
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Mol. Biosci. , 20 August 2021
Sec. Biophysics
Volume 8 - 2021 | https://doi.org/10.3389/fmolb.2021.724603
This article is part of the Research Topic Structural and Dynamic Aspects of Protein Function and Allostery View all 18 articles
Cholesterol as an allosteric modulator of G protein-coupled receptor (GPCR) function is well documented. This quintessential mammalian lipid facilitates receptor–ligand interactions and multimerization states. Functionally, this introduces a complicated mechanism for the homeostatic modulation of GPCR signaling. Chemokine receptors are Class A GPCRs responsible for immune cell trafficking through the binding of endogenous peptide ligands. CCR3 is a CC motif chemokine receptor expressed by eosinophils and basophils. It traffics these cells by transducing the signal stimulated by the CC motif chemokine primary messengers 11, 24, and 26. These behaviors are close to the human immunoresponse. Thus, CCR3 is implicated in cancer metastasis and inflammatory conditions. However, there is a paucity of experimental evidence linking the functional states of CCR3 to the molecular mechanisms of cholesterol–receptor cooperativity. In this vein, we present a means to combine codon harmonization and a maltose-binding protein fusion tag to produce CCR3 from E. coli. This technique yields ∼2.6 mg of functional GPCR per liter of minimal media. We leveraged this protein production capability to investigate the effects of cholesterol on CCR3 function in vitro. We found that affinity for the endogenous ligand CCL11 increases in a dose-dependent manner with cholesterol concentration in both styrene:maleic acid lipid particles (SMALPs) and proteoliposomes. This heightened receptor activation directly translates to increased signal transduction as measured by the GTPase activity of the bound G-protein α inhibitory subunit 3 (Gαi3). This work represents a critical step forward in understanding the role of cholesterol-GPCR allostery in regulation of signal transduction.
G protein-coupled receptors (GPCRs) are integral membrane proteins comprising a canonical seven-transmembrane alpha helical architecture (Rosenbaum et al., 2009). In response to external stimuli, this helical bundle undergoes a conformational change that is recognized by an intracellular heterotrimeric G protein (Kim et al., 2013). This molecular recognition event leads to an exchange of bound GDP for GTP in the G protein, triggering dissociation of the α and βγ subunits (Figure 1) (Sullivan et al., 1987). The α subunit then acts as an effector to influence downstream events such as modulation of adenylate cyclase functionality (Federman et al., 1992), while the βγ subunit can trigger cleavage of phosphatidylinositol-(4,5)-bisphosphate (PIP2) (Katz et al., 1992) and ion channel activation (Pegan et al., 2005; Nishida et al., 2007).
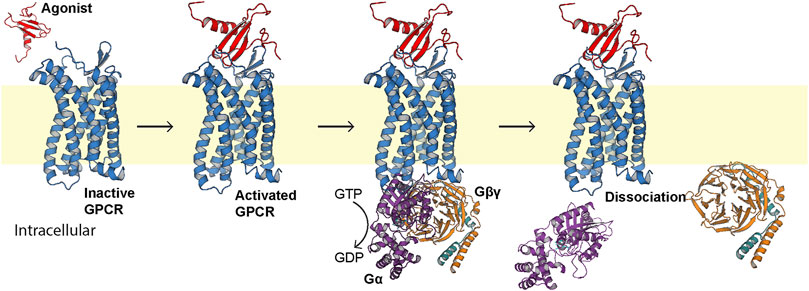
FIGURE 1. Generalized first step of GPCR signal transduction. The extracellular agonist (red, PDB ID 1EOT) (Crump et al., 1998) binds to the orthosteric pocket of the GPCR (blue) to elicit a conformational change recognized intracellularly by the heterotrimeric G protein (PDB ID 1GP2) (Wall et al., 1995). The G protein binds, exchanges bound GDP for GTP, and the α subunit (purple) dissociates from the βγ subunits (orange and teal, respectively).
Functional interplay between lipid constituents and membrane proteins is well documented. We previously reported that the bacterial K+ channel KirBac1.1 orders the membrane (Borcik et al., 2019) and that the membrane activates structural transitions and associated biological functions (Amani et al., 2020; Borcik et al., 2020). Like K+ channels, GPCRs are regulated by lipids through both direct allosteric interactions and changes to membrane mechanical and thermodynamic properties (Botelho et al., 2006). Perhaps the most widely studied of these functional lipids is cholesterol (Jafurulla et al., 2019), although allosteric effects of phosphoserines (Dawaliby et al., 2016), sphingolipids (Chattopadhyay, 2014), phosphoinositols like PIP2 (Yen et al., 2018), and the binding synergy between multiple lipid species (Xu et al., 2021) are of increasing interest. A canonical Class A GPCR cholesterol consensus motif (CCM) of (R,K)4.39–4.43–(W,Y)4.50–(I,V,L)4.46–(F,Y)2.41 has been identified in the β2 adrenergic receptor (β2AR), following the Weinstein–Ballesteros numbering convention (Ballesteros and Weinstein, 1995; Hanson et al., 2008). However, only 21% of Class A receptors contain this sequence (Hanson et al., 2008), and this motif is conspicuously absent in chemokine receptors (Legler et al., 2017). Furthermore, it is observed that such cholesterol binding motifs are not necessarily occupied even when present (Marlow et al., 2021). Cholesterol has, nevertheless, still been implicated as an allosteric modulator of chemokine receptor function (Zhukovsky et al., 2013; Pluhackova et al., 2016; Legler et al., 2017; Gahbauer et al., 2018; Calmet et al., 2020).
Chemokine receptors are a subclass of Class A GPCRs. They trigger cellular trafficking of immune cells in response to chemotactic cytokine (chemokine) ligands (Stone et al., 2017). Perhaps the most well-known chemokine receptors are CC motif chemokine receptor 5 (CCR5) and CXC motif chemokine receptor 4 (CXCR4), which act as coreceptors for HIV infection (Deng et al., 1996; Ma et al., 1998). Although the CCM identified in β2AR is absent from chemokine receptors, experimental evidence indicates receptor–lipid interactions, often driving receptor dimerization (Pluhackova et al., 2016; Legler et al., 2017; Gahbauer et al., 2018; Calmet et al., 2020). However, there are many outstanding questions regarding the direct and indirect influence of cholesterol on function in vivo. Much of what is known is derived from crystal structure co-crystallization and molecular dynamics (MD) simulations. While these studies are foundational, the conclusions are not definitive and experimental functional data are needed.
CC motif chemokine receptor 3 (CCR3, Figure 2A) is a Class A GPCR mainly expressed by eosinophils. Like all chemokine receptors, its primary messengers are endogenous peptide ligands. Specifically, CCL11, CCL24, and CCL26 (eotaxins 1, 2, and 3, respectively; CCL11 is depicted in Figure 2B) activate CCR3 and trigger chemotaxis of the expressing cell (Ge et al., 2015). This occurs through the G protein inhibitory α subunit (Gαi, Figure 2C), which triggers downstream inhibition of adenylate cyclase (Kitaura et al., 1996). It is, however, unclear as to which of the 3 Gαi subunits is primarily involved in the signaling cascade or if there is significant promiscuity in vivo between CCR3 and Gαis 1, 2, and 3. Moreover, the influence of direct lipid allostery on CCR3-G protein coupling and signal transduction is undocumented.
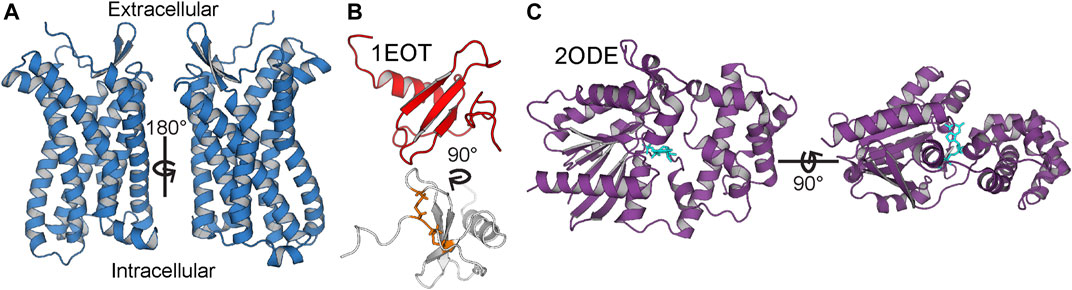
FIGURE 2. Anatomy of CCR3, CCL11, and Gαi3. (A) CCR3 homology model is marked by the canonical seven-transmembrane helical architecture. (B) Solution state NMR structure of CCL11 (PDB ID 1EOT) (Crump et al., 1998) shows the structural disulfide bonds in orange (bottom). (C) Gαi3 (purple) with bound GDP (cyan) from the crystal structure of Gαi3 bound to the regulator of G protein signaling 8, RGS8 (PDB ID 2ODE) (Soundararajan et al., 2008).
Given its role in leukocyte trafficking, CCR3 provokes inflammatory conditions such as asthma (Gauvreau et al., 2018), rheumatoid arthritis (Katschke et al., 2001), and eosinophilic esophagitis (Dunn et al., 2020). Furthermore, CCR3 is correlated with heightened invasive potential of metastatic liver (Jin et al., 2017), prostate (Ishida et al., 2018), and kidney cancers (Johrer et al., 2005). It is also a coreceptor for some strains of HIV (He et al., 1997). As a result, CCR3 is an attractive therapeutic target. However, comparatively little is known about the structural biology of CCR3 with respect to other chemokine receptors such as CCR5 and CXCR4. CCR3’s natural agonists help regulate the relative monomer–dimer higher-order oligomer populations in vivo (Song et al., 2018), but the influence of cholesterol on this interaction is unknown. Therefore, investigation of the structure–function relationship and the lipid agency is an attractive and necessary long-term goal.
In pursuit of this goal, we implemented a codon harmonization scheme we previously reported to positively influence heterologous membrane protein yield (van Aalst et al., 2020). This technique is called DNA codon usage for measured base optimization, or DUMB optimization (DO). Codon harmonization aims to site-specifically modify the codons comprising the heterologous construct to match the codon usage frequency of the native organism more closely in order to improve cotranslational folding (Angov et al., 2008; Rodriguez et al., 2018). This increases the fraction of protein that is properly folded, membrane-inserted, and functional by slowing translation through the introduction of targeted rare codons. Through implementation of DUMB optimization and incorporation of a maltose-binding protein (MBP) solubility tag (Ge et al., 2015) in our construct, we report the heterologous production of CCR3 after tag removal at yields of ∼2.5–2.8 mg/L from M9 minimal media (Bhate et al., 2013). CCL11 and Gαi3 were DUMB optimized as a matter of course in expression optimization, attaining ∼2.6 ± 0.3 and 15.1 ± 0.3 mg/L, respectively, for each, from M9 minimal media. We describe the positive cooperativity between membrane cholesterol and CCR3 binding affinity to its endogenous ligand CCL11, quantified via a fluorescence polarization assay (Rossi and Taylor, 2011) in lipid environments of increasing cholesterol content. We then confirm that this cholesterol-induced modulation of ligand affinity translates to increased signal transduction, measured via coupling to and activation of Gαi3 GTPase. This is, to our knowledge, the first experimental evidence of cholesterol-receptor interactions and their effect on ligand affinity and the efficacy of signal transduction catalyzed by CCR3.
The human CCR3 amino acid sequence was obtained from the UniProt database (P51677). The human CCL11 amino acid sequence was obtained from the UniProt database (P51671) and truncated to residues 24–97 to remove the propeptide. The human G α inhibitory 3 (Gαi3) amino acid sequence was obtained from the UniProt database (P08754). The amino acid sequences were reverse engineered into fully optimized (FO) DNA sequences according to E. coli codon usage, serving as a platform for the application of DUMB optimization. Here, the FO constructs use codons to transcribe the proteins that correspond to only the most abundantly found tRNAs in the expression vector.
The native human CCR3 DNA sequence was obtained from the European Nucleotide Archive (ENA, Sequence: U51241) and codon-harmonized according to DUMB optimization (DO) for expression in E. coli, as previously described (van Aalst et al., 2020). Briefly, codons were replaced in the expression sequence to match the codon usage frequencies found within the native sequence as E. coli codon usage permitted. Substitution was performed such that no alternative codons in the host system were chosen with a relative usage below 5% of the native usage frequency. Human and E. coli codon usage frequencies from the Graphical Codon Usage Analyzer (http://gcua.schoedl.de/) were used in designing the DO gene sequence (Fuhrmann et al., 2004). For more information on the codon harmonization process, see reference van Aalst et al., 2020. %MinMax analysis (Rodriguez et al., 2018) of the native, FO, and DO sequences visualizes the extent of deoptimization (Figure 3). The native human Gαi3 (Sequence: J03005.1) and human CCL11 (D49372.1) gene sequences were also obtained from the ENA and codon-harmonized in the same way as a matter of course in the optimization process.
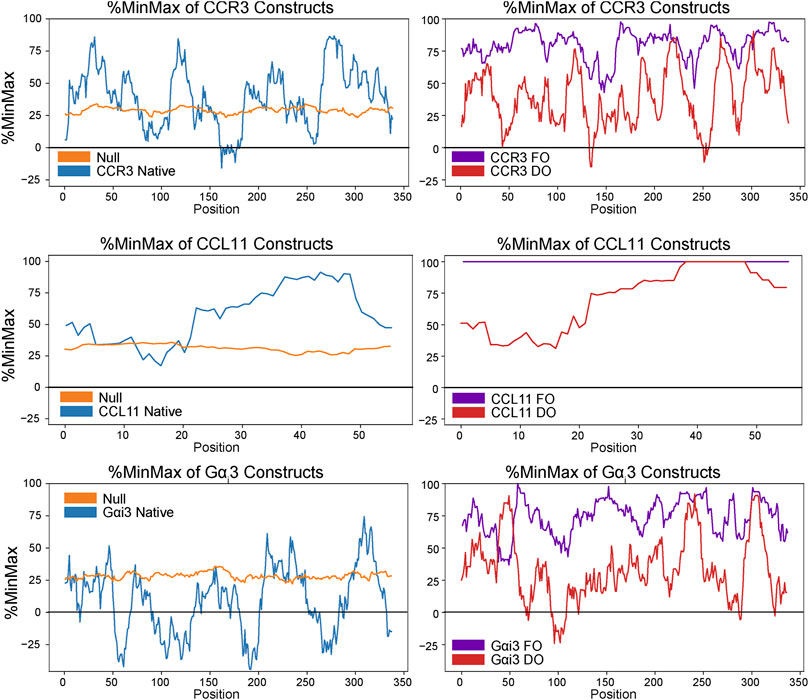
FIGURE 3. %MinMax (Rodriguez et al., 2018) of native sequences compared to optimized heterologous sequences (FO) and constructs codon-harmonized through DUMB optimization (DO). In general, %MinMax of the DO sequences (red) more closely resemble the native values (blue) than the FO values (purple).
CCR3, CCL11, and Gαi3 gene sequences were ordered from GeneArt (Thermo Fisher). The expression vector pMAL-p4x was ordered from Addgene. 5′ EcoRI and 3’ Hind III restriction enzymes (Thermo Scientific) were used to clone each sequence into the vector downstream of MalE. This resulted in an N–maltose-binding protein (MBP)-8x His tag–TEV site–CCL11–C construct, in the case of CCL11. For CCR3, the initial FO construct that was described (Ge et al., 2015) was DUMB optimized, and a GGGGS 4x repeat between the TEV site and the N terminus of CCR3 was added to promote cleavage (Chen et al., 2013). This resulted in an N–maltose-binding protein (MBP)-8x His tag–TEV site–(GGGGS)4–CCR3–C construct. The restriction enzymes NcoI and BamHI (Thermo Scientific) were used to insert the Gαi3 sequence into the expression vector pET-28a (+) (Novus Biologicals). This resulted in an N–Met-Gly–8x His–TEV site–Gαi3–C construct. In all cases, gene insertion and sequence conservation were verified by sequencing.
E. coli C43 (DE3) cells harboring the pMAL-p4x-CCR3 plasmid were grown in M9 minimal media containing 2 mM MgSO4, 0.1 mM CaCl2, 100 μg/ml ampicillin, 3 ml of 100x minimum essential vitamin stock, 96.22 mM Na2HPO4, 44.1 mM K2HPO4, 17.1 mM NaCl, 5 g glucose per L (0.5% w/v), 3.75 g NH4Cl per L (0.375% w/v), and 20 ml of Solution C (Supplementary Table S1) per L. Cultures were grown at 220 rpm and 37°C until an OD600 of ∼1.0 was reached. The cultures were cooled to 18°C and induced with 1 mM IPTG for 24 h. Cell cultures were then pelleted at 5,500 rpm for 10 min and stored at −80°C for future use.
Cell pellets were removed from storage at −80°C and resuspended in 5 ml of lysis cocktail per g of cells (20 mM HEPES, pH 8.0, 150 mM KCl, 0.02% NaN3, 10 mM MgSO4, 0.2 mg/ml lysozyme, 0.2 mg/ml RNase A, 1 mM phenylmethylsulfonyl fluoride (PMSF), and 1 mM benzamidine). PIERCE EDTA-free protease inhibitor tablets (Thermo Fisher) were added at one tablet per 6 g of cells. Cells were lysed via homogenization, and protein was extracted from membranes via addition of n-Dodecyl-β-D-Maltoside (DDM, Anatrace) and cholesteryl hemisuccinate Tris-salt (CHS, Anatrace) to final concentrations of 20 and 2 mM, respectively. Extraction took place overnight at 4°C with rocking. The solution was centrifuged at 125,000 g for 40 min at 4°C to remove cell debris. The supernatant was filtered through a 0.22-μm PES bottle top filter and loaded onto a 5-ml nickel affinity column (NAC, GE Healthcare Life Sciences) equilibrated in wash buffer (20 mM HEPES, pH 8.0, 150 mM KCl, 10 mM imidazole, 0.02% NaN3, 2.5 mM DDM, and 0.25 mM CHS). The column was then treated with five column volumes of wash buffer before elution with five column volumes of elution buffer (20 mM HEPES, pH 8.0, 150 mM KCl, 250 mM imidazole, 0.02% NaN3, 2.5 mM DDM, and 0.25 mM CHS).
Eluted protein was transferred into cleavage buffer (20 mM HEPES, pH 8.0, 150 mM KCl, 1 mM EDTA, 1 mM DDM, 0.1 mM CHS, and 0.5 mM DTT) using a desalting column (GE Healthcare Life Sciences) equilibrated with cleavage buffer. Tobacco etch virus (TEV) protease (Kapust et al., 2001) was added from glycerol stocks at a typical ratio of 1 mg TEV to 3 mg MBP-CCR3 with 1 mM DTT and set to rock overnight at 4°C. Samples were then transferred into wash buffer to remove EDTA and subjected to the NAC. Flow-through containing cleaved CCR3 was collected and the elution peak containing TEV, the MBP tag, and uncleaved MBP-CCR3 was discarded. The flow-through was then concentrated to ∼2 mg/ml using an Amicon Stirred Cell with Ultracel 30 kDa Ultrafiltration Discs (Millipore) before loading onto a HiLoad 16/600 Superdex Prep grade 200 column (GE Healthcare Life Sciences) equilibrated in exchange buffer. Cleaved CCR3 fractions were pooled, concentration was determined using optical density at 280 nm, and samples were stored at 4°C for future use.
pMAL-p4x harboring CCL11 was cultured in M9 minimal media at 220 rpm and 37°C until an OD600 of ∼0.8 was reached. The cultures were cooled to 18°C and induced with 0.5 mM IPTG for 24 h. Cell aliquots were centrifuged at 5.5 k rpm for 10 min, and the resulting pellets were stored at −80°C for future use.
Cell pellets were resuspended in CCL11 wash buffer (20 mM HEPES, pH 7.5, 300 mM KCl, 10 mM Imidazole, and 0.02% NaN3) supplemented with 0.2 mg/ml lysozyme, 0.2 mg/ml RNase A, 1 mM PMSF, 1 mM benzamidine, and 1 PIERCE EDTA-free protease inhibitor tablet per 6 g of cells. Cells were lysed via triplicate passage through a homogenizer. Cell debris was centrifuged at 17,000 rpm for 30 min followed by clarification of the lysate using a 0.22-μm PES bottle top filter. The clarified lysate was loaded onto an NAC preequilibrated in CCL11 wash buffer, washed five times with the same, and eluted with five column volumes of CCL11 elution buffer (20 mM HEPES, pH 7.5, 300 mM KCl, 250 mM imidazole, and 0.02% NaN3). The elution peak was then transferred back into CCL11 wash buffer using a desalting column for the reverse NAC.
The TEV was added in a typical ratio of 1 mg TEV per 4 mg MBP-CCL11 and set to cleave at 4 °C with rocking for 1 h. The cleavage mixture was then passed through the NAC, and the flow-through containing cleaved CCL11 was collected. This was concentrated to ∼5 ml using an Amicon Stirred Cell with Ultracel 3 kDa Ultrafiltration Discs (Millipore) and loaded onto a HiLoad 16/600 Superdex Prep grade 75 column equilibrated in CCL11 exchange buffer (20 mM HEPES, pH 7.5, 300 mM KCl, 1 mM EDTA, and 0.02% NaN3). The CCL11 elution fractions were collected, and concentration was determined via optical density at 280 nm.
E. coli BL21 codon + (DE3) cells harboring the pET-28a (+) Gαi3 plasmid were grown in M9 minimal media at 220 rpm and 37°C until an OD600 of ∼1.0 was reached. The cultures were cooled to 20°C and induced with 0.5 mM IPTG for 24 h. Cell cultures were then pelleted at 5,500 rpm for 10 min and stored at −80°C for future use.
Cell pellets were resuspended in Gαi3 wash buffer (20 mM HEPES, pH 7.5, 150 mM KCl, 10 mM Imidazole, 10 μM GDP, 2.5 mM MgCl2, and 0.02% NaN3) supplemented with 0.2 mg/ml lysozyme, 0.2 mg/ml RNase A, 1 mM PMSF, 1 mM benzamidine, and 1 PIERCE EDTA-free protease inhibitor tablet per 6 g of cells. Cells were lysed via triplicate passage through a homogenizer. Cell debris was centrifuged at 17,000 rpm for 30 min, followed by clarification of the lysate using a 0.22-μm PES bottle top filter. The clarified lysate was loaded onto an NAC preequilibrated in wash buffer, washed five times with the same, and eluted with five column volumes of elution buffer (20 mM HEPES, pH 7.5, 150 mM KCl, 250 mM Imidazole, 10 μM GDP, 2.5 mM MgCl2, and 0.02% NaN3). The elution peak was then loaded onto a HiLoad 16/600 Superdex Prep grade 75 column equilibrated in Gαi3 exchange buffer (20 mM HEPES, pH 7.5, 150 mM KCl, 10 μM GDP, 2.5 mM MgCl2, and 0.02% NaN3). The elution fractions were collected, and concentration was determined via optical density at 280 nm.
Samples were combined at a ratio of 1:1 with 2x Laemmli buffer (20% glycerol, 125 mM Tris HCl pH 6.8, 4% sodium dodecyl sulfate (SDS), 0.02% bromophenol blue) for denaturation (Laemmli, 1970). Samples were then loaded into a Mini-PROTEAN TGX precast any-kD 10-well gel (Bio-Rad) with a Precision Plus Dual Standard protein ladder (Bio-Rad). The gel was run in running buffer (192 mM glycine, 25 mM, 0.1% SDS) for 53 min to remove loading dye at 400 mA and 150 V on a PowerPac Basic module (Bio-Rad). The gel was removed from the casing and stained in staining buffer (20% methanol and 10% acetic acid with 1 mg/ml Coomassie R250) with orbital rotation at 69 rpm until the gel was no longer visible. The gel was then destained in destaining buffer (20% methanol and 10% acetic acid).
CCR3 was buffer-exchanged into CCR3 CD buffer (5 mM Tris-HCl, pH 8.0, 50 mM sodium acetate, 1 mM DDM, and 0.1 mM CHS) and concentrated to 0.3 mg/ml (7.2 μM). Immediately prior to analysis, buffer and CCR3-containing samples were diluted 4x to allow for data acquisition. Spectra were acquired using a J-815 CD spectrophotometer (JASCO Co., Easton, MD, United States) at 22°C in the spectral range of 180–260 nm at a rate of 1 nm/sec and a path length of 0.1 cm. 10 spectra each of the diluted buffer blank and CCR3 samples were recorded and averaged. Background spectra of the buffer were acquired identically and subtracted from the experimental data. Spectral fitting and secondary structure analysis for CCR3 were carried out using the DichroWeb (Whitmore and Wallace, 2004; Whitmore and Wallace, 2008) server using the K2D algorithm (Andrade et al., 1993). Presented secondary structure percentages from experimental data were calculated using DichroWeb analysis and compared to a model of CCR3. This model was generated by submitting the full-length CCR3 sequence to the Baker laboratory ROBETTA comparative modeling server (Song et al., 2013), using the CCR5 crystal structure PDB 4MBS (Tan et al., 2013a) as the template because of the high sequence similarity. The model was truncated to residues 23–317, and all truncated residues and the GGGGS4x linker were assumed to be random coils for percent secondary structure calculations. In addition, the CCL11 structure 1EOT (Crump et al., 1998) was docked to the CCR3 model using HADDOCK (van Zundert et al., 2016) to visualize the CCL11-bound CCR3 model observed throughout this work. Residue–residue interaction restraints upon drive docking were derived from information available at the GPCR Database (Duchesnes et al., 2006; Millard et al., 2014; Pandy-Szekeres et al., 2018). CCL11 was buffer-exchanged into 10 mM sodium phosphate buffer, pH 7.5. Data were acquired at a concentration of 18.14 μM and processed in the same manner as CCR3. Bestsel was used to fit CCL11 spectral data and predict the percent secondary structure from the experimental data to compare to the published structure 1EOT (Crump et al., 1998; Micsonai et al., 2015; Micsonai et al., 2018; Micsonai et al., 2021). Gαi3 was buffer-exchanged into 10 mM sodium phosphate buffer, pH 7.5, and 10 μM GDP. Spectra were acquired in the same way as before at a protein concentration of 1.85 μM. Data were fit using DichroWeb with the Contin-LL algorithm (Provencher and Glöckner, 1981) and reference set 4 (Sreerama and Woody, 2000). The extrapolated secondary structure from the experimental fit was compared to the crystal structure of activated Gαi3 in complex with RGS10 (PDB ID 2IHB) (Soundararajan et al., 2008). All non-crystallizing residues were assumed to be random coils for percent secondary structure calculations.
1-pamlitoyl-2-oleoyl-phosphatidylcholine (POPC or PC, Avanti Polar Lipids) and cholesterol (Sigma) were solvated in chloroform at 10 mg/ml, and then PC-only and 10, 20, 30, 40, and 60% cholesterol (mol%) mixtures were formed, blown down under a N2 stream, and dried overnight in vacuo to produce a lipid film. Dried films were evenly divided (one aliquot for a protein-free control) and then solvated in non-detergent buffer (NDB, 20 mM HEPES pH 8.0, 150 mM KCl, 0.02% NaN3, and 1 mM EDTA) supplemented with 25 mM 3-[(3-Cholamidopropyl) dimethylammonio]-1-propanesulfonate (CHAPS, Anatrace) using mild sonication at 5 mg lipid/ml. Solvated films were then set on the benchtop for 3–5 h before the addition of protein, added at a ratio of 1 mg of protein per 4 mg of lipids. An equal volume of exchange buffer was added to protein-free (PF) samples. For samples reconstituted in the presence of CCL11, the agonist was added at a molar ratio of 5:1 CCL11:CCR3, or an equal volume of CCL11 exchange buffer was added to control samples. All samples were set to anneal for 3 h on the benchtop, during which Bio-Bead SM-2 Resin (Bio-Rad) was prepared by 3x degassing washes with methanol, followed by 3x washes with DI water and resuspension in NDB. A double portion of Bio-Beads (∼60 mg) was added to each sample before nutation at room temperature. Samples were nutated in this way for 48–72 h, with ∼30 mg Bio-Beads being added twice daily until the detergent was completely removed, evidenced by increased turbidity and loss of detergent bubbles upon manual agitation. Bio-Beads were removed by centrifugation at 500 rpm using PIERCE columns to collect fully formed proteoliposomes. To form lipid particles (SMALPs) from proteoliposomal samples, 3:1 pre-hydrolyzed styrene:maleic acid (SMA) was added at 3 mg SMA per 1 mg of lipids dropwise with inversion to each sample to facilitate polymer disc formation (Lipodisq, Thermo Fisher). Proteoliposomal samples typically turned clear within moments of SMA addition. All samples were nutated overnight to ensure SMALP formation.
CCL11 in CCL11 exchange buffer was concentrated to >2 mg/ml and incubated with a 4x molar excess of fluorescin isothiocyanate (FITC, Invitrogen) for 1 h at room temperature. This was achieved in the dark, using an orbital shaker at 150 rpm and a pH value of 7.5 to facilitate labeling of the N terminus with the fluorophore. Following this, the sample was diluted to 1–2 ml with the same buffer and exchanged back into CCL11 exchange buffer sans excess FITC using a HiPrep 26/10 desalting column. A Lowry assay (DC protein assay, Bio-Rad) was performed to gauge protein concentrations. Fluorescence polarization assays were performed at room temperature using a Biotek Synergy NEO2 fluorescent plate reader equipped with a fluorescein filter (Biotek Instruments, Winooski, VT, United States). Fluorescence polarization was calculated automatically using the instrument as follows (Rossi and Taylor, 2011):
where I|| is the observed parallel intensity, I⊥ is the observed perpendicular intensity, and P is polarization. Static concentrations of 100 nM FITC-CCL11 and 0.1 µg/µl bovine serum albumin (BSA, Thermo Scientific) for nonspecific binding were added to each assay well. CCR3 was added to the desired concentration. Protein-free SMALPs or proteoliposomes were then added to balance out the lipid/SMA material such that the concentrations were equivalent across all wells. NDB was then added to fill to 30 µl. The concentration of FITC-CCL11, BSA, and lipids/SMA were static across all conditions and replicates. Data were normalized by subtracting the lowest zero-point (no CCR3) value in a curve from each read to bring all curves within the same reference frame.
Gαi3 hydrolyzes GTP when bound to and activated by CCR3. Unhydrolyzed GTP is enzymatically converted to ATP and then to luminescence via luciferase. GTP turnover was thus quantified using a modified protocol of the GTPase-Glo™ assay (Promega) (Mondal et al., 2015) at room temperature for all steps, with a reaction incubation time of 2 h in all cases. After incubation, reconstituted GTPase-Glo™ reagent was added and incubated for 30 min at room temperature. Detection reagent was added, followed by an additional 5–10 min of incubation. Luminescence was read using a Cytation 3 multimode reader (Biotek Instruments, Winooski, VT, United States). Intrinsic GTPase activity of Gαi3 was analyzed using 2.5 μM apo-Gαi3 in 20 mM HEPES (pH 7.5), 150 mM KCl, 5 mM MgCl2, 20 mM EDTA, 0.1 mM TCEP, 10 μM GDP, and 1 or 4 μM GTP. Preliminary CCL11-induced CCR3 activation of Gαi3 was analyzed in assay buffer (20 mM HEPES, pH 7.5, 150 mM KCl, 1 mM EDTA, 0.1 mM TCEP, 10 μM GDP, and 4 μM GTP) containing 5 mM MgCl2, 1 mM DDM, and 0.1 mM CHS. Agonist-driven GTPase activity was analyzed in SMALPs formed from POPC with 0–30% cholesterol in assay buffer with no additives. CCR3 samples reconstituted in the presence of CCL11 were analyzed in assay buffer plus 5 mM MgCl2. Assays in detergent, SMALPs, and proteoliposomes were performed with 5 μM CCL11, 1 μM CCR3, and 1 μM Gαi3. Relative light units (RLUs) of all assay runs were blank corrected by subtracting the average of three blank replicates (buffer with no GTP and background luminescence) from each replicate. % Hydrolysis was calculated from raw data as follows:
The total yield of functional, folded protein is the main bottleneck in the study of GPCRs. Here, we introduced a maltose-binding protein (MBP) fusion tag to aid in protein folding and solubility, as previously described (Ge et al., 2015). Furthermore, we employed codon harmonization, a method to optimize heterologous plasmid DNA sequences (Angov et al., 2008). We previously showed our codon harmonization strategy, dubbed DNA codon usage for measured base optimization, or DUMB optimization (DO), which dramatically increased both the yield and activity of an exogenously expressed chimeric membrane protein (van Aalst et al., 2020). Results of %MinMax analysis of proteins in this work show the codon usage of the fully optimized and codon-harmonized constructs as compared to native usage (Figure 3). Codon usage is presented as a sliding window of 21 codons to visualize the extent of optimization or deoptimization of each gene sequence to compare native usage (blue) to a random reverse transcription control (orange), FO constructs (purple), and DO constructs (red). These techniques facilitate the production of 2.6 ± 0.2 mg/L (n = 3, ± σ) of full length and functional WT CCR3 (Figure 2A), after proteolytic cleavage of the MBP tag, from M9 minimal media. It is expected that an even greater yield would be observed if the expression cultures were grown in rich media. CCL11 and Gαi3 were DUMB optimized as a matter of course in workflow optimization and produced yields of ∼2.6 ± 0.3 and 15.1 ± 0.3 mg/L, respectively, from minimal media.
An apparent band slightly above the 37-kDa marker is visible in the SDS-PAGE analysis of CCR3, consistent with the predicted molecular weight of 42.4 kDa with the linker sequence in our construct (Figure 4A) after MBP cleavage and size exclusion chromatography (SEC) elution (Supplementary Figure S1A). This band conforms to previously reported SDS-PAGE analysis of similar CCR3 constructs in which CCR3 was observed to form SDS-resistant dimers (Wang et al., 2013; Ge et al., 2015). This could explain the faint bands observed at roughly 80, 120, and 160 kDa. Full gel images are available in the Supplementary Material (Supplementary Figure S2). Circular dichroism (CD) analysis reveals deep wells at 208 and 220 nm, consistent with a highly α-helical protein (Figure 4B). Experimental CD data were fit using DichroWeb (Whitmore and Wallace, 2004; Whitmore and Wallace, 2008). The K2D algorithm (Andrade et al., 1993) was selected over the Contin-LL algorithm (Provencher and Glöckner, 1981) paired with the SMP180 reference set (Abdul-Gader et al., 2011) due to differences in the normalized RMSD (NRMSD, 0.122 vs. 0.365, respectively). NRMSD values between 0.1 and 0.2 generally suggest similarity between experimental and fit secondary structures, whereas values greater than 0.2 indicate little resemblance (Mao et al., 1982). Predicted secondary structures derived from DichroWeb also show good agreement between experimental and model data when model truncated residues are assumed to be random coils (Figure 4C). Interestingly, the small observed β-sheet amount in the model (Figure 2A), derived from the CCR5 crystal structure template (Tan et al., 2013b), is conserved in the experimental CD spectrum according to the fitting.
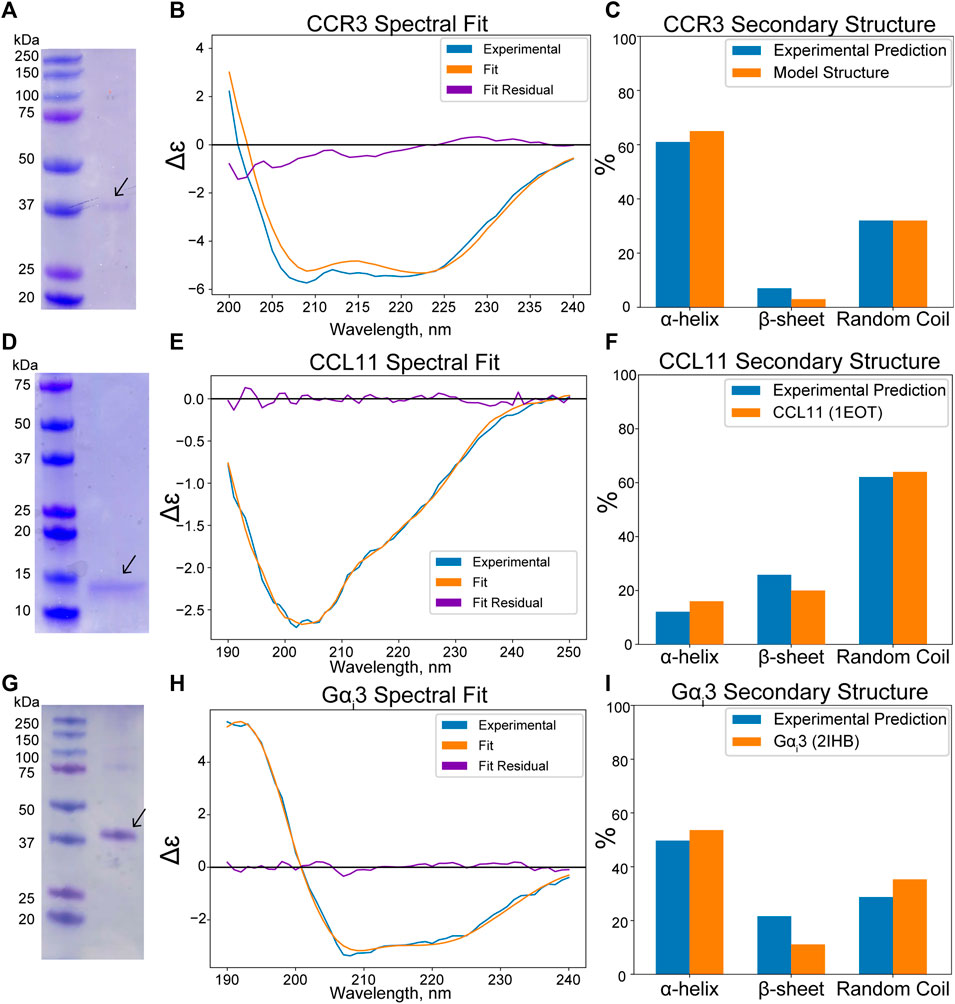
FIGURE 4. (Wavelength, nm x axis label in panel H) was cut off. (A) SDS-PAGE analysis of CCR3 reveals a ∼37-kDa band, consistent with the predicted molecular weight of 42.4 kDa. (B) Experimental CD spectra of CCR3 (blue) compared to the DichroWeb (Whitmore and Wallace, 2004) experimental fit (orange) and the residual fit (purple). (C) Quantitative assessment compared to the CCR3 model suggests a high degree of helical content. (D) SDS-PAGE analysis of CCL11 reveals an apparent molecular weight of ∼14 kDa, consistent with previous observations. (E) CD spectra of CCL11 (blue) compared to the fit (orange) and the residual fit (purple). The spectrum was fit using Bestsel (Micsonai et al., 2015; Micsonai et al., 2018; Micsonai et al., 2021). (F) Bestsel-predicted percent secondary structure for the CCL11 construct compared to the NMR structure (PDB ID 1EOT) (Crump et al., 1998). (G) SDS-PAGE analysis of Gαi3 shows good agreement with the predicted construct molecular weight of 42.7 kDa. (H) CD spectra of Gαi3 (blue) compared to the DichroWeb experimental fit (orange) and the residual fit (purple). (I) Comparison of the experimentally predicted % secondary structure to that found in the crystal structure (PDB ID 2IHB) (Soundararajan et al., 2008).
CCL11 (Figure 2B) is also produced as an MBP fusion construct in E. coli to facilitate formation of the structural disulfide bonds in the more oxidizing periplasmic space. While this has a detrimental effect on yield, it consistently produces properly folded protein. An apparent band of ∼14 kDa is observed for our CCL11 construct after MBP tag removal and subsequent SEC elution (Figure 4D; Supplementary Figure S1B). This is larger than the expected 8.4 kDa but is consistent with previously reported CCL11 constructs (Mingqing Wang, 2014). CD analysis of our construct confirms the conservation of the typical chemokine fold (Figure 4E, NRMSD 0.01858). The secondary structure distributions predicted from the fit using Bestsel (Micsonai et al., 2015; Micsonai et al., 2018; Micsonai et al., 2021) are consistent with the solution state NMR structure 1EOT (Crump et al., 1998) (Figure 4F).
The Gαi3 subunit (Figure 2C) is expressed as an N terminal His-tagged construct. The theoretical molecular weight of our construct after SEC elution (Supplementary Figure S1C) is ∼42.7 kDa, consistent with the apparent SDS-PAGE band (Figure 4G). The CD spectrum was fit using DichroWeb using the Contin-LL algorithm (Provencher and Glöckner, 1981) and spectral reference set 4 (Sreerama and Woody, 2000) (Figure 4H, NRMSD 0.045). Comparison of the DichroWeb fit–derived secondary structure to the crystal structure 2IHB (Soundararajan et al., 2008) shows relatively good agreement when accounting for possible minute differences between the inactive, GDP-bound state and the active state found in the crystal structure (Figure 4I).
GPCR structures often contain cholesterol binding motifs, but it is unclear how bilayer cholesterol impacts chemokine receptor function. Thus, we employed fluorescence polarization to measure the affinity of CCR3 for CCL11 as a function of bilayer cholesterol concentration. We attached the fluorophore fluorescein isothiocyanate (FITC) to the N terminus of CCL11, which unbound is rapidly tumbling in solution, polarizing light to a lower extent (Figure 5A). After CCL11 is bound, the decreased molecular tumbling increases polarization. Titration of cholesterol from 0 to 30% (mol%) into 1-pamlitoyl-2-oleoyl-phosphatidylcholine (POPC and PC) membrane styrene:maleic acid lipid particles (SMALPs) shows a drastic decrease in Kd (Figure 5B). A Kd of 30 ± 10 nM at 30% cholesterol, or a ∼5-fold decrease from pure POPC, is quite significant.
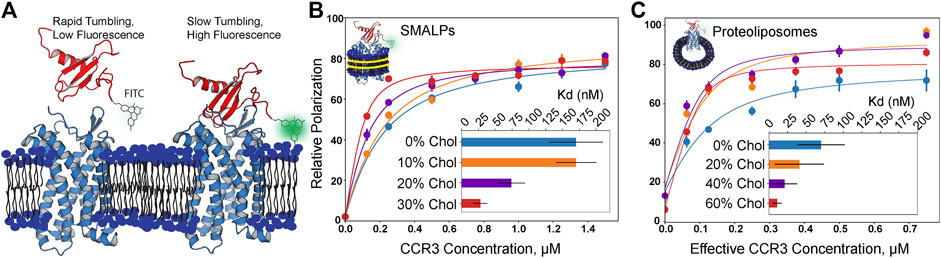
FIGURE 5. Fluorescence polarization assay results in increasing cholesterol content (mol% in PC). (A) Visualization of apo- and CCR3-bound CCL11. FITC-labeled CCL11 is rapidly tumbling in solution, and thus, the probe polarizes light to a lesser degree. Upon binding CCR3, tumbling is slowed, polarizing a greater fraction of light and giving rise to a larger polarization value. CCL11 (PDB ID 1EOT) (Crump et al., 1998) was docked to CCR3 using HADDOCK (van Zundert et al., 2016) to visualize the bound state leading to slower tumbling. (B) Membrane cholesterol content is positively correlated with increased ligand affinity in SMALPs. (C) Increased observed affinity is conserved in proteoliposomes. Concentration is halved to correct for receptor orientation (Supplementary Figure S3). Points indicate the mean ± SEM for three replicates, each read three times. Error bars may fall within the size of the points. Data were normalized by subtracting the lowest zero-point (no CCR3) value in a curve from each read to bring all curves within the same reference frame.
In order to gauge the effects of cholesterol at higher concentrations, we turned to proteoliposomes, as the membrane rigidity imparted by higher cholesterol content inhibits SMALP formation (Scheidelaar et al., 2015; Dörr et al., 2016) (Figure 5C). It is clear that the same trend is observed; however, the experimental error increased for the measurements in proteoliposomes. We corrected the effective CCR3 concentration to account for receptor orientation intractability by halving the total concentration to remove the statistical average of the receptor with the orthosteric site that is facing into the proteoliposome and thus inaccessible to the ligand. The listed receptor concentrations in proteoliposomes reflect the input concentrations that were halved during curve fitting (Figure 5C; Supplementary Figure S3). Measured Kd values were 20 ± 20 nM in 40% cholesterol and 11 ± 6 nM in 60% cholesterol, following the same inverse trend between Kd and cholesterol content as that seen in SMALPs. However, the Kd values at the same cholesterol concentrations were also lower than the corresponding measurement in SMALPs. Membrane curvature and lateral pressure in the proteoliposomal samples may play a role in GPCR function (Jones et al., 2020). We hypothesize both curvature and lateral pressure are lost in SMALPs, which could contribute to the observed Kd discrepancies between the two conditions. Increasing cholesterol presence did still lead to decreasing Kd in SMALPs, suggesting that curvature and lateral pressure are not the only phenomena responsible for the observed modulation of ligand affinity. Thus, we conclude that cholesterol is a direct positive allosteric effector of CCR3–ligand affinity.
We next investigated how the observed relationship between cholesterol and CCL11 affinity impacts G protein coupling to CCR3. Heightened activation of CCR3 should lead to increased receptor–G protein coupling. This, in turn, should drive nucleotide exchange in the Gαi3 binding pocket with CCR3 functioning as the guanine nucleotide exchange factor (GEF). The remaining GTP is then converted to a luminescent signal after conversion to ATP (Figure 6A) (Ford et al., 1994; Ford and Leach, 1998a; Ford and Leach, 1998b). In the absence of a suitable GEF, 20 mM EDTA can be used to stimulate intrinsic activity, verifying construct activity (Supplementary Figure S4). We next verified that our CCR3 and Gαi3 constructs are able to couple in detergent, using Mg2+ to stabilize the nucleotide-bound state of Gαi3 (Supplementary Figure S5). We considered this an important step as little information is available concerning which Gαi subunits CCR3 activates. Upon showing the ability for our constructs to couple, we investigated the effects of cholesterol on this interaction, hypothesizing that the dose-dependent modulation of agonist affinity would directly translate to Gαi3 coupling, activation, and GTP hydrolysis.
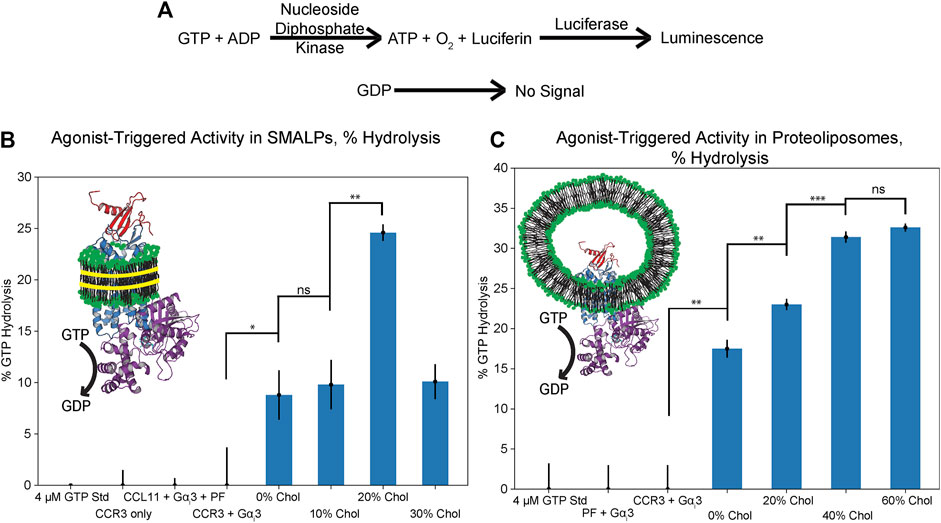
FIGURE 6. GTPase assay results in SMALPs and proteoliposomes of increasing cholesterol content (mol% in PC), presented as % GTP hydrolyzed over the course of 2 h. (A) Schematic representation of GTP hydrolysis translation to the luminescent signal (Mondal et al., 2015). (B) Cholesterol enhances GTP hydrolysis in SMALPs but dose dependence could not be verified, likely due to inhibition of disc formation. (C) Cholesterol dose dependence of GTP hydrolysis is verified through coreconstitution of CCL11 with CCR3 into proteoliposomes. Cartoons depict CCL11 (red, 1EOT) (Crump et al., 1998) bound to CCR3 (blue) reconstituted in lipids (green headgroups) formed into either SMALPs with SMA (left, yellow) or proteoliposomes (right) with Gαi3 (purple, 2ODE) (Soundararajan et al., 2008) bound to CCR3. Points indicate the mean ± standard deviation for three replicates. Bars indicate statistical significance based on the Student’s t-test: *p < 0.05, **p < 0.01, ***p < 0.001, ns is not statistically significant.
We first attempted to analyze CCL11-driven GTPase activity of Gαi3 in a SMALP environment as both the extracellular agonist orthosteric pocket and the intracellular G protein docking site would be solvent exposed (Figure 6B; Supplementary Figure S6A). While it is clear that cholesterol increases the extent of hydrolysis, dose dependence could not be verified, and the formation and integrity of the cholesterol-containing SMALPs is in question. This is likely due to the rigidity imparted by increasing the cholesterol concentration, which is documented to inhibit copoloymer intercalation within the membrane (Scheidelaar et al., 2015; Dörr et al., 2016). Unlike in the ligand-binding assay, CCR3 immobilized in SMA-resistant cholesterol proteoliposomes is unable to function in this assay regardless of orientation. This may then explain the puzzling decrease in GTPase activity between 20 and 30% datasets. Furthermore, SMA has a relatively narrow range of compatibility with common biochemical assay components such as divalent cations. Thus, CCL11-driven GTPase activity of Gαi3 in SMALPs could not be verified in the presence of Mg2+, which also adversely influences Gαi3 GTPase activity (Mondal et al., 2015).
To overcome this challenge, we reconstituted CCR3 in the presence of the peptide agonist. CCR3 was reconstituted into proteoliposomes with 0, 20, 40, and 60% cholesterol content with CCL11 present in the buffer at a molar ratio of 1:5 CCR3:CCL11. The GTPase assay was repeated in the presence of 5 mM Mg2+ (Figure 6C; Supplementary Figure S6B). Statistical significance between the GTP hydrolysis signal in 0, 20, and 40% cholesterol is clear evidence of cholesterol dose-dependent modulation of function. Furthermore, GTPase activity in 40 vs. 60% cholesterol proteoliposomes is approximately equivalent, within error, which we also observed in the ligand-binding assay. Together, this corroborates our hypothesis that the dose-dependent modulation of agonist affinity directly translates to receptor activation, Gαi3 coupling, GTP hydrolysis, and thus signal transduction.
Typically, the total exogenous yield of functional GPCRs confounds functional studies. Thus, GPCR production continues to be an area of innovation (Abiko et al., 2021; Mulry et al., 2021). Here, we implemented two techniques, codon harmonization and an MBP fusion tag, to facilitate the production of a functional human GPCR from E. coli. Codon harmonization is underutilized ostensibly due to varying degrees of experimental success. This is likely a result of the sheer number of variables that contribute to protein expression and folding (Quax et al., 2015). The use of an MBP fusion tag has also gained popularity, and successful implementation has been observed in a few cases (Bertin et al., 1992; Yeliseev et al., 2007; Serrano-Vega et al., 2008; Egloff et al., 2014; Beckner et al., 2020). Lack of widespread adoption of MBP tags for heterologous GPCR expression is, in our opinion, due to the difficulty of tag removal, as the TEV is known to be inhibited by detergents (Mohanty et al., 2003). This study represents an important step in the successful implementation of a dual heterologous expression strategy that we hypothesize will be of use both in our own future experiments and the wider GPCR structural biology community.
It was shown that the titration of membrane cholesterol increases the affinity of CCR3 for its endogenous ligand CCL11 and that this observation directly translates to agonist-driven Gαi3 GTP hydrolysis as a proxy for signal transduction. We hypothesize that the functional interplay between cholesterol, ligand affinity, and G protein docking is due to cholesterol-driven conformational sampling inhibition. Such observations were made previously, where cholesterol was hypothesized to constrain GPCR conformational selection to states with higher ligand affinity (Weis and Kobilka, 2018). Cholesterol’s influence on chemokine receptor ligand affinity has, however, been shown sparingly (Babcock et al., 2003; Calmet et al., 2020) and typically not dose-dependently. Such observations are likely conserved in chemokine receptors; therefore, we conclude that cholesterol enhances CCR3 ligand binding through a direct allosteric mechanism that is directly translated to Gαi3 coupling and GTP hydrolysis. Thus, our data indicate that cholesterol is a positive allosteric modulator of CCR3 signal transduction.
Although similar fluorescence experiments have been reported (Kawamura et al., 2014; Purvanov et al., 2018; Matti et al., 2020), the use of a recombinant, fluorescently labeled endogenous ligand is nontrivial and may function as a useful tool in future experiments involving in vitro mimicry of native biological interactions. Cholesterol as a modulator of agonist-driven GTP hydrolysis of a chemokine receptor is novel to this work. Implementation of SMALP technology is also nontrivial, although the buffer component and lipid incompatibilities still leave room for improvement. Zwitterionic SMALP polymers have broader compatibility with common biochemical reagents and may serve as a starting place in future works (Fiori et al., 2020). This may be the reason this GTPase assay is not more widely adopted in lipid environments, although some experimental evidence is adopted in high-density lipoprotein particles (nanodiscs) (Staus et al., 2019; Huang et al., 2021), and mixed micelles (Gregorio et al., 2017) are evident.
We have shown that Gαi3–CCR3 coupling is possible in vitro, but what role this might play in vivo is open to speculation. The main cellular function of Gαi subunits is as intracellular Ca2+ ion concentration and adenylate cyclase effectors (Peleg et al., 2002). However, an additional function as modulators of G protein-gated inwardly rectifying K+ channels (GIRKs) is well documented (Peleg et al., 2002; Rubinstein et al., 2007). Gαi3, specifically, was originally named Gk due to its role in the stimulation of GIRK function (Codina et al., 1987; Codina et al., 1988). A mounting body of evidence now suggests some interplay between chemokine expression and nervous system physiology, with CXCL12, the predominant endogenous ligand of CXCR4, strongly implicated in this phenomenon (Guyon, 2014). Further evidence implicates CXCL12 and CCL5 in the activation of GIRK functionality in vivo (Picciocchi et al., 2014), the latter of which is a ligand for CCR3 (Zhang et al., 2015). Thus, there is precedence for chemokine receptor–driven modulation of GIRK functionality. Although predominantly expressed by eosinophils, studies have identified expression of CCR3 in neurons, astrocytes, and microglia (Cho and Miller, 2002; Banisadr et al., 2005). It has been implicated in the pathogenesis of neuroinflammatory conditions such as Parkinson’s disease (Moghadam-Ahmadi et al., 2020) and Alzheimer’s disease (Sui et al., 2019), as well as multiple sclerosis and HIV-associated dementia (Banisadr et al., 2005). Of note, GIRK channels play a direct role in Parkinson’s pathophysiology (Mayfield et al., 2015), and upregulation of serum CCL5 is correlated with disease severity (Tang et al., 2014). Typically, upregulation of CCL5 is related to CCR5 function, but there may be a role for CCL5 and other chemokines triggering CCR3–Gαi3 coupling in neurodegenerative disorders that is certainly worth exploring.
The original contributions presented in the study are included in the article/Supplementary Material; further inquiries can be directed to the corresponding author.
EA designed and performed all experiments in coordination with BW. EA drafted the article. EA and BW contributed to subsequent revisions.
This work was funded by the NIH grant 1R35GM124979 (Maximizing Investigators’ Research Award (MIRA) R35) awarded to BW.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We would like to thank Matthew Dominguez, Isaac Scott, and Bryan Sutton (TTU Health Sciences Center) for aid in the use of a spectrometer to acquire the CD spectra. We would like to thank Michael P. Latham (Texas Tech University) for the generous gift of the TEV protease plasmid and for the use of a fluorescence plate reader in the fluorescence polarization ligand-binding assay. We would like to thank Shane Scoggin and Naima Moustaid-Moussa (Texas Tech University) for providing the use of a luminescent plate reader in the GTP hydrolysis assay.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmolb.2021.724603/full#supplementary-material
Abdul-Gader, A., Miles, A. J., and Wallace, B. A. (2011). A Reference Dataset for the Analyses of Membrane Protein Secondary Structures and Transmembrane Residues Using Circular Dichroism Spectroscopy. Bioinformatics 27, 1630–1636. doi:10.1093/bioinformatics/btr234
Abiko, L. A., Rogowski, M., Gautier, A., Schertler, G., and Grzesiek, S. (2021). Efficient Production of a Functional G Protein-Coupled Receptor in E. coli for Structural Studies. J. Biomol. NMR 75, 25–38. doi:10.1007/s10858-020-00354-6
Amani, R., Borcik, C. G., Khan, N. H., Versteeg, D. B., Yekefallah, M., Do, H. Q., et al. (2020). Conformational Changes upon Gating of KirBac1.1 into an Open-Activated State Revealed by Solid-State NMR and Functional Assays. Proc. Natl. Acad. Sci. USA 117, 2938–2947. doi:10.1073/pnas.1915010117
Andrade, M. A., Chacón, P., Merelo, J. J., and Morán, F. (1993). Evaluation of Secondary Structure of Proteins from UV Circular Dichroism Spectra Using an Unsupervised Learning Neural Network. Protein Eng. Des. Sel 6, 383–390. doi:10.1093/protein/6.4.383
Angov, E., Hillier, C. J., Kincaid, R. L., and Lyon, J. A. (2008). Heterologous Protein Expression Is Enhanced by Harmonizing the Codon Usage Frequencies of the Target Gene with Those of the Expression Host. PLoS One 3, e2189: doi:10.1371/journal.pone.0002189
Babcock, G. J., Farzan, M., and Sodroski, J. (2003). Ligand-independent Dimerization of CXCR4, a Principal HIV-1 Coreceptor. J. Biol. Chem. 278, 3378–3385. doi:10.1074/jbc.M210140200
Ballesteros, J. A., and Weinstein, H. (1995). [19] Integrated Methods for the Construction of Three-Dimensional Models and Computational Probing of Structure-Function Relations in G Protein-Coupled Receptors. Receptor Mol. Biol., 366–428. doi:10.1016/s1043-9471(05)80049-7
Banisadr, G., Rostène, W., Kitabgi, P., and Parsadaniantz, S. (2005). Chemokines and Brain Functions. Cdtia 4, 387–399. doi:10.2174/1568010054022097
Beckner, R. L., Zoubak, L., Hines, K. G., Gawrisch, K., and Yeliseev, A. A. (2020). Probing Thermostability of Detergent-Solubilized CB2 Receptor by Parallel G Protein-Activation and Ligand-Binding Assays. J. Biol. Chem. 295, 181–190. doi:10.1074/jbc.RA119.010696
Bertin, B., Freissmuth, M., Breyer, R. M., Schütz, W., Strosberg, A. D., and Marullo, S. (1992). Functional Expression of the Human Serotonin 5-HT1A Receptor in Escherichia coli. Ligand Binding Properties and Interaction with Recombinant G Protein Alpha-Subunits. J. Biol. Chem. 267, 8200–8206. doi:10.1016/s0021-9258(18)42427-1
Bhate, M. P., Wylie, B. J., Thompson, A., Tian, L., Nimigean, C., and McDermott, A. E. (2013). Preparation of Uniformly Isotope Labeled KcsA for Solid State NMR: Expression, Purification, Reconstitution into Liposomes and Functional Assay. Protein Expr. Purif. 91, 119–124. doi:10.1016/j.pep.2013.07.013
Borcik, C. G., Versteeg, D. B., Amani, R., Yekefallah, M., Khan, N. H., and Wylie, B. J. (2020). The Lipid Activation Mechanism of a Transmembrane Potassium Channel. J. Am. Chem. Soc. 142, 14102–14116. doi:10.1021/jacs.0c01991
Borcik, C. G., Versteeg, D. B., and Wylie, B. J. (2019). An Inward-Rectifier Potassium Channel Coordinates the Properties of Biologically Derived Membranes. Biophysical J. 116, 1701–1718. doi:10.1016/j.bpj.2019.03.023
Botelho, A. V., Huber, T., Sakmar, T. P., and Brown, M. F. (2006). Curvature and Hydrophobic Forces Drive Oligomerization and Modulate Activity of Rhodopsin in Membranes. Biophysical J. 91, 4464–4477. doi:10.1529/biophysj.106.082776
Calmet, P., Cullin, C., Cortès, S., Vang, M., Caudy, N., Baccouch, R., et al. (2020). Cholesterol Impacts Chemokine CCR5 Receptor Ligand‐binding Activity. FEBS J. 287, 2367–2385. doi:10.1111/febs.15145
Chattopadhyay, A. (2014). GPCRs: Lipid-dependent Membrane Receptors that Act as Drug Targets. Adv. Biol. 2014, 1–12. doi:10.1155/2014/143023
Chen, X., Zaro, J. L., and Shen, W.-C. (2013). Fusion Protein Linkers: Property, Design and Functionality. Adv. Drug Deliv. Rev. 65, 1357–1369. doi:10.1016/j.addr.2012.09.039
Cho, C., and Miller, R. J. (2002). Chemokine Receptors and Neural Function. J. Neurovirol. 8, 573–584. doi:10.1080/13550280290101003
Codina, J., Olate, J., Abramowitz, J., Mattera, R., Cook, R. G., and Birnbaumer, L. (1988). Alpha I-3 cDNA Encodes the Alpha Subunit of Gk, the Stimulatory G Protein of Receptor-Regulated K+ Channels. J. Biol. Chem. 263, 6746–6750. doi:10.1016/s0021-9258(18)68706-x
Codina, J., Yatani, A., Grenet, D., Brown, A., and Birnbaumer, L. (1987). The Alpha Subunit of the GTP Binding Protein Gk Opens Atrial Potassium Channels. Science 236, 442–445. doi:10.1126/science.2436299
Crump, M. P., Rajarathnam, K., Kim, K.-S., Clark-Lewis, I., and Sykes, B. D. (1998). Solution Structure of Eotaxin, a Chemokine that Selectively Recruits Eosinophils in Allergic Inflammation. J. Biol. Chem. 273, 22471–22479. doi:10.1074/jbc.273.35.22471
Dawaliby, R., Trubbia, C., Delporte, C., Masureel, M., Van Antwerpen, P., Kobilka, B. K., et al. (2016). Allosteric Regulation of G Protein-Coupled Receptor Activity by Phospholipids. Nat. Chem. Biol. 12, 35–39. doi:10.1038/nchembio.1960
Deng, H., Liu, R., Ellmeier, W., Choe, S., Unutmaz, D., Burkhart, M., et al. (1996). Identification of a Major Co-receptor for Primary Isolates of HIV-1. Nature 381, 661–666. doi:10.1038/381661a0
Dörr, J. M., Scheidelaar, S., Koorengevel, M. C., Dominguez, J. J., Schäfer, M., van Walree, C. A., et al. (2016). The Styrene-Maleic Acid Copolymer: a Versatile Tool in Membrane Research. Eur. Biophys. J. 45, 3–21. doi:10.1007/s00249-015-1093-y
Duchesnes, C. E., Murphy, P. M., Williams, T. J., and Pease, J. E. (2006). Alanine Scanning Mutagenesis of the Chemokine Receptor CCR3 Reveals Distinct Extracellular Residues Involved in Recognition of the Eotaxin Family of Chemokines. Mol. Immunol. 43, 1221–1231. doi:10.1016/j.molimm.2005.07.015
Dunn, J. L. M., Shoda, T., Caldwell, J. M., Wen, T., Aceves, S. S., Collins, M. H., et al. (2020). Esophageal Type 2 Cytokine Expression Heterogeneity in Eosinophilic Esophagitis in a Multisite Cohort. J. Allergy Clin. Immunol. 145, 1629–1640.e1624. doi:10.1016/j.jaci.2020.01.051
Egloff, P., Hillenbrand, M., Klenk, C., Batyuk, A., Heine, P., Balada, S., et al. (2014). Structure of Signaling-Competent Neurotensin Receptor 1 Obtained by Directed Evolution inEscherichia Coli. Proc. Natl. Acad. Sci. USA 111, E655–E662. doi:10.1073/pnas.1317903111
Federman, A. D., Conklin, B. R., Schrader, K. A., Reed, R. R., and Bourne, H. R. (1992). Hormonal Stimulation of Adenylyl Cyclase through Gi-Protein βγ Subunits. Nature 356, 159–161. doi:10.1038/356159a0
Fiori, M. C., Zheng, W., Kamilar, E., Simiyu, G., Altenberg, G. A., and Liang, H. (2020). Extraction and Reconstitution of Membrane Proteins into Lipid Nanodiscs Encased by Zwitterionic Styrene-Maleic Amide Copolymers. Sci. Rep. 10, 9940. doi:10.1038/s41598-020-66852-7
Ford, S. R., and Leach, F. R. (1998a). Bioluminescent Assay of the Guanylates. Methods Mol. Biol. 102, 55–68. doi:10.1385/0-89603-520-4:55
Ford, S. R., and Leach, F. R. (1998b). Improvements in the Application of Firefly Luciferase Assays. Methods Mol. Biol. 102, 3–20. doi:10.1385/0-89603-520-4:3
Ford, S. R., Vaden, V. R., Booth, J. L., Hall, M. S., Webster, J. J., and Leach, F. R. (1994). Bioluminescent Determination of 0.1 Picomole Amounts of Guanine Nucleotides. J. Biolumin. Chemilumin. 9, 251–265. doi:10.1002/bio.1170090403
Fuhrmann, M., Hausherr, A., Ferbitz, L., Schödl, T., Heitzer, M., and Hegemann, P. (2004). Monitoring Dynamic Expression of Nuclear Genes in Chlamydomonas Reinhardtii by Using a Synthetic Luciferase Reporter Gene. Plant Mol. Biol. 55, 869–881. doi:10.1007/s11103-004-2150-610.1007/s11103-005-2150-1
Gahbauer, S., Pluhackova, K., and Böckmann, R. A. (2018). Closely Related, yet Unique: Distinct Homo- and Heterodimerization Patterns of G Protein Coupled Chemokine Receptors and Their fine-tuning by Cholesterol. Plos Comput. Biol. 14, e1006062. doi:10.1371/journal.pcbi.1006062
Gauvreau, G. M., FitzGerald, J. M., Boulet, L. P., Watson, R. M., Hui, L., Villineuve, H., et al. (2018). The Effects of a CCR3 Inhibitor, AXP1275, on Allergen-Induced Airway Responses in Adults with Mild-To-Moderate Atopic Asthma. Clin. Exp. Allergy 48, 445–451. doi:10.1111/cea.13114
Ge, B., Wang, M., Li, J., Liu, J., and Huang, F. (2015). Maltose Binding Protein Facilitates Functional Production of Engineered Human Chemokine Receptor 3 in Escherichia coli. Process Biochem. 50, 285–293. doi:10.1016/j.procbio.2014.12.001
Gregorio, G. G., Masureel, M., Hilger, D., Terry, D. S., Juette, M., Zhao, H., et al. (2017). Single-molecule Analysis of Ligand Efficacy in β2AR-G-protein Activation. Nature 547, 68–73. doi:10.1038/nature22354
Guyon, A. (2014). CXCL12 Chemokine and its Receptors as Major Players in the Interactions between Immune and Nervous Systems. Front. Cel. Neurosci. 8, 65. doi:10.3389/fncel.2014.00065
Hanson, M. A., Cherezov, V., Griffith, M. T., Roth, C. B., Jaakola, V.-P., Chien, E. Y. T., et al. (2008). A Specific Cholesterol Binding Site Is Established by the 2.8 Å Structure of the Human β2-Adrenergic Receptor. Structure 16, 897–905. doi:10.1016/j.str.2008.05.001
He, J., Chen, Y., Farzan, M., Choe, H., Ohagen, A., Gartner, S., et al. (1997). CCR3 and CCR5 Are Co-receptors for HIV-1 Infection of Microglia. Nature 385, 645–649. doi:10.1038/385645a0
Huang, S. K., Pandey, A., Tran, D. P., Villanueva, N. L., Kitao, A., Sunahara, R. K., et al. (2021). Delineating the Conformational Landscape of the Adenosine A2A Receptor during G Protein Coupling. Cell 184, 1884–1894. e1814. doi:10.1016/j.cell.2021.02.041
Ishida, Y., Kido, A., Akahane, M., Kishi, S., Tsukamoto, S., Fujii, H., et al. (2018). Mesenchymal Stem Cells Up-Regulate the Invasive Potential of Prostate Cancer Cells via the eotaxin-3/CCR3 axis. Pathol. - Res. Pract. 214, 1297–1302. doi:10.1016/j.prp.2018.06.012
Jafurulla, M., Aditya Kumar, G., Rao, B. D., and Chattopadhyay, A. (2019). A Critical Analysis of Molecular Mechanisms Underlying Membrane Cholesterol Sensitivity of GPCRs. Cholesterol Modulation Protein Funct. Sterol Specificity Indirect Mech. 1115, 21–52. doi:10.1007/978-3-030-04278-3_2
Jin, L., Liu, W.-R., Tian, M.-X., Jiang, X.-F., Wang, H., Zhou, P.-Y., et al. (2017). CCL24 Contributes to HCC Malignancy via RhoB- VEGFA-VEGFR2 Angiogenesis Pathway and Indicates Poor Prognosis. Oncotarget 8, 5135–5148. doi:10.18632/oncotarget.14095
Jöhrer, K., Zelle-Rieser, C., Perathoner, A., Moser, P., Hager, M., Ramoner, R., et al. (2005). Up-regulation of Functional Chemokine Receptor CCR3 in Human Renal Cell Carcinoma. Clin. Cancer Res. 11, 2459–2465. doi:10.1158/1078-0432.CCR-04-0405
Jones, A. J. Y., Gabriel, F., Tandale, A., and Nietlispach, D. (2020). Structure and Dynamics of GPCRs in Lipid Membranes: Physical Principles and Experimental Approaches. Molecules 25, 4729. doi:10.3390/molecules25204729
Kapust, R. B., Tözsér, J., Fox, J. D., Anderson, D. E., Cherry, S., Copeland, T. D., et al. (2001). Tobacco Etch Virus Protease: Mechanism of Autolysis and Rational Design of Stable Mutants with Wild-type Catalytic Proficiency. Protein Eng. 14, 993–1000. doi:10.1093/protein/14.12.993
Katschke, K. J., Rottman, J. B., Ruth, J. H., Qin, S., Wu, L., LaRosa, G., et al. (2001). Differential Expression of Chemokine Receptors on Peripheral Blood, Synovial Fluid, and Synovial Tissue Monocytes/macrophages in Rheumatoid Arthritis. Arthritis Rheum., 44. 2-N: CO, 1022–1032. doi:10.1002/1529-0131(200105)44:5<1022::AID-ANR181>3.010.1002/1529-0131(200105)44:5<1022::aid-anr181>3.0.co;2-n
Katz, A., Wu, D., and Simon, M. I. (1992). Subunits βγ of Heterotrimeric G Protein Activate β2 Isoform of Phospholipase C. Nature 360, 686–689. doi:10.1038/360686a0
Kawamura, T., Stephens, B., Qin, L., Yin, X., Dores, M. R., Smith, T. H., et al. (2014). A General Method for Site Specific Fluorescent Labeling of Recombinant Chemokines. PLoS One 9, e81454. doi:10.1371/journal.pone.0081454
Kim, T. H., Chung, K. Y., Manglik, A., Hansen, A. L., Dror, R. O., Mildorf, T. J., et al. (2013). The Role of Ligands on the Equilibria between Functional States of a G Protein-Coupled Receptor. J. Am. Chem. Soc. 135, 9465–9474. doi:10.1021/ja404305k
Kitaura, M., Nakajima, T., Imai, T., Harada, S., Combadiere, C., Tiffany, H. L., et al. (1996). Molecular Cloning of Human Eotaxin, an Eosinophil-Selective CC Chemokine, and Identification of a Specific Eosinophil Eotaxin Receptor, CC Chemokine Receptor 3. J. Biol. Chem. 271, 7725–7730. doi:10.1074/jbc.271.13.7725
Laemmli, U. K. (1970). Cleavage of Structural Proteins during the Assembly of the Head of Bacteriophage T4. Nature 227, 680–685. doi:10.1038/227680a0
Legler, D. F., Matti, C., Laufer, J. M., Jakobs, B. D., Purvanov, V., Uetz-von Allmen, E., et al. (2017). Modulation of Chemokine Receptor Function by Cholesterol: New Prospects for Pharmacological Intervention. Mol. Pharmacol. 91, 331–338. doi:10.1124/mol.116.107151
Ma, Q., Jones, D., Borghesani, P. R., Segal, R. A., Nagasawa, T., Kishimoto, T., et al. (1998). Impaired B-Lymphopoiesis, Myelopoiesis, and Derailed Cerebellar Neuron Migration in CXCR4- and SDF-1-Deficient Mice. Pnas 95, 9448–9453. doi:10.1073/pnas.95.16.9448
Mao, D., Wachter, E., and Wallace, B. A. (1982). Folding of the Mitochondrial Proton Adenosine Triphosphatase Proteolipid Channel in Phospholipid Vesicles. Biochemistry 21, 4960–4968. doi:10.1021/bi00263a020
Marlow, B., Kuenze, G., Li, B., Sanders, C. R., and Meiler, J. (2021). Structural Determinants of Cholesterol Recognition in Helical Integral Membrane Proteins. Biophysical J. 120, 1592–1604. doi:10.1016/j.bpj.2021.02.028
Matti, C., D'Uonnolo, G., Artinger, M., Melgrati, S., Salnikov, A., Thelen, S., et al. (2020). CCL20 Is a Novel Ligand for the Scavenging Atypical Chemokine Receptor 4. J. Leukoc. Biol. 107, 1137–1154. doi:10.1002/JLB.2MA0420-295RRR
Mayfield, J., Blednov, Y. A., and Harris, R. A. (2015). Behavioral and Genetic Evidence for GIRK Channels in the CNS. Int. Rev. Neurobiol. 123, 279–313. doi:10.1016/bs.irn.2015.05.016
Micsonai, A., Bulyáki, É., and Kardos, J. (2021). BeStSel: From Secondary Structure Analysis to Protein Fold Prediction by Circular Dichroism Spectroscopy. Methods Mol. Biol. 2199, 175–189. doi:10.1007/978-1-0716-0892-0_11
Micsonai, A., Wien, F., Bulyáki, É., Kun, J., Moussong, É., Lee, Y.-H., et al. (2018). BeStSel: a Web Server for Accurate Protein Secondary Structure Prediction and Fold Recognition from the Circular Dichroism Spectra. Nucleic Acids Res. 46, W315–W322. doi:10.1093/nar/gky497
Micsonai, A., Wien, F., Kernya, L., Lee, Y.-H., Goto, Y., Réfrégiers, M., et al. (2015). Accurate Secondary Structure Prediction and Fold Recognition for Circular Dichroism Spectroscopy. Proc. Natl. Acad. Sci. USA 112, E3095–E3103. doi:10.1073/pnas.1500851112
Millard, C. J., Ludeman, J. P., Canals, M., Bridgford, J. L., Hinds, M. G., Clayton, D. J., et al. (2014). Structural Basis of Receptor Sulfotyrosine Recognition by a CC Chemokine: the N-Terminal Region of CCR3 Bound to CCL11/eotaxin-1. Structure 22, 1571–1581. doi:10.1016/j.str.2014.08.023
Mingqing Wang, B. G., Yang, Qiuxia., Xiaoyong, Jiang., and Huang, Fang. (2014). High-level Production of Biologically Active Chemokines in Escherichia coli. 4 ed.
Moghadam-Ahmadi, A., Khorramdelazad, H., Hassanshahi, G., Shahsavari, S., Moadab, A., and Vakilian, A. (2020). Eotaxins and C-C Chemokine Receptor Type 3 in Parkinson's Disease. Acta Neurol. Belg. 120, 589–594. doi:10.1007/s13760-018-01061-8
Mohanty, A. K., Simmons, C. R., and Wiener, M. C. (2003). Inhibition of Tobacco Etch Virus Protease Activity by Detergents. Protein Expr. Purif. 27, 109–114. doi:10.1016/s1046-5928(02)00589-2
Mondal, S., Hsiao, K., and Goueli, S. A. (2015). A Homogenous Bioluminescent System for Measuring GTPase, GTPase Activating Protein, and Guanine Nucleotide Exchange Factor Activities. ASSAY Drug Dev. Tech. 13, 444–455. doi:10.1089/adt.2015.643
Mulry, E., Ray, A. P., and Eddy, M. T. (2021). Production of a Human Histamine Receptor for NMR Spectroscopy in Aqueous Solutions. Biomolecules 11, 632. doi:10.3390/biom11050632
Nishida, M., Cadene, M., Chait, B. T., and MacKinnon, R. (2007). Crystal Structure of a Kir3.1-prokaryotic Kir Channel Chimera. Embo J. 26, 4005–4015. doi:10.1038/sj.emboj.7601828
Pándy-Szekeres, G., Munk, C., Tsonkov, T. M., Mordalski, S., Harpsøe, K., Hauser, A. S., et al. (2018). GPCRdb in 2018: Adding GPCR Structure Models and Ligands. Nucleic Acids Res. 46, D440–D446. doi:10.1093/nar/gkx1109
Pegan, S., Arrabit, C., Zhou, W., Kwiatkowski, W., Collins, A., Slesinger, P. A., et al. (2005). Cytoplasmic Domain Structures of Kir2.1 and Kir3.1 Show Sites for Modulating Gating and Rectification. Nat. Neurosci. 8, 279–287. doi:10.1038/nn1411
Peleg, S., Varon, D., Ivanina, T., Dessauer, C. W., and Dascal, N. (2002). Gαi Controls the Gating of the G Protein-Activated K+ Channel, GIRK. Neuron 33, 87–99. doi:10.1016/s0896-6273(01)00567-0
Picciocchi, A., Šiaučiūnaiteė-Gaubard, L., Petit-Hartlein, I., Sadir, R., Revilloud, J., Caro, L., et al. (2014). C-terminal Engineering of CXCL12 and CCL5 Chemokines: Functional Characterization by Electrophysiological Recordings. PLoS One 9, e87394. doi:10.1371/journal.pone.0087394
Pluhackova, K., Gahbauer, S., Kranz, F., Wassenaar, T. A., and Böckmann, R. A. (2016). Dynamic Cholesterol-Conditioned Dimerization of the G Protein Coupled Chemokine Receptor Type 4. Plos Comput. Biol. 12, e1005169. doi:10.1371/journal.pcbi.1005169
Provencher, S. W., and Gloeckner, J. (1981). Estimation of Globular Protein Secondary Structure from Circular Dichroism. Biochemistry 20, 33–37. doi:10.1021/bi00504a006
Purvanov, V., Matti, C., Samson, G. P. B., Kindinger, I., and Legler, D. F. (2018). Fluorescently Tagged CCL19 and CCL21 to Monitor CCR7 and ACKR4 Functions. Ijms 19, 3876. doi:10.3390/ijms19123876
Quax, T. E. F., Claassens, N. J., Söll, D., and van der Oost, J. (2015). Codon Bias as a Means to Fine-Tune Gene Expression. Mol. Cel 59, 149–161. doi:10.1016/j.molcel.2015.05.035
Rodriguez, A., Wright, G., Emrich, S., and Clark, P. L. (2018). %MinMax: A Versatile Tool for Calculating and Comparing Synonymous Codon Usage and its Impact on Protein Folding. Protein Sci. 27, 356–362. doi:10.1002/pro.3336
Rosenbaum, D. M., Rasmussen, S. G. F., and Kobilka, B. K. (2009). The Structure and Function of G-Protein-Coupled Receptors. Nature 459, 356–363. doi:10.1038/nature08144
Rossi, A. M., and Taylor, C. W. (2011). Analysis of Protein-Ligand Interactions by Fluorescence Polarization. Nat. Protoc. 6, 365–387. doi:10.1038/nprot.2011.305
Rubinstein, M., Peleg, S., Berlin, S., Brass, D., and Dascal, N. (2007). Gαi3primes the G Protein-Activated K+channels for Activation by Coexpressed Gβγ in intactXenopusoocytes. J. Physiol. 581, 17–32. doi:10.1113/jphysiol.2006.125864
Scheidelaar, S., Koorengevel, M. C., Pardo, J. D., Meeldijk, J. D., Breukink, E., and Killian, J. A. (2015). Molecular Model for the Solubilization of Membranes into Nanodisks by Styrene Maleic Acid Copolymers. Biophysical J. 108, 279–290. doi:10.1016/j.bpj.2014.11.3464
Serrano-Vega, M. J., Magnani, F., Shibata, Y., and Tate, C. G. (2008). Conformational Thermostabilization of the 1-adrenergic Receptor in a Detergent-Resistant Form. Proc. Natl. Acad. Sci. 105, 877–882. doi:10.1073/pnas.0711253105
Song, Y., DiMaio, F., Wang, R. Y.-R., Kim, D., Miles, C., Brunette, T., et al. (2013). High-resolution Comparative Modeling with RosettaCM. Structure 21, 1735–1742. doi:10.1016/j.str.2013.08.005
Song, Y., Ge, B., Lao, J., Wang, Z., Yang, B., Wang, X., et al. (2018). Regulation of the Oligomeric Status of CCR3 with Binding Ligands Revealed by Single-Molecule Fluorescence Imaging. Biochemistry 57, 852–860. doi:10.1021/acs.biochem.7b00676
Soundararajan, M., Willard, F. S., Kimple, A. J., Turnbull, A. P., Ball, L. J., Schoch, G. A., et al. (2008). Structural Diversity in the RGS Domain and its Interaction with Heterotrimeric G Protein -subunits. Proc. Natl. Acad. Sci. 105, 6457–6462. doi:10.1073/pnas.0801508105
Sreerama, N., and Woody, R. W. (2000). Estimation of Protein Secondary Structure from Circular Dichroism Spectra: Comparison of CONTIN, SELCON, and CDSSTR Methods with an Expanded Reference Set. Anal. Biochem. 287, 252–260. doi:10.1006/abio.2000.4880
Staus, D. P., Wingler, L. M., Pichugin, D., Prosser, R. S., and Lefkowitz, R. J. (2019). Detergent- and Phospholipid-Based Reconstitution Systems Have Differential Effects on Constitutive Activity of G-Protein-Coupled Receptors. J. Biol. Chem. 294, 13218–13223. doi:10.1074/jbc.AC119.009848
Stone, M., Hayward, J., Huang, C. Z. E. H., and E. Huma, Z. (2017). Mechanisms of Regulation of the Chemokine-Receptor Network. Ijms 18, 342. doi:10.3390/ijms18020342
Sui, Y., Zhang, Y., Dong, C., Xu, B., and Sun, X. (2019). The Small Molecular CCR3 Antagonist YM344031 Attenuates Neurodegenerative Pathologies and Improves Learning and Memory Performance in a Mouse Model of Alzheimer's Disease. Brain Res. 1719, 1–10. doi:10.1016/j.brainres.2019.05.022
Sullivan, K. A., Miller, R. T., Masters, S. B., Beiderman, B., Heideman, W., and Bourne, H. R. (1987). Identification of Receptor Contact Site Involved in Receptor-G Protein Coupling. Nature 330, 758–760. doi:10.1038/330758a0
Tan, Q., Zhu, Y., Li, J., Chen, Z., Han, G. W., Kufareva, I., et al. (2013a). Structure of the CCR5 Chemokine Receptor-HIV Entry Inhibitor Maraviroc Complex. Science 341, 1387–1390. doi:10.1126/science.1241475
Tan, Q., Zhu, Y., Li, J., Chen, Z., Han, G. W., Kufareva, I., et al. (2013b). Structure of the CCR5 Chemokine Receptor-HIV Entry Inhibitor Maraviroc Complex. Science 341, 1387–1390. doi:10.1126/science.1241475
Tang, P., Chong, L., Li, X., Liu, Y., Liu, P., Hou, C., et al. (2014). Correlation between Serum RANTES Levels and the Severity of Parkinson's Disease. Oxidative Med. Cell Longevity 2014, 1–4. doi:10.1155/2014/208408
van Aalst, E., Yekefallah, M., Mehta, A. K., Eason, I., and Wylie, B. (2020). Codon Harmonization of a Kir3.1-KirBac1.3 Chimera for Structural Study Optimization. Biomolecules 10, 430. doi:10.3390/biom10030430
van Zundert, G. C. P., Rodrigues, J. P. G. L. M., Trellet, M., Schmitz, C., Kastritis, P. L., Karaca, E., et al. (2016). The HADDOCK2.2 Web Server: User-Friendly Integrative Modeling of Biomolecular Complexes. J. Mol. Biol. 428, 720–725. doi:10.1016/j.jmb.2015.09.014
Wall, M. A., Coleman, D. E., Lee, E., Iñiguez-Lluhi, J. A., Posner, B. A., Gilman, A. G., et al. (1995). The Structure of the G Protein Heterotrimer Giα1β1γ2. Cell 83, 1047–1058. doi:10.1016/0092-8674(95)90220-1
Wang, M., Ge, B., Li, R., Wang, X., Lao, J., and Huang, F. (2013). Milligram Production and Biological Activity Characterization of the Human Chemokine Receptor CCR3. PLoS One 8, e65500. doi:10.1371/journal.pone.0065500
Weis, W. I., and Kobilka, B. K. (2018). The Molecular Basis of G Protein-Coupled Receptor Activation. Annu. Rev. Biochem. 87, 897–919. doi:10.1146/annurev-biochem-060614-033910
Whitmore, L., and Wallace, B. A. (2004). DICHROWEB, an Online Server for Protein Secondary Structure Analyses from Circular Dichroism Spectroscopic Data. Nucleic Acids Res. 32, W668–W673. doi:10.1093/nar/gkh371
Whitmore, L., and Wallace, B. A. (2008). Protein Secondary Structure Analyses from Circular Dichroism Spectroscopy: Methods and Reference Databases. Biopolymers 89, 392–400. doi:10.1002/bip.20853
Xu, P., Huang, S., Zhang, H., Mao, C., Zhou, X. E., Cheng, X., et al. (2021). Structural Insights into the Lipid and Ligand Regulation of Serotonin Receptors. Nature 592, 469–473. doi:10.1038/s41586-021-03376-8
Yeliseev, A., Zoubak, L., and Gawrisch, K. (2007). Use of Dual Affinity Tags for Expression and Purification of Functional Peripheral Cannabinoid Receptor. Protein Expr. Purif. 53, 153–163. doi:10.1016/j.pep.2006.12.003
Yen, H.-Y., Hoi, K. K., Liko, I., Hedger, G., Horrell, M. R., Song, W., et al. (2018). PtdIns(4,5)P2 Stabilizes Active States of GPCRs and Enhances Selectivity of G-Protein Coupling. Nature 559, 423–427. doi:10.1038/s41586-018-0325-6
Zhang, Q., Qin, J., Zhong, L., Gong, L., Zhang, B., Zhang, Y., et al. (2015). CCL5-Mediated Th2 Immune Polarization Promotes Metastasis in Luminal Breast Cancer. Cancer Res. 75, 4312–4321. doi:10.1158/0008-5472.CAN-14-3590
Keywords: GPCR, CCR3, chemokine receptor, CCL11, cholesterol, G protein, lipid allostery, codon harmonization, GPCR
Citation: van Aalst E and Wylie BJ (2021) Cholesterol Is a Dose-Dependent Positive Allosteric Modulator of CCR3 Ligand Affinity and G Protein Coupling. Front. Mol. Biosci. 8:724603. doi: 10.3389/fmolb.2021.724603
Received: 13 June 2021; Accepted: 14 July 2021;
Published: 20 August 2021.
Edited by:
Vincenzo Venditti, Iowa State University, United StatesReviewed by:
Oskar Engberg, University Hospital Leipzig, GermanyCopyright © 2021 van Aalst and Wylie. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Benjamin J. Wylie, YmVuamFtaW4uai53eWxpZUB0dHUuZWR1
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.