
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
BRIEF RESEARCH REPORT article
Front. Mol. Biosci. , 10 January 2023
Sec. Molecular Diagnostics and Therapeutics
Volume 9 - 2022 | https://doi.org/10.3389/fmolb.2022.1106963
This article is part of the Research Topic Microbiome in the Growth and Management of Different Types of Cancer View all 5 articles
 Sheetanshu Saproo1
Sheetanshu Saproo1 Shashanka S. Sarkar1
Shashanka S. Sarkar1 Vishakha Gautam2
Vishakha Gautam2 Chingmei W. Konyak1
Chingmei W. Konyak1 Gouri Dass1
Gouri Dass1 Arpita Karmakar1
Arpita Karmakar1 Mansi Sharma1
Mansi Sharma1 Gaurav Ahuja2
Gaurav Ahuja2 Anand Gupta3
Anand Gupta3 Juhi Tayal4
Juhi Tayal4 Anurag Mehta4
Anurag Mehta4 Srivatsava Naidu1*
Srivatsava Naidu1*Oral squamous cell carcinoma (OSCC) is the second leading cause of cancer-related morbidity and mortality in India. Tobacco, alcohol, poor oral hygiene, and socio-economic factors remain causative for this high prevalence. Identification of non-invasive diagnostic markers tailored for Indian population can facilitate mass screening to reduce overall disease burden. Saliva offers non-invasive sampling and hosts a plethora of markers for OSCC diagnosis. Here, to capture the OSCC-specific salivary RNA markers suitable for Indian population, we performed RNA-sequencing of saliva from OSCC patients (n = 9) and normal controls (n = 5). Differential gene expression analysis detected an array of salivary RNAs including mRNAs, long non-coding RNAs, transfer-RNAs, and microRNAs specific to OSCC. Computational analysis and functional predictions identified protein kinase c alpha (PRKCA), miR-6087, miR-449b-5p, miR-3656, miR-326, miR-146b-5p, and miR-497-5p as potential salivary indicators of OSCC. Notably, higher expression of PRKCA, miR-6087 and miR-449b-5p were found to be associated with therapeutic resistance and poor survival, indicating their prognostic potential. In addition, sequencing reads that did not map to the human genome, showed alignments with microbial reference genomes. Metagenomic and statistical analysis of these microbial reads revealed a remarkable microbial dysbiosis between OSCC patients and normal controls. Moreover, the differentially abundant microbial taxa showed a significant association with tumor promoting pathways including inflammation and oxidative stress. Summarily, we provide an integrated landscape of OSCC-specific salivary RNAs relevant to Indian population which can be instrumental in devising non-invasive diagnostics for OSCC.
Oral squamous cell carcinoma (OSCC) accounts for about 90% of all oral cancers and ranks sixth in global cancer incidence (D'Cruz et al., 2018). India registers one-third of the global OSCC cases (Borse et al., 2020) and is the second leading cause of cancer-related morbidity and mortality in the country (https://gco.iarc.fr/). Despite significant progress in the therapies, the overall survival remains dismal at 20% (Veluthattil et al., 2019), and the projected incidence rates of OSCC in India are alarmingly high (Sharma et al., 2018). Tobacco (various forms), alcohol, betel quid, human papillomavirus, poor oral hygiene, malnutrition continue to be major etiological factors for OSCC (Rivera, 2015). Notably, OSCC prevalence is relatively higher in Indian rural communities, and low-income strata due to the over exposure to risk factors, lack of disease awareness and inadequate quality care (Su et al., 2021). Community level screening aided with non-invasive diagnosis can result in early therapeutic interventions, improved prognosis, and survival of OSCC patients.
Liquid biopsy (LB) is an emerging diagnostic paradigm which allows capturing tumor-shed diagnostic molecules enriched in bodily fluids such as urine, blood, or saliva non-invasively (De Rubis et al., 2019). LB coupled to high-throughput technologies identified a diverse repertoire of potential tumor markers including proteins, nucleic acids, metabolites, and circulating tumor cells in various bodily fluids (Alix-Panabieres and Pantel, 2021). Notably, the clinical utility of LB-derived biomarkers holds promise for diagnosis, disease stratification, and monitoring prognosis of a broad spectrum of cancers, non-invasively (Larson et al., 2021).
Saliva directly interacts with OSCC tumor therefore considered an ideal non-invasive source for OSCC biomarkers. Recent surge in the discovery of salivary biomarkers for OSCC assessment collectively reinforced the clinical importance of this approach (Nagler, 2009). Independent studies have identified potential salivary biomarkers including proteins (Yu et al., 2016), metabolites (Song et al., 2020), circulating tumor DNA (ctDNA) (Cristaldi et al., 2019), extracellular vesicles (Gai et al., 2018), and microRNAs (miRNAs) (Menini et al., 2021), that may have diagnostic utility for OSCC. Intriguingly, qualitative, and quantitative changes in the oral microbiota have been correlated to OSCC and appears to hold potential for OSCC diagnosis (Yang et al., 2018; Zhang et al., 2019).
Salivary profilings to detect OSCC-specific RNAs, and oral microbial analyses related to OSCC have been performed previously (Yakob et al., 2014). However, these studies predominantly represent western world and other ethnic groups outside India. Notably, studies indicated that cancer biomarkers are heterogenous and show intra and inter-ethnic variations (Jing et al., 2014; Sokolenko and Imyanitov, 2018; Hirko et al., 2022). Moreover, recent salivary proteomic studies (Csosz et al., 2017; Singh et al., 2020) to identify OSCC markers, highlighted the importance of population-specific diagnostic markers. Nevertheless, a comprehensive investigation of OSCC-specific salivary RNAs appropriate to Indian ethnicity remains undetermined. In this study, we performed RNA-sequencing (RNA-seq) on saliva samples of OSCC patients and normal controls. We report for the first time, a combined OSCC-specific salivary RNAs and oral microbial dysbiosis which can be instrumental in devising non-invasive diagnostics for OSCC.
Unstimulated saliva was collected from clinically confirmed OSCC patients (n = 9) and normal control (NC) (n = 5) with no clinical evidence of OSCC. Saliva collection and RNA isolation was conducted as described previously (Pandit et al., 2013). Briefly, subjects were requested to refrain from drinking or eating for at least an hour before the collection of saliva. Approximately, 5 mL of saliva was collected from subjects into pre-chilled RNase-free conical tubes, and 5 μL of 20 U/μL RNase inhibitor (Ambion) was added immediately to prevent RNA degradation. Saliva was centrifuged at 2,600 g for 15 min at 4°C to remove cells and other debris, and the collected cell free saliva (CFS) was used for total RNA isolation by the TRIzol method (Ambion). Before saliva collection, informed consent was obtained from subjects, and all related protocols were approved by the Institute review board, Rajiv Gandhi Cancer Institute and Research Centre, New Delhi. The sample collection complied the Declaration of Helsinki.
RNA quantity and quality were analyzed on Bioanalyzer (Agilent 2,100). Small RNA-seq libraries were prepared using the Ion total RNA-seq v2.0 kit (Thermo Fisher Scientific) as per manufacturer’s protocol. To size-select the desired RNA products, Magnetic Bead Cleanup Module (Thermo Fisher Scientific) was used twice with the same sample. During the first bead binding, magnetic beads captured larger RNA species such as mRNA and lncRNA. During the second binding and with increased ethanol concentration, desired small RNA products (miRNA and other small RNA) in the supernatant re-bound to the magnetic beads. After washing the beads, the desired RNA products were eluted with pre-heated (80°C) Nuclease-free Water. Purified and size-selected RNA was converted into cDNA, and the barcodes and adaptors were ligated to the libraries. Library size distribution was verified by Bioanalyzer using a high-sensitivity DNA kit (Agilent) (Supplementary Figure S1). The libraries were diluted to 100 picomolar final concentration, and an equimolar pool was taken for clonal amplification. Template preparation for libraries on ionospheres was achieved using OneTouch 2 protocols (Thermo Fisher Scientific). The Ion 540 kit OT2 (Thermo Fisher Scientific) system in single-end mode with 200 bp chemistry was used for small RNA library sequencing.
Cutadapt (v1.8.1) algorithm was used to trim the 3′adaptor of the sequencing reads and assess the read quality. Sequencing reads were aligned to reference human genome (hg19) using Bowtie2 genome aligner (v2.2.5). Subsequently, mapped reads were screened for annotations using miRBase (v22) for miRNAs, and Gencode (v19) for other RNAs. Mapped sequencing reads were quantified using the Featurecount module (v1.4.6) of the R-package. For taxonomic classification of microbial reads, One Codex (Minot et al., 2015) algorithm was used. Total microbial diversity was calculated using the Shannon diversity index (read count ≥5, p-value ≤.05 was considered significant). The mean proportion of the sequencing reads aligned to bacterial taxa was calculated using Statistical analysis of taxonomic and functional profiles (STAMP) (v2.1.3) (Parks et al., 2014), p-value ≤.05. The differentially abundant taxa (p ≤ .05) were analyzed for associated metabolic pathways and gene ontologies (GO) using the Kyoto Encyclopedia of Genes and Genomes (KEGG) database (https://www.kegg.jp). The GOs associated with the metabolic functions were analyzed and visualized using Cytoscape (v3.8.2).
We used expression profiles of CFS RNAs from 5 NC and 9 OSCC samples. Since the gene expression matrix of the bodily fluids is often noisy, we selected only those genes that possess >5 read counts in at least 2 samples. Differential gene expression analysis was done using the DESeq2 web application (https://yanli.shinyapps.io/DEApp) Differentially expressed (DE) genes were further filtered based on their log2 fold change (FC) ≥1 or ≤ -1, p-value <.05 for the downstream analysis. The volcano plots were built on the DESeq output using the GraphPad Prism 8.2.1.
To predict the functions of DE mRNAs and miRNAs target genes, we performed pathway enrichment analysis (cancer only) using the PANTHER (http://www.pantherdb.org) with a false discovery rate (FDR) < .05. The predicted pathways and pathway-related genes were analyzed and visualized as a network using Cytoscape (Shannon et al., 2003) (v3.8.2). Target genes of the differentially abundant miRNAs were predicted using Target Scan (v7.2) with a cumulative weighted context ++ score cut-off of -.4. The gene set enrichment analysis (GSEA) was conducted using iDEP.951 web application (http://bioinformatics.sdstate.edu/idep/) using Molecular Signatures (MSig) and oncogenic signatures reference gene sets. The GSEA was visualized as a bubble plot using SRplots (www.bioinformatics.com.cn/plot_basic_gopathway_bubbleplot).
The expression profile of DE genes was analyzed using Head and Neck Squamous cell carcinoma (HNSCC) dataset available on The Cancer Genome Atlas (TCGA) portal. The protein expression data was retrieved from HNSCC dataset available on Proteomics Data Commons (PDC) portal. Survival analysis was performed using Kaplan-Meier method on Km plotter web tool (https://kmplot.com/analysis/). The correlation of drug response with respect to DE genes was determined by Receiver Operating Characterization (ROC) method (https://www.rocplot.org). The DE miRNAs-mediated drug resistance pathways were predicted and visualized using ROC plotter (https://www.rocplot.org/mirna/index).
The statistical significance of DE genes was calculated using nominal p-value in DESeq2 web application. The significance in the difference between Shannon diversities of NC and OSCC was calculated using Student’s t-test. The enrichment in microbial taxa and metabolic pathways was calculated using STAMP statistical tool.
To identify the salivary RNAs that segregate OSCC patients from normal, we performed small RNA-seq of the saliva sampled from OSCC patients and NC (the clinicopathological characteristics are described in Supplementary File S1). Approximately, 27 % of reads from the NC and 20 % from OSCC aligned to hg19 (Alignment statistics are detailed in Supplementary File S2). Among the mapped reads, RNA biotypes including mRNAs, miRNAs, long intergenic non-coding RNAs (lincRNAs), small nuclear RNAs (snRNAs), transfer RNAs (tRNAs), and pseudogenes were detected in both OSCC and NC (Figure 1A). DESeq analysis of the sequencing data yielded a total of 166 RNAs that were DE between OSCC and NC, of which 145 were upregulated and 21 were downregulated, with a log2 FC ≥ 1 or ≤ 1, p-value <.05 (Figure 1B; Supplementary File S3). Individual DE analysis of the RNA biotypes revealed 113 mRNAs (96 upregulated, and 17 downregulated) between OSCC and NC, (Figure 1C, top 30 DE mRNAs are shown in box). In addition, 7 miRNAs were found to be upregulated in OSCC compared to NC (Figure 1D) and 8 novel lincRNAs (5 upregulated, and 3 downregulated) (Figure 1E) are DE in OSCC. Notably, 35 novel tRNAs were found to be upregulated in OSCC (Figure 1F, top 10 tRNAs are shown in box). Other RNA species did not show any significant DE (Data not shown).
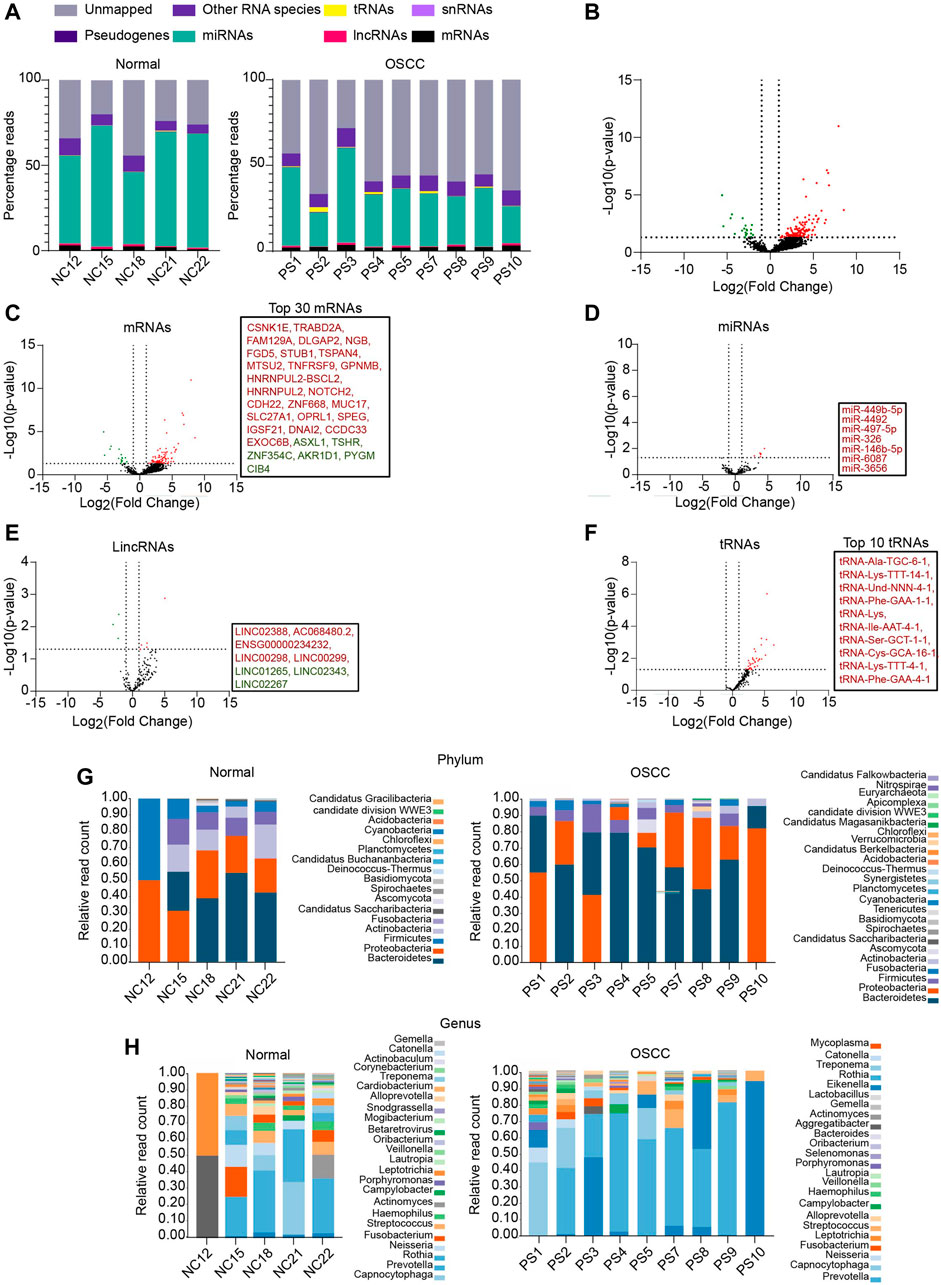
FIGURE 1. RNA profiling of salivary samples. (A) Read distribution of host salivary RNAs among normal and OSCC. (B) Volcano plots representing total altered transcripts in normal and OSCC (log2fold change ≥1 or ≤ -1, p ≤ .05). Volcano plots showing DE of (C) mRNAs, (D) miRNAs (E) lincRNAs, (F) tRNAs, respectively. Text boxes include names of top 30 mRNAs, all DE miRNAs, all DE lincRNAs, and top 10 tRNAs respectively. Relative read count of bacterial phyla (G) and genera (H) in normal and OSCC (cut off, ≥5 reads in at least 2 samples). NC = Normal control, PS = Patient sample.
Salivary transcriptomes are enriched with oral microbial RNAs (Ostheim et al., 2020). Therefore, to examine whether the sequencing reads unmapped to hg19 were of microbial origin, we aligned these sequencing reads to the microbial genomes on ONE CODEX microbial analysis platform. Approximately, 9.76% of reads from OSCC, and 3.29% of reads from NC were aligned to annotated microbial genomes. Metagenomic analysis revealed a diverse range of bacterial phyla (Figure 1G) and genera (Figure 1H) in OSCC and NC samples. A large proportion of reads remain unmapped, we speculate that these reads plausibly belong to the other uncharacterized metagenomes whose sequences are unavailable to date. Summarily, we report an array of salivary RNAs that significantly segregate OSCC from NC, and RNAs annotated to oral microbiome.
The functional associations of DE mRNAs were examined by GSEA using gene sets of canonical oncogenic pathways available on MSig database. DE mRNAs were significantly (p < .05, FDR ≤.01) associated with multiple oncogenic signature pathways including BMl1, AKT, MTOR, KRAS, AKT, IL2, PGF and VEGF and p53 (Figure 2A, Supplementary File S4). In addition, GO analysis showed a significant association (p < .05) of mRNAs such as PRKCA, NOTCH2, ZAK3, CDH22, ERBB4, CSNK1E, PCDH15, CDHR3, ROCK1, and ADCY2 with signaling pathways that are known to be dysregulated in OSCC (Figure 2B). Since PRKCA was found to be associated with multiple oncogenic pathways, we further analysed its diagnostic and prognostic relevance using OSCC TCGA data. Notably, expression analysis revealed a significant upregulation of PRKCA at mRNA (Figure 2C) and protein levels (Figure 2D) in OSCC tumors compared to adjacent normal control (NAT). Survival analysis indicated that OSCC patients with higher expression of PRKCA were relatively correlated with low survival (Figure 2E). PRKCA activates signaling pathways involved in chemotherapeutic drug resistance (He et al., 2022), this agrees with our GO analysis, therefore, we examined the plausible role of PRKCA in therapeutic resistance of OSCC. Importantly, patients with elevated expression of PRKCA showed resistance to immune checkpoint inhibitors including anti-PD1, anti-PDL1 and anti-CTLA-4 for OSCC therapy, Figure 2F shows a combined immune checkpoint therapy response with respect to PRKCA expression in OSCC patients. Taken together, salivary PRKCA expression could be a potential diagnostic, prognostic, and therapeutic marker of OSCC.
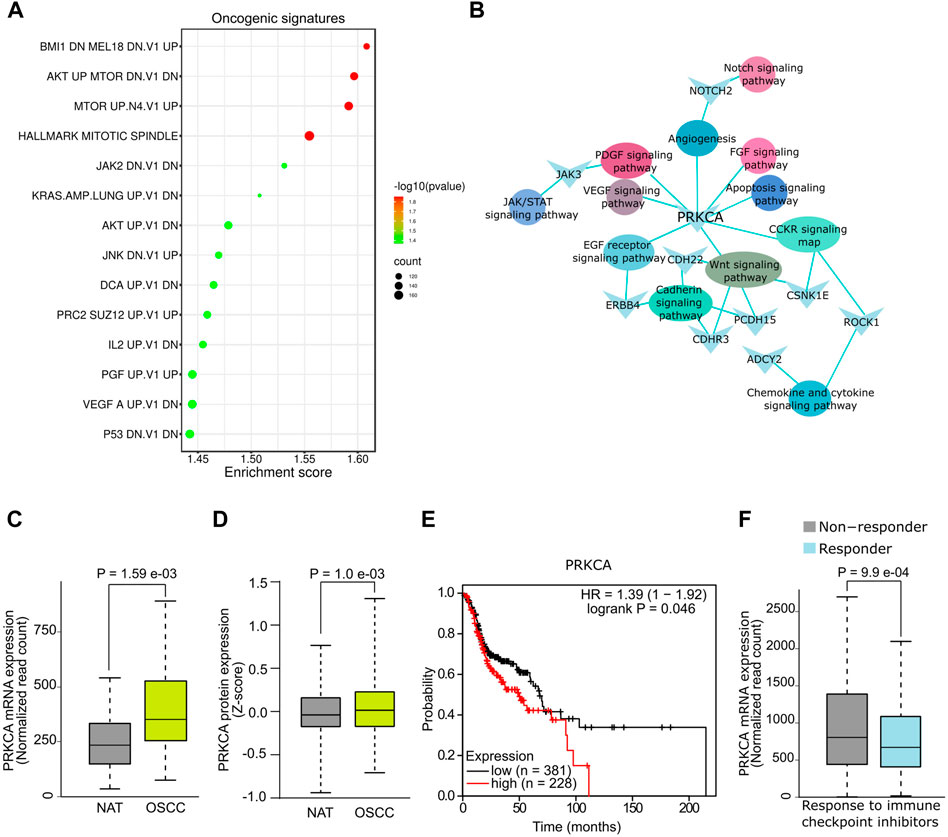
FIGURE 2. Functional analysis of salivary mRNAs. (A) GSEA showing association of DE mRNAs with oncogenic gene sets. (B) Functional networks showing DE hub mRNAs associated with multiple cancer pathways. DE of PRKCA at mRNA (C) and protein (D) levels in OSCC. (E) Survival plot showing reduced overall survival with high PRKCA expression. (F) Response of OSCC patients with high PRKCA expression towards immune checkpoint inhibitors.
The putative targets of DE-miRNAs were predicted using the Targetscan (Supplementary File S5) and subjected to functional enrichment analysis. The putative targets of miR-6087, miR-449b-5p, miR-3656, miR-326, miR-146b-5p and miR-497-5p were found to be significantly (p < .05) enriched in hallmark pathways of cancer (Figure 3A). Since the targets of DE miRNAs were associated with therapeutic resistance including p38 MAPK, EMT, RAS-PI3K, we investigated their putative role in chemo-resistance. Notably, patients with higher expression of miR-6087, miR-449-5p, miR-326 and miR-3656 showed no-response to platinum and/or taxane based drugs, compared to patients with lower expression of these miRNAs (Figures 3B–E). Conversely, the expression of miR-146b-5p and miR-497-5p was relatively lower in non-responders to platinum and taxane compounds (Figures 3F,G). Collectively, miR-6087, miR-449b-5p, miR-3656, miR-326, miR-146b-5p, and miR-497-5p can serve as diagnostic and drug resistance markers for OSCC.
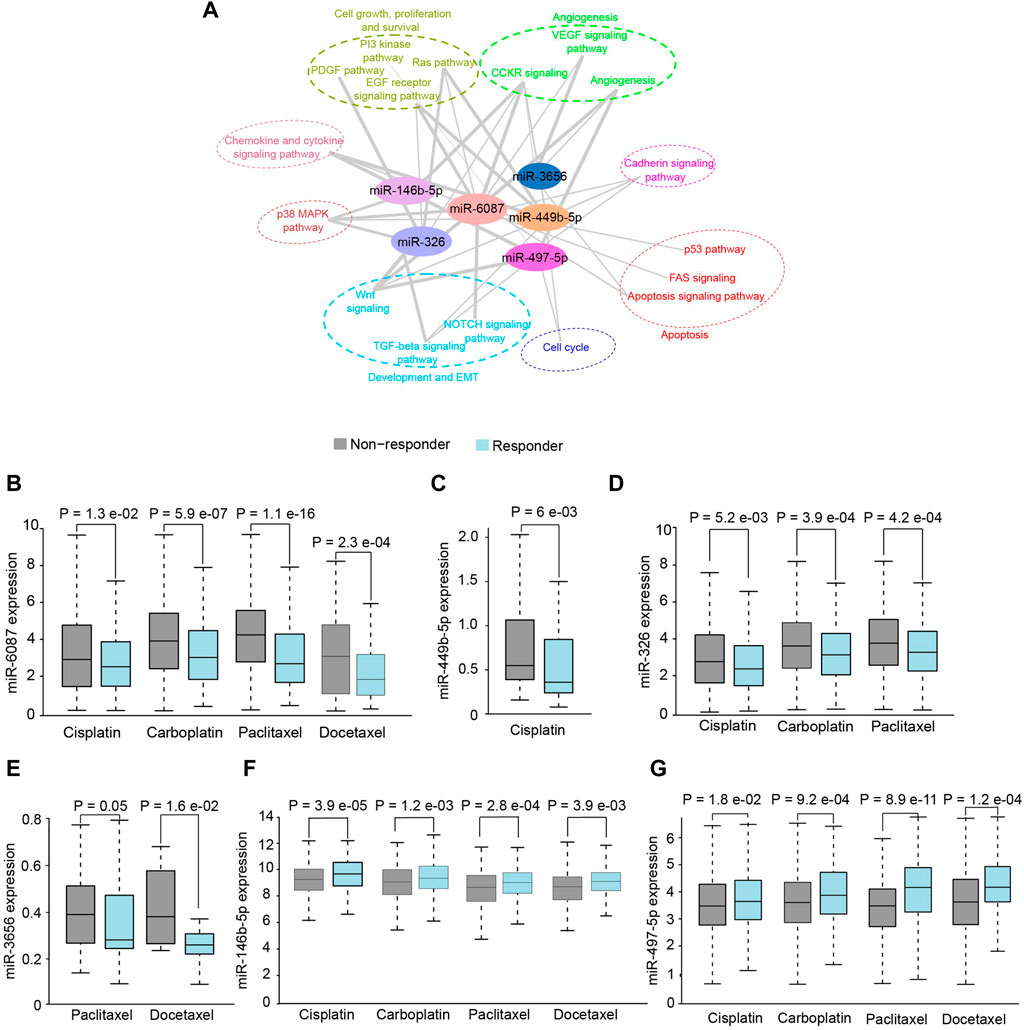
FIGURE 3. Functional analysis of salivary miRNAs. (A) Network analysis depicting association of differentially expressed miRNAs with cancer pathways. (B–G) Response of OSCC patients towards chemotherapeutics drugs with respect to higher expression of miR-6087, miR-449b-5p, miR-326, miR-3656, miR-146b-5p and miR-497-5p, respectively.
Recently, oral microbial dysbiosis is strongly associated with the incidence and progression of OSCC (Lee et al., 2017). Since a significant proportion of our sequencing reads mapped to microbial genomes, we measured the differences in microbial composition between OSCC and NC. Shannon diversity index showed a significantly higher microbial diversity in OSCC compared to NC (p < .05) (Figure 4A). Differential enrichment of microbial types by STAMP analysis revealed that phyla Firmicutes, and Actinobacteria were significantly enriched in NC (p < .05), and Bacteroidetes were significantly enriched in OSCC (p < .05) (Figure 4B). Among the 15 genera, Prevotella was significantly enriched in OSCC (p < .05), and Neisseria and Haemophilus were enriched in the NC group (Figure 4C). GO analysis of differentially abundant microbial taxa revealed associations with pathways including inflammation, ROS generation, cell proliferation, amino acid metabolism, nucleic acid metabolism, and carbohydrate metabolism suggesting a plausible role in carcinogenesis (Figure 4D). Summarily, our investigation not only identified prominent microbial dysbiosis between OSCC and NC, but also highlights the importance of analysing microbial sequences in salivary RNA-seq studies.
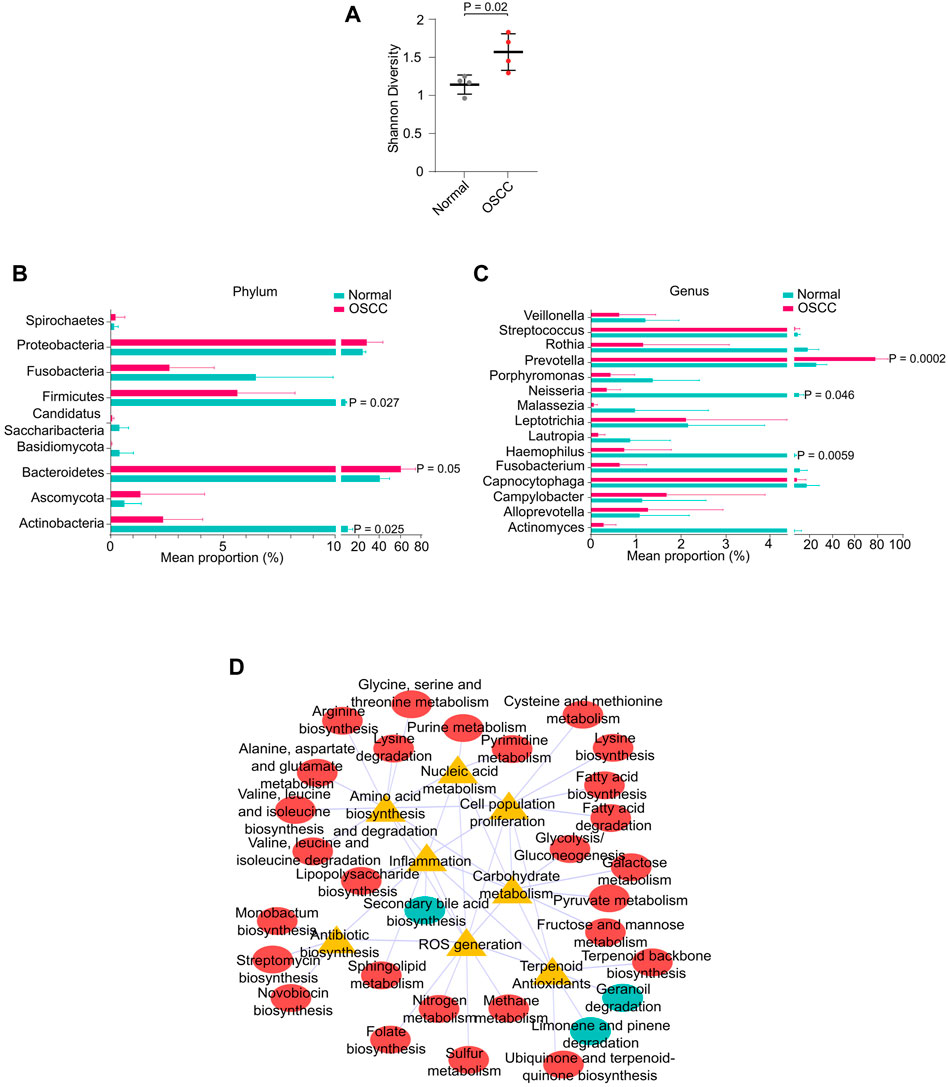
FIGURE 4. Oral microbial dysbiosis in OSCC. (A) Shannon diversity index showing microbial diversity between normal and OSCC. Mean proportion of microbial phyla (B) or genus (C) between normal and OSCC. (D) Interaction network between metabolic pathways and functional ontologies.
The concept of LB is an emerging non-invasive or minimally invasive diagnostic paradigm of precision oncology. The cancer-specific molecular signatures detectable in bodily fluids faithfully surrogate various hallmarks of cancer, including tumor-heterogeneity (Snow et al., 2019; Liebs et al., 2021). The utility of saliva for OSCC diagnosis is gaining importance. In this study, we have identified OSCC-specific salivary RNAs of human and oral microbiome from Indian subjects. Notably, the combined detection of OSCC-specific RNAs and oral microbial dysbiosis from individual saliva samples minimizes the ambiguity in understanding the role of oral dysbiosis in OSCC.
GSEA revealed significant associations of DE mRNAs with oncogenic gene set signatures including BMI1, AKT, MTOR, JAK2, KRAS, JNK, PGF, VEGF, and P53. Notably, dysregulated BMI1 pathway has previously been associated with poor survival of OSCC patients (Hu et al., 2018). In addition, upregulated AKT/mTOR pathway activity has been attributed to chemo/radio-resistance and poor prognosis in OSCC (Harsha et al., 2020). Interestingly, the genes associated with the mutant p53 pathway were found to be enriched in our OSCC dataset. The upregulation of mutant p53 has previously been associated with lymph node metastasis in OSCC patients (Li et al., 2004). In addition, JAK pathway promotes the migration potential of OSCC cells (Chuang et al., 2014). Notably, among the upregulated mRNAs, Casein kinase 1 isoform epsilon (CSNK1E), Tetraspanin-4 (TSPAN4), NOTCH2, NOTCH2NL, and Protein kinase C Alpha (PRKCA) have previously been shown to have role in OSCC pathogenesis (Hirano et al., 2009; Lin et al., 2014; Porcheri et al., 2019; Xie et al., 2020). In addition, the higher expression of TSPAN4 and CSNK1E in OSCC tumors has been shown to have diagnostic and prognostic potential (Hirano et al., 2009; Lin et al., 2014). Further, Notch signaling components has shown to be upregulated and oncogenic in OSCC (Porcheri et al., 2019). In line with these studies, it is highly plausible that the enrichment of these mRNAs in our study mirrors their expression in OSCC tumor.
Functional analysis of DE mRNAs revealed an association with oncogenic pathways of OSCC including, Wnt, cadherin, and CCKR. Upregulation of Wnt pathway components expression, and hyperactive Wnt signaling have been linked to an aggressive phenotype, poor therapeutic response, and metastasis of OSCC (Xie et al., 2020). Moreover, the crosstalk between EGFR and Wnt pathways appears to have diagnostic and prognostic value for OSCC (Lee et al., 2010). Therefore, the salivary abundance of mRNAs associated with these pathways can be potentially exploited for diagnosis and prognosis of OSCC. Notably, the salivary upregulation of PRKCA mimics its expression in OSCC tumors and correlated with low survival and therapy resistance. The upregulation of PRKCA has shown to have implications in OSCC (Ozaki-Honda et al., 2017), and attributed to chemoresistance by the phosphorylation of BCL2 in leukemia (Jiffar et al., 2004). Our analysis revealed that the increased expression of PRKCA was associated with resistance to immune checkpoint inhibitors, which are currently being used as advanced therapeutics in OSCC. Immunotherapy has been reported to improve the overall survival in OSCC; however, the resistance to immunotherapy remains a major challenge (Dos Santos et al., 2021). Taken together, PRKCA, can serve as a potential salivary indicator for survival and immunotherapy efficacy of OSCC.
The DE of miRNAs in bodily fluids has shown to have immense potential as biomarkers for cancer diagnosis, stratification, and drug efficacy (Hayes et al., 2014). The predicted targets of miRNAs in our study were enriched in OSCC-relevant Wnt signaling, cadherin signaling chemokine, and cytokine signaling pathways. Among the significantly DE-miRNAs, miR-3656 and miR-4492 (Schneider et al., 2018), miR-497 (Hu et al., 2016), and miR-146b-5p (Scapoli et al., 2010) have previously been shown to be overexpressed in OSCC tumors and implicated as putative diagnostic markers. Therefore, it is highly plausible that the DE-miRNAs were sourced from primary tumor, and may hold diagnostic potential. The role of novel miRNAs, miR-6087, miR-449b-5p, and miR-326 reported here, in OSCC pathogenesis remains elusive. The aberrant expression of miRNAs has previously been associated with multi-drug resistance in OSCC (Li et al., 2019; Cheng et al., 2021). Notably, DE of the identified miRNAs significantly correlated with chemotherapeutic drug response. The drug resistance mechanism of miRNAs operates through targeting of drug-resistance genes (Si et al., 2019). Downregulation of TP73, putative target of miR-6087, has been reported to induce chemoresistance in multiple cancer types, independent of p53 (Irwin et al., 2003). Moreover, downregulation of pro-apoptotic gene BAX, putative target of miR-6087, has been reported to induce chemoresistance in a p53 dependent manner (McCurrach et al., 1997). Further, MDM4, a putative target of miR-6087 is reported to increase chemoresistance in breast cancer (Lam et al., 2010). The higher expression of miR-6087 in OSCC, can therefore, contribute to chemoresistance in both p53 dependent and independent manner. Our analysis revealed that increased expression of miR-146b-5p and miR-497-5p increases chemosensitivity in OSCC. In line with this, the putative targets of miR-146b-5p including TRAF6 and NOVA1 have been shown to increase chemoresistance in OSCC and hepatocellular carcinoma, respectively (Wu et al., 2018; Cao et al., 2021). Additionally, the putative targets of miR-497-5p including PSMD7, CCND3, SMURF2, and WNT7A increase chemoresistance in gastric cancer, acute myeloid leukemia, lung cancer and OSCC, respectively (Tian et al., 2018; Ray et al., 2020; Smith et al., 2021; Wang et al., 2021). Owing to these observations, it is highly plausible that miR-146b-5p and miR-497-5p may impart chemosensitivity in OSCC by targeting these mRNAs. Collectively, the identified miRNAs can further be explored for their potential as diagnostic markers and therapeutic targets in larger cohorts of OSCC.
The OSCC-specific dysbiosis of phyla Firmicutes, Bacteroidetes, Actinobacteria, and genus Prevotella identified in this study supports the previous studies (Lee et al., 2017; Zhang et al., 2019). Here we report a novel salivary dysbiosis of genus Neisseria and Haemophilus. The functional relevance of oral microbes in OSCC operates through various mechanisms, including the release of genotoxic substances, carcinogens, or onco-metabolites, inflammation, and ROS-mediated toxicity (Sami et al., 2020). Accordingly, the association of microbial types enriched in OSCC with GOs such as inflammation, reactive oxygen species generation, cell proliferation, and nucleic acid, amino acid metabolism strongly suggest their potential role in the carcinogenesis of OSCC.
In summary, the OSCC-specific RNA signatures and oral microbial dysbiosis identified in this study can have diagnostic utilities tailored for Indian population. However, further screening in large cohorts is necessary to evaluate the true diagnostic potential of the identified RNAs. Nevertheless, this study strongly advocates the LB coupled RNA-seq approach for developing RNA-based non-invasive diagnostics for OSCC and contributes to the emerging Salivaomics.
The RNA-seq data presented in this study is available at GEO accession ID—GSE176077, and the analysis files are available in the Supplementary Material.
The studies involving human participants were reviewed and approved by Institute review board, Rajiv Gandhi cancer institute and research center—New Delhi (RGCIRC/IRB/290/2019). The patients/participants provided their written informed consent to participate in this study.
SS collected samples, performed RNA work, prepared libraries, analysed data and contributed in manuscript drafting. SSS performed metagenomic analysis and contributed in manuscript drafting. CK and GD contributed to sample collection and RNA quality checks, AK and MS contributed to data analysis, VG performed differential gene expression analysis under GA supervision, JT and AM identified patients and facilitated sample collection, SN conceived and designed the study, acquired funding, contributed to data analysis, and drafted the manuscript.
The study was funded by the Indian Institute of Technology Ropar.
We are grateful to Biorepository at Rajiv Gandhi Cancer Institute and Research Centre, New Delhi, India, for facilitating saliva collection. We also profusely thank Dr. Sudhakar Sahoo, Cancer Research United Kingdom Manchester, for the critical comments on the manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmolb.2022.1106963/full#supplementary-material
Alix-Panabieres, C., and Pantel, K. (2021). Liquid biopsy: From discovery to clinical application. Cancer Discov. 11, 858–873. doi:10.1158/2159-8290.CD-20-1311
Borse, V., Konwar, A. N., and Buragohain, P. (2020). Oral cancer diagnosis and perspectives in India. Sens. Int. 1, 100046. doi:10.1016/j.sintl.2020.100046
Cao, Y., Zhang, F., Wang, H., Bi, C., Cui, J., Liu, F., et al. (2021). LncRNA MALAT1 mediates doxorubicin resistance of hepatocellular carcinoma by regulating miR-3129-5p/Nova1 axis. Mol. Cell. Biochem. 476, 279–292. doi:10.1007/s11010-020-03904-6
Cheng, Y., Li, S., Gao, L., Zhi, K., and Ren, W. (2021). The molecular basis and therapeutic aspects of cisplatin resistance in oral squamous cell carcinoma. Front. Oncol. 11, 761379. doi:10.3389/fonc.2021.761379
Chuang, J. Y., Huang, Y. L., Yen, W. L., Chiang, I. P., Tsai, M. H., and Tang, C. H. (2014). Syk/JNK/AP-1 signaling pathway mediates interleukin-6-promoted cell migration in oral squamous cell carcinoma. Int. J. Mol. Sci. 15, 545–559. doi:10.3390/ijms15010545
Cristaldi, M., Mauceri, R., Di Fede, O., Giuliana, G., Campisi, G., and Panzarella, V. (2019). Salivary biomarkers for oral squamous cell carcinoma diagnosis and follow-up: Current status and perspectives. Front. Physiol. 10, 1476. doi:10.3389/fphys.2019.01476
Csosz, E., Labiscsak, P., Kallo, G., Markus, B., Emri, M., Szabo, A., et al. (2017). Proteomics investigation of OSCC-specific salivary biomarkers in a Hungarian population highlights the importance of identification of population-tailored biomarkers. PLoS One 12, e0177282. doi:10.1371/journal.pone.0177282
D'cruz, A. K., Vaish, R., and Dhar, H. (2018). Oral cancers: Current status. Oral Oncol. 87, 64–69. doi:10.1016/j.oraloncology.2018.10.013
De Rubis, G., Rajeev Krishnan, S., and Bebawy, M. (2019). Liquid biopsies in cancer diagnosis, monitoring, and prognosis. Trends Pharmacol. Sci. 40, 172–186. doi:10.1016/j.tips.2019.01.006
Dos Santos, L. V., Abrahao, C. M., and William, W. N. (2021). Overcoming resistance to immune checkpoint inhibitors in Head and Neck squamous cell carcinomas. Front. Oncol. 11, 596290. doi:10.3389/fonc.2021.596290
Gai, C., Camussi, F., Broccoletti, R., Gambino, A., Cabras, M., Molinaro, L., et al. (2018). Salivary extracellular vesicle-associated miRNAs as potential biomarkers in oral squamous cell carcinoma. BMC Cancer 18, 439. doi:10.1186/s12885-018-4364-z
Harsha, C., Banik, K., Ang, H. L., Girisa, S., Vikkurthi, R., Parama, D., et al. (2020). Targeting AKT/mTOR in oral cancer: Mechanisms and advances in clinical trials. Int. J. Mol. Sci. 21, 3285. doi:10.3390/ijms21093285
Hayes, J., Peruzzi, P. P., and Lawler, S. (2014). MicroRNAs in cancer: Biomarkers, functions and therapy. Trends Mol. Med. 20, 460–469. doi:10.1016/j.molmed.2014.06.005
He, S., Li, Q., Huang, Q., and Cheng, J. (2022). Targeting protein kinase C for cancer therapy. Cancers (Basel) 14, 1104. doi:10.3390/cancers14051104
Hirano, C., Nagata, M., Noman, A. A., Kitamura, N., Ohnishi, M., Ohyama, T., et al. (2009). Tetraspanin gene expression levels as potential biomarkers for malignancy of gingival squamous cell carcinoma. Int. J. Cancer 124, 2911–2916. doi:10.1002/ijc.24297
Hirko, K. A., Rocque, G., Reasor, E., Taye, A., Daly, A., Cutress, R. I., et al. (2022). The impact of race and ethnicity in breast cancer-disparities and implications for precision oncology. BMC Med. 20, 72. doi:10.1186/s12916-022-02260-0
Hu, J., Xu, J. F., and Ge, W. L. (2016). MiR-497 enhances metastasis of oral squamous cell carcinoma through SMAD7 suppression. Am. J. Transl. Res. 8, 3023–3031.
Hu, Q., Wu, T., Chen, X., Li, H., Du, Z., Hao, Y., et al. (2018). The poor outcome of second primary oral squamous cell carcinoma is attributed to Bmi1 upregulation. Cancer Med. 7, 1056–1069. doi:10.1002/cam4.1348
Irwin, M. S., Kondo, K., Marin, M. C., Cheng, L. S., Hahn, W. C., and Kaelin, W. G. (2003). Chemosensitivity linked to p73 function. Cancer Cell. 3, 403–410. doi:10.1016/s1535-6108(03)00078-3
Jiffar, T., Kurinna, S., Suck, G., Carlson-Bremer, D., Ricciardi, M. R., Konopleva, M., et al. (2004). PKC alpha mediates chemoresistance in acute lymphoblastic leukemia through effects on Bcl2 phosphorylation. Leukemia 18, 505–512. doi:10.1038/sj.leu.2403275
Jing, L., Su, L., and Ring, B. Z. (2014). Ethnic background and genetic variation in the evaluation of cancer risk: A systematic review. PLoS One 9, e97522. doi:10.1371/journal.pone.0097522
Lam, S., Lodder, K., Teunisse, A. F., Rabelink, M. J., Schutte, M., and Jochemsen, A. G. (2010). Role of Mdm4 in drug sensitivity of breast cancer cells. Oncogene 29, 2415–2426. doi:10.1038/onc.2009.522
Larson, M. H., Pan, W., Kim, H. J., Mauntz, R. E., Stuart, S. M., Pimentel, M., et al. (2021). Author correction: A comprehensive characterization of the cell-free transcriptome reveals tissue- and subtype-specific biomarkers for cancer detection. Nat. Commun. 12, 2553. doi:10.1038/s41467-022-30329-0
Lee, C. H., Hung, H. W., Hung, P. H., and Shieh, Y. S. (2010). Epidermal growth factor receptor regulates beta-catenin location, stability, and transcriptional activity in oral cancer. Mol. Cancer 9, 64. doi:10.1186/1476-4598-9-64
Lee, W. H., Chen, H. M., Yang, S. F., Liang, C., Peng, C. Y., Lin, F. M., et al. (2017). Bacterial alterations in salivary microbiota and their association in oral cancer. Sci. Rep. 7, 16540. doi:10.1038/s41598-017-16418-x
Li, Y., St John, M. A., Zhou, X., Kim, Y., Sinha, U., Jordan, R. C., et al. (2004). Salivary transcriptome diagnostics for oral cancer detection. Clin. Cancer Res. 10, 8442–8450. doi:10.1158/1078-0432.CCR-04-1167
Li, Y., Wang, M., Zhu, X., Cao, X., Wu, Y., and Fang, F. (2019). Prognostic significance of CIP2A in esophagogastric junction adenocarcinoma: A study of 65 patients and a meta-analysis. Dis. Markers 2019, 2312439. doi:10.1155/2019/2312439
Liebs, S., Eder, T., Klauschen, F., Schütte, M., Yaspo, M. L., Keilholz, U., et al. (2021). Applicability of liquid biopsies to represent the mutational profile of tumor tissue from different cancer entities. Oncogene 40, 5204–5212. doi:10.1038/s41388-021-01928-w
Lin, S. H., Lin, Y. M., Yeh, C. M., Chen, C. J., Chen, M. W., Hung, H. F., et al. (2014). Casein kinase 1 epsilon expression predicts poorer prognosis in low T-stage oral cancer patients. Int. J. Mol. Sci. 15, 2876–2891. doi:10.3390/ijms15022876
Mccurrach, M. E., Connor, T. M., Knudson, C. M., Korsmeyer, S. J., and Lowe, S. W. (1997). bax-deficiency promotes drug resistance and oncogenic transformation by attenuating p53-dependent apoptosis. Proc. Natl. Acad. Sci. U. S. A. 94, 2345–2349. doi:10.1073/pnas.94.6.2345
Menini, M., De Giovanni, E., Bagnasco, F., Delucchi, F., Pera, F., Baldi, D., et al. (2021). Salivary micro-RNA and oral squamous cell carcinoma: A systematic review. J. Pers. Med. 11, 101. doi:10.3390/jpm11020101
Minot, S. S., Samuel, N. K., and Greenfield, N. B. (2015). One Codex: A sensitive and accurate data platform for genomic microbial identification. bioRxiv.
Nagler, R. M. (2009). Saliva as a tool for oral cancer diagnosis and prognosis. Oral Oncol. 45, 1006–1010. doi:10.1016/j.oraloncology.2009.07.005
Ostheim, P., Tichy, A., Sirak, I., Davidkova, M., Stastna, M. M., Kultova, G., et al. (2020). Overcoming challenges in human saliva gene expression measurements. Sci. Rep. 10, 11147. doi:10.1038/s41598-020-67825-6
Ozaki-Honda, Y., Seki, S., Fujiwara, M., Matsuura, M., Fujita, S., Ikeda, H., et al. (2017). Prognostic prediction of oral squamous cell carcinoma by E-cadherin and N-cadherin expression in overall cells in tumor nests or tumor cells at the invasive front. Cancer Microenviron. 10, 87–94. doi:10.1007/s12307-017-0201-1
Pandit, P., Cooper-White, J., and Punyadeera, C. (2013). High-yield RNA-extraction method for saliva. Clin. Chem. 59, 1118–1122. doi:10.1373/clinchem.2012.197863
Parks, D. H., Tyson, G. W., Hugenholtz, P., and Beiko, R. G. (2014). Stamp: Statistical analysis of taxonomic and functional profiles. Bioinformatics 30, 3123–3124. doi:10.1093/bioinformatics/btu494
Porcheri, C., Meisel, C. T., and Mitsiadis, T. (2019). Multifactorial contribution of Notch signaling in Head and Neck squamous cell carcinoma. Int. J. Mol. Sci. 20, 1520. doi:10.3390/ijms20061520
Ray, P., Raghunathan, K., Ahsan, A., Allam, U. S., Shukla, S., Basrur, V., et al. (2020). Ubiquitin ligase SMURF2 enhances epidermal growth factor receptor stability and tyrosine-kinase inhibitor resistance. J. Biol. Chem. 295, 12661–12673. doi:10.1074/jbc.RA120.013519
Sami, A., Elimairi, I., Stanton, C., Ross, R. P., and Ryan, C. A. (2020). The role of the microbiome in oral squamous cell carcinoma with insight into the microbiome-treatment Axis. Int. J. Mol. Sci. 21, 8061. doi:10.3390/ijms21218061
Scapoli, L., Palmieri, A., Lo Muzio, L., Pezzetti, F., Rubini, C., Girardi, A., et al. (2010). MicroRNA expression profiling of oral carcinoma identifies new markers of tumor progression. Int. J. Immunopathol. Pharmacol. 23, 1229–1234. doi:10.1177/039463201002300427
Schneider, A., Victoria, B., Lopez, Y. N., Suchorska, W., Barczak, W., Sobecka, A., et al. (2018). Tissue and serum microRNA profile of oral squamous cell carcinoma patients. Sci. Rep. 8, 675. doi:10.1038/s41598-017-18945-z
Shannon, P., Markiel, A., Ozier, O., Baliga, N. S., Wang, J. T., Ramage, D., et al. (2003). Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504. doi:10.1101/gr.1239303
Sharma, S., Satyanarayana, L., Asthana, S., Shivalingesh, K. K., Goutham, B. S., and Ramachandra, S. (2018). Oral cancer statistics in India on the basis of first report of 29 population-based cancer registries. J. Oral Maxillofac. Pathol. 22, 18–26. doi:10.4103/jomfp.JOMFP_113_17
Si, W., Shen, J., Zheng, H., and Fan, W. (2019). The role and mechanisms of action of microRNAs in cancer drug resistance. Clin. Epigenetics 11, 25. doi:10.1186/s13148-018-0587-8
Singh, P., Verma, J. K., and Singh, J. K. (2020). Validation of salivary markers, IL-1β, IL-8 and Lgals3bp for detection of oral squamous cell carcinoma in an Indian population. Sci. Rep. 10, 7365. doi:10.1038/s41598-020-64494-3
Smith, C. C., Viny, A. D., Massi, E., Kandoth, C., Socci, N. D., Rapaport, F., et al. (2021). Recurrent mutations in cyclin D3 confer clinical resistance to FLT3 inhibitors in acute myeloid leukemia. Clin. Cancer Res. 27, 4003–4011. doi:10.1158/1078-0432.CCR-20-3458
Snow, A., Chen, D., and Lang, J. E. (2019). The current status of the clinical utility of liquid biopsies in cancer. Expert Rev. Mol. Diagn 19, 1031–1041. doi:10.1080/14737159.2019.1664290
Sokolenko, A. P., and Imyanitov, E. N. (2018). Molecular diagnostics in clinical oncology. Front. Mol. Biosci. 5, 76. doi:10.3389/fmolb.2018.00076
Song, X., Yang, X., Narayanan, R., Shankar, V., Ethiraj, S., Wang, X., et al. (2020). Oral squamous cell carcinoma diagnosed from saliva metabolic profiling. Proc. Natl. Acad. Sci. U. S. A. 117, 16167–16173. doi:10.1073/pnas.2001395117
Su, Y. F., Chen, Y. J., Tsai, F. T., Li, W. C., Hsu, M. L., Wang, D. H., et al. (2021). Current insights into oral cancer diagnostics. Diagn. (Basel) 11, 1287. doi:10.3390/diagnostics11071287
Tian, J., Cui, X., Feng, Y., and Gu, L. (2018). Inhibition of WNT7A-beta-catenin signaling pathway sensitizes oral squamous cell carcinoma to cisplatin. Int. J. Clin. Exp. Pathol. 11, 4926–4933.
Veluthattil, A. C., Sudha, S. P., Kandasamy, S., and Chakkalakkoombil, S. V. (2019). Effect of hypofractionated, palliative radiotherapy on quality of life in late-stage oral cavity cancer: A prospective clinical trial. Indian J. Palliat. Care 25, 383–390. doi:10.4103/IJPC.IJPC_115_18
Wang, J., Liu, R., Mo, H., Xiao, X., Xu, Q., and Zhao, W. (2021). Deubiquitinase PSMD7 promotes the proliferation, invasion, and cisplatin resistance of gastric cancer cells by stabilizing RAD23B. Int. J. Biol. Sci. 17, 3331–3342. doi:10.7150/ijbs.61128
Wu, Y. H., Wu, W. S., Lin, L. C., Liu, C. S., Ho, S. Y., Wang, B. J., et al. (2018). Bortezomib enhances radiosensitivity in oral cancer through inducing autophagy-mediated TRAF6 oncoprotein degradation. J. Exp. Clin. Cancer Res. 37, 91. doi:10.1186/s13046-018-0760-0
Xie, J., Huang, L., Lu, Y. G., and Zheng, D. L. (2020). Roles of the Wnt signaling pathway in Head and Neck squamous cell carcinoma. Front. Mol. Biosci. 7, 590912. doi:10.3389/fmolb.2020.590912
Yakob, M., Fuentes, L., Wang, M. B., Abemayor, E., and Wong, D. T. (2014). Salivary biomarkers for detection of oral squamous cell carcinoma - current state and recent advances. Curr. Oral Health Rep. 1, 133–141. doi:10.1007/s40496-014-0014-y
Yang, C. Y., Yeh, Y. M., Yu, H. Y., Chin, C. Y., Hsu, C. W., Liu, H., et al. (2018). Oral microbiota community dynamics associated with oral squamous cell carcinoma staging. Front. Microbiol. 9, 862. doi:10.3389/fmicb.2018.00862
Yu, J. S., Chen, Y. T., Chiang, W. F., Hsiao, Y. C., Chu, L. J., See, L. C., et al. (2016). Saliva protein biomarkers to detect oral squamous cell carcinoma in a high-risk population in Taiwan. Proc. Natl. Acad. Sci. U. S. A. 113, 11549–11554. doi:10.1073/pnas.1612368113
Zhang, L., Liu, Y., Zheng, H. J., and Zhang, C. P. (2019a). The oral microbiota may have influence on oral cancer. Front. Cell. Infect. Microbiol. 9, 476. doi:10.3389/fcimb.2019.00476
Keywords: non-invasive diagnostics, salivary RNAs, oral microbiota, oral squamous cell carcinoma (OSCC), liquid biospsy
Citation: Saproo S, Sarkar SS, Gautam V, Konyak CW, Dass G, Karmakar A, Sharma M, Ahuja G, Gupta A, Tayal J, Mehta A and Naidu S (2023) Salivary protein kinase C alpha and novel microRNAs as diagnostic and therapeutic resistance markers for oral squamous cell carcinoma in Indian cohorts. Front. Mol. Biosci. 9:1106963. doi: 10.3389/fmolb.2022.1106963
Received: 24 November 2022; Accepted: 28 December 2022;
Published: 10 January 2023.
Edited by:
Georgina Gonzalez-Avila, Instituto Nacional de Enfermedades Respiratorias-México (INER), MexicoReviewed by:
Xiaozhi Lv, Southern medical University, ChinaCopyright © 2023 Saproo, Sarkar, Gautam, Konyak, Dass, Karmakar, Sharma, Ahuja, Gupta, Tayal, Mehta and Naidu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Srivatsava Naidu, c3JpdmF0c2F2YS5uYWlkdUBpaXRycHIuYWMuaW4=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.