- 1Laboratory of Genomic Integrity, National Institute of Child Health and Human Development, National Institutes of Health, Bethesda, MD, United States
- 2New England Biolabs Incorporated, Ipswich, MA, United States
Y-family DNA polymerases (pols) consist of six phylogenetically separate subfamilies; two UmuC (polV) branches, DinB (pol IV, Dpo4, polκ), Rad30A/POLH (polη), and Rad30B/POLI (polι) and Rev1. Of these subfamilies, DinB orthologs are found in all three domains of life; eubacteria, archaea, and eukarya. UmuC orthologs are identified only in bacteria, whilst Rev1 and Rad30A/B orthologs are only detected in eukaryotes. Within eukaryotes, a wide array of evolutionary diversity exists. Humans possess all four Y-family pols (pols η, ι, κ, and Rev1), Schizosaccharomyces pombe has three Y-family pols (pols η, κ, and Rev1), and Saccharomyces cerevisiae only has polη and Rev1. Here, we report the cloning, expression, and biochemical characterization of the four Y-family pols from the lower eukaryotic thermophilic fungi, Thermomyces lanuginosus. Apart from the expected increased thermostability of the T. lanuginosus Y-family pols, their major biochemical properties are very similar to properties of their human counterparts. In particular, both Rad30B homologs (T. lanuginosus and human polɩ) exhibit remarkably low fidelity during DNA synthesis that is template sequence dependent. It was previously hypothesized that higher organisms had acquired this property during eukaryotic evolution, but these observations imply that polι originated earlier than previously known, suggesting a critical cellular function in both lower and higher eukaryotes.
Introduction
The Y-family DNA polymerases are responsible for copying damaged DNA during DNA replication in a process called translesion synthesis (TLS) (Sale et al., 2012). These enzymes are highly specialized in order to accommodate different structural DNA distortions caused by a wide variety of DNA lesions. The Y-family is divided into six phylogenetically distinct subfamilies: two UmuC (polV) branches; Rad30A/POLH (polη); Rad30B/POLI (polι); and DinB (pol IV, Dpo4, polκ); and Rev1 (Ohmori et al., 2001). Across the different domains of life, Y-family polymerase subfamilies are found in various combinations. For example, UmuC orthologs are only detected in Gram-positive and Gram-negative bacteria, whereas Rev1 and Rad30A/B orthologs are only detected in eukaryotes. The DinB subfamily is the most evolutionarily conserved, having members scattered throughout all three domains of life from unicellular bacteria to humans. However, differences in the distribution of Y-family DNA pols are present within each kingdom. For example, the eukaryote Saccharomyces cerevisiae (S. cerevisiae) contains neither a POLK nor a POLI gene. Indeed, it was originally assumed that polι was expressed only in higher eukaryotes. However, next generation whole genome sequencing has revealed that polι orthologs are actually distributed throughout the whole Eukaryota domain. One example is the thermophilic fungus, Thermomyces lanuginosus (T. lanuginosus) which possesses all four eukaryotic Y-family subfamilies much like humans, in contrast to its fungal relatives, S. cerevisiae and Schizosaccharomyces pombe (S. pombe). Is there logic in such seemingly random distribution of polι? Using phylogenetic analysis and comparing the biochemical characterization of Y-family pols from different species, we hoped to shed some light on this question.
The Y-family DNA pols are classically described as specializing in TLS activity that arises from their capacious active sites that accommodate DNA lesions which would otherwise obstruct the processive confined active sites of A- and B-family replicative pols (Sale et al., 2012). Each Y-family polymerase is tailored to process different lesions, leaving behind unique errors after gaining access to the replication fork (Yang and Gao, 2018). Polymerases belonging to the same subfamily often specialize in targeting the same type of DNA lesions. For example, DinB/polκ orthologs are very adept at bypassing minor groove DNA adducts (e.g., N2-dG adducts) (Liu et al., 2014; Basu et al., 2017; Jha and Ling, 2018; Stern et al., 2019). Although, the archaeal ortholog, Dpo4, is notably less efficient at bypassing bulky aromatic lesions than its eukaryotic ortholog polκ (Avkin et al., 2004; Ling et al., 2004b; Choi et al., 2006). Along with a common preference for bulky aromatics, DinB pols also share a propensity for template slippage that increases deletion events in the mutation spectra (Kokoska et al., 2002).
Eukaryotic Rad30 orthologs polη and polι are similar in sequence but exhibit very different TLS properties. Human and S. cerevisiae polη are exceptionally efficient at bypassing a thymine-thymine cyclobutane pyrimidine dimer (CPD) (Johnson et al., 2000c; Masutani et al., 2000; McCulloch et al., 2004). Although polι can insert nucleotides opposite CPDs, the efficiency is substantially lower than polη-catalyzed TLS (Tissier et al., 2000a; Vaisman et al., 2003). Polι also has a unique feature; it misincorporates dG opposite dT, 3- to 10- fold more frequently than the correct base dA (Johnson et al., 2000b; Tissier et al., 2000b; Zhang et al., 2000c; McIntyre, 2020). These dG:dT misinsertions arise from polι′s remarkably large aliphatic side chains in the finger domain, unlike any other Y-family pols (Kirouac and Ling, 2009; Makarova and Kulbachinskiy, 2012). Despite the extremely low fidelity of polι on a template dT, it is moderately accurate when incorporating opposite other target bases. The highest fidelity is found opposite the template A, where polι inserts the correct base dT with error rate of 10–4 (Johnson et al., 2000b; Tissier et al., 2000b; Zhang et al., 2000c).
The unique feature of eukaryotic Rev1 orthologs is their efficiency at bypassing both damaged guanines and abasic sites using a deoxycytidyl transferase mechanism that limits Rev1 to exclusively incorporate dC (Nelson et al., 1996). This is achieved by displacing the DNA lesion from the active site entirely and instead using a protein sidechain (R324 in S. cerevisiae and R357 in H. sapiens) as the “template” which base pairs solely with dC (Nair et al., 2005, 2011; Weaver et al., 2020).
In this manuscript, we describe the identification, purification and characterization of thermostable eukaryotic orthologs of polη, polι, polκ, and Rev1 from a thermophilic, multicellular fungal species, T. lanuginosus. Biochemical characterization of TLS DNA pols η, ι, κ, and Rev1 include determination of the enzyme’s fidelity, processivity, thermostability, metal ion requirements, and TLS specificity during bypass of CPDs, abasic sites, and benzo[a] pyrene diol epoxide (BPDE) adducts. Our findings serve as basis for comparative analysis of the properties of proteins from different species, thus providing an important insight into the functional evolution of the Y-family polymerases.
Materials and Methods
Bacterial Plasmids
Plasmids used in this study are described in Table 1. Where noted, bacteria were grown on LB agar plates containing 20 μg/ml chloramphenicol; 25 μg/ml zeocin; 30 μg/ml kanamycin; 20 μg/ml spectinomycin; or 100 μg/ml ampicillin.
Identification and Cloning of Y-Family Orthologs From T. lanuginosus
At the onset of this investigation, the complete genomic sequence of T. lanuginosus was not yet available. Therefore, in order to identify and clone the various Y-family polymerase orthologs from T. lanuginosus, we used several PCR-based procedures. Initially, we employed a degenerate PCR approach by first generating protein homology comparisons for known fungal polη, polι, polκ, and Rev1 proteins. Regions of conserved amino acid stretches within these four polymerase families were identified and degenerate PCR primers were designed based on selected conserved amino acid regions. Purified T. lanuginosus genomic DNA was purchased from DSMZ (Leibniz Institute DSMZ-German Collection of Microorganisms and Cell Cultures GmbH) (DSM 10635). PCR and nested-PCR were performed utilizing this T. lanuginosus genomic DNA and amplicons of gene segments encoding these four Y-family polymerases were cloned and sequenced. Based on the cloned DNA sequences that encoded regions of the Y-family polymerase genes, gene-specific PCR primers were designed. Additional PCR reactions were performed either using genomic DNA or DNA from a T. lanuginosus cDNA library, kindly provided by the Fungal Genomics Project at Concordia University (Adrian Tsang; Director of the Center for Functional and Structural Genomics). PCR techniques employed included RACE PCR (cDNA library), Flanking-sequence PCR (Sorensen et al., 1993) utilizing degenerate biotinylated primers and over-lapping exon PCR (genomic DNA). Full-length genes were amplified with gene-specific primers with the addition of an NdeI site at the 5′ end and either a BamHI or BglII (POLK) site at the 3′ end and cloned into pGEM vectors for sequencing. The POLH, POLK and REV1 genes were subcloned into the NdeI to BamHI sites of pET22b+ for protein expression. An E. coli codon optimized version of the POLI gene was subsequently synthesized by Genscript (Piscataway, NJ) and cloned into the pJM871 low expression vector (Frank et al., 2012) from NdeI to BamHI.
Subsequently, the full T. lanuginosus SSBP genomic sequence was published and released (https://mycocosm.jgi.doe.gov/Thelan1/Thelan1.home.html) (McHunu et al., 2013) (GenBank assembly accession GCA_000315315935.1), and we were able to confirm that our clones do indeed encode the T. lanuginosus polη, polι, polκ, and Rev1 proteins. The POLH gene is encoded on contig00146 (Genbank accession ANHP01000280) from nucleotide 84213–86129. The POLI gene is encoded on contig00250 (Genbank accession ANHP01000232) from nucleotide 126087–127864 with two introns. The POLK gene is encoded on contig00025 (Genbank accession ANHP01000258) from nucleotide 58089–60067 with one intron. The Rev1 gene is encoded on contig00203 (Genbank accession ANHP01000194) from nucleotide 112065–115607 with one intron. The Joint Genome Institute MycoCosm database designations for theses proteins is as follows: polη – jgi|Thelan1|4409|TLAN_03021-R0; polι – jgi|Thelan1|3308|TLAN_02476-R0; polκ – jgi |Thelan1|3900|TLAN_01802-R0; Rev1 – jgi|Thelan1|3900|TLAN_01802-R0.
Phylogenetic Tree Construction
A multiple sequence alignment of the newly described T. lanuginosus polι protein and polι proteins from various other organisms was performed using the ClustalW algorithm in MacVector (version 15.5.4). Known polι protein sequences were obtained from Genbank protein records or identified by BLAST homology searches of Genbank genomic sequence records. Genbank accession numbers are indicated within the legend of Figure 1. The alignment was exported from MacVector in the Nexus sequence file format. This Nexus file was imported into the SplitsTree4 (version 4.14.4) and an unrooted phylogenetic tree was generated by setting the distance method to BioNJ.
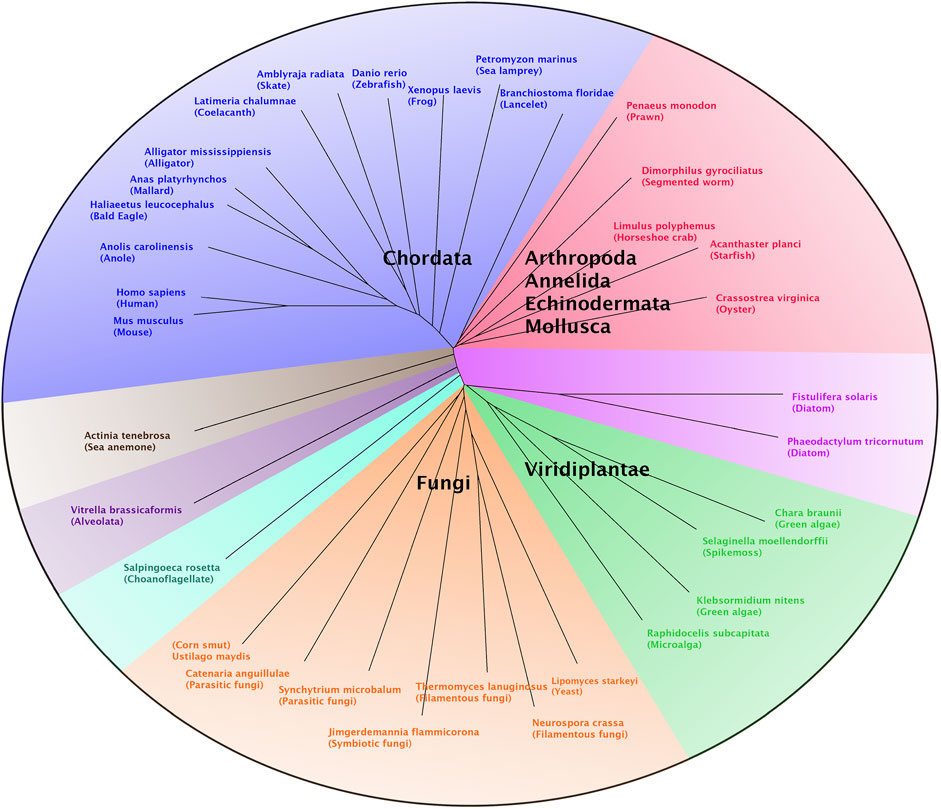
FIGURE 1. Phylogenetic analysis of polι. A multiple sequence alignment of polι proteins from a broad range of eukaryotic organisms, including T. lanuginosus polι and human polι, was performed using the ClustalW algorithm in MacVector (version 15.5.4). This alignment was exported from MacVector as a Nexus file which was then imported into SplitsTree4 (version 4.14.4) to generate an unrooted phylogenetic tree by setting the distance method to BioNJ. Most of the polι proteins cluster into several groups comprised of chordates (blue), lower animals (red), lower plants (green), fungi (orange) and diatoms (pink). Polι protein sequences for each organism were obtained from the following Genbank sequence files: Acanthaster planci (XP_022107640), Actinia tenebrosa (XP_031566011), Alligator mississippiensis (XP_006269716), Amblyraja radiata (XP_032889300), Anas platyrhynchos (XP_027302772), Anolis carolinensis (XP_016850726), Branchiostoma floridae (XP_035668094), Catenaria anguillulae (ORZ32752), Chara braunii (GBG75489), Crassostrea virginica (XP_022331447), Danio rerio (NP_001017834), Dimorphilus gyrociliatus (CAD5123533), Fistulifera solaris (GAX13589), Haliaeetus leucocephalus (XP_010581340), Homo sapiens (NP_001338561), Jimgerdemannia flammicorona (RUP43481), Klebsormidium nitens (GAQ87748), Latimeria chalumnae (XP_005994715), Limulus polyphemus (XP_022236329), Lipomyces starkeyi (ODQ70434), Mus musculus (NP_036102), Neurospora crassa (CAD70389), Penaeus monodon (XP_037797824), Petromyzon marinus (XP_032824552), Phaeodactylum tricornutum (XP_002178064), Raphidocelis subcapitata (GBF90329), Salpingoeca rosetta (XP_004998634), Selaginella moellendorffii (XP_024528028), Synchytrium microbalum (XP_031023241), T. lanuginosus (this study and ANHP01000232), Ustilago maydis (XP_011387208), Vitrella brassicaformis (CEL97102) and Xenopus laevis (XP_018100075).
Purification of the T. lanuginosus DNA Polη
E.coli strain RW644 [F− dcm ompT hsdS(rB− mB−) gal λ(DE3) ΔumuDC596:ermGT ΔdinB61:ble ΔaraD-polB:Ω Δ(gpt-proA)62] harboring pJM596 was grown overnight in 20 ml LB media containing 100 μg/ml ampicillin at 37°C. The overnight culture was then transferred into 1L fresh LB-ampicillin and grown at 37°C until an OD600 ∼0.5. At this point, IPTG was added to a final concentration of 1 mM to induce expression of T. lanuginosus DNA polη and grown for an additional 2 h before cells were harvested by centrifugation. The cell pellet was resuspended in 15 ml lysis buffer (50 mM Tris pH 8.0, 150 mM NaCl, 10 mM β-mercaptoethanol), sonicated, and cleared by ultracentrifugation at 45,000 rpm in a Beckman 50.2 Ti rotor for 45 min. Ammonium sulfate (45% saturation, 0.27 g/ml) was slowly added to the cleared lysate and stirred at 4°C for 30 min. Precipitated proteins were harvested by centrifugation and the resulting pellet resuspended in a buffer “A” (50 mM Tris pH 8.0, 20 mM NaCl, 10 mM β-mercaptoethanol, 20% v/v glycerol) and dialyzed overnight at 4°C against 1L of the same buffer. The dialyzed protein suspension was applied to a 5 ml HiTrap DEAE FF column (Cytiva, cat#17515401) and bound proteins were eluted with a 20–500 mM linear gradient of NaCl. Fractions containing polη were pooled and dialyzed against buffer “B” (10 mM sodium phosphate, pH 7.0,10 mM β-mercaptoethanol, 20% v/v glycerol) and applied to a 5 ml Bio-Scale™ Mini CHT™ Type II cartridge (BioRad, cat#7324332) and eluted with a 10–300 mM linear gradient of sodium phosphate in buffer B. Peak polη-containing fractions were pooled, aliquoted and stored at −80°C.
Purification of the T. lanuginosus DNA Polκ-
T. lanuginosus DNA polκ was expressed in RW644 harboring pJM682 and purified using the same protocol as described above for polη.
Purification of the T. lanuginosus DNA Polι
Our attempts to purify untagged T. lanuginosus polɩ using the above protocol for polη and polκ proved to be unsuccessful. We therefore decided to purify an N-terminal His-tagged version of T. lanuginosus polɩ. To do so, plasmid pJM966 was introduced into the E.coli SHuffle® T7 Express strain [F’ lac, pro, lacIq/Δ(ara-leu)7697 araD139 fhuA2 lacZ:T7 gene1 Δ(phoA)PvuII phoR ahpC galE galK λatt:pNEB3-r1-cDsbC (SpecR, lacIq) ΔtrxB rpsL150(StrR) Δgor Δ(malF)3] (New England Biolabs, cat#C3029J). Expression, cell harvest, lysis, and clearance were performed as described above for polη and polκ and the cleared lysate was dialyzed overnight at 4°C against 1L of buffer “C” (20 mM Sodium Phosphate, pH 7.5, 500 mM NaCl, 20 mM imidazole, 10 mM β-mercaptoethanol, 10% v/v glycerol) The dialysed lysate was applied to a 5 ml HisTrap HP column (Cytiva, cat#17524701) and eluted with a linear 20 mM to 1 M imidazole gradient. Fractions containing polι were pooled and dialyzed against 1L of buffer “D” (10 mM Sodium Phosphate, pH 7.5, 300 mM NaCl, 10 mM β-mercaptoethanol, 20% v/v glycerol) and applied to a 5 ml Bio-Scale™ Mini CHT™ Type II cartridge (BioRad, cat#7324332) and eluted with a 10–300 mM linear gradient of sodium phosphate. Peak polι-containing fractions were pooled, aliquoted and stored at −80°C.
Purification of T. lanuginosus Rev1
RW644 harboring pRARE2 (expressing tRNAs for seven rare codons; AGA, AGG, AUA, CUA, GGA, CCC, and CGG) was transformed with pJM863. The strain was grown overnight in 60 ml LB media containing 100 μg/ml ampicillin at 37°C. The overnight culture was then transferred into 3L fresh LB-ampicillin (1.5 L each in 2 x 4L flasks) and grown at 37 °C until an OD600 ∼0.5. At this point, IPTG was added to a final concentration of 1 mM to induce expression of T. lanuginosus Rev1 and grown for an additional 2 h before cells were harvested by centrifugation. The cell pellet was resuspended in 60 ml lysis buffer (50 mM Tris pH 7.5, 0.5 mM EDTA, 1 mM DTT, 20% glycerol), sonicated, and cleared by ultracentrifugation at 45,000 rpm in a Beckman 50.2 Ti rotor for 45 min. Ammonium sulfate (35% saturation, 0.2 g/ml) was slowly added to the cleared lysate and stirred at 4°C for 30 min. Precipitated proteins were harvested by centrifugation and the resulting pellet resuspended in a buffer “E” (10 mM Sodium phosphate pH 7.0, 25 mM NaCl) and dialyzed overnight against 2L of the same buffer at 4°C. The dialysed lysate was applied to a 20 ml HiPrep Heparin FF 16/10 column (Cytiva, cat#28936549) and eluted with a 25–400 mM NaCl linear gradient. Fractions containing Rev1 were pooled and applied to a HiPrep 16/60 Sephacryl S-200 HR size exclusion column (Cytiva, Cat#17116601) equilibrated in Phosphate Buffered Saline (PBS, pH6.8). Rev1 containing fractions were pooled and applied to a 5 ml Phosphocellulose P11 column (Whatman), equilibrated with buffer “F” (20 mM KPO4 pH 7.0, 100 μM EDTA, 10% glycerol, 1 mM DTT, and 100 mM KCl) and eluted with a 200–800 mM KCl linear gradient. Rev1-containing fractions were pooled, concentrated, and aliquoted for storage at −80°C.
DNA Templates
Undamaged synthetic oligonucleotide primers and an 48 bp abasic site-containing template were synthesized by Lofstrand Laboratories (Gaithersburg, MD) using standard technique and PAGE purified. A synthetic abasic site, dSpacer, was purchased from Glen Research (Sterling, VA) and incorporated into the oligonucleotide template using standard techniques by Lofstrand Laboratories. The 7.2 kb M13mp18 circular template was purchased from New England Biolabs (Ipswich, MA). The cis-syn CPD containing oligonucleotide was synthesized by TriLink BioTechnologies (San Diego, CA) and has been described previously (Boudsocq et al., 2004). The synthesis of a template with BPDE-dA adduct was described previously (Frank et al., 2002). The sequence of each primer-template pair is given in the legend of the respective figures. Radiolabeled primers were labeled with [γ-32P]ATP using T4 polynucleotide kinase by Lofstrand Laboratories (Gaithersburg, MD). The fluorescent primer containing a 5′-Fluorescein (6-FAM), 5′-6FAM-ATGGTACGGACGTGCTT-3′, was synthesized by Lofstrand Laboratories. The template strand complementary to the fluorescent primer used in Figure 7B–D had the following sequence: 5′-ATTAACGAATGAAGCACGTCCGTACCATCG-3′, whereby the underlined base indicates the identity of the templating base. For Figure 7E, the sequences of the three template strands used to investigate damaged and undamaged bypass were also complementary to the fluorescent primer and are as follows: dG: 5′-ATTAACGAATGAAGCACGTCCGTACCATCG-3'; abasic site (X): 5′-ATTAACGAATXAAGCACGTCCGTACCATCG-3'; and CPD: 5′-TCGATACTGGTACTAATGATTAACGAATTAAGCACGTCCGTACCATCG-3'. All primers were annealed to the unlabeled templates at a ratio of 1:2 by heating the primer and template together at 95°C in annealing buffer (0.1 M Tris-HCl, pH 8.0, 10 mg/ml BSA, 14.2 mM β-mercaptoethanol) and allowing the mixture to slowly cool to room temperature.
Radiolabeled in vitro Primer Extension
Standard 10 μl reactions contained 50 mM Tris-HCl, pH 7.5, 100 μM each dNTP, 10 mM DTT, 10 nM primer-template DNA, unless specified otherwise in the legend, and supplemented with 4 mM MgCl2 for pols η- and κ-catalyzed reactions and 4 mM MnCl2 for polι-catalyzed reactions. Reactions were carried out at 37°C for 10 min except in thermostability and processivity experiments, where the temperature and duration varied as noted in the figure legends. Reactions were terminated by the addition of 10 μl of 95% Formamide, 17.5 mM EDTA, 0.025% Xylene cyanol, and 0.025% Bromophenol blue, heated at 95°C for 5 min, and briefly chilled on ice. Aliquots containing 5 μl of the samples were separated on 15% 8 M Urea polyacrylamide gels and visualized by PhosphorImager analysis.
6-FAM in vitro Primer Extension
For in vitro polymerase assays with T. lanuginosus Rev1, 12 μl reactions containing 50 mM Tris-HCl, pH 7.5, 10% glycerol w/v, 1 mM β-mercaptoethanol, 100 nM primer-template DNA, 5 nM T. lanuginosus Rev1, and 5 mM MgCl2 (with the exception of the Me2+ activation experiment) were initiated with 100 μM dNTP. Reactions were carried out at 37°C for 10 min except in the thermostability and processivity experiments, where the temperature and duration of experiments varied, respectively, as detailed in the figure legends. Reactions were terminated by the addition of 12 μl of 10 M Urea, 100 mM EDTA, 0.2% Xylene cyanol, and 0.02% Bromophenol blue, heated at 95°C for 5 min. Aliquots containing 12 μl of the samples were separated on 22% 8 M Urea polyacrylamide gels (Triple Wide Mini-Vertical gel system, C.B.S. Scientific) and visualized using 5′-fluorescein (6-FAM) fluorescence with a Typhoon FLA 7000 (GE Healthcare).
Results and Discussion
Identification of Y-Family Polymerases in T. lanuginosus
Based on previous investigations and observations, it has been noted that the genomes of higher eukaryotes, such as humans and mice, possess genes for the four known eukaryotic Y-family pols, whereas, the model organism S. cerevisiae only possesses genes for two of the eukaryotic Y-family pols, RAD30 and REV1. Based on this observation, it has been suggested the genes encoding the closely related polη and polι proteins are paralogs of each other that diverged during evolution sometime after the origination of fungi. However, BLAST homology searches revealed that the genomes of filamentous fungi such as Neurospora crassa and Aspergillus nidulans and even the corn smut Ustilago maydis harbor a POLI gene, indicating to us that the genetic and biochemical characteristics of translesion synthesis in some “higher” fungi could be quite akin to that in higher eukaryotes such as humans. This notion of conserved functionality provided the impetus to embark on our efforts to identify and clone the four eukaryotic Y-family pols from the fungi, T. lanuginosus. Since T. lanuginosus is thermophilic, we reasoned the Y-family pols from this organism would exhibit enhanced stability during purification and biochemical analysis (see below).
In order to identify and clone the T. lanuginosus Y-family pols, protein sequence similarity comparisons were performed using known polη, polι, polκ, and Rev1 protein sequences, including other fungal sequences. Initially, degenerate PCR primers were designed based on highly conserved regions of protein homology between the various proteins within the four Y-family polymerase groups. Degenerate PCR was performed using T. lanuginosus genomic DNA and amplicons were cloned and sequenced. Subsequently, sequence of cloned regions of the Y-family polymerase genes were used to design gene-specific primers. A combination of various PCR techniques was then employed, for example degenerate PCR, nested PCR, RACE PCR, over-lapping exon PCR and flanking-sequence PCR using either genomic DNA or cDNA library DNA, to generate full-length sequences of each of the T. lanuginosus POLH, POLI, POLK and REV1 genes. Full-length genes were then subcloned into protein expression vectors to facilitate purification of the four Y-family DNA pols. After we completed cloning the Y-family pol genes, the T. lanuginosus genome was sequenced and published allowing us to confirm that our cloned sequences do indeed encode the correct T. lanuginosus proteins. Sequence comparisons between T. lanuginosus and human Y-family pols reveal modest similarities that are higher than those observed between S.cerevisiae and human homologs. T. lanuginosus and human polη have 22% identical and 15% similar residues (compare to S. cerevisiae versus human at 16% identical and 15% similar residues). T. lanuginosus and human Rev1 have 24% identical and 15% similar residues (compare to S. cerevisiae versus human at 16% identical and 16% similar residues). Additionally, T. lanuginosus and human polκ have 20% identical and 14% similar residues, and T. lanuginosus and human polι have 24% identical and 16% similar residues.
Phylogenetic analysis of the Y-Family Polymerase poli
The observation that the four eukaryotic Y-family pols are present in many filamentous fungi suggested to us that the divergence of POLI and POLH is even more ancient than the divergence of the fungi and metazoan (animals) groups. This supposition prompted us to perform a more in-depth phylogenetic analysis on the origins of the polι protein. We executed an exhaustive search for polι protein sequences within the Genbank database, including the fungi and metazoan groups, all other groups with clades containing various “lower” eukaryotes, and the plant kingdom. Although the POLI gene was absent from many “lower” or “simpler” eukaryotic organisms than fungi, as well as higher plants, we did find several examples of lower eukaryotes and simpler plants that do in fact encode a POLI gene.
A ClustalW alignment was performed using a group of sequenced POLI genes from organisms that include simple eukaryotes, lower plants, fungi and metazoans, all of which also possess genes for POLH, POLK and REV1. The unrooted phylogenetic tree of this alignment (Figure 1) reveals that the polι proteins cluster, for the most part, within several groups that include a chordate group, a lower animal grouping, including arthropods, annelids, echinoderms and mollusks, a fungi group, and a lower plant group. Polι proteins from other organisms that did not fit into these groups include a protein from a sea anemone (a Cnidarian), two proteins from diatom organisms and proteins from an Alveolate and a Choanoflagellate, which are examples of very simple single-celled eukaryotes. Interestingly, our analysis further supports the perception that the POLI gene is quite commonly found in fungal species, including some yeasts, and even other lower eukaryotic organisms and therefore substantiates our supposition that the evolutionary “split” between POLH and POLI may have preceded the emergence of fungi.
During this phylogenetic analysis of polι, one particularly noteworthy and unexpected observation was that POLI genes were quite frequently identified in some organisms within a particular group and absent in other organisms within that group. This finding holds true from the simplest eukaryotes to higher eukaryotes. For example, many protists do not possess a POLI gene such as Metamonads (e.g., diplomonads, i.e., Giardia lamblia and parabasalids, i.e., Trichomonas vaginalis), Euglenozoans (e.g., trypanosomatids, i.e., Trypanosoma brucei and Leishmania donovani) and Amoebozoans (e.g., Entamoeba histolytica). While other groups of protists do have a POLI gene such as Heteroloboseas (e.g., Naegleria gruberi) and Alveolates [e.g., Vitrella brassicaformis (Colpodellida clade)]. In direct contrast, other species of Alveolates in the Apicomplexans clade (e.g., Cryptosporidium parvum) and the Ciliophorans clade (ciliates) (e.g., Paramecium tetraurelia) do not have a POLI gene. In addition, some Stramenopiles have a POLI gene (e.g., diatoms, i.e., Phaeodactylum tricornutum and Fistulifera solaris) and some do not (e.g., diatoms, i.e., Thalassiosira oceanica and e.g., water mould, i.e., Saprolegnia parasitica). Thus, it seems the presence, or absence, of a POLI gene in the genome of any specific organism is quite indiscriminate.
This unpredictable “hit or miss” trait of POLI holds true in plants, fungi, and metazoans as well. For example, we found POLI genes in the genomes of lower plant forms such as microalga, green algae, and spikemoss but were unable to find POLI genes in any higher vascular plants. A similar situation exists in fungi. No POLI genes were found in lower fungal forms such as Microsporidia and Glomeromycota. Likewise, in Saccharomycotina (true yeasts) (e.g., S. cerevisiae) no POLI genes were found. Some fungi in the Taphrinomycotina subphylum, such as Saitoella complicate, have POLI genes, while the closely related fission yeasts, Schizosaccharomyces species, do not. Furthermore, POLI genes were found in other yeast forms (e.g., Lipomyces starkeyi), filamentous fungi, parasitic, and symbiotic fungi, and corn smut (Figure 1). However, we found no POLI genes in Basidiomycota which include club fungi and mushrooms.
Although most organisms within the various groups of animals, from the Choanoflagellates (considered to be the closest living relatives of the animals) up to the mammals, possess a POLI gene, we found it remarkable that there are numerous examples of higher eukaryotes organisms that do not possess a POLI gene. For example, we found no POLI genes in most Platyzoa (i.e., Platyhelminthes, e.g., flat worms), Rotifera (i.e., wheel animals), and Annelids (i.e., segmented worms), with the notable exception of Dimorphilus gyrociliatus which does harbor a POLI gene (Figure 1). While, many types of Mollusks do possess POLI genes, we found no POLI gene in octopi. Apparently, Arthopods such as spiders, scorpions, sea spiders, centipedes, millipedes and tardigrades (water bears) do not have POLI, but horseshoe crabs do. With a few exceptions, the majority of crustaceans and insects have POLI. Lastly, we found that the vast majority of chordates from the lancelet to humans do indeed possess a POLI gene (Figure 1). However, there are some extraordinary exceptions. We found that the tunicates (sea squirts), which are lower chordates, lack a POLI gene. Thus far, only a single fish species, Esox lucius (northern pike), was identified as lacking POLI. While alligators possess a POLI gene, the Australian saltwater crocodile (Crocodylus porosus) and the gharial (Gavialis gangeticus) both lack POLI. Most astoundingly, the majority of bird species, including Galliformes (e.g., chickens, turkeys, grouse, quail, partridges and pheasants), Bucerotiformes (hornbills), Upupiformes (hoopoes) and the vast majority of Passeriformes (e.g., sparrow, finch, tit, crow, canary, rifleman, and starlings) which includes more than half of all bird species, lack a POLI gene.
This phylogenetic analysis provides a “snapshot” of the evolution of POLI at this given moment in time. At some time in the past, the polη and polι paralogs diverged from one another due to a need to have two biochemically distinct translesion synthesis polymerases. The biochemical properties of both human polη and polι are well characterized. However, unlike polη which is defective in the human Xeroderma Pigmentosum Variant syndrome (Masutani et al., 1999), the cellular function of polι remains enigmatic (McIntyre, 2020; Vaisman and Woodgate, 2020). Therefore, the evolutionary pressure that led to this division of polη and polι remains a mystery. Perhaps this seemingly arbitrary “hit or miss” distribution of POLI genes in eukaryotic organisms can provide clues as to the cellular functioning of polι. Evaluation of the biological and biochemical translesion synthesis properties of polη and polι from closely related lower eukaryotes, one of which possesses a POLI gene and the other that does not, may provide hints as to when, and why, these two paralogs diverged and the cellular functions of polι.
Expression and Purification of T. lanuginosus Polymerases η, ι, κ, and Rev1
All four Y-family pols from T. lanuginosus were expressed in E. coli, but their expression levels, solubility, and ease of purification varied. Both pols η and κ were well-expressed in E. coli and readily purified by conventional protocols including ammonium sulfate precipitation and ion-exchange chromatography (Figure 2). Rev1 was initially poorly expressed (unpublished observations), but expression increased dramatically in the presence of the pRARE2 plasmid coding for rare E.coli tRNAs. T. lanuginosus Rev1 was also purified by conventional methods including ammonium sulfate precipitation, size-exclusion and ion-exchange chromatography (Figure 2). T. lanuginosus polι was the most problematic of the four Y-family polymerases to purify, requiring codon optimization for E.coli expression and the addition of an N-terminal Histidine tag. Subsequently, the protein was purified by affinity- and ion exchange-chromatography (Figure 2). Once the polymerases were purified to >95% homogeneity, they were characterized for metal ion requirement, ability to bypass lesions, fidelity, thermostability, and processivity.
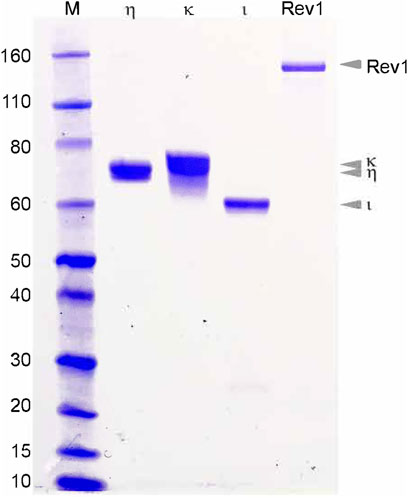
FIGURE 2. Purified T. languinosus polymerases η, κ, ι and Rev1. Two to five μg of the final purified polymerases were separated on a 12% SDS-PAGE gel. Visual analysis suggests each polymerase is greater than 95% pure.
Activation of T. lanuginosus Polymerases η, κ, ι by Mn2+ and Mg2+
DNA polymerases are known to require divalent cations to catalyze the nucleotidyl transfer reaction (Bessman et al., 1958; Mildvan and Loeb, 1979; Loeb and Kunkel, 1982; Vashishtha et al., 2016). Although it is generally believed that Mg2+ is the activating co-factor in vivo due to its cellular abundance, other divalent metal cations such as Mn2+, Co2+, Ni2+, Zn2+ have the ability to substitute for Mg2+ under certain conditions (Mildvan and Loeb, 1979; Loeb and Kunkel, 1982). Different metal co-factors can affect the activity and fidelity of DNA polymerases, promote TLS, and in the case of X-family enzymes, increase polymerization efficiency (Martin et al., 2013). In rare cases, Mn2+ appears to serve as a natural metal activator of DNA polymerases. This has been shown for X-family polymerases λ and μ and Y-family polι which exhibit great preference for Mn2+ over Mg2+ as the activating metal ion (Dominguez et al., 2000; Frank and Woodgate, 2007; Garcia-Diaz et al., 2007). In particular, human polι was inhibited by MgCl2 present at physiological concentrations and exhibited peak activity at low MnCl2 levels (0.05–0.25 mM). Furthermore, at optimal concentrations, Mn2+ ions improved the fidelity of polι-catalyzed nucleotide incorporation opposite template dT.
We were interested in determining the optimal concentration and type of divalent metal ion that is required for activation of T. lanuginosus pols η, κ, and ι. We assessed the primer extension activity of these polymerases in the presence of Mg2+ and Mn2+ at concentrations ranging from 62.5 μM to 8 mM (Figure 3). Polη exhibited greatest activity in reactions with 4–8 mM Mg2+ and 0.5–8 mM Mn2+ (Figure 3A). Reactions catalyzed by polκ were most efficient at 1–8 mM Mg2+ and 0.25–8 mM Mn2+ (Figure 3C). Polι was most active in the presence of ∼4 mM Mn2+ (Figure 3B).
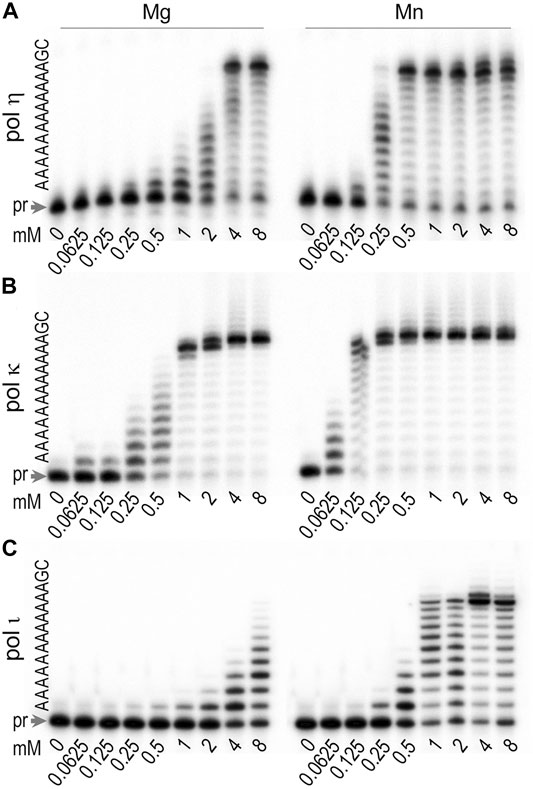
FIGURE 3. Determination of metal ion requirements (type and concentration) for optimal activity of (A) polη (B) polκ, and (C) polι. The extension of 32P-labeled primer 5′-CGATGGTACGGACGTGCTT-3′ hybridized to a template 3′-GCTACCATGCCTGCACGAAAAAAAAAAAAGC-5′ was carried out for 10 min at 37°C in the presence of 100 μM dNTPs mixture and with increasing concentrations of MgCl2 or MnCl2, ranging from 0.0625 to 8 mM. Concentrations of enzymes were 0.17 pM for polη, 0.32 pM for polκ, and 0.15 pM for polι. The sequence of the template immediately downstream of the primer (pr) is shown on the left-hand side of each gel pair.
As shown in Figure 3, Mn2+ appears to be a more conducive co-factor than Mg2+ in reactions with all three T. lanuginosus pols, as they all were active over a wider range of Mn2+ and exhibited greater activity at the same concentration of Mn2+ vs. Mg2+. However, there are some interesting differences seen between the reported metal ion requirements for human and T. lanuginosus polι. Human polι exhibits highest activity at 0.05–0.2 mM MnCl2 (Frank and Woodgate, 2007), whereas T. lanuginosus enzyme required much higher concentrations of Mn2+ with a sharp peak of activity at ∼4 mM MnCl2. In contrast, while human polι was inhibited by MgCl2 at concentrations >1 mM, T. lanuginosus polι required at least 0.5 mM Mg2+ and its activity gradually increased with concentrations up to 8 mM.
Because Mn2+ is generally known to decrease replication fidelity, we used 4 mM Mg2+ for the subsequent studies with polη and polκ. In contrast, further characterization of polι was performed in the presence of 4 mM Mn2+.
Processivity of T. lanuginosus Polymerases η, ι and κ
We next examined the processivity of T. lanuginosus pols η, ι, and κ (Figure 4A). As with the human enzymes, T. lanuginosus polη and polκ were much more processive than polι and incorporated ∼6 and ∼35 bases, respectively, in a single binding event after only 1 min. In contrast, polι incorporated ∼10–11 bases in a distributive manner. It then exhibited a strong pause after encountering two adjacent template dTs located 11 and 12 nucleotides from the 5′ end of the primer. Presumably this is due to misincorproration of dG opposite the dTs and its subsequent poor extension (Vaisman et al., 2001). These results indicate that the processivity of T. lanuginosus pols η, ι, and κ are therefore similar to their human counterparts (Washington et al., 1999; Ohashi et al., 2000a; Tissier et al., 2000b).
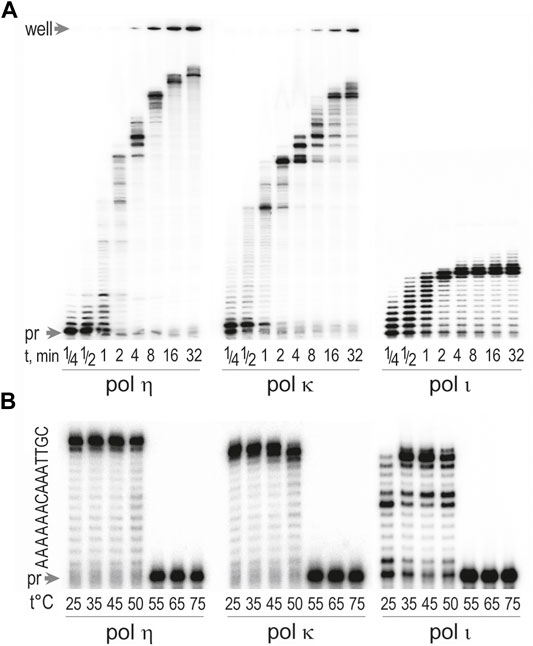
FIGURE 4. Thermostability and processivity of Thermomyces lanuginosus pols η, κ, and ι. Concentrations of enzymes were 0.17 pM for pol η, 0.32 pM for pol κ, and 0.15 pM for pol ι. (A) Processivity of pols η, κ, and ι were assayed in reactions containing 100 μM dNTPs and 4 mM MnCl2 for polι or MgCl2 for polη and polκ. Extension of 32P labeled primer 5′ TATTTATCCCAATCCAAATAAGAAACGA-3′ hybridized to the circular single-stranded 7.2 kb M13mp18 bacteriophage was at 37°C for various times ranging from 0.25 to 32 min. Reactions containing 100 μM dNTPs and 4 mM MnCl2 for reactions with polι and MgCl2 for reactions with polη and polκ. (B) Thermostability was assayed by primer extension reactions using DNA template generated by annealing of a32P-labeled primer 5′-TATTTATCCCAATCCAAATAAGAAACGA-3′ with an undamaged DNA template 5′-CGTTAAACAAAAAATCGTTTCTTATTTGGATTGGGATAAATA-3'. Reactions were carried out for 10 min at various temperatures ranging from 25 to 75°C and contained 100 μM dNTPs and 4 mM Mn2+ to activate polι or Mg2+ to activate polη and polκ.
Thermostability of T. lanuginosus Polymerases
T. lanuginosus is a thermophilic fungus with a natural habitat of growing in organic soils, e.g., decomposing vegetable matter, composts, and animal excrements, at an optimal temperature between 48 and 52°C (Tsiklinsky, 1899; Maheshwari et al., 2000; Singh et al., 2003).The ability of this fungus to live in such an environment suggests that its proteins possess enhanced thermostability (Maheshwari et al., 2000). We therefore examined the optimum temperature for the T. lanuginosus pols η, κ, and ι by incubating primer extension reactions at temperatures ranging between 25 and 75°C. All three polymerases exhibited robust activity between 25 and 50°C, but there was sharp decline in activity at temperatures greater than 50°C (Figure 4B).
Polη, Polι, and polκ-dependent Lesion Bypass Past Abasic Sites, CPDs, and benzo[a] Pyrene Adducts
The Y-family polymerases are best characterized by their ability to catalyze translesion replication past a variety of DNA lesions, in a manner distinct from the cell’s conventional replicases (Sale et al., 2012; Vaisman and Woodgate, 2017). We were therefore interested in assessing the ability and fidelity of T. lanuginosus pols η, κ, ι and to carry out translesion synthesis across a cis-syn CPD, an abasic site, and a benzo[a]pyrene adduct.
Apurinic/apyrimidinic (or abasic) sites are generated as a result of spontaneous or enzymatically-induced base loss and are considered the most frequent form of DNA damage (Lindahl, 1993; Boiteux and Guillet, 2004). An estimated 10,000 abasic sites occur per human cell per day under physiological conditions (Loeb, 1989). It is therefore not surprising that the ability of polη to copy DNA templates containing this noninstructional lesion has been extensively investigated. What is surprising, is that studies performed by various groups with human and S. cerevisiae polη produced conflicting results, ranging from those stating that “hPolη has the highest abasic lesion bypass efficiency” among human Y-family DNA pols and “is a major pol involved in abasic site bypass” (Choi et al., 2010; Sherrer et al., 2011; Patra et al., 2015; Thompson and Cortez, 2020) to those which dismiss human, or S. cerevisiae polη, as contributing to in vivo abasic site bypass because of “negligible nucleotide insertion opposite the abasic site and primer extension past the lesion” (Haracska et al., 2001; Pages et al., 2008). Somewhere in between these polar extremes are studies suggesting that polη catalyzes insertion and/or extension steps of abasic site bypass efficiently enough to ensure its involvement in TLS in vivo, but not as a major player (Zhang et al., 2000a; Masutani et al., 2000; Zhao et al., 2004; Sherrer et al., 2011).
We show here that the efficiency and fidelity of T. lanuginosus polη during TLS of abasic sites is reminiscent of properties of human and yeast polη uncovered in the third group of studies described above (Zhang et al., 2000a; Masutani et al., 2000; Zhao et al., 2004). In particular, in standing-start primer extensions T. lanuginosus polη inserted dA and dG opposite the abasic site with moderate efficiency, but further primer elongation was largely obstructed (Figure 5A). However, minuscule extension of the primer with terminal dA opposite the abasic site occurred by incorporation of an additional dA when the base 5′ to the abasic site was dT (Figure 5A, incorporation of dA). It should be noted that base selectivity of polη on the template with an abasic site is similar to its moderate fidelity on the undamaged DNA. As shown in Figure 5A, polη preferentially incorporates a correct dA opposite the undamaged dT although substantial misinsertion of dG and low level of dC and dT misincorporation can also be seen.
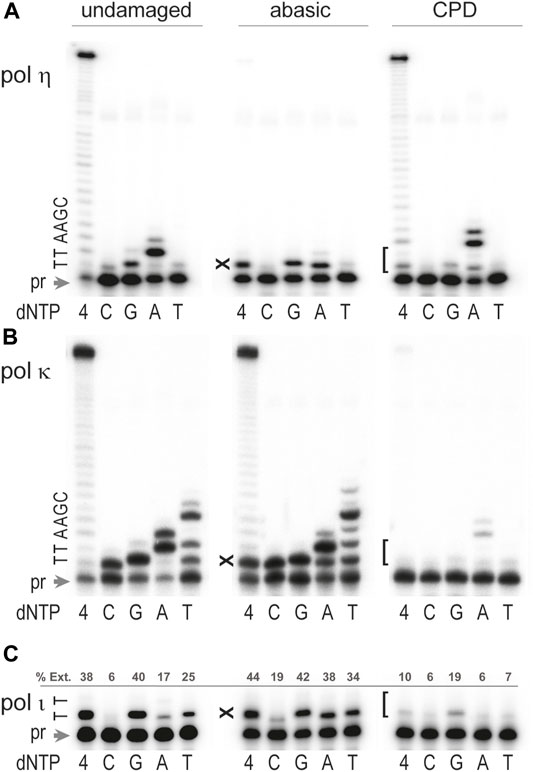
FIGURE 5. Bypass of abasic and CPD site by Thermomyces lanuginosus polymerases. The ability to bypass abasic site and CPD by (A) polη, (B) polκ, and (C) polι was assayed by measuring extension of 32P labeled primer 5′-CGATGGTACGGACGTGCTT-3′ hybridized to the undamaged or damaged DNA template. 5′-TCGATACTGGTACTAATGATTAACGAA-TN-AAGCACGTCCGTACCATCG-3'. The underlined bold TN sequence stands for the undamaged TT, TT-CPD, or TX, where X is an abasic site, pr is unextended primer. Reactions contained 100 μM each individual nucleotide (dC, dG, dA, dT) or mixture of all four dNTPs as indicated in the Figure, and 4 mM Mn2+ for reactions catalyzed by polι or Mg2+ for reactions with polη and polκ. Primer extension reactions were carried out at 37°C for 10 min. Concentrations of enzymes were 0.17 pM for pol η, 0.32 pM for polκ, and 0.15 pM for polι. The sequence of the template immediately downstream of the primer (pr) is shown on the left-hand side of each gel pair. “X” denotes the position of the abasic site and “[” indicates position of the T-T CPD. For panel C, primer extension (%Ext.) was quantified using ImageJ software and reported as the percentage of product density relative to total density for each reaction.
As with polη, there is no consensus regarding the ability of the DinB orthologs (polκ) to utilize DNA templates containing abasic sites (Strauss, 1991; Ohashi et al., 2000b; Zhang et al., 2000b; Boudsocq et al., 2001; Gerlach et al., 2001; Choi et al., 2010; Sherrer et al., 2011), which is at least partly explained by the strong effect of the surrounding sequence context on the bypass mechanism, base selectivity, and efficiency of insertion and extension steps of TLS. Thus, it has been shown that human polκ tolerates an abasic site much better when the next (5′) template base is dT, and the nucleotide preferentially incorporated opposite the lesion is dA. Furthermore, when primer/template slippage is possible, bypass often occurs through a stabilized misalignment mechanism leading to 1– or 2-base deletions (Boudsocq et al., 2001; Ling et al., 2004a). Similarly, T. lanuginosus polκ-catalyzed TLS of the abasic site was quite efficient when the 5′ template was dT and dA was preferentially incorporated opposite the lesion, although the other three nucleotides were also inserted at substantial levels (Figure 5B). The low fidelity of polκ during abasic site bypass is very similar to its highly error-prone nucleotide incorporation on undamaged DNA. The fact that the major product of T. lanuginosus polκ primer extension reactions in the presence of dT was generated by addition of four nucleotides is consistent with the aforementioned human polκ template-slippage mechanism (Mukherjee et al., 2013). Therefore, the resulting main product would contain two mismatches and total primer elongation by four bases because the DNA template used in the current study has two dA’s next to dT. When the DNA template was not susceptible to primer/template slippage (reactions with dC, dG, and dA) only one wrong nucleotide was incorporated opposite the abasic site.
Human polι has the capacity to insert deoxynucleotides opposite an abasic site, however further extension is limited (Zhang et al., 2000c; Vaisman et al., 2002; Nair et al., 2009; Choi et al., 2010; Sherrer et al., 2011). T. lanuginosus polι behaved in a similar manner, i.e., a single nucleotide was readily inserted opposite the abasic site with no indication of further extension (Figure 5C). Furthermore, akin to human polι, T. lanuginosus polι incorporates dG, dA, and dT with similar efficiency. Interestingly, when undamaged templates were used, T. lanuginosus polι was even more inaccurate than the human polymerase, i.e., not only dG, but also dT, was inserted opposite template dT more often than the correct dA (Figure 5C).
The most common ultraviolet light photoproduct is a cis-syn cyclobutane pyrimidine dimer (CPD). Unlike the ambiguity of TLS of an abasic site, there is no question as to which TLS polymerase bypasses a CPD most efficiently. Indeed, it is well established that polη can bypass a CPD with the same efficiency and accuracy as undamaged template dT’s (Johnson et al., 2000c; Masutani et al., 2000; McCulloch et al., 2004). Most other pols halt synthesis either immediately before the lesion, or opposite the first template dT of the CPD, as their active site is unable to accommodate the covalently joined dT’s of the CPD. Similar to human and S. cerevisiae polη, T. lanuginosus polη efficiently bypassed the CPD lesion (Figure 5A). Like polymerases purified from other species, T. lanuginosus polη was even more accurate while inserting nucleotides opposite the damaged bases than on undamaged DNA templates (Figure 5A).
The pols in the DinB branch of the Y-family (polκ) are unable to replicate past UV-induced cis-syn CPD dimers (Johnson et al., 2000a; Ohashi et al., 2000b; Zhang et al., 2000b) with the one reported exception of archaeal Sulfolobus solfataricus Dpo4, whose properties resemble eukaryotic polη (Boudsocq et al., 2001). Interestingly, the ability of T. lanuginosus polκ to copy a CPD-containing template resembles the behavior of its human orthologs rather than the thermophilic archaeal enzyme, i.e., T. lanuginosus polκ was virtually blocked by the CPD lesion in reactions with all four dNTPs although a very low level of the correct incorporation of two dAs opposite the damaged bases was detected in reactions where only dATP was present (Figure 5B).
It has been reported that human polι can bypass a CPD. Depending on the sequence context, it incorporates anywhere between 1–5 nucleotides, albeit with low efficiency (Tissier et al., 2000a; Vaisman et al., 2003). T. lanuginosus polι was much less efficient compared to the human enzyme. In fact, CPD bypass was barely detectible (Figure 5C). As with undamaged template dT, the preferred nucleotide inserted opposite the 3′-dT of the CPD was dG (Figure 5C).
Benzopyrene diol epoxides (BPDEs) are the highly carcinogenic metabolites of the environmental pollutant benzo[a]pyrene, found in tobacco smoke and automobile exhaust. In mammalian cells, various BPDE stereoisomers covalently bind to DNA forming adducts at the N2 and N6 position of guanine and adenine, respectively. Previous studies demonstrated that efficiency and fidelity of TLS past BPDE by different pols not only depends on the DNA base to which the adduct is linked, but is also often modulated by the stereochemistry of the adduct (Zhang et al., 2002a; Chiapperino et al., 2002; Frank et al., 2002; Rechkoblit et al., 2002; Suzuki et al., 2002; Jia et al., 2008). For example, in general, human polκ is almost completely stalled by BPDE-dA adducts. Under similar reaction conditions, it accurately and efficiently bypasses most of the BPDE-dG stereoisomers (Zhang et al., 2002a; Rechkoblit et al., 2002; Suzuki et al., 2002; Jia et al., 2008), although the cis-S-BPDE-dG represents a significant block for the polymerase (Suzuki et al., 2002). In contrast, polι is much more efficient and accurate while incorporating nucleotides opposite BPDE-dA than opposite BPDE-dG (Frank et al., 2002). Furthermore, independent of adduct stereochemistry, the correct dT is inserted by polι equally efficiently opposite various BPDE-dA isomers. On the other hand, the ability of human polη to replicate past the BPDE is strongly affected by the adduct conformation and damaged base, i.e., it readily traverses through the trans-S-BPDE-dA, but not through the isomeric trans-R- adduct, or through the BPDE-dG isomers (Chiapperino et al., 2002; Rechkoblit et al., 2002). In the current study with T. lanuginosus pols, we used DNA templates with a trans-S-BPDE-dA adduct. Reactions with polη and polκ contained Mg2+ or Mn2+, whereas polι was tested only in the presence of Mn2+.
Similar to the human enzyme, T. lanuginosus polη was quite efficient, but rather inaccurate, while incorporating nucleotides opposite the trans-S-BPDE-dA (Figures 6A,C) especially when reactions were carried out in the presence of Mn2+ which significantly boosted TLS efficiency without noticeably reducing fidelity (Figure 6C). Nucleotide incorporation opposite the lesion by polι was at least as efficient as by polη (Figure 6E), but it was less accurate and further primer extension was inhibited (Figure 6E). We have previously found that fidelity of human polι was influenced by the local sequence context of the BPDE-dA lesion (Frank et al., 2002), and when the template bases 5′ to the lesion were GAC, uncharacteristically accurate insertion of dT opposite the BPDE-dA was observed. In contrast, it appears that on the same template, T. lanuginosus polι keeps its general unfaithful nature. Finally, T. lanuginosus polκ when activated by Mg2+ barely showed any activity on the BPDE-dA-adducted DNA (Figure 6B). However, when Mg2+ was substituted by an equimolar concentration of Mn2+ (Figure 6D), T. lanuginosus polκ was able to utilize this template adding 1–2 nucleotides to the primer. Incorporation of all four nucleotides in the presence of 4 mM Mn2+occurred with similar efficiency, making synthesis by polκ highly error-prone (Figure 6D).
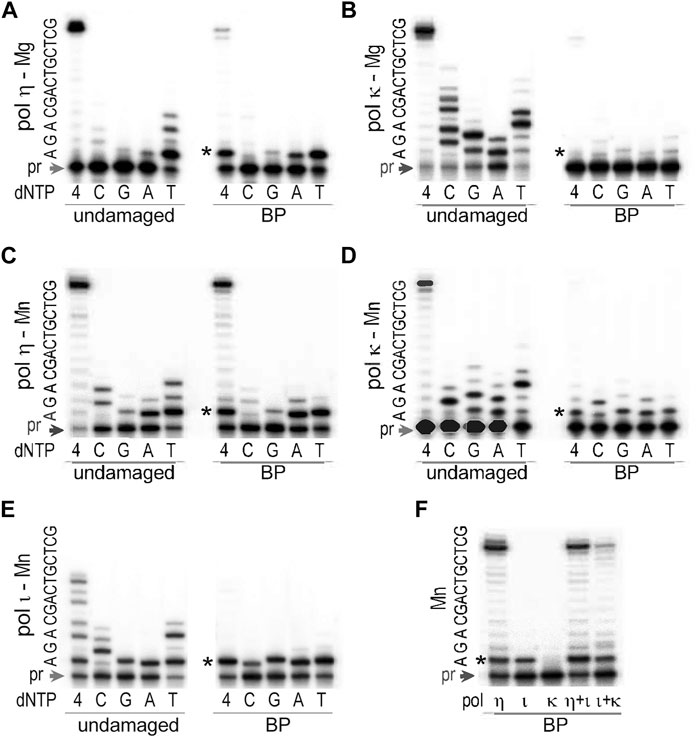
FIGURE 6. TLS past trans-S-BPDE-dA by T. lanuginosus pols. The ability to bypass BPDE-dA was assayed for (A) polη in presence of 4 mM Mg2+, (B) polκ in presence of 4 mM Mg2+, (C) polη in presence of 4 mM Mn2+, (D) polκ in presence of 4 mM Mn2+, (E) polι in presence of 4 mM Mn2+, and (F) individual, or mixture of various pols in 4 mM Mn2+. The substrate used in these assays was made by annealing of the 32P labeled primer 5′-CACTGCAGACTCTAAA-3′ and either an undamaged or BPDE-containing template 5′- GCTCGTCAGCAGATTTAGAGTCTGCAGTG-3′, where the underlined bold A stands for the undamaged, or BPDE-modified dA. Reactions contained 100 μM each individual nucleotide (dC, dG, dA, dT) or mixture of all four dNTPs as indicated in the figure and were carried out at 37°C for 10 min. Concentrations of enzymes were 0.17 pM for polη, 0.32 pM for polκ, and 0.15 pM for polι. The sequence of the template immediately downstream of the primer (pr) is shown on the left-hand side of each gel pair. The star (*) indicates position of the adduct.
While polκ itself was incapable of extending primers by more than one nucleotide after insertion opposite the BPDE-dA even in the presence of Mn2+, it was able to assist polι to bypass the lesion and elongate primers to the end of the template (Figure 6F). Nevertheless, TLS catalyzed by the combined action of polι and polκ was still less efficient than by polη alone (Figure 6F).
Characterization of T. lanuginosus Rev1
S. cerevisiae and human Rev1 function as a dCMP transferase, incorporating strictly dCMP opposite abasic sites and dG-lesions (Nelson et al., 1996; Lin et al., 1999; Masuda et al., 2001). dCMP transferase activity is instructed by Rev1’s conserved active site side chain arginine residue via a protein-templated mechanism (Nair et al., 2005; Weaver et al., 2020). We hypothesized that the conservation of the functional arginine and Rev1 consensus sequence (Figure 7A) would make T. lanuginosis Rev1 a bonafide dCMP transferase. Therefore, the DNA synthesis activity of T. lanuginosus Rev1 was characterized in vitro opposite damaged and undamaged DNA. We found that T. lanuginosus Rev1 could utilize both Mn2+ and Mg2+, but reasoned that Mg2+ was a more likely candidate for physiological utilization given its wider efficacy in comparison to Mn2+ (Figure 7B). Similar to the activities of S. cerevisiae and human Rev1, DNA synthesis by T. lanuginosus Rev1 was not processive, whereby it inserted only once opposite a templating dG and stalled at the following dT base (Figure 7C). As expected, T. lanuginosus Rev1 had a similar thermostability to the other T. lanuginosus Y-family pols (Figure 7D). The well-characterized dCMP transferase activity of Rev1 ortholog was evaluated opposite undamaged, as well as abasic site- and CPD-containing templates (Figure 7E). The ability for T. lanuginosus Rev1 to perform DNA synthesis was efficient only in the presence of dCMP opposite undamaged, or abasic site templates, akin to S. cerevisiae and human REV1 (Zhang et al., 2002b; Nair et al., 2011; Weerasooriya et al., 2014). In summary, we found that the DNA synthesis properties of T. lanuginosus Rev1 are comparable with its higher eukaryotic ortholog.
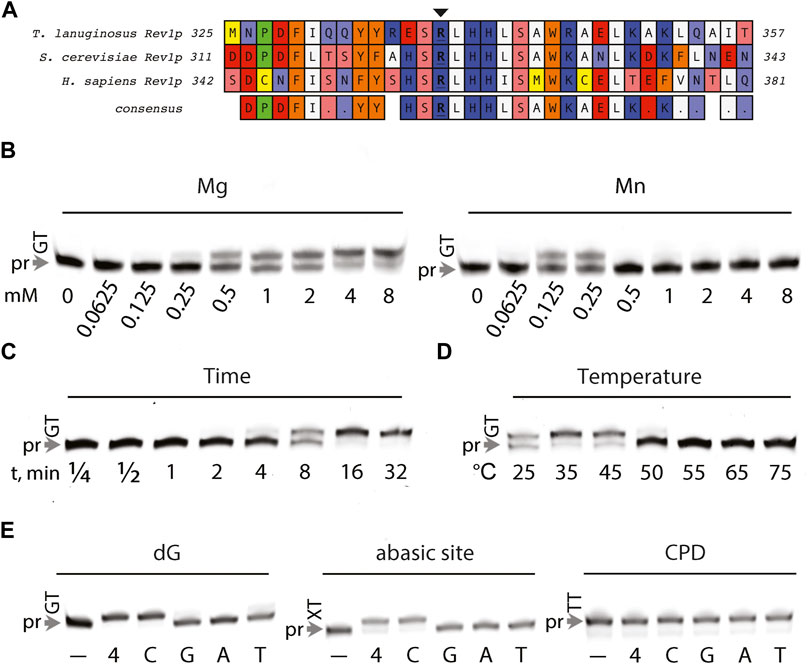
FIGURE 7. Characterization of DNA synthesis by T. lanuginosis Rev1. (A) Sequence alignment between REV1 orthologs (Thermomyces lanuginosus, Saccharomyces cerevisiae, and Homo sapiens), illustrating the conserved arginine residue (indicated with an arrow) required for protein-templated DNA synthesis (B) Serial 1:1 dilutions of Mg2+ and Mn2+ metal ions present within 6-FAM primer extension reactions, starting at 8 mM Me2+. (C) Processivity was qualitatively assessed by varying the duration (minutes) of 6-FAM primer extension reactions. (D) Thermostability was evaluated by monitoring DNA synthesis activity at varied temperature (25–75°C). (E) Lesion bypass activity for undamaged (dG), abasic site, and CPD template base/lesion identities was examined in the presence of either none (–), 100 µM dNTPs (4), 100 µM dCTP (C), 100 µM dGTP (G), 100 µM dATP (A), 100 µM dTTP (T).
Concluding Remarks
In this paper, we describe the cloning, purification, and biochemical characterization of Y-family pols from the thermophilic fungus T. lanuginosus. T. lanuginosis contains all four eukaryotic Y-family orthologs that each generally demonstrate biochemical properties similar to the properties of higher eukaryotic orthologs with the exception of the expected thermostability of the T. lanuginosis Y-family polymerases. Interestingly, while T. lanuginosis polι exhibited biochemical properties akin to the human enzyme, its metal ion requirement was significantly different. Whereas human polι is activated by very low levels of Mn2+ (250 μM) and inhibited by high levels of Mg2+ (>1 mM) (Frank and Woodgate, 2007), T. lanuginosis polι required 4–8 mM Mn2+ or Mg2+ respectively, for maximal activity in vitro.
The conservation of Y-family polymerase activities between lower and higher eukaryotic organisms suggests that TLS activity may be more essential to cellular viability than previously appreciated. Further structure-function studies of these polymerases, as well as identification and characterization of other Y-family orthologs from yet unsequenced species would help clarify the seemingly random ortholog dispersion among eukaryotes and how their presence/absence might provide evolutionary advantages in the face of different environmental stressors.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Author Contributions
Conceptualization: JM and RW. Data curation: AV, JM, MS, and SA. Formal analysis: AV, JM, and MS. Funding acquisition: RW. Investigation: AV, JM, MS, and SA. Visualization: AV, JM, and MS. Writing-original draft, Review and editing: AV, JM, MS, SA, TE, and RW.
Funding
Funding for this work was provided by the National Institute of Child Health and Human Development/National Institutes of Health Intramural Research Program (RW).
Conflict of Interest
TE is employed by New England Biolabs, Inc., a company that commercializes enzyme reagents, including DNA polymerases.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We would like to thank Dominic Quiros (NIH) for assistance with the early exploratory phase of Rev1 purification. We also thank Francine B. Perler for her work on purifying T. lanuginosus polι.
Abbreviations
Pols, polymerases; TLS, translesion synthesis; CPD, cyclobutane pyrimidine dimer; BPDE, benzo[a] pyrene diol epoxide; S. cerevisiae, Saccharomyces cerevisiae; T. lanuginosus, Thermomyces lanuginosus; S. pombe, Schizosaccharomyces pombe.
References
Avkin, S., Goldsmith, M., Velasco-Miguel, S., Geacintov, N., Friedberg, E. C., and Livneh, Z. (2004). Quantitative Analysis of Translesion DNA Synthesis across a Benzo[a]pyrene-Guanine Adduct in Mammalian Cells. J. Biol. Chem. 279, 53298–53305. doi:10.1074/jbc.M409155200
Basu, A. K., Pande, P., and Bose, A. (2017). Translesion Synthesis of 2′-Deoxyguanosine Lesions by Eukaryotic DNA Polymerases. Chem. Res. Toxicol. 30, 61–72. doi:10.1021/acs.chemrestox.6b00285
Bessman, M. J., Lehman, I. R., Simms, E. S., and Kornberg, A. (1958). Enzymatic Synthesis of Deoxyribonucleic Acid. II. General Properties of the Reaction. J. Biol. Chem. 233, 171–177. doi:10.1016/s0021-9258(19)68049-x
Boiteux, S., and Guillet, M. (2004). Abasic Sites in DNA: Repair and Biological Consequences in Saccharomyces cerevisiae. DNA Repair 3, 1–12. doi:10.1016/j.dnarep.2003.10.002
Boudsocq, F., Iwai, S., Hanaoka, F., and Woodgate, R. (2001). Sulfolobus solfataricus P2 DNA Polymerase IV (Dpo4): an Archaeal DinB-like DNA Polymerase with Lesion-Bypass Properties Akin to Eukaryotic Polη. Nucleic Acids Res. 29, 4607–4616. doi:10.1093/nar/29.22.4607
Boudsocq, F., Kokoska, R. J., Plosky, B. S., Vaisman, A., Ling, H., Kunkel, T. A., et al. (2004). Investigating the Role of the Little Finger Domain of Y-Family DNA Polymerases in Low Fidelity Synthesis and Translesion Replication. J. Biol. Chem. 279, 32932–32940. doi:10.1074/jbc.m405249200
Chiapperino, D., Kroth, H., Kramarczuk, I. H., Sayer, J. M., Masutani, C., Hanaoka, F., et al. (2002). Preferential Misincorporation of Purine Nucleotides by Human DNA Polymerase η Opposite Benzo[a]pyrene 7,8-Diol 9,10-Epoxide Deoxyguanosine Adducts. J. Biol. Chem. 277, 11765–11771. doi:10.1074/jbc.m112139200
Choi, J.-Y., Angel, K. C., and Guengerich, F. P. (2006). Translesion Synthesis across Bulky N2-Alkyl Guanine DNA Adducts by Human DNA Polymerase κ. J. Biol. Chem. 281, 21062–21072. doi:10.1074/jbc.M602246200
Choi, J.-Y., Lim, S., Kim, E.-J., Jo, A., and Guengerich, F. P. (2010). Translesion Synthesis across Abasic Lesions by Human B-Family and Y-Family DNA Polymerases α, δ, η, ι, κ, and REV1. J. Mol. Biol. 404, 34–44. doi:10.1016/j.jmb.2010.09.015
Dominguez, O., Ruiz, J. F., Lain De Lera, T., Garcia-Diaz, M., Gonzalez, M. A., Kirchhoff, T., et al. (2000). DNA Polymerase Mu (Pol μ), Homologous to TdT, Could Act as a DNA Mutator in Eukaryotic Cells. EMBO J. 19, 1731–1742. doi:10.1093/emboj/19.7.1731
Frank, E. G., and Woodgate, R. (2007). Increased Catalytic Activity and Altered Fidelity of Human DNA Polymerase ι in the Presence of Manganese. J. Biol. Chem. 282, 24689–24696. doi:10.1074/jbc.m702159200
Frank, E. G., Sayer, J. M., Kroth, H., Ohashi, E., Ohmori, H., Jerina, D. M., et al. (2002). Translesion Replication of Benzo[a]pyrene and Benzo[c]phenanthrene Diol Epoxide Adducts of Deoxyadenosine and Deoxyguanosine by Human DNA Polymerase ι. Nucleic Acids Res. 30, 5284–5292. doi:10.1093/nar/gkf643
Frank, E. G., McDonald, J. P., Karata, K., Huston, D., and Woodgate, R. (2012). A Strategy for the Expression of Recombinant Proteins Traditionally Hard to Purify. Anal. Biochem. 429, 132–139. doi:10.1016/j.ab.2012.07.016
Garcia-Diaz, M., Bebenek, K., Krahn, J. M., Pedersen, L. C., and Kunkel, T. A. (2007). Role of the Catalytic Metal during Polymerization by DNA Polymerase λ. DNA Repair 6, 1333–1340. doi:10.1016/j.dnarep.2007.03.005
Gerlach, V. L., Feaver, W. J., Fischhaber, P. L., and Friedberg, E. C. (2001). Purification and Characterization of Polκ, a DNA Polymerase Encoded by the Human DINB1 Gene. J. Biol. Chem. 276, 92–98. doi:10.1074/jbc.m004413200
Haracska, L., Washington, M. T., Prakash, S., and Prakash, L. (2001). Inefficient Bypass of an Abasic Site by DNA Polymerase η. J. Biol. Chem. 276, 6861–6866. doi:10.1074/jbc.m008021200
Jha, V., and Ling, H. (2018). Structural Basis for Human DNA Polymerase κ to Bypass Cisplatin Intrastrand Cross-Link (Pt-GG) Lesion as an Efficient and Accurate Extender. J. Mol. Biol. 430, 1577–1589. doi:10.1016/j.jmb.2018.04.023
Jia, L., Geacintov, N. E., and Broyde, S. (2008). The N-Clasp of Human DNA Polymerase κ Promotes Blockage or Error-free Bypass of Adenine- or Guanine-Benzo[a]pyrenyl Lesions. Nucleic Acids Res. 36, 6571–6584. doi:10.1093/nar/gkn719
Johnson, R. E., Prakash, S., and Prakash, L. (2000a). The Human DINB1 Gene Encodes the DNA Polymerase Polθ. Proc. Natl. Acad. Sci. 97, 3838–3843. doi:10.1073/pnas.97.8.3838
Johnson, R. E., Washington, M. T., Haracska, L., Prakash, S., and Prakash, L. (2000b). Eukaryotic Polymerases ι and ζ Act Sequentially to Bypass DNA Lesions. Nature 406, 1015–1019. doi:10.1038/35023030
Johnson, R. E., Washington, M. T., Prakash, S., and Prakash, L. (2000c). Fidelity of Human DNA Polymerase η. J. Biol. Chem. 275, 7447–7450. doi:10.1074/jbc.275.11.7447
Kirouac, K. N., and Ling, H. (2009). Structural Basis of Error-Prone Replication and Stalling at a Thymine Base by Human DNA Polymerase ι. EMBO J. 28, 1644–1654. doi:10.1038/emboj.2009.122
Kokoska, R. J., Bebenek, K., Boudsocq, F., Woodgate, R., and Kunkel, T. A. (2002). Low Fidelity DNA Synthesis by a Y Family DNA Polymerase Due to Misalignment in the Active Site. J. Biol. Chem. 277, 19633–19638. doi:10.1074/jbc.m202021200
Lin, W., Xin, H., Zhang, Y., Wu, X., Yuan, F., and Wang, Z. (1999). The Human REV1 Gene Codes for a DNA Template-dependent dCMP Transferase. Nucleic Acids Res. 27, 4468–4475. doi:10.1093/nar/27.22.4468
Lindahl, T. (1993). Instability and Decay of the Primary Structure of DNA. Nature 362, 709–715. doi:10.1038/362709a0
Ling, H., Boudsocq, F., Woodgate, R., and Yang, W. (2004a). Snapshots of Replication through an Abasic Lesion. Mol. Cel 13, 751–762. doi:10.1016/s1097-2765(04)00101-7
Ling, H., Sayer, J. M., Plosky, B. S., Yagi, H., Boudsocq, F., Woodgate, R., et al. (2004b). Crystal Structure of a Benzo[a]pyrene Diol Epoxide Adduct in a Ternary Complex with a DNA Polymerase. Proc. Natl. Acad. Sci. 101, 2265–2269. doi:10.1073/pnas.0308332100
Liu, Y., Yang, Y., Tang, T.-S., Zhang, H., Wang, Z., Friedberg, E., et al. (2014). Variants of Mouse DNA Polymerase κ Reveal a Mechanism of Efficient and Accurate Translesion Synthesis Past a Benzo[a]pyrene dG Adduct. Proc. Natl. Acad. Sci. USA 111, 1789–1794. doi:10.1073/pnas.1324168111
Loeb, L. A., and Kunkel, T. A. (1982). Fidelity of DNA Synthesis. Annu. Rev. Biochem. 51, 429–457. doi:10.1146/annurev.bi.51.070182.002241
Loeb, L. A. (1989). Endogenous Carcinogenesis: Molecular Oncology into the Twenty-First Century--Presidential Address. Cancer Res. 49, 5489–5496.
Maheshwari, R., Bharadwaj, G., and Bhat, M. K. (2000). Thermophilic Fungi: Their Physiology and Enzymes. Microbiol. Mol. Biol. Rev. 64, 461–488. doi:10.1128/MMBR.64.3.461-488.2000
Makarova, A. V., and Kulbachinskiy, A. V. (2012). Structure of Human DNA Polymerase ι and the Mechanism of DNA Synthesis. Biochem. Mosc. 77, 547–561. doi:10.1134/S0006297912060016
Martin, M. J., Garcia-Ortiz, M. V., Esteban, V., and Blanco, L. (2013). Ribonucleotides and Manganese Ions Improve Non-Homologous End Joining by Human Polµ. Nucleic Acids Res. 41, 2428–2436. doi:10.1093/nar/gks1444
Masuda, Y., Takahashi, M., Tsunekuni, N., Minami, T., Sumii, M., Miyagawa, K., et al. (2001). Deoxycytidyl Transferase Activity of the Human REV1 Protein Is Closely Associated with the Conserved Polymerase Domain. J. Biol. Chem. 276, 15051–15058. doi:10.1074/jbc.m008082200
Masutani, C., Kusumoto, R., Yamada, A., Dohmae, N., Yokoi, M., Yuasa, M., et al. (1999). The XPV (Xeroderma Pigmentosum Variant) Gene Encodes Human DNA Polymerase η. Nature 399, 700–704. doi:10.1038/21447
Masutani, C., Kusumoto, R., Iwai, S., and Hanaoka, F. (2000). Mechanisms of Accurate Translesion Synthesis by Human DNA Polymerase η. EMBO J. 19, 3100–3109. doi:10.1093/emboj/19.12.3100
McCulloch, S. D., Kokoska, R. J., Masutani, C., Iwai, S., Hanaoka, F., and Kunkel, T. A. (2004). Preferential Cis-Syn Thymine Dimer Bypass by DNA Polymerase η Occurs with Biased Fidelity. Nature 428, 97–100. doi:10.1038/nature02352
McHunu, N. P., Permaul, K., Abdul Rahman, A. Y., Saito, J. A., Singh, S., and Alam, M. (2013). Xylanase Superproducer: Genome Sequence of a Compost-Loving Thermophilic Fungus, Thermomyces Lanuginosus Strain SSBP. Genome Announc 1, e00388–13. doi:10.1128/genomeA.00388-13
McIntyre, J. (2020). Polymerase ι - an Odd Sibling Among Y Family Polymerases. DNA Repair 86, 102753. doi:10.1016/j.dnarep.2019.102753
Mildvan, A. S., Loeb, L. A., and Wu, C.-W. (1979). The Role of Metal Ions in the Mechanisms of DNA and RNA Polymerase. CRC Crit. Rev. Biochem. 6, 219–244. doi:10.3109/10409237909102564
Mukherjee, P., Lahiri, I., and Pata, J. D. (2013). Human Polymerase κ Uses a Template-Slippage Deletion Mechanism, but Can Realign the Slipped Strands to Favour Base Substitution Mutations over Deletions. Nucleic Acids Res. 41, 5024–5035. doi:10.1093/nar/gkt179
Nair, D. T., Johnson, R. E., Prakash, L., Prakash, S., and Aggarwal, A. K. (2005). Rev1 Employs a Novel Mechanism of DNA Synthesis Using a Protein Template. Science 309, 2219–2222. doi:10.1126/science.1116336
Nair, D. T., Johnson, R. E., Prakash, L., Prakash, S., and Aggarwal, A. K. (2009). DNA Synthesis across an Abasic Lesion by Human DNA Polymerase ι. Structure 17, 530–537. doi:10.1016/j.str.2009.02.015
Nair, D. T., Johnson, R. E., Prakash, L., Prakash, S., and Aggarwal, A. K. (2011). DNA Synthesis across an Abasic Lesion by Yeast REV1 DNA Polymerase. J. Mol. Biol. 406, 18–28. doi:10.1016/j.jmb.2010.12.016
Nelson, J. R., Lawrence, C. W., and Hinkle, D. C. (1996). Deoxycytidyl Transferase Activity of Yeast REV1 Protein. Nature 382, 729–731. doi:10.1038/382729a0
Ohashi, E., Bebenek, K., Matsuda, T., Feaver, W. J., Gerlach, V. L., Friedberg, E. C., et al. (2000a). Fidelity and Processivity of DNA Synthesis by DNA Polymerase κ, the Product of the Human DINB1 Gene. J. Biol. Chem. 275, 39678–39684. doi:10.1074/jbc.M005309200
Ohashi, E., Ogi, T., Kusumoto, R., Iwai, S., Masutani, C., Hanaoka, F., et al. (2000b). Error-Prone Bypass of Certain DNA Lesions by the Human DNA Polymerase Kappa. Genes Dev. 14, 1589–1594. doi:10.1101/gad.14.13.1589
Ohmori, H., Friedberg, E. C., Fuchs, R. P. P., Goodman, M. F., Hanaoka, F., Hinkle, D., et al. (2001). The Y-Family of DNA Polymerases. Mol. Cel 8, 7–8. doi:10.1016/s1097-2765(01)00278-7
Pages, V., Johnson, R. E., Prakash, L., and Prakash, S. (2008). Mutational Specificity and Genetic Control of Replicative Bypass of an Abasic Site in Yeast. Proc. Natl. Acad. Sci. 105, 1170–1175. doi:10.1073/pnas.0711227105
Patra, A., Zhang, Q., Lei, L., Su, Y., Egli, M., and Guengerich, F. P. (2015). Structural and Kinetic Analysis of Nucleoside Triphosphate Incorporation Opposite an Abasic Site by Human Translesion DNA Polymerase η. J. Biol. Chem. 290, 8028–8038. doi:10.1074/jbc.M115.637561
Rechkoblit, O., Zhang, Y., Guo, D., Wang, Z., Amin, S., Krzeminsky, J., et al. (2002). trans-Lesion Synthesis Past Bulky Benzo[a]pyrene Diol Epoxide N2-dG and N6-dA Lesions Catalyzed by DNA Bypass Polymerases. J. Biol. Chem. 277, 30488–30494. doi:10.1074/jbc.m201167200
Sale, J. E., Lehmann, A. R., and Woodgate, R. (2012). Y-family DNA Polymerases and Their Role in Tolerance of Cellular DNA Damage. Nat. Rev. Mol. Cel Biol 13, 141–152. doi:10.1038/nrm3289
Sherrer, S. M., Fiala, K. A., Fowler, J. D., Newmister, S. A., Pryor, J. M., and Suo, Z. (2011). Quantitative Analysis of the Efficiency and Mutagenic Spectra of Abasic Lesion Bypass Catalyzed by Human Y-Family DNA Polymerases. Nucleic Acids Res. 39, 609–622. doi:10.1093/nar/gkq719
Singh, S., Madlala, A. M., and Prior, B. A. (2003). Thermomyces Lanuginosus: Properties of Strains and Their Hemicellulases. FEMS Microbiol. Rev. 27, 3–16. doi:10.1016/S0168-6445(03)00018-4
Sørensen, A. B., Duch, M., Jørgensen, P., and Pedersen, F. S. (1993). Amplification and Sequence Analysis of DNA Flanking Integrated Proviruses by a Simple Two-step Polymerase Chain Reaction Method. J. Virol. 67, 7118–7124. doi:10.1128/JVI.67.12.7118-7124.1993
Stern, H. R., Sefcikova, J., Chaparro, V. E., and Beuning, P. J. (2019). Mammalian DNA Polymerase κ Activity and Specificity. Molecules 24, 2805. doi:10.3390/molecules24152805
Strauss, B. S. (1991). The ‘A Rule’ of Mutagen Specificity: A Consequence of DNA Polymerase Bypass of Non-instructional Lesions? BioEssays 13, 79–84. doi:10.1002/bies.950130206
Suzuki, N., Ohashi, E., Kolbanovskiy, A., Geacintov, N. E., Grollman, A. P., Ohmori, H., et al. (2002). Translesion Synthesis by Human DNA Polymerase κ on a DNA Template Containing a Single Stereoisomer of dG-(+)- or dG-(−)-Anti-N2-BPDE (7,8-Dihydroxy-Anti-9,10-Epoxy-7,8,9,10-Tetrahydrobenzo[a]pyrene). Biochemistry 41, 6100–6106. doi:10.1021/bi020049c
Thompson, P. S., and Cortez, D. (2020). New Insights into Abasic Site Repair and Tolerance. DNA Repair 90, 102866. doi:10.1016/j.dnarep.2020.102866
Tissier, A., Frank, E. G., McDonald, J. P., Iwai, S., Hanaoka, F., and Woodgate, R. (2000a). Misinsertion and Bypass of Thymine-Thymine Dimers by Human DNA Polymerase ι. EMBO J. 19, 5259–5266. doi:10.1093/emboj/19.19.5259
Tissier, A., McDonald, J. P., Frank, E. G., and Woodgate, R. (2000b). Polι, a Remarkably Error-Prone Human DNA Polymerase. Genes Dev. 14, 1642–1650. doi:10.1101/gad.14.13.1642
Tsiklinsky, P. (1899). Sur les Mucedinees Thermophiles. Paris: Libraires de L'academie de Medecine, 500–504.
Vaisman, A., and Woodgate, R. (2017). Translesion DNA Polymerases in Eukaryotes: what Makes Them Tick? Crit. Rev. Biochem. Mol. Biol. 52, 274–303. doi:10.1080/10409238.2017.1291576
Vaisman, A., and Woodgate, R. (2020). Mysterious and Fascinating: DNA Polymerase Ɩ Remains Enigmatic 20 Years after its Discovery. DNA Repair 93, 102914. doi:10.1016/j.dnarep.2020.102914
Vaisman, A., Tissier, A., Frank, E. G., Goodman, M. F., and Woodgate, R. (2001). Human DNA Polymerase ι Promiscuous Mismatch Extension. J. Biol. Chem. 276, 30615–30622. doi:10.1074/jbc.m102694200
Vaisman, A., Frank, E. G., McDonald, J. P., Tissier, A., and Woodgate, R. (2002). Polι-Dependent Lesion Bypass In Vitro. Mutat. Res./Fundam. Mol. Mech. Mutagen. 510, 9–22. doi:10.1016/s0027-5107(02)00248-8
Vaisman, A., Frank, E. G., Iwai, S., Ohashi, E., Ohmori, H., Hanaoka, F., et al. (2003). Sequence Context-dependent Replication of DNA Templates Containing UV-Induced Lesions by Human DNA Polymerase ι. DNA Repair 2, 991–1006. doi:10.1016/s1568-7864(03)00094-6
Vashishtha, A. K., Wang, J., and Konigsberg, W. H. (2016). Different Divalent Cations Alter the Kinetics and Fidelity of DNA Polymerases. J. Biol. Chem. 291, 20869–20875. doi:10.1074/jbc.R116.742494
Washington, M. T., Johnson, R. E., Prakash, S., and Prakash, L. (1999). Fidelity and Processivity of Saccharomyces cerevisiae DNA Polymerase η. J. Biol. Chem. 274, 36835–36838. doi:10.1074/jbc.274.52.36835
Weaver, T. M., Cortez, L. M., Khoang, T. H., Washington, M. T., Agarwal, P. K., and Freudenthal, B. D. (2020). Visualizing Rev1 Catalyze Protein-Template DNA Synthesis. Proc. Natl. Acad. Sci. USA 117, 25494–25504. doi:10.1073/pnas.2010484117
Weerasooriya, S., Jasti, V. P., and Basu, A. K. (2014). Replicative Bypass of Abasic Site in Escherichia coli and Human Cells: Similarities and Differences. PLoS One 9, e107915. doi:10.1371/journal.pone.0107915
Yang, W., and Gao, Y. (2018). Translesion and Repair DNA Polymerases: Diverse Structure and Mechanism. Annu. Rev. Biochem. 87, 239–261. doi:10.1146/annurev-biochem-062917-012405
Zhang, Y., Yuan, F., Wu, X., Rechkoblit, O., Taylor, J. S., Geacintov, N. E., et al. (2000a). Error-prone Lesion Bypass by Human DNA Polymeraseη. Nucleic Acids Res. 28, 4717–4724. doi:10.1093/nar/28.23.4717
Zhang, Y., Yuan, F., Wu, X., Wang, M., Rechkoblit, O., Taylor, J. S., et al. (2000b). Error-free and Error-Prone Lesion Bypass by Human DNA Polymerase κ In Vitro. Nucleic Acids Res. 28, 4138–4146. doi:10.1093/nar/28.21.4138
Zhang, Y., Yuan, F., Wu, X., and Wang, Z. (2000c). Preferential Incorporation of G Opposite Template T by the Low-Fidelity Human DNA Polymerase ι. Mol. Cel Biol 20, 7099–7108. doi:10.1128/mcb.20.19.7099-7108.2000
Zhang, Y., Wu, X., Guo, D., Rechkoblit, O., and Wang, Z. (2002a). Activities of Human DNA Polymerase κ in Response to the Major Benzo[a]pyrene DNA Adduct: Error-free Lesion Bypass and Extension Synthesis from Opposite the Lesion. DNA Repair 1, 559–569. doi:10.1016/s1568-7864(02)00055-1
Zhang, Y., Wu, X., Rechkoblit, O., Geacintov, N. E., Taylor, J. S., and Wang, Z. (2002b). Response of Human REV1 to Different DNA Damage: Preferential dCMP Insertion Opposite the Lesion. Nucleic Acids Res. 30, 1630–1638. doi:10.1093/nar/30.7.1630
Keywords: thermostable fungi, Y-family DNA polymerases, phylogenetic analysis, translesion DNA synthesis, DNA polymerase eta (polη), DNA polymerase iota (polι), DNA polymerase kappa (polκ), Rev1
Citation: Vaisman A, McDonald JP, Smith MR, Aspelund SL, Evans TC and Woodgate R (2021) Identification and Characterization of Thermostable Y-Family DNA Polymerases η, ι, κ and Rev1 From a Lower Eukaryote, Thermomyces lanuginosus. Front. Mol. Biosci. 8:778400. doi: 10.3389/fmolb.2021.778400
Received: 16 September 2021; Accepted: 19 October 2021;
Published: 03 November 2021.
Edited by:
Janice Pata, Wadsworth Center, United StatesReviewed by:
Penny J. Beuning, Northeastern University, United StatesPierre Barraud, Expression Génétique Microbienne, France
Copyright © 2021 Vaisman, McDonald, Smith, Aspelund, Evans and Woodgate. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Roger Woodgate, d29vZGdhdGVAbWFpbC5uaWguZ292
†ORCID: Alexandra Vaisman, orcid.org0000-0002-2521-1467; John P. McDonald, orcid.org/0000-0003-2482-148X; Mallory R. Smith, orcid.org/0000-0003-1450-7825; Sender L. Aspelund, orcid.org/0000-0003-0726-4028; Thomas C. Evans Jr, orcid.org/0000-0001-5406-0146; Roger Woodgate, orcid.org/0000-0002-2521-1467
‡Present address: Sender L. Aspelund, Novavax, Inc., Gaithersburg, MD 20878, United States
§The authors share first authorship
 Alexandra Vaisman1†§
Alexandra Vaisman1†§ Roger Woodgate
Roger Woodgate