
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Mol. Biosci. , 25 February 2022
Sec. Cellular Biochemistry
Volume 9 - 2022 | https://doi.org/10.3389/fmolb.2022.834020
This article is part of the Research Topic Genetically Mobile Elements Repurposed by Nature and Biotechnologists View all 10 articles
RmInt1 is a group II intron encoding a reverse transcriptase protein (IEP) lacking the C-terminal endonuclease domain. RmInt1 is an efficient mobile retroelement that predominantly reverse splices into the transient single-stranded DNA at the template for lagging strand DNA synthesis during host replication, a process facilitated by the interaction of the RmInt1 IEP with DnaN at the replication fork. It has been suggested that group II intron ribonucleoprotein particles bind DNA nonspecifically, and then scan for their correct target site. In this study, we investigated RmInt1 binding sites throughout the Sinorhizobium meliloti genome, by chromatin-immunoprecipitation coupled with next-generation sequencing. We found that RmInt1 binding sites cluster around the bidirectional replication origin of each of the three replicons comprising the S. meliloti genome. Our results provide new evidence linking group II intron mobility to host DNA replication.
Group II introns are considered to be ancient genetic elements present in the genomes of Eubacteria, Archaebacteria, and the organelles of some eukaryotes (Ferat and Michel, 1993; Toro, 2003; Lambowitz and Zimmerly, 2004). They have attracted considerable interest due to their role in driving eukaryotic evolution as the putative ancestor of spliceosomal introns, telomerase, and non-LTR retroelements (Martin and Koonin, 2006; Lambowitz and Belfort, 2015; Zimmerly and Semper, 2015; Novikova and Belfort, 2017; Haack and Toor, 2020), but their properties have also been exploited in the development of powerful biotechnological tools (Mohr et al., 2013; Enyeart et al., 2014; Zhao et al., 2018; Belfort and Lambowitz, 2019).
Group II introns are self-splicing RNAs and mobile retroelements generally consisting of a structurally conserved RNA and a multidomain reverse transcriptase protein (the intron-encoded protein, IEP), which interact with each other to form a ribonucleoprotein (RNP) particle facilitating intron excision and mobility reactions (Lambowitz et al., 1999; Saldanha et al., 1999; Lambowitz and Zimmerly, 2011). The ribozyme consists of six double-helical RNA domains docked around the complex domain I to form a Y-shaped structure (Agrawal et al., 2016; Qu et al., 2016). The group II intron IEP is encoded by domain IV and typically consists of four functional domains: A reverse transcriptase (RT), a maturase (X), a DNA-binding domain (D) and an endonuclease (En) domain (San Filippo and Lambowitz, 2002). Under physiological conditions, both the RT and X domains form contacts with several intron RNA domains (DIV, DII, DIII and DVI) to promote intron folding and splicing (Wank et al., 1999; Zhao and Pyle, 2017). Intron excision occurs by means of two transesterification reactions resulting in ligated exons and several forms of excised introns (lariat, circular, and linear, depending on biological determinants and host environment, Monat and Cousineau, 2020). After splicing, the IEP remains bound to the excised lariat RNA, forming the RNP complex, which performs the mobility reaction via an RNA intermediate (Cousineau et al., 1998; Martínez-Abarca et al., 2004).
Group II introns are mobile through a mechanism known as target-primed reverse transcription (TRPT) (Yang et al., 1998; Lambowitz and Zimmerly, 2011). Intron RNPs are widely thought to bind DNA nonspecifically before scanning the DNA for an intron-less target locus (retrohoming) or low-frequency ectopic sequences (retrotransposition) (Aizawa et al., 2003). Homing-site recognition involves an interaction of the C-terminal DNA-binding domain of the IEP with a small number of specific bases in the distal 5′and 3′exon regions of the DNA target site (Guo et al., 1997; Singh and Lambowitz, 2001), but principally via three base-pairing interactions between the intron and exon binding sites (EBS1/IBS1, EBS2/IBS2, and EBS3/IBS3 or δ-δ′) largely responsible for DNA target specificity (Figure 1; Mohr et al., 2000; Jiménez-Zurdo et al., 2003). The intron RNA can then insert itself into one strand of a DNA target site by reverse splicing, and the En domain of the IEP cleaves the opposite strand. Retrohoming proceeds by reverse transcription of the inserted intron RNA to generate a cDNA, using the 3′-end generated by IEP cleavage, with the cellular machinery then resolving incorporation into the host genome (Coros et al., 2008; Coros et al., 2009; Nisa-Martínez et al., 2016). However, almost half the bacterial IEPs lack En domains (Novikova et al., 2014), and the corresponding RNPs must therefore invade their single-stranded DNA targets and then make use of alternative priming strategies for the reverse transcription reaction (Ichiyanagi et al., 2002; Zhong and Lambowitz, 2003; Martínez-Abarca et al., 2004).
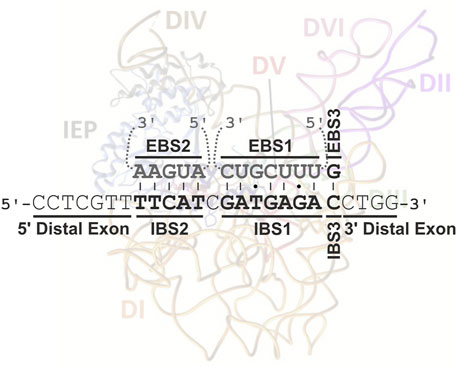
FIGURE 1. Interaction between RmInt1 and its natural DNA target in ISRm2011-2. The intron binding sites (IBSs) in the DNA target (in bold) base-pair with the exon binding sites (EBSs) in the intron RNA (in gray). These base-pairings extend from positions −13 to +1 of the intron insertion site. An intron RNP complex, adapted from Novikova and Belfort, 2017, is also shown, to illustrate the distal interaction of the IEP at positions G-16, T-15 and G+4.
One of the best characterized type IIB introns is RmInt1, an intron of the IIB3/D class found in Sinorhizobium meliloti (Martínez-Abarca et al., 1998). Despite the absence of an En domain, RmInt1 is an efficient mobile element that integrates into the ISRm2011-2 insertion sequence, which is highly abundant and present in diverse Rhizobium species (Martínez-Abarca et al., 2000; Fernández-López et al., 2005). Two retrohoming pathways have been described for RmInt1 mobility: A preferred pathway in which the intron RNA reverse splices intro single-stranded DNA at the replication fork using the nascent lagging DNA strand to prime reverse transcription, and another, less efficient pathway involving retrohoming into the leading strand template (Martínez-Abarca et al., 2004). Moreover, the colonization of the S. meliloti genome by the RmInt1 intron is also biased towards RNA insertion into the lagging DNA strand template, even though target sites located on the leading strand template are sometimes invaded, albeit less frequently (Nisa-Martínez et al., 2007). A recent work demonstrated that RmInt1 homing site selection and reverse splicing into the target locus seem to be facilitated during DNA replication by interaction of the IEP with a replicative protein, DnaN, the β-sliding clamp (García-Rodríguez et al., 2019). Finally, this intron has been reprogrammed to disrupt both plasmid-borne and chromosomal genes with a high level of efficiency (García-Rodríguez et al., 2011; García-Rodríguez et al., 2014).
Looking to the potential use of RmInt1 as a biotechnological tool, it is crucial to describe off-target insertions that could have highly deleterious effects on the host cell. Here, we investigated the potential binding of RmInt1 RNPs throughout S. meliloti genomic DNA in vivo by chromatin-immunoprecipitation coupled to next-generation sequencing (ChIP-Seq). We observed a preferential binding around the origin of replication, slightly biased through the template for the lagging strand synthesis during replication. This preference could be related to chromatin accessibility, but we cannot rule out it could be due to the presence of DnaN at the replication fork.
S. meliloti RMO17 (Villadas et al., 1995) harboring the different plasmids was cultured in TY medium supplemented with kanamycin (200 μg ml−1) at 30°C. Rhizobial bacteria were transformed by triparental mating, with E. coli containing the different plasmids. Chromatin immunoprecipitation requires the addition of formaldehyde, which is highly toxic to cells. We therefore assessed the permeability of the cells to formaldehyde and their response to the addition of this chemical before initiating the experiment (Davis et al., 2011). Cultures were grown to an OD600 of 0.4, when they were split between two flasks, with formaldehyde (1%) added to one, and a mock treatment (37 mM phosphate buffer) added to the other. Growth was monitored by measuring the OD600 at various time points. Growth had stopped after 25 min for the cells incubated with formaldehyde, whereas the bacteria in the mock treatment flask continued to grow exponentially (Supplementary Figure S1). We therefore limited the duration of treatments with this crosslinker to 20 min.
In the ChIP experiments, three plasmid constructs were used that express the RmInt1 Flag-IEP or Flag-tagged RNPs under the constitutive kanamycin resistance gene promoter: A plasmid (pKG4_FlagIEP) expressing RmInt1 RNPs tagged with a 3xFLAG epitope at the N-terminus of IEP, a similar plasmid construct (pKG_FlagIEP) expressing the 3xFLAG N-tagged IEP but lacking the ribozyme component of the intron, and the pKGEMA4 plasmid (Nisa-Martínez et al., 2007) expressing untagged active RNPs as a negative control (Figure 2). Previous works using pKGEMA4-derivative plasmids have never shown any negative effect in cell viability (Nisa-Martínez et al., 2007; García-Rodríguez et al., 2014). pKG4_FlagIEP was obtained by inserting a fragment consisting of two annealed oligonucleotides with cohesive ends containing a 3xFLAG epitope sequence into an engineered pKGEMA4 plasmid (pKGEMA4NB, containing an NdeI site at the ATG of the IEP): 5′3xflag (5′-CTAGTGGAAACAGGATGGACTACAAAGACCATGACGGTGATTATAAAGATCATGACATCGATTACAAGGATGACGATGACAAGCA-3′) and 3′3xflag (5′-TATGCTTGTCATCGTCATCCTTGTAATCGATGTCATGATCTTTATAATCACCGTCATGGTCTTTGTAGTCCATCCTGTTTCCA-3′). pKG_FlagIEP was obtained by deleting the ΔORF from pKG4_FlagIEP by cleavage at the SacI restriction sites flanking the ribozyme. Biochemical activities were assayed as previously described (Supplementary Figure S2) (García-Rodríguez et al., 2019).
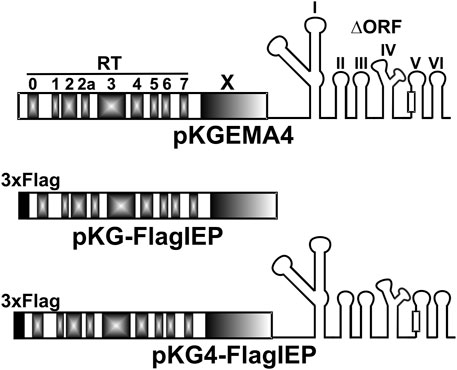
FIGURE 2. Intron constructs used in the ChIP-Seq experiments. pKGEMA4 contains the wild-type IEP sequence followed by the intron ribozyme with a partial deletion in domain IV, and gives rise to biochemically active RNPs (Nisa-Martínez et al., 2007). Different domains in the IEP are shown in gray. The square in domain V represents the catalytic triad. pKG-FlagIEP encodes the N-tagged IEP with the 3xFLAG epitope (black square) but no intron RNA. Functional RNPs are also produced from pKG4-FlagIEP, but with a 3xFLAG epitope label at the N-terminal end of the IEP.
ChIP experiments were performed as previously described (Spencer et al., 2003; O’Geen et al., 2010; Bonocora and Wade, 2015; Pini et al., 2015). Briefly, cells in early exponential growth phase (50 ml, OD600 of 0.4) were crosslinked by incubation in 1% formaldehyde in 10 mM sodium phosphate (pH 7.6) for 20 min at 30°C on an orbital shaker. Triplicate cultures were set up. Crosslinking was stopped by rapid cooling of the cells on ice, followed by the addition of cold 125 mM glycine. The cultures were then incubated for 5 minutes at room temperature with shaking. The cells were washed three times in phosphate-buffered saline (PBS) and fast-frozen on liquid nitrogen. Cultures were lysed by incubation with 0.4 mg ml−1 lysozyme in IP buffer (50 mM Tris-HCl pH 7.5, 1 mM EDTA, 150 mM NaCl, and EDTA-free protease inhibitor [Roche]). We obtained 0.2-0.5 kb DNA fragments by shearing the DNA by sonication (Branson Digital Sonicator 450) on ice, with 27 bursts of 20 s each (50% duty) at 10% amplitude. The resulting lysates were cleared by centrifugation at 10,000 rpm for 15 min at 4°C. At this point, we reserved 1/20 of the cell lysate as the input sample. Anti-FLAG® M2 magnetic beads (Sigma-Aldrich) were added to the rest of lysate, which was then incubated overnight at 4°C on a rotary shaker. ChIP samples were washed four times in TBS buffer (50 mM Tris-HCl pH 7.5, 150 mM NaCl) for 5 min each, at 4°C. The protein/DNA complexes were eluted in 500 μL freshly prepared ChIP elution buffer (50 mM Tris-HCl pH 7.5, 1 mM EDTA, 150 mM NaCl, supplemented with 150 ng μL−1 3xFLAG peptide) for 1 h at 4°C. The input and ChIP samples were incubated overnight at 65°C to reverse the crosslinking. We monitored the immunoprecipitation efficiency by dot-blot using anti-FLAG antibodies (data not shown). The samples were then successively treated with 100 μg of RNaseA for 2 h at 45°C and 100 μg of proteinase K for 2 h at 55°C. The DNA was purified by phenol extraction followed by ethanol precipitation.
A total of 18 samples were studied. Nine DNA samples were control (input) samples and the remaining samples corresponded with immunoprecipitated DNA. In turn, each group of nine samples came from bacterial cultures carrying three different plasmid constructs: A plasmid that expresses non-tagged, functional RNPs (pKGEMA4); a vector that produces 3xFLAG-tag IEP (pKG_FlagIEP); or, finally, a construct which generates 3xFLAG-tagged, active RNPs (pKG4_FlagIEP) (Figure 2). Experiments were performed in triplicates.
Subsequently, DNA samples were processed by standard protocols and sequenced at the IPBLN Genomics Unit on an Illumina Nextseq500 with 75-bp single-end reads. Finally, a total of 30 million reads were obtained (on average). Quality assessment and samples alignment were performed using the miARma-Seq software (Andrés-León et al., 2016). In detail, miARma-seq contains all the required software to process most type of NGS samples (Figure 3). In the first step, fastqc V.0.11.5 was applied to gather the overall sequence quality and to identify possible adapter accumulation (Andrews, 2010). The Per base sequence quality and the Per sequence quality scores showed that most of the reads quality were above 30. The Per base sequence content displayed a proportional distribution of the four nucleotides along the 75 bp, besides, no Illumina adapter was found in the adapter content section of the fastqc report. However, we use minion (Davis et al., 2013), a software that predict possible adapter sequences by reading the first nucleotides of the reads searching for a consensus sequence. In our case no consensus sequence was found. Next, we use Cutadapt (Martin, 2011) to filter those reads having a quality score below 30.
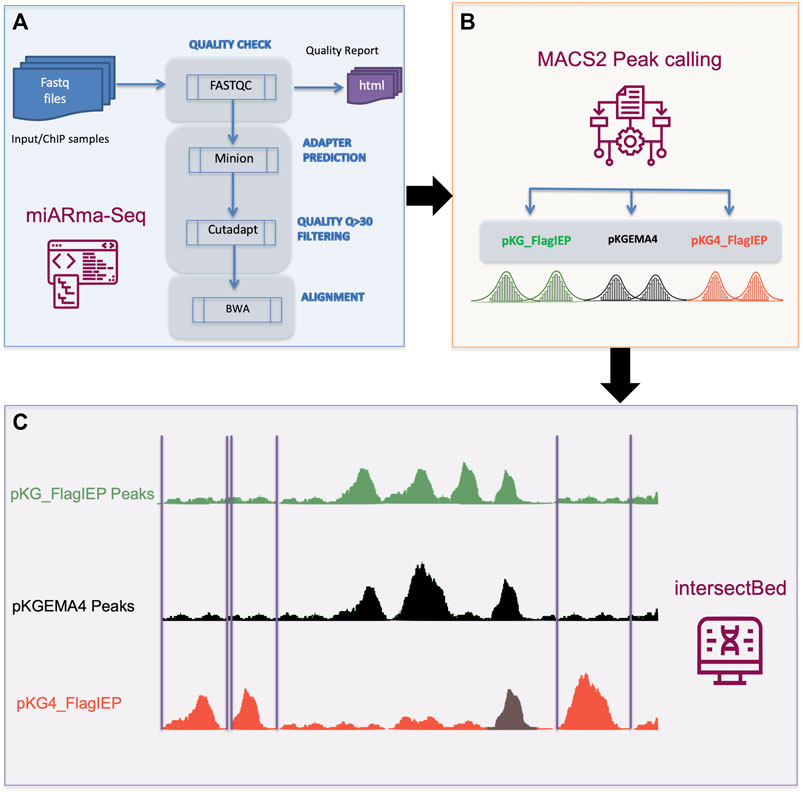
FIGURE 3. ChIP-Seq data analysis pipeline. (A) Quality assessment and samples alignment were performed using the miARma-Seq software. Fastqc 0.11.5 was used to study the overall sequence quality and to identify possible adapter accumulation. Furthermore, we applied minion to identify any possible adapter sequences. As none were found, we used cutadapt to select reads having a Phred score > 30. Next, those sequences were aligned using BWA. (B) Resulting files were processed with MACS2 to identify statically reliable peaks (FDR < 0.001) per each group of samples. (C) IntersectBed from bedtools were used to identify unique peaks in pKG4_FlagIEP by removing common peaks in pKG_FlagIEP or pKGEMA4.
Finally, an average of 27 M (>90%) reads per sample were used for the next step. We then aligned all the samples with the Sinorhizobium meliloti strain RMO17 reference genome from GenBank (accession numbers CP009144 to CP009146) using default parameters in Burrows-Wheeler Aligner software (Li and Durbin, 2010). On average we obtained an alignment percentage of reads against the reference genome above 97.53%. Aligned files were processed with MACS2 (Zhang et al., 2008) to identify statically reliable peaks. To achieve this, we analyze the three inputs and the three ChIP samples simultaneously in a single MACS2 execution, instead of separated running of the replicates and then combine common peaks. By running the samples together, we can achieve more reliable peaks even with moderately low enrichment (Wilbanks and Facciotti, 2010). Besides, the common parameter in MACS2 used in each of the three group were -f BAM --nomodel --keep-dup all -g 6.7e6. Only peaks statistically significant [LOG10 (q value) > 3 (FDR < 0.001)] were considered.
The resulting sets of enriched peaks obtained for each of the three plasmid constructs were analyzed with IntersectBed (belonging to the Bedtools suite; Quinlan and Hall, 2010) to obtain the set of common or unique peaks in each set. Once the overlapping peaks between samples from different constructs were removed, we focus to describe the unique peaks in the pKG4_Flag-IEP samples. This was accomplished in two steps: first, intersectBed -a pKG4_FlagIEP -b pKG_FlagIEP -v > file1.bed; and next, intersectBed -a file1.bed -b pKGEMA4 -v > UniquePeaks_ pKG4_FlagIEP.bed). The final bed file contains 321 statistically reliable peaks FDR< 0.001 not shared neither by FlagIEP nor by pKGEMA4.
The raw sequencing data have been deposited in the SRA database with BioProject accession number PRJNA779974.
Chromatin immunoprecipitation followed by Illumina high-throughput next-generation sequencing (ChIP-Seq) has already been successfully used to identify transcription factors providing a high-resolution snapshot of protein/DNA interactions (Spencer et al., 2003; Park, 2009; Furey, 2012; Bonocora and Wade, 2015). We aim to identify the regions of the S. meliloti genome binding RmInt1 RNPs in vivo using ChIP-Seq analyses, by introducing various plasmid constructs into RMO17, an intron-less S. meliloti strain (Figure 2). pKG4-FlagIEP encoded 3xFLAG-tagged active RNPs (Supplementary Figure S2; Reinoso-Colacio et al., 2015); pKG-FlagIEP expressed the FLAG-tagged IEP but lacking the ribozyme component of the intron; finally, as an IP control, pKGEMA4 encoded untagged functional RNPs (Nisa-Martínez et al., 2007). We constructed 18 libraries corresponding to the input and ChIP samples in triplicate, and we obtained a total of 508,880,560 high-quality reads, which we then mapped back onto the S. meliloti RMO17 genome (Supplementary Table S1), reaching around 300-fold genome coverage for each individual library. We found 574 potential binding sites corresponding to the construct producing active RNPs (pKG4_FlagIEP); 276 enriched regions for the construct expressing the IEP alone (pKG_FlagIEP); and, 428 peaks in the untagged control (pKGEMA4) (Supplementary Table S2). Then, we filtered out peaks resulting from IEP interactions in the absence of the ribozyme by comparing the binding sites identified when the 3xFLAG IEP was introduced alone (pKG_FlagIEP) to the peaks observed when the labelled protein was present together with the intron RNA forming active RNPs (pKG4_FlagIEP). Further filtering was performed by removing any peaks found in non-tagged functional RNPs (pKGEMA4), which resulted in 321 distinct DNA fragments appearing only in the FLAG-tagged, functional RNP (pKG4_FlagIEP) output data (Figure 3).
Most ChIP-seq reports consider peaks to be significant for an enrichment ≥ 1.5-2-fold (Myers et al., 2015; Šmídová et al., 2019), and the binding of some transcription factors, generally to specific motifs, results in a 10- to several hundred-fold enrichment (Martínez-Granero et al., 2014). However, the majority of the RmInt1 RNPs binding DNA sequences identified in our study (77%) showed a signal-to-noise ratio [S/N] ranging 1.08-1.14 (Supplementary Figure S3A). A 19% of the identified peaks exhibited a fold-enrichment ratio falling between 1.14 and 1.18, and only a few regions (5) presented enrichment ratios beyond 1.18 up to 1.23. ChIP-seq enrichment after immunoprecipitation is influenced by the strength of the interaction. Furthermore, low-enrichment signals in ChIP-seq experiments may reflect indirect binding to a third counterpart bound to DNA (Furey, 2012). The low ChIP signals in our samples may, therefore, reflect weak interactions of the RmInt1 RNPs with the genomic DNA, or may suggest that DNA binding is dependent on other interacting factors, for instance, DnaN (García-Rodríguez et al., 2019).
S. meliloti RMO17 genome contains 13 copies of the natural homing site of RmInt1, ISRm2011-2 (Toro et al., 2014). However, we were unable to detect significant enrichment relative to the input DNA around any of the 13 copies of the insertion sequence by qPCR on ChIP samples (Supplementary Figure S4A). Similar numbers of reads were recruited for all libraries when we considered the full-length sequence of ISRm2011-2 (Supplementary Figure S4B). Nevertheless, some differences at the IS intron insertion site were observed in the immunoprecipitated samples. We scanned the ChIP-seq libraries for three 25 nt sequences: the RmInt1 insertion site (5′-CCTCGTTTTCATCGATGAGACCTGG-3′), a sequence located 50 nt upstream (5′-CGAACGGGAGCGGCCCGACGTCGCC-3′) or a sequence located 50 nt downstream (5′-ACTGGTGGGCTACGCCCCCTTCGGC-3′). We determined the number of sequences contained in each library that aligned with the above described regions, normalizing by the total number of sequences in the corresponding library, as a means of comparing data for different libraries (Supplementary Figure S4B). Interestingly, a significant bias towards a decreased number of recruited sequences containing the insertion site was observed when active RNPs were present, probably due to intron insertion during retrohoming process. Thus, the binding of RmInt1 RNPs to DNA may not be restricted to specific sequence motifs/sequences in agreement with a previously suggested nonspecific attachment of the intron RNPs to DNA (Aizawa et al., 2003).
S. meliloti RMO17 has a tripartite genome comprising a 3.65 Mb chromosome, and two symbiotic megaplasmids: pSymA (1.47 Mb) and pSymB (1.61 Mb) (Toro et al., 2014). We identified 321 regions displaying some binding to RmInt1 active RNPs, ranging from 200–800 bp in length (Supplementary Figure S3B; Supplementary Table S2). Most of the enriched peaks corresponded to chromosomal regions (266), with only a minority of the identified sequences mapping to the megaplasmid pSymA (11) and the pSymB chromid (44). The precise coordination of genome replication, chromosome segregation and cell division is essential for population fitness, and bacteria with multipartite genomes must conserve one copy of each replicon per cell cycle (Frage et al., 2016). Differences in the proportions of regions potentially bound by RmInt1 RNPs cannot, therefore, simply be attributed to differences in copy number or replicon size. They may instead reflect differences in replicon accessibility.
We then considered possible bias in associations with respect to genome architecture. Most of the binding regions identified were located in annotated genes (51), with a certain preference toward the 3′-end of the coding sequence (Figure 4A). We also identified bound DNA fragments in intergenic regions (5), and others extending to the 5′ end of the downstream gene (11), the 3′ end of the upstream gene (27), or to both these positions (37). Again, we observed a higher number of peaks comprising the 3′-end of the gene and the downstream IR compared to the regions containing 5′CDS + IR. Although a certain bias could be considered, more evidences need to be obtained in order to establish a believable correlation between intron biology and RmInt1 RNP binding to DNA at the 3′-end of genes.
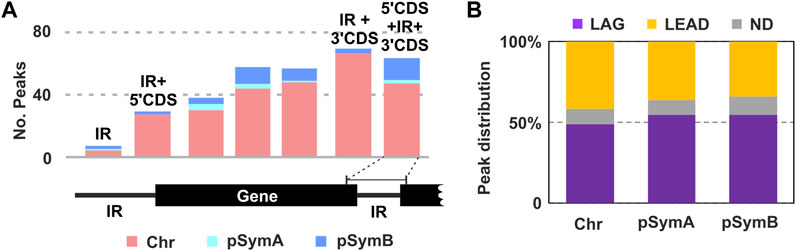
FIGURE 4. Result of the RmInt1 ChIP-Seq analysis. (A) Genomic localization of the identified RmInt1 binding sites. Bar plot showing the number of peaks found in either the coding sequence of a gene (CDS) or the intergenic regions (IR). Counts are itemized by replicon: pink corresponds to binding sites identified in the chromosome; sky blue corresponds to the pSymA megaplasmid; and, the darker blue corresponds to the pSymB chromid. IR + 3′CDS is not included in the 5′CDS+IR+3′CDS. (B) The proportions of enriched DNA sequences localizing at the template for the lagging (LAG, purple) and leading (LEAD, yellow) strands at the replication fork are shown as a percentage in a stacked bar graph. Data are presented by replicon. ND, non-determined (gray).
Previous studies have reported a preferred insertion site of the RmInt1 ribozyme on the template for lagging strand synthesis during DNA replication (Martínez-Abarca et al., 2004). We wondered whether the binding of RmInt1 RNPs displayed a similar bias genome-wide. According to the annotations of the origin and termination of replication in the genome sequence (Lato and Golding, 2020), we calculated, where possible, the most probable orientation of the binding sequences with respect to the movement of the replication fork (Figure 4B). In general, a certain preference for RmInt1 RNP binding to the template for lagging strand synthesis was detected in all three replicons, consistent with the intron colonization bias observed in nature (Nisa-Martínez et al., 2007). Since DNA transcription and replication are spatiotemporally coordinated in bacteria, head-on collisions between protein machineries during lagging strand synthesis cause replication arrest (Rocha, 2004). Thus, it is tempting to speculate that the increased availability of single-stranded DNA favors binding of En- intron RNPs.
Next, we wondered whether RmInt1 RNP binding displayed positional preferences in the genome. Then, the 321 enriched regions identified in our study were mapped in S. meliloti RMO17 genome. Remarkably, we observed a clear, biased distribution around the replication origins in each of the replicons: The chromosome, pSymA and pSymB (Figure 5). A similar result has been reported for insertion of the mobile Lactococcus lactis Ll.LtrB intron at sites clustered near the bidirectional oriC in the E. coli genome (Zhong et al., 2003). Moreover, in E. coli, the location of the wild-type Ll.LtrB retrotranspose around the origin and terminus for chromosomal DNA replication is influenced by growth conditions (Coros et al., 2005). This behavior is not exclusive to group II introns; insertion sequences and transposons also use mechanisms that coordinate transposition and DNA replication (Hu and Derbyshire, 1998; Peters and Craig, 2001).
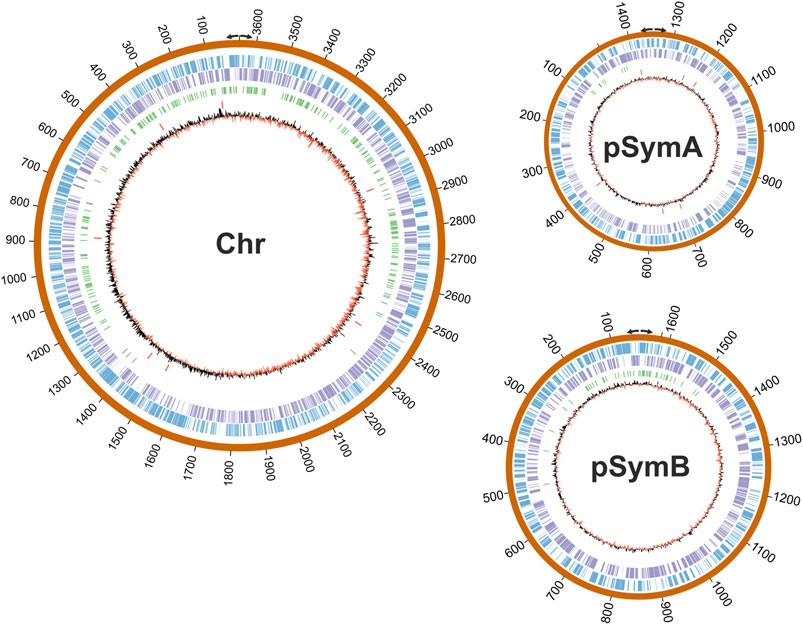
FIGURE 5. Mapping of the RmInt1 binding sites in the S. meliloti RMO17 genome. The three replicons are drawn to scale with CIRCOS. The outer track in blue corresponds to the forward annotated genes (top strand) and the purple circle corresponds to the reverse orientation genes (bottom strand). Identified ChIP-Seq peaks are shown in green and the 13 copies of the natural target sequence of RmInt1 (ISRm20011-2) are indicated using red bars. The skewed GC plot is shown in red/black lines. Divergent arrows indicate the replication origin of each plasmid according to the data published by Lato and Golding (2020).
It had been described that fast-growing bacteria requires simultaneous rounds of replication to achieve their growth rates (Rocha, 2004). Thus, regions close to the replication origin may be overrepresented relative to the regions distal to the replication origin. S. meliloti could be considered a moderately fast-growing bacteria with an optimal duplication time around 140 min (De Nisco et al., 2014). If the replication fork moves at about 600-1,000 nt s−1 (Rocha, 2004), duplication time should be enough for the completion of the chromosome synthesis in exponential growing S. meliloti cell cultures. As mentioned, experiments of segregation timing suggest that DNA replication occurs only once per cell cycle (Frage et al., 2016). Since we observed a bias of the RmInt1 RNPs around the origin of replication, we aim to discard this bias is due to differences in DNA abundance along the replicon. In that sense, we have calculated the coverage for each sample and performed a t-test of the mean coverage in control (input) against immunoprecipitated samples. This calculation gave a non-significant p-value > 0.05 when considered either the whole replicon or only the first 1,000 nt from the replication origin, indicating that the distribution of DNA reads is similar in both kind of samples (data not shown).
Many host factors, mostly related to the replication machinery, contribute to the retrohoming of group II introns (Smith et al., 2005; Coros et al., 2008; Zhao et al., 2008; Coros et al., 2009; Yao et al., 2013; Nisa-Martínez et al., 2016). One recent study revealed that the RmInt1 IEP and active RNPs interact with DnaN (β-sliding clamp), a replicative protein that forms part of the DNA polymerase III complex (García-Rodríguez et al., 2019). An analysis of S. meliloti DnaN-mCherry dynamics revealed a strict spatiotemporal localization directly connected with the order of segregation of the three bacterial replicons (Frage et al., 2016). The bidirectional replication of the chromosome occurs first, followed by the megaplasmid pSymA and then by the chromid pSymB (De Nisco et al., 2014; Frage et al., 2016). DnaN-mCherry was found to disperse before the onset of a new cell division event. By contrast to the polar location of the chromosome origin of replication, the replication origins of the megaplamids are subpolar after the completion of segregation. On the other hand, fluorescence microscopy has shown that RmInt1 IEP-EGFP RNPs are mostly dispersed throughout the cell, but that a small proportion localize at several foci within exponentially growing S. meliloti RMO17 cells (Nisa-Martínez et al., 2013). Similar results were described for GFP fusions of the Ll.ltrB group II intron (Zhao and Lambowitz, 2005; Beauregard et al., 2006; Zhao et al., 2008). We therefore suggest that interaction with DnaN may control the polar localization of RmInt1 RNPs, and that the subpolar position of the origins of megaplamids influences the binding events around these origins. The preferential location of binding sites around the origin of replication may be related to replicon segregation times.
Our results provide new evidences that support a connection between intron functionality and host DNA replication. The nonspecific binding of RmInt1 RNPs to S. meliloti genomic DNA showed a preference for the template of the lagging strand during replication. Moreover, we observed preferential binding around the origin of replication. These preferences may be related to chromatin accessibility, or the interaction of the RNPs with host replicative proteins, but also binding can be conducted by the intron RNPs distribution into the bacterial cells during DNA replication. These observations need to be considered when reprogrammed introns are used for whole genome knockout approaches.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/, PRJNA779974.
Conceptualization, NT; Methodology, MDM-S, FMG-R, and EA-L; Experiments and formal analyses, MDM-S; Writing original draft, MDM-S; Writing review and editing, all authors; Supervision and funding acquisition, NT.
This work was supported by grant BIO2017-82244-P from the MCIN /AEI/10.13039/501100011033/FEDER “Una manera de hacer Europa” and grant PID2020-113207GB-I00 from the MCIN/ AEI/10.13039/501100011033. EA-L was recipient of a postdoctoral fellowship from regional Andalusian Government (2020_DOC_00541).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We thank Ascensión Martos and Jose Maria del Arco for technical assistance and Francisco Martínez-Abarca for helpful discussions. Library preparation and Illumina sequencing were carried out at the IPBLN Genomics Facility (CSIC, Granada, Spain), and we thank Alicia Barroso del Jesus for her assistance.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmolb.2022.834020/full#supplementary-material
Agrawal, R. K., Wang, H.-W., and Belfort, M. (2016). Forks in the Tracks: Group II Introns, Spliceosomes, Telomeres and beyond. RNA Biol. 13, 1218–1222. doi:10.1080/15476286.2016.1244595
Aizawa, Y., Xiang, Q., Lambowitz, A. M., and Pyle, A. M. (2003). The Pathway for DNA Recognition and RNA Integration by a Group II Intron Retrotransposon. Mol. Cel 11, 795–805. doi:10.1016/s1097-2765(03)00069-8
Andrés-León, E., Núñez-Torres, R., and Rojas, A. M. (2016). miARma-Seq: A Comprehensive Tool for miRNA, mRNA and circRNA Analysis. Sci. Rep. 6, 25749. doi:10.1038/srep25749
Andrews, S. (2010). FastQC: A Quality Control Tool for High Throughput Sequence Data. Available online at: http://www.bioinformatics.babraham.ac.uk/projects/fastqc fastQC version 0.11.5..
Beauregard, A., Chalamcharla, V. R., Piazza, C. L., Belfort, M., and Coros, C. J. (2006). Bipolar Localization of the Group II Intron Ll.LtrB Is Maintained in Escherichia Coli Deficient in Nucleoid Condensation, Chromosome Partitioning and DNA Replication. Mol. Microbiol. 62, 709–722. doi:10.1111/j.1365-2958.2006.05419.x
Belfort, M., and Lambowitz, A. M. (2019). Group II Intron RNPs and Reverse Transcriptases: From Retroelements to Research Tools. Cold Spring Harb. Perspect. Biol. 11, a032375. doi:10.1101/cshperspect.a032375
Bonocora, R. P., and Wade, J. T. (2015). “ChIP-Seq for Genome-Scale Analysis of Bacterial DNA-Binding Proteins,” in Bacterial Transcriptional Control: Methods and Protocols, Methods in Molecular Biology. Editors I. Artsimovitch, and T. J. Santangelo (NY: Springer Science+Business Media), 1276, 327–340. doi:10.1007/978-1-4939-2392-2_20
Coros, C. J., Landthaler, M., Piazza, C. L., Beauregard, A., Esposito, D., Perutka, J., et al. (2005). Retrotransposition Strategies of the Lactococcus Lactis Ll.LtrB Group II Intron Are Dictated by Host Identity and Cellular Environment. Mol. Microbiol. 56, 509–524. doi:10.1111/j.1365-2958.2005.04554.x
Coros, C. J., Piazza, C. L., Chalamcharla, V. R., and Belfort, M. (2008). A Mutant Screen Reveals RNase E as a Silencer of Group II Intron Retromobility in Escherichia Coli. RNA 14, 2634–2644. doi:10.1261/rna.1247608
Coros, C. J., Piazza, C. L., Chalamcharla, V. R., Smith, D., and Belfort, M. (2009). Global Regulators Orchestrate Group II Intron Retromobility. Mol. Cel 34, 250–256. doi:10.1016/j.molcel.2009.03.014
Cousineau, B., Smith, D., Lawrence-Cavanagh, S., Mueller, J. E., Yang, J., Mills, D., et al. (1998). Retrohoming of a Bacterial Group II Intron: Mobility via Complete Reverse Splicing, Independent of Homologous DNA Recombination. Cell 94, 451–462. doi:10.1016/s0092-8674(00)81586-x
Davis, S. E., Mooney, R. A., Kanin, E. I., Grass, J., Landick, R., and Ansari, A. Z. (2011). Mapping E. Coli RNA Polymerase and Associated Transcription Factors and Identifying Promoters Genome-Wide. Methods Enzymol. 498, 449–471. doi:10.1016/B978-0-12-385120-8.00020-6
Davis, M. P., van Dongen, S., Abreu-Goodger, C., Bartonicek, N., and Enright, A. J. (2013). Kraken: A Set of Tools for Quality Control and Analysis of High-Throughput Sequence Data. Methods 63, 41–49. doi:10.1016/j.ymeth.2013.06.027
De Nisco, N. J., Abo, R. P., Wu, C. M., Penterman, J., and Walker, G. C. (2014). Global Analysis of Cell Cycle Gene Expression of the Legume SymbiontSinorhizobium Meliloti. Proc. Natl. Acad. Sci. USA 111, 3217–3224. doi:10.1073/pnas.1400421111
Enyeart, P. J., Mohr, G., Ellington, A. D., and Lambowitz, A. M. (2014). Biotechnological Applications of mobile Group II Introns and Their Reverse Transcriptases: Gene Targeting, RNA-Seq, and Non-Coding RNA Analysis. Mobile DNA 5, 2. doi:10.1186/1759-8753-5-2
Ferat, J.-L., and Michel, F. (1993). Group II Self-Splicing Introns in Bacteria. Nature 364, 358–361. doi:10.1038/364358a0
Fernández-López, M., Muñoz-Adelantado, E., Gillis, M., Willems, A., and Toro, N. (2005). Dispersal and Evolution of the Sinorhizobium Meliloti Group II RmInt1 Intron in Bacteria that Interact with Plants. Mol. Biol. Evol. 22, 1518–1528. doi:10.1093/molbev/msi144
Frage, B., Döhlemann, J., Robledo, M., Lucena, D., Sobetzko, P., Graumann, P. L., et al. (2016). Spatiotemporal Choreography of Chromosome and Megaplasmids in theSinorhizobium Meliloticell Cycle. Mol. Microbiol. 100, 808–823. doi:10.1111/mmi.13351
Furey, T. S. (2012). ChIP-Seq and beyond: New and Improved Methodologies to Detect and Characterize Protein-DNA Interactions. Nat. Rev. Genet. 13, 840–852. doi:10.1038/nrg3306
Galibert, F., Finan, T. M., Long, S. R., Pühler, A., Abola, P., Ampe, F., et al. (2001). The Composite Genome of the Legume Symbiont Sinorhizobium Meliloti. Science 293, 668–672. doi:10.1126/science.1060966
García-Rodríguez, F. M., Hernández-Gutiérrez, T., Díaz-Prado, V., and Toro, N. (2014). Use of the Computer-Retargeted Group II Intron RmInt1 of Sinorhizobium Melilotifor Gene Targeting. RNA Biol. 11, 391–401. doi:10.4161/rna.28373
García-Rodríguez, F. M., Neira, J. L., Marcia, M., Molina-Sánchez, M. D., and Toro, N. (2019). A Group II Intron-Encoded Protein Interacts with the Cellular Replicative Machinery through the β-Sliding Clamp. Nucleic Acids Res. 47, 7605–7617. doi:10.1093/nar/gkz468
García-Rodríguez, F. M., Barrientos-Durán, A., Díaz-Prado, V., Fernández-López, M., and Toro, N. (2011). Use of RmInt1, a Group IIB Intron Lacking the Intron-Encoded Protein Endonuclease Domain, in Gene Targeting. Appl. Environ. Microbiol. 77, 854–861. doi:10.1128/AEM.02319-10
Guo, H., Zimmerly, S., Perlman, P. S., and Lambowitz, A. M. (1997). Group II Intron Endonucleases Use Both RNA and Protein Subunits for Recognition of Specific Sequences in Double-Stranded DNA. EMBO J. 16, 6835–6848. doi:10.1093/emboj/16.22.6835
Haack, D. B., and Toor, N. (2020). Retroelement Origins of Pre‐mRNA Splicing. WIREs RNA 11, e1589. doi:10.1002/wrna.1589
Hu, W.-Y., and Derbyshire, K. M. (1998). Target Choice and Orientation Preference of the Insertion Sequence IS 903. J. Bacteriol. 180, 3039–3048. doi:10.1128/JB.180.12.3039-3048.1998
Ichiyanagi, K., Beauregard, A., Lawrence, S., Smith, D., Cousineau, B., and Belfort, M. (2002). Retrotransposition of the Ll.LtrB Group II Intron Proceeds Predominantly via Reverse Splicing into DNA Targets. Mol. Microbiol. 46, 1259–1272. doi:10.1046/j.1365-2958.2002.03226.x
Jiménez-Zurdo, J. I., García-Rodríguez, F. M., Barrientos-Durán, A., and Toro, N. (2003). DNA Target Site Requirements for Homing In Vivo of a Bacterial Group II Intron Encoding a Protein Lacking the DNA Endonuclease Domain. J. Mol. Biol. 326, 413–423. doi:10.1016/s0022-2836(02)01380-3
Lambowitz, A. M., and Belfort, M. (2015). Mobile Bacterial Group II Introns at the Crux of Eukaryotic Evolution. Microbiol. Spectr. 3 (1), MDNA3-0050-2014. doi:10.1128/microbiolspec.MDNA3-0050-2014
Lambowitz, A. M., Caprara, M. G., Zimmerly, S., and Perlman, P. S. (1999). “Group I and Group II Ribozymes as RNPs: Clues to the Past and Guides to the Future,” in The RNA World. Editors R. F. Gesteland, R. F. Cech, and J. F. Atkins. 2nd ed. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press), 451–485.
Lambowitz, A. M., and Zimmerly, S. (2011). Group II Introns: mobile Ribozymes that Invade DNA. Cold Spring Harbor Perspect. Biol. 3, a003616. doi:10.1101/cshperspect.a003616
Lambowitz, A. M., and Zimmerly, S. (2004). Mobile Group II Introns. Annu. Rev. Genet. 38, 1–35. doi:10.1146/annurev.genet.38.072902.091600
Lato, D. F., and Golding, G. B. (2020). Spatial Patterns of Gene Expression in Bacterial Genomes. J. Mol. Evol. 88, 510–520. doi:10.1007/s00239-020-09951-3
Li, H., and Durbin, R. (2010). Fast and Accurate Long-Read Alignment with Burrows-Wheeler Transform. Bioinformatics 26, 589–595. doi:10.1093/bioinformatics/btp698
Martin, M. (2011). Cutadapt Removes Adapter Sequences from High-Throughput Sequencing Reads. EMBnet j. 17, 10–12. doi:10.14806/ej.17.1.200
Martin, W., and Koonin, E. V. (2006). Introns and the Origin of Nucleus-Cytosol Compartmentalization. Nature 440, 41–45. doi:10.1038/nature04531
Martinez-Abarca, F., Barrientos-Durán, A., Fernández-López, M., and Toro, N. (2004). The RmInt1 Group II Intron Has Two Different Retrohoming Pathways for Mobility Using Predominantly the Nascent Lagging Strand at DNA Replication forks for Priming. Nucleic Acids Res. 32, 2880–2888. doi:10.1093/nar/gkh616
Martínez-Abarca, F., García-Rodríguez, F. M., and Toro, N. (2000). Homing of a Bacterial Group II Intron with an Intron-Encoded Protein Lacking a Recognizable Endonuclease Domain. Mol. Microbiol. 35, 1405–1412. doi:10.1046/j.1365-2958.2000.01804.x
Martínez-Abarca, F., Zekri, S., and Toro, N. (1998). Characterization and Splicing In Vivo of a Sinorhizobium Meliloti Group II Intron Associated with Particular Insertion Sequences of the IS630-Tc1/IS3 Retroposon Superfamily. Mol. Microbiol. 28, 1295–1306. doi:10.1046/j.1365-2958.1998.00894.x
Martínez-Granero, F., Redondo-Nieto, M., Vesga, P., Martín, M., and Rivilla, R. (2014). AmrZ Is a Global Transcriptional Regulator Implicated in Iron Uptake and Environmental Adaption in P. Fluorescens F113. BMC Genomics 15, 237. doi:10.1186/1471-2164-15-237
Mohr, G., Smith, D., Belfort, M., and Lambowitz, A. M. (2000). Rules for DNA Target-Site Recognition by a Lactococcal Group II Intron Enable Retargeting of the Intron to Specific DNA Sequences. Genes Dev. 14, 559–573. doi:10.1101/gad.14.5.559
Mohr, S., Ghanem, E., Smith, W., Sheeter, D., Qin, Y., King, O., et al. (2013). Thermostable Group II Intron Reverse Transcriptase Fusion Proteins and Their Use in cDNA Synthesis and Next-Generation RNA Sequencing. RNA 19, 958–970. doi:10.1261/rna.039743.113
Monat, C., and Cousineau, B. (2020). The circle to Lariat Ratio of the Ll.LtrB Group II Intron from Lactococcus Lactis Is Greatly Influenced by a Variety of Biological Determinants In Vivo. PLoS One 15, e0237367. doi:10.1371/journal.pone.0237367
Myers, K. S., Park, D. M., Beauchene, N. A., and Kiley, P. J. (2015). Defining Bacterial Regulons Using ChIP-Seq. Methods 86, 80–88. doi:10.1016/j.ymeth.2015.05.022
Nisa-Martínez, R., Jiménez-Zurdo, J. I., Martínez-Abarca, F., Muñoz-Adelantado, E., and Toro, N. (2007). Dispersion of the RmInt1 Group II Intron in the Sinorhizobium Meliloti Genome upon Acquisition by Conjugative Transfer. Nucleic Acids Res. 35, 214–222. doi:10.1093/nar/gkl1072
Nisa-Martínez, R., Laporte, P., Jiménez-Zurdo, J. I., Frugier, F., Crespi, M., and Toro, N. (2013). Localization of a Bacterial Group II Intron-Encoded Protein in Eukaryotic Nuclear Splicing-Related Cell Compartments. PLoS One 8, e84056. doi:10.1371/journal.pone.0084056
Nisa-Martínez, R., Molina-Sánchez, M. D., and Toro, N. (2016). Host Factors Influencing the Retrohoming Pathway of Group II Intron RmInt1, Which Has an Intron-Encoded Protein Naturally Devoid of Endonuclease Activity. PLoS ONE 11, e0162275. doi:10.1371/journal.pone.0162275
Novikova, O., and Belfort, M. (2017). Mobile Group II Introns as Ancestral Eukaryotic Elements. Trends Genet. 33, 773–783. doi:10.1016/j.tig.2017.07.009
Novikova, O., Smith, D., Hahn, I., Beauregard, A., and Belfort, M. (2014). Interaction between Conjugative and Retrotransposable Elements in Horizontal Gene Transfer. Plos Genet. 10, e1004853. doi:10.1371/journal.pgen.1004853
O’Geen, H., Frietze, S., and Farnham, P. J. (2010). Using ChIP-Seq Technology to Identify Targets of Zinc finger Transcription Factors. Methods Mol. Biol. 649, 437–455. doi:10.1007/978-1-60761-753-2_27
Park, P. J. (2009). ChIP-seq: Advantages and Challenges of a Maturing Technology. Nat. Rev. Genet. 10, 669–680. doi:10.1038/nrg2641
Peters, J. E., and Craig, N. L. (2001). Tn7: Smarter Than We Thought. Nat. Rev. Mol. Cel. Biol. 2, 806–814. doi:10.1038/35099006
Pini, F., De Nisco, N. J., Ferri, L., Penterman, J., Fioravanti, A., Brilli, M., et al. (2015). Cell Cycle Control by the Master Regulator CtrA in Sinorhizobium Meliloti. Plos Genet. 11, e1005232. doi:10.1371/journal.pgen.1005232
Qu, G., Kaushal, P. S., Wang, J., Shigematsu, H., Piazza, C. L., Agrawal, R. K., et al. (2016). Structure of a Group II Intron in Complex with its Reverse Transcriptase. Nat. Struct. Mol. Biol. 23, 549–557. doi:10.1038/nsmb.3220
Quinlan, A. R., and Hall, I. M. (2010). BEDTools: A Flexible Suite of Utilities for Comparing Genomic Features. Bioinformatics 26, 841–842. doi:10.1093/bioinformatics/btq033
Reinoso-Colacio, M., García-Rodríguez, F. M., García-Cañadas, M., Amador-Cubero, S., Pérez, J. L. G., and Toro, N. (2015). Localization of a Bacterial Group II Intron-Encoded Protein in Human Cells. Sci. Rep. 5, 12716. doi:10.1038/srep12716
Rocha, E. P. C. (2004). The Replication-Related Organization of Bacterial Genomes. Microbiology 150, 1609–1627. doi:10.1099/mic.0.26974-0
Saldanha, R., Chen, B., Wank, H., Matsuura, M., Edwards, J., and Lambowitz, A. M. (1999). RNA and Protein Catalysis in Group II Intron Splicing and Mobility Reactions Using Purified Components. Biochemistry 38, 9069–9083. doi:10.1021/bi982799l
San Filippo, J., and Lambowitz, A. M. (2002). Characterization of the C-Terminal DNA-Binding/DNA Endonuclease Region of a Group II Intron-Encoded Protein. J. Mol. Biol. 324, 933–951. doi:10.1016/s0022-2836(02)01147-6
Singh, N. N., and Lambowitz, A. M. (2001). Interaction of a Group II Intron Ribonucleoprotein Endonuclease with its DNA Target Site Investigated by DNA Footprinting and Modification Interference. J. Mol. Biol. 309, 361–386. doi:10.1006/jmbi.2001.4658
Šmídová, K., Ziková, A., PospíšilSchwarz, J. M., Schwarz, M., Bobek, J., and Vohradsky, J. (2019). DNA Mapping and Kinetic Modeling of the HrdB Regulon in Streptomyces Coelicolor. Nucleic Acids Res. 47, 621–633. doi:10.1093/nar/gky1018
Smith, D., Zhong, J., Matsuura, M., Lambowitz, A. M., and Belfort, M. (2005). Recruitment of Host Functions Suggests a Repair Pathway for Late Steps in Group II Intron Retrohoming. Genes Dev. 19, 2477–2487. doi:10.1101/gad.1345105
Spencer, V., Sun, J.-M., Li, L., and Davie, J. R. (2003). Chromatin Immunoprecipitation: A Tool for Studying Histone Acetylation and Transcription Factor Binding. Methods 31, 67–75. doi:10.1016/S1046-2023(03)00089-6
Toro, N. (2003). Bacteria and Archaea Group II Introns: Additional Mobile Genetic Elements in the Environment. Environ. Microbiol. 5, 143–151. doi:10.1046/j.1462-2920.2003.00398.x
Toro, N., Martínez-Abarca, F., and Nisa-Martínez, R. (2014). Complete Genome Sequence of the RmInt1 Group II Intronless Sinorhizobium Meliloti Strain RMO17. Genome Announc 2, e01001–14. doi:10.1128/genomeA.01001-14
Villadas, P. J., Velazquez, E., Martinez-Molina, E., and Toro, N. (1995). Identification of Nodule-Dominant Rhizobium Meliloti Strains Carrying pRmeGR4b-Type Plasmid within Indigenous Soil Populations by PCR Using Primers Derived from Specific DNA Sequences. FEMS Microbiol. Ecol. 17, 161–168. doi:10.1111/j.1574-6941.1995.tb00139.x
Wank, H., SanFilippo, J., Singh, R. N., Matsuura, M., and Lambowitz, A. M. (1999). A Reverse Transcriptase/Maturase Promotes Splicing by Binding at its Own Coding Segment in a Group II Intron RNA. Mol. Cel 4, 239–250. doi:10.1016/s1097-2765(00)80371-8
Wilbanks, E. G., and Facciotti, M. T. (2010). Evaluation of Algorithm Performance in ChIP-Seq Peak Detection. PLoS ONE 5, e11471. doi:10.1371/journal.pone.0011471
Yang, J., Mohr, G., Perlman, P. S., and Lambowitz, A. M. (1998). Group II Intron Mobility in Yeast Mitochondria: Target DNA-Primed Reverse Transcription Activity of aI1 and Reverse Splicing into DNA Transposition Sites In Vitro. J. Mol. Biol. 282, 505–523. doi:10.1006/jmbi.1998.2029
Yao, J., Truong, D. M., and Lambowitz, A. M. (2013). Genetic and Biochemical Assays Reveal a Key Role for Replication Restart Proteins in Group II Intron Retrohoming. Plos Genet. 9, e1003469. doi:10.1371/journal.pgen.1003469
Zhang, Y., Liu, T., Meyer, C. A., Eeckhoute, J., Johnson, D. S., Bernstein, B. E., et al. (2008). Model-based Analysis of ChIP-Seq (MACS). Genome Biol. 9, R137. doi:10.1186/gb-2008-9-9-r137
Zhao, C., Liu, F., and Pyle, A. M. (2018). An Ultraprocessive, Accurate Reverse Transcriptase Encoded by a Metazoan Group II Intron. RNA 24, 183–195. doi:10.1261/rna.063479.117
Zhao, C., and Pyle, A. M. (2017). Structural Insights into the Mechanism of Group II Intron Splicing. Trends Biochem. Sci. 42, 470–482. doi:10.1016/j.tibs.2017.03.007
Zhao, J., and Lambowitz, A. M. (2005). A Bacterial Group II Intron-Encoded Reverse Transcriptase Localizes to Cellular Poles. Proc. Natl. Acad. Sci. 102, 16133–16140. doi:10.1073/pnas.0507057102
Zhao, J., Niu, W., Yao, J., Mohr, S., Marcotte, E. M., and Lambowitz, A. M. (2008). Group II Intron Protein Localization and Insertion Sites Are Affected by Polyphosphate. Plos Biol. 6, e150. doi:10.1371/journal.pbio.0060150
Zhong, J., Karberg, M., and Lambowitz, A. M. (2003). Targeted and Random Bacterial Gene Disruption Using a Group II Intron (Targetron) Vector Containing a Retrotransposition-Activated Selectable Marker. Nucleic Acids Res. 31, 1656–1664. doi:10.1093/nar/gkg248
Zhong, J., and Lambowitz, A. M. (2003). Group II Intron Mobility Using Nascent Strands at DNA Replication forks to Prime Reverse Transcription. EMBO J. 22, 4555–4565. doi:10.1093/emboj/cdg433
Keywords: group II intron, RmInt1, IEP, ribonucleoprotein (RNP), ChIP-seq analysis
Citation: Molina-Sánchez MD, García-Rodríguez FM, Andrés-León E and Toro N (2022) Identification of Group II Intron RmInt1 Binding Sites in a Bacterial Genome. Front. Mol. Biosci. 9:834020. doi: 10.3389/fmolb.2022.834020
Received: 12 December 2021; Accepted: 07 February 2022;
Published: 25 February 2022.
Edited by:
Marlene Belfort, Albany State University, United StatesReviewed by:
Hengyi Xu, University of Texas at Austin, United StatesCopyright © 2022 Molina-Sánchez, García-Rodríguez, Andrés-León and Toro. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Nicolás Toro, bmljb2xhcy50b3JvQGVlei5jc2ljLmVz
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.