
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Mol. Biosci. , 10 February 2022
Sec. Cellular Biochemistry
Volume 9 - 2022 | https://doi.org/10.3389/fmolb.2022.838148
This article is part of the Research Topic Advances in Sulfation Pathways Research View all 13 articles
Sulfation is poorly understood in most invertebrates and a potential role of sulfation in the regulation of developmental and physiological processes of these organisms remains unclear. Also, animal model system approaches did not identify many sulfation-associated mechanisms, whereas phosphorylation and ubiquitination are regularly found in unbiased genetic and pharmacological studies. However, recent work in the two nematodes Caenorhabditis elegans and Pristionchus pacificus found a role of sulfatases and sulfotransferases in the regulation of development and phenotypic plasticity. Here, we summarize the current knowledge about the role of sulfation in nematodes and highlight future research opportunities made possible by the advanced experimental toolkit available in these organisms.
Sulfation, sulfonation or sulfoconjugation, is an essential and ubiquitous biochemical reaction that modifies a wide range of xenobiotic and endogenous molecules. From bacteria to plants and animals, perturbation of the molecular processes that regulate the biologic ratio of sulfated and unconjugated substrates is likely to alter cellular and organismal physiology (Leung et al., 2016; Langford et al., 2017; Gunal et al., 2019). The sulfation pathways are catalyzed by two types of enzymes, the sulfotransferases (sulfuryltransferases) and the sulfatases. The sulfotransferases transfer a sulfuryl group (SO3) from the universal donor PAPS (3′-phosphoadenosine 5′-phosphosulfate) to the hydroxyl or amino group of numerous compounds (Strott, 2002; Coughtrie, 2016). The presence of the charged sulfate group changes the physiochemical properties (water solubility and conformation) of the acceptor molecules and has therefore a major role in the detoxification and elimination of various xenobiotics (Strott, 2002; Leung et al., 2016). Multiple endogenous compounds including carbohydrates, lipids, proteins and hormone precursors (steroids) are also modified by sulfotransferases. In their case, sulfation has significant influence on the biological activity of the modified molecules and consequently in multiple biological processes (for details see (Strott, 2002; Mueller et al., 2015; Leung et al., 2016)).
The sulfatases, on the other hand, hydrolyze the sulfate ester bonds to the unconjugated form of the substrate (Hanson et al., 2004). These hydrolytic enzymes are part of the alkaline phosphatase superfamily and share a post-translational modification that greatly enhances enzymatic activity: a cysteine or serine in the catalytic center is converted to a formylglycine (Schmidt et al., 1995; Dierks et al., 1997; Dierks et al., 1998a; Dierks et al., 1998b). This review elaborates on the players, molecular mechanisms and roles of the sulfation pathways in Caenorhabditis elegans and Pristionchus pacificus. We especially highlight the use of these nematode model organisms to investigate the poorly understood relevance of the sulfation pathways in organismal development. Finally, the emerging topics and questions on sulfation in nematodes are discussed.
Nematodes, or roundworms, are one of the largest animal phyla and are found in all ecosystems: marine, fresh-water and soil (van den Hoogen et al., 2019). In addition, many nematode species are parasites of plants, animals, life stock or humans, resulting in their economic importance for agriculture, breeding and health (Lee et al., 2002). Most of these parasites are hard to culture in the laboratory, whereas some free-living species are among the most rapidly reproducing and most easily culturable species of all metazoans. One example is Caenorhabditis elegans, which was introduced as a potential model organism by Sydney Brenner in the 1960ies (Brenner, 1974). Building on a unique set of life history traits and a growing functional toolkit, the ‘C. elegans research community’ has grown rapidly to make fundamental discoveries in all areas of the life sciences. C. elegans can be grown on small agar plates in the laboratory, feeds on Escherichia coli and completes its life cycle in 3 days at 20 °C (Figure 1A). This species reproduces primarily by selfing of hermaphrodites, which produce around 200–300 sperm during larval development before switching to a female fate with oocyte production as adults. Consequently, the progenies of hermaphrodites are genetically identical and the resulting isogenic propagation represents a unique advantage among all animal model organisms. Original unbiased forward genetic screens have identified many mutants essential for animal survival, egg-laying, neurogenesis or life span (Wood and Researchers, 1988). With RNA interference and CRISPR/Cas9-based genome engineering being implemented, large scale reverse genetic screens have been performed aiming at the analysis of the knock-down or knock-out of all genes in the genome (Fire et al., 1998; Dickinson and Goldstein, 2016). Indeed, when the C. elegans genome was fully sequenced, the 100 Mb genome was shown to encode around 20,000 genes with several thousands of them being functionally characterized (Consortium, 1998). Together, these features made C. elegans one of the most important study systems in modern biology.

FIGURE 1. Nematode development and plastic traits. (A) Schematic life cycle of Caenorhabditis elegans and Pristionchus pacificus, including four postembryonic larval (juvenile) stages separated by molts. Worms can either directly develop into a reproductive adult if conditions are favorable and food is available (symbolized by plates with food), or indirectly through a stress-resistant and long-lived dauer larval stage if environmental conditions are unfavorable. (B) Predatory behavior of P. pacificus (Ppa): the eurystomatous morph of the worm is able to devour C. elegans (Ce) larvae. (C) Differential interference contrast (DIC) image depicting the mouth-form of eurystomatous (Eu) and stenostomatous (St) worms. The eurystomatous morph has two teeth (asterisks) and a wide buccal cavity whereas the stenostomatous morph shows only one tooth in a narrow buccal cavity (arrows). Bar = 10 μm.
The related nematode Pristionchus pacificus shares many of these features with C. elegans and similarly builds on a short generation time (4 days, 20°C), self-fertilizing hermaphroditism, forward and reverse genetic tools and a fully sequenced genome (Sommer and Sternberg, 1996; Dieterich et al., 2008; Han et al., 2020). P. pacificus and C. elegans, like many nematodes, can undergo direct and indirect development (Figure 1A). Under favorable environmental conditions and the availability of food, they undergo direct development, rapidly proceeding through four juvenile (larval) stages to become adults. In contrast, in the absence of food and other unfavorable circumstances, they arrest in a long-lived and stress-resistant “dauer” stage, which is adapted to survive harsh environmental conditions (Figure 1A) (for review see (Sommer and Mayer, 2015)). Besides these conserved features, P. pacificus differs from C. elegans with regard to several morphological and organismal features that turned out to be under the control of sulfation-mediated processes. Specifically, while P. pacificus can be cultured on E. coli as single food source indefinitely, it is a potential predator of other nematodes (Figure 1B). It forms teeth-like denticles in its mouth that are unknown in C. elegans or most other free-living nematodes (Figure 1C) (Bento et al., 2010). These teeth come in two alternative mouth-forms, representing an example of developmental (phenotypic) plasticity (Sommer, 2020). Worms can either form a “eurystomatous” (Eu) morph with two teeth and a wide buccal cavity, or, alternatively, a “stenostomatous” (St) morph with a single tooth and a narrow opening (Figure 1C) (Ragsdale et al., 2013). While both morphs can feed on bacteria, only the Eu form is a potential predator with associated self-recognition systems preventing the killing of kin (Lightfoot et al., 2019). The role of sulfation enzymes, the sulfatase eud-1 and the sulfotransferase seud-1/sult-1, in the regulation of mouth-form plasticity were discovered through unbiased forward genetic screens and will be described below (Ragsdale et al., 2013; Bui et al., 2018; Namdeo et al., 2018).
Eukaryotes express cytosolic and membrane-bound sulfotransferases. Cytosolic sulfotransferases (termed SULTs) sulfate xenobiotics and small molecules like neurotransmitters and hormones. The membrane-bound sulfotransferases reside in the Golgi and mediate the post-translational sulfation of endogenous macromolecules (proteins, lipids and glycosaminoglycans) in the secretory pathway (Bojarova and Williams, 2008; Leung et al., 2016). Despite low sequence conservation, sulfotransferases possess a conserved protein structure and catalytic properties, which include the binding to the PAPS cofactor. However, sulfotransferases show broad substrate specificity and may sulfate an unlimited number of substrates (Mueller et al., 2015; Langford et al., 2017). The human genome contains 13 cytoplasmic and 39 membrane-bound sulfotransferases (Langford et al., 2017; Gunal et al., 2019). Based on published literature and Wormbase (version WS282), Table 1 lists the cytosolic and membrane-bound sulfotransferases currently identified in C. elegans (36), P. pacificus (18) and Strongyloides ratti (48), a common gastro-intestinal parasitic worm of the rat (Streit, 2008).
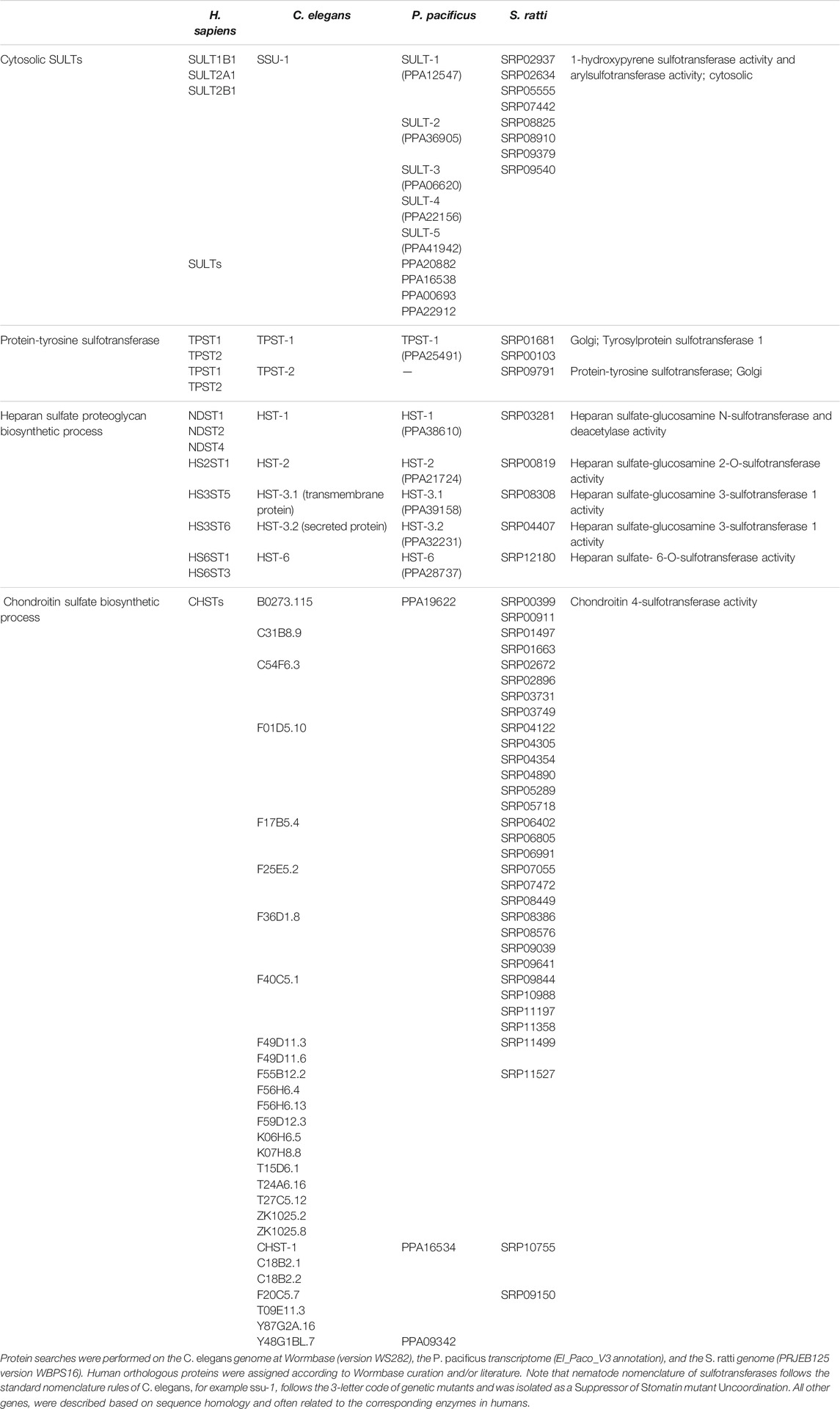
TABLE 1. Cytosolic and membrane-bound sulfotransferases in nematodes and corresponding human orthologues.
ssu-1 (suppressor of stomatin mutant uncoordination) is the only member of the cytosolic sulfotransferase gene family present in the genome of C. elegans (Carroll et al., 2006; Hattori et al., 2006; Mizuguchi et al., 2009). Orthologous proteins include the human SULT1B1, SULT2A1 and SULT2B1 enzymes (Wormbase curation). This sulfotransferase is expressed throughout development, particularly in embryos and adult stages, but shows increased levels in dauer larvae. Recombinant protein is able to sulfate simple phenol substrates, such as bisphenol A (Hattori et al., 2006). Expression of SSU-1 has also been detected in the amphid sensory neurons located in the head of the worm where it is hypothesized to modify endocrine signals (Carroll et al., 2006). A recent study observed that mutant ssu-1 animals failed to arrest development in response to osmotic stress. The authors suggest that in the sensory neurons SSU-1 sulfates a ligand of the nuclear hormone receptor NHR-1 to activate a transcriptional response to osmotic stress and induce developmental arrest (Burton et al., 2018).
Five paralogs of C. elegans SSU-1 are found in P. pacificus, termed SULT-1 to SULT-5 (Table 1) (Namdeo et al., 2018). CRISPR/Cas9-mediated knockout of these cytosolic sulfotransferases and a genetic screen for suppressors of an all-stenostomatous mutant of P. pacificus has identified a role for SULT-1 (PPA12547) in the regulation of mouth-form plasticity (see details below) (Bui et al., 2018; Namdeo et al., 2018). Additional SULTs can also be found in P. pacificus (4) and S. ratti (8). Their biological role remains unknown.
The genomes of nematodes also contain genes encoding TPSTs (Table 1) (Namdeo et al., 2018), integral membrane glycoproteins of the trans-Golgi network that post translationally sulfate tyrosine residues of soluble and membrane proteins of the secretory pathway. Sulfation is the most common modification of tyrosine residues and regulates biological activity and correct cellular processing (Monigatti et al., 2006; Stone et al., 2009; Yang et al., 2015). Two types of tyrosylprotein sulfotransferases (TPST1 and TPST2) with distinct substrate preference are present in animals; P. pacificus lacks the second TPST gene and S. ratti encodes three TPST enzymes (Table 1) (Beisswanger et al., 1998; Ouyang and Moore, 1998; Mizuguchi et al., 2009; Namdeo et al., 2018). In C. elegans, TPST-1 activity is required for cuticle organization as it controls collagen secretion (Kim et al., 2005; Kim et al., 2010).
Sulfotransferases also extensively modify carbohydrates/sugars along the secretory pathway. Carbohydrate sulfoforms function on the cell surface and in the extracellular matrix where they provide structural support and mediate communication between cells and the surrounding environment (Bowman and Bertozzi, 1999). Sulfation of carbohydrates creates structural variety, enables electrostatic interactions and generates ligands for specific receptors. The biological significance of the sulfation of carbohydrates is therefore broad as it regulates multiple processes, such as organ development, extracellular signaling, and inflammation, among others (Bowman and Bertozzi, 1999; Strott, 2002). Carbohydrate sulfotransferases are integral membrane enzymes of the Golgi apparatus that sulfate glycans attached to lipids and proteins (Bowman and Bertozzi, 1999; Strott, 2002). Recognizable carbohydrate sulfotransferases in nematodes modify the scaffold of proteoglycans (glycosaminoglycan chains constituted by repeating disaccharide units attached to core proteins) to produce variable sulfoforms. Two groups of carbohydrate sulfotransferases are present in C. elegans, P. pacificus and S. ratti: heparan sulfate (HS) and chondroitin sulfate (CS) sulfotransferases (Table 1). Sequence homology searches revealed 28 CS-6-O-sulfotransferases-like genes (CHST) in C. elegans (Mizuguchi et al., 2009), with three orthologs in P. pacificus (Namdeo et al., 2018) and 32 in S. ratti (Table 1). Five HS-sulfotransferases (HSTs) are also encoded in the genomes of the three nematodes (Mizuguchi et al., 2009; Namdeo et al., 2018) (Table 1).
a) Biological roles of CS-sulfotransferases in C. elegans
Chondroitin sulfate chains are composed of disaccharide units of glucuronic acid (GlcUA) and iduronic acid-N-acetylgalactosamine (GalNAc) heterogeneously sulfated (Sugahara et al., 2003). Sulfation of CS occurs on the 4- and 6-hydroxyl groups of GalNAc and on the 2-hydroxyl group of GlcUA. CS is the main structural component of the extracellular matrix in the brain and cartilage. In addition, CS sulfoforms regulate cell signaling through specific receptor-ligand binding influencing processes such as neuronal and skeletal development, and infection by pathogens (Bowman and Bertozzi, 1999; Sugahara et al., 2003).
Nematodes produce vast amounts of non-sulfated chondroitin (Yamada et al., 1999; Toyoda et al., 2000; Olson et al., 2006; Lawrence et al., 2008), which in C. elegans is crucial for cell division during embryogenesis and vulva morphogenesis (Herman et al., 1999; Hwang et al., 2003; Mizuguchi et al., 2003). Mutations or RNAi depletion of the nematode chondroitin synthase gene are proposed to decrease chondroitin hydration activity controlling tissue osmotic pressure (structural role) and/or disrupt cell signaling (Hwang et al., 2003; Mizuguchi et al., 2003). Although the presence of CS in C. elegans has been controversial, several studies detected different sulfoforms in the worms (Schimpf et al., 1999; Beeber and Kieras, 2002; Dierker et al., 2016). Recently, the first CS sulfotransferase enzyme has also been characterized in C. elegans, CHST-1 (Table 1) (Dierker et al., 2016), suggesting that sulfate CS has a biological role also in nematodes. Recombinant CHST-1 utilizes PAPS to sulfate a chondroitin substrate, and the levels of 4-O-sulfation in chondroitin are dramatically reduced in the chst-1 mutant strain (Dierker et al., 2016). Whether the 26 other putative CS-sulfotransferases present in C. elegans genome encode for functional sulfotransferases with effects on CS sulfation and worm biology remains unclear, similar to the function of CS sulfotransferases in other nematodes.
b) Biological roles of HS-sulfotransferases in C. elegans
HS molecules are linear glycosaminoglycan polysaccharides composed of glucuronic acid (GlcUA) and iduronic acid-N-acetylglucosamine (GlcNAc) repeat units covalently bound to a protein core. As components of extracellular matrices and cell surfaces, HS function in a wide range of cellular processes, are essential in development and homeostasis, and have several implications in disease (Bowman and Bertozzi, 1999; Strott, 2002; Lindahl and Li, 2009). Three different types of HS are present in cells: syndecans (transmembrane proteins), glycosylphosphatidylinositol (GPI)-anchored proteins (e.g., glypicans) and secreted molecules (e.g., perlecan, agrin, and collagen XVIII) (Lindahl and Li, 2009). The genome of C. elegans contains single orthologues of these major proteoglycans (syndecan/F57C7.3, glypican/F59D12.4, perlecan (unc-52) and agrin/F41G3.8) (Rogalski et al., 1993; Hutter et al., 2000; Ackley et al., 2001; Bulow and Hobert, 2004; Minniti et al., 2004; Gumienny et al., 2007). After synthesis in the Golgi complex, heparan sulfate polymers are highly modified by deacetylation, sulfation and epimerization catalyzed by specific HS modifying enzymes at selective positions. The bifunctional enzyme N-deacetylase/N-sulfotransferases (Ndst) removes the acetyl group from GlcNAc and substitutes it with a sulfuryl group (Wei et al., 1993). The reactions catalyzed by Ndst allow the subsequent epimerization of glucuronic acid to iduronic acid and sulfation of HS, as the other enzymes recognize the N-sulfate groups on the molecule (Habuchi, 2000). The HS C-5 glucuronyl-epimerase is responsible for epimerization, while the HS 2-O, HS 6-O, HS 3-O sulfotransferases add SO3 at the C2 hydroxyl group of hexuronic acids and the C6 or C3 of glucosamine units, respectively (Habuchi, 2000; Turnbull et al., 2001; Lindahl and Li, 2009). Orthologues of all of these enzymes are present in nematodes (Table 1). These modifications increase the structural diversity of HS and influence cellular processes such as cell migration, axon guidance and pathogen infection as they are binding sites for multiple ligands, receptors, growth factors, enzymes, extracellular matrix proteins and adhesion proteins found in pathogens. These interactions are specific and perform regulatory roles (Turnbull et al., 2001; Lindahl and Li, 2009).
Work in C. elegans has increased the understanding of the relationships among HS modifications and specific proteoglycans in defined biological processes. The HS-modifying enzymes have differential effects in the development of the nervous system. For example, the modifications catalyzed by C-5 glucuronyl-epimerase (hse-5), HS 2-O (hst-2) and HS 6-O (hst-6)-sulfotransferases have been described to differentially regulate specific signaling pathways controlling axonal development (Bulow and Hobert, 2004). Efficient Robo, Ephrin and possibly Integrin-associated signaling pathways are dependent on distinct HS modifications to properly guide axon patterning in specific cellular contexts. These signaling pathways act in concert with precise modifications on components of the extracellular matrix to organize the complexity of the nervous system (Bulow and Hobert, 2004). In addition, the activity of HST-6 has been shown to modulate the function of anosmin (Kallman protein, kal-1), a secreted neural cell adhesion molecule with roles in axonal growth (Bulow et al., 2002). Parallel HS modifications also participate in the migration of C. elegans hermaphrodite specific neurons (HSNs), the pair of neurons that coordinates contraction of the vulval muscles to initiate egg laying (Desai et al., 1988; Kinnunen et al., 2005; Kinnunen, 2014). The migration of these neurons that are born in the posterior towards the mid body region is supported by HST-6-dependent sulfation of syndecan and HST-2-mediated sulfation of glypican. Genetic elimination of hst-2 (expressed in the muscle and hypodermis) and hst-6 (predominantly expressed in neurons) causes severe defects in the migration of the HSNs (Kinnunen, 2014). RNAi of the hst-2 gene in C. elegans also causes aberrant morphology of the gonad and defects in egg laying, suggesting that sulfated HS chains are required for normal development and function of the reproductive tissues (Turnbull et al., 2003; Kinnunen et al., 2005).
The function of HST-3 enzymes has likewise been investigated in C. elegans. In loss-of-function mutants of hst-3.1 and hst-3.2, worm viability and overall neuronal development are not affected. Yet, HS-3-O-sulfation has a specialized role in HSNs. Together with KAL-1, both enzymes are important for the branching of neurites in these neurons (Tecle et al., 2013). Forward genetic screens, have also identified hst-6 and hst-3.2 as modifiers of kal-1-dependent neurite branching of AIY neurons, a left/right pair of interneurons with axons that innervate the nerve ring in the head of the worm (Diaz-Balzac et al., 2014). Thus, different HS modifications regulate kal-1-dependent branching in distinct tissues. As KAL-1/anosmin is the underlying gene causing hereditary Kalmman syndrome in humans (Franco et al., 1991; Legouis et al., 1991), these studies highlight the importance of using nematode model systems to understand the role of HS in development and disease-associated processes.
Mating behavior between hermaphrodites and males is also controlled by sulfation of HS molecules. During mating, the male will press its tail against the body of the hermaphrodite and move backwards in search for the vulva (Liu and Sternberg, 1995). Abnormal response to mate contact has been observed in hst-2, hst-3.1 and hse-5 mutant worms, suggesting that HS modifications are required for the response to hermaphrodite contact by the male worm (Lazaro-Pena et al., 2018). Specifically, 3-O-sulfation of glypicans regulates synaptogenesis in neurons controlling male mating behavior, the B-type ray sensory neurons (Liu and Sternberg, 1995; Koo et al., 2011; Lazaro-Pena et al., 2018).
Finally, HS modifications have also been shown to regulate cilia structure in C. elegans (Acker et al., 2021). The cilia protrusions are organized in highly compartmentalized microtubule-based domains and disruption of their composition and structure results in ciliopathies (Reiter and Leroux, 2017). Null alleles of hst-3.1 show defects in the localization of complement factor H (CFH) to the cilia of mechanosensory neurons. A potential interaction of CFH with modified HS appears to control cilia structural organization. HST-3.1 is proposed to modify the HS chain of proteoglycans that recruit CFH in the mechanosensory neurons. In the absence of this modification, CFH is not properly localized in the cilia and the animals show defects in mechanosensory neuron function (Acker et al., 2021). The role of HS modifications in cilia structure is another example of how studies in nematodes increase our knowledge on disease associated processes, since CFH is a major risk factor for age-related macular degeneration and blindness in humans (Edwards et al., 2005; Hageman et al., 2005; Haines et al., 2005; Klein et al., 2005; Zareparsi et al., 2005).
In summary, the differential modification of molecules of HS provide a “code” that mediates the selective interaction with a set of signaling factors and molecules that define the migration, patterning and function of different neurons (Bulow and Hobert, 2004; Lindahl and Li, 2009; Kinnunen, 2014).
From prokaryotes to eukaryotes sulfatases are highly conserved in sequence, structure and mechanism of action (Hanson et al., 2004; Ghosh, 2007). They share a highly conserved N-terminal region containing two consensus sulfatase motifs. The first motif is characterized by the conserved sequence C/SxPxRxxxxTG (x is any amino acid) and is crucial to generate the post-translationally modified active-site aldehyde residue, the α-formylglycine (FGly; 2-amino-3-oxypropanic acid) (Knaust et al., 1998; Dierks et al., 1999). The second sulfatase signature motif is a 12-mer sequence GY/VxS/T-xxxGKxxH. The Lys and His residues are part of the active-site and required for catalysis (Waldow et al., 1999). The C-terminal region of sulfatases is the most diverse; however, it is proposed that this region provides substrate specificity (Hanson et al., 2004). Although a large number of eukaryotic sulfatases are active on small aromatic sulfates (aryl substrates) in vitro and thus classified as arylsulfatases, many sulfatases show strong substrate specificity, specific subcellular location and optimal activity under defined conditions (Hanson et al., 2004). Sulfatase genes encode proteins with a wide range of substrate specificity but little functional redundancy, and deficiency in a single sulfatase can lead to unique disorders and developmental defects in animals (Langford et al., 2017).
Eukaryotic sulfatases are targeted to the secretory pathway. In the endoplasmic reticulum (ER), the sulfatases are modified by glycosylation and FGly is generated (Dierks et al., 1997; Dierks et al., 1998a; Hanson et al., 2004). They can then remain in the ER, be secreted, or transported to other subcellular compartments, such as the Golgi complex and the lysosome (Table 2) (Hanson et al., 2004). The steroid sulfatase (STS, human arylsulfatase C) is integrated into the membrane of the ER or Golgi apparatus (Willemsen et al., 1988; Hernandez-Guzman et al., 2003) where it plays an important role in the regulation of endocrine responses and xenobiotic metabolism. STS is the main enzyme involved in steroid desulfation and regulates hormone levels by removing sulfate from precursors, such as estrone sulfate and dehydroepiandrosterone (DHEAS), and originating active steroid hormones (for review see (Mueller et al., 2015)). Lysosome-located sulfatases (human arylsulfatases A and B) participate in the catabolism of defective and unwanted sulfated glycolipids (sulfatides) and proteoglycans that are imported into this organelle by endocytosis. Lysosome storage disorders are therefore a consequence of lysosome-sulfatase deficiency (Langford et al., 2017). The secreted sulfatases, known as Sulfs, remove the 6-O-sulfate groups of the glucosamine units of HS regulating multiple developmental processes (Hanson et al., 2004; Bojarova and Williams, 2008; Langford et al., 2017).
Nematodes also contain multiple sulfatase genes: three in C. elegans and S. ratti, and five in P. pacificus (Table 2). Sequence-wise Wormbase indicates that SUL-1 proteins are orthologues of the secreted SULF-1 and SULF-2 sulfatases that act on HS. In contrast, SUL-2 proteins show similarity with the human arylsulfatases A, B that participate in the degradation of cerebroside-3-sulfate and keratan/dermatan sulfate or CS, respectively, the steroid sulfatase (STS) and N-acetylglucosamine-6-sulfatase (GALNS) (Ragsdale et al., 2013; Perez-Jimenez et al., 2021), another lysosome-resident protein involved in the catabolism of HS (Schlotawa et al., 2020). SUL-3 proteins are most similar to the lysosome sulfatase human arylsulfatase B that participates in the degradation of keratan/dermatan sulfate and CS, or the potentially secreted arylsulfatases I and J with currently uncharacterized substrates. The biological function, localization and substrates of the nematode sulfatases are largely unexplored. To date, efforts to understand desulfation in worms have been restricted to C. elegans SUL-2 and P. pacificus EUD-1 (SUL-2 paralog). The role of EUD-1 in the regulation of mouth-form and predation is discussed below.
C. elegans SUL-2, the worm steroid sulfatase (Perez-Jimenez et al., 2021), is expressed in sensory neurons of the head of the worm where it is proposed to regulate the levels of steroid hormones. Worms with loss-of-function mutation of sul-2 or treated with the steroid sulfatase (STS) inhibitor STX64 have higher levels of sulfate steroid hormones and increased longevity. The authors of the study hypothesize that sulfated steroid hormones produced in the gonads of the worm may act in response to environmental cues, such as nutrient availability, to alter neurotransmission and promote longevity. Furthermore, higher levels of sulfated hormones as a result of reduced SUL-2 activity, or treatment of worms with sulfated steroid hormones, reduced protein aggregation and proteotoxic stress in models of Parkinson and Huntington neurodegenerative disorders. These data indicate that inhibitors of STS activity and sulfated steroid hormones have great potential in the treatment of aging-related diseases (Perez-Jimenez et al., 2021). In the same study, genetic ablation of sul-1 and sul-3 had no effects on lifespan.
The post-translational modification that enhances sulfatase activity is catalyzed by an endoplasmic reticulum (ER)-resident protein, the FGE-generating enzyme (FGE, sulfatase modifying factor 1, SUMF1) (Dierks et al., 1998a; Cosma et al., 2003; Dierks et al., 2003; Appel and Bertozzi, 2015). The FGly modification is essential for sulfatase activity (Dierks et al., 1998b; Recksiek et al., 1998) and is able to greatly stimulate catalytic proficiency, which can reach a rate enhancement (kcat/kuncat) of 1026 for alkylsulfates (most efficient enzymes known to date) (Edwards et al., 2012). In humans, lack of FGly modification in sulfatases due to mutations in FGE that alter its activity causes multiple sulfatase deficiency (Schmidt et al., 1995; Schlotawa et al., 2020). FGE orthologues are found in prokaryotes and eukaryotes. Yet, homology searches indicate that FGE homologs are not present in plants (which also lack sulfatases), nematodes and fungi, although these last two groups of organisms contain cysteine-type sulfatase genes (Landgrebe et al., 2003; Appel and Bertozzi, 2015). This observation is suggestive of a different FGly-generating system in nematodes and fungi with a yet elusive enzyme. Alternatively, as FGly modification has not been studied so far in these groups of organisms, it remains unknown if their sulfatases require this post-translational modification to be active.
The sulfatase-encoding gene eud-1 was the first gene identified to regulate mouth-form plasticity in P. pacificus (Ragsdale et al., 2013). In the P. pacificus wild type strain PS312, the majority of animals express the predatory Eu mouth-form. Therefore, genetic screens aimed at identifying eurystomatous form defective (eud) mutants that would only generate St animals. From a screen of less than 4,000 haploid genomes, 17 mutants with a Eud phenotype were isolated. Of these, four were dominant with heterozygous animals having a mutant all-St phenotype and all of them turned to be alleles of the same gene, eud-1. The cloning of eud-1 not only revealed this gene to encode a sulfatase but also that all four mutations reduce or eliminate gene function (Ragsdale et al., 2013). These reduction-of-function mutations and the associated dominant phenotype indicate this gene to be haploinsufficient and to act as a developmental switch. Indeed, eud-1 mutants can be rescued by the overexpression of a wild type copy of eud-1, whereas overexpression of the mutant version of eud-1 will not cause a Eu phenotype in wild type animals (Ragsdale et al., 2013). It should be noted that such dominant reduction-of-function mutants are extremely rare with only one example known in C. elegans after more than 50 years of forward genetic screening, whereas gain-of-function mutants with a dominant phenotype are quite common (Hodgkin, 2005).
Further characterization revealed that EUD-1 might act as a sulfatase. Ppa-EUD-1, like Cel-SUL-2, is most similar to the human arylsulfatases A, GALNS and STS (Table 2). Application of sulfate and phosphate ions in the worm diet, which are known inhibitors of arylsulfatases, mimicked the eud-1 phenotype (Ragsdale et al., 2013). As indicated above, the P. pacificus genome contains more sulfatase genes than the C. elegans genome and eud-1 is indeed one of the genes resulting from lineage-specific gene duplications. While the arylsulfatase Cel-sul-2 is a single copy gene on chromosome IV, there are three sul-2-related genes in P. pacificus (Table 2). The ancestral gene is also located on chromosome IV and is termed Ppa-sul-2.1. The two additional copies, eud-1 and Ppa-sul-2.2.1, are located on the X chromosome with an interesting head to head organization and a separation by 7 kb of potential regulatory sequence (Figure 2A). Ppa-sul-2.2.1 and Ppa-sul-2.1 were soon shown to play no role in mouth-form specification (Ragsdale and Ivers, 2016). However, the full complexity of the eud-1 locus was only revealed through several additional experiments. First, the conserved histone-acetyltransferase Ppa-lsy-12 is required for eud-1 expression suggesting that eud-1 expression and mouth-form regulation are subject to epigenetic control (Serobyan et al., 2016). Second, an antisense-eud-1 message exists at the locus that acts as a positive regulator of eud-1 (Serobyan et al., 2016). Finally, two more enzyme encoding genes are part of the eud-1 locus. These genes encode for N-acetylglucosaminidases (nag) and nag-1 and nag-2 are located adjacent to sul-2.2.1 and eud-1, respectively (Figure 2A). Inactivation of both genes results in the opposite phenotype of eud-1 mutants: all animals have the Eu mouth-form even in liquid culture conditions that induce the St morph in wild type animals (Sieriebriennikov et al., 2018). Interesting, nag-1, nag-2 and eud-1 are expressed in a small set of different sensory neurons suggesting that they play a key role in environmental perception (Figure 2B). Importantly, eud-1 is epistatic over nag-1 and nag-2 as a quadruple knockout of the complete multigene locus has an all-St phenotype similar to eud-1 single mutants (Sieriebriennikov et al., 2018). Thus, the sulfatase EUD-1 plays a crucial role in mouth-form regulation as the central developmental switch. First, it is involved in the sensing of the environment and second, it integrates different environmental inputs to guide final decision making of mouth-form. It is important to note that while these genetic experiments clearly show a key role of a sulfatase in regulating a plastic developmental decision, the exact biochemical function and the potential substrates remain currently unknown. It has been speculated based on the physical proximity of sulfatases and N-acetylglucosaminidases that they might act on the extracellular matrix of neurons. However, no experimental support of this hypothesis is currently available and future studies have to shed light on the actual substrates of EUD-1.
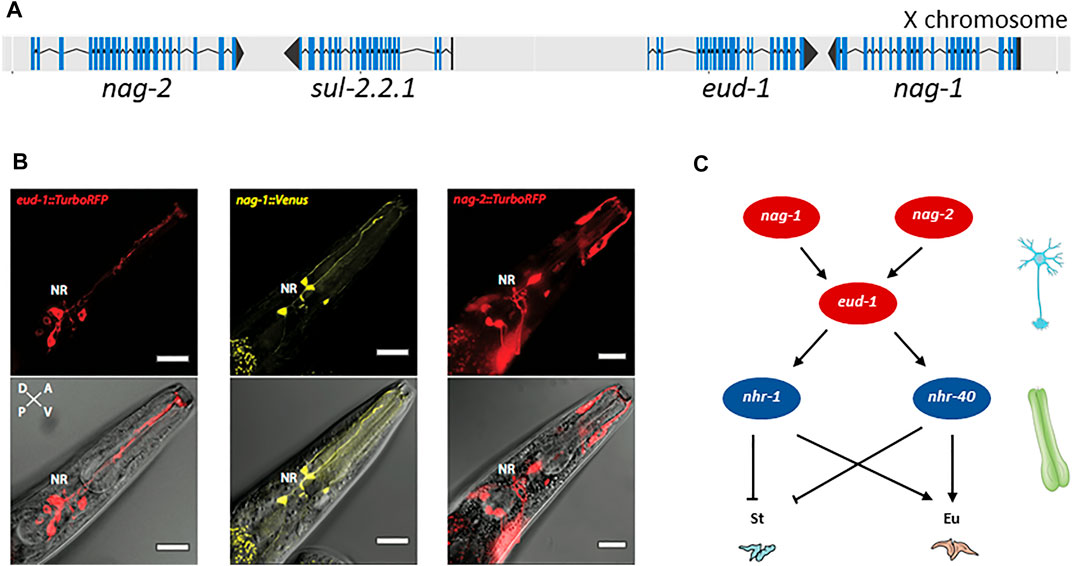
FIGURE 2. Genes involved in mouth-form dimorphism. (A) The switch gene eud-1 is located on the X chromosome of P. pacificus in an inverted tandem configuration (head-to-head orientation) with its paralog sul-2.2.1. In the same gene cluster, the two sulfatase genes are surrounded by a pair of inverted and duplicated genes encoding the α-N-acetylglucosaminidases NAG-1 and NAG-2. Blue lines indicate the coding sequence (CDS); black lines represent the untranslated regions (UTRs) of the gene. Figure adapted from (Sieriebriennikov et al., 2018). (B) eud-1, nag-1 and nag-2 are expressed in distinct sensory neurons. Depicted are head sections of worms overexpressing fluorescent eud-1, nag-1 or nag-2 transcriptional reporters. Bottom: overlay of a DIC image with the TurboRFP (red fluorescent protein) or Venus fluorescence. Top: the same without the DIC image. Bar = 20 μm. (C) Partial genetic network involved in mouth-form plasticity indicating some of the key players, the α-N-acetylglucosaminidases nag-1 and nag-2, the sulfatase-encoding gene eud-1 and the nuclear hormone receptors nhr-40 and nhr-1. nag-1, nag-2 and eud-1 are expressed in sensory neurons whereas nhr-40 and nhr-1 expression is mainly detected in the pharyngeal muscle cells, which are thought to secrete the structural components of the teeth. St: stenostomatous; Eu: eurystomatous.
Further genetic studies have identified a complex gene regulatory network (GRN) to control mouth-form plasticity with eud-1 representing the key developmental switch (Figure 2C) (Sieriebriennikov et al., 2020). The identification of this GRN was in large parts based on the strong all-St phenotype of the eud-1 mutant itself, which allowed unbiased genetic screens for suppressors resulting in all-Eu animals.
First, three suppressor alleles with an all-Eu phenotype were shown to result from mutations in the nuclear hormone receptor Ppa-nhr-40 (Kieninger et al., 2016) and were later on shown to result from gain-of-function mutations (Sieriebriennikov et al., 2020). In contrast, the loss-of-function phenotype of Ppa-nhr-40 is all-St and thus, similar to the eud-1 mutant phenotype (Sieriebriennikov et al., 2020). Also, loss-of-function mutations in another nuclear hormone receptor, Ppa-nhr-1, have an all-St phenotype, but with additional morphological defects in teeth formation unknown from other mutants (Sieriebriennikov et al., 2020). These studies would be compatible with the EUD-1 protein regulating the sulfation state of potential ligands of nuclear hormone receptors. Note that while nematodes have undergone a substantial expansion of nuclear hormone receptors with more than 250 genes in P. pacificus and C. elegans, only one ligand has been identified: the steroid hormone dafachronic acid that regulates the nuclear hormone receptor DAF-12 in multiple nematode species (Motola et al., 2006; Ogawa et al., 2009). Thus, the ligands of nematode nuclear hormone receptors remain little understood and similarly, nothing is known about a potential role of sulfation in the regulation of such ligands.
Second, the same genetic screen for eud-1 suppressors resulted in the identification of a sulfotransferase, mutations in which also produced an all-Eu phenotype and thus were named seud-1 (suppressor of eud-1) (Bui et al., 2018). Transgenic overexpression of seud-1 revealed the sulfotransferase to be dosage-dependent, similar to eud-1 (Bui et al., 2018). When mutants of eud-1 and seud-1 were used to produce all types of homozygous and heterozygous combinations, a graduation of phenotypes was observed. For example, animals with one copy of the sulfotransferase seud-1 and two copies of the sulfatase eud-1 were Eu (Bui et al., 2018). Besides the work of Bui and co-workers, an independent study resulted in the identification of the same sulfotransferase. A pharmacological screen found bisphenol A to induce the St mouth phenotype in wild type animals (Namdeo et al., 2018). As already indicated above, in C. elegans the sulfotransferase ssu-1 sulfates bisphenol A (Hattori et al., 2006). Indeed, a systematic screen of the five sulfotransferases of P. pacificus that are most similar in sequence to Cel-ssu-1, identified a single gene, named sult-1, to control mouth-form plasticity (Namdeo et al., 2018). Note that these independent studies have named the same sulfotransferase gene as seud-1 and sult-1. Importantly, the expression of SEUD-1/SULT-1 (pharyngeal muscle cells) is distinct from EUD-1 (sensory neurons) and therefore, it is possible that two independent sulfation processes regulate mouth-form plasticity (Bui et al., 2018; Namdeo et al., 2018).
Completely independent insight pointing towards the role of sulfated molecules in the biology of P. pacificus nematodes came from studies in analytical chemistry. The predatory behavior of P. pacificus is thought to be of importance in the context of the ecology of this nematode. While P. pacificus is a soil nematode, it is most reliably found in association with adult scarab beetles, i.e. cock chafers, stag beetles and dung beetles. On the living beetle, nematodes are found in the dauer stage, the arrested and stress-resistant, alternative larval stage that can survive long periods of harmful conditions. It is important to note that several, unrelated nematode species can be found on beetles and also other insects, and many of them wait for their vector to die. Indeed, after the beetles’ death back in the soil, bacteria and fungi will grow on the carcass. These food signals will cause the exit of the nematodes from the dauer stage and they rapidly mature to feed and reproduce on this time-limited food source. Indeed, recent studies in P. pacificus have indicated the dynamics and succession of nematodes on beetle carcasses in the context of the microbiome of the insect cadaver (Meyer et al., 2017; Renahan et al., 2021; Renahan and Sommer, 2021). The predatory behavior of Eu animals of P. pacificus is thought to serve a dual role: on the one hand, it allows supplementation of its bacterial diet by preying on nematodes; on the other hand, it will result in the elimination of competitors for the limited bacterial food available. This phenomenon is known as intraguild predation and represents an important concept in ecology (Polis and Holt, 1992; Quach and Chalasani, 2020).
While there is no obvious evidence that P. pacificus and C. elegans share the same ecosystem, a study by Liu and co-workers indicated a defensive response of C. elegans when it is exposed to P. pacificus predators (Liu et al., 2018). Subsequent studies revealed the behavioral details of this predator-prey competition (Quach and Chalasani, 2020). Strikingly, the original defensive response of C. elegans larvae towards P. pacificus is caused by predator-secreted sulfolipids. Specifically, fractionation of the P. pacificus secretome and analytical chemistry identified several (w-1)-branched-chain sulfolipids to induce the escape behavior in C. elegans prey (Liu et al., 2018). These findings are consistent with previous studies that showed an avoidance behavior of C. elegans against sodium dodecyl sulfate (SDS) (Hilliard et al., 2004). Importantly, the production and secretion of branched chain fatty acids is specific to P. pacificus and likely its relatives (Liu et al., 2018). The presence of multiple secreted sulfolipids in P. pacificus might open the door to several independent functions in the organism. For example, it might well be that branched chain fatty acids can serve as ligands for nuclear hormone receptors although no functional studies are yet available to support this hypothesis. In conclusion, genetic and chemical studies provide strong support for the role of sulfation in the development and the behavior of this nematode.
The literature described in this review highlights the significance of sulfation and desulfation pathways during development of different nematodes. From neuronal organization, to the regulation of phenotypic traits and behavior, sulfatases and sulfotransferases appear to modify a wide range of substrates also in nematodes. Despite the work done so far, we still possess a limited understanding on the localization, substrates and regulation of the panoply of sulfation-associated enzymes present in worms. Many of the sulfotransferases and sulfatases remain uncharacterized; their substrates are unknown, we have no understanding of their biological significance or subcellular localization, and we lack defined knowledge on their pattern of expression. Moreover, specific sulfated molecules, such as sulfolipids, act as signaling molecules between animals (Quach and Chalasani, 2020). However, we still do not understand the molecular mechanisms that generate, interpret, and execute, or the universality of this communication pathway.
An exciting research area in sulfation relates to the regulation of the enzymatic activity by post-translational modifications, such as FGly and glycosylation. As mentioned above, nematodes lack a recognizable FGE. Future studies that aim at detecting this unique modification in the proteome of nematodes, the corresponding modifying enzyme and its mechanism of action have great potential to elucidate alternative systems that introduce the FGly modification in eukaryotic sulfatases.
The significance of sulfation pathways in parasitic nematodes, like S. ratti, has never been addressed. With the development of specific sulfatase inhibitors, it is therefore of great importance to study their impact on the infection and ability of the parasite to invade the host. A deeper understanding on the sulfation players and their functions in these pathogenic worms may lead to improved therapeutic strategies that aim at eliminating the pathogen from their natural host. We are therefore looking forward to use nematodes in the quest to obtain a comprehensive understanding of the mechanisms and roles that sulfation plays in organismal development.
All authors listed have made a substantial, direct, and intellectual contribution to the work and approved it for publication.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We thank the members of the Sommer laboratory, in particular Radhika Sharma and Alejandra Schäfer, for inspiring discussions, and Wen-Sui Lo and Alejandra Schäfer for help with bioinformatic analysis of nematode sulfatase and sulfotransferase genes. We are grateful to Bogdan Sieriebriennikov for the fluorescence images depicted in Figure 2B.
Acker, N., Smith, H., Devine, C., Oltjen, S. L., Tsiropoulou, S., Smit-Mcbride, Z., et al. (2021). A Complement Factor H Homolog, Heparan Sulfation, and Syndecan Maintain Inversin Compartment Boundaries in C. elegans Cilia. Proc. Natl. Acad. Sci. U S A. 118. doi:10.1073/pnas.2016698118
Ackley, B. D., Crew, J. R., Elamaa, H., Pihlajaniemi, T., Kuo, C. J., and Kramer, J. M. (2001). The NC1/endostatin Domain of Caenorhabditis elegans Type XVIII Collagen Affects Cell Migration and Axon Guidance. J. Cel Biol 152, 1219–1232. doi:10.1083/jcb.152.6.1219
Appel, M. J., and Bertozzi, C. R. (2015). Formylglycine, a post-translationally Generated Residue with Unique Catalytic Capabilities and Biotechnology Applications. ACS Chem. Biol. 10, 72–84. doi:10.1021/cb500897w
Beeber, C., and Kieras, F. J. (2002). Characterization of the Chondroitin Sulfates in Wild Type Caenorhabditis elegans. Biochem. Biophysical Res. Commun. 293, 1374–1376. doi:10.1016/s0006-291x(02)00399-6
Beisswanger, R., Corbeil, D., Vannier, C., Thiele, C., Dohrmann, U., Kellner, R., et al. (1998). Existence of Distinct Tyrosylprotein Sulfotransferase Genes: Molecular Characterization of Tyrosylprotein Sulfotransferase-2. Proc. Natl. Acad. Sci. 95, 11134–11139. doi:10.1073/pnas.95.19.11134
Bento, G., Ogawa, A., and Sommer, R. J. (2010). Co-option of the Hormone-Signalling Module Dafachronic Acid-DAF-12 in Nematode Evolution. Nature 466, 494–497. doi:10.1038/nature09164
Bojarová, P., and Williams, S. J. (2008). Sulfotransferases, Sulfatases and Formylglycine-Generating Enzymes: a Sulfation Fascination. Curr. Opin. Chem. Biol. 12, 573–581. doi:10.1016/j.cbpa.2008.06.018
Bowman, K. G., and Bertozzi, C. R. (1999). Carbohydrate Sulfotransferases: Mediators of Extracellular Communication. Chem. Biol. 6, R9–R22. doi:10.1016/s1074-5521(99)80014-3
Brenner, S. (1974). The Genetics of Caenorhabditis elegans. Genetics 77, 71–94. doi:10.1093/genetics/77.1.71
Bui, L. T., Ivers, N. A., and Ragsdale, E. J. (2018). A Sulfotransferase Dosage-Dependently Regulates Mouthpart Polyphenism in the Nematode Pristionchus Pacificus. Nat. Commun. 9, 4119. doi:10.1038/s41467-018-05612-8
Bulow, H. E., Berry, K. L., Topper, L. H., Peles, E., and Hobert, O. (2002). Heparan Sulfate Proteoglycan-dependent Induction of Axon Branching and Axon Misrouting by the Kallmann Syndrome Gene Kal-1. Proc. Natl. Acad. Sci. 99, 6346–6351. doi:10.1073/pnas.092128099
Bülow, H. E., and Hobert, O. (2004). Differential Sulfations and Epimerization Define Heparan Sulfate Specificity in Nervous System Development. Neuron 41, 723–736. doi:10.1016/s0896-6273(04)00084-4
Burton, N. O., Dwivedi, V. K., Burkhart, K. B., Kaplan, R. E. W., Baugh, L. R., and Horvitz, H. R. (2018). Neurohormonal Signaling via a Sulfotransferase Antagonizes Insulin-like Signaling to Regulate a Caenorhabditis elegans Stress Response. Nat. Commun. 9, 5152. doi:10.1038/s41467-018-07640-w
Carroll, B. T., Dubyak, G. R., Sedensky, M. M., and Morgan, P. G. (2006). Sulfated Signal from ASJ Sensory Neurons Modulates Stomatin-dependent Coordination in Caenorhabditis elegans. J. Biol. Chem. 281, 35989–35996. doi:10.1074/jbc.m606086200
Consortium, C. E. S. (1998). Genome Sequence of the Nematode C. elegans: a Platform for Investigating Biology. Science 282, 2012–2018. doi:10.1126/science.282.5396.2012
Cosma, M. P., Pepe, S., Annunziata, I., Newbold, R. F., Grompe, M., Parenti, G., et al. (2003). The Multiple Sulfatase Deficiency Gene Encodes an Essential and Limiting Factor for the Activity of Sulfatases. Cell 113, 445–456. doi:10.1016/s0092-8674(03)00348-9
Coughtrie, M. W. H. (2016). Function and Organization of the Human Cytosolic Sulfotransferase (SULT) Family. Chemico-Biological Interactions 259, 2–7. doi:10.1016/j.cbi.2016.05.005
Desai, C., Garriga, G., Mclntire, S. L., and Horvitz, H. R. (1988). A Genetic Pathway for the Development of the Caenorhabditis elegans HSN Motor Neurons. Nature 336, 638–646. doi:10.1038/336638a0
Díaz-Balzac, C. A., Lázaro-Peña, M. I., Tecle, E., Gomez, N., and Bülow, H. E. (2014). Complex Cooperative Functions of Heparan Sulfate Proteoglycans Shape Nervous System Development in Caenorhabditis elegans. G3 (Bethesda) 4, 1859–1870. doi:10.1534/g3.114.012591
Dickinson, D. J., and Goldstein, B. (2016). CRISPR-based Methods for Caenorhabditis elegans Genome Engineering. Genetics 202, 885–901. doi:10.1534/genetics.115.182162
Dierker, T., Shao, C., Haitina, T., Zaia, J., Hinas, A., and Kjellén, L. (2016). Nematodes Join the Family of Chondroitin Sulfate-Synthesizing Organisms: Identification of an Active Chondroitin Sulfotransferase in Caenorhabditis elegans. Sci. Rep. 6, 34662. doi:10.1038/srep34662
Dierks, T., Lecca, M. R., Schlotterhose, P., Schmidt, B., and Von Figura, K. (1999). Sequence Determinants Directing Conversion of Cysteine to Formylglycine in Eukaryotic Sulfatases. EMBO J. 18, 2084–2091. doi:10.1093/emboj/18.8.2084
Dierks, T., Lecca, M. R., Schmidt, B., and Von Figura, K. (1998a). Conversion of Cysteine to Formylglycine in Eukaryotic Sulfatases Occurs by a Common Mechanism in the Endoplasmic Reticulum. FEBS Lett. 423, 61–65. doi:10.1016/s0014-5793(98)00065-9
Dierks, T., Miech, C., Hummerjohann, J., Schmidt, B., Kertesz, M. A., and Von Figura, K. (1998b). Posttranslational Formation of Formylglycine in Prokaryotic Sulfatases by Modification of Either Cysteine or Serine. J. Biol. Chem. 273, 25560–25564. doi:10.1074/jbc.273.40.25560
Dierks, T., Schmidt, B., Borissenko, L. V., Peng, J., Preusser, A., Mariappan, M., et al. (2003). Multiple Sulfatase Deficiency Is Caused by Mutations in the Gene Encoding the Human Cα-Formylglycine Generating Enzyme. Cell 113, 435–444. doi:10.1016/s0092-8674(03)00347-7
Dierks, T., Schmidt, B., and Von Figura, K. (1997). Conversion of Cysteine to Formylglycine: a Protein Modification in the Endoplasmic Reticulum. Proc. Natl. Acad. Sci. 94, 11963–11968. doi:10.1073/pnas.94.22.11963
Dieterich, C., Clifton, S. W., Schuster, L. N., Chinwalla, A., Delehaunty, K., Dinkelacker, I., et al. (2008). The Pristionchus Pacificus Genome Provides a Unique Perspective on Nematode Lifestyle and Parasitism. Nat. Genet. 40, 1193–1198. doi:10.1038/ng.227
Edwards, A. O., Ritter, R., Abel, K. J., Manning, A., Panhuysen, C., and Farrer, L. A. (2005). Complement Factor H Polymorphism and Age-Related Macular Degeneration. Science 308, 421–424. doi:10.1126/science.1110189
Edwards, D. R., Lohman, D. C., and Wolfenden, R. (2012). Catalytic Proficiency: the Extreme Case of S-O Cleaving Sulfatases. J. Am. Chem. Soc. 134, 525–531. doi:10.1021/ja208827q
Fire, A., Xu, S., Montgomery, M. K., Kostas, S. A., Driver, S. E., and Mello, C. C. (1998). Potent and Specific Genetic Interference by Double-Stranded RNA in Caenorhabditis elegans. Nature 391, 806–811. doi:10.1038/35888
Franco, B., Guioli, S., Pragliola, A., Incerti, B., Bardoni, B., Tonlorenzi, R., et al. (1991). A Gene Deleted in Kallmann's Syndrome Shares Homology with Neural Cell Adhesion and Axonal Path-Finding Molecules. Nature 353, 529–536. doi:10.1038/353529a0
Ghosh, D. (2007). Human Sulfatases: a Structural Perspective to Catalysis. Cell. Mol. Life Sci. 64, 2013–2022. doi:10.1007/s00018-007-7175-y
Gumienny, T. L., Macneil, L. T., Wang, H., De Bono, M., Wrana, J. L., and Padgett, R. W. (2007). Glypican LON-2 Is a Conserved Negative Regulator of BMP-like Signaling in Caenorhabditis elegans. Curr. Biol. 17, 159–164. doi:10.1016/j.cub.2006.11.065
Günal, S., Hardman, R., Kopriva, S., and Mueller, J. W. (2019). Sulfation Pathways from Red to green. J. Biol. Chem. 294, 12293–12312. doi:10.1074/jbc.rev119.007422
Habuchi, O. (2000). Diversity and Functions of Glycosaminoglycan Sulfotransferases. Biochim. Biophys. Acta (Bba) - Gen. Subjects 1474, 115–127. doi:10.1016/s0304-4165(00)00016-7
Hageman, G. S., Anderson, D. H., Johnson, L. V., Hancox, L. S., Taiber, A. J., Hardisty, L. I., et al. (2005). From the Cover: A Common Haplotype in the Complement Regulatory Gene Factor H (HF1/CFH) Predisposes Individuals to Age-Related Macular Degeneration. Proc. Natl. Acad. Sci. 102, 7227–7232. doi:10.1073/pnas.0501536102
Haines, J. L., Hauser, M. A., Schmidt, S., Scott, W. K., Olson, L. M., Gallins, P., et al. (2005). Complement Factor H Variant Increases the Risk of Age-Related Macular Degeneration. Science 308, 419–421. doi:10.1126/science.1110359
Han, Z., Lo, W.-S., Lightfoot, J. W., Witte, H., Sun, S., and Sommer, R. J. (2020). Improving Transgenesis Efficiency and CRISPR-Associated Tools through Codon Optimization and Native Intron Addition in Pristionchus Nematodes. Genetics 216, 947–956. doi:10.1534/genetics.120.303785
Hanson, S. R., Best, M. D., and Wong, C.-H. (2004). Sulfatases: Structure, Mechanism, Biological Activity, Inhibition, and Synthetic Utility. Angew. Chem. Int. Ed. 43, 5736–5763. doi:10.1002/anie.200300632
Hattori, K., Inoue, M., Inoue, T., Arai, H., and Tamura, H.-o. (2006). A Novel Sulfotransferase Abundantly Expressed in the Dauer Larvae of Caenorhabditis elegans. J. Biochem. 139, 355–362. doi:10.1093/jb/mvj041
Herman, T., Hartwieg, E., and Horvitz, H. R. (1999). Sqv Mutants of Caenorhabditis elegans Are Defective in Vulval Epithelial Invagination. Proc. Natl. Acad. Sci. 96, 968–973. doi:10.1073/pnas.96.3.968
Hernandez-Guzman, F. G., Higashiyama, T., Pangborn, W., Osawa, Y., and Ghosh, D. (2003). Structure of Human Estrone Sulfatase Suggests Functional Roles of Membrane Association. J. Biol. Chem. 278, 22989–22997. doi:10.1074/jbc.m211497200
Hilliard, M. A., Bergamasco, C., Arbucci, S., Plasterk, R. H., and Bazzicalupo, P. (2004). Worms Taste Bitter: ASH Neurons, QUI-1, GPA-3 and ODR-3 Mediate Quinine Avoidance in Caenorhabditis elegans. EMBO J. 23, 1101–1111. doi:10.1038/sj.emboj.7600107
Hutter, H., Vogel, B. E., Plenefisch, J. D., Norris, C. R., Proenca, R. B., Spieth, J., et al. (2000). Conservation and novelty in the Evolution of Cell Adhesion and Extracellular Matrix Genes. Science 287, 989–994. doi:10.1126/science.287.5455.989
Hwang, H.-Y., Olson, S. K., Esko, J. D., and Robert Horvitz, H. (2003). Caenorhabditis elegans Early Embryogenesis and Vulval Morphogenesis Require Chondroitin Biosynthesis. Nature 423, 439–443. doi:10.1038/nature01634
Kieninger, M. R., Ivers, N. A., Rödelsperger, C., Markov, G. V., Sommer, R. J., and Ragsdale, E. J. (2016). The Nuclear Hormone Receptor NHR-40 Acts Downstream of the Sulfatase EUD-1 as Part of a Developmental Plasticity Switch in Pristionchus. Curr. Biol. 26, 2174–2179. doi:10.1016/j.cub.2016.06.018
Kim, T. H., Hwang, S. B., Jeong, P.-Y., Lee, J., and Cho, J. W. (2005). Requirement of Tyrosylprotein Sulfotransferase-A for Proper Cuticle Formation in the nematodeC. Elegans. FEBS Lett. 579, 53–58. doi:10.1016/j.febslet.2004.11.044
Kim, T. H., Kim, D. H., Nam, H. W., Park, S. Y., Shim, J., and Cho, J. W. (2010). Tyrosylprotein Sulfotransferase Regulates Collagen Secretion in Caenorhabditis elegans. Mol. Cell 29, 413–418. doi:10.1007/s10059-010-0049-4
Kinnunen, T., Huang, Z., Townsend, J., Gatdula, M. M., Brown, J. R., Esko, J. D., et al. (2005). Heparan 2-O-Sulfotransferase, Hst-2, Is Essential for normal Cell Migration in Caenorhabditis elegans. Proc. Natl. Acad. Sci. 102, 1507–1512. doi:10.1073/pnas.0401591102
Kinnunen, T. K. (2014). Combinatorial Roles of Heparan Sulfate Proteoglycans and Heparan Sulfates in Caenorhabditis elegans Neural Development. PLoS One 9, e102919. doi:10.1371/journal.pone.0102919
Klein, R. J., Zeiss, C., Chew, E. Y., Tsai, J.-Y., Sackler, R. S., Haynes, C., et al. (2005). Complement Factor H Polymorphism in Age-Related Macular Degeneration. Science 308, 385–389. doi:10.1126/science.1109557
Knaust, A., Schmidt, B., Dierks, T., von Bülow, R., and Von Figura, K. (1998). Residues Critical for Formylglycine Formation And/or Catalytic Activity of Arylsulfatase A. Biochemistry 37, 13941–13946. doi:10.1021/bi9810205
Koo, P. K., Bian, X., Sherlekar, A. L., Bunkers, M. R., and Lints, R. (2011). The Robustness of Caenorhabditis elegans Male Mating Behavior Depends on the Distributed Properties of ray Sensory Neurons and Their Output through Core and Male-specific Targets. J. Neurosci. 31, 7497–7510. doi:10.1523/jneurosci.6153-10.2011
Landgrebe, J., Dierks, T., Schmidt, B., and Von Figura, K. (2003). The Human SUMF1 Gene, Required for Posttranslational Sulfatase Modification, Defines a New Gene Family Which Is Conserved from Pro- to Eukaryotes. Gene 316, 47–56. doi:10.1016/s0378-1119(03)00746-7
Langford, R., Hurrion, E., and Dawson, P. A. (2017). Genetics and Pathophysiology of Mammalian Sulfate Biology. J. Genet. Genomics 44, 7–20. doi:10.1016/j.jgg.2016.08.001
Lawrence, R., Olson, S. K., Steele, R. E., Wang, L., Warrior, R., Cummings, R. D., et al. (2008). Evolutionary Differences in Glycosaminoglycan fine Structure Detected by Quantitative Glycan Reductive Isotope Labeling. J. Biol. Chem. 283, 33674–33684. doi:10.1074/jbc.m804288200
Lázaro-Peña, M. I., Díaz-Balzac, C. A., Bülow, H. E., and Emmons, S. W. (2018). Synaptogenesis Is Modulated by Heparan Sulfate in Caenorhabditis elegans. Genetics 209, 195–208. doi:10.1534/genetics.118.300837
Lee, D. W., Choo, H. Y., Kaya, H. K., Lee, S. M., Smitley, D. R., Shin, H. K., et al. (2002). Laboratory and Field Evaluation of Korean Entomopathogenic Nematode Isolates against the oriental Beetle Exomala Orientalis (Coleoptera: Scarabaeidae). J. Econ. Entomol. 95, 918–926. doi:10.1093/jee/95.5.918
Legouis, R., Hardelin, J.-P., Levilliers, J., Claverie, J.-M., Compain, S., Wunderle, V., et al. (1991). The Candidate Gene for the X-Linked Kallmann Syndrome Encodes a Protein Related to Adhesion Molecules. Cell 67, 423–435. doi:10.1016/0092-8674(91)90193-3
Leung, A. W. Y., Backstrom, I., and Bally, M. B. (2016). Sulfonation, an Underexploited Area: from Skeletal Development to Infectious Diseases and Cancer. Oncotarget 7, 55811–55827. doi:10.18632/oncotarget.10046
Lightfoot, J. W., Wilecki, M., Rödelsperger, C., Moreno, E., Susoy, V., Witte, H., et al. (2019). Small Peptide-Mediated Self-Recognition Prevents Cannibalism in Predatory Nematodes. Science 364, 86–89. doi:10.1126/science.aav9856
Lindahl, U., and Li, J. p. (2009). Chapter 3 Interactions between Heparan Sulfate and Proteins-Design and Functional Implications. Int. Rev. Cel Mol Biol 276, 105–159. doi:10.1016/s1937-6448(09)76003-4
Liu, K. S., and Sternberg, P. W. (1995). Sensory Regulation of Male Mating Behavior in Caenorhabditis elegans. Neuron 14, 79–89. doi:10.1016/0896-6273(95)90242-2
Liu, Z., Kariya, M. J., Chute, C. D., Pribadi, A. K., Leinwand, S. G., Tong, A., et al. (2018). Predator-secreted Sulfolipids Induce Defensive Responses in C. elegans. Nat. Commun. 9, 1128. doi:10.1038/s41467-018-03333-6
Meyer, J. M., Baskaran, P., Quast, C., Susoy, V., Rödelsperger, C., Glöckner, F. O., et al. (2017). Succession and Dynamics ofPristionchusnematodes and Their Microbiome during Decomposition ofOryctes Borbonicuson La Réunion Island. Environ. Microbiol. 19, 1476–1489. doi:10.1111/1462-2920.13697
Minniti, A. N., Labarca, M., Hurtado, C., and Brandan, E. (2004). Caenorhabditis elegans Syndecan (SDN-1) Is Required for normal Egg Laying and Associates with the Nervous System and the Vulva. J. Cel Sci 117, 5179–5190. doi:10.1242/jcs.01394
Mizuguchi, S., Dejima, K., and Nomura, K. (2009). Sulfation and Related Genes in Caenorhabditis elegans. Tigg 21, 179–191. doi:10.4052/tigg.21.179
Mizuguchi, S., Uyama, T., Kitagawa, H., Nomura, K. H., Dejima, K., Gengyo-Ando, K., et al. (2003). Chondroitin Proteoglycans Are Involved in Cell Division of Caenorhabditis elegans. Nature 423, 443–448. doi:10.1038/nature01635
Monigatti, F., Hekking, B., and Steen, H. (2006). Protein Sulfation Analysis-A Primer. Biochim. Biophys. Acta (Bba) - Proteins Proteomics 1764, 1904–1913. doi:10.1016/j.bbapap.2006.07.002
Motola, D. L., Cummins, C. L., Rottiers, V., Sharma, K. K., Li, T., Li, Y., et al. (2006). Identification of Ligands for DAF-12 that Govern Dauer Formation and Reproduction in C. elegans. Cell 124, 1209–1223. doi:10.1016/j.cell.2006.01.037
Mueller, J. W., Gilligan, L. C., Idkowiak, J., Arlt, W., and Foster, P. A. (2015). The Regulation of Steroid Action by Sulfation and Desulfation. Endocr. Rev. 36, 526–563. doi:10.1210/er.2015-1036
Namdeo, S., Moreno, E., Rodelsperger, C., Baskaran, P., Witte, H., and Sommer, R. J. (2018). Two Independent Sulfation Processes Regulate Mouth-form Plasticity in the Nematode Pristionchus Pacificus. Development 145. doi:10.1242/dev.166272
Ogawa, A., Streit, A., Antebi, A., and Sommer, R. J. (2009). A Conserved Endocrine Mechanism Controls the Formation of Dauer and Infective Larvae in Nematodes. Curr. Biol. 19, 67–71. doi:10.1016/j.cub.2008.11.063
Olson, S. K., Bishop, J. R., Yates, J. R., Oegema, K., and Esko, J. D. (2006). Identification of Novel Chondroitin Proteoglycans in Caenorhabditis elegans: Embryonic Cell Division Depends on CPG-1 and CPG-2. J. Cel Biol 173, 985–994. doi:10.1083/jcb.200603003
Ouyang, Y.-B., and Moore, K. L. (1998). Molecular Cloning and Expression of Human and Mouse Tyrosylprotein Sulfotransferase-2 and a Tyrosylprotein Sulfotransferase Homologue in Caenorhabditis elegans. J. Biol. Chem. 273, 24770–24774. doi:10.1074/jbc.273.38.24770
Pérez-Jiménez, M. M., Monje-Moreno, J. M., Brokate-Llanos, A. M., Venegas-Calerón, M., Sánchez-García, A., Sansigre, P., et al. (2021). Steroid Hormones Sulfatase Inactivation Extends Lifespan and Ameliorates Age-Related Diseases. Nat. Commun. 12, 49. doi:10.1038/s41467-020-20269-y
Polis, G. A., and Holt, R. D. (1992). Intraguild Predation: The Dynamics of Complex Trophic Interactions. Trends Ecol. Evol. 7, 151–154. doi:10.1016/0169-5347(92)90208-s
Quach, K. T., and Chalasani, S. H. (2020). Intraguild Predation between Pristionchus Pacificus and Caenorhabditis elegans: a Complex Interaction with the Potential for Aggressive Behaviour. J. Neurogenet. 34, 404–419. doi:10.1080/01677063.2020.1833004
Ragsdale, E. J., and Ivers, N. A. (2016). Specialization of a Polyphenism Switch Gene Following Serial Duplications in Pristionchus Nematodes. Evolution 70, 2155–2166. doi:10.1111/evo.13011
Ragsdale, E. J., Müller, M. R., Rödelsperger, C., and Sommer, R. J. (2013). A Developmental Switch Coupled to the Evolution of Plasticity Acts through a Sulfatase. Cell 155, 922–933. doi:10.1016/j.cell.2013.09.054
Recksiek, M., Selmer, T., Dierks, T., Schmidt, B., and Von Figura, K. (1998). Sulfatases, Trapping of the Sulfated Enzyme Intermediate by Substituting the Active Site Formylglycine. J. Biol. Chem. 273, 6096–6103. doi:10.1074/jbc.273.11.6096
Reiter, J. F., and Leroux, M. R. (2017). Genes and Molecular Pathways Underpinning Ciliopathies. Nat. Rev. Mol. Cel Biol 18, 533–547. doi:10.1038/nrm.2017.60
Renahan, T., Lo, W. S., Werner, M. S., Rochat, J., Herrmann, M., and Sommer, R. J. (2021). Nematode Biphasic 'boom and Bust' Dynamics Are Dependent on Host Bacterial Load while Linking Dauer and Mouth‐form Polyphenisms. Environ. Microbiol. 23, 5102–5113. doi:10.1111/1462-2920.15438
Renahan, T., and Sommer, R. J. (2021). Nematode Interactions on Beetle Hosts Indicate a Role of Mouth-form Plasticity in Resource Competition. Front. Ecol. Evol. doi:10.3389/fevo.2021.752695
Rogalski, T. M., Williams, B. D., Mullen, G. P., and Moerman, D. G. (1993). Products of the Unc-52 Gene in Caenorhabditis elegans Are Homologous to the Core Protein of the Mammalian Basement Membrane Heparan Sulfate Proteoglycan. Genes Dev. 7, 1471–1484. doi:10.1101/gad.7.8.1471
Schimpf, J., Sames, K., and Zwilling, R. (1999). Proteoglycan Distribution Pattern during Aging in the Nematode Caenorhabditis elegans: an Ultrastructural Histochemical Study. Histochem. J. 31, 285–292. doi:10.1023/a:1003761817109
Schlotawa, L., Adang, L. A., Radhakrishnan, K., and Ahrens-Nicklas, R. C. (2020). Multiple Sulfatase Deficiency: A Disease Comprising Mucopolysaccharidosis, Sphingolipidosis, and More Caused by a Defect in Posttranslational Modification. Int. J. Mol. Sci. 21. doi:10.3390/ijms21103448
Schmidt, B., Selmer, T., Ingendoh, A., and Figurat, K. v. (1995). A Novel Amino Acid Modification in Sulfatases that Is Defective in Multiple Sulfatase Deficiency. Cell 82, 271–278. doi:10.1016/0092-8674(95)90314-3
Serobyan, V., Xiao, H., Namdeo, S., Rödelsperger, C., Sieriebriennikov, B., Witte, H., et al. (2016). Chromatin Remodelling and Antisense-Mediated Up-Regulation of the Developmental Switch Gene Eud-1 Control Predatory Feeding Plasticity. Nat. Commun. 7, 12337. doi:10.1038/ncomms12337
Sieriebriennikov, B., Prabh, N., Dardiry, M., Witte, H., Röseler, W., Kieninger, M. R., et al. (2018). A Developmental Switch Generating Phenotypic Plasticity Is Part of a Conserved Multi-Gene Locus. Cel Rep. 23, 2835–2843. doi:10.1016/j.celrep.2018.05.008
Sieriebriennikov, B., Sun, S., Lightfoot, J. W., Witte, H., Moreno, E., Rödelsperger, C., et al. (2020). Conserved Nuclear Hormone Receptors Controlling a Novel Plastic Trait Target Fast-Evolving Genes Expressed in a Single Cell. Plos Genet. 16, e1008687. doi:10.1371/journal.pgen.1008687
Sommer, R. J., and Mayer, M. G. (2015). Toward a Synthesis of Developmental Biology with Evolutionary Theory and Ecology. Annu. Rev. Cel Dev. Biol. 31, 453–471. doi:10.1146/annurev-cellbio-102314-112451
Sommer, R. J. (2020). Phenotypic Plasticity: From Theory and Genetics to Current and Future Challenges. Genetics 215, 1–13. doi:10.1534/genetics.120.303163
Sommer, R. J., and Sternberg, P. W. (1996). Apoptosis and Change of Competence Limit the Size of the Vulva Equivalence Group in Pristionchus Pacificus: a Genetic Analysis. Curr. Biol. 6, 52–59. doi:10.1016/s0960-9822(02)00421-9
Stone, M. J., Chuang, S., Hou, X., Shoham, M., and Zhu, J. Z. (2009). Tyrosine Sulfation: an Increasingly Recognised post-translational Modification of Secreted Proteins. New Biotechnol. 25, 299–317. doi:10.1016/j.nbt.2009.03.011
Streit, A. (2008). Reproduction inStrongyloides(Nematoda): a Life between Sex and Parthenogenesis. Parasitology 135, 285–294. doi:10.1017/s003118200700399x
Strott, C. A. (2002). Sulfonation and Molecular Action. Endocr. Rev. 23, 703–732. doi:10.1210/er.2001-0040
Sugahara, K., Mikami, T., Uyama, T., Mizuguchi, S., Nomura, K., and Kitagawa, H. (2003). Recent Advances in the Structural Biology of Chondroitin Sulfate and Dermatan Sulfate. Curr. Opin. Struct. Biol. 13, 612–620. doi:10.1016/j.sbi.2003.09.011
Tecle, E., Diaz-Balzac, C. A., and Bülow, H. E. (2013). Distinct 3-O-Sulfated Heparan Sulfate Modification Patterns Are Required for kal-1−Dependent Neurite Branching in a Context-dependent Manner in Caenorhabditis elegans. G3 (Bethesda) 3, 541–552. doi:10.1534/g3.112.005199
Toyoda, H., Kinoshita-Toyoda, A., and Selleck, S. B. (2000). Structural Analysis of Glycosaminoglycans inDrosophila and Caenorhabditis elegans and Demonstration that Tout-Velu, a Drosophila Gene Related to EXT Tumor Suppressors, Affects Heparan Sulfate In Vivo. J. Biol. Chem. 275, 2269–2275. doi:10.1074/jbc.275.4.2269
Turnbull, J., Drummond, K., Huang, Z., Kinnunen, T., Ford-Perriss, M., Murphy, M., et al. (2003). Heparan Sulphate Sulphotransferase Expression in Mice and Caenorhabditis elegans. Biochem. Soc. Trans. 31, 343–348. doi:10.1042/bst0310343
Turnbull, J., Powell, A., and Guimond, S. (2001). Heparan Sulfate: Decoding a Dynamic Multifunctional Cell Regulator. Trends Cel Biol. 11, 75–82. doi:10.1016/s0962-8924(00)01897-3
Van Den Hoogen, J., Geisen, S., Routh, D., Ferris, H., Traunspurger, W., Wardle, D. A., et al. (2019). Soil Nematode Abundance and Functional Group Composition at a Global Scale. Nature 572, 194–198. doi:10.1038/s41586-019-1418-6
Waldow, A., Schmidt, B., Dierks, T., von Bülow, R., and Von Figura, K. (1999). Amino Acid Residues Forming the Active Site of Arylsulfatase A. J. Biol. Chem. 274, 12284–12288. doi:10.1074/jbc.274.18.12284
Wei, Z., Swiedler, S. J., Ishihara, M., Orellana, A., and Hirschberg, C. B. (1993). A Single Protein Catalyzes Both N-Deacetylation and N-Sulfation during the Biosynthesis of Heparan Sulfate. Proc. Natl. Acad. Sci. 90, 3885–3888. doi:10.1073/pnas.90.9.3885
Willemsen, R., Kroos, M., Hoogeveen, A. T., Van Dongen, J. M., Parenti, G., Van Der Loos, C. M., et al. (1988). Ultrastructural Localization of Steroid Sulphatase in Cultured Human Fibroblasts by Immunocytochemistry: a Comparative Study with Lysosomal Enzymes and the Mannose 6-phosphate Receptor. Histochem. J. 20, 41–51. doi:10.1007/bf01745968
Wood, W. B., and Researchers, C. O. C. E. (1988). The Nematode Caenorhabdtis Elegans. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press.
Yamada, S., Van Die, I., Van Den Eijnden, D. H., Yokota, A., Kitagawa, H., and Sugahara, K. (1999). Demonstration of Glycosaminoglycans inCaenorhabditis Elegans. FEBS Lett. 459, 327–331. doi:10.1016/s0014-5793(99)01286-7
Yang, Y.-S., Wang, C.-C., Chen, B.-H., Hou, Y.-H., Hung, K.-S., and Mao, Y.-C. (2015). Tyrosine Sulfation as a Protein post-translational Modification. Molecules 20, 2138–2164. doi:10.3390/molecules20022138
Keywords: nematodes, sulfatases, Caenorhabditis elegans, Pristionchus pacificus, sulfotransferases, eud-1, developmental switch, developmental plasticity
Citation: Igreja C and Sommer RJ (2022) The Role of Sulfation in Nematode Development and Phenotypic Plasticity. Front. Mol. Biosci. 9:838148. doi: 10.3389/fmolb.2022.838148
Received: 20 December 2021; Accepted: 24 January 2022;
Published: 10 February 2022.
Edited by:
Jon Wolf Mueller, University of Birmingham, United KingdomReviewed by:
Paul Alexander Foster, University of Birmingham, United KingdomCopyright © 2022 Igreja and Sommer. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Catia Igreja, Y2F0aWEuaWdyZWphQHR1ZWJpbmdlbi5tcGcuZGU=; Ralf J. Sommer, cmFsZi5zb21tZXJAdHVlYmluZ2VuLm1wZy5kZQ==
†ORCID: Catia Igreja, orcid.org/0000-0003-3563-1788; Ralf J. Sommer, orcid.org/0000-0003-1503-7749
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.