- 1Department of Immunology, School of Medical Sciences, Universiti Sains Malaysia, Kubang Kerian, Malaysia
- 2Department of Otorhinolaryngology, Head and Neck Surgery, School of Medical Sciences, Universiti Sains Malaysia, Kubang Kerian, Malaysia
- 3Hospital Universiti Sains Malaysia, Kubang Kerian, Malaysia
- 4Department of Pathology, School of Medical Sciences, Universiti Sains Malaysia, Kubang Kerian, Malaysia
The IL-4/IL-13 axis is involved in the pathogenesis of allergic rhinitis (AR). In this study, we investigated the serum cytokines levels of IL-4, IL-5, IL-6, and IL-13 in AR patients, and the transcript expression levels of their receptors (i.e. IL4R, IL5RA, IL6R, and IL13RA1) in nasal epithelial cells of AR patients versus non-allergic controls. Nasal epithelial cells and blood samples of non-allergic controls (n = 30) and AR patients (n = 30) were collected to examine mRNA expression and serum cytokines levels, respectively. Bioinformatics analyses of IL-4/IL-13 receptor heterodimer association with tight junction (TJ) and JAK/STAT signaling genes were conducted in a gene expression profiling (GEP) dataset (GSE44037) of AR patients (n = 12) and healthy controls (n = 6). Serum IL-4, IL-5, IL-6 or IL-13 levels, and IL13RA1 transcript expression were significantly higher in AR patients compared with non-allergic controls. IL-4 and IL-13 serum levels were positively correlated with IL13RA1 expression in AR patients but not in non-allergic controls. In the GEP dataset (GSE44037), six TJ (CLDN4, CLDN7, CLDN12, CLDN15, TJP1, and TJP2) genes’ expressions were negatively correlated, respectively, with IL-4Rα/IL-13Rα1 heterodimeric receptor expression in AR patients and not in control samples. These six TJ genes contributed to the significant enrichment of tight junction Gene Ontology (GO ID: 0070160). Lastly, STATs DNA binding motif analysis showed that each of these TJ genes contains STATs binding consensus sequence within intronic and intergenic regions. Our results suggest that increased IL-4/IL-13 serum cytokines levels may contribute to decreased TJs expression via IL-4Rα/IL-13Rα1 heterodimeric receptor in nasal epithelium of AR patients.
Introduction
Allergic rhinitis (AR) is a common chronic inflammatory disease with high prevalence across different regions of the globe (Greiner et al., 2011; Pawankar, 2014; Sani et al., 2019; Tan et al., 2020). AR is characterized by aberrantly high Th2 cytokines levels such as interleukin-4 (IL-4), IL-5, IL-6, and IL-13 (Nur Husna et al., 2021a). IL-4 and IL-13 are key pathogenic Th2 cytokines in AR where they activate B cells to synthesize IgE, induce goblet cell hyperplasia, trigger airway hyperresponsiveness, and mucus hypersecretion (Wills-Karp and Finkelman, 2008; Sahoo et al., 2016; Bezerra Barros et al., 2020). IL-5 exerts pleiotropic effects on eosinophils by promoting their maturation, activation, survival, migration from bloodstream, and recruitment to airways in allergic disease (Abo et al., 2019; Boberg et al., 2020). IL-6 is a growth and differentiation factor for T and B cells, and it promotes the production of IgE (Gubernatorova et al., 2018).
Th2 cytokines modulate their signal through binding with their specific receptors that activate downstream signaling pathways. IL-4 and IL-13 are structurally similar, multifunctional peptides, and share a functional signaling receptor chain. IL-4 binds to two receptors i.e. the type I (composed of IL-4Rα and common γ-chain) and type II (composed of IL-4Rα and IL-13Rα1) IL-4 receptors. Binding of IL-4 with its type I receptor comprises of IL-4Rα and γc chain leads to activation of Janus kinase 1 (JAK1) and JAK3, respectively. For type II receptor, binding of IL-13 with IL-13Rα1 subunit activates TYK2/JAK2 (Bieber, 2020; Nguyen et al., 2020) that phosphorylates the tyrosine residues at the cytoplasmic tail of IL-4Rα that in turn serves as docking sites for signal transducer and activator of transcription 6 (STAT6) (Junttila, 2018; Gasparini et al., 2020). This activates IL-4 and IL-13 responsive genes in the subsequent signaling pathway of allergic responses.
Receptor for IL-5 is IL-5Rα where it relies on the β subunit to mediate the biological activities of IL-5. IL-5 binds to a ligand-specific α subunit associated with the common βc receptor subunit (common β subunit). Subsequent signaling pathways are activated including JAK/STAT, MAPK, PI3K, and NF-κB important in regulating the activities of eosinophils (Pelaia et al., 2018; Kandikattu et al., 2019). On the other hand, IL-6 binds with its cognate receptor IL-6Rα, and the complex associates with gp130 initiating their dimerization and consequent activation of JAK kinases. These mediate the phosphorylation of specific tyrosine residues on the gp130 cytoplasmic tail, which in turn acts as docking sites for STAT3 (and STAT1) SH2 domains, leading to JAK-mediated STAT3 phosphorylation and dimerization that further regulates transcription of target genes (Camporeale and Poli, 2012; Ham et al., 2018; Bin Dhuban et al., 2019).
Multiple studies have indicated the disruption of nasal epithelial barrier as the underlying cause of AR (Takano et al., 2005; Steelant et al., 2018; Nur Husna et al., 2021a). It has been proposed that Th2 cytokines signal through their respective receptors on nasal epithelial cells to suppress expression of TJs (Steelant et al., 2018; Nur Husna et al., 2021a). However, there is a lack of literature on the expression of interleukin receptors in nasal epithelial cells of AR patients. Therefore, our study was undertaken to investigate the serum cytokine levels of IL-4, IL-5, IL-6, and IL-13, the transcript expression levels of IL4R, IL5RA, IL6R, and IL13RA1 in AR patients versus non-allergic controls, and the correlation between IL-4Rα/IL-13Rα1 heterodimeric receptor (each subunit encoded by IL4R and IL13RA1 transcripts) with TJs and JAK/STAT signaling genes in AR patients and healthy controls.
Materials and Methods
Study Population
We previously recruited 30 AR patients and 30 non-allergic control subjects between March 2019 and July 2019 who attended the Hospital Universiti Sains Malaysia (HUSM) for our TJ genes expression project (Nur Husna et al., 2021b). Briefly, inclusion criteria for AR patients included a diagnosis of moderate/severe AR, positive skin prick test (SPT) to house dust mite (HDM) allergen and ≥18 years old, while non-allergic controls did not have signs or symptoms of allergy, no personal and immediate family history of allergic diseases, negative SPT to HDM allergen and ≥18 years old. The clinico-demographical characteristics of the recruited subjects are presented in Supplementary Table S1. Nasal brushing to collect nasal epithelial cells and venous blood were taken from each participant. All subjects involved in this study provided written and signed informed consent. The protocols were approved by the Human Research Ethics Committee of Universiti Sains Malaysia (JEPeM) (approved ethics code: USM/JEPeM/18060273). The samples were labeled anonymously, and all data were recorded and analyzed anonymously. All procedures were conducted in accordance with our institutional ethical standards and regulations, and with the 1964 Declaration of Helsinki and its later updates.
Blood Samples Collection
Five ml of peripheral blood samples were collected from AR patients and non-allergic controls into a 6 ml plain blood tube. The blood was stored for 1 hour at room temperature to allow samples to clot. The blood was then centrifuged at 3,500xg for 4 min to obtain the serum after separation from fibrinogen and cells. Serum samples were collected and stored at −80°C until further use.
Nasal Epithelial Cells Collection, RNA Extraction and Reverse Transcription-PCR (RT-PCR)
Cytology brush (Citotest Labware Co., Ltd., Haimen City, China) was used to collect nasal epithelial cells by fully inserting the brush into the nostrils and rubbed a few times against the medial and superior side of the inferior nasal meatus, using rotatory and linear movements. RNA was extracted from the samples using the RNeasy Mini Kit (Qiagen, Hilden, and Germany) and reverse-transcribed using iScript Reverse Transcription (RT) Supermix for RT-qPCR (Bio-Rad, Philadelphia, PA, United Sates) according to manufacturer’s protocols.
Quantitative PCR (qPCR)
qPCR was conducted using iTaq Universal SYBR Green Supermix (Bio-Rad, Philadelphia, PA, United Sates) and primers (Integrated DNA Technologies, Singapore) were designed by using the NCBI Primer-BLAST (https://www.ncbi.nlm.nih.gov/tools/primer-blast/) as presented in Supplementary Table S2. For every gene investigated, at least one primer (forward or reverse) was designed to encompass an exon-exon junction to avoid potential genomic DNA amplification, and the BLAST results of each primer was assessed to confirm the absence of alignment with genes other than the gene of interest. The Mx3005P qPCR thermal cycler (Agilent Technologies, CA, United Sates) was used in the qPCR reaction. Each qPCR reaction mixture constituted the following: 1) 10 μl of iTaq Universal SYBR Green Supermix (2x) for a final concentration of 1x; 2) 2 μl (400 nM) each for forward and reverse primer; 3) cDNA template at a final concentration of 50 ng in 20 μl; 4) Nuclease-free water added into a final volume of 20 μl qPCR reaction was subsequently conducted for 40 cycles with the following thermal profile: 1) Polymerase activation step at 95°C (25 s); 2) Denaturation step at 95°C (5 s); 3) Annealing and extension steps at 60°C (20 s). The relative transcript levels of every target gene in each sample was calculated using the 2−ΔΔCt formula in which ΔΔCt = [(Ct sample–Ct control)—ΔCt1].
Measurement of Cytokines Levels With Magnetic Luminex® Assay
The levels of Th2 cytokines (IL-4, IL-5, IL-6, and IL-13) were measured using Magnetic Luminex® Assay (R&D System, Minneapolis, United Sates). Prior to measurement of Th2 cytokines, the serum sample was thawed and centrifuged at 16,000xg for 4 min. The serum was diluted to a 2-fold dilution (i.e. 75 μl of sample plus 75 μl of calibrator diluent RD6-52) and was mixed thoroughly. Standards, controls and samples were added per well. The diluted microparticle cocktail was resuspended by inversion and vortexing. A total of 50 μl of the microparticle cocktail was added into each well of the microplate. The microplate was securely covered with a foil plate sealer. The microplate was incubated for 2 h at room temperature on a horizontal orbital microplate shaker (0.12” orbit) set at 800 ± 50 rpm. The microplate was then washed using magnetic bead washer (BioTek Instruments Inc., Winooski, United Sates). The magnet was applied to the bottom of the microplate for 1 min before removing the liquid. Each well was filled with 100 μl washing buffer for 1 min before discarding. The washing steps were repeated three times.
A total of 50 μl of diluted biotin-antibody cocktail was then added into all wells. The microplate was securely covered again with a foil plate sealer and incubated for 1 h at room temperature on the shaker (800 ± 50 rpm). The washing steps were repeated before 50 μl of diluted streptavidin-PE was added, the microplate securely covered with a foil plate sealer and incubated for 30 min at room temperature on the shaker set at 800 ± 50 rpm. The washing steps were repeated before microparticles were resuspended in 100 μl of washing buffer and incubated for 2 min on the shaker (800 ± 50 rpm). The microplate was subsequently read within 90 min using the Luminex® 200™ analyzer (Luminex, Austin, United Sates).
Correlation Analysis of Th2 Cytokine Receptors With TJ and JAK/STAT Signaling Pathway Genes
A microarray gene expression profiling (GEP) dataset (GSE44037) of nasal epithelial cells derived from 18 adults (>18 years old) comprising of 12 AR and six healthy controls (Wagener et al., 2013) were obtained from the Gene Expression Omnibus database. The similarities and differences between our AR cohort of patients with those of GSE44037 are as follows: 1) Similarities: Above 18 years old; Moderate/severe AR; Allergic status was assessed by SPT; AR patients were sensitized to at least one allergen; AR patients did not use any anti-allergy medications for at least 1 month before samples were collected; 2) Differences: Our cohort was sensitized to HDM allergens while GSE44037 dataset was sensitized to common allergens; AR patients with SPT wheal size of ≥4 mm in our cohort was considered as positive SPT instead of SPT wheal size of >3 mm.
In this GEP dataset, Pearson correlation values were calculated according to the expression of IL-4 and IL-13 receptor heterodimer [i.e. IL4R (probe ID: 203233_PM_at) and IL13RA1 (201887_PM_at) subunits], IL-5 receptor heterodimer [i.e. IL5RA (211516_PM_at) and CSF2RB (205159_PM_at) subunits], or IL-6 receptor heterodimer [i.e. IL6R (205945_PM_at) and IL6ST (204863_PM_s_at) subunits] against the total number of probes (n = 41,796) present in the microarray platform. These were conducted in AR patients (n = 12) or healthy controls (n = 6) separately. Then, presence of TJ and JAK/STAT signaling genes with significant (p < 0.05) Pearson correlation values (i.e. r > |0.603| for 12 AR subjects; r > |0.812| for six healthy control subjects) with both receptor subunit in each receptor heterodimer were shortlisted and demonstrated in Pearson correlation scatter plots. The TJ genes or desmosomal genes examined were claudins (CLDN1, CLDN2, CLDN3, CLDN4, CLDN5, CLDN6, CLDN7, CLDN8, CLDN9, CLDN10, CLDN11, CLDN12, CLDN13, CLDN14, CLDN15, CLDN16, CLDN17, CLDN18, CLDN19, CLDN20, CLDN21, CLDN22, and CLDN23), junctional adhesion molecules (JAMs; JAM1, JAM2, and JAM3), desmogleins (DSG1, DSG2, DSG3, and DSG4), desmocollins (DSC1, DSC2, and DSC3), cadherins (CDH1, CDH2, CDH3, and CDH12), and zonula occludens (ZO; TJP1, TJP2, and TJP3). The genes involved in JAK/STAT signaling examined were JAK1, JAK2, JAK3, STAT1, STAT2, STAT3, STAT6, and LTK (TYK1 gene name) and TYK2. Gene Ontology (GO) enrichment analysis of genes negatively correlated with IL4R and IL13RA1 expression was conducted using the ToppGene database as described previously (Chen et al., 2009; Brown et al., 2016).
Statistical Analysis
Data were analyzed using Student’s t-test (for normally distributed data) and Mann-Whitney U test (for not normally distributed data) to determine the difference of gene expression between AR and non-allergic control groups. Shapiro-Wilk test was conducted to assess the normality of the data. The relationship between two variables with continuous data was examined with Pearson correlation. All analyses were conducted using GraphPad Prism v6.07 (GraphPad Software Inc., CA, United Sates). All p-values were two-tailed and values < 0.05 were considered statistically significant.
Results
Sensitization of the AR Patients to HDM Allergens
In this study, we tested the sensitization to HDM allergens of Dermatophagoides pteronyssinus (D. pteronyssinus), Dermatophagoides farinae (D. farinae), and Blomia tropicalis (B. tropicalis). Only 10% (n = 3) of the AR patients were monosensitized to B. tropicalis and none of the patients were monosensitized to the other two HDMs. The rest of the 27 AR patients were either sensitized to two (n = 7/30; 23.3%) or all three (n = 20/30; 66.7%) of the HDM allergens (Figure 1).
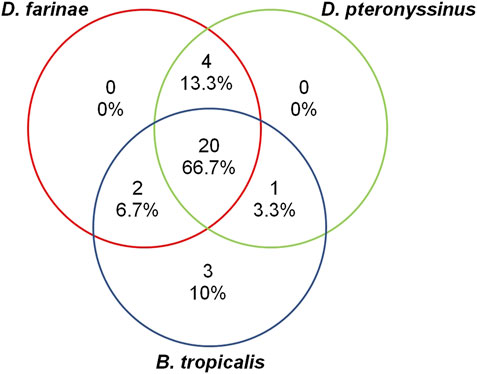
FIGURE 1. The proportion of AR patients (total n = 30) sensitized to HDM allergens D. farinae, D. pteronyssinus, and B. tropicalis.
Serum IL-4, IL-5, IL-6, and IL-13 Levels in AR Patients and Non-allergic Controls
The median age of the non-allergic control and AR groups were 25.50 (range: 20–54) and 28.00 (24–54) years old, respectively. The proportion of female subjects was 73.3% (n = 22) and 66.7% (n = 20) in non-allergic control and AR group, respectively. For the complete demographic and clinical characteristics of both groups of subjects, readers are directed to our recent publication (Nur Husna et al., 2021b). The serum levels of IL-4 (p = 0.0001), IL-5 (p = 0.0043), IL-6 (p = 0.0371) or IL-13 (p < 0.0001) were significantly higher in AR patients compared with non-allergic controls (Figures 2A–D).
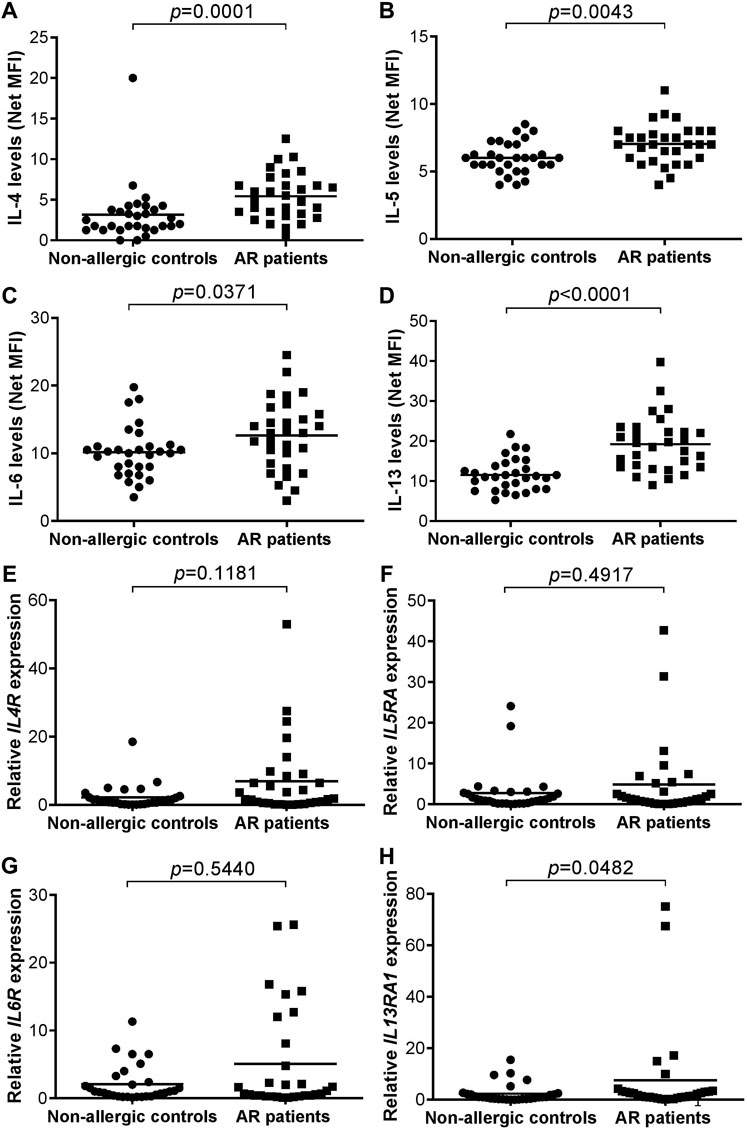
FIGURE 2. (A–D) Serum levels of IL-4 (A), IL-5 (B), IL-6 (C), and IL-13 (D) in non-allergic controls (n = 30) versus AR patients (n = 30). Bar represents mean (E–H) Relative IL4R (E), IL5RA (F), IL6R (G), and IL13RA1 (H) expression in non-allergic controls (n = 30) versus AR patients (n = 30). Bar represents mean.
Expression of IL4R, IL5RA, IL6R, and IL13RA1 Transcripts in AR Patients and Non-allergic Controls
No significant difference was observed in the expression of IL4R (p = 0.118), IL5RA (p = 0.492) or IL6R (p = 0.544) transcript in AR patients compared with non-allergic controls (Figures 2E–G). The expression of IL13RA1 (p = 0.048) transcript was borderline significantly higher in nasal epithelial cells of AR patients compared with non-allergic controls due to two outlier AR cases (Figure 2H).
Correlation of Serum Th2 Cytokine Levels With Their Receptor’s Expression Levels in AR Patients and Non-allergic Controls
The serum levels of IL-4 or IL-13 were positively and significantly associated with the expression of IL13RA1 transcripts in AR patients (r = 0.4296, p = 0.0178 and r = 0.4200, p = 0.0208, respectively) (Supplementary Figure S1). Such relationship was not observed in non-allergic controls (r = −0.1969, p = 0.2970 and r = −0.3071, and p = 0.0988, respectively) (Supplementary Figure S2). The rest of the comparisons including serum IL-4 with IL4R, serum IL-5 with IL5RA, serum IL-6 with IL6R, or serum IL-13 with IL4R did not yield a significant relationship in AR patients (Supplementary Figure S1) or non-allergic controls (Supplementary Figure S2).
Association of IL4R and IL13RA1 Expression With TJ and JAK/STAT Signaling Pathway Genes in AR Patients and Healthy Controls
Correlation analysis of IL-4/IL-13 receptor heterodimer with TJs or JAK/STAT signaling genes in AR patients (n = 12) derived from the GSE44037 GEP dataset showed that a total of 1,382 probes representing 1,088 annotated genes were positively correlated (r > 0.603, p < 0.05) with both IL4R and IL13RA1 expression (Supplementary Table S3). These 1,088 genes consisted of the JAK/STAT signaling genes STAT2 and STAT3, as well as certain TJ genes including CLDN11, JAM3, and CDH3. The expression of other JAK/STAT signaling genes (i.e,. STAT6 and TYK2) was also positively correlated with IL4R but not with IL13RA1 expression levels (Figure 3A). On the other hand, a total of 1,646 probes representing 1,325 annotated genes were negatively correlated (r < −0.603, p < 0.05) with both IL4R and IL13RA1 expression (Supplementary Table S3). In these 1,325 genes, several TJ genes were implicated including CLDN4, CLDN7, CLDN12, CLDN15, TJP1, and TJP2, while no JAK/STAT genes were involved. Other TJ genes also showed significant negative correlation with either IL4R (CLDN9, CLDN16 and CLDN19) or IL13RA1 (CLDN6, CLDN10, CLDN23, and TJP3) (Figure 3A).
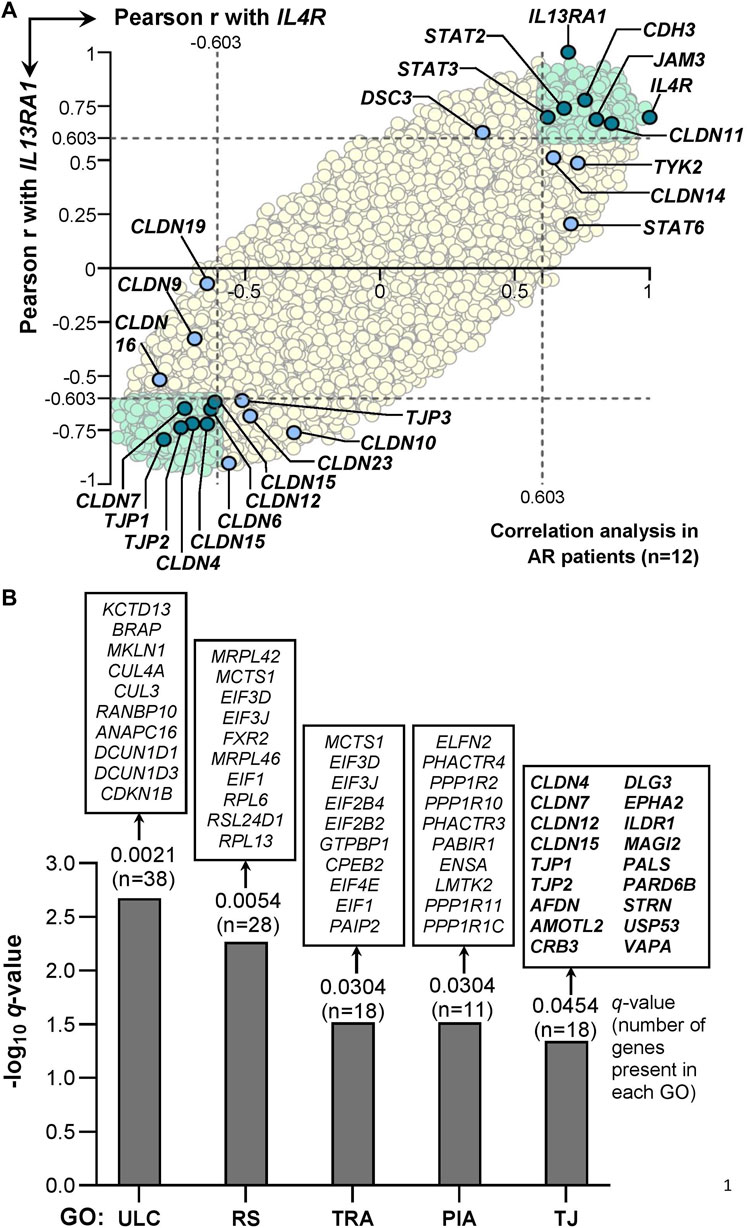
FIGURE 3. Correlation of IL-4 receptor heterodimer subunits expression (i.e., IL4R or IL13RA1) with 20,541 annotated genes in AR patients derived from GSE44037 dataset. (A) Pearson correlation of the genes expression levels with IL4R or IL13RA1 expression levels in AR patients (n = 12). The Pearson r value ± 0.603 is used as the cut-off (i.e. the dotted lines) to define positive (r > 0.603) or negative (r < −0.603) correlation with IL4R or IL13RA1 as this cut-off represents significant (i.e. p < 0.05) Pearson correlation for a sample size of 12. All genes significantly correlated with both IL4R and IL13RA1 expression levels are highlighted in light green. TJ, desmosomal or JAK/STAT signaling genes with significant correlation with both IL4R and IL13RA1 expression levels are highlighted in teal. TJ, desmosomal or JAK/STAT signaling genes with significant correlation with either IL4R or IL13RA1 expression levels only are highlighted in light blue. The rest of the background genes are highlighted in yellow. (B) Gene Ontology (GO) enrichment analysis of genes inversely associated with IL4R and IL13RA1 expression (GSE44037 dataset). Ten representative genes are displayed on top of each bar, and all 18 genes contributed to the enrichment of TJ ontology (GO ID: 0070160) are shown. ULC: Ubiquitin ligase complex; RS: Ribosomal subunit; TRA: Translation regulator activity; PIA: Phosphatase inhibitor activity; TJ: Tight junction.
In healthy controls (n = 6) derived from the same GEP dataset, none of the genes present in the GEP platform demonstrated significant association with both IL4R and IL13RA1 expression levels (Supplementary Figure S3). In terms of desmosomal genes, only DSG4 was negatively correlated with IL13RA1 expression and none of TJ and JAK/STAT genes examined showed significant correlation with either IL4R or IL13RA1 expression (Supplementary Figure S3). Interestingly, we observed that none of the TJ or JAK/STAT signaling genes presented with significant correlation with IL-5 receptor heterodimer (i.e. IL5RA and CSF2RB) or IL-6 receptor heterodimer (i.e. IL6R and IL6ST) expression in AR or healthy control subjects (Supplementary Figures S3, S5).
Gene Ontology Enrichment and STATs Consensus Sequence Analysis
To validate whether TJ ontology was enriched, we conducted GO enrichment analysis of genes negatively correlated with both IL4R and IL13RA1 expression (r < −0.603, p < 0.05). Six collective groups of GOs were enriched consisting of protein complexes, ribosome components, transcription and translation processes, phosphatases, TJ and other cellular components GOs (Supplementary Table S4). A representative GO from each of the collective group (excluding other cellular components group) was presented in Figure 3B. The TJ ontology (GO ID: 0070160) was enriched (p = 0.0010, q = 0.0454) where 18 of 134 genes annotated in the ontology were present including CLDN4, CLDN7, CLDN12, CLDN15, TJP1, and TJP2 (Figure 3B).
Activation of IL-4Rα/IL-13Rα1 heterodimeric receptor by their cytokines (IL-4 and IL-13) activates the JAK/STAT signaling pathway that leads to the activation of the transcription factor STAT6. IL-4-activated STAT6 has been shown to function as transcriptional repressor (Czimmerer et al., 2018), while IL-13-activated STAT6 enhanced permeability of epithelial cells by altering the expression of several TJ genes (Lin et al., 2019). STAT6 is capable of directly repressing transcription of genes via binding to intronic and intergenic regions of target genes (Takaki et al., 2008; Czimmerer et al., 2018). Taken together, we were interested to assess whether these six TJ genes contain STAT6 (and STATs collectively) consensus binding sequence within their genomic sequences as this may imply that STAT6 is capable of binding and regulating or suppressing their expression. The genomic sequences investigated were retrieved from UCSC Genome Browser (GRCh38/hg38) database.
STAT proteins target and bind the palindromic consensus sequence 5′-TTC(N)2-4GAA-3′ where N2-4 denotes spacer nucleotides comprising of two (N2), three (N3) or four (N4) nucleotides (Kraus et al., 2003; Goenka and Kaplan, 2011; Li et al., 2016). STAT6 differs from other STAT proteins whereby its binding element comprising of either N3 or N4 site (i.e,. 5′-TTC(N)3/4GAA-3′), and it preferentially binds N4 over N3 site (i.e. 5′-TTC(N)4GAA-3′), as well as preferential binding the regions within the first two introns over either upstream of transcription start site or downstream of transcription end site (Elo et al., 2010; Li et al., 2016). Thus, we examined for the presence of 5′-TTC(N)2–4GAA-3′ motifs within the following DNA regions of the six TJ genes: 5 kb upstream of the first exon, the first two introns and 5 kb downstream of the last exon. We observed that each gene contains at least five and two 5′-TTC(N)2-4GAA-3′ and 5′-TTC(N)3/4GAA-3′ motifs, respectively, within these regions (Table 1). In terms of STAT6 preferential motif and regions i.e,. 5′-TTC(N)4GAA-3′ within the first two introns, four TJ genes contain at least one of such motif in their first two introns i.e., CLDN7 (n = 1), CLDN12 (n = 2), TJP1 (n = 7), and TJP2 (n = 11) (Table 1). For reference, the complete list of the DNA sequences examined and the presence of STATs binding motifs for the six TJ genes are presented in Supplementary Table S5.
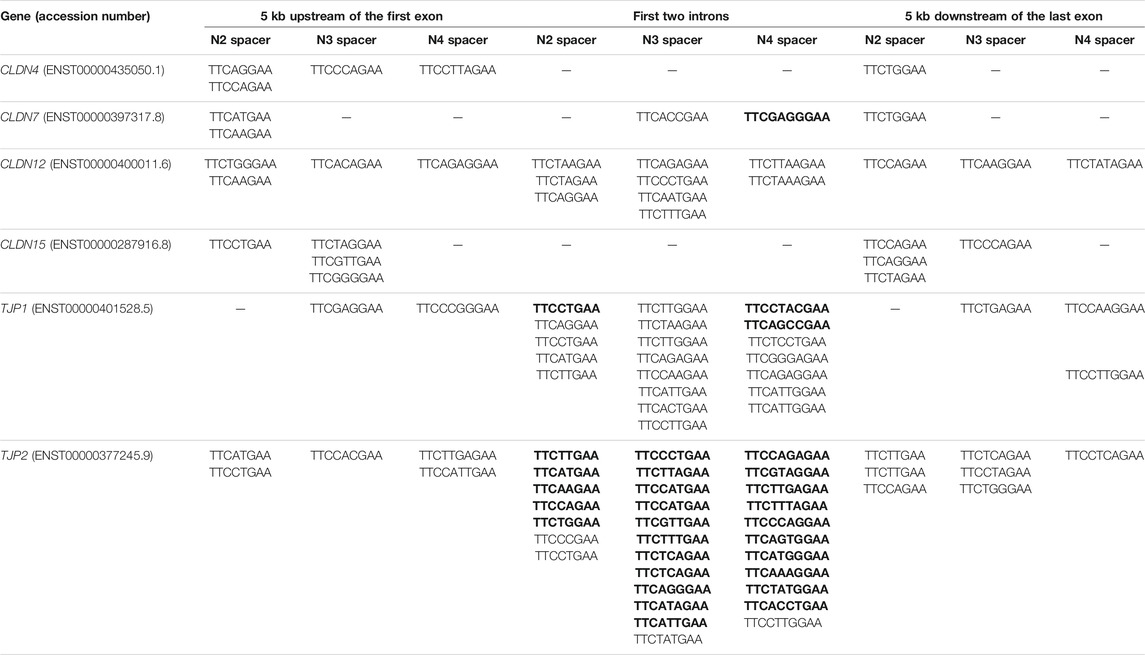
TABLE 1. Presence of STATs consensus sequence 5′-TTC(N)2-4GAA-3′ in DNA regions of TJ genes. Motif within intron 1 is in bold.
A graphical representation of the IL-4/IL-13 axis cascade and the potential pathways involving STAT6 and the TJ genes implicated in this study is presented in Figure 4. Finally, the protein-protein interaction network between IL-4/IL-13 axis and STAT6, or between TJ proteins is presented in Supplementary Figures 6A, 6B, respectively.
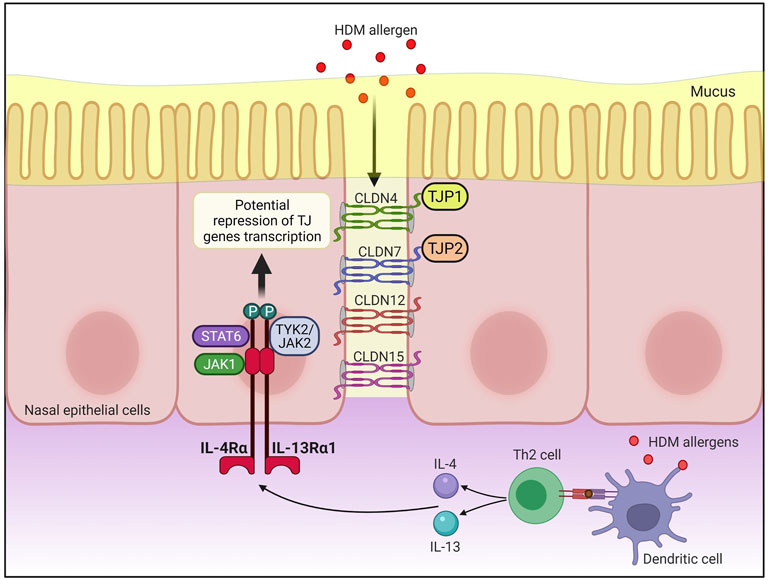
FIGURE 4. Antigen presenting cells (APCs) such as dendritic cells uptake, process and present peptides from allergens on the major histocompatibility complex (MHC) class II molecule. The antigen complex and the MHC class II molecule serve as a ligand for T cell receptors on naïve CD4+ T cells, resulting in differentiation of naïve CD4+ T cells into allergen-specific Th2 cell. Subsequently, IL-4 and IL-13 are produced by activated Th2 cells, allowing IL-4 and IL-13 to bind to IL-4Rα/IL-13Rα1 heterodimeric receptor. This activates Janus kinase 1 (JAK1) on IL-4Rα and TYK2/JAK2 on IL-13Rα1 which phosphorylates the tyrosine residues at the cytoplasmic tail of IL-4Rα that in turn serves as docking sites for signal transducer and activator of transcription 6 (STAT6). Activated STAT6 may then repress the transcription of CLDN4, CLDN7, CLDN12, CLDN15, TJP1, and TJP2 expression in the nasal epithelial cells of AR patients. CLDN, claudin; HDM, house dust mite; JAK, Janus kinase; STAT6, signal transducer, and activator of transcription 6; Th2, T helper 2; TJP, tight junction protein; TJs, tight junctions; TYK, tyrosine kinase. Created with BioRender.com.
Discussion
Th2 cytokines enable a continuous inflammation in the nasal mucosa and infiltrate within the sinonasal microenvironment that alter the composition of epithelial tight junctions (TJs) (Capaldo and Nusrat, 2009; London et al., 2016). All AR patients in this study presented with systemic atopy as defined by positive SPT to at least one of the examined aeroallergens (Rondón et al., 2014). Hence, analysis of serum Th2 cytokines levels, instead of local (nasal) levels of Th2 cytokines, was investigated to corroborate with the systemic manifestation. Significantly higher serum IL-4 and IL-5 levels were observed in AR patients compared to control group (p < 0.01) (Bi et al., 2018). In a nasal allergen challenge (NAC) study of AR patients sensitized with grass pollen, the frequency of IL-4+CD4+ T cells were significantly increased at sixth hour after NAC when compared to the control day (Shamji et al., 2015). IL-5 expression was decreased in AR patients with reduced nasal symptoms and infiltration of eosinophils into the nasal mucosa (Sato et al., 2016). AR mice models also demonstrated a significantly higher serum levels of IL-4, IL-5, and IL-13 than in the control group (Cheng et al., 2021). Furthermore, IL-5 was secreted from resident cells in response to ex vivo allergen challenge in the AR group but not in the non-allergic group, indicating that local presentation of antigen to resident allergen-specific Th2 cells represents early events of AR pathogenesis (Skrindo et al., 2015). In terms of IL-6, small nucleotide polymorphism (SNP) affecting IL-6 (rs1800795) was linked with an increased risk of AR (Zhao et al., 2016) and positively associated with the severity of AR (Zhao et al., 2018). These findings are comparable with our results where we demonstrated significantly higher levels of IL-4, IL-5, IL-6, and IL-13 in serum of AR patients compared with non-allergic controls.
We also examined the expression of IL4R, IL5RA, IL6R, and IL13RA1 in nasal epithelial cells of AR patients and non-allergic controls. However, only IL13RA1 showed a significant increase in nasal epithelial cells of AR patients compared with non-allergic controls. In our previous study, we demonstrated a significant decrease in the mRNA expression of TJ proteins such as occludin, CLDN3 and CLDN7 in nasal epithelial cells of AR patients compared with non-allergic controls (Nur Husna et al., 2021b), and the samples that we used were the same as the current study. These suggest that IL13RA1 might be involved in the disruption of TJs as IL-13Rα1 is a high-affinity heterodimer receptor that binds IL-4 and IL-13, and both of these cytokines are important factors for decreased nasal epithelial barrier integrity (Wise et al., 2014; Steelant et al., 2016; Wawrzyniak et al., 2017).
Our correlation analysis of GEP dataset demonstrated that the expression profiles of IL4R and IL13RA1 were positively correlated with JAK/STAT signaling genes, and inversely correlated with several claudins and ZO (i.e. TJP1, TJP2, and TJP3 transcripts) expression levels in AR patients but not in healthy controls. The TJ GO was also enriched in the group of genes negatively associated with both IL4R and IL13RA1 expression. This includes CLDN7 and TJP1 which we and other groups had previously shown that their expression was significantly downregulated in AR patients compared with non-allergic controls (Lee et al., 2016; Nur Husna et al., 2021b). Nasal epithelial barrier disruption induced by the activation of IL-4 or IL-13 receptors has been reported to occur in AR. Firstly, pertaining to IL-4, pre-treatment of Calu-3 (human airway epithelial cells) with anti-IL-4Rα monoclonal antibody (mAb) suppressed the effects of IL-4 and prevented IL-4-induced disruption of epithelial barrier. This was shown to occur via decreased ZO-1 expression, and pretreatment of HDM-challenged mice with anti-IL-4 mAb prevented the loss of ZO-1 expression (Steelant et al., 2018). In terms of IL-13, overexpression of miR-143 in IL-13-stimulated nasal epithelial cells (NECs) from AR patients suppressed the production of IL-13-induced inflammatory cytokines, and miR-143 rendered these effects by directly targeting and repressing IL13RA1 transcript expression (Teng et al., 2015). Treatment of human NECs with the cytokine suppressed the expression of TJ molecules including ZO-1 and CLDN3 (Huang et al., 2020). Moreover, correlation analysis of our cohort of AR and non-allergic control groups showed that serum IL-4 and IL-13 levels were positively associated with IL13RA1 expresion.
Six of the seven TJ genes (CLDN4, CLDN6, CLDN7, CLDN12, TJP1, and TJP2), which demonstrated negative association with both IL4R and IL13RA1 expression levels, have been implicated in AR such as significantly decreased expression in nasal epithelium of AR patients compared with non-allergic controls (Brandner, 2016; Siti Sarah et al., 2020; Nur Husna et al., 2021a). Moreover, our correlation analysis observation corroborates with the increased expression of Tjp2, Cldn7, and Cldn15 in epithelial cells of Stat6−/− mice (Lin et al., 2019). Our literature search did not yield publications implicating CLDN11, JAM3 or CDH3 in AR. Nevertheless, JAM3 mRNA expression was downregulated in chronic rhinosinusitis patients with nasal polyps versus healthy controls (Cornet et al., 2019). The positive association of the three TJ genes, particularly CLDN11 and JAM3, with both IL4R and IL13RA1 expression levels in AR patients remains unexplained and it represents fertile grounds for future investigations and validation.
Binding of IL-4 or IL-13 to their receptor complex, IL-4Rα and IL-13Rα1, induces the JAK/STAT signaling pathway where phosphorylation of transcription factors particularly STAT6 occurs and its subsequent translocation to nucleus to regulate genes expression. Direct transcriptional repression by STAT6 has been reported and this is achieved through STAT6 binding to intronic and intergenic regions (Takaki et al., 2008; Czimmerer et al., 2018). Our STAT6 DNA binding motif analysis showed that CLDN7, CLDN12, TJP1, and TJP2 contain STAT6 binding consensus sequence (5′-TTC(N)4GAA-3′) within its preferential binding regions i.e., the first two introns (Elo et al., 2010; Li et al., 2016). It remains to be elucidated mechanistically whether IL-4 and IL-13 signaling pathway through their receptors, IL-4Rα and IL-13Rα1, may induce epithelial barrier disruption by suppressing TJs expression via STAT6 transcriptional repression activities in AR patients.
We acknowledge the limitations of the study as follows: 1) The AR patients included in this study were sensitized to HDMs only. As the AR patients’ recruitment period in this study was conducted during the classical pollen season, we were unable to rule out the contribution of pollens sensitization to the observations in our study. However, HDM-induced AR is the most common factor causing allergic sensitization among AR patients in Malaysia (Ho et al., 1995; Liam et al., 2002). In particular, over 90% of Malaysian AR patients were positive to D. pteronyssinus sensitization while less than 10% of the AR patients were positive to either pollen species (i.e., Bermuda grass or Acacia sp.) (Liam et al., 2002); 2) The mRNA expression levels of the interleukin receptors investigated in this study, particularly IL13RA1, require confirmation at the protein levels in AR patients versus non-allergic controls; 3) Our study is dependent on GEP dataset to draw presumptions between IL-4 and IL-13 with their cognate receptors (IL-4Rα/IL-13Rα1 heterodimeric receptor) and TJ or desmosomal genes, and GEP does not necessarily correlate with functional proteins. Nonetheless, the GEP dataset was derived from human samples and our correlation observations in AR patient samples were compared with those of healthy controls; 4) Our study focused on serum cytokine levels to better mirror systemic atopy as presented by our cohort of AR patients, however this was at the expense of not measuring nasal cytokine levels. Hence, it remains unknown if the effects on the nasal epithelial integrity in our AR patients were also reflected by changes in the nasal Th2 cytokine levels.
Treatment with anti-IL-4Rα mAb is a novel strategy for AR patients. Dupilumab is a fully human anti-IL-4Rα mAb that blocks both IL-4 and IL-13 signaling (Rabe et al., 2018). In a recent randomized, double-blind and placebo-controlled phase IIb clinical trial of dupilumab in perennial AR patients, improved responses were observed in these patients in terms of nasal symptoms (Weinstein et al., 2018), highlighting that the IL-4/IL-13 axis as a novel therapeutic target in the disease.
In summary, we have demonstrated significant elevation of IL-4, IL-5, IL-6, and IL-13 serum levels in AR patients, and increased expression of IL13RA1 transcripts in nasal epithelial cells of AR patients compared with non-allergic controls. Together with the correlation analyses of TJs and JAK/STAT signaling genes expression with IL4R and IL13RA1, our findings suggest that IL-4/IL-13 axis may deregulate nasal epithelial barrier integrity by suppressing TJs expression in AR patients, and this warrants future investigation and validation.
Data Availability Statement
The original contributions presented in the study are included in the article's Supplementary Material, further inquiries can be directed to the corresponding author.
Ethics Statement
The studies involving human participants were reviewed and approved by the USM/JEPeM/18060273. The patients/participants provided their written informed consent to participate in this study.
Author Contributions
HTTT, NSMA and KKW conceived the study and recruited research grants. SMNH and NMS recruited the subjects and their clinico-demographical data. SMNH conducted the experiments. SETS co-supervised the qPCR experiments. KKW guided the molecular experiments, performed and interpreted the bioinformatics analysis. SMNH and KKW designed the study, performed the data analysis, generated figures and tables, conducted literature searches and wrote the manuscript. KKW revised the manuscript. All authors read and approved the final manuscript.
Funding
This work was supported by Universiti Sains Malaysia (USM) Grants comprising of the Research University Grant (1001/PPSP/8012349) awarded to KKW and Research University Grant (1001. PPSP.8012285) awarded to NSMA.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We would like to thank staffs of Department of Immunology, Universiti Sains Malaysia, for their kind assistance.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmolb.2022.819772/full#supplementary-material
References
Abo, H., Flannigan, K. L., Geem, D., Ngo, V. L., Harusato, A., and Denning, T. L. (2019). Combined IL-2 Immunocomplex and Anti-IL-5 mAb Treatment Expands Foxp3(+) Treg Cells in the Absence of Eosinophilia and Ameliorates Experimental Colitis. Front. Immunol. 10, 459. doi:10.3389/fimmu.2019.00459
Bezerra Barros, G. C., Paiva Ferreira, L. K. D., Ferreira, L. A. M. P., Mozzini Monteiro, T., Alves, A. F., Pereira, R. d. A., et al. (2020). 4-Carvomenthenol Ameliorates the Murine Combined Allergic Rhinitis and Asthma Syndrome by Inhibiting IL-13 and Mucus Production via p38MAPK/NF-κB Signaling Pathway axis. Int. Immunopharmacology 88, 106938. doi:10.1016/j.intimp.2020.106938
Bi, J., Hu, Y., Peng, Z., Liu, H., and Fu, Y. (2018). Changes and Correlations of Serum Interleukins, Adhesion Molecules and Soluble E-Selectin in Children with Allergic Rhinitis and Asthma. Pak J. Med. Sci. 34, 1288–1292. doi:10.12669/pjms.345.15334
Bieber, T. (2020). Interleukin‐13: Targeting an Underestimated Cytokine in Atopic Dermatitis. Allergy 75, 54–62. doi:10.1111/all.13954
Bin Dhuban, K., Bartolucci, S., d'Hennezel, E., and Piccirillo, C. A. (2019). Signaling through Gp130 Compromises Suppressive Function in Human FOXP3(+) Regulatory T Cells. Front. Immunol. 10, 1532. doi:10.3389/fimmu.2019.01532
Boberg, E., Johansson, K., Malmhäll, C., Calvén, J., Weidner, J., and Rådinger, M. (2020). Interplay between the IL-33/ST2 Axis and Bone Marrow ILC2s in Protease Allergen-Induced IL-5-Dependent Eosinophilia. Front. Immunol. 11, 1058. doi:10.3389/fimmu.2020.01058
Brandner, J. M. (2016). Importance of Tight Junctions in Relation to Skin Barrier Function. Curr. Probl. Dermatol. 49, 27–37. doi:10.1159/000441541
Brown, P. J., Wong, K. K., Felce, S. L., Lyne, L., Spearman, H., Soilleux, E. J., et al. (2016). FOXP1 Suppresses Immune Response Signatures and MHC Class II Expression in Activated B-cell-like Diffuse Large B-Cell Lymphomas. Leukemia 30, 605–616. doi:10.1038/leu.2015.299
Camporeale, A., and Poli, V. (2012). IL-6, IL-17 and STAT3: A Holy trinity in Auto-Immunity? Front. Biosci. 17, 2306–2326. doi:10.2741/4054
Capaldo, C. T., and Nusrat, A. (2009). Cytokine Regulation of Tight Junctions. Biochim. Biophys. Acta (Bba) - Biomembranes 1788, 864–871. doi:10.1016/j.bbamem.2008.08.027
Chen, J., Bardes, E. E., Aronow, B. J., and Jegga, A. G. (2009). ToppGene Suite for Gene List Enrichment Analysis and Candidate Gene Prioritization. Nucleic Acids Res. 37, W305–W311. doi:10.1093/nar/gkp427
Cheng, K.-J., Zhou, M.-L., Liu, Y.-C., Wang, C., and Xu, Y.-Y. (2021). The Role of CD40 in Allergic Rhinitis and Airway Remodelling. Mediators Inflamm. 2021, 6694109. doi:10.1155/2021/6694109
Cornet, M. E., Kostamo, K., Rinia, A. B., Zwinderman, A. H., van Egmond, D., de Groot, E. J. J., et al. (2019). Novel Roles for Nasal Epithelium in the Pathogenesis of Chronic Rhinosinusitis with Nasal Polyps. Rhinology 57, 169–179. doi:10.4193/Rhin18.128
Czimmerer, Z., Daniel, B., Horvath, A., Rückerl, D., Nagy, G., Kiss, M., et al. (2018). The Transcription Factor STAT6 Mediates Direct Repression of Inflammatory Enhancers and Limits Activation of Alternatively Polarized Macrophages. Immunity 48, 75–90. doi:10.1016/j.immuni.2017.12.010
Elo, L. L., Järvenpää, H., Tuomela, S., Raghav, S., Ahlfors, H., Laurila, K., et al. (2010). Genome-wide Profiling of Interleukin-4 and STAT6 Transcription Factor Regulation of Human Th2 Cell Programming. Immunity 32, 852–862. doi:10.1016/j.immuni.2010.06.011
Gasparini, G., Cozzani, E., and Parodi, A. (2020). Interleukin-4 and Interleukin-13 as Possible Therapeutic Targets in Systemic Sclerosis. Cytokine 125, 154799. doi:10.1016/j.cyto.2019.154799
Goenka, S., and Kaplan, M. H. (2011). Transcriptional Regulation by STAT6. Immunol. Res. 50, 87–96. doi:10.1007/s12026-011-8205-2
Greiner, A. N., Hellings, P. W., Rotiroti, G., and Scadding, G. K. (2011). Allergic Rhinitis. The Lancet 378, 2112–2122. doi:10.1016/S0140-6736(11)60130-X
Gubernatorova, E. O., Gorshkova, E. A., Namakanova, O. A., Zvartsev, R. V., Hidalgo, J., Drutskaya, M. S., et al. (2018). Non-redundant Functions of IL-6 Produced by Macrophages and Dendritic Cells in Allergic Airway Inflammation. Front. Immunol. 9, 2718. doi:10.3389/fimmu.2018.02718
Ham, S., Lima, L. G., Chai, E. P. Z., Muller, A., Lobb, R. J., Krumeich, S., et al. (2018). Breast Cancer-Derived Exosomes Alter Macrophage Polarization via gp130/STAT3 Signaling. Front. Immunol. 9, 871. doi:10.3389/fimmu.2018.00871
Ho, T. M., Murad, S., Kesavapillai, R., and Singaram, S. P. (1995). Prevalence of Allergy to Some Inhalants Among Rhinitis Patients in Malaysia. Asian Pac. J. Allergy Immunol. 13, 11–16.
Huang, Z.-Q., Liu, J., Ong, H. H., Yuan, T., Zhou, X.-M., Wang, J., et al. (2020). Interleukin-13 Alters Tight Junction Proteins Expression Thereby Compromising Barrier Function and Dampens Rhinovirus Induced Immune Responses in Nasal Epithelium. Front. Cel Dev. Biol. 8, 572749. doi:10.3389/fcell.2020.572749
Junttila, I. S. (2018). Tuning the Cytokine Responses: An Update on Interleukin (IL)-4 and IL-13 Receptor Complexes. Front. Immunol. 9, 888. doi:10.3389/fimmu.2018.00888
Kandikattu, H. K., Upparahalli Venkateshaiah, S., and Mishra, A. (2019). Synergy of Interleukin (IL)-5 and IL-18 in Eosinophil Mediated Pathogenesis of Allergic Diseases. Cytokine Growth Factor. Rev. 47, 83–98. doi:10.1016/j.cytogfr.2019.05.003
Kraus, J., Börner, C., and Höllt, V. (2003). Distinct Palindromic Extensions of the 5'‐TTC…GAA‐3' Motif Allow STAT6 Binding In Vivo. FASEB j. 17, 304–306. doi:10.1096/fj.02-0482fje
Lee, H.-J., Kim, B., Im, N.-R., Lee, D. Y., Kim, H. K., Lee, S. H., et al. (2016). Decreased Expression of E-Cadherin and ZO-1 in the Nasal Mucosa of Patients with Allergic Rhinitis: Altered Regulation of E-Cadherin by IL-4, IL-5, and TNF-Alpha. Am. J. Rhinol Allergy 30, 173–178. doi:10.2500/ajra.2016.30.4295
Li, J., Rodriguez, J. P., Niu, F., Pu, M., Wang, J., Hung, L.-W., et al. (2016). Structural Basis for DNA Recognition by STAT6. Proc. Natl. Acad. Sci. USA 113, 13015–13020. doi:10.1073/pnas.1611228113
Liam, C.-K., Loo, K.-L., Wong, C. M.-M., Lim, K.-H., and Lee, T.-C. (2002). Skin Prick Test Reactivity to Common Aeroallergens in Asthmatic Patients with and without Rhinitis. Respirology 7, 345–350. doi:10.1046/j.1440-1843.2002.00409.x
Lin, Y., Li, B., Yang, X., Liu, T., Shi, T., Deng, B., et al. (2019). Non-Hematopoietic STAT6 Induces Epithelial Tight junction Dysfunction and Promotes Intestinal Inflammation and Tumorigenesis. Mucosal Immunol. 12, 1304–1315. doi:10.1038/s41385-019-0204-y
London, N. R., Tharakan, A., and Ramanathan, M. (2016). The Role of Innate Immunity and Aeroallergens in Chronic Rhinosinusitis. Adv. Otorhinolaryngol. 79, 69–77. doi:10.1159/000445132
Nguyen, J. K., Austin, E., Huang, A., Mamalis, A., and Jagdeo, J. (2020). The IL-4/IL-13 axis in Skin Fibrosis and Scarring: Mechanistic Concepts and Therapeutic Targets. Arch. Dermatol. Res. 312, 81–92. doi:10.1007/s00403-019-01972-3
Nur Husna, S. M., Siti Sarah, C. O., Tan, H.-T. T., Md. Shukri, N., Mohd Ashari, N. S., and Wong, K. K. (2021). Reduced Occludin and Claudin-7 Expression Is Associated with Urban Locations and Exposure to Second-Hand Smoke in Allergic Rhinitis Patients. Sci. Rep. 11, 1245. doi:10.1038/s41598-020-79208-y
Nur Husna, S. M., Tan, H.-T. T., Md Shukri, N., Mohd Ashari, N. S., and Wong, K. K. (2021). Nasal Epithelial Barrier Integrity and Tight Junctions Disruption in Allergic Rhinitis: Overview and Pathogenic Insights. Front. Immunol. 12, 663626. doi:10.3389/fimmu.2021.663626
Pawankar, R. (2014). Allergic Diseases and Asthma: A Global Public Health Concern and a Call to Action. World Allergy Organ. J. 7, 12. doi:10.1186/1939-4551-7-12
Pelaia, C., Vatrella, A., Bruni, A., Terracciano, R., and Pelaia, G. (2018). Benralizumab in the Treatment of Severe Asthma: Design, Development and Potential Place in Therapy. Drug Des. Devel Ther. 12, 619–628. doi:10.2147/DDDT.S155307
Rabe, K. F., Nair, P., Brusselle, G., Maspero, J. F., Castro, M., Sher, L., et al. (2018). Efficacy and Safety of Dupilumab in Glucocorticoid-Dependent Severe Asthma. N. Engl. J. Med. 378, 2475–2485. doi:10.1056/NEJMoa1804093
Rondón, C., Campo, P., Zambonino, M. A., Blanca-Lopez, N., Torres, M. J., Melendez, L., et al. (2014). Follow-up Study in Local Allergic Rhinitis Shows a Consistent Entity Not Evolving to Systemic Allergic Rhinitis. J. Allergy Clin. Immunol. 133, 1026–1031. doi:10.1016/j.jaci.2013.10.034
Sahoo, A., Wali, S., and Nurieva, R. (2016). T Helper 2 and T Follicular Helper Cells: Regulation and Function of Interleukin-4. Cytokine Growth Factor. Rev. 30, 29–37. doi:10.1016/j.cytogfr.2016.03.011
Sani, M. M., Ashari, N. S. M., Abdullah, B., Wong, K. K., Musa, K. I., Mohamud, R., et al. (2019). Reduced CD4+ Terminally Differentiated Effector Memory T Cells in Moderate-Severe House Dust Mites Sensitized Allergic Rhinitis Patients. Asian Pac. J. Allergy Immunol. 37, 138–146. doi:10.12932/AP-191217-0220
Sato, J., Konno, N., Murakami, M., Uede, T., and Himi, T. (2016). Adenovirus-Mediated ICOSIg Gene Therapy in a Presensitized Murine Model of Allergic Rhinitis. Adv. Otorhinolaryngol. 77, 59–66. doi:10.1159/000441876
Shamji, M. H., Bellido, V., Scadding, G. W., Layhadi, J. A., Cheung, D. K. M., Calderon, M. A., et al. (2015). Effector Cell Signature in Peripheral Blood Following Nasal Allergen challenge in Grass Pollen Allergic Individuals. Allergy 70, 171–179. doi:10.1111/all.12543
Siti Sarah, C. O., Md Shukri, N., Mohd Ashari, N. S., and Wong, K. K. (2020). Zonula Occludens and Nasal Epithelial Barrier Integrity in Allergic Rhinitis. PeerJ 8, e9834. doi:10.7717/peerj.9834
Skrindo, I., Ballke, C., Gran, E., Johansen, F.-E., Baekkevold, E. S., and Jahnsen, F. L. (2015). IL-5 Production by Resident Mucosal Allergen-specific T Cells in an Explant Model of Allergic Rhinitis. Clin. Exp. Allergy 45, 1296–1304. doi:10.1111/cea.12543
Steelant, B., Farré, R., Wawrzyniak, P., Belmans, J., Dekimpe, E., Vanheel, H., et al. (2016). Impaired Barrier Function in Patients with House Dust Mite-Induced Allergic Rhinitis Is Accompanied by Decreased Occludin and Zonula Occludens-1 Expression. J. Allergy Clin. Immunol. 137, 1043–1053. doi:10.1016/j.jaci.2015.10.050
Steelant, B., Seys, S. F., Van Gerven, L., Van Woensel, M., Farré, R., Wawrzyniak, P., et al. (2018). Histamine and T Helper Cytokine-Driven Epithelial Barrier Dysfunction in Allergic Rhinitis. J. Allergy Clin. Immunol. 141, 951–963. doi:10.1016/j.jaci.2017.08.039
Takaki, H., Ichiyama, K., Koga, K., Chinen, T., Takaesu, G., Sugiyama, Y., et al. (2008). STAT6 Inhibits TGF-β1-Mediated Foxp3 Induction through Direct Binding to the Foxp3 Promoter, Which Is Reverted by Retinoic Acid Receptor. J. Biol. Chem. 283, 14955–14962. doi:10.1074/jbc.M801123200
Takano, K.-i., Kojima, T., Go, M., Murata, M., Ichimiya, S., Himi, T., et al. (2005). HLA-DR- and CD11c-Positive Dendritic Cells Penetrate beyond Well-Developed Epithelial Tight Junctions in Human Nasal Mucosa of Allergic Rhinitis. J. Histochem. Cytochem. 53, 611–619. doi:10.1369/jhc.4A6539.2005
Tan, L., Qiu, T., Xiang, R., Cao, C., Deng, Y., Tao, Z., et al. (2020). Down-Regulation of Tet2 Is Associated with Foxp3 TSDR Hypermethylation in Regulatory T Cell of Allergic Rhinitis. Life Sci. 241, 117101. doi:10.1016/j.lfs.2019.117101
Teng, Y., Zhang, R., Liu, C., Zhou, L., Wang, H., Zhuang, W., et al. (2015). miR-143 Inhibits Interleukin-13-Induced Inflammatory Cytokine and Mucus Production in Nasal Epithelial Cells from Allergic Rhinitis Patients by Targeting IL13Rα1. Biochem. Biophysical Res. Commun. 457, 58–64. doi:10.1016/j.bbrc.2014.12.058
Wagener, A. H., Zwinderman, A. H., Luiten, S., Fokkens, W. J., Bel, E. H., Sterk, P. J., et al. (2013). The Impact of Allergic Rhinitis and Asthma on Human Nasal and Bronchial Epithelial Gene Expression. PLoS One 8, e80257. doi:10.1371/journal.pone.0080257
Wawrzyniak, P., Wawrzyniak, M., Wanke, K., Sokolowska, M., Bendelja, K., Rückert, B., et al. (2017). Regulation of Bronchial Epithelial Barrier Integrity by Type 2 Cytokines and Histone Deacetylases in Asthmatic Patients. J. Allergy Clin. Immunol. 139, 93–103. doi:10.1016/j.jaci.2016.03.050
Weinstein, S. F., Katial, R., Jayawardena, S., Pirozzi, G., Staudinger, H., Eckert, L., et al. (2018). Efficacy and Safety of Dupilumab in Perennial Allergic Rhinitis and Comorbid Asthma. J. Allergy Clin. Immunol. 142, 171–177. doi:10.1016/j.jaci.2017.11.051
Wills-Karp, M., and Finkelman, F. D. (2008). Untangling the Complex Web of IL-4- and IL-13-mediated Signaling Pathways. Sci. Signal. 1, pe55. doi:10.1126/scisignal.1.51.pe55
Wise, S. K., Laury, A. M., Katz, E. H., Den Beste, K. A., Parkos, C. A., and Nusrat, A. (2014). Interleukin-4 and Interleukin-13 Compromise the Sinonasal Epithelial Barrier and Perturb Intercellular junction Protein Expression. Int. Forum Allergy Rhinology 4, 361–370. doi:10.1002/alr.21298
Zhao, H., Li, H., Zhou, C., Shi, W., Zhang, J., and Hao, Y. (2018). Aqueous Nasal spray of Chitosan Oligosaccharide Ameliorates Perennial Allergic Rhinitis by Affecting Serum Levels of Interleukin-6 and Interleukin-10. Int. J. Clin. Exp. Med. 11, 539–548.
Keywords: allergic rhinitis, IL-4, IL4R, IL-13, IL13RA1, tight junction
Citation: Nur Husna SM, Md Shukri N, Tuan Sharif SE, Tan HTT, Mohd Ashari NS and Wong KK (2022) IL-4/IL-13 Axis in Allergic Rhinitis: Elevated Serum Cytokines Levels and Inverse Association With Tight Junction Molecules Expression. Front. Mol. Biosci. 9:819772. doi: 10.3389/fmolb.2022.819772
Received: 22 November 2021; Accepted: 08 February 2022;
Published: 17 March 2022.
Edited by:
Paolo Colombo, National Research Council (CNR), ItalyReviewed by:
Laura La Paglia, Institute for High Performance Computing and Networking (ICAR), ItalyStefanie Gilles, Technical University of Munich, Germany
Copyright © 2022 Nur Husna, Md Shukri, Tuan Sharif, Tan, Mohd Ashari and Wong. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Kah Keng Wong, a2Foa2VuZ0B1c20ubXk=
 Siti Muhamad Nur Husna
Siti Muhamad Nur Husna Norasnieda Md Shukri
Norasnieda Md Shukri Sharifah Emilia Tuan Sharif
Sharifah Emilia Tuan Sharif Hern Tze Tina Tan1
Hern Tze Tina Tan1 Noor Suryani Mohd Ashari
Noor Suryani Mohd Ashari Kah Keng Wong
Kah Keng Wong