- 1Department of Genetic Engineering and Biotechnology, Shahjalal University of Science and Technology, Sylhet, Bangladesh
- 2Department of Biotechnology, Yeungnam University, Gyeongsan, South Korea
- 3Department of Genetic Engineering and Biotechnology, Jagannath University, Dhaka, Bangladesh
The present global COVID-19 pandemic caused by the noble pleomorphic severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has created a vulnerable situation in the global healthcare and economy. In this pandemic situation, researchers all around the world are trying their level best to find suitable therapeutics from various sources to combat against the SARS-CoV-2. To date, numerous bioactive compounds from different sources have been tested to control many viral diseases. However, microbial metabolites are advantageous for drug development over metabolites from other sources. We herein retrieved and reviewed literatures from PubMed, Scopus and Google relevant to antiviral microbial metabolites by searching with the keywords “antiviral microbial metabolites,” “microbial metabolite against virus,” “microorganism with antiviral activity,” “antiviral medicine from microbial metabolite,” “antiviral bacterial metabolites,” “antiviral fungal metabolites,” “antiviral metabolites from microscopic algae’ and so on. For the same purpose, the keywords “microbial metabolites against COVID-19 and SARS-CoV-2” and “plant metabolites against COVID-19 and SARS-CoV-2” were used. Only the full text literatures available in English and pertinent to the topic have been included and those which are not available as full text in English and pertinent to antiviral or anti-SARS-CoV-2 activity were excluded. In this review, we have accumulated microbial metabolites that can be used as antiviral agents against a broad range of viruses including SARS-CoV-2. Based on this concept, we have included 330 antiviral microbial metabolites so far available to date in the data bases and were previously isolated from fungi, bacteria and microalgae. The microbial source, chemical nature, targeted viruses, mechanism of actions and IC50/EC50 values of these metabolites are discussed although mechanisms of actions of many of them are not yet elucidated. Among these antiviral microbial metabolites, some compounds might be very potential against many other viruses including coronaviruses. However, these potential microbial metabolites need further research to be developed as effective antiviral drugs. This paper may provide the scientific community with the possible secret of microbial metabolites that could be an effective source of novel antiviral drugs to fight against many viruses including SARS-CoV-2 as well as the future viral pandemics.
Introduction
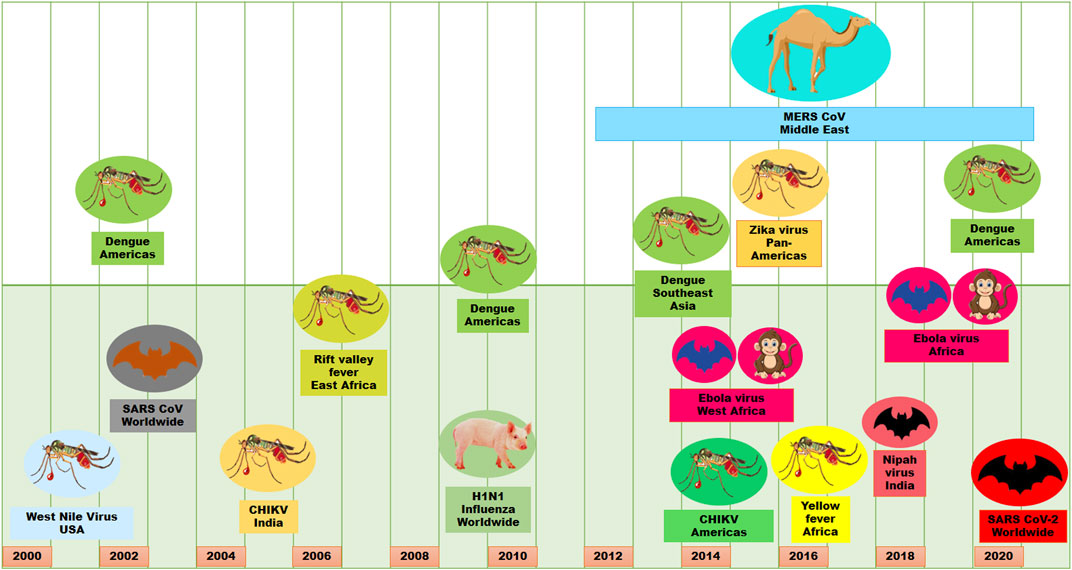
Viral infections are one of the major causes of morbidity and mortality in the world. It is very catastrophic due to the complexity, diversity, obligatory intracellular parasitic nature and pleomorphic character of viruses. These properties of viruses make it very difficult to counteract viral effects and transmission, which ultimately causes epidemics and/or pandemics (Graham et al., 2013; Meganck and Baric, 2021). Although the deadly influenza outbreak occurred in 1918, in the last 2 decades of the present century, there have been several viral epidemics or pandemics in humans (Figure 1). These viral epidemics or pandemics were caused with influenza A virus (H1N1), severe acute respiratory syndrome coronavirus (SARS-CoV), Middle East respiratory syndrome coronavirus (MERS-CoV), dengue virus (DENV), Zika virus (ZIKV), Ebola virus (EBOV), chikungunya virus (CHIKV), Henipavirus (HeV, NiV) and the recent severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) (Meganck and Baric, 2021). Moreover, human immunodeficiency virus (HIV) is life-threatening since its discovery in 1982. Some other viruses such as Crimean–Congo hemorrhagic fever virus, Herpes simplex virus, Hepatitis viruses, Rabies virus, Hantaviruses have caused outbreaks or have outbreak potential. Therefore, the increase of migration, global travel, and urbanization have made viruses outbreaks a crucial challenge for public health, especially when vaccines and antiviral therapies are still not available (Neiderud, 2015).
Viruses having a genome either RNA or DNA utilize the molecular apparatus of the host cells for their replication and cause several ailments (Tapparel et al., 2013; Cohen, 2016). Viral infections can be controlled by prophylactic strategy and/or drug therapy. However, for being obligatory intracellular parasite, most of the metabolic pathways involved in the viral replication are the same as in the host cells. From this point of view, it is difficult to design an appropriate treatment to attack the virus without triggering adverse events on the host. These aspects further highlight the main peculiarity of viruses (specificity, affinity, and self-defense mechanisms) and the difficulties of antiviral chemotherapy. Therefore, it is necessary to discover and identify new antiviral agents, which should possess primarily an adequate selectivity, power, in vivo stability profile and low toxicity (Akram et al., 2018).
Many natural and synthetic drugs having antiviral activity were considerably less effective when tested in virus-infected animal models (Martinez et al., 2015; Takizawa and Yamasaki, 2018; Mukherjee, 2019). Moreover, extraction of the natural products from the plants and the chemical synthesis of synthetic drugs have safety and economic concerns. Furthermore, conventional drugs become failed against viral infections and the onset of specific viral resistances against these drugs is a common phenomenon (Linnakoski et al., 2018; Mulwa and Stadler, 2018; Ma et al., 2020). Therefore, researchers need to search for alternative source of safe and economically cost-effective antiviral natural products. In this context, microbial metabolites might be a promising source of antiviral agents. Microorganisms are natural flora of the environment that play significant role in plenty of processes, and therefore, their metabolites have great potential to be used for antiviral treatment without severe side-effects (Cheung et al., 2014). In fact, microbial metabolites have already been a subject of intense research for the treatment of certain virus-mediated diseases (Berdy, 2005), and currently, there is an emerging trend in biotechnology for therapeutic applications of microbial metabolites as antiviral agents (Yasuhara-Bell et al., 2010a; Pham et al., 2019; Goris et al., 2021; Lobo-Galo et al., 2021). Several microbial metabolites have been demonstrated to offer promising antiviral activity against numerous DNA and RNA viruses (Tong et al., 2012; Linnakoski et al., 2018; Mulwa and Stadler, 2018). The whole world has been fighting against the current COVID-19 pandemic for more than one and a half years. As there is no newly developed specific approved drug, only repurposed drugs are used as the supportive treatment of the stormy COVID-19 caused by SARS-CoV-2 (Hakim et al., 2021), which has caused total death of 4,374,234 in the world as on August 15, 2021. Cases and death of COVID-19 is going on ceaselessly globally. As the trend of the history, more viral epidemics and/or pandemics may outbreak in the future. Therefore, it is essential to discover drugs with broad spectrum activity against SARS-CoV-2 including other catastrophic viruses. Screening and identification of natural compounds from microbial metabolites may be particularly important for drug discovery against the coronavirus alike SARS-CoV-2 as well as other viruses having potential outbreaks in the future.
This review focuses on microbial metabolites, which have shown activity against various viral pathogens. In addition, the current state of this research topic is briefly discussed, and gaps in the research are identified. Furthermore, the targets for antiviral therapeutic development and the advantages of microbial metabolites are briefly discussed. Finally, this review attempts to offer alternative conceptual framework for drug discovery for treatment of COVID-19 and alike future viral pandemics and/or epidemics.
Targets of Microbial Metabolites for Therapeutic Development
Despite of having different biology for infection, viruses share some basic steps for their replication (Figure 2A). The basic steps for viral replication include 1) viral attachment to host cells (host-viral interaction), 2) viral penetration into host cells, 3) viral uncoating into the cytoplasm, 4) viral genome replication and transcription, 5) viral protein translation and assembly, and 6) viral progeny release (Meganck and Baric, 2021). Due to having limited numbers of own coding genes, viruses must depend on the host machinery for accomplishment of viral lifecycle. The fundamental steps involved in viral lifecycle are associated with viral infection as well as pathogenesis and represent important targets for therapeutic development. The infection or the pathogenesis starts with the viral entry into the host cells (Ryu, 2017; Thaker et al., 2019). The prerequisite for viral entry is its binding on the cell surface. Viral proteins on the capsid or envelope interact with the specific receptor, which can be proteins, glycans and/or lipids in the host cell. For instance, the spike protein S of SARS-CoV-1 and SARS-CoV-2 interact with the angiotensin-converting enzyme 2 (ACE2) as the receptor expressed on the surface of the target cells (Lim et al., 2016; Fung and Liu, 2019; Chen et al., 2020; Hoffmann et al., 2020; Ou et al., 2020; Rahman et al., 2020; Walls et al., 2020; Yan et al., 2020). The interaction between the viral protein and host receptor facilitate the viral uptake often through endocytic pathways or through fusion at the plasma membrane (Millet and Whittaker, 2018; Milewska et al., 2020). Viruses escape the endosome by uncoating and the genomic material is released into the cytoplasm.
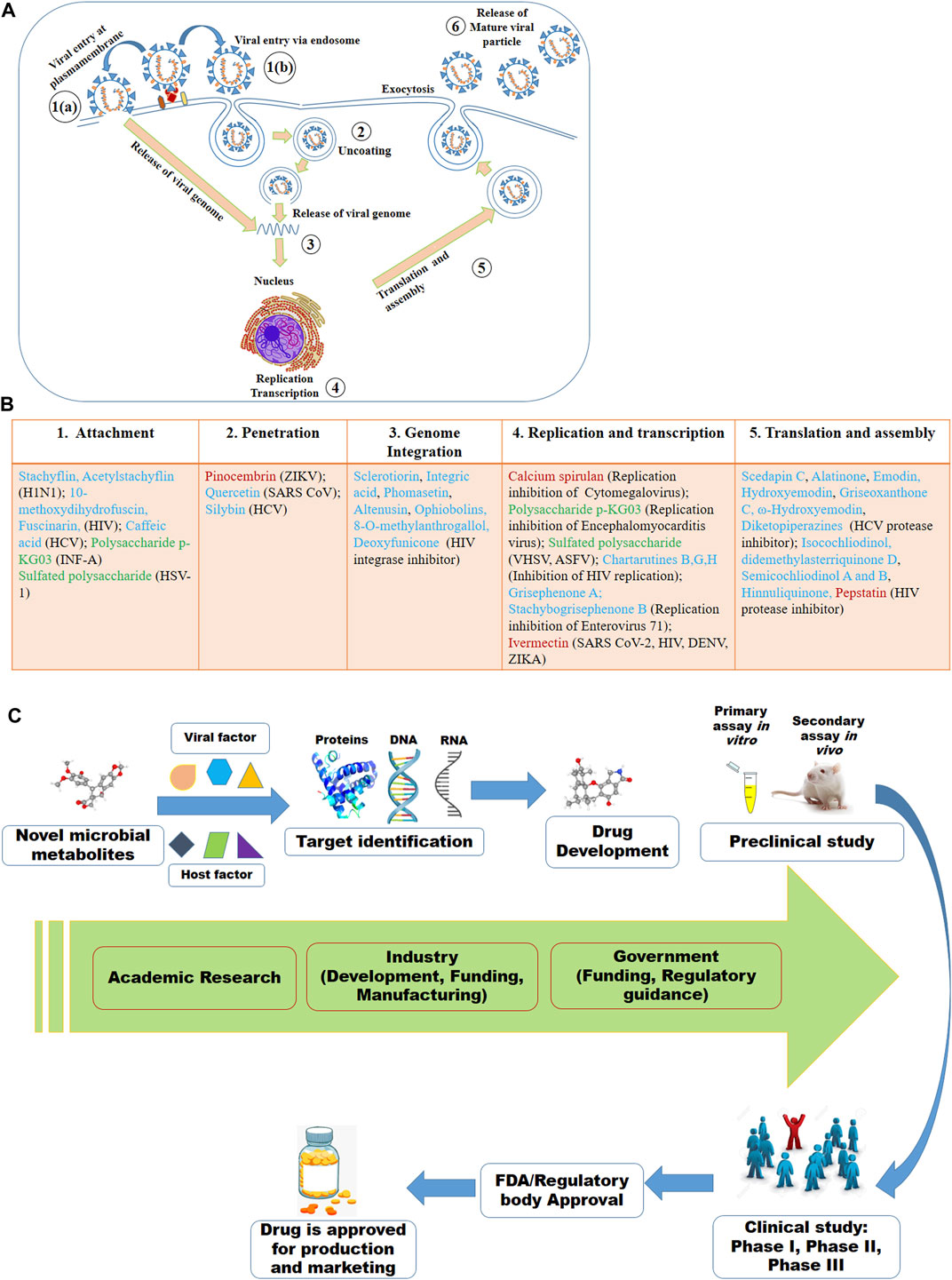
FIGURE 2. Viral lifecycle (A) and the proposed mode of actions of some of the antiviral microbial secondary metabolites (MSM) (B) listed in this review. The numbers in (A) denote the steps usually targeted by MSM. In (B), some of the antiviral MSM inhibiting targeted stages of viral lifecycle are listed. Cyan, red and green colors indicate the metabolites isolated from fungi, bacteria and microalgae, respectively. (C) Antiviral drug development strategy based on the viral and host factors.
Replication of DNA viruses is performed by using DNA dependent DNA polymerase. DNA viruses can integrate their genomes into the host genome and cause recurrent problem. RNA viruses replicate their genomes either by RNA-dependent RNA synthesis, or by RNA-dependent DNA synthesis (reverse transcription) which is followed by DNA replication and transcription. The genetic material of single-stranded positive sense RNA (ssRNA+) viruses is like mRNA which is directly translated by the host cell. The negative sense RNA (ssRNA−) viruses carry RNA that is complementary to mRNA and must be turned into ssRNA+ using RNA polymerase before translation. All positive sense RNA viruses like poliovirus, hepatitis C virus, dengue virus, ZIKV, SARS-coronavirus can arrange specialized membranous structures by remodeling host membranes where the viral genome is replicated (Cameron et al., 2009; Paul and Bartenschlager, 2013). Due to lack of RNA polymerase proofreading ability, RNA viruses have very high rate of mutation compared to DNA viruses, which eventually renders enhanced virulence and evolvability (Duffy, 2018).
Although all viruses utilize the host apparatus system for translation, viral translation is regulated differently from the host cell (Jan et al., 2016). Viral proteins and genomic materials are assembled to form the virion. The final stage of viral replication is the release of the new virions produced in the host organism. The new virions are then able to infect nearby cells and repeat the replication cycle. Some viruses are released when the host cell dies, while other viruses without directly killing the cell can leave infected cells by budding through the membrane (Lodish et al., 2000; Risco et al., 2014). The essential molecular elements involved in each of these steps in the viral lifecycle can be targeted by microbial metabolites as therapeutics.
The microbial metabolites may target either the viral or the host factors that are associated with viral pathogenesis or the completion of the viral lifecycle or viral replication (Figure 2B). The viral factors might be viral proteins associated with the binding of viruses to cells, viral protease, viral translation or others (Anderson et al., 1996; Klemm et al., 2020; Chen C. C. et al., 2021). Host-factors might be receptor on the cell surface, endocytosis, host proteases and kinases, and others (Inoue et al., 2007; Ivanov, 2008; Raj et al., 2013; Zhou et al., 2015; Kalil et al., 2021). However, the viral and the host factors associated with the viral pathogenesis and its lifecycle or replication may vary based on the viruses even of the same family. For instance, while the spike protein S of SARS-CoV-1 and SARS-CoV-2 bind with the ACE2 receptor, the S protein of MERS-CoV binds to dipeptidyl peptidase 4 (DPP4) receptor (Raj et al., 2013; Lim et al., 2016; Hoffmann et al., 2020; Rahman et al., 2020; Walls et al., 2020; Yan et al., 2020). Here, viral S protein may serve as the drug target for all these three SARS viruses, however, ACE2 might be the target for the earlier two SARS viruses and the DPP4 might be for the MERS-CoV. Similarly, a serine protease named TMPRSS2 found to be essential for the activation of hemagglutinin (HA), the key step for initiating the viral infection by the H7N9 variant of H1N1, may be an important therapeutic target. The HA activation was failed in H7N9 virus when the TMRSS2 was knocked out in the mice (Tarnow et al., 2014).
Despite the viral life cycle, a number of factors regulate the host response towards certain viral infections (Fung and Liu, 2019; Azad et al., 2021; Hakim et al., 2021). The inaugural stages of diseases include the viral phase with the appearance of symptoms. However, with the progresses of the disease, the viral phase is replaced by the host inflammatory phase, which controls viral replication usually by damaging the host cells (Peiris et al., 2003). Antiviral therapeutics are active during the viral phase or viral life cycle after which these drugs become ineffective (Widagdo et al., 2017). Treatment options for controlling inflammatory damage during inflammatory phase usually include steroids as immunomodulatory and anti-inflammatory drugs (Yang J.-W. et al., 2020). In the ongoing pandemic, the hospitalized patients with COVID-19 are being treated with the corticosteroid dexamethasone (Group, 2021). Again baricitinib, a kinase inhibitor in the JAK/STAT signaling pathway, has been approved for COVID-19 treatment, which lowers cytokine release that is a hallmark in SARS-CoV-2 infection (Stebbing et al., 2020; Hakim et al., 2021; Kalil et al., 2021). Nevertheless, the interferon (IFN) alpha and beta activates the JAK/STAT signaling pathway that in turn triggers the synthesis of a number of antiviral gene products (Chiang and Liu, 2019). Therefore, any essential event involved in the viral phase and/or the host inflammatory phase might be an important target for treatment of the respective viral disease with microbial metabolites.
Microbial Metabolites as Potential Antiviral Candidates
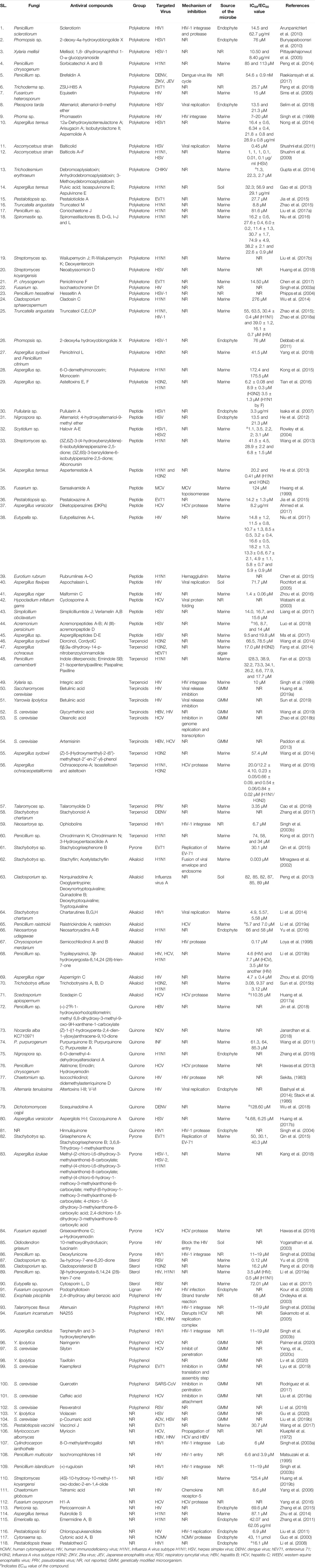
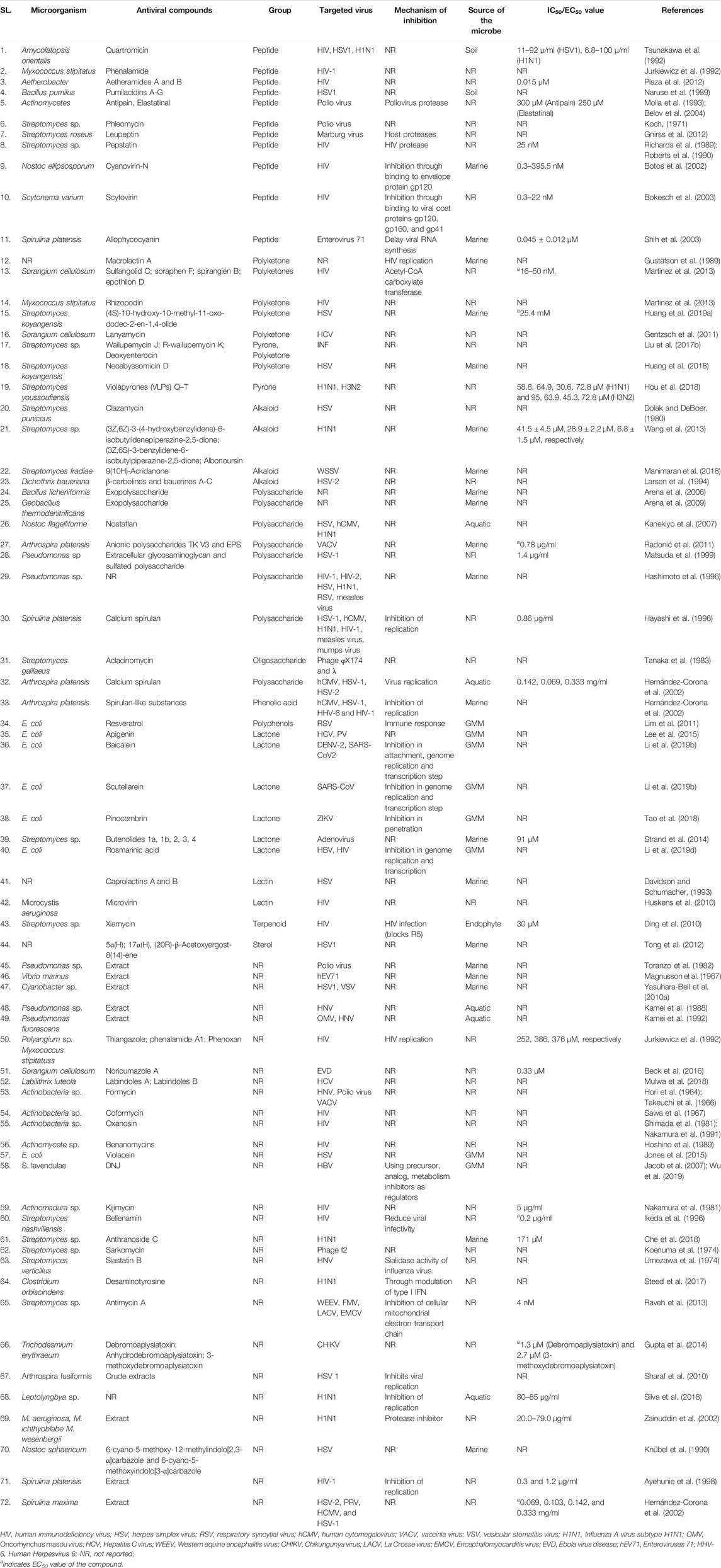
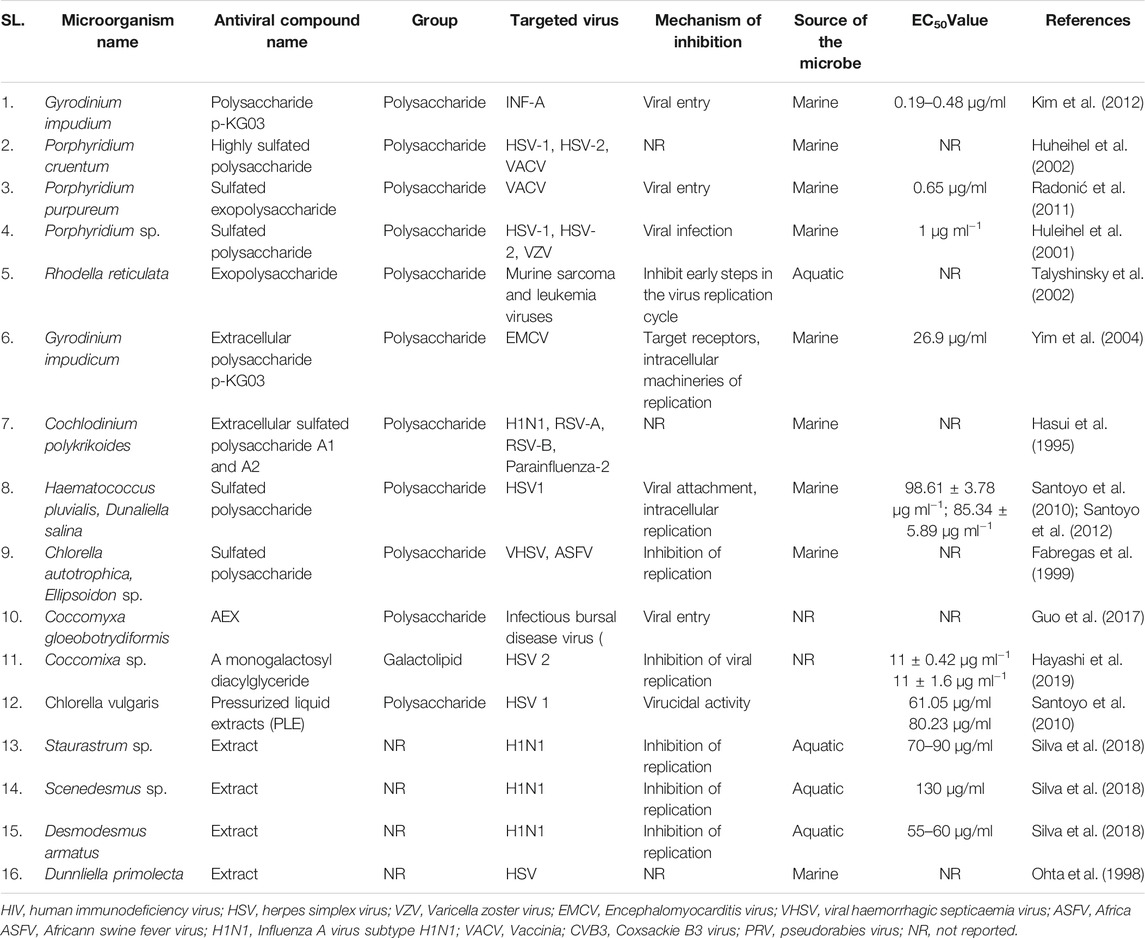
Microbial metabolites are being used as important therapeutics for treatment of infections in health and agriculture arena (Demain, 2007; Raihan et al., 2021). For being advantageous over chemically synthesized and non-microbial natural products, research and development programs are continuously adopting approaches based on microbial products for the development of novel drugs. Microbial secondary metabolites (MSMs) have been being used as easy and reliable sources for the synthesis of new pharmaceuticals and therapeutics against different types of pathogens including viruses, bacteria, fungi and parasites (Demain, 2007; Selim et al., 2018). Many microorganisms such as bacteria, fungi, actinomycetes and microalgae from numerous sources have a variety of secondary metabolites like quinones, terpenoids, lignans, alkaloids, peptides, polysaccharides, lactones, polyketide, xanthone, ester, and so on having diverse antiviral activities (Selim et al., 2018; Pan et al., 2019). Several classes of such MSMs have been used as antiviral agents. From the literatures reported previously, only the antiviral metabolites from fungi, bacteria and microalgae have been listed in the present review (Tables 1–3). Fungi from different sources are the major reservoir of antiviral metabolites followed by bacteria and microalgae. Most of the MSMs were isolated from microorganisms of the marine source (Figure 3). The MSMs clusters to different groups (Figure 4) having different mechanism of actions against viruses. Although the mechanism of actions of most of the antiviral microbial metabolites are not yet elucidated, that of a few microbial metabolites has been reported (Tables 1–3). Elucidation of mode of actions and pharmacological properties of novel antiviral microbial bioactive metabolites may lead to the development of drugs for treating human diseases developed by catastrophic viral agents.
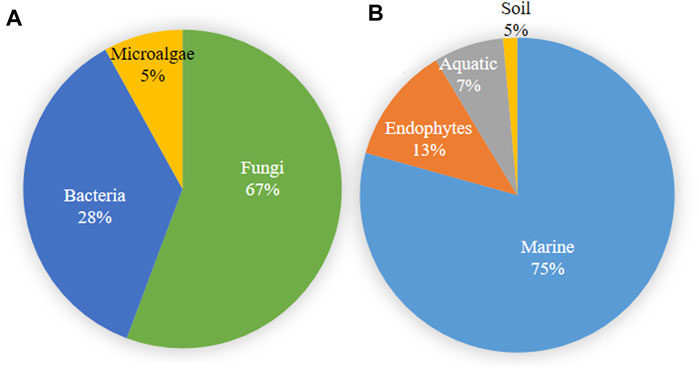
FIGURE 3. Microbial source of antiviral compounds (A) and the source of microorganisms producing antiviral compounds (B).
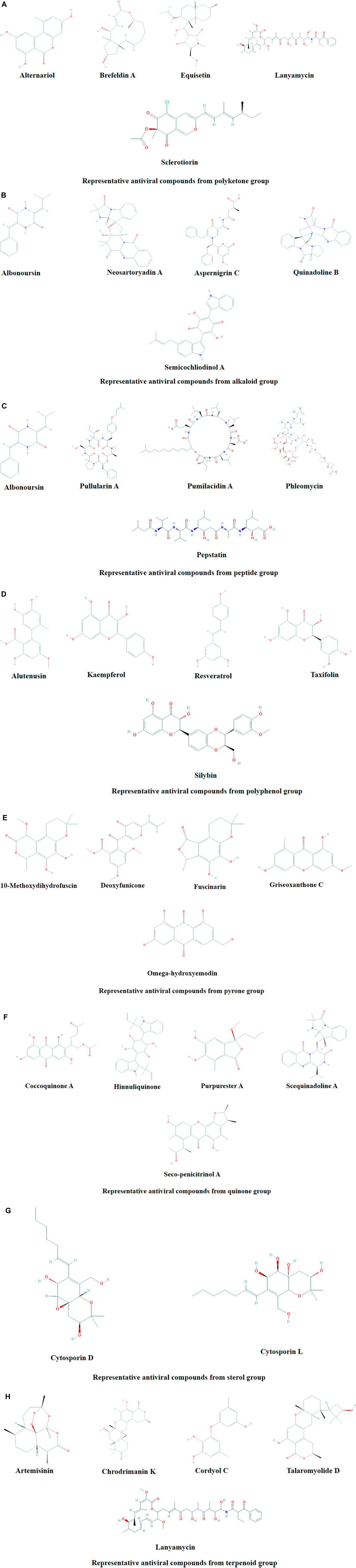
FIGURE 4. Representative antiviral compounds from polyketone (A), alkaloid (B), peptide (C), polyphenol (D), pyrone (E), quinone (F), sterol (G), and terpenoid (H) groups.
Polysaccharides
Microbial polysaccharides (MPS), the biopolymers produced through microbial metabolic process, are widely found in bacteria, fungi and algae (Tables 1–3). The antiviral metabolites so far reported from algae are MPS (Table 3). However, bacteria and fungi produced a variety of MSM including MPS (Tables 1, 2). The advantages of MPS over plant polysaccharides include lack of seasonal, geographical, pest and diseases restriction; wide variety of sources as well as short production time (Chen and Huang, 2018). Some MPS are linear (cellulose, chitin, chitosan, pullulan, alginate, curdlan) and some are branched (dextran, levan, xanthan, scleroglucan, and in lesser degree gellan). Neutral (dextran, levan, pullulan, cellulose, scleroglucan and curdlan), anionic (alginate, xanthan, gellan), and cationic (chitin and chitosan) properties of these linear and branched MPS may make them suitable against a variety of viruses (Steed et al., 2017). Due to having diversified structural properties, the antiviral mechanisms of MPS are complex and diverse, and thus suitable for a variety of applications (Steed et al., 2017; Liu et al., 2020). The antiviral mechanisms of MPS include the inhibition of events involved in viral life cycle (attachment of virus to the host cell, penetration, genetic material and protein synthesis) and the improvement of the host immunity (Liu et al., 2020). However, the antiviral mechanism of many MPS is not yet known.
Recently, studies on derivatives of MPS are given priorities because chemical modification generates enhanced or new activities to MPS (Chen and Huang, 2018). The most common derivatives are sulfonated, phosphorylated and selenizated. The derivatives of MPS having lower or no toxicity even at higher concentrations offer broad prospects for treatment of viral diseases (Saha et al., 2012; Chen and Huang, 2018; Liu et al., 2020). The bioactive sulfated polysaccharide, p-KG03, obtained from Gyrodinium impudicum showed antiviral activity (EC50 = 26.9 µg/ml) against encephalomyocarditis virus (Yim et al., 2004) and inhibited H1N1 with an EC50 value of 0.19–0.48 μg/ml through interfering the viral entry into the host cell (Kim et al., 2012). Another sulfated polysaccharide isolated from red microalgae Porphyridium sp. showed impressive antiviral activity against Herpes simplex viruses types 1 and 2 (HSV 1, 2) and Varicella zoster virus (VZV) with IC50 1 μg/ml (Huleihel et al., 2001). However, the same polysaccharide isolated from Haematococcus pluvialis showed similar inhibition rate against HSV-1 with IC50 75 µg/ml concentration (Santoyo et al., 2012). Furthermore, a number of MPS obtained from various microalgae and bacteria showed promising antiviral activity with unknown mechanism of action against numerous viruses such as HIV1, HSV-1, HSV-2, Vaccina virus, Murine sarcoma and leukemia viruses, Influenza A and B viruses, RSV-A, RSV-B, parainfluenza-2, VHSV, ASFV, hCMV, VACV mentioned in Tables 2, 3.
Peptides
Antiviral peptides (AVPs) obtained from natural sources are amphipathic and cationic nature. In addition, their hydrophobicity make them the promising drug candidate against enveloped viruses (Agarwal and Gabrani, 2020). The AVPs are reported from bacteria and fungi, however, not yet from algae (Tables 1, 2). Advantages of naturally produced microbial AVPs include high specificity and effectiveness, low toxicity and peptidase biodegradability, and low molecular weight (Boas et al., 2019). The AVPs can act at various stages of the viral life cycle through the suppression of viral gene expression. They can further prevent viral infection by many ways including inhibiting the viral particle or by competing for the receptor molecule in the host cell membrane and consequent adsorption, suppression of topoisomerase-mediated DNA-binding, DNA relaxation and formation of covalent complex (Galdiero et al., 2013; Heydari et al., 2021). Some of them can show activity by membrane destabilization of the virus (Rowley et al., 2004; Porotto et al., 2010). However, the mode of actions of most of the bacterial and fungal AVPs remains elusive (Tables 1, 2).
Sansalvamide A, a cyclic depsipeptide, isolated from marine Fusarium spp. showed antiviral activity against a poxvirus, molluscum contagiosum virus (MCV) by inhibiting the virus-encoded type-1 topoisomerase which is essential for MCV replication (Hwang et al., 1999). Simplicilliumtide J, a cyclic peptide, isolated from a deep sea derived fungal strain Simplicillium obclavatum EIODSF 020 and its analogues Verlamelin A and B showed very promising anti-HSV-1 activity with IC50 values of 15.6 μM (Liang et al., 2017). The cyclodipeptide diketopiperazines (DKPs) obtained from endophytic fungus Aspergillus versicolor exhibited anti-HSV activity through inhibition of NS3/4A protease with the IC50 value 8.2 μg/ml (Ahmed et al., 2017).
Alkaloids
Alkaloids are structurally diverse secondary metabolites which have many therapeutic applications including antiviral activity (Cushnie et al., 2014). Most of the alkaloids used as therapeutics to treat human diseases are natural products of plants although plants are unreliable, low-yielding, expensive and unstable source (Bradley et al., 2020). However, several recent studies showed that a number of fungi produce alkaloids as an MSM acting against pathogenic microbes including viruses (Table 1) (Peng et al., 2019; Sadahiro et al., 2020; Raihan et al., 2021). Nevertheless, despite the potentiality, bacterial and algal sources for alkaloids are not yet reported. Although the mechanisms of all microbial alkaloids are not yet known (Table 1), a number of studies report that alkaloids inhibit DNA polymerase, Topoisomerase, reverse transcriptase and protein synthesis (Thawabteh et al., 2019; Bleasel and Peterson, 2020; Wink, 2020), and deactivate the viral infection by acting as DNA intercalator (Croaker et al., 2016). Six indole alkaloids isolated from mangrove derived fungus Cladosporium sp. PJX-41 showed antiviral activity against H1N1 with IC50 values 82–89 μM (Peng et al., 2013). Stachyflin, a sesquiterpenoidal alkaloid, obtained from Stachybotrys sp. RF-7260 by solid state fermentation showed a promising antiviral activity in vitro against H1N1 with IC50 value 0.003 μM (Minagawa et al., 2002). Three new isoindolinone-type alkaloids named chartarutines B, G, and H isolated from sponge derived fungus Stachybotrys chartarum has been shown as antiviral agents to inhibit replication of HIV-1 with the IC50 value 4.9–5.6 mM (Li et al., 2014). Recently, it has been shown that two aminosulfonyl group containing alkaloids named Scedapin C and scequinadoline A extracted from marine-derived fungus Scedosporium apiospermum, displayed significant anti-HCV activity by inhibiting HCV protease with the EC50 values 110.35 and 128.60 μM, respectively (Huang L.-H. et al., 2017). Huang Z. et al. (2017) further showed that a deep-sea-derived fungus Aspergillus versicolor SCSIO 41502 produced Aspergilols H and I which displayed anti-HCV activity with EC50 values 4.68 and 6.25 μM, respectively (Huang Z. et al., 2017).
Polyketones
Many polyketides (derived from polyketones) isolated from microorganisms such as fungi and bacteria have been shown to inhibit the viral infection in a various way (Tables 1, 2). However, mechanisms of actions of most of the polyketones mentioned in this paper have to be elucidated. A group of polyketides are capable to inhibit viral replication. Two of such polyketides named as Alternariol and Balticolid isolated from Pleospora tarda and Ascomycetous strain exhibited potent antiviral activity with IC50 value 13.5 μM and 0.01 mg/ml, respectively (Shushni et al., 2011; Selim et al., 2018). While these polyketones inhibit viral replication, Sclerotiorin, another polyketone isolated from an endophyte Penicillium sclerotiorum essentially interferes with HIV-1 integrase and protease—two essential enzymes for maintaining the life cycle of the virus inside the host cell (Arunpanichlert et al., 2010). Furthermore, a group of polyketides namely sulfangolid C, soraphen F, spirangien B and epothilon D isolated from Sorangium cellulosum protects against HIV by interacting with the Acetyl-CoA carboxylate transferase enzyme (Martinez et al., 2013). Martinez et al. (2013) further found that Rhizopodin, derived from M. stipitatus is a potential antiviral agent although the mechanism of inhibition of the compound has not been elucidated. Another study found that marine microbe Phoma sp. produced Phomasetin which inhibited the HIV integrase, rendering it a potential drug compound against HIV (Singh et al., 1999). In fact, most of the microbial polyketides have been isolated till date is from the marine microorganisms. However, several fungi obtained from other sources are also reported to produce antiviral compounds having promising activity against DENV, ZIKV, Influenza virus, HCV and others (Table 1).
Terpenoids
Terpenoids are one of the most abundant natural aromatic compounds mostly found in plants. However, some microorganisms can synthesize terpenoids (Yamada et al., 2015). Furthermore, microbial strains can be engineered to produce such terpenoids that have antiviral activities (Ma et al., 2020). The properties and medicinal uses of terpenoids are being continuously investigated by researchers for anticancer, antioxidant, antiviral, and anti-atherosclerotic activities (Nazaruk and Borzym-Kluczyk, 2015). Based on the number of carbon atoms, terpenoids are of different types (Wang et al., 2018). Different modes of actions of different terpinoids make them important against viral infection. For instance, ochraceopone A, isoasteltoxin, and asteltoxin obtained from antarctic fungus Aspergillus ochraceopetaliformis exhibited antiviral activities against the H1N1 and H3N2 influenza viruses by inhibiting viral growth through their protease suppression with IC50 values of >20.0/12.2 ± 4.10, 0.23 ± 0.05/0.66 ± 0.09, and 0.54 ± 0.06/0.84 ± 0.02 μM, respectively (Wang et al., 2016). Three sesquiterpenes named as (Z)-5-(Hydroxymethyl)-2-(6′)-methylhept-2′-en-2′-yl)-phenol, diorcinol, cordyol C were extracted from sponge-associated fungus Aspergillus sydowii which showed anti H3N2 activity with IC50 values of 57.4, 66.5 and 78.5 μM, respectively (Wang et al., 2014). In addition, a terpenoid compound called xiamycin derived from a bacterial endophyte (Streptomyces sp.) acts as anti-HIV agent through prohibition of beta-chemokine receptor CCR5 with IC50 value of > 30 μM (Ding et al., 2010). This class of metabolites can be produced in engineered fungi such Saccharomyces cerevisiae and Yarrowia. Lipolytica (Ma et al., 2020). Oleanolic acid is such a terpenoid produced from genetically modified S. cerevisiae, which inhibited genome replication and transcription of HCV (Zhao et al., 2018a). Another metabolite named betulinic acid produced from both S. cerevisiae and Y. lipolytica showed promising anti-HIV activity by inhibiting viral release from the host cell (Huang H. et al., 2019; Sun et al., 2019). Furthermore, a lot of terpinoids derived from fungi exhibited antiviral activity against numerous viruses such as H3N2, hEV71, H1N1, HBV, HIV, PRV, and DENV (Table 1).
Quinone
Quinones are aromatic organic compounds and found ubiquitously in prokaryotes and eukaryotes. Quinones act through inhibition of electron transport as well as uncoupling of oxidative phosphorylation (Obach and Kalgutkar, 2018). Furthermore, they can act as inducers of reactive oxygen species and bioreductive alkylators of biomolecules, and suppress DNA function by interpolation into DNA (Roa-Linares et al., 2019). Quinones are used as antioxidant, antimicrobial, anticancer, anti-inflammatory, antitumor agents (El-Najjar et al., 2011; Teng et al., 2020). The coccoquinone A, an anthraquinone derivative, obtained from Aspergillus versicolor function as an anti-HSV agent with the EC50 value 6.25 µM (Huang Z. et al., 2017). Furthermore, 4-hydroxymethyl-quinoline isolated from myxobacteria Labilithrix luteola exhibited antiviral activity against HCV (Mulwa et al., 2018). Moreover, Alatinone, Emodin, and Hydroxyemodin, isolated from red alga Liagora viscida derived endophytic fungi Penicillium chrysogenum showed antiviral activity against HCV through inhibition HCV protease (Hawas et al., 2013). A citrinin dimer, seco-penicitrinol A obtained by coculturing of two marine algal-derived endophytic fungal strains Aspergillus sydowii and Penicillium citrinum showed inhibitory activity towards influenza neuraminidase in vitro with an IC50 value 24.7 µM (Yang et al., 2018). An anthraquinone derivatives called (‒)-2′R-1-hydroxyisorhodoptilometrin obtained from marine fungi Penicillium sp. OUCMDZ acted as an antiviral agent against HBV (Jin et al., 2018). Furthermore, some other promising antiviral quinone type compounds have been listed in Table 1.
Sterols
Sterols, also known as steroid alcohols, found ubiquitously in numerous plant, animals as well as microorganisms are considered as common natural bioactive compounds (Hisham Shady et al., 2021). These natural compounds inhibit viral infection through suppression of lipid dependent viral attachment to the host (Hisham Shady et al., 2021). A highly oxygenated sterol compound called Cladosporisteroid B isolated from a sponge-derived fungus Cladosporium sp. acted as an antiviral agent against H3N2 with an IC50 value 16.2 µM (Pang et al., 2018). Another new compound named 3α-hydroxy-7-ene-6,20-dione containing a rare 3α-OH configuration and synthesized by the fungus Cladosporium sp. showed antiviral activity against the respiratory syncytial virus (RSV) with the IC50 value of 0.12 µM (Yu et al., 2018). Furthermore, an ergostane analogous metabolite named 3β-hydroxyergosta-8, 14, 24 (28)-trien-7-one isolated from the marine Penicillium sp. displayed broad-spectrum antiviral activities against HIV and H1N1 with the IC50 value of 3.5 and 0.5 µM, respectively (Li et al., 2019c).
Pyrone
Pyrones, found as two isomers namely 2-pyrone and 4-pyrone, are comprised of an unsaturated six-membered ring with one oxygen atom and a ketone functional group (Teng et al., 2020). An endophytic Fusarium equiseti isolated from a marine brown alga Padina pavonica, secretes various extracellular metabolites in different media compositions (Hawas et al., 2016). When this endophytic fungus was cultivated in biomalt-peptone medium, it produced 12 known metabolites of diketopeprazines and anthraquinones which were very potent anti-HCV (HCV protease inhibitor) agent with an IC50 from 19 to 77 μM, and the most potent anti-HCV compound in this condition was Griseoxanthone C with IC50 value of 19.8 μM (Hawas et al., 2016). However, the same fungus released nine different types of anti-HCV agents with IC50 value of 10–37 μM in the presence of Czapek’smedia, and the most potent anti-HCV compound was ω-hydroxyemodin with IC50 value of 10.7 μM (Hawas et al., 2016). “One strain many compounds” (OSMAC) has been proposed as a very effective approach to discover novel bioactive compounds (Pan et al., 2019). With the OSMAC approach, a coastal saline soil-derived fungus Aspergillus iizukae produces different antiviral compounds namely Methyl-(2-chloro-l,6-dihydroxy-3-methylxanthone)-8-carboxylate; methyl-(4-chloro-l,6-dihydroxy-3-methylxanthone)-8-carboxylate; methyl-(4-chloro-6-hydroxy-1-methoxy-3-methylxanthone)-8-carboxylate; methyl-(6-hydroxy-1-methoxy-3-methylxanthone)-8-carboxylate; 4-chloro-1,6-dihydroxy-3-methylxanthone-8-carboxylic acid; and 2,4-dichloro-1,6-dihydroxy-3-methylxanthone-8-carboxylic acid (Kang et al., 2018). Among these compounds, methyl-(4-chloro-l,6-dihydroxy-3-methylxanthone)-8-carboxylate exhibits strong antiviral activities against H1N1, HSV-1, and HSV-2 with IC50 values 44.6, 21.4, and 76.7 µM, respectively. However, the other compounds show week antiviral activity (Kang et al., 2018). A marine bacteria Streptomyces youssoufiensis can produce antiviral violapyrones (VLPs) Q–T through heterologous expression of the type III polyketide synthase (PKS) gene VioA (Hou et al., 2018). The antimicrobial activity of violapyrones mainly depends on the modification of 4-OH (methylation/non-methylation) (Teng et al., 2020). The compound showed antiviral activity in methylated condition but it showed anti-MRSA (Methicillin-resistant Staphylococcus aureus) activity in non-methylated condition with losing antiviral activity. The results support the notion that methylation at 4-OH of these compounds enhanced anti-virus activity but reduced anti-MRSA activity (Hou et al., 2018).
Polyphenol
Polyphenols or phenolic compounds are one of the prominent bioactive compounds found as secondary metabolites in plants and microorganisms (Othman et al., 2019; Carpine and Sieber, 2021). For instance, a soil fungus Exophiala pisciphila produces a novel dimeric 2,4-dihydroxy alkyl benzoic acid which exhibits anti-HIV activity by inhibiting integrase, a most crucial enzyme for HIV pathogenesis and is one of the most promising drug targets for anti-retroviral therapy (Ondeyka et al., 2003). Some antiviral polyphenol compounds have been produced through genetically engineered Saccharomyces cerevisiae, E. coli, Penicillium brevicompactum, Streptomyces avermitilis, Streptomyces lavendulae, and Yarrowia lipolytica (Ma et al., 2020). These prominent bioactive compounds exhibit antiviral activities through numerous mechanisms such as inhibition of viral attachment, penetration, genome replication and transcription as well as translation and viral assembly (Tables 1, 2) (Ma et al., 2020).
Lectin, Lipid, Lignan
A unique 95 amino acid long antiviral lectin obtained from a cyanobacterium Scytonema varium inhibits HIV attachment to the host cell through binding with the viral coat proteins gp120, gp160, and gp41 with EC50 values ranging from 0.3 to 22 nM (Bokesch et al., 2003). In addition, two prominent antiviral compounds namely cyanovirin-N and agglutinin obtained from cyanobacterium Nostoc ellipsosporum and Oscillatoria agardhii, respectively act as anti-HIV agents. The former compound inhibits viral attachment by binding with gp120 and the later one inhibits viral replication (Boyd et al., 1997; Sato et al., 2007). Furthermore, a glycolipid derived from cyanobacterium showed remarkable antiviral activity against HIV-1 (Gustafson et al., 1989). Phenylpropanoid units containing compound such as podophyllotoxin of endophytic Fusarium oxysporum isolated from Juniperus recurva showed anti-HIV activity (Kour et al., 2008). Furthermore, lots of bioactive compounds show antiviral activity against various viruses such as Human cytomegalovirus; HIV, H1N1, HSV, DENV, Enterovirus 71, ZIKV, RSV, HBV, HCV, Western equine encephalitis virus, and Pseudorabies virus (Tables 1, 2).
Potential Microbial Metabolites Against SARS-CoV-2
No newly developed specific drug has been approved by the WHO, FDA or any other global regulatory body to treat SARS-CoV-2. However, some drugs for other diseases have been approved for emergency usage during the pandemic situation (Hakim et al., 2021). For instance, the microbial-derived anti-parasitic drug ivermectin (Patridge et al., 2016) has been approved by the FDA to treat COVID-19 patients. Nevertheless, the time was not also enough to discover specific drug against SARS-CoV-2. However, research is going on globally to find drug against SARS-CoV-2 either from microbial or plant sources. A semisynthetic pentacyclic sixteen-membered lactone obtained from the soil bacterium Streptomyces avermitilis, has been found in vitro as inhibitor of SARS-CoV-2 replication (Caly et al., 2020). To find anti-SARS-CoV-2 drug from either microbial or plant sources, mostly in silico studies have been done. In silico screening, molecular docking, ADMET (Absorption, Distribution, Metabolism, Elimination, and Toxicity) prediction and molecular dynamic simulation (MDS) carried out by a number of studies predicted several phytocompounds as the potential inhibitors of SARS-CoV-2 and could be candidates to the discovery of novel drugs for the treatment of COVID-19 (Basu et al., 2020; Bhuiyan et al., 2020; Choudhary et al., 2020; Prasanth et al., 2020; Puttaswamy et al., 2020; Zhang et al., 2020; Chandra et al., 2021; Prasanth et al., 2021; Sankar et al., 2021). A study screened six potential candidates (Citriquinochroman, Holyrine B, Proximicin C, Pityriacitrin B, (+)-Anthrobenzoxoconone, and Penimethavone A) as anti-SARS-CoV-2 from >24,000 natural microbial compounds (Sayed et al., 2020). Docking andMDS analysis suggests that these microbial metabolites are potential inhibitor of protease involved in the host-SARS-CoV-2 interaction. However, experimental validation is required for the hypothesis derived from the in silico studies of plant and microbial metabolites.
Since the outbreaks of SARS in 2002/2003, MERS in 2012 and the COVID-19 pandemic in 2019/2020 (all caused by β-coronaviruses), different antiviral natural compounds have been tested against coronaviruses, such as remdesivir, ribavirin or herbacetin (Cherian et al., 2020). A numbers of microbial metabolites have been discussed in the aforementioned section to show antiviral activity including viral respiratory infections. Most of the microbial metabolites listed in Tables 1–3 are experimentally reported. Some of these metabolites especially those that show activity against viral respiratory infection can be potential for repurposing drugs against SARS-CoV-2. However, it would be worth for the researchers to elucidate the mechanism of actions of all antiviral microbial metabolites. Therefore, it will be interesting to perform docking and MDS of these microbial metabolites against proteins of SARS-CoV-2 and/or humans to predict their mechanism of actions, and finally experimentally validate the prediction of the in silico study. Metabolites from probiotic bacteria and/or gut microflora have been suggested to prevent viral respiratory infections including COVID-19 (Chen J. et al., 2021; Gautier et al., 2021). Probiotic bacterial metabolites such as butyrate, desaminotyrosine, and secondary bile acid may be transported to the lung via the circulation and could prevent viral respiratory infections by inhibiting viral replication or improving the immune response against viruses (Tiwari et al., 2020; Gautier et al., 2021). However, extensive studies are required to conclude the benefits of metabolites from probiotic bacteria and/or gut micro flora in COVID-19.
Advantages of the Microbial Source for Antiviral Metabolites
Currently, researchers are focusing on natural bioactive compounds to control viral infections that are considered as the main cause for human death worldwide (Akram et al., 2018). They are designing natural broad-spectrum antiviral agents by targeting a common pathway but essential for functions in many viruses (Vigant et al., 2015). The sources of natural bioactive compounds are plants, animals and microorganisms. However, as the leading producers of essential natural bioactive compounds, microorganisms are preferred more. Microorganisms are advantageous over other natural sources such as plants and animals due to their certain unique characteristics. Most of microorganisms are available as a wide range of genetically specified strains, fast growth, high density, high production rate, efficient secretion, easy handling and propagate, and can be easily manipulated (Singh et al., 2017). Microorganisms in general act as the source of essential natural product having the advantage of viable and sustainable production of secondary metabolites by large scale fermentation with reasonable cost (Waites et al., 2009; Sun X. et al., 2015). Furthermore, microorganisms can be grown at large amount in a small space such as in a fermenter under a wide range of environmental conditions for production of MSM of versatile groups. However, plants and animals need large space and longer period for cultivation, and are not amicable to versatile environmental conditions and/or metabolic engineering is technically challenging to plants and animals (Tatsis and O’Connor, 2016).
Metabolic and genetic engineering can easily be applied to microorganisms. Genomic information of a microbe makes it easy to apply metabolic engineering to scale up the production and/or modify the natural bioactive compound (Ma et al., 2020). Modified natural bioactive compounds may be suitable to get rid of drug resistance of viruses with their high genetic variability, and microbes are the most preferable candidates in this case (Lin et al., 2014). Furthermore, metabolic engineering to contrive the microbial cellular metabolic machinery and the fermentation technology to scale up the production has introduced a low-cost microbial system for large scale production of many natural bioactive compounds including antiviral agents (Liu and Nielsen, 2019; Pham et al., 2019; Ma et al., 2020). For instance, Violacein is a bis-indole pigment produced by several Gram-negative bacterial species by the vioABCDE operon (Choi et al., 2015). Due to antimicrobial (antibacterial, antiviral and antifungal) properties, this compound has become an interesting target for metabolic engineering strategy. Recently, the Y. lipolytica chassis strain was engineered for increased production of this compound. Introduction of five genes of bacterial vioABCDE operon and overexpression of endogenous anthranilate synthase 2 and 3 of Y. lipolytica increased violacein production 2.9 fold in comparison with the control (Zheng et al., 2020). Thus, heterologous synthesis of many antiviral compounds in genetically engineered microbes which are safer and economically beneficial offers some significant advantages over plant extraction and chemical synthesis (Ma et al., 2020). However, expression of the biosynthetic pathways for production of particular compounds in microbial factories may not be cost-effective sometimes due to mainly complexity of the pathways involving a number of enzymatic steps (Pandey et al., 2016; Yang D. et al., 2020). Introduction of a number of foreign proteins in a single microbial cell may lead to unwanted interaction between genetic factors and overload of the cell capacity, resulting in decreased microbial growth and low yields of the metabolite (Johnston et al., 2020). In this case, coculturing might be a highly promising approach to overcome these complexities with high yield. Furthermore, recombinant DNA technology used for large scale industrial production of bioactive compounds is feasible in microbial systems. The advancement of recombinant DNA technology has opened new windows for development of bioactive natural products and biologics (Pham et al., 2019; Ma et al., 2020). However, the choice of microbial host cells is very crucial for production of natural and recombinant products. Different tools and strategies for engineering host cells as microbial cell factories for production of natural bioactive compounds and recombinant products have been discussed elsewhere (Pham et al., 2019; Ma et al., 2020).
Future Prospects and Conclusion
The microbial source and system for antiviral natural bioactive compounds is attracting the researchers due to its advantages over plant and animal sources. Consequently, the demand of antiviral microbial metabolites is gradually increasing because the plant extraction and chemical synthesis cannot meet the global demand due to environmental, longer time and economic concerns. Microbial fermentation technology and metabolic and genetic engineering in microbial cells provide an alternate for scalable synthesis of these compounds. The global market value for MSM including antiviral agents was 277 billion USD in 2015, which is predicted to be 400 USD by 2025 (Park et al., 2019). Again, about 77% of FDA approved antimicrobial agents are produced from microbial sources, indicating microbial bioactive compounds as the pivotal source of antimicrobial drugs (Patridge et al., 2016). Therefore, antiviral microbial metabolites may pose great possibility in the field of pharmaceutical research and commercialization in near future. However, the vast diversity of antiviral microbial natural products yet requires extensive research and evaluation to find out the specific bioactive compounds with desired medicinal properties. Hence, from selection of appropriate microorganisms to formulation of drugs from their metabolites is a long term, expensive process that deserves relentless efforts and continuous exploration (Park et al., 2019).
Despite of some drawbacks such as final product purification and structural identification, microbial metabolite is still the unparalleled source of plenty of novel antiviral drug compounds (Park et al., 2019; Ma et al., 2020; Yi et al., 2020). Advancement of OMIC sciences (genomics, proteomics, metabolomics and so on) and gene based molecular approaches such genome editing, protein engineering and mutagenesis may offer more convenient drug design. Metabolomics being an emerging area in OMICs play pivotal roles in screening of lead compound, identifying drug target and assess bioactivity, potentiality and toxicity of the metabolites. Therefore, metabolomics in addition to proteomics that allows the structural and functional evaluation of the protein or antigenic compound targeted for the drug might be a great demand now-a-days in the term of drug designing and pharmacological research (Jain, 2004; Wishart, 2016). Furthermore, the most recent genome editing tool known as CRISPR (Clustered Regularly Interspaced Short Palindromic Repeats) can also be implemented in order to make desired change in the genome, especially while designing recombinant proteins in microbial cells to explore novel antiviral drugs (Liu et al., 2016). Similar site-specific gene editing by Zinc-finger nucleases (ZFNs) and transcription activator like effector nucleases (TALENs) possess great potentiality to be used in therapeutic purpose (Gaj et al., 2013). Therefore, OMICs and gene editing approaches collectively can be feasible in achieving the desired goal in screening and modifying microbial metabolites for antiviral drugs. Another efficient approach is microbial genome mining which comes with an outstanding opportunity of evaluating activity of the silent gene and discovering novel metabolites with the assistance of the information from genome sequencing (Bachmann et al., 2014). It also enables the understanding of biochemical pathways taking place inside the microbial cell, thus allowing the potential antiviral drug compounds to be discovered and analyzed (Fields et al., 2017; Xia, 2017). Furthermore, in the near future, metabolic engineering will contribute a lot to the discovery and development of antiviral drugs from microbial metabolites. Microbial system is becoming popular for expressing heterologous antiviral bioactive compounds. However, it paves challenges to the researchers to design and express the multiple enzymatic pathways involved in biosynthesis of antiviral bioactive compounds.
A wide array of plant-based secondary metabolites show promising antiviral activity against coronaviruses (Bhuiyan et al., 2020). Microbial biotechnology may contribute to large scale production of antiviral plant secondary metabolites or to get novel pharmaceutically active metabolites. However, many of the antiviral microbial metabolites included in this study are synthesized by endophytes. Therefore, the promising plant-based metabolites can be achieved through the screening of endophytic organisms of the targeted plant because various endophytic bacteria and fungi have the ability to produce the same or similar compounds as their host plants (Xu et al., 2009; Gouda et al., 2016; Raihan et al., 2021). For example, taxol, a billion dollar anticancer drug, initially produced by Taxus brevifolia and now it is produced from its endophyte Taxomyces andreanae (Stierle et al., 1993). Similarly, camptothecin, podophyllotoxin, hypericin and azadirachtin, are produced both by the endophyte and its host plant (Kusari and Spiteller, 2011; Bhalkar et al., 2016). Therefore, metabolites of endophytic microorganisms could be an emerging source of antiviral bioactive compounds (Schulz et al., 2002; Xu et al., 2009; Gouda et al., 2016; Raihan et al., 2021). Finally, researchers should pay attention to research with microbial metabolites using the approaches aforementioned to combat against catastrophic viral infections including COVID-19 and potential outbreaks of future viral pandemic and/or epidemics. For this, it is necessary to adopt initiatives to conduct systematic longitudinal studies by applying available and newly discovered microbial metabolites against catastrophic viruses including SARS-CoV-2.
Author Contributions
Concept and design: TR and AKA; whole draft manuscript writing: TR; partial draft manuscript writing: MFR, PR, and SC; Data collection and analysis, figures preparation: TR and AKA; critical review and suggestion for editing: K-HB; Data interpretation, compilation, supervision and editing of the whole manuscript: AKA. All authors contributed to the article and approved the submitted version.
Funding
This work was partially supported by grants in aid from the Research Centre, Shahjalal University of Science and Technology, Sylhet, Bangladesh (No. LS/2020/1/17).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Agarwal, G., and Gabrani, R. (2020). Antiviral Peptides: Identification and Validation. Int. J. Pept. Res. Ther. 18, 1–20. doi:10.1007/s10989-020-10072-0
Ahmed, E., Rateb, M., Abou El-Kassem, L., and Hawas, U. W. (2017). Anti-HCV Protease of Diketopiperazines Produced by the Red Sea Sponge-Associated Fungus Aspergillus versicolor. Appl. Biochem. Microbiol. 53 (1), 101–106. doi:10.1134/S0003683817010021
Akram, M., Tahir, I. M., Shah, S. M. A., Mahmood, Z., Altaf, A., Ahmad, K., et al. (2018). Antiviral Potential of Medicinal Plants against HIV, HSV, Influenza, Hepatitis, and Coxsackievirus: a Systematic Review. Phytother. Res. 32 (5), 811–822. doi:10.1002/ptr.6024
Anderson, K. P., Fox, M. C., Brown-Driver, V., Martin, M. J., and Azad, R. F. (1996). Inhibition of Human Cytomegalovirus Immediate-Early Gene Expression by an Antisense Oligonucleotide Complementary to Immediate-Early RNA. Antimicrob. Agents Chemother. 40 (9), 2004–2011. doi:10.1128/AAC.40.9.2004
Arena, A., Gugliandolo, C., Stassi, G., Pavone, B., Iannello, D., Bisignano, G., et al. (2009). An Exopolysaccharide Produced by Geobacillus Thermodenitrificans Strain B3-72: Antiviral Activity on Immunocompetent Cells. Immunol. Lett. 123 (2), 132–137. doi:10.1016/j.imlet.2009.03.001
Arena, A., Maugeri, T. L., Pavone, B., Iannello, D., Gugliandolo, C., Bisignano, G., et al. (2006). Antiviral and Immunoregulatory Effect of a Novel Exopolysaccharide from a marine Thermotolerant Bacillus Licheniformis. Int. Immunopharmacol. 6 (1), 8–13. doi:10.1016/j.intimp.2005.07.004
Arunpanichlert, J., Rukachaisirikul, V., Sukpondma, Y., Phongpaichit, S., Tewtrakul, S., Rungjindamai, N., et al. (2010). Azaphilone and Isocoumarin Derivatives from the Endophytic Fungus Penicillium sclerotiorum PSU-A13. Chem. Pharm. Bull. 58 (8), 1033–1036. doi:10.1248/cpb.58.1033
Ayehunie, S., Belay, A., Baba, T. W., and Ruprecht, R. M. (1998). Inhibition of HIV-1 Replication by an Aqueous Extract of Spirulina Platensis (Arthrospira Platensis). J. Acquir. Immune. Defic. Syndr. Hum. Retrovirol. 18, 7–12. doi:10.1097/00042560-199805010-00002
Azad, A. K., Raihan, T., Ahmed, J., Hakim, A., Emon, T. H., Chowdhury, P. A., et al. (2021). Human Aquaporins: Functional Diversity and Potential Roles in Infectious and Non-infectious Diseases. Front. Genet. 12, 344. doi:10.3389/fgene.2021.654865
Bachmann, B. O., Van Lanen, S. G., and Baltz, R. H. (2014). Microbial Genome Mining for Accelerated Natural Products Discovery: Is a Renaissance in the Making? J. Ind. Microbiol. Biotechnol. 41 (2), 175–184. doi:10.1007/s10295-013-1389-9
Bashyal, B. P., Wellensiek, B. P., Ramakrishnan, R., Faeth, S. H., Ahmad, N., Gunatilaka, A. L., et al. (2014). Altertoxins with Potent Anti-HIV Activity from Alternaria Tenuissima QUE1Se, a Fungal Endophyte of Quercus Emoryi. Bioorg. Med. Chem. 22 (21), 6112–6116. doi:10.1016/j.bmc.2014.08.039
Basu, A., Sarkar, A., and Maulik, U. (2020). Molecular Docking Study of Potential Phytochemicals and Their Effects on the Complex of SARS-CoV2 Spike Protein and Human ACE2. Sci. Rep. 10 (1), 1–15. doi:10.1038/s41598-020-74715-4
Beck, S., Henß, L., Weidner, T., Herrmann, J., Müller, R., Chao, Y. K., et al. (2016). Identification of Entry Inhibitors of Ebola Virus Pseudotyped Vectors from a Myxobacterial Compound Library. Antivir. Res. 132, 85–91. doi:10.1016/j.antiviral.2016.05.017
Belov, G. A., Lidsky, P. V., Mikitas, O. V., Egger, D., Lukyanov, K. A., Bienz, K., et al. (2004). Bidirectional Increase in Permeability of Nuclear Envelope upon Poliovirus Infection and Accompanying Alterations of Nuclear Pores. J. Virol. 78 (18), 10166–10177. doi:10.1128/JVI.78.18.10166-10177.2004
Berdy, J. (2005). Bioactive Microbial Metabolites. J. Antibiot. Res. 58 (1), 1–26. doi:10.1038/ja.2005.1
Bhalkar, B. N., Patil, S. M., and Govindwar, S. P. (2016). Camptothecine Production by Mixed Fermentation of Two Endophytic Fungi from Nothapodytes Nimmoniana. Fungal Biol. 120 (6-7), 873–883. doi:10.1016/j.funbio.2016.04.003
Bhuiyan, F. R., Howlader, S., Raihan, T., and Hasan, M. (2020). Plants Metabolites: Possibility of Natural Therapeutics against the Covid-19 Pandemic. Front. Med. (Lausanne) 7, 444. doi:10.3389/fmed.2020.00444
Bleasel, M. D., and Peterson, G. M. (2020). Emetine, Ipecac, Ipecac Alkaloids and Analogues as Potential Antiviral Agents for Coronaviruses. Pharmaceuticals 13 (3), 51. doi:10.3390/ph13030051
Boas, L. C. P. V., Campos, M. L., Berlanda, R. L. A., De Carvalho Neves, N., and Franco, O. L. (2019). Antiviral Peptides as Promising Therapeutic Drugs. Cell Mol. Life Sci. 76 (18), 3525–3542. doi:10.1007/s00018-019-03138-w
Bokesch, H. R., O'keefe, B. R., Mckee, T. C., Pannell, L. K., Patterson, G. M., Gardella, R. S., et al. (2003). A Potent Novel Anti-HIV Protein from the Cultured Cyanobacterium Scytonema Varium. Biochemistry 42, 2578–2584. doi:10.1021/bi0205698
Botos, I., O'keefe, B. R., Shenoy, S. R., Cartner, L. K., Ratner, D. M., Seeberger, P. H., et al. (2002). Structures of the Complexes of a Potent Anti-HIV Protein Cyanovirin-N and High Mannose Oligosaccharides. J. Biol. Chem. 277, 34336–34342. doi:10.1074/jbc.M205909200
Boyd, M. R., Gustafson, K. R., Mcmahon, J. B., Shoemaker, R. H., O'keefe, B. R., Mori, T., et al. (1997). Discovery of Cyanovirin-N, a Novel Human Immunodeficiency Virus-Inactivating Protein that Binds Viral Surface Envelope Glycoprotein Gp120: Potential Applications to Microbicide Development. Antimicrob. Agents Chemother. 41 (7), 1521–1530. doi:10.1128/AAC.41.7.1521
Bradley, S. A., Zhang, J., and Jensen, M. K. (2020). Deploying Microbial Synthesis for Halogenating and Diversifying Medicinal Alkaloid Scaffolds. Front. Bioeng. Biotechnol. 8, 1240. doi:10.3389/fbioe.2020.594126
Bunyapaiboonsri, T., Yoiprommarat, S., Srikitikulchai, P., Srichomthong, K., and Lumyong, S. (2010). Oblongolides from the Endophytic Fungus Phomopsis Sp. BCC 9789. J. Nat. Prod. 73 (1), 55–59. doi:10.1021/np900650c
Caly, L., Druce, J. D., Catton, M. G., Jans, D. A., and Wagstaff, K. M. (2020). The FDA-Approved Drug Ivermectin Inhibits the Replication of SARS-CoV-2 In Vitro. Antivir. Res 178, 104787. doi:10.1016/j.antiviral.2020.104787
Cameron, C. E., Götte, M., and Raney, K. D. (2009). Viral Genome Replication. US: Springer. doi:10.1007/b135974
Cao, X., Shi, Y., Wu, X., Wang, K., Huang, S., Sun, H., et al. (2019). Talaromyolides A–D and Talaromytin: Polycyclic Meroterpenoids from the Fungus Talaromyces Sp. CX11. Org. Lett. 21 (16), 6539–6542. doi:10.1021/acs.orglett.9b02466
Carpine, R., and Sieber, S. (2021). Antibacterial and Antiviral Metabolites from Cyanobacteria: Their Application and Their Impact on Human Health. Curr. Biotechnol. 3, 65–81. doi:10.1016/j.crbiot.2021.03.001
Chandra, A., Chaudhary, M., Qamar, I., Singh, N., and Nain, V. (2021). In Silico identification and Validation of Natural Antiviral Compounds as Potential Inhibitors of SARS-CoV-2 Methyltransferase. J. Biomol. Struct. Dyn. 15, 1–11. doi:10.1080/07391102.2021.1886174
Che, Q., Qiao, L., Han, X., Liu, Y., Wang, W., Gu, Q., et al. (2018). Anthranosides A-C, Anthranilate Derivatives from a Sponge-Derived Streptomyces Sp. CMN-62. 20. Org. Lett. 20 (17), 5466–5469. doi:10.1021/acs.orglett.8b02382
Chen, C. C., Yu, X., Kuo, C. J., Min, J., Chen, S., Wu, S., et al. (2021a). Overview of Antiviral Drug Candidates Targeting Coronaviral 3C like Main Proteases. FEBS J. 2021, 15696. doi:10.1111/febs.15696
Chen, F., and Huang, G. (2018). Preparation and Immunological Activity of Polysaccharides and Their Derivatives. Int. J. Biol. Macromol. 112, 211–216. doi:10.1016/j.ijbiomac.2018.01.169
Chen, J., Hall, S., and Vitetta, L. (2021b). Altered Gut Microbial Metabolites Could Mediate the Effects of Risk Factors in COVID 19. Rev. Med. Virol. 3, e2211. doi:10.1002/rmv.2211
Chen, S., Wang, J., Wang, Z., Lin, X., Zhao, B., Kaliaperumal, K., et al. (2017). Structurally Diverse Secondary Metabolites from a Deep-Sea-Derived Fungus Penicillium chrysogenum SCSIO 41001 and Their Biological Evaluation. Fitoterapia 117, 71–78. doi:10.1016/j.fitote.2017.01.005
Chen, X., Si, L., Liu, D., Proksch, P., Zhang, L., Zhou, D., et al. (2015). Neoechinulin B and its Analogues as Potential Entry Inhibitors of Influenza Viruses, Targeting Viral Hemagglutinin. Eur. J. Med. Chem. 93, 182–195. doi:10.1016/j.ejmech.2015.02.006
Chen, Y., Guo, Y., Pan, Y., and Zhao, Z. J. (2020). Structure Analysis of the Receptor Binding of 2019-nCoV. Biochem. Biophys. Res. Commun. 525 (1), 135–140. doi:10.1016/j.bbrc.2020.02.071
Cherian, S. S., Agrawal, M., Basu, A., Abraham, P., Gangakhedkar, R. R., and Bhargava, B. (2020). Perspectives for Repurposing Drugs for the Coronavirus Disease 2019. Indian J. Med. Res. 151 (2-3), 160. doi:10.4103/ijmr.IJMR_585_20
Cheung, R. C. F., Wong, J. H., Pan, W. L., Chan, Y. S., Yin, C. M., Dan, X. L., et al. (2014). Antifungal and Antiviral Products of marine Organisms. Appl. Microbiol. Biotechnol. 98 (8), 3475–3494. doi:10.1007/s00253-014-5575-0
Chiang, H.-S., and Liu, H. M. (2019). The Molecular Basis of Viral Inhibition of IRF-And STAT-dependent Immune Responses. Front. Immunol. 9, 3086. doi:10.3389/fimmu.2018.03086
Choi, S. Y., Yoon, K.-H., Lee, J. I., and Mitchell, R. J. (2015). Violacein: Properties and Production of a Versatile Bacterial Pigment. Biomed. Res. Int. 2015, 465056. doi:10.1155/2015/465056
Choudhary, M. I., Shaikh, M., Tul-Wahab, A., and Ur-Rahman, A. (2020). In Silico identification of Potential Inhibitors of Key SARS-CoV-2 3CL Hydrolase (Mpro) via Molecular Docking, MMGBSA Predictive Binding Energy Calculations, and Molecular Dynamics Simulation. PLoS One 15 (7), e0235030. doi:10.1371/journal.pone.0235030
Cohen, F. S. (2016). How Viruses Invade Cells. Biophys. J. 110 (5), 1028–1032. doi:10.1016/j.bpj.2016.02.006
Croaker, A., King, G. J., Pyne, J. H., Anoopkumar-Dukie, S., and Liu, L. (2016). Sanguinaria canadensis: Traditional Medicine, Phytochemical Composition, Biological Activities and Current Uses. Int. J. Mol. Sci. 17 (9), 1414. doi:10.3390/ijms17091414
Cushnie, T. T., Cushnie, B., and Lamb, A. J. (2014). Alkaloids: an Overview of Their Antibacterial, Antibiotic-Enhancing and Antivirulence Activities. Int. J. Antimicrob. Agents 44 (5), 377–386. doi:10.1016/j.ijantimicag.2014.06.001
Davidson, B. S., and Schumacher, R. W. (1993). Isolation and Synthesis of Caprolactins A and B, New Caprolactams from a marine Bacterium. Tetrahedron 49, 6569–6574. doi:10.1016/S0040-4020(01)81825-1
Debbab, A., Aly, A. H., and Proksch, P. (2011). Bioactive Secondary Metabolites from Endophytes and Associated marine Derived Fungi. Fungal Divers. 49 (1), 1–12. doi:10.1007/s13225-011-0114-0
Demain, A. L. (2007). “Microbial Secondary Metabolism: a New Theoretical Frontier for Academia, a New Opportunity for Industry,” in Ciba Foundation Symposium 171 Secondary Metabolites: Their Function and Evolution: Secondary Metabolites: Their Function and Evolution: Ciba Foundation Symposium 171 (Wiley Online Library), 3–23. doi:10.1002/9780470514344.ch2
Ding, L., Münch, J., Goerls, H., Maier, A., Fiebig, H.-H., Lin, W.-H., et al. (2010). Xiamycin, a Pentacyclic Indolosesquiterpene with Selective Anti-HIV Activity from a Bacterial Mangrove Endophyte. Bioorg. Med. Chem. Lett. 20 (22), 6685–6687. doi:10.1016/j.bmcl.2010.09.010
Dolak, L. A., and DeBoer, C. (1980). Clazamycin B Is Antibiotic 354. J. Antibiot. 33 (1), 83–84. doi:10.7164/antibiotics.33.83
Duffy, S. (2018). Why Are RNA Virus Mutation Rates So Damn High? Plos Biol. 16 (8), e3000003. doi:10.1371/journal.pbio.3000003
El-Najjar, N., Gali-Muhtasib, H., Ketola, R. A., Vuorela, P., Urtti, A., Vuorela, H., et al. (2011). The Chemical and Biological Activities of Quinones: Overview and Implications in Analytical Detection. Phytochem. Rev. 10 (3), 353–370. doi:10.1007/s11101-011-9209-1
Fabregas, J., Garcıa, D., Fernandez-Alonso, M., Rocha, A., Gómez-Puertas, P., Escribano, J., et al. (1999). In Vitro inhibition of the Replication of Haemorrhagic Septicaemia Virus (VHSV) and African Swine Fever Virus (ASFV) by Extracts from marine Microalgae. Antivir. Res. 44 (1), 67–73. doi:10.1016/S0166-3542(99)00049-2
Fan, Y., Wang, Y., Liu, P., Fu, P., Zhu, T., Wang, W., et al. (2013). Indole-diterpenoids with Anti-h1n1 Activity from the Aciduric Fungus Penicillium camemberti OUCMDZ-1492. J. Nat. Prod. 76 (7), 1328–1336. doi:10.1021/np400304q
Fang, W., Lin, X., Zhou, X., Wan, J., Lu, X., Yang, B., et al. (2014). Cytotoxic and Antiviral Nitrobenzoyl Sesquiterpenoids from the marine-derived Fungus Aspergillus ochraceus Jcma1F17. Med. Chem. Comm. 5 (6), 701–705. doi:10.1039/C3MD00371J
Fields, F. R., Lee, S. W., and Mcconnell, M. J. (2017). Using Bacterial Genomes and Essential Genes for the Development of New Antibiotics. Biochem. Pharmacol. 134, 74–86. doi:10.1016/j.bcp.2016.12.002
Fung, T. S., and Liu, D. X. (2019). Human Coronavirus: Host-Pathogen Interaction. Annu. Rev. Microbiol. 73, 529–557. doi:10.1146/annurev-micro-020518-115759
Gaj, T., Gersbach, C. A., and Barbas, C. F. (2013). ZFN, TALEN, and CRISPR/Cas-based Methods for Genome Engineering. Trends Biotechnol. 31 (7), 397–405. doi:10.1016/j.tibtech.2013.04.004
Galdiero, S., Falanga, A., Tarallo, R., Russo, L., Galdiero, E., Cantisani, M., et al. (2013). Peptide Inhibitors against Herpes Simplex Virus Infections. J. Pept. Sci. 19 (3), 148–158. doi:10.1002/psc.2489
Gao, H., Guo, W., Wang, Q., Zhang, L., Zhu, M., Zhu, T., et al. (2013). Aspulvinones from a Mangrove Rhizosphere Soil-Derived Fungus Aspergillus terreus Gwq-48 with Anti-influenza A Viral (H1N1) Activity. Bioorg. Med. Chem. Lett. 23 (6), 1776–1778. doi:10.1016/j.bmcl.2013.01.051
Gautier, T., Gall, D.-L., Sweidan, A., Tamanai-Shacoori, Z., Jolivet-Gougeon, A., Loréal, O., et al. (2021). Next-generation Probiotics and Their Metabolites in COVID-19. Microorganisms 9, 941. doi:10.3390/microorganisms9050941
Gentzsch, J., Hinkelmann, B., Kaderali, L., Irschik, H., Jansen, R., Sasse, F., et al. (2011). Hepatitis C Virus Complete Life Cycle Screen for Identification of Small Molecules with Pro- or Antiviral Activity. Antivir. Res. 89 (2), 136–148. doi:10.1016/j.antiviral.2010.12.005
Gnirss, K., Kühl, A., Karsten, C., Glowacka, I., Bertram, S., Kaup, F., et al. (2012). Cathepsins B and L Activate Ebola but Not Marburg Virus Glycoproteins for Efficient Entry into Cell Lines and Macrophages Independent of TMPRSS2 Expression. Virology 424, 3–10. doi:10.1016/j.virol.2011.11.031
Goris, T., Pérez Valero, Á., Martínez, I., Yi, D., Fernández Calleja, L., San León, D., et al. (2021). Repositioning Microbial Biotechnology against COVID 19: the Case of Microbial Production of Flavonoids. Microb. Biotechnol. 14 (1), 94–110. doi:10.1111/1751-7915.13675
Gouda, S., Das, G., Sen, S. K., Shin, H. S., and Patra, J. K. (2016). Endophytes: a Treasure House of Bioactive Compounds of Medicinal Importance. Front. Microbiol. 7, 1538. doi:10.3389/fmicb.2016.01538
Graham, R. L., Donaldson, E. F., and Baric, R. S. (2013). A Decade after SARS: Strategies for Controlling Emerging Coronaviruses. Nat. Rev. Microbiol. 11 (12), 836–848. doi:10.1038/nrmicro3143
Group, R. C. (2021). Dexamethasone in Hospitalized Patients with COVID-19. New Engl. J. Med. 384 (8), 693–704. doi:10.1056/NEJMoa2021436
Gu, Y., Ma, J., Zhu, Y., Ding, X., and Xu, P. (2020). Engineering Yarrowia lipolytica as a Chassis for De Novo Synthesis of Five Aromatic-Derived Natural Products and Chemicals. ACS Synth. Biol. 9 (8), 2096–2106. doi:10.1021/acssynbio.0c00185
Guo, B., Dai, J.-R., Ng, S., Huang, Y., Leong, C., Ong, W., et al. (2000). Cytonic Acids A and B: Novel Tridepside Inhibitors of hCMV Protease from the Endophytic Fungus Cytonaema Species. J. Nat. Prod. 63 (5), 602–604. doi:10.1021/np990467r
Guo, Q., Shao, Q., Xu, W., Rui, L., Sumi, R., Eguchi, F., et al. (2017). Immunomodulatory and Anti-IBDV Activities of the Polysaccharide AEX from Coccomyxa Gloeobotrydiformis. Mar. Drugs 15 (2), 36. doi:10.3390/md15020036
Gupta, D. K., Kaur, P., Leong, S. T., Tan, L. T., Prinsep, M. R., Chu, J. J., et al. (2014). Anti-Chikungunya Viral Activities of Aplysiatoxin-Related Compounds from the marine Cyanobacterium. Trichodesmium Erythraeum. Mar. Drugs 12 (1), 115–127. doi:10.3390/md12010115
Gustafson, K., Roman, M., and Fenical, W. (1989). The Macrolactins, a Novel Class of Antiviral and Cytotoxic Macrolides from a Deep-Sea marine Bacterium. J. Am. Chem. Soc. 111 (19), 7519–7524. doi:10.1021/ja00201a036
Hakim, A., Hasan, M., Hasan, M., Lokman, S. M., Azim, K. F., Raihan, T., et al. (2021). Major Insights in Dynamics of Host Response to SARS-CoV-2: Impacts and Challenges. Front.Microb. 2384, 1. doi:10.3389/fmicb.2021.637554
Hashimoto, K., Kodama, E., Mori, S., Watanabe, J., Baba, M., Okutani, K., et al. (1996). Antiviral Activity of a Sulphated Polysaccharide Extracted from the marine Pseudomonas and marine Plant Dinoflagellata against Human Immunodeficiency Viruses and Other Enveloped Viruses. Antivir. Chem. Chemother. 7, 189–196. doi:10.1177/095632029600700403
Hasui, M., Matsuda, M., Okutani, K., and Shigeta, S. (1995). In Vitro antiviral Activities of Sulfated Polysaccharides from a marine Microalga (Cochlodinium Polykrikoides) against Human Immunodeficiency Virus and Other Enveloped Viruses. Int. J. Biol. Macromol. 17 (5), 293–297. doi:10.1016/0141-8130(95)98157-T
Hawas, U. W., Al-Farawati, R., El-Kassem, A., Lamia, T., and Turki, A. J. (2016). Different Culture Metabolites of the Red Sea Fungus Fusarium Equiseti Optimize the Inhibition of Hepatitis C Virus NS3/4A Protease (HCV PR). Mar. Drugs 14 (10), 190. doi:10.3390/md14100190
Hawas, U. W., El-Halawany, A. M., and Ahmede, E. F. (2013). Hepatitis C Virus NS3-Ns4a Protease Inhibitors from the Endophytic Penicillium chrysogenum Isolated from the Red Alga Liagora Viscida. Z. Naturforsch. C. 68 (9-10), 355–366. doi:10.1515/znc-2013-9-1003
Hayashi, K., Hayashi, T., and Kojima, I. (1996). A Natural Sulfated Polysaccharide, Calcium Spirulan, Isolated from Spirulina Platensis: In Vitro and Ex Vivo Evaluation of Anti-herpes Simplex Virus and Anti-human Immunodeficiency Virus Activities. AIDS Res. Hum. Retroviruse. 12, 1463–1471. doi:10.1089/aid.1996.12.1463
Hayashi, K., Lee, J.-B., Atsumi, K., Kanazashi, M., Shibayama, T., Okamoto, K., et al. (2019). In Vitro and In Vivo Anti-herpes Simplex Virus Activity of Monogalactosyl Diacylglyceride from Coccomyxa Sp. KJ (IPOD FERM BP-22254), a green Microalga. PloS one 14, e0219305. doi:10.1371/journal.pone.0219305
He, F., Bao, J., Zhang, X.-Y., Tu, Z.-C., Shi, Y.-M., and Qi, S.-H. (2013). Asperterrestide A, a Cytotoxic Cyclic Tetrapeptide from the marine-derived Fungus Aspergillus terreus SCSGAF0162. J. Nat. Prod. 76 (6), 1182–1186. doi:10.1021/np300897v
He, J.-W., Chen, G.-D., Gao, H., Yang, F., Li, X.-X., Peng, T., et al. (2012). Heptaketides with Antiviral Activity from Three Endolichenic Fungal Strains Nigrospora sp., Alternaria Sp. And Phialophora Sp. Fitoterapia 83, 1087–1091. doi:10.1016/j.fitote.2012.05.002
Hernández-Corona, A., Nieves, I., Meckes, M., Chamorro, G., and Barron, B. L. (2002). Antiviral Activity of Spirulina Maxima against Herpes Simplex Virus Type 2. Antivir. Res. 56 (3), 279–285. doi:10.1016/S0166-3542(02)00132-8
Heydari, H., Golmohammadi, R., Mirnejad, R., Tebyanian, H., Fasihi-Ramandi, M., and Moosazadeh-Moghadam, M. (2021). Antiviral Peptides against Coronaviridae Family: A Review. Peptides 139, 170526. doi:10.1016/j.peptides.2021.170526
Hisham Shady, N., Youssif, K. A., Sayed, A. M., Belbahri, L., Oszako, T., Hassan, H. M., et al. (2021). Sterols and Triterpenes: Antiviral Potential Supported by In-Silico Analysis. Plants 10, 41. doi:10.3390/plants10010041
Hoffmann, M., Kleine-Weber, H., Schroeder, S., Krüger, N., Herrler, T., Erichsen, S., et al. (2020). SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell 181, 271–280. e278. doi:10.1016/j.cell.2020.02.052
Hori, M., Takita, T., Koyama, G., Tadeuchi, T., and Umezawa, H. (1964). A New Antibiotic, Formycin. J. Antibiot. Ser. A 17 (3), 96–99. doi:10.11554/antibioticsa.17.3_96
Hoshino, H., Seki, J., and Takeuchi, T. (1989). New Antifungal Antibiotics, Benanomicins A and B Inhibit Infection of T-Cell with Human Immunodeficiency Virus (HIV) and Syncytium Formation by HIV. J. Antibiot. 42 (2), 344–346. doi:10.7164/antibiotics.42.344
Hou, L., Wang, S., Huang, H., Li, H., Wang, W., Li, W., et al. (2018). Generation of Methylated Violapyrones with Improved Anti-influenza A Virus Activity by Heterologous Expression of a Type III PKS Gene in a marine Streptomyces Strain. Bioorg. Med. Chem. Lett. 28 (17), 2865–2868. doi:10.1016/j.bmcl.2018.07.029
Huang, H., Song, Y., Li, X., Wang, X., Ling, C., Qin, X., et al. (2018). Abyssomicin Monomers and Dimers from the marine-derived Streptomyces Koyangensis SCSIO 5802. J. Nat. Prod. 81 (8), 1892–1898. doi:10.1021/acs.jnatprod.8b00448
Huang, H., Song, Y., Zang, R., Wang, X., and Ju, J. (2019a). Octyl Substituted Butenolides from marine-derived Streptomyces Koyangensis. Nat. Prod. Res. 1-6. doi:10.1080/14786419.2019.1686368
Huang, J., Zha, W., An, T., Dong, H., Huang, Y., Wang, D., et al. (2019b). Identification of RoCYP01 (CYP716A155) Enables Construction of Engineered Yeast for High-Yield Production of Betulinic Acid. Appl. Microbiol. Biotechnol. 103 (17), 7029–7039. doi:10.1007/s00253-019-10004-z
Huang, L.-H., Xu, M.-Y., Li, H.-J., Li, J.-Q., Chen, Y.-X., Ma, W.-Z., et al. (2017a). Amino Acid-Directed Strategy for Inducing the marine-derived Fungus Scedosporium Apiospermum F41–1 to Maximize Alkaloid Diversity. Org. Lett. 19 (18), 4888–4891. doi:10.1021/acs.orglett.7b02238
Huang, Z., Nong, X., Ren, Z., Wang, J., Zhang, X., Qi, S., et al. (2017b). Anti-HSV-1, Antioxidant and Antifouling Phenolic Compounds from the Deep-Sea-Derived Fungus Aspergillus versicolor SCSIO 41502. Bioorg. Med. Chem. Lett. 27 (4), 787–791. doi:10.1016/j.bmcl.2017.01.032
Huheihel, M., Ishanu, V., Tal, J., and Arad, S. M. (2002). Activity of Porphyridium Sp. Polysaccharide against Herpes Simplex Viruses In Vitro and In Vivo. J. Biochem. Biophys. Methods 50 (2-3), 189–200. doi:10.1016/S0165-022X(01)00186-5
Huleihel, M., Ishanu, V., Tal, J., and Arad, S. M. (2001). Antiviral Effect of Red Microalgal Polysaccharides on Herpes Simplex and Varicella Zoster Viruses. J. Appl. Phycol. 13 (2), 127–134. doi:10.1023/A:1011178225912
Huskens, D., Férir, G., Vermeire, K., Kehr, J. C., Balzarini, J., Dittmann, E., et al. (2010). Microvirin, a Novel Alpha(1,2)-mannose-specific Lectin Isolated from Microcystis Aeruginosa, Has Anti-HIV-1 Activity Comparable with that of Cyanovirin-N but a Much Higher Safety Profile. J. Biol. Chem. 285, 24845–24854. doi:10.1074/jbc.M110.128546
Hwang, Y., Rowley, D., Rhodes, D., Gertsch, J., Fenical, W., Bushman, F., et al. (1999). Mechanism of Inhibition of a Poxvirus Topoisomerase by the marine Natural Product Sansalvamide A. Mol. Pharmacol. 55 (6), 1049–1053. doi:10.1124/mol.55.6.1049
Ikeda, R., Haraguchi, Y., Ikeda, Y., Kondo, S., Takeuchi, T., Hoshino, H., et al. (1996). Inhibition of Human Immunodeficiency Virus Type 1 Infectivity by a New Amine Bellenamine. Antivir. Res. 29 (2-3), 163–173. doi:10.1016/0166-3542(95)00828-4
Inoue, Y., Tanaka, N., Tanaka, Y., Inoue, S., Morita, K., Zhuang, M., et al. (2007). Clathrin-dependent Entry of Severe Acute Respiratory Syndrome Coronavirus into Target Cells Expressing ACE2 with the Cytoplasmic Tail Deleted. J. Virol. 81 (16), 8722–8729. doi:10.1128/JVI.00253-07
Isaka, M., Berkaew, P., Intereya, K., Komwijit, S., and Sathitkunanon, T. (2007). Antiplasmodial and Antiviral Cyclohexadepsipeptides from the Endophytic Fungus Pullularia Sp. BCC 8613. Tetrahedron 63, 6855–6860. doi:10.1016/j.tet.2007.04.062
Ivanov, A. I. (2008). Pharmacological Inhibition of Endocytic Pathways: Is it Specific Enough to Be Useful? Exocytosis and endocytosis 440, 15–33. doi:10.1007/978-1-59745-178-9_2
Jacob, J. R., Mansfield, K., You, J. E., Tennant, B. C., and Kim, Y. H. (2007). Natural Iminosugar Derivatives of 1-deoxynojirimycin Inhibit Glycosylation of Hepatitis Viral Envelope Proteins. J. Microbiol. 45, 431–440.
Jain, K. K. (2004). “Applications of Proteomics Technologies for Drug Discovery,” in Proteomics: Biomedical and Pharmaceutical Applications (Springer), 201–227. doi:10.1007/1-4020-2323-5_9
Jan, E., Mohr, I., and Walsh, D. (2016). A Cap-To-Tail Guide to mRNA Translation Strategies in Virus-Infected Cells. Annu. Rev. Virol. 3, 283–307. doi:10.1146/annurev-virology-100114-055014
Janardhan, A., Kumar, A. P., Viswanath, B., Gopal, D. S., and Narasimha, G. (2018). Antiviral and Larvicidal Properties of Novel Bioactive Compounds Produced from marine Actinomycetes. Russ. J. Mar. Biol. 44 (5), 424–428. doi:10.1134/S106307401805005X
Jia, Y.-L., Guan, F.-F., Ma, J., Wang, C.-Y., and Shao, C.-L. (2015). Pestalotiolide A, a New Antiviral Phthalide Derivative from a Soft Coral-Derived Fungus Pestalotiopsis Sp. Nat. Prod. Sci. 21 (4), 227–230. doi:10.20307/nps.2015.21.4.227
Jin, Y., Qin, S., Gao, H., Zhu, G., Wang, W., Zhu, W., et al. (2018). An Anti-HBV Anthraquinone from Aciduric Fungus Penicillium Sp. OUCMDZ-4736 under Low pH Stress. Extremophiles 22, 39–45. doi:10.1007/s00792-017-0975-6
Johnston, T. G., Yuan, S.-F., Wagner, J. M., Yi, X., Saha, A., Smith, P., et al. (2020). Compartmentalized Microbes and Co-cultures in Hydrogels for On-Demand Bioproduction and Preservation. Nat. Commun. 11 (1), 1–11. doi:10.1038/s41467-020-14371-4
Jones, J. A., Vernacchio, V. R., Lachance, D. M., Lebovich, M., Fu, L., Shirke, A. N., et al. (2015). ePathOptimize: a Combinatorial Approach for Transcriptional Balancing of Metabolic Pathways. Sci. Rep. 5 (1), 1–10. doi:10.1038/srep11301
Jurkiewicz, E., Jansen, R., Kunze, B., Trowitzsch-Kienast, W., Forche, E., Reichenbach, H., et al. (1992). Three New Potent HIV-1 Inhibitors from Myxobacteria. Antivir. Chem. Chemother. 3 (4), 189–193. doi:10.1177/095632029200300401
Kalil, A. C., Patterson, T. F., Mehta, A. K., Tomashek, K. M., Wolfe, C. R., Ghazaryan, V., et al. (2021). Baricitinib Plus Remdesivir for Hospitalized Adults with Covid-19. New Engl. J. Med. 384 (9), 795–807. doi:10.1056/NEJMoa2031994
Kamei, Y., Yoshimizu, M., Ezura, Y., and Kimura, T. (1988). Effects of Environmental Water on the Infectivities of Infectious Hematopoietic Necrosis Virus (IHNV) and Infectious Pancreatic Necrosis Virus (IPNV). J. Appl. Ichthyol. 4 (1), 37–47. doi:10.1111/j.1439-0426.1988.tb00546.x
Kamei, Y., Yoshimizu, M., Ezura, Y., and Kimura, T. (1992). “Isolation and Characterization of Antiviral Substance against Salmonid Viruses, 46NW-04A Produced by an Aquatic Bacterium, Pseudomonas Fluorescens 46NW-04,” in Proceedings of the Oji International Symposium on Salmonid Diseases (Sapporo, Japan: Hokkaido University Press), 293–300.
Kanekiyo, K., Hayashi, K., Takenaka, H., Lee, J. B., and Hayashi, T. (2007). Anti-herpes Simplex Virus Target of an Acidic Polysaccharide, Nostoflan, from the Edible Blue-green Alga Nostoc Flagelliforme. Biol. Pharm. Bull. 30 (8), 1573–1575. doi:10.1248/bpb.30.1573
Kang, H.-H., Zhang, H.-B., Zhong, M.-J., Ma, L.-Y., Liu, D.-S., Liu, W.-Z., et al. (2018). Potential Antiviral Xanthones from a Coastal saline Soil Fungus Aspergillus iizukae. Mar. Drugs 16 (11), 449. doi:10.3390/md16110449
Kim, M., Yim, J. H., Kim, S.-Y., Kim, H. S., Lee, W. G., Kim, S. J., et al. (2012). In Vitro inhibition of Influenza A Virus Infection by marine Microalga-Derived Sulfated Polysaccharide P-KG03. Antivir. Res. 93 (2), 253–259. doi:10.1016/j.antiviral.2011.12.006
Klemm, T., Ebert, G., Calleja, D. J., Allison, C. C., Richardson, L. W., Bernardini, J. P., et al. (2020). Mechanism and Inhibition of the Papain like Protease, PLpro, of SARS CoV 2. EMBO J. 39 (18), e106275. doi:10.15252/embj.2020106275
Kluepfel, D., Bagli, J., Baker, H., Charest, M.-P., Kudelski, A., Sehgal, S. N., et al. (1972). Myriocin, a New Antifungal Antibiotic from Myriococcum Albomyces. J. Antibiot. 25 (2), 109–115. doi:10.7164/antibiotics.25.109
Knübel, G., Larsen, L. K., Moore, R. E., Levine, I. A., and Patterson, G. M. (1990). Cytotoxic, Antiviral Indolocarbazoles from a Blue-green Alga Belonging to the Nostocaceae. J. Antibiot. (Tokyo) 43, 1236–1239. doi:10.7164/antibiotics.43.1236
Koch, G. (1971). Differential Effect of Phleomycin on the Infectivity of Poliovirus and Poliovirus-Induced Ribonucleic Acids. J. Virol. 8 (1), 28–34. doi:10.1128/jvi.8.1.28-34.1971
Koenuma, M., Kinashi, H., and Otake, N. (1974). An Improved Screening Method for Antiphage Antibiotics and Isolation of Sarkomycin and its Relatives. J. Antibiot. 27 (10), 801–804. doi:10.7164/antibiotics.27.801
Kong, F.-D., Ma, Q.-Y., Huang, S.-Z., Wang, P., Wang, J.-F., Zhou, L.-M., et al. (2017). Chrodrimanins K–N and Related Meroterpenoids from the Fungus Penicillium Sp. SCS-KFD09 Isolated from a marine Worm, Sipunculus Nudus. J. Nat. Prod. 80 (4), 1039–1047. doi:10.1021/acs.jnatprod.6b01061
Kong, F., Zhao, C., Hao, J., Wang, C., Wang, W., Huang, X., et al. (2015). New α-glucosidase Inhibitors from a marine Sponge-Derived Fungus, Aspergillus Sp. OUCMDZ-1583. RSC Adv. 5, 68852–68863. doi:10.1039/c5ra11185d
Kour, A., Shawl, A. S., Rehman, S., Sultan, P., Qazi, P. H., Suden, P., et al. (2008). Isolation and Identification of an Endophytic Strain of Fusarium Oxysporum Producing Podophyllotoxin from Juniperus Recurva. World J. Microbiol. Biotechnol. 24 (7), 1115–1121. doi:10.1007/s11274-007-9582-5
Kusari, S., and Spiteller, M. (2011). Are We Ready for Industrial Production of Bioactive Plant Secondary Metabolites Utilizing Endophytes? Nat. Prod. Rep. 28 (7), 1203–1207. doi:10.1039/C1NP00030F
Larsen, L. K., Moore, R. E., and Patterson, G. M. (1994). Beta-Carbolines from the Blue-green Alga Dichothrix Baueriana. J. Nat. Prod. 57, 419–421. doi:10.1021/np50105a018
Lee, H., Kim, B. G., Kim, M., and Ahn, J. H. (2015). Biosynthesis of Two Flavones, Apigenin and Genkwanin, in Escherichia coli. J. Microbiol. Biotechnol. 25, 1442–1448. doi:10.4014/jmb.1503.03011
Li, E., Tian, R., Liu, S., Chen, X., Guo, L., Che, Y., et al. (2008). Pestalotheols A−D, Bioactive Metabolites from the Plant Endophytic Fungus Pestalotiopsis theae. J. Nat. Prod. 71 (4), 664–668. doi:10.1021/np700744t
Li, J., Hu, Y., Hao, X., Tan, J., Li, F., Qiao, X., et al. (2019a). Raistrickindole A, an Anti-HCV Oxazinoindole Alkaloid from Penicillium raistrickii IMB17-034. J. Nat. Prod. 82 (5), 1391–1395. doi:10.1021/acs.jnatprod.9b00259
Li, J., Tian, C., Xia, Y., Mutanda, I., Wang, K., and Wang, Y. (2019b). Production of Plant-specific Flavones Baicalein and Scutellarein in an Engineered E. coli from Available Phenylalanine and Tyrosine. Metab. Eng. 52, 124–133. doi:10.1016/j.ymben.2018.11.008
Li, J., Wang, Y., Hao, X., Li, S., Jia, J., Guan, Y., et al. (2019c). Broad-spectrum Antiviral Natural Products from the marine-derived Penicillium Sp. IMB17-046. Molecules 24, 2821. doi:10.3390/molecules24152821
Li, M., Schneider, K., Kristensen, M., Borodina, I., and Nielsen, J. (2016). Engineering Yeast for High-Level Production of Stilbenoid Antioxidants. Sci. Rep. 6 (1), 1–8. doi:10.1038/srep36827
Li, Y., Liu, D., Cen, S., Proksch, P., and Lin, W. (2014). Isoindolinone-type Alkaloids from the Sponge-Derived Fungus Stachybotrys chartarum. Tetrahedron 70, 7010–7015. doi:10.1016/j.tet.2014.07.047
Li, Z., Wang, X., and Zhang, H. (2019d). Balancing the Non-linear Rosmarinic Acid Biosynthetic Pathway by Modular Co-culture Engineering. Metab. Eng. 54, 1–11. doi:10.1016/j.ymben.2019.03.002
Liang, X., Nong, X.-H., Huang, Z.-H., and Qi, S.-H. (2017). Antifungal and Antiviral Cyclic Peptides from the Deep-Sea-Derived Fungus Simplicillium Obclavatum EIODSF 020. J. Agric. Food Chem. 65 (25), 5114–5121. doi:10.1021/acs.jafc.7b01238
Liao, H.-X., Sun, D.-W., Zheng, C.-J., and Wang, C.-Y. (2017). A New Hexahydrobenzopyran Derivative from the Gorgonian-Derived Fungus Eutypella Sp. Nat. Prod. Res. 31 (14), 1640–1646. doi:10.1080/14786419.2017.1285301
Lim, C. G., Fowler, Z. L., Hueller, T., Schaffer, S., and Koffas, M. A. (2011). High-yield Resveratrol Production in Engineered Escherichia coli. Appl. Environ. Microbiol. 77 (10), 3451–3460. doi:10.1128/AEM.02186-10
Lim, Y. X., Ng, Y. L., Tam, J. P., and Liu, D. X. (2016). Human Coronaviruses: a Review of Virus-Host Interactions. Dis. (Basel, Switzerland) 4, 26. doi:10.3390/diseases4030026
Lin, L.-T., Hsu, W.-C., and Lin, C.-C. (2014). Antiviral Natural Products and Herbal Medicines. J. Tradit. Complement. Med. 4 (1), 24–35. doi:10.4103/2225-4110.124335
Linnakoski, R., Reshamwala, D., Veteli, P., Cortina-Escribano, M., Vanhanen, H., Marjomäki, V., et al. (2018). Antiviral Agents from Fungi: Diversity, Mechanisms and Potential Applications. Front. Microbiol. 9, 2325. doi:10.3389/fmicb.2018.02325
Liu, F.-A., Lin, X., Zhou, X., Chen, M., Huang, X., Yang, B., et al. (2017a). Xanthones and Quinolones Derivatives Produced by the Deep-Sea-Derived Fungus Penicillium Sp. SCSIO Ind16f01. Mol. 22, 1999. doi:10.3390/molecules22121999
Liu, H., Chen, Z., Zhu, G., Wang, L., Du, Y., Wang, Y., et al. (2017b). Phenolic Polyketides from the marine Alga-Derived Streptomyces Sp. OUCMDZ-3434. Tetrahedron 73, 5451–5455. doi:10.1016/j.tet.2017.07.052
Liu, L., Bruhn, T., Guo, L., Götz, D. C., Brun, R., Stich, A., et al. (2011). Chloropupukeanolides C–E: Cytotoxic Pupukeanane Chlorides with a Spiroketal Skeleton from Pestalotiopsis Fici. Chem. Eur. J. 17, 2604–2613. doi:10.1002/chem.201003129
Liu, L., Liu, H., Zhang, W., Yao, M., Li, B., Liu, D., et al. (2019a). Engineering the Biosynthesis of Caffeic Acid in Saccharomyces cerevisiae with Heterologous Enzyme Combinations. Engineering 5, 287–295. doi:10.1016/j.eng.2018.11.029
Liu, Q., Yu, T., Li, X., Chen, Y., Campbell, K., Nielsen, J., et al. (2019b). Rewiring Carbon Metabolism in Yeast for High Level Production of Aromatic Chemicals. Nat. Commun. 10 (1), 1–13. doi:10.1038/s41467-019-12961-5
Liu, X., Yang, Y., Zhang, W., Sun, Y., Peng, F., Jeffrey, L., et al. (2016). Expression of Recombinant Protein Using Corynebacterium Glutamicum: Progress, Challenges and Applications. Crit. Rev. Biotechnol. 36 (4), 652–664. doi:10.3109/07388551.2015.1004519
Liu, Y., and Nielsen, J. (2019). Recent Trends in Metabolic Engineering of Microbial Chemical Factories. Curr. Opin. Biotechnol. 60, 188–197. doi:10.1016/j.copbio.2019.05.010
Liu, Z.-H., Niu, F.-J., Xie, Y.-X., Xie, S.-M., Liu, Y.-N., Yang, Y.-Y., et al. (2020). A Review: Natural Polysaccharides from Medicinal Plants and Microorganisms and Their Anti-herpetic Mechanism. Biomed. Pharmacother. 129, 110469. doi:10.1016/j.biopha.2020.110469
Lobo-Galo, N., Gálvez-Ruíz, J.-C., Balderrama-Carmona, A. P., Silva-Beltrán, N. P., and Ruiz-Bustos, E. (2021). Recent Biotechnological Advances as Potential Intervention Strategies against COVID-19. 3 Biotech. 11 (2), 11–21. doi:10.1007/s13205-020-02619-1
Lodish, H., Berk, A., Zipursky, S. L., Matsudaira, P., Baltimore, D., Darnell, J., et al. (2000). “Viruses: Structure, Function, and Uses,” in Molecular Cell Biology (New York: W. H. Freeman).
Loya, S., Reshef, V., Mizrachi, E., Silberstein, C., Rachamim, Y., Carmeli, S., et al. (1998). The Inhibition of the Reverse Transcriptase of HIV-1 by the Natural Sulfoglycolipids from Cyanobacteria: Contribution of Different Moieties to Their High Potency. J. Nat. Prod. 61 (7), 891–895. doi:10.1021/np970585j
Luo, M., Zang, R., Wang, X., Chen, Z., Song, X., Ju, J., et al. (2019). Natural Hydroxamate-Containing Siderophore Acremonpeptides A–D and an Aluminum Complex of Acremonpeptide D from the marine-derived Acremonium Persicinum SCSIO 115. J. Nat. Prod. 82 (9), 2594–2600. doi:10.1021/acs.jnatprod.9b00545
Lv, X., Wu, Y., Tian, R., Gu, Y., Liu, Y., Li, J., et al. (2020). Synthetic Metabolic Channel by Functional Membrane Microdomains for Compartmentalized Flux Control. Metab. Eng. 59, 106–118. doi:10.1016/j.ymben.2020.02.003
Lyu, X., Zhao, G., Ng, K. R., Mark, R., and Chen, W. N. (2019). Metabolic Engineering of Saccharomyces cerevisiae for De Novo Production of Kaempferol. J. Agric. Food Chem. 67 (19), 5596–5606. doi:10.1021/acs.jafc.9b01329
Ma, J., Gu, Y., and Xu, P. (2020). A Roadmap to Engineering Antiviral Natural Products Synthesis in Microbes. Curr. Opin. Biotechnol. 66, 140–149. doi:10.1016/j.copbio.2020.07.008
Ma, X., Nong, X.-H., Ren, Z., Wang, J., Liang, X., Wang, L., et al. (2017). Antiviral Peptides from marine Gorgonian-Derived Fungus Aspergillus Sp. SCSIO 41501. Tetrahedron Lett. 58 (12), 1151–1155. doi:10.1016/j.tetlet.2017.02.005
Magnusson, S., Gundersen, K., Brandberg, A., and Lycke, E. (1967). Marine Bacteria and Their Possible Relation to the Virus Inactivation Capacity of Sea Water. Acta Pathol. Microbiol. Scand. 71 (2), 274–280. doi:10.1111/j.1699-0463.1967.tb05164.x
Manimaran, M., Rajkumar, T., Vimal, S., Taju, G., Abdul Majeed, S., Sahul Hameed, A. S., et al. (2018). Antiviral Activity of 9(10H)-Acridanone Extracted from marine Streptomyces Fradiae Strain VITMK2 in Litopenaeus Vannamei Infected with white Spot Syndrome Virus. Aquaculture 488, 66–73. doi:10.1016/j.aquaculture.2018.01.032
Martinez, J. P., Hinkelmann, B., Fleta-Soriano, E., Steinmetz, H., Jansen, R., Diez, J., et al. (2013). Identification of Myxobacteria-Derived HIV Inhibitors by a High-Throughput Two-step Infectivity Assay. Microb. Cell Fact. 12 (1), 1–9. doi:10.1186/1475-2859-12-85
Martinez, J., Sasse, F., Brönstrup, M., Diez, J., and Meyerhans, A. (2015). Antiviral Drug Discovery: Broad-Spectrum Drugs from Nature. Nat. Prod. Rep. 32 (1), 29–48. doi:10.1039/C4NP00085D
Matsuda, M., Shigeta, S., and Okutani, K. (1999). Antiviral Activities of marine pseudomonas Polysaccharides and Their Oversulfated Derivatives. Mar. Biotechnol. 1 (1), 68–73. doi:10.1007/pl00011753
Matsuzaki, K., Ikeda, H., Masuma, R., Tanaka, H., and Omura, S. (1995). Isochromophilones I and Ii, Novel Inhibitors against Gp120-Cd4 Binding Produced by Penidllium Multicolor Fo-2338 I. Screening, Taxonomy, Fermentation, Isolation and Biological Activity. J. Antibiot. 48 (7), 703–707. doi:10.7164/antibiotics.48.703
Meganck, R. M., and Baric, R. S. (2021). Developing Therapeutic Approaches for Twenty-First-century Emerging Infectious Viral Diseases. Nat. Med. 27 (3), 401–410. doi:10.1038/s41591-021-01282-0
Milewska, A., Kula-Pacurar, A., Wadas, J., Suder, A., Szczepanski, A., Dabrowska, A., et al. (2020). Replication of SARS-CoV-2 in Human Respiratory Epithelium. J. Virol. 94 (15), e00957–20. doi:10.1128/JVI.00957-20
Millet, J. K., and Whittaker, G. R. (2018). Physiological and Molecular Triggers for SARS-CoV Membrane Fusion and Entry into Host Cells. Virology 517, 3–8. doi:10.1016/j.virol.2017.12.015
Minagawa, K., Kouzuki, S., Yoshimoto, J., Kawamura, Y., Tani, H., Iwata, T., et al. (2002). Stachyflin and Acetylstachyflin, Novel Anti-influenza A Virus Substances, Produced by Stachybotrys Sp. RF-7260 I. Isolation, Structure Elucidation and Biological Activities. J. Antibiot. 55 (2), 155–164. doi:10.7164/antibiotics.55.155
Molla, A., Hellen, C. U., and Wimmer, E. (1993). Inhibition of Proteolytic Activity of Poliovirus and Rhinovirus 2A Proteinases by Elastase-specific Inhibitors. J. Virol. 67 (8), 4688–4695. doi:10.1128/jvi.67.8.4688-4695.1993
Mukherjee, P. K. (2019). Antiviral Evaluation of Herbal Drugs. Quality Control and Evaluation of Herbal Drugs. Elsevier, 599–628. doi:10.1016/B978-0-12-813374-3.00016-8
Mulwa, L. S., Jansen, R., Praditya, D. F., Mohr, K. I., Wink, J., Steinmann, E., et al. (2018). Six Heterocyclic Metabolites from the Myxobacterium Labilithrix Luteola. Molecules 23, 542. doi:10.3390/molecules23030542
Mulwa, L. S., and Stadler, M. (2018). Antiviral Compounds from Myxobacteria. Microorganisms 6, 73. doi:10.3390/microorganisms6030073
Nakamura, H., Yagisawa, N., Shimada, N., Takita, T., Umezawa, H., Iitaka, Y., et al. (1981). The X-ray Structure Determination of Oxanosine. J. Antibiot. 34 (9), 1219–1221. doi:10.7164/antibiotics.34.1219
Nakamura, M., Ohno, T., Kunimoto, S., Naganawa, H., and Takeuchi, T. (1991). Kijimicin: an Inhibitor of Human Immunodeficiency Virus in Acutely and Chronically Infected Cells. J. Antibiot. 44 (5), 569–571. doi:10.7164/antibiotics.44.569
Naruse, N., Tenmyo, O., Tomita, K., Konishi, M., Miyaki, T., Kawaguchi, H., et al. (1989). Lanthiopeptin, a New Peptide Antibiotic. J. Antibiot. 42 (6), 837–845. doi:10.7164/antibiotics.42.837
Nazaruk, J., and Borzym-Kluczyk, M. (2015). The Role of Triterpenes in the Management of Diabetes Mellitus and its Complications. Phytochem. Rev. 14 (4), 675–690. doi:10.1007/s11101-014-9369-x
Neiderud, C.-J. (2015). How Urbanization Affects the Epidemiology of Emerging Infectious Diseases. Infect. Ecol. Epid. 5 (1), 27060. doi:10.3402/iee.v5.27060
Niu, S., Liu, D., Shao, Z., Proksch, P., and Lin, W. (2017). Eutypellazines A–M, Thiodiketopiperazine-type Alkaloids from Deep Sea Derived Fungus Eutypella Sp. MCCC 3A00281. RSC Adv. 7, 33580–33590. doi:10.1039/C7RA05774A
Niu, S., Si, L., Liu, D., Zhou, A., Zhang, Z., Shao, Z., et al. (2016). Spiromastilactones: A New Class of Influenza Virus Inhibitors from Deep-Sea Fungus. Eur. J. Med. Chem. 108, 229–244. doi:10.1016/j.ejmech.2015.09.037
Nong, X.-H., Wang, Y.-F., Zhang, X.-Y., Zhou, M.-P., Xu, X.-Y., Qi, S.-H., et al. (2014). Territrem and Butyrolactone Derivatives from a marine-derived Fungus Aspergillus terreus. Mar. Drugs 12 (12), 6113–6124. doi:10.3390/md12126113
Obach, R. S., and Kalgutkar, A. (2018). “Reactive Electrophiles and Metabolic Activation,” in Comprehensive Toxicology. 3rd Edition (Elsevier), 295–331. doi:10.1016/B978-0-12-801238-3.64290-3
Ohta, S., Ono, F., Shiomi, Y., Nakao, T., Aozasa, O., Nagate, T., et al. (1998). Anti-herpes Simplex Virus Substances Produced by the marine green Alga, Dunaliella Primolecta. J. Appl. Psychol. 10, 349–356. doi:10.1023/A:1008065226194
Ondeyka, J. G., Zink, D. L., Dombrowski, A. W., Polishook, J. D., Felock, P. J., Hazuda, D. J., et al. (2003). Isolation, Structure and HIV-1 Integrase Inhibitory Activity of Exophillic Acid, a Novel Fungal Metabolite from Exophiala Pisciphila. J. Antibiot. (Tokyo) 56 (12), 1018–1023. doi:10.7164/antibiotics.56.1018
Othman, L., Sleiman, A., and Abdel-Massih, R. M. (2019). Antimicrobial Activity of Polyphenols and Alkaloids in Middle Eastern Plants. Front. Microbiol. 10, 911. doi:10.3389/fmicb.2019.00911
Ou, J., Zhou, Z., Dai, R., Zhang, J., Zhao, S., Wu, X., et al. (2020). V367F Mutation in SARS-CoV-2 Spike RBD Emerging During the Early Transmission Phase Enhances Viral Infectivity Through Increased Human ACE2 Receptor Binding Affinity. J. Virol. 95 (16). doi:10.1128/JVI.00617-21
Paddon, C. J., Westfall, P. J., Pitera, D. J., Benjamin, K., Fisher, K., Mcphee, D., et al. (2013). High-level Semi-synthetic Production of the Potent Antimalarial Artemisinin. Nature 496, 528–532. doi:10.1038/nature12051
Palmer, C. M., Miller, K. K., Nguyen, A., and Alper, H. S. (2020). Engineering 4-Coumaroyl-CoA Derived Polyketide Production in Yarrowia Lipolytica through a β-oxidation Mediated Strategy. Metab. Eng. 57, 174–181. doi:10.1016/j.ymben.2019.11.006
Pan, R., Bai, X., Chen, J., Zhang, H., and Wang, H. (2019). Exploring Structural Diversity of Microbe Secondary Metabolites Using OSMAC Strategy: a Literature Review. Front. Microbiol. 10, 294. doi:10.3389/fmicb.2019.00294
Pandey, R. P., Parajuli, P., Koffas, M. A., and Sohng, J. K. (2016). Microbial Production of Natural and Non-natural Flavonoids: Pathway Engineering, Directed Evolution and Systems/synthetic Biology. Biotechnol. Adv. 34 (5), 634–662. doi:10.1016/j.biotechadv.2016.02.012
Pang, X., Lin, X., Wang, J., Liang, R., Tian, Y., Salendra, L., et al. (2018). Three New Highly Oxygenated Sterols and One New Dihydroisocoumarin from the marine Sponge-Derived Fungus Cladosporium Sp. SCSIO41007. Steroids 129, 41–46. doi:10.1016/j.steroids.2017.12.001
Park, S. R., Yoon, Y. J., Pham, J. V., Yilma, M. A., Feliz, A., Majid, M. T., et al. (2019). A Review of the Microbial Production of Bioactive Natural Products and Biologics. Front. Microbiol. 10, 1404. doi:10.3389/fmicb.2019.01404
Patridge, E., Gareiss, P., Kinch, M. S., and Hoyer, D. (2016). An Analysis of FDA-Approved Drugs: Natural Products and Their Derivatives. Drug Discov. Today 21 (2), 204–207. doi:10.1016/j.drudis.2015.01.009
Paul, D., and Bartenschlager, R. (2013). Architecture and Biogenesis of Plus-Strand RNA Virus Replication Factories. World J. Virol. 2 (2), 32. doi:10.5501/wjv.v2.i2.32
Peiris, J. S. M., Chu, C.-M., Cheng, V. C.-C., Chan, K., Hung, I., Poon, L. L., et al. (2003). Clinical Progression and Viral Load in a Community Outbreak of Coronavirus-Associated SARS Pneumonia: a Prospective Study. The Lancet 361, 1767–1772. doi:10.1016/S0140-6736(03)13412-5
Peng, F., Hou, S.-Y., Zhang, T.-Y., Wu, Y.-Y., Zhang, M.-Y., Yan, X.-M., et al. (2019). Cytotoxic and Antimicrobial Indole Alkaloids from an Endophytic Fungus Chaetomium Sp. SYP-F7950 of Panax Notoginseng. RSC Adv. 9, 28754–28763. doi:10.1039/C9RA04747F
Peng, J., Lin, T., Wang, W., Xin, Z., Zhu, T., Gu, Q., et al. (2013). Antiviral Alkaloids Produced by the Mangrove-Derived Fungus Cladosporium Sp. PJX-41. J. Nat. Prod. 76 (6), 1133–1140. doi:10.1021/np400200k
Peng, J., Zhang, X., Du, L., Wang, W., Zhu, T., Gu, Q., et al. (2014). Sorbicatechols A and B, Antiviral Sorbicillinoids from the marine-derived Fungus Penicillium chrysogenum PJX-17. J. Nat. Prod. 77 (2), 424–428. doi:10.1021/np400977e
Pham, J. V., Yilma, M. A., Feliz, A., Majid, M. T., Maffetone, N., Walker, J. R., et al. (2019). A Review of the Microbial Production of Bioactive Natural Products and Biologics. Front. Microbiol. 10, 1404. doi:10.3389/fmicb.2019.01404
Phipps, R. K., Petersen, B. O., Christensen, K. B., Duus, J. Ø., Frisvad, J. C., Larsen, T. O., et al. (2004). Hesseltin A, a Novel Antiviral Metabolite from Penicillium Hesseltinei. Org. Lett. 6 (20), 3441–3443. doi:10.1021/ol048824z
Pittayakhajonwut, P., Suvannakad, R., Thienhirun, S., Prabpai, S., Kongsaeree, P., Tanticharoen, M., et al. (2005). An Anti-herpes Simplex Virus-type 1 Agent from Xylaria Mellisii (BCC 1005). Tetrahedron Lett. 46 (8), 1341–1344. doi:10.1016/j.tetlet.2004.12.110
Plaza, A., Garcia, R., Bifulco, G., Martinez, J. P., Hütel, S., Sasse, F., et al. (2012). Aetheramides A and B, Potent HIV-Inhibitory Depsipeptides from a Myxobacterium of the New Genus “Aetherobacter”. Org. Lett. 14 (11), 2854–2857. doi:10.1021/ol3011002
Porotto, M., Yokoyama, C. C., Palermo, L. M., Mungall, B., Aljofan, M., Cortese, R., et al. (2010). Viral Entry Inhibitors Targeted to the Membrane Site of Action. J. Virol. 84 (13), 6760–6768. doi:10.1128/JVI.00135-10
Prasanth, D., Murahari, M., Chandramohan, V., Bhavya, G., Lakshmana Rao, A., Panda, S. P., et al. (2021). In-silico Strategies of Some Selected Phytoconstituents from Melissa Officinalis as SARS CoV-2 Main Protease and Spike Protein (COVID-19) Inhibitors. Mol. Simul 2021, 1–14. doi:10.1080/08927022.2021.1880576
Prasanth, D., Murahari, M., Chandramohan, V., Panda, S. P., Atmakuri, L. R., Guntupalli, C., et al. (2020). In Silico identification of Potential Inhibitors from Cinnamon against Main Protease and Spike Glycoprotein of SARS CoV-2. J. Biomol. Struct. Dyn. 39 (13), 4618–4632. doi:10.1080/07391102.2020.1779129
Puttaswamy, H., Gowtham, H. G., Ojha, M. D., Yadav, A., Choudhir, G., Raguraman, V., et al. (2020). In Silico studies Evidenced the Role of Structurally Diverse Plant Secondary Metabolites in Reducing SARS-CoV-2 Pathogenesis. Sci. Rep. 10 (1), 1–24. doi:10.1038/s41598-020-77602-0
Qin, C., Lin, X., Lu, X., Wan, J., Zhou, X., Liao, S., et al. (2015). Sesquiterpenoids and Xanthones Derivatives Produced by Sponge-Derived Fungus Stachybotry Sp. HH1 ZSDS1F1-2. J. Antibiot. 68 (2), 121–125. doi:10.1038/ja.2014.97
Radonić, A., Thulke, S., Achenbach, J., Kurth, A., Vreemann, A., König, T., et al. (2011). Anionic Polysaccharides from Phototrophic Microorganisms Exhibit Antiviral Activities to Vaccinia Virus. J. Antivir. Antiretrovir. 2, 51–55. doi:10.4172/jaa.1000023
Raekiansyah, M., Mori, M., Nonaka, K., Agoh, M., Shiomi, K., Matsumoto, A., et al. (2017). Identification of Novel Antiviral of Fungus-Derived Brefeldin A against Dengue Viruses. Trop. Med. Int. Health 45 (1), 1–7. doi:10.1186/s41182-017-0072-7
Rahman, M., Hoque, M., Islam, R., Akter, S., Rubayet-Ul-Alam, A. S. M., Siddique, M., et al. (2020). Epitope-based Chimeric Peptide Vaccine Design against S, M and E Proteins of SARS-CoV-2 Etiologic Agent of Global Pandemic COVID-19: an In Silico Approach. Peer J. 8, e9572. doi:10.7717/peerj.9572
Raihan, T., Azad, A. K., Ahmed, J., Shepon, M. R., Dey, P., Chowdhury, N., et al. (2021). Extracellular Metabolites of Endophytic Fungi from Azadirachta indica Inhibit Multidrug-Resistant Bacteria and Phytopathogens. Future Microbiol. 16, 557–576. doi:10.2217/fmb-2020-0259
Raj, V. S., Mou, H., Smits, S. L., Dekkers, D. H., Müller, M. A., Dijkman, R., et al. (2013). Dipeptidyl Peptidase 4 Is a Functional Receptor for the Emerging Human Coronavirus-EMC. Nature 495, 251–254. doi:10.1038/nature12005
Raveh, A., Delekta, P. C., Dobry, C. J., Peng, W., Schultz, P. J., Blakely, P. K., et al. (2013). Discovery of Potent Broad Spectrum Antivirals Derived from marine Actinobacteria. PLoS One 8, e82318. doi:10.1371/journal.pone.0082318
Richards, A. D., Roberts, R., Dunn, B. M., Graves, M. C., and Kay, J. (1989). Effective Blocking of HIV-1 Proteinase Activity by Characteristic Inhibitors of Aspartic Proteinases. FEBS Lett. 247 (1), 113–117. doi:10.1016/0014-5793(89)81251-7
Risco, C., De Castro, I. F., Sanz-Sánchez, L., Narayan, K., Grandinetti, G., Subramaniam, S., et al. (2014). Three-dimensional Imaging of Viral Infections. Annu. Rev. Virol. 1, 453–473. doi:10.1146/annurev-virology-031413-085351
Roa-Linares, V. C., Miranda-Brand, Y., Tangarife-Castaño, V., Ochoa, R., García, P. A., Castro, M., et al. (2019). Anti-herpetic, Anti-dengue and Antineoplastic Activities of Simple and Heterocycle-Fused Derivatives of Terpenyl-1, 4-naphthoquinone and 1, 4-anthraquinone. Molecules 24, 1279. doi:10.3390/molecules24071279
Roberts, N. A., Martin, J. A., Kinchington, D., Broadhurst, A. V., Craig, J. C., Duncan, I. B., et al. (1990). Rational Design of Peptide-Based HIV Proteinase Inhibitors. Science 248, 358–361. doi:10.1126/science.2183354
Rochfort, S., Ford, J., Ovenden, S., Wan, S. S., George, S., Wildman, H., et al. (2005). A Novel Aspochalasin with HIV-1 Integrase Inhibitory Activity from Aspergillus flavipes. J. Antibiot. 58 (4), 279–283. doi:10.1038/ja.2005.34
Rodriguez, A., Strucko, T., Stahlhut, S. G., Kristensen, M., Svenssen, D. K., Forster, J., et al. (2017). Metabolic Engineering of Yeast for Fermentative Production of Flavonoids. Bioresour. Technol. 245, 1645–1654. doi:10.1016/j.biortech.2017.06.043
Rowley, D. C., Kelly, S., Jensen, P., and Fenical, W. (2004). Synthesis and Structure–Activity Relationships of the Halovirs, Antiviral Natural Products from a marine-derived Fungus. Bioorg. Med. Chem. 12 (18), 4929–4936. doi:10.1016/j.bmc.2004.06.044
Ryu, W.-S. (2017). “Virus Life Cycle,” in Molecular Virology of Human Pathogenic Viruses (Elsevier), 31–45. doi:10.1016/B978-0-12-800838-6.00003-5
Sadahiro, Y., Kato, H., Williams, R. M., and Tsukamoto, S. (2020). Irpexine, an Isoindolinone Alkaloid Produced by Coculture of Endophytic Fungi, Irpex Lacteus and Phaeosphaeria Oryzae. J. Nat. Prod. 83 (5), 1368–1373. doi:10.1021/acs.jnatprod.0c00047
Saha, S., Navid, M. H., Bandyopadhyay, S. S., Schnitzler, P., and Ray, B. (2012). Sulfated Polysaccharides from Laminaria Angustata: Structural Features and In Vitro Antiviral Activities. Carbohydr. Polym. 87 (1), 123–130. doi:10.1016/j.carbpol.2011.07.026
Sakamoto, H., Okamoto, K., Aoki, M., Kato, H., Katsume, A., Ohta, A., et al. (2005). Host Sphingolipid Biosynthesis as a Target for Hepatitis C Virus Therapy. Nat. Chem. Biol. 1 (6), 333–337. doi:10.1038/nchembio742
Sankar, M., Ramachandran, B., Pandi, B., Mutharasappan, N., Ramasamy, V., Prabu, P. G., et al. (2021). In Silico screening of Natural Phytocompounds towards Identification of Potential lead Compounds to Treat COVID-19. Front. Mol. Bio 8, 637122. doi:10.3389/fmolb.2021.637122
Santoyo, S., Jaime, L., Plaza, M., Herrero, M., Rodriguez-Meizoso, I., Ibañez, E., et al. (2012). Antiviral Compounds Obtained from Microalgae Commonly Used as Carotenoid Sources. J. Appl. Phycol. 24, 731–741. doi:10.1007/s10811-011-9692-1
Santoyo, S., Plaza, M., Jaime, L., Ibanez, E., Reglero, G., Senorans, F. J., et al. (2010). Pressurized Liquid Extraction as an Alternative Process to Obtain Antiviral Agents from the Edible Microalga Chlorella Vulgaris. J. Appl. Phycol. 24 (4), 731–741. doi:10.1007/s10811-011-9692-1
Sato, Y., Okuyama, S., and Hori, K. (2007). Primary Structure and Carbohydrate Binding Specificity of a Potent Anti-HIV Lectin Isolated from the Filamentous Cyanobacterium Oscillatoria Agardhii. J. Biol. Chem. 282 (15), 11021–11029. doi:10.1074/jbc.M701252200
Sawa, T., Fukagawa, Y., Homma, I., Takeuchi, T., and Umezawa, H. (1967). Mode of Inhibition of Coformycin on Adenosine Deaminase. J. Antibiot. (Tokyo) 20, 227–231.
Sayed, A. M., Alhadrami, H. A., El-Gendy, A. O., Shamikh, Y. I., Belbahri, L., Hassan, H. M., et al. (2020). Microbial Natural Products as Potential Inhibitors of SARS-CoV-2 Main Protease (Mpro). Microorganisms 8, 970. doi:10.3390/microorganisms8070970
Schulz, B., Boyle, C., Draeger, S., Römmert, A.-K., and Krohn, K. (2002). Endophytic Fungi: a Source of Novel Biologically Active Secondary Metabolites. Mycol. Res. 106 (9), 996–1004. doi:10.1017/S0953756202006342
Sekita, S. (1983). Isocochliodinol and Neocochliodinol, Bis (3-Indolyl)-Benzoquinones from Chaetomium Spp. Chem. Pharm. Bull. 31 (9), 2998–3001. doi:10.1248/cpb.31.2998
Selim, K. A., Elkhateeb, W. A., Tawila, A. M., El-Beih, A. A., Abdel-Rahman, T. M., El-Diwany, A. I., et al. (2018). Antiviral and Antioxidant Potential of Fungal Endophytes of Egyptian Medicinal Plants. Fermentation 4, 49. doi:10.3390/fermentation4030049
Sharaf, M., Amara, A., Aboul-Enein, A., Helmi, S., Ballot, A., Astani, A., et al. (2010). Molecular Authentication and Characterization of the Antiherpetic Activity of the Cyanobacterium Arthrospira Fusiformis. Pharmazie 65, 132–136.
Shih, S. R., Tsai, K. N., Li, Y. S., Chueh, C. C., and Chan, E. C. (2003). Inhibition of Enterovirus 71-induced Apoptosis by Allophycocyanin Isolated from a Blue-green Alga Spirulina Platensis. J. Med. Virol. 70, 119–125. doi:10.1002/jmv.10363
Shimada, N., Yagisawa, N., Naganawa, H., Takita, T., Hamada, M., Takeuchi, T., et al. (1981). Oxanosine, a Novel Nucleoside from Actinomycetes. J. Antibiot. 34 (9), 1216–1218. doi:10.7164/antibiotics.34.1216
Shushni, M. A., Mentel, R., Lindequist, U., and Jansen, R. (2009). Balticols A–F, New Naphthalenone Derivatives with Antiviral Activity, from an Ascomycetous Fungus. Chem. Biodivers. 6 (2), 127–137. doi:10.1002/cbdv.200800150
Shushni, M. A., Singh, R., Mentel, R., and Lindequist, U. (2011). Balticolid: a New 12-membered Macrolide with Antiviral Activity from an Ascomycetous Fungus of marine Origin. Mar. Drugs 9 (5), 844–851. doi:10.3390/md9050844
Silva, T., Hamerski, L., Walter, J., Siqueira, J. E., Santos, A., Santos, J. a. M., et al. (2018). Inhibitory Effect of Microalgae and Cyanobacteria Extracts on Influenza Virus Replication and Neuraminidase Activity. Peer J. 6, e5716. doi:10.7717/peerj.5716
Sims, J. W., Fillmore, J. P., Warner, D. D., and Schmidt, E. W. (2005). Equisetin Biosynthesis in Fusarium heterosporum. Chem. Commun. 2, 186–188. doi:10.1039/B413523G
Singh, R., Kumar, M., Mittal, A., and Mehta, P. K. (2017). Microbial Metabolites in Nutrition, Healthcare and Agriculture. 3 Biotech. 7, 15. doi:10.1007/s13205-016-0586-4
Singh, S. B., Jayasuriya, H., Dewey, R., Polishook, J. D., Dombrowski, A. W., Zink, D. L., et al. (2003a). Isolation, Structure, and HIV-1-Integrase Inhibitory Activity of Structurally Diverse Fungal Metabolites. J. Ind. Microbiol. Biotechnol. 30 (12), 721–731. doi:10.1007/s10295-003-0101-x
Singh, S. B., Ondeyka, J. G., Tsipouras, N., Ruby, C., Sardana, V., Schulman, M., et al. (2004). Hinnuliquinone, a C2-Symmetric Dimeric Non-peptide Fungal Metabolite Inhibitor of HIV-1 Protease. Biochem. Biophys. Res. Commun. 324 (1), 108–113. doi:10.1016/j.bbrc.2004.08.234
Singh, S. B., Zink, D. L., Bills, G. F., Teran, A., Silverman, K. C., Lingham, R. B., et al. (2003b). Four Novel Bis-(naphtho-γ-Pyrones) Isolated from Fusarium Species as Inhibitors of HIV-1 Integrase. Bioorg. Med. Chem. Lett. 13 (4), 713–717. doi:10.1016/S0960-894X(02)01057-0
Singh, S. B., Zink, D., Polishook, J., Valentino, D., Shafiee, A., Silverman, K., et al. (1999). Structure and Absolute Stereochemistry of HIV-1 Integrase Inhibitor Integric Acid. A Novel Eremophilane Sesquiterpenoid Produced by a Xylaria Sp. Tetrahedron Lett. 40 (50), 8775–8779. doi:10.1016/S0040-4039(99)01878-X
Stack, M. E., Mazzola, E. P., Page, S. W., Pohland, A. E., Highet, R. J., Tempesta, M. S., et al. (1986). Mutagenic Perylenequinone Metabolites of Alternaria alternata: Altertoxins I, II, and III. J. Nat. Prod. 49 (5), 866–871. doi:10.1021/np50047a017
Stebbing, J., Krishnan, V., De Bono, S., Ottaviani, S., Casalini, G., Richardson, P. J., et al. (2020). Mechanism of Baricitinib Supports Artificial Intelligence Predicted Testing in COVID 19 Patients. EMBO Mol. Med. 12 (8), e12697. doi:10.15252/emmm.202012697
Steed, A. L., Christophi, G. P., Kaiko, G. E., Sun, L., Goodwin, V. M., Jain, U., et al. (2017). The Microbial Metabolite Desaminotyrosine Protects from Influenza through Type I Interferon. Science 357, 498–502. doi:10.1126/science.aam5336
Stierle, A., Strobel, G., and Stierle, D. (1993). Taxol and Taxane Production by Taxomyces Andreanae, an Endophytic Fungus of Pacific Yew. Science 260, 214–216. doi:10.1126/science.8097061
Strand, M., Carlsson, M., Uvell, H., Islam, K., Edlund, K., Cullman, I., et al. (2014). Isolation and Characterization of Anti-adenoviral Secondary Metabolites from marine Actinobacteria. Mar. Drugs 12 (2), 799–821. doi:10.3390/md12020799
Sun, J., Zhang, C., Nan, W., Li, D., Ke, D., Lu, W., et al. (2019). Glycerol Improves Heterologous Biosynthesis of Betulinic Acid in Engineered Yarrowia Lipolytica. Chem. Eng. Sci. 196, 82–90. doi:10.1016/j.ces.2018.10.052
Sun, X., Shen, X., Jain, R., Lin, Y., Wang, J., Sun, J., et al. (2015a). Synthesis of Chemicals by Metabolic Engineering of Microbes. Chem. Soc. Rev. 44 (11), 3760–3785. doi:10.1039/C5CS00159E
Sun, Y.-L., Wang, J., Wang, Y.-F., Zhang, X.-Y., Nong, X.-H., Chen, M.-Y., et al. (2015b). Cytotoxic and Antiviral Tetramic Acid Derivatives from the Deep-Sea-Derived Fungus Trichobotrys Effuse DFFSCS021. Tetrahedron 71, 9328–9332. doi:10.1016/j.tet.2015.10.010
Takeuchi, T., Iwanaga, J., Aoyagi, T., and Umezawa, H. (1966). Antiviral Effect of Formycin and Formycin B. J. Antibiot. (Tokyo) 19, 286–287.
Takizawa, N., and Yamasaki, M. (2018). Current Landscape and Future Prospects of Antiviral Drugs Derived from Microbial Products. J. Antibiot. 71 (1), 45–52. doi:10.1038/ja.2017.115
Talyshinsky, M. M., Souprun, Y. Y., and Huleihel, M. M. (2002). Anti-viral Activity of Red Microalgal Polysaccharides against Retroviruses. Cancer Cell Int 2 (1), 1–7. doi:10.1186/1475-2867-2-8
Tanaka, A., Sen, K., Morita, J., and Komano, T. (1983). Phage Inactivation by Aclacinomycin A and its Analogues. J. Antibiot. 36 (9), 1242–1244. doi:10.7164/antibiotics.36.1242
Tao, S., Qian, Y., Wang, X., Cao, W., Ma, W., Chen, K., et al. (2018). Regulation of ATP Levels in Escherichia coli Using CRISPR Interference for Enhanced Pinocembrin Production. Microb. Cell Fact. 17 (1), 1–13. doi:10.1186/s12934-018-0995-7
Tapparel, C., Sobo, K., Constant, S., Huang, S., Van Belle, S., Kaiser, L., et al. (2013). Growth and Characterization of Different Human Rhinovirus C Types in Three-Dimensional Human Airway Epithelia Reconstituted In Vitro. Virology 446, 1–8. doi:10.1016/j.virol.2013.06.031
Tarnow, C., Engels, G., Arendt, A., Schwalm, F., Sediri, H., Preuss, A., et al. (2014). TMPRSS2 Is a Host Factor that Is Essential for Pneumotropism and Pathogenicity of H7N9 Influenza A Virus in Mice. J. Virol. 88 (9), 4744–4751. doi:10.1128/JVI.03799-13
Tatsis, E. C., and O'connor, S. E. (2016). New Developments in Engineering Plant Metabolic Pathways. Curr. Opin. Biotechnol. 42, 126–132. doi:10.1016/j.copbio.2016.04.012
Teng, Y.-F., Xu, L., Wei, M.-Y., Wang, C.-Y., Gu, Y.-C., Shao, C.-L., et al. (2020). Recent Progresses in marine Microbial-Derived Antiviral Natural Products. Arch. Pharm. Res. 1-15. doi:10.1007/s12272-020-01286-3
Thaker, S. K., Ch’ng, J., and Christofk, H. R. (2019). Viral Hijacking of Cellular Metabolism. BMC Biol. 17 (1), 1–15. doi:10.1186/s12915-019-0678-9
Thawabteh, A., Juma, S., Bader, M., Karaman, D., Scrano, L., Bufo, S. A., et al. (2019). The Biological Activity of Natural Alkaloids against Herbivores, Cancerous Cells and Pathogens. Toxins 11, 656. doi:10.3390/toxins11110656
Tian, Y.-Q., Lin, X.-P., Wang, Z., Zhou, X.-F., Qin, X.-C., Kaliyaperumal, K., et al. (2016). Asteltoxins with Antiviral Activities from the marine Sponge-Derived Fungus Aspergillus Sp. SCSIO Xws02f40. Mol. 21, 34. doi:10.3390/molecules21010034
Tiwari, S. K., Dicks, L. M., Popov, I. V., Karaseva, A., Ermakov, A. M., Suvorov, A., et al. (2020). Probiotics at War against Viruses: what Is Missing from the Picture? Front. Microb. 11, 1877. doi:10.3389/fmicb.2020.01877
Tong, J., Trapido-Rosenthal, H., Wang, J., Wang, Y., Li, Q. X., Lu, Y., et al. (2012). Antiviral Activities and Putative Identification of Compounds in Microbial Extracts from the Hawaiian Coastal Waters. Mar. Drugs 10 (3), 521–538. doi:10.3390/md10030521
Toranzo, A. E., Barja, J. L., and Hetrick, F. M. (1982). Antiviral Activity of Antibiotic-Producing marine Bacteria. Can. J. Microbiol. 28 (2), 231–238. doi:10.1139/m82-031
Tsunakawa, M., Tenmyo, O., Tomita, K., Naruse, N., Kotake, C., Miyaki, T., et al. (1992). Quartromicin, a Complex of Novel Antiviral Antibiotics Production, Isolation, Physico-Chemical Properties and Antiviral Activity. J. Antibiot. 45 (2), 180–188. doi:10.7164/antibiotics.45.180
Umezawa, H., Aoyagi, T., Komiyama, T., Morishima, H., and Hamada, M. (1974). Purification and Characterization of a Sialidase Inhibitor, Siastatin, Produced by Streptomyces. J. Antibiot. (Tokyo) 27, 963–969. doi:10.7164/antibiotics.27.963
Vigant, F., Santos, N. C., and Lee, B. (2015). Broad-spectrum Antivirals against Viral Fusion. Nat. Rev. Microbiol. 13 (7), 426–437. doi:10.1038/nrmicro3475
Waites, M. J., Morgan, N. L., Rockey, J. S., and Higton, G. (2009). Industrial Microbiology: An Introduction. John Wiley & Sons.
Walls, A. C., Park, Y.-J., Tortorici, M. A., Wall, A., Mcguire, A. T., Veesler, D., et al. (2020). Structure, Function, and Antigenicity of the SARS-CoV-2 Spike Glycoprotein. Cell 181 (2), 281–292. e6. doi:10.1016/j.cell.2020.02.058
Wang, C., Liwei, M., Park, J.-B., Jeong, S.-H., Wei, G., Wang, Y., et al. (2018). Microbial Platform for Terpenoid Production: Escherichia coli and Yeast. Front. Microbiol. 9, 2460. doi:10.3389/fmicb.2018.02460
Wang, C., Su, X., Sun, M., Zhang, M., Wu, J., Xing, J., et al. (2019). Efficient Production of Glycyrrhetinic Acid in Metabolically Engineered Saccharomyces cerevisiae via an Integrated Strategy. Microb. Cell Fact. 18 (1), 1–15. doi:10.1186/s12934-019-1138-5
Wang, H., Wang, Y., Wang, W., Fu, P., Liu, P., Zhu, W., et al. (2011). Anti-influenza Virus Polyketides from the Acid-Tolerant Fungus Penicillium purpurogenum JS03-21. Nat. Prod. 74 (9), 2014–2018. doi:10.1021/np2004769
Wang, J.-F., Liang, R., Liao, S.-R., Yang, B., Tu, Z.-C., Lin, X.-P., et al. (2017). Vaccinols J–S, Ten New Salicyloid Derivatives from the marine Mangrove-Derived Endophytic Fungus Pestalotiopsis Vaccinii. Fitoterapia 120, 164–170. doi:10.1016/j.fitote.2017.06.013
Wang, J.-F., Lin, X.-P., Qin, C., Liao, S.-R., Wan, J.-T., Zhang, T.-Y., et al. (2014). Antimicrobial and Antiviral Sesquiterpenoids from Sponge-Associated Fungus, Aspergillus sydowii ZSDS1-F6. J. Antibiot. 67 (8), 581–583. doi:10.1038/ja.2014.39
Wang, J., Wei, X., Qin, X., Tian, X., Liao, L., Li, K., et al. (2016). Antiviral Merosesquiterpenoids Produced by the Antarctic Fungus Aspergillus ochraceopetaliformis SCSIO 05702. J. Nat. Prod. 79 (1), 59–65. doi:10.1021/acs.jnatprod.5b00650
Wang, P., Xi, L., Liu, P., Wang, Y., Wang, W., Huang, Y., et al. (2013). Diketopiperazine Derivatives from the marine-derived Actinomycete Streptomyces Sp. FXJ7.328. Mar. Drugs 11, 1035–1049. doi:10.3390/md11041035
Watashi, K., Hijikata, M., Hosaka, M., Yamaji, M., and Shimotohno, K. (2003). Cyclosporin A Suppresses Replication of Hepatitis C Virus Genome in Cultured Hepatocytes. Hepatology 38, 1282–1288. doi:10.1053/jhep.2003.50449
Widagdo, W., Okba, N. M., Raj, V. S., and Haagmans, B. L. (2017). MERS-coronavirus: From Discovery to Intervention. One Health 3, 11–16. doi:10.1016/j.onehlt.2016.12.001
Wink, M. (2020). Potential of DNA Intercalating Alkaloids and Other Plant Secondary Metabolites against SARS-CoV-2 Causing COVID-19. Diversity 12, 175. doi:10.3390/d12050175
Wishart, D. S. (2016). Emerging Applications of Metabolomics in Drug Discovery and Precision Medicine. Nat. Rev. Drug Discov. 15 (7), 473. doi:10.1038/nrd.2016.32
Wu, D.-L., Li, H.-J., Smith, D. R., Jaratsittisin, J., Xia-Ke-Er, X.-F.-K.-T., Ma, W.-Z., et al. (2018). Polyketides and Alkaloids from the marine-derived Fungus Dichotomomyces Cejpii F31-1 and the Antiviral Activity of Scequinadoline A against Dengue Virus. Mar. Drugs 16 (7), 229. doi:10.3390/md16070229
Wu, G., Sun, X., Yu, G., Wang, W., Zhu, T., Gu, Q., et al. (2014). Cladosins A–E, Hybrid Polyketides from a Deep-Sea-Derived Fungus, Cladosporium Sphaerospermum. J. Nat. Prod. 77 (2), 270–275. doi:10.1021/np400833x
Wu, H., Guo, Y., Chen, L., Chen, G., and Liang, Z. (2019). A Novel Strategy to Regulate 1-deoxynojirimycin Production Based on its Biosynthetic Pathway in Streptomyces Lavendulae. Front. Microbiol. 10, 1968. doi:10.3389/fmicb.2019.01968
Xia, X. (2017). Bioinformatics and Drug Discovery. Curr. Top. Med. Chem. 17 (15), 1709–1726. doi:10.2174/1568026617666161116143440
Xu, L.-L., Han, T., Wu, J.-Z., Zhang, Q.-Y., Zhang, H., Huang, B.-K., et al. (2009). Comparative Research of Chemical Constituents, Antifungal and Antitumor Properties of Ether Extracts of Panax Ginseng and its Endophytic Fungus. Phytomedicine 16, 609–616. doi:10.1016/j.phymed.2009.03.014
Yamada, Y., Kuzuyama, T., Komatsu, M., Shin-Ya, K., Omura, S., Cane, D. E., et al. (2015). Terpene Synthases Are Widely Distributed in Bacteria. Proc. Natl. Acad. Sci. 112 (3), 857–862. doi:10.1073/pnas.1422108112
Yan, R., Zhang, Y., Li, Y., Xia, L., Guo, Y., and Zhou, Q. (2020). Structural Basis for the Recognition of SARS-CoV-2 by Full-Length Human ACE2. Science 367, 1444–1448. doi:10.1126/science.abb2762
Yang, D., Park, S. Y., Park, Y. S., Eun, H., and Lee, S. Y. (2020a). Metabolic Engineering of Escherichia coli for Natural Product Biosynthesis. Trends Biotechnol. 38 (7), 745–765. doi:10.1016/j.tibtech.2019.11.007
Yang, J.-W., Yang, L., Luo, R.-G., and Xu, J.-F. (2020b). Corticosteroid Administration for Viral Pneumonia: COVID-19 and beyond. Clin. Microbiol. Infect. 26 (9), 1171–1177. doi:10.1016/j.cmi.2020.06.020
Yang, J., Liang, J., Shao, L., Liu, L., Gao, K., Zhang, J.-L., et al. (2020c). Green Production of Silybin and Isosilybin by Merging Metabolic Engineering Approaches and Enzymatic Catalysis. Metab. Eng. 59, 44–52. doi:10.1016/j.ymben.2020.01.007
Yang, L. Y., Lin, J., Zhou, B., Liu, Y. G., and Zhu, B. Q. (2016). H1-A, a Compound Isolated from Fusarium Oxysporum Inhibits Hepatitis C Virus (HCV) NS3 Serine Protease. Chin. J. Nat. Med. 14, 299–302. doi:10.1016/S1875-5364(16)30031-0
Yang, S.-Q., Li, X.-M., Li, X., Li, H.-L., Meng, L.-H., Wang, B.-G., et al. (2018). New Citrinin Analogues Produced by Coculture of the marine Algal-Derived Endophytic Fungal Strains Aspergillus sydowii EN-534 and Penicillium citrinum EN-535. Phytochem. Lett. 25, 191–195. doi:10.1016/j.phytol.2018.04.023
Yang, S.-W., Mierzwa, R., Terracciano, J., Patel, M., Gullo, V., Wagner, N., et al. (2006). Chemokine Receptor CCR-5 Inhibitors Produced by Chaetomium globosum. J. Nat. Prod. 69 (7), 1025–1028. doi:10.1021/np060121y
Yasuhara-Bell, J., Yang, Y., Barlow, R., Trapido-Rosenthal, H., and Lu, Y. (2010a). In Vitro evaluation of marine-microorganism Extracts for Anti-viral Activity. Virol. J. 7 (1), 1–11. doi:10.1186/1743-422X-7-182
Yi, M., Lin, S., Zhang, B., Jin, H., and Ding, L. (2020). Antiviral Potential of Natural Products from marine Microbes. Eur. J. Med. Chem. 1, 112790. doi:10.1016/j.ejmech.2020.112790
Yim, J. H., Kim, S. J., Ahn, S. H., Lee, C. K., Rhie, K. T., Lee, H. K., et al. (2004). Antiviral Effects of Sulfated Exopolysaccharide from the marine Microalga Gyrodinium Impudicum Strain KG03. Mar. Biotechnol. 6 (1), 17–25. doi:10.1007/s10126-003-0002-z
Yoganathan, K., Rossant, C., Ng, S., Huang, Y., Butler, M. S., Buss, A. D., et al. (2003). 10-Methoxydihydrofuscin, Fuscinarin, and Fuscin, Novel Antagonists of the Human CCR5 Receptor from Oidiodendron Griseum. J. Nat. Prod. 66 (8), 1116–1117. doi:10.1021/np030146m
Yu, G., Zhou, G., Zhu, M., Wang, W., Zhu, T., Gu, Q., et al. (2016). Neosartoryadins A and B, Fumiquinazoline Alkaloids from a Mangrove-Derived Fungus Neosartorya udagawae HDN13-313. Org. Lett. 18 (2), 244–247. doi:10.1021/acs.orglett.5b02964
Yu, M.-L., Guan, F.-F., Cao, F., Jia, Y.-L., and Wang, C.-Y. (2018). A New Antiviral Pregnane from a Gorgonian-Derived Cladosporium Sp. Fungus. Nat. Prod. Res. 32 (11), 1260–1266. doi:10.1080/14786419.2017.1342086
Zainuddin, E. N., Mundt, S., Wegner, U., and Mentel, R. (2002). Cyanobacteria a Potential Source of Antiviral Substances against Influenza Virus. Med. Microbio. Immunol. 191, 181–182. doi:10.1007/s00430-002-0142-1
Zhang, D.-H., Wu, K.-L., Zhang, X., Deng, S.-Q., and Peng, B. (2020). In Silico screening of Chinese Herbal Medicines with the Potential to Directly Inhibit 2019 Novel Coronavirus. J. Integr. Med. 18 (2), 152–158. doi:10.1016/j.joim.2020.02.005
Zhang, D., Tao, X., Chen, R., Liu, J., Li, L., Fang, X., et al. (2015). Pericoannosin A, a Polyketide Synthase–Nonribosomal Peptide Synthetase Hybrid Metabolite with New Carbon Skeleton from the Endophytic Fungus Periconia Sp. Org. Lett. 17 (17), 4304–4307. doi:10.1021/acs.orglett.5b02123
Zhang, G., Sun, S., Zhu, T., Lin, Z., Gu, J., Li, D., et al. (2011). Antiviral Isoindolone Derivatives from an Endophytic Fungus Emericella Sp. Associated with Aegiceras corniculatum. Phytochemistry 72, 1436–1442. doi:10.1016/j.phytochem.2011.04.014
Zhang, P., Li, Y., Jia, C., Lang, J., Niaz, S.-I., Li, J., et al. (2017). Antiviral and Anti-inflammatory Meroterpenoids: Stachybonoids A–F from the Crinoid-Derived Fungus Stachybotrys chartarum 952. RSC Adv. 7, 49910–49916. doi:10.1039/C7RA09859F
Zhang, S.-P., Huang, R., Li, F.-F., Wei, H.-X., Fang, X.-W., Xie, X.-S., et al. (2016). Antiviral Anthraquinones and Azaphilones Produced by an Endophytic Fungus Nigrospora Sp. From Aconitum Carmichaeli. Fitoterapia 112, 85–89. doi:10.1016/j.fitote.2016.05.013
Zhao, Y., Fan, J., Wang, C., Feng, X., and Li, C. (2018a). Enhancing Oleanolic Acid Production in Engineered Saccharomyces cerevisiae. Bioresour. Technol. 257, 339–343. doi:10.1016/j.biortech.2018.02.096
Zhao, Y., Liu, D., Proksch, P., Zhou, D., and Lin, W. (2018b). Truncateols O-V, Further Isoprenylated Cyclohexanols from the Sponge-Associated Fungus Truncatella Angustata with Antiviral Activities. Phytochemistry 155, 61–68. doi:10.1016/j.phytochem.2018.07.017
Zhao, Y., Si, L., Liu, D., Proksch, P., Zhou, D., Lin, W., et al. (2015). Truncateols A–N, New Isoprenylated Cyclohexanols from the Sponge-Associated Fungus Truncatella Angustata with Anti-h1n1 Virus Activities. Tetrahedron 71, 2708–2718. doi:10.1016/j.tet.2015.03.033
Zheng, M., Gao, Y., Wang, G., Song, G., Liu, S., Sun, D., et al. (2020). Functional Exhaustion of Antiviral Lymphocytes in COVID-19 Patients. Cell. Mol. Immunol. 17 (5), 533–535. doi:10.1038/s41423-020-0402-2
Zhou, X., Fang, W., Tan, S., Lin, X., Xun, T., Yang, B., et al. (2016). Aspernigrins with Anti-HIV-1 Activities from the marine-derived Fungus Aspergillus niger SCSIO Jcsw6F30. Bioorg. Med. Chem. Lett. 26 (2), 361–365. doi:10.1016/j.bmcl.2015.12.005
Zhou, Y., Vedantham, P., Lu, K., Agudelo, J., Carrion, R., Nunneley, J. W., et al. (2015). Protease Inhibitors Targeting Coronavirus and Filovirus Entry. Antivir. Res. 116, 76–84. doi:10.1016/j.antiviral.2015.01.011
Keywords: antiviral, microbial metabolites, pandemic, SARS-CoV-2, COVID-19
Citation: Raihan T, Rabbee MF, Roy P, Choudhury S, Baek K-H and Azad AK (2021) Microbial Metabolites: The Emerging Hotspot of Antiviral Compounds as Potential Candidates to Avert Viral Pandemic Alike COVID-19. Front. Mol. Biosci. 8:732256. doi: 10.3389/fmolb.2021.732256
Received: 28 June 2021; Accepted: 23 August 2021;
Published: 07 September 2021.
Edited by:
Mahbuba Rahman, Qatar Biomedical Research Institute, QatarReviewed by:
Lolo Wal Marzan, University of Chittagong, BangladeshLukman Sarker, Innovate Phytoceuticals Inc., Canada
Copyright © 2021 Raihan, Rabbee, Roy, Choudhury, Baek and Azad. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Abul Kalam Azad, ZGFrYXphZC1idGNAc3VzdC5lZHU=
 Topu Raihan
Topu Raihan Muhammad Fazle Rabbee
Muhammad Fazle Rabbee Puja Roy
Puja Roy Swapnila Choudhury
Swapnila Choudhury Kwang-Hyun Baek
Kwang-Hyun Baek Abul Kalam Azad
Abul Kalam Azad