
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
EDITORIAL article
Front. Mol. Biosci., 13 July 2022
Sec. Structural Biology
Volume 9 - 2022 | https://doi.org/10.3389/fmolb.2022.948326
This article is part of the Research TopicNucleic Acid Polymerases: The Two-Metal-Ion Mechanism and BeyondView all 11 articles
Editorial on the Research Topic
Nucleic Acid Polymerases: The Two-Metal-Ion Mechanism and Beyond
Nucleic acid polymerases are essential for all forms of life, performing diverse functions from genome replication and repair to the transcription of a wide variety of RNAs. Although these enzymes differ widely in substrate specificity, efficiency, accuracy, and evolutionary origin, they all catalyze the same nucleotidyltransferase reaction. This eBook on “Nucleic Acid Polymerases: The Two-Metal-Ion Mechanism and Beyond” highlights both the similarities and differences among these enzymes.
The two-metal-ion catalytic mechanism for polymerases was proposed in 1993 by Thomas A. Steitz (Steitz, 1993), based on structural studies of the 3′-5′ exonuclease active site of the Klenow fragment of E. coli DNA polymerase I (Beese and Steitz, 1991; Beese et al., 1993) and mutagenesis of the polymerase active site (Polesky et al., 1992). Structural support for this mechanism came over the next several years, when crystal structures were determined with primer-template DNA and dNTP poised for catalysis at the polymerase active sites of several different DNA polymerases and HIV-1 reverse transcriptase (Pelletier et al., 1994; Doublié et al., 1998; Huang et al., 1998; Li et al., 1998). These and subsequent structures show that polymerases have two absolutely conserved aspartate residues that coordinate two divalent cations in the polymerase active site (Figure 1A), demonstrating that the two-metal-ion catalytic mechanism is also applicable to DNA synthesis (Brautigam and Steitz, 1998).
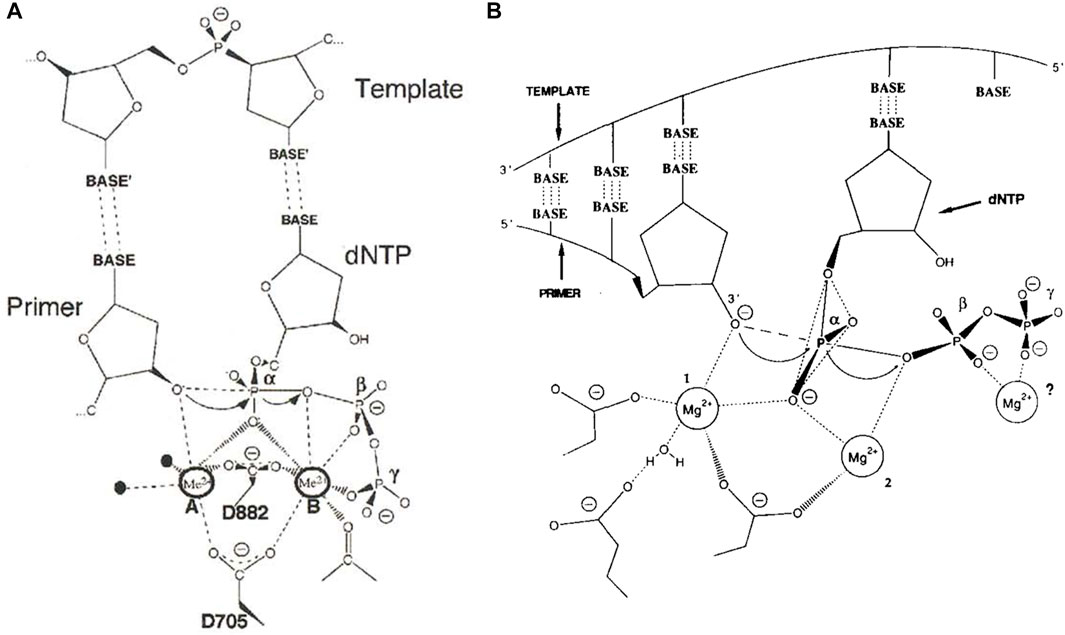
FIGURE 1. The two-metal-ion catalytic mechanism for nucleic acid polymerases (A) Two universally conserved aspartate residues (numbering is for the Klenow fragment of E. coli DNA polymerase I) coordinate two divalent cations, usually Mg2+, that facilitate deprotonation of the 3′-hydroxyl of the primer and stabilize the negative charges on the phosphates of the nucleotide and formation of the pentacovalent transition state. Figure is based on the structure of T7 DNA polymerase (Doublié et al., 1998) and is reproduced from Brautigam and Steitz (Brautigam and Steitz, 1998) with permission (B) The original mechanism proposed for polymerization based on the structure of the 3′-to-5′ exonuclease active site of Klenow fragment (Beese and Steitz, 1991). The possibility of a third metal ion binding to the beta and gamma phosphates was considered. Figure reproduced from Steitz (Steitz, 1993) with permission.
More recently, however, a three-metal-ion polymerase mechanism has been proposed based on time-resolved crystallographic studies of translesion and repair DNA polymerases eta and beta, respectively (Nakamura et al., 2012; Gao and Yang, 2016). In fact, Thomas Steitz had initially considered that a third metal ion might be involved (Figure 1B). In the first article in this eBook, Wang and Konigsberg review the effects of pH and Mg2+ concentration on high-fidelity DNA polymerase activity and argue that the three-metal-ion mechanism may not be universal to all polymerases, suggesting instead that the third metal ion stabilizes pyrophosphate binding after catalysis and thus slows product release (Wang and Konigsberg, 2022).
The review of viral RNA-directed RNA polymerases by Gong highlights the complexities of the nucleotide incorporation cycle, including conformational changes that accompany nucleotide binding and pyrophosphate release, polymerase translocation along the template that is required for processive synthesis, and events outside of the standard catalytic cycle that can impact the fidelity of replication (Gong, 2021).
Carvajal-Maldonado provide a comprehensive review of the other catalytic activities that are frequently associated with DNA polymerases: 3′-5′ exonuclease proofreading that increases replication fidelity, structure-specific 5′-nuclease activity required for Okazaki fragment maturation during lagging strand synthesis, 5′dRP lyase activity required in the base excision repair pathway, and 3′-end-trimming and single-strand extension involved in double-strand break repair (Carvajal-Maldonado et al., 2021).
Kaszubowski and Trakselis focus on the challenges of coordinating multiple polymerases during translesion synthesis where a high-fidelity replicative DNA polymerase encounters DNA damage and is replaced by one of a number of possible specialized enzymes with lower fidelity that allow replication of damaged DNA (Kaszubowski and Trakselis, 2021). The review compares passive and active mechanisms for the handoff of DNA between the polymerases and discusses the role of the sliding clamp processivity factor.
Nucleic acid synthesis is a highly dynamic process and the review by Millar emphasizes how single-molecule Fluorescent Energy Transfer techniques have been able to elucidate the conformational changes that occur during the E. coli DNA polymerase I nucleotide incorporation cycle and as the DNA transitions between the polymerase and nuclease active sites–processes that are difficult to resolve using more static structural methods (Millar, 2022).
The original research articles in this eBook emphasize how much there still is to learn about the wide variety of polymerases. Frey et al. use X-ray crystallography and molecular dynamics simulation to describe new non-nucleoside inhibitors of HIV-1 reverse transcriptase that are effective to mutants that are resistant to previously designed compounds (Frey et al., 2022). This work highlights the importance of polymerases as drug targets but also emphasizes the importance of understanding the entire nucleotide incorporation cycle, both kinetically and structurally, in the drug development process.
The work by Park et al. demonstrates that mitochondrial DNA polymerase gamma is capable of efficiently bypassing a CPD lesion at physiological concentrations of Mn2+ (Park et al., 2022). This ability is specific to polymerase gamma, not a general property of A-family DNA polymerases, underscoring the diversity of polymerases and emphasizing the role of cellular conditions in regulating activity.
Vaisman et al. address the evolutionary diversity of the translesion DNA polymerases. Biochemical characterization of the four Y-family polymerases (eta, iota, kappa and Rev1) from a lower eukaryote shows that their major properties are very similar to those of their human homologs (Vaisman et al., 2021). This work indicates that polymerase iota evolved earlier than previous sequence analysis had suggested, raising the question of what critical role this enzyme plays in both lower and higher eukaryotes.
In the final research article in this volume, Park et al. present two structures of phage RB69 DNA polymerase in open binary and partially closed ternary complexes that are distinct from previous structures of this enzyme (Park et al., 2021). Since these structures exist in a single crystal form, they suggest that initial binding of the correct incoming nucleotide and the second divalent metal ion are much weaker than expected.
The various articles in this issue demonstrate that, despite decades of seminal work on polymerases in replication and transcription, there are still many unknowns that require future research. We hope this issue will inspire graduate students and postdocs to devote their research to studying these fascinating processes that are fundamental to all life.
We wish to dedicate this special polymerase issue to the memory of Tom Steitz who was a mentor to all of us, both directly (JDP and YWY) and indirectly (IL). His pioneering structural work, insights into catalytic mechanism, and deep appreciation of the connections between biological structure and function continue to inspire our own research. We miss him deeply.
All authors have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
The authors are supported by grants awarded from the NIH to JP (R01GM080573) and to YWY (R01AI134611) and by DBT/Wellcome Trust India Alliance Intermediate Fellowship (IA/I/20/1/504905) awarded to IL.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Beese, L. S., Derbyshire, V., and Steitz, T. A. (1993). Structure of DNA Polymerase I Klenow Fragment Bound to Duplex DNA. Science 260, 352–355. doi:10.1126/science.8469987
Beese, L. S., and Steitz, T. A. (1991). Structural Basis for the 3′-5′ Exonuclease Activity of Escherichia coli DNA Polymerase I: a Two Metal Ion Mechanism. EMBO J. 10, 25–33. doi:10.1002/j.1460-2075.1991.tb07917.x
Brautigam, C. A., and Steitz, T. A. (1998). Structural and Functional Insights provided by Crystal Structures of DNA Polymerases and Their Substrate Complexes. Curr. Opin. Struct. Biol. 8, 54–63. doi:10.1016/s0959-440x(98)80010-9
Carvajal-Maldonado, D., Drogalis Beckham, L., Wood, R. D., and Doublié, S. (2021). When DNA Polymerases Multitask: Functions beyond Nucleotidyl Transfer. Front. Mol. Biosci. 8, 815845. doi:10.3389/fmolb.2021.815845
Doublié, S., Tabor, S., Long, A. M., Richardson, C. C., and Ellenberger, T. (1998). Crystal Structure of a Bacteriophage T7 DNA Replication Complex at 2.2 Å Resolution. Nature 391, 251–258. doi:10.1038/34593
Frey, K. M., Bertoletti, N., Chan, A. H., Ippolito, J. A., Bollini, M., Spasov, K. A., et al. (2022). Structural Studies and Structure Activity Relationships for Novel Computationally Designed Non-nucleoside Inhibitors and Their Interactions With HIV-1 Reverse Transcriptase. Front. Mol. Biosci. 9, 805187. doi:10.3389/fmolb.2022.805187
Gao, Y., and Yang, W. (2016). Capture of a Third Mg 2+ Is Essential for Catalyzing DNA Synthesis. Science 352, 1334–1337. doi:10.1126/science.aad9633
Gong, P. (2021). Within and beyond the Nucleotide Addition Cycle of Viral RNA-Dependent RNA Polymerases. Front. Mol. Biosci. 8, 822218. doi:10.3389/fmolb.2021.822218
Huang, H., Chopra, R., Verdine, G. L., and Harrison, S. C. (1998). Structure of a Covalently Trapped Catalytic Complex of HIV-1 Reverse Transcriptase: Implications for Drug Resistance. Science 282, 1669–1675. doi:10.1126/science.282.5394.1669
Kaszubowski, J. D., and Trakselis, M. A. (2021). Beyond the Lesion: Back to High Fidelity DNA Synthesis. Front. Mol. Biosci. 8, 811540. doi:10.3389/fmolb.2021.811540
Li, Y., Korolev, S., and Waksman, G. (1998). Crystal Structures of Open and Closed Forms of Binary and Ternary Complexes of the Large Fragment of Thermus Aquaticus DNA Polymerase I: Structural Basis for Nucleotide Incorporation. EMBO J. 17, 7514–7525. doi:10.1093/emboj/17.24.7514
Millar, D. P. (2022). Conformational Dynamics of DNA Polymerases Revealed at the Single-Molecule Level. Front. Mol. Biosci. 9, 826593. doi:10.3389/fmolb.2022.826593
Nakamura, T., Zhao, Y., Yamagata, Y., Hua, Y.-j., and Yang, W. (2012). Watching DNA Polymerase η Make a Phosphodiester Bond. Nature 487, 196–201. doi:10.1038/nature11181
Park, J., Baruch-Torres, N., Iwai, S., Herrmann, G. K., Brieba, L. G., and Yin, Y. W. (2022). Human Mitochondrial DNA Polymerase Metal Dependent UV Lesion Bypassing Ability. Front. Mol. Biosci. 9, 808036. doi:10.3389/fmolb.2022.808036
Park, J., Youn, H.-S., An, J. Y., Lee, Y., Eom, S. H., and Wang, J. (2021). Structure of New Binary and Ternary DNA Polymerase Complexes from Bacteriophage RB69. Front. Mol. Biosci. 8, 704813. doi:10.3389/fmolb.2021.704813
Pelletier, H., Sawaya, M. R., Kumar, A., Wilson, S. H., and Kraut, J. (1994). Structures of Ternary Complexes of Rat DNA Polymerase β, a DNA Template-Primer, and ddCTP. Science 264, 1891–1903. doi:10.1126/science.7516580
Polesky, A. H., Dahlberg, M. E., Benkovic, S. J., Grindley, N. D., and Joyce, C. M. (1992). Side Chains Involved in Catalysis of the Polymerase Reaction of DNA Polymerase I from Escherichia coli. J. Biol. Chem. 267, 8417–8428. doi:10.1016/s0021-9258(18)42461-1
Steitz, T. A. (1993). DNA- and RNA-Dependent DNA Polymerases. Curr. Opin. Struct. Biol. 3, 31–38. doi:10.1016/0959-440X(93)90198-T
Vaisman, A., McDonald, J. P., Smith, M. R., Aspelund, S. L., Evans, T. C., and Woodgate, R. (2021). Identification and Characterization of Thermostable Y-Family DNA Polymerases η, ι, κ and Rev1 from a Lower Eukaryote, Thermomyces Lanuginosus. Front. Mol. Biosci. 8, 778400. doi:10.3389/fmolb.2021.778400
Keywords: DNA polymerase, RNA polymerase, reverse transcriptase, cellular polymerase, viral polymerase
Citation: Pata JD, Yin YW and Lahiri I (2022) Editorial: Nucleic Acid Polymerases: The Two-Metal-Ion Mechanism and Beyond. Front. Mol. Biosci. 9:948326. doi: 10.3389/fmolb.2022.948326
Received: 19 May 2022; Accepted: 16 June 2022;
Published: 13 July 2022.
Edited and reviewed by:
Adrian Goldman, University of Helsinki, FinlandCopyright © 2022 Pata, Yin and Lahiri. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Janice D. Pata, amFuaWNlLnBhdGFAaGVhbHRoLm55Lmdvdg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.