- 1Henan Key Laboratory of Helicobacter pylori, Microbiota and Gastrointestinal Cancer, Marshall Medical Research Center, Fifth Affiliated Hospital of Zhengzhou University, Zhengzhou, China
- 2Research Center on Aging, Centre Intégré Universitaire de Santé et Services Sociaux de l'Estrie-Centre Hospitalier Universitaire de Sherbrooke, Sherbrooke, QC, Canada
- 3Department of History of Science and Scientific Archeology, University of Science and Technology of China, Hefei, China
- 4School of Environmental and Life Sciences, The University of Newcastle, Callaghan, NSW, Australia
Long noncoding RNAs (lncRNAs) possess the potential for therapeutic targeting to treat many disorders, including cancers. Several RNA-based therapeutics (ASOs and small interfering RNAs) have gained FDA approval over the past decade. And with their potent effects, lncRNA-based therapeutics are of emerging significance. One important lncRNA target is LINC-PINT, with its universalized functions and relationship with the famous tumor suppressor gene TP53. Establishing clinical relevance, much like p53, the tumor suppressor activity of LINC-PINT is implicated in cancer progression. Moreover, several molecular targets of LINC-PINT are directly or indirectly used in routine clinical practice. We further associate LINC-PINT with immune responses in colon adenocarcinoma, proposing the potential utility of LINC-PINT as a novel biomarker of immune checkpoint inhibitors. Collectively, current evidence suggests LINC-PINT can be considered for use as a diagnostic/prognostic marker for cancer and several other diseases.
Introduction
Sequencing of the human genome has revealed that ∼20,000 traditional genes encode protein molecules, occupying just 2% of the total genetic landscape (Salzberg, 2018; Viggiano, 2022). The remaining genome consists of non-protein-coding regions, including numerous ‘RNA only’ genes, among which there is a large but diverse group called long non-coding RNAs or lncRNAs for short (Elkon and Agami, 2017; Dietlein et al., 2022; Paloviita and Vuoristo, 2022). Subsequent analyses have shown that lncRNAs fulfill various regulatory roles in healthy and cancerous tissues (Geng et al., 2018; Bi et al., 2019; Farzaneh et al., 2022a; Farzaneh et al., 2022b; Farzaneh et al., 2022c; Nasrolahi et al., 2022). As such, their expression and function are often altered in disease states (Kleinbrink et al., 2021; Yang et al., 2021; Anderson and Anderson, 2022; Dietlein et al., 2022; Erdogan et al., 2022) with selected examples now being used as diagnostic markers for various diseases including cancers (Li et al., 2019; Deng and Zou, 2022; Najafi et al., 2022; Zhang et al., 2022). Among these, one particular lncRNA called LINC-PINT (P53 Induced long Non-coding Transcript) appears exceptional because of its universalized functions and relationship to the famous tumor suppressor gene P53 (He et al., 2021; Bukhari et al., 2022). Indeed, like P53, LINC-PINT was first proposed as an oncogene but later revealed as a tumor suppressor in human cancers.
LINC-PINT contributes to a variety of biological processes impacting cancer cell growth and metastasis, with involvement in processes ranging from DNA damage responses to cell senescence and apoptosis (Simchovitz et al., 2020; He et al., 2021; Wang et al., 2021; Xiang et al., 2021; Zhang et al., 2021; Bukhari et al., 2022). It has also been shown to be an essential regulator of many other diseases, including autoimmune diseases (Wang and Zhao, 2020; Salviano-Silva et al., 2021). These functions arise through interactions affecting epigenetic regulation or direct interactions with other biomolecules, including proteins and other lncRNAs (Xu et al., 2019; Bukhari et al., 2022). Some alternatively spliced variants of LINC-PINT also act as host transcripts for circular RNA (circRNA) generation, and these can serve as sources for the translation of short functional peptides (Xiang et al., 2021). More than 100 alternatively spliced variants of LINC-PINT have been identified (Bukhari et al., 2022). Still, to date, only a few of these have been thoroughly studied—a feature earning the name PINTology to describe the current and future studies involving LINC-PINT.
Therapeutic importance of lncRNA LINC-PINT
Cancer cells exhibit variability formally termed tumor heterogeneity, and this is a well-known explanation behind the failure of cancer therapy where resistant cells selectively survive repeated drug treatments (Brutovsky, 2022; Mahinfar et al., 2022). Contributing to this problem is also the lack of suitable diagnostic, therapeutic and prognostic biomarkers (Najafi, 2022). Hence, optimizing treatment outcomes for any disease is only possible with full knowledge of the underlying mechanisms. In this regard, recent studies involving lncRNAs have substantially contributed to understanding the cancer (Li et al., 2022b; Singh et al., 2022; Zhong et al., 2022). However, exploiting the potential of lncRNAs as biomarkers and therapeutic targets is just the beginning (Mirzaei et al., 2022). For example, lncRNAs associated with immune checkpoints (Li et al., 2022a; Jiang et al., 2022; Wen et al., 2022) have been used as disease biomarkers in the new wave of cancer immunotherapy that is currently revolutionizing oncology practice (Figure 1A). Nevertheless, small successes in the clinical setting are expected to help build confidence in lncRNAs as biomarkers and produce the momentum needed for their broader applications.
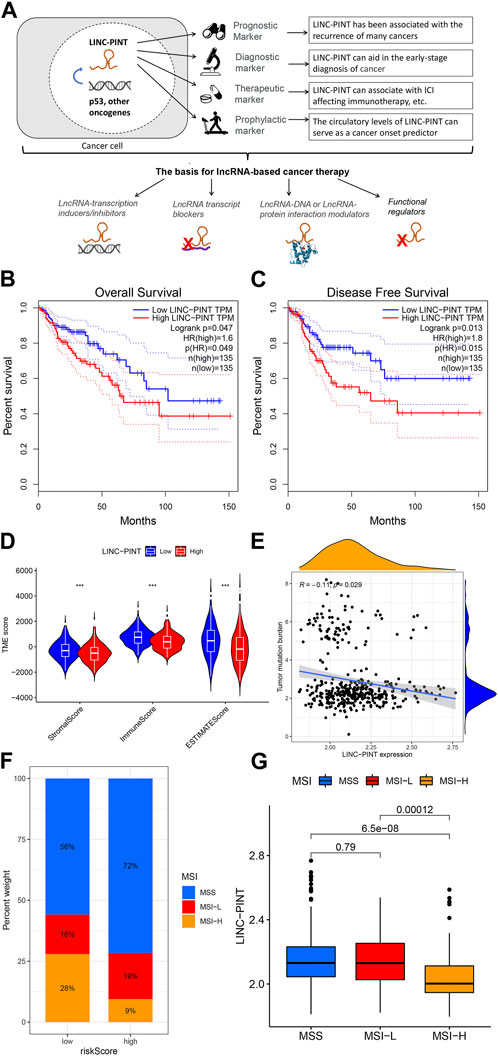
FIGURE 1. Clinical potentials of LINC-PINT. (A) Overview of the clinical applications of LINC-PINT. In short, it can be used as a prognostic marker to ascertain the course of cancer progression based on its expression level. It can also be used as a diagnostic marker to aid the early detection of ontological transformation. The third type of application, already in pre-clinical testing, predicts response to cancer therapy. Finally, less explored but equally important is the potential of LINC-PINT-based studies to determine the susceptibility of healthy subjects to cancer. These applications can be used, in turn, to tailor and administer suitable lncRNA-based cancer therapies. Correlation of LINC-PINT expression with (B) Overall survival and (C) Disease-free survival in Colon Adenocarcinoma (D) Association of LINC-PINT expression with immune scores, including stromal, immune, and ESTIMATE scores, (E) Tumor Mutation Burden, and (F–G) microsatellite instability in colon adenocarcinoma patients.
Although the tumor-suppressive function and downregulation of LINC-PINT in different cancers have been thoroughly established (He et al., 2021; Bukhari et al., 2022), its clinical use remains extremely limited. It is interesting to note that many of the vital protein targets of LINC-PINT involved in cell cycle regulation are already widely used in the diagnostic setting (Caputo et al., 2014; Xu et al., 2019). Furthermore, some of these are cancer treatments with inhibitors of LINC-PINT’s targets now being used in treating melanoma (George et al., 2019). As a starting point, the most appropriate clinical application of LINC-PINT would be as a diagnostic or predictive marker for specific cancers such as esophageal cancer, where LINC-PINT has been associated with cancer recurrence (Zhang et al., 2019; Rong et al., 2021). Indeed, the abnormal expression of LINC-PINT reflects many vital predictors of cancer outcomes such as lymph node metastasis, tumor size, and differentiation status (Figure 1A). Notably, the levels of LINC-PINT released by tumor cells into the blood could also be used to measure therapeutic responses, as specific anti-cancer drugs induce the re-expression of LINC-PINT. Other positive associations with neuropathy along with pemphigus foliaceous, diabetes, and arthritis may also be exploited, suggesting LINC-PINT as a multifaceted target for cancers and other diseases.
Data collection
The complete gene expression profiles and clinical information of 473 colon cancer patients were downloaded from the TCGA online database (https://portal.gdc.cancer.gov/). The R software package “maftools” was used to calculate each sample’s tumor mutation burden (TMB). The microsatellite instability (MSI) score and groupings of the “TCGA-COAD” samples were downloaded using “TCGAbiolinks,” including: “MSI -H,” “MSI-L,” and “MSS.”
Survival and immune function analyses
We analyzed the overall survival and disease-free survival of LINC-PINT in COAD within the online database GEPIA (http://gepia.cancer-pku.cn/). The Estimation of Stromal and Immune Cells in Malignant Tumors using the Expression Data (ESTIMATE) algorithm was used to determine the unique properties of the transcriptional profiles to infer the tumor cellularity as the tumor purity. The relationship of LINC-PINT expression, Stromal Score, and Immune Score with ESTIMATEScore was plotted as a violin plot using the “violin” package in the R program. Furthermore, we analyzed the correlations between different LINC-PINT expression groups, TMB and MSI with data presented as boxplots and scatterplots using “ggplot2”.
Relationship between LINC-PINT and immune responses in colon adenocarcinoma (COAD)
We assessed the survival of patients and immune function associated with LINC-PINT expression in COAD tissues from the TCGA resource. Notably, patients whose tumors expressed low levels of LINC-PINT showed better overall and disease-free survival (Figure 1B, C). Subsequently, LINC-PINT showed a strong association with immune indicators, Tumor Mutation Burden (TMB), and Microsatellite Instability (MSI) in the COAD tumor microenvironment. COAD cases exhibiting higher expression of LINC-PINT had significantly lower immune scores, including stromal, immune, and ESTIMATE scores (Figure 1D), meaning that patients with low expression of LINC-PINT have a better immune response. Additionally, TMB and MSI have been used as prognostic markers for many cancers, especially for those receiving immunotherapy (Rizzo et al., 2021); thus, these can be used as predictive biomarkers for the efficacy of immunotherapy. Patients with low expression of LINC-PINT showed significantly higher TMB and MSI scores (Figures 1E–G), reflecting that LINC-PINT may be used as a novel biomarker of immune checkpoint inhibitors (ICIs).
Clinical features of LINC-PINT in COAD
Additionally, we took advantage of the clinical characteristics and other clinical information of the 388 COAD cases available within the TCGA database. We stratified patients based on LINC-PINT expression in their primary tumors, dividing the samples into high-expression and low-expression groups. The relationship between LINC-PINT and the clinical features of COAD was then studied using the Wilcoxon rank sum test using p < 0.05 to denote statistical significance. Table 1 compares the differences between the high and low-expression LINC-PINT groups wherein high LINC-PINT expression showed a significantly higher proportion of tumor metastasis index (M1) cases compared to the low-expression group (p = 0.0357). Similarly, the boxplot analysis showed that the expression level of LINC-PINT was higher in patients with metastases (Figure 2A) (p = 0.003).
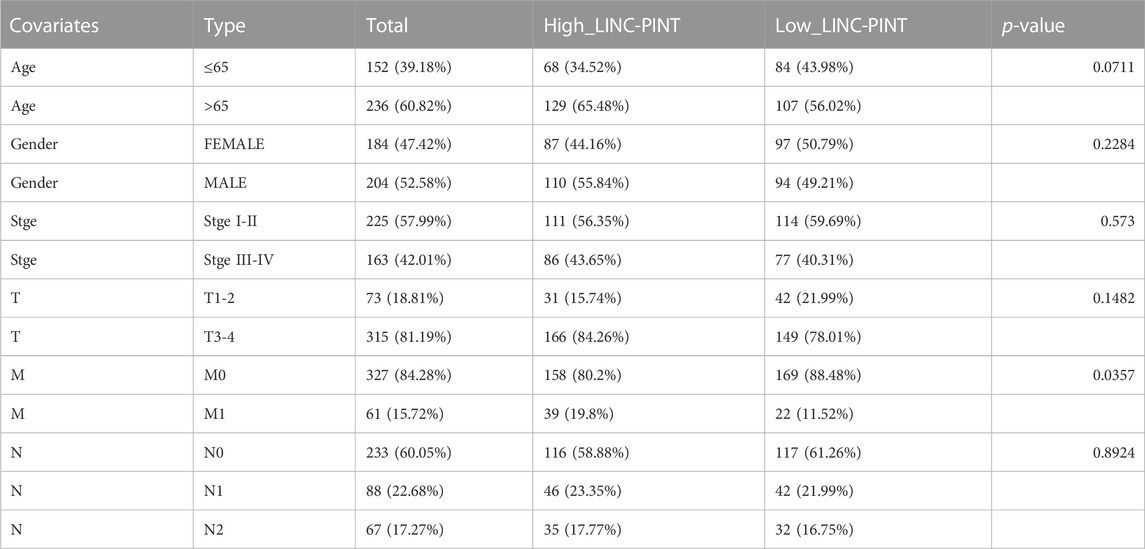
TABLE 1. Clinical features of the LINC-PINT in COAD. Out of the total of 473 COAD samples, we filtered out the complete clinical information of 388 cases from TCGA, and based on the LINC-PINT expression, the patients were divided into high and low-expression groups. Furthermore, patients were categorized based on their clinical characteristics (age, gender, T, N, M, stage, etc.) and compared the distribution of LINC-PINT high and low-expression samples in different groups. The proportion of M0 and M1 in the high-expression group was 80.2% and 19.8%, whereas in the low-expression group M0 and M1 proportion was observed as 88.48% and 11.52%, respectively. The distributions of metastasis-related clinical features were also significantly different than the control.
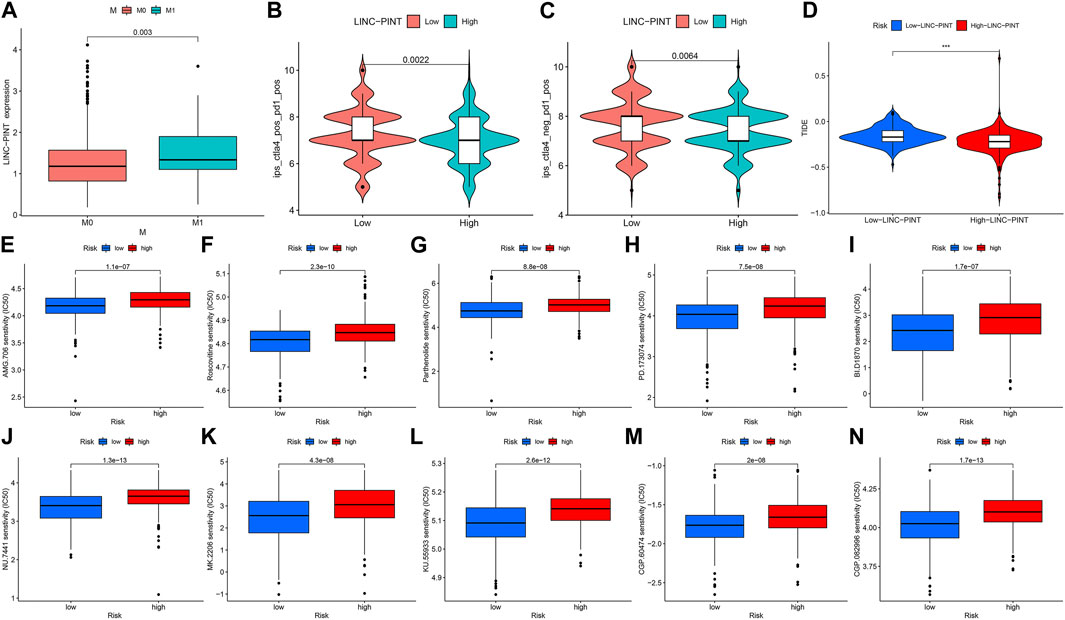
FIGURE 2. Relationship between LINC-PINT and clinical characteristics of patients with COAD. (A) Boxplot of differences in LINC-PINT expression levels between metastatic and non-metastatic groups of COAD patients. M0: non-metastasis; M1: tumor metastasis. (B, C) The immunophenotype scores in the low-LINC-PINT group and high-LINC-PINT group. (D) The TIDE score distribution between high- and low- LINC-PINT groups. (E–N) Drug sensitivity analysis of multiple chemotherapeutic drugs in the high LINC-PINT expression group and the low LINC-PINT expression group, respectively.
Furthermore, we validated the immune efficacy of LINC-PINT expression via the TCIA database. The IPS values were calculated based on immunogenicity from the TCIA database and then analyzed against LINC-PINT expression, finding higher effectiveness of immunotherapy in the low-risk group (p < 0.001) (Figures 2B, C). Moreover, Tumor Immune Dysfunction and Exclusion (TIDE) algorithm predicts ICB response and evaluates immune escape ability (http://tide.dfci.harvard.edu/). Therefore, we used the TIDE database to verify the relationship between risk scores and immune checkpoint inhibitors (ICIs) and found that the high LINC-PINT expression group has lower TIDE scores (Figure 2D). Together, this suggests that patients with high LINC-PINT expression have a lower possibility of immune escape than patients with low LINC-PINT expression, so they are more suitable for Immune Checkpoint Inhibitors (ICI) treatment.
The relationship between LINC-PINT and anticancer therapies
In addition, we also analyzed the relationship between the expression of LINC-PINT and tumor sensitivity to immunotherapy and chemotherapy drugs. IPS-PD1/PD-L1 blocker and IPS-CTLA4 blocker data on COAD from the TCGA were obtained and analyzed using The Cancer Immunome Atlas database (TCIA, https://tcia.at/home) to predict the patients’ response to ICI as high- and low-risk groups. To explore the efficacy of chemotherapeutic drugs and their relationship with PINT, we used the pRRophetic package. Using the Wilcoxon test, we screened the chemotherapeutic drugs significantly different in the LINC-PINT expression group and displayed the IC50 distribution of different drugs in the two groups of samples through a boxplot (pFilter = 0.0001). We chose the top 10 chemotherapy drugs having highly significant differences in different risk groups and displayed the IC50 distribution of different drugs in the two groups of samples by boxplot (pFilter = 0.0001) (Figure 2E–N). The results showed that COAD patients with different expressions of LINC-PINT have apparent differences in sensitivity to various chemotherapeutic drugs, particularly patients whose tumors exhibit low LINC-PINT expression are more sensitive to chemotherapy.
The future of lncRNA LINC-PINT
Compared to most lncRNAs, extensive literature exists for LINC-PINT, but a key question is whether this truly reflects most or, if not all, the underlying molecular mechanisms involved. In particular, the relationship between the functions of LINC-PINT in cancers versus non-malignant diseases needs to be better explored. For example, in viral hepatitis leading to liver cancer development, LINC-PINT foremost contributes to inflammation during viral infection and then later, to the pathogenic properties of the cancer cells (Khatun et al., 2021a; Khatun et al., 2021b), but whether the underlying processes involved are the same in both disease stages is not clear. Consequently, more intensive patient-based clinical studies are required to dissect these points. Furthermore, the large number of splice variants complicates this task, and it will be necessary to identify which variants share functions versus those that contribute to different functions. It is worth mentioning that at least one LINC-PINT variant produces small peptides, but this phenomenon’s frequency needs to be clarified. As functional products, these peptides may themselves serve as diagnostic molecules or treatment targets. It would be similarly valuable to ascertain more knowledge of the natural activators or inhibitors of LINC-PINT, which could be used to open potential new doors in cancer therapeutics. And to end on a sobering note, the number of lncRNA genes may rival or exceed the number of coding genes with LINC-PINT just one amongst thousands. However, there is the expectation that the pioneering studies involving LINC-PINT will provide important precedents for the clinical applications of lncRNAs.
Discussion
LncRNAs are crucial regulators of various cellular pathways specifically cancer-associated pathways and use different mediators to function (Peng et al., 2017; Kopp and Mendell, 2018; Ali and Grote, 2020), while the majority of lncRNAs are functionally uncharacterized (Li et al., 2017; Lorenzi et al., 2021). Specifically, the regulation of lncRNAs occurs during specific occasions, such as development and cell growth (Sun et al., 2018). Mechanistically, lncRNAs bind with DNA, RNA, and proteins for transcriptional, post-transcriptional, and post-translational regulations (Schmitt et al., 2016; Hull et al., 2021; Yue et al., 2021). LncRNAs play vital roles in cancer initiation and progression or inhibition by affecting various pathways and expression of genes (Zhuo and Kang, 2017; Statello et al., 2021), showing that lncRNAs are either onco-suppressor or onco-promoter.
Similarly, LINC-PINT is a crucial lncRNA that mainly acts as an onco-suppressor in various cancers, but its clinical use is almost neglected. However, studies have suggested it as a potential biomarker for cancer prognosis (Ghafouri-Fard et al., 2021; He et al., 2021; Ghafouri-Fard et al., 2022). Through analysis of colon cancer data from the TCGA data, we determined an enticing relationship between LINC-PINT expression, prognosis and patients’ survival. Interestingly, patients with low expression of LINC-PINT showed better survival and significantly higher TMB and MSI scores together with better responses to chemotherapy, thus supporting the clinical importance of LINC-PINT, especially its use as a biomarker for monitoring responses to immunotherapy and chemotherapy.
Conclusion
It is now commonly known that lncRNAs are key players in cancer regulation, a feature that promotes their application as target-based therapies in cancer and other relevant diseases. Interestingly, this promise has now been realized with several RNA-based therapeutics being recently approved by the FDA although their integration into the healthcare system still requires extensive clinical trials. It is well known that LINC-PINT has universalized tumor suppressor functions and a relationship to the tumor suppressor gene TP53 and several LINC-PINT targets are commonly used in clinical practice (directly or indirectly). Based on these facets, LINC-PINT emerges as a priority target for studying the clinical potential of lncRNAs (PINTology), especially for cancer therapeutics. However, the direct application of LINC-PINT is still limited due to the lack of clinical trials and clinicians’ confidence. Nevertheless, the current evidence proposes applications as a diagnostic marker for cancer and several other diseases with alterations in LINC-PINT circulatory levels providing a readily accessible platform to develop appropriate tests.
Author contributions
IB, PZ, and YM conceptualized the study, IB and MK collected data and analyzed it, IB wrote the first draft, RFT thoroughly revised the manuscript and analyzed data. FL and BS performed bioinformatics analyses and prepared figures, PZ and YM supervised the overall work and reviewed the manuscript. All authors have read the final version of the manuscript and approved it for publication.
Funding
This work was supported by Zhengzhou Major Collaborative Innovation Project (No.18XTZX12003); Key projects of discipline construction in Zhengzhou University (No. XKZDJC202001); National Key Research and development program in China (No.2020YFC2006100); Medical service capacity improvement project of Henan Province in China (grant number Yu Wei Medicine [2017] No.66).
Acknowledgments
We gratefully acknowledge the role of our host institutes in this study, and we also thank our colleagues who are listed as authors of this manuscript for their support and encouragement.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Ali, T., and Grote, P. (2020). Beyond the RNA-dependent function of LncRNA genes. Elife 9, e60583. doi:10.7554/eLife.60583
Anderson, K. M., and Anderson, D. M. (2022). LncRNAs at the heart of development and disease. Mamm. Genome 33, 354–365. doi:10.1007/s00335-021-09937-6
Bi, X., Guo, X. H., Mo, B. Y., Wang, M. L., Luo, X. Q., Chen, Y. X., et al. (2019). LncRNA PICSAR promotes cell proliferation, migration and invasion of fibroblast-like synoviocytes by sponging miRNA-4701-5p in rheumatoid arthritis. EBioMedicine 50, 408–420. doi:10.1016/j.ebiom.2019.11.024
Brutovsky, B. (2022). Scales of cancer evolution: Selfish genome or cooperating cells? Cancers (Basel) 14, 3253. doi:10.3390/cancers14133253
Bukhari, I., Khan, M. R., Hussain, M. A., Thorne, R. F., Yu, Y., Zhang, B., et al. (2022). PINTology: A short history of the lncRNA LINC-PINT in different diseases. Wiley Interdiscip. Rev. RNA 13, e1705. doi:10.1002/wrna.1705
Caputo, E., Miceli, R., Motti, M. L., Tate, R., Fratangelo, F., Botti, G., et al. (2014). AurkA inhibitors enhance the effects of B-RAF and MEK inhibitors in melanoma treatment. J. Transl. Med. 12, 216. doi:10.1186/s12967-014-0216-z
Deng, M., and Zou, W. (2022). Noncoding RNAs: Novel targets for opioid tolerance. Curr. Neuropharmacol. 21. doi:10.2174/1570159X21666221129122932
Dietlein, F., Wang, A. B., Fagre, C., Tang, A., Besselink, N. J. M., Cuppen, E., et al. (2022). Genome-wide analysis of somatic noncoding mutation patterns in cancer. Science 376, eabg5601. doi:10.1126/science.abg5601
Elkon, R., and Agami, R. (2017). Characterization of noncoding regulatory DNA in the human genome. Nat. Biotechnol. 35, 732–746. doi:10.1038/nbt.3863
Erdogan, I., Sweef, O., and Akgul, B. (2022). Long noncoding RNAs in human cancer and apoptosis. Curr. Pharm. Biotechnol. doi:10.2174/1389201023666220624094950
Farzaneh, M., Ghasemian, M., Ghaedrahmati, F., Poodineh, J., Najafi, S., Masoodi, T., et al. (2022a). Functional roles of lncRNA-TUG1 in hepatocellular carcinoma. Life Sci. 308, 120974. doi:10.1016/j.lfs.2022.120974
Farzaneh, M., Najafi, S., Dari, M. A. G., Sheykhi-Sabzehpoush, M., Dayer, D., Cheraghzadeh, M., et al. (2022b). Functional roles of long noncoding RNA MALAT1 in gynecologic cancers. Clin. Transl. Oncol. 25, 48–65. doi:10.1007/s12094-022-02914-8
Farzaneh, M., Najafi, S., Sheykhi-Sabzehpoush, M., Nezhad Dehbashi, F., Anbiyaee, O., Nasrolahi, A., et al. (2022c). The stem cell-specific long non-coding RNAs in leukemia. Clin. Transl. Oncol. 25, 345–351. doi:10.1007/s12094-022-02952-2
Geng, H., Bu, H. F., Liu, F., Wu, L., Pfeifer, K., Chou, P. M., et al. (2018). In inflamed intestinal tissues and epithelial cells, interleukin 22 signaling increases expression of H19 long noncoding RNA, which promotes mucosal regeneration. Gastroenterology 155, 144–155. doi:10.1053/j.gastro.2018.03.058
George, J., Nihal, M., Singh, C. K., and Ahmad, N. (2019). 4'-Bromo-resveratrol, a dual Sirtuin-1 and Sirtuin-3 inhibitor, inhibits melanoma cell growth through mitochondrial metabolic reprogramming. Mol. Carcinog. 58, 1876–1885. doi:10.1002/mc.23080
Ghafouri-Fard, S., Khoshbakht, T., Hussen, B. M., Taheri, M., and Samsami, M. (2022). Emerging role of non-coding RNAs in the regulation of Sonic Hedgehog signaling pathway. Cancer Cell Int. 22, 282. doi:10.1186/s12935-022-02702-y
Ghafouri-Fard, S., Shirvani-Farsani, Z., Hussen, B. M., and Taheri, M. (2021). The critical roles of lncRNAs in the development of osteosarcoma. Biomed. Pharmacother. 135, 111217. doi:10.1016/j.biopha.2021.111217
He, T., Yuan, C., and Zhao, C. (2021). Long intragenic non-coding RNA p53-induced transcript (LINC-PINT) as a novel prognosis indicator and therapeutic target in cancer. Biomed. Pharmacother. 143, 112127. doi:10.1016/j.biopha.2021.112127
Hull, R., Mbita, Z., and Dlamini, Z. (2021). Long non-coding RNAs (LncRNAs), viral oncogenomics, and aberrant splicing events: Therapeutics implications. Am. J. Cancer Res. 11, 866–883.
Jiang, H., Sun, J., Liu, F., Wu, X., and Wen, Z. (2022). An immune-related long noncoding RNA pair as a new biomarker to predict the prognosis of patients in breast cancer. Front. Genet. 13, 895200. doi:10.3389/fgene.2022.895200
Khatun, M., Sur, S., Steele, R., Ray, R., and Ray, R. B. (2021a). Inhibition of long noncoding RNA linc-pint by hepatitis C virus in infected hepatocytes enhances lipogenesis. Hepatology 74, 41–54. doi:10.1002/hep.31656
Khatun, M., Zhang, J., Ray, R., and Ray, R. B. (2021b). Hepatitis C virus evades interferon signaling by suppressing long noncoding RNA linc-pint involving C/EBP-β. J. Virol. 95, e0095221. doi:10.1128/JVI.00952-21
Kleinbrink, E. L., Gomez-Lopez, N., Ju, D., Done, B., Goustin, A. S., Tarca, A. L., et al. (2021). Gestational age dependence of the maternal circulating long non-coding RNA transcriptome during normal pregnancy highlights antisense and pseudogene transcripts. Front. Genet. 12, 760849. doi:10.3389/fgene.2021.760849
Kopp, F., and Mendell, J. T. (2018). Functional classification and experimental dissection of long noncoding RNAs. Cell 172, 393–407. doi:10.1016/j.cell.2018.01.011
Li, F., Wen, H., Bukhari, I., Liu, B., Guo, C., Ren, F., et al. (2022a). Relationship between CNVs and immune cells infiltration in gastric tumor microenvironment. Front. Genet. 13, 869967. doi:10.3389/fgene.2022.869967
Li, S., Xie, X., Peng, F., Du, J., and Peng, C. (2022b). Regulation of temozolomide resistance via lncRNAs: Clinical and biological properties of lncRNAs in gliomas (Review). Int. J. Oncol. 61, 101. doi:10.3892/ijo.2022.5391
Li, Y., Xu, J., Shao, T., Zhang, Y., Chen, H., and Li, X. (2017). RNA function prediction. Methods Mol. Biol. 1654, 17–28. doi:10.1007/978-1-4939-7231-9_2
Li, Y., Zhao, J., Yu, S., Wang, Z., He, X., Su, Y., et al. (2019). Extracellular vesicles long RNA sequencing reveals abundant mRNA, circRNA, and lncRNA in human blood as potential biomarkers for cancer diagnosis. Clin. Chem. 65, 798–808. doi:10.1373/clinchem.2018.301291
Lorenzi, L., Chiu, H. S., Avila Cobos, F., Gross, S., Volders, P. J., Cannoodt, R., et al. (2021). The RNA Atlas expands the catalog of human non-coding RNAs. Nat. Biotechnol. 39, 1453–1465. doi:10.1038/s41587-021-00936-1
Mahinfar, P., Mansoori, B., Rostamzadeh, D., Baradaran, B., Cho, W. C., and Mansoori, B. (2022). The role of microRNAs in multidrug resistance of glioblastoma. Cancers (Basel) 14, 3217. doi:10.3390/cancers14133217
Mirzaei, S., Paskeh, M. D. A., Okina, E., Gholami, M. H., Hushmandi, K., Hashemi, M., et al. (2022). Molecular landscape of LncRNAs in prostate cancer: A focus on pathways and therapeutic targets for intervention. J. Exp. Clin. Cancer Res. 41, 214. doi:10.1186/s13046-022-02406-1
Najafi, S. (2022). Circular RNAs as emerging players in cervical cancer tumorigenesis; A review to roles and biomarker potentials. Int. J. Biol. Macromol. 206, 939–953. doi:10.1016/j.ijbiomac.2022.03.103
Najafi, S., Khatami, S. H., Khorsand, M., Jamali, Z., Shabaninejad, Z., Moazamfard, M., et al. (2022). Long non-coding RNAs (lncRNAs); roles in tumorigenesis and potentials as biomarkers in cancer diagnosis. Exp. Cell Res. 418, 113294. doi:10.1016/j.yexcr.2022.113294
Nasrolahi, A., Azizidoost, S., Radoszkiewicz, K., Najafi, S., Ghaedrahmati, F., Sheykhi-Sabzehpoush, M., et al. (2022). Long non-coding RNAs involved in retinoblastoma. J. Cancer Res. Clin. Oncol. 149, 401–421. doi:10.1007/s00432-022-04398-z
Paloviita, P., and Vuoristo, S. (2022). The non-coding genome in early human development - recent advancements. Semin. Cell Dev. Biol. 131, 4–13. doi:10.1016/j.semcdb.2022.02.010
Peng, W. X., Koirala, P., and Mo, Y. Y. (2017). LncRNA-mediated regulation of cell signaling in cancer. Oncogene 36, 5661–5667. doi:10.1038/onc.2017.184
Rizzo, A., Ricci, A. D., and Brandi, G. (2021). PD-L1, TMB, MSI, and other predictors of response to immune checkpoint inhibitors in biliary tract cancer. Cancers 13, 558. doi:10.3390/cancers13030558
Rong, H., Chen, B., Ma, K., Wei, X., Peng, J., and Zhu, J. (2021). Downregulation of lncRNA LINC-PINT participates in the recurrence of esophageal squamous cell carcinoma possibly by interacting miRNA-21. Cancer Biother Radiopharm. 36, 273–279. doi:10.1089/cbr.2019.3167
Salviano-Silva, A., Farias, T. D. J., Bumiller-Bini, V., Castro, M. S., Lobo-Alves, S. C., Busch, H., et al. (2021). Genetic variability of immune-related lncRNAs: Polymorphisms in LINC-PINT and LY86-AS1 are associated with pemphigus foliaceus susceptibility. Exp. Dermatol 30, 831–840. doi:10.1111/exd.14275
Salzberg, S. L. (2018). Open questions: How many genes do we have? BMC Biol. 16, 94. doi:10.1186/s12915-018-0564-x
Schmitt, A. M., Garcia, J. T., Hung, T., Flynn, R. A., Shen, Y., Qu, K., et al. (2016). An inducible long noncoding RNA amplifies DNA damage signaling. Nat. Genet. 48, 1370–1376. doi:10.1038/ng.3673
Simchovitz, A., Hanan, M., Yayon, N., Lee, S., Bennett, E. R., Greenberg, D. S., et al. (2020). A lncRNA survey finds increases in neuroprotective LINC-PINT in Parkinson's disease substantia nigra. Aging Cell 19, e13115. doi:10.1111/acel.13115
Singh, D., Assaraf, Y. G., and Gacche, R. N. (2022). Long non-coding RNA mediated drug resistance in breast cancer. Drug Resist Updat 63, 100851. doi:10.1016/j.drup.2022.100851
Statello, L., Guo, C. J., Chen, L. L., and Huarte, M. (2021). Gene regulation by long non-coding RNAs and its biological functions. Nat. Rev. Mol. Cell Biol. 22, 96–118. doi:10.1038/s41580-020-00315-9
Sun, Q., Tripathi, V., Yoon, J. H., Singh, D. K., Hao, Q., Min, K. W., et al. (2018). MIR100 host gene-encoded lncRNAs regulate cell cycle by modulating the interaction between HuR and its target mRNAs. Nucleic Acids Res. 46, 10405–10416. doi:10.1093/nar/gky696
Viggiano, E. (2022). Molecular research in medical genetics. Int. J. Mol. Sci. 23, 6625. doi:10.3390/ijms23126625
Wang, J., and Zhao, Q. (2020). LncRNA LINC-PINT increases SOCS1 expression by sponging miR-155-5p to inhibit the activation of ERK signaling pathway in rheumatoid arthritis synovial fibroblasts induced by TNF-α. Int. Immunopharmacol. 84, 106497. doi:10.1016/j.intimp.2020.106497
Wang, Y. H., Guo, Z., An, L., Zhou, Y., Xu, H., Xiong, J., et al. (2021). LINC-PINT impedes DNA repair and enhances radiotherapeutic response by targeting DNA-PKcs in nasopharyngeal cancer. Cell Death Dis. 12, 454. doi:10.1038/s41419-021-03728-2
Wen, H., Li, F., Bukhari, I., Mi, Y., Guo, C., Liu, B., et al. (2022). Comprehensive analysis of colorectal cancer immunity and identification of immune-related prognostic targets. Dis. Markers 2022, 7932655. doi:10.1155/2022/7932655
Xiang, X., Fu, Y., Zhao, K., Miao, R., Zhang, X., Ma, X., et al. (2021). Cellular senescence in hepatocellular carcinoma induced by a long non-coding RNA-encoded peptide PINT87aa by blocking FOXM1-mediated PHB2. Theranostics 11, 4929–4944. doi:10.7150/thno.55672
Xu, Y., Wang, H., Li, F., Heindl, L. M., He, X., Yu, J., et al. (2019). Long non-coding RNA LINC-PINT suppresses cell proliferation and migration of melanoma via recruiting EZH2. Front. Cell Dev. Biol. 7, 350. doi:10.3389/fcell.2019.00350
Yang, W., Zhao, P., Liu, Y., Cao, P., Ji, X., Gao, Y., et al. (2021). Transcriptome analysis of lncRNA expression patterns in human congenital lung malformations. BMC Genomics 22, 861. doi:10.1186/s12864-021-08204-x
Yue, J., Wu, Y., Qiu, L., Zhao, R., Jiang, M., and Zhang, H. (2021). LncRNAs link cancer stemness to therapy resistance. Am. J. Cancer Res. 11, 1051–1068.
Zhang, C., Gong, C., Li, J., and Tang, J. (2021). Downregulation of long non-coding RNA LINC-PINT serves as a diagnostic and prognostic biomarker in patients with non-small cell lung cancer. Oncol. Lett. 21, 210. doi:10.3892/ol.2021.12471
Zhang, D., Xue, J., and Peng, F. (2022). The regulatory activities of MALAT1 in the development of bone and cartilage diseases. Front. Endocrinol. (Lausanne) 13, 1054827. doi:10.3389/fendo.2022.1054827
Zhang, L., Chen, J., Wang, L., Chen, L., Du, Z., Zhu, L., et al. (2019). Linc-PINT acted as a tumor suppressor by sponging miR-543 and miR-576-5p in esophageal cancer. J. Cell Biochem. 120, 19345–19357. doi:10.1002/jcb.28699
Zhong, C., Xie, Z., Zeng, L. H., Yuan, C., and Duan, S. (2022). MIR4435-2HG is a potential pan-cancer biomarker for diagnosis and prognosis. Front. Immunol. 13, 855078. doi:10.3389/fimmu.2022.855078
Keywords: PINTology, LncRNA, LINC-PINT, Colon adenocarcinoma (CAC), Cancer biomarkers, Cancer therapy, Immune checkpoint inhibitors
Citation: Bukhari I, Khan MR, Li F, Swiatczak B, Thorne RF, Zheng P and Mi Y (2023) Clinical implications of lncRNA LINC-PINT in cancer. Front. Mol. Biosci. 10:1097694. doi: 10.3389/fmolb.2023.1097694
Received: 14 November 2022; Accepted: 24 February 2023;
Published: 17 March 2023.
Edited by:
William C. Cho, QEH, Hong Kong SAR, ChinaReviewed by:
Sajad Najafi, Shahid Beheshti University of Medical Sciences, IranNiaz Muhammad, Minhaj University Lahore, Pakistan
Abdullah Shah, Shaheed Benazir Bhutto University, Pakistan
Copyright © 2023 Bukhari, Khan, Li, Swiatczak, Thorne, Zheng and Mi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Pengyuan Zheng, cHl6aGVuZ0B6enUuZWR1LmNu; Yang Mi, eWFuZ21pMTk4QHp6dS5lZHUuY24=; Rick Francis Thorne, cmlja2Z0aG9ybmVAZ21haWwuY29t
 Ihtisham Bukhari
Ihtisham Bukhari Muhammad Riaz Khan
Muhammad Riaz Khan Fazhan Li1
Fazhan Li1 Rick Francis Thorne
Rick Francis Thorne