- 1Department of Urology, Peking University First Hospital and Institute of Urology, Peking University, Beijing, China
- 2National Urological Cancer Center, Beijing, China
- 3Beijing Key Laboratory of Urogenital Diseases (male) Molecular Diagnosis and Treatment Center, Beijing, China
- 4The Guangdong and Shenzhen Key Laboratory of Male Reproductive Medicine and Genetics, Peking University Shenzhen Hospital, Institute of Urology of Shenzhen PKU‐HKUST Medical Center, Shenzhen, China
To investigate the underlying molecular mechanism of tripartite motif-containing 58 (TRIM58) in the development of clear cell renal cell carcinoma (ccRCC), we explored TRIM58 expression and methylation in tumor tissues and the association with clinicopathological features and prognosis of tissue samples; Moreover, we examined the direct gene transcription of TRIM58-specific DNA demethyltransferase (TRIM58-TET1) by the CRISPR-dCas9 fused with the catalytic domain of TET1 and the biological functions in RCC cells. In this study, we demonstrate that TRIM58 is frequently downregulated by promoter methylation in ccRCC tissues, associated significantly with tumor nuclear grade and poor patient survival. TRIM58-TET1 directly induces demethylation of TRIM58 CpG islands, and activates TRIM58 transcription in RCC cell lines. Besides, DNA demethylation of TRIM58 by TRIM58-TET1 significantly inhibits cell proliferation and migration Overall, our results demonstrate that TRIM58 is inactivated by promoter methylation, associates with tumor nuclear grade and poor survival, and TRIM58 DNA demethylation could directly activate TRIM58 transcription and inhibit cell proliferation and migration in RCC cell lines.
Introduction
Renal cell carcinoma (RCC) is one of the most common malignancies of the urinary system and accounts for approximately 2–3% of adult malignant solid tumors. The incidence of this disease has been steadily increasing in recent decades, partially because of increase in life expectancy and advance in medical imaging techniques (Ferlay et al., 2015; Bray et al., 2018). RCC lacks specific clinical manifestations, and approximately 20–30% of patients were reported to be initially diagnosed with distant metastasis (Alt et al., 2011). The response rate to targeted therapy and immunotherapy for metastatic patients is only approximately 30%. With the emergence of drug resistance, the vast majority of patients will eventually die from tumor progression (Linehan and Ricketts, 2014). Clear cell renal cell carcinoma (ccRCC) is the most common pathological subtype of RCC (accounting for 75–80%) (Moch, 2013). Therefore, it is of great significance to further investigate the molecular mechanism underlying ccRCC occurrence and development, which will benefit future research in discovering effective biomarkers and therapeutic targets.
Genome-wide epigenetic alterations are associated with human tumorigenesis. The silence of tumor suppressor genes (TSGs) by aberrant promoter methylation happens frequently and is recognized as a hallmark of the occurrence and development of cancers, including RCC (Sharma et al., 2010; Klutstein et al., 2016). Many critical TSGs have been reported to be inactivated by promoter methylation in RCC, such as APAF-1, RASSF1A, SFRP, CADM2, CDKN2A, DKK1, IRF8, and SOX7 (Morrissey et al., 2001; Christoph et al., 2006; Hirata et al., 2011; He et al., 2013; Ricketts et al., 2014; Zhang et al., 2014; Ricketts et al., 2018; Wang et al., 2019), among others. TRIM58 (tripartite motif-containing 58), a member of the TRIM family, which is located at 1q44, encodes a protein that exhibits E3 ubiquitin ligase activity. It was found to participate in a wide range of physiological processes, such as innate immunity, cell proliferation, and DNA damage repair, as well as many genetic diseases and cancers (Hatakeyama, 2011). Studies have revealed that the TRIM58 gene is hypermethylated and downregulated in a variety of malignancies including lung cancer, liver cancer, and pancreatic ductal adenocarcinoma (Tao et al., 2011; Kajiura et al., 2017; Xu et al., 2019). Moreover, TRIM58 protein can inhibit proliferation and invasion in gastrointestinal cancer cells, indicating the tumor-suppressive role in such cancers (Liu M. et al., 2018; Liu et al., 2020). However, the direct methylation–expression pattern and biological function of TRIM58 in ccRCC remains unclear.
To dissect the functional significance of DNA methylation events in human cancer, CRISPR–dCas9 has been used for targeted epigenome editing by fusion with epigenome modifying enzymes such as Dnmt3a (a DNA methyltransferase enzyme) and TET (catalyze active DNA demethylation) (Sasaki and Matsui, 2008; Morita et al., 2016; Vojta et al., 2016; Xu et al., 2016). A catalytically inactive dead Cas9 (dCas9)-based system fused to the catalytic domain of TET1 (TET1CD) that hydroxylates specific loci. This system has been designed to activate site-specific demethylation in the genome DNA (Yin and Xu, 2016; Gallego-Bartolome et al., 2018; Liu X. S. et al., 2018). Here, a TRIM58-specific DNA demethyltransferase (TRIM58-TET1) was used to directly activate TRIM58 transcription and the subsequent functional effects.
In this study, we investigated whether TRIM58 was a newly identified tumor suppressor gene with aberrant promoter methylation for ccRCC. To get this conclusion, we explored TRIM58 expression and methylation status in primary tumors, as well as its relationship with clinicopathological characteristics and survival. We also examined its tumor-suppressive functions by TRIM58-TET1 in RCC cells.
Results
TRIM58 was Frequently Silenced and Promoter Methylated in ccRCC Samples
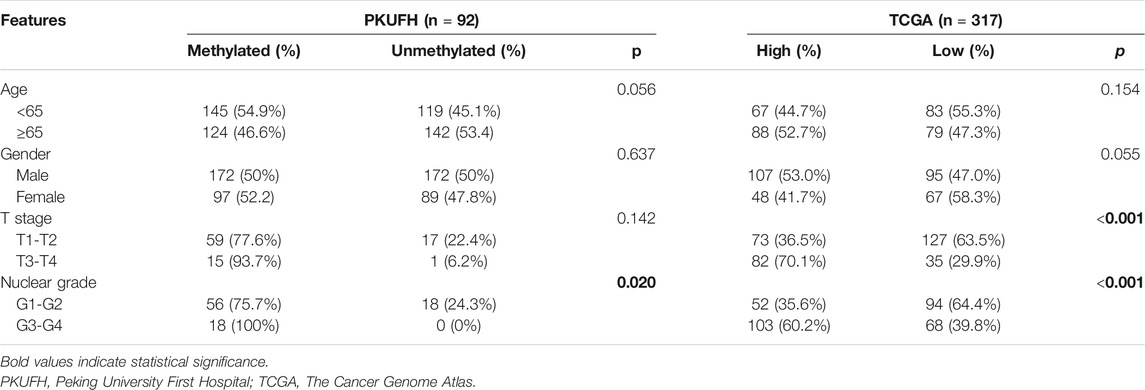
To evaluate TRIM58 expression in ccRCC tissues, the mRNA transcription level of TRIM58 was examined by qRT-PCR in ccRCC samples compared to that in adjacent non-malignant renal tissues from our center. Compared with the adjacent non-malignant renal tissues, TRIM58 was significantly decreased in 15 paired renal tumor tissues (p < 0.001, Figure 1A). TRIM58 protein expression levels were also decreased in ccRCC samples according to IHC, as compared to those in adjacent non-malignant renal tissue (Figure 1B). We next validated the expression pattern of TRIM58 in KIRC using the TCGA database. According to RNA-sequence data from 530 KIRC patients, KIRC exhibited significantly decreased expression of TRIM58 mRNA compared to that in normal renal tissues (p < 0.001, Figure 1C).
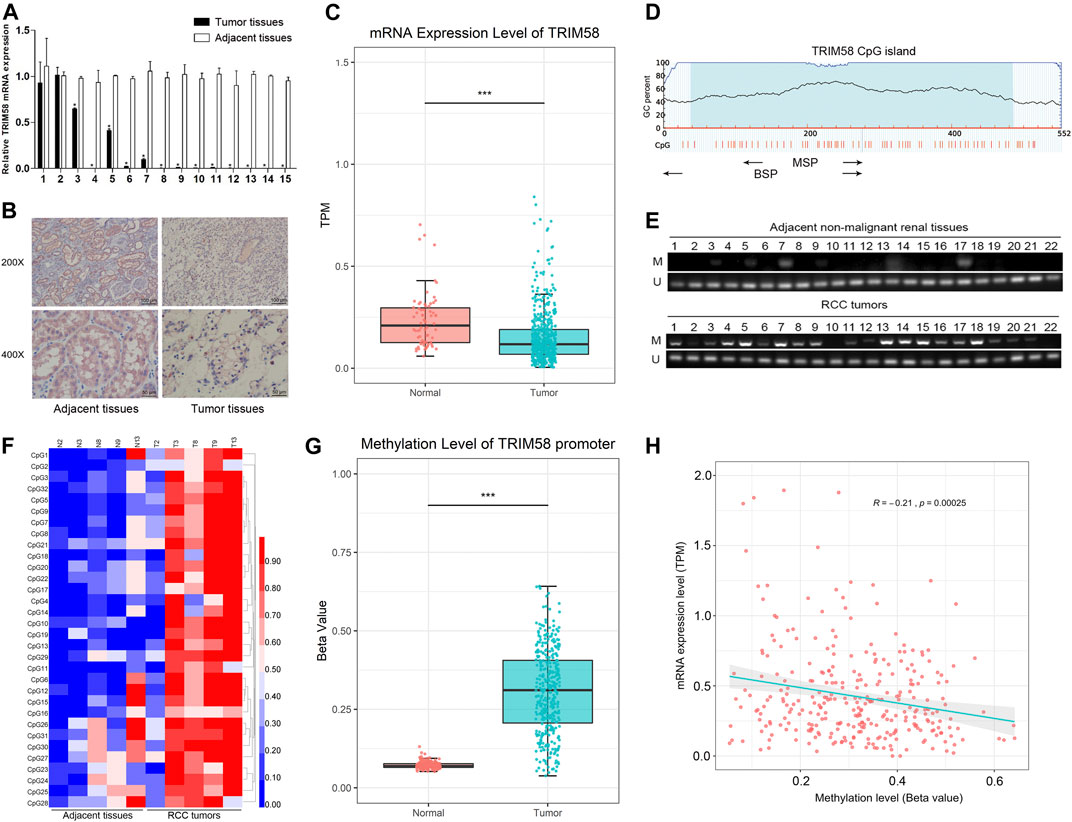
FIGURE 1. TRIM58 is frequently silenced and hypermethylated in ccRCC samples. (A) The relative mRNA transcription level of TRIM58 in 15 paired renal tumor tissues and the adjacent non-malignant renal tissues; (B) Representative IHC staining for TRIM58 in human ccRCC cancer (right panels) and adjacent normal renal tissues (left panels); (C) mRNA transcription level of TRIM58 in KIRC from TCGA database; (D) Primers targeting TRIM58 promoter region designed for MSP and BGS; (E) MSP analysis in 92 ccRCC samples and 22 adjacent non-malignant renal tissues from our center; (F) BGS analysis on TRIM58 promoter 32 CpG sites of five ccRCC samples with paired adjacent tissues; (G) Methylation level of 317 KIRC patients from UALCAN; (H) Correlation analysis between the expression and methylation of TRIM58.
It is generally known that DNA methylation in the promoter region may directly in-activate gene transcription. By exploring the UCSC Genome Browser, we found there is a CpG island near the TRIM58 promoter region. It is attractive that if there are any change of DNA methylation status of TRIM58 promoter region in ccRCC. TRIM58 methylation status was analyzed in 92 ccRCC samples and 22 adjacent non-malignant renal tissues from our center. Primers targeting this region were designed for MSP and BGS (Figure 1D). MSP analysis showed that methylation of the TRIM58 promoter could be detected in 80.4% (74/92) of ccRCC samples but in only 18.2% (4/22) of adjacent non-malignant renal tissues (p < 0.001, Figure 1E). As expected, BGS analysis of five ccRCC samples with paired adjacent non-malignant tissues demonstrated that TRIM58 methylation levels were greater in ccRCC tissues based on 32 CpG sites (Figure 1F). Meanwhile, we analyzed the methylation data of 317 KIRC patients from UALCAN and found TRIM58 promoter methylation level in KIRC was significantly higher than that in normal renal tissue (p < 0.001, Figure 1G). To investigate the relationship between the expression and promoter methylation of TRIM58, we performed correlation analysis. The results showed a significant negative correlation between mRNA expression and promoter methylation levels for this gene (R = −0.21, p < 0.001, Figure 1H).
TRIM58 Hypermethylation and Low Expression was Associated With Clinicopathological Features and Poor Prognosis in ccRCC Samples
We evaluated the relationship between TRIM58 methylation and clinicopathological features of RCC patients from our center. According to their TRIM58 methylation status of tumor tissue samples, 92 KIRC patients were allocated into a methylated group and a non-methylated group, with their corresponding clinicopathological characteristics analyzed. Among the 92 patients, 63 were males and 29 were females, with an average age of 57.28 ± 12.42 years. We found that the TRIM58 promoter methylation was significantly associated with nuclear grade. Nuclear grade was higher in the methylated group compared to the unmethylated group (p = 0.02). Moreover, methylation of the TRIM58 promoter was not significantly associated with patient age, sex, and T stage (Table 1). UALCAN patients were then divided into high and low methylation groups according to the median TRIM58 methylation level. Chi-square test showed that the methylation of TRIM58 was significantly associated with T stage and nuclear grade in ccRCC; patients with a high methylation level had higher T stages (p < 0.001) and nuclear grades (p < 0.001, Table 1).
To further explore the correlation between TRIM58 DNA methylation inactivation and the prognosis of KIRC patients, the median mRNA expression level of TRIM58 was separately set as the cut-off value using the TCGA database. Survival analysis indicated significant differences between high and low expression groups in terms of both overall survival (OS) and disease-free survival (DFS); patients with high TRIM58 expression had a longer OS (p = 0.025, Supplementary Figure S1A) and DFS (p = 0.02, Supplementary Figure S1B). In addition, patients from UALCAN with a high TRIM58 methylation level had significant worse OS (p = 0.014, Supplementary Figure S1C) and DFS (p < 0.001, Supplementary Figure S1D) than patients with low methylation. These results indicate that TRIM58 methylation inactivation is correlated with the poor prognosis of KIRC.
Methylation of TRIM58 Promoter Correlated With Its Downregulation in RCC Cell Lines
To evaluate the expression level of TRIM58 in RCC cell lines, we examined mRNA and protein expression in RCC cell lines (786-O, 769-P, ACHN and OS-RC-2). The TRIM58 expression was significantly reduced and even silenced, when compared to that in HK-2 normal human kidney proximal tubular epithelial cells using qRT-PCR (Figure 2A) and Western Blot (Figure 2B). We then examined the methylation status of the TRIM58 promoter in those cells. MSP results showed that the RCC cell lines, especially 786-O and OS-RC-2, exhibited DNA hypermethylation in the promoter region of TRIM58 compared to HK-2 cells (Figure 2C).
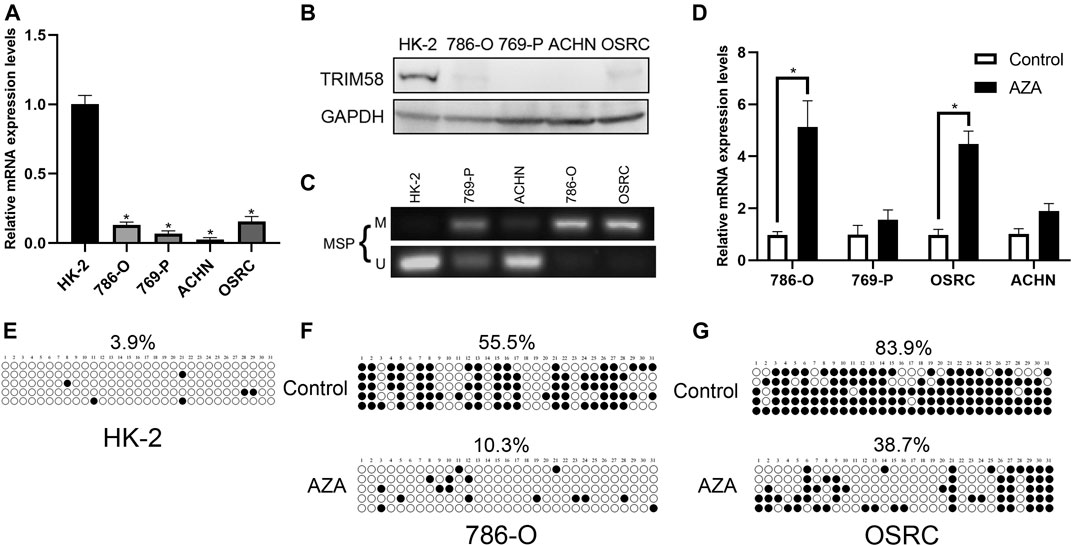
FIGURE 2. TRIM58 methylation correlates with its downregulation in RCC cell lines. (A, B) TRIM58 expression in RCC cell lines (786-O, 769-P, ACHN and OSRC) and HK-2 normal cells using qRT-PCR (A) and Western Blot (B); (C) MSP for methylation status of the TRIM58 promoter; (D) TRIM58 mRNA expression level after treating with the demethylation drug 5-AZA; (E) Methylation pattern of TRIM58 in HK-2 cells; (F, G) TRIM58 promoter CpG site methylation after treating with 5-AZA in 786-O and OS-RC-2 cells.
To determine whether TRIM58 expression was directly repressed by DNA methylation, we examined the TRIM58 mRNA expression level and DNA methylation status in RCC cell lines after treating them with the demethylation drug 5-AZA. The TRIM58 mRNA levels were restored after the treatments (Figure 2D), accompanied with a decrease of TRIM58 promoter CpG site methylation from ∼55.5% to ∼10.3% in 786-O, and from ∼83.9% to ∼38.7% in OS-RC-2, respectively (Figures 2E–G). These results suggested that TRIM58 promoter methylation mediates its decreased expression in RCC cells.
TRIM58 DNA Demethylation Directly Activated Gene Transcription
To explore whether DNA demethylation is essential for re-activation of TRIM58, TRIM58 promoter-specific DNA demethyltransferase (TRIM58-TET1) leading by four guide RNAs (sgRNA1-4) using CRISPR-dCas9 systems (Figure 3A) was transfected transiently into 786-O and OS-RC-2 RCC cell lines. Then, methylation of CpG islands within the TRIM58 promoter region was detected using bisulfite-sequencing. As expected, TRIM58-TET1 induced TRIM58 DNA demethylation in both 786-O and OS-RC-2 cells (Figures 3B,C) and significantly activated TRIM58 gene transcription and protein expression (Figures 3D,E). Taken together, these results indicated that TRIM58 promoter region demethylation could directly activate gene transcription and expression.
FIGURE 3. TRIM58 targeted demethylation directly activates gene transcription. (A) Contruction of TRIM58-TET1 using CRISPR-dCas9 systems with 4 gRNAs targeting TRIM58 promoter region and the catalytic domain of DNA hydroxymethylase; (B, C) Bisulfite clone-sequencing results from 786-O and OS-RC-2 cells transiently transfected with TRIM58-TET1; (D, E) TRIM58 gene transcription (D) and protein expression (E) following TRIM58-TET1 transfection.
TRIM58 DNA Demethylation Suppressed Cell Proliferation and Migration in RCC Cells
To characterize the biological behaviors of cancer cells following TRIM58-specific activation by DNA demethylation, we determined whether TRIM58 demethylation inhibited RCC cell proliferation. As shown in Figures 4A,B, we found that TRIM58 demethylation in 786-O and OS-RC-2 cells significantly inhibited cell proliferation by CCK-8 assay. Next, we evaluated the migration ability of TRIM58 demethylation in RCC cell lines by transwell migration assays and wound healing. The migration ability of 786-O and OS-RC-2 cells was significantly decreased following TRIM58 demethylation (Figures 4C,D). The wound-healing assays showed that TRIM58 demethylation in both RCC cells closed the wound much slower than controls (Figures 4E–H). Taken together, these results implied that TRIM58 DNA demethylation reduced the proliferation and migration phenotypes of RCC cells.
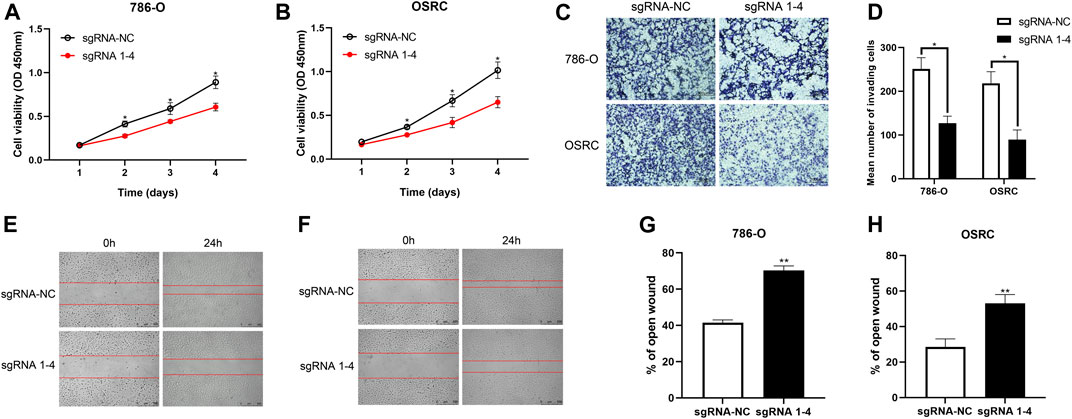
FIGURE 4. Proliferation and migration assays carried out with RCC cells transient transfected with TRIM58-TET1 vector. (A, B) CCK-8 assay for cell proliferation of TRIM58-TET1 transient transfected 786-O and OS-RC-2 cells; (C, D) Transwell assays for the migration ability of 786-O and OS-RC-2 cells; (E–H) The wound-healing assays for TRIM58 demethylation in both RCC cells.
Discussion
Methylation-mediated silencing of tumor-suppressor genes is a critical event in the occurrence and development of various malignancies, including ccRCC(Malouf et al., 2016; Morris and Latif, 2017; Joosten et al., 2018). In the previous series of research, we identified some tumor-specifically methylated genes in RCC, such as DLC1, DLEC1, IRF8, ADAMTS18 (Zhang et al., 2007; Zhang et al., 2010; Zhang et al., 2014). TRIM58, a member of the TRIM family, has been reported to play a tumor suppressive role and to be regulated by DNA methylation in lung cancer, colorectal cancer, and gastric cancer (Kajiura et al., 2017; Liu M. et al., 2018; Liu et al., 2020). However, its DNA methylation status and biological function in ccRCC remained unclear. In the present study, we preliminarily verified that TRIM58 is silenced and hypermethylated in ccRCC based on the TCGA database. The methylation level of TRIM58 was strongly associated with nuclear grade in both our samples and database, as well as with tumor stage in database. Although patients with higher T stages had higher methylation rates of TRIM58 than patients with lower T stages in our center, no statistical difference was found between them. We speculated that this was because of the small sample size of high T stage patients. In addition, ccRCC patients with high TRIM58 expression and hypomethylation levels of TRIM58 have better OS and DFS.
In vitro, we confirmed the methylation of TRIM58 in several RCC cell lines. RCC cells with low TRIM58 expression were found to be hypermethylated and the demethylation drug 5-AZA upregulated the expression of TRIM58, suggesting that the low expression of TRIM58 in ccRCC was probably induced by promoter methylation. We found TRIM58 was frequently inactivated and methylated in RCC cell lines and in primary RCC tissues. Thus, we investigated whether TRIM58 demethylation is capable of activating transcription. To target specific epigenetic alterations in cancer cells, we selected the CRISPR-dCas9 system. CRISPR-dCas9 was derived from the CRISPR-Cas9 system, and has been used in many fields such as gene regulation, epigenetic regulation and high throughput screening. dCas9 lacks nuclease activity but maintains its ability to bind both the sgRNA and targeted DNA. Several groups have developed modified versions of Cas9 for applications that go beyond genome editing (Bikard et al., 2013; Chavez et al., 2015; Kardooni et al., 2018). Here, we used dCas9-TET1 and gRNAs targeting the TRIM58 gene promoter to induce DNA demethylaton and activate transcription.
In lung squamous cell carcinoma, TRIM58 methylation is associated with eight prognostic genes, and may be used as a potential prognostic biomarker (Zhang et al., 2018). In gastric cancer, TRIM58 may function as a tumor suppressor and potentially suppress the tumor growth (Liu et al., 2020). In early-stage lung adenocarcinoma, TRIM58 was robustly silenced by hypermethylation and the restoration of TRIM58 expression in cell lines inhibited cell growth in vitro and in vivo (Kajiura et al., 2017). In the present study, we have provided evidence to demonstrate that TRIM58-TET1 induced demethylation could directly inhibit proliferation and migration of cancer cells in vitro. These facts strongly implicate the inactivation of TRIM58 by DNA methylation as a possible promoter of proliferation and migration in RCC.
Materials and Methods
Patients and Tissue Specimen Collection
92 primary clear cell renal cell carcinoma (ccRCC) samples and 22 adjacent non-malignant renal tissues with patients’ informed consent were obtained from the Urology Department of Peking University First Hospital (PKUFH), Beijing, China. This study followed the Helsinki declaration and was approved by the Institutional Ethical Review Board of PKUFH. Samples were collected immediately in the operating room after surgical removal and were stored in liquid nitrogen after rapid freezing in liquid nitrogen. The pathological diagnosis was made by professional urological pathologists.
Further, mRNA expression and clinical information for 530 kidney renal clear cell carcinoma (KIRC) patients with available RNA-sequence data were downloaded from The Cancer Genome Atlas (TCGA). Methylation data (Illumina Human Methylation 450K BeadChip) of 317 KIRC patients were downloaded from UALCAN (http://ualcan.path.uab.edu/).
Cell Lines Culture and Demethylation Treatment
RCC cell lines (786-O, 769-P, ACHN and OS-RC-2) were used in this study. HK-2 human kidney proximal tubular epithelial cells were used as normal controls. These cell lines were purchased from the American Type Culture Collection (ATCC, Manassas, VA, United States) and National Infrastructure of Cell Line Resource, China. Cell lines were routinely cultured in RPMI 1640 or DMEM, which was supplemented with 10% fetal bovine serum (Invitrogen, Carlsbad, CA, United States) and incubated in a 5% CO2 environment at 37°C. These cells were split to a low density (30% confluence) for 12 h before drug treatment, then incubated with 5-aza-2′-deoxycytidine (5-Aza; Sigma, St. Louis, MO, United States) at a concentration of 10 μM in the optional medium, which was exchanged every 24 h. After 5-Aza treatment for 72 h, the cells were harvested for further analysis.
Construction of Vectors and Transfection
The sequences of the TRIM58 gene promoter region containing the CpG islands were extracted using the UCSC genome browser. The CRISPR dCas9 plasmid dCas9-Tet1CD (#84475) and pgRNA-humanized (#44248) were purchased from Addgene. Four gRNAs that target the TRIM58 promoter were cloned into pgRNA-humanized and listed in Table 2. All designed gRNAs were subjected to MEGABLAST (https://blast.ncbi.nlm.nih.gov/Blast.cgi) to avoid mismatch to unexpected genes in the human genome.
DNA and RNA Extraction
Total RNA and genomic DNA of primary RCC tissues and cell lines were extracted using an RNA-easy Isolation Reagent (Vazyme Biotech, Nanjing, China) and TIANamp Genomic DNA Kits (TIANGEN, Shanghai, China) according to their instructions, respectively, as previously described (Zhang et al., 2014; Wang et al., 2019).
Bisulfite Treatment and Promoter Methylation Analysis
The bisulfite modification of genomic DNA was performed using the EZ DNA Methylation-Gold™ Kit (Zymo Research, Menlo Park, CA, United States). Methylation specific PCR (MSP) and bisulfite genomic sequencing (BGS) were used to analyze methylation status of TRIM58 promoter region. The primers for TRIM58 used for MSP and BGS were listed in Table 2. For BGS, PCR products were subcloned into the fast-T1 clone vector (Vazyme Biotech, Nanjing, China) and 8–10 colonies were randomly selected and sequenced.
Quantitative RT-PCR, Western Blot and Immunohistochemistry Staining of TRIM58 Expression
cDNA was synthesized using HiScript III RT SuperMix for qPCR (Vazyme Biotech, Nanjing, China). qRT-PCR was performed using spectrophotometry (ABI Prism 7500TM instrument, Applied Biosystems) with Universal SYBR Green qPCR Master Mix (Vazyme Biotech, Nanjing, China). Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was used as reference gene. Primers used for TRIM58 and GAPDH qRT-PCR were listed in Table 2.
Total protein was extracted by KeyGEN Bio TECH protein extraction kit (KGP1100) and separated on 10% SDS-PAGE and transferred onto nitrocellulose membrane. After blocking, blots were immunostained with primary antibodies and secondary antibodies respectively as previously described (Wang et al., 2017). The antibodies were as follows: TRIM58 (ab254786, Abcam, 1:500); GAPDH (0494-1-AP, proteinteach, 1:10000).
Immunohistochemistry staining was performed using a primary antibody of TRIM58 (ab254786, Abcam) at a 1:300 dilution following a protocol described previously (Liu et al., 2015). All photographs were taken randomly and measured using Image Pro Plus (Media Cybernetics, Rockville, MD, United States).
Wound-Healing Assay
The cell motility was assessed by scratch wound healing assay. 786-O and OS-RC-2 cells (2–3×106 per well) were plated in a 6-well plate for 1 day and then transfected with vectors for 24 h. The cell layers were washed with PBS after carefully scratching by sterile tips. After incubation for 0 and 24 h, photos were taken. The assays were performed in triplicate.
Transwell Migration Assay
The 786-O and OS-RC-2 cells suspended in 150 uL serum-free medium (2×105 cells/mL) were placed on the upper layer of a cell permeable membrane. Following another 24–48 h incubation, the cells migrated through the membrane were stained with 1% Crystal Violet and counted.
Statistical Analysis
When comparing two groups of measurement data, t test was used for data conforming to normal distribution, whereas a Wilcoxon test was used for data not conforming to normal distribution, and the measurement data were expressed as the mean ± standard deviation (SD). A Chi-square test was used to analyze comparisons between groups for enumeration data. Pearson correlation analysis was used to investigate the relationship between mRNA expression levels and methylation levels of TRIM58. The Kaplan–Meier method was used for survival analysis, and a log-rank test was applied for comparations between groups. R packages used in this study included “GDCRNATools,” “clusterProfiler,” “org.Hs.eg.db,” “tidyr,” “dplyr,” “ggplot2,” “ggsignif,” “survival,” and “survimier.” Annotation gene sets used in GSEA were hallmark gene sets from the Molecular Signatures Database (MSigDB). All statistical analyses were performed and visualized using RStudio (Version1.2.1335, Boston, MA, United States), GSEA (Version4.0, UC San Diego and Broad Institute, United States) 23, Medcalc (Version16.8, Ostend, Belgium), and GraphPad Prism (Version 8.0, GraphPad, Inc., La Jolla, CA, United States). A two-tailed p < 0.05 was considered statistically significant.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
Ethics Statement
The studies involving human participants were reviewed and approved by the Ethics Committee of Peking University First Hospital. The patients/participants provided their written informed consent to participate in this study.
Author Contributions
YG, CC, and AL designed the project and wrote the manuscript. YG, CC, AL, HS, GK, BM and QuZ performed experiments and data analysis. QiZ and YG supervised the project and provided financial support for the project. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by grants from the National Natural Science Foundation of China to Qi.Z. (No. 82072826 and 81872088).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmolb.2021.655126/full#supplementary-material.
References
Alt, A. L., Boorjian, S. A., Lohse, C. M., Costello, B. A., Leibovich, B. C., and Blute, M. L. (2011). Survival after complete surgical resection of multiple metastases from renal cell carcinoma. Cancer 117, 2873–2882. doi:10.1002/cncr.25836
Bikard, D., Jiang, W., Samai, P., Hochschild, A., Zhang, F., and Marraffini, L. A. (2013). Programmable repression and activation of bacterial gene expression using an engineered CRISPR-Cas system. Nucleic Acids Res. 41, 7429–7437. doi:10.1093/nar/gkt520
Bray, F., Ferlay, J., Soerjomataram, I., Siegel, R. L., Torre, L. A., and Jemal, A. (2018). Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 68, 394–424. doi:10.3322/caac.21492
Chavez, A., Scheiman, J., Vora, S., Pruitt, B. W., Tuttle, M., RIE, P., et al. (2015). Highly efficient Cas9-mediated transcriptional programming. Nat. Methods 12, 326–328. doi:10.1038/nmeth.3312
Christoph, F., Weikert, S., Kempkensteffen, C., Krause, H., Schostak, M., Köllermann, J., et al. (2006). Promoter hypermethylation profile of kidney cancer with new proapoptotic p53 target genes and clinical implications. Clin. Cancer Res. 12, 5040–5046. doi:10.1158/1078-0432.CCR-06-0144
Ferlay, J., Soerjomataram, I., Dikshit, R., Eser, S., Mathers, C., Rebelo, M., et al. (2015). Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int. J. Cancer 136, E359–E386. doi:10.1002/ijc.29210
Gallego-Bartolome, J., Gardiner, J., Liu, W., Papikian, A., Ghoshal, B., Kuo, H. Y., et al. (2018). Targeted DNA demethylation of the arabidopsis genome using the human TET1 catalytic domain. Proc. Natl. Acad. Sci. U.S.A. 115, E2125–E2134. doi:10.1073/pnas.1716945115
He, W., Li, X., Xu, S., Ai, J., Gong, Y., Gregg, J. L., et al. (2013). Aberrant methylation and loss of CADM2 tumor suppressor expression is associated with human renal cell carcinoma tumor progression. Biochem. Biophys. Res. Commun. 435, 526–532. doi:10.1016/j.bbrc.2013.04.074
Hirata, H., Hinoda, Y., Nakajima, K., Kawamoto, K., Kikuno, N., Ueno, K., et al. (2011). Wnt antagonist DKK1 acts as a tumor suppressor gene that induces apoptosis and inhibits proliferation in human renal cell carcinoma. Int. J. Cancer 128, 1793–1803. doi:10.1002/ijc.25507
Joosten, S. C., Smits, K. M., Aarts, M. J., Melotte, V., Koch, A., Tjan-Heijnen, V. C., et al. (2018). Epigenetics in renal cell cancer: mechanisms and clinical applications. Nat. Rev. Urol. 15, 430–451. doi:10.1038/s41585-018-0023-z
Kajiura, K., Masuda, K., Naruto, T., Kohmoto, T., Watabnabe, M., Tsuboi, M., et al. (2017). Frequent silencing of the candidate tumor suppressor TRIM58 by promoter methylation in early-stage lung adenocarcinoma. Oncotarget 8, 2890–2905. doi:10.18632/oncotarget.13761
Kardooni, H., Gonzalez-Gualda, E., Stylianakis, E., Saffaran, S., Waxman, J., and Kypta, R. M. (2018). CRISPR-mediated reactivation of DKK3 expression attenuates TGF-β signaling in prostate cancer. Cancers 10 (6), 165. doi:10.3390/cancers10060165
Klutstein, M., Nejman, D., Greenfield, R., and Cedar, H. (2016). DNA methylation in cancer and aging. Cancer Res. 76, 3446–3450. doi:10.1158/0008-5472.CAN-15-3278
Linehan, W. M., and Ricketts, C. J. (2014). Decade in review-kidney cancer: discoveries, therapies and opportunities. Nat. Rev. Urol. 11, 614–616. doi:10.1038/nrurol.2014.262
Liu, M., Zhang, X., Cai, J., Li, Y., Luo, Q., Wu, H., et al. (2018). Downregulation of TRIM58 expression is associated with a poor patient outcome and enhances colorectal cancer cell invasion. Oncol. Rep. 40, 1251–1260. doi:10.3892/or.2018.6525
Liu, Q., Jin, J., Ying, J., Sun, M., Cui, Y., Zhang, L., et al. (2015). Frequent epigenetic suppression of tumor suppressor gene glutathione peroxidase 3 by promoter hypermethylation and its clinical implication in clear cell renal cell carcinoma. Int. J. Mol. Sci. 16, 10636–10649. doi:10.3390/ijms160510636
Liu, X., Long, Z., Cai, H., Yu, S., and Wu, J. (2020). TRIM58 suppresses the tumor growth in gastric cancer by inactivation of β-catenin signaling via ubiquitination. Cancer Biol. Ther. 21, 203–212. doi:10.1080/15384047.2019.1679554
Liu, X. S., Wu, H., Krzisch, M., Wu, X., Graef, J., Muffat, J., et al. (2018). Rescue of fragile x syndrome neurons by DNA methylation editing of the FMR1 gene. Cell 172, 979–992. doi:10.1016/j.cell.2018.01.012
Malouf, G. G., Su, X., Zhang, J., Creighton, C. J., Ho, T. H., Lu, Y., et al. (2016). DNA methylation signature reveals cell ontogeny of renal cell carcinomas. Clin. Cancer Res. 22, 6236–6246. doi:10.1158/1078-0432.CCR-15-1217
Moch, H. (2013). An overview of renal cell cancer: pathology and genetics. Semin. Cancer Biol. 23, 3–9. doi:10.1016/j.semcancer.2012.06.006
Morita, S., Noguchi, H., Horii, T., Nakabayashi, K., Kimura, M., Okamura, K., et al. (2016). Targeted DNA demethylation in vivo using dCas9-peptide repeat and scFv-TET1 catalytic domain fusions. Nat. Biotechnol. 34, 1060–1065. doi:10.1038/nbt.3658
Morris, M. R., and Latif, F. (2017). The epigenetic landscape of renal cancer. Nat. Rev. Nephrol. 13, 47–60. doi:10.1038/nrneph.2016.168
Morrissey, C., Martinez, A., Zatyka, M., Agathanggelou, A., Honorio, S., Astuti, D., et al. (2001). Epigenetic inactivation of the RASSF1A 3p21.3 tumor suppressor gene in both clear cell and papillary renal cell carcinoma. Cancer Res. 61, 7277–7281.
Ricketts, C. J., De Cubas, A. A., Fan, H., Smith, C. C., Lang, M., Reznik, E., et al. (2018). The cancer genome atlas comprehensive molecular characterization of renal cell carcinoma. Cell Rep. 23, 313–326. doi:10.1016/j.celrep.2018.03.075
Ricketts, C. J., Hill, V. K., and Linehan, W. M. (2014). Tumor-specific hypermethylation of epigenetic biomarkers, including SFRP1, predicts for poorer survival in patients from the TCGA kidney renal clear cell carcinoma (KIRC) project. PLoS One 9, e85621. doi:10.1371/journal.pone.0085621
Sasaki, H., and Matsui, Y. (2008). Epigenetic events in mammalian germ-cell development: reprogramming and beyond. Nat. Rev. Genet. 9, 129–140. doi:10.1038/nrg2295
Sharma, S., Kelly, T. K., and Jones, P. A. (2010). Epigenetics in cancer. Carcinogenesis 31, 27–36. doi:10.1093/carcin/bgp220
Tao, R., Li, J., Xin, J., Wu, J., Guo, J., Zhang, L., et al. (2011). Methylation profile of single hepatocytes derived from hepatitis B virus-related hepatocellular carcinoma. PLoS One 6, e19862. doi:10.1371/journal.pone.0019862
Vojta, A., Dobrinić, P., Tadić, V., Bočkor, L., Korać, P., Julg, B., et al. (2016). Repurposing the CRISPR-Cas9 system for targeted DNA methylation. Nucleic Acids Res. 44, 5615–5628. doi:10.1093/nar/gkw159
Wang, L., Cui, Y., Sheng, J., Yang, Y., Kuang, G., Fan, Y., et al. (2017). Epigenetic inactivation of HOXA11, a novel functional tumor suppressor for renal cell carcinoma, is associated with RCC TNM classification. Oncotarget 8, 21861–21870. doi:10.18632/oncotarget.15668
Wang, L., Fan, Y., Zhang, L., Li, L., Kuang, G., Luo, C., et al. (2019). Classic SRY-box protein SOX7 functions as a tumor suppressor regulating WNT signaling and is methylated in renal cell carcinoma. Faseb J. 33, 254–263. doi:10.1096/fj.201701453RR
Xu, R., Xu, Q., Huang, G., Yin, X., Zhu, J., Peng, Y., et al. (2019). Combined analysis of the aberrant epigenetic alteration of pancreatic ductal adenocarcinoma. Biomed. Res. Int. 2019, 9379864. doi:10.1155/2019/9379864
Xu, X., Tao, Y., Gao, X., Zhang, L., Li, X., Zou, W., et al. (2016). A CRISPR-based approach for targeted DNA demethylation. Cell Discov. 2, 16009. doi:10.1038/celldisc.2016.9
Yin, X., and Xu, Y. (2016). Structure and function of TET enzymes. Adv. Exp. Med. Biol. 945, 275–302. doi:10.1007/978-3-319-43624-1_12
Zhang, Q., Ying, J., Li, J., Fan, Y., Poon, F. F., Ng, K. M., et al. (2010). Aberrant promoter methylation of DLEC1, a critical 3p22 tumor suppressor for renal cell carcinoma, is associated with more advanced tumor stage. J. Urol. 184, 731–737. doi:10.1016/j.juro.2010.03.108
Zhang, Q., Ying, J., Zhang, K., Li, H., Ng, K. M., Zhao, Y., et al. (2007). Aberrant methylation of the 8p22 tumor suppressor gene DLC1 in renal cell carcinoma. Cancer Lett. 249, 220–226. doi:10.1016/j.canlet.2006.08.019
Zhang, Q., Zhang, L., Li, L., Wang, Z., Ying, J., Fan, Y., et al. (2014). Interferon regulatory factor 8 functions as a tumor suppressor in renal cell carcinoma and its promoter methylation is associated with patient poor prognosis. Cancer Lett. 354, 227–234. doi:10.1016/j.canlet.2014.07.040
Keywords: clear cell renal cell carcinoma, TRIM58, DNA methylation, engineered demethyltransferase, CRISPR-dCas9
Citation: Gan Y, Cao C, Li A, Song H, Kuang G, Ma B, Zhang Q and Zhang Q (2021) Silencing of the TRIM58 Gene by Aberrant Promoter Methylation is Associated with a Poor Patient Outcome and Promotes Cell Proliferation and Migration in Clear Cell Renal Cell Carcinoma. Front. Mol. Biosci. 8:655126. doi: 10.3389/fmolb.2021.655126
Received: 18 January 2021; Accepted: 27 January 2021;
Published: 16 March 2021.
Edited by:
Yuchen Liu, Shenzhen University, ChinaCopyright © 2021 Gan, Cao, Li, Song, Kuang, Ma, Zhang and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Qian Zhang, emhhbmdxaWFuYmptdUAxMjYuY29t
†These authors have contributed equally to this work.
 Ying Gan
Ying Gan Congcong Cao
Congcong Cao Aolin Li
Aolin Li Haifeng Song
Haifeng Song Guanyu Kuang1,2,3
Guanyu Kuang1,2,3 Quan Zhang
Quan Zhang Qian Zhang
Qian Zhang