- 1National Health Commission Key Laboratory for Medical Virology and Viral Diseases, Beijing, China
- 2National Institute for Viral Disease Control and Prevention, China CDC, Beijing, China
Rotavirus (RV) is an important pathogen causing acute gastroenteritis in young humans and animals. Attachment to the host receptor is a crucial step for the virus infection. The recent advances in illustrating the interactions between RV and glycans promoted our understanding of the host range and epidemiology of RVs. VP8*, the distal region of the RV outer capsid spike protein VP4, played a critical role in the glycan recognition. Group A RVs were classified into different P genotypes based on the VP4 sequences and recognized glycans in a P genotype-dependent manner. Glycans including sialic acid, gangliosides, histo-blood group antigens (HBGAs), and mucin cores have been reported to interact with RV VP8*s. The glycan binding specificities of VP8*s of different RV genotypes have been studied. Here, we mainly discussed the structural basis for the interactions between RV VP8*s and glycans, which provided molecular insights into the receptor recognition and host tropism, offering new clues to the design of RV vaccine and anti-viral agents.
Introduction
Rotavirus (RV), belonging to the Reoviridae, is an important pathogen leading to acute gastroenteritis (AGE) in children under 5 years old and caused ∼200,000 deaths worldwide each year (Tate et al., 2016; Bányai et al., 2018). RV genome contained 11 segments of double-stranded RNA, encoding 6 structural proteins (VP) and 6 non-structural proteins (Estes and Greenberg, 2013). The RV capsid has three layers consisting of a core layer formed by VP2, an intermediate layer formed by VP6, and an outer layer formed by VP4 and VP7 (Figure 1). Based on the antigenic and molecular characteristics of VP6, RVs are currently classified into nine groups/species (A-I) and a further tentative group J (Banyai et al., 2017). Groups A, B, C, and H RVs have been identified in human infections, while other groups only cause diseases in animal species (Matthijnssens et al., 2012; Banyai et al., 2017). Among these, group A RVs (RVAs) are the most widely prevalent in humans and the leading cause of severe AGE worldwide.
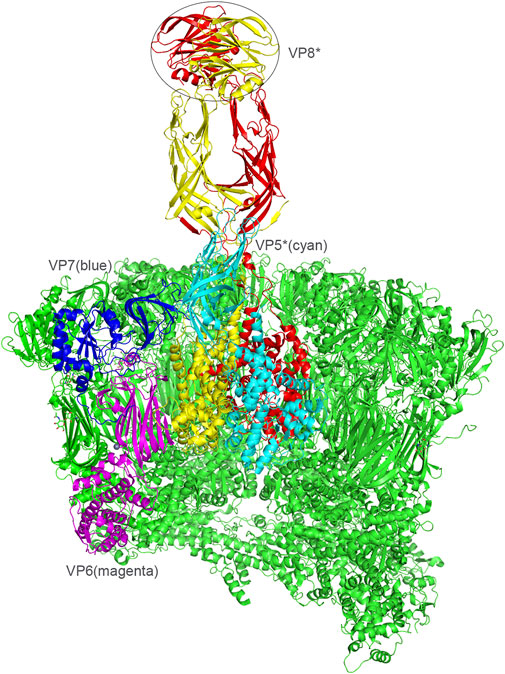
FIGURE 1. Presentation of the RV viral proteins of the middle and outer layers based on the rotavirus particle structure (PDB: 4V7Q). VP6, magenta. VP7, blue. VP4, red/yellow. VP5, cyan. VP8*, the distal domain of VP4 was shown in a circle.
VP7 is a glycoprotein and VP4 is protease-sensitive (Estes and Greenberg, 2013). VP4 extending from the VP7 shell formed the major spike protein contributing to the viral attachment and penetration (Figure 1) (Dormitzer et al., 2002b). RV was classified into G and P genotypes based on VP7 and VP4, respectively, (Matthijnssens et al., 2011). To date, no less than 37 G and 51 P genotypes of RVAs have been identified (https://rega.kuleuven.be/cev/viralmetagenomics/virus-classification). Different combinations of G and P genotypes have been reported in human infections whereas G9P[8], G1P[8], G3P[8], G2P[4], G8P[8], are the widely prevalent RVAs (Lestari et al., 2020). There is a great genetic and strain diversity of RVs, contributed by point mutations, gene rearrangement, and genetic assortment between co-circulating strains. Furthermore, interspecies transmission between human and animal RVs has been reported in different genotypes (Mukherjee et al., 2011). Though two licensed RV vaccines are effective and widely used in many countries all over the world (Anh et al., 2011; Wang et al., 2013), how effective the vaccines will be as the genetic alteration of the prevalent RVs remains unknown.
VP4 can be cleaved into two subunits, VP5* and VP8* (Larralde et al., 1991). VP8*, located at the distal terminal of the spike, is responsible for the virus-ligand interaction while VP5* facilitates the host cell penetration through the conformation rearrangement and membrane fusion (Figure 1) (Settembre et al., 2011). VP8* has been identified to interact with specific glycans in a P genotype dependent manner (Huang et al., 2012). Previously, 35 P genotypes were classified into five genogroups based on the VP8* sequences (Figure 2A) (Liu et al., 2012). The widely prevalent RVA P genotypes in humans such as P[8], P[4], P[6], and a rare P[19] genotype were classified in P[II] genogroup. P[9], P[14], and P[25] were grouped in P[III]. P[11] identified mainly in infants made P[IV] genogroup. The rare genotypes, P[17], P[30], P[31], and P[35] constituted P[V]. The remaining 23 genotypes, including P[3], P[7], all belonged to P[I] genogroup. We also constructed the phylogenetic tree of the 50 P genotypes using the VP8* sequences (Figure 2B). It is noticed that there are more lineages besides the former five branches. It was proposed that six new genogroups should be included, such as P[46]/P[29], P[49], P[5], P[28]/P[50], P[45]/P[37], P[20]/P[16] (Figure 2B). Here, we delineated recent advances in the structural basis for glycan recognition of RV VP8*s.
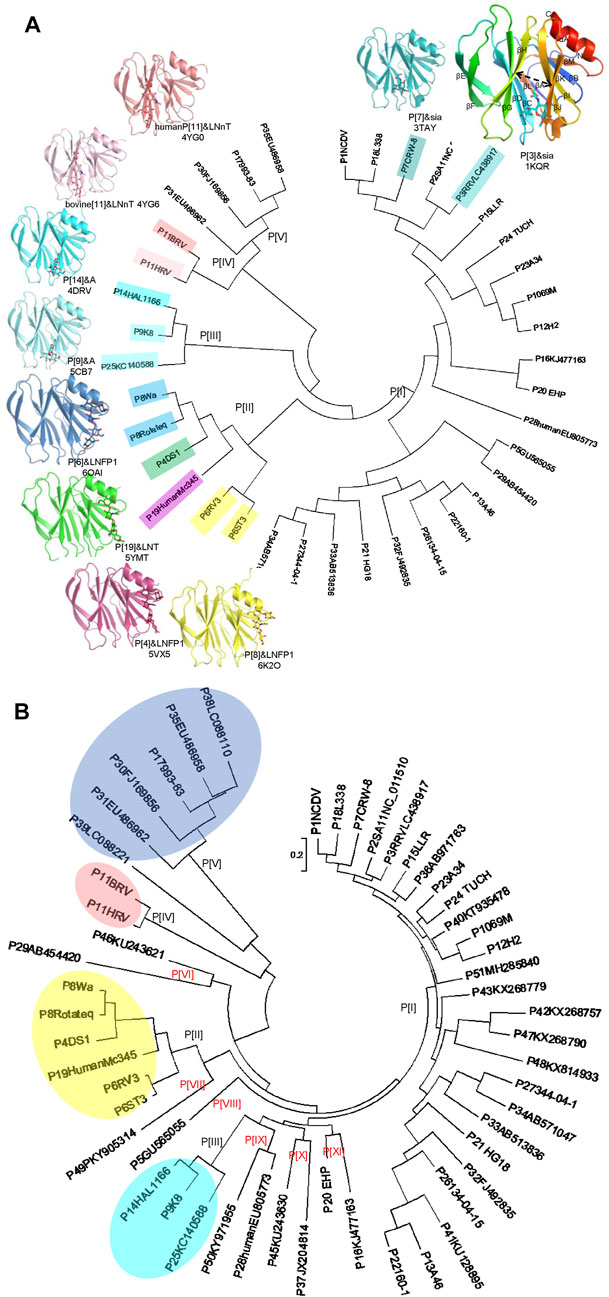
FIGURE 2. (A) Crystal structures of rotavirus VP8*s. Phylogenetic analysis of VP8*s of 35 RV P genotypes (circular dendrogram) was conducted by MEGA6 using the neighbor-joining method. The crystal structures of VP8*s in complex with glycans are presented. P[3], Rhesus rotavirus RRV in complex with Sia, rainbow, PDB ID: 1KQR. P[7], CRW-7 in complex with sia, teal, PDB ID: 3TAY. P[14], human rotavirus HAL1166 in complex with A-type HBGA, cyan, PDB ID: 4DRV. P[9], human rotavirus K8 in complex with A-type HBGA, Aquamarine, PDB ID: 5CB7. P[11], human neonatal rotavirus N155 in complex with type II tetrasaccharide LNnT, salmon, PDB ID: 4YG0. P[11], bovine rotavirus B223 in complex with type II tetrasaccharide LNnT, lightpink, PDB ID: 4YG6. P[6], human rotavirus RV3 in complex with LNFP1, skyblue, PDB ID: 6OAI. P[19], human rotavirus MC345 in complex with LNT, green, PDB ID: 5YMT. P[4], human rotavirus DS-1 in complex with LNFP1, warmpink, PDB ID: 5VX5. P[8], human rotavirus Rotateq in complex with LNFP1, yellow, PDB ID: 6K2O. (B) Phylogenetic analysis of VP8*s of 50 RV P genotypes (circular dendrogram) was conducted by MEGA6 using the neighbor-joining method. The GeneBank number: P[1]NCDV, AB119636; P[2]SA11, NC_011510; P[3]RRV, LC438917; P[4]DS1, CAD62680; P[5], GU565055; P[6]ST3, L33895; P[6]RV3, ADD31861; P[7]CRW-8, UniProtKB:P0C6Y8; P[8]Rotateq, GU565044; P[8]Wa, L34161; P[9]K8, D90260; P[10]69M, M60600; P[11]HRV, UniProtKB: B6RGK2; P[11]BRV, M92986; P[12]H2, D13397; P[13]A46, AY050274; P[14]HAL1166, L20875; P[15]LLR, JQ013506; P[16], KJ477163; P[17]993-83, D16352; P[18]L338, D13399; P[19]HumanMc345, D38054; P[20]EHP, U08424; P[21]HG18, AF237665; P[22]160-1, AF526374; P[23]A34, AY174094; P[24] TUCH, AY596189; P[25], KC140588; P[26], DQ061053; P[27], DQ242615; P28human, EU805773; P[29], AB454420; P[30], FJ169856; P[31], EU486962; P[32], FJ492835; P[33], AB513836; P[34], AB571047; P[35], EU486958; P[36], AB971763; P[37], JX204814; P[38], LC088110; P[39], LC088221; P[40], KT935478; P[41], KU128895; P[42], KX268757; P[43], KX268779; P[45], KU243630; P[46], KU243621; P[47], KX268790; P[48], KX814933; P[49], PKY905314; P[50], KY971955; P[51], MH285840.
Crystal Structures of RV VP8*s
The structures of VP8*s of ten genotypes spanning P[I] to P[IV] genogroups have been determined, including VP8*s of human and animal RVs. NMR and X-ray crystallography studies revealed that VP8* possessed a typical galectin-like fold with a two twisted β-sheets, βH and βK, separated by a shallow cleft (Dormitzer et al., 2002b) (Figure 2A). VP8* structures of animal RV strains, a porcine P[6] RV z84, a porcine P[7] RV CRW-8, a rhesus P[3] RV RRV, canine P[3] rotavirus strain K9, and bovine P[11] RV B223 have been determined (Dormitzer et al., 2002b; Blanchard et al., 2007; Mishra et al., 2018; Sun et al., 2018a) (Hu et al., 2015). The VP8* structures of human P[4] DS-1, P[8] Wa/Rotateq, P[6] RV-3, P[19] MC345, P[14] HAL1166, P[11] N155, P[9] K8, P[25] CAU12-2 are clear (Figure 2A) (Monnier et al., 2006; Blanchard et al., 2007; Hu et al., 2012; Hu et al., 2015; Yu et al., 2015; Sun et al., 2016; Liu et al., 2017; Sun et al., 2018a; Hu et al., 2018; Li et al., 2021).
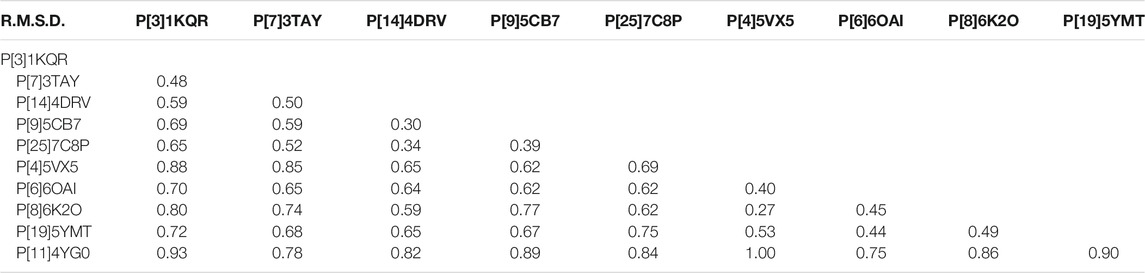
P[8] and P[4] RVs are the most prevalent P genotypes in human infection (Lestari et al., 2020). P[6] RVs mainly circulate in humans and pigs (Nyaga et al., 2018). P[19], P[14], P[9], P[25] RVs are less common in humans and show evidence of cross-species transmission (Liu et al., 2012). P[11] RVs mainly infect neonates (Liu et al., 2013). The structural comparison showed that the galectin-like fold is conserved among these VP8* structures (Figure 2A). However, the widths of the cleft between the two β-sheets are different. Human P[8], P[4], P[6], P[19], P[11] VP8*s possessed a relatively wider cleft with the width of 9.3 Å (Ångstrom), 9.0 Å, 9.2 Å, 9.2 Å, 8.6 Å, respectively (Figure 3). Meanwhile, animal P[3] and P[7] VP8* possessed a narrower cleft with the width of 7.6 Å, 6.9 Å, respectively (Figure 3). Human P[14], P[9], P[25] RV VP8*s all possessed a relatively narrow cleft of 7.1 Å (Figure 3). As previously reported (Venkataram Prasad et al., 2014; Ramani et al., 2016), it was proposed that the deletion of residue 136 and the amino acid change at position 101 may in a certain part influence the width of the cleft based on the sequence alignment and structural analysis (Figure 3). P[3]/P[7]/P[14]/P[9]/P[25] VP8*s with narrow cleft had the R101 and 136T, while P[4]/P[6]/P[8]/P[19] with the F/V/I101 and 136 deletion possessed a wider cleft (Figure 3). Consistent with the phylogenic analysis of VP8* sequences, P[3] VP8* are structurally close to P[7] VP8* with the root mean square deviation (RMSD) value of 0.48 Å (Table 1), belonging to P[I] genogroup; P[8], P[4], P[6], and P[19] VP8*s classified in P[II] genogroup exhibited similar structural characteristics with the RMSD values ranging from 0.27 Å to 0.53 Å (Table 1); P[14], P[9], and P[25] VP8*s of P[III] genogroup presented alike conformation with RMSD value of 0.30 Å, 0.34 Å, and 0.39 Å (Table 1); P[11] VP8* grouped in P[IV] genogroup are distinct to all the other VP8* structures with the RMSD value above 0.75 Å (0.75–1.00 Å) (Table 1). These indicated that the VP8* structures possessed structural conformation in a genogroup dependent manner.
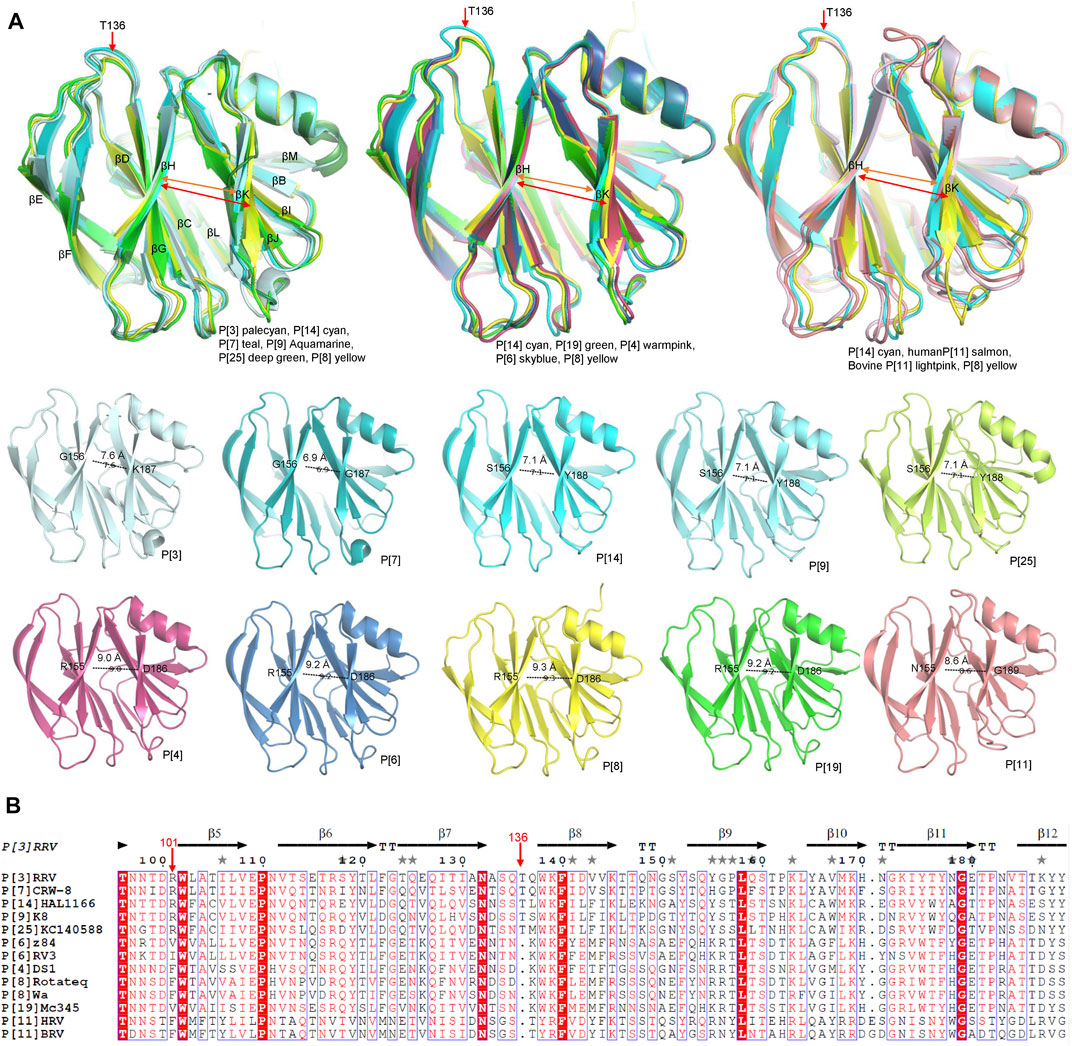
FIGURE 3. (A) Structural analysis of the VP8* structures. Superimposition of the VP8* structures to compare the width of the cleft between two β-sheets (βH and βK). The width of the cleft in VP8*s of different P genotype are measured. P[3], P[7], P[9], P[14], P[11], P[19], P[4], P[6], P[8] VP8*s are colored as described in Figure.2A and P[25] is colored deep green (PDB ID: 7C8P). (B) The structural based sequence alignment was constructed using Clustal Omega and displayed with the ESPript 3.0 (https://espript.ibcp.fr/ESPript/cgi-bin/ESPript.cgi). Residue 101 and 136 were pointed with red arrows.
Glycans Recognized by Rotaviruses
Some animal RVs were reported to recognize terminal sialic acids (SAs) (Fukudome et al., 1989; Isa et al., 1997). N-acetylneuraminic acid (Neu5Ac) and N-glycolylneuraminic acid (Neu5Gc) are the most common neuraminic acids in nature and widely expressed on the surface of most mammalian cells. Several RV strains of the P[I] genogroup, including Nebraska calf diarrhea virus (NCDV, cattle, P[1]), rhesus rotavirus (RRV, monkey, P[3]), SA11 (monkey, P[2]), porcine rotavirus strain (OSU, pig, P[7]) were sialidase sensitive and showed distinct preference for Neu5Ac or Neu5Gc (Rolsma et al., 1998; Dormitzer et al., 2002a; Isa et al., 2006). The infection of porcine P[7] RV CRW-8 could be efficiently inhibited by the ganglioside GM3Gc glycan (Blanchard et al., 2007). Canine K9 P[3] preferentially bound to Neu5Gc (Mishra et al., 2018). In addition, nuclear magnetic resonance (NMR) and cell infection studies showed that the ganglioside GM1, which lacks the terminal sialic acid but with branched sialic acid, could be a possible ligand for some human RVs, including human P[8] and P[6] (Haselhorst et al., 2009). VP8*s of human P[8] Wa and P[6] RV-3 were identified to bind to GM1 by Saturation transfer difference NMR (STD-NMR) (Fleming et al., 2014).
Later studies revealed that most animal RVs and human RVs are SA independent (Ciarlet and Estes, 1999). Recently, some human RV genotypes are found to recognize histo-blood group antigens (HBGAs) (Huang et al., 2012), indicating that HBGAs are important cell attachment factors for RVs. HBGAs are a group of carbohydrates (Yamamoto, 1994), distributing abundantly on mucosal epithelia. HBGAs also existed as free oligosaccharides in body fluids, such as saliva, milk, blood, and intestinal content. HBGAs are synthesized by sequential addition of monosaccharides to precursor disaccharides by different glycosyltransferase. The glycosyltransferases are encoded by three major gene families, secretor, Lewis, and ABO families encoding FUT2, FUT3, and A/B enzymes, respectively.
RV VP8*s recognized HBGAs in a genotype dependent manner. The P[8] and P[4] RVs that are widely prevalent in humans interacted with mucin cores, lewis b, and type I HBGA, including H type-1 antigen (H1), H1 precursor, lacto-N-tetraose (LNT), Lacto-N-fucopentaose I (LNFP1) (Huang et al., 2012; Rey et al., 2019; Sun et al., 2020). Human P[6] and P[19] bound to H1, whereas porcine P[6] and P[19] did not (Sun et al., 2018a; Li et al., 2018). P[14] was less common in human. Human P[14] VP8* was found to specifically recognize A type HBGA (Hu et al., 2012). Human P[9] and P[25], clustered in P[III] genogroup together with P[14] also bound to A type HBGA (Liu et al., 2012). Human P[11] RVs that mainly infected neonates interacted with type I and type II precursors, while bovine P[11] RVs only bound to type II precursor (Liu et al., 2013; Ramani et al., 2013; Hu et al., 2015). VP8* of human P[28] in P[I] genogroup was found to bind H1 HBGAs (Zhao et al., 2020). Bovine P[5] WC3 and its mono-reassortant G4P[5] recognized both sialic acid and the α-Gal HBGA (Alfajaro et al., 2019). Interestingly, the α-Gal epitope of the HBGA family was reported to be a ligand for bovine norovirus Newbury2 (Cho et al., 2018), indicating a common feature of the infection of certain bovine RVs and norovirus.
Mucins are large glycoproteins containing a protein core and a high number of O-linked oligosaccharides (Jensen et al., 2010). Mucin cores were reported to be recognized by several RV P genotypes. Human P[8] and P[4] RVs recognized the disaccharide core structure (GlcNAcβ1-6GalNAc) of mucin cores 2, 4, and 6 (Liu et al., 2016). Porcine P[6] (z84) VP8* interacted with mucin core 2, while human P[6] (5311142) VP8* did not bind to mucin core 2 (Sun et al., 2018a). Human and porcine P[19] bound to mucin core 2 (Sun et al., 2018a). A rare genotype P[10] VP8* also interacted with mucin core 2 (Pang et al., 2018). These results indicated that mucin cores especially mucin core 2 may play an important role in the RV infection and interspecies transmission.
The glycan binding specificity influenced the host tropism and prevalence of RVs. Animal RVs could recognize sialic acid and mucin core 2, whereas human RVs bound to HBGAs and mucin cores. The RVs that can infect both human and porcine such as P[6] and P[19] showed distinct glycan binding preference. Porcine P[6] and P[19] VP8*s recognized mucin core 2, while human P[6] and P[19] VP8*s interacted with the H1 HBGA, indicating an evolutionary path from animal to human. P[11] RV VP8* recognized type I and type II precursors that are developmentally regulated in neonates, consistent with the fact that P[11] RVs are mainly identified in neonates. P[8] and P[4] infections were mainly identified in secretors and lewis positive children (Nordgren et al., 2014), consistent with that P[8]/P[4] RVs could interact with H1 HBGA and lewis antigen.
Structural Basis for the Interactions of VP8* and Glycans
The crystal structures of VP8*and VP8*/glycans are listed in table 2. Rhesus rotavirus (RRV) VP8* with sialic acid was first determined revealing a glycan binding site consisting of R101, Y155, 187-190 KYYS (Dormitzer et al., 2002b) (Figure 4A). The glycan binding site located at one corner of the cleft between two β-sheets (βH/βK) and appeared to be an open-shallow groove. Y188 and S190 form one rim of the groove; Y155 constitutes the other rim; R101, V144, K187 and Y189 side chains make the base part. R101 was proved to be vital for the sialic acid binding (Kraschnefski et al., 2009). CRW-8 VP8* interacted with Neu5Acα2Me using the same residues except H155 and G187 (Blanchard et al., 2007) and bound to ganglioside GM3 glycans by the same pattern (Yu et al., 2011). CRW-8 and RRV VP8* binding to the Neu5Gcα2Me were determined (Yu et al., 2012) (Figure 4B), illustrating that residue 157 of VP8* influenced the glycan preference. CRW-8 VP8* with S157 showed reduced binding affinity for Neu5Gc compared to that with P157.
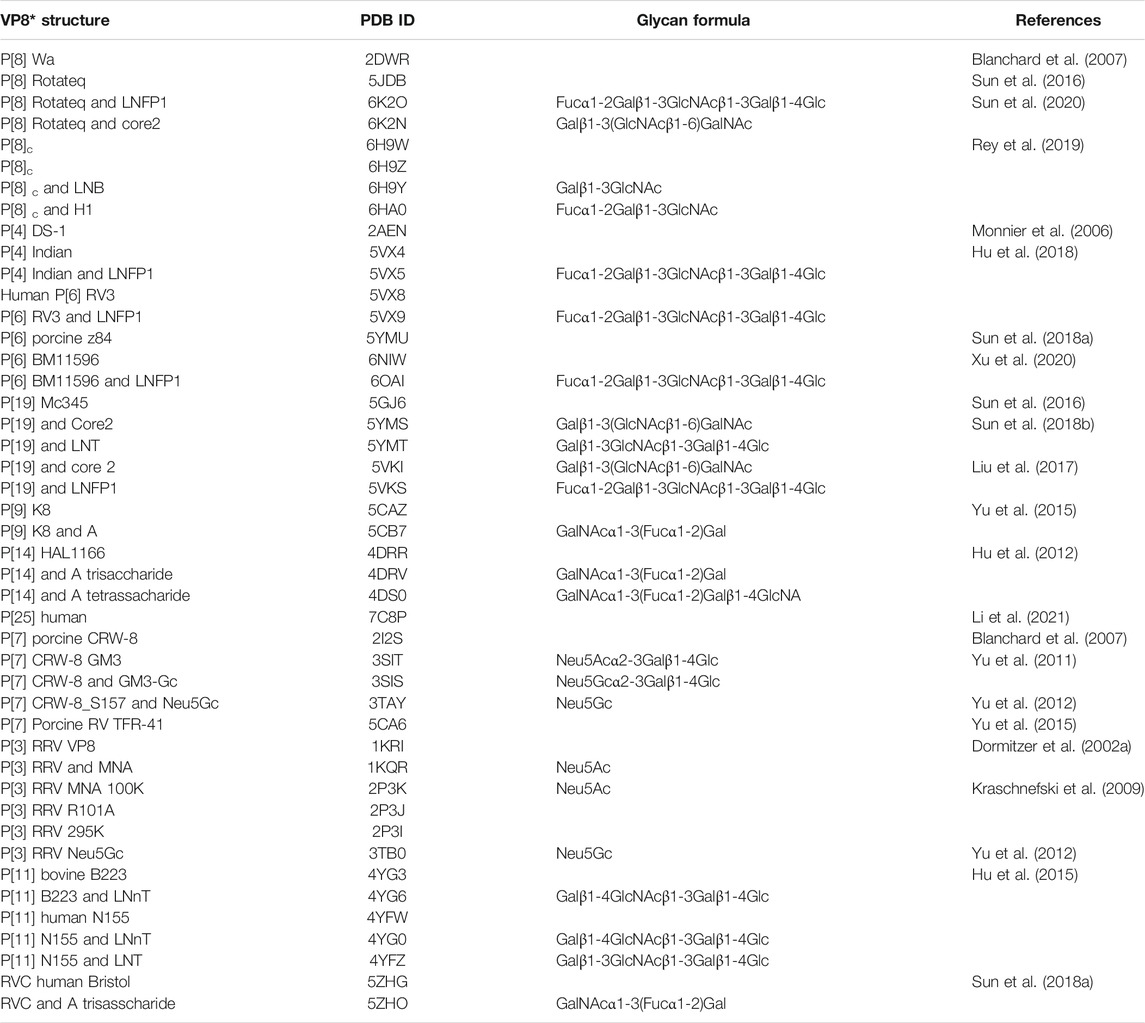
TABLE 2. Summary of VP8* and VP8*-glycan structures. The PDB ID, glycan formula, and references are included. Gal, yellow; GlcNAc, blue; GalNAc, green; Glc, magenta; Fuc, cyan.
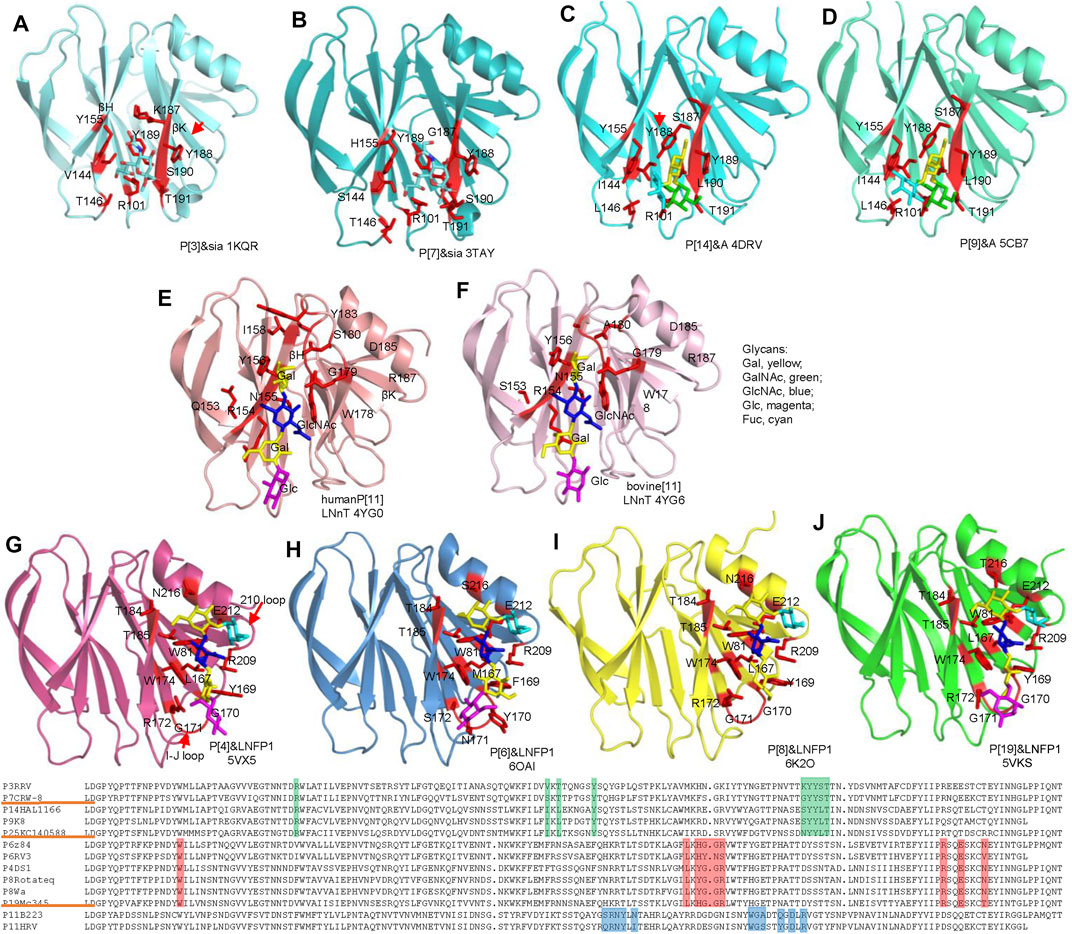
FIGURE 4. Structural analysis of different RV VP8*s in complex with glycans. VP8*s are shown as cartoon presentation. Glycans and amino acids participating the interactions are exhibited as sticks. RRV P[3] (A), CRW-8 P[7] (B), HAL1166 P[14] (C), K8 P[9] (D), human P[11] (E), bovine P[11] (F), P[4] Indian (G), RV-3 P[6] (H), Rotateq P[8] (I), human P[19] (J), VP8*s are colored as that in Figure.2A. The moieties of Gal, GalNAc, GlcNAc, Glc, Fuc are colored yellow, green, blue, magenta, cyan, respectively. Sequence alignment was done by DNAMAN. The residues involved in the glycan binding in P[3]/P[7]/P[14]/P[9], P[4]/P[6]/P[8]/P[19], P[11] VP8*s are colored green, red, blue, respectively.
The structural basis of P[14] interacting with A type HBGA has been illustrated (Hu et al., 2012) (Figure 4C). The width of the cleft between the two β-sheets is narrower than the cleft in the human VP8*, similar to that in the VP8* of the animal strains (Figure 3). P[14] VP8* bound to the A type HBGA using the same glycan binding site as P[3] VP8*. However, the structural features of the glycan binding site of P[14] VP8* is that of P[3] VP8*. The amino acid residues involved the A type HBGA binding were R101, I144, L146, Y155, S187, Y188, Y189, and L190 (Figure 4C). The orientation of Y188 was different to Y188 in P[3] VP8* and would cause steric hindrance to the binding of sialic acid, indicating that the subtle changes of the VP8* could accommodate distinct glycans. The terminal GalNAc (green) and Gal (yellow) of the HBGA contributed to all the interactions, whereas the proximal moiety Fuc (cyan) project out from the surface and did not make any direct contacts (Hu et al., 2012). Human P[9] was identified to recognize A type HBGA using the same glycan binding site as that of P[14], consisting of R101, I144, L146, Y155, S187, Y188, Y189, L190, and T191 (Figure 4D). The amino acids possessed identical conformation as those in P[14] VP8*, providing further evidence that they belong to the same genogroup.
P[11] VP8*, clustered in P[IV] genogroup, possessed a quite different conformation comparing to other VP8*. Human P[11] VP8* interacted with type I and type II precursors using a distinct glycan binding site consisting of N153, R154, N155, Y156, I158, W178, G179, S180, Y183, D185, and R187 (Hu et al., 2015) (Figure 4E). The type I tetrasaccharide lacto-N-tetraose (LNT) and type II tetrasaccharide lacto-N-neotetraose (LNnT) binding site was expansive and spanned almost the entire length of the cleft between βH and βK (Figure 4E). Bovine P[11] only interacted with type II precursor and recognized LNnT with residues of S153, R154, N155, Y156, W178, G179, A180, D185, and R187 (Figure 4F).
Human P[4]/P[6]/P[8]/P[19] VP8*s belonging to P[II] genogroup all interacted with type I HBGA (Figures 4G–J). Human P[4] and P[6] VP8*s with LNFP1 have been determined separately (Hu et al., 2018; Xu et al., 2020) (Figures 4G,H). The type I HBGAs located at a novel site consisting of the β-strand K, I-J loop, and 210 loop. Residues W81, L167, YGGR 169-172, W174, T184, T185, R209, E212, and N216 are involved in the glycan binding of human P[4] VP8* (Figure 4G). The LNFP1 binding site in human P[6] VP8* was at the same location but formed by different amino acids of W81, M167, FYNS 169-172, W174, T184, T185, R209, E212, and S216. The LNFP1 binding site in P[8] was identical to that in P[4] VP8* (Figures 4I,G), consistent with that they are close in the phylogenetic analysis. GlcNAc of LNFP1 contributed to the main interactions. Gal also participated in the interactions, whereas FucI was not involved in direct interactions (Figure 4I). The glycan binding site of P[19] VP8* is composed of W81, L167, YGGR 169-172, W174, T184, T185, R209, E212, and T216 (Figure 4J). Complex structures of P[19] VP8* with LNT and LNFP1 have been determined separately. The moieties of LNT, Gal1 (yellow), GlcNAc2 (blue), Gal3 (yellow), Glc4 (magenta) all participated in the interactions. LNFP1 located at the same place and almost overlap with the LNT. Fucose (cyan) of LNFP1 did not contribute to the binding. Superimposition of the complex structures of P[4]/P[6]/P[8]/P[19] VP8*s with LNFP1 revealed that LNFP1 overlapped at the same place in a similar conformation (Figure 5A). Only the orientation of the moiety of Glc at the reducing end displayed some variation (Figure 5A), revealing the exquisite of the interactions of different VP8*s and glycans.
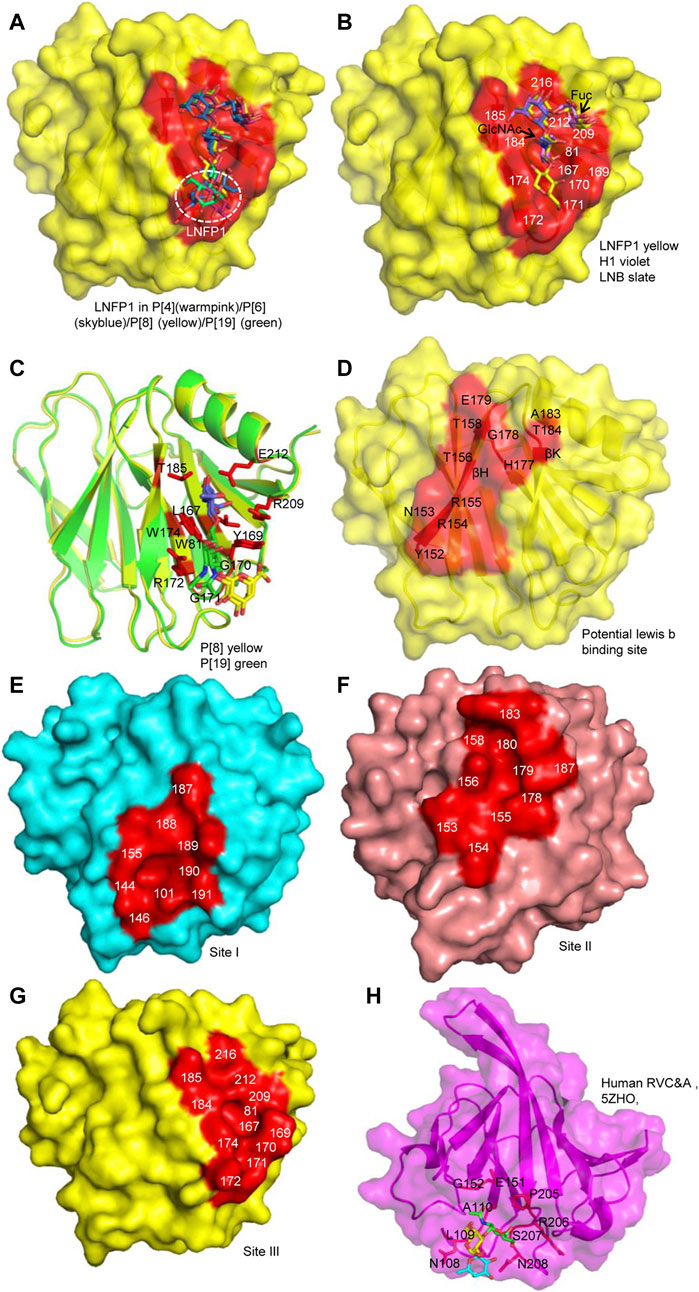
FIGURE 5. Comparison of the glycan binding sites. (A) The superimposition of LNFP1 that bound to P[4]/P[6]/P[8]/P[19] VP8*s. (B) Superimposition of type 1 HBGAs (H1, LNB, LNFP1) that located at the same glycan binding site of P[8] VP8*. (C) The interactions between P[8]/P[19] VP8* and mucin core 2. (D) The potential lewis b binding site in P[8] VP8* based on NMR. (E–G) Surface presentation of the three distinct glycan binding sites in RVA VP8*s. The residues constitute the glycan binding site were labeled. P[14] (cyan, 4DRV), human P[11] (salmon, 4YG0), P[8] (yellow, 6K2O) VP8*s are presented as models. (H) The presentation of the glycan binding site of human group C RV (RVC) VP8* (magenta, PDB ID: 5ZHO). Type A trisaccharide was shown as stick. The residues involved in the glycan binding site are labeled.
The complex structures of P[8] VP8* with different type I HBGAs (H type-1 antigen, H1; H1 precursor lacto-N-biose, LNB; Lacto-N-fucopentaose I, LNFP1) were determined (Rey et al., 2019; Sun et al., 2020). LNB and H1 located at the same site of LNFP1 (Figure 5B). LNB and H1 interacted with VP8* using similar mechanism. L-fucose of H1 was projected out and did not make direct interactions with VP8*. However, the surface plasmon resonance (SPR) assay showed that P[8]c VP8* bound more intensively to H1 (affinity constants KD = 27.9 ± 0.7 μM) compared to LNB (KD = 52.1 ± 4.3 μM) (Rey et al., 2019), implying that H1 L-fucose contributes to the glycan binding. LNFP1 overlapped exquisitely with the H1 and LNB moieties. P[8] VP8* interacted with different H1 glycans in a same site but with different binding affinity, indicating that the glycan forms may influence the RV attachment.
The interactions between P[8]/P[19] VP8* and mucin core 2 have been illustrated (Liu et al., 2017; Sun et al., 2018a; Sun et al., 2020). Mucin core 2 interacted with VP8*s at the same site as the type I HBGAs with slightly difference. GlcNAc (blue), GalNAc (green), and Gal (yellow) all participated in the interactions (Figure 5C) (Sun et al., 2020). P[8]/P[19] VP8* bound to mucin core 2 using the same pattern with residues of W81, L167, Y/HGGR 169-172, W174, T185, R209, and E212 (Figure 5C), revealing that RV VP8* can accommodate different glycans using the same residues.
P[8] and P[4] VP8*s were also reported to interact with lewis b (leb) (Huang et al., 2012; Ma et al., 2015). A recent paper elucidated the molecular mechanism for the recognition of P[8] VP8* to leb based on nuclear magnetic resonance (NMR) spectroscopy-based titration experiments and NMR-derived high ambiguity driven docking (HADDOCK) method (Xu et al., 2020). Unlike the H1 binding site composed of an α-helix and a β-sheet (referred as βα binding site), P[8] and P[4] VP8*s were identified to bind leb HBGA in another pocket consisting of the edge of two β-sheets (named ββ binding site) (Figure 5D). The potential lewis b binding site is proposed to be formed by residues of Y152, N153, R154, R155, T156, T158, H177, G178, E179, A183, and T184 (Figure 5D). Further investigations such as X-ray crystallization are needed to verify the glycan binding.
According to the crystal structures of VP8* and glycans, three glycan binding sites are identified in different RV VP8*s so far (Figures 5E–G). The first one is comprised of residues 101, 144, 146, 155, 187-191 locating in P[3]/P[7]/P[14]/P[9] (Figure 5E). The second one is that of P[11] RV VP8* (Figure 5F). The third one includes residues of 81, 167, 169-172, 174, 184, 185, 209, 212, 216 and is relatively conserved in P[II] genogroup RVs (Figure 5G). Based on the understanding of the importance of VP8* in RV-host interactions, the VP8*-based subunit vaccine has been explored. One VP8* vaccine has shown a good ability to induce neutralizing antibodies in immunized mice (Xue et al., 2015). VP8* was also presented on norovirus P particle to construct a P24-VP8* nanoparticle (Ramesh et al., 2019). A chimeric VP8* with T-cell epitope P2 exhibited better effect in inducing antibodies and protection than vaccines without P2 epitope. Polyvalent P2-VP8* vaccine candidates containing VP8*s of P[4], P[6] and P[8] are under trial (Groome et al., 2020). The VP8*-based vaccines probably be promising alternatives for future vaccines.
Other Group Rotaviruses
Group/species C rotaviruses (RVCs) have been identified as important pathogens of acute gastroenteritis in children, family-based outbreaks, as well as animal infections (Joshi et al., 2017; Vlasova et al., 2017). Human RVC VP8* was found to recognize A type HBGAs (Sun et al., 2018b). The complex structure of human RVC VP8* and type A trisaccharide exhibited that human RVC VP8* possessed a completely different glycan binding site compared to RVA VP8*s (Figure 5H). Human RVC bound to type A trisaccharide (GalNAcα1-3(Fucα1-2)Gal) using a pocket consisting of N108, L209, A110, E151, G152, P205, R206, S207, and N208 (Sun et al., 2018b). Both GalNAc and Fuc of the type A HBGA participated in the interactions, while Gal had no direct contact with the RVC VP8*.
Human group B (RVB) and group H rotavirus (RVH) caused outbreaks in China in the 1980s and mainly infected adults. Infections of human RVB and RVH have constantly reported in some areas such as Southeast Asia (Chen et al., 1985; Yang et al., 2004; Jiang et al., 2008; Joshi et al., 2019). The receptor binding specificity of human RVB and RVH is unclear. Whether they recognize sialic acid as some animal RVAs, HBGAs as human RVAs or other glycans still need further investigation.
Conclusion Remarks
Some animal RVs recognized sialic acid, such as P[3], P[7]. Some animal RVs were reported to bind sialic acid and αGal, such as bovine P[5] RVs (Alfajaro et al., 2019). The identification that some human RV VP8*s recognized HBGAs has provided new insights into the infection and transmission of RVs. So far, the interactions between VP8*s of human P[4], P[6], P[8], P[19], P[11], P[14], P[9], P[25] and HBGAs have been illustrated. In total, three distinct glycan binding sites were identified in different RVs based on crystallography. VP8* of the widely prevalent RV genotypes P[8], P[4], P[6], and a rare genotype P[19] VP8* possessed a conserved glycan binding pocket. Structural analysis revealed that the same glycan binding pocket could interact with different glycans exquisitely, such as that P[8] VP8* could accommodate H1, LNB, LNFP1, and mucin core 2. VP8*s of human RVA P[14], P[9], P[25] and human RVC all interacted with A-type HBGA, which may in a part restricted the prevalence of these RVs. The functions of these glycans, such as sialic acid, HBGAs, mucin cores in the RV infection or cross-species transmission still need more studies to clarify. Structural biology has significantly contributed to our understanding of the interaction between RV and glycans. However, the complexity and variety of glycan recognition of RV VP8*s indicated host-pathogen co-evolution with the structural and functional adaptation of RV to host glycan polymorphisms. More efforts exploring the structural basis for the VP8*-glycan interactions are necessary to fully understand the role of glycans in RV infection and transmission, which will facilitate the development of novel RV vaccines and anti-viral agents.
Author Contributions
ZD designed and directed the project. XS performed the analysis, constructed the figures and wrote the manuscript. DL designed and modified the figures. ZD and XS revised the manuscript.
Funding
This research was supported by grants from the National Science and Technology Major Project (ZDZX-2018ZX10201002-009-003), the National Natural Science Foundation of China (NSFC) (No. 21934005), and National Science and Technology Major Project (2018ZX10711-001).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Alfajaro, M. M., Kim, J.-Y., Barbé, L., Cho, E.-H., Park, J.-G., Soliman, M., et al. (2019). Dual Recognition of Sialic Acid and αGal Epitopes by the VP8* Domains of the Bovine Rotavirus G6P[5] WC3 and of its Mono-Reassortant G4P[5] RotaTeq Vaccine Strains. J. Virol. 93. doi:10.1128/jvi.00941-19
Anh, D. D., Carlos, C. C., Thiem, D. V., Hutagalung, Y., Gatchalian, S., Bock, H. L., et al. (2011). Immunogenicity, Reactogenicity and Safety of the Human Rotavirus Vaccine RIX4414 (Rotarix) Oral Suspension (Liquid Formulation) when Co-administered with Expanded Program on Immunization (EPI) Vaccines in Vietnam and the Philippines in 2006-2007. Vaccine 29 (11), 2029–2036. doi:10.1016/j.vaccine.2011.01.018
Bányai, K., Estes, M. K., Martella, V., and Parashar, U. D. (2018). Viral Gastroenteritis. The Lancet 392 (10142), 175–186. doi:10.1016/S0140-6736(18)31128-0
Bányai, K., Kemenesi, G., Budinski, I., Földes, F., Zana, B., Marton, S., et al. (2017). Candidate New Rotavirus Species in Schreiber's Bats, Serbia. Infect. Genet. Evol. 48, 19–26. doi:10.1016/j.meegid.2016.12.002
Blanchard, H., Yu, X., Coulson, B. S., and von Itzstein, M. (2007). Insight into Host Cell Carbohydrate-Recognition by Human and Porcine Rotavirus from crystal Structures of the Virion Spike Associated Carbohydrate-Binding Domain (VP8*). J. Mol. Biol. 367 (4), 1215–1226. doi:10.1016/j.jmb.2007.01.028
Chen, G.-M., Hung, T., Bridger, J. C., and McCrae, M. A. (1985). Chinese Adult Rotavirus is a Group B Rotavirus. The Lancet. 326 (8464), 1123–1124. doi:10.1016/s0140-6736(85)90710-x
Cho, E.-H., Soliman, M., Alfajaro, M. M., Kim, J.-Y., Seo, J.-Y., Park, J.-G., et al. (2018). Bovine Nebovirus Interacts with a Wide Spectrum of Histo-Blood Group Antigens. J. Virol. 92 (9), e02160. doi:10.1128/JVI.02160-17
Ciarlet, M., and Estes, M. K. (1999). Human and Most Animal Rotavirus Strains Do Not Require the Presence of Sialic Acid on the Cell Surface for Efficient Infectivity. J. Gen. Virol. 80 (Pt 4), 943–948. doi:10.1099/0022-1317-80-4-943
Dormitzer, P. R., Sun, Z.-Y. J., Blixt, O., Paulson, J. C., Wagner, G., and Harrison, S. C. (2002a). Specificity and Affinity of Sialic Acid Binding by the Rhesus Rotavirus VP8* Core. J. Virol. 76 (20), 10512–10517. doi:10.1128/jvi.76.20.10512-10517.2002
Dormitzer, P. R., Sun, Z. Y., Wagner, G., and Harrison, S. C. (2002b). The Rhesus Rotavirus VP4 Sialic Acid Binding Domain Has a Galectin Fold with a Novel Carbohydrate Binding Site. EMBO J. 21 (5), 885–897. doi:10.1093/emboj/21.5.885
Estes, M. K., and Greenberg, H. B. (2013). “Rotaviruses.” in: Fields Virology, 6th ed., Editors D. M Knipe, and P. M Howley (Philadelphia, PA, Wolters Kluwer Health/Lippincott Williams & Wilkins), 1347–1401.
Fleming, F. E., Bohm, R., Dang, V. T., Holloway, G., Haselhorst, T., Madge, P. D., et al. (2014). Relative Roles of GM1 Ganglioside, N-Acylneuraminic Acids, and 2 1 Integrin in Mediating Rotavirus Infection. J. Virol. 88 (8), 4558–4571. doi:10.1128/JVI.03431-13
Fukudome, K., Yoshie, O., and Konno, T. (1989). Comparison of Human, Simian, and Bovine Rotaviruses for Requirement of Sialic Acid in Hemagglutination and Cell Adsorption. Virology 172 (1), 196–205. doi:10.1016/0042-6822(89)90121-9
Groome, M. J., Fairlie, L., Morrison, J., Fix, A., Koen, A., Masenya, M., et al. (2020). Safety and Immunogenicity of a Parenteral Trivalent P2-VP8 Subunit Rotavirus Vaccine: a Multisite, Randomised, Double-Blind, Placebo-Controlled Trial. Lancet Infect. Dis. 20 (7), 851–863. doi:10.1016/s1473-3099(20)30001-3
Haselhorst, T., Fleming, F. E., Dyason, J. C., Hartnell, R. D., Yu, X., Holloway, G., et al. (2009). Sialic Acid Dependence in Rotavirus Host Cell Invasion. Nat. Chem. Biol. 5 (2), 91–93. doi:10.1038/nchembio.134
Hu, L., Crawford, S. E., Czako, R., Cortes-Penfield, N. W., Smith, D. F., Le Pendu, J., et al. (2012). Cell Attachment Protein VP8* of a Human Rotavirus Specifically Interacts with A-type Histo-Blood Group Antigen. Nature 485 (7397), 256–259. doi:10.1038/nature10996
Hu, L., Ramani, S., Czako, R., Sankaran, B., Yu, Y., Smith, D. F., et al. (2015). Structural Basis of Glycan Specificity in Neonate-specific Bovine-Human Reassortant Rotavirus. Nat. Commun. 6, 8346. doi:10.1038/ncomms9346
Hu, L., Sankaran, B., Laucirica, D. R., Patil, K., Salmen, W., Ferreon, A. C. M., et al. (2018). Glycan Recognition in Globally Dominant Human Rotaviruses. Nat. Commun. 9 (1), 2631. doi:10.1038/s41467-018-05098-4
Huang, P., Xia, M., Tan, M., Zhong, W., Wei, C., Wang, L., et al. (2012). Spike Protein VP8* of Human Rotavirus Recognizes Histo-Blood Group Antigens in a Type-specific Manner. J. Virol. 86 (9), 4833–4843. doi:10.1128/jvi.05507-11
Isa, P., Arias, C. F., and López, S. (2006). Role of Sialic Acids in Rotavirus Infection. Glycoconj J. 23 (1-2), 27–37. doi:10.1007/s10719-006-5435-y
Isa, P., López, S., Segovia, L., and Arias, C. F. (1997). Functional and Structural Analysis of the Sialic Acid-Binding Domain of Rotaviruses. J. Virol. 71 (9), 6749–6756. doi:10.1128/JVI.71.9.6749-6756.1997
Jensen, P. H., Kolarich, D., and Packer, N. H. (2010). Mucin-type O-Glycosylation - Putting the Pieces Together. FEBS J. 277 (1), 81–94. doi:10.1111/j.1742-4658.2009.07429.x
Jiang, S., Ji, S., Tang, Q., Cui, X., Yang, H., Kan, B., et al. (2008). Molecular Characterization of a Novel Adult Diarrhoea Rotavirus Strain J19 Isolated in China and its Significance for the Evolution and Origin of Group B Rotaviruses. J. Gen. Virol. 89 (Pt 10), 2622–2629. doi:10.1099/vir.0.2008/001933-0
Joshi, M. S., Jare, V. M., and Gopalkrishna, V. (2017). Group C Rotavirus Infection in Patients with Acute Gastroenteritis in Outbreaks in Western India between 2006 and 2014. Epidemiol. Infect. 145 (2), 310–315. doi:10.1017/S0950268816002363
Joshi, M. S., Lole, K. S., Barve, U. S., Salve, D. S., Ganorkar, N. N., Chavan, N. A., et al. (2019). Investigation of a Large Waterborne Acute Gastroenteritis Outbreak Caused by Group B Rotavirus in Maharashtra State, India. J. Med. Virol. 91 (10), 1877–1881. doi:10.1002/jmv.25523
Kraschnefski, M. J., Bugarcic, A., Fleming, F. E., Yu, X., von Itzstein, M., Coulson, B. S., et al. (2009). Effects on Sialic Acid Recognition of Amino Acid Mutations in the Carbohydrate-Binding Cleft of the Rotavirus Spike Protein. Glycobiology 19 (3), 194–200. doi:10.1093/glycob/cwn119
Larralde, G., Li, B. G., Kapikian, A. Z., and Gorziglia, M. (1991). Serotype-specific Epitope(s) Present on the VP8 Subunit of Rotavirus VP4 Protein. J. Virol. 65 (6), 3213–3218. doi:10.1128/jvi.65.6.3213-3218.1991
Lestari, F. B., Vongpunsawad, S., Wanlapakorn, N., and Poovorawan, Y. (2020). Rotavirus Infection in Children in Southeast Asia 2008-2018: Disease burden, Genotype Distribution, Seasonality, and Vaccination. J. Biomed. Sci. 27 (1), 66. doi:10.1186/s12929-020-00649-8
Li, D., Wang, M., Qi, J., Zhang, Q., Wang, H., Pang, L., et al. (2021). Human Group A Rotavirus P[25] VP8* Specifically Binds to A-type Histo-Blood Group Antigen. Virology 555, 56–63. doi:10.1016/j.virol.2020.12.016
Li, Z., Gao, C., Zhang, Y., Palma, A. S., Childs, R. A., Silva, L. M., et al. (2018). O-glycome Beam Search Arrays for Carbohydrate Ligand Discovery. Mol. Cell Proteomics 17 (1), 121–133. doi:10.1074/mcp.RA117.000285
Liu, Y., Huang, P., Jiang, B., Tan, M., Morrow, A. L., and Jiang, X. (2013). Poly-LacNAc as an Age-specific Ligand for Rotavirus P[11] in Neonates and Infants. PLoS One 8 (11), e78113. doi:10.1371/journal.pone.0078113
Liu, Y., Huang, P., Tan, M., Liu, Y., Biesiada, J., Meller, J., et al. (2012). Rotavirus VP8*: Phylogeny, Host Range, and Interaction with Histo-Blood Group Antigens. J. Virol. 86 (18), 9899–9910. doi:10.1128/jvi.00979-12
Liu, Y., Ramelot, T. A., Huang, P., Liu, Y., Li, Z., Feizi, T., et al. (2016). Glycan Specificity of P[19] Rotavirus and Comparison with Those of Related P Genotypes. J. Virol. 90 (21), 9983–9996. doi:10.1128/JVI.01494-16
Liu, Y., Xu, S., Woodruff, A. L., Xia, M., Tan, M., Kennedy, M. A., et al. (2017). Structural Basis of Glycan Specificity of P[19] VP8*: Implications for Rotavirus Zoonosis and Evolution. Plos Pathog. 13 (11), e1006707. doi:10.1371/journal.ppat.1006707
Ma, X., Li, D.-d., Sun, X.-m., Guo, Y.-q., Xiang, J.-y., Wang, W.-h., et al. (2015). Binding Patterns of Rotavirus Genotypes P[4], P[6], and P[8] in China with Histo-Blood Group Antigens. PLoS One 10 (8), e0134584. doi:10.1371/journal.pone.0134584
Matthijnssens, J., Ciarlet, M., McDonald, S. M., Attoui, H., Bányai, K., Brister, J. R., et al. (2011). Uniformity of Rotavirus Strain Nomenclature Proposed by the Rotavirus Classification Working Group (RCWG). Arch. Virol. 156 (8), 1397–1413. doi:10.1007/s00705-011-1006-z
Matthijnssens, J., Otto, P. H., Ciarlet, M., Desselberger, U., Van Ranst, M., and Johne, R. (2012). VP6-sequence-based Cutoff Values as a Criterion for Rotavirus Species Demarcation. Arch. Virol. 157 (6), 1177–1182. doi:10.1007/s00705-012-1273-3
Mishra, R., Yu, X., Kishor, C., Holloway, G., Lau, K., von Itzstein, M., et al. (2018). Specificity and Affinity of Neuraminic Acid Exhibited by Canine Rotavirus Strain K9 Carbohydrate-Binding Domain (VP8*). J. Mol. Recognit 31, e2718. doi:10.1002/jmr.2718
Monnier, N., Higo-Moriguchi, K., Sun, Z.-Y. J., Prasad, B. V. V., Taniguchi, K., and Dormitzer, P. R. (2006). High-resolution Molecular and Antigen Structure of the VP8* Core of a Sialic Acid-independent Human Rotavirus Strain. J. Virol. 80 (3), 1513–1523. doi:10.1128/JVI.80.3.1513-1523.2006
Mukherjee, A., Ghosh, S., Bagchi, P., Dutta, D., Chattopadhyay, S., Kobayashi, N., et al. (2011). Full Genomic Analyses of Human Rotavirus G4P[4], G4P[6], G9P[19] and G10P[6] Strains from North-eastern India: Evidence for Interspecies Transmission and Complex Reassortment Events. Clin. Microbiol. Infect. 17 (9), 1343–1346. doi:10.1111/j.1469-0691.2010.03383.x
Nordgren, J., Sharma, S., Bucardo, F., Nasir, W., Günaydın, G., Ouermi, D., et al. (2014). Both Lewis and Secretor Status Mediate Susceptibility to Rotavirus Infections in a Rotavirus Genotype-dependent Manner. Clin. Infect. Dis. 59, 1567–1573. doi:10.1093/cid/ciu633
Nyaga, M. M., Tan, Y., Seheri, M. L., Halpin, R. A., Akopov, A., Stucker, K. M., et al. (2018). Whole-genome Sequencing and Analyses Identify High Genetic Heterogeneity, Diversity and Endemicity of Rotavirus Genotype P[6] Strains Circulating in Africa. Infect. Genet. Evol. 63, 79–88. doi:10.1016/j.meegid.2018.05.013
Pang, L.-l., Wang, M.-x., Sun, X.-m., Yuan, Y., Qing, Y., Xin, Y., et al. (2018). Glycan Binding Patterns of Human Rotavirus P[10] VP8* Protein. Virol. J. 15 (1), 161. doi:10.1186/s12985-018-1065-9
Ramani, S., Cortes-Penfield, N. W., Hu, L., Crawford, S. E., Czako, R., Smith, D. F., et al. (2013). The VP8* Domain of Neonatal Rotavirus Strain G10P[11] Binds to Type II Precursor Glycans. J. Virol. 87 (13), 7255–7264. doi:10.1128/jvi.03518-12
Ramani, S., Hu, L., Venkataram Prasad, B. V., and Estes, M. K. (2016). Diversity in Rotavirus-Host Glycan Interactions: A "Sweet" Spectrum. Cell Mol. Gastroenterol. Hepatol. 2 (3), 263–273. doi:10.1016/j.jcmgh.2016.03.002
Ramesh, A., Mao, J., Lei, S., Twitchell, E., Shiraz, A., Jiang, X., et al. (2019). Parenterally Administered P24-VP8* Nanoparticle Vaccine Conferred Strong Protection against Rotavirus Diarrhea and Virus Shedding in Gnotobiotic Pigs. Vaccines 7 (4), 177. doi:10.3390/vaccines7040177
Rey, F. A., Gozalbo-Rovira, R., Ciges-Tomas, J. R., Vila-Vicent, S., Buesa, J., Santiso-Bellón, C., et al. (2019). Unraveling the Role of the Secretor Antigen in Human Rotavirus Attachment to Histo-Blood Group Antigens. PLOS Pathog. 15 (6), e1007865. doi:10.1371/journal.ppat.1007865
Rolsma, M. D., Kuhlenschmidt, T. B., Gelberg, H. B., and Kuhlenschmidt, M. S. (1998). Structure and Function of a Ganglioside Receptor for Porcine Rotavirus. J. Virol. 72 (11), 9079–9091. doi:10.1128/jvi.72.11.9079-9091.1998
Settembre, E. C., Chen, J. Z., Dormitzer, P. R., Grigorieff, N., and Harrison, S. C. (2011). Atomic Model of an Infectious Rotavirus Particle. EMBO J. 30 (2), 408–416. doi:10.1038/emboj.2010.322
Sun, X., Dang, L., Li, D., Qi, J., Wang, M., Chai, W., et al. (2020). Structural Basis of Glycan Recognition in Globally Predominant Human P[8] Rotavirus. Virol. Sin. 35 (2), 156–170. doi:10.1007/s12250-019-00164-7
Sun, X., Li, D., Peng, R., Guo, N., Jin, M., Zhou, Y., et al. (2016). Functional and Structural Characterization of P[19] Rotavirus VP8* Interaction with Histo-blood Group Antigens. J. Virol. 90 (21), 9758–9765. doi:10.1128/JVI.01566-16
Sun, X., Li, D., Qi, J., Chai, W., Wang, L., Wang, L., et al. (2018a). Glycan Binding Specificity and Mechanism of Human and Porcine P[6]/P[19] Rotavirus VP8*s. J. Virol. 92 (14), e00538–00518. doi:10.1128/JVI.00538-18
Sun, X., Wang, L., Qi, J., Li, D., Wang, M., Cong, X., et al. (2018b). Human Group C Rotavirus VP8*s Recognize Type A Histo-Blood Group Antigens as Ligands. J. Virol. 92 (11). doi:10.1128/JVI.00442-18
Tate, J. E., Burton, A. H., Boschi-Pinto, C., and Parashar, U. D.World Health Organization–Coordinated Global Rotavirus Surveillance Network (2016). Global, Regional, and National Estimates of Rotavirus Mortality in Children <5 Years of Age, 2000-2013. Clin. Infect. Dis. 62 (Suppl. 2), S96–S105. doi:10.1093/cid/civ1013
Venkataram Prasad, B., Shanker, S., Hu, L., Choi, J.-M., Crawford, S. E., Ramani, S., et al. (2014). Structural Basis of Glycan Interaction in Gastroenteric Viral Pathogens. Curr. Opin. Virol. 7, 119–127. doi:10.1016/j.coviro.2014.05.008
Vlasova, A., Amimo, J., and Saif, L. (2017). Porcine Rotaviruses: Epidemiology, Immune Responses and Control Strategies. Viruses 9 (3), 48. doi:10.3390/v9030048
Wang, F. T., Mast, T. C., Glass, R. J., Loughlin, J., and Seeger, J. D. (2013). Effectiveness of an Incomplete RotaTeq (RV5) Vaccination Regimen in Preventing Rotavirus Gastroenteritis in the United States. Pediatr. Infect. Dis. J. 32 (3), 278–283. doi:10.1097/INF.0b013e318275328f
Xu, S., Ahmed, L. U., Stuckert, M. R., McGinnis, K. R., Liu, Y., Tan, M., et al. (2020). Molecular Basis of P[II] Major Human Rotavirus VP8* Domain Recognition of Histo-Blood Group Antigens. Plos Pathog. 16 (3), e1008386. doi:10.1371/journal.ppat.1008386
Xue, M., Yu, L., Che, Y., Lin, H., Zeng, Y., Fang, M., et al. (2015). Characterization and Protective Efficacy in an Animal Model of a Novel Truncated Rotavirus VP8 Subunit Parenteral Vaccine Candidate. Vaccine 33 (22), 2606–2613. doi:10.1016/j.vaccine.2015.03.068
Yamamoto, F. I. (1994). Review: Recent Progress in the Molecular Genetic Study of the Histo-Blood Group ABO System. Immunohematology 10 (1), 1–7.
Yang, J.-H., Kobayashi, N., Wang, Y.-H., Zhou, X., Li, Y., Zhou, D.-J., et al. (2004). Phylogenetic Analysis of a Human Group B Rotavirus WH-1 Detected in China in 2002. J. Med. Virol. 74 (4), 662–667. doi:10.1002/jmv.20222
Yu, X., Coulson, B. S., Fleming, F. E., Dyason, J. C., von Itzstein, M., and Blanchard, H. (2011). Novel structural insights into rotavirus recognition of ganglioside glycan receptors. J. Mol. Biol. 413 (5), 929–939. doi:10.1016/j.jmb.2011.09.005
Yu, X., Dang, V. T., Fleming, F. E., von Itzstein, M., Coulson, B. S., and Blanchard, H. (2012). Structural Basis of Rotavirus Strain Preference toward N-Acetyl- or N-Glycolylneuraminic Acid-Containing Receptors. J. Virol. 86 (24), 13456–13466. doi:10.1128/JVI.06975-11
Yu, X., Mishra, R., Holloway, G., von Itzstein, M., Coulson, B. S., and Blanchard, H. (2015). Substantial Receptor-Induced Structural Rearrangement of Rotavirus VP8*: Potential Implications for Cross-Species Infection. Chembiochem 16 (15), 2176–2181. doi:10.1002/cbic.201500360
Keywords: rotavirus, VP8* structure, glycan binding specificity, sialic acid, histo-blood group antigens, mucin cores
Citation: Sun X, Li D and Duan Z (2021) Structural Basis of Glycan Recognition of Rotavirus. Front. Mol. Biosci. 8:658029. doi: 10.3389/fmolb.2021.658029
Received: 24 January 2021; Accepted: 21 June 2021;
Published: 08 July 2021.
Edited by:
Yan Liu, Imperial College London, United KingdomReviewed by:
Jacques Le Pendu, Institut National de la Santé et de la Recherche Médicale (INSERM), FranceMario Milani, Biophysics Institute, National Research Council (CNR), Italy
Copyright © 2021 Sun, Li and Duan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhaojun Duan, emhhb2p1bmRAMTI2LmNvbQ==
 Xiaoman Sun1,2
Xiaoman Sun1,2 Zhaojun Duan
Zhaojun Duan