- 1Laboratory of Molecular Neurobiology, Neuroscience Institute, Medical Academy, Lithuanian University of Health Sciences, Kaunas, Lithuania
- 2Laboratory of Molecular Neurooncology, Neuroscience Institute, Medical Academy, Lithuanian University of Health Sciences, Kaunas, Lithuania
Background: Natural non-coding antisense transcripts (ncNATs) are long non-coding RNAs (lncRNA) transcribed from the opposite strand of a separate protein coding or non-coding gene. As such, ncNATs can increase overlapping mRNA (and the coded protein) levels by stabilizing mRNA, absorbing inhibitory miRNAs and protecting the mRNA from degradation, or conversely decrease mRNA (or protein) levels by directing the mRNA towards degradation or inhibiting protein translation. Recently, growing numbers of ncNATs were shown to be dysregulated in cancerous cells, however, actual impact of ncNATs on cancer progression remains largely unknown. We therefore investigated gene expression levels of natural antisense lncRNA CHROMR (Cholesterol Induced Regulator of Metabolism RNA) and its sense protein coding gene PRKRA (Protein Activator of Interferon Induced Protein Kinase EIF2AK2) in gliomas. Next, we checked CHROMR effect on the survival of glioma patients.
Methods: We performed RNA-seq on post-surgical tumor samples from 26 glioma patients, and normal brain tissue. Gene expression in TPM values were extracted for CHROMR and PRKRA genes. These data were validated using the TCGA and GTEx gene expression databases.
Results: The gene expression level of ncNAT lncRNA CHROMR in glioma tissue was significantly higher compared to healthy brain tissue, while the expression of its sense counterpart protein coding PRKRA mRNA did not differ between glioma and healthy samples. Survival analysis showed lower survival rates in patients with low mRNA PRKRA/lncRNA CHROMR gene expression ratio compared to high ratio showing a link between lncRNA CHROMR and glioma patient survival prognosis.
Conclusion: Here we show that elevated levels of lncRNA CHROMR (i.e., low ratio of mRNA PRKRA/lncRNA CHROMR) is associated with poor prognosis for glioma patients.
1 Introduction
Natural antisense transcripts (NATs) are coding or non-coding RNA (ncRNA) sequences that are transcribed from the opposite strand of non-protein coding (nc) or protein coding (pc) genes and are complementary to or overlap with either protein-coding or non-coding transcript (Latgé et al., 2018; Santos et al., 2022). NATs can modulate the expression of their sense gene pairs (sense/antisense pairing) or of several related genes and are implicated in a broad variety of biological and pathological processes, including oncogenic progression (Guttman et al., 2009; Sun et al., 2017; Zhao et al., 2020; Krappinger et al., 2021). Most NATs are ncRNAs, among them long non-coding RNAs (lncRNAs), therefore most paired transcripts are composed of nc/nc or nc/pc pairs (Latgé et al., 2018). Regarding genomic position relative to its paired transcript, cis-NATs pairs are transcribed from the opposite strand of the same genomic locus and displays perfect RNA/RNA sequence complementarity with the opposite strand transcript (if there are no RNA modifications). Trans-NATs pairs are transcribed from different genomic loci with imperfect sequence complementarity (Latgé et al., 2018). Among cis-NATs, the estimated proportion of protein coding genes with ncNAT expression is thought to be up to 30%–50% in humans (Guttman et al., 2009; Latgé et al., 2018).
At the structural level ncNATs such as long non-coding RNAs may be multi-exonic, alternatively spliced, 5’capped, and 3’polyadenylated (Halley et al., 2013; Latgé et al., 2018). RNA polymerase II is responsible for the transcription of most lncRNAs, and its expression is controlled by promoters and enhancers, which can be induced by external stimuli (Guttman et al., 2009). NATs can drive important physiological events exerting its biological effects at small amounts (Yoshigai et al., 2014). It was shown that NAT transcription varies among cell and tissue types, and its expression is closely associated with paired sense gene (Lin et al., 2015; Zhao et al., 2020). Functionally antisense lncRNAs either promote or suppress cell proliferation, migration, and therapeutic resistance in tumors, and affect multiple stages of gene expression from epigenetic, transcriptional, to post-transcriptional, and translational modulations (Liu et al., 2021). Among the potential clinical applications of NATs, the non-coding/coding transcript pairs are of high interest for treatment (Latgé et al., 2018). However, the biological and clinical importance of NATs remains under investigation with many questions to be answered.
Gliomas account for up to 80% of malignant brain tumors, glioblastomas (GBM) being the most aggressive and malignant type (Rong et al., 2022). Despite progress in standard care of GBM patients, the outcome remains critically poor, ranging from 14.6 to 20.5 months, shortened to 8.5 months from diagnosis in elderly patients (Stupp et al., 2017). Due to the high mortality rates, caused by subpar treatment efficiency, and late detection of the tumor, the elucidation of molecular mechanisms of this disease is urgently needed.
To date, several studies have been performed to investigate the role of non-coding NATs in the context of glioma pathogenesis. Several lncRNAs, located on the antisense strand of a paired protein-coding gene (and often named after the protein coding gene, including an ‘–AS’ ending) have been linked to glioma: high levels of PTB-AS (Zhu et al., 2019), ZEB1-AS1 (Meng et al., 2018), HMMR-AS1 (Bao et al., 2021) promoted migration and proliferation in glioma, negatively impacting patient survival rates, while ST7-AS1 (Sheng et al., 2021) was associated with prolonged patient survival acting as a tumor suppressor. It is thus readily apparent that lncRNAs acting as ncNATs are critical regulators of gene expression playing an important role in gliomas, impacting patient survival rates. Thus, further inquiry into known and novel ncNATs could provide new opportunities for glioma detection and treatment.
Long non-coding RNA CHROMR (Cholesterol Induced Regulator of Metabolism RNA) (a.k.a. AC009948.5, CHROME and PRKRA-AS1) is found to be involved in controlling cholesterol homeostasis in humans, by sponging inhibitory micro RNAs (miRNAs) miR-27b, miR-33a, miR-33b and miR-128 (Hennessy et al., 2019). In addition, recent data, has linked CHROMR with several types of cancer (Bai et al., 2022; Luo et al., 2022; Wang et al., 2022), antiviral response (van Solingen et al., 2022) and autoimmune disease (Teimuri et al., 2018). Based on the location of CHROMR in the Human genome with respect to the protein-coding gene, in GENECODE 7 annotation (Derrien et al., 2012) it is categorized as biotype “non-coding natural antisense transcript”, as the 3′ end of CHROMR overlaps with the 3′ end of its paired sense gene PRKRA. However, there is no published evidence of CHROMR antisense regulation of its paired coding gene PRKRA in gliomas yet.
PRKRA (Protein Activator of Interferon Induced Protein Kinase EIF2AK2) encodes the ‘Protein Activator of PKR’ (PACT) protein, which is the activator of EIF2AK2 (‘RNA-activated protein kinase R’ (PKR), encoded by Eukaryotic Translation Initiation Factor 2 Alpha Kinase 2) gene. Under cellular stress, the PACT protein is phosphorylated, then binds to PKR, which leads to eIF2α (Eukaryotic Initiation Factor 2 alpha) phosphorylation, subsequently a halt to protein synthesis and leads to cell apoptosis (Patel et al., 2000). Conversely TRBP (TAR-RNA Binding Protein), encoded by the TARBP2 (Trans-Activation-Responsive RNA-binding Protein) gene, binds to PKR and inhibits its’ ability to phosphorylate eIF2α (Daher et al., 2001). PACT also binds with TRBP and Dicer in the RNA-induced silencing complex (RISC), where it is important to produce small interfering RNA (siRNA) and process miRNA (Lee et al., 2006; Kok et al., 2007).
In this study, we investigated the expression profiling of the most poorly understood ncRNAs species—non-coding natural antisense transcripts (ncNATs) and identified significantly upregulated expression of antisense lncRNA CHROMR in glioma as compared to normal brain tissue using RNA-seq. We found that highly upregulated CHROMR in gliomas is associated with poor patient survival in TCGA samples. Further, CHROMR regulatory role on its paired protein coding sense gene PRKRA mRNA was checked in the same patient sample. PRKRA mRNA/CHROMR gene expression ratio was significantly lower in tumorous tissues than in normal brain samples and was associated with shorter patient survival.
2 Materials and methods
2.1 Patient samples
In total 26 post-surgical glioma samples: 9 grade-II diffuse astrocytomas, IDH mutant (here referred as Low-Grade Gliomas—LGG), and 17 grade-IV Glioblastomas, IDH wild-type (GBM) were collected between 2012 and 2021 at the Hospital Lithuanian University of Health Sciences “Kauno Klinikos”, Department of Neurosurgery. Samples were snap-frozen and stored in liquid nitrogen immediately after tumor resection. Tumor diagnosis was confirmed, and tumors were classified according to the WHO classification system (2016 fourth edition update 3) (Louis et al., 2016) by the Pathology Department at Lithuanian University of Health Sciences “Kauno Klinikos”. Written consent of participation in the study was obtained from all patients before tumor resection. Permission for scientific research on glioma patients’ tissue and clinical data was obtained from the Kaunas Regional Biomedical Research Ethics Committee—No, BE-2-26-2021.
2.2 RNA isolation
100 mg of snap-frozen tissue was used to isolate total RNA, utilizing TRIzol™ reagent (Invitrogen, cat. No 15596026). The RNA integrity was estimated by a native agarose gel (1.5%) electrophoresis, evaluating 28S and 18S RNA bands, which were visible around 5k and 2k nucleotide marker point (Invitrogen™, cat. No. AM7150). The approximate quantity of the extracted RNA was measured by the NanoDrop™ 2000 spectrophotometer (Thermo Scientific™, cat. No. ND-2000). Human normal brain tissue RNA (1 mg/mL) was obtained from Invitrogen (cat. No. AM7962).
2.3 Poly-A enrichment
On average, 84.6 µg of total RNA was enriched for poly-A tailed RNAs using Dynabeads™ mRNA DIRECT™ purification kit (Invitrogen™, cat. No. 61012). Next, the poly-A enriched RNA was precipitated overnight at −80°C in a precipitation buffer (final concentrations: 10% of 3 M sodium acetate (pH 5.52) (Sigma-Aldrich, cat. No. 32319-500G-R), 100 μg/mL of glycogen (Thermo Scientific™, cat. No. R0551), and 2,5 vol of pure ethanol. Finally, precipitated RNA was resuspended in 15 µL of RNase-free water and evaluated using the RNA 6000 Pico kit (Agilent, cat. No. 5067-1513) on 2100 Bioanalyzer instrument (Agilent, cat. No. G2939BA).
2.4 Direct RNA sequencing
Sequencing libraries were made from 600 ng of poly-A enriched RNA, applying Direct RNA sequencing kit (Oxford Nanopore Technologies (ONT), cat. No. SQK-RNA002) and following manufacturer’s protocol (ver. DRS_9080_v2_revO_14 August 2019). The only correction to this protocol was made by replacing the original Reverse Transcription Adapter (RTA) to a barcoded RTA, designed by Hyeshik Chang1. All samples were divided into groups of 4, except for two glioblastoma samples, which were grouped together. Samples in each group were individually barcoded during library preparation and sequenced together. Pooling of barcoded samples (50 ng each) was done after the reverse transcription reaction (after step 8 in the protocol). Next, purified libraries were quantified with Qubit™ dsDNA HS assay kit (Invitrogen™, cat. No. Q32851) on a Qubit™ four Fluorometer (Invitrogen™, cat. No. Q33238). Finally, 200 ng of libraries were sequenced for approximately 48 h on a R9.4.1 flow cell (ONT, cat. No. FLO-MIN106D) using MinION or MinION Mk1C sequencers.
2.5 Processing of sequencing data, NATs selection criteria
Fast5 files were base-called with high accuracy mode on using ONT’s Guppy software (Version v5.0.11) applying a configuration file for dRNA-seq (“rna_r9.4.1_70bps_hac.cfg”). Base-called fast5 files were processed with a Poreplex software (Version 0.5) to extract fastq files for each barcoded sample. Next, the fastq files were aligned to the full Ensembl’s reference transcriptome (GRCh38. p13; Version 105; cDNA + ncRNA) (Cunningham et al., 2022) using Minimap2 software (Settings: ax map-ont -p 0 -N 10; Version 2.17-r941) (Li, 2018). Aligned SAM files were sorted and indexed with Samtools (Version 1.15.1) (Danecek et al., 2021) and quantified using NanoCount (Settings: -min_alignment_length 50 --min_query_fraction_aligned 0.5 --sec_scoring_value alignment_score --sec_scoring_threshold 0.95 --convergence_target 0.005 --max_em_rounds 100; Version v1.0.0. post6) (Gleeson et al., 2022). A list of all currently identified NATs was downloaded from the Guigó Lab (Derrien et al., 2012) and merged with NanoCount’s output. Of all overlapping intronic antisense lncRNA/mRNA pairs, the CHROMR and PRKRA pair had the highest (18/26%–69%) number of samples with both transcripts detected in our data. Our analysis included only these samples (GBM n = 11; LGG n = 7). All the computational work was done on a GenomeDK server, using CentosOS (Release 7.9.2009) and Miniconda virtual environment (Anaconda Software Distribution, V ersion 22.9.0).
2.6 External datasets used for analysis
Publicly available LGG and GBM data was obtained from The Cancer Genome Atlas (TCGA) dbGaP study accession phs0001782. Publicly available healthy brain tissue data was obtained from the Genotype-Tissue Expression portal (GTEx) dbGaP accession phs000424. v93. Publicly available proteomic data of healthy brain and glioma was obtained from The Human Protein Atlas version 21.1, Ensembl version 103.384, (Uhlén et al., 2015).
2.7 Statistical analyses
Statistical calculations and graphical preparations were done on the GraphPad Prism (Version 9.4.1 (681)) software. The non-parametric ANOVA Kruskal-Wallis test with Dunn’s multiple comparisons test was used for ratio comparisons. Survival analysis performed using Kaplan-Meier survival curve comparison analysis.
3 Results
3.1 Antisense lncRNA CHROMR and sense protein coding PRKRA are differentially expressed in brain gliomas
Natural antisense lncRNA CHROMR (ENST00000453026.7) according to NATs annotation in GENECODE v7 database (Derrien et al., 2012) belongs to cis overlapping antisense lncRNA transcribed at the same genomic loci as its paired protein coding gene. According to the orientation with respect to the sense protein coding gene PRKRA (ENST00000325748.9), lncRNA CHROMR could be classified as tail-to-tail overlapping intronic NAT (Figure 1A). We therefore decided to check the expression levels of both genes as well as their possible link with the survival of glioma patients.
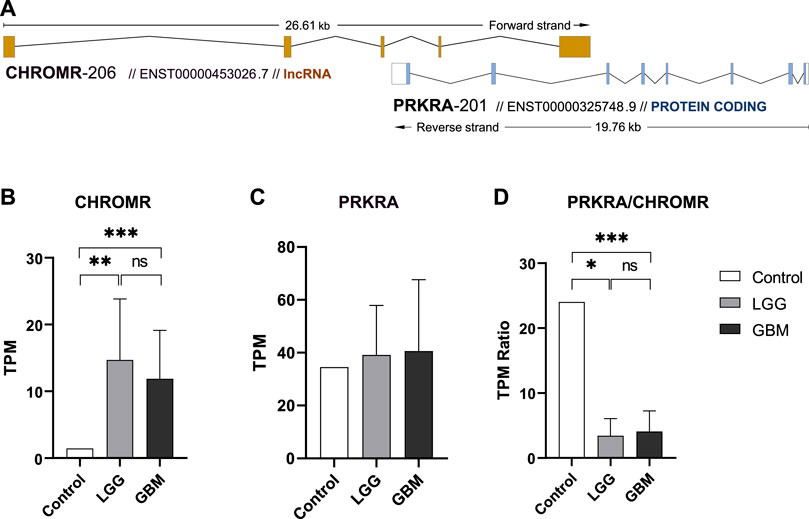
FIGURE 1. ncNAT CHROMR and its overlapped protein coding gene PRKRA expression in glioma and normal brain tissue. (A) Schematic view of lncRNA CHROMR-206 (ENST00000453026.7, + strand) and mRNA PRKRA-201 (ENST00000325748.9,—strand) transcript genomic overlap as annotated in Ensembl genome browser. Exons are depicted as colored boxes. (B–D). Our in-house RNAseq data of healthy brain (control), LGG and GBM samples for lncRNA CHROMR (B), mRNA PRKRA (C) genes and mRNA PRKRA/lncRNA CHROMR expression ratio (D). N (Control, LGG, GBM) = 1; 7; 11, respectively, one-sample t-test. *—p < 0.05; **—p < 0.01, ***—p < 0.001.
We extracted lncRNA CHROMR and mRNA PRKRA gene expression data from our in-house glioma patient RNA-seq dataset. Here we found that LGG and GBM samples had more than 3.0 log2-fold (3.35 and 3.04, respectively) significantly higher expression of lncRNA CHROMR than healthy brain (Figure 1B), while expression of protein coding PRKRA mRNA did not seem to differ between healthy brain and glioma samples (Figure 1C). This reduced the mRNA PRKRA/lncRNA CHROMR ratio in LGG and GBM samples by a log2-fold change of 2.81 and 2.57, respectively as compared to normal brain tissue (Figure 1D), which suggests a high expression of lncRNA CHROMR is characteristic of brain gliomas. Interestingly, protein-level data obtained from the Human Protein Atlas database suggests levels of PRKRA encoded protein PACT are lower in gliomas as compared to normal brain tissue (Supplementary Figure S1), but the low number of samples and qualitative nature of the data raise doubts about the validity of such results.
To validate these findings, we obtained publicly available data through the GTEx and TCGA databases for normal brain and glioma samples respectively, for comparison with our in-house generated data. Owing to possible TPM (Transcripts Per Million) differences due to sequencing depth variation between our and database samples, comparisons were focused on the mRNA PRKRA/lncRNA CHROMR ratio (Figures 2A,B). This should theoretically eliminate any effect of technical variation on compared results.
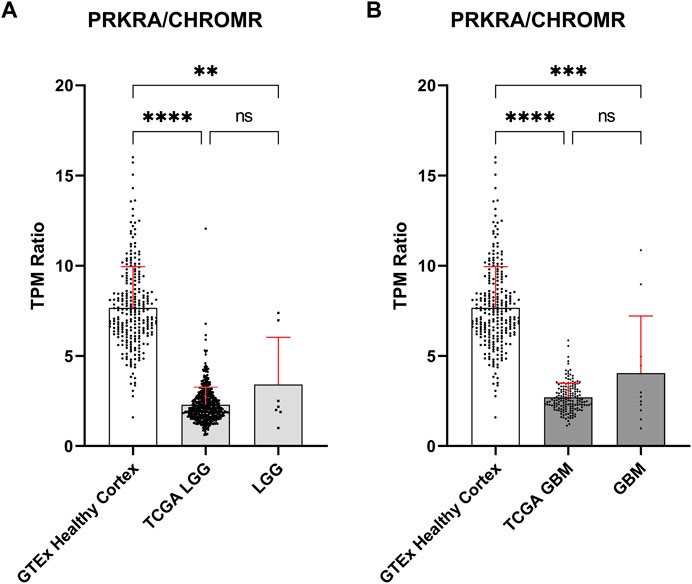
FIGURE 2. Comparison of mRNA PRKRA/lncRNA CHROMR gene expression ratio in LGG and GBM. (A,B) Comparing mRNA PRKRA/lncRNA CHROMR expression ratio of GTEx database Healthy Cortex samples with LGG (A) and GBM samples (B). N (GTEx Healthy Cortex, TCGA LGG, TCGA GBM) = 255; 532; 168; respectively. LGG and GBM here denotes our in-house RNA-seq data. Data is represented as mean with standard deviation. Kruskal-Walli’s test with Dunn’s multiple comparison test. ns—p > 0.05; ** - p < 0.01; *** - p < 0.001, **** - p < 0.0001.
The PRKRA/CHROMR ratio was consistent between glioma samples: we did not find a statistically significant difference for PRKRA/CHROMR ratio between TCGA patient and our patient samples. There was a statistically significant difference for PRKRA/CHROMR ratio when comparing pathological (both LGG and GBM) and healthy (brain cortex) brain tissue expression, confirming our previous observation from our in-house RNA-seq data. (Figures 2A,B). This confirms that a lower mRNA PRKRA/lncRNA CHROMR ratio (and thus a high expression of lncRNA CHROMR) is a feature of brain glioma.
3.2 PRKRA/CHROMR gene expression ratio has prognostic value in brain glioma
In order to evaluate the effect of the mRNA PRKRA/lncRNA CHROMR on glioma patient outcome, we performed survival analysis. Here we show that patients (LGG + GBM) with a high PRKRA/CHROMR ratio (above the third quartile) had a higher survival rate than those below the third quartile (Hazard Ratio = 0.2), though our data did not reach statistical significance (Log-rank test, p = 0.0834) (Figure 3A). Comparing high (above median) and low (below median) expression of lncRNA CHROMR (Supplementary Figure S2A) and mRNA PRKRA (Supplementary Figure S2B) individually, we could not find a statistically significant hazard ratio either, possibly due to the relatively low number of patients in our cohort.
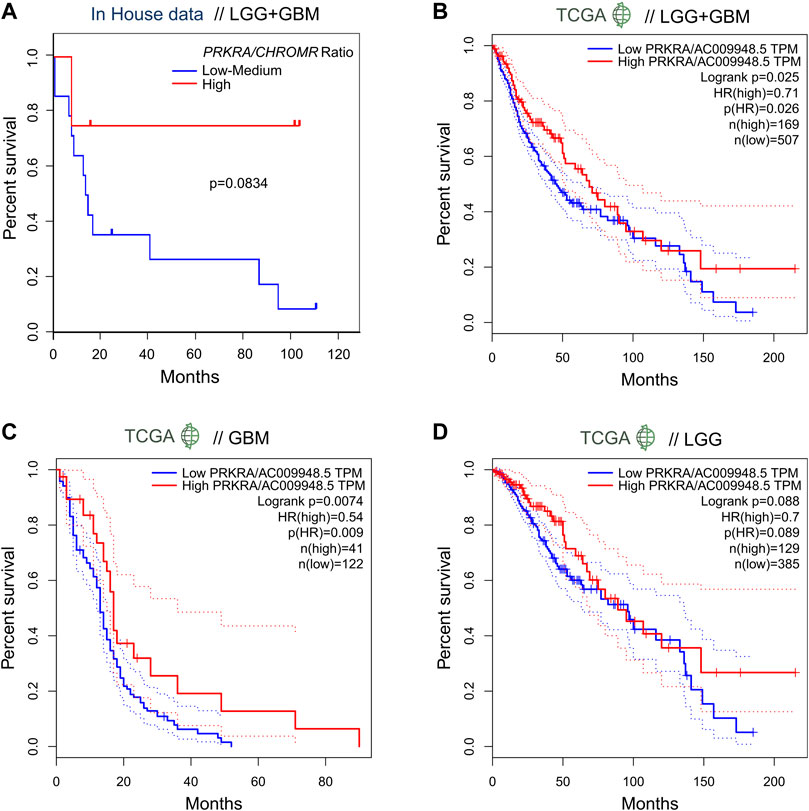
FIGURE 3. Expression ratio of mRNA PRKRA/lncRNA CHROMR affects survival ratio of LGG and GBM patients. (A) Comparison of survival probability for pooled LGG and GBM patients with High mRNA PRKRA/lncRNA CHROMR expression Ratio (above third quartile) and Low-medium expression Ratio (under third quartile). (B–D) GEPIA tool generated patient survival curves comparing mRNA PRKRA/lncRNA CHROMR High and Low gene expression (in TPM) ratio groups in pooled LGG and GBM datasets (B) and separately (C,D), dotted lines indicating 95% Confidence Interval of the survival curve. N (TCGA LGG, TCGA GBM) = 514; 163; respectively. Mantel-Cox (Log-rank) test. Statistically significant - p < 0.05. HR denotes hazard ratio.
To overcome this, we checked the survival rates of glioma patients from the TCGA database using the GEPIA online tool5 (Tang et al., 2017). When provided with analogous cutoff inputs (above the third quartile versus below the third quartile), we found a statistically significant Hazard Ratio of 0.71 for mRNA PRKRA/lncRNA CHROMR (Figure 3B) and 1.4 for lncRNA CHROMR (Supplementary Figure S2C), but not PRKRA (Supplementary Figure S2F) in LGG + GBM samples.
Interestingly, we were able to observe a difference between LGG and GBM patient survival according to our genes of interest. Unlike the combined LGG + GBM dataset, the effect of PRKRA/CHROMR ratio did not reach statistical significance in TCGA LGG patient outcomes (p = 0.088) (Figure 3D). However, the TCGA LGG patients had a significantly higher hazard ratio for high lncRNA CHROMR expression by itself (HR = 1.7, p = 0.0033) (Supplementary Figure S2D). No effect of PRKRA (Supplementary Figure S2G) could be observed on the survival of LGG patients.
When analyzing TCGA GBM samples, however, we saw that only the PRKRA/CHROMR ratio (Figure 3C), but not CHROMR (Supplementary Figure S2E) or PRKRA (Supplementary Figure S2H) separately, had prognostic value: GBM patients who had a high PRKRA/CHROMR ratio had a significantly lower hazard ratio (HR = 0.54, p = 0.0074). These results highlight that high lncRNA CHROMR is detrimental to glioma patient survival. However, it has to be emphasized that for GBM, both PRKRA and CHROMR have to be taken into account, as CHROMR alone is not informative in patient survival prognosis.
4 Discussion
Natural antisense transcripts (NATs) are coding or non-coding RNA sequences transcribed from the opposite direction from the same genomic locus as overlapped genes and seem to play important roles in pathological processes (Latgé et al., 2018; Santos et al., 2022) through various molecular mechanisms (Zhao et al., 2020; Krappinger et al., 2021; Molias et al., 2021). Only in recent years due to evolvement of high-throughput strand-specific sequencing technologies lowly expressed NATs (to which antisense long non-coding RNAs belong) attracted attention as candidate biomarkers and therapeutic targets in cancer. Here we focus on the interesting role of natural antisense lncRNA CHROMR in glioma.
Some NATs are able to bind histones or recruit histone-modifying proteins to their genomic locus, exerting either a negative or positive epigenetic regulatory effect on neighboring genes (Magistri et al., 2012). There is also evidence of NATs exerting transcriptional interference, whereby transcription of the NAT obstructs transcription of the overlapped gene (Stojic et al., 2016). Other NATs act outside the nucleus, interacting with overlapped gene mRNA, which can stabilize the mRNA (Zhu et al., 2019) or facilitate alternative splicing (Yin et al., 2017). Additionally, NATs can capture, and ‘sponge’ miRNA intended to target the sense mRNA (Zeng et al., 2019). Therefore, it is evident that NATs have multiple avenues of effect and are no doubt important in many if not most processes in the cell.
The fact that PRKRA/CHROMR ratio, but not the genes separately, had an effect on survival of glioblastoma patients, suggests that CHROMR may indeed have some NAT-associated interaction with the overlapped PRKRA gene in glioblastoma, but not low-grade glioma. Since glioblastoma is the most aggressive and malignant type of brain tumor (Rong et al., 2022), associated with high mortality rates and poor treatment efficiency (Stupp et al., 2017), such findings could prove useful in elucidating the molecular mechanisms and/or improving available treatments for glioblastomas.
Though it could be expected that lncRNA overlapping sense mRNA would modulate the levels of overlapped gene mRNA, such an effect could not be observed regarding lncRNA CHROMR and protein coding PRKRA. Indeed, it has been shown that CHROMR shows a weak interaction with the PRKRA genomic locus and that CHROMR expression does not impact the expression of the PRKRA gene (van Solingen et al., 2022) at least in the context of macrophages. However, lncRNA are known for their ability to modulate gene activity without impacting levels of mRNA, so it is not yet conclusive that CHROMR does not interact with PRKRA. Interestingly, CHROMR has been shown to have histone binding affinity in macrophages (van Solingen et al., 2022), which could still be an avenue for PRKRA regulation, without binding to PRKRA mRNA. It is also known that CHROMR can act as a sponge for various miRNAs (Hennessy et al., 2019; Wang et al., 2022), but it would be difficult to attribute miRNA sponging to observed effects on glioma. For example, miR-27b, which CHROMR seems to be able to sponge (Hennessy et al., 2019) has both been linked with glioma progression (Chen et al., 2011; Liu et al., 2015; Miao et al., 2020) and glioma repression (Chang et al., 2020; ZHAN et al., 2021). Therefore, alternate means of PRKRA-CHROMR interaction are possible, but further experiments would need to be done to elucidate the mechanism.
CHROMR has also been implicated in elevated risk for stomach adenocarcinoma (Luo et al., 2022), lung adenocarcinoma (Bai et al., 2022), increased resistance to chemotherapy in Lymphoma (Wang et al., 2022), but also in antiviral response to Influenza and COVID-19 infections (van Solingen et al., 2022) and autoimmune disease, such as multiple sclerosis (Teimuri et al., 2018). CHROMR expression is different when comparing lymphoid cancer (high) with myeloid cancer (low) cell lines (Yang et al., 2022). Our results suggest that increased CHROMR expression could be linked to development of malignancy and poorer prognosis for brain glioma patients.
Protein coding PRKRA is important for normal brain function. Mutations of the PRKRA gene are responsible for the DYT-PRKRA (DYT16) dystonia (Camargos et al., 2008; Burnett et al., 2020; Vaughn et al., 2022), characterized by death of neurons in specific brain regions, causing involuntary movements. This effect comes from aberrant and increased stress response (Vaughn et al., 2022). Perhaps there is aberrant PRKRA protein function in glioma cells, resulting in a reduced ability to respond to pro-apoptotic signals, that could be caused by the observed high CHROMR expression in gliomas. This theory would, however, require further experiments to prove.
PRKRA has been implicated in conferring chemoresistance to mucinous ovarian cancer cells (Hisamatsu et al., 2019) and in antiviral response (Vaughn et al., 2022). It is still to be determined if PRKRA acts in a similar fashion in glioma as it does the mucinous ovarian cancer cells, as gliomas are notoriously resistant to chemotherapeutic agents. It has been shown, however, that endoplasmic reticulum stress makes glioma cells more susceptible to chemotherapeutic agents through PERK signaling (Xipell et al., 2016), which like PRKRA, acts via phosphorylation of eIF2α. It could mean PRKRA dependent chemoresistance works through a different pathway, but whether PRKRA even has a role in glioma chemoresistance is still to be demonstrated. It is also not known if and how CHROMR may impact PRKRA dependent chemoresistance.
Glioma cells are able to evade the anti-tumor immune response due to the cancer’s immunosuppressive repertoire (Chen et al., 2021). Since PRKRA is known to play a role in antiviral response (Vaughn et al., 2022), perhaps our observations of high levels of CHROMR in glioma could play a role in inhibiting PRKRA related antiviral (which is related to anti-tumor) response. This would, however, require functional experiments to prove.
Both protein coding PRKRA and lncRNA CHROMR seem to have diverse functions in different tissues and different diseases, therefore need to be investigated at a tissue and disease specific context. Here we report a link between glioma patient survival and the lncRNA CHROMR gene, which could be linked with the overlapped PRKRA gene in glioblastomas. However, further experiments will have to be performed in order to elucidate the mechanism of action and other players involved in the CHROMR-Glioma link.
Data availability statement
The dataset presented in the study are deposited in the NCBI Gene Expression Omnibus repository (https://www.ncbi.nlm.nih.gov/geo/), accession number GSE219250.
Ethics statement
Permission for scientific research on glioma patients’ tissue and clinical data was obtained from the Kaunas Regional Biomedical Research Ethics Committee, No: BE-2-26-2021. The patients/participants provided their written informed consent to participate in this study.
Author contributions
DŠ, DS, and PV: generation of publication outline. DŠ: extraction of database data, statistical analysis, preparation of publication and figures. DS: collection of patient data, preparation of publication. GS: sample preparation, data analysis. RS: bioinformatic analysis, data generation. PV: data analysis. AT: collection of patient clinical samples and data. All authors contributed equally to the preparation of the manuscript.
Funding
This project was funded by the Lithuanian Research Council, project No. S-MIP-20-51.
Acknowledgments
The results here are in part based upon data generated by The Cancer Genome Atlas (TCGA), The Genotype-Tissue Expression (GTEx) Project and The Human Protein Atlas.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmolb.2023.1101953/full#supplementary-material
Footnotes
1https://github.com/hyeshik/poreplex.
3https://gtexportal.org/home/datasets.
4https://www.proteinatlas.org/about/download.
References
Bai, J., Li, H., Chen, X., Chen, L., Hu, Y., Liu, L., et al. (2022). LncRNA-AC009948.5 promotes invasion and metastasis of lung adenocarcinoma by binding to miR-186-5p. Front. Oncol. 12. doi:10.3389/fonc.2022.949951
Bao, X., Peng, Y., Shen, J., and Yang, L. (2021). Sevoflurane inhibits progression of glioma via regulating the HMMR antisense RNA 1/microRNA-7/cyclin dependent kinase 4 axis. Bioengineered 12, 7893–7906. doi:10.1080/21655979.2021.1976712
Burnett, S. B., Vaughn, L. S., Sharma, N., Kulkarni, R., and Patel, R. C. (2020). Dystonia 16 (DYT16) mutations in PACT cause dysregulated PKR activation and eIF2α signaling leading to a compromised stress response. Neurobiol. Dis. 146, 105135. doi:10.1016/j.nbd.2020.105135
Camargos, S., Scholz, S., Simón-Sánchez, J., Paisán-Ruiz, C., Lewis, P., Hernandez, D., et al. (2008). DYT16, a novel young-onset dystonia-parkinsonism disorder: Identification of a segregating mutation in the stress-response protein PRKRA. Lancet Neurol. 7, 207–215. doi:10.1016/S1474-4422(08)70022-X
Chang, K., Wang, G., Lou, J., Hao, S., Lv, R., Duan, D., et al. (2020). lncRNA TTN-AS1 upregulates RUNX1 to enhance glioma progression via sponging miR-27b-3p. Oncol. Rep. 44, 1064–1074. doi:10.3892/or.2020.7684
Chen, H., Li, M., Guo, Y., Zhong, Y., He, Z., Xu, Y., et al. (2021). Immune response in glioma’s microenvironment. Innov. Surg. Sci. 5, 20190001. doi:10.1515/iss-2019-0001
Chen, L., Li, H., Han, L., Zhang, K., Wang, G., Wang, Y., et al. (2011). Expression and function of miR-27b in human glioma. Oncol. Rep. 26, 1617–1621. doi:10.3892/or.2011.1458
Cunningham, F., Allen, J. E., Allen, J., Alvarez-Jarreta, J., Amode, M. R., Armean, I. M., et al. (2022). Ensembl 2022. Nucleic Acids Res. 50, D988–D995. doi:10.1093/nar/gkab1049
Daher, A., Longuet, M., Dorin, D., Bois, F., Segeral, E., Bannwarth, S., et al. (2001). Two dimerization domains in the trans-activation response RNA-binding protein (TRBP) individually reverse the protein kinase R inhibition of HIV-1 long terminal repeat expression. J. Biol. Chem. 276, 33899–33905. doi:10.1074/jbc.M103584200
Danecek, P., Bonfield, J. K., Liddle, J., Marshall, J., Ohan, V., Pollard, M. O., et al. (2021). Twelve years of SAMtools and BCFtools. Gigascience 10, giab008. doi:10.1093/gigascience/giab008
Derrien, T., Johnson, R., Bussotti, G., Tanzer, A., Djebali, S., Tilgner, H., et al. (2012). The GENCODE v7 catalog of human long noncoding RNAs: Analysis of their gene structure, evolution, and expression. Genome Res. 22, 1775–1789. doi:10.1101/gr.132159.111
Gleeson, J., Leger, A., Prawer, Y. D. J., Lane, T. A., Harrison, P. J., Haerty, W., et al. (2022). Accurate expression quantification from nanopore direct RNA sequencing with NanoCount. Nucleic Acids Res. 50, E19. doi:10.1093/nar/gkab1129
Guttman, M., Amit, I., Garber, M., French, C., Lin, M. F., Feldser, D., et al. (2009). Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature 458, 223–227. doi:10.1038/nature07672
Halley, P., Khorkova, O., and Wahlestedt, C. (2013). Natural antisense transcripts as therapeutic targets. Drug Discov. Today Ther. Strateg. 10, e119–e125. doi:10.1016/j.ddstr.2013.03.001
Hennessy, E. J., van Solingen, C., Scacalossi, K. R., Ouimet, M., Afonso, M. S., Prins, J., et al. (2019). The long noncoding RNA CHROME regulates cholesterol homeostasis in primate. Nat. Metab. 1, 98–110. doi:10.1038/s42255-018-0004-9
Hisamatsu, T., McGuire, M., Wu, S. Y., Rupaimoole, R., Pradeep, S., Bayraktar, E., et al. (2019). PRKRA/PACT expression promotes chemoresistance of mucinous ovarian cancer. Mol. Cancer Ther. 18, 162–172. doi:10.1158/1535-7163.MCT-17-1050
Kok, K. H., Ng, M. H. J., Ching, Y. P., and Jin, D. Y. (2007). Human TRBP and PACT directly interact with each other and associate with dicer to facilitate the production of small interfering RNA. J. Biol. Chem. 282, 17649–17657. doi:10.1074/jbc.M611768200
Krappinger, J. C., Bonstingl, L., Pansy, K., Sallinger, K., Wreglesworth, N. I., Grinninger, L., et al. (2021). Non-coding natural antisense transcripts: Analysis and application. J. Biotechnol. 340, 75–101. doi:10.1016/j.jbiotec.2021.08.005
Latgé, G., Poulet, C., Bours, V., Josse, C., and Jerusalem, G. (2018). Natural antisense transcripts: Molecular mechanisms and implications in breast cancers. Int. J. Mol. Sci. 19, 123. doi:10.3390/ijms19010123
Lee, Y., Hur, I., Park, S. Y., Kim, Y. K., Mi, R. S., and Kim, V. N. (2006). The role of PACT in the RNA silencing pathway. EMBO J. 25, 522–532. doi:10.1038/sj.emboj.7600942
Li, H. (2018). Minimap2: Pairwise alignment for nucleotide sequences. Bioinformatics 34, 3094–3100. doi:10.1093/bioinformatics/bty191
Lin, S., Zhang, L., Luo, W., and Zhang, X. (2015). Characteristics of antisense transcript promoters and the regulation of their activity. Int. J. Mol. Sci. 17, 9. doi:10.3390/ijms17010009
Liu, B., Xiang, W., Liu, J., Tang, J., Wang, J., Liu, B., et al. (2021). The regulatory role of antisense lncRNAs in cancer. Cancer Cell Int. 21, 459. doi:10.1186/s12935-021-02168-4
Liu, C., Liang, S., Xiao, S., Lin, Q., Chen, X., Wu, Y., et al. (2015). MicroRNA-27b inhibits Spry2 expression and promotes cell invasion in glioma U251 cells. Oncol. Lett. 9, 1393–1397. doi:10.3892/ol.2015.2865
Louis, D. N., Perry, A., Reifenberger, G., von Deimling, A., Figarella-Branger, D., Cavenee, W. K., et al. (2016). The 2016 world Health organization classification of tumors of the central nervous system: A summary. Acta Neuropathol. 131, 803–820. doi:10.1007/s00401-016-1545-1
Luo, L., Li, L., Liu, L., Feng, Z., Zeng, Q., Shu, X., et al. (2022). A necroptosis-related lncRNA-based signature to predict prognosis and probe molecular characteristics of stomach adenocarcinoma. Front. Genet. 13. doi:10.3389/fgene.2022.833928
Magistri, M., Faghihi, M. A., St Laurent, G., and Wahlestedt, C. (2012). Regulation of chromatin structure by long noncoding RNAs: Focus on natural antisense transcripts. Trends Genet. 28, 389–396. doi:10.1016/j.tig.2012.03.013
Meng, L., Ma, P., Cai, R., Guan, Q., Wang, M., and Jin, B. Z. (2018). Long noncoding RNA ZEB1-AS1 promotes the tumorigenesis of glioma cancer cells by modulating the miR-200c/141-ZEB1 axis. Am. J. Transl. Res. 10, 3395–3412. Available at: /pmc/articles/PMC6291700/ (Accessed November 16, 2022).
Miao, W., Li, N., Gu, B., Yi, G., Su, Z., and Cheng, H. (2020). MiR-27b-3p suppresses glioma development via targeting YAP1. Biochem. Cell Biol. 98, 466–473. doi:10.1139/bcb-2019-0300
Molias, F. B., Sim, A., Leong, K. W., An, O., Song, Y., Ng, V. H. E., et al. (2021). Antisense RNAs influence promoter usage of their counterpart sense genes in cancer. Cancer Res. 81, 5849–5861. doi:10.1158/0008-5472.CAN-21-1859
Patel, C., Handy, I., GoldsmiTh, T., and Patel, R. C. (2000). PACT, a stress-modulated cellular activator of interferon-induced double-stranded RNA-activated protein kinase, PKR. J. Biol. Chem. 275, 37993–37998. doi:10.1074/jbc.M004762200
Rong, L., Li, N., and Zhang, Z. (2022). Emerging therapies for glioblastoma: Current state and future directions. J. Exp. Clin. Cancer Res. 41. doi:10.1186/S13046-022-02349-7
Santos, F., Capela, A. M., Mateus, F., Nóbrega-pereira, S., and de Jesus, B. B. (2022). Non-coding antisense transcripts: Fine regulation of gene expression in cancer. Comput. Struct. Biotechnol. J. 20, 5652–5660. doi:10.1016/j.csbj.2022.10.009
Sheng, J., He, X., Yu, W., Chen, Y., Long, Y., Wang, K., et al. (2021). p53-targeted lncRNA ST7-AS1 acts as a tumour suppressor by interacting with PTBP1 to suppress the Wnt/β-catenin signalling pathway in glioma. Cancer Lett. 503, 54–68. doi:10.1016/j.canlet.2020.12.039
Stojic, L., Niemczyk, M., Orjalo, A., Ito, Y., Ruijter, A. E. M., Uribe-Lewis, S., et al. (2016). Transcriptional silencing of long noncoding RNA GNG12-AS1 uncouples its transcriptional and product-related functions. Nat. Commun. 7, 10406. doi:10.1038/ncomms10406
Stupp, R., Taillibert, S., Kanner, A., Read, W., Steinberg, D. M., Lhermitte, B., et al. (2017). Effect of tumor-treating fields plus maintenance temozolomide vs maintenance temozolomide alone on survival in patients with glioblastoma: A randomized clinical trial. JAMA 318, 2306–2316. doi:10.1001/jama.2017.18718
Sun, Y., Li, D., Zhang, R., Peng, S., Zhang, G., Yang, T., et al. (2017). Strategies to identify natural antisense transcripts. Biochimie 132, 131–151. doi:10.1016/j.biochi.2016.11.006
Tang, Z., Li, C., Kang, B., Gao, G., Li, C., and Zhang, Z. (2017). Gepia: A web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 45, W98–W102. doi:10.1093/nar/gkx247
Teimuri, S., Hosseini, A., Rezaenasab, A., Ghaedi, K., Ghoveud, E., Etemadifar, M., et al. (2018). Integrative analysis of lncRNAs in Th17 cell lineage to discover new potential biomarkers and therapeutic targets in autoimmune diseases. Mol. Ther. Nucleic Acids 12, 393–404. doi:10.1016/j.omtn.2018.05.022
Uhlén, M., Fagerberg, L., Hallström, B. M., Lindskog, C., Oksvold, P., Mardinoglu, A., et al. (2015). Proteomics. Tissue-based map of the human proteome. Science 347, 1260419, doi:10.1126/science.1260419
van Solingen, C., Cyr, Y., Scacalossi, K. R., de Vries, M., Barrett, T. J., de Jong, A., et al. (2022). Long noncoding RNA CHROMR regulates antiviral immunity in humans. Proc. Natl. Acad. Sci. U. S. A. 119, e2210321119. doi:10.1073/pnas.2210321119
Vaughn, L. S., Frederick, K., Burnett, S. B., Sharma, N., Bragg, D. C., Camargos, S., et al. (2022). DYT-PRKRA mutation P222L enhances PACT’s stimulatory activity on type I interferon induction. Biomolecules 12. doi:10.3390/BIOM12050713
Wang, M., Miao, Z., Cen, H., He, J., and Wei, C. (2022). Long non-coding RNA (LncRNA) CHROMR promotes the expression of the CNNM1 gene by adsorbing hsa-miR-1299 to obtain drug resistance in diffuse large B lymphoma cells. Transl. Cancer Res. 11, 1362–1371. doi:10.21037/tcr-22-1087
Xipell, E., Aragón, T., Martínez-Velez, N., Vera, B., Idoate, M. A., Martínez-Irujo, J. J., et al. (2016). Endoplasmic reticulum stress-inducing drugs sensitize glioma cells to temozolomide through downregulation of MGMT, MPG, and Rad51. Neuro Oncol. 18, 1109–1119. doi:10.1093/neuonc/now022
Yang, L. R., Lin, Z. Y., Hao, Q. G., Li, T. T., Zhu, Y., Teng, Z. W., et al. (2022). The prognosis biomarkers based on m6A-related lncRNAs for myeloid leukemia patients. Cancer Cell Int. 22, 10. doi:10.1186/s12935-021-02428-3
Yin, J., Luo, W., Zeng, X., Zeng, L., Li, Z., Deng, X., et al. (2017). UXT-AS1-induced alternative splicing of UXT is associated with tumor progression in colorectal cancer. Am. J. Cancer Res. 7, 462. 472. Available at: /pmc/articles/PMC5385636/ (Accessed January 10, 2023).
Yoshigai, E., Hara, T., Inaba, H., Hashimoto, I., Tanaka, Y., Kaibori, M., et al. (2014). Interleukin-1β induces tumor necrosis factor-α secretion from rat hepatocytes. Hepatol. Res. 44, 571–583. doi:10.1111/hepr.12157
Zhan, X., Lei, C., and Yang, L. (2021). Sevoflurane inhibits cell proliferation and migration of glioma by targeting the miR-27b/VEGF axis. Mol. Med. Rep. 23, 408. doi:10.3892/mmr.2021.12047
Zeng, T., Ni, H., Yu, Y., Zhang, M., Wu, M., Wang, Q., et al. (2019). BACE1-AS prevents BACE1 mRNA degradation through the sequestration of BACE1-targeting miRNAs. J. Chem. Neuroanat. 98, 87–96. doi:10.1016/j.jchemneu.2019.04.001
Zhao, S., Zhang, X., Chen, S., and Zhang, S. (2020). Natural antisense transcripts in the biological hallmarks of cancer: Powerful regulators hidden in the dark. J. Exp. Clin. Cancer Res. 39, 187. doi:10.1186/s13046-020-01700-0
Keywords: ncNATs, lncRNA, CHROMR, PRKRA, glioma, LGG, GBM, survival
Citation: Širvinskas D, Steponaitis G, Stakaitis R, Tamašauskas A, Vaitkienė P and Skiriutė D (2023) Antisense lncRNA CHROMR is linked to glioma patient survival. Front. Mol. Biosci. 10:1101953. doi: 10.3389/fmolb.2023.1101953
Received: 18 November 2022; Accepted: 21 February 2023;
Published: 06 March 2023.
Edited by:
Tominori Kimura, Ritsumeikan University, JapanReviewed by:
Mikio Nishizawa, Ritsumeikan University, JapanTokifumi Odaka, Kansai Medical University, Japan
Copyright © 2023 Širvinskas, Steponaitis, Stakaitis, Tamašauskas, Vaitkienė and Skiriutė. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Paulina Vaitkienė, cGF1bGluYS52YWl0a2llbmVAbHNtdS5sdA==
 Dovydas Širvinskas
Dovydas Širvinskas Giedrius Steponaitis
Giedrius Steponaitis Rytis Stakaitis
Rytis Stakaitis Arimantas Tamašauskas2
Arimantas Tamašauskas2 Paulina Vaitkienė
Paulina Vaitkienė Daina Skiriutė
Daina Skiriutė