- Department of Microbiology, Cornell University, Ithaca, NY, United States
Antibiotics and other agents that perturb the synthesis or integrity of the bacterial cell envelope trigger compensatory stress responses. Focusing on Bacillus subtilis as a model system, this mini-review summarizes current views of membrane structure and insights into how cell envelope stress responses remodel and protect the membrane. Altering the composition and properties of the membrane and its associated proteome can protect cells against detergents, antimicrobial peptides, and pore-forming compounds while also, indirectly, contributing to resistance against compounds that affect cell wall synthesis. Many of these regulatory responses are broadly conserved, even where the details of regulation may differ, and can be important in the emergence of antibiotic resistance in clinical settings.
Introduction: Membrane Homeostasis and Its Modulation in Response to Stress
The cell envelope is a multilayered outer barrier that protects the cell from a changing environment. Cell envelope stress responses (CESRs) are regulatory pathways that sense threats and mount a protective response, often involving modification of lipopolysaccharides (in Gram-negative bacteria), teichoic acids (Gram-positive bacteria), peptidoglycan, and the inner membrane (Helmann, 2016; Radeck et al., 2017; Mitchell and Silhavy, 2019). Here, we focus on Bacillus subtilis as a Gram-positive model for the role of CESRs in membrane homeostasis.
The cell membrane is a dynamic, fluid mosaic comprising a lipid bilayer and associated proteins (Figure 1). In B. subtilis, the major lipid species are phospholipids, glucolipids, and the lipoteichoic acids (LTA) (Salzberg and Helmann, 2008; Nickels et al., 2017). The membrane proteome includes proteins for transport and signaling, as well as membrane synthesis, remodeling, and protection. As the innermost and last line of defense, the cell membrane is critical for viability. In B. subtilis, for example, collapsing the proton motive force activates autolysins resulting in rapid cell lysis (Jolliffe et al., 1981). Membrane-active compounds such as detergents, antimicrobial peptides, and pore-forming compounds often trigger stress responses that modify the lipidome and membrane proteome to confer resistance. Membrane stress responses can modify the cell membrane, by (i) modulating the length, branching, and saturation of the fatty acid (FA) acyl chains, (ii) altering membrane lipid composition, or (iii) synthesizing proteins that modify or protect the membrane (Table 1).
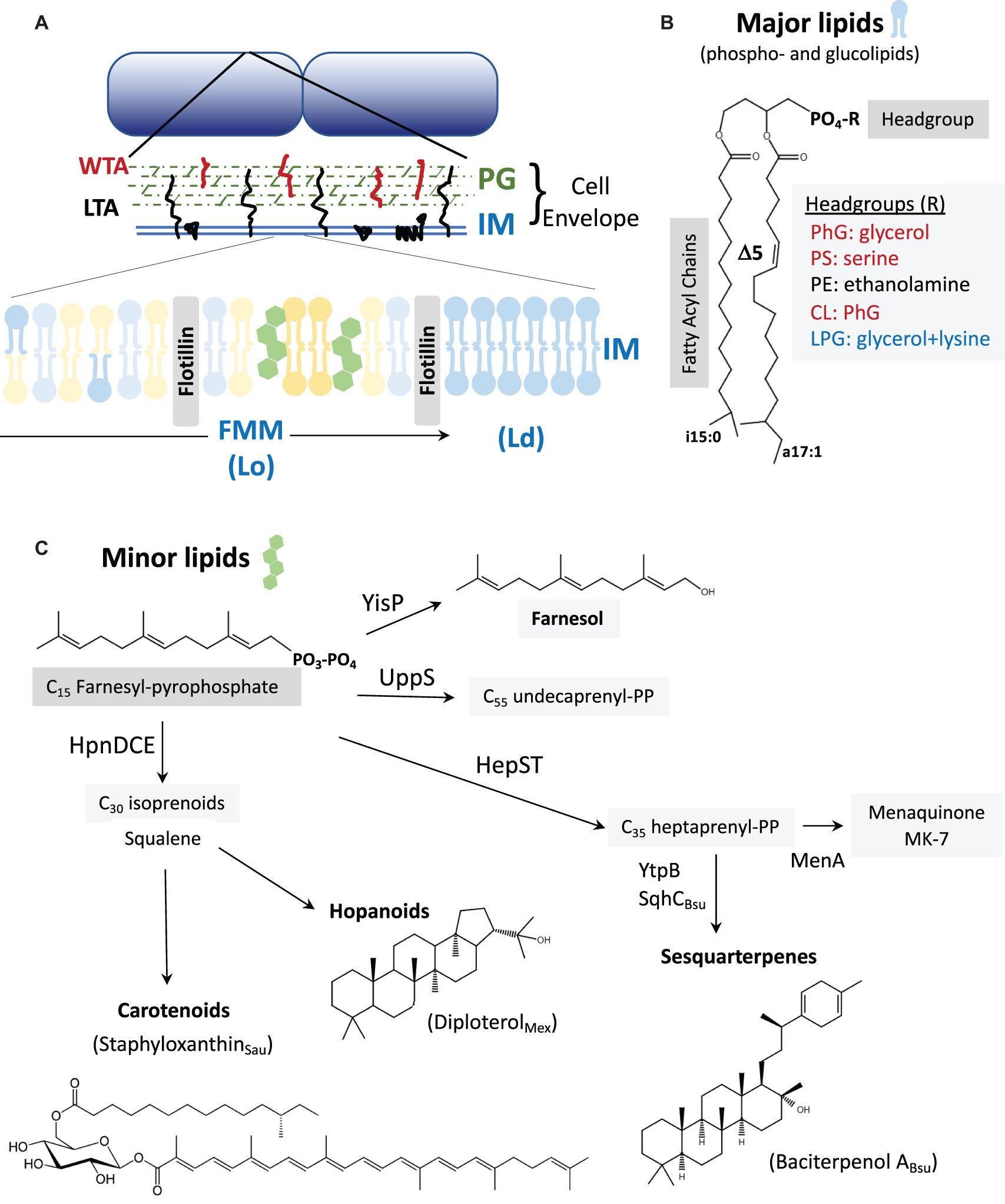
Figure 1. (A) The cell envelope: Bacillus subtilis is surrounded by a cell envelope comprised of a thick peptidoglycan (PG) layer and an inner membrane (IM). The membrane-associated lipoteichoic acid (LTA) and PG-linked wall teichoic acid (WTA) are abundant anionic polymers in the envelope (Rajagopal and Walker, 2017). The IM contains lateral microheterogeneity in the form of functional membrane microdomains (FMMs), regions of liquid-ordered (Lo) membrane together with associated proteins such as flotillins (Lopez and Koch, 2017). These are flanked by regions of higher fluidity characterized as liquid-disordered (Ld). (B) Major membrane lipids: Major membrane lipids include phospholipids and glucolipids (Nickels et al., 2017). Phospholipids (shown) vary in their FA chains, which are largely branched in B. subtilis. Shown here are a C15 iso-FA and a C17 Δ5 (unsaturated) anteiso-FA. Other FA chain lengths (including straight chains), and the positioning of the FA chains on the 1 and 2 positions of glycerol can vary. Variations in the phospholipid headgroups modulate surface charge (red are anionic, blue cationic, and black net neutral). Glucolipids are generally neutral lipids with one or more sugar residues in place of the phosphate shown. (C) Minor membrane lipids: Many of the minor lipids in the membrane are isoprenoids and are derived from the C15 intermediate farnesyl-pyrophosphate (FPP). FPP is a precursor for undecaprenyl-PP (for PG synthesis) and for the C35 intermediate heptaprenyl-PP. The latter is a precursor for the electron carrier menaquinone (MK-7) and sesquarterpenes including baciterpenol A and its derivatives (sporulenes) (Bosak et al., 2008; Takigawa et al., 2010; Sato et al., 2011; Sato, 2013). Two FPP can also be coupled in a multistep reaction by HpnDCE to generate C30 squalene (Pan et al., 2015; van der Donk, 2015), which can be processed into carotenoids [such as staphyloxanthin from S. aureus; (Garcia-Fernandez et al., 2017; Foster, 2019)] or cyclized by squalene-hopene cyclases to generate polycyclic compounds (hopanoids) (Saenz et al., 2015; Belin et al., 2018). In B. subtilis, FPP can also be dephosphorylated by YisP to generate the alcohol farnesol (Bell and Chappell, 2014; Feng et al., 2014).
The Regulation of FA Synthesis During Growth
Most bacteria utilize a type II FA synthase that catalyzes repeated cycles of acyl chain elongation (Parsons and Rock, 2013). The committed step, catalyzed by acetyl-CoA carboxylase (ACC), generates malonyl-CoA and then malonyl-ACP to serve in FA chain initiation by FabH and elongation by FabF. B. subtilis has two isoforms of FabH, and both preferentially synthesize branched chain FAs (BCFAs) (Choi et al., 2000; Kingston et al., 2011). Acylation of glycerol-3-phosphate by the PlsX/PlsY/PlsC acyltransferase system with long chain FAs generates phosphatidic acid, the precursor to all other phospholipids (Yao and Rock, 2013).
FapR is the key transcriptional regulator of membrane lipid synthesis in B. subtilis and clinically relevant pathogens such as Staphylococcus aureus, Bacillus anthracis, and Listeria monocytogenes (Schujman et al., 2003; Fujita et al., 2007; Albanesi et al., 2013; Machinandiarena et al., 2020), and modulates the overall rate of membrane synthesis in response to precursor availability. B. subtilis FapR represses genes for FA and phospholipid synthesis, and this repression is relieved by allosteric interactions with malonyl-CoA or malonyl-ACP (Schujman et al., 2006; Martinez et al., 2010).
As a branchpoint enzyme, ACC is often under complex regulation (Zhang and Rock, 2009; Salie and Thelen, 2016; Machinandiarena et al., 2020). In B. subtilis, ACC is regulated in part by YqhY, a conserved DUF322/Asp23 protein which is highly expressed and often encoded together with ACC subunits as part of an accB-accC-yqhY operon (Todter et al., 2017). The namesake, S. aureus Asp23, is a membrane-associated protein originally linked to alkaline shock (Petersen et al., 2020). Loss of Asp23/YqhY causes cell wall stress and poor growth (Muller et al., 2014; Todter et al., 2017). In B. subtilis, yqhY null mutants acquire suppressors that decrease ACC activity, but this selective pressure is alleviated in medium supplemented with acetate (Todter et al., 2017). We suggest that ACC-dependent depletion of acetyl-CoA may contribute to wall stress by negatively affecting synthesis of UDP-N-acetylglucosamine needed for peptidoglycan synthesis. A key challenge for future research will be to understand the precise role of YqhY/Asp23 proteins and how they control ACC activity to balance FA synthesis with other cellular needs.
Modulating FA Composition for Homeoviscous Adaptation
Tuning of FA composition provides one way in which the cell can optimize membrane properties in response to a changing environment. Even under non-stressed conditions, B. subtilis membranes contain ∼7 distinct FAs varying in length from C14 to C18 (indicating the number of carbon atoms) and include both branched (∼24% iso and 66% anteiso) and straight chain (∼10%) FAs (Kingston et al., 2011). Since membrane phospholipids and glucolipids each contain 2 FA chains, the lipidome contains a complex mix of species (Figure 1B), with a preponderance containing one C15 and one C17 FA chain (Kingston et al., 2011).
Modifications of FAs are important for regulating membrane fluidity in a process known as homeoviscous adaptation (de Mendoza, 2014; Ernst et al., 2016). In B. subtilis, temperature downshift induces a FA desaturase (Des) controlled by the DesKR two-component system (TCS) (Abriata et al., 2017). Des modifies existing membrane lipids, and is thereby suited for rapid adaptation. DesK is one of the better understood TCS sensors, with both kinase and phosphatase activity (Abriata et al., 2017; Fernandez et al., 2019). DesK lacks an extracellular sensor domain, but has multiple transmembrane segments that sense changes in the membrane physical state. DesK phosphorylates the DesR response regulator, which induces des, encoding a FA Δ5 desaturase (Altabe et al., 2003). The resultant unsaturated FAs increase bilayer fluidity, which restores DesK phosphatase activity in a negative feedback loop (de Mendoza, 2014). Longer term adaptation to low temperatures relies on an isoleucine-dependent switch to primarily anteiso-FAs (Weber et al., 2001). Since anteiso-FAs perturb the lateral interactions between adjacent lipids to a greater extent than iso-FAs (Figure 1B), this shift increases membrane fluidity (Kingston et al., 2011). This shift may result from a cold-dependent change in FabH activity (Beranova et al., 2008; Saunders et al., 2016).
Membranes must also adapt to conditions that increase fluidity. In B. subtilis, the ECF σ factor σW is activated by detergents, antibiotics, and bacteriocins active on the membrane (Cao et al., 2002b; Pietiainen et al., 2005; Butcher and Helmann, 2006; Helmann, 2006, 2016). A σW promoter within the fabHA-fabF operon plays a major role in homeoviscous adaptation (Kingston et al., 2011). Activation of σW leads to a decrease in FabHA levels, resulting in increased reliance on FabHB and an increase in straight chain FAs (from ∼10 to 30%). Elevated expression of the FabF elongation enzyme leads to increased FA chain length. The combined effect is a membrane with longer acyl chains and less BCFA. This increased membrane rigidity serves to protect cells against detergents and antimicrobial peptides (Kingston et al., 2011). The activation of σW is controlled by regulated proteolysis of its membrane-bound anti-σW factor (RsiW) (Schobel et al., 2004; Ellermeier and Losick, 2006; Devkota et al., 2017). However, the mechanisms by which membrane stressors trigger σW activation remain unclear.
To better understand the role of FA heterogeneity in controlling membrane properties, it would be desirable to study bacteria with chemically simple membranes. This has been achieved in B. subtilis by feeding exogenous FAs to cells with de novo FA synthesis blocked by cerulenin and a mutation to inhibit FA degradation (Nickels et al., 2020). Growth can be rescued with only two FA species: a straight-chain C16 FA (high melting) and an anteiso C15 FA (low melting). Even with only these two FA species, four distinct arrangements are possible upon acylation of glycerol-3-phosphate to generate phosphatidic acid. Cells compensate for this reduced FA complexity by altering the distribution of phospholipid headgroups, a modest induction of the DesRK system, apparent downregulation of the σW stress response, and an increase in isoprenoid lipids (Nickels et al., 2020). These results highlight the remarkable adaptability of bacterial membranes, and the interconnection between diverse stress responses.
Overview of Membrane Lipid Composition and Synthesis
One of the persistent challenges in membrane biology is to define the roles of the diverse constituent lipids (Sohlenkamp and Geiger, 2016; Dowhan et al., 2019; Chwastek et al., 2020). Although membranes have a complex and adaptable composition (the lipidome), cells are remarkably resilient to genetic alterations that remove lipid species. Because of its single membrane and ease of genetic manipulation, B. subtilis presents an attractive model system (Nickels et al., 2017). The B. subtilis lipidome comprises ∼70% phospholipids and ∼30% neutral glucolipids. The major phospholipids are phosphatidylglycerol (PhG) and phosphatidylethanolamine (PE), with minor contributions from cardiolipin and lysylphosphatidylglycerol (LPG). Variations in phospholipid headgroup size and charge modulate membrane properties (Figure 1B). Membranes also contain LTA anchored to neutral glucolipids, which together with peptidoglycan-linked wall teichoic acid (WTA) can account for up to 60% of the dry weight of the cell wall (Rajagopal and Walker, 2017; Sumrall et al., 2020). However, LTA fractionates with the wall during membrane lipid extraction, and is often not considered in lipidome measurements.
The only essential phospholipid in B. subtilis is PhG. Remarkably, the membrane can be simplified to contain close to 100% PhG with no glucolipids. Despite a greatly simplified membrane, such mutants can grow rapidly, albeit with a highly abnormal coiled filament morphology (Salzberg and Helmann, 2008). Genetic perturbations of membrane composition can lead to resistance to cationic antimicrobial peptides (CAMP). For example, gain-of-function mutations in mprF, encoding the LPG synthase/flippase, can confer daptomycin resistance possibly by reducing surface charge (Ernst et al., 2018; Ernst and Peschel, 2019). Consistently, mprF null mutants have increased daptomycin sensitivity and overexpression decreases sensitivity in B. subtilis (Hachmann et al., 2009). Daptomycin resistance also results from pgsA mutations that decrease PhG levels (Hachmann et al., 2011; Peleg et al., 2012).
In addition to the dominant phospholipids and glucolipids, membranes contain numerous other lipid species. Most prominent are the isoprenoid lipids synthesized by polymerization of C5 isoprene units (Figure 1C). The key intermediate farnesyl-PP (C15) can be joined (head-to-head) to generate squalene (C30) (Pan et al., 2015; van der Donk, 2015), a precursor of cholesterol and other sterols in eukaryotes and of structurally related hopanoid lipids in many bacteria. One major hopanoid is diploterol (Figure 1C), with five fused rings that can be further modified in a variety of ways (Belin et al., 2018). Farnesyl-PP can also be extended by UppS, which sequentially adds eight isopentenyl units to generate undecaprenyl-PP, the C55 carrier lipid that supports cell wall synthesis (Figure 1C). Alternatively, the HepST complex can extend farnesyl-PP to generate heptaprenyl (C35)-PP, an isoprenoid used as a lipid anchor for menaquinone (MK-7), the electron carrier for respiration. In B. subtilis, this same precursor can be processed to polycyclic C35-sesquarterpenoids, which may be functionally similar to C30 hopanoids (Bosak et al., 2008; Takigawa et al., 2010; Sato et al., 2011; Sato, 2013). This process is initiated by YtpB, which generates tetraprenyl-β-curcumene, and then SqhC (a homolog of squalene-hopene cyclases) to generate the C35 tetracyclic product known as baciterpenol A (Sato, 2013). Although initially described in spores, and named “sporulenes” (Bosak et al., 2008), these sesquarterpenoids are found in vegetative cells (Takigawa et al., 2010). Finally, heptaprenyl-PP can be coupled to glycerol-1-phosphate by PcrB, and then further processed by an unidentified phosphatase and the YvoF acetyltranferase to generate an ether linked lipid of unknown function (Linde et al., 2016).
Lateral Heterogeneity and Functional Membrane Microdomains
In eukaryotes, cholesterol is associated with the generation of functional membrane microdomains (FMM), also called lipid rafts. These regions have relatively low membrane fluidity (a liquid-ordered, or Lo phase) and are associated with flotillins. B. subtilis also encodes flotillin homologs, regulated by σW (Huang et al., 1999; Wiegert et al., 2001). These proteins, subsequently renamed FloA and FloT, are implicated in FMM formation (Figure 1A). The notion of FMMs in bacteria received strong impetus from the finding that yisP mutants, lacking a putative squalene synthase, was defective in biofilm formation (Lopez and Kolter, 2010). Together with the finding of a punctate localization for FloT, and chemical inhibition studies with compounds that affect sterol synthesis, this led to the proposal that bacteria harbor FMMs (Bramkamp and Lopez, 2015; Wagner et al., 2017). However, subsequent work revealed that YisP is a farnesyl-PP phosphatase that generates farnesol (Figure 1C), rather than squalene (Feng et al., 2014). Moreover, farnesol itself complements the biofilm defect of the yisP mutant, suggesting that this long chain alcohol may have an ordering effect on FMMs provided in other systems by hopanoids or carotenoids (Bell and Chappell, 2014; Feng et al., 2014). B. subtilis FMMs are enriched in flotillins (FloA and FloT) and their associated signaling complexes, with FMM formation apparently stabilized by farnesol (YisP product). No role for the C35 isoprenoid lipids has yet been demonstrated in biofilm formation (Lopez and Kolter, 2010) or in FMM formation or function.
Lateral heterogeneity, including FMMs, is likely a feature of most bacterial membranes. However, the lipid species that are required to form FMMs are still poorly understood, but likely include carotenoids, hopanoids, and other polycyclic isoprenoid lipids (Lopez and Koch, 2017). Hopanoids are structurally diverse and fulfill a broad range of functions in bacterial membranes (Belin et al., 2018). The hopanoid diplopterol (Figure 1C) orders saturated lipids and glycolipids in the outer membrane of Methylobacterium extorquens, and deficient mutants are impaired in multidrug transport (Saenz et al., 2015). Hopanoids and other polycyclic isoprenoids are present in many Gram-positive bacteria as well, suggestive of a role in the plasma membrane. In methicillin-resistant S. aureus, the carotenoid staphyloxanthin (Figure 1C) colocalizes in FMMs with FloA, and disruption of these domains with isoprenoid synthesis inhibitors interferes with the function of the penicillin-binding protein required for β-lactam resistance (PBP2a) (Garcia-Fernandez et al., 2017; Foster, 2019). The formation and function of FMMs, in both the inner (plasma) and outer membrane, remains an important area for future research.
Cell Envelope Stress Responses That Modulate Lipid Composition
Bacteria generally have a negatively charged membrane, which contributes to their susceptibility to CAMPs, bacteriocins, and antimicrobials. In B. subtilis, membrane composition and properties are regulated by ECF σ factors (Eiamphungporn and Helmann, 2008; Kingston et al., 2013; Helmann, 2016). Because of their overlapping activation and promoter recognition properties, these CESRs are intertwined and referred to as an σECF stress response (Mascher et al., 2007). In B. subtilis, activation of σX reduces the net negative charge of the membrane by increasing zwitterionic PE levels (Cao and Helmann, 2004; Ho and Ellermeier, 2019). The net negative charge of the cell wall can be further reduced by D-alanylation of teichoic acids, activated by σX (Cao and Helmann, 2004; Ho and Ellermeier, 2019) and σV, a lysozyme-responsive CESR (Guariglia-Oropeza and Helmann, 2011; Ho et al., 2011; Ho and Ellermeier, 2019). In S. aureus, surface membrane charge is modified by the induction of mprF by the GraRS TCS, thereby increasing LPG levels (Falord et al., 2011; Yang et al., 2012). In B. anthracis, the membrane-active compound targocil activates the EdsRS TCS, which induces expression of a cardiolipin synthase (Laut et al., 2020). Thus, many different stimuli can trigger changes in the membrane lipidome.
Bacillus subtilis σECF factors also control other membrane-related functions, although the effects are not yet understood. For example, σM activates the ytpAB operon. The YtpA lysophospholipase cleaves FAs from the 2 position of phospholipids resulting in a lysophospholipid (bacilysocin) suggested to function as an antibiotic (Tamehiro et al., 2002). However, it is unclear if bacilysocin is ever released at levels sufficient to serve as an antibiotic, and it may instead modify membrane properties or be an intermediate in lipid remodeling. As noted above, YtpB initiates synthesis of baciterpenol (Figure 1C; Bosak et al., 2008; Sato et al., 2011; Sato, 2013). Genetic studies have revealed only modest phenotypes for ytpAB mutants, including effects on antibiotic sensitivity, sporulation, and germination (Kingston et al., 2014; Sayer et al., 2019). In the case of ytpB, the observed phenotype (bacitracin sensitivity) was due to the accumulation of the substrate (heptaprenyl-PP) rather than a loss of baciterpenol (Kingston et al., 2014).
Genetic perturbations of membrane composition can also trigger CESRs. For example, deletion of LTA synthases induces σECF factors. An ltaS mutation upregulates σM, which then activates expression of the alternate LTA synthase LtaSa. The absence of both ltaS and ltaSa leads to activation of additional σECF factors (Hashimoto et al., 2013). The depletion of PhG, a building block of LTA, also activates σM and to a lesser extent σV (Hashimoto et al., 2009; Seki et al., 2019). The effects of mutations that affect glucolipids have been particularly challenging to understand. Glucolipids produced by UgtP are important membrane lipids and also serve as the lipid anchor of LTA. ugtP mutants lacking glucolipids are shorter and rounder, have abnormal localization of MreB, and altered assembly of FtsZ (Weart et al., 2007). Whether this abnormal morphology is due, in part, to the loss of glucolipids is unclear (Matsuoka, 2018). Mutation of ugtP activates a σECF stress response and can be suppressed by production of monoglycosyldiacylglycerol (MGlcDG) using a heterologous synthase. Since this product does not function as an LTA anchor lipid, this suggests that it is the loss of glucolipids that induces the σECF response (Matsuoka et al., 2016). The mechanistic basis for activation of σECF factors in the absence of glucolipids is unclear, but at least for σV does not require intramembrane proteolysis of the anti-σ factor (Seki et al., 2019). One hypothesis is that glucolipids might regulate folding and function of intramembrane proteins (Matsuoka, 2018).
Cell Envelope Stress Responses That Function Through Membrane Proteins
In addition to modulating lipid composition, CESRs also induce proteins that function in membrane protection and remodeling. In B. subtilis, these proteins include two flotillin homologs (FloA, FloT), two members of the phage shock protein family (LiaH, PspA), as well as antibiotic specific detoxification modules. The roles of these proteins in stabilizing and repairing the membrane are increasingly appreciated, although the precise mechanisms remain controversial.
Flotillins and Modulation of Membrane Fluidity
Flotillins are members of the widely conserved stomatin, prohibitin, flotillin, and HflK/C (SPFH) domain proteins. Flotillins localize to FMMs and are thought to be required for FMM function. In S. aureus, FloA colocalizes with staphyloxanthin in FMMs (Garcia-Fernandez et al., 2017; Foster, 2019). In other systems, flotillins and FMMs are associated with flagellar function and chemotaxis (Padilla-Vaca et al., 2019; Takekawa et al., 2019), type VII secretion (Mielich-Suss et al., 2017), signaling (Wagner et al., 2017), and interaction with the host during infection (Hutton et al., 2017). Ongoing efforts strive to track the mobility, oligomerization state, and interaction partners of flotillins in living cells.
Bacillus subtilis FloA and FloT are oligomeric, integral membrane proteins implicated in the formation of FMMs (Lopez and Kolter, 2010; Bach and Bramkamp, 2013; Bramkamp and Lopez, 2015; Lopez and Koch, 2017), and regulated by σW (Huang et al., 1999; Cao et al., 2002a). FloA and FloT are thought to help partition the membrane into low fluidity FMM regions that are spatially distinct from more fluid regions. A direct role for flotillins in FMM formation has been challenged, however, since B. subtilis FloA and FloT do not always colocalize, and form separated foci of ∼100 nm in diameter that appear spatially distinct from FMMs (Dempwolff et al., 2016). Counter-intuitively, flotillins appear to be required for regions of increased fluidity (RIFs), which are the counterpart to the FMMs. A lack of flotillins leads to a decrease in membrane fluidity and a concomitant reduction in activity of the MreB-directed elongasome complex that synthesizes peptidoglycan. This loss of membrane fluidity can be chemically complemented with fluidizing agents such as benzoyl alcohol (Zielinska et al., 2020).
Flotillins also functionally interact with DynA, a constitutively expressed dynamin homolog (Dempwolff et al., 2012; Dempwolff and Graumann, 2014). Dynamins are membrane-associated GTPases implicated in membrane remodeling, fusion and fission, and lipid mixing (Guo and Bramkamp, 2019). DynA may help repair damaged membrane regions, and contribute to resistance against antibiotics that bind membrane components, including nisin, bacitracin, and daptomycin (Sawant et al., 2016). Our understanding of flotillins and dynamins, and their roles in bacterial physiology is still incomplete and rapidly evolving.
Phage-Shock Proteins Protect Membrane Integrity
Cell envelope stress responses also support membrane stability through induction of PspA proteins, including two paralogs in B. subtilis: PspA an LiaH. Originally defined as part of the phage-shock protein response in Escherichia coli (Kobayashi et al., 2007; Flores-Kim and Darwin, 2016), PspA proteins comprise a conserved family including the vesicle-inducing protein in plastids (VIPP1/IM30) and mammalian ESCRT III (Thurotte et al., 2017; Liu et al., 2020). PspA proteins have a conserved N-terminal amphipathic helix required for membrane binding (McDonald et al., 2015, 2017), which seems to depend on anionic lipid content and regions with unfavorable packing geometries creating stored curvature elastic stress (McDonald et al., 2015). Structural studies reveal that VIPP1 forms oligomeric rings of various symmetries that stack together to form domes (Saur et al., 2017; Gupta et al., 2020). These rings are dynamic, and are thought to stabilize membranes during budding, tubulation, and fusion (Thurotte et al., 2017; Gutu et al., 2018; Junglas and Schneider, 2018). However, the role of these oligomeric structures has been questioned (Siebenaller et al., 2019). An alternative model suggests that these rings dissociate, and the resultant intrinsically disordered monomers interact with the membrane surface to form a protective protein “carpet” to stabilize the membrane and suppress proton leakage (Junglas et al., 2020).
Although PspA proteins are assumed to function in membrane protection and repair, their regulation differs markedly (Manganelli and Gennaro, 2017). B. subtilis PspA is regulated by σW (Wiegert et al., 2001; Cao et al., 2002a), whereas the paralog LiaH is regulated by the LiaRS TCS (Mascher et al., 2004; Jordan et al., 2006). Both paralogs localize to the membrane in response to stress and protect against membrane-damaging antibiotics (Wolf et al., 2010; Kingston et al., 2013; Dominguez-Escobar et al., 2014; Popp et al., 2020). In the case of LiaH, membrane association is mediated by interaction with the integral membrane protein LiaI (Dominguez-Escobar et al., 2014). LiaH may also protect the membrane against proton leakage during the export of proteins through the twin-arginine translocation (TAT) system (Hou et al., 2018; Bernal-Cabas et al., 2020). While the B. subtilis LiaRS regulon is rather limited in scope (Jordan et al., 2006; Wolf et al., 2010), LiaRS orthologs (e.g., S. aureus VraRS) play an important role in stress resistance in many Gram-positive pathogens, and mutations in these regulators are associated with clinical antibiotic resistance (Tran et al., 2016). In Mycobacterium tuberculosis, the PspA ortholog is also under control of the ECF σ factor σE (Datta et al., 2015), whereas E. coli pspA requires the σ54 RNAP and PspF activator (Joly et al., 2010; Flores-Kim and Darwin, 2016). A common theme in these systems is that PspA-like proteins are often regulated by a specific CESR; they can accumulate to high levels in stressed cells, and they seem to protect the membrane against disruptions that can dissipate the proton gradient (Manganelli and Gennaro, 2017).
Antibiotic Specific Detoxification Modules
Bacillus subtilis, like many soil bacteria, can synthesize a wide range of antimicrobial compounds and also encodes diverse resistance mechanisms (Stein, 2005; Caulier et al., 2019). Many antimicrobial peptides induce the B. subtilis LiaRS stress response that protects cells through induction of LiaH. Induction of σW also leads to expression of the SppA membrane-localized protease and its regulatory protein SppI, which function to clear the membrane of embedded peptides to protect against lantibiotics (Kingston et al., 2013; Henriques et al., 2020). Other antimicrobial peptides induce specific detoxification machinery, often including ABC transporters that either export the peptide antibiotic or disassemble membrane-bound peptide complexes (Staron et al., 2011; Dintner et al., 2014).
A prototype for such systems is the BceRS TCS, which regulates the bacitracin-specific induction of the BceAB ABC transporter (Radeck et al., 2016; Piepenbreier et al., 2020). Bacitracin is a peptide antibiotic made by Bacillus spp. that inhibits cell wall synthesis by binding to undecaprenyl-PP. The BceAB system appears to act in disassembly of bacitracin complexes to confer resistance (Kobras et al., 2020). In addition, BceAB interacts with the BceRS TCS to allow sensing of bacitracin (Ohki et al., 2003; Dintner et al., 2014; Fritz et al., 2015; Koh et al., 2020). The BceRS-AB system provides a first line of defense against bacitracin, with higher levels of antibiotic activating the protective responses mediated by the LiaRS and σECF regulons (Radeck et al., 2016). The detailed study of the B. subtilis bacitracin stress response has provided lessons relevant to the understanding of other antibiotic detoxification modules. Similar genetic modules, encoding both TCS and ABC transporter/sensors have been described for several other antimicrobial peptides (Revilla-Guarinos et al., 2014). Since induction can be quite specific, these systems provide a basis for antibiotic-inducible gene expression systems (Wolf and Mascher, 2016).
Bacillus subtilis also encodes and responds to many other secondary metabolites that can induce membrane stress (Caulier et al., 2019). For example, the toxic peptide YydF∗ is encoded by the yydFGHIJ operon, together with a radical-SAM epimerase (YydG), protease (YydH), and ABC transporter (YydIJ). Transposon insertions in the presumptive efflux pump lead to the upregulation of the LiaRS stress system (Butcher et al., 2007). Subsequent studies revealed that YydF is post-translationally processed to convert two L-amino acids to D-amino acids (Benjdia et al., 2017). The resulting epipeptide, YydF∗, induces LiaRS-regulated LiaH and the FloT flotillin (Popp et al., 2020). The modified YydF∗ peptide kills B. subtilis cells by dissipating the membrane potential via membrane permeabilization. The associated concomitant decrease in membrane fluidity together with increased membrane permeabilization induces liaIH (Popp et al., 2020). YydF∗ peptides are likely synthesized by a variety of Gram-positive organisms including Enterococcus, Staphylococcus, and Streptococcus spp. as well as members of the human microbiome (Benjdia et al., 2017).
Bacillus subtilis also has CESRs induced by polyketide and polyene-type antimicrobials. For example, Streptomyces spp. produce linear polyketides (linearmycins) that depolarize the membrane (Stubbendieck and Straight, 2015, 2017; Stubbendieck et al., 2018). Linearmycins strongly activate the LnrJK TCS that regulates an ABC transporter, LnrLMN (Stubbendieck and Straight, 2017; Revilla-Guarinos et al., 2020). This ABC transporter also provides resistance against other polyenes, including the anti-fungal amphotericin (Revilla-Guarinos et al., 2020).
Outlook
Here we provide a brief overview of the diverse ways in which CESRs help modify and protect the membrane in response to environmental threats (Table 1). This is a rapidly evolving field, and the impact of membrane composition on cell physiology is still mysterious. We have much to learn about the synthesis and roles of minor lipids (sesquarterpenes, ether lipids, lysophospholipids). There is a growing need to reconcile current models of lipid rafts, and the role that isoprenoid lipids and flotillins play in their formation. The activities of the VIPP1/IM30/PspA family of proteins in membrane repair and protection, and in particular the specific role of different oligomeric states, are still debated. Finally, the mechanisms by which diverse CESRs sense membrane perturbations are largely unknown, although considerable progress has been made in the specific cases of the DesK sensor kinase (Abriata et al., 2017), flux-sensing by peptide detoxification modules (Koh et al., 2020), and the lysozyme-mediated induction of the σV protein (Ho and Ellermeier, 2019). The overall picture is of the cell membrane as a complex and adaptable assemblage of many different lipid and protein species that still has many secrets to reveal.
Author Contributions
Both authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
Funding
This work was funded by the National Institutes of Health under award number R35GM122461 to JH. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We appreciate helpful comments from our Cornell colleagues, Thorsten Mascher, James Saenz, and Diego de Mendoza.
References
Abriata, L. A., Albanesi, D., Dal Peraro, M., and de Mendoza, D. (2017). Signal sensing and transduction by histidine kinases as unveiled through studies on a temperature sensor. Acc. Chem. Res. 50, 1359–1366. doi: 10.1021/acs.accounts.6b00593
Albanesi, D., Reh, G., Guerin, M. E., Schaeffer, F., Debarbouille, M., Buschiazzo, A., et al. (2013). Structural basis for feed-forward transcriptional regulation of membrane lipid homeostasis in Staphylococcus aureus. PLoS Pathog. 9:e1003108. doi: 10.1371/journal.ppat.1003108
Altabe, S. G., Aguilar, P., Caballero, G. M., and de Mendoza, D. (2003). The Bacillus subtilis acyl lipid desaturase is a delta5 desaturase. J. Bacteriol. 185, 3228–3231. doi: 10.1128/jb.185.10.3228-3231.2003
Bach, J. N., and Bramkamp, M. (2013). Flotillins functionally organize the bacterial membrane. Mol. Microbiol. 88, 1205–1217. doi: 10.1111/mmi.12252
Belin, B. J., Busset, N., Giraud, E., Molinaro, A., Silipo, A., and Newman, D. K. (2018). Hopanoid lipids: from membranes to plant-bacteria interactions. Nat. Rev. Microbiol. 16, 304–315. doi: 10.1038/nrmicro.2017.173
Bell, S. A., and Chappell, J. (2014). Now playing: farnesol in the biofilm. Chem. Biol. 21, 1421–1422. doi: 10.1016/j.chembiol.2014.11.001
Benjdia, A., Guillot, A., Ruffie, P., Leprince, J., and Berteau, O. (2017). Post-translational modification of ribosomally synthesized peptides by a radical SAM epimerase in Bacillus subtilis. Nat. Chem. 9, 698–707. doi: 10.1038/nchem.2714
Beranova, J., Jemiola-Rzeminska, M., Elhottova, D., Strzalka, K., and Konopasek, I. (2008). Metabolic control of the membrane fluidity in Bacillus subtilis during cold adaptation. Biochim. Biophys. Acta 1778, 445–453. doi: 10.1016/j.bbamem.2007.11.012
Bernal-Cabas, M., Miethke, M., Antelo-Varela, M., Aguilar Suarez, R., Neef, J., Schon, L., et al. (2020). Functional association of the stress-responsive LiaH protein and the minimal TatAyCy protein translocase in Bacillus subtilis. Biochim. Biophys. Acta Mol. Cell Res. 1867:118719. doi: 10.1016/j.bbamcr.2020.118719
Bosak, T., Losick, R. M., and Pearson, A. (2008). A polycyclic terpenoid that alleviates oxidative stress. Proc. Natl. Acad. Sci. U.S.A. 105, 6725–6729. doi: 10.1073/pnas.0800199105
Bramkamp, M., and Lopez, D. (2015). Exploring the existence of lipid rafts in bacteria. Microbiol. Mol. Biol. Rev. 79, 81–100. doi: 10.1128/mmbr.00036-14
Butcher, B. G., and Helmann, J. D. (2006). Identification of Bacillus subtilis σW-dependent genes that provide intrinsic resistance to antimicrobial compounds produced by Bacilli. Mol. Microbiol. 60, 765–782. doi: 10.1111/j.1365-2958.2006.05131.x
Butcher, B. G., Lin, Y. P., and Helmann, J. D. (2007). The yydFGHIJ operon of Bacillus subtilis encodes a peptide that induces the LiaRS two-component system. J. Bacteriol. 189, 8616–8625. doi: 10.1128/jb.01181-07
Cao, M., and Helmann, J. D. (2004). The Bacillus subtilis extracytoplasmic-function σX factor regulates modification of the cell envelope and resistance to cationic antimicrobial peptides. J. Bacteriol. 186, 1136–1146. doi: 10.1128/jb.186.4.1136-1146.2004
Cao, M., Kobel, P. A., Morshedi, M. M., Wu, M. F., Paddon, C., and Helmann, J. D. (2002a). Defining the Bacillus subtilis σW regulon: a comparative analysis of promoter consensus search, run-off transcription/macroarray analysis (ROMA), and transcriptional profiling approaches. J. Mol. Biol. 316, 443–457. doi: 10.1006/jmbi.2001.5372
Cao, M., Wang, T., Ye, R., and Helmann, J. D. (2002b). Antibiotics that inhibit cell wall biosynthesis induce expression of the Bacillus subtilis σW and σM regulons. Mol. Microbiol. 45, 1267–1276. doi: 10.1046/j.1365-2958.2002.03050.x
Caulier, S., Nannan, C., Gillis, A., Licciardi, F., Bragard, C., and Mahillon, J. (2019). Overview of the Antimicrobial Compounds Produced by Members of the Bacillus subtilis Group. Front. Microbiol. 10:302.
Choi, K. H., Heath, R. J., and Rock, C. O. (2000). beta-ketoacyl-acyl carrier protein synthase III (FabH) is a determining factor in branched-chain fatty acid biosynthesis. J. Bacteriol. 182, 365–370. doi: 10.1128/jb.182.2.365-370.2000
Chwastek, G., Surma, M. A., Rizk, S., Grosser, D., Lavrynenko, O., Rucinska, M., et al. (2020). Principles of membrane adaptation revealed through environmentally induced bacterial lipidome remodeling. Cell Rep. 32:108165. doi: 10.1016/j.celrep.2020.108165
Datta, P., Ravi, J., Guerrini, V., Chauhan, R., Neiditch, M. B., Shell, S. S., et al. (2015). The Psp system of Mycobacterium tuberculosis integrates envelope stress-sensing and envelope-preserving functions. Mol. Microbiol. 97, 408–422. doi: 10.1111/mmi.13037
de Mendoza, D. (2014). Temperature sensing by membranes. Annu. Rev. Microbiol. 68, 101–116. doi: 10.1146/annurev-micro-091313-103612
Dempwolff, F., and Graumann, P. L. (2014). Genetic links between bacterial dynamin and flotillin proteins. Commun. Integr. Biol. 7:e970972. doi: 10.4161/cib.29578
Dempwolff, F., Schmidt, F. K., Hervas, A. B., Stroh, A., Rosch, T. C., Riese, C. N., et al. (2016). Super resolution fluorescence microscopy and tracking of bacterial flotillin (Reggie) paralogs provide evidence for defined-sized protein microdomains within the bacterial membrane but absence of clusters containing detergent-resistant proteins. PLoS Genet. 12:e1006116. doi: 10.1371/journal.pgen.1006116
Dempwolff, F., Wischhusen, H. M., Specht, M., and Graumann, P. L. (2012). The deletion of bacterial dynamin and flotillin genes results in pleiotrophic effects on cell division, cell growth and in cell shape maintenance. BMC Microbiol. 12:298. doi: 10.1186/1471-2180-12-298
Devkota, S. R., Kwon, E., Ha, S. C., Chang, H. W., and Kim, D. Y. (2017). Structural insights into the regulation of Bacillus subtilis σW activity by anti-sigma RsiW. PLoS One 12:e0174284. doi: 10.1371/journal.pone.0174284
Dintner, S., Heermann, R., Fang, C., Jung, K., and Gebhard, S. (2014). A sensory complex consisting of an ATP-binding cassette transporter and a two-component regulatory system controls bacitracin resistance in Bacillus subtilis. J. Biol. Chem. 289, 27899–27910. doi: 10.1074/jbc.m114.596221
Dominguez-Escobar, J., Wolf, D., Fritz, G., Hofler, C., Wedlich-Soldner, R., and Mascher, T. (2014). Subcellular localization, interactions and dynamics of the phage-shock protein-like Lia response in Bacillus subtilis. Mol. Microbiol. 92, 716–732. doi: 10.1111/mmi.12586
Dowhan, W., Vitrac, H., and Bogdanov, M. (2019). Lipid-assisted membrane protein folding and topogenesis. Protein J. 38, 274–288. doi: 10.1007/s10930-019-09826-7
Eiamphungporn, W., and Helmann, J. D. (2008). The Bacillus subtilis σM regulon and its contribution to cell envelope stress responses. Mol. Microbiol. 67, 830–848. doi: 10.1111/j.1365-2958.2007.06090.x
Ellermeier, C. D., and Losick, R. (2006). Evidence for a novel protease governing regulated intramembrane proteolysis and resistance to antimicrobial peptides in Bacillus subtilis. Genes Dev. 20, 1911–1922. doi: 10.1101/gad.1440606
Ernst, C. M., and Peschel, A. (2019). MprF-mediated daptomycin resistance. Int. J. Med. Microbiol. 309, 359–363. doi: 10.1016/j.ijmm.2019.05.010
Ernst, C. M., Slavetinsky, C. J., Kuhn, S., Hauser, J. N., Nega, M., Mishra, N. N., et al. (2018). Gain-of-function mutations in the phospholipid flippase MprF confer specific daptomycin resistance. mBio 9:e01659-18.
Ernst, R., Ejsing, C. S., and Antonny, B. (2016). Homeoviscous adaptation and the regulation of membrane lipids. J. Mol. Biol. 428, 4776–4791. doi: 10.1016/j.jmb.2016.08.013
Falord, M., Mader, U., Hiron, A., Debarbouille, M., and Msadek, T. (2011). Investigation of the Staphylococcus aureus GraSR regulon reveals novel links to virulence, stress response and cell wall signal transduction pathways. PLoS One 6:e21323. doi: 10.1371/journal.pone.0021323
Feng, X., Hu, Y., Zheng, Y., Zhu, W., Li, K., Huang, C. H., et al. (2014). Structural and functional analysis of Bacillus subtilis YisP reveals a role of its product in biofilm production. Chem. Biol. 21, 1557–1563. doi: 10.1016/j.chembiol.2014.08.018
Fernandez, P., Porrini, L., Albanesi, D., Abriata, L. A., Dal Peraro, M., de Mendoza, D., et al. (2019). Transmembrane prolines mediate signal sensing and decoding in Bacillus subtilis DesK histidine kinase. mBio 10:e02564-19.
Flores-Kim, J., and Darwin, A. J. (2016). The phage shock protein response. Annu. Rev. Microbiol. 70, 83–101. doi: 10.1146/annurev-micro-102215-095359
Foster, T. J. (2019). Can beta-lactam antibiotics be resurrected to combat MRSA? Trends Microbiol. 27, 26–38. doi: 10.1016/j.tim.2018.06.005
Fritz, G., Dintner, S., Treichel, N. S., Radeck, J., Gerland, U., Mascher, T., et al. (2015). A New way of sensing: need-based activation of antibiotic resistance by a flux-sensing mechanism. mBio 6:e00975.
Fujita, Y., Matsuoka, H., and Hirooka, K. (2007). Regulation of fatty acid metabolism in bacteria. Mol. Microbiol. 66, 829–839. doi: 10.1111/j.1365-2958.2007.05947.x
Garcia-Fernandez, E., Koch, G., Wagner, R. M., Fekete, A., Stengel, S. T., Schneider, J., et al. (2017). Membrane microdomain disassembly inhibits MRSA antibiotic resistance. Cell 171, 1354–1367.e1320.
Guariglia-Oropeza, V., and Helmann, J. D. (2011). Bacillus subtilis σV confers lysozyme resistance by activation of two cell wall modification pathways, peptidoglycan O-acetylation and D-alanylation of teichoic acids. J. Bacteriol. 193, 6223–6232. doi: 10.1128/jb.06023-11
Guo, L., and Bramkamp, M. (2019). Bacterial dynamin-like protein DynA mediates lipid and content mixing. FASEB J. 33, 11746–11757. doi: 10.1096/fj.201900844rr
Gupta, T. K., Klumpe, S., Gries, K., Heinz, S., Wietrzynski, W., Ohnishi, N., et al. (2020). Structural basis for VIPP1 oligomerization and maintenance of thylakoid membrane integrity. BioRxiv [Preprint]. doi: 10.1101/2020.08.11.243204
Gutu, A., Chang, F., and O’Shea, E. K. (2018). Dynamical localization of a thylakoid membrane binding protein is required for acquisition of photosynthetic competency. Mol. Microbiol. 108, 16–31. doi: 10.1111/mmi.13912
Hachmann, A. B., Angert, E. R., and Helmann, J. D. (2009). Genetic analysis of factors affecting susceptibility of Bacillus subtilis to daptomycin. Antimicrob. Agents Chemother. 53, 1598–1609. doi: 10.1128/aac.01329-08
Hachmann, A. B., Sevim, E., Gaballa, A., Popham, D. L., Antelmann, H., and Helmann, J. D. (2011). Reduction in membrane phosphatidylglycerol content leads to daptomycin resistance in Bacillus subtilis. Antimicrob. Agents Chemother. 55, 4326–4337. doi: 10.1128/aac.01819-10
Hashimoto, M., Seki, T., Matsuoka, S., Hara, H., Asai, K., Sadaie, Y., et al. (2013). Induction of extracytoplasmic function sigma factors in Bacillus subtilis cells with defects in lipoteichoic acid synthesis. Microbiology (Reading) 159, 23–35. doi: 10.1099/mic.0.063420-0
Hashimoto, M., Takahashi, H., Hara, Y., Hara, H., Asai, K., Sadaie, Y., et al. (2009). Induction of extracytoplasmic function sigma factors in Bacillus subtilis cells with membranes of reduced phosphatidylglycerol content. Genes Genet. Syst. 84, 191–198. doi: 10.1266/ggs.84.191
Helmann, J. D. (2006). Deciphering a complex genetic regulatory network: the Bacillus subtilis σW protein and intrinsic resistance to antimicrobial compounds. Sci. Prog. 89, 243–266. doi: 10.3184/003685006783238290
Helmann, J. D. (2016). Bacillus subtilis extracytoplasmic function (ECF) sigma factors and defense of the cell envelope. Curr. Opin. Microbiol. 30, 122–132. doi: 10.1016/j.mib.2016.02.002
Henriques, G., McGovern, S., Neef, J., Antelo-Varela, M., Gotz, F., Otto, A., et al. (2020). SppI forms a membrane protein complex with sppa and inhibits its protease activity in Bacillus subtilis. mSphere 5:e00724-20.
Ho, T. D., and Ellermeier, C. D. (2019). Activation of the extracytoplasmic function sigma factor σV by lysozyme. Mol. Microbiol. 112, 410–419. doi: 10.1111/mmi.14348
Ho, T. D., Hastie, J. L., Intile, P. J., and Ellermeier, C. D. (2011). The Bacillus subtilis extracytoplasmic function sigma factor σV is induced by lysozyme and provides resistance to lysozyme. J. Bacteriol. 193, 6215–6222. doi: 10.1128/jb.05467-11
Hofler, C., Heckmann, J., Fritsch, A., Popp, P., Gebhard, S., Fritz, G., et al. (2016). Cannibalism stress response in Bacillus subtilis. Microbiology (Reading) 162, 164–176. doi: 10.1099/mic.0.000176
Hou, B., Heidrich, E. S., Mehner-Breitfeld, D., and Bruser, T. (2018). The TatA component of the twin-arginine translocation system locally weakens the cytoplasmic membrane of Escherichia coli upon protein substrate binding. J. Biol. Chem. 293, 7592–7605. doi: 10.1074/jbc.ra118.002205
Huang, X., Gaballa, A., Cao, M., and Helmann, J. D. (1999). Identification of target promoters for the Bacillus subtilis extracytoplasmic function sigma factor, σW. Mol. Microbiol. 31, 361–371. doi: 10.1046/j.1365-2958.1999.01180.x
Hutton, M. L., D’Costa, K., Rossiter, A. E., Wang, L., Turner, L., Steer, D. L., et al. (2017). A Helicobacter pylori homolog of eukaryotic flotillin is involved in cholesterol accumulation, epithelial cell responses and host colonization. Front. Cell Infect. Microbiol. 7:219.
Jolliffe, L. K., Doyle, R. J., and Streips, U. N. (1981). The energized membrane and cellular autolysis in Bacillus subtilis. Cell 25, 753–763. doi: 10.1016/0092-8674(81)90183-5
Joly, N., Engl, C., Jovanovic, G., Huvet, M., Toni, T., Sheng, X., et al. (2010). Managing membrane stress: the phage shock protein (Psp) response, from molecular mechanisms to physiology. FEMS Microbiol. Rev. 34, 797–827. doi: 10.1111/j.1574-6976.2010.00240.x
Jordan, S., Junker, A., Helmann, J. D., and Mascher, T. (2006). Regulation of LiaRS-dependent gene expression in Bacillus subtilis: identification of inhibitor proteins, regulator binding sites, and target genes of a conserved cell envelope stress-sensing two-component system. J. Bacteriol. 188, 5153–5166. doi: 10.1128/jb.00310-06
Junglas, B., and Schneider, D. (2018). What is Vipp1 good for? Mol. Microbiol. 108, 1–5. doi: 10.1111/mmi.13924
Junglas, B., Orru, R., Axt, A., Siebenaller, C., Steinchen, W., Heidrich, J., et al. (2020). IM30 IDPs form a membrane-protective carpet upon super-complex disassembly. Commun. Biol. 3:595.
Kingston, A. W., Liao, X., and Helmann, J. D. (2013). Contributions of the σW, σM and σX regulons to the lantibiotic resistome of Bacillus subtilis. Mol. Microbiol. 90, 502–518. doi: 10.1111/mmi.12380
Kingston, A. W., Subramanian, C., Rock, C. O., and Helmann, J. D. (2011). A σW-dependent stress response in Bacillus subtilis that reduces membrane fluidity. Mol. Microbiol. 81, 69–79. doi: 10.1111/j.1365-2958.2011.07679.x
Kingston, A. W., Zhao, H., Cook, G. M., and Helmann, J. D. (2014). Accumulation of heptaprenyl diphosphate sensitizes Bacillus subtilis to bacitracin: implications for the mechanism of resistance mediated by the BceAB transporter. Mol. Microbiol. 93, 37–49. doi: 10.1111/mmi.12637
Kobayashi, R., Suzuki, T., and Yoshida, M. (2007). Escherichia coli phage-shock protein A (PspA) binds to membrane phospholipids and repairs proton leakage of the damaged membranes. Mol. Microbiol. 66, 100–109. doi: 10.1111/j.1365-2958.2007.05893.x
Kobras, C. M., Piepenbreier, H., Emenegger, J., Sim, A., Fritz, G., and Gebhard, S. (2020). BceAB-type antibiotic resistance transporters appear to act by target protection of cell wall synthesis. Antimicrob. Agents Chemother. 64:e02241-19.
Koh, A., Gibbon, M. J., Van der Kamp, M. W., Pudney, C. R., and Gebhard, S. (2020). Conformation control of the histidine kinase BceS of Bacillus subtilis by its cognate ABC-transporter facilitates need-based activation of antibiotic resistance. Mol. Microbiol. 115, 157–174. doi: 10.1111/mmi.14607
Lamsa, A., Liu, W. T., Dorrestein, P. C., and Pogliano, K. (2012). The Bacillus subtilis cannibalism toxin SDP collapses the proton motive force and induces autolysis. Mol. Microbiol. 84, 486–500. doi: 10.1111/j.1365-2958.2012.08038.x
Laut, C. L., Perry, W. J., Metzger, A. L., Weiss, A., Stauff, D. L., Walker, S., et al. (2020). Bacillus anthracis responds to targocil-induced envelope damage through EdsRS activation of cardiolipin synthesis. mBio 11:e03375-19.
Linde, M., Peterhoff, D., Sterner, R., and Babinger, P. (2016). Identification and characterization of heptaprenylglyceryl phosphate processing enzymes in Bacillus subtilis. J. Biol. Chem. 291, 14861–14870. doi: 10.1074/jbc.m115.711994
Liu, J., Tassinari, M., Souza, D. P., Naskar, S., Noel, J. K., Bohuszewicz, O., et al. (2020). Bacterial Vipp1 and PspA are members of the ancient ESCRT-III membrane-remodelling superfamily. bioRxiv [Preprint]. doi: 10.1101/2020.08.13.249979
Lopez, D., and Koch, G. (2017). Exploring functional membrane microdomains in bacteria: an overview. Curr. Opin. Microbiol. 36, 76–84. doi: 10.1016/j.mib.2017.02.001
Lopez, D., and Kolter, R. (2010). Functional microdomains in bacterial membranes. Genes Dev. 24, 1893–1902. doi: 10.1101/gad.1945010
Machinandiarena, F., Nakamatsu, L., Schujman, G. E., de Mendoza, D., and Albanesi, D. (2020). Revisiting the coupling of fatty acid to phospholipid synthesis in bacteria with FapR regulation. Mol. Microbiol. 114:14574.
Manganelli, R., and Gennaro, M. L. (2017). Protecting from envelope stress: variations on the phage-shock-protein theme. Trends Microbiol. 25, 205–216. doi: 10.1016/j.tim.2016.10.001
Martinez, M. A., Zaballa, M. E., Schaeffer, F., Bellinzoni, M., Albanesi, D., Schujman, G. E., et al. (2010). A novel role of malonyl-ACP in lipid homeostasis. Biochemistry 49, 3161–3167. doi: 10.1021/bi100136n
Mascher, T., Hachmann, A. B., and Helmann, J. D. (2007). Regulatory overlap and functional redundancy among Bacillus subtilis extracytoplasmic function sigma factors. J. Bacteriol. 189, 6919–6927. doi: 10.1128/jb.00904-07
Mascher, T., Zimmer, S. L., Smith, T. A., and Helmann, J. D. (2004). Antibiotic-inducible promoter regulated by the cell envelope stress-sensing two-component system LiaRS of Bacillus subtilis. Antimicrob. Agents Chemother. 48, 2888–2896. doi: 10.1128/aac.48.8.2888-2896.2004
Matsuoka, S. (2018). Biological functions of glucolipids in Bacillus subtilis. Genes Genet. Syst. 92, 217–221. doi: 10.1266/ggs.17-00017
Matsuoka, S., Seki, T., Matsumoto, K., and Hara, H. (2016). Suppression of abnormal morphology and extracytoplasmic function sigma activity in Bacillus subtilis ugtP mutant cells by expression of heterologous glucolipid synthases from Acholeplasma laidlawii. Biosci. Biotechnol. Biochem. 80, 2325–2333. doi: 10.1080/09168451.2016.1217147
McDonald, C., Jovanovic, G., Ces, O., and Buck, M. (2015). Membrane stored curvature elastic stress modulates recruitment of maintenance proteins PspA and Vipp1. mBio 6:e001188-15.
McDonald, C., Jovanovic, G., Wallace, B. A., Ces, O., and Buck, M. (2017). Structure and function of PspA and Vipp1 N-terminal peptides: insights into the membrane stress sensing and mitigation. Biochim. Biophys. Acta Biomembr. 1859, 28–39. doi: 10.1016/j.bbamem.2016.10.018
Mielich-Suss, B., Wagner, R. M., Mietrach, N., Hertlein, T., Marincola, G., Ohlsen, K., et al. (2017). Flotillin scaffold activity contributes to type VII secretion system assembly in Staphylococcus aureus. PLoS Pathog. 13:e1006728. doi: 10.1371/journal.ppat.1006728
Mitchell, A. M., and Silhavy, T. J. (2019). Envelope stress responses: balancing damage repair and toxicity. Nat. Rev. Microbiol. 17, 417–428. doi: 10.1038/s41579-019-0199-0
Muller, M., Reiss, S., Schluter, R., Mader, U., Beyer, A., Reiss, W., et al. (2014). Deletion of membrane-associated Asp23 leads to upregulation of cell wall stress genes in Staphylococcus aureus. Mol. Microbiol. 93, 1259–1268.
Nickels, J. D., Chatterjee, S., Mostofian, B., Stanley, C. B., Ohl, M., Zolnierczuk, P., et al. (2017). Bacillus subtilis lipid extract, a branched-chain fatty acid model membrane. J. Phys. Chem. Lett. 8, 4214–4217.
Nickels, J. D., Poudel, S., Chatterjee, S., Farmer, A., Cordner, D., Campagna, S. R., et al. (2020). Impact of fatty-acid labeling of Bacillus subtilis membranes on the cellular lipidome and proteome. Front. Microbiol. 11:914.
Ohki, R., Giyanto, K. T., Masuyama, W., Moriya, S., Kobayashi, K., and Ogasawara, N. (2003). The BceRS two-component regulatory system induces expression of the bacitracin transporter, BceAB, in Bacillus subtilis. Mol. Microbiol. 49, 1135–1144. doi: 10.1046/j.1365-2958.2003.03653.x
Padilla-Vaca, F., Vargas-Maya, N. I., Elizarraras-Vargas, N. U., Rangel-Serrano, A., Cardoso-Reyes, L. R., Razo-Soria, T., et al. (2019). Flotillin homologue is involved in the swimming behavior of Escherichia coli. Arch. Microbiol. 201, 999–1008. doi: 10.1007/s00203-019-01670-8
Pan, J. J., Solbiati, J. O., Ramamoorthy, G., Hillerich, B. S., Seidel, R. D., Cronan, J. E., et al. (2015). Biosynthesis of squalene from farnesyl diphosphate in bacteria: three steps catalyzed by three enzymes. ACS Cent. Sci. 1, 77–82. doi: 10.1021/acscentsci.5b00115
Parsons, J. B., and Rock, C. O. (2013). Bacterial lipids: metabolism and membrane homeostasis. Prog. Lipid Res. 52, 249–276. doi: 10.1016/j.plipres.2013.02.002
Peleg, A. Y., Miyakis, S., Ward, D. V., Earl, A. M., Rubio, A., Cameron, D. R., et al. (2012). Whole genome characterization of the mechanisms of daptomycin resistance in clinical and laboratory derived isolates of Staphylococcus aureus. PLoS One 7:e28316. doi: 10.1371/journal.pone.0028316
Petersen, I., Schluter, R., Hoff, K. J., Liebscher, V., Bange, G., Riedel, K., et al. (2020). Non-invasive and label-free 3D-visualization shows in vivo oligomerization of the staphylococcal alkaline shock protein 23 (Asp23). Sci. Rep. 10:125.
Piepenbreier, H., Sim, A., Kobras, C. M., Radeck, J., Mascher, T., Gebhard, S., et al. (2020). From modules to networks: a systems-level analysis of the bacitracin stress response in Bacillus subtilis. mSystems 5:e00687-19.
Pietiainen, M., Gardemeister, M., Mecklin, M., Leskela, S., Sarvas, M., and Kontinen, V. P. (2005). Cationic antimicrobial peptides elicit a complex stress response in Bacillus subtilis that involves ECF-type sigma factors and two-component signal transduction systems. Microbiology (Reading) 151, 1577–1592. doi: 10.1099/mic.0.27761-0
Popp, P. F., Benjdia, A., Strahl, H., Berteau, O., and Mascher, T. (2020). The epipeptide YydF intrinsically triggers the cell envelope stress response of bacillus subtilis and causes severe membrane perturbations. Front. Microbiol. 11:151.
Radeck, J., Fritz, G., and Mascher, T. (2017). The cell envelope stress response of Bacillus subtilis: from static signaling devices to dynamic regulatory network. Curr. Genet. 63, 79–90. doi: 10.1007/s00294-016-0624-0
Radeck, J., Gebhard, S., Orchard, P. S., Kirchner, M., Bauer, S., Mascher, T., et al. (2016). Anatomy of the bacitracin resistance network in Bacillus subtilis. Mol. Microbiol. 100, 607–620. doi: 10.1111/mmi.13336
Rajagopal, M., and Walker, S. (2017). Envelope structures of gram-positive bacteria. Curr. Top. Microbiol. Immunol. 404, 1–44. doi: 10.1007/82_2015_5021
Revilla-Guarinos, A., Durr, F., Popp, P. F., Doring, M., and Mascher, T. (2020). Amphotericin B specifically induces the two-component system LnrJK: development of a novel whole-cell biosensor for the detection of amphotericin-like polyenes. Front. Microbiol. 11:2022.
Revilla-Guarinos, A., Gebhard, S., Mascher, T., and Zuniga, M. (2014). Defence against antimicrobial peptides: different strategies in Firmicutes. Environ. Microbiol. 16, 1225–1237. doi: 10.1111/1462-2920.12400
Saenz, J. P., Grosser, D., Bradley, A. S., Lagny, T. J., Lavrynenko, O., Broda, M., et al. (2015). Hopanoids as functional analogues of cholesterol in bacterial membranes. Proc. Natl. Acad. Sci. U.S.A. 112, 11971–11976. doi: 10.1073/pnas.1515607112
Salie, M. J., and Thelen, J. J. (2016). Regulation and structure of the heteromeric acetyl-CoA carboxylase. Biochim. Biophys. Acta 1861, 1207–1213.
Salzberg, L. I., and Helmann, J. D. (2008). Phenotypic and transcriptomic characterization of Bacillus subtilis mutants with grossly altered membrane composition. J. Bacteriol. 190, 7797–7807. doi: 10.1128/jb.00720-08
Sato, T. (2013). Unique biosynthesis of sesquarterpenes (C35 terpenes). Biosci. Biotechnol. Biochem. 77, 1155–1159. doi: 10.1271/bbb.130180
Sato, T., Yoshida, S., Hoshino, H., Tanno, M., Nakajima, M., and Hoshino, T. (2011). Sesquarterpenes (C35 terpenes) biosynthesized via the cyclization of a linear C35 isoprenoid by a tetraprenyl-beta-curcumene synthase and a tetraprenyl-beta-curcumene cyclase: identification of a new terpene cyclase. J. Am. Chem. Soc. 133, 9734–9737. doi: 10.1021/ja203779h
Saunders, L. P., Sen, S., Wilkinson, B. J., and Gatto, C. (2016). Insights into the mechanism of homeoviscous adaptation to low temperature in branched-chain fatty acid-containing bacteria through modeling FabH kinetics from the foodborne pathogen Listeria monocytogenes. Front. Microbiol. 7:1386.
Saur, M., Hennig, R., Young, P., Rusitzka, K., Hellmann, N., Heidrich, J., et al. (2017). A Janus-faced IM30 ring involved in thylakoid membrane fusion is assembled from IM30 tetramers. Structure 25, 1380–1390.e1385.
Sawant, P., Eissenberger, K., Karier, L., Mascher, T., and Bramkamp, M. (2016). A dynamin-like protein involved in bacterial cell membrane surveillance under environmental stress. Environ. Microbiol. 18, 2705–2720. doi: 10.1111/1462-2920.13110
Sayer, C. V., Barat, B., and Popham, D. L. (2019). Identification of L-Valine-initiated-germination-active genes in Bacillus subtilis using Tn-seq. PLoS One 14:e0218220. doi: 10.1371/journal.pone.0218220
Schobel, S., Zellmeier, S., Schumann, W., and Wiegert, T. (2004). The Bacillus subtilis σW anti-sigma factor RsiW is degraded by intramembrane proteolysis through YluC. Mol. Microbiol. 52, 1091–1105. doi: 10.1111/j.1365-2958.2004.04031.x
Scholz, R., Vater, J., Budiharjo, A., Wang, Z., He, Y., Dietel, K., et al. (2014). Amylocyclicin, a novel circular bacteriocin produced by Bacillus amyloliquefaciens FZB42. J. Bacteriol. 196, 1842–1852. doi: 10.1128/jb.01474-14
Schujman, G. E., Guerin, M., Buschiazzo, A., Schaeffer, F., Llarrull, L. I., Reh, G., et al. (2006). Structural basis of lipid biosynthesis regulation in Gram-positive bacteria. EMBO J. 25, 4074–4083. doi: 10.1038/sj.emboj.7601284
Schujman, G. E., Paoletti, L., Grossman, A. D., and de Mendoza, D. (2003). FapR, a bacterial transcription factor involved in global regulation of membrane lipid biosynthesis. Dev. Cell 4, 663–672. doi: 10.1016/s1534-5807(03)00123-0
Seki, T., Furumi, T., Hashimoto, M., Hara, H., and Matsuoka, S. (2019). Activation of extracytoplasmic function sigma factors upon removal of glucolipids and reduction of phosphatidylglycerol content in Bacillus subtilis cells lacking lipoteichoic acid. Genes Genet. Syst. 94, 71–80. doi: 10.1266/ggs.18-00046
Siebenaller, C., Junglas, B., and Schneider, D. (2019). Functional implications of multiple IM30 oligomeric states. Front. Plant Sci. 10:1500.
Sohlenkamp, C., and Geiger, O. (2016). Bacterial membrane lipids: diversity in structures and pathways. FEMS Microbiol. Rev. 40, 133–159. doi: 10.1093/femsre/fuv008
Staron, A., Finkeisen, D. E., and Mascher, T. (2011). Peptide antibiotic sensing and detoxification modules of Bacillus subtilis. Antimicrob. Agents Chemother. 55, 515–525. doi: 10.1128/aac.00352-10
Stein, T. (2005). Bacillus subtilis antibiotics: structures, syntheses and specific functions. Mol. Microbiol. 56, 845–857. doi: 10.1111/j.1365-2958.2005.04587.x
Stubbendieck, R. M., and Straight, P. D. (2015). Escape from lethal bacterial competition through coupled activation of antibiotic resistance and a mobilized subpopulation. PLoS Genet. 11:e1005722. doi: 10.1371/journal.pgen.1005722
Stubbendieck, R. M., and Straight, P. D. (2017). Linearmycins activate a two-component signaling system involved in bacterial competition and biofilm morphology. J. Bacteriol. 199:JB.00186-17.
Stubbendieck, R. M., Brock, D. J., Pellois, J. P., Gill, J. J., and Straight, P. D. (2018). Linearmycins are lytic membrane-targeting antibiotics. J. Antibiot. (Tokyo) 71, 372–381. doi: 10.1038/s41429-017-0005-z
Sumrall, E. T., Keller, A. P., Shen, Y., and Loessner, M. J. (2020). Structure and function of Listeria teichoic acids and their implications. Mol. Microbiol. 113, 627–637. doi: 10.1111/mmi.14472
Takekawa, N., Isumi, M., Terashima, H., Zhu, S., Nishino, Y., Sakuma, M., et al. (2019). Structure of Vibrio FliL, a new stomatin-like protein that assists the bacterial flagellar motor function. mBio 10:e00292-19.
Takigawa, H., Sugiyama, M., and Shibuya, Y. (2010). C35-Terpenes from Bacillus subtilis KSM 6-10. J. Nat. Prod. 73, 204–207.
Tamehiro, N., Okamoto-Hosoya, Y., Okamoto, S., Ubukata, M., Hamada, M., Naganawa, H., et al. (2002). Bacilysocin, a novel phospholipid antibiotic produced by Bacillus subtilis 168. Antimicrob. Agents Chemother. 46, 315–320. doi: 10.1128/aac.46.2.315-320.2002
Thurotte, A., Brüser, T., Mascher, T., and Schneider, D. (2017). Membrane chaperoning by members of the PspA/IM30 protein family. Commun. Integrat. Biol. 10:e1264546. doi: 10.1080/19420889.19422016.11264546
Todter, D., Gunka, K., and Stulke, J. (2017). The highly conserved Asp23 family protein YqhY plays a role in lipid biosynthesis in Bacillus subtilis. Front. Microbiol. 8:883.
Tran, T. T., Miller, W. R., Shamoo, Y., and Arias, C. A. (2016). Targeting cell membrane adaptation as a novel antimicrobial strategy. Curr. Opin. Microbiol. 33, 91–96. doi: 10.1016/j.mib.2016.07.002
van der Donk, W. A. (2015). Bacteria do it differently: an alternative path to squalene. ACS Cent. Sci. 1, 64–65. doi: 10.1021/acscentsci.5b00142
Vinayavekhin, N., Mahipant, G., Vangnai, A. S., and Sangvanich, P. (2015). Untargeted metabolomics analysis revealed changes in the composition of glycerolipids and phospholipids in Bacillus subtilis under 1-butanol stress. Appl. Microbiol. Biotechnol. 99, 5971–5983. doi: 10.1007/s00253-015-6692-0
Wagner, R. M., Kricks, L., and Lopez, D. (2017). Functional membrane microdomains organize signaling networks in bacteria. J. Membr. Biol. 250, 367–378. doi: 10.1007/s00232-016-9923-0
Weart, R. B., Lee, A. H., Chien, A. C., Haeusser, D. P., Hill, N. S., and Levin, P. A. (2007). A metabolic sensor governing cell size in bacteria. Cell 130, 335–347. doi: 10.1016/j.cell.2007.05.043
Weber, M. H., Klein, W., Muller, L., Niess, U. M., and Marahiel, M. A. (2001). Role of the Bacillus subtilis fatty acid desaturase in membrane adaptation during cold shock. Mol. Microbiol. 39, 1321–1329. doi: 10.1111/j.1365-2958.2001.02322.x
Wiegert, T., Homuth, G., Versteeg, S., and Schumann, W. (2001). Alkaline shock induces the Bacillus subtilis σW regulon. Mol. Microbiol. 41, 59–71. doi: 10.1046/j.1365-2958.2001.02489.x
Wolf, D., and Mascher, T. (2016). The applied side of antimicrobial peptide-inducible promoters from Firmicutes bacteria: expression systems and whole-cell biosensors. Appl. Microbiol. Biotechnol. 100, 4817–4829. doi: 10.1007/s00253-016-7519-3
Wolf, D., Kalamorz, F., Wecke, T., Juszczak, A., Mader, U., Homuth, G., et al. (2010). In-depth profiling of the LiaR response of Bacillus subtilis. J. Bacteriol. 192, 4680–4693. doi: 10.1128/jb.00543-10
Wormann, M. E., Corrigan, R. M., Simpson, P. J., Matthews, S. J., and Grundling, A. (2011). Enzymatic activities and functional interdependencies of Bacillus subtilis lipoteichoic acid synthesis enzymes. Mol. Microbiol. 79, 566–583. doi: 10.1111/j.1365-2958.2010.07472.x
Yamada, Y., Tikhonova, E. B., and Zgurskaya, H. I. (2012). YknWXYZ is an unusual four-component transporter with a role in protection against sporulation-delaying-protein-induced killing of Bacillus subtilis. J. Bacteriol. 194, 4386–4394. doi: 10.1128/jb.00223-12
Yang, S. J., Bayer, A. S., Mishra, N. N., Meehl, M., Ledala, N., Yeaman, M. R., et al. (2012). The Staphylococcus aureus two-component regulatory system, GraRS, senses and confers resistance to selected cationic antimicrobial peptides. Infect. Immun. 80, 74–81. doi: 10.1128/iai.05669-11
Yao, J., and Rock, C. O. (2013). Phosphatidic acid synthesis in bacteria. Biochim. Biophys. Acta 1831, 495–502. doi: 10.1016/j.bbalip.2012.08.018
Zhang, Y. M., and Rock, C. O. (2009). Transcriptional regulation in bacterial membrane lipid synthesis. J. Lipid Res. 50(Suppl), S115–S119.
Keywords: bacteria, membrane, lipid, cellular envelope, antimicrobial resistance, metabolism, Bacillus subtilis
Citation: Willdigg JR and Helmann JD (2021) Mini Review: Bacterial Membrane Composition and Its Modulation in Response to Stress. Front. Mol. Biosci. 8:634438. doi: 10.3389/fmolb.2021.634438
Received: 27 November 2020; Accepted: 13 April 2021;
Published: 11 May 2021.
Edited by:
Heidi Vitrac, University of Texas Health Science Center at Houston, United StatesReviewed by:
Turabe M. H. U. Fazil, Nanyang Technological University, SingaporeLeonel Malacrida, Universidad de la República, Uruguay
Jan Maarten Van Dijl, University Medical Center Groningen, Netherlands
Copyright © 2021 Willdigg and Helmann. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: John D. Helmann, amRoOUBjb3JuZWxsLmVkdQ==; orcid.org/0000-0002-3832-3249
 Jessica R. Willdigg
Jessica R. Willdigg John D. Helmann
John D. Helmann