- 1PG & Research, Department of Chemistry, Nehru Memorial College (Affiliated Bharathidasan University), Puthanampatti, India
- 2Department of Chemistry, M. Kumarasamy College of Engineering, Karur, India
- 3Department of Chemistry, College of Science, Deanship of Scientific Research, Taif University, Taif, Saudi Arabia
- 4South African Medical Research Council Bioinformatics Unit, South African National Bioinformatics Institute, University of the Western Cape, Cape Town, South Africa
- 5Laboratory of Computational Modeling of Drugs, South Ural State University, Chelyabinsk, Russia
This work investigated the interaction of indole with SARS-CoV-2. Indole is widely used as a medical material owing to its astounding biological activities. Indole and its derivatives belong to a significant category of heterocyclic compounds that have been used as a crucial component for several syntheses of medicine. A straightforward one-pot three-component synthesis of indole, coupled with Mannich base derivatives 1a–1j, was synthesized without a catalyst. The products were confirmed by IR, 1H-NMR, 13C-NMR, mass spectra, and elemental analysis. The indole derivatives were tested for cytotoxic activity, using three cancer cell lines and normal cell lines of Human embryonic kidney cell (HEK293), liver cell (LO2), and lung cell (MRC5) by MTT assay using doxorubicin as the standard drug. The result of cytotoxicity indole compound 1c (HepG2, LC50−0.9 μm, MCF−7, LC50−0.55 μm, HeLa, LC50−0.50 μm) was found to have high activity compared with other compounds used for the same purpose. The synthesized derivatives have revealed their safety by exhibiting significantly less cytotoxicity against the normal cell line (HEK-293), (LO2), and (MRC5) with IC50 > 100 μg/ml. Besides, we report an in silico study with spike glycoprotein (SARS-CoV-2-S). The selective molecules of compound 1c exhibited the highest docking score −2.808 (kcal/mol) compared to other compounds. This research work was successful in synthesizing a few compounds with potential as anticancer agents. Furthermore, we have tried to emphasize the anticipated role of indole scaffolds in designing and discovering the much-awaited anti-SARS CoV-2 therapy by exploring the research articles depicting indole moieties as targeting SARS CoV-2 coronavirus.
Introduction
Coronavirus has proved to be the most deadly of the 21st-century epidemics by being responsible for emergent communicable disorders. It first manifested its presence through the onset of dangerous pneumonia, started by the (SARS-CoV) infestation in 2003 (Rahman et al., 2020). In December 2019, many lung fever patients infected by a novel coronavirus were announced in Wuhan, China (Chan et al., 2020; Li et al., 2020; Zhu et al., 2020). The SARS-CoV-2 has been the cause of greater than 1.27 million deaths as of November 11, 2020 (Lu et al., 2020; Wu et al., 2020; Zhou et al., 2020). The acronym for coronavirus, namely, SARS-CoV-2, was assigned by the World Health Organization (WHO) on February 11, 2020 (Gorbalenya et al., 2020). SARS-CoV-2 has become a global health crisis involving around 212 countries (World Health Organization, 2020). Several drug mixtures are still being used.
However, the remedial outcome has been meager with secondary response (Cao et al., 2020). Adenosine triphosphate (ATP) analog was used as an antiviral drug to counter the effects of COVID-19, but more statistics are required to demonstrate its efficiency (Cohen, 2020; Holshue et al., 2020; Wang et al., 2020). On August 11, 2020, Russia became the first nation to approve a vaccine (sputnik V) to protect against infection by COVID-19 (Talha, 2020). The inherent RNA of coronaviruses and its structure information is described and discussed by other researchers (Hussain et al., 2005; Chen et al., 2020). In the biorhythms of coronaviruses, some functional and non-functional proteins are involved (Ramajayam et al., 2011; Ren et al., 2013). The emergence of drug-resistance for antiviral activity and defective antiviral drugs stimulates a great demand to develop a less toxic and more potent antiviral agent. In this regard, researchers have recently focused on naturally available indoles and their derivatives.
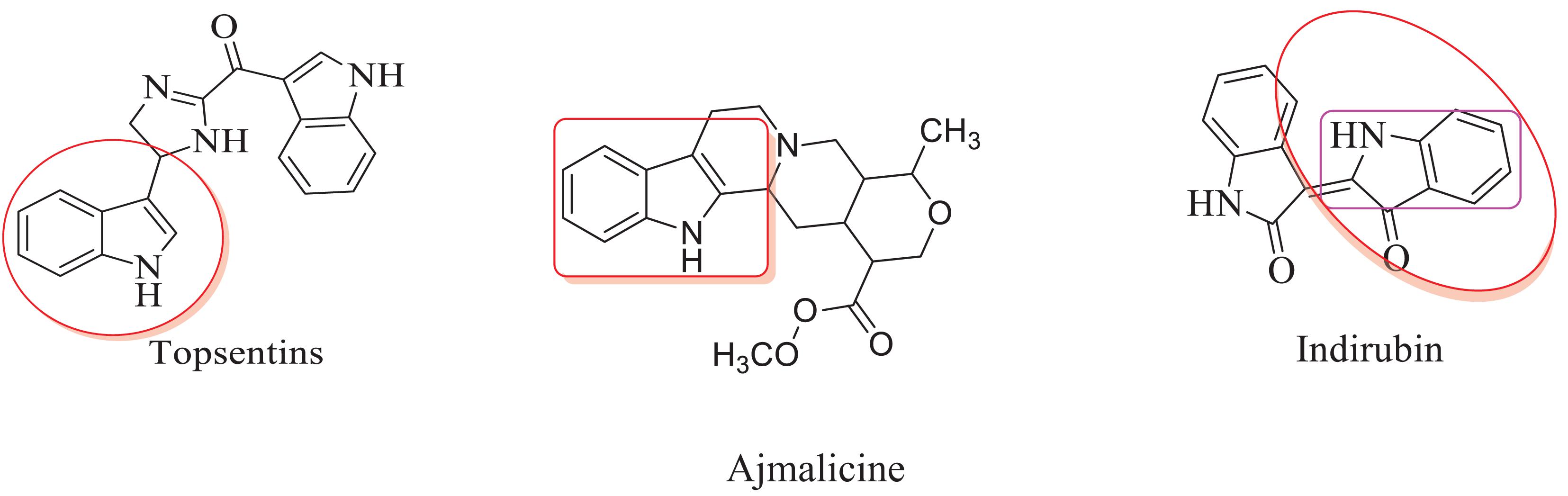
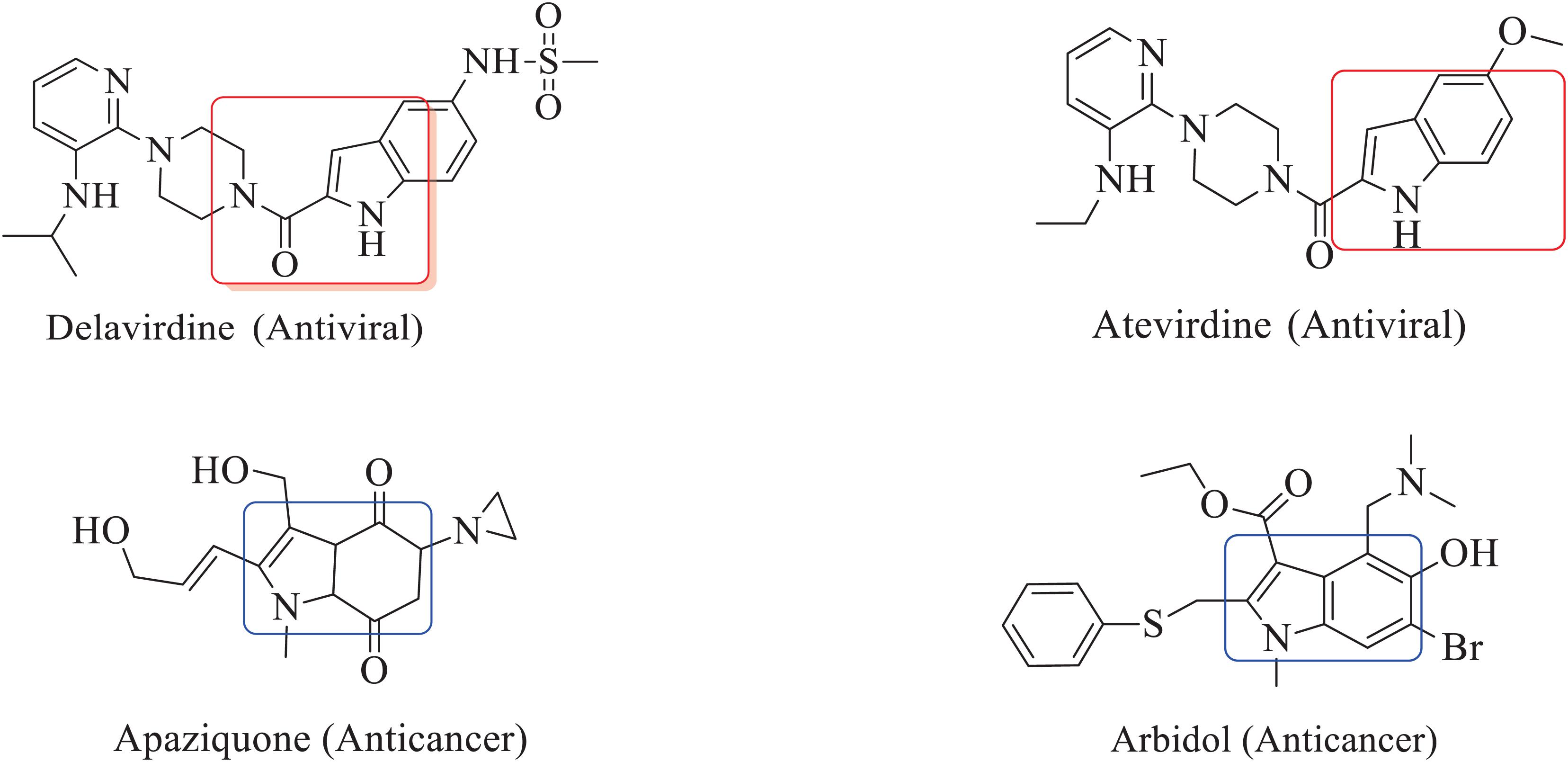
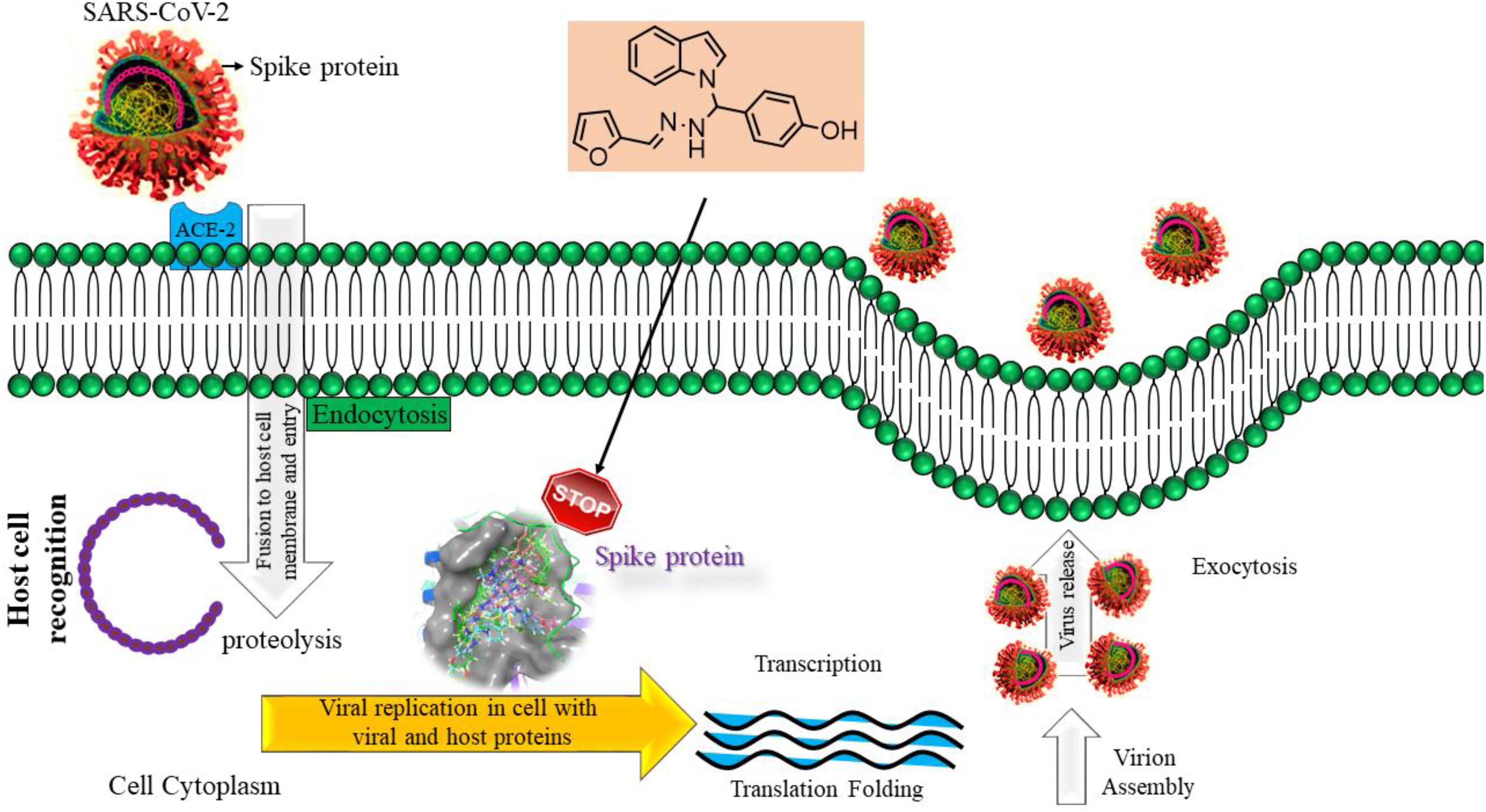
The inclusion of indole is the most significant structural modification in drug development, and it is labeled as one of the “privileged scaffolds” (Evans et al., 1988; deSa Alves et al., 2009; Welsch et al., 2010). The enlargement of a new technique for the pattern of C-N and C-C bonds that evade the pore functional group is tremendously significant in current organic chemistry (Ricci, 2008). Amino methylation is a crucial method for direct carbon–carbon and carbon-nitrogen bond-forming reactions (Hwang and Uang, 2002). Usually, amino methylation is done by the Mannich reaction using aldehyde as a methylene group source (Mannich and Krosche, 1912). Indole is perhaps the most ubiquitous motif in nature (Humphrey and Kuethe, 2006). Many natural and synthetic indole derivatives have been in great demand in medical and pharmaceutical applications since they can bind with high affinity to many receptors (Sundberg, 1970, 1996; Lounasmaa and Tolvanen, 2000; Horton et al., 2003; Gu and Hamann, 2005; Somei and Yamada, 2005; Shiri, 2012). Previously reported natural products of indole derivatives are shown in Figure 1 (Chen et al., 2019), and the biological activities of indole derivatives are offered in Figure 2 (Kumari and Singh, 2019). Indole regulates numerous aspects of microorganism physiology, including reproductive structure formation, body stability, resistance to medication, biofilm formation, and virulence (Chadha and Silakari, 2017). Based on the above properties, we prepared new indole derivatives 1a–1j via the Mannich reaction. As the indole compounds have been rigorously involved in ailments including viral infections and cancer, there exists a profound scope of exploring these multiple nuclei to curb coronaviruses (Zhang et al., 2015). Here we demonstrated that the indole moiety potently blocked the infectivity of SARS CoV-2 by targeting glycoproteins. They also potently block the enzymatic activity of SARS CoV-2 and replication of coronavirus (Hattori et al., 2021). Therefore, through this, indole derivatives developed against SARS-CoV-2 epidemics using in vitro and in silico approaches may be of immense value at this hour of global emergency and in the future.
Experimental
General
All the chemicals were purchased from Merck. The melting point was determined using an open capillary tube, and it is uncorrected. The IR spectra were recorded in KBr on a Shimadzu 8201pc (4000–400 cm–1). 1H and 13C-NMR spectra were recorded on Bruker Avance II NMR spectrometer 300 MHz with DMSO-d6 as solvent using tetramethylsilane (TMS) as an internal standard. Mass spectra were recorded using Clarus SQ8 (Perkin Elmer), and the elemental analysis (C, H, and N) was performed on a Varian EL III instrument.
General Procedure for the Synthesis of Compounds 1a–1j
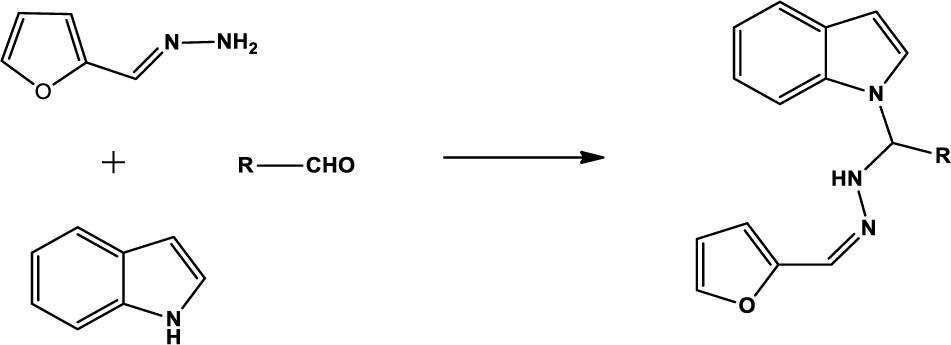
We compounded Furan-2-ylmethylenehydrzine (0.01 mol), indole (0.01 mol), and substituted aldehydes (0.01 mol) in ethanol solution to give a yellow solution with light brown color precipitate. The residue was recrystallized with ethanol. The obtained compound was purified by thin-layer chromatography (TLC). Hexane was used as eluting and solvent in TLC. All the synthesized compounds were separated by column chromatography.
(E)-1-((2furan-2ylmethylene)hydrazinyl)phenyl)methyl) 1H-indole (1a)
Light yellowish brown solid: mp 250°C; IR (KBr) (cm–1) 3440 (NH str), 3080 (CH-str Ar-ring), 1623 (C = N), 1092 (N-CH-N). 1H NMR (DMSO-d6), δ (ppm) J (Hz): 8.41 (s, 1H, CH = N), 7.99–7.79 (d, 2H, indole), 7.82–6.70 (m, 3H, furan), 7.40–6.95 (m, 9H, Ar), 7.08 (s, 1H, NH), 7.08 (s, 1H, CH); 13C NMR (DMSO-d6) δ(ppm): 150.46, 145.73, 118.96, 113.43 (4C, Furyl ring), 143.52, 128.57, 127.78, 126.95, 126.90, 125.25 (6C, Ph ring), 136.25, 126.62, 125.23, 121.69, 120.09, 118.23, 111.57, 110.76 (8C, indole ring), 135.23 (1C, C = N), 40.59 (1C, N-CH-N); EI-MS (Relative intensity %): m/z 315.14 (M++, 20); Elemental analysis: Anal.C20H17N3O: C, 76.20; H, 5.45; N, 13.35; Found C, 76.25; H, 5.55; N, 13.28.
1-((2furan-2ylmethylene)hydrazinyl)3-nitrophenyl) methyl)1H-indole (1b)
Light brown solid: mp 258°C; IR (KBr) (cm–1) 3435 (NH str), 3058 (CH-str Ar-ring), 1590 (NO2–), 1623 (C = N), 1094 (N-CH-N). 1H NMR (DMSO-d6), δ (ppm) J (Hz): 8.33–6.75 (m, 9H, Ar), 8.10 (s, 1H, CH = N), 7.95–7.82 (d, 2H, indole), 7.69–6.74 (m, 3H, furan), 7.09 (s, 1H, NH), 6.73 (s, 1H, CH); 13C NMR (DMSO-d6) δ (ppm): 150.62, 145.64, 118.96, 113.50 (4C, Furyl ring), 147.75, 135.85, 127.84, 126.69, 114.20, 114.12 (6C, Ph ring), 136.53, 126.62, 125.25, 121.62, 120.16, 118.25, 111.48, 110.70 (8C, indole ring), 135.27 (1C, C = N), 40.54 (1C, N-CH-N); EI-MS (Relative intensity %): m/z 360.12 (M+, 20); Elemental analysis: Anal.C20H16N4O3: C, 66.67; H, 4.49; N, 15.56; Found C, 66.62; H, 4.55; N, 15.48.
1-((2furan-2ylmethylene)hydrazinyl)1H-indole-1-yl)methyl)phenol (1c)
Brown solid: mp 272°C; IR (KBr) (cm–1) 3585 (OH), 3449 (NH str), 3086 (CH-str Ar-ring), 1629 (C = N), 1091 (N-CH-N). 1H NMR (DMSO-d6), δ (ppm) J (Hz): 10.24 (s, 1H, OH), 8.52 (s, 1H, CH = N), 7.95–7.52 (d, 2H, indole), 7.68–6.97 (m, 3H, furan), 7.42–6.72 (m, 9H, Ar), 7.06 (s, 1H, NH), 6.74 (s, 1H, CH); 13C NMR (DMSOd6) δ (ppm): 176.36 (1C, Ph-OH), 150.25, 145.24, 118.42, 113.47 (4C, Furyl ring), 142.18, 129.48, 129.50, 114.25, 114.22 (5C, Ph ring), 136.04, 127.89, 125.24, 121.67, 120.15, 118.29, 111.52, 110.76 (8C, indole ring), 135.29 (1C, C = N), 40.51 (1C, N-CH-N); EI-MS (Relative intensity %): m/z 331.13 (M+, 20); Elemental analysis: Anal.C20H17N3O2: C, 72.50; H, 5.18; N, 12.69; Found C, 72.45; H, 5.24; N, 12.65.
1-((4-chlorophenyl)2-furan-2ylmethylene)hydrazinyl) methyl)-1H-indole (1d)
Light brown solid: mp 260°C; IR (KBr) (cm–1) 3442 (NH str), 3082 (CH-str Ar-ring), 1626 (C = N), 1094 (N-CH-N), 818 (C-Cl). 1H NMR (DMSO-d6), δ (ppm) J (Hz): 8.11 (s, 1H, CH = N), 7.98–7.81 (d, 2H, indole), 7.84–6.74 (m, 3H, furan), 7.32–6.70 (m, 9H, Ar), 7.06 (s, 1H, NH), 6.74 (s, 1H, CH); 13C NMR (DMSO-d6) δ (ppm): 150.31, 145.35, 118.85, 113.40 (4C, Furyl ring), 136.10, 131.67, 130.50, 129.8, 129.59, 129.57 (6C, Ph ring), 136.25, 127.78, 125.26, 121.59, 120.10, 118.20, 111.59, 110.73 (8C, indole ring), 135.21 (1C, C = N), 40.50 (1C, N-CH-N); EI-MS (Relative intensity %): m/z 349.10 (M+, 20); Elemental analysis: Anal.C20H16ClN3O: C, 68.68; H, 4.62; N, 12.03; Found C, 68.65; H, 4.63; N, 12.05.
1-((2furan-2ylmethylene)hydrazinyl)4-methoxyphenyl) methyl)1H-indole (1e)
Light yellowish brown solid: mp 275°C; IR (KBr) (cm–1) 3444 (NH str), 3056 (CH-str Ar-ring), 2854 (OCH3), 1618 (C = N), 1089 (N-CH-N). 1H NMR (DMSO-d6), δ (ppm) J (Hz): 8.22 (s, 1H, CH = N), 7.96–7.86 (d, 2H, indole), 7.62–6.80 (m, 3H, furan), 7.47–6.69 (m, 9H, Ar), 7.09 (s, 1H, NH), 6.75 (s, 1H, CH). 3.86 (s, 3H, OCH3); 13C NMR (DMSO-d6) δ (ppm): 158.62 (1C, Ph, OCH3), 150.24, 145.62, 118.94, 113.48 (4C, Furyl ring), 136.51, 126.61, 125.27, 121.60, 120.18, 118.28, 111.50, 110.72 (8C, indole ring), 135.83, 127.82, 126.67, 114.18, 114.09 (5C, Ph ring), 135.29 (1C, C = N), 55.74 (1C, OCH3), 40.54 (1C, N-CH-N); EI-MS (Relative intensity %): m/z 345.15 (M+, 20); Elemental analysis: Anal.C21H19N3O2: C, 73.04; H, 5.55; N, 12.16; Found C, 70.59; H, 5.60; N, 12.30.
2-((2-furan-2-ylmethylene)hydrazinyl)(1H-indol-1-yl)methyl)phenol (1f)
Light brown solid; mp 265°C; IR (KBr) (cm–1) 3447 (NH str), 3089 (CH-str Ar-ring), 2915 (C-OCH3), 1628 (C = N), 1090 (N-CH-N). 1H NMR (DMSO-d6), δ (ppm) J (Hz): 9.82 (s, 1H, OH), 8.25 (s, 1H, CH = N), 8.22–6.83 (m, 9H, Ar), 7.97–7.81 (d, 2H, indole), 7.71–6.49 (m, 3H, furan), 7.19 (s, 1H, NH), 6.71 (s, 1H, CH); 13C NMR (DMSO-d6) δ (ppm): 148.90, 144.45, 119.08, 113.06 (4C, Furyl ring), 139.89 (1C, Ph, OH), 136.56, 126.79, 128.41, 121.07, 120.79, 119.82, 109.76, 100.24 (8C, indole ring), 131.01, 126.02, 120.42, 118.92, 116.05 (5C, Ph) 134.19 (1C, C = N), 40.49 (1C, N-CH-N); EI-MS (Relative intensity %): m/z 331.37 (M+, 20); Elemental analysis: Anal.C20H17N3O2: C, 72.50; H, 5.72; N, 12.69; Found C, 72.40; H, 5.51; N, 12.48.
4-((2-furan-2-ylmethylene)hydrazinyl)1H-indol-1-yl)methyl)-2-Methoxyphenol (1g)
Brown solid: mp 289°C; IR (KBr) (cm–1) 3442 (NH str), 3082 (CH-str Ar-ring), 2910 (C-OCH3), 1626 (C = N), 1094 (N-CH-N). 1H NMR (DMSO-d6), δ (ppm) J (Hz): 10.48 (s, 1H, OH), 8.93 (d, 1H, J = 6.1 Hz, Furyl ring), 8.16 (s, 1H, methylene), 7.92–7.80 (d, 2H, J = 7.6 Hz, indole), 7.87 (d, 1H, J = 6.7 Hz, Furyl ring), 7.20, 7.18, 6.84, 6.75 (m, 4H, indole ring), 7.09 (s, 1H, NH), 6.64 (t, 1H, Furyl ring), 6.65–6.74 (m, 2H, Ar-CH), 6.71 (s, 1H, CH), 3.85 (s, 3H, OCH3) 3.50 (s, 1H, CH); 13C NMR (DMSO-d6) δ (ppm): 150.21, 145.38, 118.82, 113.41 (4C, Furyl ring), 147.52, 146.82, 142.30, 126.37, 120.18, 115.39, 56.09 (7C, Ph ring), 136.15, 127.79, 125.23, 121.49, 120.00, 118.28, 111.52, 110.71 (8C, indole ring), 135.22 (1C, C = N), 40.51 (1C, N-CH-N); EI-MS (Relative intensity %): m/z 349.10 (M+, 20); Elemental analysis: Anal.C21H19N3O3: C, 69.75; H, 5.31; N, 11.66; Found C, 69.69; H, 5.20; N, 11.53.
4-((2-furan-2-ylmethylene)hydrazinyl)(1H-indol-1-yl)methyl)-N,N-dimethylaniline (1h)
Light brownish yellow solid; mp 270°C; IR (KBr) (cm–1) 3440 (NH str), 3085 (CH-str Ar-ring), 2946 (NH2), 1620 (C = N), 1096 (N-CH-N). 1H NMR (DMSO-d6), δ (ppm) J (Hz): 8.29 (s, 1H, CH = N), 7.93–7.83 (d, J = 7.0, 2H, indole), 7.72–6.54 (m, 3H, furan), 7.46–6.68 (m, 9H, Ar), 7.13 (s, 1H, NH), 6.76 (s, 1H, CH), 3.80–3.12 (s, 3H, N-CH3); 13C NMR (DMSO-d6) δ (ppm): 149.17, 128.13, 127.87, 127.80, 112.75, 112.72 (6C, Ph) 149.10, 144.43, 118.93, 112.67 (4C, Furyl ring), 136.57, 128.92, 128.51, 121.75, 120.77, 119.78, 109.57, 100.84, (8C, indole ring), 134.61 (1C, C = N), 41.29 (2C, N-CH3) 40.58 (1C, CH); EI-MS (Relative intensity %): m/z 331.37 (M+, 20); Elemental analysis: Anal.C22H22N4O: C, 73.42; H, 6.20; N, 15.63; Found C, 73.22; H, 6.09; N, 15.43.
2-(furan-2-ylmethylene)hydrazinyl)-3,7-dimethylocta-2,6-dien-1-yl)-1H-indole (1i)
Light brown solid; mp 280°C; IR (KBr) (cm–1) 3446 (NH str), 3080 (CH-str Ar-ring), 1620 (C = N), 1094 (N-CH-N). 1H NMR (DMSO-d6), δ (ppm) J (Hz): 8.30 (d, 1H, J = 6.2 Hz, furyl ring), 7.58–7.53 (d, 2H, J = 7.0, indole ring), 7.40, 7.31, 7.06, 6.58 (m, 4H, indole ring), 7.12 (s, 1H, NH), 6.90, 6.48 (m, 2H, Furyl ring), 6.68 (s, 1H, CH), 5.47, 5.30 (d, 2H, J = 7.7 Hz, citral), 2.00 (d, 2H, J = 6.3, citral), 2.04 (d, 2H, J = 6.0, citral), 1.82–1.71 (s, 9H, citral);13C NMR (DMSO-d6) δ (ppm): 149.98, 144.38, 118.97, 112.68 (4C, Furyl ring), 136.52, 128.91, 127.83, 121.68, 120.75, 119.81, 109.65, 100.95 (8C, indole ring), 135.52, 132.02, 123.52, 118.76, 39.45, 26.41, 24.61, 18.63, 16.10 (9C, citral) 134.62 (1C, C = N), 40.60 (1C, N-CH-N); EI-MS (Relative intensity %): m/z 361.22 (M+, 20); Elemental analysis: Anal.C23H27N3O: C, 76.43; H, 7.54; N, 11.63; Found C, 76.66; H, 7.43; N, 11.42.
1-((2-furan-2-ylmethylene)hydrazinyl)methyl)-1H-indol (1j)
Brown solid; mp 285°C; IR (KBr) (cm–1) 3442 (NH str), 3082 (CH-str Ar-ring), 1626 (C = N), 1092 (N-CH-N). 1H NMR (DMSO-d6), δ (ppm) J (Hz): 8.24 (s, 1H, CH = N), 7.78–6.59 (d, J = 6.2 Hz, 3H, Furan), 7.62–7.59 (m, 2H, indole), 7.47–6.45 (m, 9H, Ar), 7.16 (s, 1H, NH), 5.54 (d, J = 6.2Hz, 2H, CH2); 13C NMR (DMSO-d6) δ (ppm): 149.17, 145.02, 118.91, 112.62 (4C, Furyl ring), 136.49, 128.98, 127.89, 121.76, 120.72, 119.86, 109.67, 100.91 (8C, indole ring), 134.62 (1C, C = N), 40.59 (1C, CH2); EI-MS (Relative intensity %): m/z239.27 (M+, 20); Elemental analysis: Anal. C14H13N3O: C, 70.29; H, 5.49; N, 17.57; Found C, 70.35; H, 5.78; N, 17.88.
Biological Screening
Cytotoxic Activity
The cytotoxicity experiment was performed according to the United States NCI protocol, previously reported method. A detailed experimental procedure was given in Supplementary Material (Premnath et al., 2015).
Molecular Docking
Molecular docking was performed to confirm the molecular interaction with Covid-19 spike core protein to ensure the secondary biological mechanism based on the molecular pose on the binding moiety. The molecular structure of the selected ligand was drawn using Chem. Draw. Before it being considered for molecular interaction, it was 2D optimized by the energy minimization process. The 3D molecular protein crystal structure of spike glycoprotein of SARS-CoV-2 PDB ID 6WPT protein was downloaded. The protein structure was prepared using Schrodinger 12.4 software to remove water molecules and optimize the structure to become suitable to execute flexible docking. In protein preparation, hydrogen atoms were added to increase the hydrophilicity, and already existed co-crystal molecules, and missed loops were optimized (Boyd and Paull, 1995). The ligand preparation module optimized the ligand 3D structure of selected molecules to remove unwanted atomic orientation by molecular and quantum mechanics. Molecular docking, with flexible SP followed by XP, was executed. The grid-based technique, evaluation, and minimization of grid approximation procedure were followed by Premnath et al. (2016) and Muthiah et al. (2020). The confirmation of the best interactive molecule with 6WPT protein was concluded based on the G score and number of hydrogen bonds and bonding efficiency and binding energy.
Results and Discussion
Chemistry
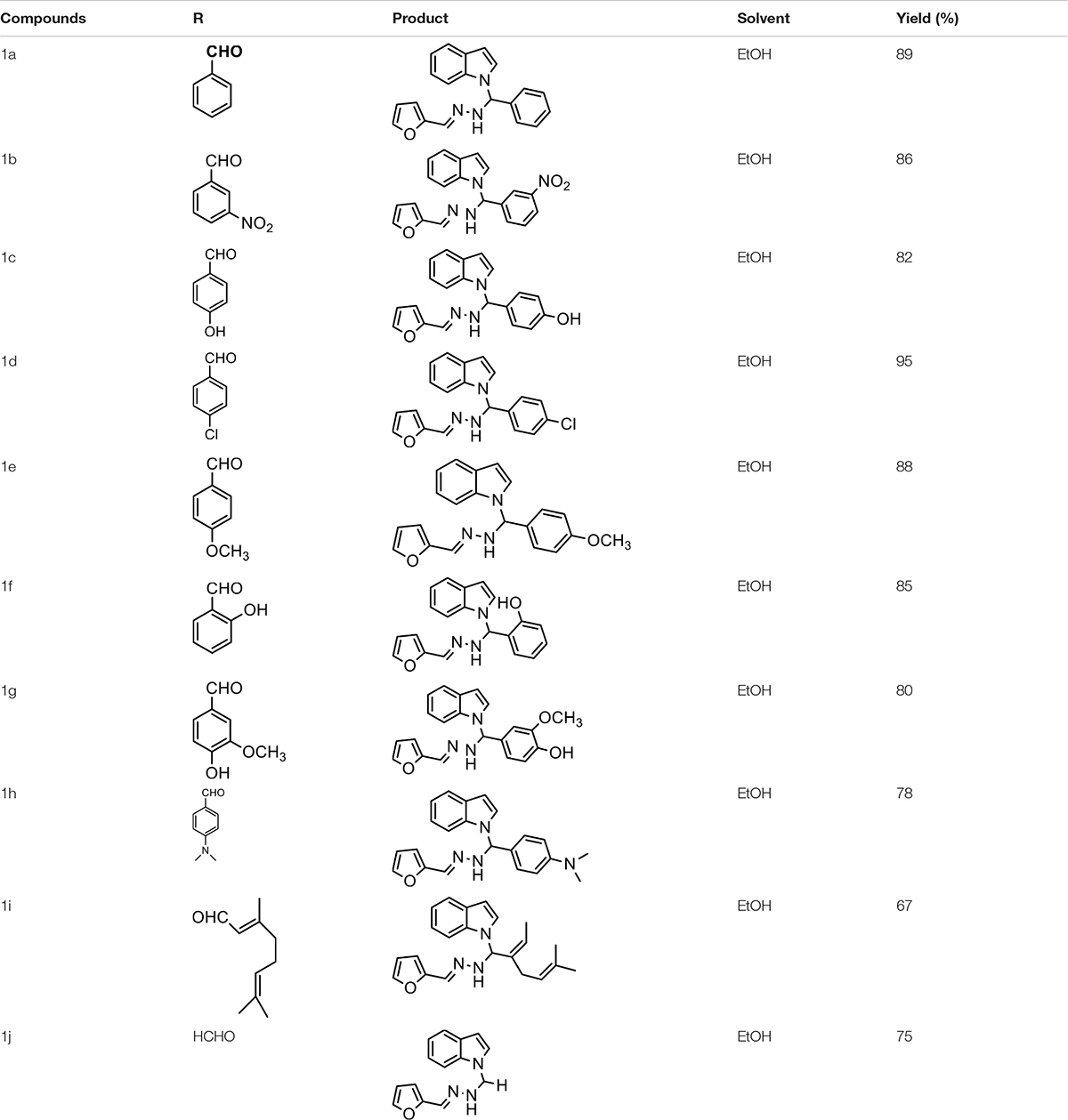
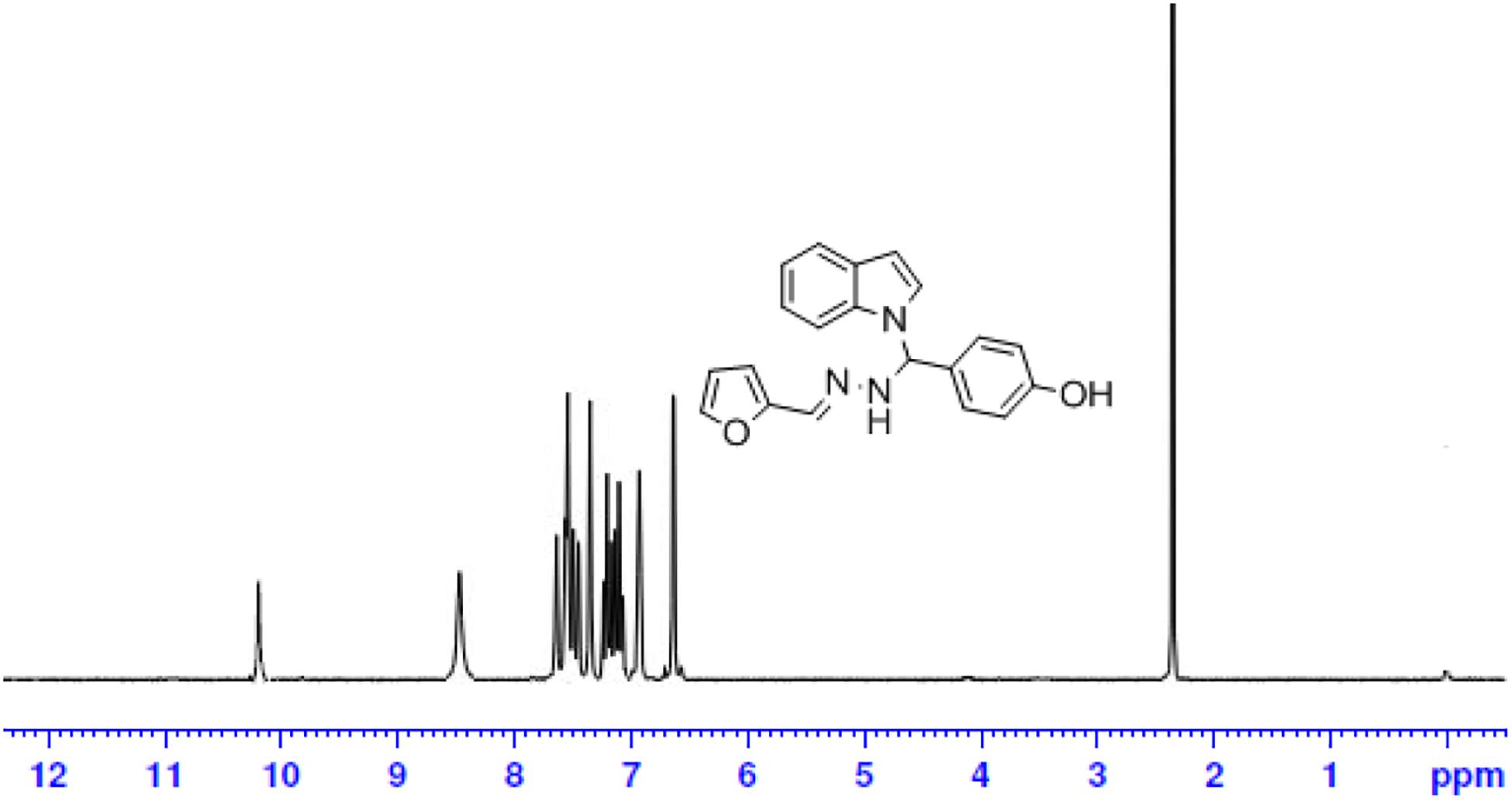
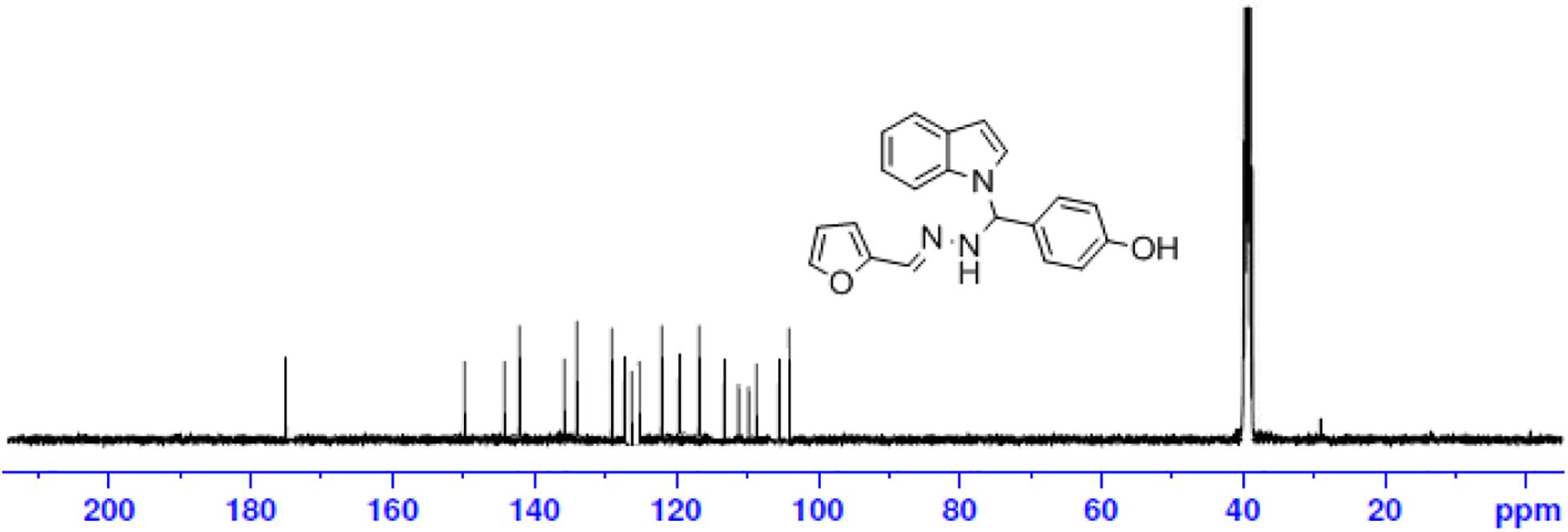
The one-pot Mannich reactions of substituted benzaldehyde, indole, and Furan-2-lymethylenehydrzine were done by reflux for 2 h using ethanol, a solvent, without any catalyst. The obtained solid 1-((2furan-2ylmethylene)hydrazinyl)phenyl)methyl)1H-indole (1a) was washed with cooled water and recrystallized using ethanol. It was purified by TLC. Hexane was used as an eluting solvent in TLC. All the synthesized compounds were separated by column chromatography. A similar procedure was carried out to synthesize the other nine compounds (1b–1j) Physicochemical data of synthesized compounds (1a–1j) are given in Table 1. The Figure 3 indicates 1H-NMR spectra of compound 1c, and Figure 4 displays 13C-NMR spectra of compound 1c. Scheme 1 represents the synthesis of compounds 1a–1j.
All the newly synthesized indole derivatives were characterized by FT-IR, which showed various functional groups. The 1H-NMR spectra of compounds (1a–1j) indicate frequency observed at 7.16–7.07 and 6.79–5.54, corresponding to the NH-CH and CH-Ph protons. The 13C -NMR spectra exhibit the peak at 144.42–118.76 and 40.60–40.53, corresponding to the NH-CH and CH-Ph carbon, respectively.
Cytotoxic Activity
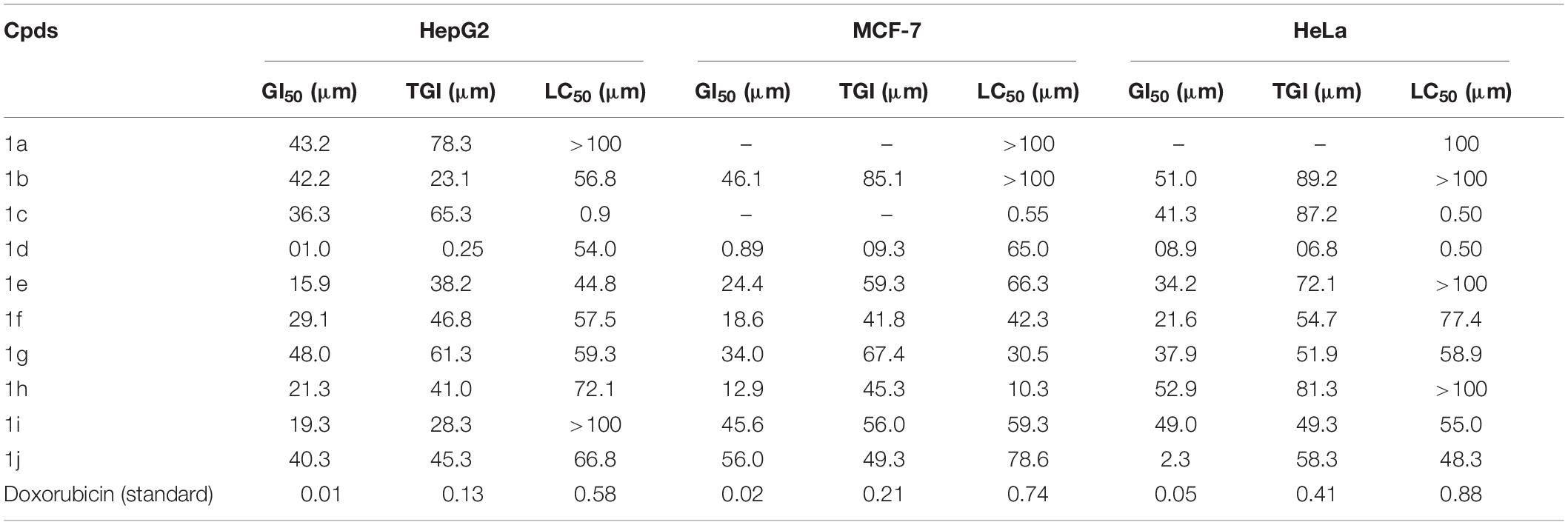
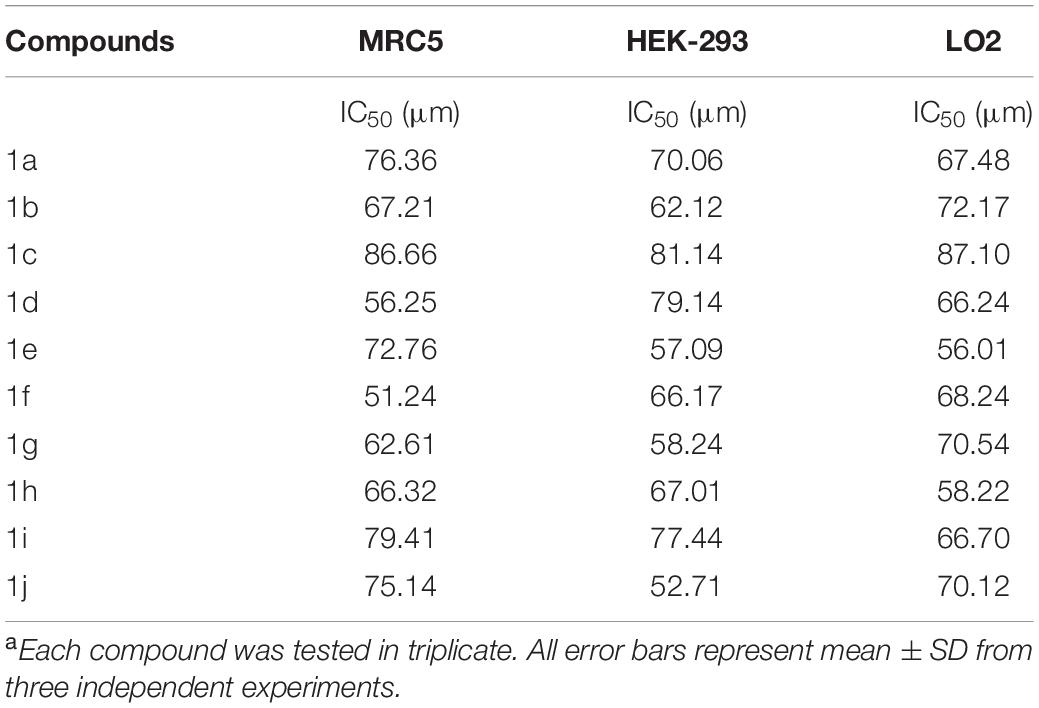
The newly prepared compounds 1a–1j are examined for their cytotoxic activity according to the United States NCI protocol, which was a previously reported method (Chadha and Silakari, 2017). The 50% growth inhibition (GI50), tumor growth inhibition (TGI), and lethal concentration (LC50) values were determined. The compounds 1c were a significant activity against (HepG2, LC50-0.9 μm, MCF-7, LC50-0.55 μm, HeLa, LC50-0.50 μm). Doxorubicin was used as a standard drug. None of the tested derivatives had shown significant activity toward the cancer cell lines. The compounds were also evaluated for their possible cytotoxicity in human embryonic kidney cells (HEK-293), lung cells (MRC-5), and liver cells (LO2) by employing MTT assay. The assay results suggested that these compounds did not significantly affect normal kidney cells’ growth (As most of the compound’s IC50 values are >100). Hence, these compounds revealed their safety for the normal cells, and the compound 1c can be taken as lead compounds for further development of more potent agents for HepG2 (Liver), MCF-7 (Breast), HeLa (Cervical) cancer cell lines. The results of cytotoxic screening of compounds (1a–1j) are shown in Table 2, and in vitro cytotoxicity of indole derivatives (1a–1j) on normal cells are shown in Table 3.
None of the tested derivatives had shown significant activity toward the cancer cell lines. The compounds were also evaluated for the possible cytotoxicity in human embryonic kidney cells (HEK-293), lung cells (MRC5), and liver cells (LO2) by employing MTT assay. The assay results suggested that these compounds did not significantly affect the growth of normal cells (as most of the compounds IC50 > 100). Hence this compounds revealed their safety for the normal cells and the compound 1c can be taken as lead compound for further development of more potential agent for HepG2 (Liver), MCF-7 (Breast), HeLa (Cervical) cancer cell lines, and in vitro cytotoxicity of indole derivatives (1a–1j) on normal cells are shown in Table 3.
Molecular Docking
PAT binds 6WPT with strong affinity via computer docking studies
Bimolecular interaction studies were used to characterize the interaction between selected drug-like molecule and protein biomolecular binding sites. The protein interaction study was executed to forecast the interactive visualization modes and binding of small molecule and their respective protein receptors. An investigation of the interactive molecular complex of ligand series disclosed very informative and important connections between the drug like molecular series and the (6WPT) protein receptor. The two-dimensional and three-dimensional protein molecular structural images were perfectly visualized using Schrodinger integrative python software to analyze the molecular interaction between the selective ligand series and protein macromolecule (6WPT) (Figure 5). The overall optimized G score for binding ligands is predicted as −2.808 kcal/mol. This is taken to indicate an expected favorable reaction. Ligand 3 (1c) were perfectly interacted and formed close molecular interactions with amino acid residues on the predicted selective binding sites of VAL367, LEU368, PHE342, GLY339, GLY112, ARG55, LEU47, ASN343, ASP115, TYR32 of receptor protein (6WPT) (XXX) during different biochemical communications of hydrogen bonding, and hydrophobic interaction (Pinto et al., 2020). The binding score of (1c) to 6WPT was moderately burly with a predictable affinity of −2.808 kcal/mol. The docking analysis characterization of novel synthesized molecules are shown in Table 4. Further, ligand series bonded with 6WPT through interactions with hydrogen bonding interaction and Pi-Pi interaction, Pi-Pi stacking interactive protein amino acids side chains of valine (Val), threonine (Thr), serine (Ser), alanine (Ala), and lysine (Lys) were analyzed and predicted important interactive bioactive binding site molecules. The molecular interaction analysis with selected small molecules of (1c) with 6WPT protein was much stronger than other series of selected ligands with a predictable affinity of −2.808 kcal/mol (Figure 5). The pathway mechanisms of spike protein interactions with highly active compound 1c are shown in Scheme 2. All the 2D structures of synthesized compounds are given in Supplementary Materials.
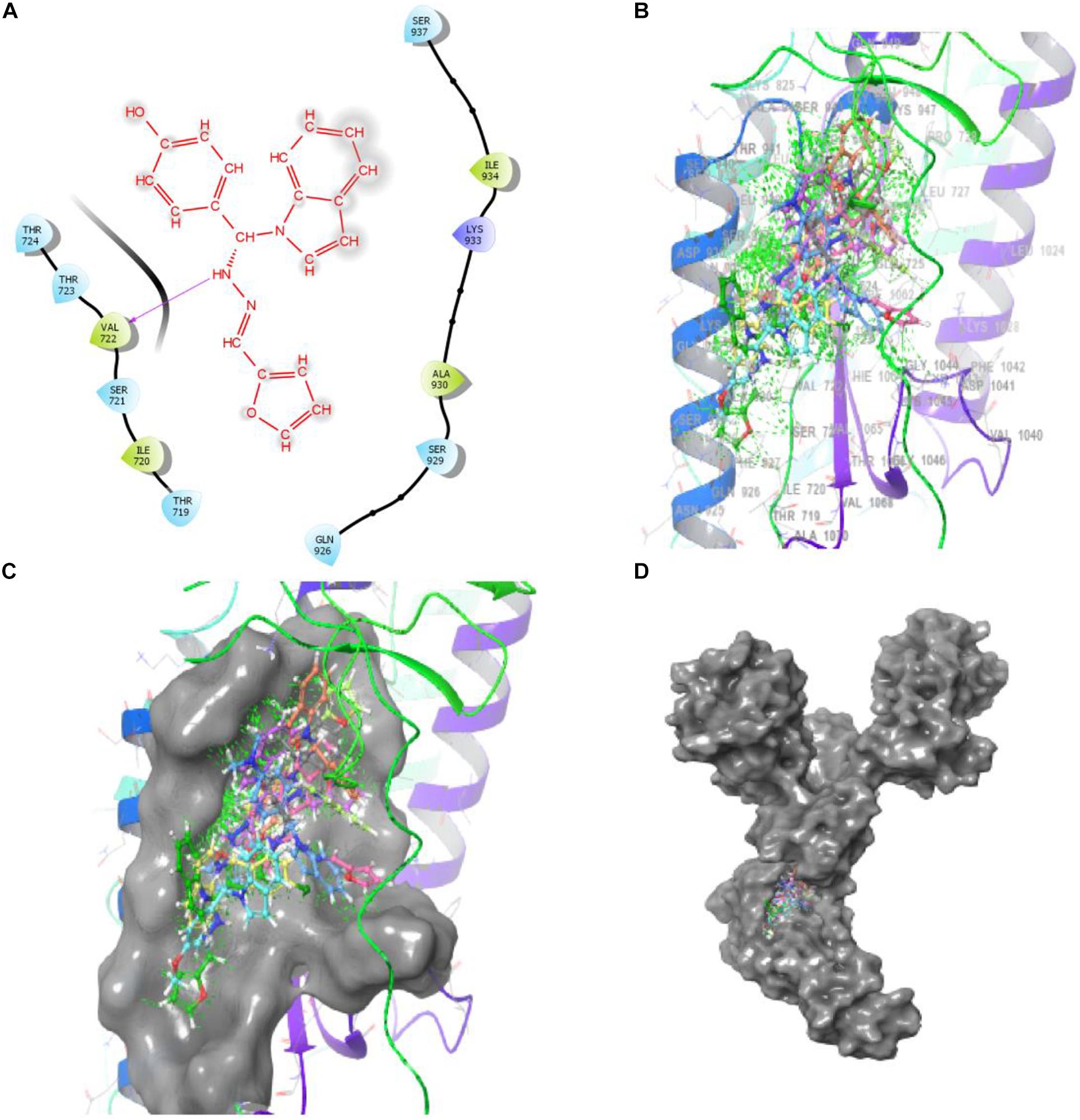
Figure 5. Compound-1c has a high affinity with 6WPT subunit. (B–D) Surface protein structure with hydrogen bonding interaction between (C) 6WPT interactive molecular pocket and ligand binding with series of ligands using a 3D molecular structure while panel (A) shows 2D binding pockets interactive sites.
Structure Activity Relationship
A structure-activity relationship analysis (SAR) was performed to find the link between the chemical structure of a dynamic molecule and its cytotoxic activity. SAR analysis makes it possible to identify the chemical group/atom that plays a critical function in modulating the cytotoxic activity of compounds within the specific system. Using the cytotoxic activity results of the indole Mannich base derivatives, preliminary SARs could be evaluated. The data of the selected indole Mannich base derivatives (1a–1j) showed that compound 1c is the most effective (HepG2, LC50-0.9 μm, MCF-7, LC50-0.55 μm, HeLa, LC50-0.50 μm) control doxorubicin.
Due to the presence of an indole ring fused to a hydroxyl benzaldehyde, it was found that the compound acquires a high cytotoxic activity against cancer cell lines. This was due to the presence of electron releasing hydroxyl group on phenyl ring attached with an indole skeleton. The rest of the compounds demonstrate feeble cytotoxic activity against all the tested cancer cell lines.
Moreover, from the docking results, it can be assumed that the docking score for indole derivatives (1a–1i) have an acceptable range except 1j compound along with essential interaction which can stabilize the compound in the active site of a protein. Compound 1j has no active site because of an absence of electron withdrawing/electron releasing group on it. From the results, the compound 1c has exhibited the highest docking score of −2.808 (Kcal/mol) compared to other compounds.
Conclusion
We have reported a facile, high-yielding, one-pot procedure for the synthesis of (1a–1j) via Mannich reaction using various kinds of protected aldehydes which was successfully employed and gave very high yields. Moreover, there were no requirements for dry solvents or protective gas atmospheres. All the newly synthesized compounds (1a–1j) were screened for in vivo cytotoxicity activities against Hep-G2 (Liver), HeLa (Cervical), and MCF-7 (Breast) cancer cell lines and normal cell lines in Human embryonic kidney cell (HEK293), liver cell (LO2), and lung cell (MRC5). Among the indole derivatives, compound 1c (HepG2, LC50-0.9 μm), (MCF-7, LC50-0.55 μm), and (HeLa, LC50-0.50 μm) was that the most active compound against the Doxorubicin standard. All other compounds were less active against the standard. The synthesized derivatives revealed a high safety level by exhibiting very low cytotoxicity against the normal cell line (HEK-293), (LO2), and (MRC5). Furthermore, we report in silico molecular docking studies against SARA-CoV-2 spike proteins and the biological characterization of the results reveal that compound 1c (−2.808 Kcal/mol) has the best multiple biological activities and can be used as a model for future derivatives based on the 1c molecular structure. It may identify the route to develop the best drug against Covid-19.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Author Contributions
PG: Organic compounds preparation; PP: Preparation of synthetic compound and chemical data analysis; KV: Manuscript editing; MA: Validation; MS: Molecular docking analysis; AI: All kinds of spectral analysis; RS: Investigation and writing original draft preparation through the contributions of all authors. All the authors contributed to the article and approved the submitted version.
Funding
MA thankfully acknowledges the Taif University researcher supporting project Number TURSP/91, Taif University, Taif, Saudi Arabia.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We wish to thank Nehru Memorial College (Affiliated Bharathidasan University), Puthanampatti, India, for taking GC-MS spectra and providing necessary facilities. We wish to thank Tamil Nadu Government for providing DST-FIST fund.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmolb.2021.637989/full#supplementary-material
References
Boyd, M. R., and Paull, K. D. (1995). Some practical considerations and applications of the national cancer institute in vitro anticancer drug discovery screen. Drug Dev. Res. 34, 91–109. doi: 10.1002/ddr.430340203
Cao, B., Wang, Y., Wen, D., Liu, W., Wang, J., Fan, G., et al. (2020). A trial of Lopinavir–Ritonavir in Adults hospitalizedwith severe Covid-19. N. Engl. J. Med. 382, 1787–1799.
Chadha, N., and Silakari, O. (2017). Indoles as therapeutics of interest in medicinal chemistry: bird’s eye view. Eur. J. Med. Chem. 134, 159–184.
Chan, J. F. W., Yuan, S., Kok, K. H., To, K. K. W., Chu, H., Yang, J., et al. (2020). A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person-to-person transmission: a study of a family cluster. Lancet 395, 514–523. doi: 10.1016/S0140-6736(20)30154-9
Chen, X. B., Xiong, S. L., Xie, Z. X., Wang, Y. C., and Liu, W. (2019). Three-component one-pot synthesis of highly functionalized bis-indole derivatives. ACS Omega. 4, 11832–11837. doi: 10.1021/acsomega.9b01159
Chen, Y., Liu, Q., and Guo, D. (2020). Emerging coronaviruses: genome structure, replication, and pathogenesis. J. Med. Virol. 92, 418–423. doi: 10.1002/jmv.25681
deSa Alves, F. R., Barreiro, E. J., and Fraga, C. A. (2009). From nature to drug discovery: the indole scaffold as a ‘privileged structure’. Mini Rev. Med. Chem. 9, 782–793. doi: 10.2174/138955709788452649
Evans, B. E., Rittle, K. E., Bock, M. G., DiPardo, R. M., Freidinger, R. M., and Whitter, W. L. (1988). Methods for drug discovery: development of potent, selective, orally effective cholecystokinin antagonists. J. Med. Chem. 31, 2235–2246. doi: 10.1021/jm00120a002
Gorbalenya, A. E., Baker, S. C., Baric, R. S., Groot, R. J., Drosten, C., Gulyaeva, A. A., et al. (2020). Severe acute respiratory syndrome-related coronavirus: the species and its viruses – a statement of the coronavirus study group. bioRxiv [preprint] doi: 10.1101/2020.02.07.937862
Gu, W., and Hamann, M. T. (2005). Indole alkaloid marine natural products: an established source of cancer drug leads with considerable promise for the control of parasitic, neurological and other diseases. Life Sci. 78, 442–453. doi: 10.1016/j.lfs.2005.09.007
Hattori, S., Higashi-Kuwata, N., Hayashi, H., Allu, S. R., Raghavaiah, J., Bulut, H., et al. (2021). A small molecule compound with an indole moiety inhibits the main protease of SARS-CoV-2 and blocks virus replication. Nat. Commun. 12:668.
Holshue, M. L., DeBolt, C., Lindquist, S., Lofy, K. H., Wiesman, J., Bruce, H., et al. (2020). First case of 2019 novel coronavirus in the United States. N. Engl. J. Med. 382, 929–936. doi: 10.1056/NEJMoa2001191
Horton, D. A., Bourne, G., and Smythe, T. (2003). The combinatorial synthesis of bicyclic privileged structures or privileged substructures. Chem. Rev. 103, 893–930. doi: 10.1021/cr020033s
Humphrey, G. R., and Kuethe, T. J. (2006). Practical methodologies for the synthesis of indoles. Chem. Rev. 106, 2875–2911. doi: 10.1021/cr0505270
Hussain, S., Pan, J., Chen, Y., Yang, Y., Xu, J., Peng, Y., et al. (2005). Identification of novel subgenomic RNAs and noncanonical transcription initiation signals of severe acute respiratory syndrome coronavirus. J. Virol. 79, 5288–5295. doi: 10.1128/JVI.79.9.5288-5295.2005
Hwang, D. R., and Uang, B. J. (2002). A modified Mannich-type reaction catalyzed by VO(acac)(2). Org. Lett. 4, 463–466. doi: 10.1021/ol017229j
Kumari, A., and Singh, R. K. (2019). Medicinal chemistry of indole derivatives: current to future therapeutic prospectives. Bioorganic Chem. 89:103021. doi: 10.1016/j.bioorg.2019.103021
Li, Q. M., Xuhua, G., Peng, W., Xiaoye, W., Lei, Z., Yeqing, T., et al. (2020). Early transmission dynamics in wuhan, China, of novel coronavirus–infected Pneumonia. N. Engl. J. Med. 382, 1199–1207. doi: 10.1056/NEJMoa2001316
Lounasmaa, M., and Tolvanen, A. (2000). Simple indole alkaloids and those with a non-rearranged monoterpenoid unit. Nat. Prod. Rep. 17, 175–191. doi: 10.1039/A809402K
Lu, R., Zhao, X., Li, J., and Niu, P. (2020). Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet 395, 565–574. doi: 10.1016/S0140-6736(20)30251-8
Mannich, C., and Krosche, W. (1912). Uebere in Kondensations produktaus Formaldehyd, Ammoniakund Antipyrin. Arch. Pharm. 250:647. doi: 10.1002/ardp.19122500151
Muthiah, I., Karthikeyan, R., Dhanaraj, P., and Vallinayagam, S. (2020). In silico structure prediction, molecular docking and dynamic simulation studies on G Protein-Coupled Receptor 116: a novel insight into breast cancer therapy. J. Biomol. Structure Dynamics [Online ahead of print] doi: 10.1080/07391102.2020.1783365
Pinto, D., Park, Y. J., Beltramello, M., Walls, A. C., Tortorici, M. A., Bianchi, S., et al. (2020). Structural and functional analysis of a potent sarbecovirus neutralizing antibody. BioRxiv [preprint] doi: 10.1101/2020.04.07.023903
Premnath, D., Enoch, I. V. M. V., Selvalumar, P. M., Indiraleka, M., and Vennila, J. J. (2016). Design, synthesis, spectral analysis, in vitro anticancer evaluation and molecular docking studies of some fluorescent 4-Amino-2, 3-Dimethyl-1-Phenyl-3-Pyrazolin-5-One, ampyrone derivatives. Interdiscip. Sci.: Computational Life Sci. 9, 130–139.
Premnath, D., Selvakumar, P. M., Ravichandiran, P., Tamil Selvan, G., Indiraleka, M., and Jannet Vennila, J. (2015). Synthesis and spectroscopic characterization of fluorescent 4-amino anti pyrine analogues: molecular docking and invitro cytotoxicity studies. Spectrochimica Acta Part A: Mol. Biomol. Spectroscopy 153, 118–123.
Rahman, N., Basharat, Z., Yousuf, M., Castaldo, G., Rastrelli, L., and Khan, H. (2020). Virtual screening of natural products against type II transmembrane serine protease (TMPRSS2), the priming agent of coronavirus 2 (SARS-CoV-2). Molecules 25:2271. doi: 10.3390/molecules2510227
Ramajayam, R., Tan, K. P., and Liang, P. H. (2011). Recent development of 3C and 3CL protease inhibitors for anti-coronavirus and anti-picornavirus drug discovery. Biochem. Soc. Trans. 39, 1371–1375. doi: 10.1042/BST0391371
Ren, Z., Yan, L., and Zhang, N. (2013). The newly emerged SARS-Like coronavirus HCoV-EMC also has an “Achilles’ heel”: current effective inhibitor targeting a 3C-likeprotease. Protein Cell 4, 248–250. doi: 10.1007/s13238-013-2841-3
Shiri, M. (2012). Indoles in multicomponent processes (MCPs). Chem. Rev. 112, 3508–3549. doi: 10.1021/cr2003954
Somei, M., and Yamada, F. (2005). Simple indole alkaloids and those with a non-rearranged monoterpenoid unit. Nat. Prod. Rep. 22, 73–103. doi: 10.1039/b316241a
Sundberg, R. J. (1996). Indoles, academic press, San Diego. J. Med. Chem. 39:5286. doi: 10.1021/jm960712t
Wang, M., Cao, R., and Zhang, L. (2020). Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Res. 30, 269–271. doi: 10.1038/s41422-020-0282-0
Welsch, M. E., Snyder, S. A., and Stockwell, B. R. (2010). Privileged scaffolds for library Design and drug discovery. Curr. Opin. Chem. Biol. 14, 347–361. doi: 10.1016/j.cbpa.2010.02.018
World Health Organization [WHO] (2020). WHO Director-General’s opening remarks at the media briefing on COVID-19-11 March 2020. Geneva: World Health Organization.
Wu, F., Zhao, S., and Yu, B. (2020). A new coronavirus associated with human respiratory disease in China. Nature 579, 265–269. doi: 10.1038/s41586-020-2008-3
Zhang, M.-Z., Chen, Q., and Yang, G.-F. (2015). A review on recent developments of indole-containing antiviral agents. Eur. J. Med. Chem. 89, 421–441. doi: 10.1016/j.ejmech.2014.10.065
Zhou, P., Yang, X. L., and Wang, X. G. (2020). A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 579, 270–273.
Keywords: indole, Mannich base, cytotoxic activity, COVID-19, spike protein
Citation: Gobinath P, Packialakshmi P, Vijayakumar K, Abdellattif MH, Shahbaaz M, Idhayadhulla A and Surendrakumar R (2021) Synthesis and Cytotoxic Activity of Novel Indole Derivatives and Their in silico Screening on Spike Glycoprotein of SARS-CoV-2. Front. Mol. Biosci. 8:637989. doi: 10.3389/fmolb.2021.637989
Received: 04 December 2020; Accepted: 26 March 2021;
Published: 11 May 2021.
Edited by:
Agnel Praveen Joseph, Science and Technology Facilities Council, United KingdomReviewed by:
Sayan Chatterjee, University School of Biotechnology, Guru Gobind Singh Indraprastha University, IndiaEliza Wyszko, Institute of Bioorganic Chemistry (PAS), Poland
Copyright © 2021 Gobinath, Packialakshmi, Vijayakumar, Abdellattif, Shahbaaz, Idhayadhulla and Surendrakumar. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Radhakrishnan Surendrakumar, c3VyZW5kcmFrdW1hckBubWMuYWMuaW4=
 Perumal Gobinath1
Perumal Gobinath1 Radhakrishnan Surendrakumar
Radhakrishnan Surendrakumar