- 1Centre for Host-Microbiome Interactions, Faculty of Dental, Oral and Craniofacial Sciences, King’s College London, London, United Kingdom
- 2Randall Centre for Cell and Molecular Biophysics, King’s College London, London, United Kingdom
Historically proteins that form highly polymeric and filamentous assemblies have been notoriously difficult to study using high resolution structural techniques. This has been due to several factors that include structural heterogeneity, their large molecular mass, and available yields. However, over the past decade we are now seeing a major shift towards atomic resolution insight and the study of more complex heterogenous samples and in situ/ex vivo examination of multi-subunit complexes. Although supported by developments in solid state nuclear magnetic resonance spectroscopy (ssNMR) and computational approaches, this has primarily been due to advances in cryogenic electron microscopy (cryo-EM). The study of eukaryotic microtubules and bacterial pili are good examples, and in this review, we will give an overview of the technical innovations that have enabled this transition and highlight the advancements that have been made for these two systems. Looking to the future we will also describe systems that remain difficult to study and where further technical breakthroughs are required.
Introduction
In the 1930s-1950s, X-ray fibre diffraction studies of filamentous proteins by pioneers such as William Astbury, Francis Crick and Linus Pauling, laid the foundation for modern structural biology (Astbury and Street, 1932; Pauling and Corey, 1951a; Pauling and Corey, 1951b; Crick, 1952). These early studies provided new insights into the structural properties of fibrous substances such as keratin and collagen, however, they could only offer global information. With the advent of single crystal X-ray diffraction (Kendrew et al., 1958), from the 1960s structural studies of soluble proteins became the principal focus due to the delivery of atomic/subatomic resolutions. In the 1980s the first soluble protein was determined by solution state nuclear magnetic resonance (NMR) spectroscopy (Williamson et al., 1985) and with the development of recombinant protein expression systems (Itakura et al., 1977; Smith et al., 1983; Cregg et al., 1993), by the mid-1990s there became a clear exponential rise in soluble/globular macromolecular structures being deposited in the protein data bank (Berman et al., 2000). Likewise, new methods for isolating and reconstituting membrane proteins has led to significant numbers of these structures (Vinothkumar and Henderson, 2010), primarily elucidated by crystallographic methods, being deposited in the PDB since the 2000s.
Using solution NMR and X-ray crystallography we have gained significant understanding of filamentous systems through studying their lower-order subunits, however, due to technical limitations of these techniques, there has been a lack in our understanding of how these components interact and how this relates to their function. For example, there is a requirement for sample homogeneity (purity and molecular mass), high amounts of material, and for crystallography the sample must form highly ordered and relatively large crystals (generally >20–50 μm3). Early advancements in the determination of high-resolution filamentous protein structures include the crystal structure of a synthetic peptide based collagen-like fragment with a defined length, published in 1994 (Bella et al., 1994). In the late 1990s the first proteinaceous structures were determined using solid state NMR (ssNMR) spectroscopy: the helical antibiotic peptide gramicidin, integrated within a lipid bilayer (Ketchem et al., 1996), and a peptide-based pentameric transmembrane helical bundle of the acetyl choline receptor (Opella et al., 1999). This ushered in a new direction for studying highly polymeric solids and by 2003 ssNMR had been successfully implemented to determine the structure of the fd filamentous bacteriophage particle coat protein (Zeri et al., 2003). Since then, the development of smaller magic angle spinning (MAS) sample rotors that spin at higher frequencies and require less sample, advances in isotopic labelling and partial deuteration of samples, and general enhancements of signal sensitivity (Ashbrook and Hodgkinson, 2018), has led to ∼140 ssNMR-derived models being deposited in the PDB (Berman et al., 2000). Of these, ∼20% are of filamentous proteins. Furthermore, in the last 10 years, hybrid approaches incorporating ssNMR, Rosetta-based in silico modelling and/or electron microscopy has provided atomic structures for the bacterial type III secretion system needle (Loquet et al., 2012; Demers et al., 2014), the M13 bacteriophage capsid (Morag et al., 2015) and Aβ amyloid fibrils (Sgourakis et al., 2015). However, over the past decade, cryo-electron microscopy (cryo-EM) has transformed all aspects of structural biology and has become the primary driving force in providing major advancements towards routine structure determination of filamentous protein assemblies (Figure 1).
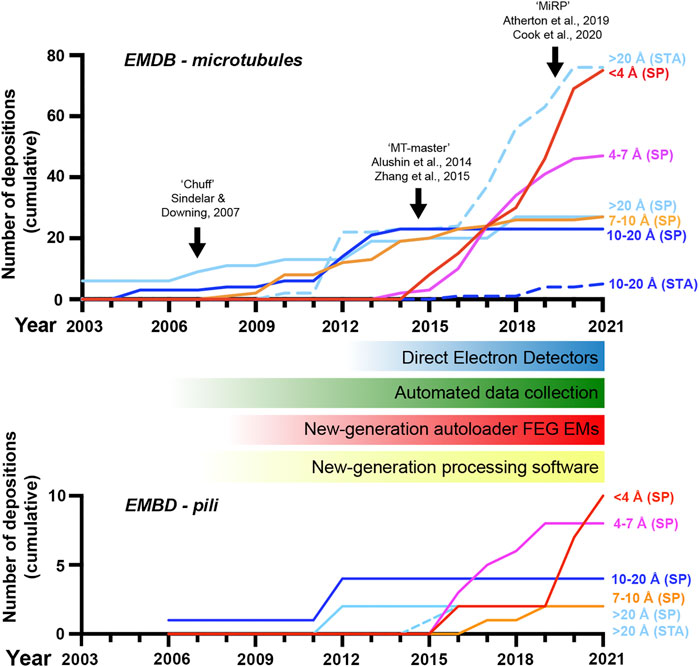
FIGURE 1. A revolution in cryo-electron microscopy of MTs and pili. Graph showing the cumulative number of electron microscopy database (EMDB) depositions over time at different resolution levels as indicated; SP, single-particle cryo-EM, STA, sub-tomogram averaging. The emergence of new general hardware and software behind the revolution in cryo-EM is shown below the MT timeline, along with black arrows indicating the introduction of several image-processing pipelines for pseudo-helical single MTs in the top graph. Included data was based on the query title:MT AND status:REL or title:pilus AND status:REL at www.emdatasource.org.
The Cryo-EM Revolution
Pre-revolution, cryo-EM produced near-atomic resolutions only in ideal sample cases; usually large macromolecules with high symmetry such as particular viral capsids. In additional preferable cases, sub-nanometer resolutions allowing secondary structure visualisation were possible, but for most targets, particularly those of low symmetry and/or small size (<200 kDa), cryo-EM was most-often limited to nm resolutions. Now, cryo-EM can routinely produce near-atomic resolution structures of even asymmetrical macromolecules of small size (currently down to around ∼50 kDa in ideal cases without a scaffold (Fan et al., 2019; Herzik et al., 2019)), with the first true atomic resolutions being reached in the last few years (Nakane et al., 2020; Yip et al., 2020). Furthermore, a significant amount of sample heterogeneity can also now be tolerated and even utilised, with snapshots of different conformational states and multimeric arrangements revealing the dynamics of macromolecular machines. Whilst remaining more challenging than single-particle cryo-EM, in the last 5 years or so cryo-electron tomography (cryo-ET) with sub-tomogram averaging has become more capable of achieving sub-nanometer resolutions in situ and near-atomic resolutions with purified macromolecular preparations (Schur, 2019; Turk and Baumeister, 2020; Pyle and Zanetti, 2021).
Core to cryo-EM’s transformation has been the introduction of direct electron detectors to replace charge-coupled devices and film, improving how faithfully transmitted electrons are recorded and allowing correction of global sample drift and local beam-induced motion (Li et al., 2013; Kuhlbrandt, 2014). Improvements to electron microscope hardware (including electron sources, stability, and energy filtration) and software (particularly increased automation in data collection) but also data processing hardware (increased computing power and improved storage) and software (in particular new Bayesian and artificial intelligence-based approaches) have also made significant contributions. The bottleneck to high-resolution cryo-EM is most often now the sample itself or the way it behaves during vitrification, however, innovation in sample preparation and data collection techniques are addressing these challenges.
In this review, we provide several examples of filamentous systems and show how our understanding has developed over the past decade due to these advances in structural biology techniques. We first discuss advancements in our understanding of microtubules (MTs) and MT complexes and then describe bacterial pili, primarily involved in adhesion. Looking to the future, we also highlight aspects that remain difficult to study and suggest where further advancements may be made.
Microtubules and Associated Proteins
MTs are tubular polymers of around 25 nm diameter built from longitudinally and laterally associated αβ-tubulin heterodimers and are a key component of the eukaryotic cytoskeleton. MTs display dynamic instability, in that they switch between polymerisation and depolymerisation phases modulated by β-tubulin’s GTPase activity, post-translational modifications and the interaction of microtubule-binding proteins. MT dynamics generate forces vital to cell division, serve as intracellular signalling platforms and provide the tracks for intracellular transport and force generation by kinesin and dynein family motor proteins (Vale, 2003; Goodson and Jonasson, 2018). While MTs can form into a number of different architectures built from 10–16 protofilaments, 3-start 13-protofilament pseudo-helices are the most commonly observed in nature (Tilney et al., 1973; Pierson et al., 1978; Wade et al., 1990). Pseudo-helical MTs are those with a discontinuity in the helical lattice known as the seam that has heterotypic (α to β-tubulin) rather than homotypic (α to α/β to β) lateral interactions between tubulin dimers.
High-Resolution Studies of MTs Using Cryo-EM
MTs were amongst the first targets to be studied by cryo-EM (Mandelkow and Mandelkow, 1985) following its development (Lepault et al., 1983; McDowall et al., 1983). Cryo-EM at this early stage already provided key advantages over EM with heavy metal stains, enabled by rapid sample vitrification in near-native conditions, yet was chiefly limited to theoretical extrapolation of 3D MT lattice architectures via analysis of real and reciprocal space patterns from 2D projections (Chretien and Wade, 1991). In the late 90s and early 2000s, 3D reconstructions of MTs with or without associated motor proteins via helical and pseudo-tomographic back-projection methods were limited to nanometre resolutions (e.g. (Arnal et al., 1996; Hoenger et al., 1998; Kikkawa et al., 2000; Kikkawa et al., 2001; Metoz et al., 1997; Nogales et al., 1999; Sosa et al., 1997a; Sosa et al., 1997b)) (Figure 1). This work gave information on the MT polymer not available from emerging crystallographic structures of αβ-tubulin or protofilament subunits (Nogales et al., 1995; Nogales et al., 1998; Lowe et al., 2001). An important and lasting shift was treating MT segments as single-particles in reference-matching approaches (Li et al., 2002). This, often combined with refinement of helical symmetrisation parameters during iterative rounds of reference-based alignment (Egelman, 2000), helped move some MT reconstructions into the sub-nanometre range allowing secondary structure identifications (Bodey et al., 2009; Sui and Downing, 2010; Alushin et al., 2012) (Figure 1).
A major challenge is differentiating between highly similar α and β-tubulin monomers during MT image processing, resulting in significant blurring of α and β-tubulin and a failure to resolve the seam in more physiologically relevant pseudo-symmetric MTs (Figure 2A). This issue was particularly prominent in studies of MTs alone, while the presence of MT-bound proteins demarcating tubulin dimers would act as fiducials during processing, alleviating the severity of the artefact, particularly for pseudo-symmetric MT architectures. Combining statistical methods for seam-finding with pseudo-symmetrical averaging approaches efficiently identified α and β-tubulin register and seam-location with MT-binding proteins acting as fiducials (Sindelar and Downing, 2007). Nevertheless, CCD and film-derived reconstructions of pseudo-helical MTs and associated proteins could not break the ∼4.5 Å resolution barrier required for visualisation of the peptide backbone and side chains (Alushin et al., 2014; Atherton et al., 2014; Fourniol et al., 2010; Maurer et al., 2012; Redwine et al., 2012; Sindelar and Downing, 2010). The introduction of direct electron detectors was central to MT reconstructions achieving near-atomic resolutions (<4.5 Å) (Figure 1). Ground-breaking work from the Nogales group, combining direct electron detector data with refined pseudo-helical processing methods achieved resolutions around 3.4 Å, allowing the authors to propose that small local nucleotide-dependent conformational changes leading to global changes in lattice compaction and twist govern dynamic instability (Zhang et al., 2015; Zhang and Nogales, 2015). This and later work to near-atomic resolutions continued to use MT binding proteins as fiducials for differentiation of α and β-tubulin in processing when working on tubulin isoforms or nucleotide or drug-induced conformational changes in the MT lattice; (Howes et al., 2018; Kellogg et al., 2017; Manka and Moores, 2018; Vemu et al., 2017; Vemu et al., 2016). However, this need for fiducials was overcome with additional image processing techniques, when near-atomic structural determination of pseudo-symmetric MTs without binding proteins in different nucleotide states was demonstrated (Zhang et al., 2018) (Figure 2A). Nevertheless, in many cases the MT-binding protein itself was the subject of structural interest, with notable examples solved near-atomic resolutions including the MT-binding regions of disease-related proteins tau (Kellogg et al., 2018) and doublecortin (Manka and Moores, 2020), minus-end binding CAMSAP (Atherton et al., 2019), MT-nucleator TPX2 (Zhang et al., 2017) and the MT-depolymerising kinesin-13 (Benoit et al., 2018) (Figure 2A).
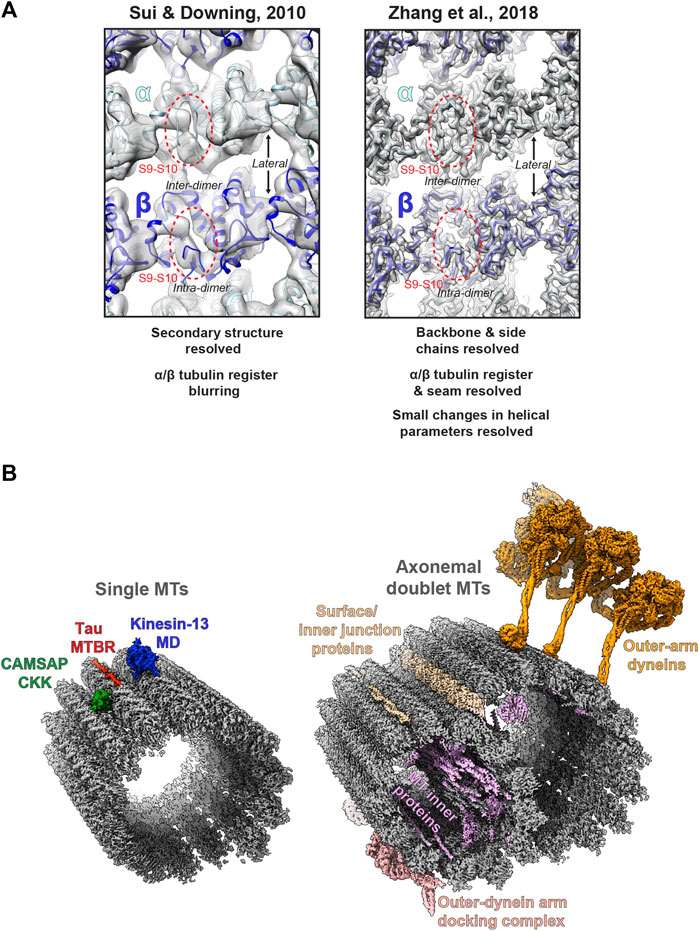
FIGURE 2. Cryo-electron microscopy of MTs (A) Images of the lumenal face of undecorated MTs, centred on an inter-dimer interface, but also showing intra-dimer interfaces and lateral interfaces between protofilaments. Grey density is shown for reconstructions of the MT alone; top, EMDB:5193 at ∼8 Å resolution (Sui and Downing, 2010); and bottom; EMDB:7973 at 3.1 Å resolution (Zhang et al., 2018), with the atomic model for the undecorated GMPCPP MT PDB:6dpu (Zhang et al., 2018) fitted into each reconstruction (α-tubulin light blue, β-tubulin dark blue). Failure to resolve differences between α and β-tubulin is a symptom of the earlier study (top), but not the more recent study (bottom), as illustrated by poor (top) or good (top) density differentiation between α and β-tubulin’s S9-S10 loop (within red dashed oval) (B) Left; exemplar cryo-EM structures from single MTs. Coloured cryo-EM densities for MT-binding proteins are shown as indicated on the single MT alone cryo-EM density map EMDB:7973 (Zhang et al., 2018) coloured grey; CAMSAP1 CKK domain (Atherton et al., 2019), MT-binding repeat (MTBR) of Tau (Kellogg et al., 2018) and the motor domain (MD) of kinesin-13 (Benoit et al., 2018). Right; exemplar cryo-EM structures from axonemal doublet MTs. Coloured cryo-EM density for MT-binding proteins are shown as indicated on the bovine tracheal cilia doublet-MT cryo-EM density map EMDB:24664 (Gui et al., 2021); bovine tracheal cilia MT inner proteins (MIPs), surface and innter junction proteins and outer-dynein arm docking complex EMDB:24664 (Gui et al., 2021) and outer-arm dynein from Tetrahymena thermophila EMDB:22677 (Rao et al., 2021).
In recent years, cryo-EM of MTs and binding partners has continued to develop (Figure 1). Near-atomic resolutions are being more readily achieved with accelerated automated and semi-automated data collection approaches (Tan et al., 2016) being adopted, allowing large amounts of data to be collected. Pipelines and methods for image processing of pseudo-symmetric MTs with or without MT-binding protein fiducials in the popular GUI-based program RELION have been introduced (Adib et al., 2019; Lacey et al., 2019; Cook et al., 2020). One challenge has been MT surface-binding proteins commonly being resolved at lower resolutions than the MT scaffold because heterogeneity in MT shape is generally amplified away from the centre of reconstructions, but also because of sample-dependent sub-stoichiometric occupancies and flexibility in the binder. In response to these challenges, new techniques have been developed to essentially subdivide MTs and bound proteins into sub-regions during processing, for example by using symmetry expansion and/or focused classification and refinement of individual MT subunits or binder sites to improve their quality (Liu et al., 2017; Debs et al., 2020; Cook et al., 2021). A recent stand-out study used focused classification methods to resolve MT inner proteins (MIPs) in the lumen of MTs extracted from Toxoplasma gondii to near-atomic resolution (Wang et al., 2021).
Although generally lagging-behind single-particle cryo-EM in resolution (Figure 1), cryo-ET can reveal 3D information on structurally heterogenous MT regions, such as lattice breaks and MT ends (Guesdon et al., 2016; Atherton et al., 2017; Gudimchuk et al., 2020). Furthermore, cryo-ET can be used to study MT architecture and organisation either in situ or in isolated ex-vivo preparations, where crowded overlapping environments render single-particle cryo-EM unsuitable (Atherton et al., 2018; McIntosh et al., 2018; Chakraborty et al., 2020). In particular cases where there are suitable repeating MT-associated sub-structures, sub-tomogram averaging (STA) can be employed to yield isotropic 3D reconstructions and improve resolution. For example, recently cryo-ET has resolved ex-vivo cytoplasmic dynein-dynactin transport teams on MTs (Chowdhury et al., 2015; Grotjahn et al., 2018), Parkinson’s disease related LRRK2-decorated MTs in cells (Watanabe et al., 2020), EB-decorated singlet MTs inside primary cilia (Kiesel et al., 2020) and revealed MT intra-lumenal F-actin in kinesore-induced cell projections (Paul et al., 2020).
Finally, alongside work on single MTs and their binding proteins, there has been a recent flurry of exciting cryo-ET/STA (Jordan et al., 2018; Zabeo et al., 2018; Owa et al., 2019; Greenan et al., 2020) and single-particle cryo-EM (Ma et al., 2019; Khalifa et al., 2020; Gui et al., 2021; Rao et al., 2021; Walton et al., 2021) work on the axonemal MT doublet structures of primary and motile cilia (Figure 2B). These studies have utilised both intact in situ and reduced membranated or de-membranated ex vivo preparations and have revealed a wealth of information on gross cilia architecture and the organisation, identities, and structure of axonemal MIPs, dynein complexes and intraflagellar transport trains (IFTs).
Studying Dynamic Interactions of Microtubules and Associated Proteins
Prior to cryo-EM’s revolution, ssNMR had provided a method of obtaining high-resolution information on drug-binding to MTs (Kumar et al., 2010). Nowadays, cryo-EM has become the tool of choice for studying the rigid interactions between MTs and their binding partners including small-molecules. However, cryo-EM struggles to resolve significant dynamics and flexible interactions due to the requirement for particle averaging and therefore ssNMR provides an ideal high-resolution method for studying the nature of these more dynamic interactions. Recent studies with labelled MT-binding domains of CAMSAPs, dynactin CAP-Gly, tau and plant companion of cellulose synthase 1 (CC1) have revealed atomic-level information on the dynamicity of their MT interfaces (Yan et al., 2015; Kadavath et al., 2018; Atherton et al., 2019; Kesten et al., 2019).
An exciting new development has been the purification of suitable amounts of isotopically labelled tubulin to produce MTs suitable for NMR studies, allowing labelled MTs and MT-binding proteins to be studied in parallel (Luo et al., 2021). In particular, this has enabled the study of the flexible and isoform-variable C-terminal tails of α and β tubulin involved in a plethora of interactions with MT-binding proteins within intact MTs (Janke, 2014; Roll-Mecak, 2020). This technical advance has now revealed the dynamic involvement of these C-terminal tails in CAMSAP-CKK domain and MAP7 MT binding at both slow and fast timescales (Luo et al., 2020) and has opened the door to further studies with a range of binding proteins.
Adhesive Bacterial Pili
The extracellular surfaces of bacteria are decorated with hair-like projections called pili or fimbriae, that are composed of smaller pilin subunits (Thanassi et al., 2012; Hospenthal et al., 2017a). Different types of pili range in their length and thickness and often have dedicated export and assembly systems that allow them to form on the bacterial surface. These include type IV-like and type V, chaperone-usher, amyloid-based, conjugative, type IV secretion, and sortase-mediated pili (Lukaszczyk et al., 2019). These pili have diverse functions including interacting with host cells during colonisation, promoting bacterial aggregation in biofilm formation, motility, conjugation, and secretion of proteins (Garnett and Matthews, 2012; Thanassi et al., 2012; Arutyunov and Frost, 2013; Berry and Pelicic, 2015). As such, these filamentous structures are often major virulence factors that drive the establishment of bacterial infection and the progression of disease. A decade ago, our primary understanding of pilus architectures was through crystallographic and solution NMR studies of monomeric pilin domains and the modelling of intact pili using low resolution (>10 Å) negative-stain and cryo-EM data (Craig et al., 2006; Salih et al., 2008; Garnett et al., 2012; Galkin et al., 2013). However, in the past 5 years ∼15 intact pilus structures have now been deposited in the PDB derived using cryo-EM data at near near-atomic resolutions (<4.5 Å) (Costa et al., 2016; Hospenthal et al., 2016; Hospenthal et al., 2017b; Spaulding et al., 2018; Filman et al., 2019; Wang et al., 2019; Zheng et al., 2019; Neuhaus et al., 2020; Shibata et al., 2020; Zheng et al., 2020; Gu et al., 2021; Pradhan et al., 2021). There has been a clear advancement in our ability to resolve high-resolution features of intact bacterial pili and this is well aligned with the introduction of direct electron detectors and processing techniques developed for filamentous helices (Figure 1). Here we will now discuss progress made in our understanding of two major classes of adhesive pili.
Type IV-like Pili
The first pilin structure was published in 1995 by John Tainer’s group; the crystal structure of an intact type IV pilus (T4P) major subunit, PilE, from Neisseria gonorrhoeae (Parge et al., 1995). In N. gonorrhea this pilus is the only known virulence factor required for infection (Kellogg et al., 1963; Swanson et al., 1987) and is important for binding host cells, other bacteria and delivering antigenic variation (Quillin and Seifert, 2018). Furthermore, the ability of the T4P to both extend and retract, coupled with its adhesive properties, provide bacteria with twitching motility and the ability to conjugate genomic material (Pelicic, 2008). The structure of PilE indicated that a long N-terminal helix could mediate pilus assembly, and the globular C-terminal region may decorate the pilus and provide specific function. In 2006 this was realised using 12.5 Å cryo-EM data to model the intact pilus (Craig et al., 2006) which displayed a tight helical packing of N-terminal helices and with a width of ∼6 nm (Figure 3). Then in 2015, cryo-ET was used to resolve the overall features of the PilA5 T4P isolated from Thermus thermophilus at 32 Å by STA (Figure 3). Strikingly, this pilus displayed a pronounced groove running along the fibre length and was much thinner at ∼3 nm (Gold et al., 2015). Over the past 5 years we have seen significant improvements in resolution first with the Neisseria meningitidis T4P using cryo-EM maps reconstructed at 6 Å (Kolappan et al., 2016) and then several other T4P models published guided by sub-nanometre resolution data from Escherichia coli, N. gonorrhoeae and Pseudomonas aeruginosa PAK (Bardiaux et al., 2019; Wang et al., 2017) (Figure 3). All these pili formed structures with widths of ∼5–6 nm. However, in the last year Vicki Gold’s group has now broken the ∼4.5 Å barrier, with cryo-EM maps for the Thermus thermophilus PilA4 and PilA5 pili at 3.2 Å and 3.5 Å, respectively, allowing backbone and sidechain details to be resolved as well as sites of glycosylation (Neuhaus et al., 2020) (Figure 3). Similar to the Neisseria-like pili, the PilA4 pilus has a width of ∼6 nm, while PilA5 is in line with the previous cryo-ET study with a width of ∼4 nm. This study has provided a clear atomic rational for the differences in overall appearance of these two classes of pili, which likely reflects their specific functions in natural transformation and twitching motility, respectively, and is based on unique inter-subunit interactions, helical parameters, and surface charge.
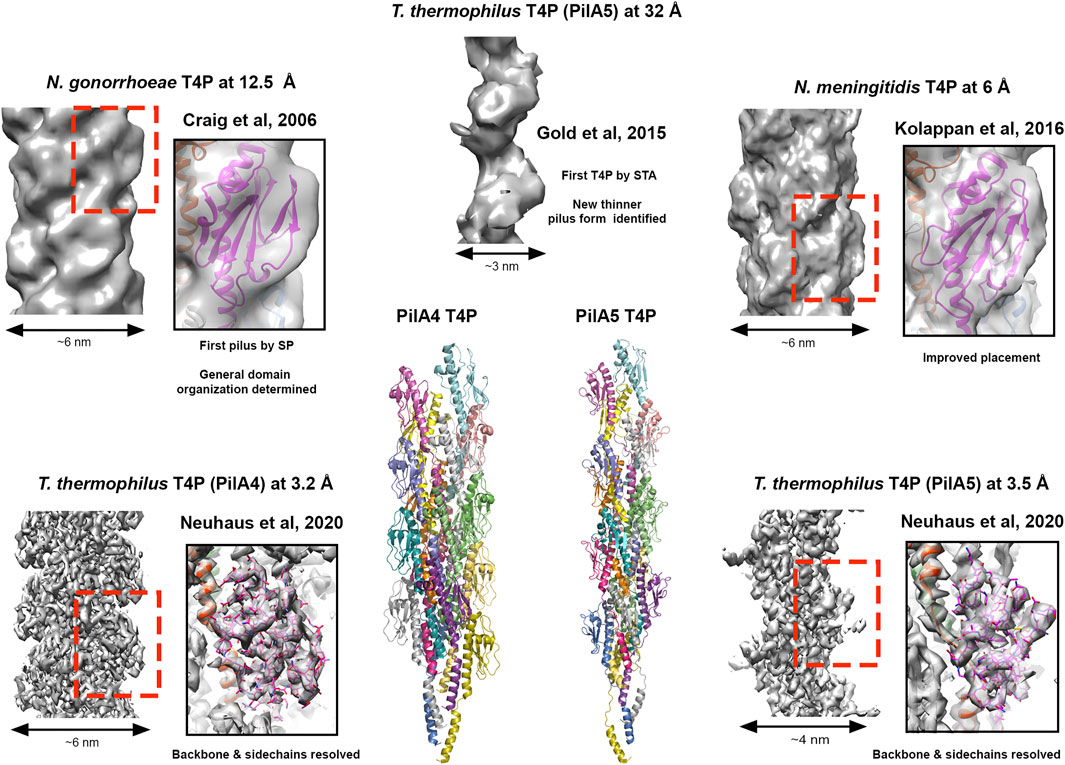
FIGURE 3. Cryo-electron microscopy of type IV pili. Images of T4P models derived from different resolutions of SP data STA data. Structures providing resolution breakthroughs are shown as example. SP Cryo-EM density and models of “thick” pili (N. meningitidis T4P at 12.5 Å (EMDB 1236; PDBID 2hil) (Craig et al., 2006), T. gonorrhoeae T4P at 6.0 Å (EMDB 8287; PDBID 5kua) (Kolappan et al., 2016) and the T. thermophilus PilA4 T4P at 3.2 Å (EMDB 10647; PDBID 6xxd) (Neuhaus et al., 2020)) and a thin “pilus” (T. thermophilus PilA5 T4P at 3.5 Å (EMDB 10648; PDBID 6xxe) (Neuhaus et al., 2020)) are shown. Cryo-EM density for the thinner T. thermophilus PilA5 T4P derived by STA at 32 Å is also presented (EMDB 3024) (Gold et al., 2015). Cartoon representation of the T. thermophilus PilA4 and PilA5 are also given as the first Cryo-EM backbone/sidechain resolved T4P structures (Neuhaus et al., 2020).
Related to the T4P, pseudo-pili have similar structures but different functions. For example, components that form the T4P system and are essential for pilus assembly are related to the type II secretion system (T2SS) which transport substrates and effectors, many of which are virulence factors, from the Gram-negative bacterial periplasm into the extracellular space (Gu et al., 2017). While the T4P system utilise ATPases that can both drive the formation and retraction of the pilus, the T2SS only contains expresses the extension ATPase, which it uses to push cargo across the bacterial outer membrane via polymerisation of the pilus via a syringe-like mechanism. In 2017 the structure of the Klebsiella oxytoca T2SS pseudo-pilus, PulG, was resolved using solution NMR and cryo-EM maps at 5.0 Å resolution, and this study has revealed that this pilus is stabilised by calcium ions and disassembles in their absence (Lopez-Castilla et al., 2017; Naskar et al., 2021). Geobacter sulfurreducens is a Gram-negative bacterium that uses surface nanowires for extracellular electron transfer. A recent cryo-EM structure of the copolymerised PilA-N/C pseudo-pilus at 3.8 Å now indicates that like the T2SS, this structure is used to push nanowires out of the bacterium (Gu et al., 2021). Furthermore, cryo-EM structures of the OmcS nanowire at 3.7 Å and 3.4 Å has uncovered a new pilus type formed through the polymerisation of OmcS hexaheme cytochromes, with hemes packed within 3.5–6.0 Å of each other to allow electron transport (Filman et al., 2019; Wang et al., 2019).
Donor-Strand Exchanged Pili
The chaperone-usher (CU) pilus assembly pathway is another well characterised system in Gram-negative bacteria with the first structure, the uropathogenic E. coli (UPEC) type 1 pilus minor FimC-FimH chaperone-adhesin complex, published at the end of the 1990s by Stefan Knight’s group. In CU systems, pilin domains consist of an incomplete Ig-like fold which lack the C-terminal strand, forming an acceptor groove, but have an additional unstructured extension at their N-terminus (Hospenthal et al., 2017a). Polymerisation proceeds through this N-terminal extension packing along the acceptor groove of an adjacent pilin subunit, which then stabilises and completes the Ig-like fold; a process called donor strand exchange. CU pili are highly variable but many consist of a major pilin subunit that makes up the majority of the fibre and then a minor pilin subunit (one or a few) at the tip, which is often an adhesin that binds carbohydrates or other receptors on the surface of host cells (e.g. E. coli type 1, P and common pili) (Garnett et al., 2012; Hospenthal et al., 2017a). However, in other CU systems, the major pilin domains that form the fibre shaft instead act as adhesive elements while the minor tip domain functions as an invasin that mediates invasion of host cells (e.g. E. coli AAF pili) (Berry et al., 2014); other arrangements also exist. Additionally, some CU pili are relatively thin (∼2–3 nm) and exist with an extended ‘beads on a string’ architecture that are relatively dynamic, with others being much thicker (∼10 nm) and forming rigid and more compacted helical arrangements.
A decade ago our understanding of CU pilus structures was primarily driven by crystallographic and NMR studies of generally monomeric or small engineered tandem subunits, however, again due to advances in cryo-EM we are now able to appreciate the functions of pilus packing through near-atomic resolution insights (Hospenthal et al., 2016; Hospenthal et al., 2017b; Spaulding et al., 2018; Zheng et al., 2019). In 2013 Lisa Craig’s group determined the global features of the enterotoxigenic E. coli (ETEC) CS1 pilus at 20 Å resolution by cryo-EM and the major CS1 pilin subunit CooA by crystallography at 1.6 Å resolution (Galkin et al., 2013). Modelling the CooA subunits into different cryo-EM maps revealed that pilins could adopt multiple orientations and structural states, resulting in different pilin packing and a dynamic pilus structure. They proposed that CS1 and other thicker class of pili may stretch in response to shear forces that they experience during colonisation. In 2015 Adam Lange’s group combined solution state NMR, ssNMR and scanning transmission electron microscopy (STEM) and determined the first atomic model of an intact UPEC type 1 pilus (Habenstein et al., 2015) while the following year, Gabriel Waksman’s group published the first near-atomic resolution structure of an intact UPEC P-pilus by cryo-EM at 3.8 Å (Hospenthal et al., 2016). This structure along with a subsequent structure of the UPEC type 1 pilus by cryo-EM at 4.2 Å was able to explain precisely how changing inter-subunit interactions within these pili can mediate spring-like properties (Hospenthal et al., 2017b; Spaulding et al., 2018). Another recent study of the ETEC CFA/I pilus by cryo-EM at 4 Å has again shown that the helical quaternary structure of the pilus is influenced by shear forces and this is likely a common function of wound CU pili (Miller et al., 2006; Zheng et al., 2019).
Within the past 5 years a new type of adhesive fibre has also been discovered in Bacteroidales, named the type V pilus (T5P) (Xu et al., 2016). These are composed of an anchor, a stalk, an adapter and a tip pilin (Hospenthal et al., 2017a). Last year the first structure of an intact T5P was determined by cryo-EM at 3.6 Å, which consisted of the polymerised FimA pilin stalk subunit from the bacterium Porphromonas gingivalis (Shibata et al., 2020). This has revealed not just a new pilus architecture but also a new mode of pilus formation. Unlike the CU pathway, T5P pilin domains contain a C-terminal extended region and when they are exported to the bacterial surface, the N-terminal strand is cleaved by the protease RgpB and released. This forms an acceptor groove that can then accept a C-terminal extension via strand exchange with an adjacent pilin subunit.
Conclusions and Perspectives
As has been discussed, structural biology of MTs and their binding proteins has advanced dramatically over the last decade, yet a number of samples and technical goals remain challenging, representing frontiers in the field. In single-particle cryo-EM of single MTs, an intriguing question is whether atomic resolutions are achievable. MTs include some intrinsic flexibility and heterogeneity, including bending and in some cases lattice breaks and even multiple seams (Cross, 2019; Debs et al., 2020). Future sample preparation, data collection and processing methods will have to reduce these variances, whilst essentially eliminating blurring of α and β-tubulin register (particularly challenging in the absence of fiducial binding proteins) in order to reach atomic resolutions. Resolving MT binding proteins with partial occupancy on the MT, or those adopting a mix of conformers remains difficult, although processing strategies including symmetry expansion, focused classifications and refinements and signal subtraction are proving fruitful (Liu et al., 2017; Debs et al., 2020; Cook et al., 2021). In some cases, rather than using the standard heterogenous tubulin purified from the brains of livestock, more homogenous sources such as purified single-isoform tubulins or tubulin from particular cell types can help improve resolve regions of tubulin isoform heterogeneity, such as the C-terminal tails and their interactions (Atherton et al., 2019; Li et al., 2020). Additionally, ssNMR is becoming a useful tool for particular cases where flexible interactions between MT-binding proteins and the MT are to be investigated.
As discussed, exciting recent data has also emerged from studies of cilia MTs that demonstrates the ability of cryo-EM to identify macromolecules de novo within in situ or ex vivo preparations (Gui et al., 2021; Kiesel et al., 2020; Wang et al., 2021). The ability to faithfully localise and identify macromolecules on MTs in situ is likely to be a prominent target in the future direction of the field. With the advent of a revolution in macromolecular structure prediction (Jumper et al., 2021) combined with expanding dataset sizes and increasing sub-tomogram averaging resolutions due to steady improvements in cryo-ET sample preparation (FIB), data-collection and image processing (Figure 1) (Turk and Baumeister, 2020; Hylton and Swulius, 2021; Pyle and Zanetti, 2021), this goal is looking more and more plausible.
On the back of cryo-EM method developments, primarily through the study of other filamentous systems such as MTs, there has also been a substantial increase in our understanding of bacterial pilus structures in recent years. This is highlighted by work shown here on type IV-like pili, CU pili and the newly characterised T5P, however, we still lack high-resolution insight of other types of pili, such as those formed from amyloids (Van Gerven et al., 2018), and lack an understanding of how minor pilin components interact within intact pilus assemblies. In many type IV pili, incorporation of minor pilins is required for pilus assembly and/or specific functions (Jacobsen et al., 2020) but it is unclear how these are distributed across the fibre or what effect these minor components have on the local pilus structure. Crystallographic and more recently cryo-EM studies have provided great insight into the atomic mechanisms of pilus biogenesis (Hospenthal et al., 2017a), however, pili often remain tethered to their secretion machinery, and there is a shortage of high-resolution observations in these states. The UPEC type 1 and P pili have become model systems to study CU pilus biogenesis and several groups have now provided snapshots of the initiation of pilus assembly at the outer membrane usher pore and the initial stages of adhesin exit into the extracellular space (Phan et al., 2011; Geibel et al., 2013; Du et al., 2018; Omattage et al., 2018; Du et al., 2021). With cryo-EM now leading the way here, the next leap will likely come once a defined number of major pilin subunits can be incorporated into these systems and then the maturation of the pilus can then be visualized in the context of secretion.
Although cryo-ET has not been used to study the CU pathway, it has been successfully implemented to elucidate the low-resolution features of T4P and T2SS pseudo-pilus assembly devices (Gold et al., 2015; Chang et al., 2016; Ghosal et al., 2019). Work on the T4P systems has revealed pilus structures emanating into the extracellular space (Gold et al., 2015; Chang et al., 2016) and with the expected future increases in cryo-ET resolution, as discussed above, in the future cryo-ET may be able to provide greater detail of how these pili form at the membrane and the role of pilus-specific assembly factors in their biogenesis (Hu et al., 2020). Cryo-EM has had a major impact on the of study filamentous systems over the past decade and it will be interesting to see how EM and its incorporation with other approaches (e.g. in silico prediction, solution/ssNMR, crystallography) will be able tackle more dynamic and heterogenous filamentous systems in the future.
Author Contributions
JG and JA wrote the manuscript. Both the authors contributed to the article and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The handling editor declared a shared affiliation with the authors at time of review.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Adib, R., Montgomery, J. M., Atherton, J., O'Regan, L., Richards, M. W., Straatman, K. R., et al. (2019). Mitotic Phosphorylation by NEK6 and NEK7 Reduces the Microtubule Affinity of EML4 to Promote Chromosome Congression. Sci. Signal. 12. doi:10.1126/scisignal.aaw2939
A. Garnett, J., and Matthews, S. (2012). Interactions in Bacterial Biofilm Development: a Structural Perspective. Curr. Protein Pept. Sci. 13, 739–755. doi:10.2174/138920312804871166
Alushin, G. M., Lander, G. C., Kellogg, E. H., Zhang, R., Baker, D., and Nogales, E. (2014). High-Resolution Microtubule Structures Reveal the Structural Transitions in αβ-Tubulin upon GTP Hydrolysis. Cell 157, 1117–1129. doi:10.1016/j.cell.2014.03.053
Alushin, G. M., Musinipally, V., Matson, D., Tooley, J., Stukenberg, P. T., and Nogales, E. (2012). Multimodal Microtubule Binding by the Ndc80 Kinetochore Complex. Nat. Struct. Mol. Biol. 19, 1161–1167. doi:10.1038/nsmb.2411
Arnal, I., Metoz, F., DeBonis, S., and Wade, R. H. (1996). Three-dimensional Structure of Functional Motor Proteins on Microtubules. Curr. Biol. 6, 1265–1270. doi:10.1016/s0960-9822(02)70712-4
Arutyunov, D., and Frost, L. S. (2013). F Conjugation: Back to the Beginning. Plasmid 70, 18–32. doi:10.1016/j.plasmid.2013.03.010
Ashbrook, S. E., and Hodgkinson, P. (2018). Perspective: Current Advances in Solid-State NMR Spectroscopy. J. Chem. Phys. 149, 040901. doi:10.1063/1.5038547
Astbury, W. T., and Street, A. (1932). X-Ray Studies of the Structure of Hair, Wool, and Related Fibres. I. General, Series A, Containing Papers of a Mathematical or Physical Character, 230. Philosophical Transactions of the Royal Society of London, 75–101. doi:10.1098/rsta.1932.0003
Atherton, J., Farabella, I., Yu, I. M., Rosenfeld, S. S., Houdusse, A., Topf, M., et al. (2014). Conserved Mechanisms of Microtubule-Stimulated ADP Release, ATP Binding, and Force Generation in Transport Kinesins. Elife 3, e03680. doi:10.7554/eLife.03680
Atherton, J., Jiang, K., Stangier, M. M., Luo, Y., Hua, S., Houben, K., et al. (2017). A Structural Model for Microtubule Minus-End Recognition and protection by CAMSAP Proteins. Nat. Struct. Mol. Biol. 24, 931–943. doi:10.1038/nsmb.3483
Atherton, J., Luo, Y., Xiang, S., Yang, C., Rai, A., Jiang, K., et al. (2019). Structural Determinants of Microtubule Minus End Preference in CAMSAP CKK Domains. Nat. Commun. 10, 5236. doi:10.1038/s41467-019-13247-6
Atherton, J., Stouffer, M., Francis, F., and Moores, C. A. (2018). Microtubule Architecture In Vitro and in Cells Revealed by Cryo-Electron Tomography. Acta Cryst. Sect D Struct. Biol. 74, 572–584. doi:10.1107/s2059798318001948
Bardiaux, B., de Amorim, G. C., Luna Rico, A., Zheng, W., Guilvout, I., Jollivet, C., et al. (2019). Structure and Assembly of the Enterohemorrhagic Escherichia coli Type 4 Pilus. Structure 27, 1082–1093. doi:10.1016/j.str.2019.03.021
Bella, J., Eaton, M., Brodsky, B., and Berman, H. M. (1994). Crystal and Molecular Structure of a Collagen-like Peptide at 1.9 Å Resolution. Science 266, 75–81. doi:10.1126/science.7695699
Benoit, M. P. M. H., Asenjo, A. B., and Sosa, H. (2018). Cryo-EM Reveals the Structural Basis of Microtubule Depolymerization by Kinesin-13s. Nat. Commun. 9, 1662. doi:10.1038/s41467-018-04044-8
Berman, H. M., Westbrook, J., Feng, Z., Gilliland, G., Bhat, T. N., Weissig, H., et al. (2000). The Protein Data Bank. Nucleic Acids Res. 28, 235–242. doi:10.1093/nar/28.1.235
Berry, A. A., Yang, Y., Pakharukova, N., Garnett, J. A., Lee, W.-c., Cota, E., et al. (2014). Structural Insight into Host Recognition by Aggregative Adherence Fimbriae of Enteroaggregative Escherichia coli. Plos Pathog. 10, e1004404. doi:10.1371/journal.ppat.1004404
Berry, J.-L., and Pelicic, V. (2015). Exceptionally Widespread Nanomachines Composed of Type IV Pilins: the Prokaryotic Swiss Army Knives. FEMS Microbiol. Rev. 39, 134–154. doi:10.1093/femsre/fuu001
Bodey, A. J., Kikkawa, M., and Moores, C. A. (2009). 9-Ångström Structure of a Microtubule-Bound Mitotic Motor. J. Mol. Biol. 388, 218–224. doi:10.1016/j.jmb.2009.03.008
Chakraborty, S., Mahamid, J., and Baumeister, W. (2020). Cryoelectron Tomography Reveals Nanoscale Organization of the Cytoskeleton and its Relation to Microtubule Curvature inside Cells. Structure 28, 991–1003. doi:10.1016/j.str.2020.05.013
Chang, Y. W., Rettberg, L. A., Treuner-Lange, A., Iwasa, J., Søgaard-Andersen, L., and Jensen, G. J. (2016). Architecture of the Type IVa Pilus Machine. Science 351, aad2001. doi:10.1126/science.aad2001
Chowdhury, S., Ketcham, S. A., Schroer, T. A., and Lander, G. C. (2015). Structural Organization of the Dynein-Dynactin Complex Bound to Microtubules. Nat. Struct. Mol. Biol. 22, 345–347. doi:10.1038/nsmb.2996
Chrétien, D., and Wade, R. H. (1991). New Data on the Microtubule Surface Lattice. Biol. Cel 71, 161–174. doi:10.1016/0248-4900(91)90062-r
Cook, A. D., Manka, S. W., Wang, S., Moores, C. A., and Atherton, J. (2020). A Microtubule RELION-Based Pipeline for Cryo-EM Image Processing. J. Struct. Biol. 209, 107402. doi:10.1016/j.jsb.2019.10.004
Cook, A. D., Roberts, A. J., Atherton, J., Tewari, R., Topf, M., and Moores, C. A. (2021). Cryo-EM Structure of a Microtubule-Bound Parasite Kinesin Motor and Implications for its Mechanism and Inhibition. J. Biol. Chem. 297, 101063. doi:10.1016/j.jbc.2021.101063
Costa, T. R. D., Ilangovan, A., Ukleja, M., Redzej, A., Santini, J. M., Smith, T. K., et al. (2016). Structure of the Bacterial Sex F Pilus Reveals an Assembly of a Stoichiometric Protein-Phospholipid Complex. Cell 166, 1436–1444. doi:10.1016/j.cell.2016.08.025
Craig, L., Volkmann, N., Arvai, A. S., Pique, M. E., Yeager, M., Egelman, E. H., et al. (2006). Type IV Pilus Structure by Cryo-Electron Microscopy and Crystallography: Implications for Pilus Assembly and Functions. Mol. Cel 23, 651–662. doi:10.1016/j.molcel.2006.07.004
Cregg, J. M., Vedvick, T. S., and Raschke, W. C. (1993). Recent Advances in the Expression of Foreign Genes in Pichia pastoris. Nat. Biotechnol. 11, 905–910. doi:10.1038/nbt0893-905
Cross, R. A. (2019). Microtubule Lattice Plasticity. Curr. Opin. Cel Biol. 56, 88–93. doi:10.1016/j.ceb.2018.10.004
Debs, G. E., Cha, M., Liu, X., Huehn, A. R., and Sindelar, C. V. (2020). Dynamic and Asymmetric Fluctuations in the Microtubule wall Captured by High-Resolution Cryoelectron Microscopy. Proc. Natl. Acad. Sci. USA 117, 16976–16984. doi:10.1073/pnas.2001546117
Demers, J.-P., Habenstein, B., Loquet, A., Kumar Vasa, S., Giller, K., Becker, S., et al. (2014). High-resolution Structure of the Shigella Type-III Secretion Needle by Solid-State NMR and Cryo-Electron Microscopy. Nat. Commun. 5, 4976. doi:10.1038/ncomms5976
Du, M., Yuan, Z., Werneburg, G. T., Henderson, N. S., Chauhan, H., Kovach, A., et al. (2021). Processive Dynamics of the Usher Assembly Platform during Uropathogenic Escherichia coli P Pilus Biogenesis. Nat. Commun. 12, 5207. doi:10.1038/s41467-021-25522-6
Du, M., Yuan, Z., Yu, H., Henderson, N., Sarowar, S., Zhao, G., et al. (2018). Handover Mechanism of the Growing Pilus by the Bacterial Outer-Membrane Usher FimD. Nature 562, 444–447. doi:10.1038/s41586-018-0587-z
Egelman, E. H. (2000). A Robust Algorithm for the Reconstruction of Helical Filaments Using Single-Particle Methods. Ultramicroscopy 85, 225–234. doi:10.1016/s0304-3991(00)00062-0
Fan, X., Wang, J., Zhang, X., Yang, Z., Zhang, J.-C., Zhao, L., et al. (2019). Single Particle Cryo-EM Reconstruction of 52 kDa Streptavidin at 3.2 Angstrom Resolution. Nat. Commun. 10, 2386. doi:10.1038/s41467-019-10368-w
Filman, D. J., Marino, S. F., Ward, J. E., Yang, L., Mester, Z., Bullitt, E., et al. (2019). Cryo-EM Reveals the Structural Basis of Long-Range Electron Transport in a Cytochrome-Based Bacterial Nanowire. Commun. Biol. 2, 219. doi:10.1038/s42003-019-0448-9
Fourniol, F. J., Sindelar, C. V., Amigues, B., Clare, D. K., Thomas, G., Perderiset, M., et al. (2010). Template-free 13-protofilament Microtubule-MAP Assembly Visualized at 8 Å Resolution. J. Cel Biol 191, 463–470. doi:10.1083/jcb.201007081
Galkin, V. E., Kolappan, S., Ng, D., Zong, Z., Li, J., Yu, X., et al. (2013). The Structure of the CS1 Pilus of Enterotoxigenic Escherichia coli Reveals Structural Polymorphism. J. Bacteriol. 195, 1360–1370. doi:10.1128/jb.01989-12
Garnett, J. A., Martinez-Santos, V. I., Saldana, Z., Pape, T., Hawthorne, W., Chan, J., et al. (2012). Structural Insights into the Biogenesis and Biofilm Formation by the Escherichia coli Common Pilus. Proc. Natl. Acad. Sci. 109, 3950–3955. doi:10.1073/pnas.1106733109
Geibel, S., Procko, E., Hultgren, S. J., Baker, D., and Waksman, G. (2013). Structural and Energetic Basis of Folded-Protein Transport by the FimD Usher. Nature 496, 243–246. doi:10.1038/nature12007
Ghosal, D., Kim, K. W., Zheng, H., Kaplan, M., Truchan, H. K., Lopez, A. E., et al. (2019). In Vivo structure of the Legionella Type II Secretion System by Electron Cryotomography. Nat. Microbiol. 4, 2101–2108. doi:10.1038/s41564-019-0603-6
Gold, V. A., Salzer, R., Averhoff, B., and Kühlbrandt, W. (2015). Structure of a Type IV Pilus Machinery in the Open and Closed State. Elife 4, e07380. doi:10.7554/eLife.07380
Goodson, H. V., and Jonasson, E. M. (2018). Microtubules and Microtubule-Associated Proteins. Cold Spring Harb Perspect. Biol. 10, a022608. doi:10.1101/cshperspect.a022608
Greenan, G. A., Vale, R. D., and Agard, D. A. (2020). Electron Cryotomography of Intact Motile Cilia Defines the Basal Body to Axoneme Transition. J. Cel Biol 219, e201907060. doi:10.1083/jcb.201907060
Grotjahn, D. A., Chowdhury, S., Xu, Y., McKenney, R. J., Schroer, T. A., and Lander, G. C. (2018). Cryo-electron Tomography Reveals that Dynactin Recruits a Team of Dyneins for Processive Motility. Nat. Struct. Mol. Biol. 25, 203–207. doi:10.1038/s41594-018-0027-7
Gu, S., Shevchik, V. E., Shaw, R., Pickersgill, R. W., and Garnett, J. A. (2017). The Role of Intrinsic Disorder and Dynamics in the Assembly and Function of the Type II Secretion System. Biochim. Biophys. Acta (Bba) - Proteins Proteomics 1865, 1255–1266. doi:10.1016/j.bbapap.2017.07.006
Gu, Y., Srikanth, V., Salazar-Morales, A. I., Jain, R., O’Brien, J. P., Yi, S. M., et al. (2021). Structure of Geobacter Pili Reveals Secretory rather Than Nanowire Behaviour. Nature 597, 430–434. doi:10.1038/s41586-021-03857-w
Gudimchuk, N. B., Ulyanov, E. V., O’Toole, E., Page, C. L., Vinogradov, D. S., Morgan, G., et al. (2020). Mechanisms of Microtubule Dynamics and Force Generation Examined with Computational Modeling and Electron Cryotomography. Nat. Commun. 11, 3765. doi:10.1038/s41467-020-17553-2
Guesdon, A., Bazile, F., Buey, R. M., Mohan, R., Monier, S., García, R. R., et al. (2016). EB1 Interacts with Outwardly Curved and Straight Regions of the Microtubule Lattice. Nat. Cel Biol 18, 1102–1108. doi:10.1038/ncb3412
Gui, M., Farley, H., Anujan, P., Anderson, J. R., Maxwell, D. W., Whitchurch, J. B., et al. (2021). De Novo identification of Mammalian Ciliary Motility Proteins Using Cryo-EM. Cell 184, 5791–5806. doi:10.1016/j.cell.2021.10.007
Habenstein, B., Loquet, A., Hwang, S., Giller, K., Vasa, S. K., Becker, S., et al. (2015). Hybrid Structure of the Type 1 Pilus of Uropathogenic Escherichia coli. Angew. Chem. Int. Ed. 54, 11691–11695. doi:10.1002/anie.201505065
Herzik, M. A., Wu, M., and Lander, G. C. (2019). High-resolution Structure Determination of Sub-100 kDa Complexes Using Conventional Cryo-EM. Nat. Commun. 10, 1032. doi:10.1038/s41467-019-08991-8
Hoenger, A., Sack, S., Thormählen, M., Marx, A., Müller, J., Gross, H., et al. (1998). Image Reconstructions of Microtubules Decorated with Monomeric and Dimeric Kinesins: Comparison with X-ray Structure and Implications for Motility. J. Cel Biol 141, 419–430. doi:10.1083/jcb.141.2.419
Hospenthal, M. K., Costa, T. R. D., and Waksman, G. (2017a). A Comprehensive Guide to Pilus Biogenesis in Gram-Negative Bacteria. Nat. Rev. Microbiol. 15, 365–379. doi:10.1038/nrmicro.2017.40
Hospenthal, M. K., Redzej, A., Dodson, K., Ukleja, M., Frenz, B., Rodrigues, C., et al. (2016). Structure of a Chaperone-Usher Pilus Reveals the Molecular Basis of Rod Uncoiling. Cell 164, 269–278. doi:10.1016/j.cell.2015.11.049
Hospenthal, M. K., Zyla, D., Costa, T. R. D., Redzej, A., Giese, C., Lillington, J., et al. (2017b). The Cryoelectron Microscopy Structure of the Type 1 Chaperone-Usher Pilus Rod. Structure 25, 1829–1838. doi:10.1016/j.str.2017.10.004
Howes, S. C., Geyer, E. A., LaFrance, B., Zhang, R., Kellogg, E. H., Westermann, S., et al. (2018). Structural and Functional Differences between Porcine Brain and Budding Yeast Microtubules. Cell Cycle 17, 278–287. doi:10.1080/15384101.2017.1415680
Hu, L. I., Yin, S., Ozer, E. A., Sewell, L., Rehman, S., Garnett, J. A., et al. (2020). Discovery of a New Neisseria Gonorrhoeae Type IV Pilus Assembly Factor, TfpC. mBio 11, e02528–20. doi:10.1128/mBio.02528-20
Hylton, R. K., and Swulius, M. T. (2021). Challenges and Triumphs in Cryo-Electron Tomography. iScience 24, 102959. doi:10.1016/j.isci.2021.102959
Itakura, K., Hirose, T., Crea, R., Riggs, A. D., Heyneker, H. L., Bolivar, F., et al. (1977). Expression in Escherichia coli of a Chemically Synthesized Gene for the Hormone Somatostatin. Science 198, 1056–1063. doi:10.1126/science.412251
Jacobsen, T., Bardiaux, B., Francetic, O., Izadi-Pruneyre, N., and Nilges, M. (2020). Structure and Function of Minor Pilins of Type IV Pili. Med. Microbiol. Immunol. 209, 301–308. doi:10.1007/s00430-019-00642-5
Janke, C. (2014). The Tubulin Code: Molecular Components, Readout Mechanisms, and Functions. J. Cel Biol 206, 461–472. doi:10.1083/jcb.201406055
Jordan, M. A., Diener, D. R., Stepanek, L., and Pigino, G. (2018). The Cryo-EM Structure of Intraflagellar Transport Trains Reveals How Dynein Is Inactivated to Ensure Unidirectional Anterograde Movement in Cilia. Nat. Cel Biol 20, 1250–1255. doi:10.1038/s41556-018-0213-1
Jumper, J., Evans, R., Pritzel, A., Green, T., Figurnov, M., Ronneberger, O., et al. (2021). Highly Accurate Protein Structure Prediction with AlphaFold. Nature 596, 583–589. doi:10.1038/s41586-021-03819-2
Kadavath, H., Cabrales Fontela, Y., Jaremko, M., Jaremko, Ł., Overkamp, K., Biernat, J., et al. (2018). The Binding Mode of a Tau Peptide with Tubulin. Angew. Chem. Int. Ed. 57, 3246–3250. doi:10.1002/anie.201712089
Kellogg, D. S., Peacock, W. L., Deacon, W. E., Brown, L., and Pirkle, C. I. (1963). Neisseria Gonorrhoeae I. J. Bacteriol. 85, 1274–1279. doi:10.1128/jb.85.6.1274-1279.1963
Kellogg, E. H., Hejab, N. M. A., Howes, S., Northcote, P., Miller, J. H., Díaz, J. F., et al. (2017). Insights into the Distinct Mechanisms of Action of Taxane and Non-taxane Microtubule Stabilizers from Cryo-EM Structures. J. Mol. Biol. 429, 633–646. doi:10.1016/j.jmb.2017.01.001
Kellogg, E. H., Hejab, N. M. A., Poepsel, S., Downing, K. H., DiMaio, F., and Nogales, E. (2018). Near-atomic Model of Microtubule-Tau Interactions. Science 360, 1242–1246. doi:10.1126/science.aat1780
Kendrew, J. C., Bodo, G., Dintzis, H. M., Parrish, R. G., Wyckoff, H., and Phillips, D. C. (1958). A Three-Dimensional Model of the Myoglobin Molecule Obtained by X-ray Analysis. Nature 181, 662–666. doi:10.1038/181662a0
Kesten, C., Wallmann, A., Schneider, R., McFarlane, H. E., Diehl, A., Khan, G. A., et al. (2019). The Companion of Cellulose Synthase 1 Confers Salt Tolerance through a Tau-like Mechanism in Plants. Nat. Commun. 10, 857. doi:10.1038/s41467-019-08780-3
Ketchem, R. R., Lee, K. C., Huo, S., and Cross, T. A. (1996). Macromolecular Structural Elucidation with Solid-State NMR-Derived Orientational Constraints. J. Biomol. NMR 8, 1–14. doi:10.1007/BF00198135
Khalifa, A. A. Z., Ichikawa, M., Dai, D., Kubo, S., Black, C. S., Peri, K., et al. (2020). The Inner junction Complex of the Cilia Is an Interaction Hub that Involves Tubulin post-translational Modifications. Elife 9, e52760. doi:10.7554/eLife.52760
Kiesel, P., Alvarez Viar, G., Tsoy, N., Maraspini, R., Gorilak, P., Varga, V., et al. (2020). The Molecular Structure of Mammalian Primary Cilia Revealed by Cryo-Electron Tomography. Nat. Struct. Mol. Biol. 27, 1115–1124. doi:10.1038/s41594-020-0507-4
Kikkawa, M., Okada, Y., and Hirokawa, N. (2000). 15 Å Resolution Model of the Monomeric Kinesin Motor, KIF1A. Cell 100, 241–252. doi:10.1016/s0092-8674(00)81562-7
Kikkawa, M., Sablin, E. P., Okada, Y., Yajima, H., Fletterick, R. J., and Hirokawa, N. (2001). Switch-based Mechanism of Kinesin Motors. Nature 411, 439–445. doi:10.1038/35078000
Kolappan, S., Coureuil, M., Yu, X., Nassif, X., Egelman, E. H., and Craig, L. (2016). Structure of the Neisseria Meningitidis Type IV Pilus. Nat. Commun. 7, 13015. doi:10.1038/ncomms13015
Kühlbrandt, W. (2014). The Resolution Revolution. Science 343, 1443–1444. doi:10.1126/science.1251652
Kumar, A., Heise, H., Blommers, M. J. J., Krastel, P., Schmitt, E., Petersen, F., et al. (2010). Interaction of Epothilone B (Patupilone) with Microtubules as Detected by Two-Dimensional Solid-State NMR Spectroscopy. Angew. Chem. Int. Edition 49, 7504–7507. doi:10.1002/anie.201001946
Lacey, S. E., He, S., Scheres, S. H., and Carter, A. P. (2019). Cryo-EM of Dynein Microtubule-Binding Domains Shows How an Axonemal Dynein Distorts the Microtubule. Elife 8, e47145. doi:10.7554/eLife.47145
Lepault, J., Booy, F. P., and Dubochet, J. (1983). Electron Microscopy of Frozen Biological Suspensions. J. Microsc. 129, 89–102. doi:10.1111/j.1365-2818.1983.tb04163.x
Li, F., Li, Y., Ye, X., Gao, H., Shi, Z., Luo, X., et al. (2020). Cryo-EM Structure of VASH1-SVBP Bound to Microtubules. Elife 9, e58157. doi:10.7554/eLife.58157
Li, H., DeRosier, D. J., Nicholson, W. V., Nogales, E., and Downing, K. H. (2002). Microtubule Structure at 8 Å Resolution. Structure 10, 1317–1328. doi:10.1016/s0969-2126(02)00827-4
Li, X., Mooney, P., Zheng, S., Booth, C. R., Braunfeld, M. B., Gubbens, S., et al. (2013). Electron Counting and Beam-Induced Motion Correction Enable Near-Atomic-Resolution Single-Particle Cryo-EM. Nat. Methods 10, 584–590. doi:10.1038/nmeth.2472
Liu, D., Liu, X., Shang, Z., and Sindelar, C. V. (2017). Structural Basis of Cooperativity in Kinesin Revealed by 3D Reconstruction of a Two-Head-Bound State on Microtubules. Elife 6, e24490. doi:10.7554/eLife.24490
López-Castilla, A., Thomassin, J.-L., Bardiaux, B., Zheng, W., Nivaskumar, M., Yu, X., et al. (2017). Structure of the Calcium-dependent Type 2 Secretion Pseudopilus. Nat. Microbiol. 2, 1686–1695. doi:10.1038/s41564-017-0041-2
Loquet, A., Sgourakis, N. G., Gupta, R., Giller, K., Riedel, D., Goosmann, C., et al. (2012). Atomic Model of the Type III Secretion System Needle. Nature 486, 276–279. doi:10.1038/nature11079
Löwe, J., Li, H., Downing, K. H., and Nogales, E. (2001). Refined Structure of αβ-tubulin at 3.5 Å Resolution 1 1Edited by I. A. Wilson. J. Mol. Biol. 313, 1045–1057. doi:10.1006/jmbi.2001.5077
Lukaszczyk, M., Pradhan, B., and Remaut, H. (2019). “The Biosynthesis and Structures of Bacterial Pili,” in Bacterial Cell Walls and Membranes. Editor A. Kuhn (Cham: Springer International Publishing), 369–413. doi:10.1007/978-3-030-18768-2_12
Luo, Y., Xiang, S., Hooikaas, P. J., van Bezouwen, L., Jijumon, A. S., Janke, C., et al. (2020). Direct Observation of Dynamic Protein Interactions Involving Human Microtubules Using Solid-State NMR Spectroscopy. Nat. Commun. 11, 18. doi:10.1038/s41467-019-13876-x
Luo, Y., Xiang, S., Paioni, A. L., Adler, A., Hooikaas, P. J., Jijumon, A. S., et al. (2021). Solid-State NMR Spectroscopy for Studying Microtubules and Microtubule-Associated Proteins. Methods Mol. Biol. 2305, 193–201. doi:10.1007/978-1-0716-1406-8_10
Ma, M., Stoyanova, M., Rademacher, G., Dutcher, S. K., Brown, A., and Zhang, R. (2019). Structure of the Decorated Ciliary Doublet Microtubule. Cell 179, 909–922. doi:10.1016/j.cell.2019.09.030
Mandelkow, E.-M., and Mandelkow, E. (1985). Unstained Microtubules Studied by Cryo-Electron Microscopy. J. Mol. Biol. 181, 123–135. doi:10.1016/0022-2836(85)90330-4
Manka, S. W., and Moores, C. A. (2020). Pseudo-repeats in Doublecortin Make Distinct Mechanistic Contributions to Microtubule Regulation. EMBO Rep. 21, e51534. doi:10.15252/embr.202051534
Manka, S. W., and Moores, C. A. (2018). The Role of Tubulin-Tubulin Lattice Contacts in the Mechanism of Microtubule Dynamic Instability. Nat. Struct. Mol. Biol. 25, 607–615. doi:10.1038/s41594-018-0087-8
Maurer, S. P., Fourniol, F. J., Bohner, G., Moores, C. A., and Surrey, T. (2012). EBs Recognize a Nucleotide-dependent Structural Cap at Growing Microtubule Ends. Cell 149, 371–382. doi:10.1016/j.cell.2012.02.049
McDowall, A. W., Chang, J.-J., Freeman, R., Lepault, J., Walter, C. A., and Dubochet, J. (1983). Electron Microscopy of Frozen Hydrated Sections of Vitreous Ice and Vitrified Biological Samples. J. Microsc. 131, 1–9. doi:10.1111/j.1365-2818.1983.tb04225.x
McIntosh, J. R., O’Toole, E., Morgan, G., Austin, J., Ulyanov, E., Ataullakhanov, F., et al. (2018). Microtubules Grow by the Addition of Bent Guanosine Triphosphate Tubulin to the Tips of Curved Protofilaments. J. Cel Biol 217, 2691–2708. doi:10.1083/jcb.201802138
Metoz, F., Arnal, I., and Wade, R. H. (1997). Tomography without Tilt: Three-Dimensional Imaging of Microtubule/motor Complexes. J. Struct. Biol. 118, 159–168. doi:10.1006/jsbi.1997.3850
Miller, E., Garcia, T., Hultgren, S., and Oberhauser, A. F. (2006). The Mechanical Properties of E. coli Type 1 Pili Measured by Atomic Force Microscopy Techniques. Biophysical J. 91, 3848–3856. doi:10.1529/biophysj.106.088989
Morag, O., Sgourakis, N. G., Baker, D., and Goldbourt, A. (2015). The NMR-Rosetta Capsid Model of M13 Bacteriophage Reveals a Quadrupled Hydrophobic Packing Epitope. Proc. Natl. Acad. Sci. USA 112, 971–976. doi:10.1073/pnas.1415393112
Nakane, T., Kotecha, A., Sente, A., McMullan, G., Masiulis, S., Brown, P. M. G. E., et al. (2020). Single-particle Cryo-EM at Atomic Resolution. Nature 587, 152–156. doi:10.1038/s41586-020-2829-0
Naskar, S., Hohl, M., Tassinari, M., and Low, H. H. (2021). The Structure and Mechanism of the Bacterial Type II Secretion System. Mol. Microbiol. 115, 412–424. doi:10.1111/mmi.14664
Neuhaus, A., Selvaraj, M., Salzer, R., Langer, J. D., Kruse, K., Kirchner, L., et al. (2020). Cryo-electron Microscopy Reveals Two Distinct Type IV Pili Assembled by the Same Bacterium. Nat. Commun. 11, 2231. doi:10.1038/s41467-020-15650-w
Nogales, E., Grayer Wolf, S., Khan, I. A., Ludueña, R. F., and Downing, K. H. (1995). Structure of Tubulin at 6.5 Å and Location of the Taxol-Binding Site. Nature 375, 424–427. doi:10.1038/375424a0
Nogales, E., Whittaker, M., Milligan, R. A., and Downing, K. H. (1999). High-resolution Model of the Microtubule. Cell 96, 79–88. doi:10.1016/s0092-8674(00)80961-7
Nogales, E., Wolf, S. G., and Downing, K. H. (1998). Structure of the αβ Tubulin Dimer by Electron Crystallography. Nature 391, 199–203. doi:10.1038/34465
Omattage, N. S., Deng, Z., Pinkner, J. S., Dodson, K. W., Almqvist, F., Yuan, P., et al. (2018). Structural Basis for Usher Activation and Intramolecular Subunit Transfer in P Pilus Biogenesis in Escherichia coli. Nat. Microbiol. 3, 1362–1368. doi:10.1038/s41564-018-0255-y
Opella, S. J., Marassi, F. M., Gesell, J. J., Valente, A. P., Kim, Y., Oblatt-Montal, M., et al. (1999). Structures of the M2 Channel-Lining Segments from Nicotinic Acetylcholine and NMDA Receptors by NMR Spectroscopy. Nat. Struct. Biol. 6, 374–379. doi:10.1038/7610
Owa, M., Uchihashi, T., Yanagisawa, H.-a., Yamano, T., Iguchi, H., Fukuzawa, H., et al. (2019). Inner Lumen Proteins Stabilize Doublet Microtubules in Cilia and Flagella. Nat. Commun. 10, 1143. doi:10.1038/s41467-019-09051-x
Parge, H. E., Forest, K. T., Hickey, M. J., Christensen, D. A., Getzoff, E. D., and Tainer, J. A. (1995). Structure of the Fibre-Forming Protein Pilin at 2.6 Å Resolution. Nature 378, 32–38. doi:10.1038/378032a0
Paul, D. M., Mantell, J., Borucu, U., Coombs, J., Surridge, K. J., Squire, J. M., et al. (2020). In Situ cryo-electron Tomography Reveals Filamentous Actin within the Microtubule Lumen. J. Cel Biol 219. doi:10.1083/jcb.201911154
Pauling, L., and Corey, R. B. (1951a). The Structure of Fibrous Proteins of the Collagen-Gelatin Group. Proc. Natl. Acad. Sci. 37, 272–281. doi:10.1073/pnas.37.5.272
Pauling, L., and Corey, R. B. (1951b). The Structure of Hair, Muscle, and Related Proteins. Proc. Natl. Acad. Sci. 37, 261–271. doi:10.1073/pnas.37.5.261
Pelicic, V. (2008). Type IV pili: e pluribus unum? Mol. Microbiol. 68, 827–837. doi:10.1111/j.1365-2958.2008.06197.x
Phan, G., Remaut, H., Wang, T., Allen, W. J., Pirker, K. F., Lebedev, A., et al. (2011). Crystal Structure of the FimD Usher Bound to its Cognate FimC-FimH Substrate. Nature 474, 49–53. doi:10.1038/nature10109
Pierson, G. B., Burton, P. R., and Himes, R. H. (1978). Alterations in Number of Protofilaments in Microtubules Assembled In Vitro. J. Cel Biol 76, 223–228. doi:10.1083/jcb.76.1.223
Pradhan, B., Liedtke, J., Sleutel, M., Lindbäck, T., Zegeye, E. D., Sullivan, O, et al. (2021). Endospore Appendages: a Novel Pilus Superfamily from the Endospores of Pathogenic Bacilli. EMBO J. 40, e106887. doi:10.15252/embj.2020106887
Pyle, E., and Zanetti, G. (2021). Current Data Processing Strategies for Cryo-Electron Tomography and Subtomogram Averaging. Biochem. J. 478, 1827–1845. doi:10.1042/bcj20200715
Quillin, S. J., and Seifert, H. S. (2018). Neisseria Gonorrhoeae Host Adaptation and Pathogenesis. Nat. Rev. Microbiol. 16, 226–240. doi:10.1038/nrmicro.2017.169
Rao, Q., Han, L., Wang, Y., Chai, P., Kuo, Y.-w., Yang, R., et al. (2021). Structures of Outer-Arm Dynein Array on Microtubule Doublet Reveal a Motor Coordination Mechanism. Nat. Struct. Mol. Biol. 28, 799–810. doi:10.1038/s41594-021-00656-9
Redwine, W. B., Hernández-López, R., Zou, S., Huang, J., Reck-Peterson, S. L., and Leschziner, A. E. (2012). Structural Basis for Microtubule Binding and Release by Dynein. Science 337, 1532–1536. doi:10.1126/science.1224151
Roll-Mecak, A. (2020). The Tubulin Code in Microtubule Dynamics and Information Encoding. Dev. Cel 54, 7–20. doi:10.1016/j.devcel.2020.06.008
Salih, O., Remaut, H., Waksman, G., and Orlova, E. V. (2008). Structural Analysis of the Saf Pilus by Electron Microscopy and Image Processing. J. Mol. Biol. 379, 174–187. doi:10.1016/j.jmb.2008.03.056
Schur, F. K. (2019). Toward High-Resolution In Situ Structural Biology with Cryo-Electron Tomography and Subtomogram Averaging. Curr. Opin. Struct. Biol. 58, 1–9. doi:10.1016/j.sbi.2019.03.018
Sgourakis, N. G., Yau, W.-M., and Qiang, W. (2015). Modeling an In-Register, Parallel "Iowa" Aβ Fibril Structure Using Solid-State NMR Data from Labeled Samples with Rosetta. Structure 23, 216–227. doi:10.1016/j.str.2014.10.022
Shibata, S., Shoji, M., Okada, K., Matsunami, H., Matthews, M. M., Imada, K., et al. (2020). Structure of Polymerized Type V Pilin Reveals Assembly Mechanism Involving Protease-Mediated Strand Exchange. Nat. Microbiol. 5, 830–837. doi:10.1038/s41564-020-0705-1
Sindelar, C. V., and Downing, K. H. (2010). An Atomic-Level Mechanism for Activation of the Kinesin Molecular Motors. Proc. Natl. Acad. Sci. 107, 4111–4116. doi:10.1073/pnas.0911208107
Sindelar, C. V., and Downing, K. H. (2007). The Beginning of Kinesin's Force-Generating Cycle Visualized at 9-Å Resolution. J. Cel Biol 177, 377–385. doi:10.1083/jcb.200612090
Smith, G. E., Summers, M. D., and Fraser, M. J. (1983). Production of Human Beta Interferon in Insect Cells Infected with a Baculovirus Expression Vector. Mol. Cel Biol 3, 2156–2165. doi:10.1128/mcb.3.12.2156-2165.1983
Sosa, H., Dias, D. P., Hoenger, A., Whittaker, M., Wilson-Kubalek, E., Sablin, E., et al. (1997a). A Model for the Microtubule-Ncd Motor Protein Complex Obtained by Cryo-Electron Microscopy and Image Analysis. Cell 90, 217–224. doi:10.1016/s0092-8674(00)80330-x
Sosa, H., Hoenger, A., and Milligan, R. A. (1997b). Three Different Approaches for Calculating the Three-Dimensional Structure of Microtubules Decorated with Kinesin Motor Domains. J. Struct. Biol. 118, 149–158. doi:10.1006/jsbi.1997.3851
Spaulding, C. N., Schreiber, H. L., Zheng, W., Dodson, K. W., Hazen, J. E., Conover, M. S., et al. (2018). Functional Role of the Type 1 Pilus Rod Structure in Mediating Host-Pathogen Interactions. Elife 7. doi:10.7554/eLife.31662
Sui, H., and Downing, K. H. (2010). Structural Basis of Interprotofilament Interaction and Lateral Deformation of Microtubules. Structure 18, 1022–1031. doi:10.1016/j.str.2010.05.010
Swanson, J., Robbins, K., Barrera, O., Corwin, D., Boslego, J., Ciak, J., et al. (1987). Gonococcal Pilin Variants in Experimental Gonorrhea. J. Exp. Med. 165, 1344–1357. doi:10.1084/jem.165.5.1344
Tan, Y. Z., Cheng, A., Potter, C. S., and Carragher, B. (2016). Automated Data Collection in Single Particle Electron Microscopy. Microscopy (Tokyo) 65, 43–56. doi:10.1093/jmicro/dfv369
Thanassi, D. G., Bliska, J. B., and Christie, P. J. (2012). Surface Organelles Assembled by Secretion Systems of Gram-Negative Bacteria: Diversity in Structure and Function. FEMS Microbiol. Rev. 36, 1046–1082. doi:10.1111/j.1574-6976.2012.00342.x
Tilney, L. G., Bryan, J., Bush, D. J., Fujiwara, K., Mooseker, M. S., Murphy, D. B., et al. (1973). Microtubules: Evidence for 13 Protofilaments. J. Cel Biol 59, 267–275. doi:10.1083/jcb.59.2.267
Turk, M., and Baumeister, W. (2020). The Promise and the Challenges of Cryo‐electron Tomography. FEBS Lett. 594, 3243–3261. doi:10.1002/1873-3468.13948
Vale, R. D. (2003). The Molecular Motor Toolbox for Intracellular Transport. Cell 112, 467–480. doi:10.1016/s0092-8674(03)00111-9
Van Gerven, N., Van der Verren, S. E., Reiter, D. M., and Remaut, H. (2018). The Role of Functional Amyloids in Bacterial Virulence. J. Mol. Biol. 430, 3657–3684. doi:10.1016/j.jmb.2018.07.010
Vemu, A., Atherton, J., Spector, J. O., Moores, C. A., and Roll-Mecak, A. (2017). Tubulin Isoform Composition Tunes Microtubule Dynamics. MBoC 28, 3564–3572. doi:10.1091/mbc.e17-02-0124
Vemu, A., Atherton, J., Spector, J. O., Szyk, A., Moores, C. A., and Roll-Mecak, A. (2016). Structure and Dynamics of Single-Isoform Recombinant Neuronal Human Tubulin. J. Biol. Chem. 291, 12907–12915. doi:10.1074/jbc.c116.731133
Vinothkumar, K. R., and Henderson, R. (2010). Structures of Membrane Proteins. Quart. Rev. Biophys. 43, 65–158. doi:10.1017/s0033583510000041
Wade, R. H., Chrétien, D., and Job, D. (1990). Characterization of Microtubule Protofilament Numbers. J. Mol. Biol. 212, 775–786. doi:10.1016/0022-2836(90)90236-f
Walton, T., Wu, H., and Brown, A. (2021). Structure of a Microtubule-Bound Axonemal Dynein. Nat. Commun. 12, 477. doi:10.1038/s41467-020-20735-7
Wang, F., Coureuil, M., Osinski, T., Orlova, A., Altindal, T., Gesbert, G., et al. (2017). Cryoelectron Microscopy Reconstructions of the Pseudomonas aeruginosa and Neisseria Gonorrhoeae Type IV Pili at Sub-nanometer Resolution. Structure 25, 1423–1435. doi:10.1016/j.str.2017.07.016
Wang, F., Gu, Y., O’Brien, J. P., Yi, S. M., Yalcin, S. E., Srikanth, V., et al. (2019). Structure of Microbial Nanowires Reveals Stacked Hemes that Transport Electrons over Micrometers. Cell 177, 361–369. doi:10.1016/j.cell.2019.03.029
Wang, X., Fu, Y., Beatty, W. L., Ma, M., Brown, A., Sibley, L. D., et al. (2021). Cryo-EM Structure of Cortical Microtubules from Human Parasite Toxoplasma Gondii Identifies Their Microtubule Inner Proteins. Nat. Commun. 12, 3065. doi:10.1038/s41467-021-23351-1
Watanabe, R., Buschauer, R., Böhning, J., Audagnotto, M., Lasker, K., Lu, T.-W., et al. (2020). The In Situ Structure of Parkinson's Disease-Linked LRRK2. Cell 182, 1508–1518. doi:10.1016/j.cell.2020.08.004
Williamson, M. P., Havel, T. F., and Wüthrich, K. (1985). Solution Conformation of Proteinase Inhibitor IIA from Bull Seminal Plasma by 1H Nuclear Magnetic Resonance and Distance Geometry. J. Mol. Biol. 182, 295–315. doi:10.1016/0022-2836(85)90347-x
Xu, Q., Shoji, M., Shibata, S., Naito, M., Sato, K., Elsliger, M.-A., et al. (2016). A Distinct Type of Pilus from the Human Microbiome. Cell 165, 690–703. doi:10.1016/j.cell.2016.03.016
Yan, S., Guo, C., Hou, G., Zhang, H., Lu, X., Williams, J. C., et al. (2015). Atomic-resolution Structure of the CAP-Gly Domain of Dynactin on Polymeric Microtubules Determined by Magic Angle Spinning NMR Spectroscopy. Proc. Natl. Acad. Sci. USA 112, 14611–14616. doi:10.1073/pnas.1509852112
Yip, K. M., Fischer, N., Paknia, E., Chari, A., and Stark, H. (2020). Atomic-resolution Protein Structure Determination by Cryo-EM. Nature 587, 157–161. doi:10.1038/s41586-020-2833-4
Zabeo, D., Heumann, J. M., Schwartz, C. L., Suzuki-Shinjo, A., Morgan, G., Widlund, P. O., et al. (2018). A Lumenal Interrupted helix in Human Sperm Tail Microtubules. Sci. Rep. 8, 2727. doi:10.1038/s41598-018-21165-8
Zeri, A. C., Mesleh, M. F., Nevzorov, A. A., and Opella, S. J. (2003). Structure of the Coat Protein in Fd Filamentous Bacteriophage Particles Determined by Solid-State NMR Spectroscopy. Proc. Natl. Acad. Sci. 100, 6458–6463. doi:10.1073/pnas.1132059100
Zhang, R., Roostalu, J., Surrey, T., and Nogales, E. (2017). Structural Insight into TPX2-Stimulated Microtubule Assembly. Elife 6. doi:10.7554/eLife.30959
Zhang, R., Alushin, G. M., Brown, A., and Nogales, E. (2015). Mechanistic Origin of Microtubule Dynamic Instability and its Modulation by EB Proteins. Cell 162, 849–859. doi:10.1016/j.cell.2015.07.012
Zhang, R., LaFrance, B., and Nogales, E. (2018). Separating the Effects of Nucleotide and EB Binding on Microtubule Structure. Proc. Natl. Acad. Sci. USA 115, E6191–E6200. doi:10.1073/pnas.1802637115
Zhang, R., and Nogales, E. (2015). A New Protocol to Accurately Determine Microtubule Lattice Seam Location. J. Struct. Biol. 192, 245–254. doi:10.1016/j.jsb.2015.09.015
Zheng, W., Andersson, M., Mortezaei, N., Bullitt, E., and Egelman, E. (2019). Cryo-EM Structure of the CFA/I Pilus Rod. Int. Union Crystallogr. J. 6, 815–821. doi:10.1107/s2052252519007966
Keywords: cryo-EM, ssNMR, filament, fibre, microtubule, pilus
Citation: Garnett JA and Atherton J (2022) Structure Determination of Microtubules and Pili: Past, Present, and Future Directions. Front. Mol. Biosci. 8:830304. doi: 10.3389/fmolb.2021.830304
Received: 07 December 2021; Accepted: 28 December 2021;
Published: 14 January 2022.
Edited by:
Annalisa Pastore, King’s College London, United KingdomReviewed by:
Louise Charlotte Serpell, University of Sussex, United KingdomCopyright © 2022 Garnett and Atherton. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: James A. Garnett, amFtZXMuZ2FybmV0dEBrY2wuYWMudWs=; Joseph Atherton, am9zZXBoLmF0aGVydG9uQGtjbC5hYy51aw==
 James A. Garnett
James A. Garnett Joseph Atherton2*
Joseph Atherton2*