
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Mol. Biosci. , 16 June 2021
Sec. Metabolomics
Volume 8 - 2021 | https://doi.org/10.3389/fmolb.2021.686110
This article is part of the Research Topic Functional Analysis of Plant-Microbe Interactions in Plant Disease Suppression and Health using Multi-Omics Techniques View all 5 articles
 Sarah F. Worsley1*
Sarah F. Worsley1* Michael C. Macey2
Michael C. Macey2 Samuel M. M. Prudence1,3
Samuel M. M. Prudence1,3 Barrie Wilkinson3
Barrie Wilkinson3 J. Colin Murrell2
J. Colin Murrell2 Matthew I. Hutchings1,3*
Matthew I. Hutchings1,3*Streptomyces species are saprophytic soil bacteria that produce a diverse array of specialized metabolites, including half of all known antibiotics. They are also rhizobacteria and plant endophytes that can promote plant growth and protect against disease. Several studies have shown that streptomycetes are enriched in the rhizosphere and endosphere of the model plant Arabidopsis thaliana. Here, we set out to test the hypothesis that they are attracted to plant roots by root exudates, and specifically by the plant phytohormone salicylate, which they might use as a nutrient source. We confirmed a previously published report that salicylate over-producing cpr5 plants are colonized more readily by streptomycetes but found that salicylate-deficient sid2-2 and pad4 plants had the same levels of root colonization by Streptomyces bacteria as the wild-type plants. We then tested eight genome sequenced Streptomyces endophyte strains in vitro and found that none were attracted to or could grow on salicylate as a sole carbon source. We next used 13CO2 DNA stable isotope probing to test whether Streptomyces species can feed off a wider range of plant metabolites but found that Streptomyces bacteria were outcompeted by faster growing proteobacteria and did not incorporate photosynthetically fixed carbon into their DNA. We conclude that, given their saprotrophic nature and under conditions of high competition, streptomycetes most likely feed on more complex organic material shed by growing plant roots. Understanding the factors that impact the competitiveness of strains in the plant root microbiome could have consequences for the effective application of biocontrol strains.
Streptomyces species are saprophytic soil bacteria that play an important ecological role as composters in soil and are prolific producers of antimicrobial compounds (van der Meij et al., 2017; Hutchings et al., 2019). The genus first evolved ca. 440 million years ago shortly after plants began to colonize land and it has been suggested that their filamentous growth and diverse secondary metabolism may have evolved to enhance their ability to colonize plant roots (Chater, 2016). Several studies have reported that streptomycetes are abundant inside the roots of the model plant Arabidopsis thaliana (Bulgarelli et al., 2012; Lundberg et al., 2012; Schlaeppi et al., 2014; Bai et al., 2015; Carvalhais et al., 2015) where they can have beneficial effects on growth (Worsley et al., 2020) while others have shown that streptomycetes can protect crop plants such as strawberry, lettuce, rice and wheat against biotic and abiotic stressors, including drought, salt stress, and pathogenic infection (Viaene et al., 2016; Newitt et al., 2019). However, different plant genotypes are associated with distinctive root-associated microbial communities and not all plant species enrich streptomycetes in their roots, a notable example being barley (Bulgarelli et al., 2015). This suggests that either specific selection mechanisms exist that enable different plant hosts to recruit particular microbial species from the soil and/or that bacterial taxa such as Streptomyces species colonize some plants better than others.
In our previous work, we reported that three Streptomyces strains isolated from the roots of A. thaliana plants can be re-introduced to new plants to promote their growth both in vitro and in soil. Furthermore, one of these strains exhibited broad spectrum antifungal activity and, when its spores were used as a seed coating, it colonized the roots of bread wheat plants and protected them against the commercially important pathogen Gaeumannomyces tritici, the causative agent of take-all disease (Worsley et al., 2020). Here we set out to test the hypothesis that streptomycetes are attracted to, and metabolize, components of the A. thaliana root exudate. Plants are known to release up to 40% of their photosynthetically fixed carbon into the surrounding soil via their roots and bacterial species in the soil have been shown to be attracted to, and be capable of metabolizing, specific resources contained within these root exudates (Haichar et al., 2012; Badri et al., 2013; Haichar et al., 2016). In turn, it is thought that the plant host might establish a beneficial root microbiome by producing particular types of root exudates that attract microbial species with desirable metabolic traits (Bais et al., 2006; Badri and Vivanco, 2009). Recent studies have further suggested that defence phytohormones, which are accumulated by plants in response to pathogenic attack, may play a key role in modulating the establishment of the normal A. thaliana root microbiome, since mutant plants that are disrupted in these pathways accumulate significantly different microbial communities (Carvalhais et al., 2013; Carvalhais et al., 2015; Lebeis et al., 2015; Liu et al., 2017). In particular, the abundance of bacteria in the A. thaliana rhizosphere and roots has been shown to correlate with concentrations of the defence phytohormone salicylic acid (Lebeis et al., 2015). Plant growth hormones such as indole-3-acetic acid have also been shown to modulate the production of Streptomyces specialized metabolites which may aid their competitive establishment in the root microbiome (Worsley et al., 2020).
Here we set out to test the hypothesis that streptomycetes are attracted by, and feed on, salicylate as well as other components of A. thaliana root exudates. We used genome sequenced Streptomyces endophyte strains that we isolated in a previous study (Worsley et al., 2020), including three which have been shown to promote A. thaliana growth in vitro and in compost, and assessed their ability to colonize mutant A. thaliana plants with altered salicylate production. Our results indicate that salicylate does not attract or feed these Streptomyces species, at least under the conditions used in our experiments. Furthermore, 13CO2 DNA stable isotope probing (DNA-SIP) of wild-type A. thaliana plants grown in compost show that Streptomyces bacteria do not feed on root exudates and are instead outcompeted by faster growing proteobacteria. We propose that streptomycetes are more likely to feed on complex organic matter that is not labeled in the 13CO2 DNA-SIP experiments. This work is an important step in understanding the role of these bacteria in the plant rhizosphere and also reveals which members of the A. thaliana microbiome feed on plant metabolites.
Root colonization by Streptomyces bacteria is affected by plant genotype. A previous study reported that A. thaliana cpr5 plants that constitutively produce salicylate have an altered root microbiota compared to wild-type plants, and that certain streptomycetes were better able to colonize these plants in vitro, due to their ability to metabolize salicylate (Lebeis et al., 2015). This suggests that salicylate might be directly responsible for recruiting these bacteria to the plant roots. To test this hypothesis, we compared root colonization efficiencies by S. coelicolor M145 and the root-associated strain Streptomyces M3 in wild-type A. thaliana Col-0 plants, cpr5 plants as well as two other genotypes, pad4 and sid2-2 plants, that are deficient in salicylate production (Table 1) (Zhou et al., 1998; Jirage et al., 1999; Wildermuth et al., 2001; Jing et al., 2007). To measure plant root colonization efficiency, we established root infection assays in which pre-germinated Streptomyces spores were used to coat A. thaliana seeds and were also added to the surrounding soil. Streptomyces strains M3 and S. coelicolor M145 (Table 1), were used because they have previously been observed to interact extensively with the A. thaliana roots using confocal microscopy (Worsley et al., 2020). Both strains have been engineered to carry the apramycin resistance (aac) gene, enabling the selective re-isolation of these strains from the roots of soil grown plants and their subsequent enumeration on agar plates containing apramycin. Colonization was measured as colony forming units (cfu) retrieved per gram of plant root tissue. This method has been used previously to assess colonization efficiency by other streptomycete strains in the plant root microbiome (Bonaldi et al., 2015; Chen et al., 2016). The results of these assays show that root colonization was significantly affected by plant genotype (Figure 1), irrespective of the Streptomyces strain used as an inoculum; plant genotype had a significant effect on the log-transformed cfu g−1 (F(3,39) = 6.17, p < 0.01), whereas the strain-genotype interaction term was insignificant (F(3,39) = 0.51, p = 0.68) in an ANOVA test. Indeed, consistent with previous findings (Lebeis et al., 2015), colonization by both strains significantly increased in the cpr5 mutant plants which constitutively make salicylate (Figure 1), compared to wild-type plants and the salicylate-deficient plants, pad4 and sid2-2 (p < 0.05 in all Tukey’s HSD tests between cpr5 and the other plant genotypes). However, we observed no significant difference in root colonization by M145 or M3 in pad4 or sid2-2 plants, compared to wild-type A. thaliana plants (p > 0.05 in Tukey’s HSD tests), suggesting that reduced quantities of salicylate do not, in turn, correspond to a reduction in colonization by streptomycetes (Figure 1). We note that the cpr5 gene has a complex role in regulating pathways involved in plant growth, immunity, and senescence (Jing et al., 2007) and that the cpr5 plants grown in our experiments were compromised in their growth compared to all the other plant genotypes tested here (Figure 1). The altered development of cpr5 plants has been observed in numerous other studies (Kirik et al., 2001; Jing et al., 2007; Brininstool et al., 2008; Jing et al., 2008; Doornbos et al., 2011; Gao et al., 2011) and gives rise to the possibility that the observed increase in colonization by Streptomyces strains M145 and M3 was linked to other aspects of the complex phenotype of the cpr5 plants and was not necessarily due to the higher levels of salicylate produced by these plants.
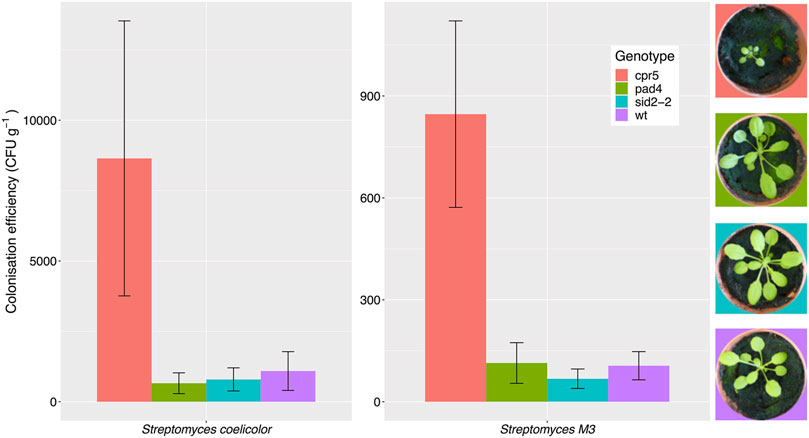
FIGURE 1. Streptomyces coelicolor M145 and the A. thaliana root endophyte strain Streptomyces M3 show increased root colonization of Arabidopsis thaliana cpr5 plants, which constitutively produce salicylic acid, compared to pad4 and sid2-2 plants, which are deficient in salicylic acid production, and wild-type (wt) Col-0 plants. Root colonization was measured as the average colony forming units (±SE) of Streptomyces bacteria per gram of root, that could be re-isolated from plants four weeks after germination. N = 6 plants per treatment. Plant growth phenotypes are shown for comparison.
To test whether the streptomycete strains used in recolonization experiments can utilize salicylate as a sole-carbon source, we grew each strain on minimal agarose medium (MM) containing either sodium citrate, or their preferred carbon sources (mannitol, maltose or sucrose). We also tested seven additional strains, four of which were also isolated from A. thaliana roots, as well as three strains of Streptomyces lydicus which are known colonizers of plant roots and were genome-sequenced in a previous study (Worsley et al., 2020). All the strains grew well on their preferred carbon source but none of the strains grew on MM plates containing salicylate as the sole carbon source (Supplementary Figure S1). This included Streptomyces M3 which had demonstrated increased levels of colonization in A. thaliana cpr5 plants (Figure 1). Thus, salicylate is not used as a carbon source by any of the Streptomyces strains used in our study, and this is supported by the absence of known SA degradation genes in the genomes of all these isolates (Supplementary Table S1). Rather than acting as a nutrient source, some root exudate molecules may recruit bacteria to the root niche by acting as chemoattractants (Rudrappa et al., 2008). In order to test whether salicylate is a chemoattractant for Streptomyces species, we grew our test strains on agar plates next to paper disks soaked in either 0.5 mM or 1 mM salicylate, or 0.1% (v/v) DMSO (solvent control) but observed no growth toward salicylate after 10 days (Supplementary Figure S2).
To test whether altered salicylate levels might indirectly benefit streptomycetes by negatively modulating the levels of the other rhizobacteria in the soil and removing competition, we established soil microcosms in deep 12-well plates, which were wetted with either sterile water or 0.5 mM salicylate. Microcosms were inoculated with spores of S. coelicolor M145-eGFP or Streptomyces M3-eGFP, both of which had colonized cpr5 plants more effectively than wild-type plants. Nine replicates of each treatment were run in parallel for each strain and the uninoculated control. Strains were recovered on apramycin-containing selective agar medium after 10 days. No apramycin resistant Streptomyces colonies were re-isolated from the control soil wells, indicating that any resulting colonies arising from the treated wells were derived from inoculated strains (Figure 2). A generalized linear model (GLM) with a negative binomial distribution demonstrated that, overall, a significantly greater number of S. coelicolor M145-eGFP colonies could be recovered from soil wells than Streptomyces M3-eGFP (p < 0.002; Figure 2). However, there was no significant effect of the different soil wetting treatments (either water or salicylate) on CFU number (p = 0.07; Figure 2). The interaction term between strain and wetting treatment was also insignificant (p = 0.20), indicating that this did not differ between the two inoculated strains. These results suggest that neither Streptomyces strain had a competitive advantage when greater concentrations of SA were present in soil. Taking these results together, we conclude that the observed increase in colonization by strains M145 and M3 in A. thaliana cpr5 plants is likely due to a pleiotropic effect of plant genotype on other aspects of plant growth rather than a direct effect of the increased presence of SA in root exudates and soil.
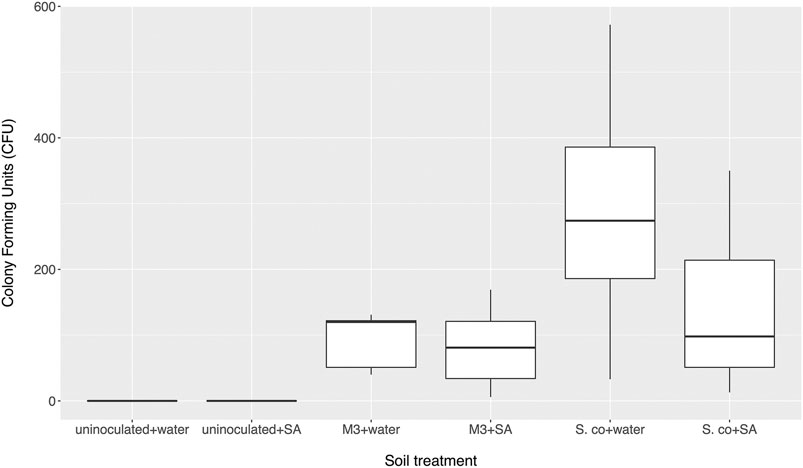
FIGURE 2. Number of colony forming units (CFU) of inoculated Streptomyces coelicolor M145 or Streptomyces M3 strains returned from soil microcosms treated with either salicylic acid (SA) or distilled water after 10 days. Control microcosms weren’t inoculated. N = 9 microcosms per treatment.
We then set out to test the hypothesis that Streptomyces bacteria can grow using a wider range of A. thaliana root exudate molecules as their sole nutrient source. We tested two S. lydicus strains, the five Streptomyces strains isolated from surface-washed A. thaliana roots (Worsley et al., 2020) and the model organism S. coelicolor M145 (Table 1). A. thaliana root exudates were collected using a small-scale hydroponics system and used to make plates with purified agarose. Agarose was used rather than agar, because streptomycetes can grow on the impurities in agar (not shown). Control plates were made using agarose and sterile water. The results show that all eight strains could grow on plates containing A. thaliana root exudates but not on the water-only control plates (Supplementary Figure S3) implying that all of the Streptomyces strains tested here can use A. thaliana root exudate as their sole food source in vitro.
Several independent studies have reported that A. thaliana plants have a relatively stable and consistent root bacterial community, which is enriched with members of the phylum Actinobacteria; this enrichment is predominantly driven by the presence of the family Streptomycetaceae (Bulgarelli et al., 2012; Lundberg et al., 2012). To test whether this was the case for plants grown in our laboratory, we repeated the bacterial 16S rRNA gene profiling using universal primers PRK341F and PRK806R (Supplementary Table S2). Plants were grown under controlled conditions in Levington F2 compost to match the conditions in our previously published study (Worsley et al., 2020) and the other experiments reported here (for chemical analysis of the Levington F2 compost, see Supplementary Table S3). The number of OTUs detected in samples was greater in the bulk soil (an average of 3,258 ± 166.28 standard deviation across the three replicates) and rhizosphere (3,630.67 ± 1,413.12) compared to the endophytic compartment (1,144 ± 76.06). The endophytic compartment also had a reduced level of diversity, with the average Shannon diversity being 6.19 ± 0.18, 6.18 ± 0.22 and 2.43 ± 0.17 in the bulk soil, rhizosphere and endosphere compartments, respectively. We found that, in agreement with published studies, Streptomycetaceae was the most abundant family of Actinobacteria in both the rhizosphere and root compartment of A. thaliana plants (Figure 3), making up 5.62% (±1.10% standard deviation) of the rhizosphere community and 1.12% (±0.50%) of the endophytic community, respectively. Actinobacteria as a whole made up 15.15% (±1.47%) and 2.87% (±0.88%) of these compartments, respectively (Figure 3 and Supplementary Figure S4). However, bacteria of the phylum Proteobacteria were found to dominate in all of the soil, rhizosphere and root compartments of the A. thaliana plants (Supplementary Figure S4) and were also significantly enriched in the endophytic compartment, relative to the surrounding soil and rhizosphere (p < 0.05 in Dunn’s multiple comparison tests between root and rhizosphere and root and soil compartments) increasing to 91.22% (±0.74% standard deviation) of the root-associated community, compared to 38.85% (±9.80%) of the rhizosphere and 35.46% (±1.97%) of the soil community, respectively.
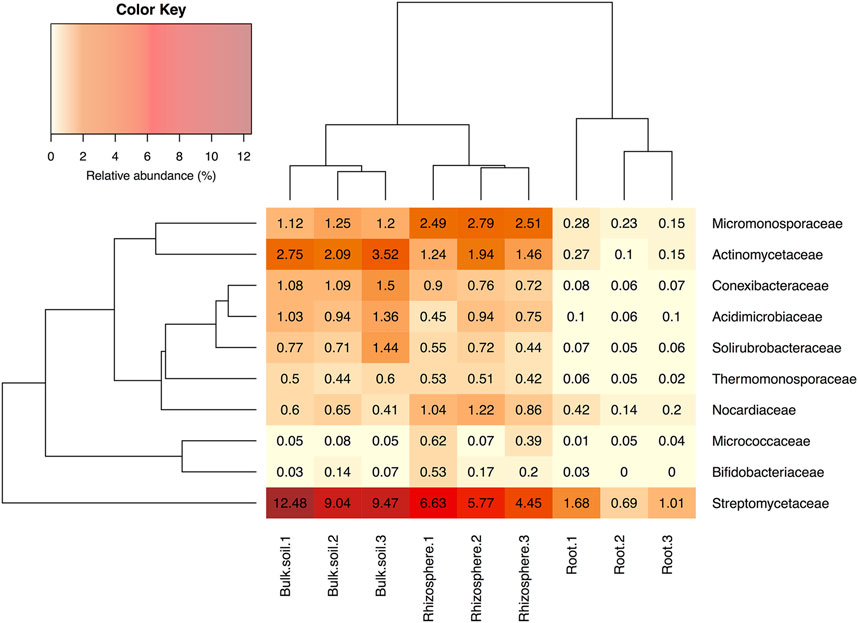
FIGURE 3. The relative abundance of actinobacterial families in soil, rhizosphere and root compartments. N = 3 replicate plants in individual pots. Streptomycetaceae was the most abundant family of Actinobacteria in all three compartments. Clustering represents Bray Curtis dissimilarities.
To test whether Streptomyces species metabolize A. thaliana root exudates we used 13CO2 DNA-SIP (Radajewski et al., 2000; Dumont and Murrell, 2005; Haichar et al., 2016). Plants incubated with 13CO2 fix the ‘heavy’ isotope of carbon which then becomes incorporated into carbon-based metabolites; many of these then leave the plant as root exudates. Bacteria feeding off labeled root exudates in the rhizosphere, or plant metabolites in the endosphere, will incorporate the 13C into their DNA as they grow and divide. By separating 12C- and 13C-labelled DNA on a cesium chloride gradient and using universal primers (Supplementary Table S2) to amplify and sequence a variable region of the 16S rRNA gene from both heavy and light fractions, we can determine which bacteria are being fed by the plant in the rhizosphere and endosphere of A. thaliana (Neufeld et al., 2007; Haichar et al., 2016). A. thaliana plants were labeled by growing them (n = 3) in Levington F2 compost for 21 days. Plants were placed in sealed growth chambers in the presence of either 12CO2 or 13CO2 (Supplementary Figure S5). Unplanted pots were also incubated with 13CO2 to account for autotrophic metabolism. Given the relatively short time frame of CO2 exposure, 13C was expected to be predominantly incorporated into plant metabolites rather than into plant cell wall material. Bacteria that incorporate 13C into their DNA must therefore either be feeding on plant metabolites or fixing 13CO2 autotrophically. After 21 days, total DNA was extracted from the rhizosphere, endosphere and unplanted soil compartments of the 12CO2 or 13CO2 incubated plants and heavy (13C) and light (12C) DNA were separated by density gradient ultracentrifugation. For the root samples, it was necessary to combine the three replicate samples into one sample for each of the CO2 treatments before ultracentrifugation, due to low DNA yields. Heavy and light DNA fractions of each gradient were determined via qPCR and used for 16S rRNA gene amplicon sequencing (Supplementary Figure S6). An average (±standard deviation) of 2,109 ± 152.42, 5,083.33 ± 204.63, 2,252.67 ± 148.35, and 4,243 ± 605.49 OTUs were detected in the 12C heavy, 12C light, 13C heavy and 13C light fractions taken from rhizosphere samples, respectively. In the endophytic fraction samples, 999, 1,072, 1,000 and 2,570 OTUs were detected in the 12C heavy, 12C light, 13C heavy and 13C light fractions, respectively.
A principal coordinates analysis, using a Bray-Curtis dissimilarity matrix of the relative abundances of genera present in each of the different rhizosphere fractions suggested that certain genera had metabolized 13C labeled host-derived resources, since 13C and 12C heavy fractions separated spatially on an ordination plot, indicating differences between the bacterial communities in these fractions (Figure 4). A permutational analysis of variance (PERMANOVA) analysis confirmed that fraction type had a significant effect on the community composition (permutations = 999, R2 = 0.80, p = <0.01).
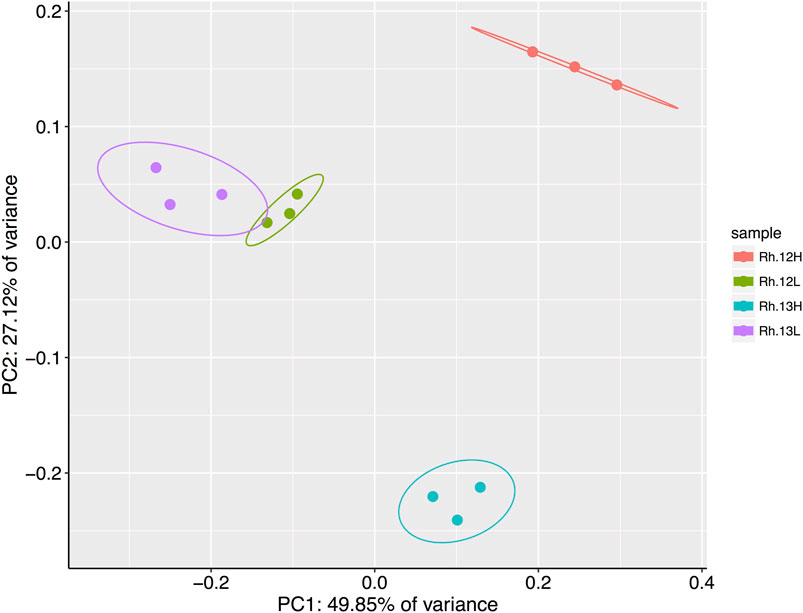
FIGURE 4. Results of a principal coordinates analysis of Bray-Curtis dissimilarities between bacterial genera present in the heavy (H) and light (L) buoyant density fractions of rhizosphere samples. Samples were taken from plants incubated under either 12CO2 (12L, 12H) or 13CO2 (13L, 13H) treatments (N = 3 replicate plants under each treatment). Principle Component 1 and 2 explain 49.85 and 27.12% of the variance in Bray–Curtis dissimilarities, respectively, between fraction types. Ellipses represent 95% confidence intervals.
Several bacterial genera were found to be enriched in the heavy vs. the light fractions of the 13CO2 unplanted control pots, suggesting that they may be capable of directly fixing CO2 from the environment. This list included bacterial genera that are known autotrophs and, thus, these genera were not considered as utilizers of root exudates if they were enriched in the heavy fractions of rhizosphere or endosphere samples taken from 13C planted pots (Supplementary Table S4). For example, several nitrifying bacteria were enriched in the heavy fractions of both the 13CO2 treated plants and the unplanted controls; this included the genera Nitrospira, Candidatus Nitrogena and Paracoccus. A diverse range of CO2 fixation strategies have been identified in nitrifying bacteria (Alfreider et al., 2018), and species in all of the aforementioned genera have been shown to use the Calvin-Benson-Bassham cycle to autotrophically fix carbon via the enzyme ribulose-1,5-bisphosphate carboxylase oxygenase (RubisCO) (Bollmann et al., 2013; Kitzinger et al., 2018; Ye et al., 2020). Genes encoding the RubisCO enzyme have also been identified in several of the other genera that were enriched in the heavy fraction of control pots, including Methylibium and Geobacter, which have both been identified as key players in CO2 fixation in rice paddies and agricultural soils (Wu et al., 2015; Xiao et al., 2021). The genus Bacteroides is also known to carry out direct carbon fixation during carbohydrate fermentation (Caldwell et al., 1969; Fischbach and Sonnenburg, 2011).
Disregarding possible autotrophs that were enriched in the heavy fractions of unplanted controls (Supplementary Table S4), a total of 28 genera showed an average of two-fold (or more) enrichment in the 13C heavy (13CH) fraction of rhizosphere samples, compared to both the 13CO2 light fraction (13CL) and 12CO2 heavy fraction (12CH), respectively, suggesting they had become labeled through the metabolism of root exudates (Supplementary Table S4; Figure 5). The majority of these taxa were in the phylum Proteobacteria (24 out of 28 genera), with only one representative each from the phyla Chloroflexi (Levilinea), Firmicutes (Pelotomaculum), Cyanobacteria (Chroococcidiopsis), and the Planctomycetes (Pirellula) (Supplementary Table S4). The most enriched genus was Pseudomonas, which demonstrated a 64-fold enrichment in relative abundance between the 13CH (8.63 ± 3.33% average relative abundance ± standard error) and 13CL (0.13 ± 0.01%) fractions and a 23-fold enrichment between the 13CH fraction compared to the 12CH control (0.37 ± 0.06%) (Figure 5).
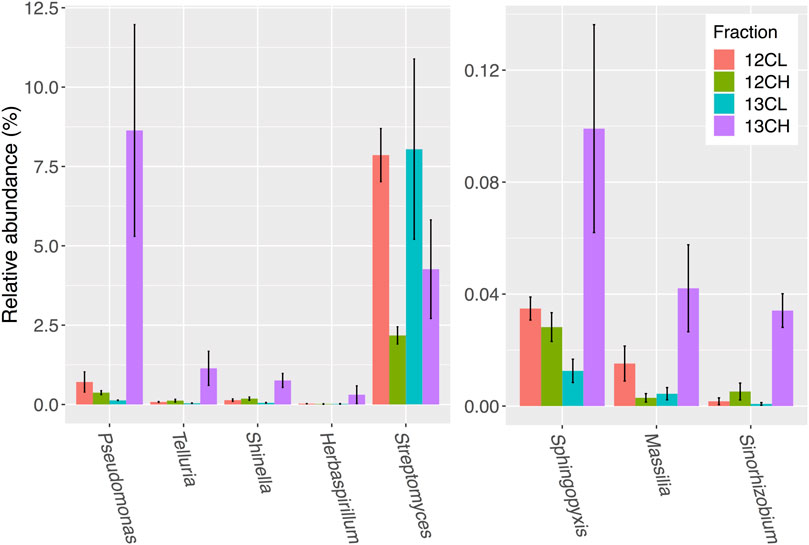
FIGURE 5. Seven bacterial genera in the rhizosphere compartment of A. thaliana plants whose relative abundance demonstrated the greatest overall enrichment in the 13CH fraction, relative to the 12CH fraction, due to the metabolism of 13C labeled root exudates. Streptomyces were not enriched but are included for comparison. 12CL/13CL and 12CH/13CH stand for buoyant density fractions representing the light (L) and heavy (H) fractions of DNA from 12CO2 (12C) or 13CO2 (13C) incubated plants, respectively. N = 3 replicate plants. Bars represent ± SE.
A total of 27 genera demonstrated more than a two-fold enrichment in the 13CH fraction of the endophytic compartment vs. 13CL and 12CH fractions (Figure 6). The majority of these genera were also Proteobacteria (21 genera in total), with three genera belonging to the phylum Planctomycetes (Blastopirellula, Pirellula and Gemmata), one to the Firmicutes (Clostridium), one to the Chloroflexi (Chloroflexus), and one to the Actinobacteria (Jatrophihabitans) (Supplementary Table S4). The most abundant genus in the 13CH fraction of the endophytic samples was Shinella, which demonstrated a 38-fold enrichment between the 13CH (26.42% relative abundance) and 13CL (0.68% relative abundance) fractions and a 23-fold enrichment between the 13CH and the 12CH control fractions (1.14%) (Figure 6). In terms of fold change, however, Pseudomonas was the most enriched genus with a 26-fold increase in abundance between 13CH (15.64% relative abundance) and 13CL (0.59% relative abundance) fractions of 13CO2 incubated plants (Figure 6). Comparing the two compartments, 15 out of the 28 genera were enriched by more than two-fold in the heavy fraction of the endosphere but not the rhizosphere samples, whereas 16 out of 29 genera were metabolizing exudates in the rhizosphere but not the endosphere. The remaining 13 genera were enriched in the heavy fractions of both the endophytic and rhizosphere compartments, suggesting that they are able to survive and make use of plant metabolites in both niches (Supplementary Table S4).
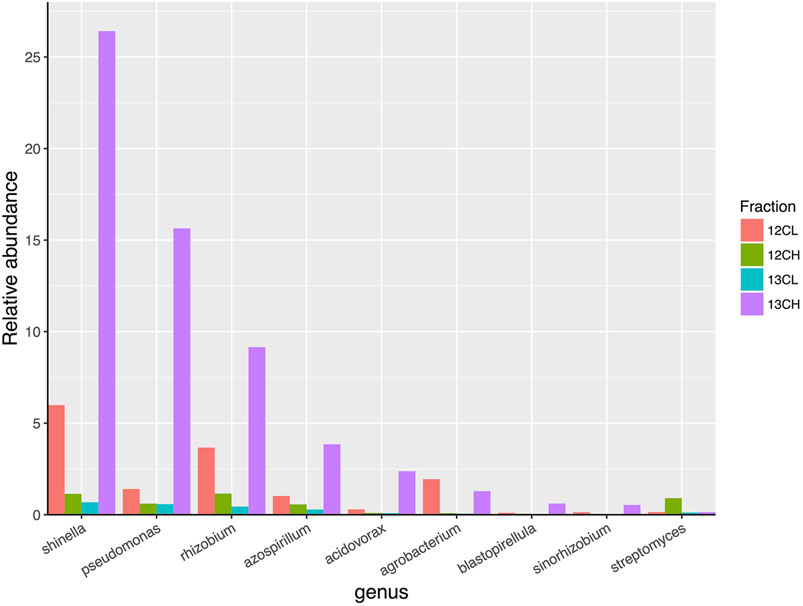
FIGURE 6. The relative abundance of eight genera of bacteria that demonstrated the greatest overall enrichment between the 13CH fraction and the 12CH fraction due to the metabolism of 13C labeled root exudates in the endophytic compartment of Arabidopsis thaliana plants. In comparison, the genus Streptomyces did not demonstrate enrichment between these fractions suggesting they did not metabolize 13C-labelled root exudates. 12CL/13CL and 12CH/13CH are the buoyant density fractions representing the light/unlabelled (L) and heavy/labeled (H) fractions of DNA from 12CO2 (12C) or 13CO2 (13C) incubated plants, respectively.
Surprisingly, there was no enrichment of Streptomyces bacteria in the 13CH fractions of either the rhizosphere or endosphere, despite being the most dominant member of the phylum Actinobacteria in both compartments (Figures 5 and 6 and Supplementary Figure S7; Supplementary Table S4). Taken together, these data suggest that, in compost, Streptomyces bacteria are outcompeted for root exudates by unicellular bacteria, particularly members of the phylum Proteobacteria. Proteobacteria were abundant in the unfractionated soil (35% average relative abundance), rhizosphere (39%) and endophytic compartment (91%), compared to Actinobacteria (present at 20, 15, and 3% in the soil, rhizosphere and root compartments, respectively) (Supplementary Figure S4) and therefore may have the upper hand at the outset of competition (Scheuring and Yu, 2012). Accordingly, genera that were found to be metabolizing the greatest amount of exudates in the roots were also found to be enriched in the endophytic compartment compared to the surrounding soil (Supplementary Figures S4 and S7).
Although typically described as free-living soil bacteria, there is increasing evidence that Streptomyces species are effective at colonizing the rhizosphere and endosphere of plant roots where they can promote plant growth and provide protection against disease (Viaene et al., 2016; Rey and Dumas, 2017; Kim et al., 2019; Newitt et al., 2019). It has even been suggested that their diverse specialized metabolism and filamentous growth may have evolved to give them a competitive advantage in this niche since it occurred ∼50 million years after plants first colonized land (Chater, 2006; 2016). Plants certainly provide a good source of food in nutritionally poor soil environments, both through the release of root exudates but also by shedding more complex organic cell wall material as roots grow and senesce (Dennis et al., 2010). There is growing evidence that root exudates enable plants to attract bacteria and shape their microbiome (Sasse et al., 2018; Zhalnina et al., 2018; Huang et al., 2019). Consistent with this, the 13CO2 DNA-SIP experiment identified several genera, particularly those in the phylum Proteobacteria, in both the rhizosphere and endosphere that became labeled with 13C due to the metabolism of A. thaliana root exudates. Many of these genera have been shown to have a positive influence on plant fitness, suggesting that root exudates may be one mechanism by which beneficial bacteria can establish and be maintained within the plant root microbiome. For example, the genus Pseudomonas was highly enriched in the heavy fractions of the rhizosphere and endosphere samples of 13C treated A. thaliana plants; this genus is well-known for producing many different bioactive and plant-growth-promoting molecules (Glick et al., 1997; Hernández-León et al., 2015; Mercado-Blanco et al., 2016; Raza et al., 2016) and certain species have been shown to promote root growth and induce systemic resistance to pathogenic infection in A. thaliana plants (Hase et al., 2003; Matilla et al., 2010; Zamioudis et al., 2013). Similarly, the genus Massilia colonizes the roots of many different plant species and has been shown to have plant growth promoting properties and antagonistic activity against phytopathogens in vitro (Ofek et al., 2012). The genus Shinella (a member of the Rhizobiaceae family) was also 13C labeled in A. thaliana rhizosphere and endosphere samples. Members of this genus have previously been isolated from the root nodules of leguminous plants in which they are capable of fixing nitrogen and enhancing plant growth (An et al., 2006; Lin et al., 2008). Other genera in the Rhizobiaceae were also labeled in the 13CO2 DNA-SIP experiment including Ensifer, Sinorhizobium and Rhizobium. Apart from inducing the formation of root nodules in leguminous plants, species in the genus Rhizobium have been shown to positively influence root development and overall plant biomass when interacting with A. thaliana plants (Zhao et al., 2017). Several of the non-proteobacterial genera that were identified as root exudate metabolisers in the rhizosphere and endosphere have been shown to be associated with plant species in other studies, although their role within the plant root microbiome is largely unknown. For example, species in the genus Jatrophihabitans (in the phylum Actinobacteria) are known to exist endophytically in several plant species and were labeled in the endosphere samples of 13C treated A. thaliana-however, these species have not been characterized in relation to plant-growth-promoting traits (Madhaiyan et al., 2013; Gong et al., 2016).
Although we confirmed in this study that Streptomyces bacteria can survive using only A. thaliana root exudates in vitro (Figure 4), 13CO2 DNA-SIP revealed that they were not 13C-labelled in our experiments and were in fact likely outcompeted for plant metabolites in both the rhizosphere and endosphere compartments by the proteobacterial genera, that were abundant and labeled in both compartments (Figure 5 and Figure 6). Many proteobacteria are thought to be “r-strategists” that have the ability to thrive and rapidly proliferate in fluctuating environments where resources are highly abundant, such as in the rhizosphere and root-associated microbiome (Philippot et al., 2013). In contrast, species of Streptomyces typically exhibit slower growth and are competitive in environments where resource availability is limited, such as in the bulk soil (van der Meij et al., 2017). Thus, it is possible that proteobacteria were able to rapidly proliferate around and within the A. thaliana plant roots and outcompete the slower growing actinobacterial species, particularly those that were already present at lower abundance. Given their diverse metabolic capabilities, it is likely that streptomycetes are able to persist at low abundance by feeding on more complex organic polymers under conditions of high competition, including older, plant-originated organic material such as root cells and mucilage that were sloughed off before 13C labeling commenced. A. thaliana also appears to exhibit a relatively small rhizosphere effect compared to other plant species (Schlaeppi et al., 2014; Bulgarelli et al., 2015), meaning that there is a weaker differentiation in terms of microenvironment and community composition between the rhizosphere and bulk soil, than there is between these two compartments and the endophytic niche, as observed in this study (Supplementary Figure S4). Thus, complex polymers may have been more readily available in the rhizosphere than exudates, particularly given the fact that the compost growth medium also contained a relatively high level of organic matter (91.08%, Supplementary Table S3).
We also tested whether Streptomyces strains S. coelicolor M145 and the A. thaliana endophyte Streptomyces M3 are specifically attracted to A. thaliana roots by a specific root exudate compound, salicylate, since a previous study suggested this molecule can modulate microbiome composition and specifically attract streptomycetes (Lebeis et al., 2015). However, we found no evidence to support this hypothesis. We found that cpr5 mutant plants, which constitutively produce and accumulate salicylate, are colonized more readily by streptomycetes than wild-type plants, but we found no difference between the levels of Streptomyces root colonization in wild-type A. thaliana and salicylate-deficient sid2-2 and pad4 plants. Our data suggest that the cpr5 effect is not directly due to salicylate, at least with the M145 and M3 strains we tested here. None of the eight Streptomyces strains we tested could use salicylate as a sole carbon source, and they did not respond chemotactically to salicylate or compete better in salicylate amended soil. We propose that the greater colonization efficiency observed in A. thaliana cpr5 plants in this and a previous study (Lebeis et al., 2015) was not due to increased levels of salicylate, but more likely due to one or more of the pleiotropic effects of the cpr5 mutation (Jing et al., 2007). The cpr5 gene encodes a putative transmembrane protein (Kirik et al., 2001) that is part of a complex regulatory signaling network; its deletion results not only in the constitutive activation of SA biosynthesis genes, but also stunted root and aerial growth, disrupted cellular organization, spontaneous cell death, early senescence in young plants, lesions on various tissue types, disrupted cell wall biogenesis, an increased production of ROS and high levels of oxidative stress (Bowling et al., 1994; Clarke et al., 2000; Kirik et al., 2001; Jing et al., 2007; Jing et al., 2008; Gao et al., 2011). Thus, it is possible that the altered cell wall composition and spontaneous cell death in crp5 plants could have resulted in easier access to plant roots by streptomycetes, particularly as these bacteria are thought to enter roots in compromised areas, such as lesions and sites of wounding (Chater, 2006; Viaene et al., 2016). We also note that although that original study (Lebeis et al., 2015) reported that salicylate could be used as a sole C and N source by a Streptomyces endophyte strain, their Supplementary Material reported that sodium citrate was included in the growth medium used in these experiments and streptomycetes can grow using sodium citrate alone (Supplementary Figure S1).
In summary, we conclude that Streptomyces bacteria do not appear to be attracted to the rhizosphere by salicylic acid and did not feed on root exudates under the conditions presented in this study, because they were outcompeted by faster growing genera in the phylum Proteobacteria. Instead, it is likely that streptomycete bacteria were able to persist in the root microbiome by utilizing the organic matter present in the soil growth medium, in addition to components of the plant cells (such as cellulose) that are sloughed off during root growth. These ubiquitous soil bacteria are likely to be present in the soil as spores or mycelia when plant seeds germinate and root growth is initiated, enabling successful transmission between plant hosts. Although not tested in this study, it would be interesting to see whether Streptomyces spores germinate in the presence of plant root exudates. A greater understanding of the factors that attract beneficial bacteria to the plant root microbiome, and maintain them there, may have significant implications for agriculture. For example, it could inform efforts to develop and deliver effective plant growth-promoting agents and ensure they are competitive within the plant root niche. Indeed, studies have shown that organic matter amendments can improve the competitiveness and overall effectiveness of streptomycete biocontrol strains, presumably by providing them with an additional source of nutrients (Newitt et al., 2019). Clearly some Streptomyces strains can colonize the endosphere of plants and confer advantages to the plant host but it is not yet clear what advantage these bacteria gain from entering the plant roots, given that they must have to evade or suppress the plant immune system and are outcompeted for plant metabolites inside or outside the roots. Future work will address these questions and test the hypothesis that soil is a mechanism for vertical transmission of Streptomyces between generations of plants.
Isolation and maintenance of Streptomyces strains. The Streptomyces strains L2, M2, M3, N1 and N2 (Table 1) were isolated from surface-washed A. thaliana roots in a previous study (Worsley et al., 2020). Genome sequences were also generated for each of these isolates, as well as for three strains of Streptomyces lydicus, one of which was isolated from the commercial biocontrol product Actinovate, while the remaining two (ATCC25470 and ATCC31975) were obtained from the American Type Culture Collection (Table 1). In the present study, Streptomyces strains were maintained on SFM agar (N1, N2, M2, M3 and S. coelicolor M145), Maltose/Yeast extract/Malt extract (MYM) agar with trace elements (L2) or ISP2 agar (S. lydicus strains) (Supplementary Table S5). Strains were spore stocked as described previously (Kieser et al., 2000).
Plasmid pIJ8660 (Supplementary Table S1) containing an optimized eGFP gene and an aac apramycin resistance marker (Sun et al., 1999) was altered using molecular cloning of the constitutive ermE* promotor to drive eGFP expression. Primers ermEKpnFwd and ermENdeRev (Supplementary Table S1) were used to amplify ermEp* from the plasmid pIJ10257 (Supplementary Table S1) (Hong et al., 2005). The PCR product was subsequently purified and then digested with the restriction enzymes KpnI and NdeI, before being ligated into the same sites in pIJ8660. The resulting plasmid, called pIJ8660/ermEp* (Supplementary Table S1), was conjugated into the Streptomyces strains M3 and S. coelicolor M145 (Table 1) as described previously (Kieser et al., 2000). Exconjugants were selected and maintained on SFM agar plates containing 50 mg mL−1 apramycin.
Plant root colonization assays in soil. Wild-type A. thaliana Col-0 as well as the mutant plant lines cpr5, pad4, and sid2-2 were obtained from the Nottingham Arabidopsis Stock Centre (Table 1). Seeds were sterilized by placing them in 70% (v/v) ethanol for 2 min, followed by 20% NaOCl for 2 min and then washing them five times in sterile dH2O. Spores of either S. coelicolor M145-eGFP or Streptomyces M3-eGFP (Table 1) were used as inoculants and were pre-germinated in 2×YT (Supplementary Table S5) at 50°C for 10 min (Kieser et al., 2000). Uninoculated 2×YT was used as a control. Surface sterilized seeds were placed into 500 μl 2×YT containing 106 spores mL−1 of pre-germinated Streptomyces spores; these were mixed for 90 min on a rotating shaker before being transferred to pots of sieved Levington F2 seed and modular compost, soaked with distilled H2O. Another 1 mL of pre-germinated spores (or 2×YT for control pots) was pipetted into the soil to a depth of approximately 2 cm below the seed. Pots were incubated at 4°C for 24 h, then grown for 4 weeks under a 12-h photoperiod. Six replicate plants were grown for each plant genotype and each Streptomyces strain.
In order to re-isolate tagged strains from the roots of the different A. thaliana genotypes we used a previously established method (Lebeis et al., 2015). Briefly, plants were taken aseptically from pots and their roots were tapped firmly to remove as much adhering soil as possible. Root material was washed twice in sterile PBS-S buffer (Supplementary Table S5) for 30 min on a shaking platform and then any remaining soil particles were removed with sterile tweezers. Cleaned roots were then transferred to 25 mL of fresh PBS-S and sonicated for 20 min to remove any residual material still attached to the root surface; this ensured that any remaining bacteria were either present in the endophytic compartment or were very firmly attached to the root surface (“the rhizoplane”). Cleaned roots were weighed, then crushed in 1 mL of sterile 10% (v/v) glycerol. 100 μl of the homogenate was plated onto three replicate soya flour plus mannitol agar plates containing 50 μg mL−1 apramycin to select for inoculated streptomycetes (Supplementary Table S5). Plates also contained 5 μg mL−1 nystatin and 100 μg mL−1 cycloheximide to inhibit fungal growth. Agar plates were incubated at 30°C for 5 days, before the number of apramycin-resistant Streptomyces colony forming units (cfu) were counted. This method has been used previously in other studies (Bonaldi et al., 2015; Chen et al., 2016) to estimate the root colonization dynamics of streptomycetes. Counts were converted to cfu per gram of root tissue and log-transformed to normalize residuals. Data were analyzed via ANOVA and post-hoc Tukey’s Honest Significant Difference (HSD) tests.
The A. thaliana endophyte Streptomyces strains L2, M2, M3, N1 and N2, as well as the S. lydicus strains ATCC25470, ATCC31975 and Actinovate (Table 1), were streaked onto minimal agarose medium plates supplemented with either their preferred carbon source as a positive control, 3.875 mM sodium citrate, or 0.5 mM salicylic acid (SA) as a carbon source (Supplementary Table S5). Preferred carbon sources were 5 g L−1 of mannitol for strains N2 and M2; 5 g L−1 maltose for strains M3, N1, S. lydicus 25470 and S. lydicus Actinovate; 5 g L−1 sucrose for L2; 10 g L−1 glucose for S. lydicus 31975. Plates with no carbon source were used as a negative control. All strains were plated in triplicate onto each type of media and incubated for seven days at 30°C before imaging. S. coelicolor M145 was not tested as it can use agarose as a sole carbon source (Stanier, 1942; Temuujin et al., 2012). Agarose was used as a gelling agent as streptomycetes can grow on impurities that are present in agar.
To identify whether the nine different streptomycetes carried homologues to known SA degradation genes, all experimentally verified pathways involving SA (or salicylate) degradation were identified using the MetaCyc database (http://www.metacyc.org/) and amino acid sequences of characterized genes involved in each of the five pathways were then retrieved from UniProt (Supplementary Table S1). Protein sequences were used to perform BLASTP searches against predicted Open Reading Frames (ORFs) for each genome-sequenced streptomycete. Previously generated sequences for N1, N2, M2, M3, L2, S. lydicus 25470, S. lydicus 31975 and S. lydicus Actinovate were used (Worsley et al., 2020) as well as the published genome sequence of S. coelicolor (Bentley et al., 2002). The results of the best hit (% identity and % query coverage) are reported.
To test whether streptomycetes grew toward SA, 4 μl of spores (106 spores mL−1) of each streptomycete (Table 1) were pipetted onto the centre of SFM agar plates. 40 μl of either 1 mM or 0.5 mM filter-sterilized SA was inoculated onto 6 mm filter paper discs (Whatman) and allowed to dry. Discs were then added to one side of the agar plate, 2 cm from the streptomycete spores. SA solutions were prepared by diluting a 100 mM stock solution to a 1 mM or 0.5 mM SA solution in PBS. The 100 mM stock solution was made by dissolving 0.138 g of SA in 2 mL of 100% DMSO before making the solution up to 10 mL with PBS. Thus, the resulting 1 and 0.5 mM solutions had a final concentration of 0.2% (v/v) and 0.1% (v/v) DMSO, respectively. To check that any observations were not due to the effects of DMSO, control plates were also run alongside the SA experiment, in which discs were soaked in 40 μl of a 0.2% DMSO (in PBS), equivalent to the final concentration of DMSO in the 1 mM SA solution. All strains and disc type pairings were plated in triplicate and were incubated for seven days at 30°C before imaging.
Levington F2 compost (4 mL) was placed into each compartment of a 12-well plate and soaked with 0.5 mL of sterile dH2O or 0.5 mM SA. Each well was then inoculated with a spore solution (107 spores mL−1) of either S. coelicolor M145-eGFP or Streptomyces M3-eGFP (Table 1), suspended in dH2O or 0.5 mM SA. Spores of eGFP-tagged M3 or M145 were used to align with in vivo plant colonization experiments (outlined above). There were nine replicate wells for each treatment. Well-plates were placed under a 12-h photoperiod for 10 days 100 mg of soil from each well was then diluted in 900 μl of water and vortexed. Serial dilutions were then plated onto SFM containing 50 μg mL−1 apramycin (for selection) in addition to 10 μg mL−1 nystatin and 100 μg mL−1 cyclohexamide (to repress fungal growth). CFU of the Streptomyces inoculum were then enumerated on 10–2 dilution plates after 4 days to assess whether SA affected the competitiveness of strains in soil. A generalized linear model (GLM) with a negative binomial distribution was generated, using the package MASS in R 3.2.3 (R Core Team, 2017), to model the effect of strain (S. coelicolor M145-eGFP or Streptomyces M3-eGFP), soil treatment (wetting with SA or dH2O), and their interaction term (soil treatment) on the number of bacterial cfu returned from soil wells. Likelihood ratio tests were used to establish the significance of terms in the model.
To test whether streptomycetes were able to utilize other compounds in root exudates more generally, root exudates were isolated from A. thaliana and used as a growth medium for the Streptomyces strains (Table 1) that we previously isolated from A. thaliana plants growing in a compost system (Worsley et al., 2020). Seeds of A. thaliana Col-0 were sterilized (as described above) and germinated on MSk agar (Supplementary Table S5). After 7 days of growth at 22°C under a photoperiod of 12 h light/12 h dark, seedlings were transferred to 12 well plates, containing 3 mL of liquid MSk (Supplementary Table S5, 0% w/v sucrose) in each well. Seedlings were then grown for a further 10 days before being washed and transferred to new wells containing 3 mL of sterile water. After 5 days plant material was removed and the liquid from each well was filter-sterilized and added to sterile agarose (0.8% w/v) to make solid growth medium plates. Spores of previously isolated Streptomyces isolates (Table 1) were streaked onto these plates and incubated for 7 days at 30°C. Agarose/water (0.8% w/v) plates were used as a control.
Seeds of A. thaliana Col-0 (Table 1) were sterilized, as described above, before being sown singly into pots containing 100 mL sieved Levington F2 compost, soaked with dH2O. These were placed in the dark at 4°C for 48 h, after which they were transferred to short-day growth conditions (8 h light/16 h dark) at 22°C for 32 days before exposure to CO2 treatments. Each plant was placed into an air-tight, transparent 1.9 L cylindrical tube. Three plants were exposed to 1,000 ppmv of 12CO2 and three plants were exposed to 1,000 ppmv of 13CO2 (99%, Cambridge isotopes, Massachusetts, United States). Three unplanted controls containing only Levington F2 compost were additionally exposed to 1,000 ppmv of 13C labeled CO2 to control for autotrophic consumption of CO2 by soil microbes. CO2 treatments took place over a period of 21 days. CO2 was manually injected into tubes every 20 min over the course of the 8 h light period, to maintain the CO2 concentration at ∼1,000 ppmv; CO2 concentration was measured using gas chromatography (see below). At the end of the light period each day, tube lids were removed to prevent the build-up of respiratory CO2. Just before the next light period, tubes were flushed with an 80%/20% (v/v) nitrogen-oxygen mix to remove any residual CO2 before replacing the lids and beginning the first injection of CO2.
The volume of CO2 to be added at each injection in the SIP experiment was determined by measuring the rate of uptake of CO2 over 20 min every 4 days. This was done using an Agilent 7890A gas chromatography instrument with a flame ionization detector and a Porapak Q (6 ft × 1/8″) HP plot/Q (30 m × 0.530 mm, 40 μm film) column with a nickel catalyst and a nitrogen carrier gas. The instrument was run with the following settings: injector temperature 250°C, detector temperature 300°C, column temperature 115°C and oven temperature 50°C. The injection volume was 100 μl and the run time was 5 min, with CO2 having a retention time of 3.4 min. Peak areas were compared to a standard curve (standards of known CO2 concentration were prepared in 120 mL serum vials that had been flushed with 80/20% nitrogen-oxygen mixture).
Two samples of root-free “bulk soil” were collected from each planted and unplanted pot; samples were snap-frozen in liquid nitrogen and stored at −80°C. Roots were then processed to collect the rhizosphere soil and endophytic samples according to a published protocol (Bulgarelli et al., 2012). For the planted pots, roots were tapped until only the soil firmly adhering to the root surface remained; the remaining soil was defined as the rhizosphere fraction. To collect this, roots were placed in 25 mL sterile, PBS-S buffer (Supplementary Table S5) and washed on a shaking platform at top speed for 30 min before being transferred to fresh PBS-S. Used PBS-S from the first washing stage was centrifuged at 1,500 x g for 15 min, and the supernatant was removed. The resulting pellet (the rhizosphere sample) was snap-frozen and stored at -80°C. The roots were then shaken in fresh PBS-S for a further 30 min before removing any remaining soil particles with sterile tweezers. Finally, the cleaned roots were transferred to fresh PBS-S and sonicated for 20 min in a sonicating water bath (Bulgarelli et al., 2012). Root samples for each plant were then snap frozen and stored at −80°C. Each root sample consisted of bacteria within the roots and those very firmly attached to the root surface (the “rhizoplane”). A modified version of the manufacturer’s protocol for the FastDNA™ SPIN Kit for Soil (MP Biomedicals) was used to extract DNA from soil, rhizosphere, and root samples. Modifications included pre-homogenization of the root material by grinding in liquid nitrogen before adding lysis buffer, an extended incubation time (10 min) in DNA matrix buffer, and elution in 150 μl of sterile water. DNA yields were quantified using a Qubit™ fluorimeter.
DNA samples from the rhizosphere, roots, and unplanted soil were subjected to caesium chloride (CsCl) density gradient separation using an established protocol (Neufeld et al., 2007). For each of the replicate rhizosphere samples from both the 12CO2 and 13CO2 incubated plants, 1.5 μg of DNA was loaded into the CsCl solution with gradient buffer. For the three unplanted soil sample replicates, 1 μg of DNA was used. For the root samples it was necessary to combine the three replicate samples for each CO2 treatment due to low DNA yields. Thus, 0.2 μg of DNA was pooled from each of the three replicates per 12CO2 and 13CO2 treatment, and the final 0.6 μg was loaded into the CsCl solution. After ultracentrifugation, the density of each fraction was measured using a refractometer (Reichert Analytical Instruments, NY, United States) in order to check for successful gradient formation. DNA was precipitated from fractions (Neufeld et al., 2007) and stored at -20°C before use as a template in qPCR and PCR reactions.
To identify fractions containing heavy (13C) and light (12C) DNA for each sample, 16S rRNA gene copy number was quantified across fractions using qPCR. Reactions were carried out in 25 μl volumes. 1 μl of template DNA (either sample DNA or standard DNA), or dH2O as a control, was added to 24 μl of reaction mix containing 12.5 μl of 2x Sybr Green Jumpstart Taq Ready-mix (Sigma Aldrich), 0.125 μl of each of the primers PRK341F and MPRK806R (Supplementary Table S2), 4 μl of 25 mM MgCl2, 0.25 μl of 20 μg μl−1 Bovine Serum Albumin (BSA, Roche), and 7 μl dH2O. Sample DNA, standards (a dilution series of the target 16S rRNA gene at known molecular quantities), and negative controls were quantified in duplicate. Reactions were run under the following conditions: 96°C for 10 min; 40 cycles of 96°C for 30 s, 52°C for 30 s, and 72°C for 1 min; 96°C for 15 s; 100 cycles at 75–95°C for 10 s, ramping 0.2°C per cycle. Reactions were performed in 96-well plates (Bio-Rad). The threshold cycle (CT) for each sample was then converted to target molecule number by comparing to CT values of a dilution series of target DNA standards. Fractions spanning the peaks in 16S rRNA gene copy number were identified and equal quantities of these were combined to create a “heavy” buoyant density (labeled) and “light” buoyant density (unlabelled) fraction for each sample, respectively. The 16S rRNA genes were amplified for each of these fractions using the Universal primers PRK341F and MPRK806R (Supplementary Table S2), and the resulting PCR product was purified and submitted for 16S rRNA gene amplicon sequencing using an Illumina MiSeq at MR DNA (Molecular Research LP), Shallowater, Texas, United States. This generated a total of 3′699′266 reads, with an average of 127,560.90 ± 85,376.35 (standard deviation) reads per sample. Sequence data were then processed at MR DNA using their custom pipeline (Dowd et al., 2008a; Dowd et al., 2008c). As part of this pipeline, pair-end sequences were merged, barcodes were trimmed, and sequences of less than 150 bp and/or with ambiguous base calls were removed. The resulting sequences were denoized, and OTUs were assigned by clustering at 97% similarity. Chimeras were removed, and OTUs were assigned taxonomies using BlastN against a curated database from GreenGenes, RDPII, and NCBI (DeSantis et al., 2006). A post processing algorithm generated best-hit files with E-values < 10e−114 and bit scores > 400. The identities of all hits were >98%. These parameters have been previously evaluated to enable reliable identification at the genus level (Dowd et al., 2008b). Plastid-like sequences and singletons were removed from downstream analysis. A total of 1′938′580 reads remained after processing, with an average of 66,847.59 ± 40,423 (standard deviation) reads per sample. OTU abundances were converted to relative abundances in all downstream analyses to control for the variation in library sizes across samples. All data received from MR DNA were then further processed and statistically analyzed using R 3.2.3 (R Core Team, 2017), using the packages tidyr and reshape (for manipulating data-frames), ggplot2 and gplots (for plotting graphs and heatmaps, respectively), vegan (for calculating Bray–Curtis dissimilarities, conducting principle coordinate and PERMANOVA analyses, calculating Shannon diversity, and for generating heatmaps) and ellipse (for plotting principal coordinate analysis). All the 16S rRNA gene amplicon sequences have been submitted to the European Nucleotide Archive (ENA) database under the study accession number PRJEB30923.
Soil pH, organic matter content (%) as well as the levels of phosphorus, potassium and magnesium (all in mg/kg), was measured by the James Hutton Institute Soil Analysis Service (Aberdeen, United Kingdom). To quantify inorganic nitrate and ammonium concentrations a KCl extraction was performed in triplicate, whereby 3 g of soil was suspended in 24 mL of 1 M KCl and incubated for 30 min, with shaking, at 250 rpm. To quantify ammonium concentration (g/kg) an indophenol blue method was used (as described in Verdouw et al., 1978). For nitrate concentration (in g/kg) vanadium (III) chloride reduction followed by chemiluminescence was used (as described in Braman and Hendrix, 1989).
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ebi.ac.uk/ena, PRJEB30923.
SW, MM, SP, BW, JM, and MH designed the research. SW, JM, and MH wrote the manuscript with comments from all other authors. SW performed the strain isolation and plant colonization experiments. SW performed the DNA stable isotope probing experiments with help from MM. SP carried out the chemical analysis of compost. SW performed all statistical analysis.
SW and SP were funded by a Natural Environment Research Council (NERC) Ph.D. studentship (NERC Doctoral Training Program Grant NE/L002582/1). MM was funded by a Biotechnology and Biological Sciences Research Council (BBSRC) Ph.D. studentship (BBSRC Doctoral Training Program grant BB/M011216/1). This work was also supported by the Norwich Research Park (NRP) through a Science Links Seed Corn Grant to MH and JM and by the NRP Earth and Life Systems Alliance (ELSA). Methods for Streptomyces culturing, genetic manipulation, plasmids and preparing growth media are available from www.actinobase.org.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The authors would like to thank Elaine Patrick for technical support and Andrew Crombie for advice on stable isotope probing.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmolb.2021.686110/full#supplementary-material
Alfreider, A., Grimus, V., Luger, M., Ekblad, A., Salcher, M. M., and Summerer, M. (2018). Autotrophic Carbon Fixation Strategies Used by Nitrifying Prokaryotes in Freshwater Lakes. FEMS Microbiol. Ecol. 94 (10), 112. doi:10.1093/femsec/fiy163
An, D.-S., Im, W.-T., Yang, H.-C., and Lee, S.-T. (2006). Shinella Granuli Gen. nov., Sp. nov., and Proposal of the Reclassification of Zoogloea Ramigera ATCC 19623 as Shinella Zoogloeoides Sp. Nov. Int. J. Syst. Evol. Microbiol. 56 (Pt 2), 443–448. doi:10.1099/ijs.0.63942-0
Badri, D. V., Chaparro, J. M., Zhang, R., Shen, Q., and Vivanco, J. M. (2013). Application of Natural Blends of Phytochemicals Derived from the Root Exudates of Arabidopsis to the Soil Reveal that Phenolic-Related Compounds Predominantly Modulate the Soil Microbiome. J. Biol. Chem. 288 (7), 4502–4512. doi:10.1074/jbc.m112.433300
Badri, D. V., and Vivanco, J. M. (2009). Regulation and Function of Root Exudates. Plant Cel Environ. 32 (6), 666–681. doi:10.1111/j.1365-3040.2009.01926.x
Bai, Y., Müller, D. B., Srinivas, G., Garrido-Oter, R., Potthoff, E., Rott, M., et al. (2015). Functional Overlap of the Arabidopsis Leaf and Root Microbiota. Nature 528 (7582), 364–369. doi:10.1038/nature16192
Bais, H. P., Weir, T. L., Perry, L. G., Gilroy, S., and Vivanco, J. M. (2006). The Role of Root Exudates in Rhizosphere Interactions with Plants and Other Organisms. Annu. Rev. Plant Biol. 57, 233–266. doi:10.1146/annurev.arplant.57.032905.105159
Bentley, S. D., Chater, K. F., Cerdeño-Tárraga, A. M., Challis, G. L., Thomson, N. R., James, K. D., et al. (2002). Complete Genome Sequence of the Model Actinomycete Streptomyces Coelicolor A3(2). Nature 417 (6885), 141–147. doi:10.1038/417141a
Bollmann, A., Sedlacek, C. J., Norton, J., Laanbroek, H. J., Suwa, Y., Stein, L. Y., et al. (2013). Complete Genome Sequence of Nitrosomonas Sp. Is79, an Ammonia Oxidizing Bacterium Adapted to Low Ammonium Concentrations. Stand. Genomic Sci. 7 (3), 469–482. doi:10.4056/sigs.3517166
Bonaldi, M., Chen, X., Kunova, A., Pizzatti, C., Saracchi, M., and Cortesi, P. (2015). Colonization of Lettuce Rhizosphere and Roots by Tagged Streptomyces. Front. Microbiol. 6, 25. doi:10.3389/fmicb.2015.00025
Bowling, S. A., Guo, A., Cao, H., Gordon, A. S., Klessig, D. F., and Dong, X. (1994). A Mutation in Arabidopsis that Leads to Constitutive Expression of Systemic Acquired Resistance. Plant Cell 6 (12), 1845–1857. doi:10.1105/tpc.6.12.1845
Braman, R. S., and Hendrix, S. A. (1989). Nanogram Nitrite and Nitrate Determination in Environmental and Biological Materials by Vanadium (III) Reduction with Chemiluminescence Detection. Anal. Chem. 61, 2715–2718. doi:10.1021/ac00199a007
Brininstool, G., Kasili, R., Simmons, L. A., Kirik, V., Hülskamp, M., and Larkin, J. C. (2008). Constitutive Expressor Of Pathogenesis-Related Genes5 Affects Cell wall Biogenesis and Trichome Development. BMC Plant Biol. 8, 58. doi:10.1186/1471-2229-8-58
Bulgarelli, D., Garrido-Oter, R., Münch, P. C., Weiman, A., Dröge, J., Pan, Y., et al. (2015). Structure and Function of the Bacterial Root Microbiota in Wild and Domesticated Barley. Cell Host & Microbe 17 (3), 392–403. doi:10.1016/j.chom.2015.01.011
Bulgarelli, D., Rott, M., Schlaeppi, K., Ver Loren van Themaat, E., Ahmadinejad, N., Assenza, F., et al. (2012). Revealing Structure and Assembly Cues for Arabidopsis Root-Inhabiting Bacterial Microbiota. Nature 488 (7409), 91–95. doi:10.1038/nature11336
Caldwell, D. R., Keeney, M., and Van Soest, P. J. (1969). Effects of Carbon Dioxide on Growth and Maltose Fermentation by Bacteroides Amylophilus. J. Bacteriol. 98 (2), 668–676. doi:10.1128/jb.98.2.668-676.1969
Carvalhais, L. C., Dennis, P. G., Badri, D. V., Kidd, B. N., Vivanco, J. M., and Schenk, P. M. (2015). Linking Jasmonic Acid Signaling, Root Exudates, and Rhizosphere Microbiomes. Mpmi 28 (9), 1049–1058. doi:10.1094/MPMI-01-15-0016-R
Carvalhais, L. C., Dennis, P. G., Badri, D. V., Tyson, G. W., Vivanco, J. M., and Schenk, P. M. (2013). Activation of the Jasmonic Acid Plant Defence Pathway Alters the Composition of Rhizosphere Bacterial Communities. PLoS One 8 (2), e56457. doi:10.1371/journal.pone.0056457
Chater, K. F. (2016). Recent Advances in Understanding Streptomyces. F1000Res 5, 2795. doi:10.12688/f1000research.9534.1
Chater, K. F. (2006). Streptomyces Inside-Out: a New Perspective on the Bacteria that Provide Us with Antibiotics. Phil. Trans. R. Soc. B 361 (1469), 761–768. doi:10.1098/rstb.2005.1758
Chen, X., Pizzatti, C., Bonaldi, M., Saracchi, M., Erlacher, A., Kunova, A., et al. (2016). Biological Control of Lettuce Drop and Host Plant Colonization by Rhizospheric and Endophytic Streptomycetes. Front. Microbiol. 7, 714. doi:10.3389/fmicb.2016.00714
Clarke, J. D., Volko, S. M., Ledford, H., Ausubel, F. M., and Dong, X. (2000). Roles of Salicylic Acid, Jasmonic Acid, and Ethylene in Cpr-Induced Resistance in Arabidopsis. The Plant Cell 12 (11), 2175–2190. doi:10.2307/3871113
Dennis, P. G., Miller, A. J., and Hirsch, P. R. (2010). Are Root Exudates More Important Than Other Sources of Rhizodeposits in Structuring Rhizosphere Bacterial Communities?. FEMS Microbiol. Ecol. 72 (3), 313–327. doi:10.1111/j.1574-6941.2010.00860.x
DeSantis, T. Z., Hugenholtz, P., Larsen, N., Rojas, M., Brodie, E. L., Keller, K., et al. (2006). Greengenes, a Chimera-Checked 16S rRNA Gene Database and Workbench Compatible with ARB. Appl. Environ. Microbiol. 72 (7), 5069–5072. doi:10.1128/AEM.03006-05
Doornbos, R. F., Geraats, B. P. J., Kuramae, E. E., Van Loon, L. C., and Bakker, P. A. H. M. (2011). Effects of Jasmonic Acid, Ethylene, and Salicylic Acid Signaling on the Rhizosphere Bacterial Community of Arabidopsis thaliana. Mpmi 24 (4), 395–407. doi:10.1094/MPMI-05-10-0115
Dowd, S. E., Callaway, T. R., Wolcott, R. D., Sun, Y., McKeehan, T., Hagevoort, R. G., et al. (2008a). Evaluation of the Bacterial Diversity in the Feces of Cattle Using 16S rDNA Bacterial Tag-Encoded FLX Amplicon Pyrosequencing (bTEFAP). BMC Microbiol. 8, 125. doi:10.1186/1471-2180-8-125
Dowd, S. E., Sun, Y., Secor, P. R., Rhoads, D. D., Wolcott, B. M., James, G. A., et al. (2008b). Survey of Bacterial Diversity in Chronic Wounds Using Pyrosequencing, DGGE, and Full Ribosome Shotgun Sequencing. BMC Microbiol. 8, 43. doi:10.1186/1471-2180-8-43
Dowd, S. E., Wolcott, R. D., Sun, Y., McKeehan, T., Smith, E., and Rhoads, D. (2008c). Polymicrobial Nature of Chronic Diabetic Foot Ulcer Biofilm Infections Determined Using Bacterial Tag Encoded FLX Amplicon Pyrosequencing (bTEFAP). PLoS One 3 (10), e3326. doi:10.1371/journal.pone.0003326
Dumont, M. G., and Murrell, J. C. (2005). Stable Isotope Probing - Linking Microbial Identity to Function. Nat. Rev. Microbiol. 3 (6), 499–504. doi:10.1038/nrmicro1162
Fischbach, M. A., and Sonnenburg, J. L. (2011). Eating for Two: How Metabolism Establishes Interspecies Interactions in the Gut. Cell Host & Microbe 10 (4), 336–347. doi:10.1016/j.chom.2011.10.002
Gao, G., Zhang, S., Wang, C., Yang, X., Wang, Y., Su, X., et al. (2011). Arabidopsis CPR5 Independently Regulates Seed Germination and Postgermination Arrest of Development through LOX Pathway and ABA Signaling. PLoS One 6 (4), e19406. doi:10.1371/journal.pone.0019406
Glick, B. R., Liu, C., Ghosh, S., and Dumbroff, E. B. (1997). Early Development of Canola Seedlings in the Presence of the Plant Growth-Promoting Rhizobacterium Pseudomonas Putida GR12-2. Soil Biol. Biochem. 29 (8), 1233–1239. doi:10.1016/S0038-0717(97)00026-6
Gong, Z.-L., Ai, M.-J., Sun, H.-M., Liu, H.-Y., Yu, L.-Y., and Zhang, Y.-Q. (2016). Jatrophihabitans Huperziae Sp. nov., an Endophytic Actinobacterium Isolated from Surface-Sterilized Tissue of the Medicinal Plant Huperzia Serrata (Thunb.). Int. J. Syst. Evol. Microbiol. 66 (10), 3972–3977. doi:10.1099/ijsem.0.001296
Haichar, F. e. Z., Heulin, T., Guyonnet, J. P., and Achouak, W. (2016). Stable Isotope Probing of Carbon Flow in the Plant Holobiont. Curr. Opin. Biotechnol. 41, 9–13. doi:10.1016/j.copbio.2016.02.023
Haichar, F. e. Z., Roncato, M.-A., and Achouak, W. (2012). Stable Isotope Probing of Bacterial Community Structure and Gene Expression in the Rhizosphere of Arabidopsis thaliana. FEMS Microbiol. Ecol. 81 (2), 291–302. doi:10.1111/j.1574-6941.2012.01345.x
Hase, S., Van Pelt, J. A., Van Loon, L. C., and Pieterse, C. M. J. (2003). Colonization of Arabidopsis Roots by Pseudomonas Fluorescens Primes the Plant to Produce Higher Levels of Ethylene upon Pathogen Infection. Physiol. Mol. Plant Pathol. 62 (4), 219–226. doi:10.1016/S0885-5765(03)00059-6
Hernández-León, R., Rojas-Solís, D., Contreras-Pérez, M., Orozco-Mosqueda, M. d. C., Macías-Rodríguez, L. I., Reyes-de la Cruz, H., et al. (2015). Characterization of the Antifungal and Plant Growth-Promoting Effects of Diffusible and Volatile Organic Compounds Produced by Pseudomonas Fluorescens Strains. Biol. Control. 81, 83–92. doi:10.1016/j.biocontrol.2014.11.011
Hong, H.-J., Hutchings, M. I., Hill, L. M., and Buttner, M. J. (2005). The Role of the Novel Fem Protein VanK in Vancomycin Resistance in Streptomyces Coelicolor. J. Biol. Chem. 280 (13), 13055–13061. doi:10.1074/jbc.M413801200
Huang, A. C., Jiang, T., Liu, Y.-X., Bai, Y.-C., Reed, J., Qu, B., et al. (2019). A Specialized Metabolic Network Selectively Modulates Arabidopsis Root Microbiota. Science 364 (6440), eaau6389. doi:10.1126/science.aau6389
Hutchings, M. I., Truman, A. W., and Wilkinson, B. (2019). Antibiotics: Past, Present and Future. Curr. Opin. Microbiol. 51, 72–80. doi:10.1016/j.mib.2019.10.008
Jing, H.-C., Anderson, L., Sturre, M. J. G., Hille, J., and Dijkwel, P. P. (2007). Arabidopsis CPR5 Is a Senescence-Regulatory Gene with Pleiotropic Functions as Predicted by the Evolutionary Theory of Senescence. J. Exp. Bot. 58 (14), 3885–3894. doi:10.1093/jxb/erm237
Jing, H.-C., Hebeler, R., Oeljeklaus, S., Sitek, B., Stühler, K., Meyer, H. E., et al. (2008). Early Leaf Senescence Is Associated with an Altered Cellular Redox Balance inArabidopsis Cpr5/old1mutants. Plant Biol. (Stuttg) 10 (Suppl. 1), 85–98. doi:10.1111/j.1438-8677.2008.00087.x
Jirage, D., Tootle, T. L., Reuber, T. L., Frost, L. N., Feys, B. J., Parker, J. E., et al. (1999). Arabidopsis thaliana PAD4 Encodes a Lipase-like Gene that Is Important for Salicylic Acid Signaling. Proc. Natl. Acad. Sci. 96 (23), 13583–13588. doi:10.1073/pnas.96.23.13583
Kieser, T., Bibb, M. J., Buttner, M. J., Chater, K. F., and Hopwood, D. A. (2000). Practical Streptomyces Genetics. Norwich: John Innes Foundation.
Kim, D.-R., Cho, G., Jeon, C.-W., Weller, D. M., Thomashow, L. S., Paulitz, T. C., et al. (2019). A Mutualistic Interaction between Streptomyces Bacteria, Strawberry Plants and Pollinating Bees. Nat. Commun. 10 (1), 4802. doi:10.1038/s41467-019-12785-3
Kirik, V., Bouyer, D., Schöbinger, U., Bechtold, N., Herzog, M., Bonneville, J.-M., et al. (2001). CPR5 Is Involved in Cell Proliferation and Cell Death Control and Encodes a Novel Transmembrane Protein. Curr. Biol. 11 (23), 1891–1895. doi:10.1016/s0960-9822(01)00590-5
Kitzinger, K., Koch, H., Lücker, S., Sedlacek, C. J., Herbold, C., Schwarz, J., et al. (2018). Characterization of the First "CandidatusNitrotoga" Isolate Reveals Metabolic Versatility and Separate Evolution of Widespread Nitrite-Oxidizing Bacteria. mBio 9 (4), 114. doi:10.1128/mBio.01186-18
Lebeis, S. L., Paredes, S. H., Lundberg, D. S., Breakfield, N., Gehring, J., McDonald, M., et al. (2015). Salicylic Acid Modulates Colonization of the Root Microbiome by Specific Bacterial Taxa. Science 349 (6250), 860–864. doi:10.1126/science.aaa8764
Lin, D. X., Wang, E. T., Tang, H., Han, T. X., He, Y. R., Guan, S. H., et al. (2008). Shinella Kummerowiae Sp. nov., a Symbiotic Bacterium Isolated from Root Nodules of the Herbal Legume Kummerowia Stipulacea. Int. J. Syst. Evol. Microbiol. 58 (Pt 6), 1409–1413. doi:10.1099/ijs.0.65723-0
Liu, H., Carvalhais, L. C., Schenk, P. M., and Dennis, P. G. (2017). Effects of Jasmonic Acid Signalling on the Wheat Microbiome Differ between Body Sites. Sci. Rep. 7, 41766. doi:10.1038/srep41766
Lundberg, D. S., Lebeis, S. L., Paredes, S. H., Yourstone, S., Gehring, J., Malfatti, S., et al. (2012). Defining the Core Arabidopsis thaliana Root Microbiome. Nature 488 (7409), 86–90. doi:10.1038/nature11237
Madhaiyan, M., Hu, C. J., Kim, S.-J., Weon, H.-Y., Kwon, S.-W., and Ji, L. (2013). Jatrophihabitans Endophyticus Gen. nov., Sp. nov., an Endophytic Actinobacterium Isolated from a Surface-Sterilized Stem of Jatropha Curcas L. Int. J. Syst. Evol. Microbiol. 63 (Pt 4), 1241–1248. doi:10.1099/ijs.0.039685-0
Matilla, M. A., Ramos, J. L., Bakker, P. A. H. M., Doornbos, R., Badri, D. V., Vivanco, J. M., et al. (2010). Pseudomonas Putida KT2440 Causes Induced Systemic Resistance and Changes in Arabidopsis Root Exudation. Environ. Microbiol. Rep. 2 (3), 381–388. doi:10.1111/j.1758-2229.2009.00091.x
Mercado-Blanco, J., Alós, E., Rey, M. D., and Prieto, P. (2016). Pseudomonas fluorescensPICF7 Displays an Endophytic Lifestyle in Cultivated Cereals and Enhances Yield in Barley. FEMS Microbiol. Ecol. 92 (8), fiw092. doi:10.1093/femsec/fiw092
Neufeld, J. D., Vohra, J., Dumont, M. G., Lueders, T., Manefield, M., Friedrich, M. W., et al. (2007). DNA Stable-Isotope Probing. Nat. Protoc. 2 (4), 860–866. doi:10.1038/nprot.2007.109
Newitt, J., Prudence, S., Hutchings, M., and Worsley, S. (2019). Biocontrol of Cereal Crop Diseases Using Streptomycetes. Pathogens 8 (2), 78. doi:10.3390/pathogens8020078
Ofek, M., Hadar, Y., and Minz, D. (2012). Ecology of Root Colonizing Massilia (Oxalobacteraceae). PLoS One 7 (7), e40117. doi:10.1371/journal.pone.0040117
Philippot, L., Raaijmakers, J. M., Lemanceau, P., and van der Putten, W. H. (2013). Going Back to the Roots: the Microbial Ecology of the Rhizosphere. Nat. Rev. Microbiol. 11 (11), 789–799. doi:10.1038/nrmicro3109
Radajewski, S., Ineson, P., Parekh, N. R., and Murrell, J. C. (2000). Stable-isotope Probing as a Tool in Microbial Ecology. Nature 403 (6770), 646–649. doi:10.1038/35001054
Raza, W., Ling, N., Liu, D., Wei, Z., Huang, Q., and Shen, Q. (2016). Volatile Organic Compounds Produced by Pseudomonas Fluorescens WR-1 Restrict the Growth and Virulence Traits of Ralstonia Solanacearum. Microbiol. Res. 192, 103–113. doi:10.1016/j.micres.2016.05.014
Rey, T., and Dumas, B. (2017). Plenty Is No Plague: Streptomyces Symbiosis with Crops. Trends Plant Sci. 22 (1), 30–37. doi:10.1016/j.tplants.2016.10.008
Rudrappa, T., Czymmek, K. J., Paré, P. W., and Bais, H. P. (2008). Root-Secreted Malic Acid Recruits Beneficial Soil Bacteria. Plant Physiol. 148 (3), 1547–1556. doi:10.1104/pp.108.127613
Sasse, J., Martinoia, E., and Northen, T. (2018). Feed Your Friends: Do Plant Exudates Shape the Root Microbiome?. Trends Plant Sci. 23 (1), 25–41. doi:10.1016/j.tplants.2017.09.003
Scheuring, I., and Yu, D. W. (2012). How to Assemble a Beneficial Microbiome in Three Easy Steps. Ecol. Lett. 15 (11), 1300–1307. doi:10.1111/j.1461-0248.2012.01853.x
Schlaeppi, K., Dombrowski, N., Oter, R. G., Ver Loren van Themaat, E., and Schulze-Lefert, P. (2014). Quantitative Divergence of the Bacterial Root Microbiota in Arabidopsis thaliana Relatives. Proc. Natl. Acad. Sci. 111 (2), 585–592. doi:10.1073/pnas.1321597111
Stanier, R. Y. (1942). Agar-decomposing Strains of the Actinomyces Coelicolor Species-Group. J. Bacteriol. 44 (5), 555–570. doi:10.1128/jb.44.5.555-570.1942
Sun, J., Kelemen, G. H., Fernández-Abalos, J. M., and Bibb, M. J. (1999). Green Fluorescent Protein as a Reporter for Spatial and Temporal Gene Expression in Streptomyces Coelicolor A3(2) This Paper Is Dedicated to the Memory of Kathy Kendrick, Whose Devotion to Understanding the Biology of Streptomyces Was Unsurpassed. Microbiology 145 (Pt 9), 2221–2227. doi:10.1099/00221287-145-9-2221
Temuujin, U., Chi, W.-J., Chang, Y.-K., and Hong, S.-K. (2012). Identification and Biochemical Characterization of Sco3487 from Streptomyces Coelicolor A3(2), an Exo- and Endo-Type -Agarase-Producing Neoagarobiose. J. Bacteriol. 194 (1), 142–149. doi:10.1128/JB.05978-11
van der Meij, A., Worsley, S. F., Hutchings, M. I., and van Wezel, G. P. (2017). Chemical Ecology of Antibiotic Production by Actinomycetes. FEMS Microbiol. Rev. 41 (3), 392–416. doi:10.1093/femsre/fux005
Verdouw, H., Van Echteld, C. J. A., and Dekkers, E. M. J. (1978). Ammonia Determination Based on Indophenol Formation with Sodium Salicylate. Water Res. 12 (6), 399–402. doi:10.1016/0043-1354(78)90107-0
Viaene, T., Langendries, S., Beirinckx, S., Maes, M., and Goormachtig, S. (2016). Streptomycesas a Plant's Best Friend?. FEMS Microbiol. Ecol. 92, fiw119. doi:10.1093/femsec/fiw119
Wildermuth, M. C., Dewdney, J., Wu, G., and Ausubel, F. M. (2001). Isochorismate Synthase Is Required to Synthesize Salicylic Acid for Plant Defence. Nature 414 (6863), 562–565. doi:10.1038/35107108
Worsley, S. F., Newitt, J., Rassbach, J., Batey, S. F. D., Holmes, N. A., Murrell, J. C., et al. (2020). Streptomyces Endophytes Promote the Growth of Arabidopsis thaliana. bioRxiv 12, 532309. doi:10.1101/532309
Wu, X., Ge, T., Wang, W., Yuan, H., Wegner, C.-E., Zhu, Z., et al. (2015). Cropping Systems Modulate the Rate and Magnitude of Soil Microbial Autotrophic CO2 Fixation in Soil. Front. Microbiol. 6, 379. doi:10.3389/fmicb.2015.00379
Xiao, K. Q., Ge, T. D., Wu, X. H., Peacock, C. L., Zhu, Z. K., Peng, J., et al. (2021). Metagenomic and 14 C Tracing Evidence for Autotrophic Microbial CO 2 Fixation in Paddy Soils. Environ. Microbiol. 23 (2), 924–933. doi:10.1111/1462-2920.15204
Ye, J., An, N., Chen, H., Ying, Z., Zhang, S., and Zhao, J. (2020). Performance and Mechanism of Carbon Dioxide Fixation by a Newly Isolated Chemoautotrophic Strain Paracoccus Denitrificans PJ-1. Chemosphere 252, 126473. doi:10.1016/j.chemosphere.2020.126473
Zamioudis, C., Mastranesti, P., Dhonukshe, P., Blilou, I., and Pieterse, C. M. J. (2013). Unraveling Root Developmental Programs Initiated by Beneficial Pseudomonas Spp. Bacteria. Plant Physiol. 162 (1), 304–318. doi:10.1104/pp.112.212597
Zhalnina, K., Louie, K. B., Hao, Z., Mansoori, N., da Rocha, U. N., Shi, S., et al. (2018). Dynamic Root Exudate Chemistry and Microbial Substrate Preferences Drive Patterns in Rhizosphere Microbial Community Assembly. Nat. Microbiol. 3 (4), 470–480. doi:10.1038/s41564-018-0129-3
Zhao, C. Z., Huang, J., Gyaneshwar, P., and Zhao, D. (2017). Rhizobium Sp. IRBG74 Alters Arabidopsis Root Development by Affecting Auxin Signaling. Front. Microbiol. 8, 2556. doi:10.3389/fmicb.2017.02556
Keywords: Streptomyces, arabidopsis, root exudates, endophytes, rhizosphere
Citation: Worsley SF, Macey MC, Prudence SMM, Wilkinson B, Murrell JC and Hutchings MI (2021) Investigating the Role of Root Exudates in Recruiting Streptomyces Bacteria to the Arabidopsis thaliana Microbiome. Front. Mol. Biosci. 8:686110. doi: 10.3389/fmolb.2021.686110
Received: 26 March 2021; Accepted: 27 May 2021;
Published: 16 June 2021.
Edited by:
Christopher Ryan Penton, Arizona State University, United StatesReviewed by:
Naeem Khan, University of Florida, United StatesCopyright © 2021 Worsley, Macey, Prudence, Wilkinson, Murrell and Hutchings. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sarah F. Worsley, cy53b3JzbGV5QHVlYS5hYy51aw==; Matthew I. Hutchings, bWF0dC5odXRjaGluZ3NAamljLmFjLnVr
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.