- Department of Pharmacy, Xuanwu Hospital, Capital Medical University, Beijing, China
As an evolutionarily phenotypic conversion program, the epithelial-mesenchymal transition (EMT) has been implicated in tumour deterioration and has facilitated the metastatic ability of cancer cells via enhancing migration and invasion. Gastric cancer (GC) remains a frequently diagnosed non-skin malignancy globally. Most GC-associated mortality can be attributed to metastasis. Recent studies have shown that EMT-related long non-coding RNAs (lncRNAs) play a critical role in GC progression and GC cell motility. In addition, lncRNAs are associated with EMT-related transcription factors and signalling pathways. In the present review, we comprehensively described the EMT-inducing lncRNA molecular mechanisms and functional perspectives of EMT-inducing lncRNAs in GC progression. Taken together, the statements of this review provided a clinical implementation in identifying lncRNAs as potential therapeutic targets for advanced GC.
Introduction
As the most frequently occurring malignancy worldwide, gastric cancer (GC) remains the fifth most diagnosed tumour and the third primary cause of tumour-related death (Sung et al., 2021). Approximately 1,089,103 people are diagnosed with GC worldwide each year, of whom about 783,000 die from this disease (Rawla and Barsouk, 2019; Yang L. et al., 2020). Asian countries, such as Japan, Mongolia, and Korea, show the highest incidence rates, with an estimated incidence rate per 100,000 of 48.1, 47.2, and 39.7, respectively (Morgan et al., 2022). GC is primarily divided into diffuse and intestinal types based on their histological characters (Lauren, 1965). Anatomically the two main types of GC are cardia and non-cardia subtypes. Additionally, the tumour, node and metastasis (TNM) system are used to assess tumour stage, including the tumour infiltration degree and size (T category), the lymph node status (N category), and the tumour distant metastasis to other organs (M category) (Amin et al., 2017). The important risk factors of the causes of GC are obesity (Kyrgiou et al., 2017), diabetes (Sona et al., 2018), smoking (Ladeiras-Lopes et al., 2008) and high salt intake (D'Elia et al., 2012). Although several risk factors are described, Helicobacter pylori infection-induced chronic inflammation is the most important known risk factor for GC (Huang et al., 1998; Uemura et al., 2001). The signs of GC in the early stage are vague and can remain undetected for years. Therefore, most GC patients are diagnosed late, often presenting tumour invasion or metastasis (Blum et al., 2013). In primary GC, the 5-year survival is 70%, whereas it is reduced to 32% amongst those with metastatic GC (Howlader N et al., 2020). In recent years, neoadjuvant chemotherapy, radiotherapy, and molecular-targeted regimen have become the mainstay of GC treatment (Digklia and Wagner, 2016; Song et al., 2017). Nevertheless, locally advanced, and metastatic GC patients still have a somber overall prognosis. Therefore, it is urgently necessary to further identify potential therapeutic targets to enhance the prognosis of patients with advanced GC.
Epithelial-mesenchymal transition (EMT) is a double-edged sword. EMT is a normal physiological process necessary for embryogenesis and wound healing. However, EMT dysregulation is a pathological process that result in cancer or fibrosis (Barriere et al., 2015; Yang J. et al., 2020). Gastrulation is an embryonic development of single layers embryo into three layers formation, of which the EMT is considered to be an important component. (Trelstad et al., 1967; Kim et al., 2017). EMT also provides a critical mechanism for the re-epithelialization of tissue which contributes to wound healing (Pastar et al., 2014). In addition, recent evidence has proven that the aberrant activation of EMT is closely linked with tumorigenesis, invasion, and metastasis of GC (Peng et al., 2014; Huang et al., 2015). As a complicated process, tumour metastasis mainly includes local invasion, intravasation, transport, and extravasation, by which malignant cells leave the primary tumour sites, sequentially colonize, and form secondary tumours at adjoining or distant organs, leading to GC-associated death (Yilmaz and Christofori, 2009). As a biological program, EMT triggers the detachment of polarized epithelial cells from neighbouring cells and converts them into mesenchymal cells, in which tumour cells forfeit epithelial polarity and transform into a mesenchymal phenotype, playing crucial roles in tumour invasion and metastasis (Kalluri and Weinberg, 2009). Epithelial markers (E-cadherin and α-catenin) and mesenchymal markers (N-cadherin, vimentin and fibronectin) are examined to determine whether cancer cells undergo the EMT (Zeisberg and Neilson, 2009). The GC patients with mesenchymal phenotype facilitate GC cell motility and metastasis and are correlated with advanced GC stage, while intestinal phenotype is mainly distributed in those at the early stage (Zheng et al., 2013). Accumulating evidence has revealed that the worst prognosis is associated with GC patients with EMT molecular subtype (Cristescu et al., 2015; Oh et al., 2018).
Non-coding RNAs (ncRNAs), including microRNAs (miRNAs), long non-coding RNAs (lncRNAs), and circular RNAs (circRNAs), can regulate the tumorigenesis of different tumours (Chan and Tay, 2018). Among these, lncRNAs dysregulation play an essential role in tumour metastasis and are conducive to the EMT (Bhan et al., 2017; Wei et al., 2020; Wang H. et al., 2021; Wang X. et al., 2021). Various lncRNAs are identified to enhance the migration, invasion of GC cells and promote the EMT process of GC (Gao et al., 2021; Li et al., 2022). Therefore, targeting lncRNAs has become a promising therapeutic regimen in patients with metastatic GC. In the present review, we summarised the regulatory functions of lncRNAs in the EMT-induced metastatic GC.
The characteristics and mechanism of lncRNAs’ action in EMT-induced metastasis
The first transcript sequence of lncRNAs was discovered in eukaryotes. LncRNAs are longer than 200 nucleotides in length and cannot be translated into proteins, and their primary structure is nucleotide sequence. Most of the lncRNAs share some similar features with messenger RNAs (mRNAs) and can be spliced, capped and polyadenylated by RNA polymerase II (Sun et al., 2018). In the past, traditional gene annotation filtered out proteins with <100 amino acids and treated them as noise (Wu et al., 2020). In recent years, research has revealed that lncRNAs contain short open reading frames (sORFs) which encode functional small peptides of approximately 100 amino acids in length using proteomics and translation technology (Zhu et al., 2018; Choi et al., 2019). These micro peptides resemble coding and noncoding genes and function as tumour regulatory factors to get involved in angiogenesis, signalling pathway transduction and metabolism in promoting cancer progression (Wu et al., 2020; Ye et al., 2020).
The functional activity of lncRNAs includes regulating transcription of neighbouring and distant genes, affecting the stability of mRNAs and interfering with signalling pathways through their crosstalk with DNA, RNA and proteins (Statello et al., 2021). Based on genomic localization and transcriptional orientation, lncRNAs are divided into intergenic lncRNAs, intronic lncRNAs, exonic lncRNAs, sense lncRNAs, and antisense lncRNAs (Ma et al., 2013). Based on the subcellular localization, lncRNAs are classified as nuclear lncRNAs and cytoplasmic lncRNAs (Kapranov et al., 2007). Nuclear lncRNAs mainly mediate the transcription (Sun et al., 2018), and cytoplasmic lncRNAs commonly participate in post-transcriptional regulation and function as miRNA sponges (Du et al., 2016).
LncRNAs are a highly heterogeneous group of epigenetic regulators that can mediate EMT-induced metastasis utilizing diverse mechanisms. First, lncRNAs can regulate gene expression at chromatin modification, transcription processing, and post-transcriptional processing (Dykes and Emanueli, 2017). LncRNAs recruit chromatin remodelling complexes to specific sites and coordinate genome activity by controlling chromosome structure and affecting histone status and DNA methylation status (Beckedorff et al., 2013; Böhmdorfer and Wierzbicki, 2015), which is significantly associated with the overall suvival and disease-specific survival of GC (Dai J. et al., 2021). After transcription, most lncRNAs are transcribed by RNA polymerase II and share some similarities with mRNAs, including 5′ end capping, 3’ end polyadenylating, splicing, and intracellular transporting (Canzio et al., 2019). Second, transcription factors (TFs) play an essential role in the development of GC. LncRNAs have been implicated in gene transcription through biding to EMT-inducing TFs (Long et al., 2017). The lncRNAs - TFs interaction directly regulates gene transcription via inducing the targeted protein degradation by phosphorylation and ubiquitination (Lamouille et al., 2014; Xiu et al., 2019). Thirdly, some identified lncRNAs shows oncogenic properties, acting as competing endogenous RNAs (ceRNAs) to sponge miRNA at the post-transcriptional level, as consequently, the interaction with targeted mRNA, thus increasiong the expression of oncogenic mRNA to facilitate the cancer occurrence and progression (Arun K. et al., 2018; Liu H. et al., 2018; Qi et al., 2020).
LncRNAs regulate EMT-induced metastasis by mediating the signalling pathways
EMT is induced by a variety of signalling pathways, including TGF-β, BMP, Wnt-β-catenin, NOTCH, Shh, and receptor tyrosine kinases, and many feedback activation/inhibition mechanisms have been demonstrated (Deshmukh et al., 2021). In the process above, lncRNAs regulate the EMT process in GC by mediating various signalling pathways, including Wnt, PI3K/AKT, Hippo, MEK/ERK, and Notch1 (Table 1).
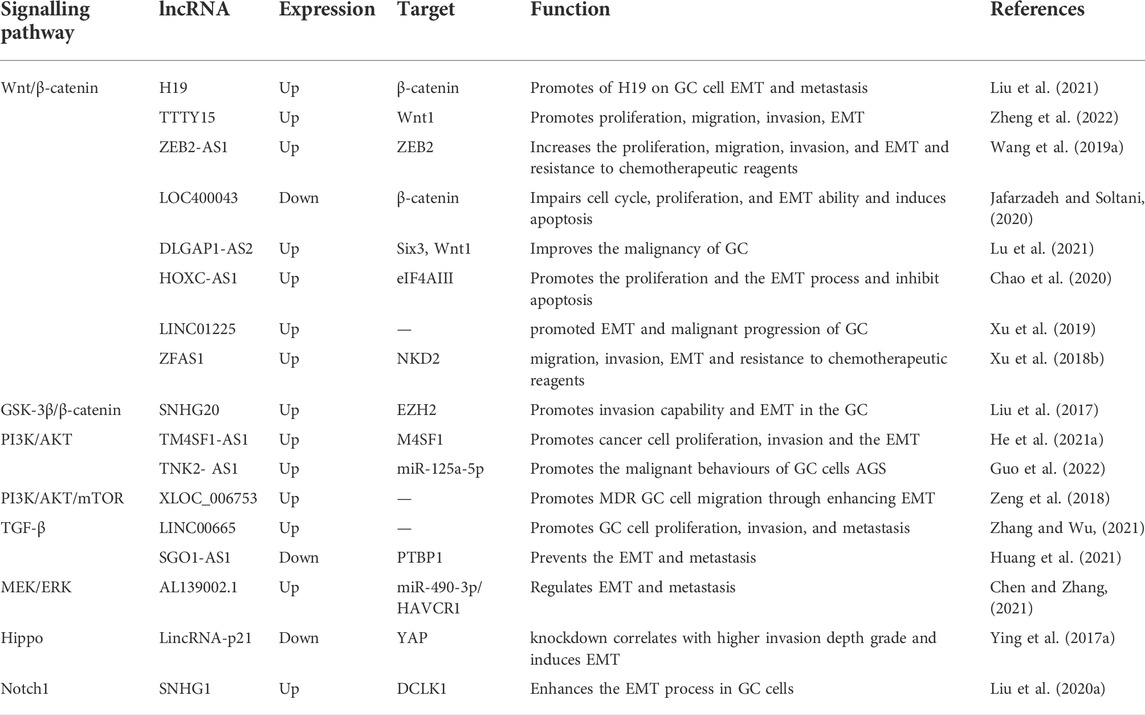
TABLE 1. LncRNAs and their targeting signalling pathways in the regulation of EMT-induced GC metastasis.
Wnt is a secretory glycoprotein that acts by autocrine or paracrine. After secretion, Wnt can bind to cell surface-specific receptors, leading to β-catenin accumulation via phosphorylating and dephosphorylating a series of downstream proteins. As a multifunctional protein, β-catenin reacts with E-cadherin at the cell junctions and is involved in the formation of adhesive bonds. Free β-catenin can reach the nucleus and get involved in gene expression, while its dysfunction or activation can trigger tumorigenesis. The primary elements of the Wnt signalling pathway consist of secreted protein Wnt family, transmembrane receptor Frizzled family, casein kinase (CK1), Dishevelled (DVL), adenomatous polyposis coli (APC), Axin, glycogen synthase kinase 3-beta (GSK3β), β-catenin and TF transcription factor T-cell factor/lymphoid enhancer-binding factor (TCF/LEF) family (MacDonald et al., 2009; Zhan et al., 2017; Albrecht et al., 2021).
LncRNA H19 is increased in different malignancies, and it functions as an oncogene. H19 is overexpressed in GC and associated with poor prognosis. H19 can transfer β-catenin into the nucleus and activate Wnt/β-catenin signalling, facilitating EMT of GC cell (Liu et al., 2021). LncRNA ViM antisense RNA 1 (VIM-AS1) is highly expressed in GC and associated with prognostic outcomes. VIM-AS1 may enhance cell migration, invasion, and EMT by mediating frizzled 1(FDZ1), and activating the Wnt/β-catenin pathway (Sun et al., 2020). The expression of lncRNA Zinc finger E-box-binding homeobox two antisense RNA 1 (ZEB2-AS1) is up-regulated in GC specimens, down-regulated of ZEB2-AS1 can suppress the proliferation, EMT, and Wnt/β-catenin signalling (Wang F. et al., 2019). LncRNA testis-specific transcript, Y-linked 15 (TTTY15) (Zheng et al., 2022), lncRNA DLGAP1 antisense RNA 2 (DLGAP1-AS2) (Lu et al., 2021), and lncRNA HOXC cluster antisense RNA 1(HOXC-AS1) (Zhou C. et al., 2020) are also overexpressed and can regulate the Wnt/β-catenin signalling pathway to promote EMT in GC.
GSK-3β is a serine/threonine-protein kinase. In the absence of Wnt signalling, phosphate groups can be added to n-terminal serine/threonine residues of β-catenin by GSK-3β. After covalent modification by β-TRCP ubiquitination, the phosphorylated β-catenin is degraded (Stamos and Weis, 2013). LncRNA small nucleolar RNA host gene 20 (SNHG20) acts as an oncogene in GC. The expressions of E-cadherin and p21 can be markedly suppressed when SNHG20 is overexpressed in MKN45 and BGC-823 cells via binding to the enhancer of zeste homolog 2 (EZH2) and mediating the GSK-3β/β-catenin signalling pathway, and SNHG20 can be a therapeutic target for GC (Liu et al., 2017).
The AKT, also called protein kinase B, signalling pathway is involved in the molecular mechanisms underlying many cancers, which plays a vital role in tumor cell proliferation, metastasis and drug resistance, lncRNAs can regulate the relative expressions of key genes in the phosphoinositide 3-kinase (PI3K)/AKT pathway (Peng et al., 2017; Lin et al., 2020). The PI3K/Akt signalling pathway can impact the EMT in various manners to alter the aggressiveness of cancer (Xu et al., 2015). GC tissues and cells exhibit increased expression of lncRNA Transmembrane four superfamily 1-antisense 1 (TM4SF1-AS1). However, the proliferation, invasion, EMT and enhanced apoptosis of cancer cells can be inhibited when TM4SF1-AS1 is depleted. The underlying mechanism is associated with the suppression of TM4SF1 and PI3K-AKT signalling pathways (He C. et al., 2021).
Transforming growth factor-β (TGF-β) family is a group of structurally related proteins, is involved many cellular functions, including EMT and migration, and many human diseases, including vascular diseases, autoimmune disorders, and carcinogenesis (Syed, 2016). LncRNA Shugoshin-like protein 1-antisense 1 (SGO1-AS1) facilitates TGF-β1/2 mRNA decay by competitively binding to the PTBP1 protein, leading to impaired TGF-β production, and preventing EMT and metastasis (Huang et al., 2021). Extracellular signal-regulated kinase (ERK) cascade can regulates proliferation, differentiation, survival, and apoptosis of cells, the mitogen extracellular signal-regulated kinase (MEK) functions as an upstream essential protein of ERK (Liu W. et al., 2015). LncRNA AL139002.1 is highly expressed in GC cells, and lncRNA AL139002.1/miR-490-3p/HAVCR1 functions critically in GC by mediating the MEK/ERK signalling (Chen and Zhang, 2021). The Hippo pathway can mainly restrict adult tissue growth and regulate cell proliferation, differentiation, and migration in developing organs. In addition, abnormal cell growth and neoplasia are observed when the Hippo pathway is dysregulated (Meng et al., 2016). LINC00649 acts as an oncogene to promote the EMT by targeting the miR-16-5p/YES-associated protein 1 (YAP1)/Hippo signalling pathway (Wang H. et al., 2021).
The Notch family consists of four highly conserved transmembrane receptors. Enzyme activity of G-secretase is required to release active regions within cells. Notch is involved in many physiological processes of embryonic development and normal cells, regulating cell growth, apoptosis, and differentiation. Notch1, a member of the Notch family, has been linked to various cancers (Gharaibeh et al., 2020). LncRNA small nucleolar RNA host gene 1(SNHG1) is overexpressed in GC and regulates the EMT process and cell migration by miR-15b/DCLK1/Notch1 axis (Liu Z. Q. et al., 2020).
LncRNAs regulate EMT-induced metastasis in GC through transcription factors
TFs also known as DNA-binding factors, control the transcription from DNA to RNA via binding to a specific DNA sequence (Latchman, 1993). TFs regulates RNA polymerase activity via interacting with two classes of surface domain: a sequence-specific DNA binding domain (i.e., zinc finger and homeodomain) and an activation domain that binds to various cofactors to recruit RNA polymerase (Bhagwat and Vakoc, 2015; Mulero et al., 2018). LncRNAs exist in both cytoplasm and nucleus and their functions are activated by two main mechanisms. In the nucleus, lncRNAs bind to TFs directly by interacting with DNA to regulate the transcription of GC metastasis-related genes (Fatima et al., 2015). In the cytoplasm, lncRNAs bind to tissue-specific protein, altering the post-translational modification to induce the protein ubiquitination and degradation (Table 2) (Liao et al., 2021).
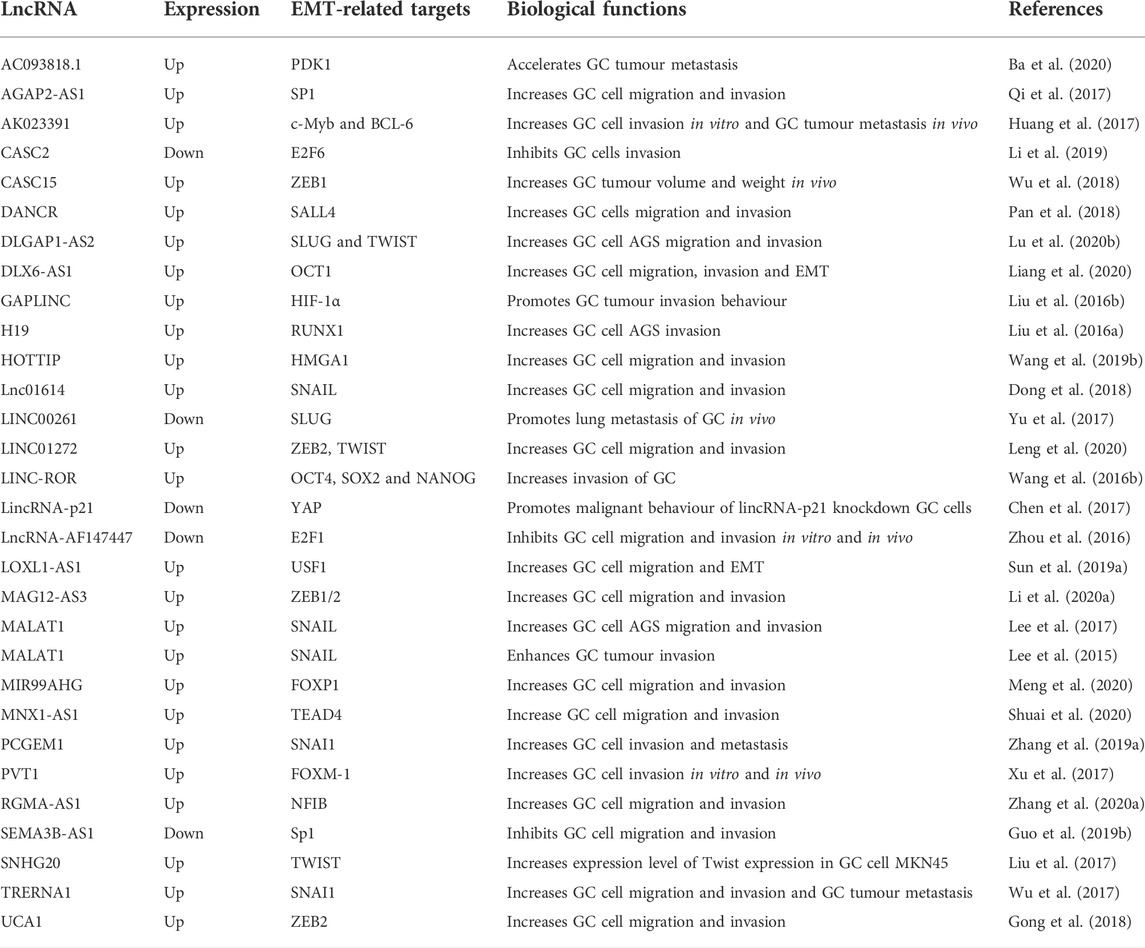
TABLE 2. LncRNAs and their associated transcription factors in the regulation of EMT-induced metastasis in gastric cancer.
LncRNAs participate in tumour progression and metastasis by modulating EMT (Xu et al., 2016). In addition, lncRNAs function critically in the induction and regulation of EMT-TFs (Pavlič et al., 2022). The loss of expressions of the cadherin family proteins remains the hallmark of EMT, which is crucial in cell-cell adherents junctions (Wang and Zhou, 2013). During EMT, decreased expression of E-cadherin translocate β-catenin to the nucleus and activates numerous notable TFs, including SNAIL, SLUG, Twist-related protein 1 (TWIST1), zinc-finger E-box-binding homeobox 1 (ZEB1), and 2 (ZEB2) (Chan and Wang, 2015; Stemmler et al., 2019).
SNAIL (also known as SNAI1), a zinc-finger transcriptional repressor, modulates EMT during tumour progression (Wang et al., 2013). It binds to E-box, an E-cadherin promoter region, which converts epithelial cells to mesenchymal cells (Villarejo et al., 2014). The overexpression of SNAIL up-regulates XBP1 (Li et al., 2015) and ALX1 (Yuan et al., 2013), subsequently inducing the activation of EMT. The Notch activity (Timmerman et al., 2004) and Wilms’ tumour one homolog (Wt1) (Martínez-Estrada et al., 2010) promote EMT through transcriptional induction of the SNAIL repressor. Additionally, overexpression of SNAIL suppresses miR-192 and miR-194 but up-regulates miR-205, let-7i, and SNORD13. Those identified changes are correlated with the initiation of SNAIL-mediated EMT in cancer cells (Przygodzka et al., 2019). Depleting lnc01614 in GC cells (SGC7901 and AGS) exhibits attenuated migration and invasion caused by decreased SNAIL expression (Dong et al., 2018). Lee and their colleagues have also confirmed that siR-MALAT1 reduces gastric tumorigenesis by inhibiting invasiveness via reduced expression of SNAIL (Lee et al., 2017). The lncRNAs TRERNA and PCGEM1 function as enhancers of SNAI1 to contribute to the metastasis of GC (Zhang J. et al., 2019).
In addition to SNAIL, SLUG is another notable SNAIL superfamily of zinc-finger TFs. It acts as a transcriptional repressor to bind to E-box to mediate the expressions of target genes responsible for the EMT (Stegmann et al., 1999). The expression of SLUG is controlled by Tbx18 and Wt1, which regulate EMT activation (Takeichi et al., 2013). Moreover, the high-mobility group AT-hook 2 (HMGA2) has a positive correlation with SLUG expression in EMT activation (Li et al., 2014). The work performed by Lu and their colleagues has demonstrated that depletion of DLGAP1-AS2 suppresses the migration and invasion of GC cells AGS via down-regulating SLUG (Lu J. et al., 2020).
As a primary helix-loop-helix TF, TWIST participates in recognizing E-box elements. Overexpression of TWIST plays an essential role in promoting EMT (Chava et al., 2019). TWIST activation is controlled by several signalling pathways, such as Akt (Tang et al., 2016) and STAT3 (Zhang et al., 2015). Overexpression of TWIST up-regulates miR-214 to facilitate the EMT process (Liu C. et al., 2018). The in vitro investigation has indicated that inhibition of LINC01272 and DLGAP1-AS2 attenuates the migration and invasion of GC cells. Exposure of GC cells to LINC01272 siRNA and DLGAP1-AS2 siRNA significantly inhibits the expression of TWIST (Lu J. et al., 2020; Leng et al., 2020).
ZEB1/2 encodes zinc finger and down-regulates E-cadherin to induce EMT in carcinomas (Bürglin and Affolter, 2016). Li and their colleagues have confirmed that the expression of ZEB1/2 is significantly inhibited in MAG12-AS3-depleted GC cells (Li D. et al., 2020). In another case, lncRNA UCA1 contributed to GC metastasis via regulating miR-203/ZEB2 axis (Gong et al., 2018).
In addition to classic EMT-TFs described above, lncRNAs also regulate some other TFs to influence the progression and metastasis of GC. For example, overexpression of forkhead box M1 (FOXM1) enhances GC cell motility, and this effect can be reversed by blocking Cath-D (Yang et al., 2017). Cytoplasmic lncRNA plasmacytoma variant translocation 1 (PVT1) is a valuable prognostic predictor in GC. High PVT1 expression promotes the invasiveness of GC cell lines through binding to FOXM1 protein which implicate in high TNM staging and lymph node metastasis (Xu et al., 2017). The lncRNA MNX1-AS1 enhances the migration and invasion of GC cells in vitro. TEA domain DNA-binding family of TF 4 (TEAD4) acts as an oncogene to mediate Hippo signalling driving cancer progression. By binding to TEAD4, MNX1-AS1 promotes tumorigenesis through up-regulating BCL2 expression (Shuai et al., 2020). Additionally, the study performed by Zhou et al. have shown that the EMT-induced lncRNA AF147447 negatively regulates the expression of E2F1 and promotes GC tumour metastasis via the miR-34c/MUC2 axis (Zhou et al., 2016).
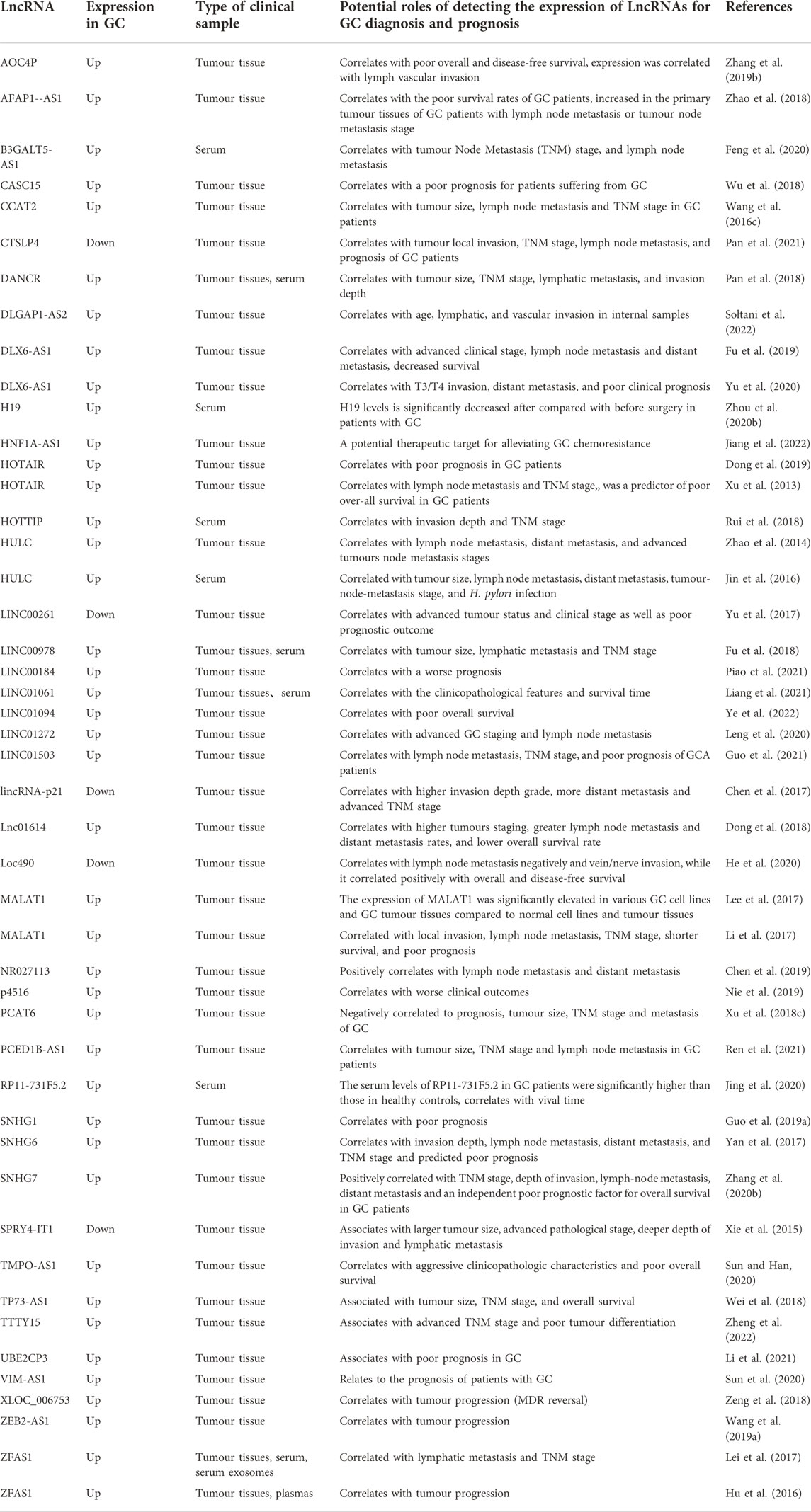
LncRNAs regulate EMT-induced metastasis in GC through sponging miRNAs
miRNAs are a group of small RNAs with approximately 22 nt in length. miRNAs bind to the complementary sequence in targeted mRNAs, leading to the degradation of targeted mRNAs via RNA-induced silencing complex (RISC). Like lncRNAs, miRNAs play a critical role in different tumours. Many miRNAs are significantly up-regulated in cancer cells, resulting in cancer development. Several miRNAs even mediate the progression of various tumours (Karagkouni et al., 2021). In the study on the regulation of gene expression, miRNA and lncRNA are vital links. One of the widely recognized types is the endogenous competition mechanism. Different from directly regulating target genes, some lncRNAs inhibit the degradation or inhibition effect of miRNAs on target genes by binding to miRNAs. Such a regulatory strategy is widely reported in GC. LncRNA metallothionein 1 J, pseudogene (MT1JP) sponges miR-92a-3p and mediates the downstream F-Box-WD Repeat-Containing Protein 7(FBXW7) gene, which in turn impacts the progression of GC (Zhang G. et al., 2018). LINC01234 functions as the ceRNA of miR-204-5p and blocks the activation of core-binding factor b (CBFB) in GC (Chen et al., 2018). Some lncRNAs that regulate the target genes and are involved in the EMT progress of GC via the competitive binding of lncRNAs and miRNAs are summarized in Table 3.
Some miRNAs can regulate the expressions of TFs, while lncRNAs can suppress the degradation of target genes by binding these miRNAs to promote the expressions of TFs, leading to the promoted EMT process of GC. SNHG1 promotes the proliferation and invasion of GC cells via modulating the miR-140/ADAM10 axis (Guo et al., 2019a). GC cell lines display markedly high expressions of lncRNAs small nucleolar RNA host gene (SNHG3) and TWIST. Depletion of SNHG3 significantly inhibits the proliferation, migration, and invasion of GC cell lines. SNHG3 acts as an endogenous sponge to reduce the expression of miR-326 and regulates the expression of TWIST by competitively binding to miR-326 (Rao et al., 2021). Overexpression of small nucleolar RNA host gene 7 (SNHG7) has been reported in most human tumors, including lung cancer, and it acts as an oncogenic lncRNA in GC and may be a promising therapeutic candidate for GC patients. SNHG7 promotes the migration and invasion of GC cells by inhibiting miR-34a (Zhang Y. et al., 2020). As a potential oncogene, small nucleolar RNA host gene 6 (SNHG6) is involved in the initiation and progression of hepatocellular carcinoma, and SNHG6 functions as an oncogene in GC cells by post-transcriptionally mediating and transcriptionally silencing miR-101-3p/ZEB1 via recruiting EZH2 to the promoter of p27 (Yan et al., 2017).
Emerging evidence has shown that EMT plays a critical role in the chemoresistance of tumor cells. The resistance of lung cancer cells to doxorubicin can be effectively reversed by inhibiting EMT (Ying Y. et al., 2017). EMT is associated with treatment resistance (Gaianigo et al., 2017), suppressing EMT may enhance the chemosensitivity. SNHG6 positively regulates B-Cell Lymphoma 2 (BCL-2) by sponging miR-1297. The DDP resistance, proliferation, and metastasis of DDP-resistant cells can be suppressed by depletion of SNHG6 (Mei et al., 2021). Exosome HOTTIP can regulate the miR-218/high-mobility group A1 (HMGA1) axis, contributing to cisplatin resistance in GC cells. HOTTIP regulates HMGA1 by acting as a ceRNA of miR-218 in GC cells. Serum exosome HOTTIP has been related to cisplatin resistance in GC patients (Wang et al., 2019b), lncRNA H19 suppresses the chemosensitivity to ADM via sponging miR-152 from TCF4 in GC cells (Jiang et al., 2020). HNF1A-AS1 promoted chemoresistance by facilitating EMT process through upregulating EIF5A2 expression by sponging of miR-30b-5p (Jiang et al., 2022).
LncASNR (apoptosis suppressing-non-coding RNA) inhibits the expression of miR-519e-5p but up-regulates fibroblast growth factor receptor 2 (FGFR2). As a receptor for FGF, FGFR2 can deliver the FGF signal to RAS-ERK and PI3K-AKT signal cascades, facilitating EMT-related migration and invasion of GC cells (Chen Z. et al., 2021). Overexpression of lncRNA HLA complex group 18 (HCG18) induced by hepatocyte nuclear factor 1 homeobox A (HNF1A) promotes GC progression by competitively binding to miR-152-3p and up-regulating DNAJB12. HNF1A can facilitate its transcription by binding to the HCG18 promoter. The DNAJB12 and cytosolic heat shock protein 70 (Hsp70) can promote the triage of nascent polytopic membrane proteins for folding or degradation by cooperating on the endoplasmic reticulum’s cytoplasmic face (Ma et al., 2020). GC tissues and cell lines show high expression of lncRNA PCED1B antisense RNA 1 (PCED1B-AS1), and its expression has been linked to the clinicopathological characteristics of GC patients. Moreover, PCED1B-AS1, as a ceRNA, up-regulates C-X-C motif chemokine receptor 1 (CXCR1) by competitively binding to miR-215-3p, leading to enhanced malignancy of GC cells, and this finding indicates that PCED1B-AS1/miR-215-3p/CXCR1 axis may be a potential mechanism involved in the progression of GC (Ren et al., 2021). CXCR1 can regulate the malignant biological behaviors of cancer cells by controlling the activation of AKT and ERK1/2 signaling pathways. Depletion of CXCR1 up-regulates E-cadherin in GC cells (Wang J. et al., 2016). Nuclear receptor subfamily two group F member 1-antisense RNA 1 (NR2F1-AS1)/miR-190a/Pleckstrin Homology Like Domain Family Member 2 (PHLDB2), a ceRNA, can facilitate the EMT process of GC cells, and PHLDB2 can enhance the expression and phosphorylation of AKT3 to promote the EMT process of GC cells (Lv et al., 2021). LINC00689/miR-526b-3p/A disintegrin and metalloproteinase domain 9 (ADAM9) participates in many biological processes, including myogenesis, fertilization, cell migration, inflammatory response, proliferation, and cell-cell interactions (Yin et al., 2020). miR-338-3p has a negative correlation with LINC00689 in GC. Homeobox A3 (HOXA3) is one target gene of miR-338-3p, and ectopic expression of LINC00689 inhibits miR-338-3p and up-regulates HOXA3 in GC cells (Lu H. et al., 2020). HOXA3 can activate EGFR/Ras/Raf/MEK/ERK signalling pathway, promoting the tumor growth of colon cancer. LINC00689 functions as a ceRNA by sponging miR-526b-3p in GC cells (Zhang X. et al., 2018).
As growth factors, cell signal transducers, and nuclear TFs, proto-oncogenes primarily regulate biological activities in normal cells. Changes in these genes affect their encoded proteins, becoming oncogenes, which drive cell proliferation and play a critical role in tumorigenesis (Kontomanolis et al., 2020). The expression of HOTAIR is often increased in GC tissues and cell lines, and a high expression of HOTAIR has been linked with poor prognosis in GC patients. HOTAIR can sponge miR-217 and inhibit its expression in GC. HOTAIR can facilitate the development of GC by up-regulating glypican-5 (GCP5) via sponging miR-217 (Dong et al., 2019). GPC5 is an oncogene and may play a critical role in regulating tumorigenesis. Early studies have confirmed that miR-217 functions as a cancer suppressor by directly targeting the GPC5 oncogene in GC (Wang et al., 2015). HOTAIR can interact with polycomb repressive complex 2 (PRC2), thereby mediating the downstream targets via epigenetic regulation. HOTAIR also binds to PRC2 to activate its target genes C-Met (HGF/C-Met/Snail pathway) and Snail via epigenetically decreasing the expression of miR34a, thereby facilitating EMT in advanced stages of GC (Liu Y. W. et al., 2015).
Depletion of LINC00649 inhibits YAP1 and releases miR-16-5p, leading to the recovery of the Hippo pathway, and some downstream oncogenes are suppressed accordingly, such as EGFR, SOX2, and OCT4, suppressing the malignant phenotypes in GC cells (Wang H. et al., 2021). YAP1 has been identified as an oncogene, which can promote the pathogenesis of multiple cancers and immunosuppression (He S. et al., 2021; Wang and Gao, 2021). LncRNA metastasis associated lung adenocarcinoma transcript 1 (MALAT1) promotes cell proliferation and invasion in GC, and its up-regulation is associated with local invasion, lymph node metastasis, and TNM stage. MALAT1 is negatively correlated with miR-1297 and functions as a molecule sponging miR-1297, antagonizing its ability to inhibit the expression of high mobility group box 2 (HMGB2) (Li et al., 2017). HMGB2 is an essential protein in carcinogenesis, and it is associated with increased proliferation, invasion, and glycolysis of GC cells (Cui et al., 2019). FAM225A (Chen N. et al., 2021), HCP5 (Gao et al., 2021), MIAT (Li X. M. et al., 2020), and UBE2CP3 (Li et al., 2021) all can promote the expressions of oncogenes.
In normal cells, there are tumor suppressor genes besides oncogenes. Tumor suppressor genes play a fundamental role in the normal growth and differentiation of cells. They protect the body from tumor invasion, block tumor growth, and promote the normal development of cells (Kontomanolis et al., 2020). LncRNA actin alpha 2, smooth muscle antisense RNA 1 (ACTA2-AS1) can suppress malignant phenotypes of GC cells as it can function as a ceRNA to bind to miR-378a-3p and antagonize the inhibitory impacts of miR-378a-3p on the expression of phosphatidylinositol-specific phospholipase C X domain containing 2 (PLCXD2) (Liu et al., 2022). PLCXD2 is linked to an enhanced risk of esophageal squamous cell carcinoma in the Han Chinese population (Wang et al., 2019c). LncRNA MRI503HG up-regulates tumour suppressor candidate 3 (TUSC3) in GC cells through sponging miR-224-5p, leading to GC progression. Depletion of ATF6 partially rescues EMT in GC cells overexpressing lncRNA MIR503HG. GC cell invasion is inhibited by overexpressing lncRNA MIR503HG, which reduces protein contents of N-cadherin and vimentin to hinder EMT in GC cells (Lin et al., 2021). Overexpression of TUSC3 impedes cell proliferation and triggers apoptosis in retinoblastoma (Kong et al., 2020).
As an essential epigenetic regulation, methylation can be described as the transfer of the active methyl group to the target chemicals catalyzed by methyltransferases, and the DNA sequence composition remains unchanged in this process. Methylation deregulation is involved in many diseases, including human cancers (Dai X. et al., 2021). Ubiquitination regulates several steps in autophagy via post-translational modification, which is a primary lysosome-mediated intracellular degradation pathway. Multiple ubiquitin chains act as selective markers to attach to protein aggregates and dysfunctional organelles, thereby accelerating the degradation in an autophagy-dependent manner (Grumati and Dikic, 2018). Overexpression of lncRNA prostate cancer-associated transcript 6(PCAT6) has been reported in GC, which facilitates the progression of GC by endogenous competition with miRNA-30 by targeting makorin ring finger protein 3 (MKRN3). MKRN3 participates in the processes of gene transcription and ubiquitination. The invasive ability of GC cells is enhanced when PCAT6 is overexpressed. The expressions of EMT-related genes at the protein level are also remarkably increased (Xu Y. et al., 2018). LncRNA The C-C motif chemokine ligand 2 (CCL2) suppresses the expression of miR-128 in GC. miR-128 mimic significantly down-regulates the expression of poly (ADP-ribose) polymerase 2 (PARP2). As a leading member of the PARP family, PARP2 possesses multiple biological functions, such as DNA repair, synthetic lethality, apoptosis, necrosis, and histone binding (Ma et al., 2022). LINC00240 can bind to miR-124-3p as a ceRNA to enhance the expression of DNA methyltransferase 3b (DNMT3B), a member of the DNMT family. DNMT3B can enhance cell proliferation, invasion, and migration of GC cells. siRNA targeting LINC00240 up-regulates E-cadherin and down-regulates vimentin in GC SGC-7901 cells and BGC-823 cells (Li Y. et al., 2020).
In addition to the widely recognized endogenous competition mechanisms, there are a few other mechanisms of lncRNAs and miRNAs. For example, miR-21 is negatively mediated by lncRNA Maternally expressed gene 3 (MEG3) and can enhance metastasis in GC. The MEG3/miR-21 axis is involved in the progression and metastasis of GC through mediating EMT (Xu G. et al., 2018).
Diagnostic and prognostic value of EMT-related LncRNAs in metastatic gastric cancer
Most GC--associated mortality is attributed to tissue metastasis. GC preferably metastasizes to the liver accounting for 48% of metastatic GC patients. Moreover, the other common sites for GC to spread include the peritoneum, lung and bone, accounting for 32%, 15% and 12% of patients with metastatic cancer, respectively (Riihimäki et al., 2016). At present, the combined chemotherapy protocols, such as FOLFOX (oxaliplatin and 5-FU/leucovorin), CAPOX (oxaliplatin and capecitabine) and FOLFIRI (irinotecan and 5-FU/leucovorin) are the most commonly regimen of chemotherapy treatment for GC. In addition, targeted drugs (Trastuzumab, Ramucirumab, Larotrectinib and Entrectinib) might be helpful in GC patients with gene over-expression and mutation. Growing evidence has shown that immunotherapy is a promising treatment for GC. FDA approved nivolumab and pembrolizumab, in combination with chemotherapy, for the treatment of patients with locally advanced or metastatic GC (Takei et al., 2022). Although the availability of numerous drugs for the treatment of GC, 39% of GC patients were found to have metastatic disease and had a poor prognosis (Dai W. et al., 2021). Thus, there is an urgent need to find potential valuable biomarkers for prognosis of patients with metastatic GC.
Carbohydrate antigen (CA) 12-5 and CA 72-4 are the most frequently used biomarkers in diagnosis of patients with GC (Matsuoka and Yashiro, 2018). In addition, carcinoembryonic antigen (CEA) and CA 19-9 act as the prognostic predictors in clinical practice, as they have not detected in the early stage of GC (Feng et al., 2017). Therefore, some novel reliable markers supporting diagnosis and prognosis of GC are needed. Previous study has demonstrated that circulating lncRNAs is detectable in plasma, and it significant increase in GC patients than that of health donors (Arita et al., 2013). Therefore, lncRNAs are adopted to diagnose tumors at early stages, and they can also predict the prognosis, metastasis risks, and recurrence after surgery (Necula et al., 2019).
As diagnostic biomarkers, some lncRNAs are differentially expressed in the serum and plasma of GC patients and normal patients. For example, using gastric juice ABHD11-AS1 as a marker, ABHD11-AS1 levels were significantly increase in early GC patients, reaching to 71.4% (Yang Y. et al., 2016). Serum B3GALT5-AS1 levels were significantly higher in GC patients than that of in normal individuals. High serum B3GALT5-AS1 levels were also associated with TNM stage and lymph node metastasis (Feng et al., 2020). The level of serum exosome lncRNA H19 in GC patients was significantly up-regulated before and after surgery when compared with that in healthy controls, and the postoperative level was significantly lower than that before operation. Preoperative lncRNA H19 levels were significantly correlated with TNM stage. The area under the ROC curve (AUC) value of exosome lncRNA H19 was significantly higher than the AUC value of cancer antigen 19-9, 72-4 and carcinoembryonic antigen (Feng et al., 2020). The AUC of exosome HOTTIP was 0.827, and its diagnostic ability was significantly higher than that of CEA, CA 19-9 and CA72-4 and exosome HOTTIP overexpression was an independent prognostic factor in GC patients (Feng et al., 2020). In addition, several studies have investigated the effects of H. pylori infection on GC progression by regulating lncRNAs expression. For example, lncRNA AF147447 decreased expression by H. pylori infection and acts as a tumour suppressor in the development of GC (Zhou et al., 2016). Li and their collegues addressed a significant associations with high expression of lncRNA51663 and FLJ46906 and increased risk of H. pylori infection-induced GC (Li N. et al., 2020). In addition, serum H19 and LINC00152 could function as potential biomarkers for GC patients with H. pylori infection due to the joint effect of H19 and LINC00152 and H. pylori infection on the increased risk of GC (Yang T. et al., 2016).
Accumulating evidence has shown that lncRNAs can act as prognostic biomarkers in predicting GC tumor size and Lauren classification, depth of invasion, Lymph node and distant metastasis, TNM stage. Highly expressed lncRNA DANCR was tested in tumour tissues and serum form GC patients than that of from healthy controls (Pan et al., 2018). Moreover, HOTAIR expression is significantly elevated in GC tissues when compared with adjacent non-cancer tissues. This study also confirmed the association of HOTAIR overexpression with poor overall survival in patients with diffuse-type GC (Petkevicius et al., 2022). Yang and their colleagues have performed study to determine the expression of lncRNA ABHD11-AS1 in gastric juice from GC patients relate to tumour size, tumour stage, Lauren type and blood CEA level (Yang Y. et al., 2016). In addition, elevated expression of lncRNA M26317 might be a potential biomarker that correlate with Lauren’s classification, lymph node and distant metastasis (Li et al., 2018). The lncRNAs RP11-119F7.4, C5orf66-AS1 and DLEU2 were differentially expressed in GC tissue and non-tumour gastric tissue, and were predominantly correlated with Lauren histologic classification of GC (Sun et al., 2015; Zhou Q. et al., 2020; Hu et al., 2022). In addition to this, some lncRNAs expressed in some specific tissue (i.e. GAS5 and H19 expressed in embryo tissue) that can be targeted using nucleic acid-based drugs, small molecule inhibitors, and gene-editing methods at different functional levels to provide various therapeutic options (Arun G. et al., 2018).
In clinical practice, the lncRNAs expression might be tested in patients’ fluid samples from whole blood, serum or plasma, and tissue samples from gastric carcinoma tumour tissue and surrounding tissues or adjacent non-cancer tissues using qRT-PCT technique, to diagnose and predict the lymphatic metastasis and distal metastasis of GC. The single lncRNAs or combined lncRNAs or combined lncRNAs with the well-established biomarkers (CA12-5, CA72-4 and CA19-9) are promising biomarkers for assessing the diagnosis and prognosis of advanced gastric carcinoma. According to present studies, lncRNAs has potential valuable of being the biomarker for patients with GC in clinical settings. Therefore, the effects of lncRNAs on diagnosis and prognosis of GC are summarized in Table 4.
Perspectives
LncRNAs play an important role in the development of GC. Some identified oncogenic lncRNAs overexpressed in gastric cancerous tissue, such as H19, HOTAIR, and MALAT1, whereas others are expressed in the tissue from gastric carcinoma at a low level, such as LincRNA-p21, LINC00261, CTLSP4 and SPRY4-IT1. LncRNAs regulate EMT process by targeting EMT-related signalling pathways directly (i.e., H19, HOTAIR, ZEB2-AS1, LincRNA-p21 and SNHG1), or function as ceRNAs (i.e., H19, HOTTIP, MALAT1, SNHG1 and SNHG6) for tumour suppressive miRNAs. Furthermore, dysfunction of lncRNAs regulates apoptosis and cell cycle in GC cell lines, for example, SNHG6 function as a positive regulator for BCL-2 gene expression. In addition, SNHG6 implicated in initiation and EMT-induced metastasis of GC by regulating ZEB1 expression. Dysregulated lncRNAs (SNGH6, HOTTIP, H19, HNF1A-AS1 and ZFAS1) exert the functional role in chemoresistance leading to enhanced EMT ability in GC cell lines and tissues. Aberrant expression of H19 is involved in progression and metastasis in numerous cancer through regulating various of targeted genes. For example, it regulates VEGF expression by competitively binding miR-138 in glioma (Liu Z. Z. et al., 2020). H19/miRNA-140 axis promotes ovarian cancer cell migration by upregulating Wnt1 expression (Wang and Gao, 2021). Additionally, H19 upregulates PFTK1 expression through targeting miR-194 in pancreatic cancer cells (Sun Y. et al., 2019). Since targeting lncRNAs are currently under development by researchers, H19 might be a promising target in the treatment of patients with advanced GC.
In addition to being a potential therapeutic target for GC, another important clinical value of lncRNAs is as a diagnostic marker (differentially expression in GC and surrounding tissues or in GC patients and health individuals) or prognostic markers (association with Lauren’s classification, TNM stage, lymph node metastasis and overall survival time). It is likely that the overexpression of serum-derived lncRNAs, such as H19, HOTTIP, DANCR and HULC, may be an early event in tumorigenesis of the GC. The upregulation of tumour tissue-derived lncRNAs (HOTAIR, MALAT1, SHNG1 and SHNG6) might be adverse prognostic factors of GC. To ensure high specificity and sensitivity of the diagnosis and prognosis of GC, the expression level of lncRNAs, both diagnostic and prognostic markers, can be used alone or in combination with existing markers. The above insights may help to provide strategies for basic research and clinical diagnosis and treatment with lncRNAs. However, more in-depth investigations are still required to verify the practicality of lncRNAs in clinical application.
Author contributions
Y-NF, B-YL, and X-ZD proposed the topic, wrote the manuscript, Y-NF, B-YL, KW, X-XL, and LZ selected the literature. Y-NF and B-YL drafted the manuscript. X-ZD reviewed and revised the manuscript. All authors have read and approved the final manuscript.
Funding
National Natural Science Foundation of China (No. 81773778), China Postdoctoral Science Foundation (2019M650775, 2020T130438) and The sixth batch of Beijing Municipal TCM experts academic experience inheritance project.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Albrecht, L. V., Tejeda-Muñoz, N., Cell, E. M. D. R. J. A. R. O., and Biology, D. (2021). Cell Biology of canonical Wnt signaling. Annu. Rev. Cell Dev. Biol. 37, 369–389. doi:10.1146/annurev-cellbio-120319-023657
Amin, M. B., Greene, F. L., Edge, S. B., Compton, C. C., Gershenwald, J. E., Brookland, R. K., et al. (2017). The Eighth Edition AJCC Cancer Staging Manual: Continuing to build a bridge from a population-based to a more "personalized" approach to cancer staging. Ca. Cancer J. Clin. 67, 93–99. doi:10.3322/caac.21388
Arita, T., Ichikawa, D., Konishi, H., Komatsu, S., Shiozaki, A., Shoda, K., et al. (2013). Circulating long non-coding RNAs in plasma of patients with gastric cancer. Anticancer Res. 33, 3185–3193.
Arun, G., Diermeier, S. D., and Spector, D. L. (2018a). Therapeutic targeting of long non-coding RNAs in cancer. Trends Mol. Med. 24, 257–277. doi:10.1016/j.molmed.2018.01.001
Arun, K., Arunkumar, G., Bennet, D., Chandramohan, S. M., Murugan, A. K., and Munirajan, A. K. (2018b). Comprehensive analysis of aberrantly expressed lncRNAs and construction of ceRNA network in gastric cancer. Oncotarget 9, 18386–18399. doi:10.18632/oncotarget.24841
Ba, M. C., Ba, Z., Long, H., Cui, S. Z., Gong, Y. F., Yan, Z. F., et al. (2020). LncRNA AC093818.1 accelerates gastric cancer metastasis by epigenetically promoting PDK1 expression. Cell Death Dis. 11, 64. doi:10.1038/s41419-020-2245-2
Barriere, G., Fici, P., Gallerani, G., Fabbri, F., and Rigaud, M. (2015). Epithelial mesenchymal transition: A double-edged sword. Clin. Transl. Med. 4, 14. doi:10.1186/s40169-015-0055-4
Beckedorff, F. C., Amaral, M. S., Deocesano-Pereira, C., and Verjovski-Almeida, S. (2013). Long non-coding RNAs and their implications in cancer epigenetics. Biosci. Rep. 33, e00061. doi:10.1042/BSR20130054
Bhagwat, A. S., and Vakoc, C. R. (2015). Targeting transcription factors in cancer. Trends Cancer 1, 53–65. doi:10.1016/j.trecan.2015.07.001
Bhan, A., Soleimani, M., and Mandal, S. S. (2017). Long noncoding RNA and cancer: A new paradigm. Cancer Res. 77, 3965–3981. doi:10.1158/0008-5472.CAN-16-2634
Blum, M. A., Takashi, T., Suzuki, A., and Ajani, J. A. (2013). Management of localized gastric cancer. J. Surg. Oncol. 107, 265–270. doi:10.1002/jso.23183
Böhmdorfer, G., and Wierzbicki, A. T. (2015). Control of chromatin structure by long noncoding RNA. Trends Cell Biol. 25, 623–632. doi:10.1016/j.tcb.2015.07.002
Bürglin, T. R., and Affolter, M. (2016). Homeodomain proteins: An update. Chromosoma 125, 497–521. doi:10.1007/s00412-015-0543-8
Canzio, D., Nwakeze, C. L., Horta, A., Rajkumar, S. M., Coffey, E. L., Duffy, E. E., et al. (2019). Antisense lncRNA transcription mediates DNA demethylation to drive stochastic protocadherin α promoter choice. Cell 177, 639–653. e15. doi:10.1016/j.cell.2019.03.008
Chan, J. J., and Tay, Y. (2018). Noncoding RNA:RNA regulatory networks in cancer. Int. J. Mol. Sci. 19, 1310. doi:10.3390/ijms19051310
Chan, S. H., and Wang, L. H. (2015). Regulation of cancer metastasis by microRNAs. J. Biomed. Sci. 22, 9. doi:10.1186/s12929-015-0113-7
Chao, Z., Na, A., Chunyan, C., and Guodong, W. (2020). lncRNA HOXC-AS1 promotes gastric cancer via binding eIF4AIII by activating Wnt/β-catenin signaling. J. Gene Med. 22, e3202. doi:10.1002/jgm.3202
Chava, S., Gayatri, M. B., and Reddy, A. B. M. (2019). “Chapter 3 - EMT contributes to chemoresistance in pancreatic cancer,” in Breaking tolerance to pancreatic cancer unresponsiveness to chemotherapy. Editor G. P. NAGARAJU (Massachusetts, US: Academic Press).
Chen, N., Zhu, X., Zhu, Y., Shi, J., Zhang, J., Tang, C., et al. (2021a). The regulatory relationship and function of LncRNA FAM225A-miR-206-ADAM12 in gastric cancer. Am. J. Transl. Res. 13, 8632–8652.
Chen, Q. F., Hu, C. F., Sun, K., and Lang, Y. P. (2019). LncRNA NR027113 promotes malignant progression of gastric carcinoma via EMT signaling pathway. Eur. Rev. Med. Pharmacol. Sci. 23, 4746–4755. doi:10.26355/eurrev_201906_18056
Chen, X., Chen, Z., Yu, S., Nie, F., Yan, S., Ma, P., et al. (2018). Long noncoding RNA LINC01234 functions as a competing endogenous RNA to regulate CBFB expression by sponging miR-204-5p in gastric cancer. Clin. Cancer Res. 24, 2002–2014. doi:10.1158/1078-0432.CCR-17-2376
Chen, Y., Wei, G., Xia, H., Yu, H., Tang, Q., and Bi, F. (2017). Down regulation of lincRNA-p21 contributes to gastric cancer development through Hippo-independent activation of YAP. Oncotarget 8, 63813–63824. doi:10.18632/oncotarget.19130
Chen, Y., and Zhang, R. (2021). Long non-coding RNA AL139002.1 promotes gastric cancer development by sponging microRNA-490-3p to regulate Hepatitis A Virus Cellular Receptor 1 expression. Bioengineered 12, 1927–1938. doi:10.1080/21655979.2021.1922329
Chen, Z., Li, Y., Tan, B., Li, F., Zhao, Q., Fan, L., et al. (2021b). Long non-coding RNA ASNR targeting miR-519e-5p promotes gastric cancer development by regulating FGFR2. Front. Cell Dev. Biol. 9, 679176. doi:10.3389/fcell.2021.679176
Choi, S. W., Kim, H. W., and Nam, J. W. (2019). The small peptide world in long noncoding RNAs. Brief. Bioinform. 20, 1853–1864. doi:10.1093/bib/bby055
Cristescu, R., Lee, J., Nebozhyn, M., Kim, K.-M., Ting, J. C., Wong, S. S., et al. (2015). Molecular analysis of gastric cancer identifies subtypes associated with distinct clinical outcomes. Nat. Med. 21, 449–456. doi:10.1038/nm.3850
Cui, G., Cai, F., Ding, Z., and Pathology, L. G. J. H. (2019). HMGB2 promotes the malignancy of human gastric cancer and indicates poor survival outcome. Hum. Pathol. 84, 133–141. doi:10.1016/j.humpath.2018.09.017
D'Elia, L., Rossi, G., Ippolito, R., Cappuccio, F. P., and Strazzullo, P. (2012). Habitual salt intake and risk of gastric cancer: A meta-analysis of prospective studies. Clin. Nutr. 31, 489–498. doi:10.1016/j.clnu.2012.01.003
Dai, J., Nishi, A., Li, Z. X., Zhang, Y., Zhou, T., You, W. C., et al. (2021a). DNA methylation signatures associated with prognosis of gastric cancer. BMC Cancer 21, 610. doi:10.1186/s12885-021-08389-0
Dai, W., Xiao, Y., Tang, W., Li, J., Hong, L., Zhang, J., et al. (2021b). Identification of an EMT-related gene signature for predicting overall survival in gastric cancer. Front. Genet. 12, 661306. doi:10.3389/fgene.2021.661306
Dai, X., Ren, T., Zhang, Y., and Nan, N. (2021c). Methylation multiplicity and its clinical values in cancer. Expert Rev. Mol. Med. 23, e2. doi:10.1017/erm.2021.4
Deshmukh, A. P., Vasaikar, S. V., Tomczak, K., Tripathi, S., Den Hollander, P., Arslan, E., et al. (2021). Identification of EMT signaling cross-talk and gene regulatory networks by single-cell RNA sequencing. Proc. Natl. Acad. Sci. U. S. A. 118, e2102050118. doi:10.1073/pnas.2102050118
Digklia, A., and Wagner, A. D. (2016). Advanced gastric cancer: Current treatment landscape and future perspectives. World J. Gastroenterol. 22, 2403–2414. doi:10.3748/wjg.v22.i8.2403
Dong, X., He, X., Guan, A., Huang, W., Jia, H., Huang, Y., et al. (2019). Long non-coding RNA Hotair promotes gastric cancer progression via miR-217-GPC5 axis. Life Sci. 217, 271–282. doi:10.1016/j.lfs.2018.12.024
Dong, Y., Wang, Z. G., and Chi, T. S. (2018). Long noncoding RNA Lnc01614 promotes the occurrence and development of gastric cancer by activating EMT pathway. Eur. Rev. Med. Pharmacol. Sci. 22, 1307–1314. doi:10.26355/eurrev_201803_14472
Du, Z., Sun, T., Hacisuleyman, E., Fei, T., Wang, X., Brown, M., et al. (2016). Integrative analyses reveal a long noncoding RNA-mediated sponge regulatory network in prostate cancer. Nat. Commun. 7, 10982. doi:10.1038/ncomms10982
Dykes, I. M., and Emanueli, C. (2017). Transcriptional and Post-transcriptional Gene Regulation by Long Non-coding RNA. Genomics, Proteomics and Bioinformatics 15, 177–186.
Fatima, R., Akhade, V. S., Pal, D., and Rao, S. M. (2015). Long noncoding RNAs in development and cancer: Potential biomarkers and therapeutic targets. Mol. Cell. Ther. 3, 5. doi:10.1186/s40591-015-0042-6
Feng, F., Tian, Y., Xu, G., Liu, Z., Liu, S., Zheng, G., et al. (2017). Diagnostic and prognostic value of CEA, CA19-9, AFP and CA125 for early gastric cancer. BMC Cancer 17, 737. doi:10.1186/s12885-017-3738-y
Feng, W., Zong, W., Li, Y., Shen, X., Cui, X., and Ju, S. (2020). Abnormally expressed long noncoding RNA B3GALT5-AS1 may serve as a biomarker for the diagnostic and prognostic of gastric cancer. J. Cell. Biochem. 121, 557–565. doi:10.1002/jcb.29296
Fu, M., Huang, Z., Zang, X., Pan, L., Liang, W., Chen, J., et al. (2018). Long noncoding RNA LINC00978 promotes cancer growth and acts as a diagnostic biomarker in gastric cancer. Cell Prolif. 51. doi:10.1111/cpr.12425
Fu, X., Tian, Y., Kuang, W., Wen, S., and Guo, W. (2019). Long non-coding RNA DLX6-AS1 silencing inhibits malignant phenotypes of gastric cancer cells. Exp. Ther. Med. 17, 4715–4722. doi:10.3892/etm.2019.7521
Gaianigo, N., Melisi, D., and Carbone, C. (2017). EMT and treatment resistance in pancreatic cancer. Cancers 9, 122. doi:10.3390/cancers9090122
Gao, M., Liu, L., Yang, Y., Li, M., Ma, Q., and Chang, Z. (2021). LncRNA HCP5 induces gastric cancer cell proliferation, invasion, and EMT processes through the miR-186-5p/wnt5a Axis under hypoxia. Front. Cell Dev. Biol. 9, 663654. doi:10.3389/fcell.2021.663654
Gharaibeh, L., Elmadany, N., Alwosaibai, K., and Alshaer, W. (2020). Notch1 in cancer therapy: Possible clinical implications and challenges. Mol. Pharmacol. 98, 559–576. doi:10.1124/molpharm.120.000006
Gong, P., Qiao, F., Wu, H., Cui, H., Li, Y., Zheng, Y., et al. (2018). LncRNA UCA1 promotes tumor metastasis by inducing miR-203/ZEB2 axis in gastric cancer. Cell Death Dis. 9, 1158. doi:10.1038/s41419-018-1170-0
Grumati, P., and Dikic, I. (2018). Ubiquitin signaling and autophagy. J. Biol. Chem. 293, 5404–5413. doi:10.1074/jbc.TM117.000117
Guo, L., Ma, H., Kong, Y., Leng, G., Liu, G., and Zhang, Y. (2022). Long non-coding RNA TNK2 AS1/microRNA-125a-5p axis promotes tumor growth and modulated phosphatidylinositol 3 kinase/AKT pathway. J. Gastroenterol. Hepatol. 37, 124–133. doi:10.1111/jgh.15683
Guo, W., Huang, J., Lei, P., Guo, L., and Li, X. (2019a). LncRNA SNHG1 promoted HGC-27 cell growth and migration via the miR-140/ADAM10 axis. Int. J. Biol. Macromol. 122, 817–823. doi:10.1016/j.ijbiomac.2018.10.214
Guo, W., Liang, X., Liu, L., Guo, Y., Shen, S., Liang, J., et al. (2019b). MiR-6872 host gene SEMA3B and its antisense lncRNA SEMA3B-AS1 function synergistically to suppress gastric cardia adenocarcinoma progression. Gastric Cancer 22, 705–722. doi:10.1007/s10120-019-00924-0
Guo, Y., Sun, P., Guo, W., and Dong, Z. (2021). Long non-coding RNA LINC01503 promotes gastric cardia adenocarcinoma progression via miR-133a-5p/VIM Axis and EMT process. Dig. Dis. Sci. 66, 3391–3403. doi:10.1007/s10620-020-06690-9
He, C., Qi, W., and Wang, Z. (2021a). Effect and mechanism of downregulating the long-chain noncoding RNA TM4SF1-AS1 on the proliferation, apoptosis and invasion of gastric cancer cells. World J. Surg. Oncol. 19, 226. doi:10.1186/s12957-021-02334-y
He, S., Zhang, H., Xiao, Z., Bhushan, S., Gao, K., and Bioengineered, W. W. J. (2021b). The interaction of TEA domain transcription factor 4 (TEAD4) and Yes-associated protein 1 (YAP1) promoted the malignant process mediated by serum/glucocorticoid regulated kinase 1 (SGK1). Bioengineered 12, 601–614. doi:10.1080/21655979.2021.1882142
He, Z., Duan, Z., Chen, L., Li, B., and Zhou, Y. (2020). Long non-coding RNA Loc490 inhibits gastric cancer cell proliferation and metastasis by upregulating RNA-binding protein Quaking. Aging 12, 17681–17693. doi:10.18632/aging.103876
Howlader, N., Noone, A. M., Krapcho, M., Miller, D., Brest, A., Yu, M., et al. (2020). SEER cancer statistics review, 1975-2017. Bethesda, MD: National Cancer Institute.
Hu, J., Wang, M., Yang, Y., Xing, Y., and Li, S. (2022). LncRNA DLEU2 silencing impedes the migration, invasion and EMT in gastric cancer cell by suppressing PI3K/AKT signaling pathway. Immunopharmacol. Immunotoxicol. 2022, 1–13. doi:10.1080/08923973.2022.2078727
Hu, Z., Fubing, W., Hao, C., Qian, T., Shili, Q., Shanshan, C., et al. (2016). Increased expression of long-noncoding RNA ZFAS1 is associated with epithelial-mesenchymal transition of gastric cancer. Aging 8, 2023–2038. doi:10.18632/aging.101048
Huang, D., Zhang, K., Zheng, W., Zhang, R., Chen, J., Du, N., et al. (2021). Long noncoding RNA SGO1-AS1 inactivates TGFβ signaling by facilitating TGFB1/2 mRNA decay and inhibits gastric carcinoma metastasis. J. Exp. Clin. Cancer Res. 40, 342. doi:10.1186/s13046-021-02140-0
Huang, J. Q., Sridhar, S., Chen, Y., and Hunt, R. H. (1998). Meta-analysis of the relationship between Helicobacter pylori seropositivity and gastric cancer. Gastroenterology 114, 1169–1179. doi:10.1016/s0016-5085(98)70422-6
Huang, L., Wu, R.-L., and Xu, A. M. (2015). Epithelial-mesenchymal transition in gastric cancer. Am. J. Transl. Res. 7, 2141–2158.
Huang, Y., Zhang, J., Hou, L., Wang, G., Liu, H., Zhang, R., et al. (2017). LncRNA AK023391 promotes tumorigenesis and invasion of gastric cancer through activation of the PI3K/Akt signaling pathway. J. Exp. Clin. Cancer Res. 36, 194. doi:10.1186/s13046-017-0666-2
Jafarzadeh, M., and Soltani, B. M. (2020). Long noncoding RNA LOC400043 (LINC02381) inhibits gastric cancer progression through regulating Wnt signaling pathway. Front. Oncol. 10, 562253. doi:10.3389/fonc.2020.562253
Ji, Z., Tang, T., Chen, M., Dong, B., Sun, W., Wu, N., et al. (2021). C-Myc-activated long non-coding RNA LINC01050 promotes gastric cancer growth and metastasis by sponging miR-7161-3p to regulate SPZ1 expression. J. Exp. Clin. Cancer Res. 40, 351. doi:10.1186/s13046-021-02155-7
Jiang, L., Zhang, Y., Su, P., Ma, Z., Ye, X., Kang, W., et al. (2022). Long non-coding RNA HNF1A-AS1 induces 5-FU resistance of gastric cancer through miR-30b-5p/EIF5A2 pathway. Transl. Oncol. 18, 101351. doi:10.1016/j.tranon.2022.101351
Jiang, X., Ding, W., Shen, W., and Jin, J. (2020). H19/miR-152-3p/TCF4 axis increases chemosensitivity of gastric cancer cells through suppression of epithelial-mesenchymal transition. Transl. Cancer Res. 9, 3915–3925. doi:10.21037/tcr-20-1736
Jin, C., Shi, W., Wang, F., Shen, X., Qi, J., Cong, H., et al. (2016). Long non-coding RNA HULC as a novel serum biomarker for diagnosis and prognosis prediction of gastric cancer. Oncotarget 7, 51763–51772. doi:10.18632/oncotarget.10107
Jing, R., Liu, S., Jiang, Y., Zong, W., Ju, S., and Cui, M. (2020). Determination of serum RP11-731F5.2 as a noninvasive biomarker for gastric cancer diagnosis and prognosis. Pathol. Res. Pract. 216, 153261. doi:10.1016/j.prp.2020.153261
Kalluri, R., and Weinberg, R. A. (2009). The basics of epithelial-mesenchymal transition. J. Clin. Invest. 119, 1420–1428. doi:10.1172/JCI39104
Kapranov, P., Cheng, J., Dike, S., Nix, D. A., Duttagupta, R., Willingham, A. T., et al. (2007). RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science 316, 1484–1488. doi:10.1126/science.1138341
Karagkouni, D., Karavangeli, A., Paraskevopoulou, M. D., and Biology, A. G. H. J. M. I. M. (2021). Characterizing miRNA-lncRNA interplay. Methods Mol. Biol. 2372, 243–262. doi:10.1007/978-1-0716-1697-0_21
Kim, D. H., Xing, T., Yang, Z., Dudek, R., Lu, Q., and Chen, Y. H. (2017). Epithelial mesenchymal transition in embryonic development, tissue repair and cancer: A comprehensive overview. J. Clin. Med. 7, E1. doi:10.3390/jcm7010001
Kong, L., Sun, Y., Chen, M., Dai, Y., Experimental, Z. L. J., and Medicine, T. (2020). Downregulation of microRNA-320a inhibits proliferation and induces apoptosis of retinoblastoma cells via targeting TUSC3. Exp. Ther. Med. 20, 9. doi:10.3892/etm.2020.9137
Kontomanolis, E. N., Koutras, A., Syllaios, A., Schizas, D., Mastoraki, A., Garmpis, N., et al. (2020). Role of oncogenes and tumor-suppressor genes in carcinogenesis: A review. Anticancer Res. 40, 6009–6015. doi:10.21873/anticanres.14622
Kyrgiou, M., Kalliala, I., Markozannes, G., Gunter, M. J., Paraskevaidis, E., Gabra, H., et al. (2017). Adiposity and cancer at major anatomical sites: Umbrella review of the literature. Bmj 356, j477. doi:10.1136/bmj.j477
Ladeiras-Lopes, R., Pereira, A. K., Nogueira, A., Pinheiro-Torres, T., Pinto, I., Santos-Pereira, R., et al. (2008). Smoking and gastric cancer: Systematic review and meta-analysis of cohort studies. Cancer Causes Control 19, 689–701. doi:10.1007/s10552-008-9132-y
Lamouille, S., Xu, J., and Derynck, R. (2014). Molecular mechanisms of epithelial–mesenchymal transition. Nat. Rev. Mol. Cell Biol. 15, 178–196. doi:10.1038/nrm3758
Lauren, P. (1965). The two histological main types of gastric carcinoma: Diffuse and SO-called intestinal-type carcinoma. AN attempt at a histo-clinical classification. Acta Pathol. Microbiol. Scand. 64, 31–49. doi:10.1111/apm.1965.64.1.31
Lee, N. K., Lee, J. H., Ivan, C., Ling, H., Zhang, X., Park, C. H., et al. (2017). MALAT1 promoted invasiveness of gastric adenocarcinoma. BMC Cancer 17, 46. doi:10.1186/s12885-016-2988-4
Lee, S. K., Lee, J. H., Lee, N. K., and Park, C. H. (2015). Sa1983 long non-coding RNA MALAT-1 promote invasion and migration of gastric cancer. Gastroenterology 148, S374–S374. doi:10.1016/s0016-5085(15)31256-7
Lei, P., Wei, L., Min, F., Zhen-Hua, H., Xia, L., Wen, Z., et al. (2017). Exosomes-mediated transfer of long noncoding RNA ZFAS1 promotes gastric cancer progression. J. Cancer Res. Clin. Oncol. 143, 991–1004. doi:10.1007/s00432-017-2361-2
Leng, X., Liu, G., Wang, S., Song, J., Zhang, W., Zhang, X., et al. (2020). LINC01272 promotes migration and invasion of gastric cancer cells via EMT. Onco. Targets. Ther. 13, 3401–3410. doi:10.2147/OTT.S242073
Li, D., She, J., Hu, X., Zhang, M., Sun, R., and Qin, S. (2021). The ELF3-regulated lncRNA UBE2CP3 is over-stabilized by RNA–RNA interactions and drives gastric cancer metastasis via miR-138-5p/ITGA2 axis. Oncogene 40, 5403–5415. doi:10.1038/s41388-021-01948-6
Li, D., Wang, J., Zhang, M., Hu, X., She, J., Qiu, X., et al. (2020a). LncRNA MAGI2-AS3 is regulated by BRD4 and promotes gastric cancer progression via maintaining ZEB1 overexpression by sponging miR-141/200a. Mol. Ther. Nucleic Acids 19, 109–123. doi:10.1016/j.omtn.2019.11.003
Li, D., Xu, M., Wang, Z., Huang, P., Huang, C., Chen, Z., et al. (2022). The EMT-induced lncRNA NR2F1-AS1 positively modulates NR2F1 expression and drives gastric cancer via miR-29a-3p/VAMP7 axis. Cell Death Dis. 13, 84. doi:10.1038/s41419-022-04540-2
Li, H., Chen, X., Gao, Y., Wu, J., Zeng, F., and Song, F. (2015). XBP1 induces snail expression to promote epithelial- to-mesenchymal transition and invasion of breast cancer cells. Cell. Signal. 27, 82–89. doi:10.1016/j.cellsig.2014.09.018
Li, J., Gao, J., Tian, W., Li, Y., and Zhang, J. (2017). Long non-coding RNA MALAT1 drives gastric cancer progression by regulating HMGB2 modulating the miR-1297. Cancer Cell Int. 17, 44. doi:10.1186/s12935-017-0408-8
Li, L., Wang, Y. Y., Mou, X. Z., Ye, Z. Y., and Zhao, Z. S. (2018). Up-regulation of long noncoding RNA M26317 correlates with tumor progression and poor prognosis in gastric cancer. Hum. Pathol. 78, 44–53. doi:10.1016/j.humpath.2018.04.008
Li, N., Ouyang, Y., Chen, S., Peng, C., He, C., Hong, J., et al. (2020b). Integrative analysis of differential lncRNA/mRNA expression profiling in Helicobacter pylori infection-associated gastric carcinogenesis. Front. Microbiol. 11, 880. doi:10.3389/fmicb.2020.00880
Li, X. M., Jiao, Y. Y., Luan, B. H., Wu, H. X., Wang, R. R., and Zhong, J. (2020c). Long non-coding RNA MIAT promotes gastric cancer proliferation and metastasis via modulating the miR-331-3p/RAB5B pathway. Oncol. Lett. 20, 355. doi:10.3892/ol.2020.12219
Li, Y., Jiang, L., Lv, S., Xu, H., Fan, Z., He, Y., et al. (2019). E2F6-mediated lncRNA CASC2 down-regulation predicts poor prognosis and promotes progression in gastric carcinoma. Life Sci. 232, 116649. doi:10.1016/j.lfs.2019.116649
Li, Y., Yan, J., Wang, Y., Wang, C., Zhang, C., and Li, G. (2020d). LINC00240 promotes gastric cancer cell proliferation, migration and EMT via the miR-124-3p/DNMT3B axis. Cell biochem. Funct. 38, 1079–1088. doi:10.1002/cbf.3551
Li, Y., Zhao, Z., Xu, C., Zhou, Z., Zhu, Z., and You, T. (2014). HMGA2 induces transcription factor Slug expression to promote epithelial-to-mesenchymal transition and contributes to colon cancer progression. Cancer Lett. 355, 130–140. doi:10.1016/j.canlet.2014.09.007
Liang, M., Yunshan, J., and Ning, W. (2022). Long non-coding RNA CCL2 promoted gastric cancer function via miR-128/PARP2 signal pathway. Bioengineered 13, 1602–1611. doi:10.1080/21655979.2021.2020548
Liang, W., Xia, B., Yan, M., Zhai, G., and Li, M. (2021). Enhanced LINC01061 levels as a serum biomarker in gastric cancer and promotion of malignant transformation. Oncol. Res. Treat. 44, 242–251. doi:10.1159/000508310
Liang, Y., Zhang, C. D., Zhang, C., and Dai, D. Q. (2020). DLX6-AS1/miR-204-5p/OCT1 positive feedback loop promotes tumor progression and epithelial-mesenchymal transition in gastric cancer. Gastric Cancer 23, 212–227. doi:10.1007/s10120-019-01002-1
Liao, Z., Nie, H., Wang, Y., Luo, J., Zhou, J., and Ou, C. (2021). The emerging landscape of long non-coding RNAs in colorectal cancer metastasis. Front. Oncol. 11, 641343. doi:10.3389/fonc.2021.641343
Lin, H., Wang, J., Wang, T., Wu, J., Wang, P., Huo, X., et al. (2021). The LncRNA mir503hg/miR-224-5p/TUSC3 signaling cascade suppresses gastric cancer development via modulating ATF6 branch of unfolded protein response. Front. Oncol. 11, 708501. doi:10.3389/fonc.2021.708501
Lin, J., Song, T., Li, C., and Mao, W. (2020). GSK-3β in DNA repair, apoptosis, and resistance of chemotherapy, radiotherapy of cancer. Biochim. Biophys. Acta Mol. Cell Res. 1867, 118659. doi:10.1016/j.bbamcr.2020.118659
Liu, C., Luo, J., Zhao, Y. T., Wang, Z. Y., Zhou, J., Huang, S., et al. (2018a). TWIST1 upregulates miR-214 to promote epithelial-to-mesenchymal transition and metastasis in lung adenocarcinoma. Int. J. Mol. Med. 42, 461–470. doi:10.3892/ijmm.2018.3630
Liu, G., Xiang, T., Wu, Q. F., and Wang, W. X. (2016a). Long noncoding RNA H19-derived miR-675 enhances proliferation and invasion via RUNX1 in gastric cancer cells. Oncol. Res. 23, 99–107. doi:10.3727/096504015X14496932933575
Liu, H., Zhang, Z., Wu, N., Guo, H., Zhang, H., Fan, D., et al. (2018b). Integrative analysis of dysregulated lncRNA-associated ceRNA network reveals functional lncRNAs in gastric cancer. Genes (Basel) 9, 303. doi:10.3390/genes9060303
Liu, J., Liu, L., Wan, J. X., and Song, Y. (2017). Long noncoding RNA SNHG20 promotes gastric cancer progression by inhibiting p21 expression and regulating the GSK-3β/β-catenin signaling pathway. Oncotarget 8, 80700–80708. doi:10.18632/oncotarget.20959
Liu, J., Wang, G., Zhao, J., Liu, X., Zhang, K., Gong, G., et al. (2021). LncRNA H19 promoted the epithelial to mesenchymal transition and metastasis in gastric cancer via activating wnt/β-catenin signaling. Dig. Dis. 40, 436–447. doi:10.1159/000518627
Liu, L., Zhao, X., Zou, H., Bai, R., Yang, K., and Tian, Z. (2016b). Hypoxia promotes gastric cancer malignancy partly through the HIF-1α dependent transcriptional activation of the long non-coding RNA GAPLINC. Front. Physiol. 7, 420. doi:10.3389/fphys.2016.00420
Liu, W., Zhang, Z., Zhang, Y., Chen, X., Guo, S., Lei, Y., et al. (2015a). THERAPYHMGB1-mediated autophagy modulates sensitivity of colorectal cancer cells to oxaliplatin via MEK/ERK signaling pathway. Cancer Biol. Ther. 16, 511–517. doi:10.1080/15384047.2015.1017691
Liu, Y. W., Sun, M., Xia, R., Zhang, E. B., Liu, X. H., Zhang, Z. H., et al. (2015b). LincHOTAIR epigenetically silences miR34a by binding to PRC2 to promote the epithelial-to-mesenchymal transition in human gastric cancer. Cell Death Dis. 6, e1802. doi:10.1038/cddis.2015.150
Liu, Z., Hu, K., Wang, X., Zhang, Y., Wang, W., and Wu, Y. (2022). LncRNA ACTA2-AS1 inhibits malignant phenotypes of gastric cancer cells. Open Med. 17, 266–279. doi:10.1515/med-2021-0406
Liu, Z. Q., He, W. F., Wu, Y. J., Zhao, S. L., Wang, L., Ouyang, Y. Y., et al. (2020a). LncRNA SNHG1 promotes EMT process in gastric cancer cells through regulation of the miR-15b/DCLK1/Notch1 axis. BMC Gastroenterol. 20, 156. doi:10.1186/s12876-020-01272-5
Liu, Z. Z., Tian, Y. F., Wu, H., Ouyang, S. Y., and Kuang, W. L. (2020b). LncRNA H19 promotes glioma angiogenesis through miR-138/HIF-1α/VEGF axis. Neoplasma 67, 111–118. doi:10.4149/neo_2019_190121N61
Long, Y., Wang, X., Youmans, D. T., and Cech, T. R. (2017). How do lncRNAs regulate transcription? Sci. Adv. 3, eaao2110. doi:10.1126/sciadv.aao2110
Lu, H., Zhang, Q., Sun, Y., Wu, D., and Liu, L. (2020a). LINC00689 induces gastric cancer progression via modulating the miR-338-3p/HOXA3 axis. J. Gene Med. 22, e3275. doi:10.1002/jgm.3275
Lu, J., Tang, Y., Zhang, W., Zhang, S., Qi, W., Ma, G., et al. (2020b). Expression and mechanism of long non-coding RNA DLGAP1-AS2 in gastric cancer. Tumor 40, 153–162.
Lu, J., Xu, Y., Xie, W., Tang, Y., Zhang, H., Wang, B., et al. (2021). Long noncoding RNA DLGAP1-AS2 facilitates Wnt1 transcription through physically interacting with Six3 and drives the malignancy of gastric cancer. Cell Death Discov. 7, 255. doi:10.1038/s41420-021-00649-z
Lv, J., Zhang, S., Liu, Y., Li, C., Guo, T., Zhang, S., et al. (2021). NR2F1-AS1/miR-190a/PHLDB2 induces the epithelial-mesenchymal transformation process in gastric cancer by promoting phosphorylation of AKT3. Front. Cell Dev. Biol. 9, 688949. doi:10.3389/fcell.2021.688949
Ma, L., Bajic, V. B., and Zhang, Z. (2013). On the classification of long non-coding RNAs. RNA Biol. 10, 925–933. doi:10.4161/rna.24604
Ma, L., Jiang, Y., and Wu, N. (2022). Long non-coding RNA CCL2 promoted gastric cancer function via miR-128/PARP2 signal pathway. Bioengineered 13, 1602–1611. doi:10.1080/21655979.2021.2020548
Ma, P., Li, L., Liu, F., and Zhao, Q. (2020). HNF1A-Induced lncRNA HCG18 facilitates gastric cancer progression by upregulating DNAJB12 via miR-152-3p. Onco. Targets. Ther. 13, 7641–7652. doi:10.2147/OTT.S253391
Macdonald, B. T., Tamai, K., and Cell, X. H. J. D. (2009). Wnt/beta-catenin signaling: Components, mechanisms, and diseases. Dev. Cell 17, 9–26. doi:10.1016/j.devcel.2009.06.016
Martínez-Estrada, O. M., Lettice, L. A., Essafi, A., Guadix, J. A., Slight, J., Velecela, V., et al. (2010). Wt1 is required for cardiovascular progenitor cell formation through transcriptional control of Snail and E-cadherin. Nat. Genet. 42, 89–93. doi:10.1038/ng.494
Matsuoka, T., and Yashiro, M. (2018). Biomarkers of gastric cancer: Current topics and future perspective. World J. Gastroenterol. 24, 2818–2832. doi:10.3748/wjg.v24.i26.2818
Mei, J., Liu, G., Li, R., Xiao, P., Yang, D., Bai, H., et al. (2021). LncRNA SNHG6 knockdown inhibits cisplatin resistance and progression of gastric cancer through miR-1297/BCL-2 axis. Biosci. Rep. 41, BSR20211885. doi:10.1042/BSR20211885
Meng, Q., Wang, X., Xue, T., Zhao, Q., Wang, W., and Zhao, K. (2020). Long noncoding RNA MIR99AHG promotes gastric cancer progression by inducing EMT and inhibiting apoptosis via miR577/FOXP1 axis. Cancer Cell Int. 20, 414. doi:10.1186/s12935-020-01510-6
Meng, Z., Moroishi, T., Genes, K.-L. G. J., and Development, (2016). Mechanisms of Hippo pathway regulation. Genes Dev. 30, 1–17. doi:10.1101/gad.274027.115
Morgan, E., Arnold, M., Camargo, M. C., Gini, A., Kunzmann, A. T., Matsuda, T., et al. (2022). The current and future incidence and mortality of gastric cancer in 185 countries, 2020–40: A population-based modelling study. eClinicalMedicine 47, 101404. doi:10.1016/j.eclinm.2022.101404,
Mulero, M. C., Shahabi, S., Ko, M. S., Schiffer, J. M., Huang, D.-B., Wang, V. Y.-F., et al. (2018). Protein cofactors are essential for high-affinity DNA binding by the nuclear factor κB RelA subunit. Biochemistry 57, 2943–2957. doi:10.1021/acs.biochem.8b00158
Necula, L., Matei, L., Dragu, D., Neagu, A. I., Mambet, C., Nedeianu, S., et al. (2019). Recent advances in gastric cancer early diagnosis. World J. Gastroenterol. 25, 2029–2044. doi:10.3748/wjg.v25.i17.2029
Nie, M.-L., Han, J., Huang, H.-C., Guo, T., Huangfu, L.-T., Cheng, X.-J., et al. (2019). The novel lncRNA p4516 acts as a prognostic biomarker promoting gastric cancer cell proliferation and metastasis. Cancer Manag. Res. 11, 5375–5391. doi:10.2147/CMAR.S201793
Oh, S. C., Sohn, B. H., Cheong, J.-H., Kim, S.-B., Lee, J. E., Park, K. C., et al. (2018). Clinical and genomic landscape of gastric cancer with a mesenchymal phenotype. Nat. Commun. 9, 1777. doi:10.1038/s41467-018-04179-8
Pan, L., Liang, W., Gu, J., Zang, X., Huang, Z., Shi, H., et al. (2018). Long noncoding RNA DANCR is activated by SALL4 and promotes the proliferation and invasion of gastric cancer cells. Oncotarget 9, 1915–1930. doi:10.18632/oncotarget.23019
Pan, T., Yu, Z., Jin, Z., Wu, X., Wu, A., Hou, J., et al. (2021). Tumor suppressor lnc-CTSLP4 inhibits EMT and metastasis of gastric cancer by attenuating HNRNPAB-dependent Snail transcription. Mol. Ther. Nucleic Acids 23, 1288–1303. doi:10.1016/j.omtn.2021.02.003
Pastar, I., Stojadinovic, O., Yin, N. C., Ramirez, H., Nusbaum, A. G., Sawaya, A., et al. (2014). Epithelialization in wound healing: A comprehensive review. Adv. Wound Care 3, 445–464. doi:10.1089/wound.2013.0473
Pavlič, A., Hauptman, N., Boštjančič, E., and Zidar, N. (2022). Long non-coding RNAs as potential regulators of EMT-related transcription factors in colorectal cancer-A systematic review and bioinformatics analysis. Cancers 14, 2280. doi:10.3390/cancers14092280
Peng, W.-X., Koirala, P., and Oncogene, Y.-Y. M. J. (2017). LncRNA-mediated regulation of cell signaling in cancer. Oncogene 36, 5661–5667. doi:10.1038/onc.2017.184
Peng, Z., Wang, C.-X., Fang, E.-H., Wang, G.-B., and Tong, Q. (2014). Role of epithelial-mesenchymal transition in gastric cancer initiation and progression. World J. Gastroenterol. 20, 5403–5410. doi:10.3748/wjg.v20.i18.5403
Petkevicius, V., Thon, C., Steponaitiene, R., Skieceviciene, J., Janciauskas, D., Jechorek, D., et al. (2022). Differential expression of long noncoding RNA HOTAIR in intestinal metaplasia and gastric cancer. Clin. Transl. Gastroenterol. 13, e00483. doi:10.14309/ctg.0000000000000483
Piao, H. Y., Guo, S., Jin, H., Wang, Y., and Zhang, J. (2021). LINC00184 involved in the regulatory network of ANGPT2 via ceRNA mediated miR-145 inhibition in gastric cancer. J. Cancer 12, 2336–2350. doi:10.7150/jca.49138
Przygodzka, P., Papiewska-Pająk, I., Bogusz-Koziarska, H., Sochacka, E., Boncela, J., and Kowalska, M. A. (2019). Regulation of miRNAs by Snail during epithelial-to-mesenchymal transition in HT29 colon cancer cells. Sci. Rep. 9, 2165. doi:10.1038/s41598-019-39200-7
Qi, F., Liu, X., Wu, H., Yu, X., Wei, C., Huang, X., et al. (2017). Long noncoding AGAP2-AS1 is activated by SP1 and promotes cell proliferation and invasion in gastric cancer. J. Hematol. Oncol. 10, 48. doi:10.1186/s13045-017-0420-4
Qi, M., Yu, B., Yu, H., and Li, F. (2020). Integrated analysis of a ceRNA network reveals potential prognostic lncRNAs in gastric cancer. Cancer Med. 9, 1798–1817. doi:10.1002/cam4.2760
Rao, J., Fu, J., Meng, C., Huang, J., Qin, X., and Zhuang, S. (2021). LncRNA SNHG3 promotes gastric cancer cells proliferation, migration, and invasion by targeting miR-326. J. Oncol. 2021, 9935410. doi:10.1155/2021/9935410
Rawla, P., and Barsouk, A. (2019). Epidemiology of gastric cancer: Global trends, risk factors and prevention. Prz. Gastroenterol. 14, 26–38. doi:10.5114/pg.2018.80001
Ren, J., Xu, N., Zhou, R., Huang, F., Zhang, H., and Li, W. (2021). Long non-coding RNA PCED1B antisense RNA 1 promotes gastric cancer progression via modulating microRNA-215-3p/C-X-C motif chemokine receptor 1 axis. Bioengineered 12, 6083–6095. doi:10.1080/21655979.2021.1971503
Riihimäki, M., Hemminki, A., Sundquist, K., Sundquist, J., and Hemminki, K. (2016). Metastatic spread in patients with gastric cancer. Oncotarget 7, 52307–52316. doi:10.18632/oncotarget.10740
Rui, Z., Yanli, Z., Xin, Z., Yongmei, Y., Xin, Z., Xiaohui, L., et al. (2018). Exosomal long noncoding RNA HOTTIP as potential novel diagnostic and prognostic biomarker test for gastric cancer. Mol. Cancer 17, 68. doi:10.1186/s12943-018-0817-x
Shuai, Y., Ma, Z., Liu, W., Yu, T., Yan, C., Jiang, H., et al. (2020). TEAD4 modulated LncRNA MNX1-AS1 contributes to gastric cancer progression partly through suppressing BTG2 and activating BCL2. Mol. Cancer 19, 6. doi:10.1186/s12943-019-1104-1
Soltani, R., Amini, M., Mazaheri Moghaddam, M., Jebelli, A., Ahmadiyan, S., Bidar, N., et al. (2022). LncRNA DLGAP1-AS2 overexpression associates with gastric tumorigenesis: A promising diagnostic and therapeutic target. Mol. Biol. Rep. 49, 6817–6826. doi:10.1007/s11033-021-07038-w
Sona, M. F., Myung, S. K., Park, K., and Jargalsaikhan, G. (2018). Type 1 diabetes mellitus and risk of cancer: A meta-analysis of observational studies. Jpn. J. Clin. Oncol. 48, 426–433. doi:10.1093/jjco/hyy047
Song, Z., Wu, Y., Yang, J., Yang, D., and Fang, X. (2017). Progress in the treatment of advanced gastric cancer. Tumour Biol. 39, 1010428317714626. doi:10.1177/1010428317714626
Stamos, J. L., and Weis, W. I. (2013). The β-catenin destruction complex. Cold Spring Harb. Perspect. Biol. 5, a007898. doi:10.1101/cshperspect.a007898
Statello, L., Guo, C.-J., Chen, L.-L., and Huarte, M. (2021). Gene regulation by long non-coding RNAs and its biological functions. Nat. Rev. Mol. Cell Biol. 22, 96–118. doi:10.1038/s41580-020-00315-9
Stegmann, K., Boecker, J., Kosan, C., Ermert, A., Kunz, J., and Koch, M. C. (1999). Human transcription factor SLUG: Mutation analysis in patients with neural tube defects and identification of a missense mutation (D119E) in the slug subfamily-defining region. Mutat. Res. 406, 63–69. doi:10.1016/s1383-5726(99)00002-3
Stemmler, M. P., Eccles, R. L., Brabletz, S., and Brabletz, T. (2019). Non-redundant functions of EMT transcription factors. Nat. Cell Biol. 21, 102–112. doi:10.1038/s41556-018-0196-y
Sun, J. G., Li, X. B., Yin, R. H., and Li, X. F. (2020). lncRNA VIM-AS1 promotes cell proliferation, metastasis and epithelial-mesenchymal transition by activating the Wnt/β-catenin pathway in gastric cancer. Mol. Med. Rep. 22, 4567–4578. doi:10.3892/mmr.2020.11577
Sun, J., Song, Y., Chen, X., Zhao, J., Gao, P., Huang, X., et al. (2015). Novel long non-coding RNA RP11-119F7.4 as a potential biomarker for the development and progression of gastric cancer. Oncol. Lett. 10, 115–120. doi:10.3892/ol.2015.3186
Sun, Q., Hao, Q., and Prasanth, K. V. (2018). Nuclear long noncoding RNAs: Key regulators of gene expression. Trends Genet. 34, 142–157. doi:10.1016/j.tig.2017.11.005
Sun, Q., Li, J., Li, F., Li, H., Bei, S., Zhang, X., et al. (2019a). LncRNA LOXL1-AS1 facilitates the tumorigenesis and stemness of gastric carcinoma via regulation of miR-708-5p/USF1 pathway. Cell Prolif. 52, e12687. doi:10.1111/cpr.12687
Sun, Y., and Han, C. (2020). Long non-coding RNA TMPO-as1 promotes cell migration and invasion by sponging MIR-140-5p and inducing SOX4-mediated EMT in gastric cancer. Cancer Manag. Res. 12, 1261–1268. doi:10.2147/CMAR.S235898
Sun, Y., Zhu, Q., Yang, W., Shan, Y., Yu, Z., Zhang, Q., et al. (2019b). LncRNA H19/miR-194/PFTK1 axis modulates the cell proliferation and migration of pancreatic cancer. J. Cell. Biochem. 120, 3874–3886. doi:10.1002/jcb.27669
Sung, H., Ferlay, J., Siegel, R. L., Laversanne, M., Soerjomataram, I., Jemal, A., et al. (2021). Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. Ca. Cancer J. Clin. 71, 209–249. doi:10.3322/caac.21660
Takei, S., Kawazoe, A., and Shitara, K. (2022). The new era of immunotherapy in gastric cancer. Cancers (Basel) 14, 1054. doi:10.3390/cancers14041054
Takeichi, M., Nimura, K., Mori, M., Nakagami, H., and Kaneda, Y. (2013). The transcription factors Tbx18 and Wt1 control the epicardial epithelial-mesenchymal transition through bi-directional regulation of Slug in murine primary epicardial cells. PLoS One 8, e57829. doi:10.1371/journal.pone.0057829
Tang, H., Massi, D., Hemmings, B. A., Mandalà, M., Hu, Z., Wicki, A., et al. (2016). AKT-ions with a TWIST between EMT and MET. Oncotarget 7, 62767–62777. doi:10.18632/oncotarget.11232
Timmerman, L. A., Grego-Bessa, J., Raya, A., Bertrán, E., Pérez-Pomares, J. M., Díez, J., et al. (2004). Notch promotes epithelial-mesenchymal transition during cardiac development and oncogenic transformation. Genes Dev. 18, 99–115. doi:10.1101/gad.276304
Trelstad, R. L., Hay, E. D., and Revel, J. D. (1967). Cell contact during early morphogenesis in the chick embryo. Dev. Biol. 16, 78–106. doi:10.1016/0012-1606(67)90018-8
Uemura, N., Okamoto, S., Yamamoto, S., Matsumura, N., Yamaguchi, S., Yamakido, M., et al. (2001). Helicobacter pylori infection and the development of gastric cancer. N. Engl. J. Med. 345, 784–789. doi:10.1056/NEJMoa001999
Villarejo, A., Cortés-Cabrera, A., Molina-Ortíz, P., Portillo, F., and Cano, A. (2014). Differential role of Snail1 and Snail2 zinc fingers in E-cadherin repression and epithelial to mesenchymal transition. J. Biol. Chem. 289, 930–941. doi:10.1074/jbc.M113.528026
Wang, F., Zhu, W., Yang, R., Xie, W., and Wang, D. (2019a). LncRNA ZEB2-AS1 contributes to the tumorigenesis of gastric cancer via activating the Wnt/β-catenin pathway. Mol. Cell. Biochem. 456, 73–83. doi:10.1007/s11010-018-03491-7
Wang, H., Di, X., Bi, Y., Sun, S., and Wang, T. (2021a). Long non-coding RNA LINC00649 regulates YES-associated protein 1 (YAP1)/Hippo pathway to accelerate gastric cancer (GC) progression via sequestering miR-16-5p. Bioengineered 12, 1791–1802. doi:10.1080/21655979.2021.1924554
Wang, H., Dong, X., Gu, X., Qin, R., Jia, H., and One, J. G. J. P. (2015). The MicroRNA-217 functions as a potential tumor suppressor in gastric cancer by targeting GPC5. PLoS One 10, e0125474. doi:10.1371/journal.pone.0125474
Wang, J., Hu, W., Wu, X., Wang, K., Yu, J., Luo, B., et al. (2016a). CXCR1 promotes malignant behavior of gastric cancer cells in vitro and in vivo in AKT and ERK1/2 phosphorylation. Int. J. Oncol. 48, 2184–2196. doi:10.3892/ijo.2016.3428
Wang, J., Lv, B., Su, Y., Wang, X., Bu, J., and Oncotargets, L. Y. J. (2019b). THERAPYExosome-mediated transfer of lncRNA HOTTIP promotes cisplatin resistance in gastric cancer cells by regulating HMGA1/miR-218 Axis. Onco. Targets. Ther. 12, 11325–11338. doi:10.2147/OTT.S231846
Wang, J., Wang, Q., Wei, B., Zhou, Y., Qian, Z., Gao, Y., et al. (2019c). Intronic polymorphisms in genes LRFN2 (rs2494938) and DNAH11 (rs2285947) are prognostic indicators of esophageal squamous cell carcinoma. BMC Med. Genet. 20, 72. doi:10.1186/s12881-019-0796-9
Wang, S., Liu, F., Deng, J., Cai, X., Han, J., and Liu, Q. (2016b). Long noncoding RNA ROR regulates proliferation, invasion, and stemness of gastric cancer stem cell. Cell. Reprogr. 18, 319–326. doi:10.1089/cell.2016.0001
Wang, X., Gong, Z., Ma, L., and Wang, Q. (2021b). LncRNA GACAT1 induces tongue squamous cell carcinoma migration and proliferation via miR-149. J. Cell. Mol. Med. 25, 8215–8221. doi:10.1111/jcmm.16690
Wang, Y., and Gao, W. J. (2021). Long non-coding RNA-H19 promotes ovarian cancer cell proliferation and migration via the microRNA-140/Wnt1 axis. Kaohsiung J. Med. Sci. 37, 768–775. doi:10.1002/kjm2.12393
Wang, Y. J., Liu, J. Z., Lv, P., Dang, Y., Gao, J. Y., and Wang, Y. (2016c). Long non-coding RNA CCAT2 promotes gastric cancer proliferation and invasion by regulating the E-cadherin and LATS2. Am. J. Cancer Res. 6, 2651–2660.
Wang, Y., Shi, J., Chai, K., Ying, X., and Zhou, B. P. (2013). The role of snail in EMT and tumorigenesis. Curr. Cancer Drug Targets 13, 963–972. doi:10.2174/15680096113136660102
Wang, Y., and Zhou, B. P. (2013). Epithelial-mesenchymal transition-A hallmark of breast cancer metastasis. Cancer Hallm. 1, 38–49. doi:10.1166/ch.2013.1004
Wei, L., Sun, J., Zhang, N., Zheng, Y., Wang, X., Lv, L., et al. (2020). Noncoding RNAs in gastric cancer: Implications for drug resistance. Mol. Cancer 19, 62. doi:10.1186/s12943-020-01185-7
Wei, Z., Yajuan, Z., Wenbin, W., Min, C., and Chunjie, M. (2018). Enhanced expression of lncRNA TP73-AS1 predicts unfavorable prognosis for gastric cancer and promotes cell migration and invasion by induction of EMT. Gene 678, 377–383. doi:10.1016/j.gene.2018.08.055
Wu, H., Hu, Y., Liu, X., Song, W., Gong, P., Zhang, K., et al. (2017). LncRNA TRERNA1 function as an enhancer of SNAI1 promotes gastric cancer metastasis by regulating epithelial-mesenchymal transition. Mol. Ther. Nucleic Acids 8, 291–299. doi:10.1016/j.omtn.2017.06.021
Wu, P., Mo, Y., Peng, M., Tang, T., Zhong, Y., Deng, X., et al. (2020). Emerging role of tumor-related functional peptides encoded by lncRNA and circRNA. Mol. Cancer 19, 22. doi:10.1186/s12943-020-1147-3
Wu, Q., Xiang, S., Ma, J., Hui, P., Wang, T., Meng, W., et al. (2018). Long non-coding RNA CASC15 regulates gastric cancer cell proliferation, migration and epithelial mesenchymal transition by targeting CDKN1A and ZEB1. Mol. Oncol. 12, 799–813. doi:10.1002/1878-0261.12187
Xie, M., Nie, F. Q., Sun, M., Xia, R., Liu, Y. W., Zhou, P., et al. (2015). Decreased long noncoding RNA SPRY4-IT1 contributing to gastric cancer cell metastasis partly via affecting epithelial-mesenchymal transition. J. Transl. Med. 13, 250. doi:10.1186/s12967-015-0595-9
Xiu, B., Chi, Y., Liu, L., Chi, W., Zhang, Q., Chen, J., et al. (2019). LINC02273 drives breast cancer metastasis by epigenetically increasing AGR2 transcription. Mol. Cancer 18, 187. doi:10.1186/s12943-019-1115-y
Xu, G., Meng, L., Yuan, D., Li, K., Zhang, Y., Dang, C., et al. (2018a). MEG3/miR-21 axis affects cell mobility by suppressing epithelial-mesenchymal transition in gastric cancer. Oncol. Rep. 40, 39–48. doi:10.3892/or.2018.6424
Xu, M. D., Wang, Y., Weng, W., Wei, P., Qi, P., Zhang, Q., et al. (2017). A positive feedback loop of lncRNA-PVT1 and FOXM1 facilitates gastric cancer growth and invasion. Clin. Cancer Res. 23, 2071–2080. doi:10.1158/1078-0432.CCR-16-0742
Xu, Q., Deng, F., Qin, Y., Zhao, Z., Wu, Z., Xing, Z., et al. (2016). Long non-coding RNA regulation of epithelial–mesenchymal transition in cancer metastasis. Cell Death Dis. 7, e2254. doi:10.1038/cddis.2016.149
Xu, W., He, L., Li, Y., Tan, Y., Zhang, F., and Hong Xu %J Bioscience, B. (2018b). Silencing of lncRNA ZFAS1 inhibits malignancies by blocking Wnt/β-catenin signaling in gastric cancer cells. Biosci. Biotechnol. Biochem. 82, 456–465. doi:10.1080/09168451.2018.1431518
Xu, W., Yang, Z., Adhesion, N. L. J. C., and Migration, (2015). A new role for the PI3K/Akt signaling pathway in the epithelial-mesenchymal transition. Cell adh. Migr. 9, 317–324. doi:10.1080/19336918.2015.1016686
Xu, Y., Sun, J. Y., Jin, Y. F., and Yu, H. (2018c). PCAT6 participates in the development of gastric cancer through endogenously competition with microRNA-30. Eur. Rev. Med. Pharmacol. Sci. 22, 5206–5213. doi:10.26355/eurrev_201808_15718
Xu, Y., Zhang, G., Zou, C., Qi, W., Gong, Z., Zhang, G., et al. (2019). Long non-coding RNA LINC01225 promotes proliferation, invasion and migration of gastric cancer via Wnt/β-catenin signalling pathway. J. Cell. Mol. Med. 23, 7581–7591. doi:10.1111/jcmm.14627
Xu, Z. Y., Yu, Q. M., Du, Y. A., Yang, L. T., Dong, R. Z., Huang, L., et al. (2013). Knockdown of long non-coding RNA HOTAIR suppresses tumor invasion and reverses epithelial-mesenchymal transition in gastric cancer. Int. J. Biol. Sci. 9, 587–597. doi:10.7150/ijbs.6339
Yan, K., Tian, J., Shi, W., Xia, H., and Zhu, Y. (2017). LncRNA SNHG6 is associated with poor prognosis of gastric cancer and promotes cell proliferation and EMT through epigenetically silencing p27 and sponging miR-101-3p. Cell. Physiol. biochem. 42, 999–1012. doi:10.1159/000478682
Yang, J., Antin, P., Berx, G., Blanpain, C., Brabletz, T., Bronner, M., et al. (2020a). Guidelines and definitions for research on epithelial–mesenchymal transition. Nat. Rev. Mol. Cell Biol. 21, 341–352. doi:10.1038/s41580-020-0237-9
Yang, L., Cui, M., Zhang, L., and Song, L. (2017). FOXM1 facilitates gastric cancer cell migration and invasion by inducing Cathepsin D. Oncotarget 8, 68180–68190. doi:10.18632/oncotarget.19254
Yang, L., Ying, X., Liu, S., Lyu, G., Xu, Z., Zhang, X., et al. (2020b). Gastric cancer: Epidemiology, risk factors and prevention strategies. Chin. J. Cancer Res. 32, 695–704. doi:10.21147/j.issn.1000-9604.2020.06.03
Yang, T., Zeng, H., Chen, W., Zheng, R., Zhang, Y., Li, Z., et al. (2016a). Helicobacter pylori infection, H19 and LINC00152 expression in serum and risk of gastric cancer in a Chinese population. Cancer Epidemiol. 44, 147–153. doi:10.1016/j.canep.2016.08.015
Yang, X.-Z., Cheng, T.-T., He, Q.-J., Lei, Z.-Y., Chi, J., Tang, Z., et al. (2018). LINC01133 as ceRNA inhibits gastric cancer progression by sponging miR-106a-3p to regulate APC expression and the Wnt/β-catenin pathway. Mol. Cancer 17, 126. doi:10.1186/s12943-018-0874-1
Yang, Y., Shao, Y., Zhu, M., Li, Q., Yang, F., Lu, X., et al. (2016b). Using gastric juice lncRNA-ABHD11-AS1 as a novel type of biomarker in the screening of gastric cancer. Tumour Biol. 37, 1183–1188. doi:10.1007/s13277-015-3903-3
Ye, M., Zhang, J., Wei, M., Liu, B., and Dong, K. (2020). Emerging role of long noncoding RNA-encoded micropeptides in cancer. Cancer Cell Int. 20, 506. doi:10.1186/s12935-020-01589-x
Ye, Y., Ge, O., Zang, C., Yu, L., Eucker, J., and Chen, Y. (2022). LINC01094 predicts poor prognosis in patients with gastric cancer and is correlated with EMT and macrophage infiltration. Technol. Cancer Res. Treat. 21, 15330338221080977. doi:10.1177/15330338221080977
Yilmaz, M., and Christofori, G. (2009). EMT, the cytoskeleton, and cancer cell invasion. Cancer Metastasis Rev. 28, 15–33. doi:10.1007/s10555-008-9169-0
Yin, G., Tian, P. R., He, A. B., Yan, W., Li, T. X., and Sun, Z. P. (2020). Lncrna linc00689 promotes the progression of gastric cancer through upregulation of adam9 by sponging mir-526b-3p. Cancer Manag. Res. 12, 4227–4239. doi:10.2147/CMAR.S231042
Ying, C., Guoqing, W., Hongwei, X., Huangfei, Y., Qiuling, T., and Feng, B. (2017a). Down regulation of lincRNA-p21 contributes to gastric cancer development through Hippo-independent activation of YAP. Oncotarget 8, 63813–63824. doi:10.18632/oncotarget.19130
Ying, Y., Qingwu, L., Mingming, X., Zhenju, S., Chaoyang, T., and Physiology, T. Z. J. C. (2017b). Emodin: One main ingredient of shufeng jiedu capsule reverses chemoresistance of lung cancer cells through inhibition of EMT. Cell. Physiol. biochem. 42, 1063–1072. doi:10.1159/000478754
Yu, L., Chun-Dong, Z., Cheng, Z., and Dong-Qiu, D. (2020). DLX6-AS1/miR-204-5p/OCT1 positive feedback loop promotes tumor progression and epithelial-mesenchymal transition in gastric cancer. Gastric Cancer 23, 212–227. doi:10.1007/s10120-019-01002-1
Yu, Y., Li, L., Zheng, Z., Chen, S., Chen, E., and Hu, Y. (2017). Long non-coding RNA linc00261 suppresses gastric cancer progression via promoting Slug degradation. J. Cell. Mol. Med. 21, 955–967. doi:10.1111/jcmm.13035
Yuan, H., Kajiyama, H., Ito, S., Yoshikawa, N., Hyodo, T., Asano, E., et al. (2013). ALX1 induces snail expression to promote epithelial-to-mesenchymal transition and invasion of ovarian cancer cells. Cancer Res. 73, 1581–1590. doi:10.1158/0008-5472.CAN-12-2377
Zeisberg, M., and Neilson, E. G. (2009). Biomarkers for epithelial-mesenchymal transitions. J. Clin. Invest. 119, 1429–1437. doi:10.1172/JCI36183
Zeng, L., Liao, Q., Zou, Z., Wen, Y., Wang, J., Liu, C., et al. (2018). Long non-coding RNA XLOC_006753 promotes the development of multidrug resistance in gastric cancer cells through the PI3K/AKT/mTOR signaling pathway. Cell. Physiol. biochem. 51, 1221–1236. doi:10.1159/000495499
Zhan, T., Rindtorff, N., and Oncogene, M. B. J. (2017). Wnt signaling in cancer. Oncogene 36, 1461–1473. doi:10.1038/onc.2016.304
Zhang, C., Guo, F., Xu, G., Ma, J., and Shao, F. (2015). STAT3 cooperates with Twist to mediate epithelial-mesenchymal transition in human hepatocellular carcinoma cells. Oncol. Rep. 33, 1872–1882. doi:10.3892/or.2015.3783
Zhang, G., Li, S., Lu, J., Ge, Y., Wang, Q., Ma, G., et al. (2018a). LncRNA MT1JP functions as a ceRNA in regulating FBXW7 through competitively binding to miR-92a-3p in gastric cancer. Mol. Cancer 17, 87. doi:10.1186/s12943-018-0829-6
Zhang, J., Jin, H. Y., Wu, Y., Zheng, Z. C., Guo, S., Wang, Y., et al. (2019a). Hypoxia-induced LncRNA PCGEM1 promotes invasion and metastasis of gastric cancer through regulating SNAI1. Clin. Transl. Oncol. 21, 1142–1151. doi:10.1007/s12094-019-02035-9
Zhang, K., Lu, C., Huang, X., Cui, J., Li, J., Gao, Y., et al. (2019b). Long noncoding RNA AOC4P regulates tumor cell proliferation and invasion by epithelial-mesenchymal transition in gastric cancer. Ther. Adv. Gastroenterol. 12, 1756284819827697. doi:10.1177/1756284819827697
Zhang, W., Zhan, F., Li, D., Wang, T., and Huang, H. (2020a). RGMB-AS1/miR-22-3p/NFIB axis contributes to the progression of gastric cancer. Neoplasma 67, 484–491. doi:10.4149/neo_2020_190418N350
Zhang, X., Liu, G., Ding, L., Jiang, T., Shao, S., Gao, Y., et al. (2018b). HOXA3 promotes tumor growth of human colon cancer through activating EGFR/Ras/Raf/MEK/ERK signaling pathway. J. Cell. Biochem. 119, 2864–2874. doi:10.1002/jcb.26461
Zhang, X., and Wu, J. (2021). LINC00665 promotes cell proliferation, invasion, and metastasis by activating the TGF-β pathway in gastric cancer. Pathol. Res. Pract. 224, 153492. doi:10.1016/j.prp.2021.153492
Zhang, Y., Yuan, Y., Zhang, Y., Cheng, L., Zhou, X., and Chen, K. (2020b). SNHG7 accelerates cell migration and invasion through regulating miR-34a-Snail-EMT axis in gastric cancer. Cell Cycle 19, 142–152. doi:10.1080/15384101.2019.1699753
Zhao, H., Zhang, K., Wang, T., Cui, J., XI, H., Wang, Y., et al. (2018). Long non-coding RNA AFAP1-antisense RNA 1 promotes the proliferation, migration and invasion of gastric cancer cells and is associated with poor patient survival. Oncol. Lett. 15, 8620–8626. doi:10.3892/ol.2018.8389
Zhao, Y., Guo, Q., Chen, J., Hu, J., Wang, S., and Sun, Y. (2014). Role of long non-coding RNA HULC in cell proliferation, apoptosis and tumor metastasis of gastric cancer: A clinical and in vitro investigation. Oncol. Rep. 31, 358–364. doi:10.3892/or.2013.2850
Zheng, H. X., Cai, Y. D., Wang, Y. D., Cui, X. B., Xie, T. T., Li, W. J., et al. (2013). Fas signaling promotes motility and metastasis through epithelial-mesenchymal transition in gastrointestinal cancer. Oncogene 32, 1183–1192. doi:10.1038/onc.2012.126
Zheng, X., Peng, B., Wu, X., Ye, J., Zhao, H., Li, Y., et al. (2022). Male-specific long non-coding RNA testis-specific transcript, Y-linked 15 promotes gastric cancer cell growth by regulating Wnt family member 1/β-catenin signaling by sponging microRNA let-7a-5p. Bioengineered 13, 8605–8616. doi:10.1080/21655979.2022.2053814
Zhou, C., An, N., Cao, C., and Wang, G. (2020a). lncRNA HOXC-AS1 promotes gastric cancer via binding eIF4AIII by activating Wnt/β-catenin signaling. J. Gene Med. 22, e3202. doi:10.1002/jgm.3202
Zhou, H., Shen, W., Zou, H., Lv, Q., and Shao, P. (2020b). Circulating exosomal long non-coding RNA H19 as a potential novel diagnostic and prognostic biomarker for gastric cancer. J. Int. Med. Res. 48, 300060520934297. doi:10.1177/0300060520934297
Zhou, Q., Li, H., Jing, J., Yuan, Y., and Sun, L. (2020c). Evaluation of C5orf66-AS1 as a potential biomarker for predicting early gastric cancer and its role in gastric carcinogenesis. Onco. Targets. Ther. 13, 2795–2805. doi:10.2147/OTT.S239965
Zhou, X., Chen, H., Zhu, L., Hao, B., Zhang, W., Hua, J., et al. (2016). Helicobacter pylori infection related long noncoding RNA (lncRNA) AF147447 inhibits gastric cancer proliferation and invasion by targeting MUC2 and up-regulating miR-34c. Oncotarget 7, 82770–82782. doi:10.18632/oncotarget.13165
Keywords: gastric cancer, epithelial-mesenchymal transition, long non-coding RNAs, signalling pathways, transcription factors, microRNAs
Citation: Feng Y-N, Li B-Y, Wang K, Li X-X, Zhang L and Dong X-Z (2022) Epithelial-mesenchymal transition-related long noncoding RNAs in gastric carcinoma. Front. Mol. Biosci. 9:977280. doi: 10.3389/fmolb.2022.977280
Received: 24 June 2022; Accepted: 29 September 2022;
Published: 12 October 2022.
Edited by:
Xuefeng Liu, The Ohio State University, United StatesReviewed by:
Ann-Kristin Östlund Farrants, Stockholm University, SwedenWilliam K. K. Wu, Chinese University of Hong Kong, China
Copyright © 2022 Feng, Li, Wang, Li, Zhang and Dong. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xian-Zhe Dong, ZG9uZ3hpYW56aGVAMTYzLmNvbQ==
†These authors have contributed equally to this work and share first authorship
 Ying-Nan Feng
Ying-Nan Feng Bo-Ya Li
Bo-Ya Li Ke Wang
Ke Wang Xiao-Xi Li
Xiao-Xi Li Lan Zhang
Lan Zhang Xian-Zhe Dong
Xian-Zhe Dong