- 1School of Behavioural and Health Sciences, Faculty of Health Sciences, Australian Catholic University, Banyo, QLD, Australia
- 2Mater Research Institute, University of Queensland, Brisbane, QLD, Australia
Sulfate is an important nutrient that modulates a diverse range of molecular and cellular functions in mammalian physiology. Over the past 2 decades, animal studies have linked numerous sulfate maintenance genes with neurological phenotypes, including seizures, impaired neurodevelopment, and behavioral abnormalities. Despite sulfation pathways being highly conserved between humans and animals, less than one third of all known sulfate maintenance genes are clinically reportable. In this review, we curated the temporal and spatial expression of 91 sulfate maintenance genes in human fetal brain from 4 to 17 weeks post conception using the online Human Developmental Biology Resource Expression. In addition, we performed a systematic search of PubMed and Embase, identifying those sulfate maintenance genes linked to atypical neurological phenotypes in humans and animals. Those findings, together with a search of the Online Mendelian Inheritance in Man database, identified a total of 18 candidate neurological dysfunction genes that are not yet considered in clinical settings. Collectively, this article provides an overview of sulfate biology genes to inform future investigations of perturbed sulfate homeostasis associated with neurological conditions.
Introduction
Sulfate is essential for healthy growth and development (Dawson, 2011). Supplied from the diet and catabolism of sulfur-containing amino acids, intracellular sulfate is transformed into 3′-phosphoadenosine 5′-phosphosulfate (PAPS) which is the universal sulfate donor for sulfate conjugation (sulfonation) to a wide range of endogenous and exogenous molecules via sulfotransferases (Mulder and Jakoby, 1990). Sulfonation biotransforms the physiological properties of molecules, including: (i) inactivation of steroids, thyroid hormone and neurotransmitters (Darras et al., 1999; Richard et al., 2001; Dawson, 2012); (ii) detoxification of xenobiotics and certain pharmaceutical drugs (McCarver and Hines, 2002); and (iii) maintenance of tissue structure and function via the sulfate content of glycosaminoglycans (Yamaguchi, 2001). Sulfatases remove sulfate from substrates to maintain the required balance of sulfonated to unconjugated substrates (Hanson et al., 2004). Disturbances in any of these sulfation pathways has the potential for adverse physiological consequences.
In recent years, interest in human sulfate biology has expanded following animal studies that show adverse phenotypes linked to sulfate biology genes (Langford et al., 2017). Neurological dysfunction is one of the most predominant phenotypes linked to disturbances of sulfate biology. This is not surprising when considering that sulfonation biotransforms numerous target molecules in the brain, including inactivation of thyroid hormone, metabolism of neurotransmitters (e.g., serotonin, noradrenaline and dopamine) and modulation of neurosteroid actions on GABAA, N-methyl-D-aspartate (NMDA) glutaminergic and ơ-opioid receptors (Rivett et al., 1982; Kríz et al., 2008). Sulfonation of proteoglycans (e.g., heparan sulfate) and cerebroside sulfate also plays an important role in maintaining the structure and function of brain tissue (Schwartz and Domowicz, 2018). Animal studies have also shown that hyposulfataemia leads to reduced sulfonation capacity and abnormal behavioral phenotypes, including impaired memory, increased anxiety and seizures in mice (Dawson et al., 2003; Dawson et al., 2004; Dawson et al., 2005). More recent studies have shown that reduced transfer of sulfate across the blood-brain barrier of neonatal mice leads to: (i) decreased heparan sulfate levels in the subventricular zone (SVZ) and rostral migratory stream; (ii) impairment in perineuronal net formation (which contains sulfonated proteoglycans) in the hippocampus and somatosensory cortex; (iii) increased neural stem cell proliferation and decreased neuronal maturation in the SVZ; (iv) seizures; and (v) impaired behavioral phenotypes, including impaired long-term spatial memory and defects in social interaction and social memory (Zhang et al., 2020). Despite the diverse roles for sulfate during neurodevelopment, most human sulfate biology genes have not yet been considered in clinical genetics.
To date, 91 genes are known to contribute to maintaining sulfate homeostasis, including those encoding: sulfate transporters; PAPS synthetases and transporters; enzymes in the pathway of sulfate generation; cytosolic and membrane-bound sulfotransferases; and sulfatases (Langford et al., 2017). However, only 24 of these genes are currently considered in clinical genetics, with 16 of those genes linked to neurological dysfunction (Table 1). Animal studies have shown additional sulfate biology genes are linked to adverse neurological phenotypes (Langford et al., 2017), suggesting that the number of clinically reportable genes is currently underestimated. This warrants further investigation into the role of sulfate biology genes in human neurophysiology.

TABLE 1. Sulfate biology genes that are either clinically reportable or not yet captured in clinical genetics.
In this study, we curated the temporal and spatial mRNA expression patterns of all known sulfate biology genes in the developing human fetal brain between 4 and 17 weeks gestation, corresponding to a critical time for nervous system formation in utero. We show moderate to abundant mRNA expression levels of 62 genes, suggesting a critical role of sulfate in early neurodevelopment. Additionally, we collated 18 sulfate biology genes implicated in atypical neurological function from the animal and human research literature which are currently not captured in clinical resources. Overall, these findings provide data for future investigations of sulfate biology genes associated with neurological dysfunction.
Sulfate Biology Gene Expression in Human Fetal Brain
We obtained data of gene expression in the human fetal brain from the online Human Developmental Biology Resource (HDBR) Expression (Lindsay et al., 2016) (https://www.hdbr.org/) from 6 November 2021 to 12 January 2022. The HDBR is one of the rare human fetal tissue databases providing gene expression under 8 weeks post-conception compared to other databases (Johnson et al., 2009; Sunkin et al., 2013), which is essential for getting a better understanding of brain development since neurulation starts after 3 weeks post conception (O'Rahilly and Müller, 1994). Taking in account that brain development is dynamic, HDBR reports anatomical regionalisation gene expression results, which is also essential for mapping the differential pattern of gene expression related to selected brain regions compared to using full brain tissues (Ono et al., 2017). Data were available for four brain regions (cerebrum, brainstem, diencephalon and cerebellum) from 4 to 17 weeks post-conception. The gene expression levels for each tissue sample were averaged for each set of technical replicates, and then quantile normalized within each set of biological replicates using limma. Finally, they are averaged for all biological replicates. Specifically, we used transcripts per million (TPM) mapped reads-normalized RNA-Seq data to plot results for each gene. Of the 91 sulfate biology genes, 16 were abundantly expressed (>100 TPM) in at least one brain region at one gestational time point, 46 genes had moderate (>10 to 100 TPM) mRNA expression, and 15 genes had lower (≥0.1 to 10 TPM) mRNA expression levels (Figures 1–4). Fourteen genes (SLC13A1, SLC26A3, SLC26A9, SULT1A4, SULT1C3, SULT2A1, SULT2B1, SULT6B1, CHST4, CHST13, GAL3ST2, HS3ST6, ARSH and ARSJ) had undetectable or negligible (<0.1 TPM) mRNA expression levels (data not shown). The limitations of these data are that mRNA levels may: (i) not be representative of encoded protein level; (ii) not be predictive of functional impact when abnormal, for example altered expression of highly-expressed genes does not necessarily result in a more severe neurodevelopmental phenotype than altered expression of lowly-expressed genes; (iii) be redundant as other family member genes may compensate for altered expression level of a particular gene; (iv) not identify any sex-specific effects as the data are derived from pooled samples, which warrants future investigations to compare sulfate biology gene expression between male and female samples. Furthermore, our focus on genes expressed in the brain doesn’t take into account any of the sulfate biology genes expressed in other tissues that may influence neurodevelopment, such as the thyroid or adrenal gland or testis that could theoretically influence neurodevelopment through endocrine effects. Nonetheless, the temporal and spatial mRNA expression profiles of sulfate biology genes reported in the present study provide information for the next phase of research into understanding the regulation of sulfate homeostasis in the developing brain.
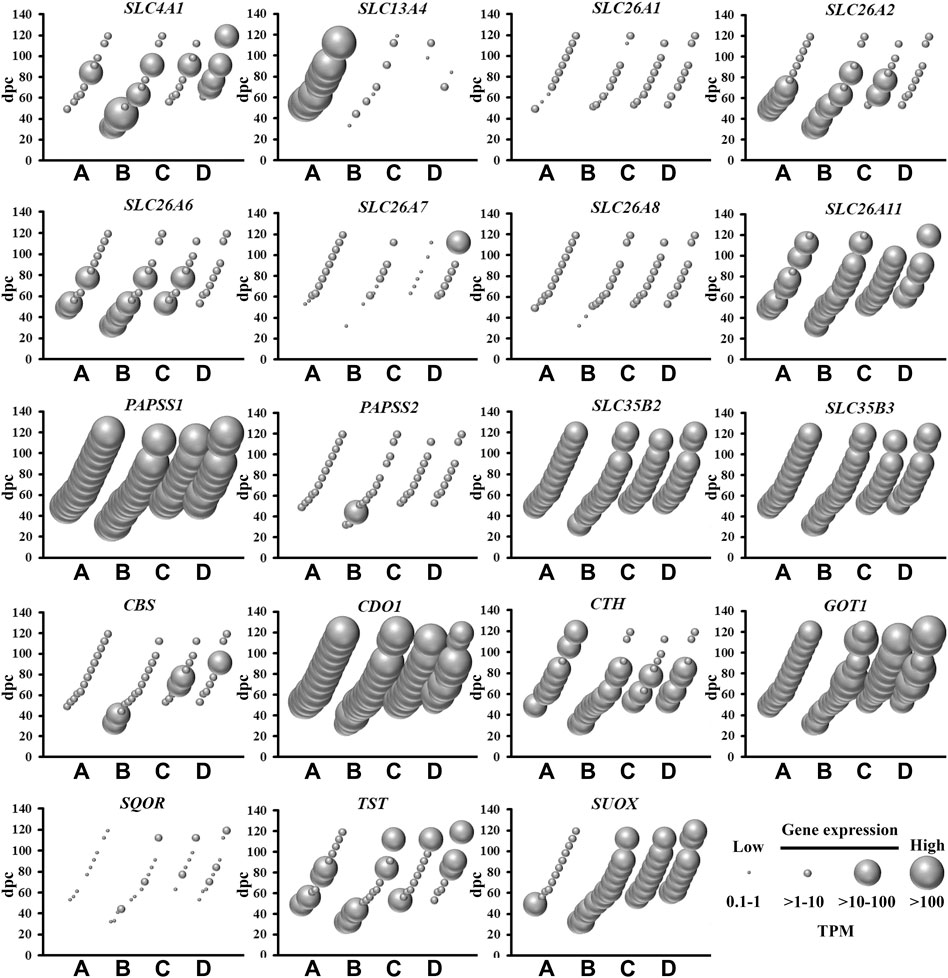
FIGURE 1. Spatial and temporal mRNA expression of sulfate transporters, PAPS synthetases and transporters, and enzymes in the pathways of sulfate generation in fetal brain regions (A) cerebrum, (B) brainstem, (C) diencephalon, and (D) cerebellum. dpc, days post-conception. TPM, transcripts per million.
Genes Involved in Sulfate and PAPS Supply
Amongst the most abundantly expressed sulfate biology genes at all gestational time points are those involved in: (i) transporting sulfate from circulation to brain across the blood brain barrier (SLC13A4) (Zhang et al., 2020); (ii) the major (CDO1) and minor (CTH) biochemical pathways of sulfate generation (Dawson et al., 2020); and (iii) synthesis (PAPSS1) and transportation (SLC35B2 and SLC35B3) of PAPS (Langford et al., 2017) (Figure 1). SLC13A4 mRNA expression was specifically expressed in the cerebrum, indicating that sulfate transport from circulation likely contributes an important role in sulfate supply in early neurodevelopment. Of great interest are the data showing abundant mRNA expression of those genes involved in pathways of sulfate generation from the sulfur-containing amino acid cysteine. Previous studies suggested that the developing fetus has negligible capacity to generate sulfate, based on low CDO1 and CTH mRNA expression levels in the liver (Gaull et al., 1972; Loriette and Chatagner, 1978). However, a more recent animal study showed the expression of Cdo1 mRNA in the developing mouse fetus from mid-gestation (Rakoczy et al., 2015). The present study shows abundant expression of CDO1, suggesting that the fetal brain most likely generates sulfate, in addition to obtaining sulfate from circulation via SLC13A4. The present study also shows that PAPSS1 is the most abundantly expressed sulfate biology gene in all four selected regions of the fetal brain, suggesting that this gene encodes for the predominant PAPS synthetase in fetal brain tissue, rather than PAPSS2 which is expressed in cartilage and linked to developmental dwarfism disorders (Bownass et al., 2019). In addition, the findings of abundant SLC35B2 and SLC35B3 mRNA expression suggest that PAPS is actively transported in the golgi of fetal brain cells for sulfonation reactions via membrane-bound sulfotransferases. Collectively, these findings of abundant mRNA expression of genes involved in sulfate supply and generation, as well as generation and transport of PAPS, suggest that sulfate has an important physiological role in early fetal neurodevelopment.
Sulfotransferase Genes
There are two classes of sulfotransferases based on their protein sub-cellular localization: (i) cytosolic sulfotransferases which sulfonate neurotransmitters, bile acids, xenobiotics and steroids; and (ii) membrane-bound sulfotransferases that sulfonate proteoglycan and lipid molecules (Gamage et al., 2006). The present study shows abundant mRNA levels of four cytosolic (Figure 2) and 22 membrane-bound (Figure 3) sulfotransferases. SULT4A1 is the most abundantly expressed cytosolic sulfotransferase mRNA in all four fetal brain regions. Whilst the substrate of SULT4A1-mediated sulfonation has not yet been identified, a recent study suggests it plays a role in the regulation of neuronal morphology and synaptic activity (Culotta et al., 2020). Abundant mRNA levels of SULT1E1 and SULT1C4, suggest that these two sulfotransferases, that sulfonate estrogenic compounds (Dawson, 2012), may regulate hormonal signalling pathways during fetal neurodevelopment. Interestingly, SULT1E1 mRNA is mainly expressed in cerebrum, which is a critical region in hormonal regulation due to its development into the thalamic and hypothalamic structures modulating endocrine body system. SULT1A1 mRNA expression was abundant in the cerebrum, with lower levels detected in the other three brain regions. SULT1A1 sulfonates phenolic compounds, and while it has a major role of Phase II metabolism in the liver (Pacifici, 2005), it is also proposed to contribute to the detoxification and clearance of drugs and toxins in the brain (Salman et al., 2009). It is intriguing to see a single burst of abundant SULT1B1, iodothyronine sulfotransferase (Stanley et al., 2005), mRNA expression at 10 weeks post-conception. This time point coincides with the early stage of neuronal migration, a neurodevelopmental process that relies on the action of thyroid hormone which is regulated by SULT1B1-mediated sulfonation (Wang et al., 1998; Morreale de Escobar et al., 2004). In addition to the cytosolic sulfotransferases, the majority of all known membrane-bound sulfotransferases were abundantly expressed (Figure 3), including: carbohydrate sulfotransferases CHST1, CHST2, CHST3, CHST8, CHST10, CHST11, CHST14, and CHST15; galactose 3-O-sulfotransferases GAL3ST3 and GAL3ST4; heparan sulfate 2-O-sulfotransferase HS2ST1; heparan sulfate 3-O-sulfotransferases HS3ST2, HS3ST4, and HS3ST5; heparan sulfate 6-O-sulfotransferases HS6ST1, HS6ST2 and HS6ST3; heparan N-deacetylase/N-sulfotransferases NDST1 and NDST; tyrosylprotein sulfotransferase TPST1; and uronyl 2-sulfotransferase UST. Overall, these findings suggest the sulfonation of numerous substrates in the developing fetal brain.
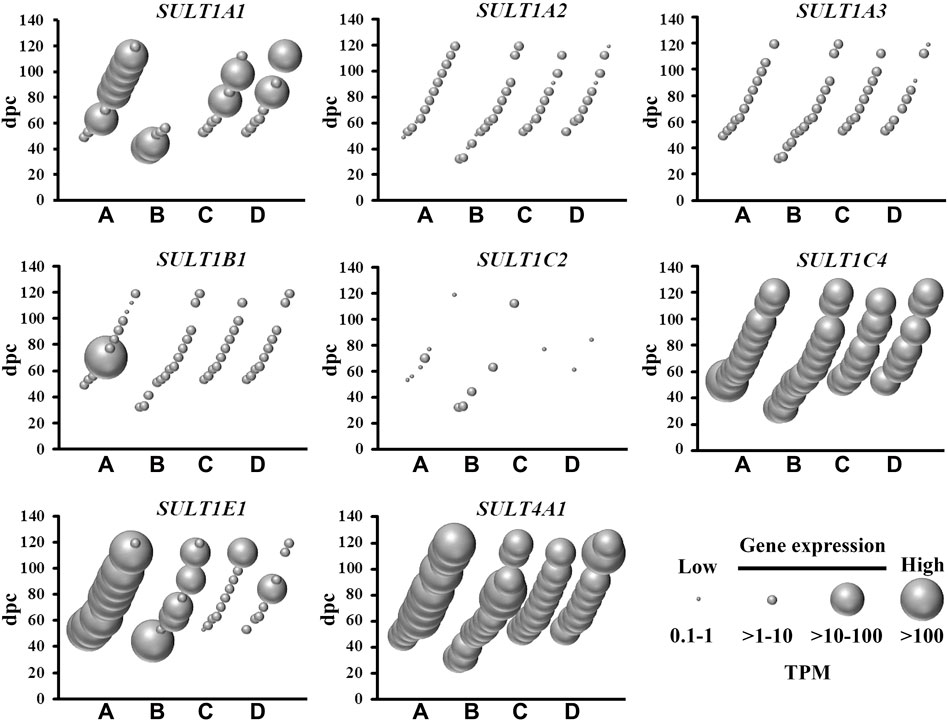
FIGURE 2. Spatial and temporal mRNA expression of cytosolic sulfotransferases in fetal brain regions (A) cerebrum, (B) brainstem, (C) diencephalon, and (D) cerebellum. dpc, days post-conception. TPM, transcripts per million.
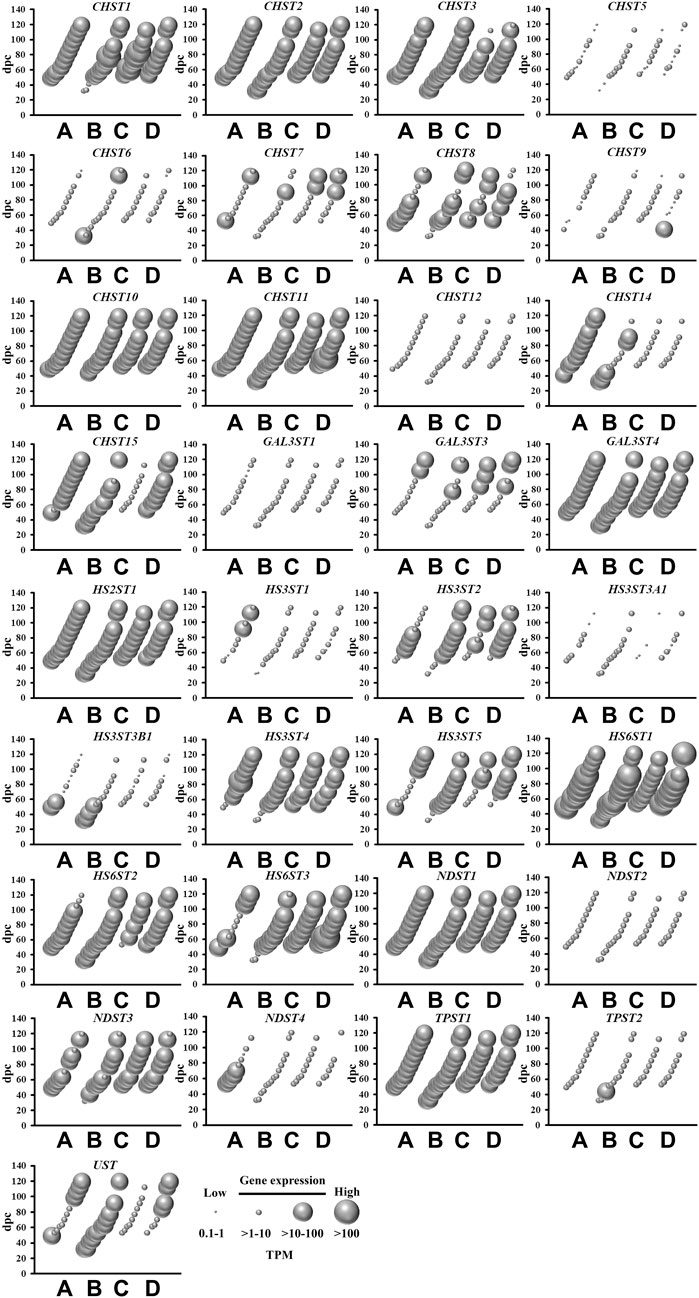
FIGURE 3. Spatial and temporal mRNA expression of membrane-bound sulfotransferases in fetal brain regions (A) cerebrum, (B) brainstem, (C) diencephalon, and (D) cerebellum. dpc, days post-conception. TPM, transcripts per million.
Sulfatase Genes
The removal of sulfate from lipids, glycosaminoglycans and steroids is mediated by sulfatases (Hanson et al., 2004), of which nine are abundantly expressed in the fetal brain (Figure 4). This is relevant to sulfatases that target a range of substrates in the central nervous system, including sulfoglycolipids (ARSA), proteoglycans (ARSB, ARSK, GNS, IDS, SULF1 and SULF2) and steroids (STS). With the exception of STS, all of these sulfatase genes were expressed in all four brain regions over the 4–17 weeks gestational period. STS mRNA level is low in cerebrum when compared to brainstem, diencephalon and cerebellum, suggesting that steroid activity is spatially confined to the latter three brain regions during early neurodevelopment. SULF1 and SULF2 mRNA, highly expressed in developing fetal brain, lead to the formation of enzyme removing 6-O-sulfate groups of proteoglycans and modulating morphogen Shh gradient during embryonic development. By modifying the Shh gradient in the embryo, the regulation of motorneuron to oligodendrocyte precursor cell fate change is significantly affected in the developing ventral spinal cord (Jiang et al., 2017). The present study also shows abundant sulfatase modifying factor 2 (SUMF2) mRNA levels in comparison to low SUMF1 mRNA levels, indicating that SUMF2 is the major sulfatase regulating factor in early fetal brain tissue. There were short bursts of increased ARSI, GALNS and SGSH mRNA expression that were spatially enriched, suggesting a localized and temporary requirement for these glycosaminoglycans metabolizing sulfatases. Taken together, the abundant mRNA expression of numerous sulfatases indicates an active process of regulating sulfate content in the developing fetal brain.
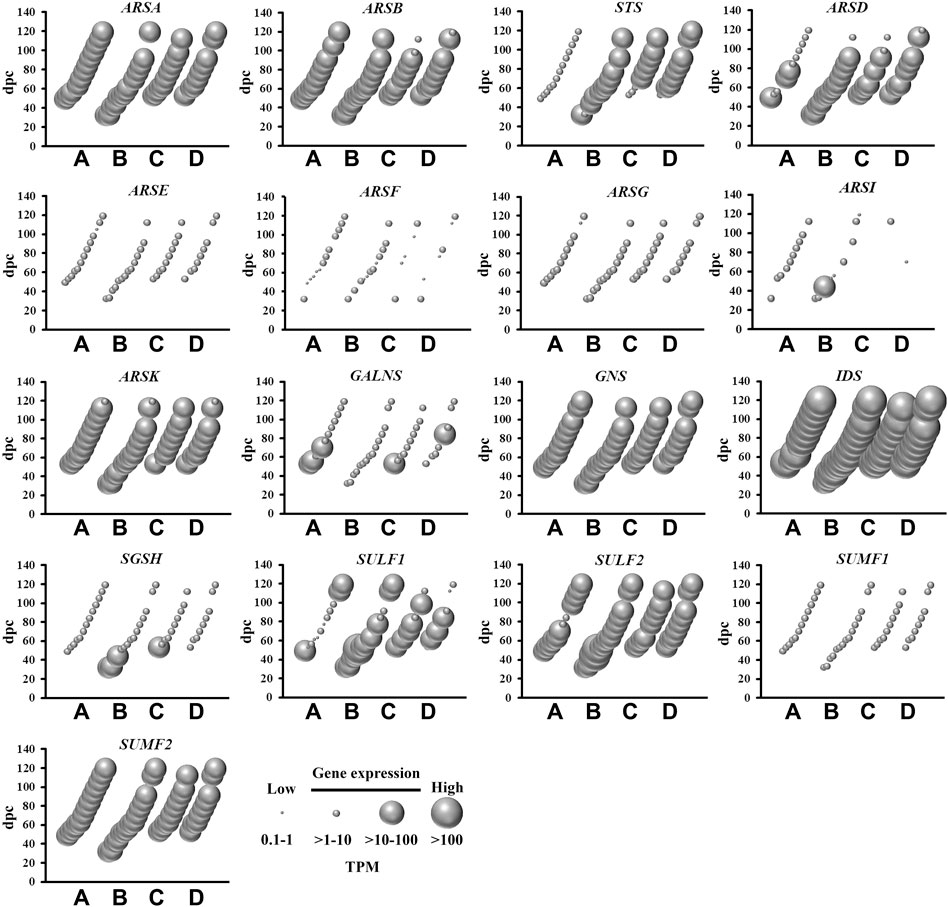
FIGURE 4. Spatial and temporal mRNA expression of sulfatases and sulfatase modifying factors in fetal brain regions (A) cerebrum, (B) brainstem, (C) diencephalon, and (D) cerebellum. dpc, days post-conception. TPM, transcripts per million.
Candidate Neurological Dysfunction Sulfate Biology Genes
Despite the mRNA expression of 62 sulfate biology genes in the fetal brain (Figures 1–4), only 12 of these genes are considered for neurological dysfunction in routine clinical genetics (Table 1). As an approach to inform clinical geneticists and potentially expand the list of clinically reportable genes, we undertook a systematic online search (10–23 September 2021). We used PubMed and Medline to identify sulfate biology genes reported to be associated with atypical neurological phenotypes in humans using the search terms: (“brain” or “cerebellum” or “cerebral” or “neurodevelop”) and (“gene” or “dna” or “gene expression regulation” or “gene expression” or “gene genetic regulation”) and (“sulphate” or “sulfate” or “SO4”). We included articles that associated any of the sulfate biology genes with neurological conditions. All references used in this study were peer-reviewed research articles published in English between January 2001 and September 2021. In addition, we used OMIM (https://www.ncbi.nlm.nih.gov/omim/?term=) to expand our search prior to 2001 and to include any research articles not captured in our literature search. Overall, our research literature and OMIM searches identified 42 research articles that report the involvement of sulfate biology genes in neurological dysfunction.
From the curated list of research articles, we identified 18 candidate neurological dysfunction genes that are not yet clinically reportable (Table 2). Ten of these genes were reported from animal studies (SLC13A1, SLC13A4, SLC35B2, CDO1, CHST12, CHST15, NDST2, NDST3, SULF2 and UST), 2 from human and animal studies (ARSG and SULF1) and 6 from human research studies (SQOR, TST, SULT4A1, CHST10, HS6ST2 and ARSK). Thirteen of these genes are abundantly expressed in the human brain between 4 and 17 weeks gestation, whereas SQOR, CHST12, NDST2 and ARSG are expressed at low level (Figures 1–4) and SLC13A1 mRNA level is negligible (data not shown). The SLC13A1 gene is primarily expressed in the proximal tubule of the kidney where it mediates sulfate reabsorption (Ullrich and Murer, 1982; Lee et al., 2005). Disruption of human SLC13A1 and mouse Slc13a1 causes renal sulfate wasting and hyposulfataemia, which is proposed to reduce sulfate supply to the brain (Dawson et al., 2003; Bowling et al., 2012). Sulfide:quinone oxidoreductase (SQOR) is an enzyme in the minor pathway of sulfate generation (Dawson, 2013) and its loss leads to excess hydrogen sulfide levels that cause encephalopathy and Leigh disease (Friederich et al., 2020). Individuals with SQOR mutations first present with coma in early childhood (Friederich et al., 2020), suggesting a physiological role for SQOR in the brain beyond the 17-week gestational end point used to assess gene expression in the present study. Increasing CHST12, NDST2 and ARSG mRNA levels in brain at developmental ages beyond 17 weeks gestation (Humphries et al., 1998; Hiraoka et al., 2000; Khateb et al., 2018), also explains their low mRNA levels presented in this study. Nonetheless, the 18 genes listed in Table 2, warrant further research to determine whether these candidate neurological dysfunction genes are to be considered in clinical genetics.
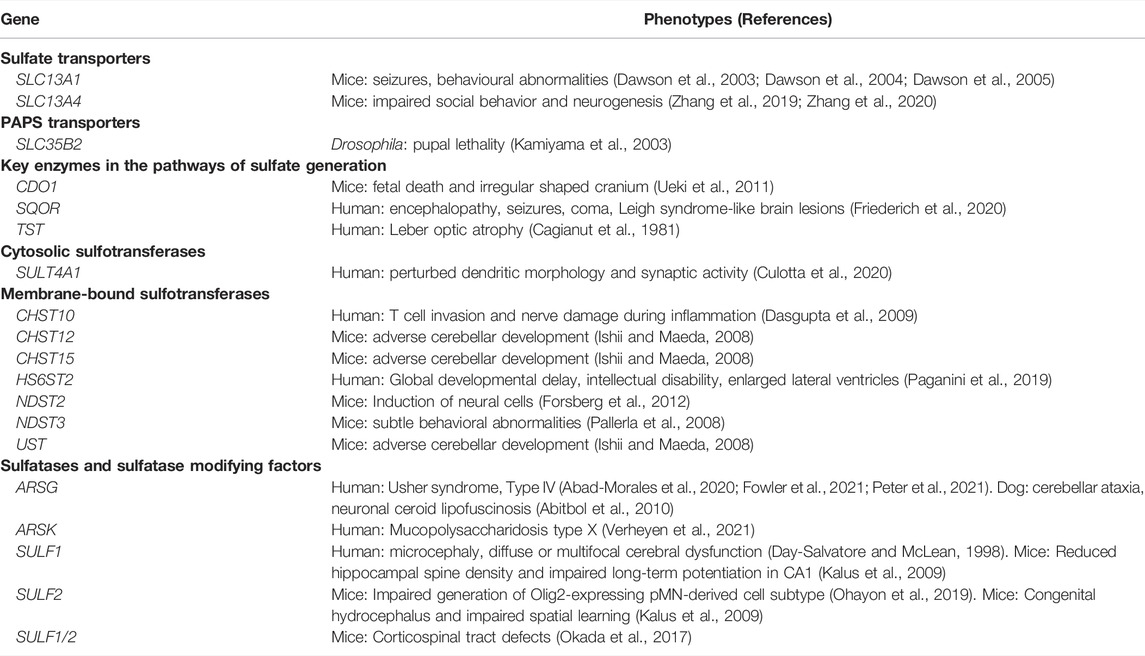
TABLE 2. Neurological phenotypes reported for sulfate biology genes that are not yet captured in clinical genetics.
Summary
In this study, we show the moderate to abundant mRNA expression of 62 sulfate biology genes in the human fetal brain between 4 and 17 weeks post-conception, a critical period in nervous system formation. This relatively large number of genes highlights the physiological relevance of sulfate in neurodevelopment and is a testament to the genetic complexity of maintaining sulfate homeostasis. However, sulfate biology is underappreciated in clinical settings, with only 12 genes routinely considered for certain neurological conditions. Our finding of an additional 18 genes reported in the research literature with neurological dysfunction, warrants future studies to validate the pathogenetics of these genes in human brain. Expanding the list of clinically reportable sulfate biology genes has the potential for improved genetic counselling, and potentially for developing therapeutic approaches towards clinical treatments. Overall, this study curated a list of sulfate biology genes involved in early brain development that provide information for future research of the physiological roles and regulation of sulfate homeostasis in human brain.
Author Contributions
This study was designed by TC, FEF and PAD. Online searches were conducted by TC and PAD and all authors contributed to the preparation of the manuscript and approved the final version.
Funding
This study was funded by the Mater Foundation. PAD is supported by a Mater Foundation Principal Research Fellowship.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We acknowledge the administrative and IT support of Mater Research Ltd. for this research that was carried out in part at the Translational Research Institute (TRI), which is supported by a grant from the Australian Government.
References
Abad-Morales, V., Navarro, R., Burés-Jelstrup, A., and Pomares, E. (2020). Identification of a Novel Homozygous ARSG Mutation as the Second Cause of Usher Syndrome Type 4. Am. J. Ophthalmol. Case Rep. 19, 100736. doi:10.1016/j.ajoc.2020.100736
Abitbol, M., Thibaud, J.-L., Olby, N. J., Hitte, C., Puech, J.-P., Maurer, M., et al. (2010). A Canine Arylsulfatase G ( ARSG ) Mutation Leading to a Sulfatase Deficiency Is Associated with Neuronal Ceroid Lipofuscinosis. Proc. Natl. Acad. Sci. U.S.A. 107 (33), 14775–14780. doi:10.1073/pnas.0914206107
Bowling, F. G., Heussler, H. S., McWhinney, A., and Dawson, P. A. (2012). Plasma and Urinary Sulfate Determination in a Cohort with Autism. Biochem. Genet. 51 (1-2), 147–153. doi:10.1007/s10528-012-9550-0
Bownass, L., Abbs, S., Armstrong, R., Baujat, G., Behzadi, G., Berentsen, R. D., et al. (2019). PAPSS2 ‐related Brachyolmia: Clinical and Radiological Phenotype in 18 New Cases. Am. J. Med. Genet. 179 (9), 1884–1894. doi:10.1002/ajmg.a.61282
Cagianut, B., Rhyner, K., Furrer, W., and Schnebli, H. P. (1981). Thiosulphate-sulphur Transferase (Rhodanese) Deficiency in Leber's Hereditary Optic Atrophy. The Lancet 318 (8253), 981–982. doi:10.1016/s0140-6736(81)91171-5
Culotta, L., Scalmani, P., Vinci, E., Terragni, B., Sessa, A., Broccoli, V., et al. (2020). SULT4A1 Modulates Synaptic Development and Function by Promoting the Formation of PSD-95/NMDAR Complex. J. Neurosci. 40 (37), 7013–7026. doi:10.1523/jneurosci.2194-19.2020
Darras, V. M., Hume, R., and Visser, T. J. (1999). Regulation of Thyroid Hormone Metabolism during Fetal Development. Mol. Cell. Endocrinol. 151 (1-2), 37–47. doi:10.1016/s0303-7207(99)00088-x
Dasgupta, S., Silva, J., Wang, G., and Yu, R. K. (2009). Sulfoglucuronosyl Paragloboside Is a Ligand for T Cell Adhesion: Regulation of Sulfoglucuronosyl Paragloboside Expression via Nuclear Factor κB Signaling. J. Neurosci. Res. 87 (16), 3591–3599. doi:10.1002/jnr.22153
Dawson, P. A., Beck, L., and Markovich, D. (2003). Hyposulfatemia, Growth Retardation, Reduced Fertility, and Seizures in Mice Lacking a Functional NaS I -1 Gene. Proc. Natl. Acad. Sci. U.S.A. 100 (23), 13704–13709. doi:10.1073/pnas.2231298100
Dawson, P. A. (2013). Role of Sulphate in Development. Reproduction 146 (3), R81–R89. doi:10.1530/REP-13-0056
Dawson, P. A., Steane, S. E., and Markovich, D. (2004). Behavioural Abnormalities of the Hyposulphataemic Nas1 Knock-Out Mouse. Behav. Brain Res. 154 (2), 457–463. doi:10.1016/j.bbr.2004.03.013
Dawson, P. A., Steane, S. E., and Markovich, D. (2005). Impaired Memory and Olfactory Performance in NaSi-1 Sulphate Transporter Deficient Mice. Behav. Brain Res. 159, 15–20. doi:10.1016/j.bbr.2004.09.020
Dawson, P. A. (2011). Sulfate in Fetal Development. Semin. Cell Developmental Biol. 22 (6), 653–659. doi:10.1016/j.semcdb.2011.03.004
Dawson, P. A. (2012). “The Biological Roles of Steroid Sulfonation,” in Steroids - from Physiology to Clinical Medicine. Editor S. M. Ostojic (Rijeka: Intech), 45–64.
Dawson, P. A., Weerasekera, S. J., Atcheson, R. J., Twomey, S. A., and Simmons, D. G. (2020). Molecular Analysis of the Human Placental Cysteine Dioxygenase Type 1 Gene. Mol. Genet. Metab. Rep. 22, 100568. doi:10.1016/j.ymgmr.2020.100568
Day-Salvatore, D., and McLean, D. (1998). Blepharophimosis, Hypoplastic Radius, Hypoplastic Left Heart, Telecanthus, Hydronephrosis, Fused Metacarpals, and ?prehensile? Halluces: A New Syndrome? Am. J. Med. Genet. 80 (4), 309–313. doi:10.1002/(sici)1096-8628(19981204)80:4<309::aid-ajmg2>3.0.co;2-j
Forsberg, M., Holmborn, K., Kundu, S., Dagälv, A., Kjellén, L., and Forsberg-Nilsson, K. (2012). Undersulfation of Heparan Sulfate Restricts Differentiation Potential of Mouse Embryonic Stem Cells. J. Biol. Chem. 287 (14), 10853–10862. doi:10.1074/jbc.M111.337030
Fowler, N. H., El-Rashedy, M. I., Chishti, E. A., Vander Kooi, C. W., and Maldonado, R. S. (2021). Multimodal Imaging and Genetic Findings in a Case of ARSG-Related Atypical Usher Syndrome. Ophthalmic Genet. 42 (3), 338–343. doi:10.1080/13816810.2021.1891552
Friederich, M. W., Elias, A. F., Kuster, A., Laugwitz, L., Larson, A. A., Landry, A. P., et al. (2020). Pathogenic Variants in SQOR Encoding Sulfide:quinone Oxidoreductase Are a Potentially Treatable Cause of Leigh Disease. Jrnl Inher Metab. Disea 43 (5), 1024–1036. doi:10.1002/jimd.12232
Gamage, N., Barnett, A., Hempel, N., Duggleby, R. G., Windmill, K. F., Martin, J. L., et al. (2006). Human Sulfotransferases and Their Role in Chemical Metabolism. Toxicol. Sci. 90 (1), 5–22. doi:10.1093/toxsci/kfj061
Gaull, G., Sturman, J. A., and Räihä, N. C. R. (1972). Development of Mammalian Sulfur Metabolism: Absence of Cystathionase in Human Fetal Tissues. Pediatr. Res. 6 (6), 538–547. doi:10.1203/00006450-197206000-00002
Hanson, S. R., Best, M. D., and Wong, C.-H. (2004). Sulfatases: Structure, Mechanism, Biological Activity, Inhibition, and Synthetic Utility. Angew. Chem. Int. Ed. 43 (43), 5736–5763. doi:10.1002/anie.200300632
Hiraoka, N., Nakagawa, H., Ong, E., Akama, T. O., Fukuda, M. N., and Fukuda, M. (2000). Molecular Cloning and Expression of Two Distinct Human Chondroitin 4-O-Sulfotransferases that Belong to the HNK-1 Sulfotransferase Gene Family. J. Biol. Chem. 275 (26), 20188–20196. doi:10.1074/jbc.M002443200
Humphries, D. E., Lanciotti, J., and Karlinsky, J. B. (1998). cDNA Cloning, Genomic Organization and Chromosomal Localization of Human Heparan Glucosaminyl N-deacetylase/N-Sulphotransferase-2. Biochem. J. 332 (Pt 2), 303–307. doi:10.1042/bj3320303
Ishii, M., and Maeda, N. (2008). Spatiotemporal Expression of Chondroitin Sulfate Sulfotransferases in the Postnatal Developing Mouse Cerebellum. Glycobiology 18 (8), 602–614. doi:10.1093/glycob/cwn040
Jiang, W., Ishino, Y., Hashimoto, H., Keino-Masu, K., Masu, M., Uchimura, K., et al. (2017). Sulfatase 2 Modulates Fate Change from Motor Neurons to Oligodendrocyte Precursor Cells through Coordinated Regulation of Shh Signaling with Sulfatase 1. Dev. Neurosci. 39 (5), 361–374. doi:10.1159/000464284
Johnson, M. H., Grossmann, T., and Kadosh, K. C. (2009). Mapping Functional Brain Development: Building a Social Brain through Interactive Specialization. Developmental Psychol. 45 (1), 151–159. doi:10.1037/a0014548
Kalus, I., Salmen, B., Viebahn, C., Figura, K. v., Schmitz, D., D'Hooge, R., et al. (2009). Differential Involvement of the Extracellular 6-O-Endosulfatases Sulf1 and Sulf2 in Brain Development and Neuronal and Behavioural Plasticity. J. Cell Mol Med 13 (11-12), 4505–4521. doi:10.1111/j.1582-4934.2008.00558.x
Kamiyama, S., Suda, T., Ueda, R., Suzuki, M., Okubo, R., Kikuchi, N., et al. (2003). Molecular Cloning and Identification of 3′-Phosphoadenosine 5′-Phosphosulfate Transporter. J. Biol. Chem. 278 (28), 25958–25963. doi:10.1074/jbc.M302439200
Khateb, S., Kowalewski, B., Bedoni, N., Damme, M., Pollack, N., Saada, A., et al. (2018). A Homozygous Founder Missense Variant in Arylsulfatase G Abolishes its Enzymatic Activity Causing Atypical Usher Syndrome in Humans. Genet. Med. 20 (9), 1004–1012. doi:10.1038/gim.2017.227
Kříž, L., Bičíková, M., and Hampl, R. (2008). Roles of Steroid Sulfatase in Brain and Other Tissues. Physiol. Res. 57 (5), 657–668. doi:10.33549/physiolres.931207
Langford, R., Hurrion, E., and Dawson, P. A. (2017). Genetics and Pathophysiology of Mammalian Sulfate Biology. J. Genet. Genomics 44, 7–20. doi:10.1016/j.jgg.2016.08.001
Lee, A., Dawson, P. A., and Markovich, D. (2005). NaSi-1 and Sat-1: Structure, Function and Transcriptional Regulation of Two Genes Encoding Renal Proximal Tubular Sulfate Transporters. Int. J. Biochem. Cell Biol. 37 (7), 1350–1356. doi:10.1016/j.biocel.2005.02.013
Lindsay, S. J., Xu, Y., Lisgo, S. N., Harkin, L. F., Copp, A. J., Gerrelli, D., et al. (2016). HDBR Expression: A Unique Resource for Global and Individual Gene Expression Studies during Early Human Brain Development. Front. Neuroanat. 10, 86. doi:10.3389/fnana.2016.00086
Loriette, C., and Chatagner, F. (1978). Cysteine Oxidase and Cysteine Sulfinic Acid Decarboxylase in Developing Rat Liver. Experientia 34 (8), 981–982. doi:10.1007/BF01915299
Martin, A. R., Williams, E., Foulger, R. E., Leigh, S., Daugherty, L. C., Niblock, O., et al. (2019). PanelApp Crowdsources Expert Knowledge to Establish Consensus Diagnostic Gene Panels. Nat. Genet. 51 (11), 1560–1565. doi:10.1038/s41588-019-0528-2
McCarver, D. G., and Hines, R. N. (2002). The Ontogeny of Human Drug-Metabolizing Enzymes: Phase II Conjugation Enzymes and Regulatory Mechanisms: Table 1. J. Pharmacol. Exp. Ther. 300 (2), 361–366. doi:10.1124/jpet.300.2.361
Morreale de Escobar, G., Obregon, M., and Escobar del Rey, F. (2004). Role of Thyroid Hormone during Early Brain Development. Eur. J. Endocrinol. 151 (Suppl. 3), U25–U37. doi:10.1530/eje.0.151u025
Mulder, G. J., and Jakoby, W. B. (1990). “Sulfation,” in Conjugation Reactions in Drug Metabolism: An Integrated Approach: Substrates, Co-substrates, Enzymes and Their Interactions in Vivo and in Vitro. Editor G. J. Mulder (London: Taylor & Francis), 107–161.
O'Rahilly, R., and Müller, F. (1994). Neurulation in the normal Human Embryo. Ciba Found. Symp. 181, 70–89. ; discussion 82-79. doi:10.1002/9780470514559.ch5
Ohayon, D., Escalas, N., Cochard, P., Glise, B., Danesin, C., and Soula, C. (2019). Sulfatase 2 Promotes Generation of a Spinal Cord Astrocyte Subtype that Stands Out through the Expression of Olig2. Glia 67 (8), 1478–1495. doi:10.1002/glia.23621
Okada, T., Keino-Masu, K., Nagamine, S., Kametani, F., Ohto, T., Hasegawa, M., et al. (2017). Desulfation of Heparan Sulfate by Sulf1 and Sulf2 Is Required for Corticospinal Tract Formation. Sci. Rep. 7 (1), 13847. doi:10.1038/s41598-017-14185-3
Ono, H., Ogasawara, O., Okubo, K., and Bono, H. (2017). RefEx, a Reference Gene Expression Dataset as a Web Tool for the Functional Analysis of Genes. Sci. Data 4, 170105. doi:10.1038/sdata.2017.105
Pacifici, G. M. (2005). Sulfation of Drugs and Hormones in Mid-gestation Human Fetus. Early Hum. Development 81 (7), 573–581. doi:10.1016/j.earlhumdev.2004.10.021
Paganini, L., Hadi, L. A., Chetta, M., Rovina, D., Fontana, L., Colapietro, P., et al. (2019). A HS6ST2 Gene Variant Associated with X‐linked Intellectual Disability and Severe Myopia in Two Male Twins. Clin. Genet. 95 (3), 368–374. doi:10.1111/cge.13485
Pallerla, S. R., Lawrence, R., Lewejohann, L., Pan, Y., Fischer, T., Schlomann, U., et al. (2008). Altered Heparan Sulfate Structure in Mice with Deleted NDST3 Gene Function. J. Biol. Chem. 283 (24), 16885–16894. doi:10.1074/jbc.M709774200
Peter, V. G., Quinodoz, M., Sadio, S., Held, S., Rodrigues, M., Soares, M., et al. (2021). New Clinical and Molecular Evidence Linking Mutations in ARSG to Usher Syndrome Type IV. Hum. Mutat. 42 (3), 261–271. doi:10.1002/humu.24150
Rakoczy, J., Lee, S., Weerasekera, S. J., Simmons, D. G., and Dawson, P. A. (2015). Placental and Fetal Cysteine Dioxygenase Gene Expression in Mouse Gestation. Placenta 36 (8), 956–959. doi:10.1016/j.placenta.2015.06.003
Richard, K., Hume, R., Kaptein, E., Stanley, E. L., Visser, T. J., and Coughtrie, M. W. H. (2001). Sulfation of Thyroid Hormone and Dopamine during Human Development: Ontogeny of Phenol Sulfotransferases and Arylsulfatase in Liver, Lung, and Brain1. J. Clin. Endocrinol. Metab. 86 (6), 2734–2742. doi:10.1210/jcem.86.6.7569
Rivett, A. J., Eddy, B. J., and Roth, J. A. (1982). Contribution of Sulfate Conjugation, Deamination, and O-Methylation to Metabolism of Dopamine and Norepinephrine in Human Brain. J. Neurochem. 39 (4), 1009–1016. doi:10.1111/j.1471-4159.1982.tb11490.x
Salman, E. D., Kadlubar, S. A., and Falany, C. N. (2009). Expression and Localization of Cytosolic Sulfotransferase (SULT) 1A1 and SULT1A3 in normal Human Brain. Drug Metab. Dispos 37 (4), 706–709. doi:10.1124/dmd.108.025767
Schwartz, N. B., and Domowicz, M. S. (2018). Proteoglycans in Brain Development and Pathogenesis. FEBS Lett. 592 (23), 3791–3805. doi:10.1002/1873-3468.13026
Stanley, E. L., Hume, R., and Coughtrie, M. W. H. (2005). Expression Profiling of Human Fetal Cytosolic Sulfotransferases Involved in Steroid and Thyroid Hormone Metabolism and in Detoxification. Mol. Cell Endocrinol. 240 (1-2), 32–42. doi:10.1016/j.mce.2005.06.003
Sunkin, S. M., Ng, L., Lau, C., Dolbeare, T., Gilbert, T. L., Thompson, C. L., et al. (2013). Allen Brain Atlas: an Integrated Spatio-Temporal portal for Exploring the central Nervous System. Nucleic Acids Res. 41, D996–d1008. doi:10.1093/nar/gks1042
Ueki, I., Roman, H. B., Valli, A., Fieselmann, K., Lam, J., Peters, R., et al. (2011). Knockout of the Murine Cysteine Dioxygenase Gene Results in Severe Impairment in Ability to Synthesize Taurine and an Increased Catabolism of Cysteine to Hydrogen Sulfide. Am. J. Physiology-Endocrinology Metab. 301 (4), E668–E684. doi:10.1152/ajpendo.00151.2011
Ullrich, K. J., and Murer, H. (1982). Sulphate and Phosphate Transport in the Renal Proximal Tubule. Phil. Trans. R. Soc. Lond. B 299, 549–558. doi:10.1098/rstb.1982.0151
Verheyen, S., Blatterer, J., Speicher, M. R., Bhavani, G. S., Boons, G.-J., Ilse, M.-B., et al. (2021). Novel Subtype of Mucopolysaccharidosis Caused by Arylsulfatase K (ARSK) Deficiency. J. Med. Genet., 16, jmedgenet–2021. doi:10.1136/jmedgenet-2021-108061
Wang, J., Falany, J. L., and Falany, C. N. (1998). Expression and Characterization of a Novel Thyroid Hormone-Sulfating Form of Cytosolic Sulfotransferase from Human Liver. Mol. Pharmacol. 53 (2), 274–282. doi:10.1124/mol.53.2.274
Yamaguchi, Y. (2001). Heparan Sulfate Proteoglycans in the Nervous System: Their Diverse Roles in Neurogenesis, Axon Guidance, and Synaptogenesis. Semin. Cell Developmental Biol. 12 (2), 99–106. doi:10.1006/scdb.2000.0238
Zhang, Z., Dawson, P. A., Piper, M., and Simmons, D. G. (2019). Postnatal N-Acetylcysteine Administration Rescues Impaired Social Behaviors and Neurogenesis in Slc13a4 Haploinsufficient Mice. EBioMedicine 43, 435–446. doi:10.1016/j.ebiom.2019.03.081
Keywords: sulfate, brain, embryological, fetal, gene expression, neurological dysfunction
Citation: Clarke T, Fernandez FE and Dawson PA (2022) Sulfation Pathways During Neurodevelopment. Front. Mol. Biosci. 9:866196. doi: 10.3389/fmolb.2022.866196
Received: 30 January 2022; Accepted: 24 March 2022;
Published: 14 April 2022.
Edited by:
Tarsis G. Ferreira, Optimvia, United StatesReviewed by:
William Davies, Cardiff University, United KingdomAntonio Rossi, University of Pavia, Italy
Copyright © 2022 Clarke, Fernandez and Dawson. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Paul A. Dawson, cGF1bC5kYXdzb25AbWF0ZXIudXEuZWR1LmF1
 Taylor Clarke
Taylor Clarke Francesca E. Fernandez1
Francesca E. Fernandez1 Paul A. Dawson
Paul A. Dawson