- 1Guangzhou Municipal and Guangdong Provincial Key Laboratory of Protein Modification and Degradation, Affiliated Cancer Hospital & Institute of Guangzhou Medical University, School of Basic Medical Sciences, Guangzhou Medical University, Guangzhou, China
- 2Department of Oncology, School of Medicine, Barbara Ann Karmanos Cancer Institute, Wayne State University, Detroit, MI, United States
- 3Departments of Pharmacology & Pathology, School of Medicine, Wayne State University, Detroit, MI, United States
- 4Department of Surgery, UT Southwestern Medical Center, Dallas, TX, United States
Characterizing mechanisms of protein homeostasis, a process of balancing between protein synthesis and protein degradation, is important for understanding the potential causes of human diseases. The ubiquitin–proteasome system (UPS) is a well-studied mechanism of protein catabolism, which is responsible for eliminating misfolded, damaged, or aging proteins, thereby maintaining quality and quantity of cellular proteins. The UPS is composed of multiple components, including a series of enzymes (E1, E2, E3, and deubiquitinase [DUB]) and 26S proteasome (19S regulatory particles + 20S core particle). An impaired UPS pathway is involved in multiple diseases, including cancer. Several proteasome inhibitors, such as bortezomib, carfilzomib, and ixazomib, are approved to treat patients with certain cancers. However, their applications are limited by side effects, drug resistance, and drug–drug interactions observed in their clinical processes. To overcome these shortcomings, alternative UPS inhibitors have been searched for in many fields. Copper complexes (e.g., CuET, CuHQ, CuCQ, CuPDTC, CuPT, and CuHK) are found to be able to inhibit a core component of the UPS machinery, such as 20S proteasome, 19S DUBs, and NPLOC4/NPL4 complex, and are proposed to be one class of metal-based anticancer drugs. In this review, we will summarize functions and applications of copper complexes in a concise perspective, with a focus on connections between the UPS and cancer.
Introduction
Protein, a complex molecule composed of amino acids, is the basic component of all living organisms. Protein homeostasis is a dynamic balance between protein synthesis and protein degradation, which is critical for maintaining healthy cell functions. In contrast, altered protein homeostasis may produce misfolded, aggregated, and mutated proteins, which are implicated in various pathological conditions and diseases, including cancer (Bastola et al., 2018). Although half-lives of different proteins may vary greatly, the turnover rate of a protein is usually related to its function or subcellular location. The ubiquitin–proteasome system (UPS) and autophagy play major roles in degradations of short-lived and long-lived proteins, respectively (Bachmair and Varshavsky, 1989; Kuma et al., 2004). Autophagy relies on formation of different membrane structures to engulf and destroy cargos (including proteins and organelles), whereas the UPS is mainly carried out to degrade proteins through a hierarchical enzymatic cascade. Both autophagy and the UPS constitute potential targets for cancer therapy. In particular, proteasome inhibitors bortezomib, carfilzomib, and ixazomib were approved by the US Food and Drug Administration (FDA) for treatment of multiple myeloma in 2003, 2012, and 2015, respectively (Manasanch and Orlowski, 2017). However, side effects and secondary drug resistance of these proteasome inhibitors limit their wide applications (Brunnert et al., 2019; Ge et al., 2020). These challenges highlight an urgent need for development of novel UPS inhibitors for cancer patients. In this mini-review, we first describe key components of the UPS (Figure 1), and then discuss potential implication of copper complexes as new UPS inhibitors in cancer treatment.
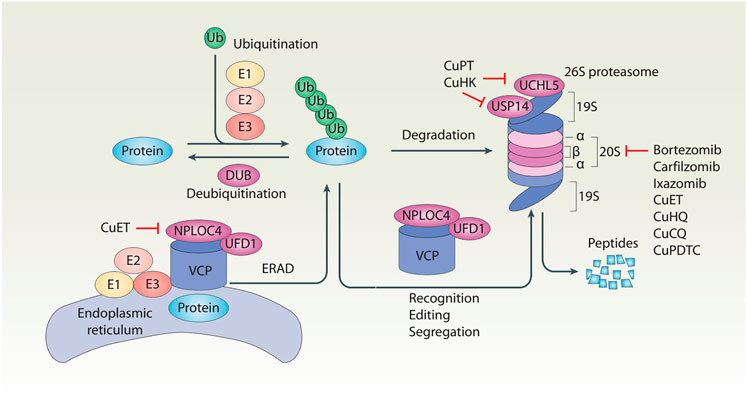
FIGURE 1. Schematic of the ubiquitin–proteasome system of protein degradation. In the ubiquitin–proteasome system, ubiquitination and deubiquitination are two reversible events that control protein levels. Generally, ubiquitination is the addition of ubiquitin (Ub) molecules to lysine residues of a protein by a cascade of enzymes (E1, E2, and E3), leading to the degradation of the substrate proteins in the 26S proteasome. In contrast, deubiquitination is catalyzed by cytosolic deubiquitinase (DUBs) or proteasomal DUBs (e.g., UCHL5 and USP14) to remove Ub from ubiquitinated proteins. In addition, the VCP–UFD1–NPLOC4 complex not only participates in the recognition, editing, and segregation of ubiquitinated substrates, but also plays a unique role in the regulation of endoplasmic reticulum-associated degradation (ERAD) by extracting substrates for degradation in the cytoplasmic proteasome.
Ubiquitin–Proteasome System: A Complexed but Coordinated System
The UPS is a highly controlled mechanism of protein degradation and turnover in cells, starting with monomeric ubiquitin, a small protein with a molecular mass of approximately 8 kDa (Ciehanover et al., 1978; Johnson et al., 1995). Ubiquitin is first activated by E1 in an adenosine triphosphate (ATP)–dependent manner. Activated ubiquitin is then conjugated by an E2 enzyme, resulting in transfer of ubiquitin to an internal lysine of the target protein via one of E3 ligases. Subsequently, through an ATP-dependent process, the polyubiquitin–protein conjugate is degraded by the 26S proteasome complex composed of two subcomplexes, the 20S proteasome and the 19S regulatory particle. Disassembly of the 26S complexes into the 20S proteasomes and the 19S regulatory particle can be caused by diminished ATP (Hoglinger et al., 2003), aspartate deficiency (Meul et al., 2020), or elevated reactive oxygen species (ROS) levels (Livnat-Levanon et al., 2014). Finally, an abnormal protein is cleaved into short peptide fragments, and the polyubiquitin chain is released and is trimmed into monomeric ubiquitin by deubiquitinating enzymes (DUBs) (Mevissen and Komander, 2017). Of note, the E3 ubiquitin ligase is responsible for substrate recognition and more than 600 putative E3 ligases determine the diversity of the UPS.
The Proteasome as a Target for Cancer Therapy
The 20S proteasome is a 700-kDa large complex formed by four stacked rings, two outer α-rings and two inner β-rings. Each α-ring or β-ring is formed by seven subunits. The constitutive active sites are β1, β2, and β5 that are responsible for caspase-like (C-like), trypsin-like (T-like), and chymotryptic activity (CT-like), respectively. The 19S regulatory particle plays essential roles in processing ubiquitylated substrates by binding, deubiquitinating, and unfolding ubiquitinated proteins (Liu and Jacobson, 2013). Substrate recognition is mediated by ubiquitin receptors (e.g., proteasome 26S subunits non-ATPase 2 [PSMD2/RPN1] and non-ATPase 4 [PSMD4/RPN10]and adhesion regulating molecule 1 [ADRM1/RPN13]) located in the 19S regulatory particle (Martinez-Fonts et al., 2020). In order to pass through a narrow channel in the 20S proteasome, domains within target proteins must be unfolded before entering. ATPases of the 19S regulatory particle exhibit conformational changes and function in gate opening, which is required for the degradation of a specific protein (Smith et al., 2011).
The proteasome has been demonstrated as a potential therapeutic target for cancer. Specially, 20S proteasome inhibitors bortezomib, carfilzomib, and ixazomib, which mainly target the CT-like catalytic subunit of the proteasome β5 subunit, are approved to treat patients with certain cancers, such as multiple myeloma. Although the precision mechanism remains obscure, several pathways are involved in proteasome inhibitor-induced tumor suppression. Proteasome inhibition results in stabilization of several key tumor suppressing proteins (e.g., NFKB inhibitor alpha [NFKBIA/IKBA] (Chen et al., 1995), bcl2-associated X, apoptosis regulator [BAX] (Li and Dou, 2000), cyclin-dependent kinase inhibitor 1A [CDKN1A/p21] (Bloom et al., 2003), cyclin-dependent kinase inhibitor 1B [CDKN1B/p27] (Pagano et al., 1995), and tumor protein p53 [TP53] (Lopes et al., 1997)), which are responsible for cell growth inhibition or cell death induction. In addition, inhibition of the proteasome causes the accumulation of unfolded and misfolded proteins, which triggers unfolded protein response (UPR), endoplasmic reticulum (ER) stress, and apoptosis (Obeng et al., 2006). However, severe side effects and drug resistance observed in clinical process of these applications demand discovery of more specific, less toxic proteasome inhibitors.
Targeting DUBs: A Novel Strategy for Cancer Therapy
DUBs can be divided into six families based on their different structures: ubiquitin C-terminal hydrolases (UCHs), ubiquitin-specific proteases (USPs), ovarian-tumor proteases (OTUs), JAMM/MPN domain–associated metallopeptidases (JAMMs), Machado–Joseph disease protein domain proteases (MJD), and monocyte chemotactic protein-induced protein (MCPIP) (Mevissen and Komander, 2017). Three DUBs are associated with the proteasome, two cysteine proteases [ubiquitin C-terminal hydrolase L5 (UCHL5/UCH37) and ubiquitin-specific peptidase 14 (USP14)], and a proteasome 26S subunit [non-ATPase 14 (PSMD14/RPN11)]. The Zn(II) metallo-protease PSMD14 is an integral subunit of the 19S regulatory particles, whereas the activities of UCHL5 and USP14 can be enhanced when they were recruited to the 19S regulatory particles (D'Arcy et al., 2015). USP14 binds to PSMD2, while UCHL5 can form a complex with PSMD4 and ADRM1 (Leggett et al., 2002; Stone et al., 2004; Qiu et al., 2006). Proteasomal DUBs may play different and overlapping functions in inhibiting ubiquitination. For example, UCHL5 or USP14 removes the polyubiquitin chain from the distal end of target protein, whereas PSMD14 cuts the entire ubiquitin chain at the base of the polyubiquitin chain during degradation process (Lam et al., 1997; Verma et al., 2002; Yao and Cohen, 2002). Binding of ubiquitinated proteins to USP14 causes 20S proteasome gate opening, thereby linking proteasomal deubiquitination to protein degradation (Peth et al., 2009). Indeed, knockdown of UCHL5 or USP14 alone increases the rate of proteasomal protein degradation but fails to affect structure and proteolytic capacity of the proteasome (Hu et al., 2005; Yao et al., 2006). However, these findings are challenged by a recent study that the loss of UCHL5 activity also impairs global protein turnover (Deol et al., 2020).
Accumulating evidence suggests a protumor role of proteasomal DUBs in different cancers. For example, UCHL5 accelerates growth of endometrial cancer by activating the WNT-catenin beta 1 (CTNNB1) pathway (Liu et al., 2020). UCHL5 positively regulates Hedgehog signaling by removing ubiquitination of Smoothened, Frizzled class receptor (SMO) protein in cancer cells (Zhou et al., 2018). In vitro and in vivo studies have shown that genetic and pharmacological inhibition of USP14 induces degradation of an androgen receptor (AR), favoring tumor suppression in AR-positive breast cancer cells or prostate cancer cells (Liao et al., 2017; Liao et al., 2018). USP14 promotes tumor growth by regulating DNA damage response or ER stress-mediated autophagy in non-small cell lung cancer cells (Moghadami et al., 2020; Sharma and Almasan, 2020). Alone this line, several compounds targeting UCHL5 and USP14 have been developed, including b-AP15 (D'Arcy et al., 2011), VLX1570 (b-AP15 analog) (Wang et al., 2016), AC17 (curcumin analog) (Zhou et al., 2013), and various metal-based complexes (Liu et al., 2014b; Chen et al., 2018a; Li et al., 2019a). Inhibition of deubiquitinating activity of USP14 and UCHL5 by b-AP15 rescues the protein level of TP53 in tumors from Tp53−/− mice (Ma et al., 2020). VLX1570 or b-AP15 inhibits tumor migration and induces apoptosis in diffuse large B-cell lymphoma, prostate cancer, and Ewing sarcoma cells (Shukla et al., 2016; Cai et al., 2017; Jiang et al., 2019). Importantly, a phase I study showed that VLX1570 has antitumor effects in patients with multiple myeloma, although two patients developed severe pulmonary toxicity (Rowinsky et al., 2020). Similar to proteasome inhibitors, efforts are needed to develop specific DUB inhibitors with greater therapeutic effects and less toxic side effects.
ER-Associated Degradation: A Quality-Controlled Degradation of Misfolded Proteins
Disruption of protein folding in ER activates UPR or ER-associated degradation (ERAD) to eliminate misfolded proteins. Unlike UPR, ERAD is a quality control process of ubiquitination of misfolded proteins in ER and subsequent proteasome degradation. Although signal and modulation of ERAD is still poorly understood, it has been shown that the valosin-containing protein (VCP/CDC48/p97)–ubiquitin recognition factor in ER-associated degradation 1 (UFD1)-NPL4 homolog, ubiquitin recognition factor (NPLOC4) complex, may play a major role in blocking activation of ERAD. VCP is a conserved chaperone-like ATPase associated with protein unfolding activities by translocating ubiquitinated substrates through its central pore, which is formed by a homohexameric, ring-shaped complex. In addition to ERAD, VCP also participates in regulation of the mitochondria-associated degradation pathway and quality control on ribosomes translating cytosolic proteins (Forster et al., 2014). VCP commonly collaborates with heterodimeric cofactors UFD1 and NPLOC4. Hyperosmotic stress induces phase separation of proteasome-containing nuclear foci (containing ubiquitinated proteins, VCP, and several proteasome-related proteins), which collectively constitute a proteolytic site (Yasuda et al., 2020). Compared to normal cells, cancer cells are characterized with upregulation of the VCP-UFD1-NPLOC4 pathway to digest false-synthesized and misfolded proteins (Tsujimoto et al., 2004; Yamamoto et al., 2004; Huiting et al., 2018; Lu et al., 2019). Thus, inhibition of the VCP-UFD1-NPLOC4 pathway may represent as a selective treatment option for cancer.
Copper Complexes as UPS Inhibitors in Cancer Treatment
Copper is an essential transition metal ion and plays an important role in maintaining normal cellular function (Linder and Hazegh-Azam, 1996). Excessive copper ions (Cu(II)) may generate ROS, which mediates oxidative damage to lipids, proteins, and DNA. Multiple types of tumors (e.g., breast and colorectal cancers) have aberrantly elevated copper levels, which promote tumor progression by increasing cell proliferation and stimulating angiogenesis and metastasis (Denoyer et al., 2015; Li, 2020; Shanbhag et al., 2020). This abnormal copper pathway in cancer can be targeted by two strategies. On one hand, a copper chelator can be used to inhibit the pro-survival effect of copper in cancer cells (Gupte and Mumper, 2009). On the other hand, using copper ionophores or copper complexes to increase intracellular copper levels could also suppress tumor growth (Wehbe et al., 2017).
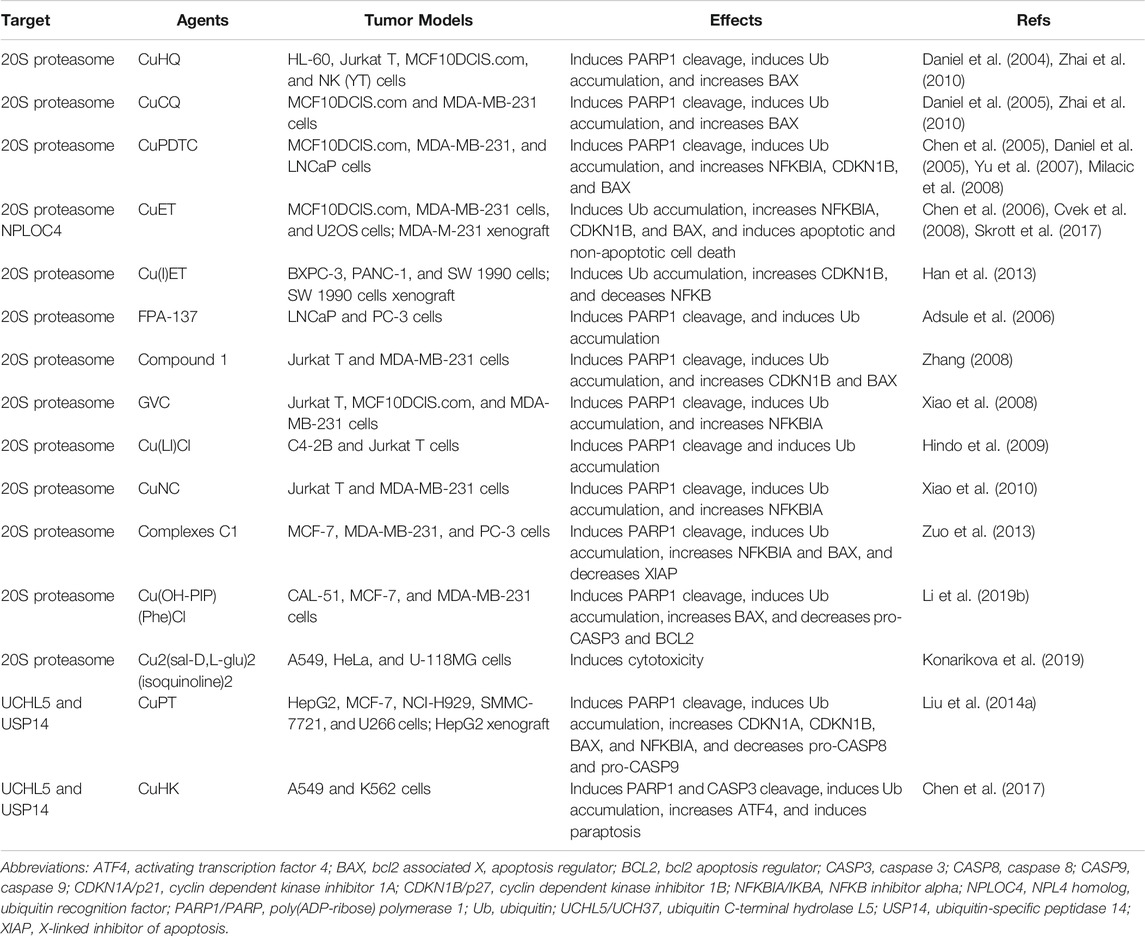
In addition to mediating oxidative damage, several lines of evidence emphasize selective role of copper complexes as potential UPS inhibitors, which contribute to their anticancer activities. First, addition of Cu(II), but not other metal ions [e.g., Zn(II), Ni(II), and Al(III)] and Cd(II), to protein samples decreases the thermal stability of ubiquitin (Milardi et al., 2007). Second, incubation with Cu(II) leads to formation of spherical ubiquitin aggregation, and this process is inhibited by Cu(II) chelation or reduction to Cu(I) (Arnesano et al., 2009). Third, Cu(II) at micromolar concentration inhibits all three kinds of proteasomes activities (Santoro et al., 2016). Fourth, in a cell-free system, Cu(II) impairs channel gating of the 20S proteasome, but does not catalyze redox reactions (Santoro et al., 2016). Fifth, Cu(II) decreases proteasome activity in HeLa cells through ROS-mediated proteasome inhibition and disassembly of the 26S proteasome (Santoro et al., 2016). Finally, several copper complexes have strong anticancer activities by inhibiting the UPS in vitro and in vivo (Table 1). In this section, we will highlight several well-studied copper complexes in tumor therapy.
CuET
Disulfiram (DSF) is used to treat patients with alcohol dependence and has been repurposed for cancer treatment (McMahon et al., 2020). DSF is easily reduced to diethyldithiocarbamate (ET), which is a strong chelator of divalent metal ions. Formation of the DSF-copper (CuDSF) complex inhibits the chymotrypsin-like activity of the purified 20S or 26S proteasome in breast cancer cells (MDA-MB-231) (Chen et al., 2006). This selective inhibition of proteasome activity has been further confirmed in malignant MCF10DCIS.com breast cells, but not in normal MCF-10A breast cells (Chen et al., 2006). Addition of Cu(II) enhances anticancer activity of DSF by converting to bis(ET)-Cu(II) complex (CuET) (Cen et al., 2004). Interestingly, ZnET also has an effect of inhibiting proteasome activity similar to CuET (Cvek et al., 2008), although it is not clear whether they have the same structural basis. The activity of CuET on the cellular 26S proteasome is higher than that on purified 20S proteasome core particles (Cvek et al., 2008), indicating that CuET may mainly target 19S regulatory particles. In addition to inducing apoptosis, CuET also triggers non-apoptotic cell death, such as paraptosis in drug-resistant prostate cancer cells (Chen et al., 2018b). The anti-cancer activity of CuET or CuIET has been proven in multiple xenograft mouse models (including, but not limited to SW 1990, MDA-MB-231, and AMO-1 cancer cells) and has good tolerance (Han et al., 2013; Skrott et al., 2017; Peng et al., 2020). Moreover, some clinical trials of the combination of DSF and copper have been completed (NCT00742911 and NCT03034135) or are in progress (NCT04265274, NCT03714555, NCT03363659, and NCT02715609), providing a potential strategy for tumor therapy.
Inhibiting VCP segregase adaptor NPLOC4 is another appealing mechanism contributing to the anticancer activity of CuET (Skrott et al., 2017). Unlike NMS873 (a well-known inhibitor of ATPase activity of VCP), CuET has no effect on this ATPase activity (Skrott et al., 2017). Isothermal calorimetry analysis reveals that CuET can directly bind to NPLOC4, leading to NPLOC4 aggregation (Skrott et al., 2017). Consequently, ectopic overexpression of NPLOC4 reverses CuET-mediated cytotoxicity (Skrott et al., 2017), although the role of VCP in this process remains unknown. Similar to proteasome inhibitors (e.g., MG132 and bortezomib), CuET induces accumulation of ubiquitylated proteins and rapid deubiquitylation of histone H2A in MCF-7 cells (Skrott et al., 2017). In another case, CuET prevents degradation of NFKBIA and Ub(G76V)-GFP (a degradation substrate for proteasome) in vitro (Skrott et al., 2017). However, different from bortezomib or MG132, CuET induces a weak accumulation of hypoxia inducible factor 1 subunit alpha (HIF1A) (Skrott et al., 2017). These findings establish overlap and difference in substrate selection between CuET and classic proteasome inhibitors.
CuHQ and CuCQ
The copper-containing compound NCI-109268 was identified as a proteasome inhibitor from a screening library of the National Cancer Institute (USA), which contains 1990 compounds (Daniel et al., 2004). Later, it was discovered that the bis(8-hydroxyquinoline)–Cu(II) complex (CuHQ) has a similar structure with NCI-109268, and its inhibitory effect on the CT-like activity is stronger than the T-like activity of proteasome (Daniel et al., 2004). CuHQ induces accumulation of ubiquitinated proteins and subsequent apoptotic death in Jurkat T cells (Daniel et al., 2004), suggesting potential anticancer activity. Moreover, increased cellular copper concentrations by a copper-enriched culture medium enhances sensitivity of prostate cancer PC-3 cells to 8-hydroxyquinoline (Daniel et al., 2004), indicating that exogenous copper can be used as a sensitizer for chemotherapeutics.
As a topical antifungal drug used clinically, clioquinol (5-chloro-7-iodo-8-hydroxyquinoline, CQ) is an analog of 8-hydroxyquinoline. CQ can form a stable complex with Cu(II), namely CuCQ, which exhibits potent cytotoxicity in human cancer cell lines (Khan et al., 2020; Wehbe et al., 2018). Mechanistically, CuCQ acts as an inhibitor of the CT-like activity of proteasome, leading to apoptotic death in MDA-MB-231 cells (Daniel et al., 2005). In particular, CQ-mediated cytoplasmic clearance of baculoviral IAP repeat containing 2 (BIRC2/CIAP1) and baculoviral IAP repeat containing 3 (BIRC3/CIAP2) contribute to CuCQ-induced apoptosis (Cater and Haupt, 2011). Similar to 8-hydroxyquinoline, copper-enriched cancer cells are also sensitive to CQ treatment (Daniel et al., 2005), further suggesting that formation of CuCQ can suppress tumor growth. Importantly, CQ inhibits the proteasome in leukemia and myeloma cells, but is not effective on normal cells (Mao et al., 2009), indicating that it has selective antitumor activity. Other analogs of CQ also inhibit the proteasomal activity and proliferation of cancer cells, such as human breast cancer (MCF10DCIS.com), ovarian cancer (A2780), and lung cancer (A549) cells (Zhai et al., 2010; Oliveri et al., 2017). However, the features recognized by these CQ analogs on the proteasome are largely unknown.
CuPDTC
The mixture of pyrrolidine dithiocarbamate (PDTC) and copper inhibits proteasome function and induces cell death in human breast cancer (MDA-MB-231) and prostate cancer (LNCaP) cells (Chen et al., 2005; Daniel et al., 2005). The involvement of bis(PDTC)-Cu(II) (CuPDTC) in inhibiting the CT-like activity of proteasome is confirmed in MDA-MB-231 cells (Milacic et al., 2008). Moreover, in MDA-MB-231 cells, several tumor-related proteins (e.g., NFKBIA and CDKN1B) are identified as proteasome targets of CuPDTC (Milacic et al., 2008). It is unclear whether PDTC analog-driven proteasome inhibitors have similar protein substrate targets for tumor suppression (Yu et al., 2007; Wang et al., 2011). In addition, CuPDTC is also engaged in induction of protein cleavage during apoptosis. For example, in apoptosis induced by chemotherapeutics, caspase or calpain family proteins usually play a parallel role to cleave poly-ADP-ribose polymerase 1 (PARP1/PARP) to impair nuclear function (Pink et al., 2000). In MDA-MB-231 cells, CuPDTC induces calpain-dependent PARP1 cleavage and apoptosis, and this process is reversed by calpastatin (a calpain inhibitor) (Milacic et al., 2008). Whether CuPDTC-induced apoptosis depends on calpain (rather than caspase) needs to be further investigated in a variety of cancer cells. At least, in cisplatin-resistant neuroblastoma cells, CuPDTC can induce cell death and cell cycle arrest by increasing TP53 protein expression, which usually leads to caspase-dependent apoptosis (Zhang et al., 2008). Interestingly, similar to CuPDTC, ZnPDTC also triggers proteasome inhibition and PARP1 cleavage (Milacic et al., 2008), raising questions about whether and how CuPDTC and ZnPDTC bind to the 20S proteasome. Further research also needs to confirm the direct molecular link between CuPDTC-induced proteasome inhibition, apoptosis induction, and structural protein cleavage.
CuPT
Pyrithione has a broad antimicrobial activity and excellent metal binding potential (Turley et al., 2000), whereas copper pyrithione (CuPT) not only has antifouling paint biocides (Almond and Trombetta, 2016), but also exerts potent anticancer activity (Liu et al., 2014a). CuPT inhibits cancer cell growth by targeting active sites of 19S DUBs (UCHL5 and USP14), leading to accumulation of both total and K48-linked ubiquitinated proteins (e.g., CDKN1A, CDKN1B, BAX, and NFKBIA) and GFPu (a surrogate proteasome substrate) (Liu et al., 2014b). Accordingly, CuPT inhibits DUB activity of the 26S proteasome in a cell-free system and can compete with UbVS (a potent inhibitor against UCHL5 and USP14) binding with UCHL5 and USP14 (Liu et al., 2014b). In contrast, although high doses may produce off-target effects, low doses of CuPT cannot prevent the CT-like activity of the 20S proteasome. The anticancer activity of CuPT is mediated by inducing apoptosis in various cancer cell lines (MCF-7, U266, and HepG2), primary monocytes from patients with acute myeloid leukemia, and xenograft mouse models (Liu et al., 2014b). These findings expand our understanding of extensive role of metal pyrithione complexes in inhibiting proteasomal DUBs (Zhao et al., 2016a; Zhao et al., 2016b; Zhao et al., 2017; Chen et al., 2018).
CuHK
Hinokitiol is a tropolone-based phenolic component isolated from Cupressaceae heartwood and its anticancer effects and antimicrobial activity have been well-documented (Inoue et al., 2020; Wu et al., 2020). In addition, the hinokitiol copper complex (CuHK) has been confirmed as an inhibitor of the 19S DUB, but not the CT-like activity of the 20S proteasome (Chen et al., 2017). This function of CuHK on DUB inhibition causes accumulation of ubiquitinated proteins and GFPu in cancer cells (A549 and K562) and HEK293 cells, respectively (Chen et al., 2017). Consequently, CuHK triggers paraptosis-like cell death (Chen et al., 2017), a caspase-independent form of regulated cell death characterized by ER and/or mitochondria dilation (Fontana et al., 2020). In particular, activating transcription factor 4 (ATF4)-mediated ER stress, but not ROS generation, favors CuHK-induced paraptosis-like cell death in A549 and K562 cells (Chen et al., 2017). It remains to be defined whether CuHK-induced paraptosis is involved in activation of ERAD machinery in ER.
Conclusion and Perspectives
The essential role of the UPS in controlling cancer biology has aroused great interest in the development of proteasome inhibitors as anticancer drugs. In recent years, tremendous progress has been made in understanding the potential of copper complexes as anticancer drugs by inhibiting multiple components and regulators of the UPS, such as 20S proteasome, 19S DUBs, and NPLOC4, although it is expected that E1, E2, and E3 might also be their targets. A main challenge in the future is to reveal precise mechanisms and specific substrate aspects of UPS inhibition by copper complexes. It is also important to profile activity and side effects of copper complexes versus other metal-containing drugs on inhibiting protein degradation and inducing different cell death modalities, toward developing them as clinically used anticancer agents. In addition, it will be interesting to distinguish between UPS-dependent and UPS-independent anticancer activities of copper complexes. A systematic and rigorous research on drug structure and function may help in addressing these issues. However, severe side effects and poor target specificity remain major obstacles for the development of copper complexes as anticancer drugs. Various nanoparticles may help provide better targeting and drug delivery of metal complexes for multiple tumor models (Sharma et al., 2018). It is also important to improve target selectivity by modifying the ligand of copper complexes and investigating the structure-activity relationship.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Author Contributions
All authors wrote the manuscript. JL and DT edited, reviewed, and approved the manuscript before submission.
Funding
XC is supported by the National Natural Science Foundation of China (81802405); JL. is supported by the National Funds for Developing Local Colleges and Universities (B16056001) and Natural Science Foundation research team of Guangdong Province (2018B030312001).
References
Adsule, S., Chen, D., Ahmed, F., Dou, Q. P., Padhye, S., et al. (2006). Novel Schiff base copper complexes of quinoline-2 carboxaldehyde as proteasome inhibitors in human prostate cancer cells. J. Med. Chem. 49, 7242–7246. doi:10.1021/jm060712l
Almond, K. M., and Trombetta, L. D. (2016). The effects of copper pyrithione, an antifouling agent, on developing zebrafish embryos. Ecotoxicology 25, 389–398. doi:10.1007/s10646-015-1597-3
Arnesano, F., Calò, V., Bonfrate, E., Ingrosso, C., Losacco, M., Pellegrino, T., et al. (2009). Copper-triggered aggregation of ubiquitin. PLoS One 4, e7052. doi:10.1371/journal.pone.0007052
Bachmair, A., and Varshavsky, A. (1989). The degradation signal in a short-lived protein. Cell 56, 1019–1032. doi:10.1016/0092-8674(89)90635-1
Bastola, P., Oien, D. B., Cooley, M., and Chien, J. (2018). Emerging cancer therapeutic targets in protein homeostasis. AAPS J. 20, 94. doi:10.1208/s12248-018-0254-1
Bloom, J., Amador, V., Bartolini, F., DeMartino, G., and Pagano, M. (2003). Proteasome-mediated degradation of p21 via N-terminal ubiquitinylation. Cell 115, 71–82. doi:10.1016/s0092-8674(03)00755-4
Brunnert, D., Stühmer, T., Kirner, S., Heiden, R., Goyal, P., Driessen, C., et al. (2019). Novel cell line models to study mechanisms and overcoming strategies of proteasome inhibitor resistance in multiple myeloma. Biochim. Biophys. Acta (BBA) - Mol. Basis Dis. 1865, 1666–1676. doi:10.1016/j.bbadis.2019.04.003
Cai, J., Liao, Y., Liu, N., Guo, Z., Chen, J., Yang, L., et al. (2017). A novel deubiquitinase inhibitor b-AP15 triggers apoptosis in both androgen receptor-dependent and -independent prostate cancers. Oncotarget 8, 63232–63246. doi:10.18632/oncotarget.18774
Cater, M. A., and Haupt, Y. (2011). Clioquinol induces cytoplasmic clearance of the X-linked inhibitor of apoptosis protein (XIAP): therapeutic indication for prostate cancer. Biochem. J. 436, 481–491. doi:10.1042/BJ20110123
Cen, D., Brayton, D., Shahandeh, B., Meyskens, F. L., and Farmer, P. J. (2004). Disulfiram facilitates intracellular Cu uptake and induces apoptosis in human melanoma cells. J. Med. Chem. 47, 6914–6920. doi:10.1021/jm049568z
Chen, D., Cui, Q. C., Yang, H., and Dou, Q. P. (2006). Disulfiram, a clinically used anti-alcoholism drug and copper-binding agent, induces apoptotic cell death in breast cancer cultures and xenografts via inhibition of the proteasome activity. Cancer Res. 66, 10425–10433. doi:10.1158/0008-5472.CAN-06-2126
Chen, D., Peng, F., Cui, Q. C., Daniel, K. G., Orlu, S., Liu, J., et al. (2005). Inhibition of prostate cancer cellular proteasome activity by a pyrrolidine dithiocarbamate-copper complex is associated with suppression of proliferation and induction of apoptosis. Front. Biosci. 10, 2932–2939. doi:10.2741/1749
Chen, W., Yang, W., Chen, P., Huang, Y., and Li, F. (2018). Disulfiram copper nanoparticles prepared with a stabilized metal ion ligand complex method for treating drug-resistant prostate cancers. ACS Appl. Mater. Inter. 10, 41118–41128. doi:10.1021/acsami.8b14940
Chen, X., Chen, J., Yang, Q., Yang, L., Xu, D., Zhang, P., et al. (2017). Hinokitiol copper complex inhibits proteasomal deubiquitination and induces paraptosis-like cell death in human cancer cells. Eur. J. Pharmacol. 815, 147–155. doi:10.1016/j.ejphar.2017.09.003
Chen, X., Chen, J., Zhang, P., Huang, Q., Zhang, X., Yang, L., et al. (2018a). Inhibition of proteasomal deubiquitinase by silver complex induces apoptosis in non-small cell lung cancer cells. Cell Physiol Biochem 49, 780–797. doi:10.1159/000493041
Chen, X., Yang, Q., Zhang, X., Zhang, P., Liao, S., He, Z., et al. (2018b). Cadmium pyrithione suppresses tumor growth in vitro and in vivo through inhibition of proteasomal deubiquitinase. Biometals 31, 29–43. doi:10.1007/s10534-017-0062-6
Chen, Z., Palombella, V. J., Melandri, F., Scherer, D., Ballard, D., and Maniatis, T. (1995). Signal-induced site-specific phosphorylation targets I kappa B alpha to the ubiquitin-proteasome pathway. Genes Develop. 9, 1586–1597. doi:10.1101/gad.9.13.1586
Ciehanover, A., Hod, Y., and Hershko, A. (1978). A heat-stable polypeptide component of an ATP-dependent proteolytic system from reticulocytes. Biochem. Biophysical Res. Commun. 81, 1100–1105. doi:10.1016/0006-291x(78)91249-4
Cvek, B., Milacic, V., Taraba, J., and Dou, Q. P. (2008). Ni(II), Cu(II), and Zn(II) Diethyldithiocarbamate Complexes Show Various Activities against the Proteasome in Breast Cancer CellsCu(II), and Zn(II) diethyldithiocarbamate complexes show various activities against the proteasome in breast cancer cells. J. Med. Chem. 51, 6256–6258. doi:10.1021/jm8007807
D'Arcy, P., Fryknäs, M., Lindsten, K., De Cesare, M., Perego, P., Sadeghi, B., et al. (2011). Inhibition of proteasome deubiquitinating activity as a new cancer therapy. Nat. Med. 17, 1636–1640. doi:10.1038/nm.2536
D'Arcy, P., Wang, X., and Linder, S. (2015). Deubiquitinase inhibition as a cancer therapeutic strategy. Pharmacol. and Ther. 147, 32–54. doi:10.1016/j.pharmthera.2014.11.002
Daniel, K. G., Gupta, P., Harbach, R. H., Guida, W. C., and Dou, Q. P. (2004). Organic copper complexes as a new class of proteasome inhibitors and apoptosis inducers in human cancer cells. Biochem. Pharmacol. 67, 1139–1151. doi:10.1016/j.bcp.2003.10.031
Daniel, K. G., Orlu, S., Cui, Q. C., Miller, F. R., and Dou, Q. P. (2005). Clioquinol and pyrrolidine dithiocarbamate complex with copper to form proteasome inhibitors and apoptosis inducers in human breast cancer cells. Breast Cancer Res. 7, R897–R908. doi:10.1186/bcr1322
Denoyer, D., Masaldan, S., La Fontaine, S., and Cater, M. A. (2015). Targeting copper in cancer therapy: “Copper that Cancer”. Metallomics 7, 1459–1476. doi:10.1039/c5mt00149h
Deol, K. K., Du, J., Bisbee, H. A., Guenette, R. G., and Strieter, E. R. (2020). Proteasome-bound UCH37/UCHL5 debranches ubiquitin chains to promote degradation. Mol. Cell 80, 796–809. doi:10.1016/j.molcel.2020.10.017
Fontana, F., Raimondi, M., Marzagalli, M., Di Domizio, A., and Limonta, P. (2020). The emerging role of paraptosis in tumor cell biology: perspectives for cancer prevention and therapy with natural compounds. Biochim. Biophys. Acta-Rev. Cancer 1873, 188338. doi:10.1016/j.bbcan.2020.188338
Forster, F., Schuller, J. M., Unverdorben, P., and Aufderheide, A. (2014). Emerging mechanistic insights into AAA complexes regulating proteasomal degradation. Biomolecules 4, 774–794. doi:10.3390/biom4030774
Ge, M., Qiao, Z., Kong, Y., Lu, H., and Liu, H. (2020). Exosomes mediate intercellular transfer of non-autonomous tolerance to proteasome inhibitors in mixed‐lineage leukemia. Cancer Sci. 111, 1279–1290. doi:10.1111/cas.14351
Gupte, A., and Mumper, R. J. (2009). Elevated copper and oxidative stress in cancer cells as a target for cancer treatment. Cancer Treat. Rev. 35, 32–46. doi:10.1016/j.ctrv.2008.07.004
Han, J., Yue, X., Chang, J., Shi, W., and Hua, Y. (2013). A binuclear complex constituted by diethyldithiocarbamate and copper(I) functions as a proteasome activity inhibitor in pancreatic cancer cultures and xenografts. Toxicol. Appl. Pharmacol. 273, 477–483. doi:10.1016/j.taap.2013.09.009
Hindo, S. S., Tomco, D., Heeg, M. J., Hryhorczuk, L., McGarvey, B. R., Dou, Q. P., et al. (2009). Metals in anticancer therapy: copper(II) complexes as inhibitors of the 20S proteasome. Eur. J. Med. Chem. 44, 4353–4361. doi:10.1016/j.ejmech.2009.05.019
Hoglinger, G. U., Michel, P. P., Medja, F., Lombès, A., Ruberg, M., Friguet, B., et al. (2003). Dysfunction of mitochondrial complex I and the proteasome: interactions between two biochemical deficits in a cellular model of Parkinson’s disease. J. Neurochem. 86, 1297–1307. doi:10.1046/j.1471-4159.2003.01952.x
Hu, M., Song, L., Jeffrey, P. D., Chenova, T. A., Wilkinson, K. D., Cohen, R. E., et al. (2005). Structure and mechanisms of the proteasome-associated deubiquitinating enzyme USP14. EMBO J. 24, 3747–3756. doi:10.1038/sj.emboj.7600832
Huiting, L. N., Zhang, G., Roderick, J., Li, B., Anderson, N., Wang, Y., et al. (2018). UFD1 contributes to MYC-mediated leukemia aggressiveness through suppression of the proapoptotic unfolded protein response. Leukemia 32, 2339–2351. doi:10.1038/s41375-018-0141-x
Inoue, Y., Murata, I., Nomura, H., Isshiki, Y., and Kanamoto, I. (2020). Evaluation of antibacterial activity expression of the hinokitiol/cyclodextrin complex against bacteria. ACS Omega 5, 27180–27187. doi:10.1021/acsomega.0c03222
Jiang, L., Wang, J., He, Q., Chen, X., Lan, X., Chen, J., et al. (2019). Proteasomal cysteine deubiquitinase inhibitor b-AP15 suppresses migration and induces apoptosis in diffuse large B cell lymphoma. J. Exp. Clin. Cancer Res. 38, 453. doi:10.1186/s13046-019-1446-y
Johnson, E. S., Ma, P. C., Ota, I. M., and Varshavsky, A. (1995). A proteolytic pathway that recognizes ubiquitin as a degradation signal. J. Biol. Chem. 270, 17442–17456. doi:10.1074/jbc.270.29.17442
Khan, R., Khan, H., Abdullah, Y., and Dou, Q. P. (2020). Feasibility of repurposing clioquinol for cancer therapy. Pra 15, 14–31. doi:10.2174/1574892815666200227090259
Konarikova, K., Frivaldskaau, J., Gbelcova, H., Sveda, M., Ruml, T., Janubova, M., et al. (2019). Schiff base Cu(II) complexes as inhibitors of proteasome in human cancer cells. Bll 120, 646–649. doi:10.4149/BLL_2019_107
Kuma, A., Matsui, M., Yamamoto, A., Nakaya, H., Yoshimori, T., Ohsumi, Y., et al. (2004). The role of autophagy during the early neonatal starvation period. Nature 432, 1032–1036. doi:10.1038/nature03029
Lam, Y. A., Xu, W., DeMartino, G. N., and Cohen, R. E. (1997). Editing of ubiquitin conjugates by an isopeptidase in the 26S proteasome. Nature 385, 737–740. doi:10.1038/385737a0
Leggett, D. S., Borodovsky, A., Crosas, B., Schmidt, M., Baker, R. T., Walz, T., et al. (2002). Multiple associated proteins regulate proteasome structure and function. Mol. Cell 10, 495–507. doi:10.1016/s1097-2765(02)00638-x
Li, B., and Dou, Q. P. (2000). Bax degradation by the ubiquitin/proteasome-dependent pathway: involvement in tumor survival and progression. Proc. Natl. Acad. Sci. 97, 3850–3855. doi:10.1073/pnas.070047997
Li, D. D., Wang, L.-Y., Dai, L.-L., Yang, Z.-B., Zhi, S., Zhang, N., et al. (2019a). Novel copper complexes that inhibit the proteasome and trigger apoptosis in triple-negative breast cancer cells. ACS Med. Chem. Lett. 10, 1328–1335. doi:10.1021/acsmedchemlett.9b00284
Li, X., Long, H., Zhang, P., Su, H., and Liu, J. (2019b). A new gold(I) complex-Au(PPh3)PT is a deubiquitinase inhibitor and inhibits tumor growth. EBioMedicine 39, 159–172. doi:10.1016/j.ebiom.2018.11.047
Li, Y. (2020). Copper homeostasis: emerging target for cancer treatment. IUBMB Life 72, 1900–1908. doi:10.1002/iub.2341
Liao, Y., Hua, X., Cai, J., Xia, X., Wang, X., Huang, H., et al. (2017). Proteasome-associated deubiquitinase ubiquitin-specific protease 14 regulates prostate cancer proliferation by deubiquitinating and stabilizing androgen receptor. Cell Death Dis 8, e2585. doi:10.1038/cddis.2016.477
Liao, Y., Liu, N., Cai, J., Guo, Z., Li, Y., Jiang, L., et al. (2018). Growth arrest and apoptosis induction in androgen receptor-positive human breast cancer cells by inhibition of USP14-mediated androgen receptor deubiquitination. Oncogene 37, 1896–1910. doi:10.1038/s41388-017-0069-z
Linder, M. C., and Hazegh-Azam, M. (1996). Copper biochemistry and molecular biology. Am. J. Clin. Nutr. 63, 797S–811S. doi:10.1093/ajcn/63.5.797
Liu, C. W., and Jacobson, A. D. (2013). Functions of the 19S complex in proteasomal degradation. Trends Biochem. Sci. 38, 103–110. doi:10.1016/j.tibs.2012.11.009
Liu, D., Song, Z., Wang, X., and Ouyang, L. (2020). Ubiquitin C-terminal hydrolase L5 (UCHL5) accelerates the growth of endometrial cancer via activating the wnt/β-catenin signaling pathway. Front. Oncol. 10, 865. doi:10.3389/fonc.2020.00865
Liu, N., Huang, H., Zhao, C., Liao, S., Yang, C., Liu, S., et al. (2014a). Clinically used antirheumatic agent auranofin is a proteasomal deubiquitinase inhibitor and inhibits tumor growth. Oncotarget 5, 5453–5471. doi:10.18632/oncotarget.2113
Liu, N., Li, X., Liao, S., Song, W., Yang, C., Zhao, C., et al. (2014b). A novel proteasome inhibitor suppresses tumor growth via targeting both 19S proteasome deubiquitinases and 20S proteolytic peptidases. Sci. Rep. 4, 5240. doi:10.1038/srep05240
Livnat-Levanon, N., Kleifeld, O., Krutauz, D., Segref, A., Rinaldi, T., Erpapazoglou, Z., et al. (2014). Reversible 26S proteasome disassembly upon mitochondrial stress. Cell Rep. 7, 1371–1380. doi:10.1016/j.celrep.2014.04.030
Lopes, U. G., Erhardt, P., Yao, R., and Cooper, G. M. (1997). p53-dependent induction of apoptosis by proteasome inhibitors. J. Biol. Chem. 272, 12893–12896. doi:10.1074/jbc.272.20.12893
Lu, B. S., Zhang, Y.-P., Guo, P.-Y., Li, W., and Liu, K.-L. (2019). Upregulation of NPL4 promotes bladder cancer cell proliferation by inhibiting DXO destabilization of cyclin D1 mRNA. Cancer Cell Int 19, 149. doi:10.1186/s12935-019-0874-2
Ma, Y. S., Yu, F., Wu, T.-M., Liu, J.-B., Zhang, Y.-J., Xia, Q., et al. (2020). Inhibition of USP14 and UCH37 deubiquitinating activity by b-AP15 as a potential therapy for tumors with p53 deficiency. Sig Transduct Target. Ther. 5, 30. doi:10.1038/s41392-020-0143-9
Manasanch, E. E., and Orlowski, R. Z. (2017). Proteasome inhibitors in cancer therapy. Nat. Rev. Clin. Oncol. 14, 417–433. doi:10.1038/nrclinonc.2016.206
Mao, X., Sprangers, R., Wang, X., Venugopal, A., Wood, T., Zhang, Y., et al. (2009). Clioquinol inhibits the proteasome and displays preclinical activity in leukemia and myeloma. Leukemia 23, 585–590. doi:10.1038/leu.2008.232
Martinez-Fonts, K., Tomita, T., Elsasser, S., Nager, A. R., Shi, Y., et al. (2020). The proteasome 19S cap and its ubiquitin receptors provide a versatile recognition platform for substrates. Nat. Commun. 11, 477, doi:10.1038/s41467-019-13906-8
McMahon, A., Chen, W., and Li, F. (2020). Old wine in new bottles: advanced drug delivery systems for disulfiram-based cancer therapy. J. Controlled Release 319, 352–359. doi:10.1016/j.jconrel.2020.01.001
Meul, T., Schmitt, S., Mayr, C. H., Mattner, L. F., Schiller, H. B., Yazgili, A. S., et al. (2020). Mitochondrial regulation of the 26S proteasome. Cell Rep. 32, 108059. doi:10.1016/j.celrep.2020.108059
Mevissen, T. E. T., and Komander, D. (2017). Mechanisms of deubiquitinase specificity and regulation. Annu. Rev. Biochem. 86, 159–192. doi:10.1146/annurev-biochem-061516-044916
Milacic, V., Giovagnini, L., Diez, A., Fregona, D., and Dou, Q. P. (2008). Pyrrolidine dithiocarbamate-zinc(II) and -copper(II) complexes induce apoptosis in tumor cells by inhibiting the proteasomal activity. Toxicol. Appl. Pharmacol. 231, 24–33. doi:10.1016/j.taap.2008.03.009
Milardi, D., Grasso, G., Magrì, A., Tabbì, G., Scintilla, S., Natile, G., et al. (2007). Ubiquitin stability and the lys 63-linked polyubiquitination site are compromised on copper binding. Angew. Chem. Int. Ed. 46, 7993–7995. doi:10.1002/anie.200701987
Moghadami, A. A., Kalantary-Charvadeh, A., Hamzavi, M., Mosayyebi, B., Sedghi, H., Ghorbani Haghjo, A., et al. (2020). Inhibition of USP14 induces ER stress-mediated autophagy without apoptosis in lung cancer cell line A549. Cell Stress and Chaperones 25, 909–917. doi:10.1007/s12192-020-01125-w
Obeng, E. A., Gutman, D. M., Harrington, W. J., Lee, K. P., and Boise, L. H. (2006). Proteasome inhibitors induce a terminal unfolded protein response in multiple myeloma cells. Blood 107, 4907–4916. doi:10.1182/blood-2005-08-3531
Oliveri, V., Milardi, D., Viale, M., Maric, I., Sgarlata, C., and Vecchio, G. (2017). Amino- and chloro-8-hydroxyquinolines and their copper complexes as proteasome inhibitors and antiproliferative agents. Metallomics 9, 1439–1446. doi:10.1039/c7mt00156h
Pagano, M., Theodoras, A., Beer-Romero, P., Del Sal, G., Chau, V., Yew, P., et al. (1995). Role of the ubiquitin-proteasome pathway in regulating abundance of the cyclin-dependent kinase inhibitor p27. Science 269, 682–685. doi:10.1126/science.7624798
Peng, Y., Meng, Y., Hu, S., Ding, J., and Zhou, W. (2020). Nanoscale copper(II)-Diethyldithiocarbamate coordination polymer as a drug self-delivery system for highly robust and specific cancer therapy. Mol. Pharmaceutics 17, 2864–2873. doi:10.1021/acs.molpharmaceut.0c00284
Peth, A., Besche, H. C., and Goldberg, A. L. (2009). Ubiquitinated proteins activate the proteasome by binding to Usp14/Ubp6, which causes 20S gate opening. Mol. Cell 36, 794–804. doi:10.1016/j.molcel.2009.11.015
Pink, J. J., Tagliarino, C., Planchon, S. M., Yang, X., Froelich, C. J., and Boothman, D. A. (2000). Activation of a cysteine protease in MCF-7 and T47D breast cancer cells during β-lapachone-mediated apoptosis. Exp. Cell Res. 255, 144–155. doi:10.1006/excr.1999.4790
Qiu, X. B., Li, C.-J., Miao, S., Wang, L., and Goldberg, A. L. (2006). hRpn13/ADRM1/GP110 is a novel proteasome subunit that binds the deubiquitinating enzyme, UCH37. EMBO J. 25, 5742–5753. doi:10.1038/sj.emboj.7601450
Rowinsky, E. K., Paner, A., Berdeja, J. G., Paba-Prada, C., Venugopal, P., Porkka, K., et al. (2020). Phase 1 study of the protein deubiquitinase inhibitor VLX1570 in patients with relapsed and/or refractory multiple myeloma. Invest. New Drugs 38, 1448–1453. doi:10.1007/s10637-020-00915-4
Santoro, A. M., Attanasio, F., Lanza, V., Pappalardo, G., Tomasello, M. F., Cunsolo, A., et al. (2016). Copper(II) ions affect the gating dynamics of the 20S proteasome: a molecular and in cell study. Sci. Rep. 6, 33444. doi:10.1038/srep33444
Shanbhag, V. C., Jasmer, K., Papageorgiou, C., Singh, K., and Petris, M. J. Copper metabolism as a unique vulnerability in cancer. Biochim. Biophys. Acta (BBA) - Mol. Cell Res. 1868, 118893, doi:10.1016/j.bbamcr.2020.118893 (2020).
Sharma, A., and Almasan, A. (2020). USP14 regulates DNA damage response and is a target for radiosensitization in non-small cell lung cancer. Ijms 21, 6383. doi:10.3390/ijms21176383
Sharma, A., Goyal, A. K., and Rath, G. (2018). Recent advances in metal nanoparticles in cancer therapy. J. Drug Target. 26, 617–632. doi:10.1080/1061186X.2017.1400553
Shukla, N., Smith, R. S., Ambati, S., Munoz, S., Merchant, M., D'Arcy, P., et al. (2016). Proteasome addiction defined in ewing sarcoma is effectively targeted by a novel class of 19S proteasome inhibitors. Cancer Res. 76, 4525–4534. doi:10.1158/0008-5472.CAN-16-1040
Skrott, Z., Andersen, K. K., Friis, S., Majera, D., Gursky, J., Ozdian, T., et al. (2017). Alcohol-abuse drug disulfiram targets cancer via p97 segregase adaptor NPL4. Nature 552, 194–199. doi:10.1038/nature25016
Smith, D. M., Fraga, H., Reis, C., Kafri, G., and Goldberg, A. L. (2011). ATP binds to proteasomal ATPases in pairs with distinct functional effects, implying an ordered reaction cycle. Cell 144, 526–538. doi:10.1016/j.cell.2011.02.005
Stone, M., Seeger, M., Bech-Otschir, D., Wallace, M., and Gordon, C. (2004). Uch2/Uch37 is the major deubiquitinating enzyme associated with the 26S proteasome in fission yeast. J. Mol. Biol. 344, 697–706. doi:10.1016/j.jmb.2004.09.057
Tsujimoto, Y., Hoshida, Y., Kono, T., Oka, T., Yamamoto, S., Nonomura, N., et al. (2004). Elevated expression of valosin-containing protein (p97) is associated with poor prognosis of prostate cancer. Clin. Cancer Res. 10, 3007–3012. doi:10.1158/1078-0432.ccr-03-0191
Turley, P. A., Fenn, R. J., and Ritter, J. C. (2000). Pyrithiones as antifoulants: environmental chemistry and preliminary risk assessment. Biofouling 15, 175–182. doi:10.1080/08927010009386308
Verma, R., Aravind, L., Oania, R., McDonald, W. H., Yates, J. R., Koonin, E. V., et al. (2002). Role of Rpn11 metalloprotease in deubiquitination and degradation by the 26S proteasome. Science 298, 611–615. doi:10.1126/science.1075898
Wang, F., Liu, X., Li, L., Wu, S., Dou, Q. P., and Yan, B. (2011). A novel dithiocarbamate analogue with potentially decreased ALDH inhibition has copper-dependent proteasome-inhibitory and apoptosis-inducing activity in human breast cancer cells. Cancer Lett. 300, 87–95. doi:10.1016/j.canlet.2010.09.010
Wang, X., Hillert, E.-K., Olofsson, M. H., Pierrou, S., Hillertz, P., Gullbo, J., et al. (2016). The proteasome deubiquitinase inhibitor VLX1570 shows selectivity for ubiquitin-specific protease-14 and induces apoptosis of multiple myeloma cells. Sci. Rep. 6, 26979. doi:10.1038/srep26979
Wehbe, M., Malhotra, A. K., Anantha, M., Lo, C., Dragowska, W. H., Dos Santos, N., et al. (2018). Development of a copper-clioquinol formulation suitable for intravenous use. Drug Deliv. Transl. Res. 8, 239–251. doi:10.1007/s13346-017-0455-7
Wehbe, M., Leung, A. W. Y., Abrams, M. J., Orvig, C., and Bally, M. B. (2017). A Perspective - can copper complexes be developed as a novel class of therapeutics? Dalton Trans. 46, 10758–10773. doi:10.1039/c7dt01955f
Wu, Y. J., Wu, L.-H., Liou, H.-P., Pangilinan, C. R., Tyan, Y.-C., and Lee, C.-H. (2020). Hinokitiol reduces tumor metastasis by inhibiting heparanase via extracellular signal-regulated kinase and protein kinase B pathway. Int. J. Med. Sci. 17, 403–413. doi:10.7150/ijms.41177
Xiao, Y., Bi, C., Fan, Y., Cui, C., Zhang, X., and Dou, Q. P. (2008). L-glutamine Schiff base copper complex as a proteasome inhibitor and an apoptosis inducer in human cancer cells. Int. J. Oncol. 33, 1073–1079.
Xiao, Y., Chen, D. I., Zhang, X., Cui, Q., Fan, Y., Bi, C., et al. (2010). Molecular study on copper-mediated tumor proteasome inhibition and cell death. Int. J. Oncol. 37, 81–87. doi:10.3892/ijo_00000655
Yamamoto, S., Hoshida, Y., Sakon, M., Kameyama, M., Imaoka, S., Sekimoto, M., et al. (2004). Expression of valosin-containing protein in colorectal carcinomas as a predictor for disease recurrence and prognosis. Clin. Cancer Res. 10, 651–657. doi:10.1158/1078-0432.ccr-1576-03
Yao, T., and Cohen, R. E. (2002). A cryptic protease couples deubiquitination and degradation by the proteasome. Nature 419, 403–407. doi:10.1038/nature01071
Yao, T., Xu, W., DeMartino, G. N., Florens, L., Swanson, S. K., Washburn, M. P., et al. (2006). Proteasome recruitment and activation of the Uch37 deubiquitinating enzyme by Adrm1. Nat. Cell Biol 8, 994–1002. doi:10.1038/ncb1460
Yasuda, S., Kaiho, A., Guo, Q., Ikeuchi, K., Endo, A., Arai, N., et al. (2020). Stress- and ubiquitylation-dependent phase separation of the proteasome. Nature 578, 296–300. doi:10.1038/s41586-020-1982-9
Yu, Z., Wang, F., Milacic, V., Li, X., Cui, Q. C., Zhang, B., et al. (2007). Evaluation of copper-dependent proteasome-inhibitory and apoptosis-inducing activities of novel pyrrolidine dithiocarbamate analogues. Int. J. Mol. Med. 20, 919–925.
Zhai, S., Cui, Q. C., Sun, Y., Dou, Q. P., and Yan, B. (2010). Tumor cellular proteasome inhibition and growth suppression by 8-hydroxyquinoline and clioquinol requires their capabilities to bind copper and transport copper into cells. J. Biol. Inorg. Chem. 15, 259–269. doi:10.1007/s00775-009-0594-5
Zhang, H., Wu, J. S., and Peng, F. (2008). Potent anticancer activity of pyrrolidine dithiocarbamate-copper complex against cisplatin-resistant neuroblastoma cells. Anticancer Drugs 19, 125–132. doi:10.1097/CAD.0b013e3282f2bdff
Zhang, X., Bi, C., Fan, Y., Cui, Q., Chen, D., Xiao, Y., et al. (2008). Induction of tumor cell apoptosis by taurine Schiff base copper complex is associated with the inhibition of proteasomal activity. Int. J. Mol. Med. 22, 677–682.
Zhao, C., Yang, C., Zang, D., Lan, X., Liao, S., Zhang, P., et al. (2017). Repurposing an antidandruff agent to treating cancer: zinc pyrithione inhibits tumor growth via targeting proteasome-associated deubiquitinases. Oncotarget 8, 13942–13956. doi:10.18632/oncotarget.14572
Zhao, C., Zang, D., Lan, X., Liao, S., Yang, C., Zhang, P., et al. (2016a). A novel nickel complex works as a proteasomal deubiquitinase inhibitor for cancer therapy. Oncogene 35, 5916–5927. doi:10.1038/onc.2016.114
Zhao, C., Zang, D., Lan, X., Liao, S., Yang, C., Zhang, P., et al. (2016b). Platinum-containing compound platinum pyrithione is stronger and safer than cisplatin in cancer therapy. Biochem. Pharmacol. 116, 22–38. doi:10.1016/j.bcp.2016.06.019
Zhou, B., Li, B., Wang, H., Liu, H., Wang, X., Qiu, X., et al. (2013). Deubiquitinase inhibition of 19S regulatory particles by 4-arylidene curcumin analog AC17 causes NF-κB inhibition and p53 reactivation in human lung cancer cells. Mol. Cancer Ther. 12, 1381–1392. doi:10.1158/1535-7163.MCT-12-1057
Zhou, Z., Pang, S., Chen, P., Jiang, W., Shan, Z., and Zhang, Q. (2018). The deubiquitinase UCHL5/UCH37 positively regulates Hedgehog signaling by deubiquitinating Smoothened. J. Mol. Cell Biol 10, 243–257. doi:10.1093/jmcb/mjx036
Keywords: copper complex, cancer, ubiquitin, proteasome, degradation
Citation: Chen X, Dou QP, Liu J and Tang D (2021) Targeting Ubiquitin–Proteasome System With Copper Complexes for Cancer Therapy. Front. Mol. Biosci. 8:649151. doi: 10.3389/fmolb.2021.649151
Received: 04 January 2021; Accepted: 12 February 2021;
Published: 13 April 2021.
Edited by:
Graça Soveral, University of Lisbon, PortugalReviewed by:
Pádraig D'Arcy, Linköping University, SwedenBenita Sjogren, Purdue University, United States
Copyright © 2021 Chen, Dou, Liu and Tang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jinbao Liu, amxpdUBnemhtdS5lZHUuY24=; Daolin Tang, ZGFvbGluLnRhbmdAdXRzb3V0aHdlc3Rlcm4uZWR1
 Xin Chen
Xin Chen Q. Ping Dou
Q. Ping Dou Jinbao Liu
Jinbao Liu Daolin Tang
Daolin Tang