- 1School of Life Science and Technology, Weifang Medical University, Weifang, China
- 2Department of Biology, Vrije Universiteit Brussel, Brussels, Belgium
- 3VIB-VUB Center for Structural Biology, Vrije Universiteit Brussel, Brussels, Belgium
- 4Structural Biology Brussels, Vrije Universiteit Brussel, Brussels, Belgium
- 5SAFE Pharmaceutical Technology Co, Ltd., Beijing, China
The endogenous plasmid pUTI89 harbored by the uropathogenic Escherichia coli (UPEC) strain UTI89 plays an important role in the acute stage of infection. The partitioning gene parB is important for stable inheritance of pUTI89. However, the function of partitioning genes located on the plasmid in pathogenesis of UPEC still needs to be further investigated. In the present study, we observed that disruption of the parB gene leads to a deficiency in biofilm formation in vitro. Moreover, in a mixed infection with the wild type strain and the parB mutant, in an ascending UTI mouse model, the mutant displayed a lower bacterial burden in the bladder and kidneys, not only at the acute infection stage but also extending to 72 hours post infection. However, in the single infection test, the reduced colonization ability of the parB mutant was only observed at six hpi in the bladder, but not in the kidneys. The colonization capacity in vivo of the parB-complemented strain was recovered. qRT-PCR assay suggested that ParB could be a global regulator, influencing the expression of genes located on both the endogenous plasmid and chromosome, while the gene parA or the operon parAB could not. Our study demonstrates that parB contributes to the virulence of UPEC by influencing biofilm formation and proposes that the parB gene of the endogenous plasmid could regulate gene expression globally.
Introduction
Uropathogenic E. coli (UPEC), a major cause of urinary tract infections (UTI) (Emori & Gaynes, 1993), generally express virulence factors such as fimbrial adhesins and iron uptake proteins. Some UPEC strains, such as UTI89, carry a large endogenous virulence plasmid pUTI89. In a previous study, we found that interruption or deletion of parB, which encodes a type Ib partitioning protein, causes loss of the endogenous plasmid at a ratio of about one percent (Song et al., 2015). However, the parA and parAB mutants could harbor the plasmid rather stably. It would be interesting to further investigate the function of parA, parB and parAB in the virulence of UPEC and what genes on the chromosome are influenced by these mutations located on the endogenous plasmid.
Plasmids are extra-chromosomal genetic elements varying in size from several thousands to hundred thousand of base pairs. Plasmids contribute to the plasticity and rearrangement of the bacterial genome by horizontal gene transfer into plasmid-free cells of the same or different species and facilitate bacterial survival in specific niches. In addition, beneficial components on plasmids can be integrated into the bacterial chromosome. The genes on the endogenous plasmids could encode iron-uptake proteins, fimbriae, hemolysin and other factors functioning in symbiosis, pathogenesis and metabolism (Imre et al., 2006).
Most of the large plasmids are harbored in a single copy and under tight control, to avoid loading metabolic burden to the host and reduce the risk of plasmid loss (de la Cueva-Mendez and Pimentel, 2007). The partitioning system is one of the mechanisms to ensure that the plasmids can be stably maintained and inherited in the host (Yao et al., 2007; Salje et al., 2010; McVicker et al., 2019). Most of the known plasmid-encoded partitioning loci include three essential components, an ATPase-like protein (ParA/SopA/ParF or StbA/ParM), a DNA-binding protein (ParB/SopB/ParG or StbB/ParR) (Mohl et al., 2001) and one or more cis-acting centromere-like sites (parS/sopC/parC) (Barilla et al., 2007; Hanai and Arai, 2015). The former two components are encoded in one operon (Ringgaard et al., 2007). The centromere-like site can be bound by the DNA-binding protein, forming a partitioning complex. It was shown that the parABS partition system is responsible for chromosome and plasmid segregation in bacteria and archaea (Guilhas et al., 2020).
According to the ATPase encoded by the parA gene, the partitioning loci were divided into two types (Gerdes et al., 2000). Type I partitioning loci (par1) can be sub-classified into type Ia and type Ib, according to the size of the Walker-type ATPase. Type II partitioning loci (par2) are actin-like ATPases. However, two types of partitioning loci can sometimes reside on an endogenous plasmid (Ebersbach and Gerdes, 2001) such as the virulence plasmid pUTI89 of uropathogenic E. coli UTI89 (Chen et al., 2006), the F-like plasmid pB171 of enteropathogenic E.coli B171 (Tobe et al., 1999) and the IncHI plasmid R27 of Salmonella typhimurium (Sherburne et al., 2000; Lawley and Taylor, 2003). The comprehension of the network including ATP, DNA and the partitioning genes is still far from final and the plasmid partitioning system has been regarded as a simple model to understand the chromosomal segregation (Howard and Gerdes, 2010). Even though we have characterized the parA/B/AB insertional mutants of UTI89 in growth and plasmid stability in vitro, the effects of mutations on biofilm formation, an important concern of bacterial virulence, and the colonization in urinary tract in vivo still need to be further deciphered.
In this study, we removed the antibiotic resistance gene replacing the parA, parB or parAB from the previously constructed mutants (Song, et al., 2015) and constructed a parB-complemented strain by inserting the parB gene and its original promoter into the nadB gene, which is naturally inactive in UTI89 (Li et al., 2012). We observed that disruption of parB leads to a deficiency in biofilm formation in vitro and a reduction in the colonization of mice in vivo, while complementation of parB in trans restores the virulence of the mutant to the level of the wild type strain. In addition, we demonstrated that the parB mutation influences the expression of genes on both plasmid and chromosome. Collectively, this study demonstrates that the endogenous plasmid gene parB contributes to the virulence of UPEC strain UTI89 and shows that the genes are globally influenced by a parB mutation. This study could be helpful to further understand the function of the partitioning genes in bacterial pathogenesis and natural plasmid maintenance.
Materials and methods
Bacterial strains and growth conditions
The bacterial strains and plasmids used in this study are listed in Table 1. E. coli DH5α was used for cloning. The following antibiotics were used at the indicated concentrations: nalidixic acid, 20 μg ml−1; kanamycin, 25 μg ml−1; chloramphenicol 25 μg ml−1, carbenicillin 100 μg ml−1, except if mentioned otherwise.
Plasmid construction
The PCR primers used for amplification of target fragments are listed in Table 2. The PCR reactions were performed using Ex Taq DNA polymerase (Takara Bio) at the appropriate annealing temperature. The plasmid pDONR221-nadBU-p060-cat was generated by the following steps. The parB with its promoter region was amplified with the primer set EcorpP060F-pP060RP1 (Table 2). The cat cassette was obtained with the primer set P1-EcorP2 (Table 2), using pKD3 as the template. Then overlapping PCR was performed with the mixture of above two fragments as template and the primer set EcorpP060F-EcorP2 (Table 2). The resulting fragment, as well as pDONR221-nadBUTI89, which was constructed as pDONR221-nadBK12 (Li et al., 2012), were digested with EcoRI. The digested products were ligated and transformed by electroporation. The positive clones were selected and then further confirmed by PCR and sequencing.
Construction of the complemented strain
Complementation of the parB mutant strain was realized by inserting parB with its promoter region into the nadB gene on the chromosome. The Red recombination system was employed to recombine the linear fragment amplified by the primer set nadBPF-nadBPR (Table 2) with pDONR221-nadBUTI89-p060-cat as the template. The complemented strain UTI89Δp060 nadB::p060cat was confirmed by PCR with the primer set linadB5-linadB6 (Table 2) and sequencing.
Motility assay
Motility assays were performed by picking fresh bacterial colonies with toothpicks onto the surface of LB plates containing 0.3% agar and incubating at 30°C for 16 h. The diameter of cloudy area of bacteria was measured. The motile capability was expressed in the cloudy area percentage of the mutants which was calculated with the formulation that the cloudy area of the wild type strain is divided by that of the transposon insertion mutant (Zdziarski et al., 2008).
Yeast agglutination test
Briefly, agglutination of yeast cells was examined by mixing 25 μl of overnight E. coli culture in LB with 25 μl of a 5% commercial baker’s yeast suspension in PBS (per litre: 10 g NaCl, 0.25 g KCl, 0.1438 g Na2HPO4, 0.25 g KH2PO4; pH 7.4). Occurrence of visible clumping was compared between mutants and wild type strains (Li et al., 2012).
Biofilm formation assay
Biofilm formation assays were carried out using crystal violet staining in 96-well microliter-plates and quantified as previously described (Merritt et al., 2005). Briefly, the bacterium was inoculated in a 5-ml minimal A medium supplemented with nicotinamide and grown to stationary phase. Then these cultures were diluted by 1:100 in minimal A medium supplemented with nicotinamide. Of each dilution 100 μl was plated in sets of three wells in a round-bottom 96 well plate (BD Falcon™ no. 353911, U bottom). The plate was covered and incubated at 30°C for 48 h. The cultures were cautiously removed using a pipette and the wells were washed twice with PBS and then stained for a conventional crystal violet assay. Briefly, biofilms were incubated in aqueous crystal violet solution (0.05% w/v) for 30 min at room temperature followed by four washes with PBS. The biofilms on the wells of the microtiter plates were photographed by a Canon digital camera. To quantify biofilm formation, 150 µl of 96% ethanol (v/v) was added to each well, incubated for 10 min at room temperature and 100 µl of the extracted solution was removed to a new 96-well plate and the optical density was determined at 595 nm.
RNA extraction and cDNA synthesis
Total RNA was extracted from the tissue using TRIzol® Reagent according to the manufacturer’s instructions (Invitrogen) and genomic DNA was removed using DNase I (Takara). Then RNA quality was determined by 2,100 Bioanalyser (Agilent) and quantified using the ND-2000 (NanoDrop Technologies). High-quality RNA (OD260/280 = 1.8–2.2, OD260/230 ≥ 2.0, RIN ≥ 6.5, 28 S:18 S ≥ 1.0, >10 μg) was used for cDNA synthesis. First-strand cDNAs were synthesized using a SuperScript III First-Strand Synthesis SuperMix Kit (Invitrogen).
Quantitative RT-PCR
Quantitative real-time PCR (qRT-PCR) was performed in a LightCycler480 II (Roche) with iQ™ SYBR Green Supermix (Bio-Rad). The reaction was in a total volume of 25 µl with 12.5 µl SYBR Green Supermix and 10-fold diluted cDNA as templates. The reaction conditions were as follows: 95°C for 3 min, 40 cycles at 95°C for 10 s, 55°C for 30 s, 72°C for 10 s, and one cycle at 50°C for 3 min. The assays were carried out in technical duplicates for three biological replicates with a no-template and a no-RT control. The housekeeping gene gapA, dnaE and dld were used for normalization. The relative expression folds were calculated using the 2ΔΔCq method (Livak and Schmittgen, 2001).
Ascending urinary tract infection mouse model
Bacteria for inoculation were grown statically for 24 h at 37°C in LB broth, collected and adjusted to OD660 = 1.5 in PBS (per litre: 10 g NaCl, 0.25 g KCl, 0.1438 g Na2HPO4, 0.25 g KH2PO4; pH 7.4). Eight-week-old female C3H/HeN mice (Harlan, Horst, Netherlands) were anesthetized by Anesketin (Eurovet, Brussels, Belgium) and Rompun (Bayer, Brussels, Belgium) (15 µl of each for one mouse) and infected via transurethral catheterization of 50 µl of bacterial suspension with 107 CFU of a mixture of equal amount of the transposon insertion mutants, specific null mutants or complemented strain and UTI89NalR, which is a spontaneous nalidixic acid resistant mutant of UTI89. At each indicated time point, mice were sacrificed by inhalation of carbon dioxide. The bladders and kidneys were immediately harvested and homogenized using a 5-ml vessel grinder and plain plunger. Serial dilutions were plated onto LB agar, with the appropriate antibiotics when necessary and the colony forming units (CFU) were enumerated.
Statistical method
The Mann-Whitney test was applied for the comparison of the data collected from the single strain assay and the one-tailed paired t-test for the comparison of the data collected from the mixed infection assay. Both analyses were performed using GraphPad software.
Ethical issues
The animal experiments were approved by the Ethical Committee for Animal Experiments of Vrije Universiteit Brussel (project number 06-219-3) and complied with all relevant national legislation and institutional policies.
Results
Insertion in parB and traV reduces biofilm formation of UTI89
In a previous study, we isolated several transposon miniTn5phoA2 (Pattery et al., 1999) insertion mutants with the insertion localized on the endogenous plasmid (Song, et al., 2015). In the present investigation on the influence of these mutations on the in vitro phenotype of UTI89, we discovered that mutant DV8163 and DV8165 (traV) show a relatively low biofilm formation on the flexible plates among the six mutants derived from DV7729 with insertions in the plasmid (Song, et al., 2015). DV8163 only formed around 20% biofilm and DV8165 around 60% (Figure 1). DV8163 harbors miniTn5phoA2 fused into the parB ORF at the end of the parB gene as described previously (Song et al., 2015). The parB gene is the downstream gene within the parAB operon in pUTI89, encoding a DNA-binding protein. The specific parB/p060 knock-out strain DV9920 demonstrated the same biofilm formation as the transposon insertion mutant DV8163 (Figure 1). The complemented strain DV10145, which was consrtucted by reintroduction of parB with its natural promoter sequence into the naturally inactivated nadB of DV9920, recoved the biofilm formation to the level of the wild type strain (Figure 1).
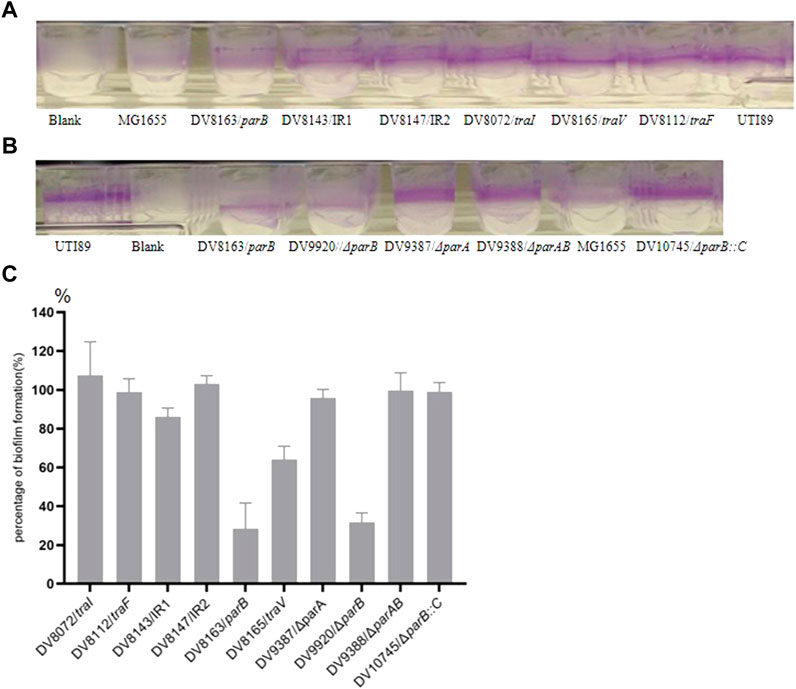
FIGURE 1. Biofilm formation by the transposon insertion mutants. (A) and (B) the biofilm formation on the wells of microtiter plates; (C). Percentage of Biofilm formation by the mutants. The percentage of biofilm formation by each mutant was calculated according to the formula, [(ODmutant-ODblank)/(ODparent strain-ODblank)]×100%. The mutants with the disrupted genes are indicated in the X-coordinate. IR = intergenic region between p017-18 open reading frames.
The mutant DV8165 carries the miniTn5phoA2 insertion at the beginning of the traV gene. The traV gene encodes an outer membrane lipoprotein (Harris et al., 2001), which functions in the assembly of F-type pili. All mutants derived from DV7729 have similar motility and agglutinate yeast cells as UTI89 (Figure 2), including the other two mutants and DV8072 with insertion in traI and DV8112 with the insertion in traF. Both DV8143 and DV8147 harboring the insertion in the intergenic region between the UTI89_p017 and UTI89_p018 at different nucleotide positions have the same phenotypes as UTI89 in the above test.
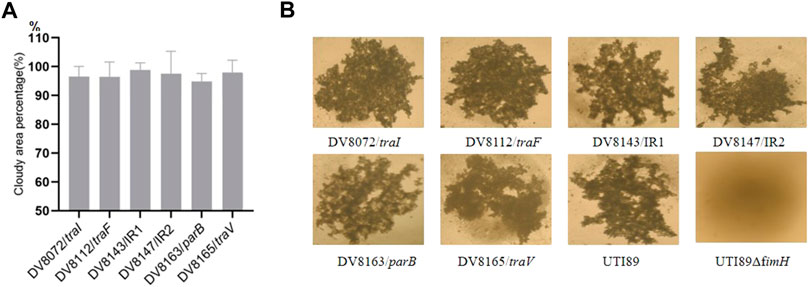
FIGURE 2. Motility and yeast agglutination assay of transponson insertional mutants. (A). The percentage of cloudy area formed by the transposon insertional mutants comparing to that formed by the wild strain. (B). Yeast agglutination by the transposon insertional mutants with the wild type strain as a positive control and fimH deletion strain as a negative control. IR = intergenic region between p017-18 open reading frames.
Reduced colonization of the parB transposon insertion mutant in mice
Since endogenous plasmids have been regarded as virulence factors in bacterial pathogens, we further investigated the consequences of these pUTI89 insertions on the bacterial colonization in the well-established ascending infection mouse model by mixed bacterial challenge. The bacterial burden in the bladder and kidneys of C3H/HeN mice was investigated at 24 h post infection. In the mixed infection, the bacterial burden of DV8163, a miniTn5phoA2 insertion in the parB gene, in the bladder is reduced in seven out of ten mice; there is no obvious difference in the other three mice. The bacterial load of DV8163 in the kidneys is reduced in eight out of ten mice and in the other two mice the load is slightly increased (Figure 3). Furthermore, comparing the number of bacterial colonies from the mice infected by mutant DV8163 and wild type strain demonstrated that the mutant DV8163 has a significantly lower bacterial burden than the wild type strain in the bladders (p = 0.0040) and kidneys (p = 0.0104) in mixed infection (Figure 3).
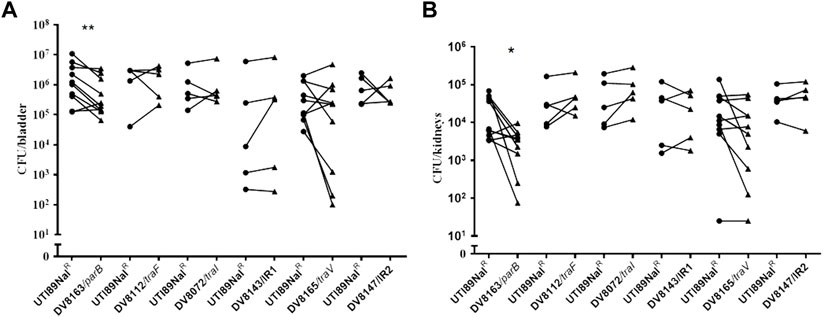
FIGURE 3. In vivo infection of mice with mutant strains. Mixed colonization of mutants and UTI89NalR in bladder (A) and kidneys (B) of mice. The mice were sacrificed at 24 h post infection. *, p < 0.05; **, p < 0.01. Circles stand for the wild type strain and the triangles represent the transposon insertion mutants, which are labeled on the X-axis respectively. One symbol represents the number of the bacteria recovered from the bladder or kidneys from one mouse. IR = intergenic region between p017-18 open reading frames.
No significant difference was detected with any of the other five pUTI89 mutants, including mutants DV8072 (traI), DV8112 (traF), DV8165 (traV), DV8143 and DV8147, in the bladder and kidneys of mice at 24 h post infection (Figure 3). The results from the mutants DV8143 and DV8147 are consistent with the single in vivo colonization test of a p017-18 mutant reported before, even though the previous test was done at six hpi (Cusumano et al., 2010).
We also tested whether plasmid-free UTI89 clones can be recovered from the mice infected with the parB mutant DV8163, which generates one percent plasmid-free bacteria in vitro (Song et al., 2015). The suspension of the grinded bladders or kidneys of mice was spread on LB agar plates without kanamycin and incubated overnight at 37°C. Then the resulting bacterial colonies were replica plated using sterile velvet to LB agar plates with kanamycin. All the tested colonies grew well, implying that all recovered bacteria colonizing the urinary tract of the mice harbored the plasmid. Given that the plasmid can be lost at a ratio of one percent in vitro, we presume that an unknown selection mechanism is exerting a pressure to retain the endogenous plasmid in vivo.
Deletion of parB causes colonization deficiency of UTI89
Since the parB::miniTn5phoA2 mutant DV8163 showed decreased colonization ability in vivo, we investigated whether the transposon insertion mutant of parB as well as the parA and parAB deletion mutants, alone or mixed with the wild type strain, is attenuated in the mouse model. Interestingly, we found that a statistically significant difference between DV9679 (UTI89 ΔparB::cat) and the DV7603 strain, a spontaneous nalidixic acid resistant mutant of UTI89, has emerged at six hpi both in the colonization in the bladder and kidneys and the difference broadened further with the elongation of the co-inoculation time span to 24 and 72 hpi. Even though the major trend is that the DV7603 strain outnumbers the mutant DV9679 at the first three time points, the mutant DV9679 occasionally colonizes slightly more efficiently than the wild type strain in the bladder and kidneys of one to three of the ten mice tested in each group (Figures 4A,B). The mutant DV9679 is outnumbered by the DV7603 strain in the bladder and kidneys of all 10 mice tested at 72 hpi (Figures 4A,B).
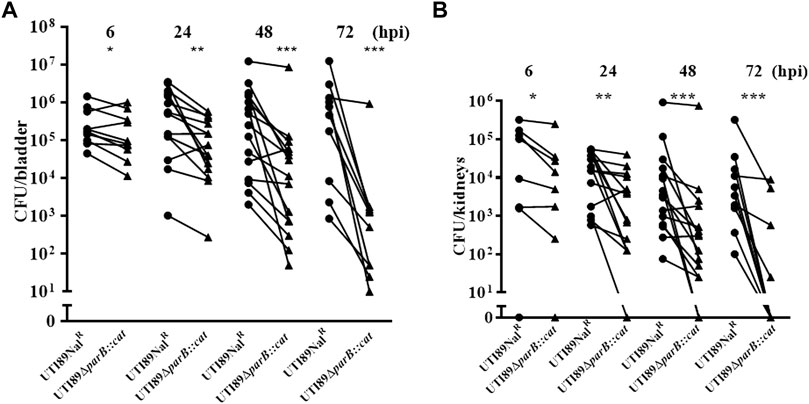
FIGURE 4. Mixed infection of mice with equal amounts of the parB mutant and the wild type strain UTI89NalR. UTI89ΔparB:cat instead of DV9679 was used for simple demonstration the colonization in bladder (A) and kidneys (B) with the deleted genes. *, p < 0.05; **, p < 0.01; ***, p < 0.001. Circles stand for the wild type strain and the triangles represent the transposon insertional mutants, which are labeled on the X-axis, respectively. One symbol represents the number of the bacteria recovered from the bladder or kidneys from one mouse.
Unexpectedly, no statistically significant difference can be detected between the parA mutant DV9383 or the parAB mutant DV9384 and the DV7603 strain either in bladders or in kidneys of the tested mice at any time after infection, including six hpi, 24 hpi, 48 hpi and 72 hpi (Supplementary Figure S1). In conclusion, deletion of parB reduces colonization ability compared to DV7603, but deletion of parA or parAB does not lead to a decrease in colonization of murine bladder or kidneys.
We tested whether the single colonization would show similar differences as in the mixed incubation test in mice. Mice infected with the DV7603 strain or the mutants DV9679, DV9383 or DV9384 were sacrificed at 6, 24, 48 and 72 hpi and the number of CFU recovered from bladder and kidneys was counted. The colonization ability of mutant strains DV9383 and DV9384, with deletions of the parA or parAB genes respectively, was not statistically significantly reduced in the bladder and kidneys at these tested time points (Supplementary Figure S2), compared to the DV7603 strain. In contrast, the mutant DV9679 with a deletion of the parB gene displayed a reduced colonization at six hpi in the bladder (Figure 5), but not at the other three time points, and not in kidneys at all four tested time points (Supplementary Figure S2).
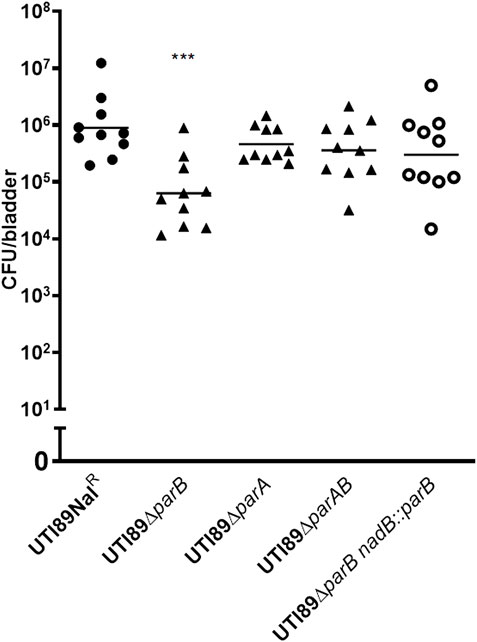
FIGURE 5. Single colonization tests in vivo. The lines in each column indicate average CFUs of bacteria recovered from the bladder. The mice were sacrificed at 6 h hours post infection as mentioned. The filled dots represent the wild type strain, the filled triangles represent the gene deletion mutants and the hollow circle represent the parB-complemented strain, which are labeled on the X-axis, respectively. One symbol represents the number of the bacteria recovered from the bladder or kidneys from one mouse. ***, p < 0.001.
Complementation of the parB mutant restores the colonization ability in mice
To exclude that the above observation would be caused by a polar effect of the parB mutation on the nearby genes and to confirm that the parB gene is a virulence factor following Koch’s postulates, we introduced the parB gene with its promoter region into the nadB gene region on the UTI89 chromosome and performed transurethral inoculation into the bladder of mice with this complemented strain DV9984, alone or together with an equal number of DV7603. The colonization by the mixture was assessed at six hpi, 24 hpi, 48 hpi and 72 hpi, both in bladder and kidneys of ten mice in each group (Figure 6). No statistically significant differences were detected at the four time points (Figure 6). The colonization ability in the urinary tract by DV9984 alone is recovered also in the bladder of mice (Figure 5). This in vivo test demonstrated that the parB gene inserted in the chromosome can complement the attenuated colonization ability in the bladder and kidneys caused by the deletion of the parB gene on the plasmid.
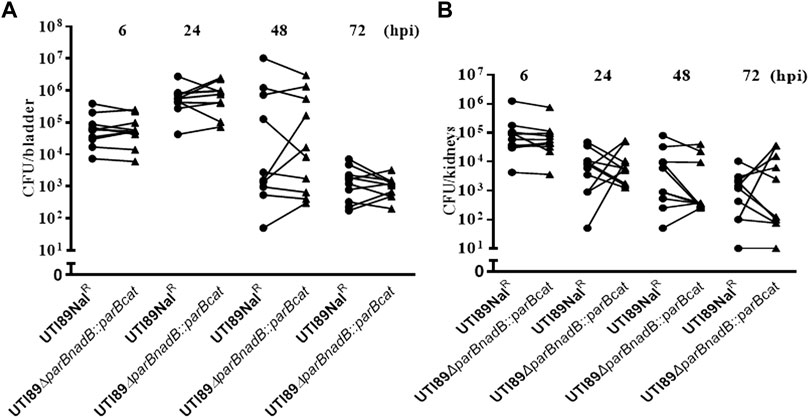
FIGURE 6. The colonization ability of parB-complemented and wild type UTI89 strains in an UTI mouse model. (A) In bladder of mice. (B) In the kidneys of mice. Circles stand for the wild type strain and the triangles represent the parB-complemented strain, which are labeled on the X-axis. One symbol represents the number of the bacteria recovered from the bladder or kidneys from one mouse.
qRT-PCR confirms the influence of deletion of parB on gene expression
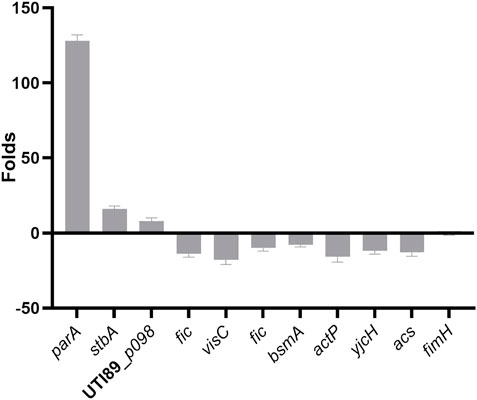
To further illustrate the effects of the parB mutation on the transcription of virulence-related genes of UTI89, RNA was extracted and expression levels were analyzed by qRT-PCR, comparing the UTI89ΔparB mutant with UTI89 at the exponential growth stage. The expression level variation between in the mutant UTI89ΔparB::cat and the wild type strain is shown in Figure 7.
We mainly focused on plasmid genes involved in plasmid stability, separation and stable inheritance and some virulence or virulence-related genes on the chromosome. Deletion of parB (UTI89_p060) causes the expression of parA, p062(stbA) encoding a plasmid segregation protein, UTI89_p098 encoding type I toxin-antitoxin system Hok family toxin increased 128, 16 and eight folds respectively. The changed expression of stbA by the deletion of parB is consistent with the previous report in the mini plasmid model (Gerdes et al., 2000).
Deletion of parB, also causes changes in gene expression on the chromosome. As a binding protein, ParB possibly binds to the regulatory regions of the affected genes or indirectly affects gene expression by influencing chromosomal genes. The gene cvpA (UTI89_C2597) (Shaffer et al., 2017) and visC (UTI89_C3292) (Floyd et al., 2016) were chosen since they are involved in biofilm formation of UPEC. Indeed, the expression level of cvpA and visC in the parB mutant is down-regulated 10–20 fold compared to the wild type strain. Fic (UTI89_C3864) encoding a cell filamentation protein involved in the synthesis of PAB or folate decreased 10-fold in the parB-deleted strain. The Fic protein was shown to be essential for growth of E. coli and involved in regulation of cell division (Komano et al., 1991). BsmA (UTI89_C4789) encoding biofilm stress and motility protein decreased 8-fold in the parB deleted strain. The bsmA mutants are altered in their ability to produce biofilms, in biofilm structure, in cell motility, and in response to pH and oxidative stress (Weber et al., 2010). ActP (UTI89_C4657) encoding acetate permease, YjcH (UTI89_C4658) encoding inner membrane protein and ACS (UTI89_C4659) encoding acetyl-CoA synthetase decreased nearly 16 folds in the parB-deleted strain. It was shown that deletion of the acs-yjcH-actP operon in E. coli not only decreases its cytotoxic level to macrophages, but also attenuates virulence and colonization capability in avian lungs in vivo for colibacillosis infection models (Zhuge et al., 2019). However, as a main virulence factors of UPEC, the expression of FimH (UTI89_C5017), the type-1 fimbrial tip adhesin, is not influenced significantly by mutation of parB.
Discussion
We systematically tested virulence contribution of the parAB operon and the single genes parA and parB by comparing the virulence of the mutants with the wild type strain. The parB gene is especially important for the bacterial colonization. It was reported that parAB enhances the stability of a mini-R plasmid more than stbAB does (Ebersbach and Gerdes, 2001). In our previous study, we confirmed that deletion of parB could cause endogenous plasmid loss (Song, et al., 2015). However, the mutation did not influence the growth of the mutant strain without antibiotic pressure (Song, et al., 2015). To our knowledge, no report has been presented yet about the function of the component genes in the partitioning operon in the pathogenesis of uropathogenic E. coli. Here, we report that the parB gene contributes to virulence of UPEC in a mouse model potentially through biofilm formation and global regulation of gene expression.
Although several mutants (traV, traI and traF) involved in pilus assembly were tested, only the traV mutation shows a reduction of biofilm formation up to 60% compared to the parental strain (Figure 1). This could result from either the subcellular location of the gene products or the potential polar effects of the transposon insertion into traV (DV8165), since many genes involved in pilus assembly are located downstream of the insertion. However, the colonization ability of the traV mutant in the mouse model is equal to the wild type strain.
These data on the colonization of the parB mutant in vivo in single strain infection in our study are consistent with a previous investigation, using plasmid-free cells or deletion mutants of other potential virulence genes on the plasmid (Cusumano et al., 2010). In that report, the plasmid-free bacterium was isolated by treatment with ethidium bromide and also carried a stbAB deletion. The colonization ability of this plasmid-free UTI89 was greatly reduced in vivo (Cusumano et al., 2010). However, in our study, we did not observe the same reduction in the bladder or kidneys with the plasmid-free strain isolated without chemical treatment and not carrying the stbAB deletion (Supplementary Figure S3). Both studies confirmed that some of the plasmid components (p028-p030 and p062-p063 in the previous study) including the parB gene are important factors for virulence in the acute colonization stage in an UTI mouse model. However, in our study the parAB (p060-p061) mutant (Supplementary material Figures S1, S2) displays a different colonization behavior from the p062-p063 (stbAB) deletion mutant (Cusumano et al., 2010), considering that both operons function in plasmid maintenance and stability. Combination of our results and the previous study that deletion of parAB causes the plasmids to be less stable than deletion of p062-p063 (stbAB) when the corresponding fragments were inserted in a mini-R plasmid (Ebersbach and Gerdes, 2001) suggests that the parAB system is not only crucial for stable plasmid inheritance, but also would have an important function in vivo colonization. So, it is reasonable to declare that the parB gene of the parAB operon plays a role in the virulence of UPEC, both in biofilm formation in vitro and in colonization in mouse.
We used qRT-PCR to investigate the impact of the parB deletion on genes encoding known virulence factors, the proteins contributing to plasmid stability, and the component essential for biofilm formation. The expression of the adhesin FimH, one of the most important virulence factors of UTI89 is not influenced by the parB mutation. Genes relevant to plasmid stability, such as parA and stbAB are upregulated, which means that these genes could be in a fluctuating way expressed and accelerate plasmid instability when parB is absent. The gene parA product could accumulate in bacterial cell since it cannot be recruited to parB-parS complex to function in proper plasmid segregation. Indeed, the interaction of the ParA and ParB components, which leads to proper separation of plasmid copies, has been extensively studied in plasmids and bacteria of the proteobacteria phylum. These studies provide the foundation for filament- and non-filament-based models of plasmid and bacterial chromosome segregation (Lioy et al., 2015). Indeed, a recent study shows that ParB binds CTP and hydrolyzes it upon interaction with centromer-like parS motifs (Osorio-Valeriano et al., 2019). The CTPase activity of ParB is critical for the partition complex formation in vivo and potentially it could mediate a regulatory link between CTP-dependent metabolic pathways and DNA segregation (Taylor et al., 2021).
Taken together, our study presented here determined that parB is also involved in the virulence of UPEC, and not only in plasmid segregation and potentially involved in the regulation of gene expression in UPEC. Our results shed light on the contribution of the partitioning gene parB to the virulence and gene regulation of UPEC and identify novel virulence genes.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
Ethics statement
The animal study was reviewed and approved by Ethical Committee for Animal Experiments of Vrije Universiteit Brussel.
Author contributions
NS, QW and ZL did the experimental work and wrote the first draft. ZL, HDG and J-PH corrected the manuscript. J-PH, HDG and ZL designed the research. J-PH supervised the work. All authors contributed to the corrections and proof of the final manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (Grant Number 31873014), by Shandong Project for Talents Introduction and Development on Youth Innovation Team of Higher Education, China, and by the China Scholarship Council—Vrije Universiteit Brussel (CSC—VUB) joint program and the LISUM (Linking Sino—European Universities through Mobility) project of the Erasmus Mundus External Cooperation Window program.
Conflict of Interest
Authors QW and ZL were employed by SAFE Pharmaceutical Technology Co., Ltd.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmolb.2022.1053888/full#supplementary-material
SUPPLEMENTARY FIGURE S1 | Mixed infection test of the parA or parAB mutant with the wild type strain UTI89NalR. UTI89ΔparA::Km instead of DV9383, UTI89ΔparAB::Km instead of DV9384 are used for simple demonstration the colonization with the deleted genes. No significant differences were observed either in bladder or in kidneys by the parA or parAB mutant comparing to the wild type strain. Circles stands for the wild type strain and the triangles represent the parB-complemented strain, which are labeled on the X-axis. One symbol represents the number of the bacteria recovered from the bladder or kidneys from one mouse.
SUPPLEMENTARY FIGURE S2 | Single colonization tests in vivo. Horizontal lines in each column indicate average CFUs of bacteria recovered from the bladder (A) or kidneys (B). The mice were sacrificed at the indicated time points post infection as mentioned in the figure. Circles standfor the wild type strain and the triangles represent the transposon insertion mutants, which are labeled on the X-axis respectively. One symbol represents the number of the bacteria recovered from the bladder or kidneys from one mouse.
SUPPLEMENTARY FIGURE S3 | Single colonization tests of UTI89 and UTI89 plasmid-free strain in vivo. No differences were observed in colonization capacity between two strains in bladder and kidneys of the infected mice. Circles stand for the wild type strain and the squares represents the plasmidfree UTI89, which are labeled on the X-axis respectively. One symbol represents the number of the bacteria recovered from the bladder or kidneys from one mouse.
References
Barilla, D., Carmelo, E., and Hayes, F. (2007). The tail of the ParG DNA segregation protein remodels ParF polymers and enhances ATP hydrolysis via an arginine finger-like motif. Proc. Natl. Acad. Sci. U. S. A. 104, 1811–1816. doi:10.1073/pnas.0607216104
Chen, S. L., Hung, C. S., Xu, J., Reigstad, C. S., Magrini, V., Sabo, A., et al. (2006). Identification of genes subject to positive selection in uropathogenic strains of Escherichia coli: A comparative genomics approach. Proc. Natl. Acad. Sci. U. S. A. 103, 5977–5982. doi:10.1073/pnas.0600938103
Cusumano, C. K., Hung, C. S., Chen, S. L., and Hultgren, S. J. (2010). Virulence plasmid harbored by uropathogenic Escherichia coli functions in acute stages of pathogenesis. Infect. Immun. 78, 1457–1467. doi:10.1128/IAI.01260-09
Datsenko, K. A., and Wanner, B. L. (2000). One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U. S. A. 97, 6640–6645. doi:10.1073/pnas.120163297
de la Cueva-Mendez, G., and Pimentel, B. (2007). Gene and cell survival: Lessons from prokaryotic plasmid R1. EMBO Rep. 8, 458–464. doi:10.1038/sj.embor.7400957
Ebersbach, G., and Gerdes, K. (2001). The double par locus of virulence factor pB171: DNA segregation is correlated with oscillation of ParA. Proc. Natl. Acad. Sci. U. S. A. 98, 15078–15083. doi:10.1073/pnas.261569598
Emori, T. G., and Gaynes, R. P. (1993). An overview of nosocomial infections, including the role of the microbiology laboratory. Clin. Microbiol. Rev. 6, 428–442. doi:10.1128/CMR.6.4.428
Floyd, K. A., Mitchell, C. A., Eberly, A. R., Colling, S. J., Zhang, E. W., DePas, W., et al. (2016). The UbiI (VisC) aerobic ubiquinone synthase is required for expression of type 1 pili, Biofilm Formation, and pathogenesis in uropathogenic Escherichia coli. J. Bacteriol. 198, 2662–2672. doi:10.1128/JB.00030-16
Gerdes, K., Moller-Jensen, J., and Bugge Jensen, R. (2000). Plasmid and chromosome partitioning: Surprises from phylogeny. Mol. Microbiol. 37, 455–466. doi:10.1046/j.1365-2958.2000.01975.x
Guilhas, B., Walter, J. C., Rech, J., David, G., Walliser, N. O., Palmeri, J., et al. (2020). ATP-driven separation of liquid phase condensates in bacteria. Mol. Cell 79, 293–303. doi:10.1016/j.molcel.2020.06.034
Hanai, R., and Arai, Y. (2015). New roles of DNA and SopB in polymerization of SopA of Escherichia coli F plasmid. J. Biochem. 157, 459–466. doi:10.1093/jb/mvv003
Harris, R. L., Hombs, V., and Silverman, P. M. (2001). Evidence that F-plasmid proteins TraV, TraK and TraB assemble into an envelope-spanning structure in Escherichia coli. Mol. Microbiol. 42, 757–766. doi:10.1046/j.1365-2958.2001.02667.x
Howard, M., and Gerdes, K. (2010). What is the mechanism of ParA-mediated DNA movement? Mol. Microbiol. 78, 9–12. doi:10.1111/j.1365-2958.2010.07316.x
Imre, A., Olasz, F., Kiss, J., and Nagy, B. (2006). A novel transposon-based method for elimination of large bacterial plasmids. Plasmid 55, 235–241. doi:10.1016/j.plasmid.2005.11.006
Komano, T., Utsumi, R., and Kawamukai, M. (1991). Functional analysis of the fic gene involved in regulation of cell division. Res. Microbiol. 142, 269–277. doi:10.1016/0923-2508(91)90040-h
Lawley, T. D., and Taylor, D. E. (2003). Characterization of the double-partitioning modules of R27: Correlating plasmid stability with plasmid localization. J. Bacteriol. 185, 3060–3067. doi:10.1128/jb.185.10.3060-3067.2003
Li, Z. L., Bouckaert, J., Deboeck, F., De Greve, H., and Hernalsteens, J. P. (2012). Nicotinamide dependence of uropathogenic Escherichia coli UTI89 and application of nadB as a neutral insertion site. Microbiology 158, 736–745. doi:10.1099/mic.0.052043-0
Lioy, V. S., Volante, A., Soberon, N. E., Lurz, R., Ayora, S., and Alonso, J. C. (2015). ParAB partition dynamics in firmicutes: Nucleoid bound ParA captures and tethers ParB-plasmid complexes. Plos One 10, e0131943. doi:10.1371/journal.pone.0131943
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25, 402–408. doi:10.1006/meth.2001.1262
McVicker, G., Hollingshead, S., Pilla, G., and Tang, C. M. (2019). Maintenance of the virulence plasmid in Shigella flexneri is influenced by Lon and two functional partitioning systems. Mol. Microbiol. 111, 1355–1366. doi:10.1111/mmi.14225
Merritt, J. H., Kadouri, D. E., and O'Toole, G. A. (2005). Growing and analyzing static biofilms. Curr. Protoc. Microbiol. 1, 1. doi:10.1002/9780471729259.mc01b01s00
Mohl, D. A., Easter, J., and Gober, J. W. (2001). The chromosome partitioning protein, ParB, is required for cytokinesis in Caulobacter crescentus. Mol. Microbiol. 42, 741–755. doi:10.1046/j.1365-2958.2001.02643.x
Mulvey, M. A., Schilling, J. D., and Hultgren, S. J. (2001). Establishment of a persistent Escherichia coli reservoir during the acute phase of a bladder infection. Infect. Immun. 69, 4572–4579. doi:10.1128/IAI.69.7.4572-4579.2001
Osorio-Valeriano, M., Altegoer, F., Steinchen, W., Urban, S., Liu, Y., Bange, G., et al. (2019). ParB-type DNA segregation proteins are CTP-dependent molecular switches. Cell 179, 1512–1524. e1515. doi:10.1016/j.cell.2019.11.015
Pattery, T., Hernalsteens, J. P., and De Greve, H. (1999). Identification and molecular characterization of a novel Salmonella enteritidis pathogenicity islet encoding an ABC transporter. Mol. Microbiol. 33, 791–805. doi:10.1046/j.1365-2958.1999.01526.x
Ringgaard, S., Ebersbach, G., Borch, J., and Gerdes, K. (2007). Regulatory cross-talk in the double par locus of plasmid pB171. J. Biol. Chem. 282, 3134–3145. doi:10.1074/jbc.M609092200
Salje, J., Gayathri, P., and Lowe, J. (2010). The ParMRC system: Molecular mechanisms of plasmid segregation by actin-like filaments. Nat. Rev. Microbiol. 8, 683–692. doi:10.1038/nrmicro2425
Shaffer, C. L., Zhang, E. W., Dudley, A. G., Dixon, B. R. E. A., Guckes, K. R., Breland, E. J., et al. (2017). Purine biosynthesis metabolically constrains intracellular survival of uropathogenic Escherichia coli. Infect. Immun. 85, 004711. doi:10.1128/IAI.00471-16
Sherburne, C. K., Lawley, T. D., Gilmour, M. W., Blattner, F. R., Burland, V., GrotbEck, E., et al. (2000). The complete DNA sequence and analysis of R27, a large IncHI plasmid from Salmonella typhi that is temperature sensitive for transfer. Nucleic Acids Res. 28, 2177–2186. doi:10.1093/nar/28.10.2177
Song, N., Xu, J., Li, Z., and Hernalsteens, J. P. (2015). Curing a large endogenous plasmid by single substitution of a partitioning gene. Plasmid 82, 10–16. doi:10.1016/j.plasmid.2015.06.001
Taylor, J. A., Seol, Y., Budhathoki, J., Neuman, K. C., and Mizuuchi, K. (2021). CTP and parS coordinate ParB partition complex dynamics and ParA-ATPase activation for ParABS-mediated DNA partitioning. Elife 10, e65651. doi:10.7554/eLife.65651
Tobe, T., Hayashi, T., Han, C. G., Schoolnik, G. K., Ohtsubo, E., and Sasakawa, C. (1999). Complete DNA sequence and structural analysis of the enteropathogenic Escherichia coli adherence factor plasmid. Infect. Immun. 67, 5455–5462. doi:10.1128/IAI.67.10.5455-5462.1999
Weber, M. M., French, C. L., Barnes, M. B., Siegele, D. A., and McLean, R. J. C. (2010). A previously uncharacterized gene, yjfO (bsmA), influences Escherichia coli biofilm formation and stress response. Microbiol. Read. 156, 139–147. doi:10.1099/mic.0.031468-0
Woodcock, D. M., Crowther, P. J., Doherty, J., JefferSon, S., Decruz, E., Noyer-WeidnerM., , et al. (1989). Quantitative evaluation of Escherichia coli host strains for tolerance to cytosine methylation in plasmid and phage recombinants. Nucleic Acids Res. 17, 3469–3478. doi:10.1093/nar/17.9.3469
Yao, S., Helinski, D. R., and Toukdarian, A. (2007). Localization of the naturally occurring plasmid ColE1 at the cell pole. J. Bacteriol. 189, 1946–1953. doi:10.1128/JB.01451-06
Zdziarski, J., Svanborg, C., Wullt, B., Hacker, J., and Dobrindt, U. (2008). Molecular basis of commensalism in the urinary tract: Low virulence or virulence attenuation? Infect. Immun. 76 (2), 695–703. doi:10.1128/IAI.01215-07
Keywords: parB, uropathogenic Escherichia coli, biofilm, virulence, regulation
Citation: Song N, De Greve H, Wang Q, Hernalsteens J-P and Li Z (2022) Plasmid parB contributes to uropathogenic Escherichia coli colonization in vivo by acting on biofilm formation and global gene regulation. Front. Mol. Biosci. 9:1053888. doi: 10.3389/fmolb.2022.1053888
Received: 26 September 2022; Accepted: 28 November 2022;
Published: 16 December 2022.
Edited by:
Han Yao, Shanghai Jiao Tong University, ChinaReviewed by:
Preeti Srivastava, Indian Institute of Technology Delhi, IndiaAdrian F. Alvarez, Institute of Cellular Physiology, National Autonomous University of Mexico, Mexico
Copyright © 2022 Song, De Greve, Wang, Hernalsteens and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jean-Pierre Hernalsteens, SmVhbi1QaWVycmUuSGVybmFsc3RlZW5zQHZ1Yi5iZQ==, Zhaoli Li, bGl6aGFvbGlAc2FmZWdscC5jb20=
 Ningning Song
Ningning Song Henri De Greve3,4
Henri De Greve3,4 Zhaoli Li
Zhaoli Li