- 1Institutes of Biomedical Sciences and Shanghai Cancer Center, Fudan University, Shanghai, China
- 2Department of Chemistry, Fudan University, Shanghai, China
- 3Beijing Advanced Innovation Center for Precision Medicine, Beihang University, Beijing, China
- 4Department of Clinical Laboratory, Guangxi Medical University Cancer Hospital, Nanning, China
- 5NHC Key Laboratory of Glycoconjugates Research, Fudan University, Shanghai, China
Glycosylation inhibition has great potential in cancer treatment. However, the corresponding cellular response, protein expression and glycosylation changes remain unclear. As a cell-permeable small-molecule inhibitor with reduced cellular toxicity, N-linked glycosylation inhibitor-1 (NGI-1) has become a great approach to regulate glycosylation in mammalian cells. Here for the first time, we applied a nascent proteomic method to investigate the effect of NGI-1 in hepatocellular carcinoma (HCC) cell line. Besides, hydrophilic interaction liquid chromatography (HILIC) was adopted for the enrichment of glycosylated peptides. Glycoproteomic analysis revealed the abundance of glycopeptides from LAMP2, NICA, and CEIP2 was significantly changed during NGI-1 treatment. Moreover, the alterations of LAMP2 site-specific intact N-glycopeptides were comprehensively assessed. NGI-1 treatment also led to the inhibition of Cathepsin D maturation and the induction of autophagy. In summary, we provided evidence that NGI-1 repressed the expression of glycosylated LAMP2 accompanied with the occurrence of lysosomal defects and autophagy.
Introduction
The role of glycosylation in modulating various aspects of protein biology is a broad research field (Ward et al., 2021; Critcher et al., 2022). O-linked glycosylation and N-linked glycosylation are the two most important forms of protein glycosylation (Sun et al., 2021). Unlike the O-linked glycosylation, N-linked glycosylation is one type of biological processes that proteins undergo during de novo synthesis (Xu et al., 2018; Karki et al., 2021; Tian et al., 2021). N-linked protein glycosylation mainly begins from endoplasmic reticulum (ER). Next, a range of complex enzymatic reactions were carried out with the help of numerous glycosidases, glycosyltransferases, transporters and regulatory proteins located in ER or Golgi apparatus (Groux-Degroote et al., 2021). Well-controlled glycosylation is very important for the maturation and transport of glycoproteins. Once the N-linked glycosylation pathway is interrupted, the unfolded and misfolded glycoproteins are easy to accumulate in the ER, resulting in ER stress (Bull and Thiede, 2012; Di Patria et al., 2022; Wang et al., 2022). Under the condition of continuous stress, the excessive accumulation of abnormally folded proteins in the ER lumen is fatal to cells, which can cause unfolded protein response (UPR) and apoptosis (Sovolyova et al., 2014; Read and Schroder, 2021). However, the detailed response events and underlying molecular mechanisms in N-glycosylation inhibition remain to be elucidated, especially in the fine process of protein de novo synthesis.
NGI-1 is developed as a novel inhibitor of N-linked glycosylation with cellular permeability, which can directly target and block the function of the oligosaccharyltransferase (OST) catalytic subunits STT3A and STT3B in the ER (Rinis et al., 2018; Lu et al., 2019; Zhu et al., 2019). Compared with traditional glycosylation inhibitor tunicamycin with obvious toxicity, NGI-1 does not cause complete inhibition of glycosylation and has low cytotoxicity (Dawson et al., 2007; Contessa et al., 2010; Alymova et al., 2022). Notably, the development of NGI-1 provides a pharmacological method to mildly regulate N-linked glycosylation in mammalian cells (Baro et al., 2019; Shu et al., 2019). The clinical value of NGI-1 in the application of oncologic therapies has recently attracted people’s attention. In lung cancer cells, NGI-1 selectively inhibits the proliferation of epidermal growth factor receptor (EGFR)-dependent cell lines by destroying the glycosylation and cell surface localization of EGFR. Many results confirmed that inhibition of glycosylation could be a promising method for the treatment of receptor tyrosine kinase-dependent cancers (Lopez-Sambrooks et al., 2016). Also, the addition of NGI-1 to tyrosine kinase inhibitor therapy is fatal to drug-resistant cell lines (Lopez Sambrooks et al., 2016). Besides, NGI-1 can sensitize multiple models of neoplasms to chemotherapy or radiation, and may overcome the limited success of conventional strategies (Wahl and Lawrence, 2019; Phillips et al., 2022). The mechanisms involved need to be further investigated to promote the clinical application of NGI-1 in tumor therapy.
Although some studies have analyzed the dynamic protein response under traditional glycosylation inhibitor treatment (Xiao et al., 2016; Shu et al., 2019), our study focused on qualitative and quantitative analyses of newly synthesized proteins. Because the nascent sub-proteome is the first to respond to perturbations in theory. Through the bioorthogonal-chemistry-based enrichment strategy, the non nascent proteome is removed, and the remaining newly synthesized proteins can be identified and quantified more easily in mass spectrometry due to the eliminated interference of non nascent proteome. Moreover, to quantify the changes of newly synthesized proteins is more reasonable to attribute the changed results to the changed condition (McClatchy et al., 2018; van Bergen et al., 2022).
Here, we performed a bioorthogonal non-canonical amino-acid tagging (BONCAT)-based quantitative proteomics method for identification and quantitation of newly synthesized proteins under the treatment of NGI-1 to enhance our mechanistic understanding of N-linked glycosylation inhibition process (Dieterich et al., 2007; Shah et al., 2022). According to the following bioinformatic analyses, the functions of the differentially expressed proteins were annotated. Then, the glycoproteome analysis was applied to identify the de novo synthesized glycoproteins in response to N-glycosylation inhibition. Functional experiments were performed to confirm the alteration of lysosome-associated membrane protein-2 (LAMP2) under the condition of glycosylation inhibition. Our study demonstrated that NGI-1 had an impact on a wide range of biological processes by regulating protein glycosylation, providing novel insights in explaining its antitumor mechanism.
Materials and Methods
Cell Culture and NGI-1 Treatment
The Huh7 and HCCLM3 cells were obtained from the Chinese Academy of Sciences. Cell culture was performed using Dulbecco’s modified Eagle’s medium (DMEM, Gibco) supplemented with 10% fetal bovine serum (FBS, Biological Industries). Briefly, cells were plated for 48 h prior to incubation with various doses or temporal gradients of NGI-1 (MCE). Briefly, 10 mM NGI-1 was dissolved in 1 ml DMSO to create a storage solution, and further diluted to 5–50 μM with culture medium. The same dosage of DMSO was used as a control. Following NGI-1 treatment, the cells were washed with phosphate buffered saline (PBS, Hyclone) and lysed on ice with RIPA buffer (Beyotime) containing Roche protease inhibitor cocktail. Then, the homogenate was sonicated for 30s and centrifuged at 4°C for 10 min at 18,000×g.
Chemical Metabolic Labeling
Cells were cultured to about 70–80% confluence and treated with l-methionine-free DMEM (Gibco, 21013024) and dialyzed FBS (Gibco) for half an hour. AHA (Cambridge Isotope Laboratories) at 500 μM was added in the absence or presence of 10 μM NGI-1 for 24 h. At the end of incubation, cells were collected and lysed with 0.5% Sodium dodecyl sulfate (SDS, Sigma-Aldrich) containing Roche protease inhibitor cocktail. The concentration of extracted protein was measured by BCA assay (Beyotime).
Click Reaction
After metabolic labeling by AHA, a click reaction was performed as previously published (Dieterich et al., 2007). Briefly, a cocktail of 500 μM alkyne-biotin reagent (Sigma-Aldrich), 1 mM Copper Sulfate (CuSO4, Sigma-Aldrich), 6 mM 3-(4-((bis((1-tert-butyl-1H-1,2,3-triazol-4-yl)methyl)amino)methyl)-1H-1,2,3-triazol-1-yl)propanol (BTTP, Click Chemistry Tools) and 6 mM Ascorbate (Sigma-Aldrich) was added to 500 μg cell lysate. The samples were incubated for 3 hours at room temperature.
Enrichment of Newly Synthesized Proteins
Newly synthesized proteins were enriched as previously described (Ma et al., 2017; Ma et al., 2018) with minor modifications. The enrichment was conducted with NeutrAvidin Agarose Resins (GenScript). The beads were washed by 6 M urea (Sigma), 20% ACN and PBS respectively. After reconstitution, the beads were reduced with 10 mM dithiothreitol (DTT) and alkylated with 25 mM iodoacetamide (IAA). Sequencing-grade modified trypsin (Promega) was utilized for on-beads digestion. The obtained peptides were desalted by Zip-Tip (Millipore) and stored at -20°C until use. For glycopeptide enrichment, the Glycopeptide Enrichment Kit (Novagen, Darmstadt, Germany) was used following the instructions.
LC-MS/MS
The desalted peptides were analyzed on Orbitrap Exploris 480 MS. Peptides were separated by a C18 column (75 μm × 500 mm column, ThermoFisher) on an Easy nLC 1,200 high-pressure liquid chromatography (HPLC) system (Thermo Fisher Scientific) operating at 300 nL/min. Buffer A (0.1% formic acid) and buffer B (0.1% formic acid in 80% ACN) were used. Peptides were separated by a linear gradient from 8% B to 23% B in 90 min followed by a linear increase to 50% B in 27 min. For data-dependent acquisition experiments, the full MS resolutions were set to 120,000 and full MS AGC target was 300%. Mass range was set to 350–1,200 Da. AGC target value for fragment spectra was set at 75% with a resolution of 15,000. Isolation width was set at 1.6 m/z. Precursor ions were fragmented by higher energy collisional dissociation with a normalized collision energy of 29%. All data were acquired in positive polarity profile mode. For intact glycopeptide analysis, the experimental instrument and conditions were described previously with minor modification (Zhang et al., 2019; Cao et al., 2021). To ensure equal amount of analyte loading, the same volume (4 μl) was loaded onto the trap column (PepMap C18, 100 μm × 2 cm) and the analytical column (PepMap C18, 75 μm × 25 cm) by using a gradient of 1–25% solvent B (solvent A: water with 0.1% FA; solvent B: ACN with 0.1% FA) in 60 min, followed by an increase to 45% B in 20 min.
Data Analysis
Peptide identification and label-free relative quantification analysis was performed in PEAKS Studio software (Bioinformatics Solutions Inc., Waterloo, Canada). PEAKS provided an unbiased quantitative workflow based on the MS1 peak area without internal standard. The protein database was from Swiss-Prot reviewed, date July 2020, with species of Homo sapiens (20,380 entries). The input parameters were: 10 ppm precursor mass tolerance, 0.02 Da fragment mass tolerance. The maximum false discovery rates for protein and peptides were set at 1%. Gene ontology (GO) analysis, including cellular component (CC), molecular function (MF), and biological process (BP), was performed through the David Functional Annotation Tool (Huang da et al., 2009). The intact glycopeptides were identified by pGlyco 3.0 (Version 2020.12.08) and quantified by pQuant (Liu et al., 2014; Zeng et al., 2021). Parameters for database search could be found in our previous study (Zhang et al., 2019; Cao et al., 2021). The default setting of 1% FDR at the glycopeptide level was used.
Western Blotting
The obtained cell proteins were resolved by SDS-PAGE. After transferring, the membranes were blocked in 5% defatted milk (Bio-Rad) and then incubated with the primary antibodies: anti-LAMP2 (1:500 dilution, Santa Cruz Biotechnology, United States), Cathepsin D (1:1,000 dilution, Cell Signaling Technology, United States), LC3B (1:1,000 dilution, Cell Signaling Technology, United States), ATG5 (1:1,000 dilution, Proteintech, United States) and β-actin (1:10,000 dilution, Multisciences, China) overnight at 4°C, followed by HRP-conjugated secondary antibodies (Multisciences, China). Immunoblots were visualized by ECL substrate (Biosharp) and Bio-Rad Image Lab software.
Lysosome Isolation and Visualization
Lysosomes were isolated by using Minute™ Lysosome Isolation Kit (Invent Biotechnologies, United States) according to instructions. For visualization of lysosomes, 50 nM LysoTracker Red (Beyotime) was added to culture medium for 1 hour incubation. After fixation with 4% paraformaldehyde, the cells were stained with DAPI (Keygen Biotech). Olympus fluorescence microscopy was used for image acquisition.
Transient Transfection and Cell Viability Assay
The small interfering RNA (siRNA) targeting human ATG5 was commercially synthesized (Sangon Biotech) as described (Shi et al., 2021). Along with scrambled RNAi oligonucleotides, the siRNA targeting AGT5 was transfected into cells via transfection reagent (BBI). Cell Counting Kit-8 (CCK8, Dojindo) was performed to confirm the influence of NGI-1 on cell viability and was repeated at least three times.
Statistical Analysis
Figures were plotted with GraphPad Prism (GraphPad Software Inc.) and Hiplot (https://hiplot.com.cn). The numeric data were showed as means ± standard deviation. Two tailed Student t-test was used for statistical comparison. p < 0.05 was considered statistically significant. N-glycan structures were drawn with GlycoWorkbench.
Results
Analysis of de novo Synthesized Proteins Under Glycosylation Inhibition
For comprehensive characterization of the protein alterations under glycosylation inhibition, we applied the small-molecule OST inhibitor NGI-1 to the de novo protein synthesis process (Figure 1A). After starvation, the cells were treated with AHA to specifically label de novo synthesized proteins for 24 h under 10 μM NGI-1 treatment or DMSO. Then the cells were harvested, lysed and reacted with alkyne-biotin, followed by enrichment via biotin-avidin affinity interaction in parallel. After incubation and thorough washing, the bound proteins were digested on beads. Ten-percent enzymolysis products were utilized for desalination, and the rest were used for glycopeptide enrichment (Figure 1A). The obtained peptides and glycopeptides were sent to LC-MS/MS for further identification and quantification. In the proteome, 5,508 newly synthesized proteins were identified, of which 4,993 proteins obtained quantitative information. The technical replicates showed good correlation (Figure 1B). As the cutoff criterion was set to fold change (FC, NGI treatment/DMSO) > 2 or <0.5, a total of 78 newly synthesized proteins (31 up- and 47 down-regulated) were differentially expressed (Figures 1C,D; Supplementary Table S1). Actin cytoskeleton organization and protein binding process were most enriched by GO-BP and GO-MF analysis, and the cytoplasm was enriched according to GO-CC analysis (Figures 2A–C). The Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis of the differentially expressed proteins indicated significant enrichment in lysosome-related pathways (Figure 2D).
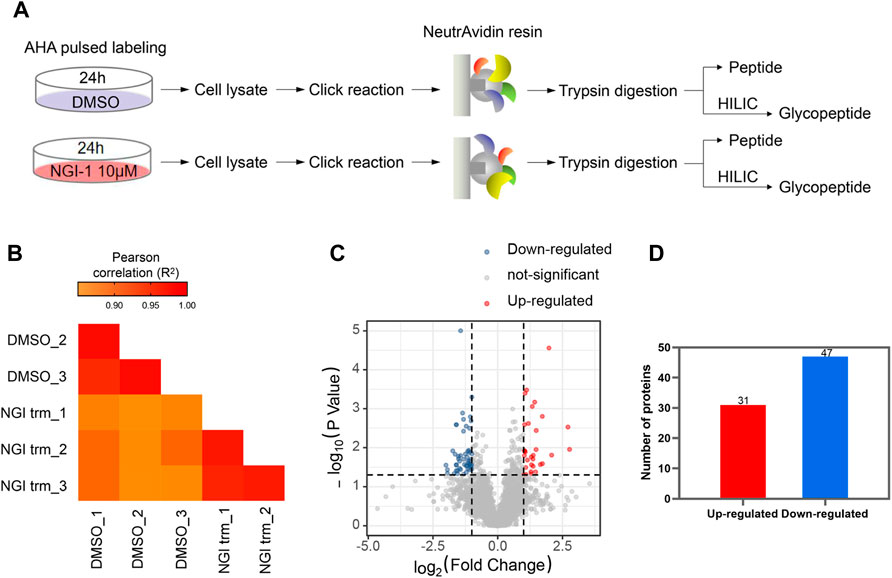
FIGURE 1. Analysis of nascent proteome under glycosylation inhibition (A) Workflow for metabolic labeling of de novo protein synthesis in NGI-1 treatment (B) The Pearson Correlation Coefficient R2 values in technical repetition (C) Proteins with statistical significance after a t-test were shown in red (up-regulated) or blue (down-regulated) on a volcano plot. The vertical lines correspond to 2.0-fold up and down (ratio between NGI treatment and DMSO), and the horizontal line represents a p-value of 0.05. Each point in the plot represents a different protein (D) A total of 31 up-regulated and 47 down-regulated de novo synthesized proteins were identified.
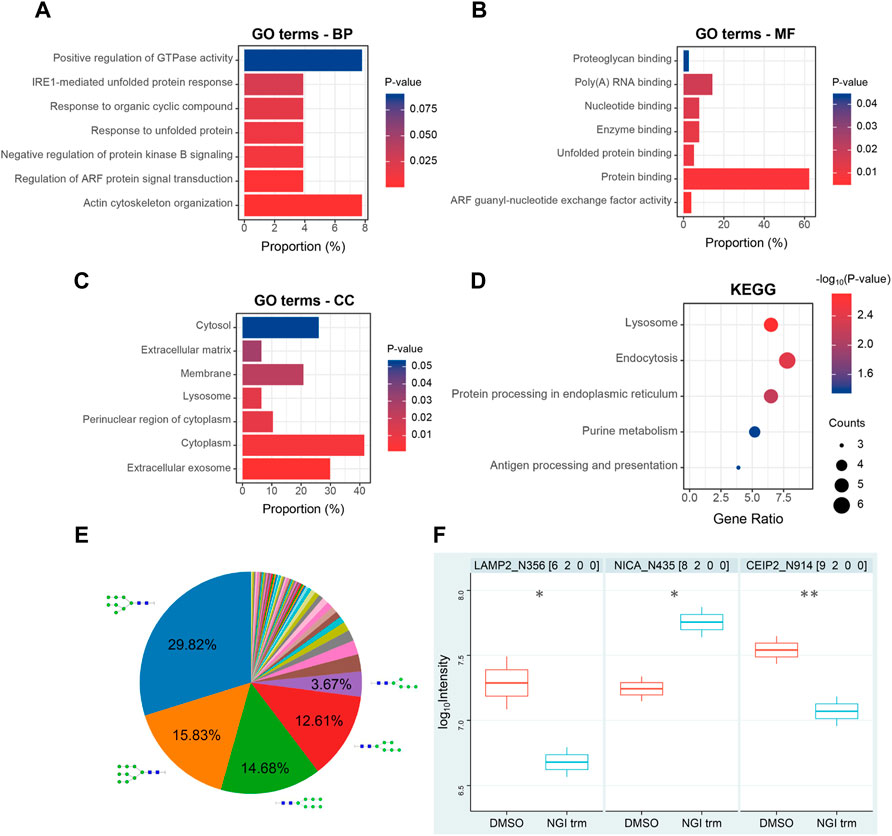
FIGURE 2. Bioinformatic analysis of nascent proteome (A–C) GO biological process (BP), molecular function (MF), and cellular component (CC) categories enriched during N-glycosylation inhibition (D) KEGG pathway analysis of differentially expressed de novo synthesized proteins (E) Distribution of the identified N-glycan compositions (F) Differentially expressed de novo synthesized intact glycopeptides in Huh7 cells upon NGI-1 treatment. p values were determined using two-tailed t-tests. *p < 0.05, **p < 0.01. The number after the protein name represents N-glycosylation site.
Analysis of Newly Synthesized Glycoproteins During NGI-1 Treatment
For convenience, a four-digit nomenclature in HNSF order was used for glycan annotation (H, Hexose; N, N-acetylhexosamine; S, Sialic acid; F, Fucose). In the glycoproteome, 436 newly synthesized N-glycopeptides were identified under the condition of NGI-1 treatment (Supplementary Table S2). Comparing the distribution of N-glycan compositions, H8N2 and H9N2 represented the most common compositions in the identification (Figure 2E). With the help of a quantitation software tool termed pQuant (Liu et al., 2014), the peak area was determined for each glycopeptide. At last 47 N-glycopeptides passed the strict filtering criteria (Zhang et al., 2019; Cao et al., 2021) and obtained quantitative information (Supplementary Table S3). Compared with the control group, LAMP2_N356 (H6N2) and CEIP2_N914 (H9N2) were significantly decreased under NGI-1 treatment (p = 0.0434 and p = 0.0049, respectively), while NICA_N435 (H8N2) was significantly increased (p = 0.0465) (Figure 2F).
NGI-1 Treatment Inhibited the Expression of Glycosylated LAMP2 and Caused Lysosomal Defects
LAMP2 is one of the major proteins on the surface of lysosomes, which is crucial for lysosomal function (Li and Pfeffer, 2016; Liu et al., 2021). As LAMP2 was significantly suppressed in proteome-wide experiment, we first investigated the potential effect of NGI-1 treatment on LAMP2. Western blotting showed that the expression of glycosylated LAMP2 in Huh7 cells was considerably reduced under NGI-1 treatment in a concentration-dependent manner (Supplementary Figure S1). To further confirm the suppression extent of LAMP2 by NGI-1, fine mapping of the N-glycosylation profile of LAMP2 was achieved based on lysosome pre-separation. Moreover, we observed that the top 10 intact glycopeptides of LAMP2 were generally decreased in varying degrees (Figure 3A and Supplementary Table S4). Among them, LAMP2_N275 (H6N5S1) exhibited the most obvious down-regulation (Figure 3B). As illustrated in Figure 4A, prolonged NGI-1 incubation led to the accumulation of lysosomal fluorescence signals. Thus, we hypothesized that glycosylation inhibition caused lysosomal dysfunction. We investigated the maturation of lysosomal protease Cathepsin D, which plays an important role in lysosomal protein degradation (Shi et al., 2021). Western blotting demonstrated that NGI-1 treatment strongly inhibited the cleavage of preprocathepsin D, thereby reducing the production of mature Cathepsin D forms at different concentrations (Figure 4B) and different durations (10 μM) (Figure 4C).
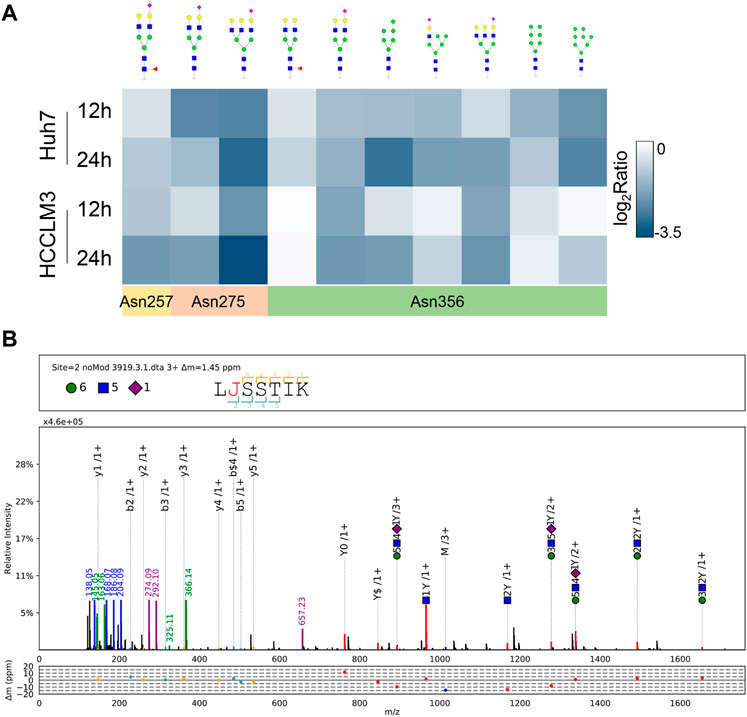
FIGURE 3. Site-specific glycan profiling of LAMP2 (A) Heat map showing site-specific intact glycopeptides from LAMP2 in Huh7 and HCCLM3 cells with NGI-1 treatment. The positions of identified N-glycosylation sites were annotated (B) pGlyco annotation of LAMP2_N275 (H6N5S1). “J” represents the glycosylation site “N”; green circle: hexose (H); blue square: N-acetylglucosamine (N); purple rhombus: sialic acid (S). The upper frame of spectrum is designed to annotate peptide sequence and glycan composition. The mass deviations of the annotated peaks are shown in the box below.
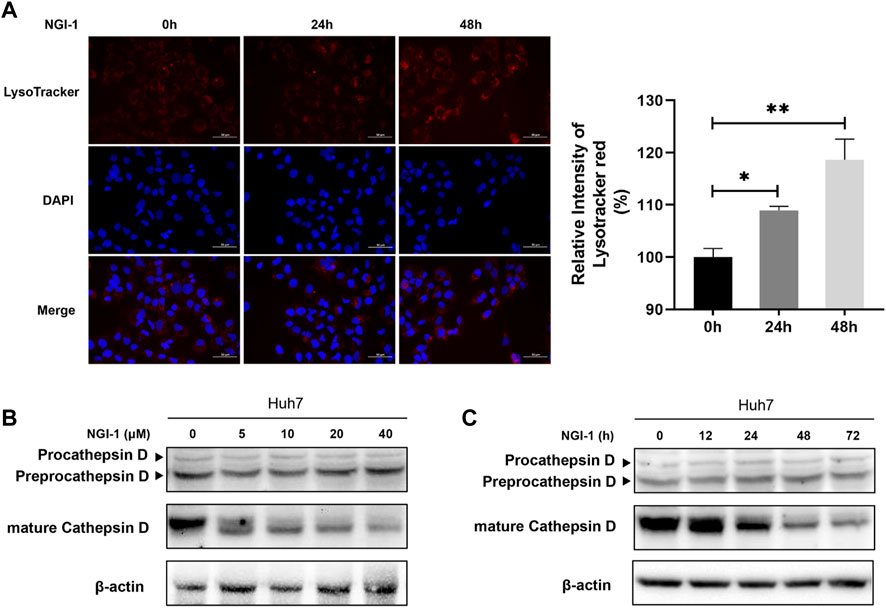
FIGURE 4. NGI-1 treatment causes lysosomal defects in Huh7 cells. (A) Huh7 cells were treated with 10 μM NGI-1 for different time (0, 24, 48 h), followed by labeling with LysoTracker Red and stained with DAPI. Relative intensity of LysoTracker Red was quantified from images (Student t-test; *p < 0.05, **p < 0.01) (B,C) Immunoblot analysis of Cathepsin D in Huh7 cells with NGI-1 treatment. Cells were treated either with different concentrations for 24 h (B) or for different durations at a concentration of 10 μM (C).
Suppression of Autophagy Occurrence Promoted NGI-1-Mediated Cytotoxicity
Have shown that NGI-1 treatment disabled LAMP2 by glycosylation inhibition, we therefore hypothesized that NGI-1 was related to chaperone-mediated autophagy which is mediated by LAMP proteins (Hubert et al., 2016; Shi et al., 2021). To solve the problem, we detected the level of autophagy along with NGI-1 treatment. The preliminary results showed that NGI-1 treatment induced a large amount of LC3-II accumulation (Figure 5A). Autophagy is an evolutionarily conserved intracellular process, which maintains homeostasis by degrading and cycling intracellular components in response to various environmental stresses (Cao et al., 2020). Thus, we speculated that the occurrence of autophagy might affect cell viability. To test the hypothesis, the autophagy essential component ATG5 was knocked down to examine the influence of NGI-1-mediated cytotoxicity. Upon NGI-1 treatment, due to the ATG5 knockdown, LC3-II accumulation was considerably decreased (Figure 5B). Under 24 or 48 h treatment, ATG5 knockdown had no effect on NGI-1-mediated decline in cell viability. With the increase of treatment time to 72 h, knocking down of ATG5 exhibited more obvious suppression on cell viability (Figure 5C). The findings suggested that inhibition of autophagy occurrence partially promoted NGI-1-mediated cytotoxicity.
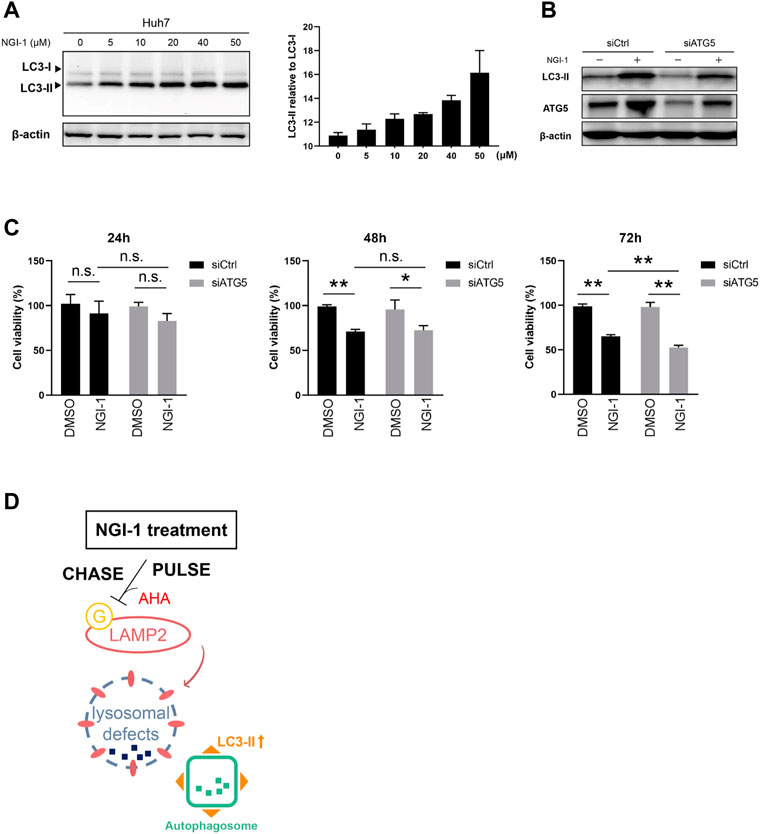
FIGURE 5. NGI-1 treatment induces autophagy occurrence in Huh7 cells. (A) Huh7 cells were treated with NGI-1 at different concentrations and LC3 expression pattern was examined by immunoblot. Endogenous LC3-II/LC3-I ratio was quantified (B) Western blotting analysis of LC3 and ATG5 expression in scrambled siRNA- or ATG5 siRNA-treated Huh7 cells incubated with DMSO or NGI-1 (10 μM) for 24 h. Actin was used as the internal control (C) Ctrl and ATG5 knockdown Huh7 cells were treated with 10 μM NGI-1 for indicated times and cell viability was determined by Cell Counting Kit-8. Data were presented as Mean ± SD from three independent experiments. *p < 0.05, **p < 0.01, n. s = not significant (D) The responses of Huh7 cells toward NGI-1 treatment.
Discussion
It is widely known that N-linked protein glycosylation is an important cotranslational and post-translational modification in biology, playing an essential part in development, organ specific function and disease (Zielinska et al., 2010; Esmail and Manolson, 2021). Tumor-related glycosylation alterations are connected with tumor progression, malignant transformation and immune evasion (Rodrigues et al., 2018; Bindeman and Fingleton, 2022). In view of the important role of glycosylation in tumor development, glycosylation inhibition has exhibited huge potential as an auxiliary means of antitumor therapies (Song et al., 2020).
Recently, a novel small-molecule OST inhibitor (NGI-1) has provided a pharmacological approach to partially disrupt N-linked glycosylation (Lopez-Sambrooks et al., 2016; Lu et al., 2019; Phillips et al., 2022). In the process of protein synthesis, a unique carbohydrate structure is transferred from a lipid carrier to the -Asn-X-Ser/Thr- (X≠Pro) sequence of the nascent peptides via the OST complex at ER membrane (Aebi, 2013). Therefore, OST represents a critical node for N-glycosylation regulation. Based on this view, NGI-1 was designed to block the function of OST catalytic subunit and induce a selective loss of cell viability (Lopez Sambrooks et al., 2018). However, little is known about de novo protein synthesis during the inhibition of glycosylation induced by NGI-1.
Combined with metabolic labeling technology and label-free quantitative proteomics, a workflow was built to characterize the de novo protein synthesis in the process of NGI-1-induced glycosylation inhibition. The structure of AHA contains methionine analogues, which enables it to be assembled into de novo synthesized proteins. Besides, the azide group of AHA could be used for the enrichment by click reaction (Dieterich et al., 2007; Wang et al., 2016; Vargas-Diaz and Altelaar, 2022). The protein alterations of nascent proteome in NGI-1-treated cells probably reflected specific stress response under N-linked glycosylation inhibition. Our data showed that, the lysosome, endocytosis and ER protein processing related pathways were disturbed as the distinct response to NGI-1 treatment. The discovery of nascent proteome provided a new perspective for comprehending the importance of glycosylation in the growth process of cell. In addition, the advantage of newly synthesized protein enrichment enabled it to be applied in characterizing the glycosylation changes that occurred during protein synthesis, which were usually not detectable. Our glycoproteomic results showed that the alteration of the oligo-mannose glycans was apparently enriched in cancer cells, which was similar to a previous study (Sun et al., 2016). Considering that the increase of high-mannose glycans is a characteristic feature of malignant transformation, this observation indicated that some caution should be used when popularizing the effects of processing inhibitors on a complex process (Li et al., 2015; Boyaval et al., 2022).
It was speculated that when NGI-1 treatment blocks the N-glycosylation synthesis pathway, the changes of related functional glycoproteins may reflect the status of ongoing biological processes. We then focused on a highly glycosylated lysosome membrane protein LAMP2, whose expression of newly synthesized protein and intact glycopeptide were both dramatically down-regulated under NGI-1 treatment. As expected, the overall expression of glycosylated LAMP2 exhibited a similar alteration trend as observed in nascent glycoproteome. LAMP2 is known as a key protein involved in the control of membrane fusion between autophagosome and lysosome. Lysosomal defects usually abolish the fusion of autophagosomes and lysosomes, resulting in excessive accumulation of autophagosomes and occurrence of human diseases (Ikeda et al., 2021; Jiang and Mizushima, 2014). A novel finding in this study was that lysosomal defects occurred along with autophagy accumulation in the NGI-1-exposed Huh7 cells (Figure 5D), suggesting that the fusion of autophagosome-lysosome was blocked up (Liu et al., 2021).
This work highlights the importance of using nascent glycoproteome to comprehensively characterize the targets of glycosylation inhibitors, so that such inhibitors can be better developed and used for basic research or clinical practice. However, due to the complexity of glycoprotein biosynthesis, many studies should be further expanded. For example, more experiments are needed to explore the pathways related to glycosylation inhibition, including glycan biosynthesis. Besides, LAMP2 has different degrees of N-glycosylation under abnormal physiological conditions. But the key sites responsible for controlling its function remain to be determined in the context of cancer.
In conclusion, a novel technique process was successfully established for characterization of de novo synthesized proteins and glycoproteins during glycosylation inhibition, which would promote the clinical application of NGI-1 in future cancer treatment. We also proposed that NGI-1 induced lysosomal defects probably by impairing the lysosomal membrane protein LAMP2.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author Contributions
HL and HS conceived and designed the research. All authors were responsible for acquisition and analysis of data; furthermore, XC, PM and YS performed the research; GY, JY and XZ provided technical assistance; CL and LZ performed the data analyses; XC and HS drafted the manuscript; HL revised and commented on the draft, and all authors read and approved the final version of the manuscript.
Funding
This work was supported by the National Key Research and Development Program of China (2017YFA0505100), NSF of China (Grants 82121004), Shanghai Projects (19ZR1403200) and Natural Science Fund Project of Guangxi Province of China (2018GXNSFAA281053).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank openbiox community and Hiplot team (https://hiplot.com.cn) for providing technical assistance and valuable tools for data analysis and visualization. The authors thank Shu Zhang, Zhongshan Hospital, Fudan University, Shanghai, China, for her generous support.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmolb.2022.899192/full#supplementary-material
Reference
Aebi, M. (2013). N-linked Protein Glycosylation in the Er. Biochim. Biophys. Acta (Bba) - Mol. Cel Res. 1833, 2430–2437. doi:10.1016/j.bbamcr.2013.04.001
Alymova, I. V., Cipollo, J. F., Parsons, L. M., Music, N., Kamal, R. P., Tzeng, W.-P., et al. (2022). Aberrant Cellular Glycosylation May Increase the Ability of Influenza Viruses to Escape Host Immune Responses through Modification of the Viral Glycome. mBio [Online ahead of print]. doi:10.1128/mbio.02983-21
Baro, M., Lopez Sambrooks, C., Quijano, A., Saltzman, W. M., and Contessa, J. (2019). Oligosaccharyltransferase Inhibition Reduces Receptor Tyrosine Kinase Activation and Enhances Glioma Radiosensitivity. Clin. Cancer Res. 25, 784–795. doi:10.1158/1078-0432.CCR-18-0792
Bindeman, W. E., and Fingleton, B. (2022). Glycosylation as a Regulator of Site-specific Metastasis. Cancer Metastasis. Rev. 41, 107–129. doi:10.1007/s10555-021-10015-1
Boyaval, F., Dalebout, H., Van Zeijl, R., Wang, W., Fariña-Sarasqueta, A., Lageveen-Kammeijer, G. S. M., et al. (2022). High-Mannose N-Glycans as Malignant Progression Markers in Early-Stage Colorectal Cancer. Cancers 14, 1552. doi:10.3390/cancers14061552
Bull, V. H., and Thiede, B. (2012). Proteome Analysis of Tunicamycin-Induced Er Stress. Electrophoresis 33, 1814–1823. doi:10.1002/elps.201100565
Cao, Q., You, X., Xu, L., Wang, L., and Chen, Y. (2020). Paqr3 Suppresses the Growth of Non-small Cell Lung Cancer Cells via Modulation of Egfr-Mediated Autophagy. Autophagy 16, 1236–1247. doi:10.1080/15548627.2019.1659654
Cao, X., Cao, Z., Shao, Y., Liu, C., Yan, G., Meng, X., et al. (2021). Analysis of Serum Paraoxonase 1 Using Mass Spectrometry and Lectin Immunoassay in Patients with Alpha-Fetoprotein Negative Hepatocellular Carcinoma. Front. Oncol. 11, 651421. doi:10.3389/fonc.2021.651421
Contessa, J. N., Bhojani, M. S., Freeze, H. H., Ross, B. D., Rehemtulla, A., and Lawrence, T. S. (2010). Molecular Imaging of N-Linked Glycosylation Suggests Glycan Biosynthesis Is a Novel Target for Cancer Therapy. Clin. Cancer Res. 16, 3205–3214. doi:10.1158/1078-0432.CCR-09-3331
Critcher, M., Hassan, A. A., and Huang, M. L. (2022). Seeing the Forest through the Trees: Characterizing the Glycoproteome. Trends Biochem. Sci. S0968, 00048. doi:10.1016/j.tibs.2022.02.007
Dawson, J. P., Bu, Z., and Lemmon, M. A. (2007). Ligand-Induced Structural Transitions in Erbb Receptor Extracellular Domains. Structure 15, 942–954. doi:10.1016/j.str.2007.06.013
Di Patria, L., Annibalini, G., Morrone, A., Ferri, L., Saltarelli, R., Galluzzi, L., et al. (2022). Defective Igf-1 Prohormone N-Glycosylation and Reduced Igf-1 Receptor Signaling Activation in Congenital Disorders of Glycosylation. Cell. Mol. Life Sci. 79, 150. doi:10.1007/s00018-022-04180-x
Dieterich, D. C., Lee, J. J., Link, A. J., Graumann, J., Tirrell, D. A., and Schuman, E. M. (2007). Labeling, Detection and Identification of Newly Synthesized Proteomes with Bioorthogonal Non-canonical Amino-Acid Tagging. Nat. Protoc. 2, 532–540. doi:10.1038/nprot.2007.52
Esmail, S., and Manolson, M. F. (2021). Advances in Understanding N-Glycosylation Structure, Function, and Regulation in Health and Disease. Eur. J. Cel Biol. 100, 151186. doi:10.1016/j.ejcb.2021.151186
Groux-Degroote, S., Foulquier, F., Cavdarli, S., and Delannoy, P. (2021). Reticular And Golgi Glycosylation: Advances And Associated Diseases. Med. Sci. (Paris) 37, 609–617. doi:10.1051/medsci/2021082
Huang, D. W., Sherman, B. T., and Lempicki, R. A. (2009). Systematic and Integrative Analysis of Large Gene Lists Using David Bioinformatics Resources. Nat. Protoc. 4, 44–57. doi:10.1038/nprot.2008.211
Hubert, V., Peschel, A., Langer, B., Gröger, M., Rees, A., and Kain, R. (2016). Lamp-2 Is Required for Incorporating Syntaxin-17 into Autophagosomes and for Their Fusion with Lysosomes. Biol. Open. 5, 1516–1529. doi:10.1242/bio.018648
Ikeda, S., Nah, J., Shirakabe, A., Zhai, P., Oka, S.-i., Sciarretta, S., et al. (2021). Yap Plays a Crucial Role in the Development of Cardiomyopathy in Lysosomal Storage Diseases. J. Clin. Invest. 131, e143173. doi:10.1172/JCI143173
Jiang, P., and Mizushima, N. (2014). Autophagy and Human Diseases. Cell. Res. 24, 69–79. doi:10.1038/cr.2013.161
Karki, R., Rimal, S., and Rieth, M. D. (2021). Predicted N‐terminal N ‐linked Glycosylation Sites May Underlie Membrane Protein Expression Patterns in Saccharomyces cerevisiae. Yeast 38, 497–506. doi:10.1002/yea.3657
Li, J., and Pfeffer, S. R. (2016). Lysosomal Membrane Glycoproteins Bind Cholesterol and Contribute to Lysosomal Cholesterol Export. Elife 5, e21635. doi:10.7554/eLife.21635
Li, T., Dong, Z.-R., Guo, Z.-Y., Wang, C.-H., Zhi, X.-T., Zhou, J.-W., et al. (2015). Mannose-mediated Inhibitory Effects of PA-MSHA on Invasion and Metastasis of Hepatocellular Carcinoma via EGFR/Akt/IκBβ/NF-κB Pathway. Liver IntInt 35, 1416–1429. doi:10.1111/liv.12644
Liu, C., Song, C.-Q., Yuan, Z.-F., Fu, Y., Chi, H., Wang, L.-H., et al. (2014). Pquant Improves Quantitation by Keeping Out Interfering Signals and Evaluating the Accuracy of Calculated Ratios. Anal. Chem. 86, 5286–5294. doi:10.1021/ac404246w
Liu, Y., Zhao, D., Peng, W., Xue, P., Jiang, X., Chen, S., et al. (2021). Atmospheric Pm2.5 Blocking up Autophagic Flux in Huvecs via Inhibiting Sntaxin-17 and Lamp2. Ecotoxicology Environ. Saf. 208, 111450. doi:10.1016/j.ecoenv.2020.111450
Lopez Sambrooks, C., Baro, M., Quijano, A., Narayan, A., Cui, W., Greninger, P., et al. (2018). Oligosaccharyltransferase Inhibition Overcomes Therapeutic Resistance to Egfr Tyrosine Kinase Inhibitors. Cancer Res. 78, 5094–5106. doi:10.1158/0008-5472.CAN-18-0505
Lopez-Sambrooks, C., Shrimal, S., Khodier, C., Flaherty, D. P., Rinis, N., Charest, J. C., et al. (2016). Oligosaccharyltransferase Inhibition Induces Senescence in Rtk-Driven Tumor Cells. Nat. Chem. Biol. 12, 1023–1030. doi:10.1038/nchembio.2194
Lu, H., Cherepanova, N. A., Gilmore, R., Contessa, J. N., and Lehrman, M. A. (2019). Targeting STT3A‐oligosaccharyltransferase with NGI‐1 Causes Herpes Simplex Virus 1 Dysfunction. FASEB j. 33, 6801–6812. doi:10.1096/fj.201802044RR
Ma, Y., McClatchy, D. B., Barkallah, S., Wood, W. W., and Yates, J. R. (2017). Hilaq: A Novel Strategy for Newly Synthesized Protein Quantification. J. Proteome Res. 16, 2213–2220. doi:10.1021/acs.jproteome.7b00005
Ma, Y., McClatchy, D. B., Barkallah, S., Wood, W. W., and Yates, J. R. (2018). Quantitative Analysis of Newly Synthesized Proteins. Nat. Protoc. 13, 1744–1762. doi:10.1038/s41596-018-0012-y
McClatchy, D. B., Ma, Y., Liem, D. A., Ng, D. C. M., Ping, P., and Yates, J. R. (2018). Quantitative Temporal Analysis of Protein Dynamics in Cardiac Remodeling. J. Mol. Cell Cardiol. 121, 163–172. doi:10.1016/j.yjmcc.2018.07.126
Phillips, A. J., Lobl, M. B., Hafeji, Y. A., Safranek, H. R., Mohr, A. M., and Mott, J. L. (2022). Glycosylation of Fgfr4 in Cholangiocarcinoma Regulates Receptor Processing and Cancer Signaling. J. Cell. Biochem. 123, 568–580. doi:10.1002/jcb.30204
Read, A., and Schröder, M. (2021). The Unfolded Protein Response: An Overview. Biology (Basel) 10, 384. doi:10.3390/biology10050384
Rinis, N., Golden, J. E., Marceau, C. D., Carette, J. E., Van Zandt, M. C., Gilmore, R., et al. (2018). Editing N-Glycan Site Occupancy with Small-Molecule Oligosaccharyltransferase Inhibitors. Cel Chem. Biol. 25, 1231–1241. doi:10.1016/j.chembiol.2018.07.005
Rodrigues, J. G., Balmaña, M., Macedo, J. A., Poças, J., Fernandes, Â., de-Freitas-Junior, J. C. M., et al. (2018). Glycosylation in Cancer: Selected Roles in Tumour Progression, Immune Modulation and Metastasis. Cell Immunol. 333, 46–57. doi:10.1016/j.cellimm.2018.03.007
Shah, S. H., Schiapparelli, L. M., Yokota, S., Ma, Y., Xia, X., Shankar, S., et al. (2022). Quantitative Boncat (Qboncat) Allows Identification of Newly Synthesized Proteins after Optic Nerve Injury. J. Neurosci [Online ahead of print]. doi:10.1523/JNEUROSCI.3100-20.2022
Shi, W., Lu, D., Wu, C., Li, M., Ding, Z., Li, Y., et al. (2021). Coibamide a Kills Cancer Cells through Inhibiting Autophagy. Biochem. Biophysical Res. Commun. 547, 52–58. doi:10.1016/j.bbrc.2021.01.112
Shu, J., Dang, L., Zhang, D., Shah, P., Chen, L., Zhang, H., et al. (2019). Dynamic Analysis of Proteomic Alterations in Response to N‐linked Glycosylation Inhibition in a Drug‐resistant Ovarian Carcinoma Cell Line. FEBS. J. 286, 1594–1605. doi:10.1111/febs.14811
Song, X., Zhou, Z., Li, H., Xue, Y., Lu, X., Bahar, I., et al. (2020). Pharmacologic Suppression of B7-H4 Glycosylation Restores Antitumor Immunity in Immune-Cold Breast Cancers. Cancer Discov. 10, 1872–1893. doi:10.1158/2159-8290.CD-20-0402
Sovolyova, N., Healy, S., Samali, A., and Logue, S. E. (2014). Stressed to Death - Mechanisms of Er Stress-Induced Cell Death. Biol. Chem. 395, 1–13. doi:10.1515/hsz-2013-0174
Sun, S., Shah, P., Eshghi, S. T., Yang, W., Trikannad, N., Yang, S., et al. (2016). Comprehensive Analysis of Protein Glycosylation by Solid-phase Extraction of N-Linked Glycans and Glycosite-Containing Peptides. Nat. Biotechnol. 34, 84–88. doi:10.1038/nbt.3403
Sun, X., Zhan, M., Sun, X., Liu, W., and Meng, X. (2021). C1GALT1 in Health and Disease (Review). Oncol. Lett. 22, 589. doi:10.3892/ol.2021.12850
Tian, Y., Zhu, Q., Sun, Z., Geng, D., Lin, B., Su, X., et al. (2021). One‐Step Enzymatic Labeling Reveals a Critical Role of O‐GlcNAcylation in Cell‐Cycle Progression and DNA Damage Response. Angew. Chem. Int. Ed. 60, 26128–26135. doi:10.1002/anie.202110053
van Bergen, W., Heck, A. J. R., and Baggelaar, M. P. (2022). Recent Advancements in Mass Spectrometry-Based Tools to Investigate Newly Synthesized Proteins. Curr. Opin. Chem. Biol. 66, 102074. doi:10.1016/j.cbpa.2021.07.001
Vargas-Diaz, D., and Altelaar, M. (2022). Automated High-Throughput Method for the Fast, Robust, and Reproducible Enrichment of Newly Synthesized Proteins. J. Proteome Res. 21, 189–199. doi:10.1021/acs.jproteome.1c00743
Wahl, D. R., and Lawrence, T. S. (2019). No Sugar Added: A New Strategy to Inhibit Glioblastoma Receptor Tyrosine Kinases. Clin. Cancer Res. 25, 455–456. doi:10.1158/1078-0432.CCR-18-2113
Wang, J., Zhang, J., Lee, Y.-M., Koh, P.-L., Ng, S., Bao, F., et al. (2016). Quantitative Chemical Proteomics Profiling of De Novo Protein Synthesis during Starvation-Mediated Autophagy. Autophagy 12, 1931–1944. doi:10.1080/15548627.2016.1196317
Wang, X., Wei, Z., Cheng, B., Li, J., He, Y., Lan, T., et al. (2022). Endoplasmic Reticulum Stress Promotes Hbv Production by Enhancing Use of the Autophagosome/Multivesicular Body Axis. Hepatology 75, 438–454. doi:10.1002/hep.32178
Ward, S., O'Sullivan, J. M., and O'Donnell, J. S. (2021). The Biological Significance of Von Willebrand Factor O-Linked Glycosylation. Semin. Thromb. Hemost. 47, 855–861. doi:10.1055/s-0041-1726373
Xiao, H., Smeekens, J. M., and Wu, R. (2016). Quantification of Tunicamycin-Induced Protein Expression and N-Glycosylation Changes in Yeast. Analyst 141, 3737–3745. doi:10.1039/c6an00144k
Xu, X.-X., Li, S.-T., Wang, N., Kitajima, T., Yoko-o, T., Fujita, M., et al. (2018). Structural and Functional Analysis of Alg1 Beta-1,4 Mannosyltransferase Reveals the Physiological Importance of its Membrane Topology. Glycobiology 28, 741–753. doi:10.1093/glycob/cwy060
Zeng, W.-F., Cao, W.-Q., Liu, M.-Q., He, S.-M., and Yang, P.-Y. (2021). Precise, Fast and Comprehensive Analysis of Intact Glycopeptides and Modified Glycans with Pglyco3. Nat. Methods 18, 1515–1523. doi:10.1038/s41592-021-01306-0
Zhang, S., Cao, X., Liu, C., Li, W., Zeng, W., Li, B., et al. (2019). N-glycopeptide Signatures of Iga2 in Serum from Patients with Hepatitis B Virus-Related Liver Diseases. Mol. Cell Proteomics 18, 2262–2272. doi:10.1074/mcp.RA119.001722
Zhu, S., Wan, W., Zhang, Y., Shang, W., Pan, X., Zhang, L.-K., et al. (2019). Comprehensive Interactome Analysis Reveals that Stt3b Is Required for N-Glycosylation of Lassa Virus Glycoprotein. J. Virol. 93. doi:10.1128/JVI.01443-19
Zielinska, D. F., Gnad, F., Wiśniewski, J. R., and Mann, M. (2010). Precision Mapping of an In Vivo N-Glycoproteome Reveals Rigid Topological and Sequence Constraints. Cell 141, 897–907. doi:10.1016/j.cell.2010.04.012
Glossary
OST Oligosaccharyltransferase
UPR Unfolded protein response
PAGE polyacrylamide gel electrophoresis
EGFR Epidermal growth factor receptor
F Fucose
BONCAT Bioorthogonal non-canonical amino-acid tagging
LAMP2 Lysosome-associated membrane protein-2
DMEM Dulbecco’s modified Eagle’s medium
FBS Fetal bovine serum
N N-acetylhexosamine
NGI-1 N-linked glycosylation inhibitor-1
AHA Azidohomoalanine
DTT Dithiothreitol
H Hexose
HPLC High-pressure liquid chromatography
GO Gene ontology
CC Cellular component
MF Molecular function
BP Biological process
S Sialic acid
SDS Sodium dodecyl sulfate
siRNA Small interfering RNA
CCK8 Cell Counting Kit-8
FC Fold change
KEGG Kyoto Encyclopedia of Genes and Genomes
HCC Hepatocellular carcinoma
IAA Iodoacetamide
LC-MS/MS Liquid chromatography-tandem mass spectrometry
PBS Phosphate buffered saline
ER Endoplasmic reticulum
HILIC Hydrophilic interaction liquid chromatography.
Keywords: hepatocellular carcinoma, nascent proteome, glycosylation, glycoproteome, LAMP2
Citation: Cao X, Meng P, Shao Y, Yan G, Yao J, Zhou X, Liu C, Zhang L, Shu H and Lu H (2022) Nascent Glycoproteome Reveals That N-Linked Glycosylation Inhibitor-1 Suppresses Expression of Glycosylated Lysosome-Associated Membrane Protein-2. Front. Mol. Biosci. 9:899192. doi: 10.3389/fmolb.2022.899192
Received: 18 March 2022; Accepted: 12 April 2022;
Published: 27 April 2022.
Edited by:
Wenjuan Zeng, Sichuan University, ChinaReviewed by:
Qiong Wang, Johns Hopkins University, United StatesDing Liu, Moderna Inc, United States
Zhixin Tian, Tongji University, China
Yong Zhang, Sichuan University, China
Copyright © 2022 Cao, Meng, Shao, Yan, Yao, Zhou, Liu, Zhang, Shu and Lu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hong Shu, c2h1aG9uZ0BneG11LmVkdS5jbg==; Haojie Lu, bHVoYW9qaWVAZnVkYW4uZWR1LmNu
†These authors have contributed equally to this work
 Xinyi Cao
Xinyi Cao Peiyi Meng2†
Peiyi Meng2† Guoquan Yan
Guoquan Yan Hong Shu
Hong Shu Haojie Lu
Haojie Lu