- 1Department of Anatomy, School of Basic Medicine, Anhui Medical University, Hefei, Anhui, China
- 2High Magnetic Field Laboratory, Hefei Institutes of Physical Science, Chinese Academy of Sciences, Science Island, Hefei, China
- 3Institutes of Physical Science and Information Technology, Anhui University, Hefei, Anhui, China
- 4Science Island Branch of Graduate School, University of Science and Technology of China, Hefei, Anhui, China
- 5School of Life Sciences, Peking University, Beijing, China
- 6Department of Biological Chemistry and Molecular Pharmacology, Harvard Medical School, Boston, MA, United States
- 7State Key Laboratory of Molecular Reaction Dynamics, Dalian Institute of Chemical Physics, Chinese Academy of Sciences, Dalian, China
- 8International Magnetobiology Frontier Research Center, Science Island, Hefei, China
Iron-sulfur proteins play essential roles in a wide variety of cellular processes such as respiration, photosynthesis, nitrogen fixation and magnetoreception. The stability of iron-sulfur clusters varies significantly between anaerobic and aerobic conditions due to their intrinsic sensitivity to oxygen. Iron-sulfur proteins are well suited to various practical applications as molecular redox sensors or molecular “wires” for electron transfer. Various technologies have been developed recently using one particular iron-sulfur protein, MagR, as a magnetic tag. However, the limited protein stability and low magnetic sensitivity of MagR hindered its wide application. Here in this study, the iron-sulfur binding site of pigeon clMagR was rationally re-designed. One such mutation, T57C in pigeon MagR, showed improved iron-sulfur binding efficiency and higher iron content, as well as prolonged thermostability. Thus, clMagRT57C can serve as a prototype for further design of more stable and sensitive magnetic toolbox for magnetogenetics in the future.
Introduction
Iron-sulfur clusters are essential cofactors consisting of ferrous (Fe2+) or ferric (Fe3+) iron and sulfide (S2-) ions and comprise the largest class of metalloproteins present in almost all organisms. The most common types of iron-sulfur clusters are rhombic [2Fe-2S], cubic [3Fe-4S] and cubic [4Fe-4S] (Kiley and Beinert, 2003; Hinton et al., 2022). Iron-sulfur clusters are usually bind to cysteine (Cys) residues through iron ions. In addition, histidine (His), aspartic acid (Asp) and glutamic acid (Glu) residues can also serve as coordination bonds to iron-sulfur clusters and exhibit unique functions (Wiley et al., 2007; Zeng et al., 2008; Gruner et al., 2011; Volbeda et al., 2019).
Electron transfer is perhaps the most obvious function which has been identified since early 1960s in photosynthesis and respiration systems (Beinert, 1960; Mortenson et al., 1962). Later, a wide variety of biological functions have emerged for these clusters, including nitrogen fixation, DNA replication and repair (Kiley and Beinert, 2003; Johnson et al., 2005; Fontecave, 2006; Mettert and Kiley, 2015; Rouault, 2015). In 2015, Qin et al. reported an iron-sulfur protein MagR (Magnetoreceptor, originally named IscA) played essential roles in animal magnetoreception through the interaction with cryptochrome (Cry) (Qin et al., 2016). The MagR/Cry-based biocompass model (Qin et al., 2016; Xie, 2022) combined the concept of both the magnetite-based mechanism (Hsu et al., 2007; Eder et al., 2012; Wiltschko and Wiltschko, 2013; Monteil and Lefevre, 2020; Schuler et al., 2020) and radical-pair based mechanism (Ritz et al., 2000; Liedvogel et al., 2007; Gegear et al., 2008; Lau et al., 2012; Wiltschko and Wiltschko, 2014; Xu et al., 2021), thus provided an solution for both polarity detection and inclination detection. The iron-sulfur cluster of MagR is required for the assembly of the MagR/Cry protein complex (Qin et al., 2016), and has been suggested to mediate the long range intermolecular electron transport chain in MagR/Cry complex (Qin et al., 2016; Xie, 2022), and contribute to the intrinsic magnetic moment of MagR and MagR/Cry complex (Guo et al., 2021). Furthermore, two different types of iron-sulfur clusters, [2Fe-2S] and [3Fe-4S], have been identified in MagR and may serve as a magnetic switch to modulate the magnetic property of MagR (Guo et al., 2021).
In pace with the growing interest in elucidating the underlying mechanism of MagR as a putative magnetoreceptor, various technologies have been developed recently using MagR as a magnetic tag (Jiang et al., 2017; Xue et al., 2020; Kang et al., 2021). Biological manipulation via magnetic fields, which is also refer to as magnetogenetics, has been a pre-eminent goal for scientists. It is especially appealing for in vivo applications since magnetic field can penetrate deep into tissues, which allow non-invasive remote modulation of biological processes possible. This is achieved by fusing a magnetic tag to a mechanically sensitive ion channel such as TRPV4, and then applying magnetic field to exert magnetic force on the channel to open up the associated channel and activate biological systems, such as neuronal functions. To engineer MagR as a suitable actuator for magnetogenetics, a rationally re-designed MagR with improved stability and higher magnetic sensitivity is required to overcome the thermal fluctuations at room temperature. Therefore, there has been a major effort to re-design a better MagR for applications, for example, a single-chain tetramer MagR was designed as a building block to increase the protein self-assembly efficiency and thus increase the magnetic sensitivity by polymerization (Yang et al., 2022). Another particular approach is to design a more stable binding site to host the iron-sulfur cluster since iron-sulfur clusters are critical for the magnetism of MagR, as described in this paper.
The stability of iron-sulfur clusters varies significantly between anaerobic and aerobic conditions due to their intrinsic sensitivity to oxygen. When they are harbored in proteins, the stability is primarily dependent on the microenvironments within a biomolecular structure, such as the oxygen accessibility to the clusters, and coordination bonds of the clusters, et al. (Meyer, 2008). Iron-sulfur clusters in some proteins (e.g., thermophilic Fd, a thermostable [2Fe-2S] ferredoxin from hyperthermophilic bacterium Aquifex aeolicus) are unusually stable for weeks even exposed in air (Mitou et al., 2003), while in many or in the majority of iron-sulfur proteins are very sensitive to oxygen and only stable for tens of seconds (e.g., nitrogenase) (Eady et al., 1972). As for pigeon (Columba livia) MagR (clMagR), the iron-sulfur cluster is normally stable for 4–5 days at room temperature (298 K) and 7 days at 4°C (277 K).
To further stabilize the iron-sulfur cluster binding in MagR, here in this study, the binding site was rationally re-designed based on the 3D structural model of MagR. Two hotspot regions located close to the iron-sulfur binding site and around E128 and T57/R58 respectively were identified. Site-directed mutagenesis were then designed aiming to stabilize the iron-sulfur cluster binding. One such mutation, T57C in clMagR, has been identified with increased iron-sulfur cluster binding half-life. The prolonged thermostability of clMagRT57C makes it suitable to serve as a prototype for further fine-tuning as a magnetic actuator for controlling biological processes in the future.
Results
E128 is not a potential ligand of iron-sulfur cluster in pigeon MagR
Three highly conserved cysteines (Cys60, Cys124, Cys126) have been identified to bind two types of iron-sulfur clusters, [2Fe-2S] and [3Fe-4S], in freshly purified pigeon MagR in aerobic conditions (Guo et al., 2021) (Figures 1A,B). A three-dimensional structural model of pigeon MagR was initially generated based on bacterial homologous IscA structure (PDB ID:1R94) as described previously (Qin et al., 2016), then fine-tuned using the MODELLER homology modeling package (Yang et al., 2022). A careful analysis of the structural model revealed a hotspot residue, glutamic acid (E128), located close (around 3.6 Å) to the iron-sulfur cluster (Figure 1C and Supplementary Figure S1), leading to the question if E128A serve as a potential ligand of iron-sulfur cluster? May or may not, would it be possible to design a more stable binding site by mutating E128 to cysteine residue for iron-sulfur cluster coordination?
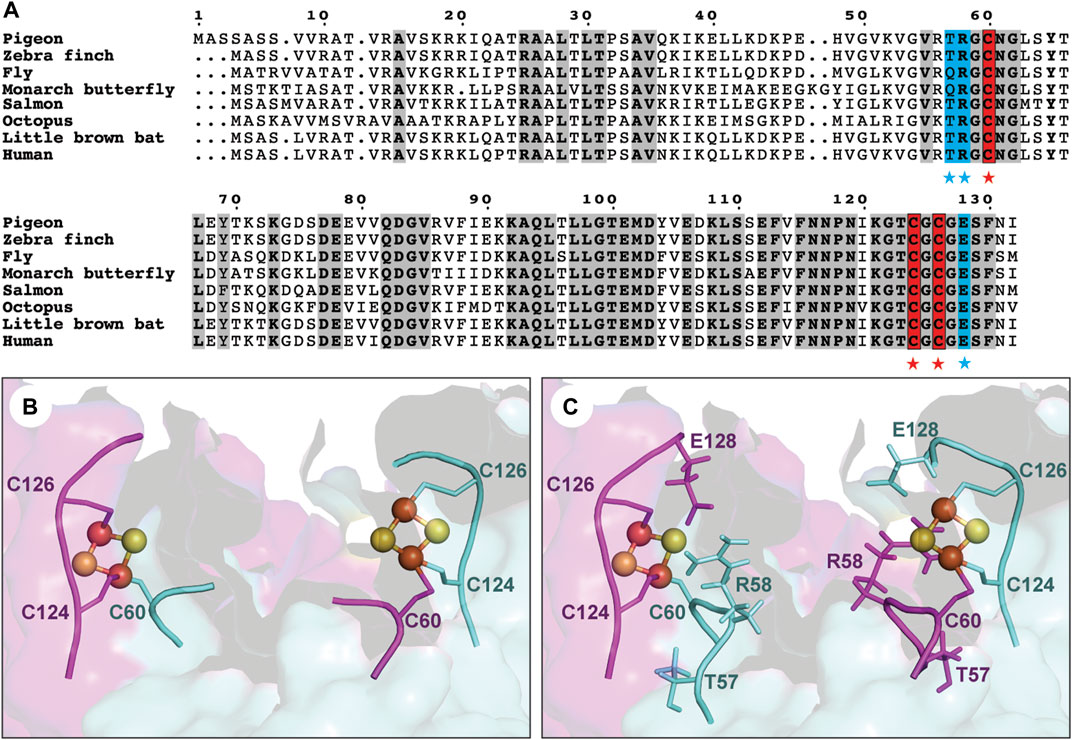
FIGURE 1. Sequence conservation and structural feature of the iron-sulfur cluster binding site of clMagR. (A) Sequence alignment of MagR from eight representative species. Three conserved cysteines (C60, C124 and C126) forming the iron-sulfur cluster binding site are highlighted in red background and marked by red stars. Residues are mutated in this study are shown in blue background and marked by blue stars. The conserved residues are shown in bold and grey background. (B) Six conserved cysteines from clMagR dimer form two iron-sulfur cluster binding sites in structural model. (C) Three nearby residues (T57, R58 and E128) located in two hotspot regions could potentially coordinate with iron-sulfur binding sites are shown in structural model.
Not only cysteine, but also histidine and glutamate acid residues could potentially coordinate [2Fe−2S] cluster binding. As previously reported in the transcription regulator RsrR, glutamic acid and histidine both served as ligands of iron-sulfur cluster and played critical roles in sensing the redox status of the cell via the facile cycling of the [2Fe−2S] cluster between +2 and +1 states (Volbeda et al., 2019). To further investigate if E128 in pigeon MagR could potentially function similarly, we performed redox cycling of clMagR protein. Briefly, we firstly measured the Ultraviolet–visible (UV-Vis) absorption spectra of as purified clMagRWT (Figure 2A, black line), followed the subsequent addition of stoichiometric hydrogen peroxide (H2O2) and incubated at 4°C for 30 min (orange line), then, adding stoichiometric sodium dithionite and incubated at 4°C for about 100 min (green line). Finally, the samples were re-added with stoichiometric hydrogen peroxide (H2O2) and incubated at 4°C for 60 min (purple line). The results showed that re-adding H2O2 after clMagRWT reduction did not re-oxidize the protein, indicating that clMagR is lacking the glutamate acid-mediated [2Fe−2S] redox cycle.
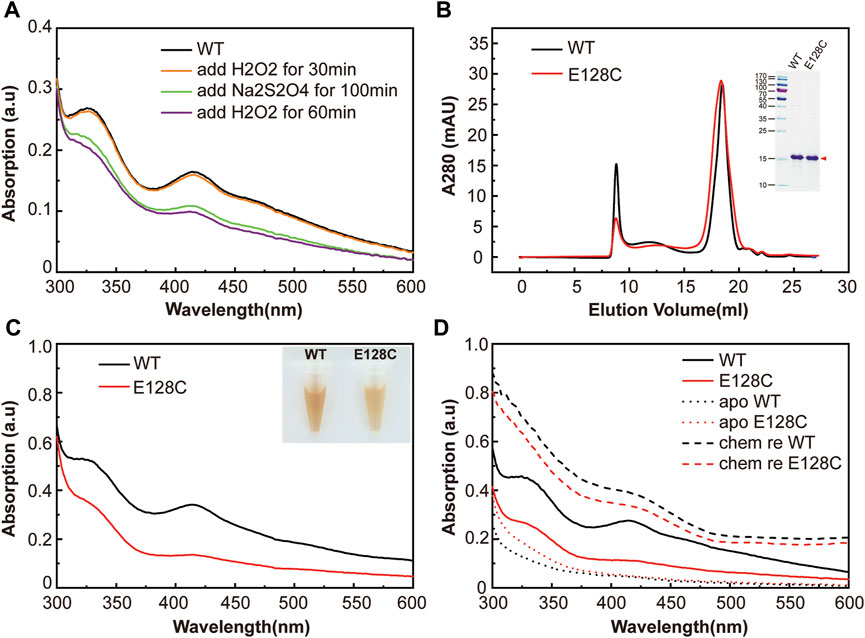
FIGURE 2. The functional role of E128 in clMagR iron-sulfur cluster binding. (A) The redox cycling experiment of clMagRWT protein monitored by UV-Vis absorption spectrum. As isolated protein (WT) was shown as black line, subsequently H2O2 oxidized protein, Na2S2O4 reduced protein and H2O2 re-oxidized protein were shown as orange, green and purple lines, respectively. (B) Size-exclusion chromatography of purified wild type clMagR protein (WT, black line) and its mutant clMagRE128C (abbreviated as E128C, red line). SDS-PAGEs of protein preparation are shown as inserts. (C) UV–Vis absorption spectrum of purified clMagR (WT) and its mutant (E128C), with the same color scheme as in (A) (D) UV-Vis absorption spectrum of chemically reconstituted clMagR (WT) and its mutant (E128C). As-isolated proteins were shown as solid lines, apo proteins with iron-sulfur cluster removal were shown as dotted lines, and chemically reconstituted proteins (labeled as chem re) were shown as dashed lines.
We then tested if substituting E128 to cysteine residue could enhance or stabilize the iron-sulfur cluster. A single amino acid substitution in clMagR (clMagRE128C) was generated to test this hypothesis, and clMagRE128A was used as a control to compare the effect on iron-sulfur binding by mutagenesis. Both mutants and wild-type proteins were expressed and purified to homogeneity. Size-exclusion chromatography and SDS-PAGE showed similar conformations and molecular weight among clMagRE128A, clMagRE128C and clMagRWT (Figure 2B and Supplementary Figure S2A), indicating the mutants preserved the correct protein folding of MagR. The purified clMagRE128C and clMagRE128A proteins showed similar characteristic brownish color compared with clMagRWT in solution (Figure 2C and Supplementary Figure S2B). Ultraviolet–visible (UV–Vis) spectrum from 300 nm to 600 nm wavelength showing absorption peaks at 320 and 420 nm, and a shoulder at 460 nm (Dailey et al., 1994; Netz et al., 2016; Guo et al., 2021), further confirmed that the iron-sulfur cluster incorporation was not affected by mutation (Figure 2C and Supplementary Figure S2B). However, lighter coloration and reduced UV-Vis absorption peaks (320 nm, 420 nm, 460 nm) of clMagRE128C protein were observed, suggesting the impaired iron-sulfur cluster binding by mutation. One possibility could be the additional cysteine (-SH) in clMagRE128C may interfere with the assembly of iron-sulfur clusters or lead to local structural instability in iron-sulfur binding site.
Wild type clMagR and its E128 mutant protein were chemically reconstituted respectively to check the iron-sulfur binding capacity in vitro (Figure 2D and Supplementary Figure S2C). Briefly, the iron-sulfur clusters of MagR were removed by incubating with sodium dithionite and EDTA, and confirmed by UV-Vis spectrum (apo clMagR, dotted line). Then, fresh ferrous ammonium sulfate (Fe(NH4)2(SO4)2) and sodium sulfide (Na2S) were added to the apo protein to reconstitute iron-sulfur clusters. The iron-sulfur cluster constitution was verified by UV-Vis spectrum (chem re clMagR, dashed line). The UV absorption of reconstituted proteins appeared to be significantly higher than that of as-isolated clMagR. The successful reconstitution of clMagR and its E128 mutants (especially clMagRE128A) suggested that E128 might not a ligand of iron-sulfur cluster and should not be involved in iron-sulfur binding directly.
Highly conserved cysteine rich motifs such as CXC motif (e.g., C124 and C126 in pigeon MagR) and CX2C motif were often found in iron-sulfur binding site. We designed clMagRG125_C126insK mutation to covert CXC motif to CX2C motif around this region (Supplementary Figure S1), but also found the iron-sulfur binding efficiency decreased as well (Supplementary Figure S3).
Taken together, we concluded that E128 was not a potential ligand of iron-sulfur cluster in clMagR, and mutating E128 to cysteine residue neither increased the binding efficiency nor affected the type of iron-sulfur clusters in clMagR.
T57C mutation improved the iron-sulfur cluster binding in pigeon MagR
Based on the structural model of clMagR, the second hotspot region was found located around the conserved C60 residue (Figure 1C). We analyzed the known iron-sulfur cluster binding protein structures from RCSB Protein Data Bank and performed sequence alignment for the CX2C motif of iron-sulfur binding sites based on known structures (PDB ID: 3ZXS, 4Z3Y, 5C4I, 4UNF, 4S23, 1DUR). We mainly focused on CXGC motif, since C60 has been identified as the ligand of iron-sulfur cluster and has a nearby glycine residue in position 59 in clMagR (Supplementary Figure S1) (Qin et al., 2016; Guo et al., 2021). Two mutants, clMagRT57C and clMagRT57_R58insC, were then designed aiming to reconstruction of an artificial CXGC motif in clMagR and to provide additional ligand for [2Fe−2S] binding. clMagRR58C was also generated as a control of clMagRT57C.
All three mutants were expressed and purified as described above. Size-exclusion chromatography and SDS-PAGE showed similar conformations and molecular weights compared with wild type clMagR (Figure 3A). Freshly purified clMagRWT protein and three mutants (clMagRT57C, clMagRR58C and clMagRT57_R58insC) all showed brown color, indicating the iron-sulfur cluster binding in purified proteins. Purified clMagRT57C showed slightly thicker color, whereas the color of clMagRR58C and clMagRT57_R58insC protein were relatively lighter compared with that of clMagRWT at the same concentration (Figure 3B and Supplementary Figure S3). Consistently, the UV-Vis absorption peaks (320 nm, 420 nm, 460 nm) of clMagRT57C were higher than those of clMagRWT, but clMagRR58C showed significantly decreased UV-Vis absorption (Figure 3B), and clMagRT57_R58insC even showed very different UV-Vis spectrum indicating the iron-sulfur cluster binding site was almost abolished (Supplementary Figure S3). After chemical reconstitution to incorporate iron-sulfur clusters, both the chemically reconstituted clMagRT57C (Figure 3C, dashed line) and clMagRWT (Figure 2D, dashed line) showed similar UV-Vis absorption, which is higher than that of clMagRR58C (Figure 3D, dashed line), indicating the iron-sulfur binding probably was impaired by R58C substitution.
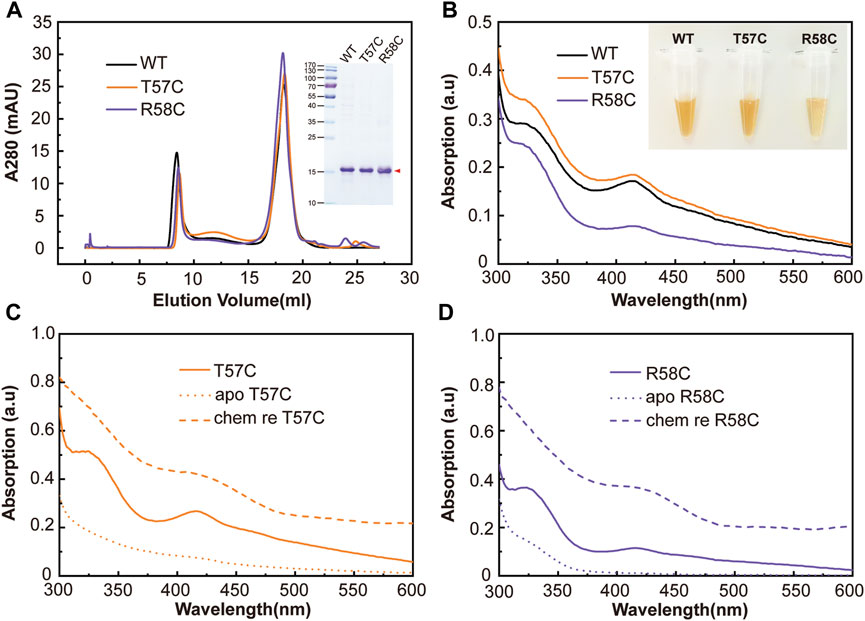
FIGURE 3. Mutations designed around C60 region in clMagR identified T57C increased iron-sulfur cluster binding. (A) Size-exclusion chromatography of purified wild type clMagR (WT, black line) and its mutants, clMagRT57C (abbreviated as T57C, orange line) and clMagRR58C (abbreviated as R58C, purple line). SDS-PAGEs of protein preparation are shown as inserts. (B) UV–Vis absorption spectrum of purified clMagR (WT) and its mutants (T57C and R58C), with the same color scheme as in (A) (C,D) UV-Vis absorption spectrum of chemically reconstituted clMagR mutants, T57C (C) and R58C (D). The data for chemically reconstituted wild type clMagR protein was shown in Figure 2D. As-isolated proteins were shown as solid lines, apo proteins with iron-sulfur cluster removal were shown as dotted lines, and chemically reconstituted proteins (labeled as chem re) were shown as dashed lines.
The total iron content of clMagRWT and its mutants was measured by Ferrozine assay, an accurate and rapid method of the quantitation of iron in biological systems (Im et al., 2013; Landry et al., 2013; de Mello Gabriel et al., 2021). It is obvious that all mutations except T57C decreased the iron content in clMagR, and T57C even showed increased iron content compared that of wild type clMagR (Figure 4A), which was consistent with the UV-Vis spectrum results and the coloration of the purified proteins as well.
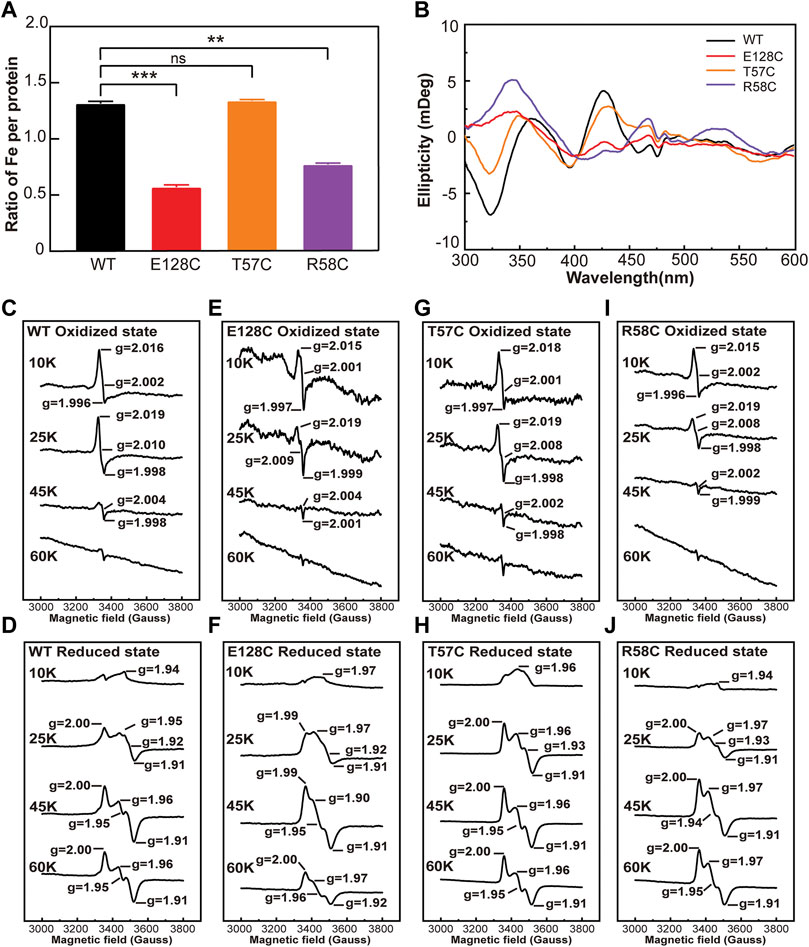
FIGURE 4. The iron content and the type of iron-sulfur cluster of wild type clMagR and its mutants. (A) Iron content of purified wild type clMagR (WT, black) and its mutants measured by Ferrozine assay and presented as the ratio of iron atoms in each protein. Student’s t-test: **, p < 0.01; ***, p < 0.001; ns, p >0.05, “ns” indicate no significant differences. Error bars: SD of three experiments. (B) Circular dichroism (CD) spectrum of wild type clMagR and its mutants. (C–J) EPR spectrum of wild type clMagR (WT, C,D) and its mutants, E128C (E,F), T57C (G,H) and R58C (I,J). EPR were measured both at oxidized (upper) status and reduced status (lower), and recorded at different temperatures (10 K, 25 K, 45 K and 60 K). Asterisks above bars indicate statistically significant differences between treatments at.
Circular dichroism (CD) spectroscopy was further applied to characterize the types of iron–sulfur cluster and their protein environments. As shown in Figure 4B and Supplementary Figure S2D, both wild type clMagR and its mutants showed distinctly positive peaks at 371 nm and 426 nm and three negative peaks at 324 nm, 396 nm, and 463 nm, respectively, suggesting the presence of [2Fe–2S] cluster (Azam et al., 2020). Since [4Fe–4S] or [3Fe–4S] clusters usually exhibit negligible CD intensity compared to [2Fe–2S], thus CD spectroscopy cannot exclude the possible existence of [4Fe–4S] or [3Fe–4S].
Electron paramagnetic resonance (EPR) spectroscopy was then used to further identify the iron-sulfur cluster types in different states of wild type clMagR protein and its mutants (Figures 4C–J and Supplementary Figure S4). The oxidized clMagRWT protein was characterized by a rhombic EPR signal with g values at g1 = 2.016, g2 = 2.002, and g3 = 1.996, and disappeared at 45 K (Figure 4C), suggesting the presence of [3Fe–4S] (Rothery et al., 2001; Hoppe et al., 2011; Pandelia et al., 2011; Liu et al., 2013; Guo et al., 2021). Moreover, after reduction with sodium dithionite, the EPR signal from the [2Fe-2S] cluster can be observed at 45K and 60K (Figure 4D) (Netz et al., 2016; Zhang et al., 2017; Guo et al., 2021), which is consistent with previous report (Guo et al., 2021) . Thus, two distinct iron–sulfur clusters, [2Fe–2S] and [3Fe–4S], were assigned by EPR spectroscopy of clMagRWT. As for the mutants, the g values of clMagRE128A, clMagRE128C, clMagRT57C and clMagRR58C were all similar with clMagRWT both in oxidized and reduced states (Figures 4E–J and Supplementary Figure S4), indicating that mutation in these positions did not affect the type of iron-sulfur cluster clMagR bound. The EPR spectral signal of clMagRT57C was stronger than that of clMagRWT at 25 K in the reduced state (Figure 4H), indicating that the iron-sulfur clusters binding in clMagRT57C might be improved at this temperature. Whether clMagRT57C has enhanced stability at ambient temperature remains unknown and further investigation is required.
Taking together, the stronger EPR signal, the thicker coloration of purified protein, the increased UV-Vis absorption and Ferrozine staining in clMagRT57C suggested that a more stabilized iron-sulfur cluster binding, thus clMagRT57C might serve as a valuable candidate for further exploration.
Prolonged iron-sulfur cluster stability in clMagRT57C
To further investigate if clMagRT57C could stabilize the iron-sulfur cluster binding at ambient temperature, UV-Vis spectrums were measured with freshly purified clMagRWT and clMagRT57C for continuous 7 days parallelly at room temperature at the same concentration (200 μM), to monitor the iron-sulfur cluster bound in proteins. The loss of iron-sulfur cluster binding in protein was shown by the decrease in UV-Vis absorption. As shown in Figures 5A,B, the absorption peaks of clMagRWT were much lower than that of clMagRT57C after 7 days. It is obvious that wild type clMagR protein is losing its iron-sulfur clusters significantly faster than clMagRT57C. By mutating T57 to cysteine residue, the iron-sulfur cluster was stabilized in bound form in clMagR protein even at room temperature.
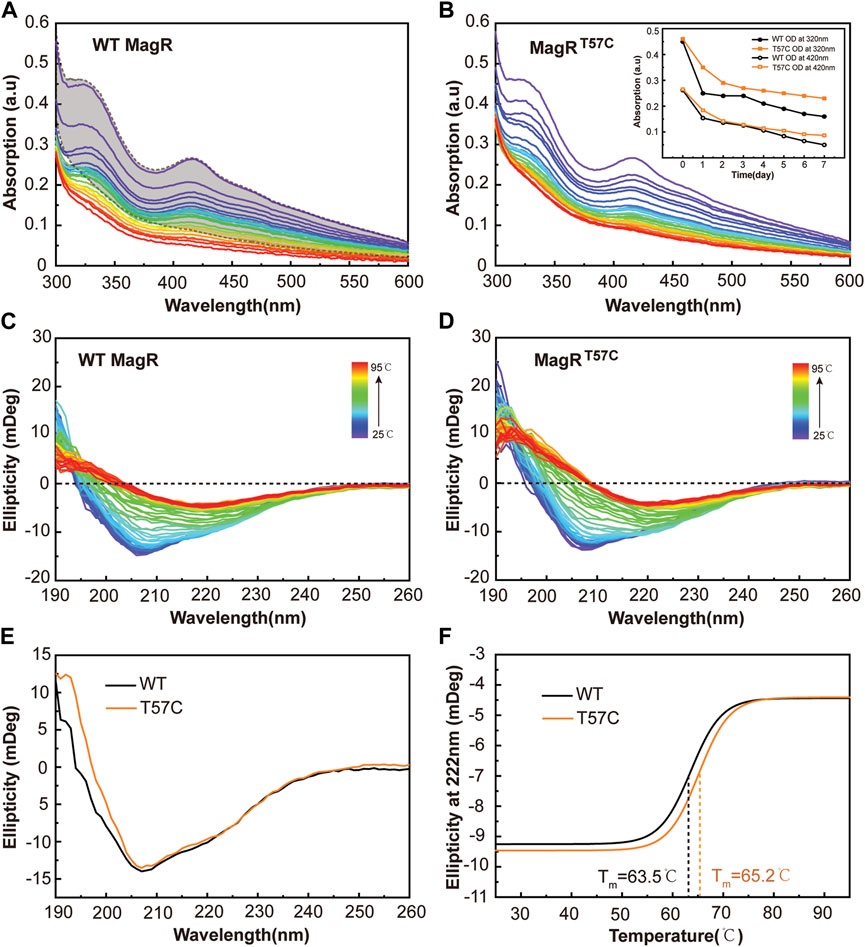
FIGURE 5. The clMagRT57C shows prolonged iron-sulfur cluster stability. (A–B) UV–Vis absorption spectrum of wild type clMagR (WT, (A) and clMagRT57C (T57C, (B) measured every 6 h at room temperature for continuous 7 days, showing time-dependent absorption decay of both proteins at different rate. The colors of the traces follow UV-Vis spectrum trend from purple (starting point) to red (ending point). The shaded area in (A) represents the range of UV–Vis absorption spectral of clMagRT57C in 7 days (as in B) as a reference for comparison. The iron-sulfur cluster dissociation curves determined by absorption at 320 nm and 420 nm from A and B are shown as inserts in (B) (C–D) Thermal stability analysis of wild type clMagR (WT, (C) and clMagRT57C (T57C, (D) measured by CD spectroscopy from 25 to 95°C. (E) Far-UV (190–260 nm) CD spectrum of wild type clMagR (WT, black line) and clMagRT57C (T57C, orange line). (F) Thermal unfolding curves of wild type clMagR (WT, black line) and clMagRT57C (T57C, orange line). Boltzmann sigmoidal fit of CD thermal denaturation curve at 222 nm depicting a melting temperature of 63.5°C for clMagRWT and 65.2°C for clMagRT57C.
To further address the underlying mechanism of how T57C mutation leads to thermostability increase in clMagR, far-UV CD spectroscopy (190–260 nm) was applied to follow the unfolding and folding of proteins as a function of temperature from 25 to 95°C at 1°C intervals (Figures 5C,D) (Kanagarajan et al., 2021; Wensien et al., 2021). The clMagRT57C mutant showed improved thermostability compared with clMagRWT in the temperature range we recorded (Figures 5C,D). A side-by-side comparison of the secondary structure of both proteins at room temperature revealed largely unchanged profile between clMagRWT and clMagRT57C (Figure 5E), indicating that site-directed mutagenesis did not disrupt the overall structures of the protein, which is consistent with the size-exclusion chromatography result (Figure 3A). Further analysis showed that melting points (Tm) of clMagRT57C was significantly higher than that of clMagRWT, suggesting the increased thermostability by mutating T57 to cysteine (Figure 5F).
Discussion
Iron-sulfur proteins attracted much attention of protein design as they are of tremendous interest for their electron transfer properties and play essential roles in various fundamental biological processes (Nanda et al., 2016). The general purpose of re-designing an iron-sulfur protein could be to increase the stability in different environments and/or the sensitivity to redox changes, to explore its applications in protein-based therapies, or serve as biosensor, et al.
Most iron-sulfur proteins can be unstable when they are accessible to oxidizing substances (Rouault and Klausner, 1996). Some proteins such as pigeon MagR which contains relatively stable iron-sulfur clusters, mostly because that the cluster is bound in a region of the protein inaccessible to solvent and oxidants. Designing a more stable iron-sulfur binding site based on native protein has been very challenging (Nanda et al., 2016). One particular approach is to bury the cluster within the hydrophobic center of the protein by substitute nearby hydrophilic residues to hydrophobic residues, thus, to make the cluster inaccessible to oxidants. However, this approach could potentially damage their inherent susceptibility to redox changes. MagR, as a putative magnetoreceptor, its sensitivity to magnetic field changes could be modulated by different types of iron-sulfur clusters bound to the protein and regulated by redox cycle as well (Guo et al., 2021) . Therefore, we chose a different approach to stabilize the iron-sulfur cluster binding by adding an additional ligand to the cluster.
We screened various mutations in two hotspots around the iron-sulfur cluster binding site in pigeon MagR and found that most mutants we tested did not affect the type of iron-sulfur clusters bound to the protein. And most mutants also showed decreased iron-sulfur cluster binding efficiency and lower iron content compared with that of wild type protein. Only one particular mutation, T57C in clMagR showed improved iron-sulfur binding efficiency and iron content, without alter the type of bound iron-sulfur cluster. More importantly, prolonged thermostability has been found with clMagRT57C, and CD spectroscopy data also reveals an overall structural stability improvement.
The ultimate goal of protein engineering of MagR is to provide a molecular tool for magnetogenetics in the future with improved stability at ambient temperature and hypersensitivity to external magnetic field changes. Previously we rationally designed a single-chain tetramer MagR (SctMagR) as a building block to facilitate MagR assembly (Yang et al., 2022), and here in this study, we designed a better iron-sulfur binding site in clMagRT57C with prolonged stability at room temperature. The work we presented in here, as well as in previous report, may serve as steady steps toward the magnetogenetics applications in the future.
Method
Expression and purification of clMagRWT and its mutant proteins
The expression vector of the pigeon (Columba livia) MagR (clMagR) was constructed as previously described (Qin, Nature Materials, 2016). The expression vectors of the clMagRE128A, clMagRE128C, clMagRG125_C126insK, clMagRT57C, clMagRR58C and clMagRT57_R58insC were obtained by site-directed mutagenesis (TIANGEN) using clMagRWT plasmid as template.
All proteins were recombinantly expressed in E. coli strain BL21 (DE3). Briefly, the protein expression was induced by 20 µM isopropyl β-D-1-thiogalactopyranoside (IPTG) overnight at 15°C (288K). Bacteria cells were harvested and resuspended in lysis buffer (20 mM Tris, 500 mM NaCl, PH 8.0) with complete protease inhibitor cocktail (Roche) and lysed by sonication on ice. The supernatant was collected after centrifugation and loaded onto a Strep-Tactin affinity column (IBA). The column was washed with washing buffer (20 mM Tris, 500 mM NaCl, pH 8.0) for approximately 30 column volumes (CV) to remove unbound proteins. Afer washing, proteins were eluted from Strep-Tactin affinity columns with elution buffer (20 mM Tris, 500 mM NaCl, 5 mM desthiobiotin, pH 8.0). PageRuler Prestained Protein Ladder (Thermo Scientific, Product# 26616) was used as the molecular weight standards for all SDS-PAGEs.
UV-Vis analysis of clMagRWT and mutant proteins
Wild type MagR and mutant proteins were prepared at 200 μM in TBS buffer (20 mM Tris, 150 mM NaCl, pH 8.0) and UV-Vis absorption measurements in the near UV visible wavelength (300–600 nm) were recorded using a nanodrop spectrophotometer (Thermo Scientific, NanoDrop OneC).
Electron paramagnetic resonance spectroscopy
X-Band (∼9.6 GHz) EPR spectra were recorded on an EMX plus 10/12 spectrometer (Bruker, Billerica, MA), equipped with Oxford ESR910 Liquid Helium cryostat. For the oxidized protein samples, 200 μL of 1 mM as-isolated purified protein were mixed with 50 μl of glycerol in TBS buffer (20 mM Tris, 150 mM NaCl, pH 8.0). For reduced protein samples, 10 mM sodium dithionite (Na2S2O4) was added to the above protein solutions. Then, the protein samples were transferred into 4 mm diameter quartz EPR tubes (Wilmad 707-SQ-250 M) and frozen in liquid nitrogen. The EPR signals of oxidized and reduced proteins were recorded at different temperatures (10 K, 25 K, 45 K, and 60 K) with a modulation amplitude of 2 G, a microwave frequency of 9.40 GHz, an incident microwave power of 2 mW, and a sweep time of 25.60 s.
Chemical reconstitution
A concentration of 400 μM purified (as-isolated) wild type clMagR (clMagRWT) or mutant proteins were incubated overnight at 4°C in TBS buffer (20 mM Tris, 150 mM NaCl, pH 8.0) with 10 mM EDTA and 10 mM sodium dithionite (Na2S2O4) to remove the bound iron-sulfur clusters. The mixture was desalted with a PD MiniTrap G-25 desalting column (GE Healthcare) and the obtained protein was labeled “apo clMagR”. Then, 5 mM DTT was added into the protein solution and incubated for 30 min at 4°C, followed by dropwise addition of fresh Fe(NH4)2(SO4)2 and Na2S both at 8-fold molar excess relative to protein. The mixture was incubated at 4°C overnight. The protein solution was then passed through a PD MiniTrap G-25 desalting column to remove unbound iron and sulfate, and the obtained protein sample labeled “chem re clMagR”.
Circular dichroism spectroscopy
Circular dichroism (CD) was applied evaluate secondary structures in protein in the far UV range (190–260 nm) and to monitor protein-bound co-factors such as iron-sulfur clusters in the near UV-Visible range (300–600 nm) in this study. As for the protein-bound iron-sulfur cluster types analysis, purified wild-type MagR protein (clMagRWT) and mutants were prepared at 100 μM in TBS buffer (20 mM Tris, 150 mM NaCl, pH 8.0) and measured in 1 cm diameter quartz cells at room temperature using a MOS-500 (Biologic) CD Spectrometer. To analyze the secondary structure and thermal stability of wild type MagR and its mutants, measurements were performed using a J-1700 CD Spectrometer (JASCO Corporation, JPN) in the Far-UV range (190–260 nm). Purified wild-type MagR protein (clMagRWT) and mutants were prepared at 10 μM protein (20 mM Na2HPO4, pH 8.0) in 1 mm quartz cuvettes at room temperature. The thermal stability of wild-type MagR protein (clMagRWT) and mutants were monitored and recorded by CD spectrum in the range of 25°C to 95°C with temperature increases at 1°C intervals. The melting temperature values were calculated by sigmoidal fitting of the thermal denaturation curve at 222 nm using Boltzmann function.
Ferrozine assay
Ferrous iron reacts with ferrozine (0.1% (w/v) ferrozine in 50% (w/v) ammonium acetate) to form an intense purple complex that can be quantified spectrophotometrically at 562 nm using a microplate reader. The iron (Fe) content in MagR and its mutant proteins was quantified by reducing Fe with hydroxylamine hydrochloride (10% (w/v) HAHCl in 1M HCl) and analyzed by ferrozine assay. Briefly, aliquots of protein and HAHCl mixture (80 μl HAHCl and 20 μl proteins at 100μM, total 100 μl) were incubated at 37°C for 30 min in the dark in a 96-well plate, then, 100 μl ferrozine was added into each well and incubated at 37°C for additional 15 min in the dark. The iron-ferrozine complex was measured at 562 nm on a microplate reader (Tecan Spark). A standard curve for ferrozine assay was generated using a series ferric chloride solution (0–500 μM) in 1 M HCl. The total iron content of MagR protein and its mutants were calculated by comparing its absorbance to that of a range of standard concentrations of equal volume that had been prepared in a way similar to that of the protein samples by linear regression analysis. There were three replicates for each protein sample. Histograms and statistical analyses were performed by using the software GraphPad Prism. Student’s t-test was used to test for differences in total iron between protein samples and considered significant at p <0.05.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding authors.
Author contributions
CX, JW, and YZ conceived the idea and designed the study. TT carried out protein purification, site-directed mutagenesis, CD spectroscopy and EPR experiments. TT and CX did data analysis. ZG helped with EPR experiments. YZ, XZ, and JZ contributed to the Ferrozine assay data analysis. FF and PZ helped with size-exclusion chromatography data analysis. SW helped with Far-UV CD data analysis. YZ and GL constructed and optimized the structure model of MagR. JW and GL provided valuable suggestions on data analysis. TT and CX wrote the paper. TC provided valuable discussions and did English editing on the manuscript. All authors commented on the manuscript.
Funding
This research was funded by the Presidential Foundation of Hefei Institutes of Physical Science, Chinese Academy of Sciences (http://english.hf.cas.cn), grants Y96XC11131 (CX), National Natural Science Foundation of China (https://www.nsfc.gov.cn/english/site_1/index.html) grants 31640001 (CX).
Acknowledgments
We thank the Steady High Magnetic Field Facilities (High Magnetic Field Laboratory, CAS) for assistance with EPR measurements, and thanks to Dr. Wei Tong and Jinxing Li for their technical support. We are grateful to the core facilities of the College of Life Sciences of the University of Science and Technology of China and the core facilities of the College of Life Sciences of Anhui University for their help in CD spectrum measurements. We also thank Huangtao Xu, Shuai Xu and Xianglong Zhao for discussions.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmolb.2022.1051943/full#supplementary-material
References
Azam, T., Przybyla-Toscano, J., Vignols, F., Couturier, J., Rouhier, N., and Johnson, M. K. (2020). The arabidopsis mitochondrial glutaredoxin GRXS15 provides [2Fe-2S] clusters for ISCA-mediated [4Fe-4S] cluster maturation. Int. J. Mol. Sci. 21 (23), E9237. doi:10.3390/ijms21239237
Beinert, H. S. (1960). Studies on succinic and DPNH dehydrogenase preparations by paramagnetic resonance (EPR) spectroscopy.
Dailey, H. A., Finnegan, M. G., and Johnson, M. K. (1994). Human ferrochelatase is an iron-sulfur protein. Biochemistry 33 (2), 403–407. doi:10.1021/bi00168a003
de Mello Gabriel, G. V., Pitombo, L. M., Rosa, L. M. T., Navarrete, A. A., Botero, W. G., do Carmo, J. B., et al. (2021). The environmental importance of iron speciation in soils: Evaluation of classic methodologies. Environ. Monit. Assess. 193 (2), 63. doi:10.1007/s10661-021-08874-w
Eady, R. R., Smith, B. E., Cook, K. A., and Postgate, J. R. (1972). Nitrogenase of Klebsiella pneumoniae. Purification and properties of the component proteins. Biochem. J. 128 (3), 655–675. doi:10.1042/bj1280655
Eder, S. H., Cadiou, H., Muhamad, A., McNaughton, P. A., Kirschvink, J. L., and Winklhofer, M. (2012). Magnetic characterization of isolated candidate vertebrate magnetoreceptor cells. Proc. Natl. Acad. Sci. U. S. A. 109 (30), 12022–12027. doi:10.1073/pnas.1205653109
Fontecave, M. (2006). Iron-sulfur clusters: Ever-expanding roles. Nat. Chem. Biol. 2 (4), 171–174. doi:10.1038/nchembio0406-171
Gegear, R. J., Casselman, A., Waddell, S., and Reppert, S. M. (2008). Cryptochrome mediates light-dependent magnetosensitivity in Drosophila. Nature 454 (7207), 1014–1018. doi:10.1038/nature07183
Gruner, I., Fradrich, C., Bottger, L. H., Trautwein, A. X., Jahn, D., and Hartig, E. (2011). Aspartate 141 is the fourth ligand of the oxygen-sensing [4Fe-4S]2+ cluster of Bacillus subtilis transcriptional regulator Fnr. J. Biol. Chem. 286 (3), 2017–2021. doi:10.1074/jbc.M110.191940
Guo, Z., Xu, S., Chen, X., Wang, C., Yang, P., Qin, S., et al. (2021). Modulation of MagR magnetic properties via iron-sulfur cluster binding. Sci. Rep. 11 (1), 23941. doi:10.1038/s41598-021-03344-2
Hinton, T. V., Batelu, S., Gleason, N., and Stemmler, T. L. (2022). Molecular characteristics of proteins within the mitochondrial Fe-S cluster assembly complex. Micron 153, 103181. doi:10.1016/j.micron.2021.103181
Hoppe, A., Pandelia, M. E., Gartner, W., and Lubitz, W. (2011). [Fe₄S₄]- and [Fe₃S₄]-cluster formation in synthetic peptides. Biochim. Biophys. Acta 1807 (11), 1414–1422. doi:10.1016/j.bbabio.2011.06.017
Hsu, C. Y., Ko, F. Y., Li, C. W., Fann, K., and Lue, J. T. (2007). Magnetoreception system in honeybees (Apis mellifera). PLoS One 2 (4), e395. doi:10.1371/journal.pone.0000395
Im, J., Lee, J., and Loffler, F. E. (2013). Interference of ferric ions with ferrous iron quantification using the ferrozine assay. J. Microbiol. Methods 95 (3), 366–367. doi:10.1016/j.mimet.2013.10.005
Jiang, M., Zhang, L., Wang, F., Zhang, J., Liu, G., Gao, B., et al. (2017). Novel application of magnetic protein: Convenient one-step purification and immobilization of proteins. Sci. Rep. 7 (1), 13329. doi:10.1038/s41598-017-13648-x
Johnson, D. C., Dean, D. R., Smith, A. D., and Johnson, M. K. (2005). Structure, function, and formation of biological iron-sulfur clusters. Annu. Rev. Biochem. 74, 247–281. doi:10.1146/annurev.biochem.74.082803.133518
Kanagarajan, S., Carlsson, M. L. R., Chakane, S., Kettisen, K., Smeds, E., Kumar, R., et al. (2021). Production of functional human fetal hemoglobin in Nicotiana benthamiana for development of hemoglobin-based oxygen carriers. Int. J. Biol. Macromol. 184, 955–966. doi:10.1016/j.ijbiomac.2021.06.102
Kang, J., Kang, D., Yeom, G., and Park, C. J. (2021). Molecular diagnostic system using engineered fusion protein-conjugated magnetic nanoparticles. Anal. Chem. 93 (50), 16804–16812. doi:10.1021/acs.analchem.1c03247
Kiley, P. J., and Beinert, H. (2003). The role of Fe–S proteins in sensing and regulation in bacteria. Curr. Opin. Microbiol. 6 (2), 181–185. doi:10.1016/s1369-5274(03)00039-0
Landry, A. P., Cheng, Z., and Ding, H. (2013). Iron binding activity is essential for the function of IscA in iron-sulphur cluster biogenesis. Dalton Trans. 42 (9), 3100–3106. doi:10.1039/c2dt32000b
Lau, J. C., Rodgers, C. T., and Hore, P. J. (2012). Compass magnetoreception in birds arising from photo-induced radical pairs in rotationally disordered cryptochromes. J. R. Soc. Interface 9 (77), 3329–3337. doi:10.1098/rsif.2012.0374
Liedvogel, M., Maeda, K., Henbest, K., Schleicher, E., Simon, T., Timmel, C. R., et al. (2007). Chemical magnetoreception: Bird cryptochrome 1a is excited by blue light and forms long-lived radical-pairs. PLoS One 2 (10), e1106. doi:10.1371/journal.pone.0001106
Liu, Y., Guo, S., Yu, R., Ji, J., and Qiu, G. (2013). HdrC2 from Acidithiobacillus ferrooxidans owns two iron-sulfur binding motifs but binds only one variable cluster between [4Fe-4S] and [3Fe-4S]. Curr. Microbiol. 66 (1), 88–95. doi:10.1007/s00284-012-0244-y
Mettert, E. L., and Kiley, P. J. (2015). How is Fe-S cluster formation regulated? Annu. Rev. Microbiol. 69, 505–526. doi:10.1146/annurev-micro-091014-104457
Meyer, J. (2008). Iron-sulfur protein folds, iron-sulfur chemistry, and evolution. J. Biol. Inorg. Chem. 13 (2), 157–170. doi:10.1007/s00775-007-0318-7
Mitou, G., Higgins, C., Wittung-Stafshede, P., Conover, R. C., Smith, A. D., Johnson, M. K., et al. (2003). An Isc-type extremely thermostable [2Fe-2S] ferredoxin from Aquifex aeolicus. Biochemical, spectroscopic, and unfolding studies. Biochemistry 42 (5), 1354–1364. doi:10.1021/bi027116n
Monteil, C. L., and Lefevre, C. T. (2020). Magnetoreception in microorganisms. Trends Microbiol. 28 (4), 266–275. doi:10.1016/j.tim.2019.10.012
Mortenson, L., Valentine, R., and Carnahan, J. (1962). An electron transport factor from Clostridiumpasteurianum. Biochem. Biophys. Res. Commun. 7, 448–452. doi:10.1016/0006-291x(62)90333-9
Nanda, V., Senn, S., Pike, D. H., Rodriguez-Granillo, A., Hansen, W. A., Khare, S. D., et al. (2016). Structural principles for computational and de novo design of 4Fe-4S metalloproteins. Biochim. Biophys. Acta 1857 (5), 531–538. doi:10.1016/j.bbabio.2015.10.001
Netz, D. J., Genau, H. M., Weiler, B. D., Bill, E., Pierik, A. J., and Lill, R. (2016). The conserved protein Dre2 uses essential [2Fe-2S] and [4Fe-4S] clusters for its function in cytosolic iron-sulfur protein assembly. Biochem. J. 473 (14), 2073–2085. doi:10.1042/BCJ20160416
Pandelia, M. E., Nitschke, W., Infossi, P., Giudici-Orticoni, M. T., Bill, E., and Lubitz, W. (2011). Characterization of a unique [FeS] cluster in the electron transfer chain of the oxygen tolerant [NiFe] hydrogenase from Aquifex aeolicus. Proc. Natl. Acad. Sci. U. S. A. 108 (15), 6097–6102. doi:10.1073/pnas.1100610108
Qin, S., Yin, H., Yang, C., Dou, Y., Liu, Z., Zhang, P., et al. (2016). A magnetic protein biocompass. Nat. Mat. 15 (2), 217–226. doi:10.1038/nmat4484
Ritz, T., Adem, S., and Schulten, K. (2000). A model for photoreceptor-based magnetoreception in birds. Biophys. J. 78 (2), 707–718. doi:10.1016/s0006-3495(00)76629-x
Rothery, R. A., Blasco, F., and Weiner, J. H. (2001). Electron transfer from heme bL to the [3Fe-4S] cluster of Escherichia coli nitrate reductase A (NarGHI). Biochemistry 40 (17), 5260–5268. doi:10.1021/bi002393k
Rouault, T. A., and Klausner, R. D. (1996). Iron-sulfur clusters as biosensors of oxidants and iron. Trends biochem. Sci. 21 (5), 174–177. doi:10.1016/s0968-0004(96)10024-4
Rouault, T. A. (2015). Mammalian iron-sulphur proteins: Novel insights into biogenesis and function. Nat. Rev. Mol. Cell Biol. 16 (1), 45–55. doi:10.1038/nrm3909
Schuler, D., Monteil, C. L., and Lefevre, C. T. (2020). Magnetospirillum gryphiswaldense. Trends Microbiol. 28 (11), 947–948. doi:10.1016/j.tim.2020.06.001
Volbeda, A., Martinez, M. T. P., Crack, J. C., Amara, P., Gigarel, O., Munnoch, J. T., et al. (2019). Crystal structure of the transcription regulator RsrR reveals a [2Fe-2S] cluster coordinated by Cys, glu, and his residues. J. Am. Chem. Soc. 141 (6), 2367–2375. doi:10.1021/jacs.8b10823
Wensien, M., von Pappenheim, F. R., Funk, L. M., Kloskowski, P., Curth, U., Diederichsen, U., et al. (2021). A lysine-cysteine redox switch with an NOS bridge regulates enzyme function. Nature 593 (7859), 460–464. doi:10.1038/s41586-021-03513-3
Wiley, S. E., Paddock, M. L., Abresch, E. C., Gross, L., van der Geer, P., Nechushtai, R., et al. (2007). The outer mitochondrial membrane protein mitoNEET contains a novel redox-active 2Fe-2S cluster. J. Biol. Chem. 282 (33), 23745–23749. doi:10.1074/jbc.C700107200
Wiltschko, R., and Wiltschko, W. (2014). Sensing magnetic directions in birds: Radical pair processes involving cryptochrome. Biosens. (Basel) 4 (3), 221–242. doi:10.3390/bios4030221
Wiltschko, R., and Wiltschko, W. (2013). The magnetite-based receptors in the beak of birds and their role in avian navigation. J. Comp. Physiol. A Neuroethol. Sens. Neural Behav. Physiol. 199 (2), 89–98. doi:10.1007/s00359-012-0769-3
Xie, C. (2022). Searching for unity in diversity of animal magnetoreception: From biology to quantum mechanics and back. Innovation. 3 (3), 100229. doi:10.1016/j.xinn.2022.100229
Xu, J., Jarocha, L. E., Zollitsch, T., Konowalczyk, M., Henbest, K. B., Richert, S., et al. (2021). Magnetic sensitivity of cryptochrome 4 from a migratory songbird. Nature 594 (7864), 535–540. doi:10.1038/s41586-021-03618-9
Xue, L., Hu, T., Guo, Z., Yang, C., Wang, Z., Qin, S., et al. (2020). A novel biomimetic magnetosensor based on magneto‐optically involved conformational variation of MagR/cry4 complex. Adv. Electron. Mat. 6 (4), 1901168. doi:10.1002/aelm.201901168
Yang, P., Cai, T., Zhang, L., Yu, D., Guo, Z., Zhang, Y., et al. (2022). A rationally designed building block of the putative magnetoreceptor MagR. Bioelectromagnetics 43 (5), 317–326. doi:10.1002/bem.22413
Zeng, J., Zhang, X., Wang, Y., Ai, C., Liu, Q., and Qiu, G. (2008). Glu43 is an essential residue for coordinating the [Fe2S2] cluster of IscR from Acidithiobacillus ferrooxidans. FEBS Lett. 582 (28), 3889–3892. doi:10.1016/j.febslet.2008.09.060
Keywords: iron-sulfur cluster, magnetoreceptor, MagR, rational design, thermostability
Citation: Tong T, Zhou Y, Fei F, Zhou X, Guo Z, Wang S, Zhang J, Zhang P, Cai T, Li G, Zhang Y, Wang J and Xie C (2022) The rational design of iron-sulfur cluster binding site for prolonged stability in magnetoreceptor MagR. Front. Mol. Biosci. 9:1051943. doi: 10.3389/fmolb.2022.1051943
Received: 23 September 2022; Accepted: 13 October 2022;
Published: 10 November 2022.
Edited by:
Guijun Wan, Nanjing Agricultural University, ChinaReviewed by:
Vitalii Zablotskii, Institute of Physics (ASCR), CzechiaJianfei Sun, Southeast University, China
Copyright © 2022 Tong, Zhou, Fei, Zhou, Guo, Wang, Zhang, Zhang, Cai, Li, Zhang, Wang and Xie. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yuebin Zhang, emhhbmd5YkBkaWNwLmFjLmNu; Junfeng Wang, anVuZmVuZ0BobWZsLmFjLmNu; Can Xie, Y2FueGllQGhtZmwuYWMuY24=
 Tianyang Tong1,2
Tianyang Tong1,2 Guohui Li
Guohui Li Can Xie
Can Xie