- 1Department of Biological Sciences, School of Science, Xi’an Jiaotong-Liverpool University, Suzhou, Jiangsu, China
- 2School of Electronic Science and Engineering, University of Electronic Science and Technology of China, Chengdu, China
- 3Department of Chemistry, Rhodes University, Grahamstown, South Africa
The remarkable rise of the current COVID-19 pandemic to every part of the globe has raised key concerns for the current public healthcare system. The spike (S) protein of SARS-CoV-2 shows an important part in the cell membrane fusion and receptor recognition. It is a key target for vaccine production. Several researchers studied the nature of this protein under various environmental conditions. In this work, we applied molecular modeling and extensive molecular dynamics simulation approaches at 0°C (273.15 K), 20°C (293.15 K), 40°C (313.15 K), and 60°C (333.15 K) to study the detailed conformational alterations in the SARS-CoV-2 S protein. Our aim is to understand the influence of temperatures on the structure, function, and dynamics of the S protein of SARS-CoV-2. The structural deviations, and atomic and residual fluctuations were least at low (0°C) and high (60°C) temperature. Even the internal residues of the SARS-CoV-2 S protein are not accessible to solvent at high temperature. Furthermore, there was no unfolding of SARS-CoV-2 spike S reported at higher temperature. The most stable conformations of the SARS-CoV-2 S protein were reported at 20°C, but the free energy minimum region of the SARS-CoV-2 S protein was sharper at 40°C than other temperatures. Our findings revealed that higher temperatures have little or no influence on the stability and folding of the SARS-CoV-2 S protein.
Introduction
The outbreaks of Severe Acute Respiratory Syndrome CoV 1 (SARS-CoV-1), Middle-East Respiratory Syndrome CoV (MERS-CoV), and Severe Acute Respiratory Syndrome CoV 2 (SARS-CoV-2) were caused by zoonotic viruses in 2003, 2012, and 2019–2020 with a fatality ratio of 10%, 35%, and 5%, respectively (Lee et al., 2003; Cheng et al., 2007; Zaki et al., 2012; de Groot et al., 2013; Reusken et al., 2013; Rothan and Byrareddy, 2020). The International Virus Classification Commission (ICTV) termed this 2019 novel CoV as SARS-CoV-2 (Chen et al., 2020; Zhu et al., 2020). SARS-CoV-2 virus spread from humans to humans, and animals to humans (Khan et al., 2020a; Khan et al., 2021a). The COVID-19-infected patient develops mild to moderate symptoms and recovers. Some patients have serious symptoms such as atypical pneumonia and chest pain (Huang et al., 2020a; Cheung et al., 2020; Lu et al., 2020; Rothan and Byrareddy, 2020). The phenomenal spread of the current COVID-19 pandemic to every part of the sphere has raised key concerns for the healthcare system. To combat this pandemic, the researchers are using all possible approaches and practices to inhibit the synthesis of crucial non-structural viral proteins, inhibit the viral replicase enzyme, inhibit the formation of viral RNA, prevent the self-assembly of viruses, or boost the human immune response against the virus.
The Spike (S) protein of SARS-CoV-2 performs a vital part in the cell membrane fusion and receptor recognition. It has two subunits such as S1 and S2. A receptor-binding domain (RBD) is present on the S1 subunit. The RBD recognizes and attaches to the host receptor angiotensin-converting enzyme 2 (ACE-2). The membrane fusion (MF) is facilitated by the S2 subunit by making 6 helical bundles through two heptad repeat (HR) domains (Huang et al., 2020b). The S protein has a size of 180–200 kDa. It has an extracellular N-terminal, a transmembrane (TM) attached to the membrane, and small intracellular C-terminal domains.
The S proteins are covered with polysaccharide for camouflage and escaping the host immune system in the course of entry (Huang et al., 2020b). The S protein of SARS-CoV-2 contains 1,273 amino acid residues. It contains a signal peptide (1–13 amino acids), an S1 subunit (14–685 residues), and an S2 subunit (686–1,273 residues). The S1 subunit has 14–305 N-terminal domain amino acids and 319–541 RBD amino acids. The S2 subunit has 788–806 fusion peptide (FP) amino acids, 912–984 HR1 amino acids, 1,163–1,213 HR2 amino acids, 1,213–1,237 TM domain amino acids, and 1,237–1,273 cytoplasm domain amino acids (Xia et al., 2020). The SARS-CoV-2 S protein lives as a sedentary precursor in native state. During the viral contagion, the proteases from target cells trigger the S protein by slicing it into two different subunits (Bertram et al., 2013), which is needed for triggering the MF domain after entry of virus into the target cells.
The SARS-CoV-2 S protein is an important target for vaccine production. In this work, we applied molecular modeling and molecular dynamics simulations approaches at 0°C (273.15 K), 20°C (293.15 K), 40°C (313.15 K), and 60°C (333.15 K) to study the detailed conformational variations in the SARS-CoV-2 S protein. It is worth noting that, experimentally, in terms of information on inactivation of viruses, data in the range 40–60°C are essential (Bertrand et al., 2012). Our findings revealed that higher temperatures have little or no influence on the SARS-CoV-2 S protein. We found that the structural deviations, atomic, and residual fluctuations were least at low (0°C) and high (60°C) temperature. The solvent accessible area plot indicated that the internal residues of SARS-CoV-2 spike protein are not exposed to solvent at high temperature. The secondary structure scheme indicated that there was no such denaturation of the SARS-CoV-2 S protein at higher temperature. The most stable conformations of the SARS-CoV-2 S protein was found at 20°C, but the free energy state region of SARS-CoV-2 spike protein was sharper at 40°C than other temperatures.
Materials and Methods
Structure Modeling of the SARS-CoV-2 S Protein
The structures of S protein of SARS-CoV-2 (PDB: 6vsb) were taken from PDB (Wrapp et al., 2020). The missing atoms in the structure of S protein were modeled using MODELLER (Webb and Sali, 2016). The complete protocols are stated in preceding publications (Khan et al., 2015; Khan et al., 2016c; Khan et al., 2017a; Khan et al., 2017b; Khan et al., 2021d). Structure analysis was performed using PDBsum (Laskowski et al., 2018) and numerous modules of MD simulations. PyMOL was used for visualization and drawing structure.
MD Simulations
MD simulations were achieved on SARS-CoV-2 spike protein at 0°C (273.15 K), 20°C (293.15 K), 40°C (313.15 K), and 60°C (333.15 K) via GROMACS 2018.2 (Van Der Spoel et al., 2005) using a standard protocol (Khan et al., 2020b; Khan et al., 2020c; Khan et al., 2021b). Na+ and Cl− ions were supplemented to neutralize the system. Absolute production phase of 100 ns was attained at 0°C (273.15 K), 20°C (293.15 K), 40°C (313.15 K), and 60°C (333.15 K). The complete MD simulation procedure is cited in prior publications (Khan et al., 2016a; Khan et al., 2016b; Durrani et al., 2020; Hassan et al., 2020; Qausain et al., 2020).
Essential Dynamics
ED was obtained for the SARS-CoV-2 S protein at 0°C (273.15 K), 20°C (293.15 K), 40°C (313.15 K), and 60°C (333.15 K). It is estimated as:
ri denotes the coordinate, ith Cα atom, N signifies the Cα atoms, and <ri > indicates time average over all configurations (Khan et al., 2020d).
Gibbs Free Energy Landscape
GFE landscape can suggest conformational variations in the SARS-CoV-2 S protein at 0°C (273.15 K), 20°C (293.15 K), 40°C (313.15 K), and 60°C (333.15 K) (Khan et al., 2016c). The GFE landscape was projected on PC1 and PC2.
kB, T, and P(PC1, PC2) denote Boltzmann constant, temperature, and normalized joint probability distribution for SARS-CoV-2 spike protein at 0°C (273.15 K), 20°C (293.15 K), 40°C (313.15 K), and 60°C (333.15 K) respectively.
Results and Discussion
Structure Analysis of Spike Protein
The SARS-CoV-2 spike protein comprises N-terminal, TM, and C-terminal segments (Bosch et al., 2003). It consists of a signal peptide (1–13 residues at the N-terminal), an S1 subunit (14–685 amino acid residues), and an S2 subunit (686–1,273 amino acid residues). The S1 is accountable for receptor attachment, and S2 is accountable for membrane fusion. The S1 subunit contains 14–305 NTD residues and 319–541 RBD residues. The S2 contains 788–806 FP amino acid residues, 912–984 HR1 residues, 1,163–1,213 HR2 residues, 1,213–1,237 TM domain residues, and 1,237–1,273 cytoplasm domain residues (Huang et al., 2020b). The residues that participated in strand, α-helix, and 3–10 helix formations are 271 (28.3%), 190 (19.8%), and 23 (2.4%), respectively. The structure of spike protein includes 18 β-hairpins, 13 β-sheets, 52 β-strands, 18 β-bulges, 29 helix-helix interactions, 22 helices, 16 γ-turns, 76 β-turns, and 12 disulfides (Figure 1).
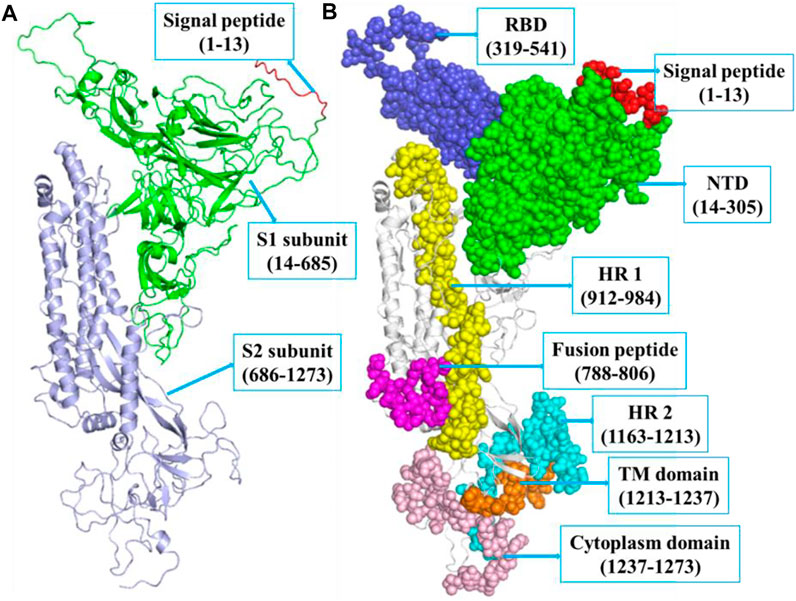
FIGURE 1. (A) The structure of SARS-CoV-2 spike protein indicating SP (red, 1–13 aa), S1 (green, 14–685 aa), and S2 (blue, 686–1,273 aa), respectively. (B) The S1 subunit includes NTD domain (green, 14–305 aa) and RBD (blue, 319–541 aa). The S2 subunit includes FP (magenta, 788–806 aa), hepta-peptide repeat sequence 1 (yellow, 912–984 aa), hepta-peptide repeat sequence 2 (cyan, 1,163–1,213 aa), TM domain (orange, 1,213–1,237 aa), and cytoplasm domain (pink, 1,237–1,273 aa), respectively.
Structural Deviations
To investigate the structural dynamics of the SARS-CoV-2 S protein, the RMSD, RMSF, and the Rg were considered throughout 100-ns MD simulations at 0°C (273.15 K), 20°C (293.15 K), 40°C (313.15 K), and 60°C (333.15 K), respectively (Kuzmanic and Zagrovic, 2010). The mean RMSD values of the SARS-CoV-2 S protein at 0, 20, 40, and 60°C were estimated to be 1.53, 2.51, 3.26, and 2.23 nm, respectively (Figure 2). It has been estimated that RMSD values, and residual and atomic fluctuations increase from 0 to 40°C. It attained a low structural deviation equilibrium at 60°C. The SARS-CoV-2 S protein is least deviated at low (0°C) and high (60°C) temperature. The average radius of gyration (Rg) values for the SARS-CoV-2 S protein at 0, 20, 40, and 60°C was found to be 4.09, 4.37, 4.32, and 3.48 nm, respectively. The Rg is described as the allotment of atoms of a molecule around its axis. The calculation of Rg is a significant indicator that is broadly used in calculating the structural activity. At different temperatures, there is a conformational change in the SARS-CoV-2 S protein that changes the radius of gyration. It was estimated that the SARS-CoV-2 S protein is tightly packed at 60°C. At 20°C–40°C, it shows high fluctuations throughout the time scale.
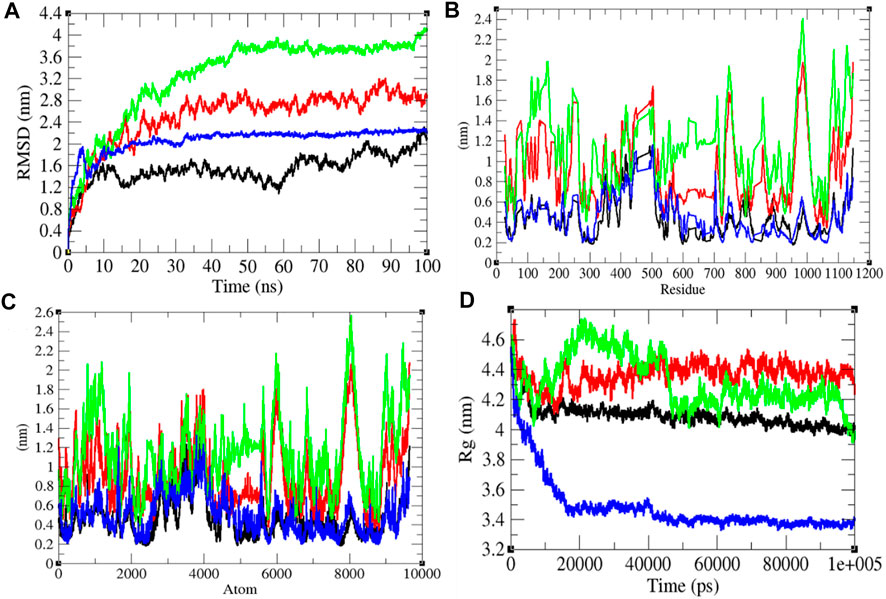
FIGURE 2. Structural dynamics. (A) RMSD plot for the SARS-CoV-2 S protein vs. time. (B) RMSF vs. residues. (C) RMSF vs. residues. (D) Rg plot vs. time. The values calculated at 0°C (black), 20°C (red), 40°C (green), and 60°C (blue), respectively.
Solvent Accessible Surface Area
It has been assumed as a significant element in molecular stability and folding analysis. The average solvent accessible surface area values for the SARS-CoV-2 S protein at 0, 20, 40, and 60°C were found to be 437.71, 439.92, 418.90, and 384.66 nm2, respectively (Figure 3). The solvation energy for the SARS-CoV-2 S protein at 0, 20, 40, and 60°C was found to be 752.14, 730.93, 668.86, and 657.95 kJ/mol/nm2, respectively. An increase in temperature from 0 to 20°C has not much effect on SASA of the SARS-CoV-2 S protein. At 40°C–60°C, the solvent accessible surface area of the SARS-CoV-2 S protein continuously decreases. This specifies that the internal residues of the SARS-CoV-2 S protein are not exposed to solvent at high temperature. This might be due to stability and compactness of the SARS-CoV-2 S protein at higher temperature. The solvation energy refers to the free-energy change during the simulations. The solvation free energy is also less at higher temperature. The solvent accessible surface area was further divided into hydrophobic and hydrophilic regions. The hydrophobic regions for the SARS-CoV-2 S protein at 0, 20, 40, and 60°C were found to be 224.22, 224.34, 221.61, and 218.62 nm2, respectively. The hydrophilic regions for the SARS-CoV-2 S protein at 0, 20, 40, and 60°C were 254.52, 256.53, 250.9, and 243.08 nm2, respectively. Both hydrophobic and hydrophilic regions are sparingly accessible to solvent at higher temperatures.
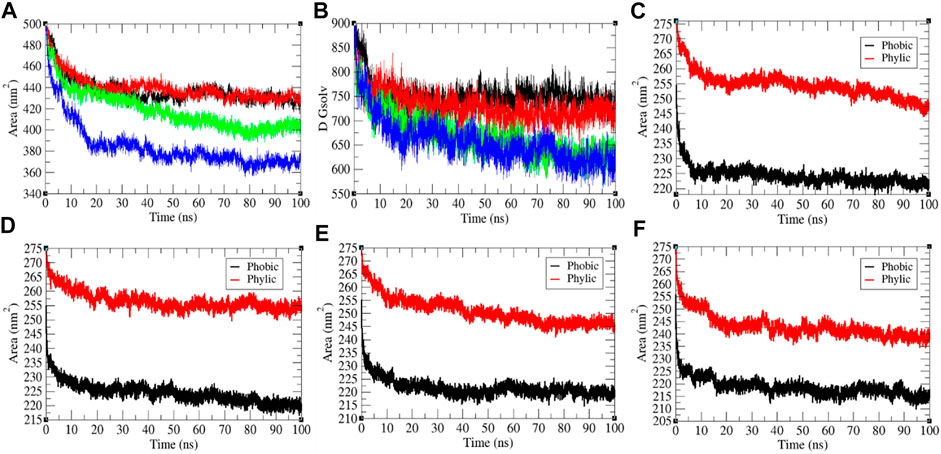
FIGURE 3. The solvent accessible surface area of the SARS-CoV-2 S protein. (A) SASA vs. time. (B) Free energy of solvation vs. time. The color codes have the same meaning as described in Figure 2. It was resolved into hydrophobic and hydrophilic regions for the SARS-CoV-2 S protein at (C) 0°C, (D) 20°C, (E) 40°C, and (F) 60°C, respectively.
Secondary Structure Analysis
The secondary structure in the SARS-CoV-2 S protein was analyzed at each period at 0, 20, 40, and 60°C (Table 1). The mean residues involved in the assembly of the SARS-CoV-2 S protein at 0, 20, 40, and 60°C were found to be 60%, 61%, 59%, and 59%, respectively (Figure 4). There was no such unfolding of the SARS-CoV-2 S protein reported at higher temperature from this analysis. The β-sheet of the SARS-CoV-2 S protein slightly unfolds from 30 to 28% at 60°C, while the α-helix (21%) remained unchanged at 60°C. The most stable conformation of the SARS-CoV-2 S protein was found at 20°C. Furthermore, we calculated the volume and density of the SARS-CoV-2 S protein at 0, 20, 40, and 60°C, respectively. Moreover, the volume of the SARS-CoV-2 S protein was found to be 176.02, 176.04, 174.71, and 172.03 nm3 at 0, 20, 40, and 60°C, respectively, while the density of the SARS-CoV-2 S protein was calculated to be 1,001.84 g/L, 1,001.72 g/L, 1,009.39 g/L, and 1,025.18 g/L at 0, 20, 40, and 60°C, respectively. The volume of the SARS-CoV-2 S protein slightly decreases and density increases at higher temperature. This might be due to different structure conformations at higher temperatures.
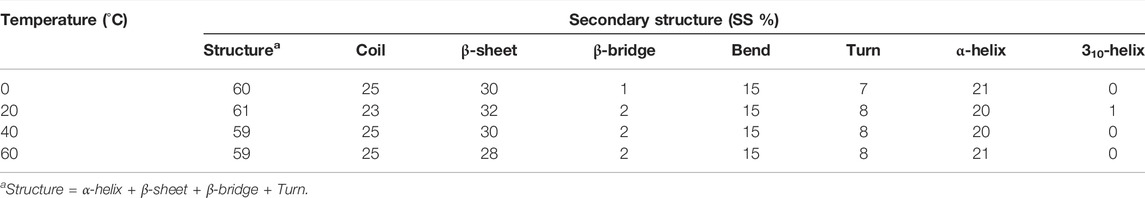
TABLE 1. Percentage of residues in SARS-CoV-2 spike protein at 0, 20, 40, and 60°C contributed in mean structure development.
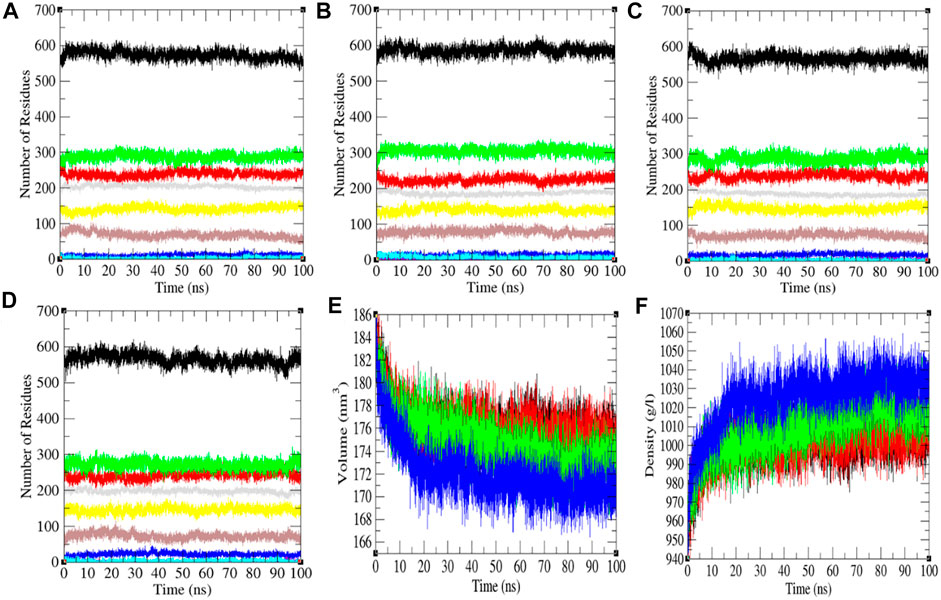
FIGURE 4. The secondary structure conformations. The secondary structure plot of the SARS-CoV-2 S protein at (A) 0°C, (B) 20°C, (C) 40°C, and (D) 60°C, respectively. (E) Volume and (F) density of SARS-CoV-2 spike protein. The color codes have the same meaning as described in Figure 2.
Hydrogen Bonding and the Mean Square Displacement
The H-bond is a noteworthy element in stabilizing the molecule. It was estimated between the main chain and side chains (M-S) of the SARS-CoV-2 S protein at 0, 20, 40, and 60°C, respectively. The mean H-bonds between M-S chains of the SARS-CoV-2 S protein were found to be 413.43, 407.89, 417.99, and 417.51 at 0, 20, 40, and 60°C, respectively (Figure 5). The strength of hydrogen bonds becomes stronger at 40–60°C. There is no sign of denaturation at higher temperatures. Furthermore, the mean square displacement (MSD) of atoms from a set of original positions of the SARS-CoV-2 S protein at 0, 20, 40, and 60°C was computed. The displacement of atoms from a set of initial positions of the SARS-CoV-2 S protein was estimated to be higher at 40°C only. In short, higher temperature has not much impact on unfolding and denaturation of the SARS-CoV-2 S protein.
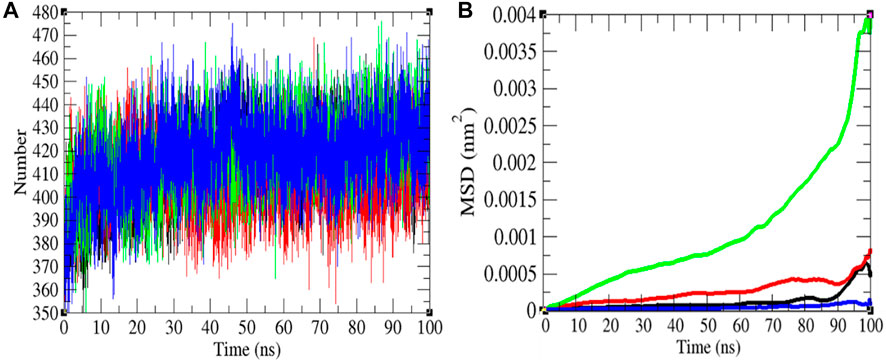
FIGURE 5. Hydrogen bonds and MSD. (A) The H-bond estimation between M-S chains of the SARS-CoV-2 S protein was calculated. The color codes have the same meaning as described in Figure 2. (B) The MSD of the SARS-CoV-2 S protein at 0°C (black), 20°C (red), 40°C (green), and 60°C (blue), respectively.
Principal Component Analysis
It shows global expansion of the SARS-CoV-2 S protein at 0, 20, 40, and 60°C. It estimates mean atomic motions of the SARS-CoV-2 S protein at 0, 20, 40, and 60°C. The eigenvalues were 8,360.61, 33,665.60, 53,083.50, and 8,911.24 nm2 for the SARS-CoV-2 S protein at 0, 20, 40, and 60°C, respectively. It was higher at 20–40°C (Figure 6). The average atomic motions in the SARS-CoV-2 S protein was highest at 40°C. The atomic motions are also related to activity in case of protein molecules. It can be assumed that at low and high environmental temperatures, the atomic motions and activity of the SARS-CoV-2 S protein are low.
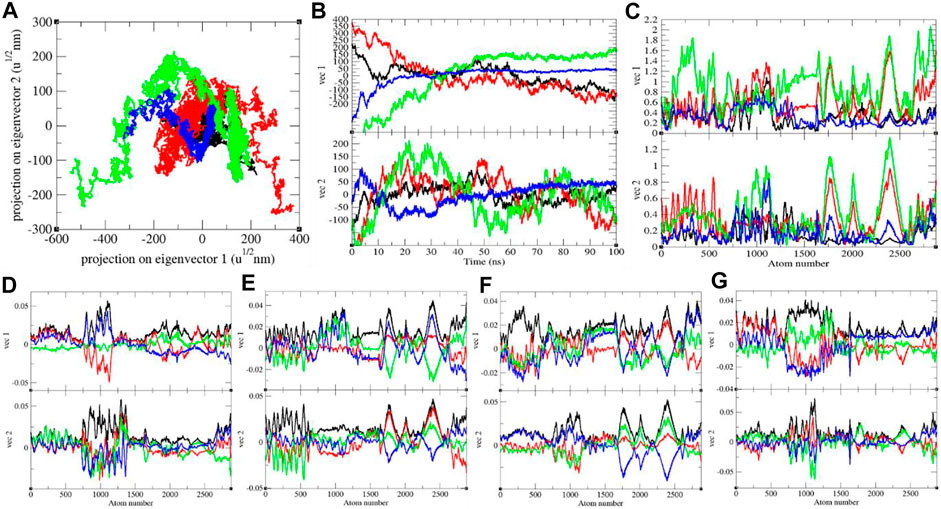
FIGURE 6. (A) The 2D projection and (B) projections of trajectories of SARS-CoV-2 spike protein. (C) Eigen RMSF. The color codes have the same meaning as described in Figure 2. The Eigen components were calculated for SARS-CoV-2 spike protein at (D) 0°C, (E) 20°C, (F) 40°C, and (G) 60°C, respectively.
GFE Landscape
The GFE landscape exhibited diverse forms for the SARS-CoV-2 S protein at 0, 20, 40, and 60°C (Figure 7). Every atomic pair covariance displays diverse frameworks in respective events. The GFE patterns are relatively similar with minor changes at 0–20°C and 40–60°C. The following GFE curve with reflective blue shade implies lower energy state. Extra blue regions describe shifts in the molecular conformation lagged by the thermodynamically new favorable areas. The GFE state in the global energy minimum section of the SARS-CoV-2 S protein at 40°C is sharper than other temperatures. This indicates that temperature slightly affects the GFE patterns in the case of the SARS-CoV-2 spike protein. The GFE landscape suggests that the temperature slightly affects the atomic motions of SARS-CoV-2 spike protein. The denaturation was not reported from the secondary structure analysis. The potential energy and the enthalpy were also calculated during the course of simulations. The potential energy was found to be −10,713,734.28 kJ/mol, −10,434,949.84 kJ/mol, −10,161,602.16 kJ/mol, and −9,893,572.08 kJ/mol at 0, 20, 40, and 60°C, respectively. The enthalpy was found to be −9,100,901.71 kJ/mol, −8,704,052.14 kJ/mol, −8,312,630.26 kJ/mol, and −7,926,537.36 kJ/mol at 0, 20, 40, and 60°C, respectively.
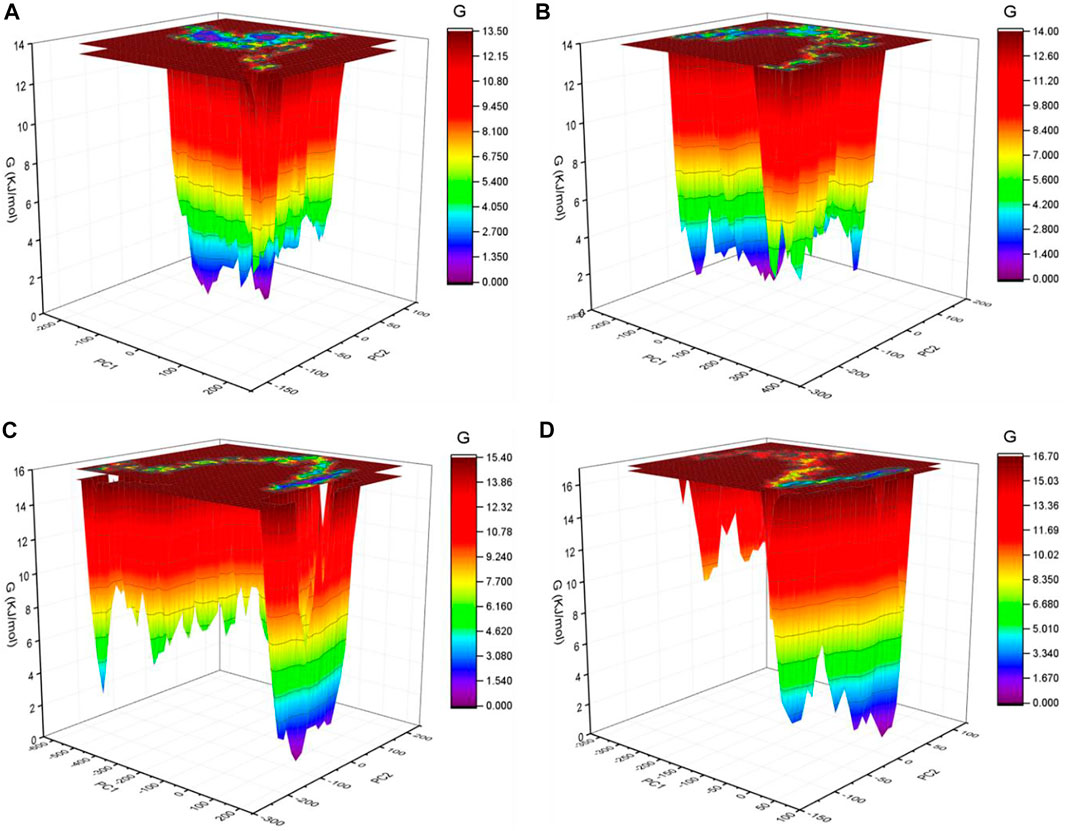
FIGURE 7. GFE landscape. The GFE landscape plot achieved for SARS-CoV-2 spike protein at (A) 0°C, (B) 20°C, (C) 40°C, and (D) 60°C, respectively.
Recently, several inhibitors and their mode of action have been demonstrated (Khan et al., 2021c; Rani et al., 2021). Edwards et al. found that the spike protein samples kept at diverse temperatures did not show any considerable denaturation, while they observed an increase in upper molecular weight bands in a sample that was kept at 37°C (Edwards et al., 2021). Kumar et al. imitated SARS-CoV-2 by polymer beads covered with the S protein of SARS-CoV-2 to investigate the effect of different temperatures on attachment of virus-imitating nano-spheres to lung tissues incubated at 33 and 37°C. They found that the existence of the RBD of S protein controlled the binding by Calu-3 airway epithelial tissues. They also found that there was no temperature correlation to binding of BSA-coated nano-spheres. Additionally, the 4–40°C temperature had no influence on S-RBD-ACE-2 ligand–receptor, and the negligible effect on the S-RBD protein structure (up to 40°C) was reported. The protein denaturation occurred at 51°C. Their outcomes suggested that 4–40°C temperature has a slight influence on SARS-CoV-2 and ACE-2 contact (Kumar et al., 2021). Zhou et al. performed MD simulations at 36–40°C to prove SARS-CoV-2 and ACE2 binding. They found that it was less stable under 40°C than under 37°C, and reduced infection rate at higher temperature (Zhou et al., 2021). Martí et al. also performed MD simulations at 298 K (24.85°C), 310 K (36.85°C), 324 K (50.85°C), 338 K (64.85°C), 358 K (84.85°C), and 373 K (99.85°C), respectively. They suggested that temperature brings structural and conformational variations in the S1 subunit and affects the RBD. Nevertheless, the influence of temperature up to 373 K was not adequate to cause a noteworthy alteration of the S protein of SARS-CoV-2 (Martí et al., 2021). Our results also suggested that the temperature has the least effect on the structure conformations of S protein of SARS-CoV-2.
Conclusion
Previously, we published several articles based on finding potential inhibitors of SARS-CoV-2. In the present work, we focused on the SARS-CoV-2 S protein as it performs a vital part in the cell membrane fusion and receptor recognition. Researchers have demonstrated the nature of the S protein of SARS-CoV-2 on diverse environmental conditions. We applied molecular modeling and extensive molecular dynamics simulations approaches at different temperatures to investigate the structural conformational of SARS-CoV-2 spike protein. There are several hypotheses proposed regarding the temperature dependence of the COVID-19 transmission. We concluded that temperature has no effect or has the least effect on the structure conformations of S protein of SARS-CoV-2. Minor changes were reported in the structure and thermodynamic properties that are mentioned in this paper.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author Contributions
Conceptualization, FK and DL. Methodology, FK. Software, FK and KL. Validation, FK and DL. Formal analysis, FK and DL. Investigation, FK and DL. Resources, FK and DL. Data curation, FK and DL. Writing—original draft preparation, FK, KL, and DL. Writing—review and editing, FK. Visualization, FK; supervision, DL. Project administration, DL. Funding acquisition, FK and DL. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the Sichuan Science and Technology Program (No. 2021YFH0093), and the China Postdoctoral Science Foundation (No. 2020M673187).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We would like to acknowledge The Centre for High Performance Computing (CHPC), South Africa.
References
Bertram, S., Dijkman, R., Habjan, M., Heurich, A., Gierer, S., Glowacka, I., et al. (2013). TMPRSS2 Activates the Human Coronavirus 229E for Cathepsin-independent Host Cell Entry and Is Expressed in Viral Target Cells in the Respiratory Epithelium. J. Virol. 87, 6150–6160. doi:10.1128/jvi.03372-12
Bertrand, I., Schijven, J. F., Sánchez, G., Wyn-Jones, P., Ottoson, J., Morin, T., et al. (2012). The Impact of Temperature on the Inactivation of Enteric Viruses in Food and Water: a Review. J. Appl. Microbiol. 112, 1059–1074. doi:10.1111/j.1365-2672.2012.05267.x
Bosch, B. J., Van Der Zee, R., De Haan, C. A. M., and Rottier, P. J. M. (2003). The Coronavirus Spike Protein Is a Class I Virus Fusion Protein: Structural and Functional Characterization of the Fusion Core Complex. J. Virol. 77, 8801–8811. doi:10.1128/jvi.77.16.8801-8811.2003
Chen, Y., Liu, Q., and Guo, D. (2020). Emerging Coronaviruses: Genome Structure, Replication, and Pathogenesis. J. Med. Virol. 92, 418–423. doi:10.1002/jmv.25681
Cheng, V. C. C., Lau, S. K. P., Woo, P. C. Y., and Yuen, K. Y. (2007). Severe Acute Respiratory Syndrome Coronavirus as an Agent of Emerging and Reemerging Infection. Clin. Microbiol. Rev. 20, 660–694. doi:10.1128/cmr.00023-07
Cheung, K. S., Hung, I. F. N., Chan, P. P. Y., Lung, K. C., Tso, E., Liu, R., et al. (2020). Gastrointestinal Manifestations of SARS-CoV-2 Infection and Virus Load in Fecal Samples from a Hong Kong Cohort: Systematic Review and Meta-Analysis. Gastroenterology 159, 81–95. doi:10.1053/j.gastro.2020.03.065
de Groot, R. J., Baker, S. C., Baric, R. S., Brown, C. S., Drosten, C., Enjuanes, L., et al. (2013). Middle East Respiratory Syndrome Coronavirus (MERS-CoV): Announcement of the Coronavirus Study Group. J. Virol. 87, 7790–7792. doi:10.1128/jvi.01244-13
Durrani, R., Khan, F. I., Ali, S., Wang, Y., and Yang, B. (2020). A Thermolabile Phospholipase B from Talaromyces marneffei GD-0079: Biochemical Characterization and Structure Dynamics Study. Biomolecules 10, 231. doi:10.3390/biom10020231
Edwards, R. J., Mansouri, K., Stalls, V., Manne, K., Watts, B., Parks, R., et al. (2021). Cold Sensitivity of the SARS-CoV-2 Spike Ectodomain. Nat. Struct. Mol. Biol. 28, 128–131. doi:10.1038/s41594-020-00547-5
Hassan, F., Khan, F. I., Song, H., Lai, D., and Juan, F. (2020). Effects of Reverse Genetic Mutations on the Spectral and Photochemical Behavior of a Photoactivatable Fluorescent Protein PAiRFP1. Spectrochimica Acta A: Mol. Biomol. Spectrosc. 228, 117807. doi:10.1016/j.saa.2019.117807
Huang, C., Wang, Y., Li, X., Ren, L., Zhao, J., Hu, Y., et al. (2020a). Clinical Features of Patients Infected with 2019 Novel Coronavirus in Wuhan, China. The Lancet 395, 497–506. doi:10.1016/s0140-6736(20)30183-5
Huang, Y., Yang, C., Xu, X.-F., Xu, W., and Liu, S.-W. (2020b). Structural and Functional Properties of SARS-CoV-2 Spike Protein: Potential Antivirus Drug Development for COVID-19. Acta Pharmacol. Sin 41, 1141–1149. doi:10.1038/s41401-020-0485-4
Khan, A., Khan, M., Saleem, S., Babar, Z., Ali, A., Khan, A. A., et al. (2020a). Phylogenetic Analysis and Structural Perspectives of RNA-dependent RNA-Polymerase Inhibition from SARs-CoV-2 with Natural Products. Interdiscip. Sci. Comput. Life Sci. 12, 335–348. doi:10.1007/s12539-020-00381-9
Khan, A., Umbreen, S., Hameed, A., Fatima, R., Zahoor, U., Babar, Z., et al. (2021a). In Silico Mutagenesis-Based Remodelling of SARS-CoV-1 Peptide (ATLQAIAS) to Inhibit SARS-CoV-2: Structural-Dynamics and Free Energy Calculations. Interdiscip. Sci. Comput. Life Sci. 13, 521–534. doi:10.1007/s12539-021-00447-2
Khan, F. I., Bisetty, K., Wei, D.-Q., and Hassan, M. I. (2016a). A pH Based Molecular Dynamics Simulations of Chitinase II Isolated from Thermomyces Lanuginosus SSBP. Cogent Biol. 2, 1168336. doi:10.1080/23312025.2016.1168336
Khan, F. I., Govender, A., Permaul, K., Singh, S., and Bisetty, K. (2015). Thermostable Chitinase II from Thermomyces Lanuginosus SSBP: Cloning, Structure Prediction and Molecular Dynamics Simulations. J. Theor. Biol. 374, 107–114. doi:10.1016/j.jtbi.2015.03.035
Khan, F. I., Gupta, P., Roy, S., Azum, N., Alamry, K. A., Asiri, A. M., et al. (2020b). Mechanistic Insights into the Urea-Induced Denaturation of Human Sphingosine Kinase 1. Int. J. Biol. Macromolecules 161, 1496–1505. doi:10.1016/j.ijbiomac.2020.07.280
Khan, F. I., Hassan, F., Ali, H., and Lai, D. (2021b). Mechanism of pH-Induced Conformational Changes in MurE Ligase Obtained from Salmonella enterica Serovar Typhi. J. Biomol. Struct. Dyn. 39, 1898–1905. doi:10.1080/07391102.2020.1739560
Khan, F. I., Hassan, F., Anwer, R., Juan, F., and Lai, D. (2020c). Comparative Analysis of Bacteriophytochrome Agp2 and its Engineered Photoactivatable NIR Fluorescent Proteins PAiRFP1 and PAiRFP2. Biomolecules 10, 1286. doi:10.3390/biom10091286
Khan, F. I., Kang, T., Ali, H., and Lai, D. (2021c). Remdesivir Strongly Binds to RNA-dependent RNA Polymerase, Membrane Protein, and Main Protease of SARS-CoV-2: Indication from Molecular Modeling and Simulations. Front. Pharmacol. 12, 710778. doi:10.3389/fphar.2021.710778
Khan, F. I., Lai, D., Anwer, R., Azim, I., and Khan, M. K. A. (2020d). Identifying Novel Sphingosine Kinase 1 Inhibitors as Therapeutics against Breast Cancer. J. Enzyme Inhib. Med. Chem. 35, 172–186. doi:10.1080/14756366.2019.1692828
Khan, F. I., Lan, D., Durrani, R., Huan, W., Zhao, Z., and Wang, Y. (2017a). The Lid Domain in Lipases: Structural and Functional Determinant of Enzymatic Properties. Front. Bioeng. Biotechnol. 5, 16. doi:10.3389/fbioe.2017.00016
Khan, F. I., Nizami, B., Anwer, R., Gu, K.-R., Bisetty, K., Hassan, M. I., et al. (2017b). Structure Prediction and Functional Analyses of a Thermostable Lipase Obtained from Shewanella Putrefaciens. J. Biomol. Struct. Dyn. 35, 2123–2135. doi:10.1080/07391102.2016.1206837
Khan, F. I., Shahbaaz, M., Bisetty, K., Waheed, A., Sly, W. S., Ahmad, F., et al. (2016b). Large Scale Analysis of the Mutational Landscape in β-glucuronidase: A Major Player of Mucopolysaccharidosis Type VII. Gene 576, 36–44. doi:10.1016/j.gene.2015.09.062
Khan, F. I., Song, H., Hassan, F., Tian, J., Tang, L., Lai, D., et al. (2021d). Impact of Amino Acid Substitutions on the Behavior of a Photoactivatable Near Infrared Fluorescent Protein PAiRFP1. Spectrochimica Acta Part A: Mol. Biomol. Spectrosc. 253, 119572. doi:10.1016/j.saa.2021.119572
Khan, F. I., Wei, D.-Q., Gu, K.-R., Hassan, M. I., and Tabrez, S. (2016c). Current Updates on Computer Aided Protein Modeling and Designing. Int. J. Biol. Macromolecules 85, 48–62. doi:10.1016/j.ijbiomac.2015.12.072
Kumar, S., Paul, A., Chatterjee, S., Pütz, S., Nehra, N., Wang, D. S., et al. (2021). Effect of Ambient Temperature on Respiratory Tract Cells Exposed to SARS-CoV-2 Viral Mimicking Nanospheres-An Experimental Study. Biointerphases 16, 011006. doi:10.1116/6.0000743
Kuzmanic, A., and Zagrovic, B. (2010). Determination of Ensemble-Average Pairwise Root Mean-Square Deviation from Experimental B-Factors. Biophysical J. 98, 861–871. doi:10.1016/j.bpj.2009.11.011
Laskowski, R. A., Jabłońska, J., Pravda, L., Vařeková, R. S., and Thornton, J. M. (2018). PDBsum: Structural Summaries of PDB Entries. Protein Sci. 27, 129–134. doi:10.1002/pro.3289
Lee, N., Hui, D., Wu, A., Chan, P., Cameron, P., Joynt, G. M., et al. (2003). A Major Outbreak of Severe Acute Respiratory Syndrome in Hong Kong. N. Engl. J. Med. 348, 1986–1994. doi:10.1056/nejmoa030685
Lu, R., Zhao, X., Li, J., Niu, P., Yang, B., Wu, H., et al. (2020). Genomic Characterisation and Epidemiology of 2019 Novel Coronavirus: Implications for Virus Origins and Receptor Binding. The Lancet 395, 565–574. doi:10.1016/s0140-6736(20)30251-8
Martí, D., Torras, J., Bertran, O., Turon, P., and Alemán, C. (2021). Temperature Effect on the SARS-CoV-2: A Molecular Dynamics Study of the Spike Homotrimeric Glycoprotein. Comput. Struct. Biotechnol. J. 19, 1848–1862. doi:10.1016/j.csbj.2021.03.037
Qausain, S., Khan, F. I., Lai, D., Hassan, M. I., Basheeruddin, M., Ahmed, N., et al. (2020). Mechanistic Insights into the Urea-Induced Denaturation of a Non-seleno Thiol Specific Antioxidant Human Peroxiredoxin 6. Int. J. Biol. Macromolecules 161, 1171–1180. doi:10.1016/j.ijbiomac.2020.05.168
Rani, J., Bhargav, A., Khan, F. I., Ramachandran, S., Lai, D., and Bajpai, U. (2021). In Silico prediction of Natural Compounds as Potential Multi-Target Inhibitors of Structural Proteins of SARS-CoV-2. J. Biomol. Struct. Dyn., 1–17. doi:10.1080/07391102.2021.1968497
Reusken, C. B., Haagmans, B. L., Müller, M. A., Gutierrez, C., Godeke, G.-J., Meyer, B., et al. (2013). Middle East Respiratory Syndrome Coronavirus Neutralising Serum Antibodies in Dromedary Camels: a Comparative Serological Study. Lancet Infect. Dis. 13, 859–866. doi:10.1016/s1473-3099(13)70164-6
Rothan, H. A., and Byrareddy, S. N. (2020). The Epidemiology and Pathogenesis of Coronavirus Disease (COVID-19) Outbreak. J. Autoimmun. 109, 102433. doi:10.1016/j.jaut.2020.102433
Van Der Spoel, D., Lindahl, E., Hess, B., Groenhof, G., Mark, A. E., and Berendsen, H. J. C. (2005). GROMACS: Fast, Flexible, and Free. J. Comput. Chem. 26, 1701–1718. doi:10.1002/jcc.20291
Webb, B., and Sali, A. (2016). Comparative Protein Structure Modeling Using MODELLER. Curr. Protoc. Protein Sci. 86, 5.6.1–5.6.37. doi:10.1002/cpps.20
Wrapp, D., Wang, N., Corbett, K. S., Goldsmith, J. A., Hsieh, C.-L., Abiona, O., et al. (2020). Cryo-EM Structure of the 2019-nCoV Spike in the Prefusion Conformation. Science 367, 1260–1263. doi:10.1126/science.abb2507
Xia, S., Zhu, Y., Liu, M., Lan, Q., Xu, W., Wu, Y., et al. (2020). Fusion Mechanism of 2019-nCoV and Fusion Inhibitors Targeting HR1 Domain in Spike Protein. Cell Mol Immunol 17, 765–767. doi:10.1038/s41423-020-0374-2
Zaki, A. M., Van Boheemen, S., Bestebroer, T. M., Osterhaus, A. D. M. E., and Fouchier, R. A. M. (2012). Isolation of a Novel Coronavirus from a Man with Pneumonia in Saudi Arabia. N. Engl. J. Med. 367, 1814–1820. doi:10.1056/nejmoa1211721
Zhou, Z., Yang, Z., Ou, J., Zhang, H., Zhang, Q., Dong, M., et al. (2021). Temperature Dependence of the SARS-CoV-2 Affinity to Human ACE2 Determines COVID-19 Progression and Clinical Outcome. Comput. Struct. Biotechnol. J. 19, 161–167. doi:10.1016/j.csbj.2020.12.005
Keywords: SARS-CoV-2, COVID-19, spike protein, MD simulations, Gibbs free energy
Citation: Khan FI, Lobb KA and Lai D (2022) The Molecular Basis of the Effect of Temperature on the Structure and Function of SARS-CoV-2 Spike Protein. Front. Mol. Biosci. 9:794960. doi: 10.3389/fmolb.2022.794960
Received: 03 November 2021; Accepted: 27 January 2022;
Published: 25 March 2022.
Edited by:
Matthew Bogyo, Stanford University, United StatesReviewed by:
Shashikant Ray, Mahatma Gandhi Central University, Motihari, IndiaMohammad Hassan Baig, Yonsei University Health System, South Korea
Copyright © 2022 Khan, Lobb and Lai. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Dakun Lai, ZGtsYWlAdWVzdGMuZWR1LmNu; Kevin A. Lobb, Sy5Mb2JiQHJ1LmFjLnph
 Faez Iqbal Khan
Faez Iqbal Khan Kevin A. Lobb
Kevin A. Lobb Dakun Lai
Dakun Lai