- 1Department of Bioinformatics, Alagappa University, Karaikudi, India
- 2Department of Animal Science, School of Life Sciences, Bharathidasan University, Tiruchirappalli, India
- 3Department of Biotechnology, Alagappa University, Karaikudi, India
SARS-CoV-2, an etiological agent of COVID-19, has been the reason for the unexpected global pandemic, causing severe mortality and imposing devastative effects on public health. Despite extensive research work put forward by scientist around globe, so far, no suitable drug or vaccine (safe, affordable, and efficacious) has been identified to treat SARS-CoV-2. As an alternative way of improvising the COVID-19 treatment strategy, that is, strengthening of host immune system, a great deal of attention has been given to phytocompounds from medicinal herbs worldwide. In a similar fashion, the present study deliberately focuses on the phytochemicals of three Indian herbal medicinal plants viz., Mentha arvensis, Coriandrum sativum, and Ocimum sanctum for their efficacy to target well-recognized viral receptor protein through molecular docking and dynamic analyses. Nucleocapsid phosphoprotein (N) of SARS-CoV-2, being a pivotal player in replication, transcription, and viral genome assembly, has been recognized as one of the most attractive viral receptor protein targets for controlling the viral multiplication in the host. Out of 127 phytochemicals screened, nine (linarin, eudesmol, cadinene, geranyl acetate, alpha-thujene, germacrene A, kaempferol-3-O-glucuronide, kaempferide, and baicalin) were found to be phenomenal in terms of exhibiting high binding affinity toward the catalytic pocket of target N-protein. Further, the ADMET prediction analysis unveiled the non-tumorigenic, noncarcinogenic, nontoxic, non-mutagenic, and nonreproductive nature of the identified bioactive molecules. Furthermore, the data of molecular dynamic simulation validated the conformational and dynamic stability of the docked complexes. Concomitantly, the data of the present study validated the anti-COVID efficacy of the bioactives from selected medicinal plants of Indian origin.
Introduction
Although a year has been completed since the unprecedented emergence of severe acute respiratory syndrome coronavirus (SARS-CoV-2), the pandemic menace prevails till date. The World Health Organization’s (WHO) Day-to-Day data till 4th February 2021 state that the morbidity rate of “coronavirus disease 2019” (COVID-19) across 220 countries is crossing 102.9 (102, 942, 987) million, and the rate of mortality is 2.29 (2,232,233 deaths). The ever-increasing infected victims as well as mortality rate alarms the dire need for early diagnosis and identification of a drug or vaccine to treat COVID patients. Owing to the lack of therapeutic choices, the WHO announces COVID-19 as a “public health emergency of international concern (PHEIC),” implying that this pandemic seeks orchestrated global action in all clinical aspects (de Wit et al., 2016; Wu C. et al., 2020a).
SARS-CoV-2 is a positive-sense single-stranded (+ss) RNA virus that belongs to the family Coronaviridae and genera Betacoronavirus. It infects a wide range of host, including human, cattle, pigs, cats, and birds. Particularly, in human, it causes various symptoms, from mild respiratory infections, fever, dyspnea, lung lesions (Li et al., 2005; Killerby et al., 2018), and enteric disease to severe life-threatening pneumonia (de Wit et al., 2016; Killerby et al., 2018; Phan et al., 2020a; Huang et al., 2020; Li et al., 2020; Parry, 2020; Riou and Althaus, 2020). As far as strategies of SARS-CoV-2 spread are concerned, the significant virulence traits including immune evasion, replication inside host, and transmission from human-to-human are the major barriers for clinicians and other healthcare workers to treat and prevent COVID-19 (Chan et al., 2020; Li et al., 2020; Prompetchara, et al., 2020).
In general, the viral replication inside the host cell involves the synthesis of proteins, namely, envelope (E), membrane/matrix (M), spike (S), and nucelocapsid phosphoprotein (N) (Brian and Baric 2005). Accordingly, in the recent days, the E, M, S, and N proteins have been targeted for antiviral drug and vaccine designing investigations. However, any mutation in the outer membrane proteins viz., S, E, and M proteins aid SARS-CoV-2 to gain drug resistance (Benvenuoto et al., 2020; Phan 2020b; Dawood, 2020; Ou et al., 2020; Pachetti et al., 2020; Yin, 2020). Therefore, the N protein has particularly been considered as an attractive drug target (Wu F. et al., 2020b; Zhou et al., 2020).
The N protein of SARS-CoV-2 is a multifunctional protein chiefly involved in viral replication (Chang et al., 2014), virulence, immunogenicity (Burbelo et al., 2020; Randad et al., 2020), and pathogenesis (Yasui et al., 2008; Gao et al., 2020). The C-terminal domain (CTD) of the N protein binds with the M protein through dimerization and creates a physical link between the viral genome and its envelope, which thereby forms the helical ribonucleoprotein (RNP) complex. This complex not only renders protection to viral genome but also modulates the host intracellular machinery, and consistently plays a regulatory role throughout the viral life cycle (Masters et al., 1990; Narayanan et al., 2003; Chang et al., 2009). Earlier studies have robustly demonstrated the contribution of the N protein in host–pathogen interactions by regulating host cell cycle, apoptosis, and actin reorganization (Hsieh et al., 2005; Surjit et al., 2006). In addition, the viral N protein inhibits interferon-β and thus facilitates SARS-CoV-2 to evade the host immune response (Kopecky-Bromberg et al., 2000; Lu et al., 2011). Therefore, such a protein that majorly contributes to the viral replication and immune evasion could be a promising target to develop therapeutic countermeasures in controlling SARS-CoV-2 and infection-mediated further havoc.
As traced to antique Indian traditional medicinal system, the consumption of plant and plant-derived natural products has shown efficient therapeutic effects against various health ailments (Alagu Lakshmi et al., 2020; Muthuramalingam et al., 2020; Vellingiri et al., 2020; Gowrishankar et al., 2021). Notably, consumption of herbal plants has been a well-recognized home remedy for common cold (Alagu Lakshmi et al., 2020; Muthuramalingam et al., 2020; Vellingiri et al., 2020; Gowrishankar et al., 2021). Against common cold, a wide spectrum of herbs with proven medicinal benefits have been used in the traditional home remedy that reinforce the immune system (Lin et al., 2014; Wang and Liu 2014; Ganjhu et al., 2015). Based on this, three herbal plants viz., Mentha arvensis, Coriandrum sativum, and Ocimum sanctum were considered in the present study.
Mentha arvensis L., an aromatic plant popularly known as menthol mint and kitchen herb (in India; The wealth of India, 2003), holds not only medicinal values but also clutches varied industrial applications viz., flavorings, food, confectionary, cosmetic, perfumery, and pharmaceutics (Kumar et al., 2012; Lal et al., 2020). M. arvensis is a natural antioxidant (Kumar and Chattopadhyay, 2007), and it has been reported to exhibit antimycotic efficacy (Yadav et al., 2006). The mint leave juice also displayed diversified health benefits as it has been administered against liver and spleen disease, diarrhea, dysentery, indigestion, asthma, and jaundice. It has been a traditional remedy for rheumatic pains, arthritis, and inflamed joints (Salin et al., 2011; de Sousa Guedes et al., 2016; de Sousa Guedes and Souza, 2017). C. sativum, exhibits myriad pharmaceutical potentials (viz., antidiabetic, antiseptic, anti-inflammatory, antihypertensive, anxiolytic, antimicrobial, anti-cancerous, antimutagenic, diuretic, cognition improvement, and antioxidant) (Hussain et al., 2018; Kačániová et al., 2020), and its inhibitory efficacy against angiotensin-converting enzyme has been envisaged as the most significant action mechanism against COVID-19 (Khan and Kumar, 2019; Wei et al., 2019; Vellingiri et al., 2020). O. sanctum (tulsi) has been the holy herb with innumerable medicinal/health benefits, deployed since ancient period. It has been well demonstrated for multifaceted therapeutic propensity viz., anti-inflammatory, antidiabetic, immunomodulatory, antifertility, anticancer, cardio and hepatoprotective, antiviral, antifungal, and antibacterial efficacies (Seth and Sharma, 2004; Prakash and Gupta, 2005; Mallikarjun et al., 2016; Yamani et al., 2016; Jamshidi and Cohen, 2017; Mousavi et al., 2018). Most importantly, tulsi leaves have been proven to show beneficial effects against bronchitis and pyrexia through boosting/strengthening cellular as well as humoral immune responses (Mukherjee et al., 2005). Although a few reports have documented the plausible anti-COVID efficacy of O. sanctum as it targets the main protease of SARS-CoV-2, the efficacy of three selected plants against SARS-CoV-2 N protein has not been explored so far. Therefore, in the current investigation, the phytochemicals of these herbal plants (M. arvensis, C. sativum, and O. sanctum) were analyzed for their potential to inhibit the N protein through an in silico approach.
Materials and Methods
Protein Selection and Active Site Prediction
The crystal structure of the SARS-CoV-2 nucleocapsid phosphoprotein essential for virion formation and replication (PDB ID: 6ZCO; 1.36 Å) was retrieved from the RCSB Protein Databank (https://www.rcsb.org/). Based on the resolution (1.36 Å), stable atomic orientation, and CTD of the N protein crystal structure, 6ZCO was selected in the present study. All crystalline water molecules and bound ligand molecules were removed, and the polar hydrogen and Gasteiger charges to protein structures for the docking simulation were also assigned, as described earlier by Afriza et al. (2018).
Ligand Selection and Preparation
The chemical structure of phytocompounds was obtained from the Pubchem database (https://pubchem.ncbi.nlm.nih.gov/) in .sdf (structure date file) format. Then the file format was converted to PDB (Protein Data Bank) coordinate file format using the Open Babel (http://openbabel.org) (Rolta et al., 2020).
Molecular Docking
Molecular docking studies were conducted using AutoDock Vina in order to predict the accuracy of binding affinity as well as ligand-binding poses into protein active sites. Initially, both the ligand and receptor were preprocessed by adding the hydrogen, to assign the charge particle, and to remove the unwanted water molecules and heteroatom, and file format conversion was done by AutoDockTools. Then the grid map was defined to the active site (Ala264, Val270, Phe274, Arg277, Gla281, Phe286, and Gly284) of the receptor, and the grid box dimension was set as 20 × 20 × 20. The default scoring function of AutoDock Vina was used to calculate the docking score, and the lowest binding energy docking poses were selected for further interaction analysis as described in an earlier study by Chen et al. (2018). Discover Studio 3.5 is used to analyze the binding pose 2D and 3D interaction analysis of the protein–ligand complexes (Shivanika et al., 2020).
ADME Prediction
The top hit compounds obtained through molecular docking studies were further screened based on their ADME (absorption, distribution, metabolism, and excretion) properties, physicochemical properties (Lipinski’s rule of five principles), pharmacokinetics (Pks), and drug-likeness properties using the Molinspiration and AdmetSAR servers (Isa et al., 2018).
Molecular Dynamic Simulation
The obtained docking results of the best docked complexes were further subjected to molecular dynamic (MD) simulation using the GROMACS 4.5.5 package with the GROMOS53a6 force field for all atoms to get a protein topology parameter. The PRODRG web server was used to analyze the topology and force filed parameter of the ligand (Zheng et al., 2014). The protein–ligand complexes were solvated in a cubic box with the water model of SPC216 and neutralized by adding -Cl counter ions. Then the energy minimization of the system was performed by using the steepest descent algorithm. To equilibrate the system with constant volume and temperature from 300 K for 100 ps, NVT ensembles followed by the NPT ensembling at a constant temperature and constant pressure for 300 K for 1 bar. Finally, MD simulations were conducted for 50 ns (Ul Qamar et al., 2019). Root mean square deviation (RMSD), hydrogen bond analysis, radius of gyration (Rg), potential energy, root mean square fluctuations (RMSFs), secondary structure analyses, and SASA were done using GROMACS. XM Grace software was used to analyze the plot of RMSD, RMSFs, hydrogen bonds, etc. (Muthumanickam et al., 2020).
Results and Discussion
Given the prominence that boosting/strengthening the immune status of an individual would be a convincing alternative to prevent COVID infectivity, we deliberately investigated three selective medicinal herbs (viz., Mentha arvensis, Coriandrum sativum, and Ocimum sanctum) against one of the most important structural proteins named nucleocapsid phosphoprotein, which is a least variable and highly conserved structure of CoV (Lin et al., 2014). Indian traditional knowledge system has a historical background with proof of concept toward curing effects against common cold. In view of that, during this COVID-19 pandemic, Ministry of AYUSH, Government of India has identified and listed diverse medicinal shrubs and herbs employed in-house as home remedies with proven efficacy to strengthen the respiratory tracks and immune system.6 Most evidently, the WHO has estimated that nearly 80% of the population in underdeveloped countries depend chiefly on traditional medicines against COVID-19. On the view of tradition-based phyto-immune boosters, the WHO has enlisted nearly 21,000 global plants of therapeutic potential; among which, around 2,500 varieties were of Indian origin.33
Indian traditional system strongly relies on the quote “Food as Medicine,” and the other armors such as balanced diet and proper physical exercise further immunize the system. Based on these Indian naturopathic values, the patients affected with respiration illness were recommended for herbal steam inhalation therapy to subset the symptoms (Amini et al., 2017; Singh et al., 2017). In substantiation with the current study, earlier reports by Alagu Lakshmi et al. (2020) had signified the scientific merit toward deploying complementary herbal medicines against COVID-19. Similarly, in an earlier report by Singh et al. (2017), they denoted the speedy improvement of patients infected with common cold viral infection upon neti treatment along with vitamins as well as minerals (Amini et al., 2017; Singh et al., 2017). In an earlier study by our group, we have demonstrated the promising effects of phytochemicals from traditional Indian herbal steam inhalation therapy against COVID-19 through an in silico approach (Gowrishankar et al., 2021). Therefore, it is anticipated that phytochemicals might possibly set forth an initial developmental step for combinatorial naturopathic therapy either as an antiviral agent or as an immune booster in order to effectively manage COVID-19.
Unlike the other studies that target different structural proteins of SARS-CoV-2, in the present study, we chose a most important viral structural protein, nucleocapsid phosphoprotein, as it is highly abundant and least variable as well as highly conserved in CoV (Lin et al., 2014). The three domains of the N protein holds three different roles viz., N-terminal binds RNA, C-terminal aids in oligomerization, and central Ser/Arg rich linker helps in phosphorylation reactions (Chang et al., 2013; Yadav et al., 2020). This strong binding of the N protein with the RNA genome creates ribonucleoprotein complex, which exclusively triggers the production of virion core and RNA-dependent RNA synthesis for replication of virus (McBride et al., 2014; Cong et al., 2020). In addition to it, N-proteins have been investigated to uphold regulatory role during infection with host, starting from actin filament reorganization to apoptosis (Surjit et al., 2006; Du et al., 2008). As the N protein involves in the replication, transcription, and viral genome assembly, it could be an attractive drug target of SARS-CoV-2 in controlling the viral multiplication in host (Yadav et al., 2020). In par with the current study, a very recent study by Yadav et al. (2020) has emphasized that the inhibition of the N protein would be a convincing approach in treating the viral disease progression. Therefore, in the present study, we intentionally made an effort to virtually substantiate the antiviral efficacy of three AYUSH, GoI enlisted immune booster Indian herbs’ (viz., Mentha arvensis, Coriandrum sativum, and Ocimum sanctum) associated compounds against the N protein of SARS-CoV-2 through an in silico approach.
Molecular Docking Studies
In order to identify the potential drug candidates for managing COVID-19, molecular docking analysis was performed for 127 phytoconstituents from three selected medicinal plants against the active site of SARS-CoV-2 RNA binding domain of nucleocapsid phosphoprotein. The results revealed that most of the phytoconstituents interacted with target protein efficiently. Further, the phytoconstituents with the highest docking affinity were assigned as potential small molecules, and their interaction analysis was studied in detail. The overall binding affinity of 127 phytoconstituents toward the target protein is tabulated in Supplementary Table S1.
Binding Mode of Phytoconstituents From Mentha arvensis Against the N Protein
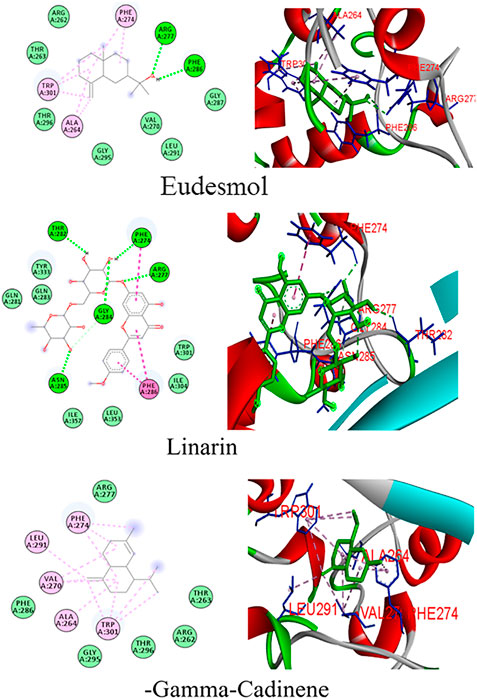
Docking simulations of major phytocompounds of Mentha arvensis against the N protein using ADT showed eudesmol as a top hit, exhibiting the highest docking score of −10.1 kcal/mol, followed by the phytochemicals linarin (−8.4 kcal/mol) and (−)−gamma−cadinene (−7.5 kcal/mol) (Table 1). The results indicated that out of sixty-six small molecules screened virtually, eudesmol, linarin, and (−)−gamma−cadinene possess the strong interactions with the N protein by executing greatest binding affinity. The amino acid residues of the N protein involved in hydrogen bond, hydrophobic, and electrostatic interactions with these ligands were also observed through docking analysis using AutoDock Vina (Figure 1).
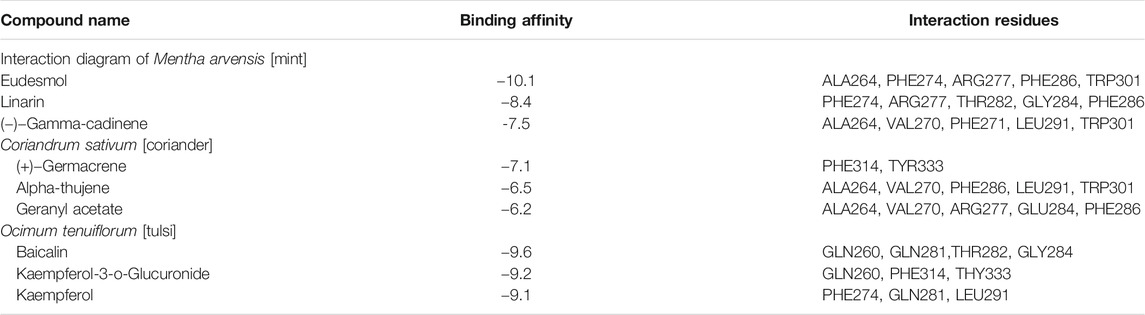
TABLE 1. List of top three hit phytochemicals from each of the three selected plants (Mentha arvensis, Coriandrum sativum, and Ocimum tenuiflorum) along with their binding energy and interaction residues against the N protein as predicted through molecular docking.
Eudesmol formed seven hydrophobic interactions (Pi-Alkyl) with the N protein, involving the amino acid residues Ala264 (3.86 and 6.06), Phe274 (5.86 and 6.01), and Trp301 (6.56, 5.25, and 7.47); two hydrogen bond interaction with Arg277 (5.33) and Phe286 (4.70) residues; and seven van der Waals interaction with Arg262, Thr263, Gly287, Val270, Leu291, Gly295, and Thr296, whereas linarin interacted with the N protein through six hydrogen bond interactions with residues viz., Phe274 (3.96), Arg277 (6.25), Thr282 (5.14), Gly284 (3.40 and 3.77), and Asn285 (3.75); three Hydrophobic interactions (Pi–Pi Stacked and Pi–Pi T-shaped) with Phe274 (4.65) and Phe286 (5.28 and 4.14) residues; and some van der Waals interactions with Gln281, Gln283, Trp301, Ile304, Tyr333, Leu353, and Ile357. (−)−Gamma−cadinene−N-protein complex showed eleven hydrophobic interactions (Pi-Alkyl) with Ala264 (5.99), Val270 (4.97, 4.96, and 6.01), Phe274 (4.47, 5.32, and 5.52,), Leu291 (4.41), and Trp301 (6.32, 7.38, 6.85, and 6.16), and six van der Waals interaction with Arg261, Thr263, Arg277, Phe286, Gly295, and Thr296. No hydrogen bonds were imputed between linarin and the binding site of the N protein (Figure 1).
Phytoconstituents From Coriandrum sativum With the N Protein
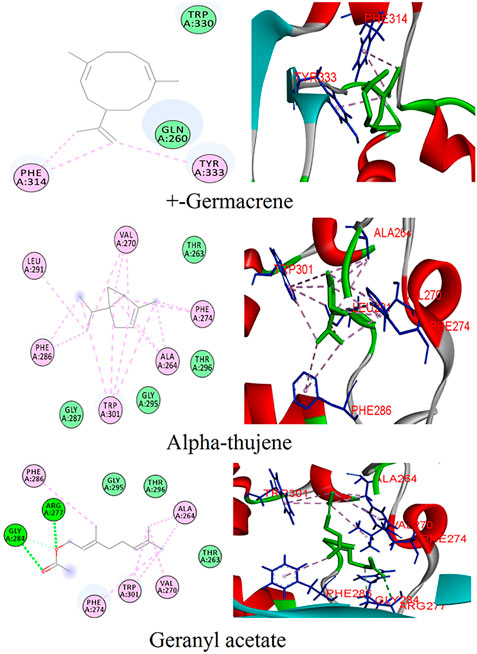
The phytoligands of Coriandrum sativum were virtually screened against the N protein, and the docked phytoligands were ranked based on a stringer filter which included bonding affinity, strength of hydrogen bonding, and other hydrophobic, electrostatic, and van der Waals interaction. Out of 38 phytoligands, the top most docking poses and binding orientation were selected. The top ranked phytoligands include (+)−germacrene A, alpha-thujene, and geranyl acetate, which bind firmly at the active site of the target protein with high binding affinity and good molecular interactions (Table 1) (Figure 2). (+)−Germacrene A was bound to the N protein with the docking score of −7.1 kcal/mol, forming three hydrophobic interactions: (Pi-Alkyl) with Phe314 (5.35 and 5.50) and Tyr333(5.63), and two van der Waals interaction with Gln260 and Trp330. Alpha-thujene bound to the active site of the N protein with a docking score of 6.4 kcal/mol, and it displayed fifteen hydrophobic interactions (Pi-Alkyl) with Ala264 (5.51, 4.65, and 4.12), Val270 (4.78, 4.93, and 4.98), Phe274 (6.66 and 6.43), Phe286 (4.65 and 4.53), Leu291 (4.67), and Trp301 (6.55, 6.11, 5.36, and 6.68), and van der Waals interaction with residues Thr263, Gly287, Gly295, and Thr296. Geranyl acetate bound effectively to the active site of the N protein with the docking score of 6.2 kcal/mol, and it formed three hydrogen bonds (two conventional and one carbon hydrogen bond) with Arg277 (5.48) and Glu284 (3.48 and 3.91), respectively. Together, geranyl acetate also extended seven hydrophobic interactions (Pi-Alkyl) with resides Ala264 (3.74 and 4.06), Val270 (5.58), Phe274 (6.61), Phe286 (4.08), and Tro301 (5.37 and 6.62), and van der Waals interaction with Thr263, Gly295, and Thr296 residues (Figure 2).
Binding Mode of Phytoconstituents From Ocimum sanctum With the N Protein
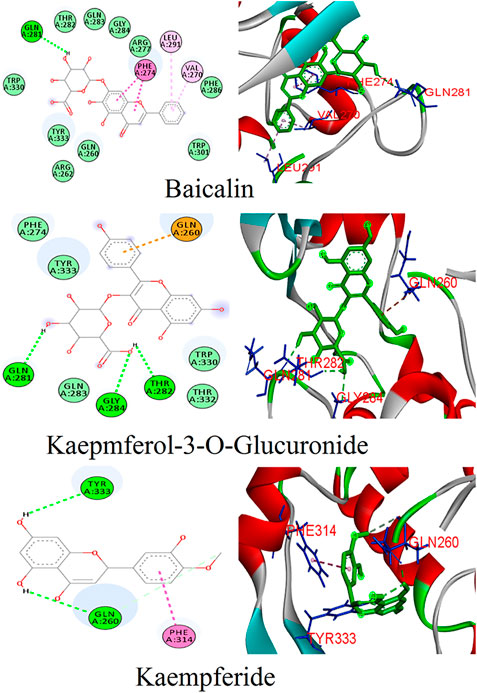
Twenty-three phytoligands from the immune booster herb Ocimum tenuiflorum were docked into the binding pocket of the N protein. After docking, three best phytoligands were selected based on their accurate binding pose and binding energy score. The top hit phytoligands were ranked in the sequence of baicalin, kaempferol-3-O-glucuronide, and kaempferide as they depicted a bonding affinity of −9.6, −9.2, and −9.1 kcal/mol, respectively (Table 1). Baicalin formed hydrogen bonds as well as hydrophobic interaction with the active site residues of receptor (Figure 3). It formed one hydrogen bond with the active site residue GLn281 (5.80); four hydrophobic interactions (Pi-alkyl, Pi–Pi stacked) with the active site residues of Val270 (5.24), Leu291 (7.32), and Phe274(4.85, 4.42); and some van der Waals interactions with resides Gln260, Arg261, Arg277, Thr282, Gln283, Gly284, Phe286, Trp301, Trp330, and Tyr333 (Figure 3). Kaempferol-3-O-glucuronide was stabilized by the three hydrogen bonds with active site residues of Gln281 (4.42), Thr282 (5.27), and Gly284 (3.73). One electrostatic interaction (Pi-cation) with the amino acid residue of Gln260 and some van der Waals interactions with amino acids residues of Phe274, Gln283, Trp330, Thr332, and Tyr333 were formed. Kaempferide was stabilized by forming three hydrogen bond interactions with active site residues of Gln260 (3.00 and 4.66) and Tyr333 (6.20), and one hydrophobic interaction (Pi–Pi stacked) with active site residue Phe314 (6.57) (Figure 3). A known antiviral drug nucleozin (which has been reported to target the N protein of influenza virus) was used as the positive control, which displayed a binding energy of −6.8 kcal mol−1 (Supplementary Table S1), and it builds one hydrogen bond with Ala264 (2.69Å); seven hydrophobic interactions (Pi–Pi stacked, Pi-alkyl, Pi-sigma, and Pi–Pi T-shaped) with amino acid residues Ala264 (4.52 Å), Val270 (3.93 Å), Phe286 (4.51 Å), Leu291 (5.33 Å), Trp301 (4.61 Å), Ile304 (5.35 Å), and Ala308 (3.66 Å); and van der Waals interaction with Arg262, Thr263, Arg277, Phe274, GLy295, Thr296, Ala305, Phe307, Leu353, and Ile357 (Supplementary Figure S1).
Overall, the docking results revealed that every docked complex formed fair number of Pi-alkyl, Pi–Pi stacked, Pi–Pi T-shaped, and Pi-cation interactions, which were largely involved in charge transfer that aid in intercalating the small molecules (drug) in the active site of the receptor (Arthur and Uzairu, 2019). The top hit phytoligands from each of three herbal plants displayed strong hydrophobic interactions and hydrogen bonds, which stabilized strong chemical bonding between phytoligands and the active site of the N protein.
Drug-Likeness and ADME/T Prediction
Physicochemical properties, biological activity, and pharmacological profiles including absorption, distribution, metabolism, elimination, and toxicity (ADMET) features were envisaged using Molinspiration and admetSAR web server. The ADMET properties are essential in current drug discovery and development process. Nowadays, computational modeling is used instead of in vitro and in vivo evaluation of ADMET properties. The ultimate goal of in silico analysis is the perfect prediction of the in vivo pharmacokinetics of a probable drug molecule. Molinspiration results unveiled that all top phytoligands have obeyed Lipinski “Rule of 5” principles such as molecular weight (MW < 500Da), high lipophilicity expressed as logP (logP < 5), hydrogen bond donors (HBD < 5), and hydrogen bond acceptors (HBA < 10) (Table 2). Pharmacological parameters viz., blood–brain barrier penetration, human intestinal absorption, CYP2D6 inhibitor, Caco-2 cell permeability, carcinogenicity, and biodegradation of top phytoligands are depicted in Table 3. The very essential of ADMET property is the aqueous solubility of a drug, predicating the rate of absorption and transport of a drug molecule in the body and Caco-2 permeability, as it is one of the most important properties to measure the rate of transport of a drug molecule across the Caco-2 cell line. In addition, the BBB is a crucial factor for drugs, as it is a physiological barrier which protects the drug molecules to cross from blood to the brain (Alagu Lakshmi et al., 2020). In the present study, the ADMET prediction analysis revealed that the top hit phytoligands have the capability to aqueous solubility, Caco-2 permeability, cross the BBB, and novel absorption in the intestine. Therefore, the phytoligands envisaged in the current investigation could plausibly be considered as drug candidates for further studies.
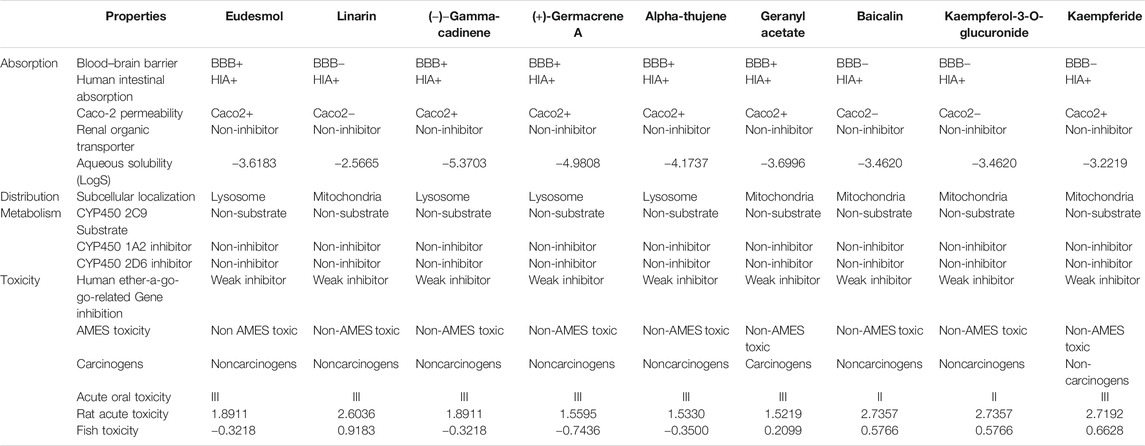
TABLE 2. Pharmacodynamic profile of top three hit phytochemicals from each of the three selected plants (Mentha arvensis, Coriandrum sativum, and Ocimum tenuiflorum).
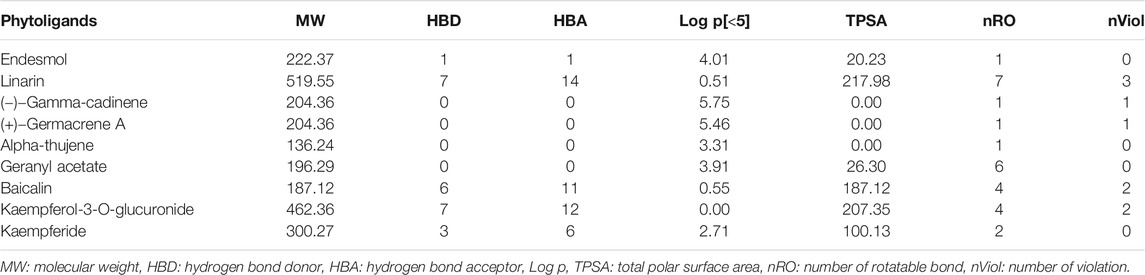
TABLE 3. In silico drug-likeness and molecular property prediction in top three hit phytochemicals from each of the three selected plants (Mentha arvensis, Coriandrum sativum, and Ocimum tenuiflorum).
Molecular Dynamic Simulation
MD simulations for protein–ligand complexes were performed for 50 ns. MD simulation is one of the attractive approaches to investigate the stability and dynamic behavior of the protein–ligand complexes in different binding poses under different physiological conditions. The observations of Cα backbone RMSD graph of docked complexes of each system suggested their stability during the simulation time period (Figure 4). Further, the RMSD graph also revealed that docked complexes were highly stable between 0.25 and 1.0 nm with minor deviations. The phytoligands viz., eudesmol, (+)−germacrene A, and baicalin acquired stability with an average RMSD of 0.5 nm, which depicted the stability of the protein–phytoligand complex in the active site of receptor. The phytoligands linarin geranyl acetate, kaempferol-3-O-glucuronide, and kaempferol (−)−gamma−cadinene showed slight deviation of RMSD around 0.65–0.8 nm. Alpha-thujene depicted more deviations at 1.0 nm; however, after 25 ns, it maintained the stability until 50 ns simulation. The RMSD analysis of the N protein with top phytoligands displayed that each phytoligand remained stable in the active site of the N protein throughout simulation (Figure 4).
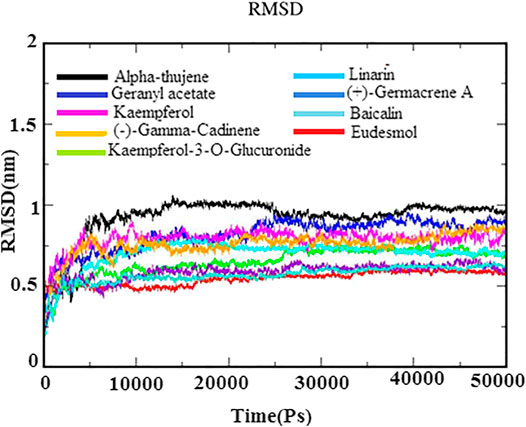
FIGURE 4. RMSD backbone plot for N-protein–inhibitor complexes (nine complexes) during 50 ns simulation as a function of timescale in ps.
The root mean square fluctuation (RMSF) graphs of Cα backbone atom was used to study the dynamic behavior of essential amino acid residues precipitated with the ligand. As shown in Figure 5, the RMSF within the range of 0.4–1.8 Å had less structural fluctuations on interacting residues. Although high fluctuations were observed between the residues of 315–330, 335–340, and 364, these regions were denoted as the loop and disordered. Hence, the fluctuation does not affect the binding of ligand into the active site of protein.
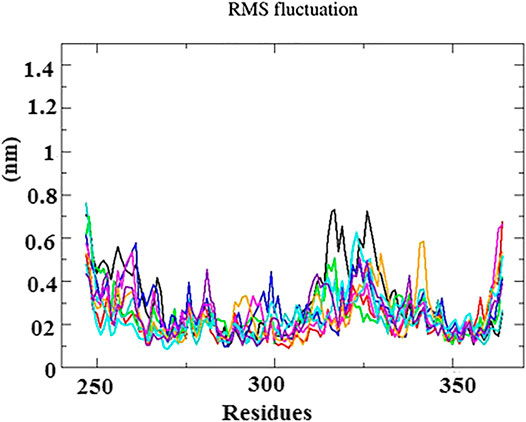
FIGURE 5. RMS fluctuation plot for N-protein–inhibitor complexes (nine complexes) during the 50 ns simulation as a function of the number of residues.
In ligand–receptor (proteins) interaction, the role of hydrogen bond is inevitable for the molecular recognition, binding stability, and backbone conformation. Therefore, in order to assess the stabilizing interaction factor between the docked complexes, the number of hydrogen bonds was calculated to investigate the nature of the H-bond at the active site of the N protein. The H-bonds were monitored throughout 50 ns of simulation and is depicted in Figure 6. A maximum number of hydrogen bonds (n = 10) were identified with the complexes of the N protein and eudesmol as well as baicalin. Next to these, (−)−gamma−cadinene showed 8 h-bonds, linarin and kaempferol-3-O-glucuronide showed 7 H-bonds, (+)−germacrene A and geranyl acetate have shown 6 H-bonds, kaempferol showed 5H-bonds, and alpha-thujene showed 3H-bonds. All the H-bonds were stable and consistent throughout the 50-ns simulation.
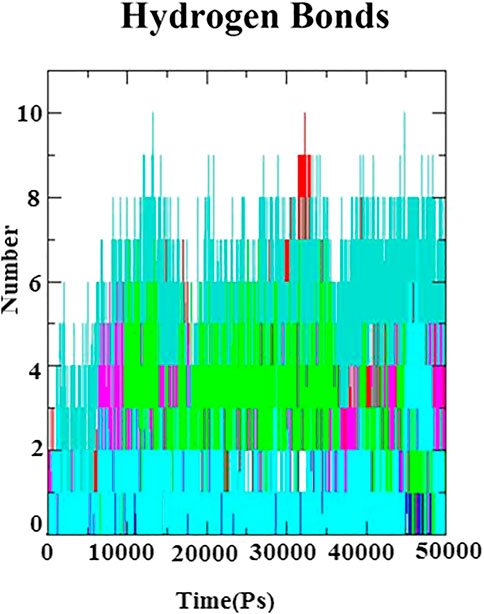
FIGURE 6. Hydrogen bond plot for N-protein–inhibitor complexes (nine complexes) during the 50 ns simulation as a function of timescale in ps.
Conclusion
Concomitantly, in the current study, we envisaged nine (linarin, eudesmol, cadinene, geranyl acetate, alpha-thujene, germacrene A, kaempferol-3-O-glucuronide, kaempferide, and baicalin) phytochemicals out of 127 screened from three Indian herbal medicinal plants (viz., Mentha arvensis, Coriandrum sativum, and Ocimum sanctum) to target the N protein by exhibiting high binding affinity toward its catalytic pocket. Although a plethora of studies have targeted several other viral proteins (viz., spike protein, main protease, and receptor protein), the present study is first of its kind in envisaging phytochemicals against the N protein of SARS-CoV-2. Moreover, the data of ADMET prediction analysis depicted the nontumorigenic, noncarcinogenic, nontoxic, nonmutagenic, and nonreproductive nature of the identified bioactive molecules. Furthermore, the molecular dynamic simulation analysis validated the conformational and dynamic stability of the docked complexes. Overall, the data of the present study virtually authenticated the anti-COVID efficacy of phytochemicals from selected medicinal herbs of Indian origin.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material; further inquiries can be directed to the corresponding authors.
Author Contributions
SM: designing the research work and writing—original draft. AK: data analysis and discussion for the original draft; PB: conceived and conceptualized the research idea, supervising, and review editing. SG: conceived and conceptualized the research idea, supervising, and review editing. SP: critical editing.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors thank DST-FIST [Grant no. SR/FST/LSI-639/2015(C)], UGC-SAP [Grant no. F.5-1/ 2018/DRS-II (SAP-II)], and DST-PURSE [Grant no. SR/ PURSE Phase 2/38 (G)] for rendering instrumentation and infrastructure facilities. SG gratefully thank the UGC for Start-Up Grant (Grant No. F.30-381/2017(BSR)/F.D Diary No. 2892), DST-SERB-EEQ Project Grant (File No.:EEQ/2020/000288), and Alagappa University for AURF (Ref: ALU: AURF Start-up Grant: 2018)). The authors acknowledge financial support rendered by RUSA 2.0 [F.24-51/2014-U, Policy (TN Multi-Gen), Department of Education, Government of India].
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmolb.2021.637329/full#supplementary-material
References
Afriza, D., Suriyah, W. H., and Ichwan, S. J. A. (2018). In Silico analysis of Molecular Interactions between the Anti-apoptotic Protein Survivin and Dentatin, Nordentatin, and Quercetin. J. Phys. Conf. Ser. 1073 (3), 032001. doi:10.1088/1742-6596/1073/3/032001
Alagu Lakshmi, S., Shafreen, R. M. B., Priya, A., and Shunmugiah, K. P. (2020). Ethnomedicines of Indian Origin for Combating COVID-19 Infection by Hampering the Viral Replication: Using Structure-Based Drug Discovery Approach. J. Biomol. Struct. Dyn. 2020, 1–16. doi:10.1080/07391102.2020.1778537
Amini, H., Hosseini, V., Schindler, C., Hassankhany, H., Yunesian, M., Henderson, S. B., et al. (2017). Spatiotemporal Description of BTEX Volatile Organic Compounds in a Middle Eastern Megacity: Tehran Study of Exposure Prediction for Environmental Health Research (Tehran SEPEHR). Environ. Pollut. 226, 219–229. doi:10.1016/j.envpol.2017.04.027
Arthur, D. E., and Uzairu, A. (2019). Molecular Docking Studies on the Interaction of NCI Anticancer Analogues with Human Phosphatidylinositol 4,5-bisphosphate 3-kinase Catalytic Subunit. J. King Saud Univ. - Sci. 31 (4), 1151–1166. doi:10.1016/j.jksus.2019.01.011
Benvenuoto, D., Demir, A. B., GiovanettiBianchi, M. M., Angeletti, S., Pascarella, S., Cauda, R., et al. (2020). Evidence for Mutations in Sars-Cov-2 Italian Isolates Potentially Affecting Virus Transmission. J. Med. Virol. 92 (10), 2232–2237. doi:10.1002/jmv.26104
Brian, D. A., and Baric, R. S. (2005). Coronavirus Genome Structure and Replication. Curr. Top. Microbiol. Immunol. 287, 1–30. doi:10.1007/3-540-26765-4_1
Burbelo, P. D., RiedoMorishima, F. X. C., Rawlings, S., Smith, D., Das, S., Strich, J. R., et al. (2020). Detection of Nucleocapsid Antibody to SARS-CoV-2 Is More Sensitive Than Antibody to Spike Protein in COVID-19 Patients. medRxiv [Epub ahead of print]. doi:10.1101/2020.04.20.20071423
Chan, J. F., Yuan, S., Kok, K. H., To, K. K., Chu, H., Yang, J., et al. (2020). A Familial Cluster of Pneumonia Associated with the 2019 Novel Coronavirus Indicating Person-To-Person Transmission: a Study of a Family Cluster. Lancet 395, 514e523. doi:10.1016/s0140-6736(20)30154-9
Chang, C. K., Chen, C. M., Chiang, M. H., Hsu, Y. L., and Huang, T. H. (2013). Transient Oligomerization of the SARS-CoV N Protein-Iimplication for Virus Ribonucleoprotein Packaging. PloS one 8 (5), e65045. doi:10.1371/journal.pone.0065045
Chang, C. K., Hou, M. H., Chang, C. F., Hsiao, C. D., and Huang, T. H. (2014). The SARS Coronavirus Nucleocapsid Protein-Fforms and Functions. Antivir. Res 103, 39–50. doi:10.1016/j.antiviral.2013.12.009
Chen, H., Fu, W., Wang, Z., Wang, X., Lei, T., Zhu, F., et al. (2019). Reliability of Docking-Based Virtual Screening for GPCR Ligands with Homology Modeled Structures: A Case Study of the Angiotensin II Type I Receptor. ACS Chem. Neurosci. 10 (1), 677–689. doi:10.1021/acschemneuro.8b00489
Cong, Y., Ulasli, M., Schepers, H., Mauthe, M., V'kovski, P., Kriegenburg, F., et al. (2020). Nucleocapsid Protein Recruitment to Replication-Transcription Complexes Plays a Crucial Role in Coronaviral Life Cycle. J. Virol. 94 (4). doi:10.1128/JVI.01925-19
Dawood, A. A. (2020). Mutated COVID-19 May Foretell a Great Risk for Mankind in the Future. New Microbes New Infect. 35, 100673. doi:10.1016/j.nmni.2020.100673
de Sousa Guedes, J. P., da Costa Medeiros, J. A., de Souza E Silva, R. S., de Sousa, J. M., da Conceição, M. L., and de Souza, E. L. (2016). The Efficacy of Mentha Arvensis L. And M. Piperita L. Essential Oils in Reducing Pathogenic Bacteria and Maintaining Quality Characteristics in Cashew, Guava, Mango, and Pineapple Juices. Int. J. Food Microbiol. 238, 183–192. doi:10.1016/j.ijfoodmicro.2016.09.005
de Sousa Guedes, J. P., and de Souza, E. L. (2018). Investigation of Damage to Escherichia coli, Listeria Monocytogenes and Salmonella Enteritidis Exposed to Mentha Arvensis L. And M. Piperita L. Essential Oils in Pineapple and Mango Juice by Flow Cytometry. Food Microbiol. 76, 564–571. doi:10.1016/j.fm.2017.09.020
de Wit, E., van Doremalen, N., Falzarano, D., and Munster, V. J. (2016). SARS and MERS: Recent Insights into Emerging Coronaviruses. Nat. Rev. Microbiol. 14 (8), 523–534. doi:10.1038/nrmicro.2016.81
Du, L., Zhao, G., Lin, Y., Chan, C., He, Y., Jiang, S., et al. (2008). Priming with rAAV Encoding RBD of SARS-CoV S Protein and Boosting with RBD-specific Peptides for T Cell Epitopes Elevated Humoral and Cellular Immune Responses against SARS-CoV Infection. Vaccine 26 (13), 1644–1651. doi:10.1016/j.vaccine.2008.01.025
Ganjhu, R. K., Mudgal, P. P., Maity, H., Dowarha, D., Devadiga, S., Nag, S., et al. (2015). Herbal Plants and Plant Preparations as Remedial Approach for Viral Diseases. Virusdisease 26 (4), 225–236. doi:10.1007/s13337-015-0276-6
Gao, T., Hu, M., Zhang, X., Li, H., Zhu, L., Liu, H., et al. (2020). Highly Pathogenic Coronavirus N Protein Aggravates Lung Injury by MASP-2-Mediated Complement Over-activation. medRxiv [Epub ahead of print]. doi:10.1101/2020.03.29.20041962
Gowrishankar, S., Muthumanickam, S., Kamaladevi, A., Karthika, C., Jothi, R., Boomi, P., et al. (2021). Promising Phytochemicals of Traditional Indian Herbal Steam Inhalation Therapy to Combat COVID-19–An In Silico Study. Food Chem. Toxicol. 148, 111966. doi:10.1016/j.fct.2020.111966
Hsieh, P. K., Chang, S. C., Huang, C., Lee, T., Hsiao, C., Kou, Y., et al. (2005). Assembly of Severe Acute Respiratory Syndrome Coronavirus RNA Packaging Signal into Virus-like Particles Is Nucleocapsid Dependent. J. Virol. 79 (22), 13848–13855. doi:10.1128/JVI.79.22.13848-13855.2005
Huang, C., Wang, Y., Li, X., Ren, L., Zhao, J., Hu, Y., et al. (2020). Clinical Features of Patients Infected with 2019 Novel Coronavirus in Wuhan, China. Lancet 395 (10223), 497–506. doi:10.1016/S0140-6736(20)30183-5
Huang, C. K., Hsu, Y. L., Chang, Y. H., Chao, F. A., Wu, M. C., Huang, Y. S., et al. (2009). Multiple Nucleic Acid Binding Sites and Intrinsic Disorder of Severe Acute Respiratory Syndrome Coronavirus Nucleocapsid Protein: Implications for Ribonucleocapsid Protein Packaging. J. Virol. 83 (5), 2255–2264. doi:10.1128/JVI.02001-08
Hussain, F., Jahan, N., Rahman, K.-u., Sultana, B., and Jamil, S. (2018). Identification of Hypotensive Biofunctional Compounds of Coriandrum Sativumand Evaluation of Their Angiotensin-Converting Enzyme (ACE) Inhibition Potential. Oxidative Med. Cell Longevity 2018, 1. doi:10.1155/2018/4643736
Isa, M. A., Majumdhar, R. S., and Haider, S. (2018). In Silico docking and Molecular Dynamics Simulation of 3-dehydroquinate Synthase (DHQS) from Mycobacterium tuberculosis. J. Mol. Model. 24 (6), 132. doi:10.1007/s00894-018-3637-4
Jamshidi, N., and Cohen, M. M. (2017). The Clinical Efficacy and Safety of Tulsi in Humans: A Systematic Review of the Literature. Evid Based Complement Alternat. Med. 2017 (2017), 9217567. doi:10.1155/2017/9217567
Kačániová, M., Galovičová, L., Ivanišová, E., Vukovic, N. L., Štefániková, J., Valková, V., et al. (2020). Antioxidant, Antimicrobial and Antibiofilm Activity of Coriander (Coriandrum Sativum L.) Essential Oil for its Application in Foods. Foods 9 (3), 282. doi:10.3390/foods9030282
Khan, M. Y., and Kumar, V. (2019). Mechanism & Inhibition Kinetics of Bioassay-Guided Fractions of Indian Medicinal Plants and Foods as ACE Inhibitors. J. Tradit. Complement. Med. 9 (1), 73–84. doi:10.1016/j.jtcme.2018.02.001
Killerby, M. E., Biggs, H. M., Haynes, A., Dahl, R. M., Mustaquim, D., Gerber, S. I., et al. (2018). Human Coronavirus Circulation in the United States 2014-2017. J. Clin. Virol. 101, 52–56. doi:10.1016/j.jcv.2018.01.019
Kopecky-Bromberg, S. A., Martínez-Sobrido, L., Frieman, M., Baric, R. A., and Palese, P. (2000). Severe Acute Respiratory Syndrome Coronavirus Open reading Frame (ORF) 3b, ORF 6, and Nucleocapsid Proteins Function as Interferon Antagonists. J. Virol. 81 (2), 548–557. doi:10.1128/JVI.01782-06
Kumar, A., and Chattopadhyay, S. (2007). DNA Damage Protecting Activity and Antioxidant Potential of Pudina Extract. Food Chem. 100 (4), 1377–1384. doi:10.1016/j.foodchem.2005.12.015
Kumar, A., Khajuria, V., and Aggarwal, S. (2012). Secondary Metabolites ofMentha Arvensisand Their Biological Activities. Anal. Chem. Lett. 2, 373–400. doi:10.1080/22297928.2012.10662623
Lal, R. K., Chanotiya, C. S., Dhawan, S. S., Gupta, P., Mishra, A., Srivastava, S., et al. (2020). Estimation of Intra-Specific Genetic Variability and Half-Sib Familyselection Using AMMI (Additive Main Effects and Multiplicative Interactions) Model in Menthol Mint (Menthaarvensis L.). J. Med. Aromat. Plants 42 (1-2), 102–113.
Li, Q., Guan, X., Wu, P., Wang, X., Zhou, L., TongRen, Y. R., et al. (2020). Early Transmission Dynamics in Wuhan, China, of Novel Coronavirus-Infected Pneumonia. N. Engl. J. Med. 382, 1199e1207. doi:10.1056/nejmoa2001316
Li, W., Shi, Z., Yu, M., Ren, W., Smith, C., Epstein, J. H., et al. (2005). Bats Are Natural Reservoirs of SARS-like Coronaviruses. Science 310 (5748), 676–679. doi:10.1126/science.1118391
Lin, S. Y., Liu, C. L., Chang, Y. M., Zhao, J., Perlman, S., and Hou, M. H. (2014). Structural Basis for the Identification of the N-Terminal Domain of Coronavirus Nucleocapsid Protein as an Antiviral Target. J. Med. Chem. 57 (6), 2247–2257. doi:10.1021/jm500089r
Lu, X., Pan, J., Tao, J., and Guo, D. (2011). SARS-CoV Nucleocapsid Protein Antagonizes IFN-β Response by Targeting Initial Step of IFN-β Induction Pathway, and its C-Terminal Region Is Critical for the Antagonism. Virus Genes 42 (1), 37–45. doi:10.1007/s11262-010-0544-x
Mallikarjun, S., Rao, A., Rajesh, G., Shenoy, R., and Pai, M. (2016). Antimicrobial Efficacy of Tulsi Leaf (Ocimum Sanctum) Extract on Periodontal Pathogens: An In Vitro Study. J. Indian Soc. Periodontol. 20 (2), 145–150. doi:10.4103/0972-124X.175177
Masters, P. S., Parker, M. M., Ricard, C. S., Duchala, C., Frana, M. F., Holmes, K. V., et al. (1990). Structure and Function Studies of the Nucleocapsid Protein of Mouse Hepatitis Virus. Adv. Exp. Med. Biol. 276, 239–246. doi:10.1007/978-1-4684-5823-7_33
McBride, R., Van Zyl, M., and Fielding, B. C. (2014). The Coronavirus Nucleocapsid Is a Multifunctional Protein. Viruses 6 (8), 2991–3018. doi:10.3390/v6082991
Mousavi, L., Salleh, R. M., and Murugaiyah, V. (2018). Phytochemical and Bioactive Compounds Identification of Ocimum Tenuiflorum Leaves of Methanol Extract and its Fraction with an Anti-diabetic Potential. Int. J. Food Properties 21, 2390–2399. doi:10.1080/10942912.2018.1508161
Mukherjee, R., Das, P. K., and Ram, G. C. (2005). Immunotherapeutic Potential of Ocimum Sanctum Linn Bovine Subclinical Mastitis. Rev. Vet. Sci. 79 (1), 37–43. doi:10.1016/j.rvsc.2004.11.001
Muthumanickam, S., Indhumathi, T., Boomi, P., Balajee, R., Jeyakanthan, J., Anand, K., et al. (2020). In Silico approach of Naringin as Potent Phosphatase and Tensin Homolog (PTEN) Protein Agonist against Prostate Cancer. J. Biomol. Struct. Dyn., 1–10. doi:10.1080/07391102.2020.1830855
Muthuramalingam, P., Jeyasri, R., Valliammai, A., Selvaraj, A., Karthika, C., Gowrishankar, S., et al. (2020). Global Multi-Omics and Systems Pharmacological Strategy Unravel the Multi-Targeted Therapeutic Potential of Natural Bioactive Molecules against COVID-19: An In Silico Approach. Genomics 112 (6), 4486–4504. doi:10.1016/j.ygeno.2020.08.003
Narayanan, K., Kim, K. H., and Makino, S. (2003). Characterization of N Protein Self-Association in Coronavirus Ribonucleoprotein Complexes. Virus. Res. 98 (2), 131–140. doi:10.1016/j.virusres.2003.08.021
Ou, J., Zhou, Z., Zhang, J., Lan, W., Zhao, S., Wu, J., et al. (2020). RBD Mutations from Circulating SARS-CoV-2 Strains Enhance the Structure Stability and Infectivity of the Spike Proteins. bioRxiv [Epub ahead of print]. doi:10.1101/2020.03.15.991844
Pachetti, M., Marini, B., Benedetti, F., Giudici, F., Mauro, E., Storici, P., et al. (2020). Emerging SARS-CoV-2 Mutation Hot Spots Include a Novel RNA-Dependent-RNA Polymerase Variant. J. Transl. Med. 18, 179. doi:10.1186/s12967-020-02344-6
Parry, J. (2020). China Coronavirus: Cases Surge as Official Admits Human to Human Transmission. BMJ 368, m236. doi:10.1136/bmj.m236
Phan, L. T., Nguyen, T. V., Luong, Q. C., Nguyen, T. V., Nguyen, H. T., Le, H. Q., et al. (2020a). Importation and Human-To-Human Transmission of a Novel Coronavirus in Vietnam. N. Engl. J. Med. 382, 872–874. doi:10.1056/NEJMc2001272
Phan, T. (2020b). Genetic Diversity and Evolution of SARS-CoV-2. Infect. Genet. Evol. 81, 104260. doi:10.1016/j.meegid.2020.104260
Prakash, P., and Gupta, N. (2005). Therapeutic Uses of Ocimum sanctum Linn (Tulsi) with a Note on Eugenol and Its Pharmacological Actions: A Short Review. Indian J. Physiol Pharmacol. 49 (2), 125–131.
Prompetchara, E., Ketloy, C., and Palaga, T. (2020). Immune Responses in COVID-19 and Potential Vaccines: Lessons Learned from SARS and MERS Epidemic. Asian Pac. J. Allergy Immunol. 38, 1–9. doi:10.12932/AP-200220-0772
Randad, P. R, Pisanic, N., Kruczynski, K., Manabe, Y. C., Thomas, D., Pekosz, A., et al. (2020). COVID-19 Serology at Population Scale: SARS-CoV-2-Specific Antibody Responses in Saliva. medRxiv [Epub ahead of print]. doi:10.1101/2020.05.24.20112300
Riou, J., and Althaus, C. L. (2020). Pattern of Early Human-To-Human Transmission of Wuhan 2019 Novel Coronavirus (2019-nCoV), December 2019 to January 2020. Euro Surveill. 25 (4), 2000058. doi:10.2807/1560-7917.ES.2020.25.4.2000058
Rolta, R., Yadav, R., Salaria, D., Trivedi, S., Imran, M., Sourirajan, A., et al. (2020). In Silico screening of Hundred Phytocompounds of Ten Medicinal Plants as Potential Inhibitors of Nucleocapsid Phosphoprotein of COVID-19: an Approach to Prevent Virus Assembly. J. Biomol. Struct. Dyn., 1–18. doi:10.1080/07391102.2020.1804457
Salin, O., Törmäkangas, L., Leinonen, M., Saario, E., Hagström, M., Ketola, R. A., et al. (2011). Corn Mint (Mentha Arvensis) Extract Diminishes Acute Chlamydia Pneumoniae Infection In Vitro and In Vivo. J. Agric. Food Chem. 59, 12836–12842. doi:10.1021/jf2032473
Shivanika, C., Kumar, D., Ragunathan, V., Pawan, T., Sumitha, A., Brindha Devi, P., et al. (2020). Molecular Docking, Validation, Dynamics Simulations, and Pharmacokinetic Prediction of Natural Compounds against the SARS-CoV-2 Main-Protease. J. Biomol. Struct. Dyn., 1–27. doi:10.1080/07391102.2020.1815584
Singh, M., Singh, M., Jaiswal, N., and Chauhan, A. (2017). Heated, Humidified Air for the Common Cold. Cochrane Database Syst. Rev. 8 (8), CD001728. doi:10.1002/14651858.CD001728.pub6
Surjit, M., Liu, B., Chow, V. T., and Lal, S. K. (2006). The Nucleocapsid Protein of Severe Acute Respiratory Syndrome-Coronavirus Inhibits the Activity of Cyclin-cyclin-dependent Kinase Complex and Blocks S Phase Progression in Mammalian Cells. J. Biol. Chem. 281 (16), 10669–10681. doi:10.1074/jbc.M509233200
The wealth of India (2003). A Dictionary of Indian Raw Materials and Industial Products. Raw Materials. New Delhi: L-M: Council of Scientific and Industrial Research, 337–346.
Ul Qamar, M. T., Maryam, A., Muneer, I., Xing, F., Ashfaq, U. A., Khan, F. A., et al. (2019). Computational Screening of Medicinal Plant Phytochemicals to Discover Potent Pan-Serotype Inhibitors against Dengue Virus. Scientific Rep. 9 (1), 1433. doi:10.1038/s41598-018-38450-1
Vellingiri, B., Jayaramayya, K., Iyer, M., Narayanasamy, A., Govindasamy, V., Giridharan, B., et al. (2020). COVID-19: A Promising Cure for the Global Panic. Sci. Total Environ. 725, 138277. doi:10.1016/j.scitotenv.2020.138277
Wang, X., and Liu, Z. (2014). Prevention and Treatment of Viral Respiratory Infections by Traditional Chinese Herbs. Chin. Med. J. 127 (7), 1344–1350.
Wei, J., Liu, Z., Zhao, Y., Zhao, L., Xue, T., and Lan, Q. (2019). Phytochemical and Bioactive Profile of Coriandrum Sativum L. Food Chem. 286, 260–267. doi:10.1016/j.foodchem.2019.01.171
Wu, C., Liu, Y., Yang, Y., Zhang, P., Zhong, W., Wang, Y., et al. (2020a). Analysis of Therapeutic Targets for SARS-CoV-2 and Discovery of Potential Drugs by Computational Methods. Acta Pharm. Sin. B. 10 (5), 766–788. doi:10.1016/j.apsb.2020.02.008
Wu, F., Zhao, S., Yu, B., Chen, Y., Wang, W., Song, Z., et al. (2020b). A New Coronavirus Associated with Human Respiratory Disease in China. Nature 579, 265–269. doi:10.1038/s41586-020-2008-3
Yadav, R., Imran, M., Dhamija, P., Suchal, K., and Handu, S. (2020). Virtual Screening and Dynamics of Potential Inhibitors Targeting RNA Binding Domain of Nucleocapsid Phosphoprotein from SARS-CoV-2. J. Biomol. Struct. Dyn., 1–16. doi:10.1080/07391102.2020.1778536
Yadav, R. S., Kumar, S., and Dikshit, A. (2006). Antifungal Properties of Essential Oil of Mentha Spicata L. Var. MSS-5. Indian J. Crop Sci. 1 (1and2), 197–200.
Yamani, H. A., Pang, E. C., Mantri, N., and Deighton, M. A. (2016). Antimicrobial Activity of Tulsi (Ocimum Tenuiflorum) Essential Oil and Their Major Constituents against Three Species of Bacteria. Front. Microbiol. 7 (7), 681. doi:10.3389/fmicb.2016.00681
Yasui, F., Kai, C., Kitabatake, M., Inoue, S., Yoneda, M., Yokochi, S., et al. (2008). Prior Immunization with Severe Acute Respiratory Syndrome (SARS)-associated Coronavirus (SARS-CoV) Nucleocapsid Protein Causes Severe Pneumonia in Mice Infected with SARS-CoV. J. Immunol. 181, 6337–6348. doi:10.4049/jimmunol.181.9.6337
Yin, C. (2020). Genotyping Coronavirus SARS-CoV-2: Methods and Implications. Genomics 112 (5), 3588–3596. doi:10.1016/j.ygeno.2020.04.016
Zheng, N., Cheng, J., Zhang, W., Li, W., Shao, X., Xu, Z., et al. (2014). Binding Difference of Fipronil with GABAARs in Fruitfly and Zebrafish: Insights from Homology Modeling, Docking, and Molecular Dynamics Simulation Studies. J. Agric. Food Chem. 62 (44), 10646–10653. doi:10.1021/jf503851z
Keywords: ADMET profiles, medicinal plants, molecular docking, molecular dynamic simulation, nucleocapsid phosphoprotein, SARS-CoV-2
Citation: Muthumanickam S, Kamaladevi A, Boomi P, Gowrishankar S and Pandian SK (2021) Indian Ethnomedicinal Phytochemicals as Promising Inhibitors of RNA-Binding Domain of SARS-CoV-2 Nucleocapsid Phosphoprotein: An In Silico Study. Front. Mol. Biosci. 8:637329. doi: 10.3389/fmolb.2021.637329
Received: 03 December 2020; Accepted: 11 February 2021;
Published: 02 July 2021.
Edited by:
Da’san Mahmoud Mousa Jaradat, Al-Balqa Applied University, JordanReviewed by:
Roopali Rajput, National Institute of Tuberculosis and Respiratory Diseases, IndiaSayan Chatterjee, Guru Gobind Singh Indraprastha University, India
Copyright © 2021 Muthumanickam, Kamaladevi, Boomi, Gowrishankar and Pandian. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Pandi Boomi, cGJvb21pMTk4M0BnbWFpbC5jb20=; Shanmugaraj Gowrishankar, Z293cmlzaGFua2FyLmFsdUBnbWFpbC5jb20=
†These authors have contributed equally to this work
 Sankar Muthumanickam1†
Sankar Muthumanickam1† Arumugam Kamaladevi
Arumugam Kamaladevi Pandi Boomi
Pandi Boomi Shanmugaraj Gowrishankar
Shanmugaraj Gowrishankar