- 1Department of Oncology, Ruijin Hospital, Shanghai Jiao Tong University School of Medicine, Shanghai, China
- 2Department of Gastrointestinal Medical Oncology, Fudan University Shanghai Cancer Center, Shanghai, China
- 3Department of Oncology, Shanghai Medical College, Fudan University, Shanghai, China
- 4Department of Oncology, Chengdu First People’s Hospital, Chengdu, China
Background: Immune checkpoint inhibitor (ICI) therapies have revolutionized the treatment of metastatic cutaneous melanoma, but have only benefitted a subset of them. Gene mutations were reported to impact the ICI therapy outcomes in metastatic melanoma but have not been fully investigated. Hence, we systematically analyzed the impact of cancer-related gene mutations on the clinical outcome in metastatic melanoma patients who underwent ICI therapies.
Methods: Publicly available discovery and validation cohorts (312 patients and 110 patients respectively, all the patients received ICI therapies) were included in this study. Cox proportional hazards regression analysis was used to assess the association of 468 cancer-related gene mutations with overall survival (OS) in the discovery cohort, and the polygenic hazard score (PHS) was constructed subsequently, and validated in the validation cohort. The Tumor Immune Estimation Resource (TIMER) online tools, which are based on The Cancer Genome Atlas database, were used to analyze the impact of gene mutations on tumor-infiltrated immune cells in melanoma samples.
Results: We found eight gene mutations that were significantly associated with the overall survival (BAP1, CARD11, IGF1R, KMT2D, PTPRD, PTPRT, ROS1, and TERT, p < 0.05, mutation frequency >0.05). The PHS, which was based on these genes, was found to effectively discriminate the subset which benefited most from ICI therapies (HR = 1·54, 95%CI, 1.25–1.95; p < 0.001). After adjusting with age, sex, ICI regimes, and tumor mutation burden (TMB), we found that PHS was an independent predictor for the outcome of ICI therapies (adjusted HR = 1.84, 95%CI, 1.22–2.79; p = 0.004). The PHS was validated in the validation cohort (log-Rank p = 0.038). Further research found that CARD11 and PTPRD mutations were significantly associated with more tumor-infiltrated immune cells in melanoma samples.
Conclusion: For the first time, we have shown that PHS can independently and effectively predict the ICI therapy outcome in metastatic melanoma, which once validated by larger research, may help the decision-making process in melanoma.
Introduction
Melanoma is notorious for its rapid progression and enormous capacity of metastasis. In 2018, over 280,000 new patients suffered from cutaneous melanoma with over 60,000 cancer-related deaths worldwide (Bray et al., 2018). In Asia, the mucosal and acral melanomas accounted for up to 58% of all melanoma types and were more aggressive compared with cutaneous melanoma (Chang et al., 2004).
Because of its intrinsic property of resistance to chemo/radiotherapy, the mainstay therapies for metastatic melanoma have been limited to high-dose interleukin-2, dacarbazine, and vemurafenib before the emergence of immune checkpoint inhibitors (ICI), (Flaherty et al., 2010), and the 5-year survival rate stayed between 15–20 % for decades, which was quite unsatisfactory (Siegel et al., 2018). The ICI treatments, including antibodies targeting programmed death-1 (PD-1), programmed death-ligand 1 (PD-L1), or cytotoxic T-lymphocyte-associated protein 4 (CTLA4), provide substantial survival benefits for metastatic melanoma (Hodi et al., 2010) (Hodi et al., 2018), especially with durable anti-tumor effects for patients who achieved complete response after receiving ICI therapies, with over 85 % having a 5-year progression-free survival rate; while for unselected patients, the 5-year survival rate was 43.2 % and 33.0 % for anti-PD-1 therapy and anti- CTLA4, respectively, according to the KEYNOTE-006 research (Robert et al., 2019). For the acral and mucosal melanomas, the pembrolizumab, an anti-PD-1 monoclonal antibody, had achieved limited efficacy in second line settings, with ORR 15.8 % and 13.3 %, respectively (Si et al., 2019). Hence, it is critical to identify sensitive predictors for the ICI outcome in metastatic melanoma.
Previous studies showed that the PD-L1 expression level was not a solid predictor for ICI therapy (Carlino et al., 2018) (Larkin et al., 2015). Tumor mutation burden (TMB) was validated to be an effective predictor for ICI treatment outcome in various cancer types, as high TMB tumors might yield more neoantigens, which were believed to trigger the tumor immunity, but this association was not significant in metastatic melanoma (Samstein et al., 2019). Recently, some research studies reported that somatic gene mutations were associated with the ICI therapy outcomes in various cancer types including metastatic melanoma (Wang et al., 2019) (Xiao et al., 2018). However, it was believed that a single somatic gene mutation was not efficient enough to detect the beneficial metastatic melanoma subgroups for ICI therapies. As reported, the 21 gene model in breast cancer prognostic prediction could effectively assess the recurrence risk and help treatment decision-making (Varga et al., 2019); it was reasonable to speculate that the polygenic mutation model might be effective to predict the ICI therapy outcomes. Hence, we systematically analyzed the association of 468 cancer-related gene mutations with the ICI therapy outcomes in a melanoma cohort consisting of 321 patients, constructed a polygenic hazard score (PHS), and validated our findings in another independent cohort, in this post-hoc analysis.
Materials and methods
Data acquisition
Two independent datasets were downloaded from the publicly available cBioPortal database (https://www.cbioportal.org), i.e., the Memorial Sloan Kettering Cancer Center Integrated Mutation Profiling of Actionable Cancer Targets (MSK-IMPACT) dataset (https://www.cbioportal.org/study/summary?id=tmb_mskcc_2018), which included 321 metastatic melanoma patients who received anti-PD-1/PD-L1 therapy, anti-CTLA-4 therapy or a combination of anti-PD-1/PD-L1 with anti-CTLA-4 therapies (Samstein et al., 2019); and the DFCI (Dana-Farber Cancer Institute) metastatic melanoma (DFCIMM) dataset (https://www.cbioportal.org/study/clinicalData?id=skcm_dfci_2015) which included 110 metastatic melanoma patients who received anti-CTLA-4 therapy alone (Van Allen et al., 2015). The MSK-IMPACT dataset contained 468 cancer-related gene-sequencing data, and the detailed gene list is included in Supplementary Table S1. DFCIMM datasets contained whole exome sequencing data. Both of them contained integral clinical and survival information.
Discovery analysis
We used the MSK-IMPACT dataset consisting of all somatic mutation data, except the synonymous mutation of the 468 cancer-related genes, to assess which gene mutation was significantly associated with the overall survival (OS). We divided the 321 metastatic melanoma patients into two groups according to a certain gene mutational status: the group of patients with certain gene mutations, and the other group of patients carrying the gene without mutations or with synonymous mutation (e.g., the TP53 mutational group and the TP53 wildtype or synonymous mutational group), and then we performed the univariate Cox proportional hazards regression analysis for these two groups to evaluate the association between gene mutation status and the OS of metastatic melanoma patients who received ICI therapies. Each of the 468 genes was assessed accordingly, and those genes with p value < 0.05 and mutation frequency >0.05 (i.e., mutation counts >321 × 5%) were selected as candidate genes for the multivariate Cox regression analysis with adjustments for age, sex, TMB, and ICI regimes (i.e., anti-PD-1/PD-L1, anti-CTLA-4, and the combination of anti-PD-1/PD-L1 with anti-CTLA-4). Kaplan–Meier survival curves were generated to visualize the results.
Construct the polygenic hazard score model
The PHS was built and used as an effective survival analysis model to predict the time to events (Tan et al., 2019) (Seibert et al., 2018). Those genes with p value < 0.05 and mutation rate >0.05 were used to construct the PHS model. The score was defined as the vector product of a patient’s mutational status (MS) (MS = 1 or 0, MS = 1 represents the patients carrying the mutational gene, MS = 0 represents the patients carrying the wildtype gene or synonymous gene) and the corresponding log hazard ratio (logHR) from the aforementioned Cox proportional hazards regression for the selected genes (see equation).
To ascertain whether the PHS was independently associated with OS or not, we subsequently performed the multivariate Cox regression analysis of PHS with adjustments for age, sex, TMB, and ICI regimes, and divided the 321 metastatic patients into three different levels (high-, intermediate-, and low-risk groups) according the PHS value, finally generating the Kaplan–Meier survival curves.
Validation analysis
We further validated this PHS mode in the DFCIMM cohort using the calculation method mentioned previously. Then, the 110 metastatic patients were also divided into three different groups according the PHS value, and Kaplan–Meier survival curves were generated at last.
Tumor-infiltrated immune cells’ assessment
In order to explore the possible mechanisms underlining the impact of those gene mutations on the survival of metastatic melanoma who received ICI therapies, we compared the differences in tumor-infiltrated immune cells between the groups of the patients carrying mutational genes and patients carrying wildtype genes from the (TCGA) dataset using TIMER online tools (https://cistrome.shinyapps.io/timer/) (Li et al., 2017), and boxplots were obtained.
All the data were analyzed by using the R language (version 3.5.1), and p values were two-sided with a significance level of 0.05.
Results
Eight genes were found in the discovery analysis
A total of 321 metastatic melanoma patients from the MSK-IMPCAT cohort who received immunotherapies were analyzed (116 patients received a combination of anti CTLA-4 and anti PD-1/PD-L1 immunotherapies, 75 and 130 patients received anti CTLA-4 and anti PD-1/PD-L1 respectively), and the general clinicopathological characteristics of these patients are presented in Table1. Meanwhile, clinical information of patients in the validation cohort (DFCIMM) is presented in Supplementary Table S2.
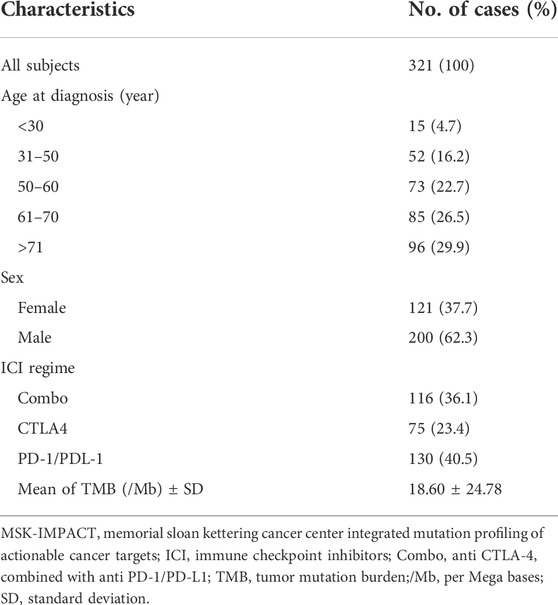
TABLE 1. Clinicopathological characteristics of the MSK-IMPACT metastatic melanoma cohort that underwent ICI treatment.
In the univariate Cox regression analysis, eight gene mutations were found to be significantly associated with OS in this cohort, i.e., BAP1, CARD11, IGF1R, KMT2D, PTPRD, PTPRT, ROS1,and TERT (HR = 1.72, 0.58, 0.47, 0.62, 0.66, 0.71, 0.68, and 0.71 respectively; p = 0.028, 0.033, 0.034, 0.034, 0.029, 0.029, 0.043, and 0.008, respectively); details are included In Table 2.
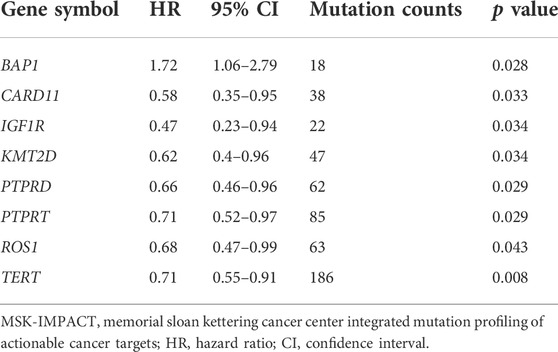
TABLE 2. These gene mutations were significantly associated with overall survival in the MSK-IMPACT metastatic melanoma cohort.
Kaplan–Meier survival curves are presented in Figure 1, and it was revealed that seven gene mutations were associated with better OS, except the BAP1 gene mutation which was associated with poorer OS.
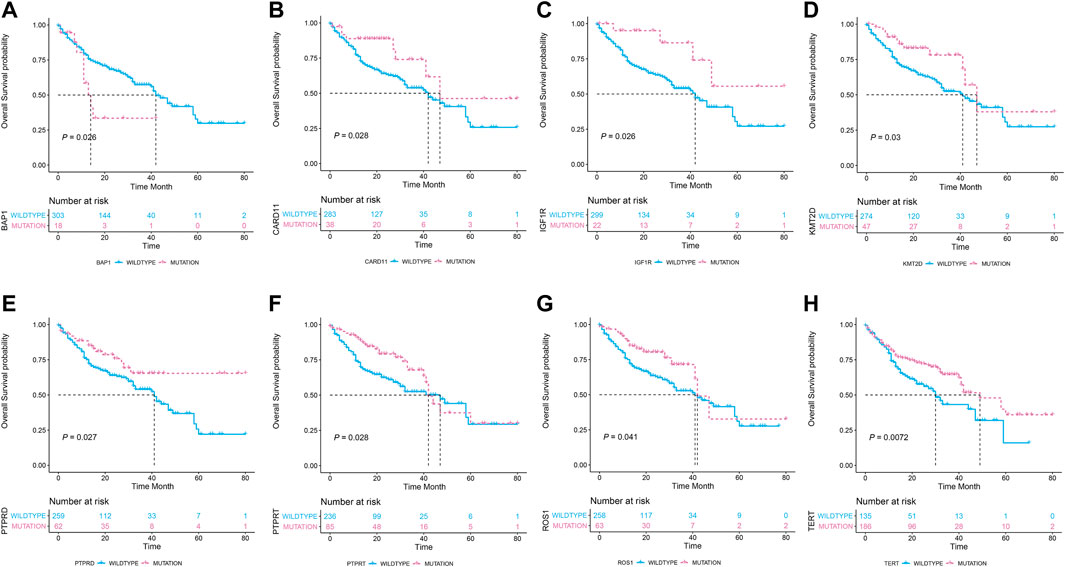
FIGURE 1. The association of the eight gene mutation statuses with the overall survival in 321 metastatic melanoma patients who received immune checkpoint inhibitor treatments. (A) Patients carrying BP1 mutations (n = 18) had poor overall survival than the wildtype counterparts (n = 303). (B–H): Patients carrying CARD11, IGF1R, KMT2D, PTPRD, PTPRT, ROS1, and TERT mutations had prolonged overall survival than their wildtype counterparts.
Polygenic hazard score model was constructed and validated
Because the OS is also affected by the age, ICI regime, and TMB, we subsequently performed the multivariate Cox regression analysis to generate the adjusted HR for each of the eight gene mutations (Supplementary Table S3). Subsequently, PHS was calculated for each of 321 patients based on the genotype regarding the eight genes’ mutational status as illustrated in the method part. After that, univariate and multivariate Cox regression analyses were performed to test the PHS model. It was revealed that, PHS can effectively predict the OS in the MSK-IMPACT cohort (i.e., the discovery cohort, HR = 1·54, 95%CI, 1.25–1.95; p < 0.001) and this prediction was independent from the TMB, age, sex, and ICI regime (adjusted HR = 1.84, 95%CI, 1.22–2.79; p = 0.004). Details are included in Table 3.
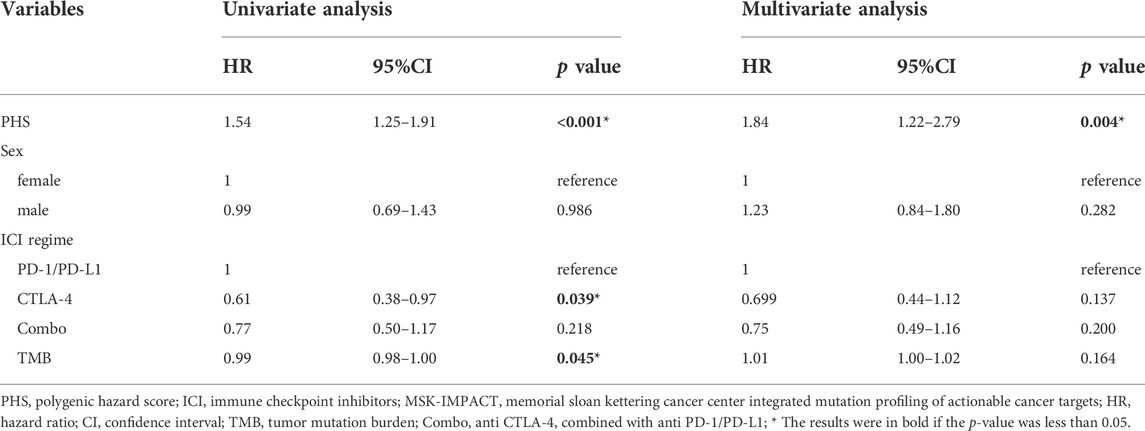
TABLE 3. Univariate and multivariate analyses of PHS associated with overall survival in the MSK-IMPACT metastatic melanoma cohort that underwent ICI treatment.
Additionally, the Kaplan–Meier survival curves of the three groups which were divided by the PHS value were presented in Figure 2A (log-Rank p < 0.001). In the DFCIMM cohort, the PHS was calculated using the same adjusted HR of those eight genes and was validated. It was shown that PHS was significantly associated with OS in 110 patients, the Kaplan–Meier survival curves of the three groups which were divided by PHS value are presented in Figure 2B (log-Rank p = 0.038).
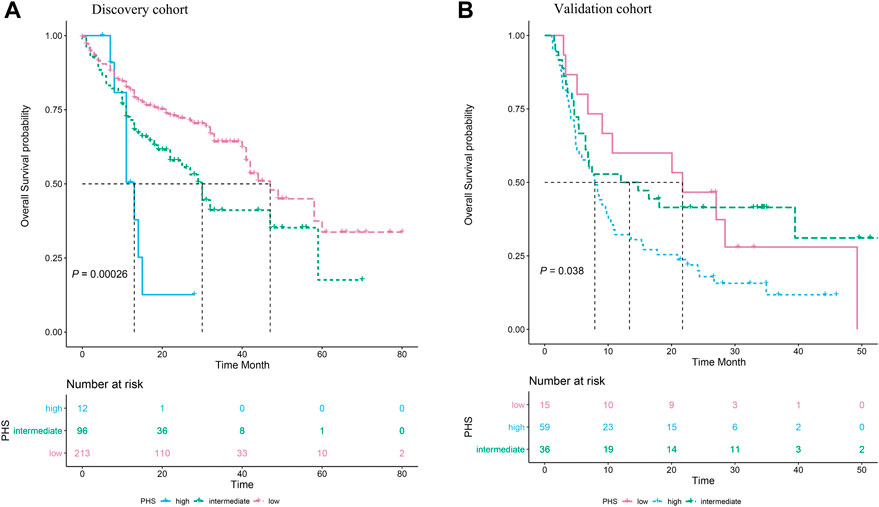
FIGURE 2. The association of polygenic hazard ratio (PHS) levels with the overall survival in metastatic melanoma patients treated with immune checkpoint inhibitors in two independent datasets. (A). Three subgroups, i.e., the high PHS, the intermediate PHS, and the low PHS, have significantly different overall survival rates in the MSK-IMPACT cohort. (B)The PHS model was successfully validated in the DFCI metastatic melanoma cohort, the group with high PHS had the poorest overall survival compared to the groups with intermediate PHS and low PHS.
CARD11 and PTPRD were associated with higher tumor-infiltrated immune cells in abundance
In those eight genes whose mutations can impact the OS in the metastatic melanoma patients, two genes, CARD11 and PTPRD, were associated with higher tumor-infiltrated immune cell abundance in skin melanoma tumor samples from the TCGA database. Boxplots in the Figure 3 showed that samples carrying CARD11 or PTPRD mutations harbored more dendric cells and CD4+ T cells. Specifically, CARD11 mutation was significantly associated with more dendric cells (p < 0.05); PTPRD mutation was significantly associated with more CD4+ T cells (p < 0.05).
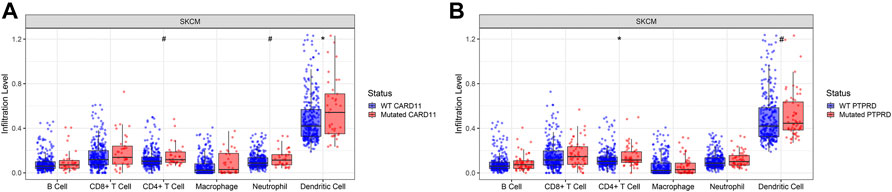
FIGURE 3. CARD11 and PTPRD genes were associated with higher tumor-infiltrated immune cell abundance in skin melanoma tumor samples from The Cancer Genome Atlas database. (A). CARD11 gene mutational status was significantly associated with more dendric cells (p < 0.05), and also associated with more CD4+ T cells and neutrophilic cells (p < 0.1). (B). PTPRD gene mutational status was significantly associated with more CD4+ T cells (p < 0.05), and was also associated with more dendric cells (p < 0.1). *: p < 0.05, #: p < 0.1.
Discussion
For ICI therapies in the metastatic melanoma, which was widely recognized as an immunogenic malignancy, the outcomes were different from patient to patient, suggesting that good biomarkers were critically needed. For patients who could not benefit from the ICI therapy, ICI therapies might result with unnecessary immune-mediated toxicities, sometimes this adverse effect could be lethal (Weber et al., 2015).
In our research, we identified eight genes which were significantly associated with longer median OS in the metastatic melanoma. Previous reports also suggested that a higher burden of gene copy number loss was associated with the anti-CTLA-4 and anti-PD-1 therapy response and the TP53 gene mutation was negatively associated with survival in the melanoma treated with anti-CTLA4 therapies (Van Allen et al., 2015; Weber et al., 2015; Li et al., 2017; Roh et al., 2017; Seibert et al., 2018; Xiao et al., 2018; Tan et al., 2019; Varga et al., 2019), However, a single gene prediction model was not effective enough to distinguish the responders or non-responders in ICI treatments.
Polygenic models have been well established in the risk prediction realm. But, for the melanoma or ICI therapies, there is no polygenic model for now. Hence, we developed this PHS model to predict the ICI therapy outcomes for the first time, and validated our PHS model in another independent cohort. Interestingly, when we performed the multivariate Cox regression model including the age, sex, ICI regimes, TMB, and PHS, it was found that PHS was still with a p value less than 0.05 while the p value of TMB changed into a larger value than 0.05, which suggested that PHS could more accurately and effectively predict the survival in metastatic melanoma under ICI treatments than TMB.
Furthermore, we found that two out of those eight gene (CARD11 and PTPRD) mutations were associated with more tumor-infiltrated immune cells in tumor tissues. This can partially explain the different outcomes after ICI treatments, because it was reported that more intratumor immune cells may lead to the benefit for ICI therapies in melanoma (Siddiqui et al., 2019). However, the other six gene mutations were not significantly associated with different tumor-infiltrated immune cell abundance. As for the CADR11 gene, researchers found that it played an important role in maintaining the normal adaptive immunity (Ma et al., 2017). Previous research also proved that deletion of the CADR11 gene (alias CARMA1) in regulatory T cells could produce IFN-γ leading to dramatical anti-tumor effects, and contributed positively to the ICI therapies (Di Pilato et al., 2019). Another study reported that CARD11 mutant skin cutaneous melanoma had higher immunogenicity compared to wildtype counterparts, and CARD11 mutation was associated with a longer OS after ICI therapy (Si et al., 2021). This was consistent with our findings, i.e., melanoma patients carrying mutational CARD11 samples survived longer than the wildtype melanoma patients after ICI therapies. As for the PTPRD gene, there were no reports about this gene in the tumor immunity area yet, but the PTPRD gene was involved in the JAK/STAT signaling which was found to be associated with tumor immunotherapy (Owen et al., 2019). PTPRD encodes a kind of protein tyrosine phosphatase which is involved in tyrosine phosphorylation. Walia et al. reported that PTPRD acts as a tumor-suppressor and was one of the most frequently mutated genes in cutaneous metastatic melanoma, mutant PTPRD loses the function of inhibiting cell proliferation and migration, some kind of mutants even promote cell migration (Walia et al., 2014).
In conclusion, we built a PHS model which can effectively and independently predict the ICI outcome in metastatic melanoma, and we successfully validated our PHS model in an independent cohort. This PHS model may serve to help the clinical decision-making for metastatic melanoma patients. However, due to the relatively small patients’ size, this PHS model may need improvements when larger cohorts are available.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material; further inquiries can be directed to the corresponding author.
Author contributions
Conception and design: LZ, TL, and XZ; Statistical analysis of data: LZ, TL, and JJ; Manuscript writing: LZ and JW; Final approval for the manuscript: All authors; Accountable for all aspects of the work: All authors.
Funding
This work was supported by the Natural Science Foundation of China (No. 81871948 to XZ).
Acknowledgments
We gratefully thank the support from the National Natural Science Foundation of China; We also gratefully thank the network convenience support from Fei Luo, a network engineer from China Merchants Securities Company.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmolb.2022.1001792/full#supplementary-material
References
Bray, F., Ferlay, J., Soerjomataram, I., Siegel, R. L., Torre, L. A., and Jemal, A. (2018). Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. Ca. Cancer J. Clin. 68 (6), 394–424. published Online First: 2018/09/13. doi:10.3322/caac.21492
Carlino, M. S., Long, G. V., Schadendorf, D., Robert, C., Ribas, A., Richtig, E., et al. (2018). Outcomes by line of therapy and programmed death ligand 1 expression in patients with advanced melanoma treated with pembrolizumab or ipilimumab in KEYNOTE-006: A randomised clinical trial. Eur. J. Cancer 101, 236–243. published Online First: 2018/08/11. doi:10.1016/j.ejca.2018.06.034
Chang, J. W., Yeh, K. Y., Wang, C. H., Yang, T. S., Chiang, H. F., Wei, F. C., et al. (2004). Malignant melanoma in taiwan: A prognostic study of 181 cases. Melanoma Res. 14 (6), 537–541. published Online First: 2004/12/04. doi:10.1097/00008390-200412000-00016
Di Pilato, M., Kim, E. Y., Cadilha, B. L., PruBmann, J. N., Nasrallah, M. N., Seruggia, D., et al. (2019). Targeting the CBM complex causes Treg cells to prime tumours for immune checkpoint therapy. Nature 570 (7759), 112–116. published Online First: 2019/05/17. doi:10.1038/s41586-019-1215-2
Flaherty, K. T., Puzanov, I., Kim, K. B., Ribas, A., McArthur, G. A., Sosman, J. A., et al. (2010). Inhibition of mutated, activated BRAF in metastatic melanoma. N. Engl. J. Med. 363 (9), 809–819. published Online First: 2010/09/08. doi:10.1056/NEJMoa1002011
Hodi, F. S., Chiarion-Sileni, V., Gonzalez, R., Grob, J. J., Rutkowski, P., Cowey, C. L., et al. (2018). Nivolumab plus ipilimumab or nivolumab alone versus ipilimumab alone in advanced melanoma (CheckMate 067): 4-year outcomes of a multicentre, randomised, phase 3 trial. Lancet. Oncol. 19 (11), 1480–1492. published Online First: 2018/10/27. doi:10.1016/s1470-2045(18)30700-9
Hodi, F. S., O'Day, S. J., McDermott, D. F., Weber, R. W., Sosman, J. A., Haanen, J. B., et al. (2010). Improved survival with ipilimumab in patients with metastatic melanoma. N. Engl. J. Med. 363 (8), 711–723. published Online First: 2010/06/08. doi:10.1056/NEJMoa1003466
Larkin, J., Chiarion-Sileni, V., Gonzalez, R., Grob, J. J., Cowey, C. L., Lao, C. D., et al. (2015). Combined nivolumab and ipilimumab or monotherapy in untreated melanoma. N. Engl. J. Med. 373 (1), 23–34. published Online First: 2015/06/02. doi:10.1056/NEJMoa1504030
Li, T., Fan, J., Wang, B., Traugh, N., Chen, Q., Liu, J. S., et al. (2017). Timer: A web server for comprehensive analysis of tumor-infiltrating immune cells. Cancer Res. 77 (21), e108–e110. published Online First: 2017/11/03. doi:10.1158/0008-5472.Can-17-0307
Ma, C. A., Stinson, J. R., Zhang, Y., Abbott, J. K., Weinreich, M. A., Hauk, P. J., et al. (2017). Germline hypomorphic CARD11 mutations in severe atopic disease. Nat. Genet. 49 (8), 1192–1201. published Online First: 2017/06/20. doi:10.1038/ng.3898
Owen, K. L., Brockwell, N. K., and Parker, B. S. (2019). JAK-STAT signaling: A double-edged sword of immune regulation and cancer progression. Cancers (Basel) 11 (12), E2002. published Online First: 2019/12/18. doi:10.3390/cancers11122002
Robert, C., Ribas, A., Schachter, J., Arance, A., Grob, J. J., Mortier, L., et al. (2019). Pembrolizumab versus ipilimumab in advanced melanoma (KEYNOTE-006): Post-hoc 5-year results from an open-label, multicentre, randomised, controlled, phase 3 study. Lancet. Oncol. 20 (9), 1239–1251. published Online First: 2019/07/28. doi:10.1016/s1470-2045(19)30388-2
Roh, W., Chen, P. L., Reuben, A., Spencer, C. N., Prieto, P. A., Miller, J. P., et al. (2017). Integrated molecular analysis of tumor biopsies on sequential CTLA-4 and PD-1 blockade reveals markers of response and resistance. Sci. Transl. Med. 9 (379), eaah3560. published Online First: 2017/03/03. doi:10.1126/scitranslmed.aah3560
Samstein, R. M., Lee, C. H., Shoushtari, A. N., Hellmann, M. D., Shen, R., Janjigian, Y. Y., et al. (2019). Tumor mutational load predicts survival after immunotherapy across multiple cancer types. Nat. Genet. 51 (2), 202–206. published Online First: 2019/01/16. doi:10.1038/s41588-018-0312-8
Seibert, T. M., Fan, C. C., Wang, Y., Zuber, V., Karunamuni, R., Parsons, J. K., et al. (2018). Polygenic hazard score to guide screening for aggressive prostate cancer: Development and validation in large scale cohorts. Bmj 360, j5757. published Online First: 2018/01/13. doi:10.1136/bmj.j5757
Si, L., Zhang, X., Shu, Y., Pan, H., Wu, D., Liu, J., et al. (2019). A phase ib study of pembrolizumab as second-line therapy for Chinese patients with advanced or metastatic melanoma (KEYNOTE-151). Transl. Oncol. 12 (6), 828–835. published Online First: 2019/04/14. doi:10.1016/j.tranon.2019.02.007
Si, Y., Lin, A., Ding, W., Meng, H., Luo, P., and Zhang, J. (2021). CARD11 alteration as a candidate biomarker of skin cutaneous melanoma treated with immune checkpoint blockade. Am. J. Transl. Res. 13 (1), 286–300. published Online First: 2021/02/03.
Siddiqui, I., Schaeuble, K., Chennupati, V., Fuertes Marraco, S. A., Calderon-Copete, S., Pais Ferreira, D., et al. Intratumoral tcf1(+)pd-1(+)cd8(+) T cells with stem-like properties promote tumor control in response to vaccination and checkpoint blockade immunotherapy. Immunity 2019;50(1):195–211.e10. doi:10.1016/j.immuni.2018.12.021published Online First: 2019/01/13]
Siegel, R. L., Miller, K. D., and Jemal, A. (2018). Cancer statistics, 2018. Ca. Cancer J. Clin. 68 (1), 7–30. published Online First: 2018/01/10. doi:10.3322/caac.21442
Tan, C. H., Bonham, L. W., Fan, C. C., Mormino, E. C., Sugrue, L. P., Broce, I. J., et al. (2019). Polygenic hazard score, amyloid deposition and Alzheimer's neurodegeneration. Brain 142 (2), 460–470. published Online First: 2019/01/29. doi:10.1093/brain/awy327
Van Allen, E. M., Miao, D., Schilling, B., Shukla, S. A., Blank, C., Zimmer, L., et al. (2015). Genomic correlates of response to CTLA-4 blockade in metastatic melanoma. Sci. (New York, NY) 350 (6257), 207–211. published Online First: 2015/09/12. doi:10.1126/science.aad0095
Varga, Z., Sinn, P., and Seidman, A. D. (2019). Summary of head-to-head comparisons of patient risk classifications by the 21-gene Recurrence Score® (RS) assay and other genomic assays for early breast cancer. Int. J. Cancer 145 (4), 882–893. published Online First: 2019/01/18. doi:10.1002/ijc.32139
Walia, V., Prickett, T. D., Kim, J. S., Gartner, J. J., Lin, J. C., Zhou, M., et al. (2014). Mutational and functional analysis of the tumor-suppressor PTPRD in human melanoma. Hum. Mutat. 35 (11), 1301–1310. published Online First: 2014/08/13. doi:10.1002/humu.22630
Wang, F., Zhao, Q., Wang, Y. N., Jin, Y., He, M. M., Liu, Z. X., et al. (2019). Evaluation of POLE and POLD1 mutations as biomarkers for immunotherapy outcomes across multiple cancer types. JAMA Oncol. 5, 1504–1506. [published Online First: 2019/08/16]. doi:10.1001/jamaoncol.2019.2963
Weber, J. S., Yang, J. C., Atkins, M. B., and Disis, M. L. (2015). Toxicities of immunotherapy for the practitioner. J. Clin. Oncol. 33 (18), 2092–2099. published Online First: 2015/04/29. doi:10.1200/jco.2014.60.0379
Keywords: immune checkpoint inhibitor, somatic gene mutation, polygenic hazard score, metastatic melanoma, tumor mutation burden
Citation: Zhao L, Luo T, Jiang J, Wu J and Zhang X (2022) Eight gene mutation-based polygenic hazard score as a potential predictor for immune checkpoint inhibitor therapy outcome in metastatic melanoma. Front. Mol. Biosci. 9:1001792. doi: 10.3389/fmolb.2022.1001792
Received: 24 July 2022; Accepted: 10 10 August 20222022;
Published: 02 September 2022.
Edited by:
Deshui Jia, Shanghai General Hospital, ChinaReviewed by:
Jiwei Zhang, Shanghai University of Traditional Chinese Medicine, ChinaDedong Cao, Renmin Hospital of Wuhan University, China
Jia Liu, Seton Hall University, United States
Copyright © 2022 Zhao, Luo, Jiang, Wu and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiaowei Zhang, ZG9uZ2Zhbmd6aGl6aXpoeHdAYWxpeXVuLmNvbQ==
†These authors have contributed equally to this work
 Liqin Zhao
Liqin Zhao Ting Luo4†
Ting Luo4† Xiaowei Zhang
Xiaowei Zhang