
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
MINI REVIEW article
Front. Mol. Biosci., 09 August 2021
Sec. Molecular Diagnostics and Therapeutics
Volume 8 - 2021 | https://doi.org/10.3389/fmolb.2021.702484
This article is part of the Research TopicSynthetic Live Biotherapeutic Products for DiseasesView all 8 articles
The microbiome is a collection of genomes from microbiota, including all microorganisms in a niche, through direct and indirect interactions with the host. Certain microorganisms can exist in areas conventionally considered to be sterile, such as the bone matrix. Osseous microbiota dysbiosis caused by host-microbiome perturbation or external infections may ultimately lead to osteomyelitis, a bone inflammatory disorder. Our review covers the current discoveries on the impact of host-microbiome on osteomyelitis and some common osseous diseases. Some studies suggest that the microbiotas from both osseous and non-osseous tissues (e.g., blood or gut) impact the pathogenicity of osteomyelitis and other osseous diseases (e.g., rheumatoid arthritis). We believe that this review will provide readers with a better understanding on the role of the microbiome to the host’s bone health.
Osteomyelitis is a bone inflammatory disease that usually results from microbial infections. In other instances, certain osteomyelitic cases are not caused by microbial infections and exhibit auto-inflammatory bone disorders, e.g., chronic nonbacterial osteomyelitis (Buch et al., 2019). The diverse forms of osteomyelitis are classified according to differentiating features, such as pathogenesis, duration of infection, localization, implant presence, anatomy, and co-morbidity (Zimmerli, 2015).
The recent development of microbiome research shows that the host microbiota interacts with the body to maintain a homeostatic balance or exacerbate the state of infection in the host. Microbiome, the collection of genomes from the microbiota, varies from the different niches in the host (e.g., oral cavity, skin, and gastrointestinal tract). While the microbiome of each niche is unique and separate, it can influence different remote areas of the host and the corresponding microbiome within that niche.
Given the tenacious nature of osteomyelitic infections, investigating the impacts of the host microbiota on osteomyelitis is critical to understanding the pathology, etiology, diagnosis, prevention, therapeutics, and prognosis of osteomyelitis. Osteomyelitis is correlated with the microbiota within the osteomyelitic bones and microbes in blood and other distant organs, including the gastrointestinal tract, via indirect interactions. Herein, we provide a review on the impact of the host-associated microbiome on osteomyelitis. This review would provide readers a better understanding of the role of the host microbiome and how the microbiome dysbiosis influences the host’s susceptibility to osteomyelitis. The review is divided into two main sections addressing direct and indirect microbiome-associated osteomyelitis.
Osteomyelitis is generally associated with microbial infections. These infectious microorganisms (Table 1) in osteomyelitic sites are commonly detected using cultivation-, microscope-, histology- and sequencing-based methods. The conventional cultivating methods mainly utilize aerobic and anaerobic conditions to cultivate microbes from osteomyelitic samples (Lavery et al., 1995; Abdulrazak et al., 2005; Van Asten et al., 2016). In contrast, the emergence of culturomics leverages multiple culture conditions combined with the rapid identification of microbes, rendering isolation of over 3,000 microbial colonies possible (Jneid et al., 2018). Nevertheless, the cultivation-based approaches are only capable of screening species that are culturable under the typical nutritional conditions, thus incapable of reflecting the actual microbial abundance and distribution. Additionally, culturomics requires laborious and time-consuming cultivating steps, limiting its practical applications to large sample sizes.
High-throughput sequencing or the next generation sequencing (NGS) is a swifter and more accurate approach that overcomes the above shortcomings and is currently widely used to quickly and comprehensively investigate microbiome. Common NGS-based methods for studying osteomyelitis comprise the 16S rRNA gene sequencing and metagenome sequencing. The 16S rRNA gene sequencing generally studies the variable regions instead of the whole gene following PCR amplification. In contrast, metagenome sequencing analyzes all microorganisms’ complete genomes in a specimen and provides microbial identity at the species level. However, metagenomics cannot identify the microbial behaviors or the metabolic states (Cassir et al., 2016). Thus, metagenomic sequencing is often coupled to other molecular approaches, e.g., transcriptomics and proteomics, to acquire complete information on the actual functions of the microbiome.
Microscopic methods have also been employed to investigate the osteomyelitic microbiome, providing the exact microbial locations and other taggable biochemical information. These approaches include scanning electron microscopy (SEM) and peptide nucleic acid fluorescent in situ hybridization (PNA-FISH) coupled to confocal laser scanning microscopy (CLSM), e.g., a combinative utilization of them shows the predominance of coccoid microorganisms (Malone et al., 2019) and the biofilm formation (Johani et al., 2019) in osteomyelitic tissues.
These various approaches applied to study the microbiome have their respective advantages and disadvantages. The cultivation-based methods can provide physiological properties and assess the potential for virulence and antimicrobial resistance at the strain level. Molecular methods generally identify more microbes than the culture-based methods (Malone et al., 2019; Zou et al., 2020), and the highly efficient NGS can quickly and accurately provide microbial diversity and distribution (Lagier et al., 2015). The advantages of these methods complement each other limitations and are often performed in tandem for better characterization of the osteomyelitic microbiome. These approaches have been successfully used to study the direct microbiome-associated osteomyelitis of diabetic foot infection and jaw bone infection, and are currently employed to study other osteomyelitic symptoms in many osseous tissues including long bones, vertebral column, clavicle, and sternum.
DFO is a severe form of infection in diabetic patients and can result in lower extremity amputation if left untended. Despite being a severe concern among diabetic patients, there are currently no universally acknowledged guidelines for DFO diagnosis or treatment. DFO is generally accompanied by elevated blood sugar concentration, compromised immunity, and concurrent vascular insufficiency (Lipsky, 1997; Markanday, 2014; Tong et al., 2015), and these complications facilitate the hematogenous or contiguous microbial infections, leading to severe inflammations. The DFO pathogenesis is mainly regulated by the DFO microbiome, which is however influenced by many factors such as demographic characteristics, personal hygiene, grade of severity, and antibiotic therapies (Jneid et al., 2017).
Nevertheless, the DFO microbiota comprises a single dominant microbial species or a complex community infecting the bone. For instance, a study on infected osteomyelitic tissues showed that a single species infection by an anaerobic, non-motile, Gram-negative bacilli Prevotella fusca is positively correlated with the duration of diabetic foot infection (Zou et al., 2020). Other studies discovered that certain Enterococcus faecalis contribute to wound recovery, whereas Staphylococcus aureus infection can exacerbate an infected wound by triggering microbial pathogenesis (Jneid et al., 2018). The wound healing properties of E. faecalis are attributed to its probiotics status and its ability to help regulate the host immune responses (Franz et al., 2011). S. aureus, the most prevalent pathogen in osteomyelitic infections (Jagodzinski et al., 2009; Peltola et al., 2010; Hatzenbuehler and Pulling, 2011; Urish and Cassat, 2020), affects its host in several approaches (Figure 1).
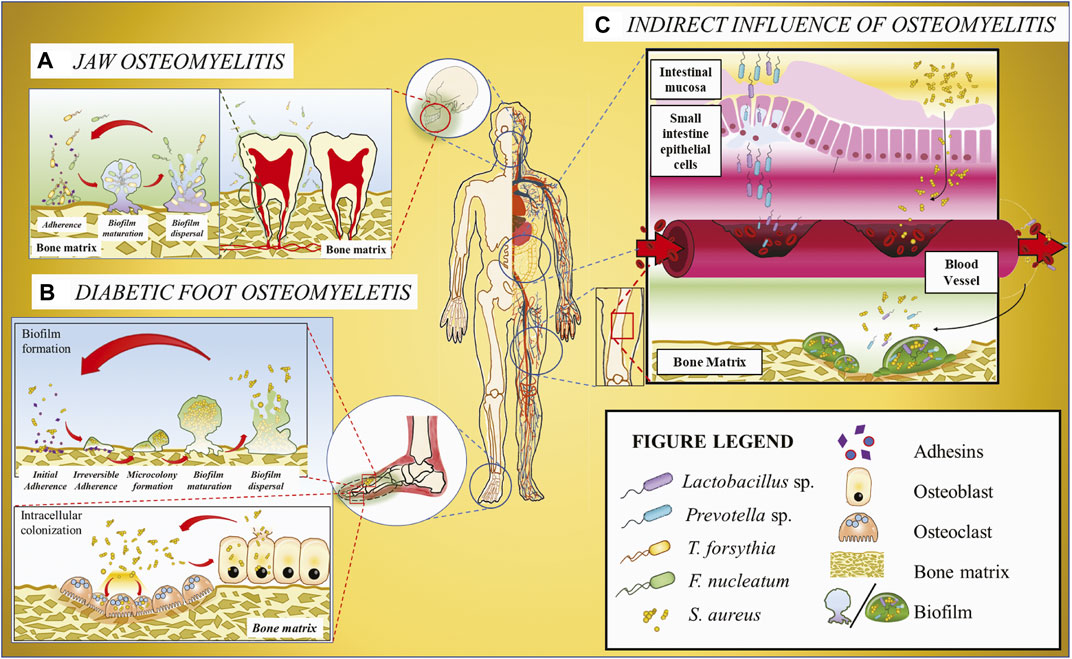
FIGURE 1. Schematic illustration of the host microbiome’s impacts on osteomyelitis. (A) Jaw osteomyelitis associated with two bacteria T. forsythia and F. nucleatum acts as a bridging bacterium to facilitate T. forsythia colonization and subsequent biofilm formation. (B) DFO involving S. aureus. As the most prevalent osteomyelitic pathogen, S. aureus may generate biofilm on the bone matrix or invade osteoclasts and osteoblasts mediated with adhesins, and the released bacteria from cells can be recovered to the original state. (C) Osteomyelitis or other osseous diseases associated with infectious microorganisms originating from gut microbiota. Some bacteria, e.g., Lactobacillus and Prevotella, translocate from gastrointestinal tract to the blood vessel across the gut epithelium barrier and then colonize in the bone matrix through the systematic circulation, ultimately resulting in osseous diseases.
S. aureus binds to the host tissues by expressing adhesins that facilitate the attachment to host extracellular matrix proteins such as collagen, fibrinogen, and fibronectin (Kdaa et al., 2020). Upon attachment, S. aureus evades the host immune cells and may form biofilms on the host tissue, increasing the microbial tolerance against any antimicrobial treatments. Previous studies have discovered Staphylococcal biofilm formation within chronic osteomyelitic bones (Gristina et al., 1985; Marrie and Costerton, 1985) and in vitro (O'Neill et al., 2007; Esteban et al., 2010). Additionally, commensal bacterial species are known to form multispecies biofilm, increasing the pathogenicity of the biofilm complex (Jneid et al., 2017). The commensal microbes within multispecies biofilm were documented in DFO specimens observed using SEM and/or CLSM-coupled PNA-FISH (Johani et al., 2019; Malone et al., 2019). The dynamic shift between the sessile and motile lifestyles of S. aureus in DFO tissues confers the pathogen resistance against non-surgical treatments, resulting in persistent chronic and acute infections.
Furthermore, S. aureus can infiltrate all types of osseous cells (osteoblasts, osteoclasts, and osteocytes) (Ellington et al., 2001; Klenerman, 2007; Reott et al., 2008; Mohamed et al., 2014; Yang et al., 2018) and then persist in a quiescent state, forming quasi-dormant small-colony variants (SCV) (Yang et al., 2018). This dormant lifestyle is less susceptible to antibiotics than the wild-type counterparts and can cause latent or recurrent infections upon release from these cells, potentially resulting in chronic infections (Proctor et al., 2006). Studies have demonstrated that the recovered SCV phenotypes were highly dynamic and could be rapidly reverted to the fully virulent wild-type form (Tuchscherr et al., 2011). These characteristics may explain the high infecting rate and recurrence of S. aureus-associated osteomyelitis.
Also, S. aureus produces a lot of factors that influence the host immune system. For example, immune evasion proteins SCIN and Efb help S. aureus evade the host immune system by inhibiting C3 convertase (Garcia et al., 2012), which belongs to the serine protease family and is necessary for innate immunity as a part of the complement system; chemotaxis inhibitory protein of S. aureus (CHIPS) inhibits neutrophil and monocyte chemotaxis toward C5a and formylated peptides by binding specifically to the C5aR and formylated peptide receptor (Postma et al., 2004); Staphylococcal protein A (SpA) can block antibody-mediated phagocytosis via binding Fcγ domain of Immunoglobulin G (Graille et al., 2000) or directly bind to osteoblasts, resulting in inhibition of osteoblastic proliferation and mineralization and even induction of apoptosis (Claro et al., 2011).
Studies of multispecies colonies have shown that certain coagulase-negative pathogenic Staphylococci such as S. epidermidis and S. lugdunensis, do not further exacerbate the wound severity compared to a single pathogen type infection (Yang et al., 2018). This unusual phenomenon agrees with the observation found in a study that showed reduced colonization of pathogenic S. aureus on the human nasal due to S. lugdunensis colonization (Zipperer et al., 2016). This inhibition of S. aureus colonization is hypothesized to be due to S. lugdunensis ability to produce lugdunin (thiazolidine-containing cyclic peptide antibiotics). The microbial interactions described above suggest intricate and subtle interactions between osteomyelitic microbes, functioning as both agonists and antagonists to regulate the host’s health.
Osteomyelitis of the jaw (i.e., maxilla and mandible) is a common disease found in patients suffering from head and neck infections (Baltensperger and Eyrich, 2009); however, there currently is no conclusive microbiome analysis of jaw osteomyelitis. There are some microbes specific to jaw osteomyelitis that have been identified (Table 1). One such example is that an Escherichia coli strain exhibiting multiple antibiotic resistances was isolated from bilateral maxillary osteomyelitis of a diabetic individual (PaDhiary et al., 2013). This strain presumptively enters the oral cavity through animal feces-contaminated water or food and then colonizes the jaw. Some soil-inhabited filamentous bacteria, actinomyces, were also discovered in maxilla osteomyelitis patients, and the entry portal is suggested to be from pulpal or periodontal infection (Gannepalli et al., 2015). Actinomyces are common commensals in the human gastrointestinal tract that may revert to be pathogenic upon infiltration of tissue layers or bones in this case. Other commonly found microbes in jaw osteomyelitic patients include Cutibacterium acnes (Park et al., 2017), a commensal strain of human skin and mucosal surface. The sequencing of C. acnes genome shows the presence of genes that are responsible for bacterial evasion from host immune system, biofilm formation, and resistance to clinical treatments such as multiple antibiotic treatment.
Similar to DFO, chronic osteomyelitis of the jaw (COMJ) pathogenesis is also associated with multispecies microbial communities. These microbial communities may exhibit antagonistic or synergistic interactions among the microbial community. A study of mandible or maxilla COMJ of twelve Brazilian patients showed three predominant commensal anaerobic strains (Parvimonas micra, Staphylococcus spp. and Fusobacterium nucleatum), indicative of the co-existence of these microbes resulting in the COMJ pathogenesis (Gaetti-Jardim Júnior et al., 2010).
Studies using NGS on COMJ provide vital information on the microbiome composition (Goda et al., 2014). It was discovered that the core microbiome comprises predominantly of the anaerobic microbes Fusobacterium nucleatum followed by Tannerella sp. and Porphyromonas sp. although the microbial populations from COMJ patient samples dramatically vary depending on the disease progression and the patient’s bone health. An in vitro study shows that F. nucleatum synergistically forms biofilms with Tannerella forsythia (Figure 1) (Sharma et al., 2005) dependent on surface contact rather than the bacterial biochemical cues; moreover, polymicrobial biofilms on a bone surface have been observed in an osteomyelitic jaw (Sedghizadeh et al., 2009). Hence, it is possible that F. nucleatum may also form biofilms with T. forsythia in COMJ. Other studies on F. nucleatum revealed that it acts an opportunistic pathogen in multispecies infections such as bacterial vaginosis (Citron, 2002), acute appendicitis (Swidsinski et al., 2011), and anaerobic bacteremia (Brook, 2010). F. nucleatum plays a crucial role in these infections by being a bridging bacterium to assist in the colonization of other bacteria, e.g., providing coaggregation conditions and an anaerobic environment for other anaerobes propagation. Thus, it is hypothesized that F. nucleatum plays a similar role in the pathogenesis and biofilm formation in the jaw bone, resulting in the development of COMJ. In addition, the multispecies biofilm of F. nucleatum with P. gingivalis and T. forthysia were found to induce severe periodontitis with massive bone resorption (Polak et al., 2009; Settem et al., 2012), suggesting that they may also synergistically form biofilms in COMJ.
The microbiome can indirectly influence osteomyelitis pathogenesis, in which the microbiota of non-osseous tissues produces biochemical signals that trigger the cells and microbes within the osteomyelitic tissue. In this section, we will discuss the impact of the gastrointestinal and serum-based microbiome on host osteomyelitis pathogenesis.
The gastrointestinal tract (GIT) is the most densely microorganism-populated region of humans or mammals, giving rise to an ecosystem comprising commensal, symbiotic, and pathogenic microorganisms. The gastrointestinal microbiota alone outnumber their host’s genes by more than 100 times, whereas the changes of these populations are regulated by many factors including diet, lifestyle and the environment (Turnbaugh et al., 2009). Currently, the gut microbiota is closely correlated with human diseases, especially autoinflammatory diseases including asthma, arthritis, colitis, diabetes, and lupus (Bach, 2002; Chervonsky, 2010; Maslowski and Mackay, 2011; Bodkhe et al., 2019; Marietta et al., 2019), and modulation of the gut microbiome can be applied to treating some autoimmune diseases (Balakrishnan and Taneja, 2018). Although there are limited studies correlating the intestinal microbiome to osseous tissues, other studies focused on the host-microbe interactions have identified three main manners through which the gastrointestinal microbiome may influence distant organs (Hernandez et al., 2016), i.e., regulation of nutritional absorption, regulation of the immune system at the gut endothelium, and translocation of microbes and/or their metabolites across the endothelial barrier into the systemic circulation.
One such example was demonstrated in turkey poults fed with rye. The rye diet increases the Lactobacillus population in the intestinal microbiota while encouraging Enterobacteriaceae translocation, leading to significant reduction in bone strength and bone mineralization (Tellez et al., 2015). This study suggests that rye disrupts the epithelial tight junctions in the intestinal tract, causing infiltration of microbes into the systemic circulation and ultimately alterations of bone mineralization. Another example looked into the dietary intake in mice and its influence on the intestinal microbiota composition (Lukens et al., 2014). The mice fed with low-fat diets had enriched Prevotella populations and lower abundance of Lactobacillus compared to mice fed with normal diets. Mice fed with high-fat diets showed changes in intestinal microbiota that inhibit the osteomyelitis development in osteomyelitis-susceptible Pstpip2cmo mice (Phillips et al., 2016). The Pstpip2cmo mouse expresses a homozygous Leu98Pro missense mutation in the Pombe Cdc15 homologous protein PSTPIP2 (proline-serine-threonine phosphatase interacting protein 2), resulting in the increased susceptibility of the mice to develop autoinflammatory diseases, bone deformities, and elevated levels of IL-1β (Hartland, 2020). Further studies revealed that these gut microbiome alterations upregulate pro-IL-1β levels, suggesting that the gut microbiome can indirectly affect osteomyelitis via regulating pro-IL-1β levels in the circulatory system.
Additionally, studies have shown variation in GIT microbiota from rodents and humans of different genders, where these changes impact the host differently. Studies on rodents revealed that female B6 mice have higher abundance of Lactobacillaceae and Bacteroides compared to males; whereas female BALB/c mice have higher abundance of Bifidobacteriaceae than males (Elderman et al., 2018). Similarly, such changes in the microbiota are observed in human patients, where microbiota isolated from elderly women has lower abundance of Bacteroidetes than elderly men (Mueller et al., 2006; Dominianni et al., 2015). This gender specific variation of the GIT microbiota primarily results from the changes in the host biochemistry, where these alterations of the microbiome affect bone formation and are correlated to the increased susceptibility of patients to various osseous diseases (e.g., osteoporosis). This increased susceptibility is likely due to the microbiome influencing various immunological-related genes (Elderman et al., 2018) and the production of sex hormones (Menon et al., 2013). Thus far, it has been understood that the microbiome in different genders are considerably complex, encompassing the hormone level changes, T-cell activation, and modifications in cytokine production (Ibáñez et al., 2019).
The perturbation of the gut microbiome is also associated with a number of other bone and joint diseases. The monocolonization of the commensal Lactobacillus bifidus in interleukin-1 receptor antagonist-knockout mice result in the spontaneous development of autoimmune T-cell-mediated arthritis. This localization resulted in a faster onset of the disease compared to normal mice (Abdollahi-Roodsaz et al., 2008). The L. bifidus-triggered arthritis results from the imbalance of TREG-TH17 cell homeostasis through TLR2–TLR4 signaling. The presence of Prevotella copri in the gut was found to trigger the onset of untreated rheumatoid arthritis. This phenomenon was discovered through sequencing rheumatoid arthritis patient’s stool samples (Scher et al., 2013), and the onset development of rheumatoid arthritis is possibly due to P. copri’s ability to dominate the intestinal microbiota.
Healthy human blood is not as sterile as previously perceived since discovering a diversified microbiome in healthy human blood (Potgieter et al., 2015; Païssé et al., 2016). These blood-borne microorganisms infiltrate the human host through infected wounds or microbial translocation from the respiratory or intestinal epithelium. These microbes then circulate the human host until finding a suitable site for localization. When some microbes reach the bones, where the capillaries are abundant but tortuous thereby containing relatively slower bloodstream, these microbes readily accumulate and ultimately block the capillaries, leading to osteonecrosis and a series of inflammatory reactions. It should be noted that although some blood microbes in the blood might be dormant due to the host immune reactions or antibiotic treatments, they could however be resuscitated at suitable conditions (Panaiotov et al., 2018) and be potentially pathogenic.
The bacterial chondronecrosis with osteomyelitis (BCO) is a common cause of lameness in commercial broiler chickens worldwide and results in substantial economic loss. BCO pathogenesis was found to be correlated to the blood microbiome. Studies indicate that the microorganisms associated with BCO may originate from the intestinal or respiratory tract microbiota, which crosses the epithelium barrier and then enters the bloodstream (Wideman and Prisby, 2013). Chickens with BCO have an increased abundance of Staphylococcus sp., Granulicatella sp., and Microbacterium sp. compared to healthy chickens (Mandal et al., 2016). A study on osteomyelitis-associated genes indicates that chickens suffering from BCO showed downregulated level of Runt-related transcription factor (RUNX2) and secreted protein acidic and cysteine rich (SPARC) genes (Paludo et al., 2017). RUNX2 is an important transcription factor that regulates the shape and differentiation of osteoblasts. Downregulation of RUNX2 blocks osteoblastic and chondrocyte differentiations, ultimately impairing the ossification process (Ma et al., 2010). SPARC is a calcium-binding matricellular glycoprotein and is involved in bone development, repair, and tissue remodeling (Orlando et al., 2013). We hypothesize that the dysbiosis of the blood microbiome results in the downregulation of RUNX2 and SPARC in BCO chickens, which further leads to osteomyelitis.
The oral cavity is the primary gateway to the human body, and the plethora of microorganisms (over 600 prokaryote species according to human oral microbiome database), whether colonizing or transiently inhabiting in the oral cavity, are very likely to translocate to different body sites. The human oral microbiome is the most extensively studied human microflora due to its relatively simple sampling process and strong correlation with common oral infectious diseases (e.g., dental caries and periodontitis). However, despite the extensive studies on the oral microbiome, there is much more to be explored as the microbiome network remains difficult to unravel due to the highly dynamic microbial compositions and complex interactions between microbe-host/microbe-microbe that impact the host biochemistry. Moreover, the variation of microbial distribution within the microenvironment of the oral cavity is also complex. For example, swabs from the center of the tongue exhibit more Streptococcus salivarius whereas the left outer part of the tongue is predominantly Haemophilus parainfluenza (Zeus et al., 2019). These changes in the microbiome composition influence the susceptibility of the host acquiring jaw osteomyelitis (discussed in Jaw Osteomyelitis section), providing an access for the oral microbiome to translocate to other parts of the human host. Despite these confounding factors, the oral microbiome may exhibit a possible role as modulatory target or biomarker in children and adolescents with chronic nonbacterial osteomyelitis (Zeus et al., 2021).
Changes in the host-microbiome are known to affect osteomyelitis pathogenesis in direct and indirect manners. The direct interaction of osteomyelitic microbiota induces inflammation by forming single- or multi-species biofilms on the surface of the bones. Some of these microbes can also burrow into the host cells forming quasi-dormant small-colony variants (SCV). These dormant microbes are protected from the host immune system or antibiotic treatments and shift to a planktonic lifestyle when there is a change in the host biochemistry. The microbiota from other niches indirectly influences osteomyelitis by altering the biochemical signals that regulate the host immune responses. Certain microbiota members can infiltrate into the host systemic circulation and then generate microbial embolism, causing tissue necrosis and chronic diseases such as bacterial chondronecrosis with osteomyelitis. Thus, it can be concluded that the various microbiomes within the patient influence the severity of the osteomyelitis. Further understanding of these microbial populations’ roles would be needed to improve the treatment of osteomyelitis and encourage rapid patient recovery by regulating the host microbiome.
Conceptualization, JC, and CH; Formal analysis-JC, AX, and YM; Investigation-JC, CQ, and CH; Resources-CQ, and CH; Writing-original draft preparation, JC, AX, and YM; Writing-review and editing, CH; Visualization-JC, and CH; Supervision-CQ, and CH; Project administration-JC; Fund acquisition-CH; All authors discussed, commented and agreed to publish this version of the manuscript.
This work was supported by the Shenzhen Institutes of Advanced Technology External Funds (DWKF20190001), National Natural Science Foundation of China’s Research Fund for International Young Scientists (22050410270), Guangdong Innovative Projects for the Characteristics of General Colleges and Universities (2018KTSCX200) and Guangdong Innovative and Entrepreneurial Research Team Program (2019ZT08Y191).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The authors wish to thank Xinyi Chen for comments on the manuscript. We also recognize the technical help provided by our undergraduate students, Yu Liqing, Cai Yanwei, and Song Zixian. The authors also wish to thank the advice provided by Professor Dai Lei and Assistant Professor Tan Yang from the Shenzhen Institute of Advanced Technology (SIAT), and Associate Professor Tang Bin from the Southern University of Science and Technology.
BCO, bacterial chondronecrosis with osteomyelitis; COMJ, chronic osteomyelitis of the jaw; CLSM, confocal laser scanning microscope; GIT, gastrointestinal tract; SCV, small-colony variants; SEM, scanning electron microscopy; PNA-FISH, peptide nucleic acid fluorescent in situ hybridization.
Abdollahi-Roodsaz, S., Joosten, L. A. B., Koenders, M. I., Devesa, I., Roelofs, M. F., Radstake, T. R. D. J., et al. (2008). Stimulation of TLR2 and TLR4 Differentially Skews the Balance of T Cells in a Mouse Model of Arthritis. J. Clin. Invest. 118, 205–216. doi:10.1172/JCI32639
Abdulrazak, A., Ibrahim Bitar, Z., Ayesh Al-Shamali, A., and Ahmed Mobasher, L. (2005). Bacteriological Study of Diabetic Foot Infections. J. Diabetes Complications 19, 138–141. doi:10.1016/j.jdiacomp.2004.06.001
Bach, J.-F. (2002). The Effect of Infections on Susceptibility to Autoimmune and Allergic Diseases. N. Engl. J. Med. 347, 911–920. doi:10.1056/NEJMra020100
Balakrishnan, B., and Taneja, V. (2018). Microbial Modulation of the Gut Microbiome for Treating Autoimmune Diseases. Expert Rev. Gastroenterol. Hepatol. 12, 985–996. doi:10.1080/17474124.2018.1517044
Baltensperger, M., and Eyrich, G. (2009). “Osteomyelitis of the Jaws: Definition and Classification,” in Osteomyelitis of the Jaws (Berlin, Heidelberg: Springer), 5–56. doi:10.1007/978-3-540-28766-7_2
Bodkhe, R., Balakrishnan, B., and Taneja, V. (2019). The Role of Microbiome in Rheumatoid Arthritis Treatment. Ther. Adv. Musculoskelet. 11, 1759720X1984463. doi:10.1177/1759720X19844632
Brook, I. (2010). The Role of Anaerobic Bacteria in Bacteremia. Anaerobe 16, 183–189. doi:10.1016/j.anaerobe.2009.12.001
Buch, K., Thuesen, A. C. B., Brøns, C., and Schwarz, P. (2019). Chronic Non-bacterial Osteomyelitis: a Review. Calcif Tissue Int. 104, 544–553. doi:10.1007/s00223-018-0495-0
Cai, Y. L., Cao, Y., Fan, X. Z., Luo, X. R., Meng, J. F., Xue, Y. M., et al. (2019). Microbiome Analysis of Diabetic Foot Osteomyelitis by Metagenome Sequencing Technology. Zhonghua Yi Xue Za Zhi 99, 2057–2061. doi:10.3760/cma.j.issn.0376-2491.2019.26.011
Cassir, N., Benamar, S., and La Scola, B. (2016). Clostridium Butyricum : from Beneficial to a New Emerging Pathogen. Clin. Microbiol. Infect. 22, 37–45. doi:10.1016/j.cmi.2015.10.014
Chervonsky, A. V. (2010). Influence of Microbial Environment on Autoimmunity. Nat. Immunol. 11, 28–35. doi:10.1038/ni.1801
Citron, D. M. (2002). Update on the Taxonomy and Clinical Aspects of the GenusFusobacterium. Clin. Infect. Dis. 35, S22–S27. doi:10.1086/341916
Claro, T., Widaa, A., O'Seaghdha, M., Miajlovic, H., Foster, T. J., O'Brien, F. J., et al. (2011). Staphylococcus aureus Protein A Binds to Osteoblasts and Triggers Signals that Weaken Bone in Osteomyelitis. Plos One 6, e18748. doi:10.1371/journal.pone.0018748
Dominianni, C., Sinha, R., Goedert, J. J., Pei, Z., Yang, L., Hayes, R. B., et al. (2015). Sex, Body Mass index, and Dietary Fiber Intake Influence the Human Gut Microbiome. PloS One 10, e0124599. doi:10.1371/journal.pone.0124599
Elderman, M., Hugenholtz, F., Belzer, C., Boekschoten, M., van Beek, A., de Haan, B., et al. (2018). Sex and Strain Dependent Differences in Mucosal Immunology and Microbiota Composition in Mice. Biol. Sex Diff. 9, 1–18. doi:10.1371/journal.pone.012459910.1186/s13293-018-0186-6
Ellington, J. K., Elhofy, A., Bost, K. L., and Hudson, M. C. (2001). Involvement of Mitogen-Activated Protein Kinase Pathways in Staphylococcus aureus Invasion of normal Osteoblasts. Infect. Immun. 69, 5235–5242. doi:10.1128/IAI.69.9.5235-5242.2001
Esteban, J., Molina-Manso, D., Spiliopoulou, I., Cordero-Ampuero, J., Fernández-Roblas, R., Foka, A., et al. (2010). Biofilm Development by Clinical Isolates of Staphylococcus spp. From Retrieved Orthopedic Prostheses. Acta Orthop. 81, 674–679. doi:10.3109/17453674.2010.537810
Franz, C. M. A. P., Huch, M., Abriouel, H., Holzapfel, W., and Gálvez, A. (2011). Enterococci as Probiotics and Their Implications in Food Safety. Int. J. Food Microbiol. 151, 125–140. doi:10.1016/j.ijfoodmicro.2011.08.014
Gaetti-Jardim Júnior, E., Fardin, A. C., Gaetti-Jardim, E. C., Castro, A. L. d., Schweitzer, C. M., and Avila-Campos, M. J. (2010). Microbiota Associated with Chronic Osteomyelitis of the Jaws. Braz. J. Microbiol. 41, 1056–1064. doi:10.1590/S1517-83822010000400025
Gannepalli, A., Ayinampudi, B. K., Baghirath, P. V., and Reddy, G. V. (2015). Actinomycotic Osteomyelitis of Maxilla Presenting as Oroantral Fistula: a Rare Case Report. Case Rep. Dentistry 2015, 1–5. doi:10.1155/2015/689240
Garcia, B. L., Ramyar, K. X., Ricklin, D., Lambris, J. D., and Geisbrecht, B. V. (2012). Advances in Understanding the Structure, Function, and Mechanism of the SCIN and Efb Families of Staphylococcal Immune Evasion Proteins. Adv. Exp. Med. Biol. 946, 113–133. doi:10.1007/978-1-4614-0106-3_7
Goda, A., Maruyama, F., Michi, Y., Nakagawa, I., and Harada, K. (2014). Analysis of the Factors Affecting the Formation of the Microbiome Associated with Chronic Osteomyelitis of the Jaw. Clin. Microbiol. Infect. 20, O309–O317. doi:10.1111/1469-0691.12400
Graille, M., Stura, E. A., Corper, A. L., Sutton, B. J., Taussig, M. J., Charbonnier, J.-B., et al. (2000). Crystal Structure of a Staphylococcus aureus Protein A Domain Complexed with the Fab Fragment of a Human IgM Antibody: Structural Basis for Recognition of B-Cell Receptors and Superantigen Activity. Proc. Natl. Acad. Sci. 97, 5399–5404. doi:10.1073/pnas.97.10.5399
Gristina, A., Oga, M., Webb, L., and Hobgood, C. (1985). Adherent Bacterial Colonization in the Pathogenesis of Osteomyelitis. Science 228, 990–993. doi:10.1126/science.4001933
Hartland, E. L. (2020). A Potential New Target for Autoinflammatory Bone Disease. J. Biol. Chem. 295, 3401–3402. doi:10.1074/jbc.H120.012867
Hatzenbuehler, J., and Pulling, T. J. (2011). Diagnosis and Management of Osteomyelitis. Am. Fam. Phys. 84, 1027–1033. PMID: 22046943
Hernandez, C. J., Guss, J. D., Luna, M., and Goldring, S. R. (2016). Links between the Microbiome and Bone. J. Bone Miner Res. 31, 1638–1646. doi:10.1002/jbmr.2887
Ibáñez, L., Rouleau, M., Wakkach, A., and Blin-Wakkach, C. (2019). Gut Microbiome and Bone. Jt. Bone Spine 86, 43–47. doi:10.1016/j.jbspin.2018.02.008
Jagodzinski, N. A., Kanwar, R., Graham, K., and Bache, C. E. (2009). Prospective Evaluation of a Shortened Regimen of Treatment for Acute Osteomyelitis and Septic Arthritis in Children. J. Pediatr. Orthop. 29, 518–525. doi:10.1097/BPO.0b013e3181ab472d
Jneid, J., Cassir, N., Schuldiner, S., Jourdan, N., Sotto, A., Lavigne, J.-P., et al. (2018). Exploring the Microbiota of Diabetic Foot Infections with Culturomics. Front. Cel. Infect. Microbiol. 8, 1–8. doi:10.3389/fcimb.2018.00282
Jneid, J., Lavigne, J. P., La Scola, B., and Cassir, N. (2017). The Diabetic Foot Microbiota: a Review. Hum. Microbiome J. 5-6, 1–6. doi:10.1016/j.humic.2017.09.002
Johani, K., Fritz, B. G., Bjarnsholt, T., Lipsky, B. A., Jensen, S. O., Yang, M., et al. (2019). Understanding the Microbiome of Diabetic Foot Osteomyelitis: Insights from Molecular and Microscopic Approaches. Clin. Microbiol. Infect. 25, 332–339. doi:10.1016/j.cmi.2018.04.036
Kdaa, C., Ilab, C., Amma, C., Hkka, C., Mtma, C., Svca, C., et al. (2020). Intracellular Staphylococcus aureus in Bone and Joint Infections: A Mechanism of Disease Recurrence, Inflammation, and Bone and Cartilage Destruction. Bone 141, 1–17. doi:10.1016/j.bone.2020.115568
Klenerman, L. (2007). A History of Osteomyelitis from the Journal of Bone and Joint Surgery. J. Bone Jt. Surg. 89-B, 667–670. doi:10.1302/0301-620X.89B5.19170
Lagier, J.-C., Hugon, P., Khelaifia, S., Fournier, P.-E., La Scola, B., and Raoult, D. (2015). The Rebirth of Culture in Microbiology through the Example of Culturomics to Study Human Gut Microbiota. Clin. Microbiol. Rev. 28, 237–264. doi:10.1128/CMR.00014-14
Lavery, L. A., Sariaya, M., Ashry, H., and Harkless, L. B. (1995). Microbiology of Osteomyelitis in Diabetic Foot Infections. J. Foot Ankle Surg. 34, 61–64. doi:10.1016/S1067-2516(09)80103-8
Lesens, O., Desbiez, F., Vidal, M., Robin, F., Descamps, S., Beytout, J., et al. (2011). Culture of Per-Wound Bone Specimens: a Simplified Approach for the Medical Management of Diabetic Foot Osteomyelitis. Clin. Microbiol. Infect. 17, 285–291. doi:10.1111/j.1469-0691.2010.03194.x
Lipsky, B. A. (1997). Osteomyelitis of the Foot in Diabetic Patients. Clin. Infect. Dis. 25, 1318–1326. doi:10.1086/516148
Lukens, J. R., Gurung, P., Vogel, P., Johnson, G. R., Carter, R. A., McGoldrick, D. J., et al. (2014). Dietary Modulation of the Microbiome Affects Autoinflammatory Disease. Nature 516, 246–249. doi:10.1038/nature13788
Ma, X.-l., Liu, Z.-p., Ma, J.-x., Han, C., and Zang, J.-c. (2010). Dynamic Expression of Runx2, Osterix and AJ18 in the Femoral Head of Steroid-Induced Osteonecrosis in Rats. Orthop. Surg. 2, 278–284. doi:10.1111/j.1757-7861.2010.00100.x
Malone, M., Fritz, B. G., Vickery, K., Schwarzer, S., Sharma, V., Biggs, N., et al. (2019). Analysis of Proximal Bone Margins in Diabetic Foot Osteomyelitis by Conventional Culture, DNA Sequencing and Microscopy. APMIS 127, 660–670. doi:10.1111/apm.12986
Mandal, R. K., Jiang, T., Al-Rubaye, A. A., Rhoads, D. D., Wideman, R. F., Zhao, J., et al. (2016). An Investigation into Blood Microbiota and its Potential Association with Bacterial Chondronecrosis with Osteomyelitis (BCO) in Broilers. Sci. Rep. 6, 1–11. doi:10.1038/srep25882
Marietta, E., Horwath, I., Balakrishnan, B., and Taneja, V. (2019). Role of the Intestinal Microbiome in Autoimmune Diseases and its Use in Treatments. Cell Immunol. 339, 50–58. doi:10.1016/j.cellimm.2018.10.005
Markanday, A. (2014). Diagnosing Diabetic Foot Osteomyelitis: Narrative Review and a Suggested 2-Step Score-Based Diagnostic Pathway for Clinicians. Proc. Open Forum Infect. Dis. 1, ofu060. doi:10.1093/ofid/ofu060
Marrie, T. J., and Costerton, J. W. (1985). Mode of Growth of Bacterial Pathogens in Chronic Polymicrobial Human Osteomyelitis. J. Clin. Microbiol. 22, 924–933. doi:10.1128/jcm.22.6.924-933.1985
Maslowski, K. M., and Mackay, C. R. (2011). Diet, Gut Microbiota and Immune Responses. Nat. Immunol. 12, 5–9. doi:10.1038/ni0111-5
Menon, R., Watson, S. E., Thomas, L. N., Allred, C. D., Dabney, A., Azcarate-Peril, M. A., et al. (2013). Diet Complexity and Estrogen Receptor β Status Affect the Composition of the Murine Intestinal Microbiota. Appl. Environ. Microbiol. 79, 5763–5773. doi:10.1128/AEM.01182-13
Mohamed, W., Sommer, U., Sommer, U., Sethi, S., Domann, E., Thormann, U., et al. (2014). Intracellular Proliferation of S. aureus in Osteoblasts and Effects of Rifampicin and Gentamicin on S. aureus Intracellular Proliferation and Survival. eCM 28, 258–268. doi:10.22203/eCM.v028a18
Mueller, S., Saunier, K., Hanisch, C., Norin, E., Alm, L., Midtvedt, T., et al. (2006). Differences in Fecal Microbiota in Different European Study Populations in Relation to Age, Gender, and Country: a Cross-Sectional Study. Appl. Environ. Microbiol. 72, 1027–1033. doi:10.1128/AEM.72.2.1027-1033.2006
O'Neill, E., Pozzi, C., Houston, P., Smyth, D., Humphreys, H., Robinson, D. A., et al. (2007). Association between Methicillin Susceptibility and Biofilm Regulation in Staphylococcus aureus Isolates from Device-Related Infections. J. Clin. Microbiol. 45, 1379–1388. doi:10.1128/JCM.02280-06
Orlando, B., Giacomelli, L., Ricci, M., Barone, A., and Covani, U. (2013). Leader Genes in Osteogenesis: a Theoretical Study. Arch. Oral Biol. 58, 42–49. doi:10.1016/j.archoralbio.2012.07.010
PaDhiary, S. K., SrivaStava, G., PanDa, S., SubuDhi, S., and LenKa, S. (2013). E.coli Associated Extensive Bilateral Maxillary Osteomyelitis: A Rare Case Report. J. Clin. Diagn. Res. 7, 2380–2832. doi:10.7860/JCDR/2013/5628.3531
Païssé, S., Valle, C., Servant, F., Courtney, M., Burcelin, R., Amar, J., et al. (2016). Comprehensive Description of Blood Microbiome from Healthy Donors Assessed by 16S Targeted Metagenomic Sequencing. Transfusion 56, 1138–1147. doi:10.1111/trf.13477
Paludo, E., Ibelli, A. M. G., Peixoto, J. O., Tavernari, F. C., Lima-Rosa, C. A. V., Pandolfi, J. R. C., et al. (2017). The Involvement of RUNX2 and SPARC Genes in the Bacterial Chondronecrosis with Osteomyelitis in Broilers. Animal 11, 1063–1070. doi:10.1017/S1751731116002433
Panaiotov, S., Filevski, G., Equestre, M., Nikolova, E., and Kalfin, R. (2018). Cultural Isolation and Characteristics of the Blood Microbiome of Healthy Individuals. Adv. Microbiol. 08, 406–421. doi:10.4236/aim.2018.85027
Park, S. N., Roh, H., Lim, Y. K., and Kook, J. K. (2017). Complete Genome Sequence of Cutibacterium Acnes KCOM 1861 Isolated from a Human Jaw Osteomyelitis Lesion. Korean J. Microbiol. 53, 126–128. doi:10.7845/kjm.2017.7016
Peltola, H., Pääkkönen, M., Kallio, P., Kallio, M. J. T., and Group, O.-S. A. S. (2010). Short- versus Long-Term Antimicrobial Treatment for Acute Hematogenous Osteomyelitis of Childhood. Pediatr. Infect. Dis. J. 29, 1123–1128. doi:10.1097/INF.0b013e3181f55a89
Phillips, F. C., Gurung, P., and Kanneganti, T.-D. (2016). Microbiota and Caspase-1/caspase-8 Regulate IL-1β-mediated Bone Disease. Gut Microbes 7, 334–341. doi:10.1080/19490976.2016.1182289
Polak, D., Wilensky, A., Shapira, L., Halabi, A., Goldstein, D., Weiss, E. I., et al. (2009). Mouse Model of Experimental Periodontitis Induced by Porphyromonas gingivalis/Fusobacterium Nucleatum infection: Bone Loss and Host Response. J. Clin. Periodontol. 36, 406–410. doi:10.1111/j.1600-051X.2009.01393.x
Postma, B., Poppelier, M. J., van Galen, J. C., Prossnitz, E. R., van Strijp, J. A. G., de Haas, C. J. C., et al. (2004). Chemotaxis Inhibitory Protein of Staphylococcus aureus Binds Specifically to the C5a and Formylated Peptide Receptor. J. Immunol. 172, 6994–7001. doi:10.4049/jimmunol.172.11.6994
Potgieter, M., Bester, J., Kell, D. B., Pretorius, E., and Danchin, A. (2015). The Dormant Blood Microbiome in Chronic, Inflammatory Diseases. Fems Microbiol. Rev. 39, 567–591. doi:10.1093/femsre/fuv013
Proctor, R. A., von Eiff, C., Kahl, B. C., Becker, K., Mcnamara, P., Herrmann, M., et al. (2006). Small colony Variants: a Pathogenic Form of Bacteria that Facilitates Persistent and Recurrent Infections. Nat. Rev. Microbiol. 4, 295–305. doi:10.1038/nrmicro1384
Reott, M. A., Ritchie-Miller, S. L., Anguita, J., and Hudson, M. C. (2008). TRAIL Expression Is Induced in Both Osteoblasts Containing intracellular Staphylococcus Aureus and Uninfected Osteoblasts in Infected Cultures. FEMS Microbiol. Lett. 278, 185–192. doi:10.1111/j.1574-6968.2007.00988.x
Scher, J. U., Sczesnak, A., Longman, R. S., Segata, N., Ubeda, C., Bielski, C., et al. (2013). Expansion of Intestinal Prevotella Copri Correlates with Enhanced Susceptibility to Arthritis. Elife 2, e01202. doi:10.7554/eLife.01202.001
Sedghizadeh, P. P., Kumar, S. K. S., Gorur, A., Schaudinn, C., Shuler, C. F., and Costerton, J. W. (2009). Microbial Biofilms in Osteomyelitis of the Jaw and Osteonecrosis of the Jaw Secondary to Bisphosphonate Therapy. J. Am. Dental Assoc. 140, 1259–1265. doi:10.14219/jada.archive.2009.0049
Settem, R. P., El-Hassan, A. T., Honma, K., Stafford, G. P., and Sharma, A. (2012). Fusobacterium Nucleatum and Tannerella Forsythia Induce Synergistic Alveolar Bone Loss in a Mouse Periodontitis Model. Infect. Immun. 80, 2436–2443. doi:10.1128/IAI.06276-11
Sharma, A., Inagaki, S., Sigurdson, W., and Kuramitsu, H. K. (2005). Synergy between Tannerella Forsythia and Fusobacterium Nucleatum in Biofilm Formation. Oral Microbiol. Immunol. 20, 39–42. doi:10.1111/j.1399-302X.2004.00175.x
Swidsinski, A., Dörffel, Y., Loening-Baucke, V., Theissig, F., Rückert, J. C., Ismail, M., et al. (2011). Acute Appendicitis Is Characterised by Local Invasion with Fusobacterium Nucleatum/necrophorum. Gut 60, 34–40. doi:10.1136/gut.2009.191320
Tellez, G., Latorre, J. D., Kuttappan, V. A., Hargis, B. M., and Hernandez-Velasco, X. (2015). Rye Affects Bacterial Translocation, Intestinal Viscosity, Microbiota Composition and Bone Mineralization in turkey Poults. PLoS One 10, e0122390. doi:10.1371/journal.pone.0122390
Tong, S. Y. C., Davis, J. S., Eichenberger, E., Holland, T. L., and Fowler, V. G. (2015). Staphylococcus aureus Infections: Epidemiology, Pathophysiology, Clinical Manifestations, and Management. Clin. Microbiol. Rev. 28, 603–661. doi:10.1128/CMR.00134-14
Tuchscherr, L., Medina, E., Hussain, M., Völker, W., Heitmann, V., Niemann, S., et al. (2011). Staphylococcus aureus Phenotype Switching: an Effective Bacterial Strategy to Escape Host Immune Response and Establish a Chronic Infection. EMBO Mol. Med. 3, 129–141. doi:10.1002/emmm.201000115
Turnbaugh, P. J., Ridaura, V. K., Faith, J. J., Rey, F. E., Knight, R., and Gordon, J. I. (2009). The Effect of Diet on the Human Gut Microbiome: a Metagenomic Analysis in Humanized Gnotobiotic Mice. Sci. Transl. Med. 1, 6ra14. doi:10.1126/scitranslmed.3000322
Urish, K. L., and Cassat, J. E. (2020). Staphylococcus aureus Osteomyelitis: Bone, Bugs, and Surgery. Infect. Immun. 88, e00932–19. doi:10.1128/IAI.00932-19
Van Asten, S. A. V., La Fontaine, J., Peters, E. J. G., Bhavan, K., Kim, P. J., and Lavery, L. A. (2016). The Microbiome of Diabetic Foot Osteomyelitis. Eur. J. Clin. Microbiol. Infect. Dis. 35, 293–298. doi:10.1007/s10096-015-2544-1
Wei, X., Pushalkar, S., Estilo, C., Wong, C., Farooki, A., Fornier, M., et al. (2012). Molecular Profiling of Oral Microbiota in Jawbone Samples of Bisphosphonate-Related Osteonecrosis of the Jaw. Oral Dis. 18, 602–612. doi:10.1111/j.1601-0825.2012.01916.x
Wideman, R. F., and Prisby, R. D. (2013). Bone Circulatory Disturbances in the Development of Spontaneous Bacterial Chondronecrosis with Osteomyelitis: a Translational Model for the Pathogenesis of Femoral Head Necrosis. Fendo 3, 1–14. doi:10.3389/fendo.2012.00183
Yang, D., Wijenayaka, A. R., Solomon, L. B., Pederson, S. M., Findlay, D. M., Kidd, S. P., et al. (2018). Novel Insights into Staphylococcus aureus Deep Bone Infections: the Involvement of Osteocytes. MBio 9, e00415–18. doi:10.1128/mBio.00415-18
Zeus, M., Janssen, S., Fischer, U., Laws, H. J., Borkhardt, A., and Oommen, P. (2019). Role of the Oral Microbiome in Chronic Non-bacterial Osteomyelitis in Children. London, UK: BMJ Publishing Group Ltd. doi:10.1136/annrheumdis-2019-eular.7875
Zeus, M., Janssen, S., Laws, H.-J., Fischer, U., Borkhardt, A., and Oommen, P. T. (2021). Results from a Pilot Study on the Oral Microbiome in Children and Adolescents with Chronic Nonbacterial Osteomyelitis. Z. Rheumatol., 1–10. doi:10.1007/s00393-021-01035-x
Zimmerli, W. (2015). “Osteomyelitis,” in Bone and Joint Infections: From Microbiology to Diagnostics and Treatment, Hoboken, NJ: Wiley Blackwell 197–203. doi:10.1002/9781118581742.ch13
Zipperer, A., Konnerth, M. C., Laux, C., Berscheid, A., Janek, D., Weidenmaier, C., et al. (2016). Human Commensals Producing a Novel Antibiotic Impair Pathogen Colonization. Nature 535, 511–516. doi:10.1038/nature18634
Keywords: microbiome, osteomyelitis, direct interaction, indirect interaction, pathogenesis
Citation: Chen J, Xiong A, Ma Y, Qin C and Ho CL (2021) Impact of the Host-Microbiome on Osteomyelitis Pathogenesis. Front. Mol. Biosci. 8:702484. doi: 10.3389/fmolb.2021.702484
Received: 29 April 2021; Accepted: 30 July 2021;
Published: 09 August 2021.
Edited by:
Shuyi Zhang, Tsinghua University, ChinaReviewed by:
Ana Cláudia Coelho, University of Trás-os-Montes and Alto Douro, PortugalCopyright © 2021 Chen, Xiong, Ma, Qin and Ho. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chun Loong Ho, aGVqbEBzdXN0ZWNoLmVkdS5jbg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.