
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Mol. Biosci. , 20 September 2022
Sec. RNA Networks and Biology
Volume 9 - 2022 | https://doi.org/10.3389/fmolb.2022.974348
This article is part of the Research Topic Construction of Transcriptional and Alternative Splicing Regulatory Networks View all 7 articles
Farmed chinese giant salamander (Andrias davidianus) was an important distinctive economically amphibian that exhibited male-biased sexual size dimorphism. Fgf9 and rspo1 genes antagonize each other in Wnt4 signal pathway to regulate mammalian gonadal differentiation has been demonstrated. However, their expression profile and function in A. davidianus are unclear. In this study, we firstly characterized fgf9 and rspo1 genes expression in developing gonad. Results showed that fgf9 expression level was higher in testes than in ovaries and increased from 1 to 6 years while rspo1 expression was higher in ovaries than in testes. In situ hybridization assay showed that both fgf9 and rspo1 genes expressed at 62 dpf in undifferentiated gonad, and fgf9 gene was mainly expressed in spermatogonia and sertoli cells in testis while strong positive signal of rspo1 was detected in granular cell in ovary. During sex-reversal, fgf9 expression was significantly higher in reversed testes and normal testes than in ovaries, and opposite expression pattern was detected for rspo1. When FH535 was used to inhibit Wnt/β-catenin pathway, expression of rspo1, wnt4 and β-catenin was down-regulated. Conversely, expression of fgf9, dmrt1, ftz-f1 and cyp17 were up-regulated. Furthermore, when rspo1 and fgf9 were knocked down using RNAi technology, respectively. We observed that female biased genes were down regulated in ovary primordial cells after rspo1 was knocked down, while the opposite expression profile was observed in testis primordial cells after fgf9 was knocked down. These results suggested that fgf9 and rspo1 played an antagonistic role to regulate sex differentiation in the process of the gonadal development and provided a foundation for further functional characterizations. The data also provided basic information for genome editing breeding to improve the Chinese giant salamander farming industry.
The Chinese giant salamander A. davidianus is the largest extant amphibian, from aquatic to terrestrial vertebrates, in world with the body length to 1.8 m (Turvey et al., 2019). Due to habitat destruction, wild population were sharply decreased. From 1998, A. davidianus was listed as endangered on the China Species Red List. Artificial propagation provide a approach to conserve A. davidianus and also generate a new industry. From 2006, commercial sale of F2 generation Chinese giant salamander has been allowed by the Chinese government. Now, commercial breeding and farming of giant salamander has became an important industry. However, male salamander grow faster than female which impeded the development of the industry.
Fibroblast growth factor-9 (fgf9) is one of the fibroblast growth factor, which plays an important role in the development of ovarian cancer, bone development, and gonadal differentiation (Blasio et al., 1995; Harada and Akita, 2020). Fgf9 was originally isolated from a human glial cell line. Based on its activity, it was named glia-activating factor (Plotnikov et al., 2001). Later, further studies found that fgf9 not only plays an important role in ovarian cancer and bone development but also plays a very important role in mammal sex determination and differentiation (Colvin et al., 2001; Behr et al., 2010). In mice, the lack of fgf9 disrupts the expression of sox9 in the XY gonads and up regulates the expression of wnt4, preventing the embryo from progressing through the male differentiation pathway and resulting in the phenomenon of sex reversal (Colvin et al., 2001). Transgenic mice with loss-of-function fgf9 exhibit male-to-female sex reversal, and gain of fgf9 copy number was found in human 46, a patient with XX sex reversal and disorders of sex development (Li et al., 2020).
Kamata et al. (2004) discovered a gene with a thrombospondin (TSP) type I structure and named it R-spondin (roof plate-specific spondin). R-spondin-1 can bind to specific receptors to activate the Wnt/β-catenin pathway to regulate cell proliferation, differentiation and apoptosis (van Meurs et al., 2011). In mice, inactivation of rspo1 by homologous recombination led to partial XX sex reversal (Zhang et al., 2011). Expression of rspo1 in various vertebrate species showed that rspo1 promote ovarian development (Binnerts and Kim, 2007; Wei et al., 2007; Bagheri-Fam et al., 2008; Kocer et al., 2008; Tomizuka et al., 2008). Human r-spondin1 (rspo1) was considered to be an activating gene in ovarian development. Rspo1 activated the Rspo1/β-catenin/Wnt signal pathway in the human and mouse reproductive system, especially for the early sex differentiation (Kim and Capel, 2006; Tomaselli et al., 2011; Biason-Lauber, 2012). In human, mutations in rspo1 gene can lead to complete female-to-male sexual reversal (Parma et al., 2006). The results suggested that rspo1 is necessary for human sex determination.
Studies shown that in mammals, the sox9/fgf9 and rspo1/wnt4 signal pathways compete with each other to reach a balance to determine sex differentiation (Piprek, 2009). When the expression of male-biased genes (sox9, fgf9) is higher than that of female-biased genes (wnt4, rspo1), the balance of sexual differentiation tilts towards the male pathway, and the gonads develop into testes. Otherwise, the gonads will develop into ovaries (Kim et al., 2006). There is an interactive positive feedback loop between fgf9 and sox9 (Kim and Capel, 2006). Studies have found that XY fgf9/wnt4 and FGFR2/Wnt4 double mutants develop testes with male somatic cells and germ cells, which indicates that the main role of FGF signal is to inhibit female-promoting genes (Jameson et al., 2012). Whether the gonads differentiate towards the testes depends not only on the expression of male genes (such as sox9) but also on the active suppression of female genes (such as wnt4) (Jameson et al., 2012). The R-spondin1/Wnt4/β-catenin pathway and foxl2 transcription factor work in a complementary manner to promote ovarian growth and inhibit testicular development (Ottolenghi et al., 2007; Maatouk et al., 2008; Nef and Vassalli, 2009). Through the scattered in-frame missense and splicing mutations of the MAP3K1 gene, the expression of WNT/β-catenin/FOXL2 activity are enhanced and the expression of sox9/fgf9/fgfr2/sry is reduced to mediate the tilting of the sex determination pathway from male to female balance (Loke et al., 2014).
In the previous study, we have determined the beginning time of sex differentiation at 98 dpf (Hu et al., 2019a) and explore the female-specific marker (Hu et al., 2019b). Additionally, we found high temperature at 28°C could induce the sex reversal from undifferentiated genetic female to male and estradiol induce the undifferentiated genetic male to female (Hu et al., 2019c). In present study, we characterized fgf9 and rspo1 expression using qRT-PCR, and determined their expression in undifferentiated and differentiated gonad by in situ hybridization. We also study antagonistic relationship of fgf9 and rspo1 genes using the Wnt/β-catenin inhibitor (FH535). Furthermore, RNAi technology was used to study the function of fgf9 and rspo1 genes.
The experimental animals (A. davidianus Blanchard, 1871; Cryptobranchidae, Caudata) were collected from the salamander breeding base at Zhejiang Yongqiang Chinese Giant Salamander Co., Ltd., (Jinhua, Zhejiang Province, China). The salamanders were killed after anaesthesia by MS222 according to Yangtze River Fisheries Research Institute Care Committee (No. 2013001). According to previous study, we found that gonad of A. davidianus began to differentiate on the 98 days after fertilization (Hu et al., 2019a). The developing stage gonads of 62 dpf (days post fertilization), 98 dpf, 130 dpf and 1-, 2-, 3-, 4-, 5-, and 6-years-old (6 years old) giant salamanders in the normal group and sex hormone induction group were collected. Two samples of each tissue were collected, and one was stored in RNAstore Reagent for RNA extraction (Tiangen, DP408); the other part was preserved in 4% paraformaldehyde (pH 7.5) to prepare tissue sections; at the same time, the muscle tissue of each individual above was collected and preserved in absolute ethanol for DNA extraction using the TIANamp Genomic DNA Kit (Tiangen, Beijing, China) including RNase A treatment according the manufacturer’s instructions (Hu et al., 2019b). The testes, ovary, liver, spleen, lung, kidney, stomach, intestine, brain and pituitary tissues were collected from 2 years old A. davidianus, and put into RNAstore Reagent, and then transferred to a refrigerator at −80°C for storage until RNA extraction.
The larvae at 55 dpf were treated by estradiol or high temperature (28°C) respectively until 8 months post-fertilization to obtain the sex reversal individual according the previous study (Hu et al., 2019c). Using tissue sections and microscopes, the physiological sex of A. davidianus samples was identified, and, the genetic sex of used A. davidianus were identified using female-specific markers adf340 developed by our laboratory (Hu et al., 2019b). The PCR system consisted of 12.5 μl 2X Master Mix (TsingKe, TSE003), 100 ng genomic DNA, 0.5 µl of each primer (10 μmol/L), and double distilled water to reach a final volume of 25 μl. The reaction conditions were as follows: 95°C for 5 min; 33 cycles of 94°C for 15 s, 60°C for 15 s, 72°C for 30 s; and 72°C for 5 min. If the sex and genotype were consistent, there was no sex reversal. If the sex and genotype were different, namely, the sex was male, and the genotype was female, the individual was considered a sex reversal male (RM) individual; if the sex was female, and the genotype was male, the individual was considered a sex reversal female (RF) individual.
According to the partial sequences of the fgf9 and rspo1 genes in the giant salamander transcriptome database from our laboratory (Hu et al., 2019c), primers were designed to amplify the 5′ or 3′ UTR (Table 1). The 5′-RACE-Ready cDNA and 3′-RACE-Ready cDNA libraries were constructed according to the SMART™ RACE kit (Clontech, 634923) instructions as templates for RACE. PCR was performed as follows: five cycles of 94°C for 30 s and 72°C for 3 min; five cycles of 94°C for 30 s, 70°C for 30 s, and 72°C for 3 min; and then 27 cycles of 94°C for 30 s, 68°C for 30 s, and 72°C for 3 min. The amplified product was electrophoresed on 1% agarose and the purified fragment was cloned into the pMD18-T vector, transfected into E. coli strain TOP10 (Invitrogen, C404003), and then sequenced.
The sequence was searched in the GenBank database using BLAST (http://www.ncbi.nlm.nih.gov/BLAST/). The protein molecular weight was deduced using the website (http://www.bio-soft.net/sms/index.html). The conserved domain was predicted with online software (http://www.ncbi.nlm.nih.gov/structure). The protein domains of fgf9 and rspo1 were analyzed by the online software SMART (http://smart.embl-heidelberg.de/smart/set_mode.cgi). The phylogenetic tree of fgf9 and rspo1 was constructed based on protein sequence by MEGA 7.0 (Kumar et al., 2016) with the NJ method (neighbor-joining method). Bootstrap values were estimated from 1,000 replications.
To detect fgf9 and rspo1 expression in gonadal tissue, two pairs of primers were designed to obtain a riboprobe (Table 1). A 468 bp cDNA fragment was amplified from the fgf9 ORF domain, and a 533 bp cDNA fragment was amplified from rspo1. Primers were designed to introduce the T7 promoter sequence in the 5′- or 3′- side of the amplicons that will later be used as riboprobes, and PCR products were purified by a QIAquick Gel Extraction Kit (QIAGEN, Cat# 28706). To obtain probes, transcription was performed using the MEGAshortscript T7 High Yield Transcription Kit (Invitrogen, AM1354) according the manufacturer’s instructions In situ hybridization was performed following the method described in previous study (Hu et al., 2016). Sections were deparaffinized, hydrated and treated with proteinase K and then hybridized using sense or antisense DIG-labeled RNA probe at 70°C for 12 h. Hybridization signals were then detected with anti-DIG (Roche, 11093274910) conjugated with POD. DAB (Beyotime, P0202) was used as substrate for POD (Hu et al., 2016).
Ovary of female at 1 year old were dissected and washed with the PBS containing 1% Penicillin-Streptomycin (P/S) (Gibco, Thermo Fisher Scientific), then the gonads were cut into small pieces and placed on a 70 μm nylon mesh (Biologix,15-1070) and gently pushed through the mesh with constantly dripping of cold DMEM/F-12 medium (Gibco, 11320082) containing 10% FBS (BioInd, 18455), 1% heparin (Gibco, A16198.03) and Penicillin-Streptomycin (P/S) (Gibco, 15140122). The collected cells were washed using DMEM medium and then centrifuged at 500 × g for 10 min at 4°C. The supernatant was discarded and the cells were collected. The cells were divided into six groups and seed in a 6-well plate (Corning) with the density of 1 × 106 cells/well. Three were set as control and the other three for FH535 treatment. The FH535 (MCE,108409-83-2) was added to the treated groups with the final concentration of 20 uM and incubate at 28°C for 3 days. The cells were collected and RNA was extracted for qRT-PCR.
According to fgf9 and rspo1 sequence, three siRNA at different sites were designed and synthesized for each gene (GenePharma, Shanghai, China, Table1). The siRNA marked by FAM was used as the negative control. The primordial gonadal cells were prepared according to the above description. The siRNA was transfected into the gonadal cells according to the manufacture’s instruction of Lipofectamine TM3000 (Thermo Fisher Scientific). Then the cells were incubated in opti-MEM medium (Gibco, Thermo Fisher Scientific) at 37°C for 6 h. After that, opti-MEM medium was removed and 2 ml of DMEM medium containing 10% FBS and P/S were added and incubated at 28°C for 48 h. The signal of green fluorescence was detected by OLYMPUS IX73. The RNA was extracted according the TRIzol method for qRT-PCR. To further detect the function of fgf9 and rspo1, the siRNA of fgf9 and rspo1 with the best inhibition effect were injection into the salamander after the sex was identified by the sex specific primers according to previous study (Hu et al., 2019b; 2019d). Sex related gene expression profile were detected in ovary and testis after rspo1/siRNA and fgf9/siRNA treatment for 72 h, respectively.
Total RNA was isolated from tissues using Trizol reagent (Invitrogen, 15596018). The cDNA was synthesized using PrimeScript™ RT reagent Kit following manufacturer’s instructions (Takara, RR047A). The expression of fgf9 and rspo1 genes in various tissues and developing gonad in A. davidianus was analyzed by quantitative real-time PCR (qRT-PCR). qRT-PCR was conducted using 2 × T5 Fast qPCR Mix (SYBR Green) (TsingKe, TSE202) following manufacturer’s instructions independently in triplicate on the QuantStudio 5 Real-Time PCR System (Applied Biosystems, California, United States). β-Actin was used as the internal control gene, cDNA was used as a template, tissue from three individuals was used, and each reaction was repeated three times. qRT-PCR was performed as follows: 95°C for 30 s; forty cycles of 95°C for 5 s, 60°C for 30 s, and 72°C for 30 s; and then 72°C for 5 min. The difference in expression was analyzed by one-way ANOVA followed by Duncan multiple comparison tests using SPSS 22.0 (IBM, New York, NY, United States). Significance was set at p < 0.05. Bars with different letters differ significantly (p < 0.05).
With RT-PCR and RACE amplification, full-length fgf9 and rspo1 cDNA were cloned from A. davidianus gonad at 1 year old (GenBank Accession No. MW685518 and MW685519). The full length of fgf9 was 1489 bp with a 5′UTR of 612 bp and a 3′UTR of 247 bp, and the full length of rspo1 was 1708 bp with a 5′UTR of 430 bp, an ORF of 786 bp, and a 3′UTR of 492 bp. The ORF of fgf9 encoded a protein of 209 aa, and that of rspo1 encoded a protein of 261 aa, with molecular weights of 23.35 and 28.76 kDa, respectively. A comparison of the selected amino acid sequences of amphibian and mammalian fgf9 showed that this gene is conserved with more than 85% identity (Supplementary Figure S1A). Based on the alignment of the amino acid sequence, a phylogenetic tree was constructed by the neighbour-joining method. The tree of fgf9 showed that A. davidianus is more similar to Cynops orientalis with 93% identity than to Xenopus laevis or Xenopus tropicalis (Supplementary Figures S1A,B). The phylogenetic tree of rspo1 showed that A. davidianus is more similar to C. orientalis with 85% identify than to Xenopus laevis or Xenopus tropicalis (Supplementary Figure S2B).
Genetic sex of the salamander were identified and salamanders without sex reversal were used to analyze the expression profile. Expression levels of the fgf9 and rspo1 genes in various tissues from 1 year old and developing gonad were detected using qRT-PCR. High expression levels of fgf9 were observed in the kidney and testis, followed by the liver, and relatively low expression levels were observed in the spleen and intestine; other tissues showed medium expression levels (Figure 1A). Expression pattern of fgf9 in developing gonad indicated that the expression of fgf9 in testes was significantly higher than that in ovaries (p < 0.05), and mRNA transcription gradually increased from 1 to 6 years of life (Figure 1B). Expression of rspo1 was observed in various tissues and significantly higher expression levels were observed in the ovary than that in other tissues (p < 0.05), medium expression levels were observed in the kidney, and low levels were observed in the remaining tested tissues (Figure 1C). In the developing gonad, rspo1 expression was significantly higher in the ovary than in the testis, with expression levels gradually increased from 1 to 5 years of life and decreasing at 6 years (Figure 1D).
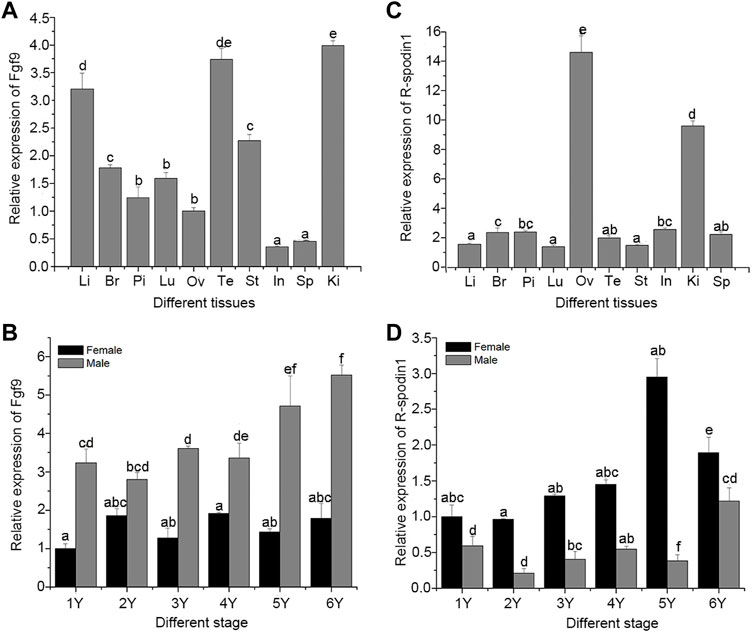
FIGURE 1. Expression of fgf9 and rspo1 gene: expression analysis of fgf9 (A) and rspo1 (B) in various tissues by qRT-PCR; expression analysis of fgf9 (C) and rspo1 (D) in developing gonads by qRT-PCR. Tissue from three individuals and each reaction was repeated three times. Bars with different letters are significantly different (p < 0.05). Li, Liver; Br, Brain; Pi, pituitary; Lu:Lung; Ov, Ovary; Te, Testis; St, Stomach; In, Intestinal; Ki, Kidney; Sp, Spleen.
To detect whether fgf9 or rspo1 expressed before gonad differentiation, In situ hybridization was conducted to identify the expression of fgf9 and rspo1 in the early developing gonad which was too small to detect by the qRT-PCR. At 62 dpf before the gonad differentiation, fgf9 and rspo1 positive signal were detected in the undifferentiated gonad of A. davidianus (Figures 2A1,A2,B1,B2). While the sense probe had no signal (Supplementary Figure S3). At 98 dpf when the gonad begin to differentiation, positive signal of fgf9 and rspo1 were also both detected in developing gonad (Figures 2A3,A4,B3,B4). At 130 dpf, expression of fgf9 and rspo1 were also detected in the gonad. When the gonadal differentiation is complete, we observed that fgf9 signal was strongly distributed in spermatogonia, spermatocyte and sertoli cells in testis while very weak signal in granular cell in ovary in the 2 years old A. davidianus (Figures 2A7,A8,A9,A10). For rspo1 gene, strong signal was detected in granular cell in ovary and weak signal in spermatogonia, spermatocyte and sertoli cells in testis (Figures 2B7,B8,B9,B10).
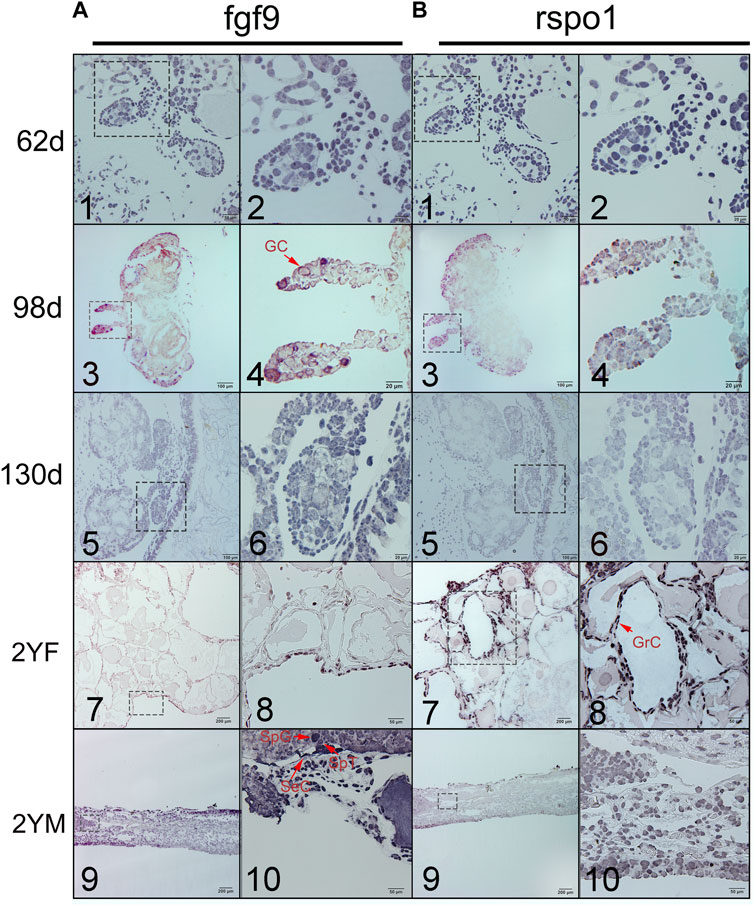
FIGURE 2. Expression of fgf9 (A) and rspo1 (B) in gonads detected by in situ hybridization. 1. Undifferentiated gonad at 62dpf; 2. Large magnification of frame area in (1); 3. Differentiating gonad at 98 dpf; 4. Large magnification of frame area in (3); 5. Differentiating gonad at 130dpf; 6. Large magnification of frame area in (5); 7: 2 year old ovary; 8. Large magnification of frame area in (7); 9. 2 year old testis; 10. Large magnification of frame area in (9). 62dpf: gonad at 62 days after fertilization; 98dpf: gonad at 98 days after fertilization; 2 YF: 2-year-old female gonad;2 YM: 2-year-old male gonad. Three samples were used per stage. Germ cell (GC), granulosa cells (GrC), sertoli cell (SeC), spermatogonia (SpG), spermatocyte (SpT).
Expression level of fgf9 and rspo1 were detected in the gonad and sex-reversal gonad at 1 year old. Expression of fgf9 was significantly higher in testis than in ovary and highest expression was observed in sex-reversal testis (Figure 3A). Conversely, in the gonads and sex-reversal gonads, the expression level of rspo1 was significantly higher in sex-reversal ovaries than in ovaries, and the rspo1 expression level was significantly higher in ovaries and sex-reversal ovaries than the testes and sex-reversal testes (p < 0.05) (Figure 3B).
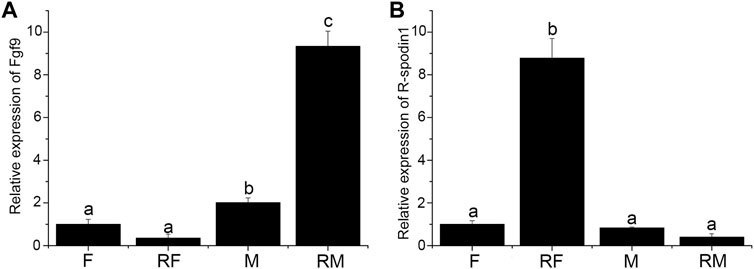
FIGURE 3. Expression of fgf9 and rspo1 in normal gonads and sex-reversal gonads by qRT-PCR. (A) Expression analysis of fgf9 mRNA in normal gonads and sex-reversal gonads by qRT-PCR; (B) expression analysis of rspo1 mRNA in normal gonads and sex-reversal gonads by qRT-PCR. The gonads were collected at 1 year old from three individuals and each reaction was repeated three times. Bars with different letters are significantly different (p < 0.05).
To provide further evidence of rspo1 in gonad development and potential antagonistic relationship of fgf9 and rspo1 genes, primary gonadal cell from 1 year old salamander ovary were collected and incubated with or without FH535. After the primary gonadal cells were treated with FH535, expression profile of sex related gene showed that cyp17, dmrt1, sf1, fgf9 expression were upregulated while rspo1, wnt4 and β-catenin expression was down-regulated comparing with the control without FH535 treatment (Figure 4). Additionally, we observed that cyp19a and frizzled2 expression profile was unchanged (Figure 4).
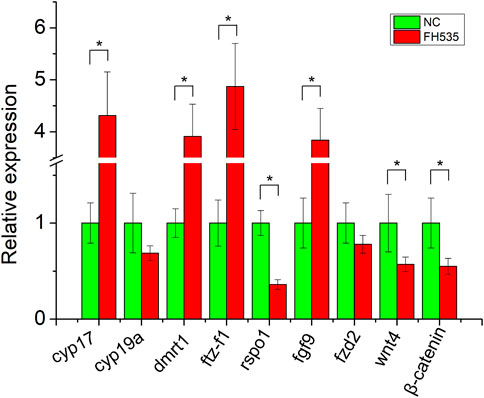
FIGURE 4. Expression profile of sex-related genes after FH535 treatment. The results from three replicates and *indicate the significantly difference (p < 0.05).
To examine the function of fgf9 and rspo1 genes in the gonad, three siRNA sites were designed according the sequence of fgf9 and rspo1, respectively. The control siRNA site marked by FAM was transfected into the gonadal cells and we found that high transfection efficiency was exhibited in ovary cell with 60% and low in testis cell with about 20% (Figure 5A). Expression level of fgf9 and rspo1 were significantly down-regulated in ovary and testis cells at site siRNA R684 and F890, respectively (Figures 5B,D). Seventy-two hours post siRNA R684 treatment in vitro and vivo, we found that expression of cyp17, foxl2, rspo1 and cyp19a were significantly down regulated while dmrt1 and ftz-f1 expression were not changed (Figures 5C,F). Seventy-two hours after siRNA F890 treatment in testis cells, expression of cyp17, fgf9, dmrt1 and ftz-f1 were significantly down regulated while cyp19a expression was up regulated (Figures 5E,G). Only foxl2 expression was not changed.
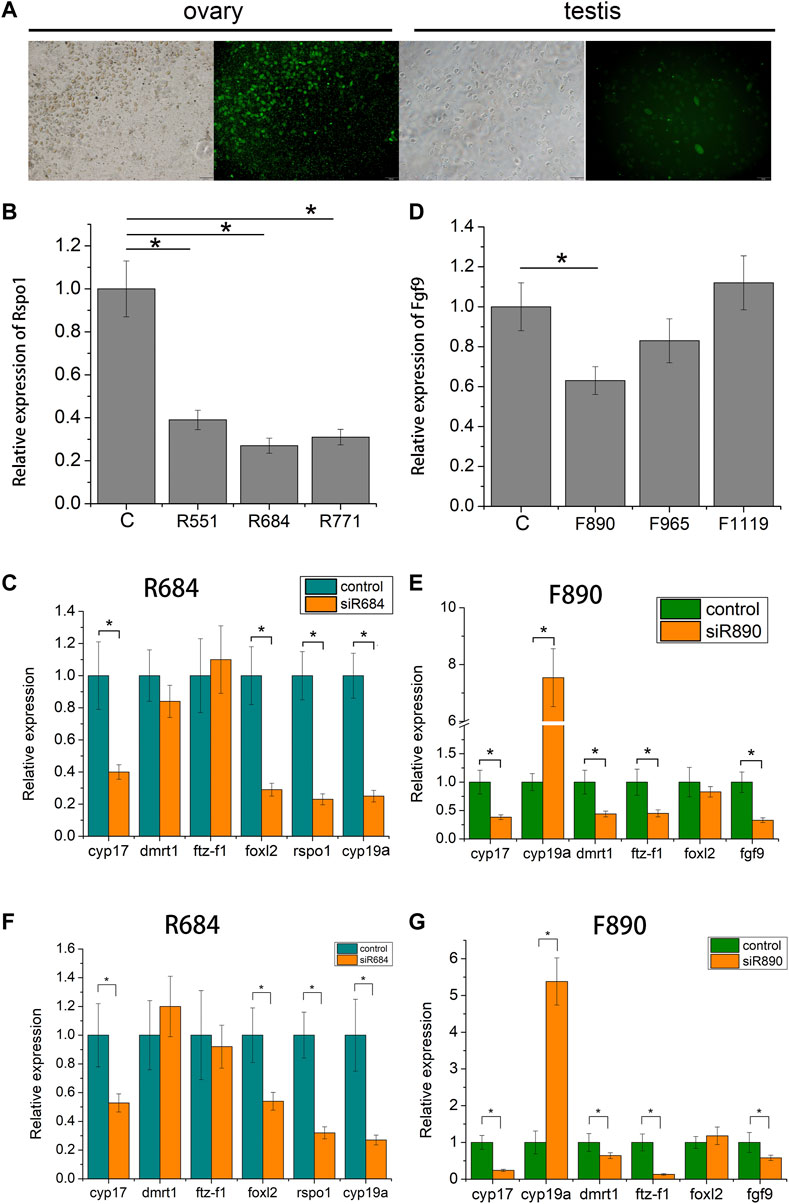
FIGURE 5. Expression profiles of sex-related gene in Andrias davidianus gonad after siRNA treatment. (A) control siRNA marked by FAM transfected into the gonadal cells; (B) Expression level of each rspo1 siRNA sites in ovary cell; (C) expression profile of sex-related genes after R684 treatment in ovary cell; (D) expression level of each fgf9 siRNA sites in testis cell; (E) expression profile of sex-related genes after F890 treatment in testis cell; (F) expression profile of sex-related genes after R684 treatment in ovary tissue; (G) expression profile of sex-related genes after F890 treatment in testis tissue. The results from three replicates and *indicate the significantly difference (p < 0.05).
Among different A. davidianus tissues, the highest expression level of fgf9 was observed in the kidney, followed by the testis and liver, and a lower level was found in other tissues. In previous studies, fgf9 has been proven to be widely expressed in various tissues in goats, mice, and cattle (Liao and Lin, 2009; Lai et al., 2016; Huang, 2020). Yamamura found that in Rana rugosa fgf9 was highly expressed in the ovary but was not detected in the testis; however, it did not show a sex-dependent expression pattern during the sex determination process. Therefore, Yamamura found fgf9 may not be a key factor in the gonadal differentiation of Rana rugosa (Yamamura et al., 2005). In this study, the expression of fgf9 increased from 1 to 6 years during gonadal development, and the expression in the testis was higher than that in the ovaries of the same stage, indicating that fgf9 plays a role in testicular development. Additionally, we found that fgf9 expression was increased during female to male sex reversal which suggested that fgf9 was associated with testis differentiation. Similar expression pattern was observed in other species. In mice, when fgf9 was knocked out, the male to female sex reversal was happen (Colvin et al., 2001). In the mouse, fgf9 is expressed in gonad prior to sex determination, Loss of Fgf9 leads to XY sex reversal (Kim et al., 2006). The results of in situ hybridization showed that fgf9 was expressed in the undifferentiated gonad of A. davidianus at 62 dpf and 98 dpf. Previous study showed that the gonads of A. davidianus began to differentiate on the 98 days after fertilization (Hu et al., 2019a), and the fgf9 gene signal was detected in the gonads of A. davidianus at 62 dpf and 98 dpf, indicating that fgf9 was expressed in A. davidianus at the early stage of gonadal differentiation and affected gonadal differentiation. The weakly positive signal in the ovarian granulosa cells of the 2-years-old A. davidianus and the strong positive signal in the testis indicate that the fgf9 gene always plays an important role in the testes of A. davidianus, but the role in the ovary is not obvious.
The relative expression of rspo1 was assessed by qRT-PCR. The results showed that the relative expression of rspo1 in A. davidianus was the highest in the ovary. The level in the ovary was significantly higher than that in other tissues. Similar rspo1 expression patters were reported in Peldiscus sinensis (Zhang, 2019). The expression of rspo1 in Danio rerio was significantly higher in adult females than in males in the kidney, gonad and muscle. During the gonadal differentiation of vertebrates, the expression of rspo1 in females was significantly higher than that in males. In the developing gonad of A. davidianus, the rspo1 expression level was significantly higher in the ovary than in the testis, with expression levels gradually increasing from 1 to 5 years and decreasing at 6 years. The results showed that rspo1 gradually played a role in ovarian development, but after sexual maturation, the level of rspo1 began to decline, and the expression level in testis began to rise, suggesting that it may play some role in sexually mature testis. However, there is no relevant research or report on the function of rspo1 in testis. The rspo1 gene can play a key role in human and mouse reproductive systems by activating rspo1/β-catenin/Wnt signal pathways, especially in early sex determination (Nam et al., 2007; Zhang et al., 2011). Wnt4 has been found to promote ovarian differentiation by inhibiting male sex differentiation (Biason-Lauber and Konrad, 2008). But the expression profile of wnt4 in the ovary was affected by changes of protein R-spondin-1 (Biason-Lauber, 2012). In the present study, we used FH535 to inhibit rspo1 expression. From the results, we observed that potential antagonistic genes fgf9 expression was upregulated and expression of sex differentiation gene were changed except cyp19a. The results suggested that fgf9 antagonizes the expression of rspo1 gene and regulate sex differentiation through rspo1/β-catenin/Wnt signal pathways. Smith found that rspo1 is a highly conserved and ancient part of the vertebrate ovarian determinant (Smith et al., 2008). The localization of rspo1 gene expression in ovarian and testicular cells was identified by in situ hybridization in A. davidianus. The results showed that rspo1 gene signals were both detected in the gonads of A. davidianus at 62 and 98 dpf, suggesting that rspo1 may play a role in gonadal differentiation and that when the gonads of A. davidianus begin to differentiate, rspo1 begins to be expressed, affecting the gonad differentiation of A. davidianus. The rspo1 gene positive signals were strongly detected in the ovarian cells in 2-years-old A. davidianus (Figure 2B7), while the positive signal was not strong in the testis (Figure 2B9), suggesting that rspo1 affects the process of gonadal differentiation into ovary and that it may also play a role in the development of testis, but this role has not yet been explained. Among the gonads and sex-reversal gonads, the expression level of rspo1 was the highest in sex-reversal ovaries, followed by ovaries, and the rspo1 expression level was significantly higher in ovaries and sex-reversal ovaries than that in testes and sex-reversal testes. The above results showed that rspo1 was highly expressed in ovaries and different tissues and during developmental stages of A. davidianus, as well as in gonads and sex-reversal gonads in A. davidianus. It can be determined that rspo1 plays an important role in ovarian differentiation and development.
In the present study, we characterized fgf9 and rspo1 genes expression in various tissues and developing gonads. In situ hybridization was used to detect the expression of fgf9 and rspo1 in undifferentiated gonad and locate the expression. Function and potential antagonistic relationship of fgf9 and rspo1 genes were studied and confirmed that potential antagonistic relationship of fgf9 and rspo1 genes in WNT4 pathway to regulate the sex differentiation in A. davidianus. Furthermore, genome editing technology would carried out for further study in future. The data provided basic information for genome editing breeding to improve the Chinese giant salamander farming industry.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/genbank/, MW685518; https://www.ncbi.nlm.nih.gov/genbank/, MW685519.
The animal study was reviewed and approved by the Yangtze River Fisheries Research Institute Care Committee (No. 2013001).
QH and HX conceived the experiments, JZ, QH, and XX performed the experiment, In situ hybridization was carried out by YZ and XX, ZL, and HT collected the samples. JZ wrote the manuscript and all of the authors reviewed the manuscript.
This work was supported by National Nature Science Foundation of China (31502155); Key Laboratory of Freshwater Aquatic Biotechnology and Breeding, Ministry of Agriculture, Heilongjiang Fisheries Research Institute, Chinese Academy of Fishery Sciences (KF-2017-06).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmolb.2022.974348/full#supplementary-material
SUPPLEMENTARY FIGURE S1 | Multiple alignment of the FGF9 amino acid sequence (A) and NJ phylogenetic tree (B).
SUPPLEMENTARY FIGURE S2 | Multiple alignment of the RSPO1 amino acid sequence (A) and the NJ phylogenetic tree (B).
SUPPLEMENTARY FIGURE S3 | Result of in situ hybridization with fgf9 (A) and rspo1 (B) sense 628 probe; no positive signal was detected.
Bagheri-Fam, S., Sim, H., Bernard, P., Jayakody, I., Taketo, M. M., Scherer, G., et al. (2008). Loss of FGFr2 leads to partial XY sex reversal. Dev. Biol. 314 (1), 71–83. doi:10.1016/j.ydbio.2007.11.010
Behr, B., Leucht, P., Longaker, M. T., and Quarto, N. (2010). Fgf-9 is required for angiogenesis and osteogenesis in long bone repair. Proc. Natl. Acad. Sci. U. S. A. 107 (26), 11853–11858. doi:10.1073/pnas.1003317107
Biason-Lauber, A., and Konrad, D. (2008). WNT4 and sex development. Sex. Dev. 2 (4-5), 210–218. doi:10.1159/000152037
Biason-Lauber, A. (2012). Wnt4, RSPO1, and FOXL2 in sex development. Semin. Reprod. Med. 30 (05), 387–395. doi:10.1055/s-0032-1324722
Binnerts, M. E., Kim, K. A., Bright, J. M., Patel, S. M., Tran, K., Zhou, M., et al. (2007). R-Spondin1 regulates Wnt signaling by inhibiting internalization of LRP6. Proc. Natl. Acad. Sci. U. S. A. 104, 14700–14705. doi:10.1073/pnas.0702305104
Blasio, A. M. Di., Carniti, C., Viganò, P., and Vignali, M. (1995). Basic fibroblast growth factor and ovarian cancer. J. Steroid Biochem. Mol. Biol. 53 (1-6), 375–379. doi:10.1016/0960-0760(95)00082-b
Colvin, J. S., Green, R. P., Schmahl, J., Capel, B., and Ornitz, D. M. (2001). Male-to-female sex reversal in mice lacking fibroblast growth factor 9. Cell 104 (6), 875–889. doi:10.1016/s0092-8674(01)00284-7
Harada, M., and Akita, K. (2020). Mouse fibroblast growth factor 9 N143T mutation leads to wide chondrogenic condensation of long bones. Histochem. Cell Biol. 153 (4), 215–223. doi:10.1007/s00418-020-01844-2
Hu, Q. M., Chang, C. F., Wang, Q. H., Tian, H. F., Qiao, Z. G., Wang, L., et al. (2019b). Genome-wide RAD sequencing to identify a sex-specific marker in Chinese giant salamander Andrias davidianus. BMC Genomics 20, 415. doi:10.1186/s12864-019-5771-5
Hu, Q. M., Meng, Y., Tian, H. F., Zhang, Y., and Xiao, H. B. (2016). Sexually dimorphic expression of foxl2 and Ftz-F1 in Chinese giant salamander Andrias davidianus. J. Exp. Zool. B Mol. Dev. Evol. 326 (6), 363–374. doi:10.1002/jez.b.22693
Hu, Q. M., Meng, Y., Wang, D., Tian, H. F., and Xiao, H. B. (2019d). Characterization and function of the T-box 1 gene in Chinese giant salamander Andrias davidianus. Genomics 111, 1351–1359. doi:10.1016/j.ygeno.2018.09.007
Hu, Q. M., Tian, H. F., Li, W., Meng, Y., Wang, Q. H., and Xiao, H. B. (2019c). Identification of critical sex-biased genes in Andrias davidianus by de novo transcriptome. Mol. Genet. Genomics 294 (2), 287–299. doi:10.1007/s00438-018-1508-4
Hu, Q. M., Tian, H. F., and Xiao, H. B. (2019a). Effects of temperature and sex steroids on sex ratio, growth, and growth-related gene expression in theChinese giant salamander Andrias davidianus. Aquat. Biol. 28, 79–90. doi:10.3354/ab00710
Huang, K. (2020). Regulation of fgf9 on the differentiation of goat intramuscular adipocytes. Animal Sci. J. 92, e13627. doi:10.1111/asj.13627
Jameson, S. A., Lin, Y. T., and Capel, B. (2012). Testis development requires the repression of Wnt4 by FGF signaling. Dev. Biol. 370 (1), 24–32. doi:10.1016/j.ydbio.2012.06.009
Kamata, T., Katsube, K., Michikawa, M., Yamada, M., Takada, S., and Mizusawa, H. (2004). R-spondin, a novel gene with thrombospondin type 1 domain, was expressed in the dorsal neural tube and affected in Wnts mutants. Biochim. Biophys. Acta 1676 (1), 51–62. doi:10.1016/j.bbaexp.2003.10.009
Kim, Y., and Capel, B. (2006). Balancing the bipotential gonad betweenalternative organ fates: A new perspective on an old problem. Dev. Dyn. 235, 2292–2300. doi:10.1002/dvdy.20894
Kim, Y., Kobayashi, A., Sekido, R., DiNapoli, L., Brennan, J., Chaboissier, M. C., et al. (2006). fgf9 and Wnt4 act as antagonistic signals to regulate mammalian sex determination. PLoS Biol. 4 (6), e187. doi:10.1371/journal.pbio.0040187
Kocer, A., Pinheiro, I., Pannetier, M., Renault, L., Parma, P., Radi, O., et al. (2008). R-spondin1 and FOXL2 act into two distinct cellular types during goat ovarian differentiation. BMC Dev. Biol. 8, 36. doi:10.1186/1471-213X-8-36
Kumar, S., Stecher, G., and Tamura, K. (2016). MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33 (7), 1870–1874. doi:10.1093/molbev/msw054
Lai, M. S., Wang, C. Y., Yang, S. H., Wu, C. C., Sun, H. S., Tsai, S. J., et al. (2016). The expression profiles of fibroblast growth factor 9 and its receptors in developing mice testes. Organogenesis 12 (2), 61–77. doi:10.1080/15476278.2016.1171448
Li, Y. H., Chen, T. M., Huang, B. M., Yang, S. H., Wu, C. C., Lin, Y. M., et al. (2020). fgf9 is a downstream target of SRY and sufficient to determine male sex fate in ex vivo XX gonad culture. Biol. Reprod. 103 (6), 1300–1313. doi:10.1093/biolre/ioaa154
Liao, B., and Lin, X. K. (2009). Research progress in fgf9 regulating gonadal differentiation. Adv. Veterinary Sci. Comp. Med. 30 (05), 80–83.
Loke, J., Pearlman, A., Radi, O., Zuffardi, O., Giussani, U., Pallotta, R., et al. (2014). Mutations in MAP3K1 tilt the balance from SOX9/FGF9 to WNT/β-catenin signaling. Hum. Mol. Genet. 23 (4), 1073–1083. doi:10.1093/hmg/ddt502
Maatouk, D. M., DiNapoli, L., Alvers, A., Parker, K. L., Taketo, M. M., and Capel, B. (2008). Stabilization of beta-catenin in XY gonads causes male-to-female sex-reversal. Hum. Mol. Genet. 17 (19), 2949–2955. doi:10.1093/hmg/ddn193
Nam, J. S., Turcotte, T. J., and Yoon, J. K. (2007). Dynamic expression of R-spondin family genes in mouse development. Gene Expr. Patterns 7 (3), 306–312. doi:10.1016/j.modgep.2006.08.006
Nef, S., and Vassalli, J. D. (2009). Complementary pathways in mammalian female sex determination. J. Biol. 8(8), 74, doi:10.1186/jbiol173
Ottolenghi, C., Pelosi, E., Tran, J., Colombino, M., Douglass, E., Nedorezov, T., et al. (2007). Loss of Wnt4 and Foxl2 leads to female-to-male sex reversal extending to germ cells. Hum. Mol. Genet. 16 (23), 2795–2804. doi:10.1093/hmg/ddm235
Parma, P., Radi, O., Vidal, V., Chaboissier, M. C., Dellambra, E., Valentini, S., et al. (2006). R-spondin1 is essential in sex determination, skin differentiation and malignancy. Nat. Genet. 38 (11), 1304–1309. doi:10.1038/ng1907
Piprek, R. P. (2009). Genetic mechanisms underlying male sex determination in mammals. J. Appl. Genet. 50 (4), 347–360. doi:10.1007/BF03195693
Plotnikov, A. N., Eliseenkova, A. V., Ibrahimi, O. A., Shriver, Z., Sasisekharan, R., Lemmon, M. A., et al. (2001). Crystal structure of fibroblast growth factor 9 reveals regions implicated in dimerization and autoinhibition. J. Biol. Chem. 276 (6), 4322–4329. doi:10.1074/jbc.M006502200
Smith, C. A., Shoemaker, C. M., Roeszler, K. N., Queen, J., Crews, D., and Sinclair, A. H. (2008). Cloning and expression of R-Spondin1 in different vertebrates suggests a conserved role in ovarian development. BMC Dev. Biol. 24 (8), 72. doi:10.1186/1471-213X-8-72
Tomaselli, S., Megiornim, F., Lin, L., Mazzilli, M. C., Gerrelli, D., Majore, S., et al. (2011). Human RSPO1/R-spondin1 is expressed during early ovary development and augments β-catenin signaling. PLoS One 6 (1), e16366. doi:10.1371/journal.pone.0016366
Tomizuka, K., Horikoshi, K., Kitada, R., Sugawara, Y., Iba, Y., Kojima, A., et al. (2008). R-spondin1 plays an essential role in ovarian development through positively regulating Wnt-4 signaling. Hum. Mol. Genet. 17 (9), 1278–1291. doi:10.1093/hmg/ddn036
Turvey, S. T., Marr, M. M., Barnes, I., Brace, S., Cunningham, A. A., Murphy, R. W., et al. (2019). Historical museum collections clarify the evolutionary history of cryptic species radiation in the world's largest amphibians. Ecol. Evol. 9 (18), 10070–10084. doi:10.1002/ece3.5257
van Meurs, J. B., Trikalinos, S. H., Ralston, M., Balcells, S., Brandi, M. L., Brixen, K., et al. (2008). Large-scale analysis of association between LRP5 and LRP6 variants and osteoporosis. JAMA 299 (11), 1277–90.
Wei, Q., Yokota, C., Semenov, M. V., Doble, B., Woodgett, J., and He, X. (2007). R-spondin1 is a high affinity ligand for LRP6 and induces LRP6 phosphorylation and beta-catenin signaling. J. Biol. Chem. 282(21), 15903–15911. doi:10.1074/jbc.M701927200
Yamamura, Y., Aoyama, S., Oshima, Y., Kato, T., Osawa, N., and Nakamura, M. (2005). Molecular cloning and expression in gonad of Rana rugosa WT1 and fgf9. Zool. Sci. 22 (9), 1045–1050. doi:10.2108/zsj.22.1045
Zhang, Y., Li, F., Sun, D., Liu, J., Liu, N., and Yu, Q. (2011). Molecular analysis shows differential expression of RSPO1 in Zebrafish (Danio rerio) gonads. Mol. Biol. Rep. 38 (1), 275–282. doi:10.1007/s11033-010-0105-3
Keywords: Andrias davidianus, FGF9, rspo1, antagonize, sex differentiation
Citation: Zhang J, Xia X, Zhu Y, Lian Z, Tian H, Xiao H and Hu Q (2022) Potential antagonistic relationship of fgf9 and rspo1 genes in WNT4 pathway to regulate the sex differentiation in Chinese giant salamander (Andrias davidianus). Front. Mol. Biosci. 9:974348. doi: 10.3389/fmolb.2022.974348
Received: 21 June 2022; Accepted: 30 August 2022;
Published: 20 September 2022.
Edited by:
Zongtao Sun, Ningbo University, ChinaReviewed by:
Linyan Zhou, Southwest University, ChinaCopyright © 2022 Zhang, Xia, Zhu, Lian, Tian, Xiao and Hu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Qiaomu Hu, aHFtdTA4MDZAMTYzLmNvbQ==
†These authors have contributed equally to this work and share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.