
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
MINI REVIEW article
Front. Mol. Biosci. , 14 December 2022
Sec. Biophysics
Volume 9 - 2022 | https://doi.org/10.3389/fmolb.2022.1070526
This article is part of the Research Topic Recent Advances in Molecular Properties of DNA-Protein Interactions, Chromatin and Their Biological Roles View all 6 articles
In the last decade, the CRISPR-Cas technology has gained widespread popularity in different fields from genome editing and detecting specific DNA/RNA sequences to gene expression control. At the heart of this technology is the ability of CRISPR-Cas complexes to be programmed for targeting particular DNA loci, even when using catalytically inactive dCas-proteins. The repertoire of naturally derived and engineered dCas-proteins including fusion proteins presents a promising toolbox that can be used to construct functional synthetic genetic circuits. Rational genetic circuit design, apart from having practical relevance, is an important step towards a deeper quantitative understanding of the basic principles governing gene expression regulation and functioning of living organisms. In this minireview, we provide a succinct overview of the application of CRISPR-dCas-based systems in the emerging field of synthetic genetic circuit design. We discuss the diversity of dCas-based tools, their properties, and their application in different types of genetic circuits and outline challenges and further research directions in the field.
Interactions of proteins and nucleic acids are the cornerstone of genome functioning in living organisms. They mediate the execution of complex genetic and epigenetic networks and circuits in bacteria, archaea, eukaryotes, and viruses. Understanding the molecular mechanisms behind living organisms’ operation now goes hand-in-hand with the advances in synthetic biology which aims to construct biological systems and their components in a bottom-up approach (Jia and Schwille, 2019). A quote by Nobel laureate Richard Feynman “What I cannot create, I do not understand” nicely illustrates this trend and the challenges ahead.
Around 20 years ago two pioneering works by Elowitz and Leibler (2000) and Gardner et al. (2000) showcased the first artificial transcriptional genetic circuits in bacteria. These circuits implemented periodic oscillations of gene expression (repressilator circuit) and persistent switching between the expression of two genes upon transient exposure to a chemical signal (toggle switch circuit). The circuits were based on bacterial DNA-binding repressor proteins and corresponding DNA operator sequences. Since then the field of genetic circuit design has been rapidly advancing [reviewed in (Xia et al., 2019)]. Transcriptional regulatory circuits based on libraries of orthogonal repressor-operator pairs (Nielsen et al., 2016), RNA interference (Bloom et al., 2014), and more recently CRISPR-dCas complexes (see below) have been constructed. The design of complex multi-layered genetic circuits usually employs abstract principles similar to those used in the construction of digital electronic circuits from elementary modules, called logic gates. These logic gates (such as AND, OR, NOT, etc.) produce discrete output signals depending on one or more input signals (in genetic circuits rates of specific gene expression can be treated as such signals). Practical applications of genetic circuits include the design of living therapeutics (Brennan, 2022) [including those for altering the human microbiome (Robinson et al., 2022) or designing advanced CAR T therapies for cancer treatment (Cho et al., 2018a)], biosensors (Del Valle et al., 2021; Nguyen et al., 2021), tissue engineering in regenerative medicine (Healy and Deans, 2019), genetic modification of crop plants (de Lange et al., 2018), etc.
The discovery of CRISPR-Cas adaptive immunity systems in bacteria and archaea (Makarova et al., 2020) and the demonstration of their application for targeted genome editing in bacteria and eukaryotes (Jinek et al., 2012; Cong et al., 2013) gave impetus to a whole new era in synthetic biology. Soon afterward it was demonstrated that catalytically dead CRISPR-Cas9 proteins can be repurposed to interfere with gene transcription (Qi et al., 2013) and used as vehicles to deliver fused moieties to the specific genome loci in a programmable manner (Xu and Qi, 2019). These discoveries spurred rapid progress in the application of CRISPR-dCas technology for the construction of transcriptional genetic circuits in recent years. In this mini-review we survey the diversity and properties of CRISPR-dCas-based systems, analyze reported and prospective applications thereof for genetic circuits design, outline challenges and further research directions in the field.
CRISPR-dCas systems are derived from wild-type CRISPR-Cas systems by inactivating their nuclease activity via point mutations while retaining their ability to bind DNA loci. In nature, CRISPR-Cas systems are represented by a very diverse group of genes (Makarova et al., 2020). For practical applications, class 2 type II (based on Cas9 effector nucleases) and type V (based on a heterogeneous family of Cas12 nucleases) are most valuable since their effector nucleases are mainly DNA binding and are represented by a single protein.
The most studied Cas9 nuclease is SpCas9 from S. pyogenes, a 158 kDa (1368 aa) protein that in nature forms a complex with two RNA molecules—CRISPR RNA and transactivating CRISPR RNA. The former encodes the spacer sequence that is used to guide target-specific SpCas9 binding. Key elements of both RNAs may be combined via a tetraloop into a single guide RNA (sgRNA), which binds to SpCas9 with a Kd of around 10 pM (Wright et al., 2015) (see Figure 1A). The process of SpCas9-sgRNA binding to the DNA is now known in detail (Wang et al., 2022). As a first step, the SpCas9-sgRNA complex finds a three-nucleotide-long protospacer adjacent motif (PAM) on the non-target DNA strand via its C-terminal domain (NGG is the optimal PAM sequence for SpCas9). Next, the “recognition” and “nuclease” lobes of Cas9 rotate, simultaneously bending the DNA and exposing the PAM-proximal bases on the target DNA strand for sgRNA invasion and further target sequence interrogation through base pairing, resulting in the formation of an R-loop structure. The formation of a full RNA-DNA duplex and its conformational kinking activates the nuclease domains (RuvC and HNH) which cleave the DNA strands. Two mutations (D10A, H841A) in SpCas9 inactivate these domains resulting in a dead-SpCas9 (SpdCas9).
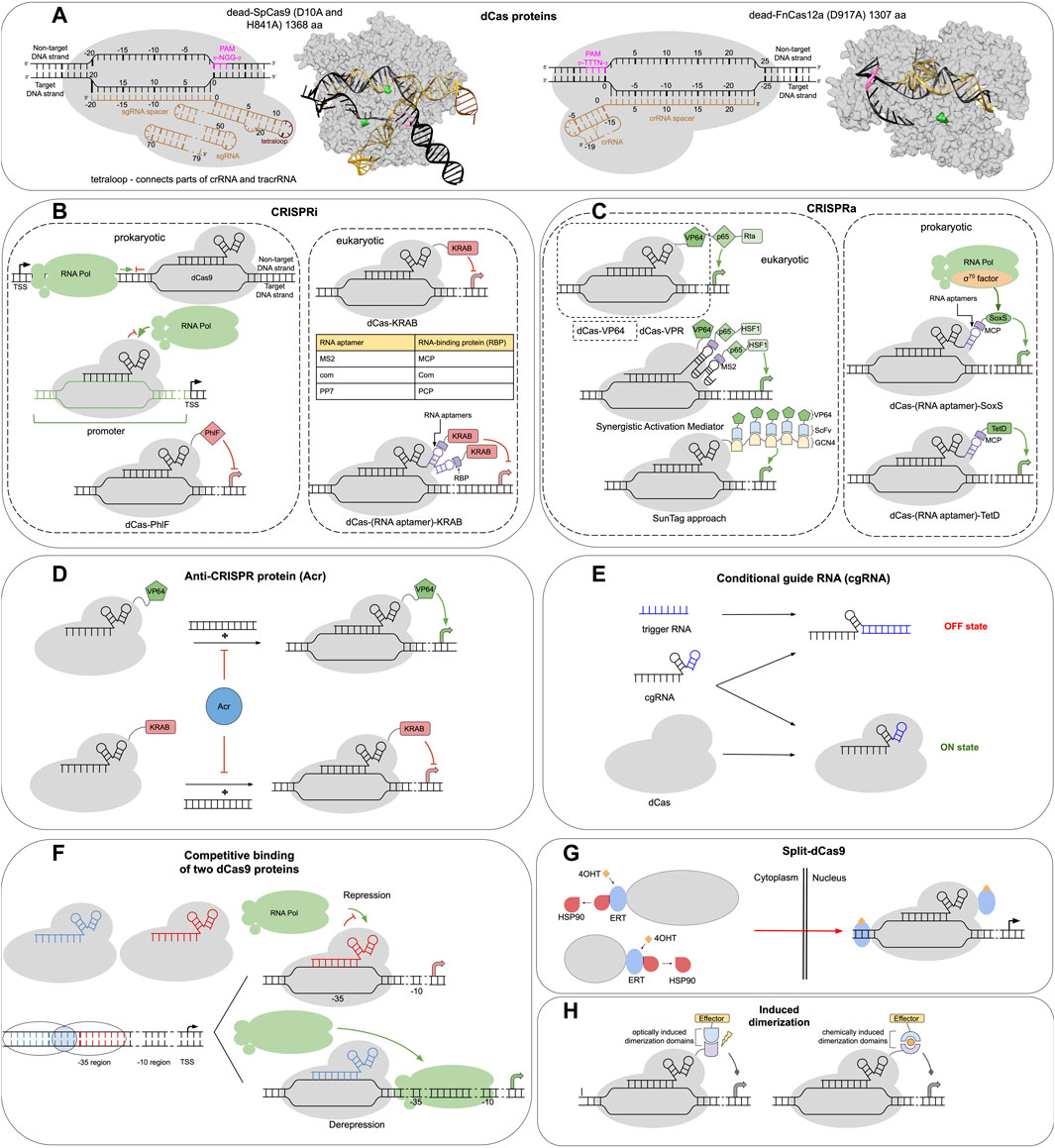
FIGURE 1. CRISPR-dCas toolbox for genetic circuit design. (A) Schematic and structural representation of the dCas proteins: dead-SpCas9 (left) and dead-FnCas12a (right). The colors of elements in the annotated schematic representations correspond to those in the structural representation. Amino-acid residues which are mutated in the dCas proteins are highlighted in green, PAM stands for protospacer adjacent motif. (B–H) dCas-based constructs that can be used for transcriptional gene expression control. (B) Principal schema of sequence-specific gene repression using CRISPR interference (CRISPRi) in prokaryotes (left) and eukaryotes (right). (C) Various approaches to sequence-specific gene activation using CRISPR-dCas systems (CRISPRa) in prokaryotes (right) and eukaryotes (left). (D) Additional regulation of CRISPRi and CRISPRa by Anti-CRISPR proteins (Acr). (E) Regulation of dCas activity via conditional sgRNA and trigger RNA. (F) Combination of two interfering CRISPR-dCas complexes for fine control of target gene expression. (G) Activation of split-dCas9 proteins fused to ERT domains of estrogen receptor (see text). (H) Additional control of sequence-specific dCas targeting activity for gene expression regulation by chemical- or light-induced dimerization domains.
The use of SpdCas9 to interfere with RNA polymerase (RNAP) binding or progression along the DNA in prokaryotes resulted in the first application of such systems for sequence-specific control of gene expression (termed CRISPRi) (see Figure 1B) (Qi et al., 2013). Qi et al. (2013) have shown that up to 300-fold gene expression repression can be achieved by targeting dCas9 to the gene promoter region (blocking RNAP binding) or the beginning of the gene coding sequence (interfering with transcription elongation). Interestingly, in the latter case, the effect is only achieved when targeting the DNA non-template strand (Qi et al., 2013). The specificity and strength of the dCas9-sgRNA complex binding to the target DNA sequence is governed by around 20 nucleotides at the 5′-end of sgRNA—the so-called spacer sequence. The 7–12 bp long PAM proximal part of the spacer (usually called the seed sequence) is especially important for binding specificity. Mismatches in this region have a severe impact on binding or cleavage, while the PAM distal region can tolerate multiple mismatches (Feng et al., 2021). Seed sequence mismatches result in a 70%–90% decrease in gene repression in CRISPRi assays (Qi et al., 2013). The overall Kd for Cas9–RNA complex binding to a bona fide DNA target has been estimated to be around 0.5 nM (Sternberg et al., 2014). For comparison, the high-affinity bacterial transcription repressor LacI binds to its cognate operator LacO in vitro with a Kd of around 10 pM (Du et al., 2019), while eukaryotic p53 dimers bind to their response elements with Kds above 5 nM (Weinberg et al., 2005).
A family of dCas9 proteins may be derived from SpCas9 orthologs in various organisms. These Cas9 proteins may vary in size (e.g., 984 aa in C. jejuni, 1629 aa in F. novicida), recognized PAM sequences (e.g., NNGRRT in S. aureus, NNNNGATT in N. meningitidis, NNARAAW in S. thermophilus), optimal spacer length (e.g., 21–24 nt in N. meningitidis), the structure of CRISPR RNA (Anzalone et al., 2020). So far dCas9 from S. thermophilus, N. meningitidis, T. denticola have been used (Brocken et al., 2018). An additional advantage of having different dCas9 orthologs at hand is the simultaneous use of several orthogonal dCas9 (i.e., binding specifically only their cognate CRISPR/sgRNAs) (Esvelt et al., 2013). For a multi-layered gene circuit (see below) this may alleviate competition between several sgRNAs for the same pool of dCas9 protein, potentially increasing circuit robustness and alleviating interference between genetic components.
Cas12 nucleases are another source of CRISPR-dCas systems. The Cas12 family is more diverse than Cas9 family and is further classified into different subtypes (e.g., Cas12a from F. novicida, E. eligens, Acidaminococcus sp., Lachnospiraceae sp. or Cas12e from Deltaproteobacteria) (Makarova et al., 2020). Cas12 nucleases usually require an AT-rich PAM site located upstream of the target region (contrary to Cas9), many lack tracrRNA and have only one nuclease domain (RuvC) which can be inactivated by a point mutation to produce dCas12 (Anzalone et al., 2020) (Figure 1A). One advantage of certain dCas12a variants is their nuclease-independent ability to process pre-crRNA into crRNA, thus many crRNAs may be simultaneously encoded by one customized CRISPR array even for CRIPSRi applications (Zetsche et al., 2017). In prokaryotes, initial dCas12a-based CRISPRi assays have demonstrated up to ∼8-fold gene expression repression (Kim et al., 2017). Interestingly, unlike dCas9, dCas12a has been shown to repress transcription elongation much better when targeted to the template DNA strand rather than the coding strand (Kim et al., 2017). Disadvantages of wild-type dCas12a versus dCas9 include a more restrictive PAM (e.g., TTTV for AsCas12a) and lower regulation efficiency, although engineered dCas12a have improved properties (see below) (Kim et al., 2017).
During the last decade many engineered Cas9, Cas12 variants, and sgRNAs have been developed for genome editing, aiming to relax PAM requirements (Collias and Beisel, 2021), reduce off-target activity, and increase on-target activity (Anzalone et al., 2020; Riesenberg et al., 2022). Simultaneously optimizing all these properties is difficult, since the relaxation of PAM requirements or reducing off-target activity often comes at a cost of the reduced on-target activity. For the purpose of dCas applications, what essentially matters is the on-target and off-target binding of dCas, which may not be directly related to the nuclease activity of its catalytically active counterpart. Since the high concentration of dCas proteins is usually required for the effective operation of genetic circuits, dCas toxicity [likely caused by its off-target binding, which may happen even without sgRNA (Jones et al., 2017)] poses an important problem, especially in bacteria (Zhang and Voigt, 2018). An original approach to alleviating dCas9 toxicity suggested by Zhang and Voigt involved reducing dCas DNA affinity by mutating its PAM-recognizing domain and simultaneously fusing it to another sequence-specific transcription factor to rescue DNA specificity and affinity of the whole construct (Zhang and Voigt, 2018) (see Figure 1B, bottom). Such a design allowed to reach ∼9,000 dCas9 molecules per E. coli cell compared to ∼500 in the wild-type case. Some other examples of dCas engineering include altering the PAM interacting domain of SpdCas9 to specifically recognize start codons of protein-coding genes (5′-CAT-3′ PAM) (Wang et al., 2021) and development of hyperdCas12a—a version of dCas12a from Lachnospiraceae bacterium with enhanced binding affinity resulting in significantly improved CRISPRi and CRISPRa outcomes in eukaryotes (Guo et al., 2022).
The versatility of dCas-systems in gene expression regulation can be significantly enhanced by combining them with other functional protein domains. For instance, in bacteria instead of transcription repression, gene activation can be achieved by fusing dCas with prokaryotic activator domains and targeting them near promoters (CRISPRa technology) (Figure 1C). A number of such activator domains have been used so far, including the omega subunit of RNAP, TetD, and SoxS activators from E. coli (Dong et al., 2018; Villegas Kcam et al., 2021). SoxS-based designs have been found to confer the best results, yielding up to a 50-fold increase in mRNA levels of activated genes, provided the SoxS domain has proper positioning and orientation relative to the promoter (Dong et al., 2018; Klanschnig et al., 2022).
In eukaryotes, the effects of bare dCas-sgRNA binding on transcription repression are limited (around two-fold repression has been reported for human cells) (Qi et al., 2013). This is likely due to the much more complex mechanisms of transcription regulation in chromatin, where nucleosomes alone may block transcription elongation unless other chromatin factors are recruited to help (Bondarenko et al., 2006; Petesch and Lis, 2012). Using dCas as a vehicle to target transcriptional effectors is a way to address this challenge and expand the toolbox toward gene activation (Figures 1B, C). To enable gene silencing in eukaryotes, linking several chromatin effectors to dCas9 proteins has been attempted [see (Gilbert et al., 2013; Xu and Qi, 2019)]. For human cells, the most efficient results (up to 15-fold repression of GFP expression in HEK-293 cells), so far, were achieved by fusing dCas9 to the KRAB domain (Krüppel associated box) of Kox1 protein (Gilbert et al., 2013; Gilbert et al., 2014), which was successfully used in the construction of complex genetic logic gates (Kim et al., 2019). KRAB interacts with KAP1 protein, which acts as a scaffold for various heterochromatin-inducing factors (HP1, SETDB1, NuRD, N-CoR1, еtc.) (Groner et al., 2010). In yeast, fusing dCas9 with Mxi1 domain, which is thought to interact with the histone deacetylase Sin3, yielded the best results in maintaining consistent gene repression states (more than 10-fold repression of GFP expression was achieved) (Gilbert et al., 2013; Gander et al., 2017).
For CRISPR-mediated activation in eukaryotes (CRISPRa) initial works relied on a VP64 transcription activator derived from the herpes simplex virus. Although moderately effective [25-fold activation reported by Gilbert et al. (2013) in human HEK293 cells], it has been successfully used in the construction of genetic circuits and AND gates in yeast (Hofmann et al., 2019). Further CRISPRa studies focused on developing more potent and more flexible activator designs. Chavez et al. (2015) found that adding a tripartite VPR activator consisting of serially connected VP64, p65, and Rta domains to the C-terminus of dCas9 resulted in more than an order of magnitude better activation than in the dCas9-VP64 case (up to a 320-fold increase in activation) (see Figure 1C). The dCas9-VPR was employed for the construction of AND gates in human HEK293T cells (Gao et al., 2016). Another highly potent but somewhat more complex CRISPRa design is the synergistic activation mediator (SAM), yielding more than 100-fold greater activation than dCas9-VP64. SAM combines dCas9-VP64 protein with a special scaffold sgRNA which incorporates two MS2 RNA hairpin aptamers that recruit exogenously introduced MCP-p650-HSF1 constructs via MS2 binding to the bacteriophage MS2 coat protein (MCP) (Konermann et al., 2015). A more detailed discussion of the effectiveness of different CRISPRa methods in eukaryotes can be found elsewhere (Shakirova et al., 2020).
The use of RNA hairpins and their corresponding protein binding domains provides a versatile platform for the modular design of dCas-based systems. Using this strategy Zalatan et al. (2015) have simultaneously implemented CRISPRa together with CRISPRi using one dCas9 protein but different sgRNAs attached to various aptamers (Figures 1B, C). Interestingly, the recruitment of VP64 domain through an MS2-MCP interaction yielded ∼18-fold better gene activation results than in the case of fusing VP64 directly to the dCas9 protein. This can be in part explained by the fact that MCP binds as a dimer to its RNA target. Such RNA-mediated scaffolding designs are not without limitations, since RNA hairpins interfere with sgRNA transcription. Routes to circumvent this problem include using Pumilio RNA-binding protein that binds to a non-hairpin structure (Cheng et al., 2016) or coupling activators via antigen-antibody interactions derived system [e.g., SunTag system (Tanenbaum et al., 2014)].
The interaction of dCas-RNA complexes with regulatory proteins or RNAs provides additional instruments to the toolbox (Figures 1D, E). AntiCRISPR proteins (Acrs), that bind and inactivate dCas were shown to practically abolish CRISPRa and CRISPRi activity of dCas-based systems. Arcs have been used in genetic circuits to construct feed-forward loops and pulse generators (Nakamura et al., 2019). An alternative way to deactivate dCas complexes is to inactivate or activate sgRNA. To this end, conditional guide RNAs (cgRNA) have been designed. They can be blocked or activated by binding a trigger RNA. The effectiveness of such activation/deactivation of up to 50% has been achieved (Hochrein et al., 2021). cgRNA may be used as a signal input in genetic logic gates (Hochrein et al., 2021). Alternatively, aptazyme, which upon binding of a small ligand (guanine), catalyzes self-cleavage and consecutive activation of sgRNA was used as a way to trigger CRISPRa. However, such a design had significant levels of gene activation even without guanine (Tang et al., 2017). Finally, competition of two dCas-sgRNA complexes that target neighboring DNA sites has been used to expand the flexibility of dCas-based circuits (Anderson and Voigt, 2021) (Figure 1F). Such a design is good for ratiometric signal processing when the relative strength of two signals is used as an input to the circuit.
Another powerful strategy used to expand dCas-based instruments’ repertoire is the use of split proteins, including those with induced dimerization and split-dCas itself (Figures 1G, H). Chemically induced dimerizers (CID), such as FKBP/FRB system induced by rapamycin (Gao et al., 2016; Bao et al., 2017), or optogenetically induced dimerizers (OID), such as the Magnet system (Kawano et al., 2015; Nihongaki et al., 2017) have been used to link dCas to transcriptional effectors. Such designs provide spatiotemporal control of gene expression, have almost no background activity when no dimerizing agent is present, and manifest efficiencies comparable to direct dCas protein fusion constructs [e.g., up to 165-fold activation in CRISPRa using VPR (Gao et al., 2016)]. Finally, the application of split-dCas proteins has been reported. In one case split-dCas domains of a dCas9-VP64 system were fused to the CID-domains of the FKBP/FRB system (Zetsche et al., 2015). However, such a design was shown to have a considerable level of background activation due to the spontaneous auto-assembly of dCas9 in vivo (Zetsche et al., 2015). To dampen auto-assembly, split-dCas parts have to be kept further apart. In eukaryotes, this was shown to be possible through the differential addition of nuclear localization and nuclear export signals to the split parts (Zetsche et al., 2015). Alternatively, fusing dCas9-split parts to ligand-binding domains from nuclear receptors was demonstrated to be effective. Nguyen et al. (2016) have fused dCas9 split parts to the ERT domains of the estrogen receptor. ERT keeps dCas in the cytoplasm due to interactions with Hsp90, unless 4-hydroxytamoxifen is added. 4-hydroxytamoxifen disrupts the interactions and allows dCas parts to enter the nucleus (Figure 1G). In the nucleus, they complement each other to form a functional complex. Such systems were shown to induce ∼100-fold gene activation, which was ∼2.5-fold less than achievable with non-split versions of dCas9-VPR systems (Nguyen et al., 2016).
Borrowed from the world of digital electronics, logic gates are a key abstraction used to describe and design genetic circuits (see Figure 2). In transcriptional genetic circuits “signals” are rates of transcription initiated at specific promoters. It is simple to see that CRISPRi technology can be straightforwardly used to implement a transcriptional inverter, also called a NOT gate. In this case transcription of sgRNA (controlled by an input promoter) results in the repression of an output gene (Figure 2A) (Didovyk et al., 2016; Tickman et al., 2022). For practical applications, however, the exact quantitative characteristics of NOT gates have to be taken into account (ON/OFF-state expression levels, transfer function shape and dynamic range).
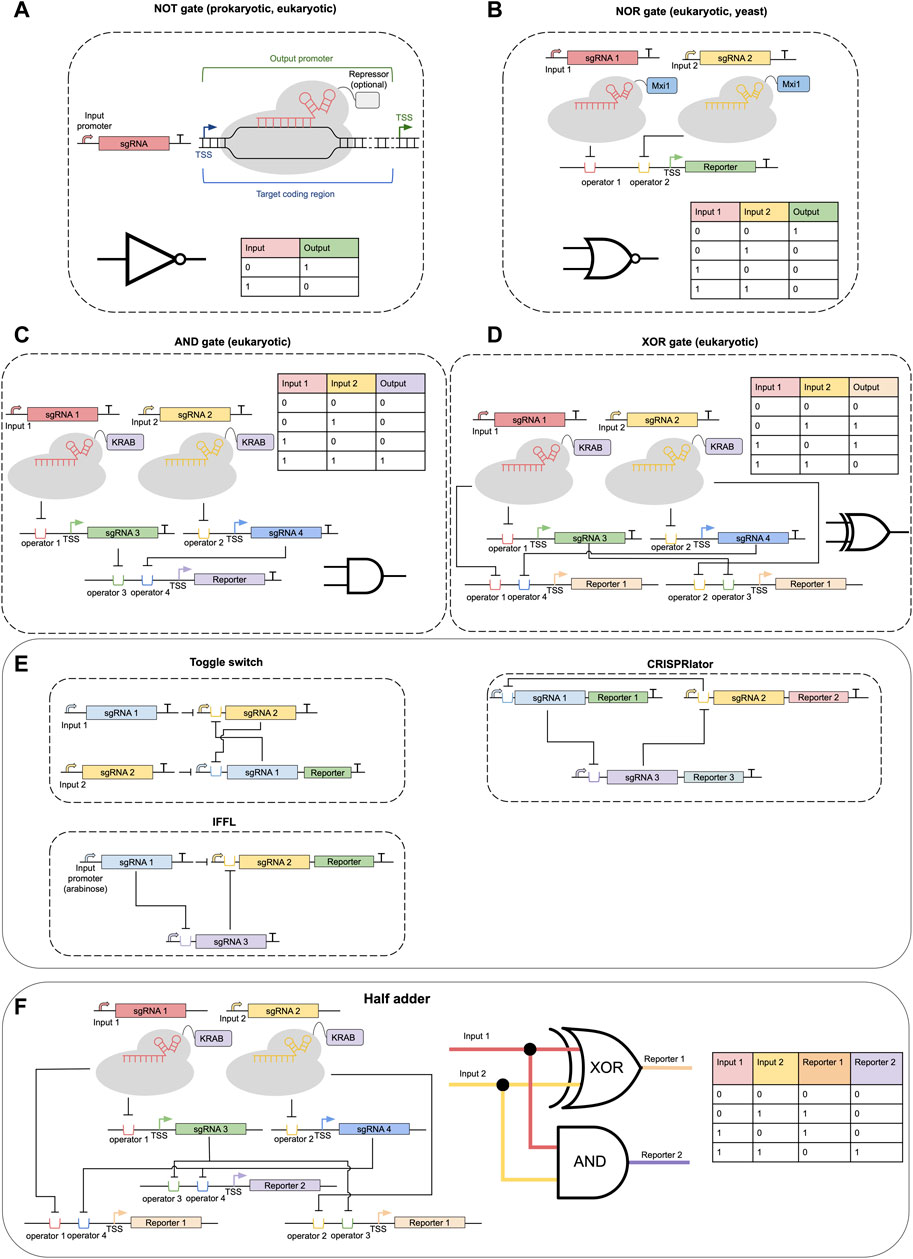
FIGURE 2. CRISPR-dCas-based genetic circuits. (A) A NOT gate based on CRISPRi approach. (B) A typical NOR gate in eukaryotes built with dCas9 fused to a Mxi1 chromatin repressor domain. (C) A variant of AND gate based on the CRISPRi approach. (D) A variant of XOR gate based on the CRISPRi approach. (E) Simple genetic circuits: toggle switch, incoherent feed-forward loop, CRISPRlator. (F) A two-layer CRISPR-dCas-based circuit implementing a half-adder device. Where appropriate truth tables and schematic representations of logic gates are shown.
Of more practical interest is the creation of gates that have two inputs (AND, OR, AND NOT = NAND, OR NOT = NOR, etc.), since they can be combined to create logic devices of arbitrary complexity. In E. coli, the first NOR gates were implemented by Nielsen and Voigt (2014). In their design, two different input promoters regulate the transcription of identical sgRNAs, which in turn target dCas9 to a single DNA locus near the promoter of an output reporter gene. In eukaryotes, when using dCas-fusions with regulatory domains, greater flexibility is allowed in terms of the exact positions that may be targeted by dCas for gene repression or activation. This allows the use of two different sgRNAs as input signals, which target dCas constructs to the two operator sequences near the promoter of an output gene (see Figure 2B). Such strategies have been successfully implemented in yeast and human HEK-193T cells (Gander et al., 2017; Kim et al., 2019). Potentially, it should be possible to create an OR gate using the CRISPRa approach in a similar way, however, it has not yet been reported in the literature to our knowledge. AND and XOR gates may be implemented as a two-layer circuit by combining NOR and two NOT gates (Figures 2C, D) (Kim et al., 2019). Design of NAND/AND gates is also possible by considering dCas expression itself as one of the transcriptional input signals (Liu et al., 2014; Hofmann et al., 2019), however, such gates are hard to use as modules in multi-layer circuits. Alternatively, a conditional sgRNA approach may be employed to create AND gates driven by only RNA input signals: The conditional sgRNA and trigger RNA that activates it (Hanewich-Hollatz et al., 2019).
One of the promises of CRISPR-dCas technology is the assumed ease to create multi-layer circuits driven by sets of orthogonal sgRNA, where the expression of one specific sgRNA regulates the expression of other sgRNA and so on (see, example circuits in Figures 2E, F). Indeed, such layering has been shown to be feasible. For example, Tickman et al. (2022) have combined CRISPRi and CRISPRa in E.coli to make three-layer systems, including feed-forward loops (FFL). Gander et al. (2017) have constructed genetic circuits in yeast based on NOR gates with up to seven layers. Santos-Moreno et al. (2020) were able recently to implement classical genetic circuits such as a bistable toggle switch and repressilator (termed CRISPRlator) in E. coli using CRISPRi (Figure 1E). And Kim et al. (2019) have developed a platform based on dCas9-KRAB and four sgRNAs that allowed the realization of various one and two-layer circuits in HEK293T mammalian cells, including a half-adder circuit (Figure 2F).
Implementation of multi-layered gene circuits is not without challenges. Some of the key requirements for an effective operation of a multi-layered genetic circuit are the orthogonality of its modules (low crosstalk between signals), compatibility of input and output signals between modules (signal thresholds and dynamic ranges, acceptable levels of transcriptional leak, shape, and quality of signal transfer functions), the robustness of genetic elements functioning with respect to their genetic context (Brophy and Voigt, 2014).
For example, the choice of orthogonal sgRNA libraries and their operator targets is not a straightforward task, although some algorithms have been suggested (Didovyk et al., 2016). An optimally designed sgRNA library should at least take into account 1) potential crosstalk due to dCas-sgRNA tolerance to base pairing mismatches especially in the 5′-end of the spacer and 2) potential off-target binding to the genomic DNA. Moreover, the effectiveness of sgRNA transcription from the respective genetic constructs and its post-transcriptional processing have to be taken into account. In prokaryotic systems, one challenge is transcriptional read through due to the short nature of sgRNAs. To mitigate this problem strong terminators should be used (Nielsen and Voigt, 2014). In eukaryotes, nuclear export signals, the 5′-cap and 3′-poly-A tail, interfere with sgRNA functioning. To mitigate these effects flanking sequences are often introduced into the constructs of sgRNAs that are cleaved after transcription. Cleavage has been achieved through ribozymes (Gander et al., 2017) or the tRNA cleavage system (Kim et al., 2019).
Context-dependent effects, which alter operator performance and gene expression (especially if promoter sequence is altered) are another problem for modular circuit design. These effects may be due to the influence of nearby sequences on promoter melting, RNA hairpins that block ribosome binding sites, or transcriptional read-through (Brophy and Voigt, 2014). In eukaryotes, the influence of chromatin structure and dynamics adds another layer of complexity. In genome-wide screens, it was found that both the dCas9-Mxi1 and dCas9-KRAB constructs strongly suppress transcription when targeted to some regions of DNA, but not others (Gilbert et al., 2013). Nucleosomes are known to impede Cas9 access to the DNA (Horlbeck et al., 2016), so their positioning might be another factor that has to be taken into account.
One other challenge of CRISPR-dCas systems is their toxicity, especially in bacteria (Cho et al., 2018b), likely originating from dCas non-specific binding. Certain engineered dCas proteins may be used to address this problem (see above) (Zhang and Voigt, 2018). We hypothesize that the use of dCas-proteins with longer and more specific PAM sequences may actually be beneficial for genetic circuit design. It might reduce toxicity by lowering off-target binding, while specific PAMs may be easily incorporated into the genetic elements of the circuit, which are anyway designed artificially.
Optimization of dose-response transfer functions and signal thresholds is another challenge. The transfer functions of dCas-based genetic elements depend on many factors, including the concentration of dCas protein, sgRNA and their target DNA site, their on-target and off-target binding efficiencies, interactions of fused effector domains with transcription and chromatin machinery, etc. In the design process, plasmid copy numbers and expression levels of individual components of the gene circuit should be carefully optimized (Nielsen and Voigt, 2014). One drawback of dCas-based transfer functions is their linear-like dose-response characteristics. In conventional transcription genetic circuits, based on repressor proteins, digital-like S-shaped dose-response curves are obtained due to cooperative DNA binding (due to repressor dimerization or DNA looping) (Brophy and Voigt, 2014). dCas-sgRNA complexes per se do not bind cooperatively to the DNA, however, additional factors, such as fusion of chromatin effector domains or depletion of dCas pool due to non-specific binding of genomic DNA (Santos-Moreno et al., 2020) may change the dose-response curve in a favorable direction. This is, for instance, needed for the design of bistable toggle switch circuits (Santos-Moreno et al., 2020).
Despite certain challenges, CRISPR-dCas-based tools offer great flexibility in designing multi-layer synthetic genetic circuits. Their versatility already allowed to create rather complex circuits in bacterial (Tickman et al., 2022), mammalian (Kim et al., 2019), yeast (Gander et al., 2017), and even plant cells (Khan et al., 2022). Such terms as “genetic programs,” “genetic computation,” “complex logic computation” and even “CRISPR/Cas9-based core processor” are now being used to refer to such circuits. We envision that further progress in optimizing and characterizing CRISPR-dCas components should be bridged with attempts at automated in silico design of such circuits with sufficient confidence. Practical applications of dCas-based genetic circuits would require connecting such circuits to effective signal input and output molecular devices. Such technologies are already being developed. For example, promising results have been already obtained in the activation of dCas by soluble extracellular factors using fusion constructs between antibodies, dCas-proteins, and proteases (Schwarz et al., 2017). Taken together, dCas-based genetic circuits are a promising technology that will likely contribute to the development of new cell-based therapeutics, diagnostic devices, sensors, epigenetic therapies, tissue engineering, and development of plants with improved properties and serve as a tool to understand genome regulation in living organisms.
AS wrote the manuscript. RN conceived and prepared figures, contributed to literature analysis. AG contributed to literature analysis. RV contributed to figure preparation and literature analysis. GG contributed to manuscript revision. All of the authors read and approved the final manuscript.
This work was funded by the Russian Ministry of Science and Higher Education grant No. 075-15-2021-1062.
This work used the infrastructure provided by the Interdisciplinary Scientific and Educational School of Moscow University “Molecular Technologies of the Living Systems and Synthetic Biology.” We thank referees for helping us to improve the quality of the manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Anderson, D. A., and Voigt, C. A. (2021). Competitive dCas9 binding as a mechanism for transcriptional control. Mol. Syst. Biol. 17 (11), e10512. doi:10.15252/msb.202110512
Anzalone, A. V., Koblan, L. W., and Liu, D. R. (2020). Genome editing with CRISPR–Cas nucleases, base editors, transposases and prime editors. Nat. Biotechnol. 38 (7), 824–844. doi:10.1038/s41587-020-0561-9
Bao, Z., Jain, S., Jaroenpuntaruk, V., and Zhao, H. (2017). Orthogonal genetic regulation in human cells using chemically induced CRISPR/Cas9 activators. ACS Synth. Biol. 6 (4), 686–693. doi:10.1021/acssynbio.6b00313
Bloom, R. J., Winkler, S. M., and Smolke, C. D. (2014). A quantitative framework for the forward design of synthetic miRNA circuits. Nat. Methods 11 (11), 1147–1153. doi:10.1038/nmeth.3100
Bondarenko, V. A., Steele, L. M., Újvári, A., Gaykalova, D. A., Kulaeva, O. I., Polikanov, Y. S., et al. (2006). Nucleosomes can form a polar barrier to transcript elongation by RNA polymerase II. Mol. Cell 24 (3), 469–479. doi:10.1016/j.molcel.2006.09.009
Brennan, A. M. (2022). Development of synthetic biotics as treatment for human diseases. Synth. Biol. 7 (1), ysac001. doi:10.1093/synbio/ysac001
Brocken, D. J. W., Tark-Dame, M., and Dame, R. T. (2018). dCas9: A versatile tool for epigenome editing. Curr. Issues Mol. Biol. 26, 15–32. doi:10.21775/cimb.026.015
Brophy, J. A. N., and Voigt, C. A. (2014). Principles of genetic circuit design. Nat. Methods 11 (5), 508–520. doi:10.1038/nmeth.2926
Chavez, A., Scheiman, J., Vora, S., Pruitt, B. W., Tuttle, M., Iyer, P. R. E., et al. (2015). Highly efficient Cas9-mediated transcriptional programming. Nat. Methods 12 (4), 326–328. doi:10.1038/nmeth.3312
Cheng, A. W., Jillette, N., Lee, P., Plaskon, D., Fujiwara, Y., Wang, W., et al. (2016). Casilio: A versatile CRISPR-cas9-pumilio hybrid for gene regulation and genomic labeling. Cell Res. 26 (2), 254–257. doi:10.1038/cr.2016.3
Cho, J. H., Collins, J. J., and Wong, W. W. (2018). Universal chimeric antigen receptors for multiplexed and logical control of T cell responses. Cell 173 (6), 1426–1438. doi:10.1016/j.cell.2018.03.038
Cho, S., Choe, D., Lee, E., Kim, S. C., Palsson, B., and Cho, B. K. (2018). High-level dCas9 expression induces abnormal cell morphology in Escherichia coli. ACS Synth. Biol. 7 (4), 1085–1094. doi:10.1021/acssynbio.7b00462
Collias, D., and Beisel, C. L. (2021). CRISPR technologies and the search for the PAM-free nuclease. Nat. Commun. 12 (1), 555. doi:10.1038/s41467-020-20633-y
Cong, L., Ran, F. A., Cox, D., Lin, S., Barretto, R., Habib, N., et al. (2013). Multiplex genome engineering using CRISPR/Cas systems. Science 339 (6121), 819–823. doi:10.1126/science.1231143
de Lange, O., Klavins, E., and Nemhauser, J. (2018). Synthetic genetic circuits in crop plants. Curr. Opin. Biotechnol. 49, 16–22. doi:10.1016/j.copbio.2017.07.003
Del Valle, I., Fulk, E. M., Kalvapalle, P., Silberg, J. J., Masiello, C. A., and Stadler, L. B. (2021). Translating new synthetic biology advances for biosensing into the earth and environmental sciences. Front. Microbiol. 11, 618373. doi:10.3389/fmicb.2020.618373
Didovyk, A., Borek, B., Hasty, J., and Tsimring, L. (2016). Orthogonal modular gene repression in Escherichia coli using engineered CRISPR/Cas9. ACS Synth. Biol. 5 (1), 81–88. doi:10.1021/acssynbio.5b00147
Dong, C., Fontana, J., Patel, A., Carothers, J. M., and Zalatan, J. G. (2018). Author Correction: Synthetic CRISPR-Cas gene activators for transcriptional reprogramming in bacteria. Nat. Commun. 9 (1), 4318. doi:10.1038/s41467-018-06909-4
Du, M., Kodner, S., and Bai, L. (2019). Enhancement of LacI binding in vivo. Nucleic Acids Res. 47 (18), 9609–9618. doi:10.1093/nar/gkz698
Elowitz, M. B., and Leibler, S. (2000). A synthetic oscillatory network of transcriptional regulators. Nature 403 (6767), 335–338. doi:10.1038/35002125
Esvelt, K. M., Mali, P., Braff, J. L., Moosburner, M., Yaung, S. J., and Church, G. M. (2013). Orthogonal Cas9 proteins for RNA-guided gene regulation and editing. Nat. Methods 10 (11), 1116–1121. doi:10.1038/nmeth.2681
Feng, H., Guo, J., Wang, T., Zhang, C., and hui, Xing X. (2021). Guide-target mismatch effects on dCas9–sgRNA binding activity in living bacterial cells. Nucleic Acids Res. 49 (3), 1263–1277. doi:10.1093/nar/gkaa1295
Gander, M. W., Vrana, J. D., Voje, W. E., Carothers, J. M., and Klavins, E. (2017). Digital logic circuits in yeast with CRISPR-dCas9 NOR gates. Nat. Commun. 8 (1), 15459. doi:10.1038/ncomms15459
Gao, Y., Xiong, X., Wong, S., Charles, E. J., Lim, W. A., and Qi, L. S. (2016). Complex transcriptional modulation with orthogonal and inducible dCas9 regulators. Nat. Methods 13 (12), 1043–1049. doi:10.1038/nmeth.4042
Gardner, T. S., Cantor, C. R., and Collins, J. J. (2000). Construction of a genetic toggle switch in Escherichia coli. Nature 403 (6767), 339–342. doi:10.1038/35002131
Gilbert, L. A., Horlbeck, M. A., Adamson, B., Villalta, J. E., Chen, Y., Whitehead, E. H., et al. (2014). Genome-Scale CRISPR-mediated control of gene repression and activation. Cell 159 (3), 647–661. doi:10.1016/j.cell.2014.09.029
Gilbert, L. A., Larson, M. H., Morsut, L., Liu, Z., Brar, G. A., Torres, S. E., et al. (2013). CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell 154 (2), 442–451. doi:10.1016/j.cell.2013.06.044
Groner, A. C., Meylan, S., Ciuffi, A., Zangger, N., Ambrosini, G., Dénervaud, N., et al. (2010). “KRAB–Zinc finger proteins and KAP1 can mediate long-range transcriptional repression through heterochromatin spreading,”PLoS Genet. Editor H. D. Madhani, 6, e1000869.
Guo, L. Y., Bian, J., Davis, A. E., Liu, P., Kempton, H. R., Zhang, X., et al. (2022). Multiplexed genome regulation in vivo with hyper-efficient Cas12a. Nat. Cell Biol. 24 (4), 590–600. doi:10.1038/s41556-022-00870-7
Hanewich-Hollatz, M. H., Chen, Z., Hochrein, L. M., Huang, J., and Pierce, N. A. (2019). Conditional guide RNAs: Programmable conditional regulation of CRISPR/Cas function in bacterial and mammalian cells via dynamic RNA nanotechnology. ACS Cent. Sci. 5 (7), 1241–1249. doi:10.1021/acscentsci.9b00340
Healy, C. P., and Deans, T. L. (2019). Genetic circuits to engineer tissues with alternative functions. J. Biol. Eng. 13, 39. doi:10.1186/s13036-019-0170-7
Hochrein, L. M., Li, H., and Pierce, N. A. (2021). High-performance allosteric conditional guide RNAs for mammalian cell-selective regulation of CRISPR/Cas. ACS Synth. Biol. 10 (5), 964–971. doi:10.1021/acssynbio.1c00037
Hofmann, A., Falk, J., Prangemeier, T., Happel, D., Köber, A., Christmann, A., et al. (2019). A tightly regulated and adjustable CRISPR-dCas9 based AND gate in yeast. Nucleic Acids Res. 47 (1), 509–520. doi:10.1093/nar/gky1191
Horlbeck, M. A., Witkowsky, L. B., Guglielmi, B., Replogle, J. M., Gilbert, L. A., Villalta, J. E., et al. (2016). Nucleosomes impede Cas9 access to DNA in vivo and in vitro. eLife 5, e12677. doi:10.7554/eLife.12677
Jia, H., and Schwille, P. (2019). Bottom-up synthetic biology: Reconstitution in space and time. Curr. Opin. Biotechnol. 60, 179–187. doi:10.1016/j.copbio.2019.05.008
Jinek, M., Chylinski, K., Fonfara, I., Hauer, M., Doudna, J. A., and Charpentier, E. (2012). A programmable dual-RNA–guided DNA endonuclease in adaptive bacterial immunity. Science 337 (6096), 816–821. doi:10.1126/science.1225829
Jones, D. L., Leroy, P., Unoson, C., Fange, D., Ćurić, V., Lawson, M. J., et al. (2017). Kinetics of dCas9 target search in Escherichia coli. Science 357 (6358), 1420–1424. doi:10.1126/science.aah7084
Kawano, F., Suzuki, H., Furuya, A., and Sato, M. (2015). Engineered pairs of distinct photoswitches for optogenetic control of cellular proteins. Nat. Commun. 6 (1), 6256. doi:10.1038/ncomms7256
Khan, M. A., Herring, G., Oliva, M., Fourie, E., Zhu, J. Y., Johnston, B., et al. (2022). CRISPRi-based circuits for genetic computation in plants. bioRxiv. doi:10.1101/2022.07.01.498372
Kim, H., Bojar, D., and Fussenegger, M. (2019). A CRISPR/Cas9-based central processing unit to program complex logic computation in human cells. Proc. Natl. Acad. Sci. U. S. A. 116 (15), 7214–7219. doi:10.1073/pnas.1821740116
Kim, S. K., Kim, H., Ahn, W. C., Park, K. H., Woo, E. J., Lee, D. H., et al. (2017). Efficient transcriptional gene repression by type V-A CRISPR-cpf1 from eubacterium eligens. ACS Synth. Biol. 6 (7), 1273–1282. doi:10.1021/acssynbio.6b00368
Klanschnig, M., Cserjan-Puschmann, M., Striedner, G., and Grabherr, R. (2022). CRISPRactivation-SMS, a message for PAM sequence independent gene up-regulation in Escherichia coli. Nucleic Acids Res. 50, 10772–10784. doi:10.1093/nar/gkac804
Konermann, S., Brigham, M. D., Trevino, A. E., Joung, J., Abudayyeh, O. O., Barcena, C., et al. (2015). Genome-scale transcriptional activation by an engineered CRISPR-Cas9 complex. Nature 517 (7536), 583–588. doi:10.1038/nature14136
Liu, Y., Zeng, Y., Liu, L., Zhuang, C., Fu, X., Huang, W., et al. (2014). Synthesizing AND gate genetic circuits based on CRISPR-Cas9 for identification of bladder cancer cells. Nat. Commun. 5 (1), 5393. doi:10.1038/ncomms6393
Makarova, K. S., Wolf, Y. I., Iranzo, J., Shmakov, S. A., Alkhnbashi, O. S., Brouns, S. J. J., et al. (2020). Evolutionary classification of CRISPR-cas systems: A burst of class 2 and derived variants. Nat. Rev. Microbiol. 18 (2), 67–83. doi:10.1038/s41579-019-0299-x
Nakamura, M., Srinivasan, P., Chavez, M., Carter, M. A., Dominguez, A. A., La Russa, M., et al. (2019). Anti-CRISPR-mediated control of gene editing and synthetic circuits in eukaryotic cells. Nat. Commun. 10 (1), 194. doi:10.1038/s41467-018-08158-x
Nguyen, D. P., Miyaoka, Y., Gilbert, L. A., Mayerl, S. J., Lee, B. H., Weissman, J. S., et al. (2016). Ligand-binding domains of nuclear receptors facilitate tight control of split CRISPR activity. Nat. Commun. 7 (1), 12009. doi:10.1038/ncomms12009
Nguyen, P. Q., Soenksen, L. R., Donghia, N. M., Angenent-Mari, N. M., de Puig, H., Huang, A., et al. (2021). Wearable materials with embedded synthetic biology sensors for biomolecule detection. Nat. Biotechnol. 39 (11), 1366–1374. doi:10.1038/s41587-021-00950-3
Nielsen, A. A. K., Der, B. S., Shin, J., Vaidyanathan, P., Paralanov, V., Strychalski, E. A., et al. (2016). Genetic circuit design automation. Science 352 (6281), aac7341. doi:10.1126/science.aac7341
Nielsen, A. A. K., and Voigt, C. A. (2014). Multi-input CRISPR/Cas genetic circuits that interface host regulatory networks. Mol. Syst. Biol. 10, 763. doi:10.15252/msb.20145735
Nihongaki, Y., Furuhata, Y., Otabe, T., Hasegawa, S., Yoshimoto, K., and Sato, M. (2017). CRISPR–Cas9-based photoactivatable transcription systems to induce neuronal differentiation. Nat. Methods 14 (10), 963–966. doi:10.1038/nmeth.4430
Petesch, S. J., and Lis, J. T. (2012). Overcoming the nucleosome barrier during transcript elongation. Trends Genet. 28 (6), 285–294. doi:10.1016/j.tig.2012.02.005
Qi, L. S., Larson, M. H., Gilbert, L. A., Doudna, J. A., Weissman, J. S., Arkin, A. P., et al. (2013). Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell 152 (5), 1173–1183. doi:10.1016/j.cell.2013.02.022
Riesenberg, S., Helmbrecht, N., Kanis, P., Maricic, T., and Pääbo, S. (2022). Improved gRNA secondary structures allow editing of target sites resistant to CRISPR-Cas9 cleavage. Nat. Commun. 13 (1), 489. doi:10.1038/s41467-022-28137-7
Robinson, C. M., Short, N. E., and Riglar, D. T. (2022). Achieving spatially precise diagnosis and therapy in the mammalian gut using synthetic microbial gene circuits. Front. Bioeng. Biotechnol. 10, 959441. doi:10.3389/fbioe.2022.959441
Santos-Moreno, J., Tasiudi, E., Stelling, J., and Schaerli, Y. (2020). Multistable and dynamic CRISPRi-based synthetic circuits. Nat. Commun. 11 (1), 2746. doi:10.1038/s41467-020-16574-1
Schwarz, K. A., Daringer, N. M., Dolberg, T. B., and Leonard, J. N. (2017). Rewiring human cellular input–output using modular extracellular sensors. Nat. Chem. Biol. 13 (2), 202–209. doi:10.1038/nchembio.2253
Shakirova, K. M., Ovchinnikova, V. Y., and Dashinimaev, E. B. (2020). Cell reprogramming with CRISPR/Cas9 based transcriptional regulation systems. Front. Bioeng. Biotechnol. 8, 882. doi:10.3389/fbioe.2020.00882
Sternberg, S. H., Redding, S., Jinek, M., Greene, E. C., and Doudna, J. A. (2014). DNA interrogation by the CRISPR RNA-guided endonuclease Cas9. Nature 507 (7490), 62–67. doi:10.1038/nature13011
Tanenbaum, M. E., Gilbert, L. A., Qi, L. S., Weissman, J. S., and Vale, R. D. (2014). A protein-tagging system for signal amplification in gene expression and fluorescence imaging. Cell 159 (3), 635–646. doi:10.1016/j.cell.2014.09.039
Tang, W., Hu, J. H., and Liu, D. R. (2017). Aptazyme-embedded guide RNAs enable ligand-responsive genome editing and transcriptional activation. Nat. Commun. 8 (1), 15939. doi:10.1038/ncomms15939
Tickman, B. I., Burbano, D. A., Chavali, V. P., Kiattisewee, C., Fontana, J., Khakimzhan, A., et al. (2022). Multi-layer CRISPRa/i circuits for dynamic genetic programs in cell-free and bacterial systems. Cell Syst. 13 (3), 215–229.e8. doi:10.1016/j.cels.2021.10.008
Villegas Kcam, M. C., Tsong, A. J., and Chappell, J. (2021). Rational engineering of a modular bacterial CRISPR–Cas activation platform with expanded target range. Nucleic Acids Res. 49 (8), 4793–4802. doi:10.1093/nar/gkab211
Wang, J., Teng, Y., Zhang, R., Wu, Y., Lou, L., Zou, Y., et al. (2021). Engineering a PAM-flexible SpdCas9 variant as a universal gene repressor. Nat. Commun. 12 (1), 6916. doi:10.1038/s41467-021-27290-9
Wang, J. Y., Pausch, P., and Doudna, J. A. (2022). Structural biology of CRISPR–Cas immunity and genome editing enzymes. Nat. Rev. Microbiol. 20, 641–656. doi:10.1038/s41579-022-00739-4
Weinberg, R. L., Veprintsev, D. B., Bycroft, M., and Fersht, A. R. (2005). Comparative binding of p53 to its promoter and DNA recognition elements. J. Mol. Biol. 348 (3), 589–596. doi:10.1016/j.jmb.2005.03.014
Wright, A. V., Sternberg, S. H., Taylor, D. W., Staahl, B. T., Bardales, J. A., Kornfeld, J. E., et al. (2015). Rational design of a split-Cas9 enzyme complex. Proc. Natl. Acad. Sci. U. S. A. 112 (10), 2984–2989. doi:10.1073/pnas.1501698112
Xia, P. F., Ling, H., Foo, J. L., and Chang, M. W. (2019). Synthetic genetic circuits for programmable biological functionalities. Biotechnol. Adv. 37 (6), 107393. doi:10.1016/j.biotechadv.2019.04.015
Xu, X., and Qi, L. S. (2019). A CRISPR–dCas toolbox for genetic engineering and synthetic biology. J. Mol. Biol. 431 (1), 34–47. doi:10.1016/j.jmb.2018.06.037
Zalatan, J. G., Lee, M. E., Almeida, R., Gilbert, L. A., Whitehead, E. H., La Russa, M., et al. (2015). Engineering complex synthetic transcriptional programs with CRISPR RNA scaffolds. Cell 160 (1), 339–350. doi:10.1016/j.cell.2014.11.052
Zetsche, B., Heidenreich, M., Mohanraju, P., Fedorova, I., Kneppers, J., DeGennaro, E. M., et al. (2017). Multiplex gene editing by CRISPR–Cpf1 using a single crRNA array. Nat. Biotechnol. 35 (1), 31–34. doi:10.1038/nbt.3737
Zetsche, B., Volz, S. E., and Zhang, F. (2015). A split-Cas9 architecture for inducible genome editing and transcription modulation. Nat. Biotechnol. 33 (2), 139–142. doi:10.1038/nbt.3149
Keywords: synthetic genetic circuits, CRISPR-dCas, dCas, CRISPRi, CRISPRa, transcription
Citation: Shaytan AK, Novikov RV, Vinnikov RS, Gribkova AK and Glukhov GS (2022) From DNA-protein interactions to the genetic circuit design using CRISPR-dCas systems. Front. Mol. Biosci. 9:1070526. doi: 10.3389/fmolb.2022.1070526
Received: 15 October 2022; Accepted: 05 December 2022;
Published: 14 December 2022.
Edited by:
Philipp Thomas, Imperial College London, United KingdomReviewed by:
James Carothers, University of Washington, United StatesCopyright © 2022 Shaytan, Novikov, Vinnikov, Gribkova and Glukhov. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: A. K. Shaytan, c2hheXRhbl9ha0BtYWlsLmJpby5tc3UucnU=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.