- 1Research Center of Molecular Diagnostics and Sequencing, Axbio Biotechnology (Shenzhen) Co., Ltd., Shenzhen, China
- 2Research Center of Molecular Diagnostics and Sequencing, Research Institute of Tsinghua University in Shenzhen, Shenzhen, China
- 3Department of Biochemistry and Molecular Biology, The Key Laboratory of Environment and Genes Related to Disease of Ministry of Education, Health Science Center, Xi’an Jiaotong University, Xi’an, China
- 4Graduate School of Heilongjiang University of Chinese Medicine, Harbin, China
- 5Department of Gastroenterology and Hepatology, Mayo Clinic, Rochester, MN, United States
Background and Objective: One of the most recent forms of programmed cell death, ferroptosis, is crucial in tumorigenesis. Ferroptosis is characterized by iron-dependent oxidative destruction of cellular membranes following the antioxidant system’s failure. However, it is unknown whether ferroptosis-related genes (FRGs) are associated with colon adenocarcinoma (COAD) metastasis, immune cell infiltration, and oxidative stress in COAD. The current study concentrated on FRGs expression in colon cancer metastasis, their relationship to immune cell infiltration (ICI), and potential pathological pathways in COAD.
Methods and Results: Clinical information and mRNA expression patterns for patients with COAD metastasis were obtained from the public TCGA database. Patients with low mRNA levels showed good overall survival than patients with high mRNA levels. The genomic-clinicopathologic nomogram was subsequently created by combining risk score and clinicopathological features. Absolute Shrinkage and Selection Operator have shown a 4 gene signature that can stratify cancer patients into high-risk versus low-risk. These four FRGs were found to be significantly linked to the overall survival of COAD patients and predicted high risk score. Next, age, stage, and PTNM were combined in univariate and multivariate cox regression models to perform a filtering procedure. The receiver operating characteristic (ROC) and calibration curves indicated that constructed signature model exhibited high prediction accuracy and clinical relevance in COAD. ARID3A showed a strong negative correlation with a wide range of immune tumour-infiltrating cells in COAD microenvironment. According to the single sample gene set enrichment analysis (ssGSEA) results, FRGs are involved in variety of pathological pathways including PI3K-AKT-mTOR pathway, reactive oxygen species (ROS) pathway, response to hypoxia pathway, and other inflammation related pathways. Moreover, dysregulation of FRGs in COAD patients showed a significance correlation with wide range of miRNAs and transcription factors (TFs).
Conclusion: We identified new diagnostic biomarkers and established prognostic models for ferroptosis related programmed cell death in COAD metastasis. FRGs may improve tumor cell survival by activating the TGFB pathway, which can stimulate ROS production, accelerates ECM breakdown, and promote tumor progression and invasion. Genes implicated in ferroptosis, as revealed by the Kaplan Meier and a genomic-clinicopathologic nomogram, are potential therapeutic targets and prognosis indications for metastasis COAD patients.
1 Introduction
1.1 Colon cancer
Colon cancer is the third most frequently diagnosed cancer in males and females worldwide, with 80, 690 (8%), 70,340 (8%) new cases and 28,400 (9%), 24,180 (8%) deaths in male and female, respectively (Siegel et al., 2022). The high incidence rate or mortality is because of the lack of early detection and the fact that it is often diagnosed in its later stages (Zhou et al., 2019). The 5-year overall survival (OS) rate for COAD patients is still low, despite the availability of various targeted medicines and immunotherapies in recent years. Therefore, studying the molecular mechanism of the occurrence and development of colorectal cancer is an important subject of clinical research. Preventive measures, such as screening and finding new therapeutic targets, are also critical to improve patients’ survival and prognosis of colon cancer.
1.2 Iron death
Various human diseases can be prevented by targeting regulated cell death, which includes necroptosis, pyroptosis, ferroptosis, entotic cell death, lysosome-dependent cell death, and autophagy-dependent cell death. Ferroptosis is iron-dependent programmed cell death, first postulated by Dixon in 2012; it differs from apoptosis, pyrolysis, and autophagy at levels of cell morphology, biochemical features, and regulation and occurs through Fe(II)-dependent lipid peroxidation to insufficient cellular reducing capacity (Dixon et al., 2012; Jiang et al., 2020). Several studies have connected ferroptosis to cancer development and progression (Xia et al., 2019; Jiang et al., 2020). Since tumor cells can maintain or acquire ferroptosis sensitivity while surviving cell death, ferroptosis therapy for cancer is gaining attention. Wei et al. found that small molecule drugs that activated p53 had potent inhibitory action against HCT116 cells by inducing ferroptosis (Wei et al., 2018). Combinatorial therapy with ferroptosis medicine and tumor necrosis factor-related apoptosis-inducing ligands led to synergistic apoptosis and growth regression of CRC (Lee et al., 2019). Thereby, ferroptosis-related genes (FRGs) are very significant in cancer patients (Jiang et al., 2015; Ou et al., 2016; Bersuker et al., 2019; Doll et al., 2019; Li et al., 2020) and could be promising therapeutic targets and prognostic indicators in colon adenocarcinoma (COAD).
1.3 Working hypothesis
Our working hypothesis was that FRGs promotes colon cancer metastasis and play a role in oxidative stress.
1.4 Study design
Based on above mentioned hypothesis we developed prognostic model, validated, and explored the mechanism by which FRGs promotes COAD progression and invasion. Our results showed that there are solid predictive genes and offered a novel, personalized approach to treating COAD.
2 Materials and methods
2.1 Data acquisition and identification of differentially expressed genes (DEGs)
Ferroptosis-related genes were derived from Ze-Xian Liu et al. RNA-sequencing expression (level 3) profiles and corresponding clinical information for COAD were downloaded from the TCGA dataset (https://portal.gdc.com). R package, version 4.0.3 was used to implement the analysis. Genes with a p-value of less than 0.05 and log FC > 1 were chosen for further investigation. Only patients who had M1 metastases and higher were considered for participation in the trial according to the inclusion criteria for patient selection.
2.2 Nomogram construction
The Cox regression analysis was conducted to determine if risk scores and relevant clinical indicators could be identified as prospective predictors of OS for COAD patients. Based on the Cox regression analysis results, a prognostic nomogram was built using the stepwise Cox regression model to predict the 1, 3, and 5-year OS of COAD patients included in the TCGA dataset. This was done to determine the probability of survival. The area under the curve was used to evaluate the nomogram’s ability to discriminate between categories (AUC). Using the calibration curve, a graph comparing the expected OS of the nomogram to the observed survival rates was constructed.
2.3 Correlation between genes and pathways in COAD
The genes found in the associated pathways were gathered and examined using the R software’s GSVA package, with the parameter method set to the ssGSEA algorithm’ as the final step. Finally, Spearman correlation was used to examine the correlation between the genes and the pathway score. The analysis methods and R packages were implemented by R version 4.0.3. p-value < 0.05 was considered statistically significant.
2.4 Regulatory network of FRGs
We predicted the potential ironoptosis-associated non-coding RNAs by performing co-expression analysis with identified iron-related genes in COAD. We predicted miRNAs that target FGRs using the GSCA-hosted TCGA database (GSCA - Gene Set Cancer Analysis (hust.edu.cn). We then utilized the knockTF database to identify FRGs-specific transcription factors (TFs) that significantly influence FGRs based on expression and ChIP-seq/motif evidence (KnockTF-Search (licpathway.net)).
3 Results
3.1 Identification of DEGs related to ferroptosis in the TCGA cohort
In total, 426 sample, 52 metastases, 333 non-metastases COAD patients and 41 normal samples from TCGA cohorts were considered for inclusion in the study (Supplementary Table S1). We investigated RNA-seq data from the TCGA dataset to determine the expression differences of ferroptosis-related genes between tumor tissues and neighboring normal tissues. Among the 25 ferroptosis-related genes that were investigated, tumor tissues and adjacent normal tissues displayed significant differences in 21 genes. CDKN1A, HSPA5, EMC2, SLC7A11, MT1G, GPX4, FANCD2, CISD1, FDFT1, SLC1A5, SAT1, TFRC, RPL8, NCOA4, LPCAT3, GLS2, CARS1, ATP5MC3, ALOX15, ACSL4, and ATL1 (Figure 1A). To be more specific the differential expressed gene in metastases and non-metastases tissues were identified. Among the above-mentioned genes, the following genes were identified FDFT1, SLC1A5, RPL8, CARS1, and ALOX15 in addition to HSPB1.
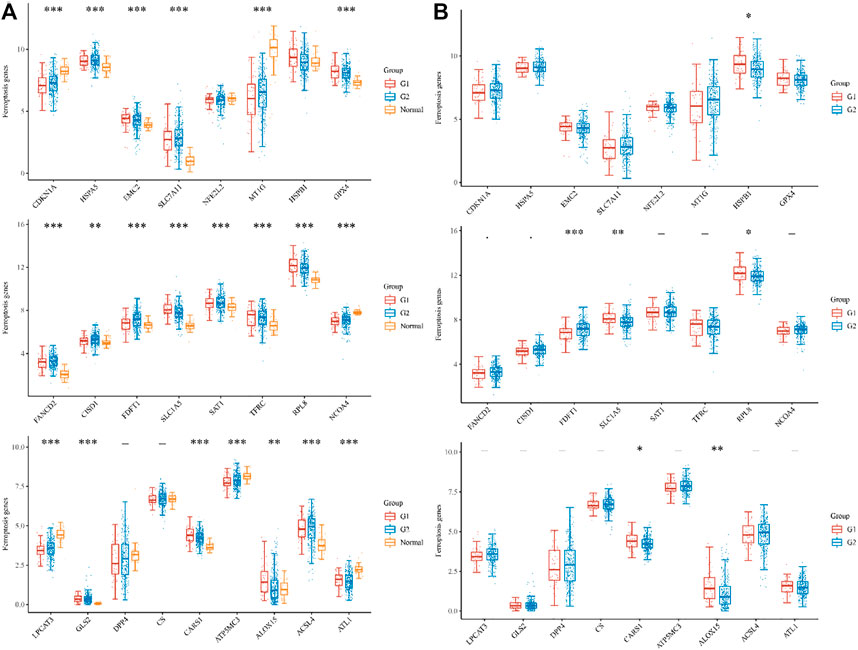
FIGURE 1. The expression distribution of ferroptosis-related mRNA in tumor tissues and normal tissues. (A) histogram shows the dysregulation of iron death in metastasis G1, non-metastases G2, and Normal samples, respectively. (B)histogram shows the number of significantly differentially expressed genes in metastasis G1, non-metastases G2, respectively. *p < 0.05, **p < 0.01, ***p < 0.001.
3.2 Prognostic genes identified in the TCGA COAD cohort
To reduce dimensionality and construct prognostic models based on Cox and lasso regression methods, iron death-related gene difference analysis results were utilized to evaluate the prognosis of these genes on CAOD tumor patients. Based on the median cut-off value, high- and low-risk patients were separated into two groups. The following formula was used to determine the risk score of the outcome =
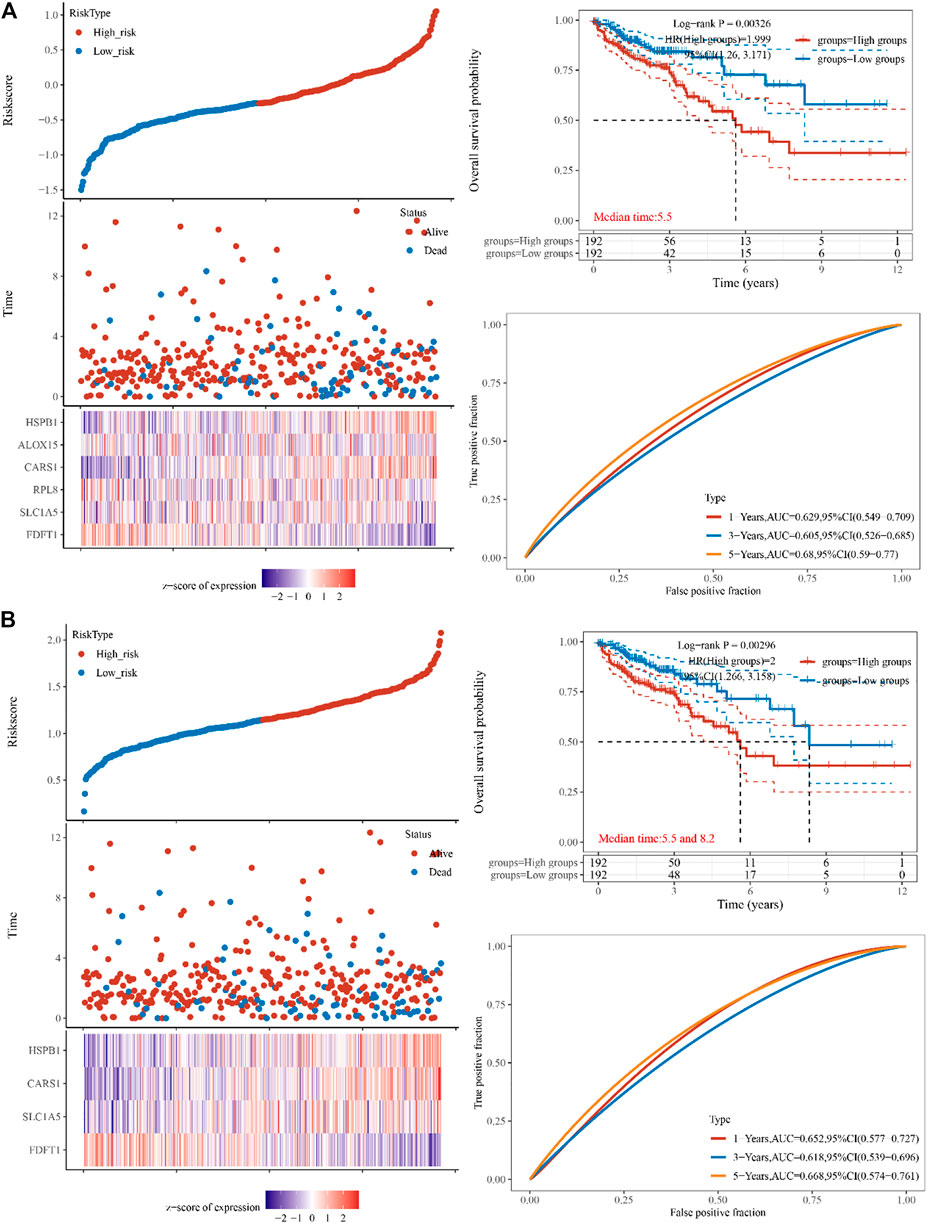
FIGURE 2. Constructing the TCGA cohort’s genes. (A) The multi-factor cox regression analysis showed six prognosis related genes in colon cancer (B) The forest analysis showed four prognosis related genes in colon cancer using 10x cross-validation (p < 0.05) The top scatters represent TCGA cohort’s risk score distribution from low to high. Kaplan-Meier survival analysis of the risk model from dataset, comparison among different groups was made by log-rank test. HR (High exp) represents the hazard ratio of the low-expression sample relatives to the high-expression sample. HR > 1 indicates the genes are a risk factor, and HR < 1 indicates the genes are a protective factor. HR (95%Cl). Heatmap is the gene expression from the signature. Prognostic performance was evaluated using the AUC of the time-dependent ROC curve analysis for OS in the TCGA cohort.
3.3 Independent prognostic value of the FRGs gene in COAD
The TCGA dataset was used to do univariate and multivariate Cox regression analysis on the identified 4 genes signature to determine whether the ability of the prognostic significant in predicting OS was independent. The effect of the 4 genes and clinical factors on prognosis was investigated with the help of the Cox regression analysis. These factors included age, pT, pM, pN, and Pathological tumor-node-metastasis (pTNM) staging. The variable was significant in the univariate Cox regression analysis and was thought to be related to prognosis (Figure 3A). Multivariate Cox regression analysis found these variables to be non-significant, except for age and pT stage and of the four discovered genes, CARS1 and HSPB1 had a hazard ratio greater than 1 (Figure 3B). Based on the independent prognostic factors for the OS of multi-factor Cox regression analysis, we developed a nomogram for predicting 1, 2, 3, and 5-year survival rates (Figures 3C, D). In the TCGA dataset, the calibration curve for the probability of 1, 2, 3, and 5-year OS exhibited the best agreement between observation and prediction. After bias adjustment, the C-index was found to be 0.703, p < 0.001 indicating a strong level of clinical diagnosis performance for the model signature genes. As a result, the identified ferroptosis-related gene impacts the prognosis of COAD and may serve as a possible diagnostic factor for COAD patients.
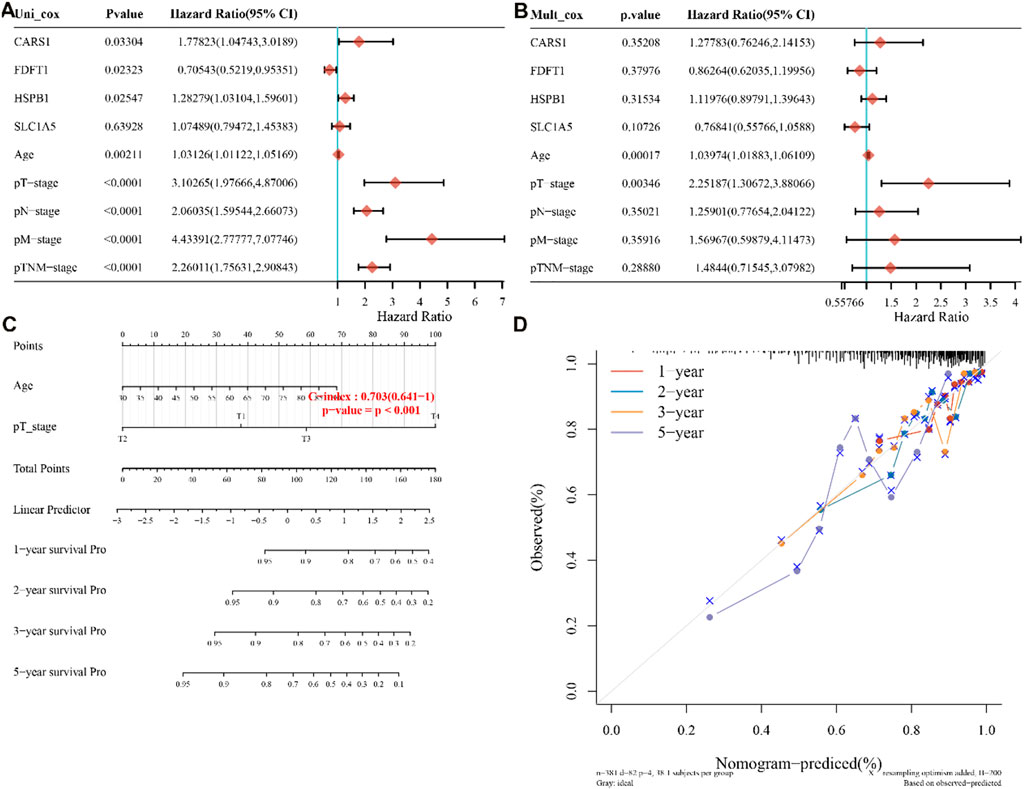
FIGURE 3. four-genes prognostic value and predictive nomogram. (A,B) Multivariate and univariate Cox regression analysis of the four genes is both available, (C) Nomogram development based on independent prognostic value for the 4-genes, (D) Predicting 1–5-year survival using a nomogram. pT,pN, pM are pathological stages where T is the stage (generally related to the size and depth of the tumor), N is lymph node metastasis, and M is distant metastasis. pTNM stands for Pathological tumor-node-metastasis (pTNM) staging.
3.4 Characterization of ferroptosis-associated molecules in COAD microenvironment
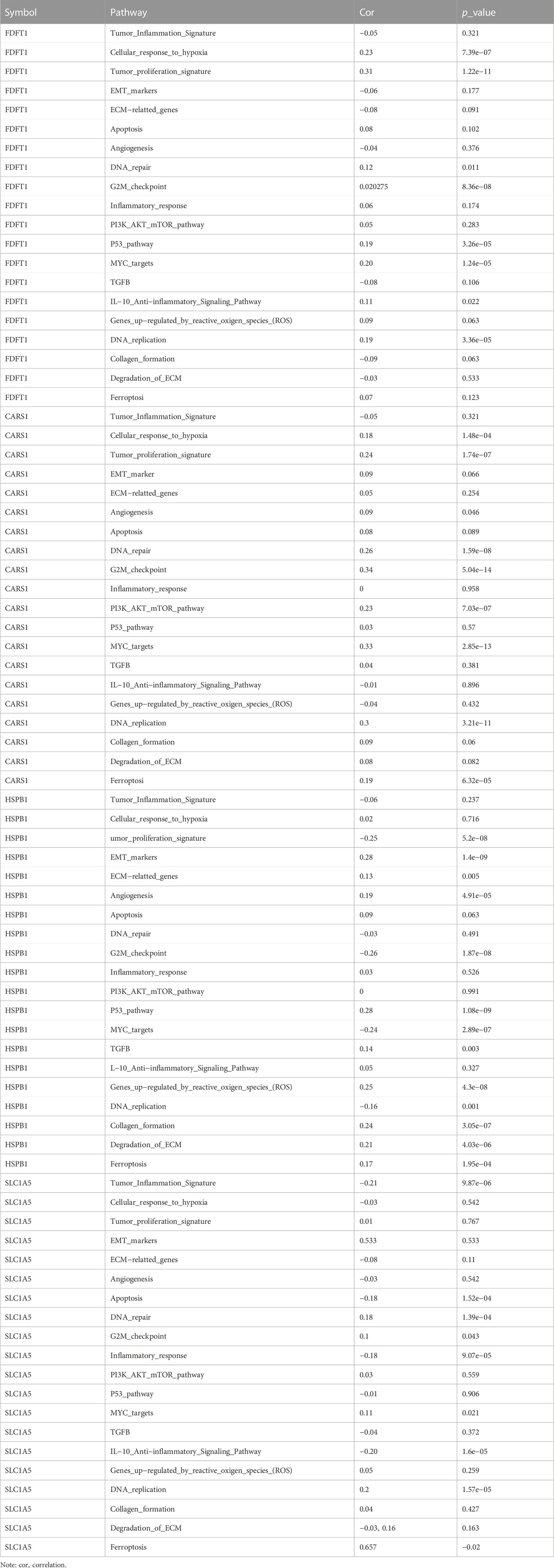
The enrichment fraction of each sample on each pathway was calculated in turn using the ssGSEA algorithm to obtain the connection between the sample and the pathway, and we then obtained the relationship between the gene and the pathway by calculating the correlation between gene expression and pathway score using spearman analysis. The tumor cell proliferation signature score was found to be positively correlated with CARS1 and FDFT1, indicating that the signature model has a high proliferation rate. Furthermore, we found a statistically significant activation of the cellular response to hypoxia signature and the G2M checkpoint in SLC1A5, CARS1, and FDFT1 (Table1).
3.5 FRGs correlate with immune cell infiltration in COAD
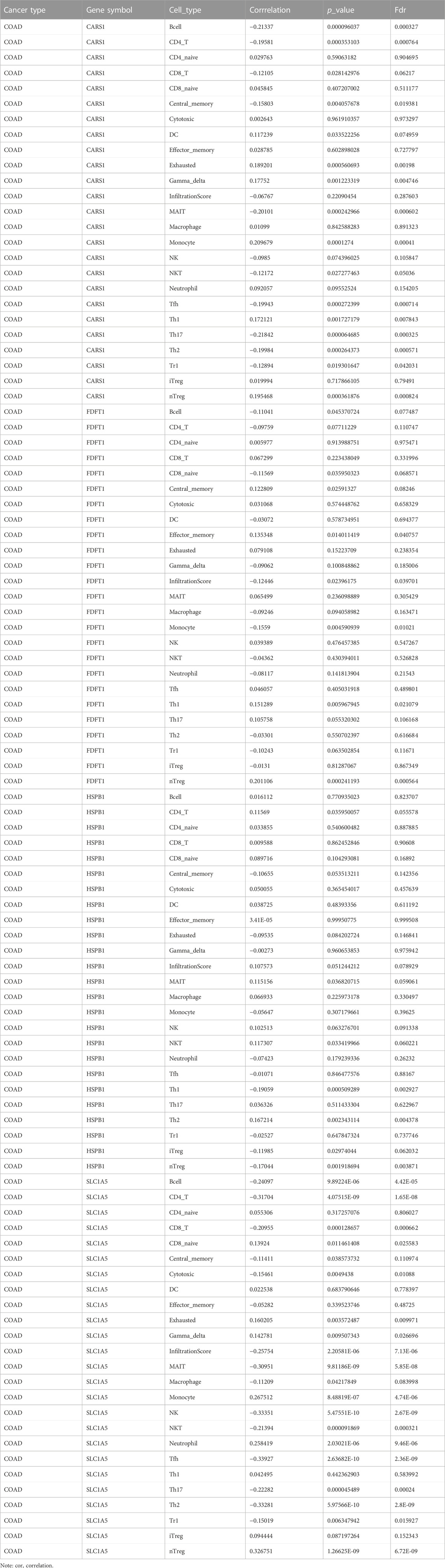
An increasing number of studies have suggested an interaction between immune response and pathophysiological processes. GSCA was performed to evaluate the correlation between FRGs and the available immune cells in COAD. FRFs correlated negatively with the infiltration of immune cells, including B cells, CD4-T, CD8-T, central memory, NKT, Tfh, Th17, Th2, and other immune cells presented in Table 2. These results indicated that dysregulation of FRGs expression is associated with worse immune cell infiltration in the COAD microenvironment.
3.5 Prediction of FRGs’ regulation network
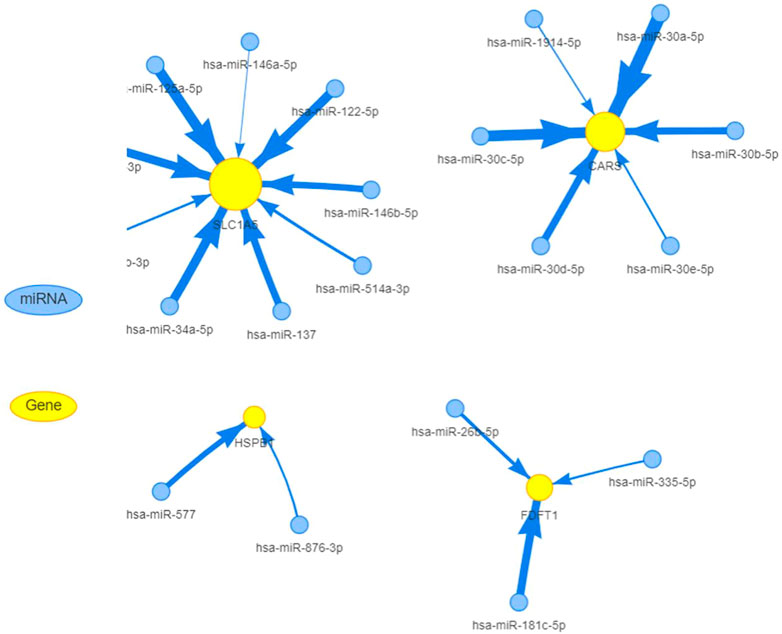
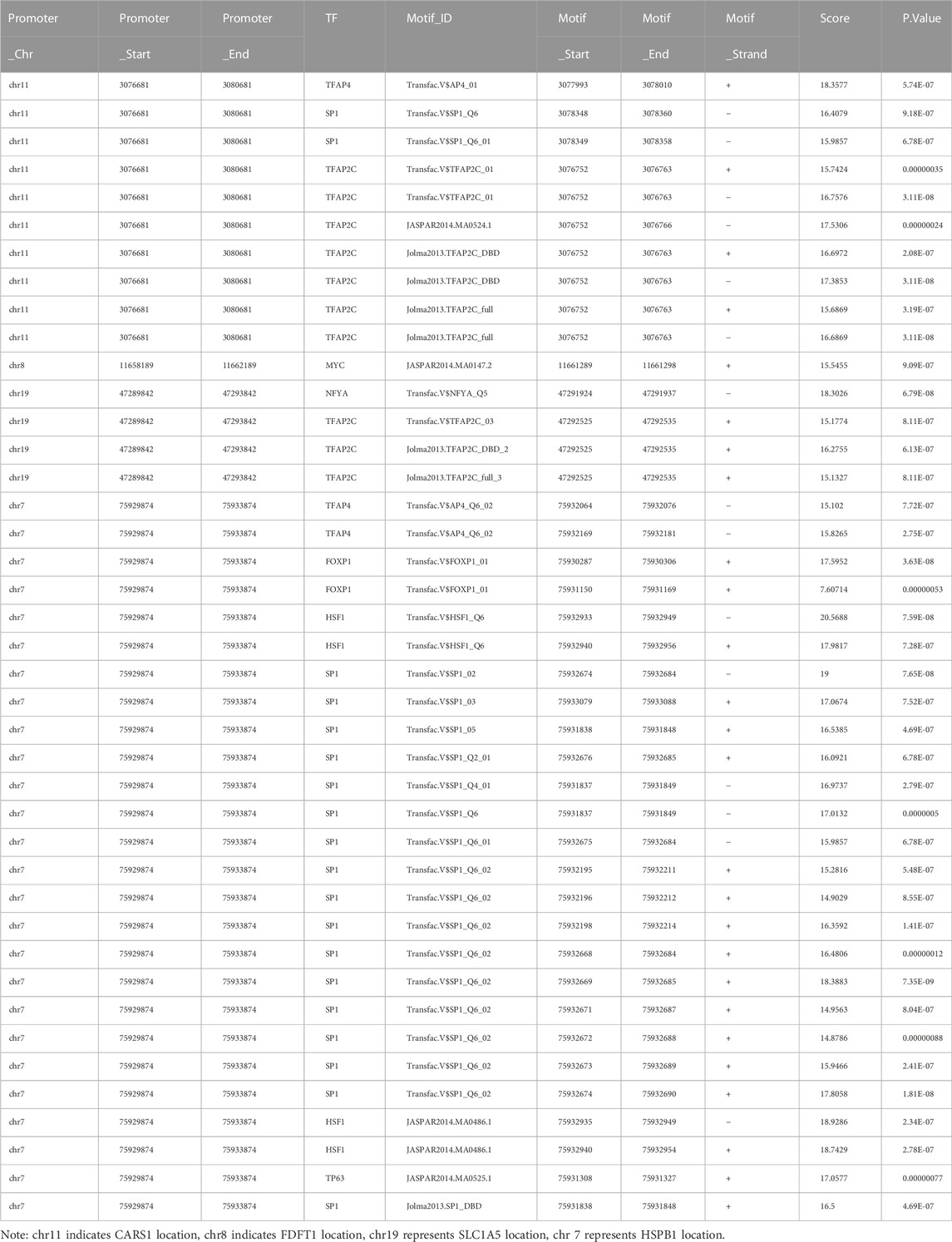
MiRNAs have been linked to the regulation of tumor-associated genes in a variety of cancer types, although their role in the deregulation of FGRs remains uncertain. We initially identified the FGRs miRNAs network in COAD using the GSCALite tool. Figure 4 depicts evidence that miRNAs target FGRs identified by the GSCALite program. Using ChIP-seq datasets, we also discovered transcription factors that may posttranscriptionally modulate FGRs expression (Table 3). Hsa miR- 37–3p has experimentally verified to target ironoptosis-related SLC1A5 in melanoma (doi: 10.1038/s41418-017-0053-8). These consistent results suggest that anticipated miRNAs may have a role in COAD as well.
4 Discussion
Colon cancer is the most prevalent primary malignant tumor that is characterized by rapid growth and treatment resistance, resulting in significant mortality and morbidity. Iron death is a pattern of cell death caused by the accumulation of active oxygen species (ROS) of iron and lipids within cells and plays a vital role in tumorigenesis and development. (Dixon, 2017; Jiang et al., 2021). A number of studies have shown that ferroptosis can be used to treat cells that have developed resistance to drugs, which emphasized the importance of ferroptosis in the treatment of CRC patients (Friedmann Angeli et al., 2019; Wang et al., 2021). However, the underlying mechanisms and biomarkers of iron death in cancer remain to be studied. In the current study, we used bioinformatics and statistical methods to screen TCGA for information relevant to ferroptosis in COAD metastasis. We also created a risk for ferroptosis, which we believe could serve as a possible diagnostic biomarker. Using the TCGA data collection, we initially evaluated DEGs ferroptosis genes in primary and next in metastasis COAD patients. According to the findings of the Kaplan Meier survival analysis four genes having a significant correlation with OS. The prognostic accuracy of these ferroptosis-related genes in COAD patients was examined using univariate and multivariate Cox regression. Patients with COAD who were classified as high risk had a lower overall survival time than those who were classified as low risk. Studies have shown that fatal lipid peroxidation [20] is a cause of ferroptotic cell death (Gao et al., 2015). The accumulation of intracellular iron resulting from the depletion of ferritin or iron transporters and the subsequent peroxidation causes the formation of lipid peroxides and iron hypertrophy (Stockwell and Jiang, 2019). Four ferroptosis-related genes (FDFT1, HSPB1, SLC1A5, and CARS1 were identified in this investigation. Two genes involved in ferroptosis and lipid metabolism, FDFT1 and HSPB1, have been related to a poor prognosis in colorectal cancer, which is congruent with our own findings in COAD metastases (Liu et al., 2021; Yang et al., 2021). In addition to being increased in CRC, HSPB1 was also revealed to be related to worse survival in the present study (Wang et al., 2012; Konda et al., 2017). However, neither CARS1 nor SLC1A5 have previously been documented in CRC, therefore our results suggest that this area requires additional investigation. One of the most important regulators of cell survival is the PI3K/Akt/mTOR pathway. ROS can activate this pathway either by oxidizing kinases directly or by oxidizing the negative phosphatase regulators of this system, including phosphatase and tensin homolog (PTEN), protein-tyrosine phosphatase 1B (PTP1B), and protein phosphatase 2 (PP2) (PP2A). The effects of above mentioned FRGs on ROS can cause an increase in activity of the PI3K/Akt/mTOR signaling cascade. Furthermore, Curcumin’s anti-cancer effects in colon cancer cells are modulated by HSPB1’s ability to induce reactive oxygen species (ROS) generation and autophagy (Liang et al., 2018). The above results appeared to support the existing results that FRGs contributed to colon cancer progression and invasion via oxidative phosphorylation. FGRs may act as ROS-inducing agents and can activate TGFB, which is known to promote tumorigenesis, angiogenesis, and metastasis. Additionally, TGFB regulates the genes responsible for inflammation, cell proliferation, differentiation, and survival. Hypoxia is known to stimulate the production of mitochondrial reactive oxygen species (mROS), which in turn increases and stabilizes hypoxia-inducible factor-1 (HIF1a), which contributes to the survival and progression of tumors by upregulating the genes that regulate tumor angiogenesis and metastasis. On the other hand, several distinct DNA repair systems collaborate to decrease the risk that this damage may lead to dangerous mutations. There is a link between FRGs and genes involved in DNA repair pathways, allowing cells to be more resistant to DNA damage. FGRs showed positive correlation with a checkpoint control pathway and enhanced the activity of genes involved in DNA replication pathway that would normally prohibit cells from initiating DNA replication when breaks are present. Previous research reported that increasing SLC7A11 and GPX4 expression in HCT116 inhibited iron-induced lipid peroxidation and protected cells from ferroptosis (Yuan et al., 2021).
The infiltration of different types of immune cells is a principal determining factor for the immune response at primary and secondary tumor sites in the tumor microenvironment. We further analyzed the influence of CARS1 level on immune cell infiltration. The levels of FRGs significantly affects the infiltration level of B cells, CD4-T, CD8-T, central memory, NKT, Tfh, Th17, Th2, and Tr1cells. These results indicated that FRGs had a key regulatory effect on the immune cells in COAD patients. This is a preliminary exploration, but we intend to follow it up with experimental confirmation and biogenesis research.
Study limitation: All the findings in this manuscript were of speculation based on the transcriptional analyses using bioinformatics analysis. In vitro and in vivo investigations should be conducted to confirm the functional involvement in COAD. In conclusion, based on bioinformatics and statistical analysis, this study revealed associations between ferroptosis-related genes (FRGs) and colon adenocarcinoma (COAD). Prognostic models for ferroptosis-related programmed cell death in COAD metastasis were developed by identifying 4 signature genes, constructing a nomogram based on univariate and multivariate Cox regression model, and analyzing related pathways, immune cell infiltration, and regulatory networks.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding authors.
Author contributions
SB conceived the idea, designed the study, and wrote the original draft. YH and II validated the results. YS, WF, YG, and HT reviewed and edited the paper. MR and SA performed statistical analysis. All authors discussed the results and contributed to the final manuscript.
Funding
The work is paid by 2021Szvup096, Shenzhen Science and Technology Innovation Strategic Research and Technology Transfer Promotion Center. This work is also supported by Grant no, LHTD20170002, Science and Technology Innovation Bureau of Nanshan district, Shenzhen.
Conflict of interest
Authors SB, YH, II, YS, WF, ZT, YW, and HT were employed by Research Center of Molecular Diagnostics and Sequencing, Axbio Biotechnology (Shenzhen) Co., Ltd.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmolb.2022.1102735/full#supplementary-material
References
Bersuker, K., Hendricks, J. M., Li, Z., Magtanong, L., Ford, B., Tang, P. H., et al. (2019). The CoQ oxidoreductase FSP1 acts parallel to GPX4 to inhibit ferroptosis. Nature 575 (7784), 688–692. doi:10.1038/s41586-019-1705-2
Dixon, S. J. (2017). Ferroptosis: Bug or feature? Immunol. Rev. 277 (1), 150–157. doi:10.1111/imr.12533
Dixon, S. J., Lemberg, K. M., Lamprecht, M. R., Skouta, R., Zaitsev, E. M., Gleason, C. E., et al. (2012). Ferroptosis: An iron-dependent form of nonapoptotic cell death. Cell 149 (5), 1060–1072. doi:10.1016/j.cell.2012.03.042
Doll, S., Freitas, F. P., Shah, R., Aldrovandi, M., da Silva, M. C., Ingold, I., et al. (2019). FSP1 is a glutathione-independent ferroptosis suppressor. Nature 575 (7784), 693–698. doi:10.1038/s41586-019-1707-0
Friedmann Angeli, J. P., Krysko, D. V., and Conrad, M. (2019). Ferroptosis at the crossroads of cancer-acquired drug resistance and immune evasion. Nat. Rev. Cancer 19 (7), 405–414. doi:10.1038/s41568-019-0149-1
Gao, M., Monian, P., and Jiang, X. (2015). Metabolism and iron signaling in ferroptotic cell death. Oncotarget 6 (34), 35145–35146. doi:10.18632/oncotarget.5671
Jiang, L., Kon, N., Li, T., Wang, S. J., Su, T., Hibshoosh, H., et al. (2015). Ferroptosis as a p53-mediated activity during tumour suppression. Nature 520 (7545), 57–62. doi:10.1038/nature14344
Jiang, M., Qiao, M., Zhao, C., Deng, J., Li, X., and Zhou, C. (2020). Targeting ferroptosis for cancer therapy: Exploring novel strategies from its mechanisms and role in cancers. Transl. Lung Cancer Res. 9 (4), 1569–1584. doi:10.21037/tlcr-20-341
Jiang, X., Stockwell, B. R., and Conrad, M. (2021). Ferroptosis: Mechanisms, biology and role in disease. Nat. Rev. Mol. Cell Biol. 22 (4), 266–282. doi:10.1038/s41580-020-00324-8
Konda, J. D., Olivero, M., Musiani, D., Lamba, S., and Di Renzo, M. F. (2017). Heat-shock protein 27 (HSP27, HSPB1) is synthetic lethal to cells with oncogenic activation of MET, EGFR and BRAF. Mol. Oncol. 11 (6), 599–611. doi:10.1002/1878-0261.12042
Lee, Y. S., Lee, D. H., Jeong, S. Y., Park, S. H., Oh, S. C., Park, Y. S., et al. (2019). Ferroptosis-inducing agents enhance TRAIL-induced apoptosis through upregulation of death receptor 5. J. Cell. Biochem. 120 (1), 928–939. doi:10.1002/jcb.27456
Li, J., Cao, F., Yin, H. L., Huang, Z. J., Lin, Z. T., Mao, N., et al. (2020). Ferroptosis: Past, present and future. Cell Death Dis. 11 (2), 88. doi:10.1038/s41419-020-2298-2
Liang, H. H., Huang, C. Y., Chou, C. W., Makondi, P. T., Huang, M. T., Wei, P. L., et al. (2018). Heat shock protein 27 influences the anti-cancer effect of curcumin in colon cancer cells through ROS production and autophagy activation. Life Sci. 209, 43–51. doi:10.1016/j.lfs.2018.07.047
Liu, Y., Guo, F., Guo, W., Wang, Y., Song, W., and Fu, T. (2021). Ferroptosis-related genes are potential prognostic molecular markers for patients with colorectal cancer. Clin. Exp. Med. 21 (3), 467–477. doi:10.1007/s10238-021-00697-w
Ou, Y., Wang, S. J., Li, D., Chu, B., and Gu, W. (2016). Activation of SAT1 engages polyamine metabolism with p53-mediated ferroptotic responses. Proc. Natl. Acad. Sci. U. S. A. 113 (44), E6806–E6812. doi:10.1073/pnas.1607152113
Siegel, R. L., Miller, K. D., Fuchs, H. E., and Jemal, A. (2022). Cancer statistics, 2016. Ca. Cancer J. Clin. 72 (1), 7–30. doi:10.3322/caac.21332
Stockwell, B. R., and Jiang, X. (2019). A physiological function for ferroptosis in tumor suppression by the immune system. Cell Metab. 30 (1), 14–15. doi:10.1016/j.cmet.2019.06.012
Wang, J. J., Liu, Y., Zheng, Y., Lin, F., Cai, G. F., and Yao, X. Q. (2012). Comparative proteomics analysis of colorectal cancer. Asian pac. J. Cancer Prev. 13 (4), 1663–1666. doi:10.7314/apjcp.2012.13.4.1663
Wang, Y., Zhao, G., Condello, S., Huang, H., Cardenas, H., Tanner, E. J., et al. (2021). Frizzled-7 identifies platinum-tolerant ovarian cancer cells susceptible to ferroptosis. Cancer Res. 81 (2), 384–399. doi:10.1158/0008-5472.CAN-20-1488
Wei, G., Sun, J., Hou, Z., Luan, W., Wang, S., Cui, S., et al. (2018). Novel antitumor compound optimized from natural saponin Albiziabioside A induced caspase-dependent apoptosis and ferroptosis as a p53 activator through the mitochondrial pathway. Eur. J. Med. Chem. 157, 759–772. doi:10.1016/j.ejmech.2018.08.036
Xia, X., Fan, X., Zhao, M., and Zhu, P. (2019). The relationship between ferroptosis and tumors: A novel landscape for therapeutic approach. Curr. Gene Ther. 19 (2), 117–124. doi:10.2174/1566523219666190628152137
Yang, C., Huang, S., Cao, F., and Zheng, Y. (2021). A lipid metabolism-related genes prognosis biomarker associated with the tumor immune microenvironment in colorectal carcinoma. BMC cancer 21 (1), 1182. doi:10.1186/s12885-021-08902-5
Yuan, Y., Ni, S., Zhuge, A., Li, B., and Li, L. (2021). Iron regulates the warburg effect and ferroptosis in colorectal cancer. Front. Oncol. 11, 614778. doi:10.3389/fonc.2021.614778
Keywords: COAD microenvironment, metastasis, ferroptosis, immune cell infiltration, TCGA
Citation: Baldi S, He Y, Ivanov I, Sun Y, Feng W, Refat M, Mohammed SAD, Adlat S, Tian Z, Wang Y, Gao Y and Tian H (2022) Novel characterization discoveries of ferroptosis-associated molecules in COAD microenvironment based TCGA data. Front. Mol. Biosci. 9:1102735. doi: 10.3389/fmolb.2022.1102735
Received: 19 November 2022; Accepted: 30 November 2022;
Published: 13 December 2022.
Edited by:
Yanqing Liu, Columbia University, United StatesReviewed by:
Jun Shang, Cincom System, Inc., United StatesYufei Liu, Huashan Hospital, Fudan University, China
Shiqiang Jin, Bristol Myers Squibb, United States
Copyright © 2022 Baldi, He, Ivanov, Sun, Feng, Refat, Mohammed, Adlat, Tian, Wang, Gao and Tian. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Salem Baldi, c2FsaW1sYWJvcmF0b3J5QGdtYWlsLmNvbQ==; Yaping Gao, Z2FveXBAdHNpbmdodWEtc3oub3Jn; Hui Tian, aHVpLnRpYW5AYXhiaW8uY24=
 Salem Baldi
Salem Baldi Yun He1
Yun He1