Abstract
Background: Autophagy is a highly regulated and evolutionarily conserved process in eukaryotes which is responsible for protein and organelle degradation. Although this process was described over 60 years ago, the selective autophagy of mitochondria (mitophagy) was recently coined in 2005. Research on the topic of mitophagy has made rapid progress in the past decade, which proposed to play critical roles in human health and disease. This study aimed to visualize the scientific outputs and research trends of mitophagy.
Methods: Articles and reviews related to the topic of mitophagy were retrieved from the Web of Science Core Collection on 30 November 2021. Two kinds of software (CiteSpace and VOSviewer) were used to perform a visualized analysis of countries/regions, institutions, authors, journals, references, and keywords.
Results: From 2005 to 2021, total 5844 publications on mitophagy were identified for final analysis. The annual number of publications grew yearly over the past 17 years. United States (N = 2025) and Chinese Academy of Sciences is the leading country and institute (N = 112) ranked by the number of publications, respectively. The most productive author was Jun Ren (N = 38) and Derek P. Narendra obtained the most co-cited times (2693 times). The journals with the highest output and the highest co-citation frequency were Autophagy (N = 208) and Journal of Biological Chemistry (co-citation: 17226), respectively. Analyses of references and keywords suggested that “mechanism of mitochondrial quality control”, “molecule and signaling pathway in mitophagy”, and “mitophagy related diseases” were research hotspots, and parkin-mediated mitophagy and its roles in skeletal muscle and inflammation-related diseases may be the frontiers of future research.
Conclusion: Although mitophagy research has flourished and attracted attention from all over the world, the regional imbalance in the development of mitophagy research was observed. Our results provided a comprehensive global research landscape of mitophagy from 2005– 2021 from a perspective of bibliometrics, which may serve as a reference for future mitophagy studies.
Introduction
As the powerhouse of the cell, mitochondria play essential roles in regulating cellular metabolism and physiology (Harper et al., 2018). Thus, the mitochondrial quality control is important to maintain the life of cells, as well as human health (Pickles et al., 2018). Mitophagy was a recently identified form of selective autophagy, during which dysfunctional or superfluous mitochondria were removed (Gustafsson and Dorn, 2019a). Although mitophagy was described firstly in yeast, it is an evolutionarily conserved mechanism that has also been identified in mammals (Priault et al., 2005; Palikaras et al., 2018). So far, at least two distinct pathways have been found to be involved in the mechanisms of mitophagy. One is the PINK1/Parkin pathway, and the other is the mitophagy receptor pathway (Gustafsson et al., 2019b); its functional roles include mitochondrial quality and quantity control, metabolic reprogramming, and differentiation (Palikaras et al., 2018). For more than a decade, significant progress has been made in the field of mitophagy and a list of diseases, such as neurodegenerative disease (Malpartida et al., 2021), cardiovascular disease (Bravo-San Pedro et al., 2017), and cancer (Panigrahi et al., 2020), were found to be closely associated with mitophagy impairment. Recently, several systematic reviews on mitophagy have been performed (Palikaras et al., 2018; Pickles et al., 2018; Onishi et al., 2021). With these reviews, we can understand the mechanism, function, and related diseases of mitophagy. However, the comprehensive knowledge of the global research status, current hotspots, and future trends of mitophagy is still lacking.
Bibliometric analysis is a powerful approach that uses literature metrics or indicators to quantitatively measure the research performance in a certain field (Deng et al., 2020; Wang et al., 2021c). It was coined more than 50 years ago (Pritchard, 1969), and now it has been widely used as a methodology for evaluating trends and frontiers from a large number of publications (Zhao et al., 2020; Hu et al., 2021). Through bibliometric analysis, influential articles, main research fields, and new research directions can be timely and comprehensively achieved by researchers (Thompson and Walker, 2015). Compared with traditional reviews, bibliometric-based analyses can provide a more comprehensive perspective on the research trends, and the data is more objective (Yan et al., 2021). With the development of bibliometrics, dozens of software tools for conducting bibliometric analysis are available for researchers (Moral-Muñoz et al., 2020). CiteSpace and VOSviewer represent the two most commonly used tools for visualization (Moral-Muñoz et al., 2020). Recently, thousands of bibliometric studies have been conducted in a range of fields (Donthu et al., 2021), while only one study reported on mitophagy so far which focused on comparing the difference in research progress in China and other developed countries (Chen et al., 2021). The global research status of mitophagy remains largely unknown.
In this study, a bibliometric-based analysis was performed to systematically evaluate the mitophagy studies from 2005 to 2021. By taking advantage of CiteSpace and VOSviewer, we comprehensively analyzed the publication output, disciplinary composition, countries/regions, institutions, publishers, funders, authors, journals, top-cited articles, references, and appeared keywords. Our results draw a visualization map of the global research landscape of mitophagy, helping researchers, especially for those who are new to this field, to deepen their understanding of the current status and future directions of mitophagy research.
Methods
Data Collection and Cleaning
Publications were extracted from the database of the Web of Science Core Collection (WoSCC) in this study. WoSCC is the world’s oldest, most widely used, and authoritative database of research publications and citations, which contains records of articles from the highest impact journals worldwide (Birkle et al., 2020). A literature search was performed within 1 day on 30 November 2021. The search formula was set as follows: Mitophagy (Topic) and 2022 (Exclude–Publication Years) and Meeting Abstracts or Editorial Materials or Early Access or Book Chapters or Corrections or Proceedings Papers or Letters or Retracted Publications or News Items or Retractions (Exclude–Document Types). A search by Topic will be searched in the title, abstract, author keywords, and keywords plus. A total of 5846 literatures were obtained and exported for full record and cited references in the format of plain text files. Using Endnote software, a total of two duplications were found. After removing duplications, a total of 5844 unique records remained, including 4434 articles and 1410 reviews (Figure 1). Research articles published original findings and reviews and are written based on existing articles. Based on these two types of documents, the impact factor (IF) of an academic journal is calculated by Clarivate. Therefore, consistent with other studies (Chen et al., 2020; Hu et al., 2021; Zhang et al., 2021), we only used articles and reviews for bibliometric analysis in this study.
FIGURE 1
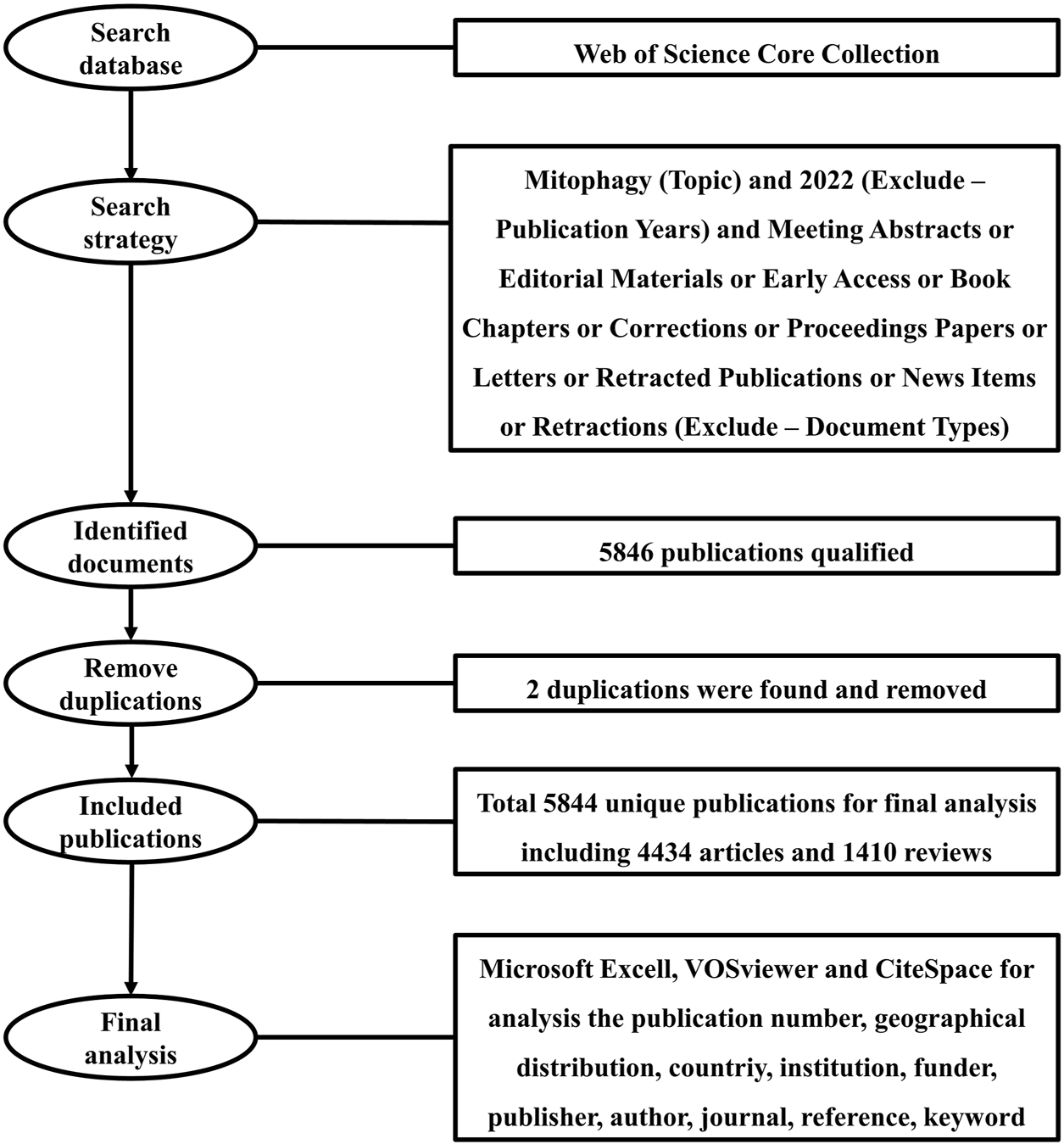
Flowchart of the methodology and design of this study.
Of note, we have cleaned the data before analysis (Wang et al., 2021b; Zhang et al., 2021). These included: 1) Publications from Taiwan and the People’s Republic of China were reclassified to China; 2) Publications from England, Scotland, Northern Ireland, and Wales were assigned to the United Kingdom; 3) Publications from Austria, Belgium, Bulgaria, Croatia, Cyprus, Czech Republic, Denmark. Estonia, Finland, France, Germany, Greece, Hungary, Ireland, Italy, Latvia, Lithuania, Luxembourg, Malta, Netherlands, Poland, Portugal, Romania, Slovakia, Slovenia, Spain, and Sweden were merged into European Union; 4) Publications from Cent S Univ and Cent South Univ merged into Cent South Univ; 5) Publications from Guangdong Med Coll and Guangdong Med Univ merged into Guangdong Med Univ; 6) Publications from Univ Texas Southwestern Med Ctr Dallas and Univ Texas SW Med Ctr Dallas merged into Univ Texas Southwestern Med Ctr Dallas.
Visualized Analysis
CiteSpace software (Drexel University, Philadelphia, PA, United States) is a freely available Java application, which was widely used for visualizing and analyzing trends and patterns in the scientific literature (Chen, 2006). It was designed by Chen (2004) and last updated on 17 January 2021 (Version 5.8. R2). Web of Science is the primary source of input data for CiteSpace. It has been widely used for visualizing bibliometric networks by other studies (Hu et al., 2021; Ma et al., 2021). In this study, CiteSpace was used to perform a bibliometric analysis of collaborations (countries/regions, institutions, authors), the timeline view of co-occurrence (keywords), and citation bursts (references and keywords). Co-occurrence analysis was used to find the number of terms where they occur together in the same article and weighted by the frequency of occurrence. Citation bursts means that the frequency of a term has soared within a certain short period. The primary parameters were set as follows: time slicing (2005–2021), years per slice (2 years), and selection criteria (g-index, k = 25). Other parameters were set according to the CiteSpace manual for different situations.
VOSviewer software is a useful tool for constructing and visualizing bibliometric networks (Van Eck and Waltman, 2010). It was developed by the Center for Science and Technology Research at Leiden University (The Netherlands) in 2007 (Van Eck and Waltman, 2007). The latest version 1.6.17 was released on 22 July 2021, and is free for download. Co-authorship networks, citation-based networks, and co-occurrence networks can be created based on data downloaded from the Web of Science. It also has been widely used for visualizing bibliometric networks by other groups (Chen et al., 2020; Xiong et al., 2021). In this study, VOSviewer was used for visualizing the co-authorship between countries/regions, institutions, authors, and the co-occurrence of keywords.
Other information such as impact factor (IF) and Journal Citation Reports (JCR) division of journals were obtained from the Web of Science website (www.webofscience.com) directly on 30 November 2021. Microsoft Office Excel 2016 was used to analyze the annual publications. WordArt (https://wordart.com/) was used to create the word cloud of the subject categories. GunnMap 2 (http://gunnmap.herokuapp.com/) was used to display the global publication distribution on a world map.
Results
Analysis of Publication Trends and Subject Categories
From the first article published on 10 June 2005 to 30 November 2021, a total of 5844 publications on mitophagy research were identified. Figure 2A shows the annual number of publications and citations worldwide. In general, the annual number of publications increased year by year. The early stage (2005–2014) saw a relatively stable growth, with a publication count of no more than 300 per year. While a steep growth was observed since 2015. Up to date (30 November 2021), the publication outputs have reached over 1100 in 2021. The number of 2020 (1032) was over 27 times that of 2010 (74). The annual number of citations were also increased yearly, and the number in 2020 (51433) was over 40 times that of 2010 (1281).
FIGURE 2
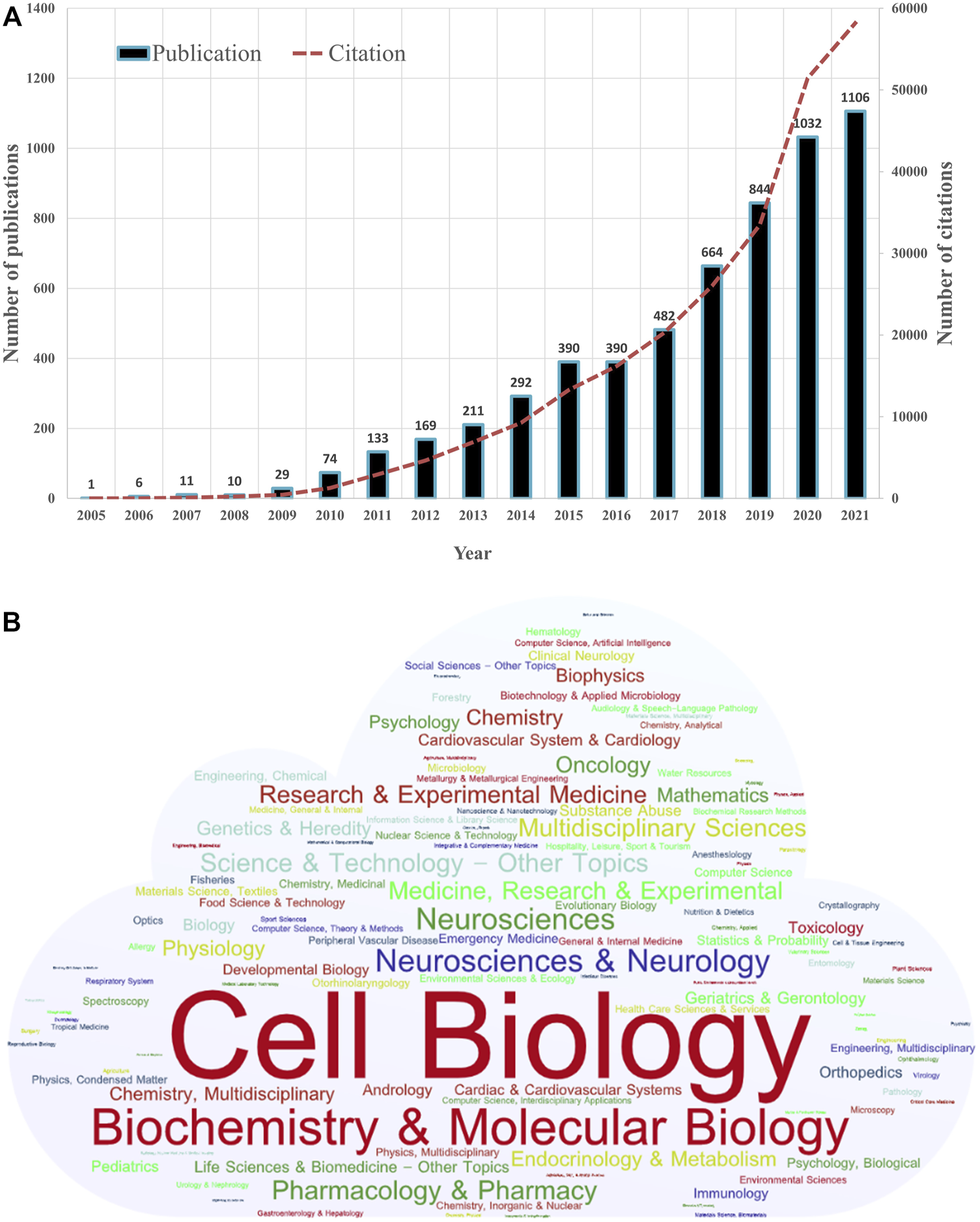
Distribution of publications on mitophagy by year and category. (A) Number of annual publications and citations of mitophagy from 2005 to 2021 globally. (B) The word cloud map of the subject categories on mitophagy (Created with https://wordart.com/).
By the analysis of CiteSpace, we identified 127 disciplinary categories involved in the field of mitophagy (Figure 2B). Through WordArt, a map of the word cloud of the subject categories was created (the more publications belong to the category, the bigger the word appears in the word cloud). Cell Biology (1916, 32.79%) is the most common category, followed by Biochemistry and Molecular Biology (1574, 26.93%), Neurosciences and Neurology (616, 10.54%), Neurosciences (566, 9.69%) and Pharmacology and Pharmacy (437, 7.48%).
Analysis of Countries and Institutions
Figure 3A shows the geographical distribution of global productivity of studies on mitophagy. A total of 76 different countries/regions covering five continents have taken part in mitophagy research. Thirteen countries/regions published only one paper, and 12 countries/regions published over 100 articles. These results indicated that mitophagy research has attracted attention from all over the world. However, research development of mitophagy in different countries/regions is uneven. Among the top 10 most prolific countries or political entities (Table 1), United States had the highest number of publications (2025, 34.65%), followed by China (1968, 33.68%), the European Union (1373, 23.49%), the United Kingdom (418, 7.15%), and Japan (364, 6.23%). These top five countries or political entities account for most of the total publications. From Figure 3B, we can see that the number of annual publications slowly increased after 2015 in the United States, the European Union, the United Kingdom, and Japan. Whereas, a relatively sharp rise was witnessed in China after 2015, especially during the year from 2017 to 2021. Therefore, we can conclude that China has been the main driver of the increase in mitophagy research since 2015. Among the top 10 most prolific institutions, six of them come from China, and the remaining institutions come from the United States (N = 2), the United Kingdom (N = 1), and Canada (N = 1). Chinese Acad Sci (112, 1.92%) is the leading institute, followed by Univ Pittsburgh (109, 1.87%), Univ Calif San Diego (92, 1.57%) and Zhejiang Univ (88, 1.51%). Of note, no institutions from the European Union appeared on the list.
TABLE 1
| Rank | Country | Year | Centrality | Count (%) | Rank | Institution (Country) | Year | Centrality | Count (%) |
|---|---|---|---|---|---|---|---|---|---|
| 1 | United States | 2006 | 0.51 | 2025 (34.65%) | 1 | Chinese Acad Sci (China) | 2010 | 0.03 | 112 (1.92%) |
| 2 | China | 2009 | 0.21 | 1968 (33.68%) | 2 | Univ Pittsburgh (United States) | 2007 | 0.13 | 109 (1.87%) |
| 3 | European Union | 2005 | 0.39 | 1373 (23.49%) | 3 | Univ Calif San Diego (United States) | 2009 | 0.05 | 92 (1.57%) |
| 4 | UK | 2009 | 0.35 | 418 (7.15%) | 4 | Zhejiang Univ (China) | 2013 | 0.04 | 88 (1.51%) |
| 5 | Japan | 2008 | 0.05 | 364 (6.23%) | 5 | Fudan Univ (China) | 2010 | 0.01 | 87 (1.49%) |
| 6 | South Korea | 2007 | 0.05 | 242 (4.14%) | 6 | Cent South Univ (China) | 2015 | 0.04 | 85 (1.45%) |
| 7 | Canada | 2006 | 0.08 | 241 (4.12%) | 7 | UCL (UK) | 2010 | 0.07 | 74 (1.27%) |
| 8 | Australia | 2007 | 0.07 | 139 (2.38%) | 8 | Shanghai Jiao Tong Univ (China) | 2013 | 0.02 | 73 (1.25%) |
| 9 | India | 2013 | 0.1 | 129 (2.21%) | 9 | Sun Yat-sen Univ (China) | 2013 | 0.03 | 68 (1.16%) |
| 10 | Russia | 2009 | 0.06 | 98 (1.68%) | 10 | McGill Univ (Canada) | 2011 | 0.03 | 67 (1.15%) |
Top 10 countries/political entities and institutions contributed to publications on mitophagy.
UK: United Kingdom. UCL: Univ College London. Year: The year in which the earliest article was published.
FIGURE 3
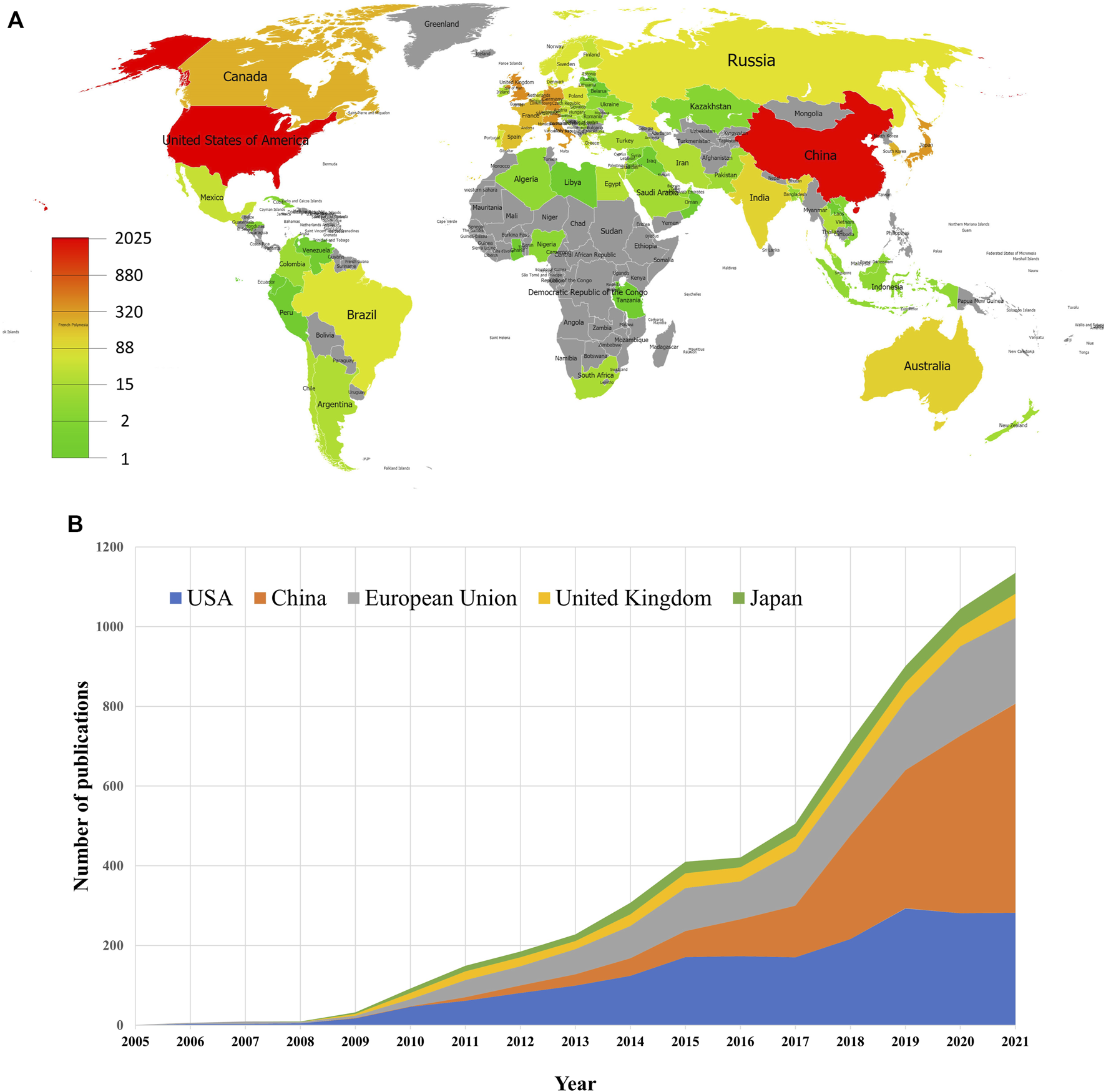
Geographical distribution of publications on mitophagy. (A) Geographical distribution map of publications on mitophagy. (B) Trends of the annual publications of mitophagy in the United States, China, European Union, United Kingdom, and Japan.
Next, network visualization of the co-authorship countries and institutions was analyzed via CiteSpace. In the cooperative network map (Figure 4), the nodes represent countries/institutions (the larger circle, the higher the number of publications); lines between the nodes represent a collaboration between two countries/institutions on the same article (the wider lines, the more frequency of collaborations); the purple ring represents active cooperation of the country/institutions (centrality≥0.10). Herein, high centrality indicates active cooperation. From Figure 4 and Table 1, we can see that United States (0.51) and Univ Pittsburgh (0.13) obtained the highest centrality among countries and institutions, respectively. In addition, many other countries or political entities, such as European Union, United Kingdom, Canada, India, Australia, Russia and Norway, also had a high centrality (purple ring). However, China and Japan do not have a high centrality, indicating their insufficient international cooperation.
FIGURE 4
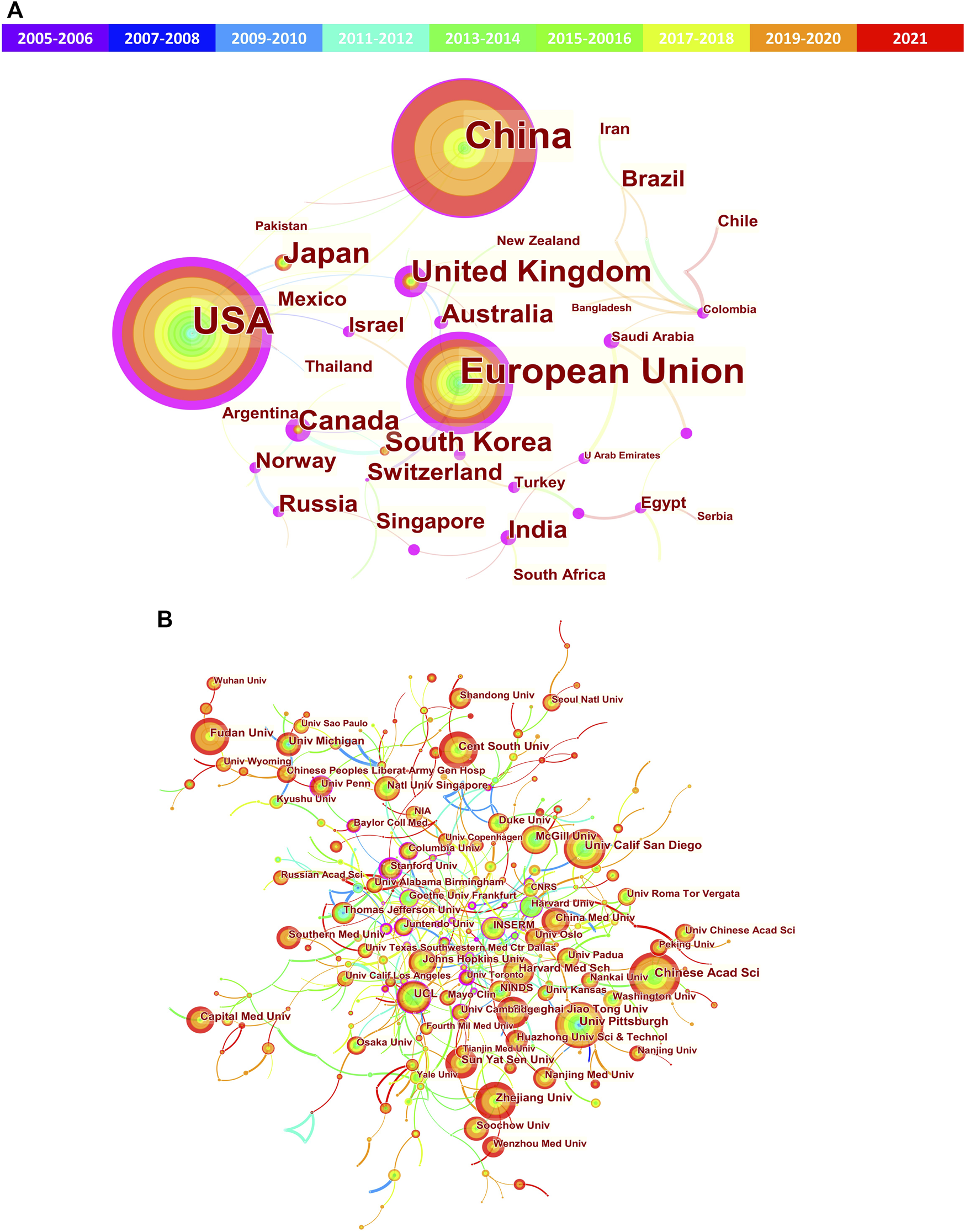
Cooperation network of the countries/regions and institutions related to mitophagy. (A) Network map of countries/regions; (B) Network map of institutions. The nodes represent countries/regions or institutions (the larger circle, the higher the number of publications); lines between the nodes represent collaboration (the wider lines, the more frequency of collaborations); the purple ring represents active cooperation (centrality≥0.10).
Analysis of Authors and Co-cited Authors
A total of 27872 authors published literature on mitophagy (data not shown). Among the top 10 prolific authors (Table 2), Jun Ren published the largest number of papers (N = 38), followed by Richard J Youle (N = 37), Roberta A Gottlieb (N = 34), and Asa B Gustafsson (N = 33). In the cooperative network map (Figure 5A), the 114 authors who published at least 10 papers and had co-authorship with others were visualized. A total of 114 terms, 12 clusters, 280 links, and a total link strength of 1106 were generated. Each node represented an author. The size of the nodes is determined by the number of publications (The higher the number, the larger the node). The same color represented the same cluster. The line between the nodes represented the co-authorship between authors (The stronger the cooperation relationship, the wider the line). The number of total link strengths reflected the total co-authorship strength between authors.
TABLE 2
| Rank | Author | Documents | Institutions (Countries) | Rank | Co-cited author | Co-citation | Institutions (Countries) | Countries |
|---|---|---|---|---|---|---|---|---|
| 1 | Jun Ren | 38 | Fudan Univ (China) | 1 | Derek P. Narendra | 2693 | NINDS (United States) | United States |
| 2 | Richard J. Youle | 37 | NINDS (United States) | 2 | Noboru Mizushima | 1446 | Univ Tokyo (Japan) | Univ Tokyo |
| 3 | Roberta A. Gottlieb | 34 | Cedars Sinai Med Ctr (United States) | 3 | Richard J. Youle | 1313 | NINDS (United States) | United States |
| 4 | Asa B. Gustafsson | 33 | Univ Calif San Diego (United States) | 4 | Hao Zhou | 1295 | Chinese Peoples Liberat Army Hosp (China) | China |
| 5 | Quan Chen | 32 | Nankai Univ (China) | 5 | Sven Geisler | 1141 | Univ Tubingen (Germany) | Germany |
| 6 | Hao Zhou | 32 | Chinese Peoples Liberat Army Hosp (China) | 6 | Michael Lazarou | 1085 | Monash Univ (Australia) | Australia |
| 7 | Daniel J. Klionsky | 31 | Univ Michigan (United States) | 7 | Gilad Twig | 1069 | Boston Univ (United States) | United States |
| 8 | Michael P. Lisanti | 30 | Thomas Jefferson Univ (United States) | 8 | Daniel J. Klionsky | 940 | Univ Michigan (United States) | United States |
| 9 | Federica Sotgia | 29 | Univ Manchester (England) | 9 | Hsiuchen Chen | 857 | Caltech (United States) | United States |
| 10 | Nobutaka Hattori | 28 | Juntendo Univ (Japan) | 10 | Noriyuki Matsuda | 800 | Tokyo Metropolitan Inst Med Sci (Japan) | Japan |
Top 10 authors and co-cited authors contributed to mitophagy research.
NINDS, NIH, national institute of neurological disorders and stroke; Caltech, California Institute of Technology.
FIGURE 5
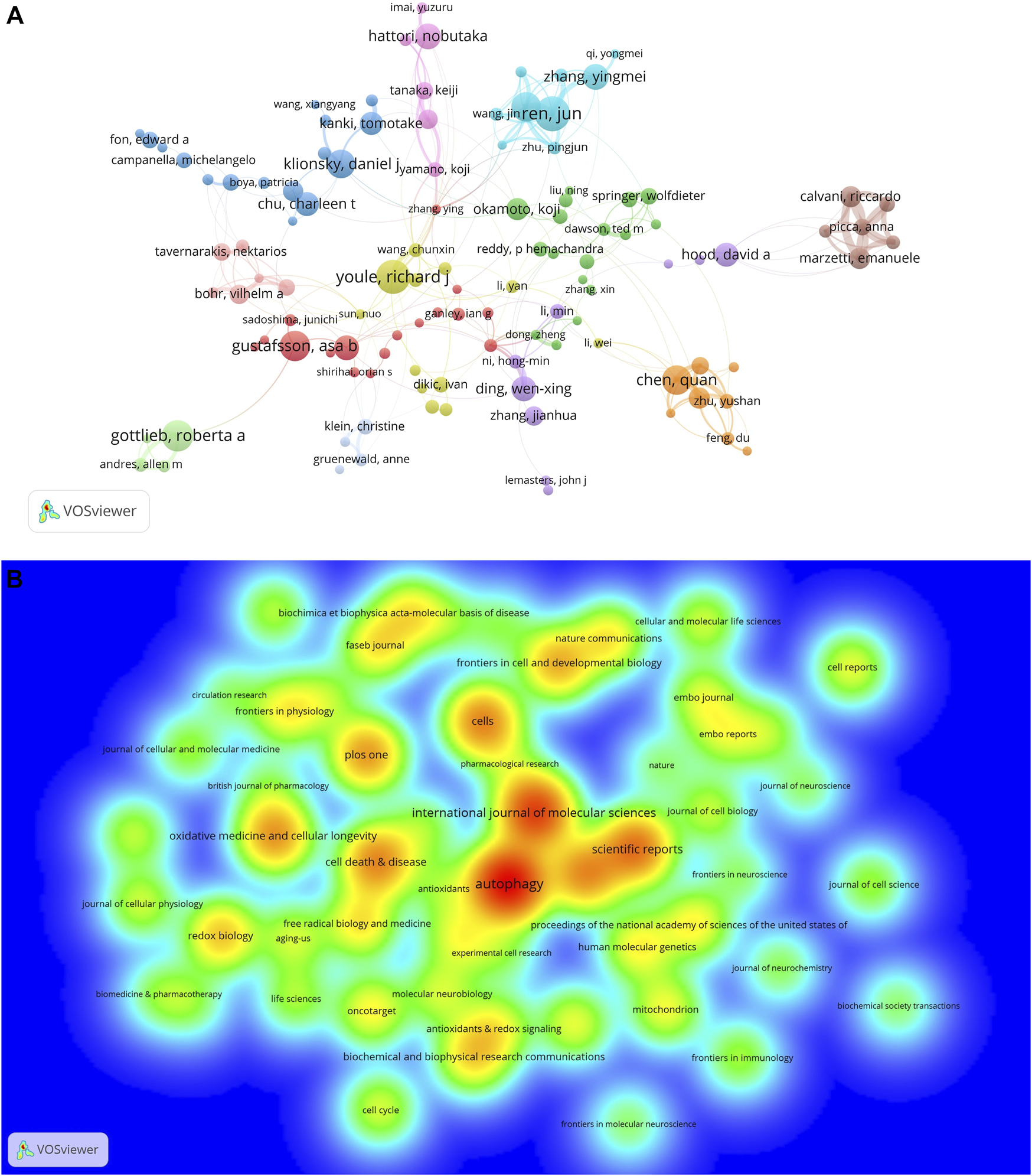
Co-authorship of authors and productive journals on mitophagy. (A) Co-authorship network of authors in the field of mitophagy. (B) Density map of top 56 most productive journals published papers on mitophagy.
Co-citation is defined as the frequency with which two documents are cited together by another or more articles at the same time, providing a way to study the specialty structure of science (Wang et al., 2021b). In this study, a total of 106127 co-cited authors were identified (data not shown). Among the top 10 most frequently co-cited authors (Table 2), Derek P Narendra (2693 times) ranked first, followed by Noboru Mizushima (1446 times) and Richard J Youle (1313 times). It is worth noting that Richard J Youle, Hao Zhou, and Daniel J Klionsky were the top 10 authors ranked by both numbers of publication and co-citation frequency.
Analysis of Journals and Co-Cited Journals
A total of 1021 academic journals had published articles on mitophagy (data not shown). The top 10 journals published with the highest number of publications were shown in Table 3. Among them, Autophagy (N = 208) most productive one and had the highest IF 2020 (16.016). Eight (80%) of the journals had an IF 2020 over 5 and 6 (60%) were located in JCR (2020) Q1 region. A density map of the top 56 productive journals (with over 20 publications) was shown in Figure 5B. The top 10 journals ranked by co-cited frequency were also shown in Table 3. We can see that the journal with the highest co-citations was Journal of Biological Chemistry (co-citation: 17226). Notably, Autophagy, Journal of Biological Chemistry, and PLoS One were the top 10 journals ranked both by a number of publications and co-cited frequency.
TABLE 3
| Rank | Journal | Count | IF (2020) | JCR Category quartile | Co-cited journal | Citation | IF (2020) | JCR Category quartile |
|---|---|---|---|---|---|---|---|---|
| 1 | Autophagy | 208 | 16.016 | Cell Biology (Q1) | Journal of Biological Chemistry | 17226 | 5.157 | Biochemistry and Molecular Biology (Q2) |
| 2 | International Journal of Molecular Sciences | 155 | 5.924 | Biochemistry and Molecular Biology (Q1); Chemistry, Multidisciplinary (Q2) | PNAS* | 13970 | 11.205 | Multidisciplinary Sciences (Q1) |
| 3 | Scientific Reports | 115 | 4.38 | Multidisciplinary Sciences (Q1) | Nature | 13427 | 49.962 | Multidisciplinary Sciences (Q1) |
| 4 | Cells | 103 | 6.6 | Cell Biology (Q2) | Autophagy | 12774 | 16.016 | Cell Biology (Q1) |
| 5 | Cell Death and Disease | 101 | 8.469 | Cell Biology (Q1) | Journal of Cell Biology | 10499 | 10.539 | Cell Biology (Q1) |
| 6 | Journal of Biological Chemistry | 97 | 5.157 | Biochemistry and Molecular Biology (Q2) | Cell | 9829 | 41.584 | Biochemistry and Molecular Biology (Q1); Cell Biology (Q1) |
| 7 | Oxidative Medicine and Cellular Longevity | 93 | 6.543 | Cell Biology (Q2) | Science | 8462 | 47.728 | Multidisciplinary Sciences (Q1) |
| 8 | PLoS One | 90 | 3.24 | Multidisciplinary Sciences (Q2) | PLoS One | 6957 | 3.24 | Multidisciplinary Sciences (Q2) |
| 9 | Frontiers In Cell and Developmental Biology | 78 | 6.684 | Cell Biology (Q2); Developmental Biology (Q1) | Human Molecular Genetics | 6528 | 6.15 | Biochemistry and Molecular Biology (Q1); Genetics and Heredity (Q1) |
| 10 | Redox Biology | 69 | 11.799 | Biochemistry and Molecular Biology (Q1) | EMBO Journal | 6285 | 11.598 | Biochemistry and Molecular Biology (Q1); Cell Biology (Q1) |
Top 10 journals and co-cited journals contributed to mitophagy research.
What are the publication patterns for mitophagy studies in general journals? In this study, we found that most articles on mitophagy were published in a cell or molecular biology journals (Table 3). Unexpectedly, the Journal of Biological Chemistry was the most frequently co-cited journal (co-citation: 17226) with the IF 2020 as 5.157 that far exceeded some high-impact journals, such as Nature (co-citation: 13427; IF 2020: 49.962), Cell (co-citation: 9829; IF 2020: 41.584), and Science (co-citation: 8462; IF 2020: 47.728). Also notably, Autophagy (IF 2020: 16.016), Journal of Biological Chemistry (IF 2020: 5.157), and PLoS One (IF 2020: 3.24) were the top 10 both productive and cited journals. These results provided some guidance to future authors deciding where to publish their work on mitophagy research.
Analysis of Publishers and Funders
According to the published time, we divided the 5844 papers into two groups. There were 644 papers that were published during 2005–2013; and 5200 papers that were published during 2014–2021. Then the publishers and funders of each group were analyzed separately to show the trends over time. The top 10 publishers and funders of each group were presented in Figure 6. During 2005–2013, Elsevier (20.50%), Springer Nature (12.58%), Taylor and Francis (11.02%), Wiley (7.45%), and Public Library Science (5.75%) were the top five largest publishers in the field of mitophagy (Figure 6A). During 2014–2021, there was a huge increase in the number of publications on Mdpi (7.93%) and Frontiers Media Sa (6.71%), which ranked fourth and fifth, respectively (Figure 6B). It is noteworthy that Elsevier (25.01%) and Springer Nature (14.56%) were always the top two publishers in the two time periods.
FIGURE 6
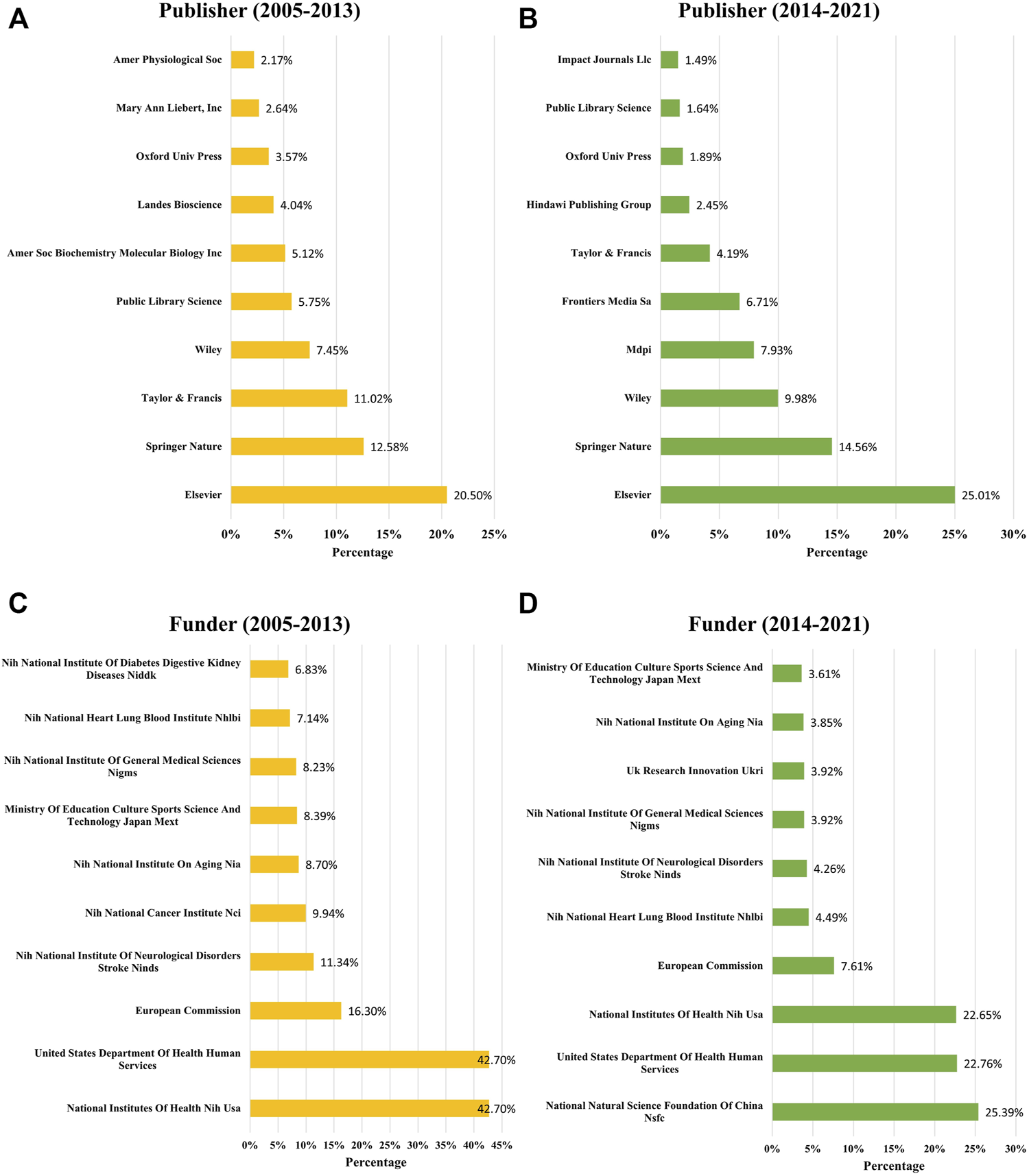
Publishers and funders changes for mitophagy research. Top 10 publishers with the largest number of articles on mitophagy during 2005–2013 (A) and 2014–2021 (B). Top 10 funders sponsored the largest number of publications during 2005–2013 (C) and 2014–2021 (D).
During 2005–2013, over 40% of papers were funded by the National Institutes Of Health, NIH, United States (42.7%) or United States Department Of Health and Human Services (42.7%), all belong to the United States (Figure 6C). Other top five funders that sponsored the largest number of publications were the European Commission (16.30%), (NIH) National Institute Of Neurological Disorders and Stroke, NINDS (11.34%), and (NIH) National Cancer Institute, NCI (9.94%). While during 2014–2021, the National Natural Science Foundation Of China, NSFC, has become one of the most important funders and sponsored the largest number of publications (25.39%) (Figure 6D). Followed by the United States Department Of Health and Human Services (22.76%), National Institutes Of Health, NIH, United States (22.65%), European Commission (7.61%), and (NIH) National Heart, Lung, and Blood Institute NHLBI (7.61%).
Top Cited Publications and Co-Cited References
The top-cited publications and co-cited references were fundamental and the basis of a certain field. In this study, the 10 most cited publications on mitophagy are listed in Table 4. All the top 10 publications were cited more than 900 times. The article (entitled A role for mitochondria in NLRP3 inflammasome activation) published in 2011 by Rongbin Zhou et al. obtained the largest number of citations (2,876). Of those, seven are articles (70%) and the rest are reviews (30%). As we can see from Table 4, all the top 10 most cited publications were published in high-impact journals before 2015. There were six papers with main themes on the functional roles of PINK1/Parkin and its related pathways in mitophagy. We suggested further reading these articles, if you wanted to get an in-depth knowledge of this field.
TABLE 4
| Rank | Title | Type | First author | Journal | Year | Citation | Major themes |
|---|---|---|---|---|---|---|---|
| 1 | A role for mitochondria in NLRP3 inflammasome activation | Article | Rongbin Zhou | Nature | 2011 | 2,876 | Reported a central role for mitophagy in the process of NLRP3 inflammasomes activation highlighting that mitochondria are essential for inflammatory response |
| 2 | Mechanisms of mitophagy | Review | Richard J Youle | Nature Reviews Molecular Cell Biology | 2011 | 1,904 | Comprehensively discussed the identified pathways that mediate mitophagy in yeast and mammalian cells and the role of mitophagy in Parkinson’s disease |
| 3 | PINK1/Parkin-mediated mitophagy is dependent on VDAC1 and p62/SQSTM1 | Article | Sven Geisler | Nature Cell Biology | 2010 | 1,763 | Reported the functional role of PINK1/Parkin-mediated mitophagy through VDAC1 ubiquitination in the development of Parkinson’s disease |
| 4 | PINK1 Is Selectively Stabilized on Impaired Mitochondria to Activate Parkin | Article | Derek P Narendra | Plos Biology | 2010 | 1,727 | Provide a novel explanation for how PINK1 and Parkin work together to protect against damaged mitochondria by promoting mitophagy |
| 5 | Phosphorylation of ULK1 (hATG1) by AMP-Activated Protein Kinase Connects Energy Sensing to Mitophagy | Article | Daniel F Egan | Science | 2011 | 1,606 | Uncovers a mechanism that AMPK directly regulates mitophagy through phosphorylating and activating ULK1 thus establishing a direct molecular link between nutrient status and cell survival |
| 6 | The ubiquitin kinase PINK1 recruits autophagy receptors to induce mitophagy | Article | Michael Lazarou | Nature | 2015 | 1216 | Shows that PINK1 induces mitophagy directly, through the phospho-ubiquitin-mediated recruitment of NDP52 and OPTN, and that these receptors have an early role in recruiting the autophagy machinery |
| 7 | PINK1 stabilized by mitochondrial depolarization recruits Parkin to damaged mitochondria and activates latent Parkin for mitophagy | Article | Noriyuki Matsuda | Journal of Cell Biology | 2010 | 1149 | Describe the mechanism underlying the functional interplay between ubiquitination catalyzed by Parkin and mitochondrial quality control regulated by PINK1 |
| 8 | PINK1-dependent recruitment of Parkin to mitochondria in mitophagy | Article | Cristofol Vives-Bauza | PNAS | 2010 | 1068 | Demonstrate that Parkin, together with PINK1, modulates mitochondrial trafficking, especially to the perinuclear region, a subcellular area associated with autophagy |
| 9 | The Roles of PINK1, Parkin, and Mitochondrial Fidelity in Parkinson’s Disease | Review | Alicia M Pickrell | Neuron | 2015 | 1042 | Summarize the functions of PINK1 and Parkin in normal cells, their molecular mechanisms of action, and the pathophysiological consequences of their loss |
| 10 | Autophagy, mitochondria and oxidative stress: cross-talk and redox signaling | Review | Jisun Lee | Biochemical Journal | 2012 | 948 | Summarize the basic mechanisms of mitophagy and the crosstalk between autophagy, redox signaling, and mitochondrial dysfunction highlighting its impact on chronic pathologies, particularly on neurodegenerative diseases |
Top 10 mitophagy-related publications with the most citations (up to 13 December 2021).
PNAS, Proceedings of the national academy of sciences of the United States of America.
Next, we constructed a cluster analysis for all the co-cited references by CiteSpace software and presented the clusters in form of a timeline view in Figure 7A. Total 14 clusters were formed: #0 mitochondrial dynamics; #1 mitochondrial quality control; #2 Parkinson’s disease; #3 signaling pathway; #4 selective autophagy; #5 mitochondrial fission; #6 neurodegenerative disease; #7 mitochondrial dysfunction; #8 heart failure; #9 skeletal muscle; #10 parkin-mediated mitophagy; #11 chronic obstructive pulmonary disease; #12 pathophysiological role; #13 colorectal cancer stress response. Among them, “parkin-mediated mitophagy” was the most recent cluster. In addition, the top 25 references with the strongest citation bursts were identified by Citespace analysis (Figure 7B). Citation bursts represent a rapid rise in citations within a certain period, which can help characterize the dynamics of a research field (Amjad et al., 2022). In this study, the first burst of reference began in 2008. Among these 25 references, four references (28%) had a burst duration until 2021. The reference with the strongest burstiness (strength = 140.19) was published in Nature Cell Biology by Geisler S et al., in 2010.
FIGURE 7
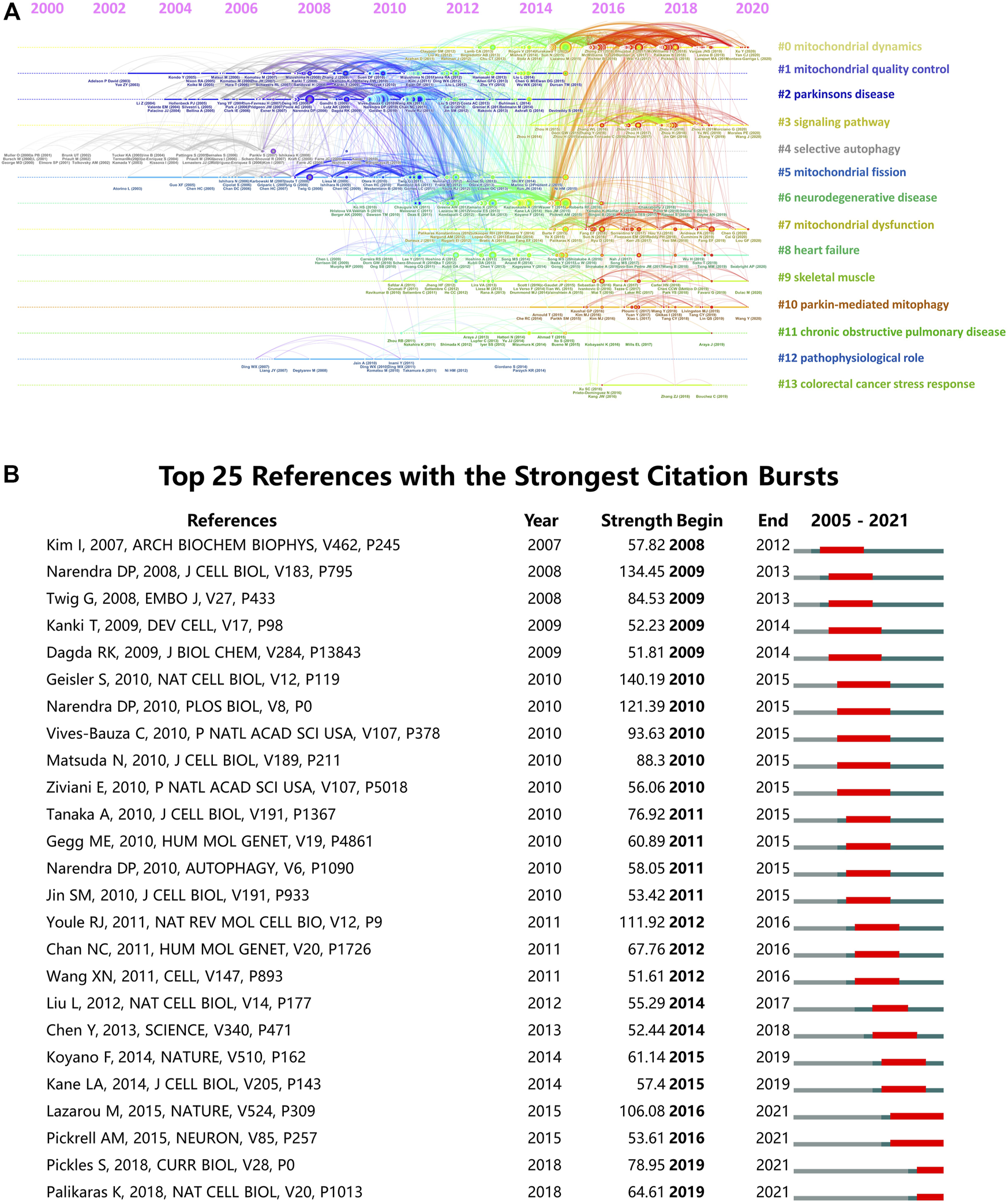
Analysis of co-cited references of the 5844 articles on mitophagy. (A) Timeline view of co-cited references. (B) Top 25 references with strongest citation bursts.
Analysis of Keywords
To investigate the research hotspots in the field of mitophagy, a total of 7506 keywords (author keywords) were extracted from the 5844 publications for co-occurrence analysis on VOSviewer and Citespace software. Table 5 shows the top 30 high-frequency keywords. Among these keywords, “mitophagy” (2333 times) was the most frequently appeared one, followed by “mitochondria” (1242 times), “autophagy” (1185 timesand), “apoptosis” (410 times), and “parkin” (391 times). Keyword cluster analysis is based on the coexistence network to simplify the keywords in a small number of clusters. Through Citespace analysis, seventeen clusters were generated and listed as fellow: #0 mitochondrial quality control; #1 oxidative stress; #2 endoplasmic reticulum; #3 Parkinson’s disease; #4 amyotrophic lateral sclerosis; #5 mitochondrial dysfunction; #6 mitochondrial DNA; #7 PGC-1 alpha; #8 reactive oxygen species; #9 selective autophagy; #10 tumor stroma; #11 mitochondrial dynamics; #12 cell death; #13 breast cancer; #14 drug resistance; #15 acute lung injury; #16 mitochondrial homeostasis (Figure 8A and Table 6). Herein, the serial numbers are sorted by cluster size, and the clusters indicated that publications are grouped into different topics according to the keywords.
TABLE 5
| Rank | Keyword | Occurrences | Rank | Keyword | Occurrences | Rank | Keyword | Occurrences |
|---|---|---|---|---|---|---|---|---|
| 1 | Mitophagy | 2333 | 11 | Mitochondrial dysfunction | 191 | 21 | ubiquitin | 101 |
| 2 | Mitochondria | 1242 | 12 | Neurodegeneration | 164 | 22 | metabolism | 83 |
| 3 | Autophagy | 1185 | 13 | Reactive oxygen species | 159 | 23 | drp1 | 77 |
| 4 | Apoptosis | 410 | 14 | Mitochondrial biogenesis | 140 | 24 | fission | 73 |
| 5 | Parkin | 391 | 15 | Inflammation | 136 | 25 | skeletal muscle | 70 |
| 6 | Oxidative stress | 352 | 16 | ROS | 124 | 26 | fusion | 68 |
| 7 | Parkinson’s disease | 302 | 17 | Mitochondrial fission | 120 | 27 | hypoxia | 68 |
| 8 | Pink1 | 275 | 18 | Alzheimer’s disease | 110 | 28 | bnip3 | 66 |
| 9 | Mitochondrial dynamics | 267 | 19 | Cancer | 101 | 29 | cell death | 65 |
| 10 | Aging | 192 | 20 | Mitochondrial quality control | 101 | 30 | ampk | 61 |
Top 30 keywords related to mitophagy.
FIGURE 8
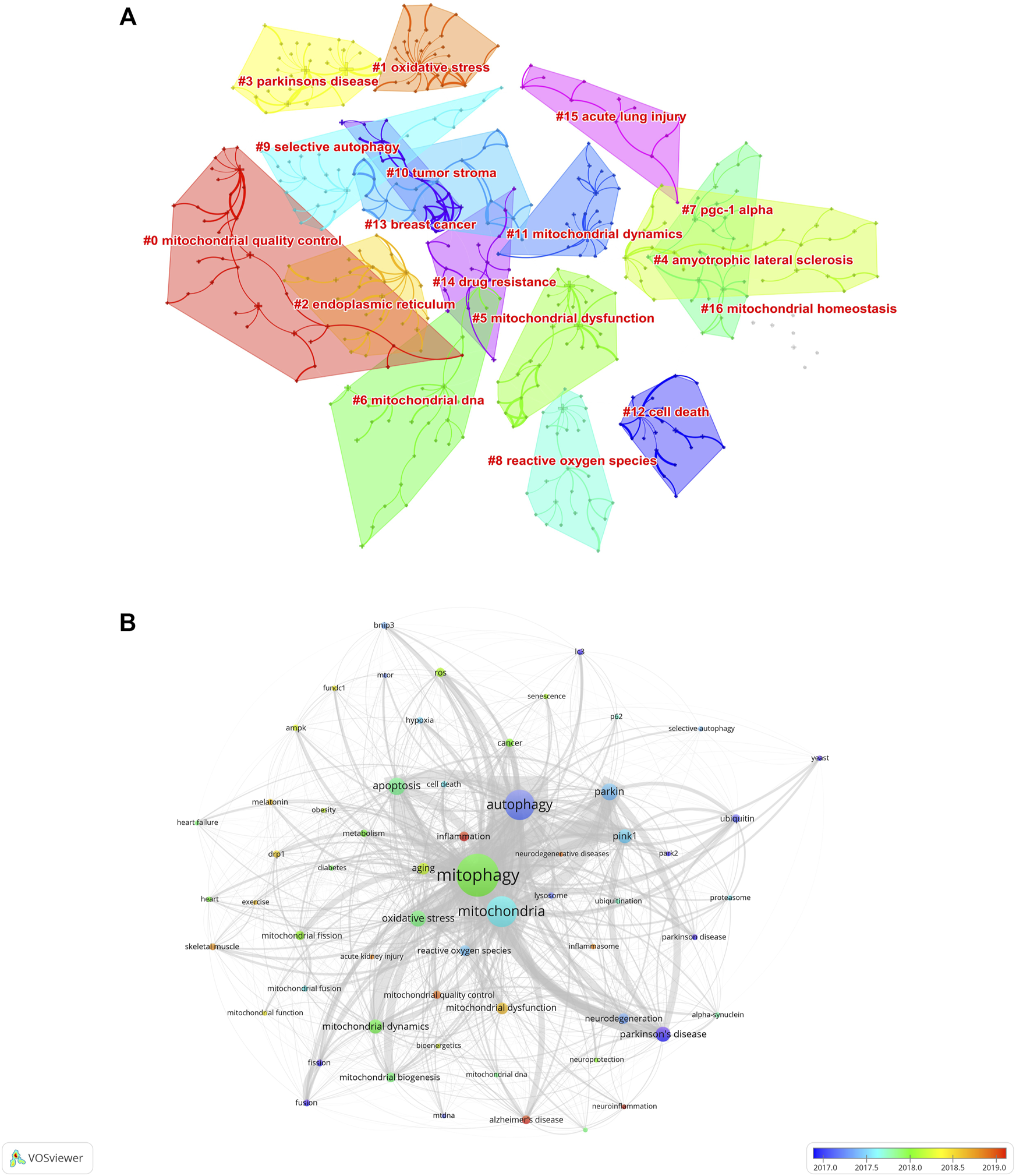
Analysis of keywords of the 5844 articles on mitophagy. (A) Clustering map of author keywords related to the research of mitophagy. (B) Author keyword overlay visualization map. The size of each circle indicates the frequency of occurrences of the author keyword. According to the color label in the lower right corner, the color of each circle indicates the average year when the keyword appeared in articles. The distance between any two circles is indicative of their co-occurrence link, and the thickness of the connecting line indicates the strength of the link.
TABLE 6
| Cluster ID | Size | Silhouette | Mean (Year) | Label | Cluster ID | Size | Silhouette | Mean (Year) | Label |
|---|---|---|---|---|---|---|---|---|---|
| 0 | 33 | 0.99 | 2015 | Mitochondrial quality control | 9 | 24 | 0.99 | 2016 | Selective autophagy |
| 1 | 30 | 0.98 | 2014 | Oxidative stress | 10 | 21 | 0.97 | 2011 | Tumor stroma |
| 2 | 30 | 0.98 | 2016 | Endoplasmic reticulum | 11 | 21 | 1.00 | 2015 | Mitochondrial dynamics |
| 3 | 29 | 0.94 | 2014 | Parkinson’s disease | 12 | 19 | 0.99 | 2014 | Cell death |
| 4 | 29 | 0.94 | 2016 | Amyotrophic lateral sclerosis | 13 | 17 | 0.99 | 2012 | Breast cancer |
| 5 | 28 | 0.93 | 2013 | Mitochondrial dysfunction | 14 | 16 | 0.89 | 2015 | Drug resistance |
| 6 | 27 | 0.98 | 2016 | Mitochondrial DNA | 15 | 12 | 0.95 | 2016 | Acute lung injury |
| 7 | 26 | 0.94 | 2016 | PGC-1 alpha | 16 | 8 | 0.98 | 2018 | Mitochondrial homeostasis |
| 8 | 25 | 1.00 | 2016 | Reactive oxygen species |
Top 17 largest clusters of keywords in the field of mitophagy.
To further visualize the research frontiers in the field of mitophagy, the keyword co-occurrence analysis was performed on VOSviewer. In the overlay visualization map (Figure 8B), different colors of author keywords were marked according to the average publication years. For example, the keywords of “yeast’, “ubiquitin”, “park2”, “fusion”, “fission”, “autophagy”, and “Parkinson’s disease/Parkinson disease” were marked in blue, indicating that they were mainly found in the early years. While keywords such as “inflammation,” “Alzheimer’s disease,” “neuroinflammation”, “mitochondrial quality control”, “neurodegenerative disease”, “acute kidney injury”, skeletal muscle”, “exercise”, “drp1”, and “melatonin” marked in yellow or red, indicating that these keywords were represented the recent major topics in the field of mitophagy and may also become the frontiers in the future.
Discussion
Nowadays, more and more research are being done around the world, and more and more scientific discoveries are being made. The biggest challenge is how to keep up with the remarkable explosion of knowledge in some active fields such as mitophagy. In this study, we took advantage of bibliometric analysis to visualize the global research landscape of mitophagy from 2005 to 2021. A total of 5844 articles were identified. Mitophagy research has flourished and attracted attention from all over the world, especially in United States, China, and the European Union. We found a list of outstanding institutions, researchers, journals, publishers, and funders which made a significant contributions to it. Research trends and hotspots were summarized and frontiers were predicted. Our findings provide a historical prospect and new insights into mitophagy.
Trend in Countries/Regions
Although the annual number of publications on mitophagy research increased year by year, a regional imbalance in the development of mitophagy research was observed in this study (Figure 2B). With no doubt, United States, China, and the European Union were on the dominant position in the field of mitophagy (Figure 2B, Figure 4A, and Table 1). This could be partially explained by a large number of researchers and institutions and substantial fundings in them (Table 1 and Figure 6C). Previous studies suggested that economic progress and interregional cooperation are two important factors for promoting the development of research in a certain field (Wagner et al., 2018; Mormina, 2019). According to the GDP (https://datacatalog.worldbank.org/search/dataset/0038130), our data showed that high-income countries were more productive than low-income countries. This was consistent with other bibliometric studies conducted on mitophagy (Chen et al., 2021) and other topics, such as ferroptosis in cancer (Li et al., 2022), ATAC-seq (Zhao et al., 2020), intestinal microbiota in obesity (Yao et al., 2018), COVID-19 (Tantengco, 2021), and Viral Hepatitis (Ornos and Tantengco, 2022). Of note, although China and United States have a roughly equal number of publications, the degree of centrality was much lower in China (0.21) than that in United States (0.51). This indicated that United States put more emphasis on international cooperation, while China was more likely to conduct studies through national collaborative research.
Knowledge Base and Landmarks
The knowledge base is the collection of co-cited references within the corresponding research community and the most cited articles are usually regarded as the landmarks (Chen et al., 2012; Zhang et al., 2021). In this bibliometric analysis of mitophagy, the top-cited articles and co-cited references were identified and reviewed as follows.
In 2005, the term mitophagy was firstly coined (Lemasters, 2005; Priault et al., 2005) for selective autophagy of mitochondria. In the early years, studies in yeast led to the discovery of several key proteins involved in mitophagy, such as Uth1p (Kissová et al., 2004), Aup1p (Tal et al., 2007), Mdm38 (Nowikovsky et al., 2007) and Atg32 (Kanki et al., 2009; Okamoto et al., 2009). In 2008, Derek demonstrated the role of Parkin protein in mediating the engulfment of mitochondria by autophagosomes and their subsequent degradation in mammalian cells (Narendra et al., 2008). And then, in 2010, four outstanding studies revealed the molecule mechanism of PINK1/Parkin-mediated mitophagy and its pathogenic mechanisms in the pathogenesis of Parkinson’s disease (Geisler et al., 2010; Matsuda et al., 2010; Narendra et al., 2010; Vives-Bauza et al., 2010). Hence, the functional links between PINK1 and Parkin as a classical mitophagy pathway were convincingly demonstrated. From then on, the role of mitophagy was massively studied and mitochondria were considered a promising therapeutical target for Parkinson’s disease (Malpartida et al., 2021). In that year, Ivana also identified the mitochondrial protein Nix (also known as BNIP3L) as a selective autophagy receptor in mammalian cells (Novak et al., 2010) and Atsushi Tanaka et al established the essential role of p97 in Parkin-mediated ubiquitination of mitofusins during mitophagy (Novak et al., 2010). These breakthroughs led to the mechanisms of mitophagy being initially established (Youle and Narendra, 2011). After that, other mechanisms such as AMPK-mediated phosphorylation of ULK1 (Egan et al., 2011), hypoxia-induced dephosphorylation of FUNDC1 (Liu et al., 2012), PINK1-dependent phosphorylation of ubiquitin (Koyano et al., 2014; Lazarou et al., 2015) were successively uncovered. In addition to the historic discovery of the functional links between mitophagy/autophagy and NLRP3 inflammasome activation (Zhou et al., 2011), these findings offer new insights into the mechanisms of mitophagy (Lee et al., 2012; Pickrell and Youle, 2015; Palikaras et al., 2018). In parallel, broader roles of mitophagy in other different diseases have attracted more and more attention after 2010 (Figure 7A).
Hotspots Evolution and Frontiers
By analyses the keywords, research hotspots and trends can be effectively obtained (Wang H. et al., 2021; Zhang et al., 2021). The research trends on mitophagy can be roughly divided into three stages. During the first phase (2005–2010), researchers have focused mostly on the discoveries of new mechanisms in yeast and the roles of mitochondrial fission and fusion in neurodegenerative diseases (Priault et al., 2005; Kissova et al., 2006; Abeliovich, 2007; Tatsuta and Langer, 2008; Chen and Chan, 2009; Munakata and Klionsky, 2010). During the second phase (2011–2015), researchers’ attention have mostly been focused on the mitochondrial stress and dynamic network and roles of mitophagy in cancer, heart failure and related diseases (Egan et al., 2011; Dang, 2012; Dutta et al., 2013; Drew et al., 2014; Chourasia et al., 2015). During the third phase (2016–2021), researchers mostly attached attention to parkin-mediated mitophagy and its roles in skeletal muscle and inflammation-related diseases (Akabane et al., 2016; Kimura et al., 2017; Kravic et al., 2018; Lin et al., 2019; Han et al., 2020; Tran and Reddy, 2021). In all, we summarized the main research hotspots below: “mechanism of mitochondrial quality control”, “molecule and signaling pathway in mitophagy” and “mitophagy-related diseases”.
Most Prolific and Influential Scientists
Top productive authors and top co-cited authors are the leading scientists in their respective fields. In this study, we found three scholars who were not only the top 10 productive authors but also the top 10 co-cited authors, namely Richard J. Youle, Hao Zhou, and Daniel J. Klionsky (Table 1). This implied their notable contributions to the field of mitophagy. Why can they be both prolific and influential? By further reading their publications, we can understand that Richard J. Youle mainly focused on the mechanisms of mitophagy (Youle and Narendra, 2011) and the roles of PINK1, Parkin, and mitochondrial fidelity in Parkinson’s Disease (Pickrell and Youle, 2015); Hao Zhou mainly focused on the role of mitophagy in cardiovascular disease, especially in cardiac microvascular ischemia/reperfusion injury (Zhou et al., 2018; Wang et al., 2020); Daniel J. Klionsky mainly focused on the selective mechanism during mitophagy (Kanki et al., 2009; Wang and Klionsky, 2011; Gatica et al., 2018). Their research directions were well matched with the research hotspots and frontiers of mitophagy. This may be one of the reasons why they can be the most important players in the research field of mitophagy.
Conclusion
The present study was hardly not without limitations. First, this study queried only the WoSCC database. High-quality articles published in journals that are not included in the WoSCC database may be missed. The combination of Web of Science with other databases, such as PubMed, Google scholar, Dimensions, and Scopus, would enable a more robust bibliometric analysis. Second, this study also comes with certain limitations inherent in CiteSpace and VOSviewer software. For instance, some keywords in the articles may be not included in the analysis due to the incomplete keyword extraction methods and the clustering analysis based on only the main information (not the full text). Third, although we have standardized some terms that have different expression types, bias may still exist if other synonyms were available.
In summary, by bibliometric analysis of 5844 publications over the previous 17 years, we found that mitophagy research has flourished and attracted attention from all over the world, especially in the United States (2025, 34.65%) and China (1968, 33.68%). A list of outstanding institutions and scholars made a significant contribution, such as Chinese Acad Sci (112, 1.92%), Univ Pittsburgh (109, 1.87%), Richard J. Youle (N = 38), and Derek P. Narendra (co-citation: 2693). Many journals played an important role in promoting the development of mitophagy research, such as Autophagy (N = 208) and Journal of Biological Chemistry (co-citation: 17226). We summarized current research hotspots (“mechanism of mitochondrial quality control”, “molecule and signaling pathway in mitophagy”, and “mitophagy related diseases”) and predicted future frontiers (“parkin-mediated mitophagy” and its roles in “skeletal muscle” and “inflammation-related diseases”). This information will provide researchers with references for future mitophagy studies.
Statements
Data availability statement
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding author.
Author contributions
GL and TX conceived the study. GL and WY collected the data. TX and HY re-examined the data. GL and TX analyzed the data and wrote the manuscript. WY, YY, YC, YL, and WZ reviewed and revised the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This project was supported by the Hunan Province Office of Education (No. 21B0052), the Hunan Clinical Research Center for Chronic Kidney Disease (No. 2019SK4009), Changsha Natural Science Foundation project (No. kq2202439), the Scientific Research Projects of the Health Commission of Hunan Province (No. 202203050006), National Natural Science Foundation of China (No. 82100788), Fellowship of China Postdoctoral Science Foundation (No. BX2021382, No. 2021M693571), Hunan Provincial Natural Science Foundation for Outstanding Youth (No. 2020JJ 2020), and Major Research and Development Program of Hunan Province (No. 2020SK2116). The funders had no role in the conduct of the study, the analysis or interpretation of data, and the preparation, review, or approval of the manuscript.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1
Abeliovich H. (2007). Mitophagy: The Life-Or-Death Dichotomy Includes Yeast. Autophagy3, 275–277. 10.4161/auto.3915
2
Akabane S. Uno M. Tani N. Shimazaki S. Ebara N. Kato H. et al (2016). PKA Regulates PINK1 Stability and Parkin Recruitment to Damaged Mitochondria through Phosphorylation of MIC60. Mol. Cell62, 371–384. 10.1016/j.molcel.2016.03.037
3
Amjad T. Shahid N. Daud A. Khatoon A. (2022). Citation Burst Prediction in a Bibliometric Network. Scientometrics127, 2773–2790. 10.1007/s11192-022-04344-3
4
Birkle C. Pendlebury D. A. Schnell J. Adams J. (2020). Web of Science as a Data Source for Research on Scientific and Scholarly Activity. Quantitative Sci. Stud.1, 363–376. 10.1162/qss_a_00018
5
Bravo-San Pedro J. M. Kroemer G. Galluzzi L. (2017). Autophagy and Mitophagy in Cardiovascular Disease. Circ. Res.120, 1812–1824. 10.1161/circresaha.117.311082
6
Chen C. (2004). Searching for Intellectual Turning Points: Progressive Knowledge Domain Visualization. Proc. Natl. Acad. Sci. U. S. A.101 Suppl 1 (Suppl. 1), 5303–5310. 10.1073/pnas.0307513100
7
Chen C. (2006). CiteSpace II: Detecting and Visualizing Emerging Trends and Transient Patterns in Scientific Literature. J. Am. Soc. Inf. Sci.57, 359–377. 10.1002/asi.20317
8
Chen C. Hu Z. Liu S. Tseng H. (2012). Emerging Trends in Regenerative Medicine: a Scientometric Analysis inCiteSpace. Expert Opin. Biol. Ther.12, 593–608. 10.1517/14712598.2012.674507
9
Chen H. Chan D. C. (2009). Mitochondrial Dynamics-Fusion, Fission, Movement, and Mitophagy-In Neurodegenerative Diseases. Hum. Mol. Genet.18, R169–R176. 10.1093/hmg/ddp326
10
Chen J. Li X. Jia Y. Xia Z. Ye J. (2021). Publication Trends on Mitophagy in the World and China: A 16-Year Bibliometric Analysis. Front. Cell Dev. Biol.9, 793772. 10.3389/fcell.2021.793772
11
Chen Y. Li Y. Guo L. Hong J. Zhao W. Hu X. et al (2020). Bibliometric Analysis of the Inflammasome and Pyroptosis in Brain. Front. Pharmacol.11, 626502. 10.3389/fphar.2020.561494
12
Chourasia A. H. Tracy K. Frankenberger C. Boland M. L. Sharifi M. N. Drake L. E. et al (2015). Mitophagy Defects Arising from BNip3 Loss Promote Mammary Tumor Progression to Metastasis. EMBO Rep.16, 1145–1163. 10.15252/embr.201540759
13
Dang C. V. (2012). Links between Metabolism and Cancer. Genes Dev.26, 877–890. 10.1101/gad.189365.112
14
Deng Z. Wang H. Chen Z. Wang T. (2020). Bibliometric Analysis of Dendritic Epidermal T Cell (DETC) Research from 1983 to 2019. Front. Immunol.11, 259. 10.3389/fimmu.2020.00259
15
Donthu N. Kumar S. Mukherjee D. Pandey N. Lim W. M. (2021). How to Conduct a Bibliometric Analysis: An Overview and Guidelines. J. Bus. Res.133, 285–296. 10.1016/j.jbusres.2021.04.070
16
Drew B. G. Ribas V. Le J. A. Henstridge D. C. Phun J. Zhou Z. et al (2014). HSP72 Is a Mitochondrial Stress Sensor Critical for Parkin Action, Oxidative Metabolism, and Insulin Sensitivity in Skeletal Muscle. Diabetes63, 1488–1505. 10.2337/db13-0665
17
Dutta D. Xu J. Kim J.-S. Dunn Jr. W. A. Leeuwenburgh C. (2013). Upregulated Autophagy Protects Cardiomyocytes from Oxidative Stress-Induced Toxicity. Autophagy9, 328–344. 10.4161/auto.22971
18
Egan D. F. Shackelford D. B. Mihaylova M. M. Gelino S. Kohnz R. A. Mair W. et al (2011). Phosphorylation of ULK1 (hATG1) by AMP-Activated Protein Kinase Connects Energy Sensing to Mitophagy. Science331, 456–461. 10.1126/science.1196371
19
Gatica D. Lahiri V. Klionsky D. J. (2018). Cargo Recognition and Degradation by Selective Autophagy. Nat. Cell Biol.20, 233–242. 10.1038/s41556-018-0037-z
20
Geisler S. Holmström K. M. Skujat D. Fiesel F. C. Rothfuss O. C. Kahle P. J. et al (2010). PINK1/Parkin-mediated Mitophagy Is Dependent on VDAC1 and p62/SQSTM1. Nat. Cell Biol.12, 119–131. 10.1038/ncb2012
21
Gustafsson Å. B. Dorn G. W. 2nd (2019b). Evolving and Expanding the Roles of Mitophagy as a Homeostatic and Pathogenic Process. Physiol. Rev.99, 853–892. 10.1152/physrev.00005.2018
22
Gustafsson Å. B. Dorn G. W. (2019a). EVOLVING AND EXPANDING THE ROLES OF MITOPHAGY AS A HOMEOSTATIC AND PATHOGENIC PROCESS. Physiol. Rev.99, 853–892. 10.1152/physrev.00005.2018
23
Han S. Jeong Y. Y. Sheshadri P. Cai Q. (2020). Mitophagy Coordination with Retrograde Transport Ensures the Integrity of Synaptic Mitochondria. Autophagy16, 1925–1927. 10.1080/15548627.2020.1810919
24
Harper J. W. Ordureau A. Heo J.-M. (2018). Building and Decoding Ubiquitin Chains for Mitophagy. Nat. Rev. Mol. Cell Biol.19, 93–108. 10.1038/nrm.2017.129
25
Hu S. Alimire A. Lai Y. Hu H. Chen Z. Li Y. (2021). Trends and Frontiers of Research on Cancer Gene Therapy from 2016 to 2020: A Bibliometric Analysis. Front. Med.8, 740710. 10.3389/fmed.2021.740710
26
Kanki T. Wang K. Cao Y. Baba M. Klionsky D. J. (2009). Atg32 Is a Mitochondrial Protein that Confers Selectivity during Mitophagy. Dev. Cell17, 98–109. 10.1016/j.devcel.2009.06.014
27
Kimura T. Isaka Y. Yoshimori T. (2017). Autophagy and Kidney Inflammation. Autophagy13, 997–1003. 10.1080/15548627.2017.1309485
28
Kissová I. Deffieu M. Manon S. Camougrand N. (2004). Uth1p Is Involved in the Autophagic Degradation of Mitochondria. J. Biol. Chem.279, 39068–39074. 10.1074/jbc.M406960200
29
Kissová I. Deffieu M. Samokhvalov V. Velours G. Bessoule J. J. Manon S. et al (2006). Lipid Oxidation and Autophagy in Yeast. Free Radic. Biol. Med.41, 1655–1661. 10.1016/j.freeradbiomed.2006.08.012
30
Koyano F. Okatsu K. Kosako H. Tamura Y. Go E. Kimura M. et al (2014). Ubiquitin Is Phosphorylated by PINK1 to Activate Parkin. Nature510, 162–166. 10.1038/nature13392
31
Kravic B. Harbauer A. B. Romanello V. Simeone L. Vögtle F.-N. Kaiser T. et al (2018). In Mammalian Skeletal Muscle, Phosphorylation of TOMM22 by Protein Kinase CSNK2/CK2 Controls Mitophagy. Autophagy14, 311–335. 10.1080/15548627.2017.1403716
32
Lazarou M. Sliter D. A. Kane L. A. Sarraf S. A. Wang C. Burman J. L. et al (2015). The Ubiquitin Kinase PINK1 Recruits Autophagy Receptors to Induce Mitophagy. Nature524,309, 314-+.10.1038/nature14893
33
Lee J. Giordano S. Zhang J. (2012). Autophagy, Mitochondria and Oxidative Stress: Cross-Talk and Redox Signalling. Biochem. J.441, 523–540. 10.1042/bj20111451
34
Lemasters J. J. (2005). Selective Mitochondrial Autophagy, or Mitophagy, as a Targeted Defense against Oxidative Stress, Mitochondrial Dysfunction, and Aging. Rejuvenation Res.8, 3–5. 10.1089/rej.2005.8.3
35
Li G. Liang Y. Yang H. Zhang W. Xie T. (2022). The Research Landscape of Ferroptosis in Cancer: A Bibliometric Analysis. Front. Cell Dev. Biol.10, 841724. 10.3389/fcell.2022.841724
36
Lin Q. Li S. Jiang N. Shao X. Zhang M. Jin H. et al (2019). PINK1-parkin Pathway of Mitophagy Protects against Contrast-Induced Acute Kidney Injury via Decreasing Mitochondrial ROS and NLRP3 Inflammasome Activation. Redox Biol.26, 101254. 10.1016/j.redox.2019.101254
37
Liu L. Feng D. Chen G. Chen M. Zheng Q. Song P. et al (2012). Mitochondrial Outer-Membrane Protein FUNDC1 Mediates Hypoxia-Induced Mitophagy in Mammalian Cells. Nat. Cell Biol.14, 177–185. 10.1038/ncb2422
38
Ma D. Yang B. Guan B. Song L. Liu Q. Fan Y. et al (2021). A Bibliometric Analysis of Pyroptosis from 2001 to 2021. Front. Immunol.12, 731933. 10.3389/fimmu.2021.731933
39
Malpartida A. B. Williamson M. Narendra D. P. Wade-Martins R. Ryan B. J. (2021). Mitochondrial Dysfunction and Mitophagy in Parkinson's Disease: From Mechanism to Therapy. Trends Biochem. Sci.46, 329–343. 10.1016/j.tibs.2020.11.007
40
Matsuda N. Sato S. Shiba K. Okatsu K. Saisho K. Gautier C. A. et al (2010). PINK1 Stabilized by Mitochondrial Depolarization Recruits Parkin to Damaged Mitochondria and Activates Latent Parkin for Mitophagy. J. Cell Biol.189, 211–221. 10.1083/jcb.200910140
41
Moral-Muñoz J. A. Herrera-Viedma E. Santisteban-Espejo A. Cobo M. J. (2020). Software Tools for Conducting Bibliometric Analysis in Science: An Up-To-Date Review. El Prof. Inf.29.
42
Mormina M. (2019). Science, Technology and Innovation as Social Goods for Development: Rethinking Research Capacity Building from Sen's Capabilities Approach. Sci. Eng. Ethics25, 671–692. 10.1007/s11948-018-0037-1
43
Munakata N. Klionsky D. J. (2010). "Autophagy Suite": Atg9 Cycling in the Cytoplasm to Vacuole Targeting Pathway. Autophagy6, 679–685. 10.4161/auto.6.6.12396
44
Narendra D. P. Jin S. M. Tanaka A. Suen D. F. Gautier C. A. Shen J. et al (2010). PINK1 Is Selectively Stabilized on Impaired Mitochondria to Activate Parkin. PLoS Biol.8, e1000298. 10.1371/journal.pbio.1000298
45
Narendra D. Tanaka A. Suen D.-F. Youle R. J. (2008). Parkin Is Recruited Selectively to Impaired Mitochondria and Promotes Their Autophagy. J. Cell Biol.183, 795–803. 10.1083/jcb.200809125
46
Novak I. Kirkin V. Mcewan D. G. Zhang J. Wild P. Rozenknop A. et al (2010). Nix Is a Selective Autophagy Receptor for Mitochondrial Clearance. EMBO Rep.11, 45–51. 10.1038/embor.2009.256
47
Nowikovsky K. Reipert S. Devenish R. J. Schweyen R. J. (2007). Mdm38 Protein Depletion Causes Loss of Mitochondrial K+/H+ Exchange Activity, Osmotic Swelling and Mitophagy. Cell Death Differ.14, 1647–1656. 10.1038/sj.cdd.4402167
48
Okamoto K. Kondo-Okamoto N. Ohsumi Y. (2009). Mitochondria-Anchored Receptor Atg32 Mediates Degradation of Mitochondria via Selective Autophagy. Dev. Cell17, 87–97. 10.1016/j.devcel.2009.06.013
49
Onishi M. Yamano K. Sato M. Matsuda N. Okamoto K. (2021). Molecular Mechanisms and Physiological Functions of Mitophagy. Embo J.40, 27. 10.15252/embj.2020104705
50
Ornos E. D. B. Tantengco O. A. G. (2022)). Research Trends, Gaps, and Prospects for Viral Hepatitis in Southeast Asia: A Bibliometric Analysis. Sci. Technol. Libr., 1–13. 10.1080/0194262x.2022.2028698
51
Palikaras K. Lionaki E. Tavernarakis N. (2018). Mechanisms of Mitophagy in Cellular Homeostasis, Physiology and Pathology. Nat. Cell Biol.20, 1013–1022. 10.1038/s41556-018-0176-2
52
Panigrahi D. P. Praharaj P. P. Bhol C. S. Mahapatra K. K. Patra S. Behera B. P. et al (2020). The Emerging, Multifaceted Role of Mitophagy in Cancer and Cancer Therapeutics. Seminars Cancer Biol.66, 45–58. 10.1016/j.semcancer.2019.07.015
53
Pickles S. Vigié P. Youle R. J. (2018). Mitophagy and Quality Control Mechanisms in Mitochondrial Maintenance. Curr. Biol.28, R170–R185. 10.1016/j.cub.2018.01.004
54
Pickrell A. M. Youle R. J. (2015). The Roles of PINK1, Parkin, and Mitochondrial Fidelity in Parkinson's Disease. Neuron85, 257–273. 10.1016/j.neuron.2014.12.007
55
Priault M. Salin B. Schaeffer J. Vallette F. M. Di Rago J.-P. Martinou J.-C. (2005). Impairing the Bioenergetic Status and the Biogenesis of Mitochondria Triggers Mitophagy in Yeast. Cell Death Differ.12, 1613–1621. 10.1038/sj.cdd.4401697
56
Pritchard A. (1969). Statistical Bibliography or Bibliometrics. J. documentation25, 348–349. 10.1007/bf01901469
57
Tal R. Winter G. Ecker N. Klionsky D. J. Abeliovich H. (2007). Aup1p, a Yeast Mitochondrial Protein Phosphatase Homolog, Is Required for Efficient Stationary Phase Mitophagy and Cell Survival. J. Biol. Chem.282, 5617–5624. 10.1074/jbc.m605940200
58
Tantengco O. A. G. (2021). Investigating the Evolution of COVID-19 Research Trends and Collaborations in Southeast Asia: A Bibliometric Analysis. Diabetes & Metabolic Syndrome Clin. Res. Rev.15, 102325. 10.1016/j.dsx.2021.102325
59
Tatsuta T. Langer T. (2008). Quality Control of Mitochondria: Protection against Neurodegeneration and Ageing. Embo J.27, 306–314. 10.1038/sj.emboj.7601972
60
Thompson D. F. Walker C. K. (2015). A Descriptive and Historical Review of Bibliometrics with Applications to Medical Sciences. Pharmacotherapy35, 551–559. 10.1002/phar.1586
61
Tran M. Reddy P. H. (2021). Defective Autophagy and Mitophagy in Aging and Alzheimer's Disease. Front. Neurosci.14, 18. 10.3389/fnins.2020.612757
62
Van Eck N. J. Waltman L. (2010). Software Survey: VOSviewer, a Computer Program for Bibliometric Mapping. Scientometrics84, 523–538. 10.1007/s11192-009-0146-3
63
Van Eck N. J. Waltman L. (2007). VOS: A New Method for Visualizing Similarities between Objects. Springer Berlin Heidelberg, 299–306.
64
Vives-Bauza C. Zhou C. Huang Y. Cui M. De Vries R. L. A. Kim J. et al (2010). PINK1-dependent Recruitment of Parkin to Mitochondria in Mitophagy. Proc. Natl. Acad. Sci. U.S.A.107, 378–383. 10.1073/pnas.0911187107
65
Wagner C. S. Whetsell T. Baas J. Jonkers K. (2018). Openness and Impact of Leading Scientific Countries. Front. Res. Metrics Anal.3. 10.3389/frma.2018.00010
66
Wang H. Deng S. Fan X. Li J. Tang L. Li Y. et al (2021a). Research Trends and Hotspots of Extracorporeal Membrane Oxygenation: A 10-Year Bibliometric Study and Visualization Analysis. Front. Med.8. 10.3389/fmed.2021.752956
67
Wang J. Toan S. Zhou H. (2020). New Insights into the Role of Mitochondria in Cardiac Microvascular Ischemia/reperfusion Injury. Angiogenesis23, 299–314. 10.1007/s10456-020-09720-2
68
Wang K. Klionsky D. J. (2011). Mitochondria Removal by Autophagy. Autophagy7, 297–300. 10.4161/auto.7.3.14502
69
Wang S. Zhou H. Zheng L. Zhu W. Zhu L. Feng D. et al (2021b). Global Trends in Research of Macrophages Associated with Acute Lung Injury over Past 10 Years: A Bibliometric Analysis. Front. Immunol.12. 10.3389/fimmu.2021.669539
70
Wang S. Zhou H. Zheng L. Zhu W. Zhu L. Feng D. et al (2021c). Global Trends in Research of Macrophages Associated with Acute Lung Injury over Past 10 Years: A Bibliometric Analysis. Front. Immunol.12, 669539. 10.3389/fimmu.2021.669539
71
Xiong J. Qi W. Liu J. Zhang Z. Wang Z. Bao J. et al (2021). Research Progress of Ferroptosis: A Bibliometrics and Visual Analysis Study. J. Healthc. Eng.2021, 2178281. 10.1155/2021/2178281
72
Yan W. T. Lu S. Yang Y. D. Ning W. Y. Cai Y. Hu X. M. et al (2021). Research Trends, Hot Spots and Prospects for Necroptosis in the Field of Neuroscience. Neural Regen. Res.16, 1628–1637. 10.4103/1673-5374.303032
73
Yao H. Wan J.-Y. Wang C.-Z. Li L. Wang J. Li Y. et al (2018). Bibliometric Analysis of Research on the Role of Intestinal Microbiota in Obesity. PeerJ6, e5091. 10.7717/peerj.5091
74
Youle R. J. Narendra D. P. (2011). Mechanisms of Mitophagy. Nat. Rev. Mol. Cell Biol.12, 9–14. 10.1038/nrm3028
75
Zhang J. Song L. Xu L. Fan Y. Wang T. Tian W. et al (2021). Knowledge Domain and Emerging Trends in Ferroptosis Research: A Bibliometric and Knowledge-Map Analysis. Front. Oncol.11, 686726. 10.3389/fonc.2021.686726
76
Zhao Y. Zhang X. Song Z. Wei D. Wang H. Chen W. et al (2020). Bibliometric Analysis of ATAC-Seq and its Use in Cancer Biology via Nucleic Acid Detection. Front. Med.7, 584728. 10.3389/fmed.2020.584728
77
Zhou H. Wang S. Hu S. Chen Y. Ren J. (2018). ER-mitochondria Microdomains in Cardiac Ischemia-Reperfusion Injury: A Fresh Perspective. Front. Physiol.9, 755. 10.3389/fphys.2018.00755
78
Zhou R. Yazdi A. S. Menu P. Tschopp J. (2011). A Role for Mitochondria in NLRP3 Inflammasome Activation. Nature469, 221–225. 10.1038/nature09663
Summary
Keywords
mitophagy, bibliometric analysis, CiteSpace, VOSviewer, visualization
Citation
Li G, Yin W, Yang Y, Yang H, Chen Y, Liang Y, Zhang W and Xie T (2022) Bibliometric Insights of Global Research Landscape in Mitophagy. Front. Mol. Biosci. 9:851966. doi: 10.3389/fmolb.2022.851966
Received
17 January 2022
Accepted
16 June 2022
Published
18 July 2022
Volume
9 - 2022
Edited by
Martin Van Der Laan, Saarland University, Germany
Reviewed by
Ralf J. Braun, Danube Private University, Austria
Raman Kumar, Guru Nanak Dev Engineering College, India
Néstor Montalván-Burbano, University of Almeria, Spain
Ourlad Alzeus Gaddi Tantengco, University of the Philippines Manila, Philippines
Updates
Copyright
© 2022 Li, Yin, Yang, Yang, Chen, Liang, Zhang and Xie.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Tingting Xie, xtt1991.good@csu.edu.cn
This article was submitted to Cellular Biochemistry, a section of the journal Frontiers in Molecular Biosciences
Disclaimer
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.