
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Mol. Biosci. , 19 October 2022
Sec. Molecular Diagnostics and Therapeutics
Volume 9 - 2022 | https://doi.org/10.3389/fmolb.2022.967510
This article is part of the Research Topic Cancer Stem Cells: Hidden Modulators of Tumor Immune Response View all 5 articles
 Mohd Imran Khan1
Mohd Imran Khan1 Taehwan Park1
Taehwan Park1 Mohammad Azhar Imran1
Mohammad Azhar Imran1 Venu Venkatarame Gowda Saralamma1
Venu Venkatarame Gowda Saralamma1 Duk Chul Lee2
Duk Chul Lee2 Jaehyuk Choi3
Jaehyuk Choi3 Mohammad Hassan Baig1*
Mohammad Hassan Baig1* Jae-June Dong1*
Jae-June Dong1*Heat shock protein 90 (Hsp90) is a molecular chaperone playing a significant role in the folding of client proteins. This cellular protein is linked to the progression of several cancer types, including breast cancer, lung cancer, and gastrointestinal stromal tumors. Several oncogenic kinases are Hsp90 clients and their activity depends on this molecular chaperone. This makes HSP90 a prominent therapeutic target for cancer treatment. Studies have confirmed the inhibition of HSP90 as a striking therapeutic treatment for cancer management. In this study, we have utilized machine learning and different in silico approaches to screen the KCB database to identify the potential HSP90 inhibitors. Further evaluation of these inhibitors on various cancer cell lines showed favorable inhibitory activity. These inhibitors could serve as a basis for future development of effective HSP90 inhibitors.
Due to their distinct cellular distribution, molecular chaperones have emerged as appealing drug targets for therapeutic research (Schopf et al., 2017; Kim et al., 2018; Li et al., 2018). Heat-shock proteins, a molecular chaperone, vary from 10 kDa to >100 kDa in size and are located across different cellular compartments. HSP90α and HSP90β in the cytoplasm, GRP94 in the endoplasmic reticulum, and TRAP-1 (tumor necrosis factor receptor-associated protein 1) in the mitochondria are different isoforms of 90 kDa HSPs (Hoter et al., 2018).
Hsp90 reportedly involves a wide range of essential cell functions (Wu et al., 2017). It plays a crucial role in the folding and stabilizing of more than 200 client proteins (Trepel et al., 2010; Azoitei et al., 2014). Hsp90 is well reported to be involved in various cancer progression by playing a prominent role in the biological functions and maintaining the conformation of several oncoproteins (Boroumand et al., 2018; Liew et al., 2022). The role of HSP90 has been thoroughly investigated and linked with progression of various cancer types, including breast cancer, lung cancer, melanoma, and gastrointestinal stromal tumors (Whitesell and Lindquist, 2005; Sherman and Multhoff, 2007; Wandinger et al., 2008; Graner, 2016; Chatterjee and Burns, 2017; Wu et al., 2017). Studies have well established that compared to normal cells, the expression level of Hsp90 is much higher in various cancer cells (Beliakoff and Whitesell, 2004; Mahalingam et al., 2009; Moser et al., 2009; Miyata et al., 2013; Sanchez et al., 2020). In cancer cells, the inhibition of Hsp90 promotes the degradation of client oncoproteins, thereby making it a significant cancer therapeutic strategy (Bhat et al., 2014; Ozgur and Tutar, 2016; Liu et al., 2019). In addition, the role of Hsp90 is reported in several other diseases (Cowen et al., 2009; Lackie et al., 2017; Wang et al., 2017). It is widely investigated as a crucial therapeutic drug target for viral, parasitic, fungal, and various neurodegenerative disorders (Ernst et al., 2014; Lackie et al., 2017).
Hsp90 is considered a prominent anticancer drug target because of its interaction with several kinases (almost 60% kinases) (Taipale et al., 2012). The inhibition of HSP90 results in the degradation of the client kinases (mediated by ubiquitination). The central role of HSP90 in these tumor mediating interactions has placed it as one of the most prominent anticancer drug targets. This has also constantly developed research interest towards developing Hsp90 targeting anticancer agents (Bhat et al., 2014; Kumalo et al., 2015). In the early 1990s, researchers identified the druggable pocket of HSP90 (Sanchez et al., 2020). Since then, many inhibitors targeting the N-terminal domain have been developed, some of which are also under clinical trials (Yuno et al., 2018). Despite so much research, there is still no HSP90 targeting molecule approved by FDA for cancer monotherapy.
The recent advancement in technology has fastened the drug discovery process. A large number of molecules have been developed using advanced computational techniques (Baig et al., 2016; Lin et al., 2020). Virtual screening is an efficient and cost-effective strategy for identifying chemical moieties and structural scaffolds potentially crucial for the binding to a target protein (Lionta et al., 2014) (Baig et al., 2016).
We used three different ML techniques on the ChEMBL activity dataset along with DUD-E decoys to increase the performance and balance out the number bias. The relevant features were selected to develop a robust model for HSP90. The best ML model was chosen on several parameters and was further utilized for screening the Korean Chemical Bank (https://chembank.org/) to identify the potential HSP90 inhibitor. Further various in silico techniques were applied to evaluate the selected compounds’ binding potential and stability against HSP90. Finally, the top compounds were experimentally validated. The outcome of this study results in identifying potent HSP90 inhibitors with novel scaffolds.
ChEMBL database (Mendez et al., 2019) was used to select the dataset of molecules having previously reported activity against HSP90 (ChEMBL ID: CHEMBL3880). The selected assay was a single protein assay designated to HSP 90-alpha. The dataset was filtered for (i). Organism: human and (ii). Binding activity type: IC50.
Further the data was preprocessed and filtered for desired information. The unique molecules were segregated to remove the redundancy. To make a classification model, molecules having IC50 activity value less than or equal to 100 nM were treated as active, and more than 500 nM were treated as inactive. Decoys were generated by the DUD-E database (Mysinger et al., 2012). Both active and inactive datasets were initially segregated in smiles format.
RDkit was used to calculate the MACCS KEYS, ECFP4, and 1D&2D descriptors. The biophysical molecular descriptors and fingerprints comprising 1385 unique properties were calculated. To reduce the complexity, improve predictive power, and reduce overfitting of the model, feature selection was performed. Finally, a total of 62 relevant features were selected with Pearson correlation coefficient more than 0.3.
ML model was built using Python, Scikit-learn (Bac et al., 2021). The dataset was divided into training and test sets in a ratio of 20:80 using the stratified method. Three ML models were built: Randomforest, XGBoost, and SVM (Support Vector Machine). The model data was preserved in the github repository (https://github.com/marinewhlae/HSP90). Hyperparameter tuning was done by the grid search cross-validation method.
The model was validated through several performance parameters, such as Accuracy, Precision, Recall, MCC (Matthews correlation coefficient), F1 score, and ROC-AUC (Receiver Operating Characteristic curve - Area Under the Curve).
Y-scrambling is one of the methods used to verify machine learning models and is called Y-randomization or Y-permutation. By randomly mixing response variables (Y data) and intentionally breaking the connection between feature variables (X data), this is an attempt to verify the accidental action of coincidence in model performance, i.e., whether the model’s performance on selected dataset was a random accidental event.
Randomly mix the Y value to create a pair mixed with the existing X value for model learning.
This process is repeated 1000 times, and the performance metric (Accuracy, MCC, F1 score) is compared with the existing learned model.
If the performance of a model learned with existing data is similar to or inferior to that of a model learned through y-scrambling, even if the performance metric of the model is high, the prediction is unreliable.
The selected ML model was used for screening the KCB. KCB is a collection of highly active 578,112 structures. The database was also pre-processed to remove the redundancy and the descriptor information required for screening purposes. The ML model was applied to the compound library, and the molecules predicted to be over 90% active were selected.
The structure of Human HSP90 in Complex with Geldanamycin was extracted from the RCSB Protein Databank (PDB ID: 1YET) (Stebbins et al., 1997). HSP90 and Geldanamycin were separated and were subjected to redock using CCDC GOLD v5.8.1 (Jones et al., 1997). The binding orientation of the redock and crystal confirmation of Geldanamycin within the binding site of HSP90 were compared. The molecules identified from the ML approach were further screened against HSP90 based on molecular docking study using CCDC GOLD. The molecules with a PLP fitness score higher than Geldanamycin were selected and further evaluated using autodock (Morris et al., 2009).
The high performing molecules in complex with the target protein were then subjected to molecular dynamic simulation studies. The thermal stability, binding affinity, and relative motion were the prime objective. Gromacs (v 2020.04 package) (Van Der Spoel et al., 2005) was used to perform the MD throughout the study. CHARMM27 force field and TIP3P water model was applied to the cubic simulation box of 1.2 Å radius. Ligand topology was generated at SwissParam. The simulation box was filled with solvent; and subsequently, the system was electro neutralized using sodium and chloride ions. The bad contacts were corrected through a steepest descent minimization algorithm fixing a maximum force and steps at 1000 kJ/mol/nm and 50,000 respectively. Two rounds of 100 ps of equilibration was performed. First, an isothermal and isochoric equilibration (NVT) was done using Particle Mesh Ewald electrostatics, followed by an isothermal and isobaric equilibration (NPT). Temperature coupling was applied to rectify the temperature differences. Finally, the production MD was performed for 100 ns using trajectories generated after NPT equilibration.
The MMPBSA.py module utilizing the AMBER software was used to appraise the Molecular Mechanic/Poisson-Boltzmann Surface Area (MM-PBSA) (Miller et al., 2012). The binding free energy between the ligand and the receptor was calculated by keeping the account for the vacuum potential energy and solvation free energy terms. Poisson–Boltzmann equation and solvent-accessible surface area (SASA) methods were harnessed to estimate polar and nonpolar energies.
This approach calculates the binding free energy (ΔG binding) based on the following equation:
where
and
where ΔGpolar represents the electrostatic and ΔGnon-polar is the nonpolar contribution to the solvation free energy.
3-(4,5-imethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT), Dimethyl sulphoxide (DMSO) were purchased from Sigma-Aldrich. RPMI-1640 medium, fetal bovine serum (FBS), penicillin/streptomycin, trypsin–EDTA, and phosphate buffer saline (PBS) were purchased from Gibco Life Technology. All the HSP90 inhibitors (Compound-1, 2, 3, and 4) were provided by the KCB, Republic of Korea. HSP90 inhibitors were prepared in DMSO at 10 mM stock solution and stored at −20°C. Human breast cancer cells MDA-MB-231 and human lung cancer cells A549 were obtained from Korean Cell Line Bank.
Human breast cancer cells (MDA-MB-231) and lung cancer cells (A549) were grown in RPMI-1640 medium with 10% heat-inactivated fetal bovine serum (FBS) and 100U/ml of penicillin and 10 μg/ml streptomycin at 37°C in a humidified atmosphere with 5% CO2 in the incubator.
Antiproliferative effects of all the compounds on the MDA-MB-231 and A549 cell line were evaluated by 3-(4,5- imethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay. In brief, MDA-MB-231 and A549 cells were seeded into 96-well plates (10,000 cells/well) and were allowed to adhere overnight. The cells were then treated with the top 4 selected HSP90 inhibitors (Compound-1, 2, 3, and 4) at various concentrations (0.321µM, 0.625µM, 1.25µM, 2.5µM, and 5 µM) for 72 h. To measure the cell viability, 10 µl of MTT (5 mg/ml) solution was added to each well, and the cells were further incubated for another 3 h at 37 °C. The supernatant was removed and subsequently 200 μl of DMSO was added to each well to dissolve the formazan product. Absorbance at 540 nm was measured using a microplate reader (Promega, Discover), and percentage of cell viability was calculated as follows: (optical density of experimental sample/optical density of control) * 100.
The use of Machine learning techniques in drug discovery has attracted much research interest (Sato et al., 2010; Carpenter and Huang, 2018; Zhu et al., 2020). ML improves the decision-making in hit discovery to retrieve accurate outcomes (Adeshina et al., 2020; Dara et al., 2021). This study aimed to build a robust classification model for screening the potential hits for HSP90. At the outset, this study incorporated a total of 354 inhibitors with reported inhibitory activities against HSP90 from the ChEMBL Bioassay database.
The primary dataset for molecular descriptors calculation contained 218 active and 136 inactive, and 518 decoy entries (Table 1). The training and test data distribution was in the ratio of 20:80. The magnitude of the biological activity range was sufficiently wide and evenly distributed between the sampling sets. Therefore, the multi-dimensional chemical property space was evenly rendered between training and test sets. In total, 1385 1D&2D, MACCS KEYS, and ECFP4 unique descriptors were calculated (Table 2). Molecular descriptors represent a molecule’s physical, chemical, or topological features that are experimentally or theoretically defined (Todeschini and Consonni, 2008; Dong et al., 2015). The descriptors were filtered using the Pearson correlation coefficient matrix. The relevant features with characteristic correlation with the continuous data of inhibitory activity and low dimensionality were considered as an independent chemical variables. The descriptors that contained a correlation value greater than 0.3 were selected. In this study, 62 high correlating features were used for the classification. A plot for the Pearson correlation coefficient matrix for adopted 1D&2D (20), MACCS KEYS (16) and ECFP4 (26) descriptors against their inhibitory activities are shown in Figure 1. A significant correlation was found between inhibitory activity and selected features.
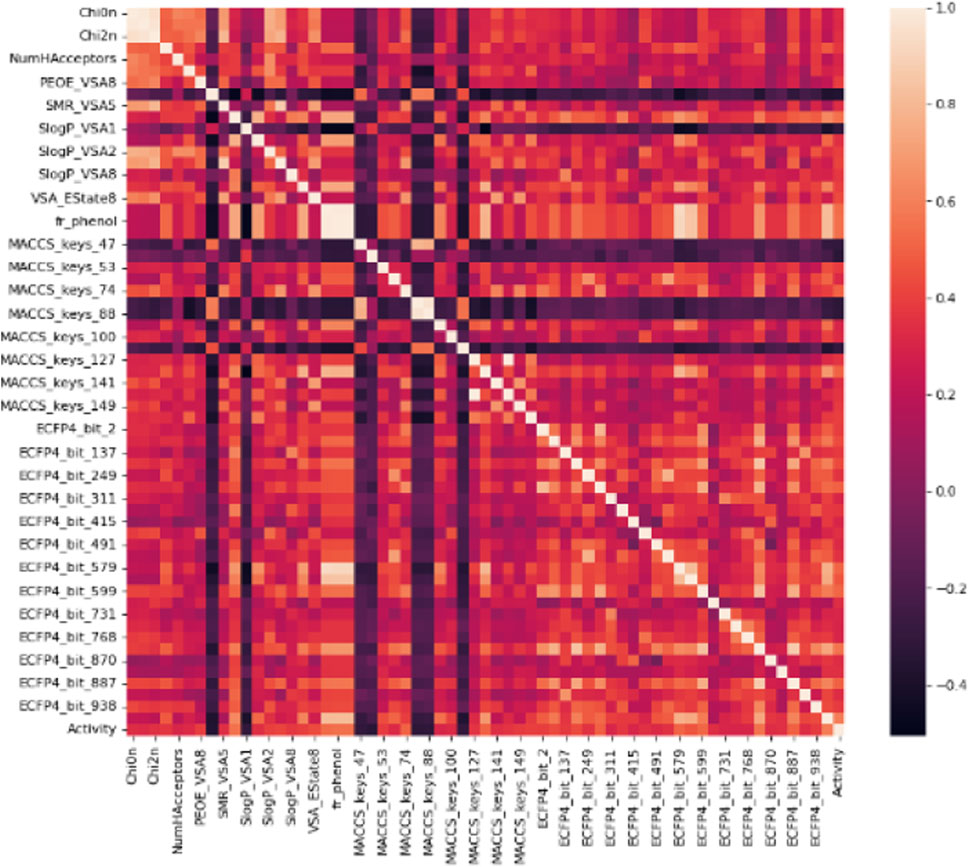
FIGURE 1. A correlation histogram of selected descriptors against the inhibitory activity. The scale represents the value of the Pearson correlation coefficient.
Machine learning models employing SVM, Random Forest, and XGBoost were generated on the identical binary dataset. The selected parameter sets were optimized to attain maximum accuracy for each classifier. Internal 10-fold cross-validation to tune the hyperparameters was performed. Data was compared to the original class label for all the classifiers to evaluate true positives, true negatives, false positives, and false negatives. The confusion matrix and the evaluated quality parameters of the classifier are illustrated in Figure 2.
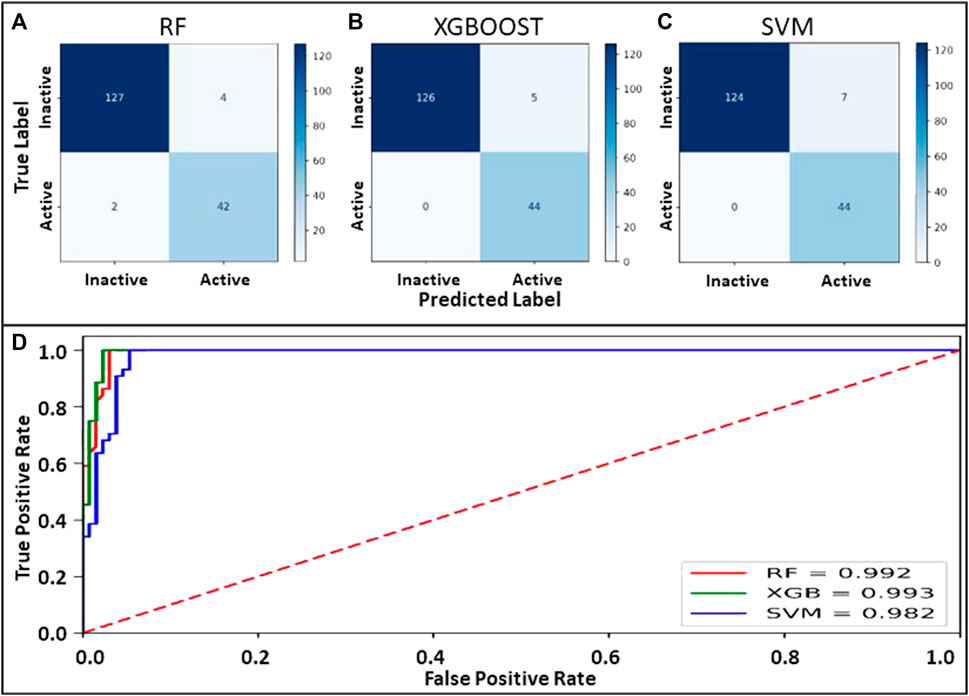
FIGURE 2. (A–C) Confusion matrix for the models prepared using RF, XGBOOST and SVM. (D): ROC curve showing the performance of binary class models.
The performance of the binary classifier was also analyzed on a comparative ROC plot drawn between the proportion of truly predicted positive classes among all actual positive classes and the proportion of incorrectly predicted negative classes among all actual negatives to encapsulate the findings in confusion matrices.
The accuracy evaluation for the internal ten-fold cross-validated SVM, RF, and XGB was computed while keeping the complexity parameter to 1.0 (Table 3). Scores for six performance parameters viz., accuracy, precision, recall, MCC, F1, and ROC-AUC for the 3 different methods were compared to each other. The ROC-AUC for the XCBoost model was significantly highest among the 3 models. Concretely, the tradeoff between statistical results of the performance parameters among the 3 classifiers establishes an XGBoost model as the best-suited binary class prediction method for our dataset.
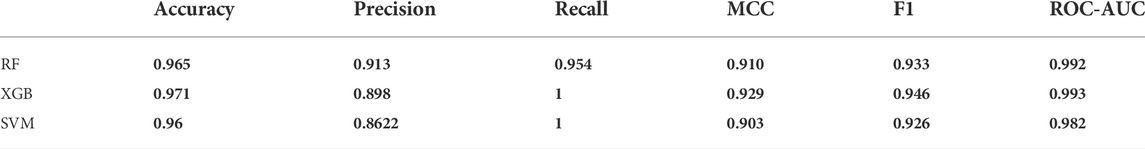
TABLE 3. The performance parameter collation for the Ramdom forest, XGBoost and Support Vector Machine models.
Although the accuracy and MCC scores indicated (Table 3) in all three models are above 0.9, demonstrating their ability to generalize outside the initial dataset characteristics, the difference lies in the results of performance for recall values. We found the lowest recall value for RF (0.954), and the highest recall value possessed by XBG and SVM models (1.0). Overall, the higher value of the five performance metric parameters, including accuracy, precision, recall, MCC, and F1, indicates XBGoost as the most accurate classifier in combination with the selected descriptors (Table 3). We performed Y-scrambling randomization test on the XGB model. The results favor the argument for the model not being a change event (Figure 3).
The test dataset with 577,939 molecules was screened on the XGBoost model. A total of 6807 molecules were predicted as active hits against HSP90 (Table 4).
The selected active molecules were further screened against HSP90 using CCDC Gold and Autodock. Finally, the top 4 molecules with binding scores from both the docking software higher than Geldanamycin were selected. The PLP fitness score from Gold, binding energy from Autodock, and Classification Probability score from the ML model for the selected compounds are shown in Table 5.
The 2D representation of the four potential molecules selected against HSP90 is shown in Figure 4. When compared with the drug geldanamycin, these molecules showed no significant structural similarity. All the molecules have an extended ring structure with molecular weights ranging from 565.840 Da (Compound 1), 533.846 Da (Compound 2), 572.779 Da (Compound 3), and 593.826 Da (Compound 4).
The stability of the selected hits in complex with HSP90 was further confirmed through molecular dynamic simulations (Liu et al., 2018; Naqvi et al., 2018). Apart from stability, we sought to evaluate the binding affinity and the effect of inhibitors on the dynamic structure of the protein. The RMSD, Hydrogen bond, and free energy plot were calculated. (Figure 5; Table 5). The RMSD values for Compounds 1, 2, and 3 were below 2Å, while compound 4 showed a slightly higher fluctuation.
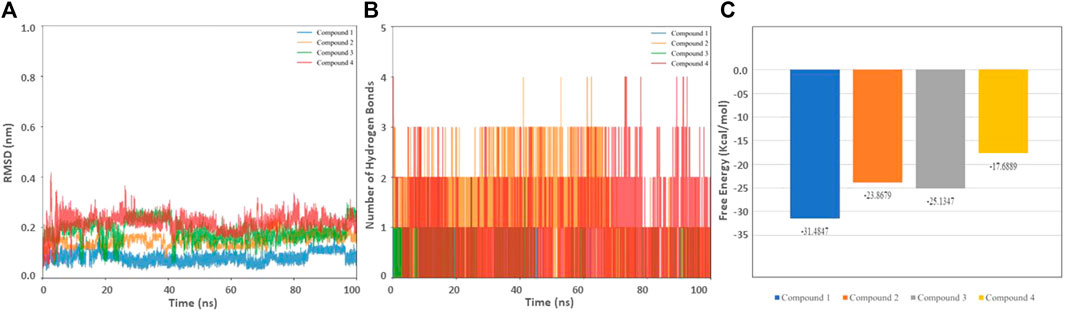
FIGURE 5. Biophysical simulation analysis of the complexes. (A) RMSD plot of HSP90 with the identified compounds (B) Hydrogen bond frequency plot (C) Binding free energy.
The principal component analysis (PCA) was carried out on the simulation trajectories to segregate, discover and evaluate the meaningful conformational changes among all the multidirectional atomic thermal fluctuations (David and Jacobs, 2014; Khan et al., 2021). The biggest eigenvectors from the analysis depict the rigorous atomic motion in the complex (Mazanetz et al., 2014). We investigated the projection of 8 eigenvectors for the PCA of HSP90 bound with Gelendamycin and four high-scoring hits (Compound1, 2, 3, and 4). The trajectory suggested different atomic motions during the simulation. The 2D projection of the trajectories in the essential subspace is projected in Figures 6A–D. The results show that the Compound 1 and 2 bound structure of HSP90 occupies a common conformational space, indicating higher complex structural stability.
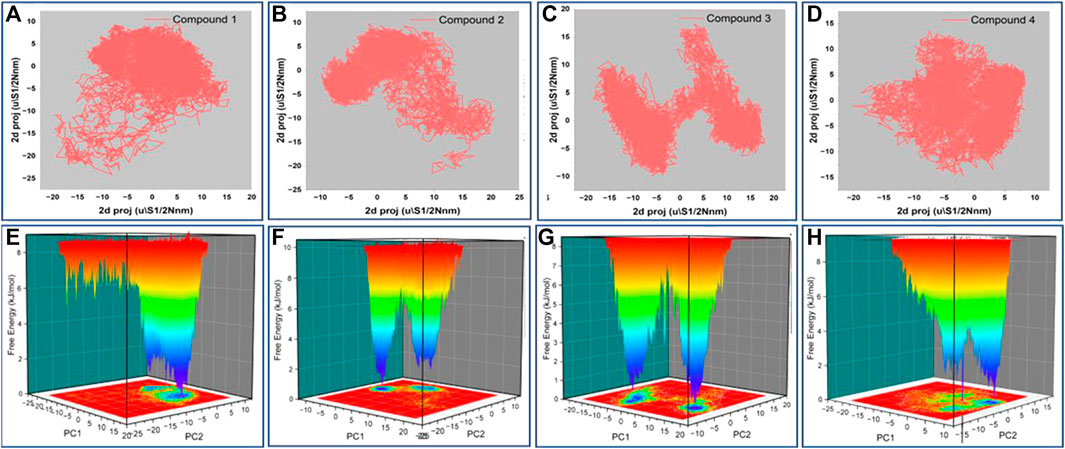
FIGURE 6. (A–D) 2D projection of the trajectory allocation for motion of the proteins in complex with compound 1,2,3 and 4 in the essential subspace, (E–H) 3D projection of the free energy landscape as a function of first two principal components.
Further, we plotted the free energy landscapes in Figures 6E–H to understand the folding pattern fluctuations among the top four selected complexes. The simulation ensembles of the four compound bound structure showed narrow peaks with distinct minima among the first two essential components. Complex with compound 1 has most of the ensembles in a narrow range of conformational space with no major state transition. These findings suggest compact packing and better stability of the compound 1 bound structure followed by 2, 3, and 4.
We also determined the stable number of hydrogen bonds stabilizing the interactions between HSP90 and the molecules. As anticipated, Compound 3 and 4 were stabilized with 3–4 stable and consistent hydrogen bonds, indicating their strong binding capability. In drug discovery, the analysis of MM-PBSA is considered an efficient strategy to account binding affinity (Genheden and Ryde, 2015). The binding free energy values for Compounds 1, 2, 3, and 4 were found to be −31.4847, −23.8679, −25.1347, and −17.6889 kcal/mol, respectively (Table 6). The selected compounds were further evaluated on different cancer lines.
Antiproliferative activity of the selected compounds was assessed in MDA-MB-231 and A549 cells by MTT assay. The inhibitory effect of these compounds against both cell lines was evaluated using the half-maximal inhibition concentration (IC50) value (a concentration required to inhibit cell growth in 50% of the cell population) at 72 h. Along with the increasing concentration of these compounds (Compound-1, 2, 3, and 4) treatments, all these inhibitors significantly decreased human breast cancer MDA-MB-231 cell and human lung cancer A549 cell proliferation in a dose-dependent manner (Figure 7). The Half inhibitory concentration (IC50) of these compounds (Compound-1, 2, 3, 4) on MDA-MB-231 was 3.62 µM, 3.13 µM, 4.5 µM, and 1.86 µM respectively, and on A549 was 4.13 µM, 6.75 µM, 4.13 µM, and 12.32 µM respectively (Table 7). 17-DMAG is a semi-synthetic geldanamycin analog that binds specifically to Hsp90’s ATP binding site and inhibits the protein folding process. The cell proliferation of MCF-7 and MDA-MB-231 is inhibited by 17-DMAG, with IC50 values of 3.11 μM and 2.16 μM, respectively (Ghadban et al., 2016). Our compounds were found to be demonstrating potency close to 17-DMAG. Overall, the selected compounds (Compound-1, 2, 3, and 4) significantly reduced cell proliferation in MDA-MB-231 and A549 cell lines with low IC50 values at 72 h (Table.7).
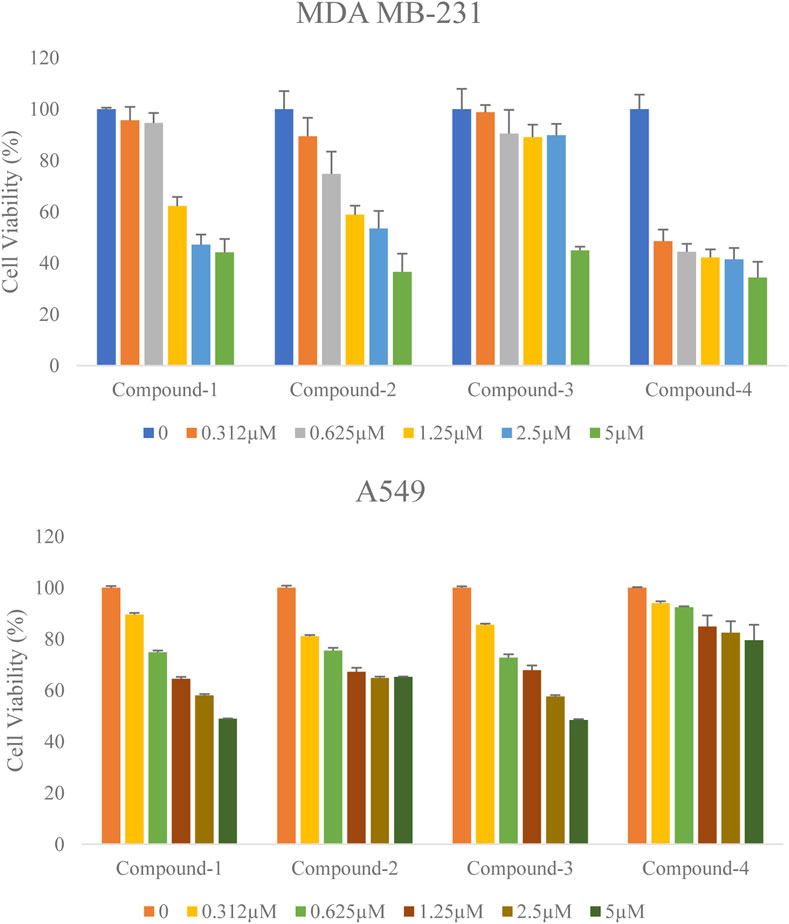
FIGURE 7. Effect of the selected compounds (Compound-1, 2, 3 and 4) on the cell viability of human breast cancer cells MDA-MB-231 and lung cancer cells A549. Both the cells were treated with various concentrations of these compounds for 72 h. Cell viability was determined by MTT assay. Percentages of viable cells were calculated by comparing treated and solvent control (DMSO) cells. Data are the mean ± S.D. of three replicates.
Heat shock protein 90 (Hsp90) is a molecular chaperone playing a significant role in the folding of client proteins. This cellular protein linkage with cancer progression makes it a viable therapeutic target. This study has utilized machine learning and different in silico approaches to identify potential HSP90 inhibitors. Further experimental validation of the selected compounds on various cancer cell lines proved their anticancer potential. Compound-1 and Compound-3 showed antiproliferative activity with IC50 below 5 µM against both the studied cancer cells. While Compound-2 and 4 also demonstrated good antiproliferative activity against MDA-MB-231 (IC50 < 3.5 µM). The finding of this work identified four novel and promising leads for further development of anticancer drugs targeting Hsp90.
The original contributions presented in the study are included in the article/Supplementary Material; further inquiries can be directed to the corresponding authors.
Study design: MHB, JJ-D, Experiment: PT, MI, MIK, VG, Validation: MI, PT, VG, DL, JC, Manuscript writing: MIK, JJD, MHB, DL.
National Research Foundation of Korea (Grant Number: NRF-2021K1A4A7A02098793).
The authors are thankful to the Korea Chemical Bank (www.chembank.org) of the Korea Research Institute of Chemical Technology for providing the chemical library used in this study.
JC was employed by the company BNJBiopharma.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Adeshina, Y. O., Deeds, E. J., and Karanicolas, J. (2020). Machine learning classification can reduce false positives in structure-based virtual screening. Proc. Natl. Acad. Sci. U. S. A. 117, 18477–18488. doi:10.1073/pnas.2000585117
Azoitei, N., Diepold, K., Brunner, C., Rouhi, A., Genze, F., Becher, A., et al. (2014). HSP90 supports tumor growth and angiogenesis through PRKD2 protein stabilization. Cancer Res. 74, 7125–7136. doi:10.1158/0008-5472.CAN-14-1017
Bac, J., Mirkes, E. M., Gorban, A. N., Tyukin, I., and Zinovyev, A. (2021). Scikit-dimension: A Python package for intrinsic dimension estimation. Entropy (Basel) 23, 1368. doi:10.3390/e23101368
Baig, M. H., Ahmad, K., Roy, S., Ashraf, J. M., Adil, M., Siddiqui, M. H., et al. (2016). Computer aided drug design: Success and limitations. Curr. Pharm. Des. 22, 572–581. doi:10.2174/1381612822666151125000550
Beliakoff, J., and Whitesell, L. (2004). Hsp90: An emerging target for breast cancer therapy. Anticancer. Drugs 15, 651–662. doi:10.1097/01.cad.0000136876.11928.be
Bhat, R., Tummalapalli, S. R., and Rotella, D. P. (2014). Progress in the discovery and development of heat shock protein 90 (Hsp90) inhibitors. J. Med. Chem. 57, 8718–8728. doi:10.1021/jm500823a
Boroumand, N., Saghi, H., Avan, A., Bahreyni, A., Ryzhikov, M., Khazaei, M., et al. (2018). Therapeutic potency of heat-shock protein-90 pharmacological inhibitors in the treatment of gastrointestinal cancer, current status and perspectives. J. Pharm. Pharmacol. 70, 151–158. doi:10.1111/jphp.12824
Carpenter, K. A., and Huang, X. (2018). Machine learning-based virtual screening and its applications to alzheimer's drug discovery: A review. Curr. Pharm. Des. 24, 3347–3358. doi:10.2174/1381612824666180607124038
Chatterjee, S., and Burns, T. F. (2017). Targeting heat shock proteins in cancer: A promising therapeutic approach. Int. J. Mol. Sci. 18, E1978. doi:10.3390/ijms18091978
Cowen, L. E., Singh, S. D., Kohler, J. R., Collins, C., Zaas, A. K., Schell, W. A., et al. (2009). Harnessing Hsp90 function as a powerful, broadly effective therapeutic strategy for fungal infectious disease. Proc. Natl. Acad. Sci. U. S. A. 106, 2818–2823. doi:10.1073/pnas.0813394106
Dara, S., Dhamercherla, S., Jadav, S. S., Babu, C. M., and Ahsan, M. J. (2021). Machine learning in drug discovery: A review. Artif. Intell. Rev. 55, 1947–1999. doi:10.1007/s10462-021-10058-4
David, C. C., and Jacobs, D. J. (2014). Principal component analysis: A method for determining the essential dynamics of proteins. Methods Mol. Biol. 1084, 193–226. doi:10.1007/978-1-62703-658-0_11
Dong, J., Cao, D. S., Miao, H. Y., Liu, S., Deng, B. C., Yun, Y. H., et al. (2015). ChemDes: An integrated web-based platform for molecular descriptor and fingerprint computation. J. Cheminform. 7, 60. doi:10.1186/s13321-015-0109-z
Ernst, J. T., Neubert, T., Liu, M., Sperry, S., Zuccola, H., Turnbull, A., et al. (2014). Identification of novel HSP90α/β isoform selective inhibitors using structure-based drug design. demonstration of potential utility in treating CNS disorders such as Huntington's disease. J. Med. Chem. 57, 3382–3400. doi:10.1021/jm500042s
Genheden, S., and Ryde, U. (2015). The MM/PBSA and MM/GBSA methods to estimate ligand-binding affinities. Expert Opin. Drug Discov. 10, 449–461. doi:10.1517/17460441.2015.1032936
Ghadban, T., Jessen, A., Reeh, M., Dibbern, J. L., Mahner, S., Mueller, V., et al. (2016). In vitro study comparing the efficacy of the water-soluble HSP90 inhibitors, 17-AEPGA and 17-DMAG, with that of the nonwater-soluble HSP90 inhibitor, 17-AAG, in breast cancer cell lines. Int. J. Mol. Med. 38, 1296–1302. doi:10.3892/ijmm.2016.2696
Graner, M. W. (2016). HSP90 and immune modulation in cancer. Adv. Cancer Res. 129, 191–224. doi:10.1016/bs.acr.2015.10.001
Hoter, A., El-Sabban, M. E., and Naim, H. Y. (2018). The HSP90 family: Structure, regulation, function, and implications in Health and disease. Int. J. Mol. Sci. 19, E2560. doi:10.3390/ijms19092560
Jones, G., Willett, P., Glen, R. C., Leach, A. R., and Taylor, R. (1997). Development and validation of a genetic algorithm for flexible docking. J. Mol. Biol. 267, 727–748. doi:10.1006/jmbi.1996.0897
Khan, M. I., Baig, M. H., Mondal, T., Alorabi, M., Sharma, T., Dong, J. J., et al. (2021). Impact of the double mutants on spike protein of SARS-CoV-2 B.1.617 lineage on the human ACE2 receptor binding: A structural insight. Viruses 13, 2295. doi:10.3390/v13112295
Kim, H. H., Hyun, J. S., Choi, J., Choi, K. E., Jee, J. G., and Park, S. J. (2018). Structural ensemble-based docking simulation and biophysical studies discovered new inhibitors of Hsp90 N-terminal domain. Sci. Rep. 8, 368. doi:10.1038/s41598-017-18332-8
Kumalo, H. M., Bhakat, S., and Soliman, M. E. (2015). Heat-shock protein 90 (Hsp90) as anticancer target for drug discovery: An ample computational perspective. Chem. Biol. Drug Des. 86, 1131–1160. doi:10.1111/cbdd.12582
Lackie, R. E., Maciejewski, A., Ostapchenko, V. G., Marques-Lopes, J., Choy, W. Y., Duennwald, M. L., et al. (2017). The hsp70/hsp90 chaperone machinery in neurodegenerative diseases. Front. Neurosci. 11, 254. doi:10.3389/fnins.2017.00254
Li, T., Jiang, H. L., Tong, Y. G., and Lu, J. J. (2018). Targeting the Hsp90-Cdc37-client protein interaction to disrupt Hsp90 chaperone machinery. J. Hematol. Oncol. 11, 59. doi:10.1186/s13045-018-0602-8
Liew, H. Y., Tan, X. Y., Chan, H. H., Khaw, K. Y., and Ong, Y. S. (2022). Natural HSP90 inhibitors as a potential therapeutic intervention in treating cancers: A comprehensive review. Pharmacol. Res. 181, 106260. doi:10.1016/j.phrs.2022.106260
Lin, X., Li, X., and Lin, X. (2020). A review on applications of computational methods in drug screening and design. Molecules 25, E1375. doi:10.3390/molecules25061375
Lionta, E., Spyrou, G., Vassilatis, D. K., and Cournia, Z. (2014). Structure-based virtual screening for drug discovery: Principles, applications and recent advances. Curr. Top. Med. Chem. 14, 1923–1938. doi:10.2174/1568026614666140929124445
Liu, X., Shi, D., Zhou, S., Liu, H., Liu, H., and Yao, X. (2018). Molecular dynamics simulations and novel drug discovery. Expert Opin. Drug Discov. 13, 23–37. doi:10.1080/17460441.2018.1403419
Liu, Y., Liu, X., Li, L., Dai, R., Shi, M., Xue, H., et al. (2019). Identification and structure-activity studies of 1, 3-Dibenzyl-2-aryl imidazolidines as novel Hsp90 inhibitors. Molecules 24, E2105. doi:10.3390/molecules24112105
Mahalingam, D., Swords, R., Carew, J. S., Nawrocki, S. T., Bhalla, K., and Giles, F. J. (2009). Targeting HSP90 for cancer therapy. Br. J. Cancer 100, 1523–1529. doi:10.1038/sj.bjc.6605066
Mazanetz, M. P., Laughton, C. A., and Fischer, P. M. (2014). Investigation of the flexibility of protein kinases implicated in the pathology of Alzheimer’s disease. Molecules. 19 (7), 9134–9159. doi:10.3390/molecules19079134
Mendez, D., Gaulton, A., Bento, A. P., Chambers, J., De Veij, M., Felix, E., et al. (2019). ChEMBL: Towards direct deposition of bioassay data. Nucleic Acids Res. 47, D930–D940. doi:10.1093/nar/gky1075
Miller, B. R., McGee, T. D., Swails, J. M., Homeyer, N., Gohlke, H., and Roitberg, A. E. (2012). MMPBSA.py: An efficient program for end-state free energy calculations. J. Chem. Theory Comput. 8, 3314–3321. doi:10.1021/ct300418h
Miyata, Y., Nakamoto, H., and Neckers, L. (2013). The therapeutic target Hsp90 and cancer hallmarks. Curr. Pharm. Des. 19, 347–365. doi:10.2174/138161213804143725
Morris, G. M., Huey, R., Lindstrom, W., Sanner, M. F., Belew, R. K., Goodsell, D. S., et al. (2009). AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J. Comput. Chem. 30 (16), 2785–2791. doi:10.1002/jcc.21256
Moser, C., Lang, S. A., and Stoeltzing, O. (2009). Heat-shock protein 90 (Hsp90) as a molecular target for therapy of gastrointestinal cancer. Anticancer Res. 29, 2031–2042.
Mysinger, M. M., Carchia, M., Irwin, J. J., and Shoichet, B. K. (2012). Directory of useful decoys, enhanced (DUD-E): Better ligands and decoys for better benchmarking. J. Med. Chem. 55, 6582–6594. doi:10.1021/jm300687e
Naqvi, A. A. T., Mohammad, T., Hasan, G. M., and Hassan, M. I. (2018). Advancements in docking and molecular dynamics simulations towards ligand-receptor interactions and structure-function relationships. Curr. Top. Med. Chem. 18, 1755–1768. doi:10.2174/1568026618666181025114157
Ozgur, A., and Tutar, Y. (2016). Heat shock protein 90 inhibition in cancer drug discovery: From chemistry to futural clinical applications. Anticancer. Agents Med. Chem. 16, 280–290. doi:10.2174/1871520615666150821093747
Sanchez, J., Carter, T. R., Cohen, M. S., and Blagg, B. S. J. (2020). Old and new approaches to target the Hsp90 chaperone. Curr. Cancer Drug Targets 20, 253–270. doi:10.2174/1568009619666191202101330
Sato, T., Honma, T., and Yokoyama, S. (2010). Combining machine learning and pharmacophore-based interaction fingerprint for in silico screening. J. Chem. Inf. Model. 50, 170–185. doi:10.1021/ci900382e
Schopf, F. H., Biebl, M. M., and Buchner, J. (2017). The HSP90 chaperone machinery. Nat. Rev. Mol. Cell Biol. 18, 345–360. doi:10.1038/nrm.2017.20
Sherman, M., and Multhoff, G. (2007). Heat shock proteins in cancer. Ann. N. Y. Acad. Sci. 1113, 192–201. doi:10.1196/annals.1391.030
Stebbins, C. E., Russo, A. A., Schneider, C., Rosen, N., Hartl, F. U., and Pavletich, N. P. (1997). Crystal structure of an hsp90-geldanamycin complex: Targeting of a protein chaperone by an antitumor agent. Cell 89, 239–250. doi:10.1016/s0092-8674(00)80203-2
Taipale, M., Krykbaeva, I., Koeva, M., Kayatekin, C., Westover, K. D., Karras, G. I., et al. (2012). Quantitative analysis of HSP90-client interactions reveals principles of substrate recognition. Cell 150, 987–1001. doi:10.1016/j.cell.2012.06.047
Todeschini, R., and Consonni, V. (2008). Handbook of molecular descriptors. Hoboken, New Jersey, US: John Wiley & Sons.
Trepel, J., Mollapour, M., Giaccone, G., and Neckers, L. (2010). Targeting the dynamic HSP90 complex in cancer. Nat. Rev. Cancer 10, 537–549. doi:10.1038/nrc2887
Van Der Spoel, D., Lindahl, E., Hess, B., Groenhof, G., Mark, A. E., and Berendsen, H. J. (2005). Gromacs: Fast, flexible, and free. J. Comput. Chem. 26, 1701–1718. doi:10.1002/jcc.20291
Wandinger, S. K., Richter, K., and Buchner, J. (2008). The Hsp90 chaperone machinery. J. Biol. Chem. 283, 18473–18477. doi:10.1074/jbc.R800007200
Wang, Y., Jin, F., Wang, R., Li, F., Wu, Y., Kitazato, K., et al. (2017). HSP90: A promising broad-spectrum antiviral drug target. Arch. Virol. 162, 3269–3282. doi:10.1007/s00705-017-3511-1
Whitesell, L., and Lindquist, S. L. (2005). HSP90 and the chaperoning of cancer. Nat. Rev. Cancer 5, 761–772. doi:10.1038/nrc1716
Wu, J., Liu, T., Rios, Z., Mei, Q., Lin, X., and Cao, S. (2017). Heat shock proteins and cancer. Trends Pharmacol. Sci. 38, 226–256. doi:10.1016/j.tips.2016.11.009
Yuno, A., Lee, M. J., Lee, S., Tomita, Y., Rekhtman, D., Moore, B., et al. (2018). Clinical evaluation and biomarker profiling of Hsp90 inhibitors. Methods Mol. Biol. 1709, 423–441. doi:10.1007/978-1-4939-7477-1_29
Keywords: cancer, machine learing, pharmacophore, Hsp90, virtual screeening
Citation: Khan MI, Park T, Imran MA, Gowda Saralamma VV, Lee DC, Choi J, Baig MH and Dong J-J (2022) Development of machine learning models for the screening of potential HSP90 inhibitors. Front. Mol. Biosci. 9:967510. doi: 10.3389/fmolb.2022.967510
Received: 13 June 2022; Accepted: 15 August 2022;
Published: 19 October 2022.
Edited by:
Mustafa Aziz Hatiboglu, Bezmiâlem Vakıf Üniversitesi, TurkeyReviewed by:
Yusuf Tutar, University of Health Sciences, TurkeyCopyright © 2022 Khan, Park, Imran, Gowda Saralamma, Lee, Choi, Baig and Dong. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jae-June Dong, czgydG9uaWdodEB5dWhzLmFj; Mohammad Hassan Baig, bWhiYWlnQHlvbnNlaS5hYy5rcg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.