- 1Institute of Bioorganic Chemistry, Polish Academy of Sciences, Poznan, Poland
- 2Department of Chemistry, The RNA Institute, University at Albany, State University of New York, Albany, NY, United States
G•U wobble base pair frequently occurs in RNA structures. The unique chemical, thermodynamic, and structural properties of the G•U pair are widely exploited in RNA biology. In several RNA molecules, the G•U pair plays key roles in folding, ribozyme catalysis, and interactions with proteins. G•U may occur as a single pair or in tandem motifs with different geometries, electrostatics, and thermodynamics, further extending its biological functions. The metal binding affinity, which is essential for RNA folding, catalysis, and other interactions, differs with respect to the tandem motif type due to the different electrostatic potentials of the major grooves. In this work, we present the crystal structure of an RNA 8-mer duplex r[UCGUGCGA]2, providing detailed structural insights into the tandem motif I (5′UG/3′GU) complexed with Ba2+ cation. We compare the electrostatic potential of the presented motif I major groove with previously published structures of tandem motifs I, II (5′GU/3′UG), and III (5′GG/3′UU). A local patch of a strongly negative electrostatic potential in the major groove of the presented structure forms the metal binding site with the contributions of three oxygen atoms from the tandem. These results give us a better understanding of the G•U tandem motif I as a divalent metal binder, a feature essential for RNA functions.
Introduction
Watson–Crick (WC) base pairs shape the RNA double-helical landscape. However, multiple non-WC interactions (Leontis et al., 2002) have been distinguished in RNA structures and implicated in various biological functions of RNA (Westhof and Fritsch, 2000; Chandrasekhar and Malathhi, 2003; Brown, 2020). The G•U wobble base pair is the most frequent among the non-WC base pairs in RNA molecules. Its existence was proposed by F. Crick over 50 years ago, alongside the hypothesis that G•U plays an important role in the decoding of messenger RNA (mRNA) codons (Crick., 1966). Since then, G•U pairs have been found in multiple classes of RNA, including messenger RNA (mRNA) (Benard et al., 1998), ribosomal RNA (rRNA) (Woese et al., 1983; Gutell et al., 1994), transfer RNA (tRNA) (Ladner et al., 1975), small nuclear RNA (Wu and Manley, 1992; Sashital et al., 2004; Sashital et al., 2007), and ribozymes [such as group I and II introns, and in hepatitis delta virus (HDV)] (Cech et al., 1981; Allain and Varani, 1995; Strobel and Cech, 1995; Been and Wickham, 1997; Nishikawa et al., 1997; Colmenarejo and Tinoco, 1999).
The G•U pairs can be accommodated within A-form RNA helices with minimal structural distortions. In a typical G•U base pair, the uracil is displaced into the major groove of the RNA helix. However, other types of G•U pairing, i.e., the bifurcated G•U pairs (non-helical regions), the tautomeric G•U pairs (all positions of the codon/anticodon triplets), and the minor groove-shifted G•U pair (tRNALys wobble position of the codon/anticodon triplet) have been observed (reviewed in Westhof et al., 2019). G•U pairs usually interact on their Watson–Crick edges, exploiting the subtleties of the G and U electronic configurations (reviewed in Westhof et al., 2019). Beyond numerous types of single G•U pairing, the regular G•U pair can also form tandems that are often present in rRNA structures (Gautheret et al., 1995). The types of G•U tandems vary in terms of their structural, electrostatic, and thermodynamic properties (Xu et al., 2007). The frequency of the different tandem motifs in rRNA follows the trend of 5′UG/3′GU (motif I) > 5′GG/3′UU (motif III) > 5′GU/3′UG (motif II) (Figure 1). The same trend applies to the thermodynamic stabilities of the G•U tandems (He et al., 1991; Gautheret et al., 1995; Wu et al., 1995; Deng and Sundaralingam, 2000).
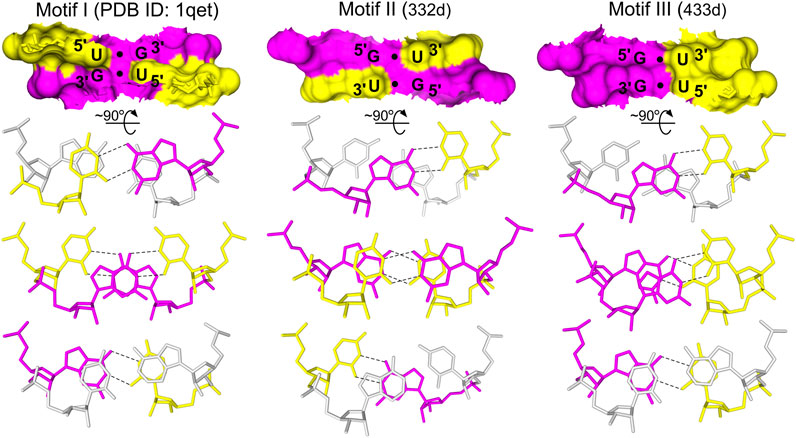
FIGURE 1. The three types of G•U tandem motifs. The tandem motifs are shown as surface representations based on the NMR and X-ray structures (Biswas and Sundaralingam, 1997; McDowell et al., 1997; Trikha et al., 1999). The stacking pattern characteristic for each motif is shown, including the base pairs flanking the G•U tandems (gray). In G•U tandems, G is magenta and U is yellow. Hydrogen bonds (black dashed lines) are presented only for G•U pairs.
The significance of the G•U base pair in RNA structures is emphasized by its high conservation (Sprinzl et al., 1996). Throughout the three domains of life, the G3•U70 pair is a major determinant of the amino acid acceptor identity of tRNAAla (Hou and Schimmel, 1988; McClain and Foss, 1988; Chong et al., 2018). The unique chemical groups of the G•U pair exposed to the major and minor grooves, its thermodynamic stability, conformational flexibility, and the non-isostericity of G•U compared to the WC pair are essential attributes for its biological functions (Varani and McClain, 2000; Ananth et al., 2013). The exocyclic N2 amino group of G exposed to the minor groove of G•U 1) mediates contacts with proteins, e.g., alanyl-tRNA synthetase (Musier-Forsyth et al., 1991; Ananth et al., 2013; Naganuma et al., 2014; Chong et al., 2018); 2) is involved in the 5′ splice site selection and stabilization of the transition state in group I self-splicing introns (Strobel and Cech, 1995; 1996); and 3) thermodynamically stabilizes interactions within the catalytic core of group II self-splicing introns (Boudvillain and Pyle, 1998; Konforti et al., 1998). The major groove of the G•U pair, due to its deep negative potential, may function as a recognition site for metals, amino acids such as R, H, K, and N, and other positively charged ligands (Varani and McClain, 2000; Chen et al., 2010; Ananth et al., 2013). The unbounded O4 carbonyl group of U is one of the G3•U70 tRNAAla elements recognized by alanyl-tRNA synthetase (Naganuma et al., 2014). The conserved G (G•U) of the catalytic triad in group II introns is involved in RNA triplex formation through its major groove edge (Keating et al., 2010; Manigrasso et al., 2020); the G chemical groups exposed to the major groove are critical for catalysis (Konforti et al., 1998). The non-isosteric nature of G•U, along with the presence of the N2 amino group in the G•U minor groove, is an important element for the Tetrahymena ribozyme catalytic activity (Strobel and Cech, 1995) and tRNAAla aminoacylation (Hou and Schimmel, 1988; Park et al., 1989; Ananth et al., 2013). Moreover, the non-isostericity of G•U plays a role in RNA tertiary interactions. The shift of U into the major groove of the G•U pair allows it to form a minor groove suitable for close contact with the WC pair of the other helix, creating the so-called along-groove packing motif (Gagnon and Steinberg, 2002; Ananth et al., 2013).
Here, we present the crystal structure of an RNA duplex containing motif I of the G•U wobble tandem. In the previous study, the major groove of G•U wobble motif I has been determined as less negative than the major groove of motifs II and III and accordingly suggested as the least favorable metal binding site among the G•U tandems (Xu et al., 2007). In this work, we observed that the 5′UG/3′GU tandem complexed with the Ba2+ cation, showing that G•U wobble motif I attracts metal ions and functions as their potent binding site. Structural analysis of the presented RNA duplex confirmed the A-form helix and indicated a few unique features of the G•U tandem.
Results and Discussion
Overall Structure Quality
The crystal of the r(UCGUGCGA)2 duplex was obtained even though an equimolar of its complementary strand UCGCACGA was used for crystallization as a hybrid duplex. The presence of a divalent cation (Ba2+), bound in the major groove (see below), might be the major driving force behind the formation of the r(UCGUGCGA)2 duplex. The crystals in the H32 space group, with one duplex in the asymmetric unit, diffracted X-rays to 2.2 Å resolution. The detailed diffraction data and model refinement statistics are listed in Table 1. There are 10 structures of RNA octanucleotides in the Protein Data Bank (PDB), determined at resolutions between 1.15 and 2.6 Å.
The obtained electron density maps allowed tracing RNA unambiguously and modeling one Ba2+ cation and two water molecules in its coordination sphere (Figure 2). The strong peak (9.9σ level) in the anomalous difference electron density map corroborated the presence of Ba2+; at the used X-ray energy of 12,400 eV, the f′′ of barium is ∼4.1e. Individual base pairs were well-defined in the maps for canonical WC pairs (Figure 3A) and for the 5′UG/3′GU wobble tandem (Figure 3B).
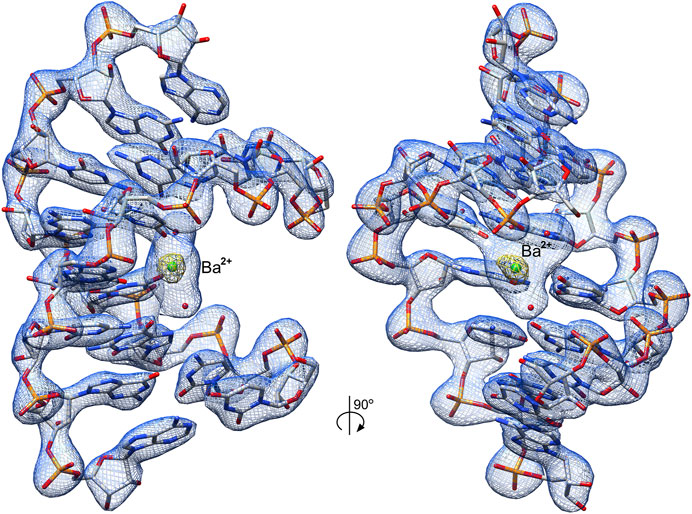
FIGURE 2. Crystal structure of the r(UCGUGCGA)2 duplex. The structure contains the 5′UG/3′GU wobble tandem constituting the binding site of Ba2+ (green ball) and two water molecules (red balls). The 2Fo–Fc electron density map (blue mesh) around the RNA duplex is contoured at the 1.5σ level. The anomalous difference electron density map (yellow mesh) is shown at the 7σ level; the strong map peak verifies the presence of Ba2+.
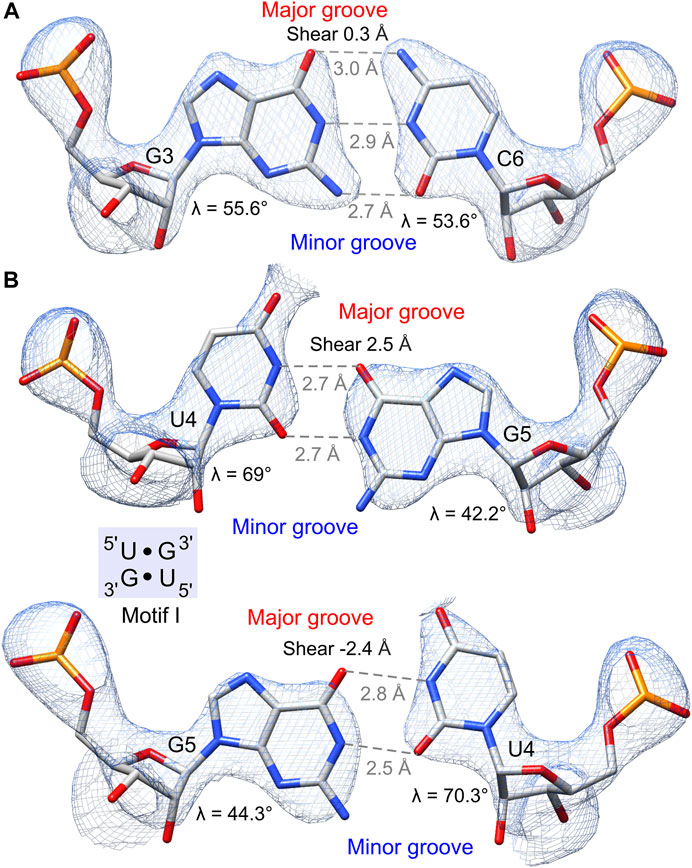
FIGURE 3. Hydrogen bonding pattern and geometry of the G•U pair and its canonical counterpart. The 2Fo–Fc electron density map (blue mesh) of wobble pairs is contoured at the 1.5σ level. (A) G–C pair preceding the 5′UG/3′GU tandem. (B) 5′UG/3′GU tandem representing motif I (light blue box). The guanosine N7, guanosine O6, and the unpaired uridine O4 carbonyl group of G•U pairs are exposed to the major groove and generate a region of negative potential. The non-hydrogen-bonded N2 amino groups of guanosines are positioned in the minor groove. Unique structural features of G•U such as shear and unequal glycosidic bond angles (λ) at G and U are indicated.
The Major Groove Created by the 5′UG/3′GU Wobble Tandem Is Optimal for Binding Divalent Cations
Ba2+ was bound in the duplex major groove by the exo-O4 atoms of U4 residues in both chains and the O6 atom of G5 (chain A) (Figures 2 and 4). Two water molecules coordinated Ba2+ in the major groove. The coordination sphere of Ba2+ was incomplete, although seemingly compatible with the most common Ba2+ coordination number of 9. More ligands of the Ba2+ cation (e.g., four more water molecules) may not be visible in the electron density maps due to the limited data resolution.
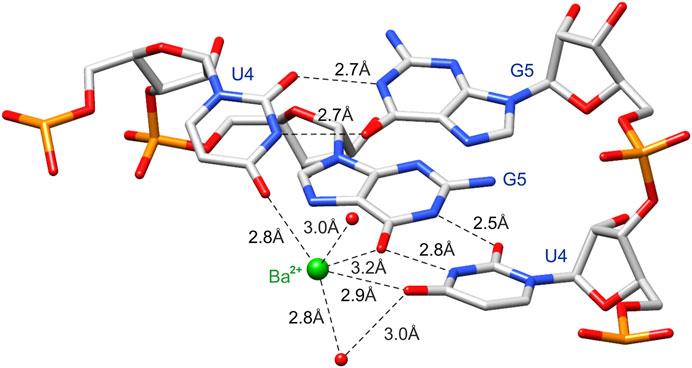
FIGURE 4. The cation binding site is created by functional groups of the 5′UG/3′GU wobble tandem [O6 carbonyl group of G5 (chain A) and O4 carbonyl group of U4 in both chains] poised into the major groove. Ba2+ and water molecules are marked as green and red balls, respectively.
The G•U wobble pair is negatively charged with the group composition of guanosine N7, guanosine O6, and the unpaired O4 carbonyl of uridine, which are exposed to the major groove forming a region of strong negative electrostatic potential (Figure 3B) (McDowell and Turner, 1996; Varani and McClain, 2000). A comparison between the surface electrostatic potential of the presented 5′UG/3′GU wobble tandem duplex and an ideal RNA helix containing the canonical 5′CG/3′GC motif showed clear centralization of the negative charge within the major groove of the wobble tandem (Figure 5). In contrast, WC pairs exposed their amino groups into the major groove and disturbed the negative electrostatic potential (Figures 3A and 5).
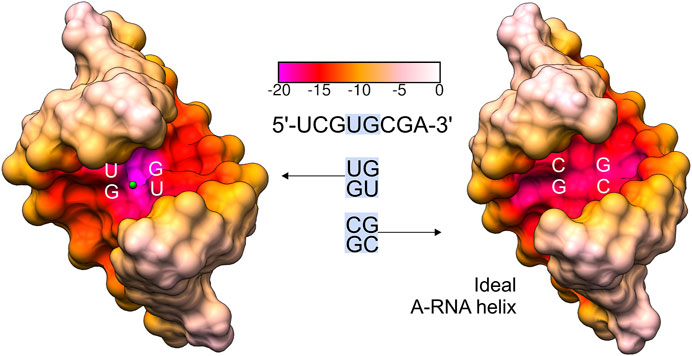
FIGURE 5. Comparison of the major groove electrostatic potential in the presented 5′UG/3′GU wobble tandem (motif I) and ideal A-helix RNA containing canonical 5′CG/3′GC counterparts (generated using Coot software) (Emsley et al., 2010). Ba2+ is shown as a green ball. The electrostatic surface was calculated using the APBS/PDB2PQR server (default settings) (Baker et al., 2001; Dolinsky et al., 2004; Jurrus et al., 2018).
The negatively polarized surface within the major groove of the G•U wobble pair has a strong affinity for divalent metal ions (Ott et al., 1993; Allain and Varani, 1995; Konforti et al., 1998). This effect is even stronger for two consecutive G•U pairs (Cate and Doudna, 1996; McDowell and Turner, 1996; Kieft and Tinoco, 1997). In the presented structure, the Ba2+ cation was located in the center of the 5′UG/3′GU tandem (Figures 2 and 4). Universally, the G•U pair is polarized, and the G side is more negative than U. In the 5′UG/3′GU wobble tandem (motif I), the two Gs belonged to different strands, and the center of the tandem was the most negative part of the duplex (Figures 4 and 5) (Xu et al., 2007).
The database of the metal ion binding sites in RNA (MeRNA) indicates that the G•U pair major groove is the most common metal binding RNA motif (Stefan et al., 2006). Divalent metal ions are usually crucial for RNA structure and function. Metal ions can counteract the repulsion between the negatively charged phosphate backbone and stabilize various RNA tertiary structures for diverse functions (Cate and Doudna, 1996; Draper, 2004; Woodson, 2005; Marcia and Pyle, 2014). For example, the capacity of the G•U pair to interact with metal ions was postulated as an important factor for RNA catalysis, i.e., partially hydrated Mg2+ ion interacting with N7 and O6 of the reverse G•U pair in the HDV ribozyme active site directly participates in RNA cleavage reactions (Chen et al., 2010).
Generally, G•U wobble pairs enhance the negative potential in the major groove. However, the stacking patterns and major groove sizes associated with various G•U tandem motifs affect the absolute electrostatic potential (Xu et al., 2007). The major groove created by a single G•U pair and the three motifs of the wobble tandem (I, II, and III) differed in widths and consequently showed diverse contributions of their phosphate backbones to electrostatic potential (formal electrostatic potential; Figure 6). The G•U tandem motif I (Figure 6A) has been proposed to exhibit less negative formal electrostatic potential than the G•U tandem of motifs II and III (Xu et al., 2007) (Figures 6B, C). The 1eka structure (Chen et al., 2000), representing the G•U tandem motif I contained a significantly extended major groove (Figure 6A); the inter-strand phosphate–phosphate distance for this helix was 19.8 Å (Xu et al., 2007). Its wide major groove caused a lower concentration of negative potential from the bases and the backbone. In effect, the negativity of the G•U pair was diffused and the major grove electrostatic potential was similar or even weaker than those of its canonical counterparts (Xu et al., 2007) (Figure 6). The major groove of the presented helix, also containing motif I wobble tandem, revealed a direct inter-strand phosphate–phosphate distance of 16.5 Å (Supplementary Table S1). Thus, the major groove width was closer to those of the G•U tandem motifs II and III (Xu et al., 2007) (Figures 5 and 6) and canonical counterparts (∼14.9 Å in an ideal A-helix; Supplementary Table S1). Moreover, analysis of the surface electrostatic potential of this duplex indicated the presence of a negatively charged region within the major groove, which was, in fact, the Ba2+ cation binding site (Figures 4 and 5).
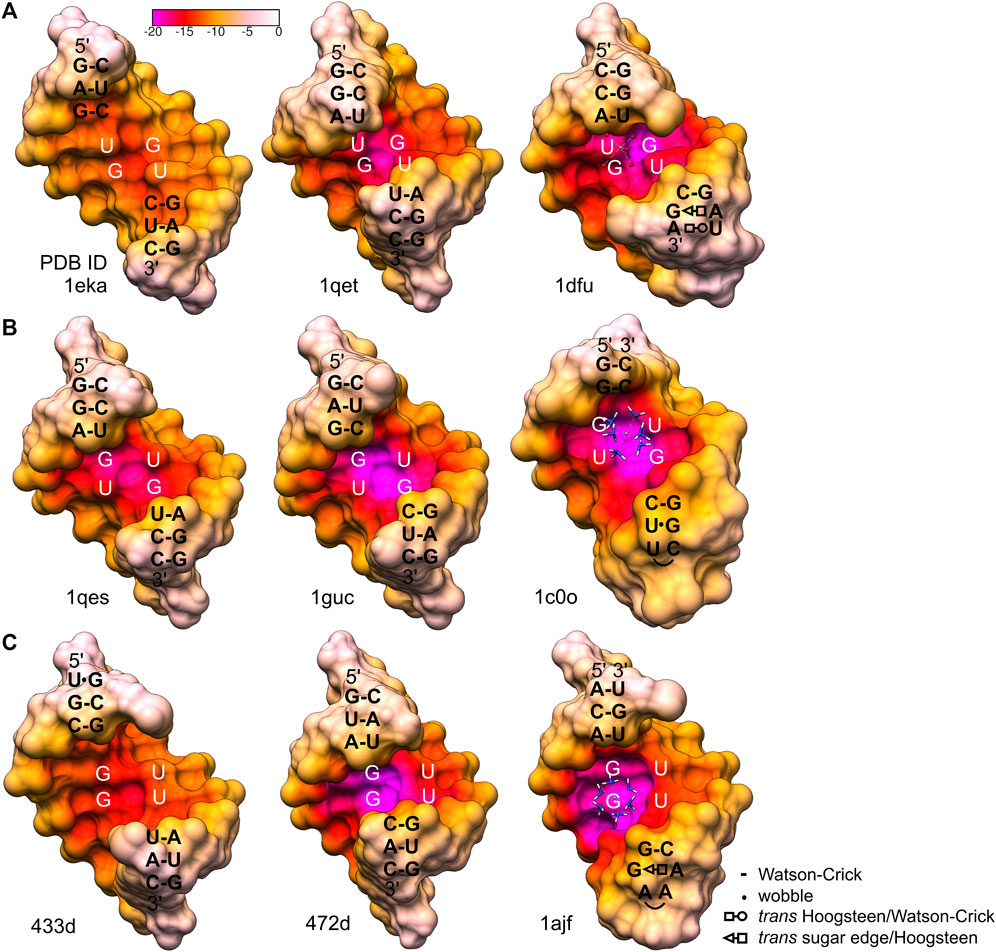
FIGURE 6. Electrostatic potential of the major grooves found in the X-ray and NMR structures containing different types of wobble tandems (McDowell and Turner, 1996; Kieft and Tinoco, 1997; McDowell et al., 1997; Colmenarejo and Tinoco, 1999; Trikha et al., 1999; Chen et al., 2000; Deng and Sundaralingam, 2000; Lu and Steitz, 2000). (A) 5′UG/3′GU wobble tandem, motif I. (B) 5′GU/3′UG wobble tandem, motif II. 1c0o shows the P5 hairpin of a group I intron complexed with [Co(NH3)6]3+. (C) 5′GU/3′GU wobble tandem, motif III. 1ajf is a model of a group I intron P5b stem–loop structure complexed with [Co(NH3)6]3+. The electrostatic surface was calculated by the APBS/PDB2PQR server using default settings (Baker et al., 2001; Dolinsky et al., 2004; Jurrus et al., 2018). The original structures 1dfu, 433d, and 1ajf were truncated to octamers to correspond to the other presented motifs. Interactions in sequences flanking the G•U tandem motifs are represented as shown in the legend, according to (Leontis et al., 2002).
Another G•U tandem motif I, found in loop E of 5S rRNA [1dfu (Lu and Steitz, 2000), truncated to the duplex of octamers], was characterized by quite negative formal electrostatic potential of the major groove that binds Mg2+ (Figure 6A). The direct inter-strand phosphate–phosphate distance within the G•U tandem in that structure was ∼15.4 Å. The general geometry of the helix in the 1dfu structure was affected by other non-WC base pairs in the neighborhood of the 5′UG/3′GU tandem (1dfu; Figure 6A). The opposite effect, which was a relatively low electrostatic potential in the major groove of motif III, can be observed in the structure 433d (Trikha et al., 1999; Xu et al., 2007) (Figure 6C). The proximity of two asymmetrical tandem G•U pairs seemed to affect the helix geometry. As a result, the major groove was wider than the ones in a standard A-form RNA helix, and the pattern of the electrostatic potential was also altered.
The geometrical dissimilarity of the G•U pair with its U•G and WC counterparts (see below) affected the stacking interactions between the G•U pair and flanking base pairs (Ananth et al., 2013). The three G•U tandem motifs were characterized by different stacking patterns: I, purine (R)–R inter-strand stacking; II, R–pyrimidine (Y) intra-strand stacking; and III, a mix of intra- and inter-strand stacking (Figures 1 and 7). The reported diverse G•U tandem motifs in identical sequential contexts (PDB IDs 1eka and 1guc, 1qet and 1qes) showed varied stacking patterns with the flanking base pairs and differed in electrostatic potential (Figures 6A, B) (Xu et al., 2007). Moreover, the flanking sequences in 5′GAG-tandem-CUC3′ and in 5′GGA-tandem-UCC3′ seemed to exert opposite effects on the absolute value of the electrostatic potential for motif I (1eka, 1qet; Figure 6A) and motif II (1guc, 1qes; Figure 6B).
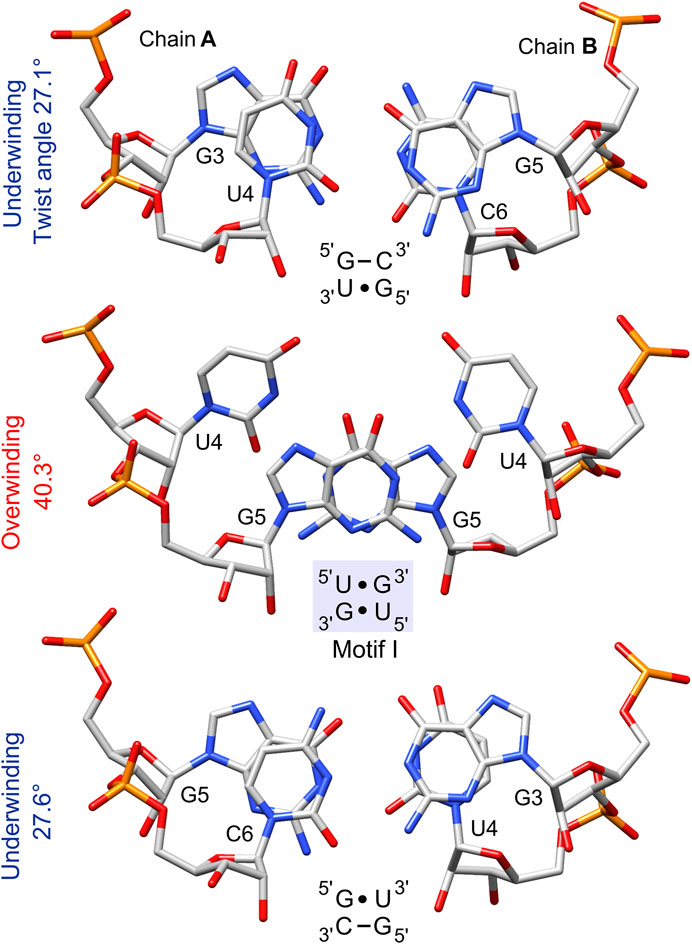
FIGURE 7. The stacking pattern within the 5′UG/3′GU tandem and between the wobble tandem pairs and flanking GC pairs. The 5′UG/3′GU wobble tandem represents motif I (light blue box), in which inter-strand stacking involves rings of guanosines, whereas unstacked uracils face each other. Local twist angle variations associated with each step are indicated.
Overall, the sequential context of the wobble G•U pair and tandems impact the helix geometry, including the base stacking pattern and the major groove width. These structural features influence the overall electrostatic potential and eventual metal recognition and binding. It has been proposed that uniformity and the absence of positively charged groups make the major groove of G•U pairs an attractive binding site for metal ions to a greater extent than formal electrostatic potential (Chin et al., 1999). Colmenarejo and Tinoco observed that Mg2+ preferentially binds to G•U motif I, whereas complex ion [Co(NH3)6]3+ favorably binds to motifs II and III (1999). Thus, the binding affinity of specific RNAs is likely a composite of multiple factors involving geometry, electrostatic potential, hydration pattern, and some quantum effects (Xu et al., 2007). It was noted that Ba2+ is not a physiological cation and that the binding of Mg2+ occurs in vivo with two conceivable outcomes which may depend on the sequential context. One involves the direct coordination of Mg2+ by similar donors, as observed in our structure or in 1dfu, where Mg2+ recognition is affected by protein (Lu and Steitz, 2000). The second possibility is the indirect binding of hydrated Mg2+, only via water-mediated interactions. In both scenarios, the binding mode of Mg2+ could also change the RNA structure, and new structures with other flanking sequences are needed to elucidate more details. Knowledge about the predisposition of the G•U pair for binding cations is also useful in biochemical applications, for example, in crystallography to facilitate experimental phasing of X-ray structures (Cate and Doudna, 1996; Kieft and Tinoco, 1997; Keel et al., 2007).
The 5′UG/3′GU Tandem in A-Helix Reveals Geometric Variations
All the calculations of the duplex structural parameters were performed using 3DNA v2.4.3 (2019apr06) (Li et al., 2019). For structural characterization of the 5′UG/3′GU tandem, unless specified otherwise, we have used simple base pairs and step parameters due to their more intuitive interpretation of structural variations in structures containing non-WC pairs. The simple parameters provide a complete qualitative description of the base-pairing geometries and their step parameters (Lu et al., 2015; Li et al., 2019). Hydrogen bonds in all WC pairs and the G•U wobble pairs of the presented duplex were in the expected range of 2.5–3.2 Å (Supplementary Table S2). The distance between the U4 (B) O2 atom and G5 (A) N2 atom in the wobble pair was 3.2 Å, indicating the presence of a weak hydrogen bond; however, the geometry was suboptimal for a stabilized interaction.
The global helix geometry resembled the A-form since the riboses revealed C3′-endo sugar puckers and the displacement from the helical axis (X-displacement) had a negative value, on average −4.46 Å, which is close to the ideal A-RNA helix, approx. −4 Å. However, a large negative X-displacement of −10.3 Å were observed at the 5′UG/3′GU step (local base-pair helical parameters; Supplementary Table S3). The individual torsion angles were within the ranges typical for the A-helix form (Table 2 and Supplementary Table S4) (Schneider et al., 1997). The average of the glycosidic dihedral angles (chi, χ) was −165 ± 3° for purines and was −163 ± 4° for pyrimidines. These values agree with the C3′-endo ribose and glycosidic bonds in anti-conformation (Table 2 and Supplementary Table S4). Similarly, the average of the delta (δ) angles of 82 ± 8° corresponded to the universal value for C3′-endo sugar conformation (81 ± 7°; Table 2 and Supplementary Table S4) (Gelbin et al., 1996; Parkinson et al., 1996; Schneider et al., 1997).
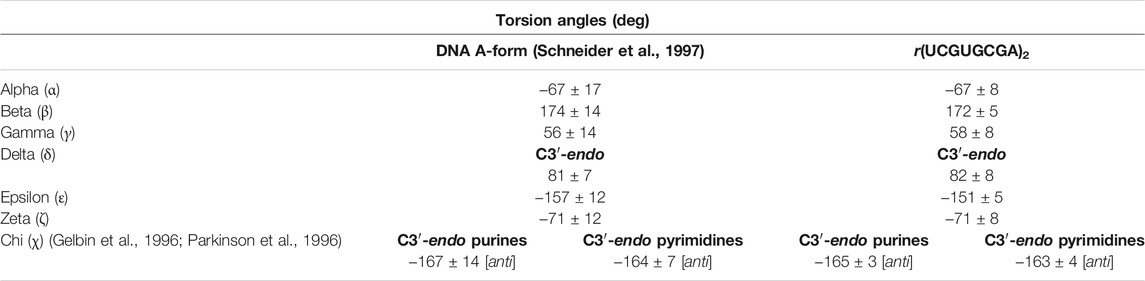
TABLE 2. Universal torsion angles for the A-helix and torsion angles (Gelbin et al., 1996; Parkinson et al., 1996; Schneider et al., 1997) in the r(UCGUGCGA)2 duplex calculated using the 3DNA server, v2.4.3-2019apr06 (Lu and Olson, 2003; Li et al., 2019).
The negative slide and positive roll values calculated for the present duplex (Table 3) are in agreement with the trend predicted for A-helix (Lu and Olson, 2003). Furthermore, the roll angles at each base-pair step alternated (Table 3). This tendency has been observed in alternating purine (R)–pyrimidine (Y) sequences in which the roll values tended to be lower at 5′R to 3′Y steps and higher at 5′Y–3′R steps (Dock-Bregeon et al., 1988; Dock-Bregeon et al., 1989; Biswas et al., 1997). The buckle angles tended to be negative for the 5′R–3′Y pairs and positive for the 5′Y–3′R pairs (Table 4).
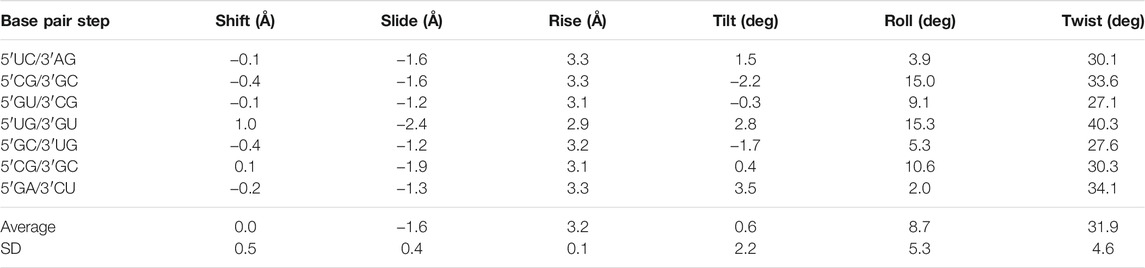
TABLE 3. Simple base-pair step parameters of the r(UCGUGCGA)2 duplex based on consecutive C1′–C1′ vectors calculated using the 3DNA server, v2.4.3-2019apr06 (Lu and Olson, 2003; Li et al., 2019)
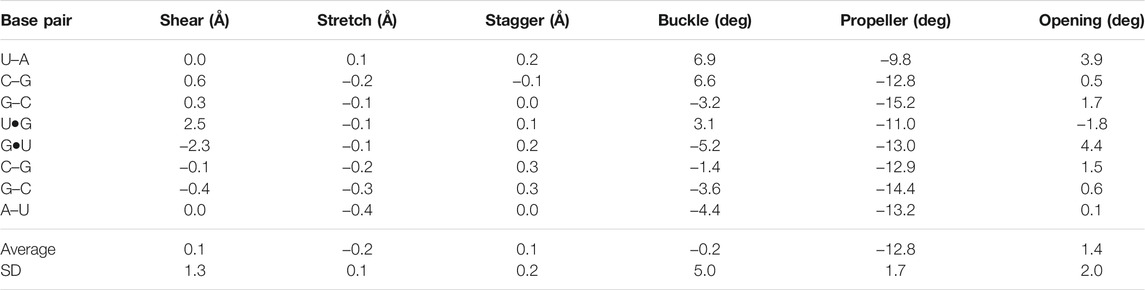
TABLE 4. Simple base-pair parameters of the r(UCGUGCGA)2 duplex based on RC8-YC6 vectors calculated using the 3DNA server, v2.4.3-2019apr06 (Lu and Olson, 2003; Li et al., 2019)
Several distinctive geometric features of the G•U wobble pair occurred in the 5′UG/3′GU tandem region. The key characteristic of the U•G and G•U wobble pairs is the shear of 2.5 and −2.4 Å, respectively (Lu and Olson, 2003) (Table 4 and Figure 3B). The displacement of the G•U pair relative to the WC counterparts provoked an asymmetry in the glycosidic bond angles (λ) subtended at the glycosyl carbon atoms C1 (Figure 3). The λ angles for the WC base pairs were nearly identical, measuring ∼54° (Figure 3A) (Varani and McClain, 2000). In wobble pairs, the λ angles measured ∼43° at G and ∼70° at U (Figure 3B and Supplementary Table S5). The above structural characteristic of the G•U pair caused its geometric dissimilarity (non-isostericity) with the WC base pairs and non-self-isostericity with the U•G pair (G•U ≠ U•G). The presence of the non-isosteric G•U contributes to the overall geometry of the RNA structures and may provide RNA recognition elements (Ananth et al., 2013).
The non-isostericity of G•U leads to the local twist angle variations (Ananth et al., 2013). The herein presented twist angle values around wobble pairs were calculated as simple base-pair step parameters in 3DNA based on the consecutive C1′–C1′ vectors (Li et al., 2019). Since the G•U tandem represents motif I, the 5′UG/3′GU tandem base-pair step had a high twist angle of 40.3°, while the steps between the wobble tandem and the flanking WC base pairs (5′GU/3′CG and 5′GC/3′UG) exhibited lower than average twist angles of 27.1° and 27.6° (Table 3 and Figure 7). The compensatory effect of the under- and overtwisting of the 5′UG/3′GU tandem and associated base pairs allowed the RNA duplex to maintain an average twist of ∼32°, which was close to the value for a typical A-RNA helix. A similar underwinding–overwinding–underwinding trend was also observed in other duplexes carrying the 5′UG/3′GU tandem motif I (e.g., PDB ID: 315d, 1eka) (Biswas et al., 1997; Chen et al., 2000).
The 5′UG/3′GU tandem was characterized by a stacking pattern typical of motif I (Figures 1, 7). At the U•G/G•U step, inter-strand stacking involved rings of guanosine, whereas unstacked uracils faced each other (Gautheret et al., 1995; Biswas et al., 1997) (Figures 1 and 7). The observed inter-strand stacking of G5 (A) and G5 (B) corresponded to overwinding (twist = 40.3°; Table 3 and Figure 7) and mostly resulted from the combination of a negative slide (−2.4 Å) and a positive shift (1 Å) (Lu and Olson, 2003) (Table 3). The strong stacking interaction between guanosines and the enhanced stacking of the U•G and G•U pairs with the flanking WC pairs (Figure 7) provided thermodynamic stability of the 5′UG/3′GU tandem—one of the major reasons making this motif the most abundant among G•U wobble tandem motifs (Gautheret et al., 1995; Wu et al., 1995; McDowell and Turner, 1996; Masquida and Westhof, 2000; Ananth et al., 2013). Comparison of the thermodynamic stability between the different wobble tandems revealed the following trend motif I > motif III > motif II (He et al., 1991; Wu et al., 1995; Deng and Sundaralingam, 2000). Moreover, the C–G pair succeeding G•U (as in the presented duplex) was thermodynamically more stable than G–C, A–U, and U–A (all after G•U) and, hence, was most frequent in the RNA structures (He et al., 1991; Ananth et al., 2013).
Conclusion
G•U wobble base pairs play a key role in RNA biology. While several structures including such pairs have been reported, this work presents the first detailed analysis based on an experimental model of the motif I G•U tandem complexed with a divalent metal cation. This is particularly important as metal binding by G•U pairs appears vital to RNA folding, catalytic functions, and RNA–protein interactions.
The metal binding site in the r(UCGUGCGA)2 duplex was formed by the G•U pair tandem in the major groove. The O6 carbonyl of G and two O4 carbonyls of U within the tandem created the coordination sphere of the metal. The G•U pairs were non-self-isosteric (G•U ≠ U•G), which is consistent with the asymmetric coordination sphere of the metal cation. It is also apparent that the distribution of the electrostatic potential within the major groove contributed to the formation of the metal binding site. The aforementioned features may provide elements for recognition by RNA binding proteins.
Materials and Methods
Crystallization Conditions and Diffraction Data Collection
The crystal of the r(UCGUGCGA)2 duplex was obtained in the presence of its complementary strand UCGCACGA. The crystals grew in the 12th condition of nucleic acid mini-screen from Hampton [10% m-phenylenediamine (MPD), 0.04 M sodium cacodylate trihydrate, pH 6.0, 0.012 M spermine tetrahydrochloride, 0.08 M potassium chloride, and 0.02 M barium chloride] in 2 weeks with size of ∼0.1 × 0.06 × 0.03 mm, using a hanging drop setting. They were cryoprotected by increasing the MPD concentration to 35%.
Diffraction data were collected at the SER-CAT beamline 22-BM at the Advanced Photon Source, Argonne National Laboratory (Lemont, IL, USA). The diffraction data were processed with the XDS package (Kabsch, 2010); the processing statistics are given in Table 1.
Determination and Refinement of the Crystal Structures
The crystal structure of the r(UCGUGCGA)2 duplex was solved by molecular replacement in PHASER (McCoy et al., 2007) using an ideal single-stranded RNA, generated in Coot (Emsley et al., 2010). The data were also processed in the H3 space group and tested for twinning using L-test and by twin refinement in Refmac5 (Murshudov et al., 2011). The estimated twin fraction was ∼20%, and Rwork/Rfree did not decrease; therefore, we decided to use data in the H32 space group. The initial model was refined initially in phenix.refine (Afonine et al., 2012) and in the final cycles in Refmac5 (Murshudov et al., 2011) with external restraints generated by the RestraintLib (Kowiel et al., 2020) server. Coot (Emsley et al., 2010) was used for manual model corrections between rounds of automatic model refinement. The presence of Ba2+ was inferred from the peak size of the electron density map, correlated with the content of the crystallization solution. The anomalous difference electron density map, calculated with Phenix (Liebschner et al., 2019), confirmed the presence of an anomalous scatterer, interpreted as Ba2+.
Determination of Structural Parameters and Electrostatic Potential
Structural parameters of the r(UCGUGCGA)2 duplex were analyzed using the 3DNA (v2.4.3-2019apr06) server (Li et al., 2019). The electrostatic potential of the r(UCGUGCGA)2 duplex and the other structures compared in this study [PDB ID: 1eka, 1qet, 1dfu, 1qes, 1guc, 1c0o, 433d, 472d, and 1ajf; ideal A-helix r(UCGCGCGA)2 generated by COOT] (Emsley et al., 2010) was calculated using the APBS/PDB2PQR server (Baker et al., 2001; Dolinsky et al., 2004; Jurrus et al., 2018) with default settings (accessed in March 2021). The results of the electrostatic potential calculations were presented using Chimera 1.13.1 software (Pettersen et al., 2004).
Data Availability Statement
Atomic coordinates and structure factors for the crystal structure of RNA duplex r(UCGUGCGA)2 in complex with Ba2+ cation have been deposited with the Protein Data Bank (PDB) under the PDB ID 7ouo.
Author Contributions
AR performed the analysis and drafted the manuscript. MR solved the structure and edited the text. YZ and SM synthesized the strands and performed the crystallization. JS designed and managed the project and funding and edited the text.
Funding
This study is supported by the National Science Foundation (NSF) (CHE-1845486 to JS).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
Diffraction data were collected at the SER-CAT beamline 22-BM at the Advanced Photon Source, Argonne National Laboratory, supported by the US Department of Energy, Office of Science, Office of Basic Energy Sciences under contract W-31-109-Eng-38.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmolb.2021.762786/full#supplementary-material
References
Afonine, P. V., Grosse-Kunstleve, R. W., Echols, N., Headd, J. J., Moriarty, N. W., Mustyakimov, M., et al. (2012). Towards Automated Crystallographic Structure Refinement with phenix.Refine. Acta Crystallogr. D Biol. Cryst. 68, 352–367. doi:10.1107/S0907444912001308
Allain, F. H.-T., and Varani, G. (1995). Divalent Metal Ion Binding to a Conserved Wobble Pair Defining the Upstream Site of Cleavage of Group I Self-Splicing Introns. Nucl. Acids Res. 23, 341–350. doi:10.1093/nar/23.3.341
Ananth, P., Goldsmith, G., and Yathindra, N. (2013). An Innate Twist between Crick's Wobble and Watson-Crick Base Pairs. RNA 19, 1038–1053. doi:10.1261/rna.036905.112
Baker, N. A., Sept, D., Joseph, S., Holst, M. J., and Mccammon, J. A. (2001). Electrostatics of Nanosystems: Application to Microtubules and the Ribosome. Proc. Natl. Acad. Sci. 98, 10037–10041. doi:10.1073/pnas.181342398
Been, M. D., and Wickham, G. S. (1997). Self-cleaving Ribozymes of Hepatitis delta Virus RNA. Eur. J. Biochem. 247, 741–753. doi:10.1111/j.1432-1033.1997.00741.x
Benard, L., Mathy, N., Grunberg-Manago, M., Ehresmann, B., Ehresmann, C., and Portier, C. (1998). Identification in a Pseudoknot of a U*G Motif Essential for the Regulation of the Expression of Ribosomal Protein S15. Proc. Natl. Acad. Sci. 95, 2564–2567. doi:10.1073/pnas.95.5.2564
Biswas, R., and Sundaralingam, M. (1997). Crystal Structure of r(GUGUGUA)dC with Tandem G·U/U·G Wobble Pairs with Strand Slippage. J. Mol. Biol. 270, 511–519. doi:10.1006/jmbi.1997.1118
Biswas, R., Wahl, M. C., Ban, C., and Sundaralingam, M. (1997). Crystal Structure of an Alternating Octamer r(GUAUGUA)dC with Adjacent G·U Wobble Pairs. J. Mol. Biol. 267, 1149–1156. doi:10.1006/jmbi.1997.0936
Boudvillain, M., and Marie Pyle, A. (1998). Defining Functional Groups, Core Structural Features and Inter-domain Tertiary Contacts Essential for Group II Intron Self-Splicing: a NAIM Analysis. EMBO J. 17, 7091–7104. doi:10.1093/emboj/17.23.7091
Brown, J. A. (2020). Unraveling the Structure and Biological Functions of RNA Triple Helices. WIREs RNA 11, e1598. doi:10.1002/wrna.1598
Cate, J. H., and Doudna, J. A. (1996). Metal-binding Sites in the Major Groove of a Large Ribozyme Domain. Structure 4, 1221–1229. doi:10.1016/s0969-2126(96)00129-3
Cech, T. R., Zaug, A. J., and Grabowski, P. J. (1981). In Vitro splicing of the Ribosomal RNA Precursor of Tetrahymena: Involvement of a Guanosine Nucleotide in the Excision of the Intervening Sequence. Cell 27, 487–496. doi:10.1016/0092-8674(81)90390-1
Chandrasekhar, K., and Malathi, R. (2003). Non-Watson Crick Base Pairs Might Stabilize RNA Structural Motifs in Ribozymes - a Comparative Study of Group-I Intron Structures. J. Biosci. 28, 547–555. doi:10.1007/BF02703330
Chen, J.-H., Yajima, R., Chadalavada, D. M., Chase, E., Bevilacqua, P. C., and Golden, B. L. (2010). A 1.9 Å Crystal Structure of the HDV Ribozyme Precleavage Suggests Both Lewis Acid and General Acid Mechanisms Contribute to Phosphodiester Cleavage. Biochemistry 49, 6508–6518. doi:10.1021/bi100670p
Chen, X., Mcdowell, J. A., Kierzek, R., Krugh, T. R., and Turner, D. H. (2000). Nuclear Magnetic Resonance Spectroscopy and Molecular Modeling Reveal that Different Hydrogen Bonding Patterns Are Possible for G·U Pairs: One Hydrogen Bond for Each G·U Pair in R(GGCGUGCC)2 and Two for Each G·U Pair in R(GAGUGCUC)2. Biochemistry 39, 8970–8982. doi:10.1021/bi992938e
Chong, Y. E., Guo, M., Yang, X.-L., Kuhle, B., Naganuma, M., Sekine, S.-i., et al. (2018). Distinct Ways of G:U Recognition by Conserved tRNA Binding Motifs. Proc. Natl. Acad. Sci. USA 115, 7527–7532. doi:10.1073/pnas.1807109115
Colmenarejo, G., and Tinoco, I. (1999). Structure and Thermodynamics of Metal Binding in the P5 helix of a Group I Intron Ribozyme 1. Edited by P. E. Wright. J. Mol. Biol. 290, 119–135. doi:10.1006/jmbi.1999.2867
Crick, F. H. C. (1966). Codon-anticodon Pairing: The Wobble Hypothesis. J. Mol. Biol. 19, 548–555. doi:10.1016/s0022-2836(66)80022-0
Deng, J., and Sundaralingam, M. (2000). Synthesis and crystal Structure of an Octamer RNA R(guguuuac)/r(guaggcac) with G·G/U·U Tandem Wobble Base Pairs: Comparison with Other Tandem G·U Pairs. Nucleic Acids Res. 28, 4376–4381. doi:10.1093/nar/28.21.4376
Dock-Bregeon, A. C., Chevrier, B., Podjarny, A., Johnson, J., De Bear, J. S., Gough, G. R., et al. (1989). Crystallographic Structure of an RNA helix: [U(UA)6A]2. J. Mol. Biol. 209, 459–474. doi:10.1016/0022-2836(89)90010-7
Dock-Bregeon, A. C., Chevrier, B., Podjarny, A., Moras, D., Debear, J. S., Gough, G. R., et al. (1988). High Resolution Structure of the RNA Duplex [U(U-A)6A]2. Nature 335, 375–378. doi:10.1038/335375a0
Dolinsky, T. J., Nielsen, J. E., Mccammon, J. A., and Baker, N. A. (2004). PDB2PQR: an Automated Pipeline for the Setup of Poisson-Boltzmann Electrostatics Calculations. Nucleic Acids Res. 32, W665–W667. doi:10.1093/nar/gkh381
Emsley, P., Lohkamp, B., Scott, W. G., and Cowtan, K. (2010). Features and Development ofCoot. Acta Crystallogr. D Biol. Cryst. 66, 486–501. doi:10.1107/S0907444910007493
Gagnon, M. G., and Steinberg, S. V. (2002). GU Receptors of Double Helices Mediate tRNA Movement in the Ribosome. RNA 8, 873–877. doi:10.1017/S135583820202602x
Gautheret, D., Konings, D., and Gutell, R. R. (1995). G.U Base Pairing Motifs in Ribosomal RNA. RNA 1, 807–814.
Gelbin, A., Schneider, B., Clowney, L., Hsieh, S.-H., Olson, W. K., and Berman, H. M. (1996). Geometric Parameters in Nucleic Acids: Sugar and Phosphate Constituents. J. Am. Chem. Soc. 118, 519–529. doi: DOI doi:10.1021/ja9528846
Gutell, R. R., Larsen, N., and Woese, C. R. (1994). Lessons from an Evolving rRNA: 16S and 23S rRNA Structures from a Comparative Perspective. Microbiol. Rev. 58, 10–26. doi:10.1128/mr.58.1.10-26.1994
He, L., Kierzek, R., Santalucia, J., Walter, A. E., and Turner, D. H. (1991). Nearest-neighbor Parameters for G.cntdot.U Mismatches: 5'GU3'/3'UG5' Is Destabilizing in the Contexts CGUG/GUGC, UGUA/AUGU, and AGUU/UUGA but Stabilizing in GGUC/CUGG. Biochemistry 30, 11124–11132. doi:10.1021/bi00110a015
Honig, B., Pyle, A. M., Chin, K., and Sharp, K. A. (1999). Calculating the Electrostatic Properties of RNA Provides New Insights into Molecular Interactions and Function. Nat. Struct. Biol. 6, 1055–1061. doi:10.1038/14940
Hou, Y.-M., and Schimmel, P. (1988). A Simple Structural Feature Is a Major Determinant of the Identity of a Transfer RNA. Nature 333, 140–145. doi:10.1038/333140a0
Jurrus, E., Engel, D., Star, K., Monson, K., Brandi, J., Felberg, L. E., et al. (2018). Improvements to the APBS Biomolecular Solvation Software Suite. Protein Sci. 27, 112–128. doi:10.1002/pro.3280
Keating, K. S., Toor, N., Perlman, P. S., and Pyle, A. M. (2010). A Structural Analysis of the Group II Intron Active Site and Implications for the Spliceosome. RNA 16, 1–9. doi:10.1261/rna.1791310
Keel, A. Y., Rambo, R. P., Batey, R. T., and Kieft, J. S. (2007). A General Strategy to Solve the Phase Problem in RNA Crystallography. Structure 15, 761–772. doi:10.1016/j.str.2007.06.003
Kieft, J. S., and Tinoco, I. (1997). Solution Structure of a Metal-Binding Site in the Major Groove of RNA Complexed with Cobalt (III) Hexammine. Structure 5, 713–721. doi:10.1016/s0969-2126(97)00225-6
Konforti, B. B., Abramovitz, D. L., Duarte, C. M., Karpeisky, A., Beigelman, L., and Pyle, A. M. (1998). Ribozyme Catalysis from the Major Groove of Group II Intron Domain 5. Mol. Cell 1, 433–441. doi:10.1016/S1097-2765(00)80043-X
Kowiel, M., Brzezinski, D., Gilski, M., and Jaskolski, M. (2020). Conformation-dependent Restraints for Polynucleotides: the Sugar Moiety. Nucleic Acids Res. 48, 962–973. doi:10.1093/nar/gkz1122
Ladner, J. E., Jack, A., Robertus, J. D., Brown, R. S., Rhodes, D., Clark, B. F., et al. (1975). Structure of Yeast Phenylalanine Transfer RNA at 2.5 A Resolution. Proc. Natl. Acad. Sci. 72, 4414–4418. doi:10.1073/pnas.72.11.4414
Leontis, N. B., Stombaugh, J., and Westhof, E. (2002). The Non-watson-crick Base Pairs and Their Associated Isostericity Matrices. Nucleic Acids Res. 30, 3497–3531. doi:10.1093/nar/gkf481
Li, S., Olson, W. K., and Lu, X.-J. (2019). Web 3DNA 2.0 for the Analysis, Visualization, and Modeling of 3D Nucleic Acid Structures. Nucleic Acids Res. 47, W26–W34. doi:10.1093/nar/gkz394
Liebschner, D., Afonine, P. V., Baker, M. L., Bunkóczi, G., Chen, V. B., Croll, T. I., et al. (2019). Macromolecular Structure Determination Using X-Rays, Neutrons and Electrons: Recent Developments in Phenix. Acta Cryst. Sect. D Struct. Biol. 75, 861–877. doi:10.1107/S2059798319011471
Lu, M., and Steitz, T. A. (2000). Structure of Escherichia coli Ribosomal Protein L25 Complexed with a 5S rRNA Fragment at 1.8-A Resolution. Proc. Natl. Acad. Sci. 97, 2023–2028. doi:10.1073/pnas.97.5.2023
Lu, X.-J., Bussemaker, H. J., and Olson, W. K. (2015). DSSR: an Integrated Software Tool for Dissecting the Spatial Structure of RNA. Nucleic Acids Res. 43, gkv716. doi:10.1093/nar/gkv716
Lu, X.-J., and Olson, W. K. (2003). 3DNA: a Software Package for the Analysis, Rebuilding and Visualization of Three-Dimensional Nucleic Acid Structures. Nucleic Acids Res. 31, 5108–5121. doi:10.1093/nar/gkg680
Manigrasso, J., Chillón, I., Genna, V., Vidossich, P., Somarowthu, S., Pyle, A. M., et al. (2020). Visualizing Group II Intron Dynamics between the First and Second Steps of Splicing. Nat. Commun. 11, 2837. doi:10.1038/s41467-020-16741-4
Marcia, M., and Pyle, A. M. (2014). Principles of Ion Recognition in RNA: Insights from the Group II Intron Structures. Rna 20, 516–527. doi:10.1261/rna.043414.113
Masquida, B., and Westhof, E. (2000). On the Wobble GoU and Related Pairs. RNA 6, 9–15. doi:10.1017/s1355838200992082
McClain, W. H., and Foss, K. (1988). Changing the Identity of a tRNA by Introducing a G-U Wobble Pair Near the 3′ Acceptor End. Science 240, 793–796. doi:10.1126/science.2452483
McCoy, A. J., Grosse-Kunstleve, R. W., Adams, P. D., Winn, M. D., Storoni, L. C., and Read, R. J. (2007). Phasercrystallographic Software. J. Appl. Cryst. 40, 658–674. doi:10.1107/S0021889807021206
McDowell, J. A., He, L., Chen, X., and Turner, D. H. (1997). Investigation of the Structural Basis for Thermodynamic Stabilities of Tandem GU Wobble Pairs: NMR Structures of (rGGAGUUCC)2 and (rGGAUGUCC)2,. Biochemistry 36, 8030–8038. doi:10.1021/bi970122c
McDowell, J. A., and Turner, D. H. (1996). Investigation of the Structural Basis for Thermodynamic Stabilities of Tandem GU Mismatches: Solution Structure of (rGAGGUCUC)2 by Two-Dimensional NMR and Simulated Annealing,. Biochemistry 35, 14077–14089. doi:10.1021/bi9615710
Murshudov, G. N., Skubák, P., Lebedev, A. A., Pannu, N. S., Steiner, R. A., Nicholls, R. A., et al. (2011). REFMAC5 for the Refinement of Macromolecular crystal Structures. Acta Crystallogr. D Biol. Cryst. 67, 355–367. doi:10.1107/S0907444911001314
Musier-Forsyth, K., Usman, N., Scaringe, S., Doudna, J., Green, R., and Schimmel, P. (1991). Specificity for Aminoacylation of an RNA helix: an Unpaired, Exocyclic Amino Group in the Minor Groove. Science 253, 784–786. doi:10.1126/science.1876835
Naganuma, M., Sekine, S.-i., Chong, Y. E., Guo, M., Yang, X.-L., Gamper, H., et al. (2014). The Selective tRNA Aminoacylation Mechanism Based on a Single GU Pair. Nature 510, 507–511. doi:10.1038/nature13440
Nishikawa, F., Fauzi, H., and Nishikawa, S. (1997). Detailed Analysis of Base Preferences at the Cleavage Site of a Trans-acting HDV Ribozyme: a Mutation that Changes Cleavage Site Specificity. Nucleic Acids Res. 25, 1605–1610. doi:10.1093/nar/25.8.1605
Ott, G. n., Arnold, L., and Limmer, S. (1993). Proton NMR Studies of Manganese Ion Binding to tRNAderived Acceptor Arm Duplexes. Nucl. Acids Res. 21, 5859–5864. doi:10.1093/nar/21.25.5859
Park, S. J., Hou, Y. M., and Schimmel, P. (1989). A Single Base Pair Affects Binding and Catalytic Parameters in the Molecular Recognition of a Transfer RNA. Biochemistry 28, 2740–2746. doi:10.1021/bi00432a056
Parkinson, G., Vojtechovsky, J., Clowney, L., Brünger, A. T., and Berman, H. M. (1996). New Parameters for the Refinement of Nucleic Acid-Containing Structures. Acta Cryst. D 52, 57–64. doi:10.1107/S0907444995011115
Pettersen, E. F., Goddard, T. D., Huang, C. C., Couch, G. S., Greenblatt, D. M., Meng, E. C., et al. (2004). UCSF Chimera? A Visualization System for Exploratory Research and Analysis. J. Comput. Chem. 25, 1605–1612. doi:10.1002/jcc.20084
Sashital, D. G., Cornilescu, G., Butcher, S. E., Brow, D. A., and Butcher, S. E. (2004). U2-U6 RNA Folding Reveals a Group II Intron-like Domain and a Four-helix Junction. Nat. Struct. Mol. Biol. 11, 1237–1242. doi:10.1038/nsmb863
Sashital, D. G., Venditti, V., Angers, C. G., Cornilescu, G., and Butcher, S. E. (2007). Structure and Thermodynamics of a Conserved U2 snRNA Domain from Yeast and Human. RNA 13, 328–338. doi:10.1261/rna.418407
Schneider, B., Neidle, S., and Berman, H. M. (1997). Conformations of the Sugar-Phosphate Backbone in Helical DNA crystal Structures. Biopolymers 42, 113–124. doi:10.1002/(sici)1097-0282(199707)42:1<113:aid-bip10>3.0.co;2-o
Sprinzl, M., Steegborn, C., Hubel, F., and Steinberg, S. (1996). Compilation of tRNA Sequences and Sequences of tRNA Genes. Nucleic Acids Res. 24, 68–72. doi:10.1093/nar/24.1.68
Stefan, L. R., Zhang, R., Levitan, A. G., Hendrix, D. K., Brenner, S. E., and Holbrook, S. R. (2006). MeRNA: a Database of Metal Ion Binding Sites in RNA Structures. Nucleic Acids Res. 34, D131–D134. doi:10.1093/nar/gkj058
Strobel, S. A., and Cech, T. R. (1996). Exocyclic Amine of the Conserved G·U Pair at the Cleavage Site of the Tetrahymena Ribozyme Contributes to 5'-Splice Site Selection and Transition State Stabilization. Biochemistry 35, 1201–1211. doi:10.1021/bi952244f
Strobel, S. A., and Cech, T. R. (1995). Minor Groove Recognition of the Conserved G⋅U Pair at the Tetrahymena Ribozyme Reaction Site. Science 267, 675–679. doi:10.1126/science.7839142
Trikha, J., Filman, D. J., and Hogle, J. M. (1999). Crystal Structure of a 14 Bp RNA Duplex with Non-symmetrical Tandem GxU Wobble Base Pairs. Nucleic Acids Res. 27, 1728–1739. doi:10.1093/nar/27.7.1728
Varani, G., and McClain, W. H. (2000). The G X U Wobble Base Pair. A Fundamental Building Block of RNA Structure Crucial to RNA Function in Diverse Biological Systems. EMBO Rep. 1, 18–23. doi:10.1093/embo-reports/kvd001
Westhof, E., and Fritsch, V. (2000). RNA Folding: beyond Watson-Crick Pairs. Structure 8, R55–R65. doi:10.1016/s0969-2126(00)00112-x
Westhof, E., Yusupov, M., and Yusupova, G. (2019). The Multiple Flavors of GoU Pairs in RNA. J. Mol. Recognit. 32, e2782. doi:10.1002/jmr.2782
Woese, C. R., Gutell, R., Gupta, R., and Noller, H. F. (1983). Detailed Analysis of the Higher-Order Structure of 16S-like Ribosomal Ribonucleic Acids. Microbiol. Rev. 47, 621–669. doi:10.1128/mr.47.4.621-669.1983
Woodson, S. A. (2005). Metal Ions and RNA Folding: a Highly Charged Topic with a Dynamic Future. Curr. Opin. Chem. Biol. 9, 104–109. doi:10.1016/j.cbpa.2005.02.004
Wu, J., and Manley, J. L. (1992). Multiple Functional Domains of Human U2 Small Nuclear RNA: Strengthening Conserved Stem I Can Block Splicing. Mol. Cell. Biol. 12, 5464–5473. doi:10.1128/mcb.12.12.5464
Wu, M., Mcdowell, J. A., and Turner, D. H. (1995). A Periodic Table of Tandem Mismatches in RNA. Biochemistry 34, 3204–3211. doi:10.1021/bi00010a009
Keywords: GU wobble pair, X-ray crystallography, RNA, structure, metal ion
Citation: Ruszkowska A, Zheng YY, Mao S, Ruszkowski M and Sheng J (2022) Structural Insights Into the 5′UG/3′GU Wobble Tandem in Complex With Ba2+ Cation. Front. Mol. Biosci. 8:762786. doi: 10.3389/fmolb.2021.762786
Received: 22 August 2021; Accepted: 24 November 2021;
Published: 13 January 2022.
Edited by:
Anna M. Kietrys, Carnegie Mellon University, United StatesReviewed by:
Blaine Mooers, University of Oklahoma Health Sciences Center, United StatesIsabel Chillón, European Molecular Biology Laboratory Grenoble, France
Copyright © 2022 Ruszkowska, Zheng, Mao, Ruszkowski and Sheng. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jia Sheng, SlNoZW5nQGFsYmFueS5lZHU=; Milosz Ruszkowski, bXJ1c3prb3dza2lAaWJjaC5wb3puYW4ucGw=
 Agnieszka Ruszkowska1
Agnieszka Ruszkowska1 Milosz Ruszkowski
Milosz Ruszkowski Jia Sheng
Jia Sheng