
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Mol. Biosci., 09 April 2021
Sec. RNA Networks and Biology
Volume 8 - 2021 | https://doi.org/10.3389/fmolb.2021.634146
This article is part of the Research TopicRNA-Protein Interactions in mRNA Translation and DecayView all 9 articles
RNA molecules are increasingly being identified as facilitating or impeding the interaction of proteins and nucleic acids, serving as so-called scaffolds or decoys. Long non-coding RNAs have been commonly implicated in such roles, particularly in the regulation of nuclear processes including chromosome topology, regulation of chromatin state and gene transcription, and assembly of nuclear biomolecular condensates such as paraspeckles. Recently, an increased awareness of cytoplasmic RNA scaffolds and decoys has begun to emerge, including the identification of non-coding regions of mRNAs that can also function in a scaffold-like manner to regulate interactions of nascently translated proteins. Collectively, cytoplasmic RNA scaffolds and decoys are now implicated in processes such as mRNA translation, decay, protein localization, protein degradation and assembly of cytoplasmic biomolecular condensates such as P-bodies. Here, we review examples of RNA scaffolds and decoys in both the nucleus and cytoplasm, illustrating common themes, the suitability of RNA to such roles, and future challenges in identifying and better understanding RNA scaffolding and decoy functions.
The cell, with its variety of cellular compartments, varying polarities, competing interactions, and differing sites of molecular synthesis poses challenges to the formation of biomolecular interactions essential to all biological processes. This necessitates ways to bring biomolecules together in a tightly regulated manner. One mechanism for this localization is the use of molecular scaffolds to enable particular interactions while inhibiting off-pathway interactions, thus increasing the efficiencies of the biological processes in which they are involved in Zappulla and Cech (2006). Whereas proteins have long been appreciated to sometimes function as scaffolds, it is becoming increasingly clear that RNA molecules can also facilitate a wide range of interactions among and between proteins and nucleic acids, in many cellular contexts.
In this review, we define an “RNA scaffold” as an RNA molecule capable of bringing together 2 or more macromolecules to form a complex with functional activity. These macromolecules may be proteins, other RNAs or DNA molecules that in the absence of the RNA scaffold do not interact or do so very poorly. By this definition, perhaps the most well-known RNA scaffold molecules are ribosomal RNAs (rRNAs), which provide the structural and catalytic core of ribosomes around which approximately 57–78 ribosomal proteins (depending on species) (Lecompte et al., 2002) assemble to generate functioning ribosomes. Beyond this, long non-coding RNAs (lncRNAs) and even messenger RNAs (mRNAs) are now increasingly being shown to perform a diverse array of scaffolding roles. While rRNAs, bound by ribosomal proteins, are generally regarded as mostly static ribonucleoprotein complexes (RNPs) (Mathis et al., 2017)- although study of “specialized ribosomes” may shift this view (Genuth and Barna, 2018) – lncRNA and mRNAs are generally thought to facilitate more transient, regulatable interactions. Thus, RNAs can scaffold both stable cellular complexes and facilitate transient macromolecular interactions.
RNA can also function as a “decoy” molecule, which we define similarly to a scaffold, except that in this case, two or more macromolecules are brought together by the RNA decoy in a complex that prevents the sequestered macromolecules from forming other interactions and functional complexes at other cellular locations. As with scaffolds, RNA decoy interactions can be transient and regulatable.
RNAs as molecular scaffolds or decoys possess several advantages over other types of biomolecules like proteins and DNA, including: (1) RNAs fold more readily than DNA into complex 3D structures by virtue of diverse secondary and tertiary structural interactions. (2) RNA abundance can be easily and rapidly increased (new transcription) or decreased (RNA decay) while using 1–2 orders of magnitude less energy than occurs for similar regulation of protein (Lynch and Marinov, 2015); indeed translation is the most energy-intensive process in a cell (Topisirovic and Sonenberg, 2011), whereas transcription (barring initiation) is largely an ATP-independent process (Imashimizu et al., 2014). (3) Unlike DNA, RNA is unencumbered with serving as a cell’s nuclear-localized permanent genetic material, thus increasing RNA’s regulatory potential. (4) Compared to proteins, RNA binds other molecules much more efficiently. For instance, 4–17 nts of RNA are capable of binding specific proteins, whereas protein-protein interaction domains typically range from tens to hundreds of amino acids (Prikryl et al., 2011; Chujo et al., 2016; Lunde et al., 2017). Additionally, RNA molecules are generally longer than most proteins (typical size of a globular protein = 5 nm; radius of gyration of mRNA = 16.8–20.8 nm) allowing greater spatial interaction potential with other biomolecules. (5) Evolution of RNA scaffolding functions likely occurs faster and is under less constraint than with protein scaffolds. This is evidenced by lower conservation of lncRNA genes versus protein coding genes (Johnsson et al., 2014), and the more complex protein folding rules and solubility issues that proteins face given their complement of 20 + distinct amino acids.
In this review, we discuss examples of many nuclear and cytoplasmic RNA scaffolds and decoys, some of which regulate relatively stable complexes, while others facilitate transient macromolecular interactions. Concepts of RNA-driven assemblies that have emerged, their physiological importance, and key remaining questions for future study will be highlighted. We will also focus on recent findings of mRNAs acting as cytoplasmic RNA scaffolds, and other means by which nascently translated proteins find their interaction partners. Finally, we discuss steps that can be taken to identify new RNA scaffolds and decoys.
RNA scaffolds can generally be classified as either “structured” or “flexible.” RNAs in the former class tend to scaffold stable complexes and are enriched in secondary/tertiary RNA structure, while RNAs in the latter class often aid more transient macromolecular interactions and tend to be less structured.
In this category, the RNP, composed of the RNA scaffold along with its partner proteins, is sufficiently stable in structure that it can be assessed by X-ray crystallography or by high resolution cryo-electron microscopy (cryo-EM). Zappulla and Cech (Zappulla and Cech, 2006; Zappulla, 2020) have suggested that structured RNPs can further be sub-categorized based on the degree to which the RNA or its protein binding partners drive the overall structure of the RNP.
(i) Structure determined mostly by RNA: In this category, the RNP structure is determined in large part by the folded RNA. An example of this is the ribosome, in which disordered tails or internal loops of proteins constituting the RNP are ordered only after they bind to folded rRNA (Brodersen et al., 2002; Klein et al., 2004). The rRNA structure is so robust that even after degrading most ribosomal proteins (∼95%) from purified ribosomes, rRNA structure and catalytic peptidyl transferase activity still largely persist (Noller et al., 1992). Both sequences and secondary structures of rRNA are highly conserved.
(ii) Structure determined mostly by protein: The RNP has a specific structure that is determined in large part by previously established protein-proteins interactions that constitute the RNP, evident in small nuclear RNPs (snRNPs) and small nucleolar RNPs (snoRNPs). Several snRNP and snoRNP proteins can be crystallized in heteromeric complexes in the absence of their RNA component (Achsel et al., 1998; Deckert et al., 2006; Rashid et al., 2006). That said, during the splicing process, certain snRNPs undergo significant structural re-arrangements dependent on the RNA component; indeed RNA base-pairing driven interactions are essential to spliceosome catalysis (Matera and Wang, 2014; Galej, 2018).
The rest of this review will focus on flexible RNA scaffolds, which nucleate RNPs that lack a rigid structure. In this case, the primary role of the RNA is typically to maintain spatial proximity of macromolecular components, and aid interactions that are often (though not always) more transient in nature. Such RNAs often lack strong sequence conservation and can tolerate large deletions or insertions, but remain functional (Zappulla and Cech, 2006). Conversely, certain structures or subdomains within the flexible RNA scaffold can be of critical functional importance; these can often be transferred in a modular-like fashion to other regions of the RNA and retain functional activity (Stuckenholz et al., 2003; Zappulla et al., 2005; Zappulla and Cech, 2006).
Most known RNA scaffolds and decoys are defined as lncRNAs, which are a heterogenous group of RNAs > 200 nucleotides in length, and which generally lack long conserved open reading frames, though some may encode short peptides (Slavoff et al., 2013). lncRNAs are transcribed from diverse genomic locations including intergenic regions, and from within protein-coding gene elements including introns, exons, promoters, 5′ and 3′ untranslated regions (UTRs); both sense and antisense lncRNA transcription, with respect to the protein coding gene is observed (Khandelwal et al., 2015). Most lncRNAs are produced by RNA polymerase II, and thus like mRNAs are 5′ methyl-guanosine capped, spliced and 3′ polyadenylated, although the efficiency of these events, particularly splicing, is often lower for lncRNAs (Melé et al., 2017; Mukherjee et al., 2017; Rinn and Chang, 2020). LncRNAs that lack features of mRNA also exist including enhancer-derived ncRNAs (unstable, non-polyadenylated) and other lncRNAs with non-canonical 3′ ends (Zhang Y. et al., 2014; Villegas and Zaphiropoulos, 2015; Li W. et al., 2016). RNA polymerase I, III and, in plants, RNA polymerase V, also generate lncRNAs (Bohmdorfer et al., 2016; Sun et al., 2018). Most lncRNAs localize to the nucleus, possibly due to specific sequence motifs that interact with nuclear RNA binding proteins (Zhang B. et al., 2014; Shukla et al., 2018; Rinn and Chang, 2020), and are typically expressed in low quantities, although their expression ranges from zero to exceeding abundant housekeeping mRNAs (Derrien et al., 2012). LncRNAs tend to exhibit more specific temporal, developmentally regulated and cell-type specific expression patterns than most protein coding genes, suggesting important regulatory functions, as well as tight regulation of lncRNA transcription and decay (Derrien et al., 2012; Gloss and Dinger, 2016). LncRNAs with scaffold or decoy functions impact diverse aspects of nuclear cell biology, select examples of which we discuss below (Figure 1 and Table 1).
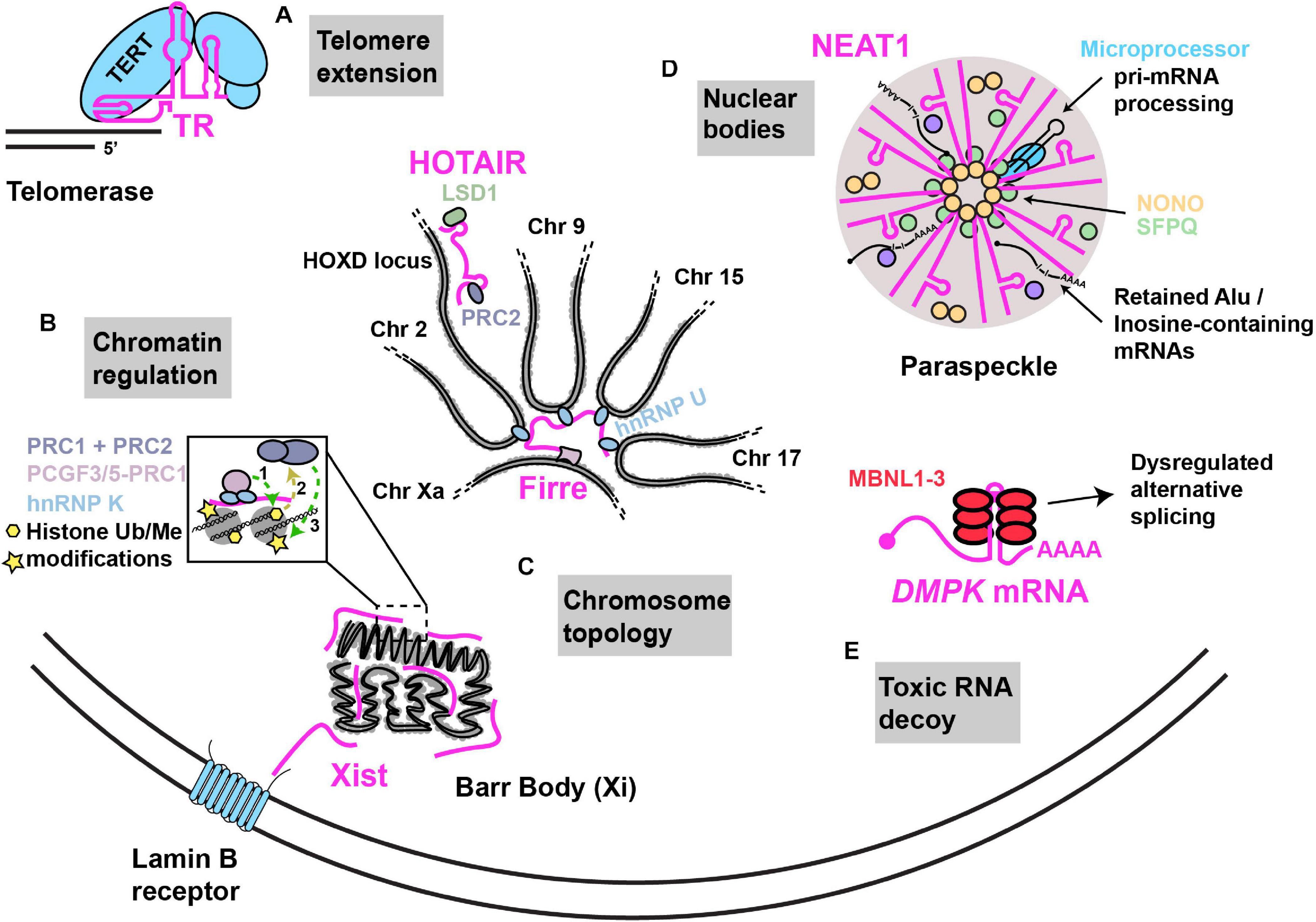
Figure 1. Examples of nuclear RNA scaffolds and decoys. Scaffold and decoy RNAs are depicted in pink. (A) Telomere extension: the TR lncRNA scaffolds and provides the telomeric repeat template for the Telomerase complex that consists of the reverse transcriptase protein (TERT) along with other accessory proteins that function in telomere extension. (B) Chromatin regulation: Xist lncRNA establishes X chromosome inactivation by (1) hnRNP K initiated recruitment of; non-canonical PRC1 complex (Ub ligase) (2), whose activities recruit (3) canonical PRC1 and PRC2 (methyltransferases). HOTAIR recruits PRC2 and LSD1 (demethylase) to modify chromatin at numerous gene loci, including the HOXD locus. (C) Chromosome topology: (i) Firre lncRNA expressed from the active X chromosome, establishes chromosome territories by bringing together loci across multiple different chromosomes with the help of interaction with hnRNP U. (ii) Xist lncRNA can interact with the Lamin B receptor, resulting in recruitment of the inactivated X chromosome (“Barr body”) to the nuclear lamina. (D) Nuclear bodies: NEAT1–2 lncRNA acts as an RNA scaffold driving the assembly of nuclear paraspeckles via its interaction with NONO, SFPQ and several other proteins. NEAT1–2 facilitates NONO-Microprocessor interactions to aid in pri-miRNA processing. Paraspeckles also sequester Inosine-modified RNAs, preventing their export. (E) Toxic RNA decoy: CUG repeat expansions in the 3′UTR of DMPK sequester Muscleblind-like (MBNL) proteins, thus impairing alternative splicing.
An elegant example of an RNP featuring a flexible RNA scaffold is the telomerase enzyme that functions in the maintenance of genome stability by counteracting the loss of DNA at chromosomal ends following rounds of DNA replication (Figure 1). Telomerase is scaffolded by the telomerase RNA (TR; also named telomerase RNA component; TERC), a lncRNA that functions both as a template in the reverse transcriptase-driven extension and maintenance of telomeric DNA, and which binds and spatially organizes various telomerase complex proteins including the telomerase reverse transcriptase (TERT), and several other species-specific accessory proteins (Chan and Blackburn, 2004; Wu et al., 2017). RNA structural modeling in different species suggests that TR usually harbors 3 long arms that each bind to specific proteins of the telomerase complex (Zappulla and Cech, 2006; Zhang et al., 2011; Zappulla, 2020). Each arm consists of double-helical regions with several internal loops and bulges, features that provide flexibility to RNAs in solution (Nowakowski and Tinoco, 1997). Another conserved feature of TR is the presence of a template/pseudoknot domain (“t/PK”) which contains the template used for replicating telomeric sequence, and a pseudoknot implicated in aiding TERT catalysis, template positioning and telomerase assembly (Chen and Greider, 2005; Theimer et al., 2005; Qiao and Cech, 2008; Wang et al., 2016). TR length ranges significantly between species from 150nts in ciliates to 3kb in certain yeast species. Furthermore, large stretches of TR can be deleted or domains transferred to other parts of TR without significant loss of function suggesting that TR structural flexibility allows maintenance of function (Livengood et al., 2002; Zappulla and Cech, 2006).
Telomerase complex assembly, and particularly recruitment to telomeres is often transient, and typically regulated in a cell-cycle dependent manner (Vasianovich and Wellinger, 2017). In yeast, assembly of the active telomerase enzyme depends on binding of a particular subunit (Est3) to the reverse transcriptase, which in turn must already be bound (along with another subunit, Est1) to the TR. Such assembly only occurs in G2/M, and while protein interaction domains of telomerase subunits are known, the underlying regulation of this process remains unclear (Tucey and Lundblad, 2014). Interestingly, in humans, TR and TERT remain stably associated throughout the cell cycle based on competitor interaction studies, but a key telomerase subunit (Telomerase Cajal body protein 1; TCAB1) decreases interaction with the TR during M phase. Finally, single-molecule imaging of TERT in mammalian cells revealed that during S-phase, telomerase mostly exists in a freely diffusing nuclear state, and only transiently interacts with telomeric ends (Schmidt et al., 2016). Specifically, TERT exhibited ∼2400 telomere encounters in an 8 hr S-phase, with only rare long-lasting telomerase-telomere interactions of ∼3.7 mins observed in about 4% of cells.
Many lncRNAs associate with chromatin, and can enact either widespread or local changes in chromatin conformation that can regulate chromosome topology, localization and the process of transcription (Werner and Ruthenburg, 2015).
The most studied lncRNA modifier of chromosome topology is Xist (X-inactive specific transcript), a 17 kb lncRNA that mediates X chromosome inactivation (XCI), a method of sex-chromosome gene dosage compensation where one X chromosome in every female cell is almost completely transcriptionally silenced at random (Chow et al., 2005). Following transcription, Xist lncRNA coats and is tethered to the entire future inactivated X chromosome (“Xi”) by an AT-rich DNA interacting protein called hnRNP U (Hasegawa et al., 2010). Ultimately, Xist promotes condensation of the Xi into a compact structure termed the Barr body, which is repositioned either to the nuclear lamina, or to peri nucleolar regions (Andrulis et al., 1998; Zhang et al., 2007; Rego et al., 2008); in both cases, these regions are enriched in and likely help maintain transcriptionally inactive heterochromatin (Clemson et al., 1996; Padeken and Heun, 2014). Xist harbors several conserved, structured tandem-repeat regions, named A to F, that are implicated in Xist association with the Xi, Xist spreading across Xi, and Xist association with transcriptional silencing complexes (Wutz et al., 2002; Pintacuda et al., 2017b). Although many Xist structural domains and protein interactions have been described (Chu et al., 2015; Smola et al., 2016), one intriguing example is interaction of a region of Xist overlapping it’s 5′ localized F-repeat with the Lamin B receptor (LBR), a trans-membrane protein embedded in the inner nuclear membrane (Chen et al., 2016). This interaction induces Xi localization to the nuclear lamina, which may facilitate Xist spreading on Xi and transcriptional silencing due to chromosome immobilization (Chen et al., 2016; Figure 1).
Another repeat-containing lncRNA that affects chromosome topology is Firre, which unlike Xist, appears to be transcribed predominantly from the active X chromosome (“Xa”) (Fang et al., 2020) and can associate in trans with a region of the Xi. Firre expression also promotes Xi positioning at the nuclear lamina and near the nucleolus in a manner dependent on the zinc finger transcription factor CTCF (Deng et al., 2015; Fang et al., 2020). Firre also helps preserve H3K27me3, a repressive chromatin mark, via Polycomb repressive complex 2 targeting (PRC2) to Xi (Yang et al., 2015).
Firre also regulates inter-autosomal interactions by interacting via its repeat domains with hnRNP U (Figure 1). This drives formation of nuclear domains in proximity to Firre’s genomic locus on the Xa, which induces co-localization of loci on chromosomes 2, 9, 15, and 17 (Hacisuleyman et al., 2014). Both deletion of the Firre locus and hnRNP U knockdown result in loss of colocalization of the trans-chromosomal interacting loci (Hacisuleyman et al., 2014). Firre null cells also exhibit > 1000 genes with significant changes in gene expression as assessed by RNA-seq (Hacisuleyman et al., 2014), while Firre KO mice exhibit close to 4000 differentially expressed genes across multiple tissues, and exhibit a specific defect in hematopoiesis (Lewandowski et al., 2019). Thus, Firre appears to act as a scaffold that initiates and organizes specific chromosomal domains within the nucleus through specific chromatin interactions, which in turn regulates diverse gene expression programs (Nakagawa and Hirano, 2014).
LncRNAs can regulate transcriptional activity either by regulating chromatin modification and thus conformation, or by directly affecting transcription. This is achieved by lncRNAs recruiting factors such as histone modifying complexes, DNA methyltransferases, transcription factors and other DNA-binding transcriptional regulators to specific genomic loci. This recruitment process may involve a lncRNA binding to a specific loci, or an entire chromosome (as discussed for Xist), in cis or in trans, by base pairing with other RNAs, by the lncRNA binding directly to DNA, or by RNA-protein interactions (Long et al., 2017).
Numerous Xist-interacting proteins have been identified by various cross linking and mass-spectrometry approaches (McHugh et al., 2015; Almeida et al., 2017; Brockdorff, 2017; Pintacuda et al., 2017a), many of which are chromatin modifiers, which likely help initiate and maintain XCI. In one study, amongst other Xist directly interacting proteins including hnRNP U and LBR, a transcriptional repressor named SHARP (SMRT and HDAC associated repressor protein) was identified as an Xist binder which in turn recruits SMRT (silencing mediator of retinoid and thyroid receptor) and histone deacetylase 3 (HDAC3) (Chu et al., 2015; McHugh et al., 2015). More recent work has identified a 600 nt region of the Xist RNA overlapping the B repeat, termed the Xist RNA Polycomb interaction domain (XR-PID), which is bound by the RNA-binding protein (RBP) hnRNP K (Pintacuda et al., 2017a). This promotes recruitment of a non-canonical Polycomb chromatin-modifying complex (PCGF3/5-PRC1) which ubiquitinates Histone H2A. This serves as a recruitment signal for other PRC1 complexes and the PRC2 Polycomb complex, which promotes H3K27me3 repressive modification (Almeida et al., 2017), a feature of XCI (Wutz, 2011; Figure 1). Interestingly, PRC2 Xi recruitment is inhibited in SHARP or HDAC3 knockdown cells (McHugh et al., 2015), though understanding of how histone deacetylation and ubiquitination co-operate in PRC1 and 2 recruitment, in an Xist dependent manner, remains unclear. In summary, Xist harbors a complex, cis-acting RNA-scaffolding function, involving interactions with several chromatin modifiers and DNA binding proteins.
RNA scaffolds can also drive recruitment of chromatin modifiers in trans at specific gene loci; this was first reported for the 2.2 kb human HOX antisense intergenic RNA (HOTAIR), which is encoded on chromosome 12 at the HOXC locus (Rinn et al., 2007). HOTAIR appears capable of simultaneously binding PRC2 via a 5′ region and Lysine-specific demethylase 1 (LSD1) via a 3′ region (Tsai et al., 2010; Figure1). Chromatin-IP analysis (ChIP) revealed that depletion of HOTAIR results in striking transcriptional activation of a 40 kb region of HOXD locus on chromosome 2 owing to loss of PRC2-mediated H3K27me3 and gain of LSD1-regulated H3K4me2. HOTAIR depletion also alters these chromatin modifications, and PRC2 and/or LSD1 binding, at hundreds of other genes (Tsai et al., 2010). Besides implications for a role in development, given the nature of HOX gene function in embryogenesis (although this is controversial (Selleri et al., 2016)) HOTAIR is also overexpressed and may affect pathology in various cancers (Hajjari and Salavaty, 2015).
LncRNAs can also directly affect transcription by interacting with transcription factors and the transcriptional apparatus. For instance, in breast cancer cells, HOTAIR scaffolds a complex involving LSD1 (Amente et al., 2010a, b) and HBXIP, which is a co-activator and binding partner of the oncogenic transcription factor c-Myc. Ultimately, c-Myc recruitment of the HBXIP/HOTAIR/LSD complex transcriptionally activates c-Myc target genes which in turn drives c-Myc-mediated oncogenesis (Li Y. et al., 2016). Many other lncRNAs interact with chromatin modifying proteins to regulate chromatin modification, or interact with transcription factors, resulting in either transcriptional activation or repression. We guide readers to these excellent reviews (Rinn and Chang, 2012; Rutenberg-Schoenberg et al., 2016; Kopp and Mendell, 2018).
lncRNAs can function as RNA scaffolds for nuclear bodies (Chujo et al., 2016), which are membrane-less liquid-like organelles rich in protein and RNAs. These include nucleoli, paraspeckles, Cajal bodies and Polycomb bodies (Mao et al., 2011). Very generally, nuclear bodies can act as sites of biogenesis, maturation, storage, and sequestration of specific RNPs or regulators of nuclear RNA processes. Some nuclear bodies harbor specific lncRNAs implicated in their scaffolding which tend to be characterized by repetitive sequences. These may either concentrate biomolecules in a nuclear body, and thus facilitate a molecular process, or sequester in a seemingly inert state specific biomolecules away from other cellular locations, often altering gene expression mechanisms in the process. Thus, nuclear body RNA scaffolds can also sometimes be considered as decoys.
Nuclear paraspeckles, which are implicated in regulating transcription, splicing, RNA processing and export, are scaffolded by the nuclear-enriched abundant transcript 1 (NEAT1) lncRNA (Bond and Fox, 2009; Figure 1). Paraspeckles are nucleated at NEAT1 loci, particularly following transcription of the NEAT1_2 (22.7 kb) lncRNA isoform; a second isoform, NEAT1_1 (3.7 kb) has a more supplementary role in paraspeckle formation (Clemson et al., 2009; Naganuma et al., 2012). NEAT1 is essential for the formation and maintenance of paraspeckles based on knockdown and over-expression studies. Several RBPs localize in paraspeckles (Naganuma et al., 2012; Kawaguchi et al., 2015), including NONO, PSPC1, and SFPQ; all 3 of these are part of the Drosophila Behavior/Human Splicing family of proteins and function in numerous steps of nuclear mRNA metabolism including transcription, splicing, processing and export (Knott et al., 2016). Careful truncation analyses, RBP-NEAT1 tethering assays and microscopy have identified that the middle domain of NEAT1–2 contains functionally redundant repetitive sequences that bind NONO and SFPQ. A dimerization domain of NONO also drives paraspeckle assembly, a process that exhibits phase transition-like mechanisms similar to other RNP membrane less bodies (Boeynaems et al., 2018; Yamazaki et al., 2018). Consistent with the importance of the central NEAT1_2 region, electron and super-resolution microscopy indicates that the middle region of NEAT1–2 locates to the inner core of paraspeckles, whereas 5′ and 3′ ends of NEAT1_2 are located in the outer shell (Souquere and Pierron, 2015; West et al., 2016).
NEAT1 and paraspeckles regulate gene expression in diverse ways through both scaffolding and decoy functions. As a scaffolding example, NONO-SFPQ heterodimeric complexes bind to and enhance processing of select pri-miRNAs by the Microprocessor complex, which conducts the first step in miRNA processing (Jiang et al., 2017). NEAT1 binds both NONO-SFPQ dimers (presumably in the central region) and the Microprocessor complex at a 3′ stem loop motif (also implicated in regulating NEAT1_2 stability (Yamazaki et al., 2018)). Importantly, knockdown or knockout of NEAT1 strongly impedes NONO interactions with Microprocessor, and impairs pri-miRNA processing, indicating NEAT1 scaffolds Microprocessor interactions with NONO-SFPQ to aid the production of mature miRNA (Jiang et al., 2017; Figure 1).
A decoy function of paraspeckles and NEAT1 is sequestration, and thus limiting export of mRNAs harboring inverted repeat Alu elements in their 3′ untranslated region (UTR; Alu elements are transposable elements that account for 11% of the human genome, and which likely derive from 7SL ncRNA (Deininger, 2011); discussed later). This feature makes RNAs targets for Adenosine-to Inosine (A to I) editing by Adenosine deaminase enzymes (Athanasiadis et al., 2004). Paraspeckles are enriched in A to I hyper-edited RNA (Zhang and Carmichael, 2001), owing to such modified RNA being bound by NONO, and several such mRNAs are retained in the nucleus (or specifically, where examined microscopically, in paraspeckles themselves). This depends on the mRNAs harboring a modified Alu element in their 3′UTR, as well as the presence of NEAT1 expression and paraspeckle formation (Prasanth et al., 2005; Chen et al., 2008; Chen and Carmichael, 2009; Osenberg et al., 2009). Such mRNA sequestration can be regulated; for example, mCAT2 mRNA is released from paraspeckles following a stress-activated 3′UTR cleavage event (Prasanth et al., 2005) and may in turn regulate innate immune and inflammatory responses (Lowenstein and Padalko, 2004; Pisani and Baron, 2019).
Another decoy function of NEAT1 is sequestration of transcription regulating factors. In response to either proteasomal inhibition or viral infection, NEAT1 RNA abundance increases > 4 fold, causing significant paraspeckle enlargement and sequestration of SFPQ. This titrates SFPQ away from specific promoters, altering mRNA expression in a context dependent manner (Hirose et al., 2014; Imamura et al., 2014). In NEAT1 knockdown or knockout contexts, failure to transcriptionally regulate these mRNAs correctly following proteasomal inhibition or viral infection leads to increased cell death and impaired innate immune responses, respectively (Hirose et al., 2014; Imamura et al., 2014). Thus, regulation of NEAT1 expression and paraspeckle formation may play a key role in mediating stress-inducible gene expression responses by sequestration of both mRNA and proteins.
While repetitive sequences often are of functional benefit to lncRNA scaffolds (Duszczyk et al., 2011; Yamazaki et al., 2018), aberrant repeat expansion in mRNAs is implicated in playing decoy functions that contribute to various neurodegenerative diseases. To date, 10 neurodegenerative diseases including Myotonic dystrophy type 1 and 2, Fragile X-associated tremor ataxia syndrome, Amyotrophic Lateral Sclerosis (ALS), Huntington’s disease-like 2 and a variety of other Ataxia conditions feature repetitive sequence expansions in regions including UTRs, introns, promoter and variant exon sequences (Wojciechowska and Krzyzosiak, 2011; Swinnen et al., 2020). Myotonic dystrophy type 1, caused by expansion of a CTG repeat in the 3′UTR of the DMPK gene represents the clearest example of an RNA decoy disease (Mastroyiannopoulos et al., 2010; Figure 1). This expansion results in the formation of a stable hairpin conformation that leads to accumulation of RNA nuclear foci (Taneja et al., 1995; Davis et al., 1997; Tian et al., 2000); why such RNAs fail to export is still unclear (Smith et al., 2007; Mastroyiannopoulos et al., 2010). In turn, these RNA foci sequester specific RBPs, particularly muscleblind-like (MBNL) family proteins MBNL1, MBNL2, and MBNL3 (Miller, 2000; Mankodi et al., 2001). These proteins regulate alternative splicing in a tissue-specific manner, and splicing defects are observed in various mouse models and patients (Swinnen et al., 2020). Other means of repeat expansion toxicity, including repeat-associated translation of repeat sequences via a non-canonical translation mechanism, and loss of protein expression (owing to poor transcription or enhanced mRNA decay) are also proposed mechanisms in many of the above-mentioned diseases (Swinnen et al., 2020).
Finally, the nucleolus is also scaffolded by at least two distinct RNA molecules. In humans, nucleoli form due to juxtaposition of rDNA arrays, located on five chromosomes in strongly euchromatic regions, to typically form 1–3 nucleoli per cell (Savino et al., 2001). The essential rRNA transcription factor UBF1 binds to and maintains rDNA arrays in an open chromatin state which initiates nucleoli formation (Grob et al., 2014). This involves rRNA transcription itself, which confers spatiotemporal regulation on nucleoli formation, whose assembly varies during embryogenesis and cell cycle progression (Hernandez-verdun, 2011; Berry et al., 2015; Falahati et al., 2016). However, a long-stranding puzzle in the field was that RNAP II inhibition strongly dispersed nucleoli into multiple small foci, even though RNAP II activity is negligible in nucleoli (Carmo-Fonseca, 2015). An answer to this conundrum came from sequencing of nucleolar RNA fractions, and subsequent RNA fluorescence in situ (FISH) experiments, which identified a strong enrichment of Alu repeat-containing RNAs (AluRNAs) in nucleoli that mostly derive from introns and subsequent pre-mRNA splicing of multiple genes (Caudron-Herger et al., 2015). Knockdown and ectopic expression of AluRNAs confirmed a clear role in stimulating assembly of normal-sized nucleoli, which involves a scaffolding function whereby AluRNAs bind nucleolin and nucleophosmin, two abundant structural proteins of the nucleolus that harbor low-complexity domains (possibly aiding phase-transition like properties of the nucleolus), and which also bind UBF1 (Li et al., 1996; Hisaoka et al., 2010). Thus, trans acting AluRNA scaffolds appear to “glue” small nucleolar bodies together (Caudron-Herger et al., 2015, 2016). Critically, depletion of AluRNAs or impairment of RNAP II transcription strongly reduces rRNA transcription, whereas AluRNA overexpression enhanced rRNA transcription, arguing that AluRNA scaffolding facilitates nucleolar functions in rRNA production (Caudron-Herger et al., 2015, 2016). Whether other nucleolar functions (Iarovaia et al., 2019), including roles in telomerase (Fu and Collins, 2007) and signal recognition particle assembly (Massenet, 2019) (see below) are impacted by AluRNA-driven scaffolding is unknown but worthy of future study.
Several examples of flexible cytoplasmic RNA scaffolds and decoys have recently come to light, particularly involving cytoplasmic lncRNAs and mRNAs. These, together with a long-known ribosome-associated flexible RNA scaffold are now discussed (Figure 2 and Table 1).
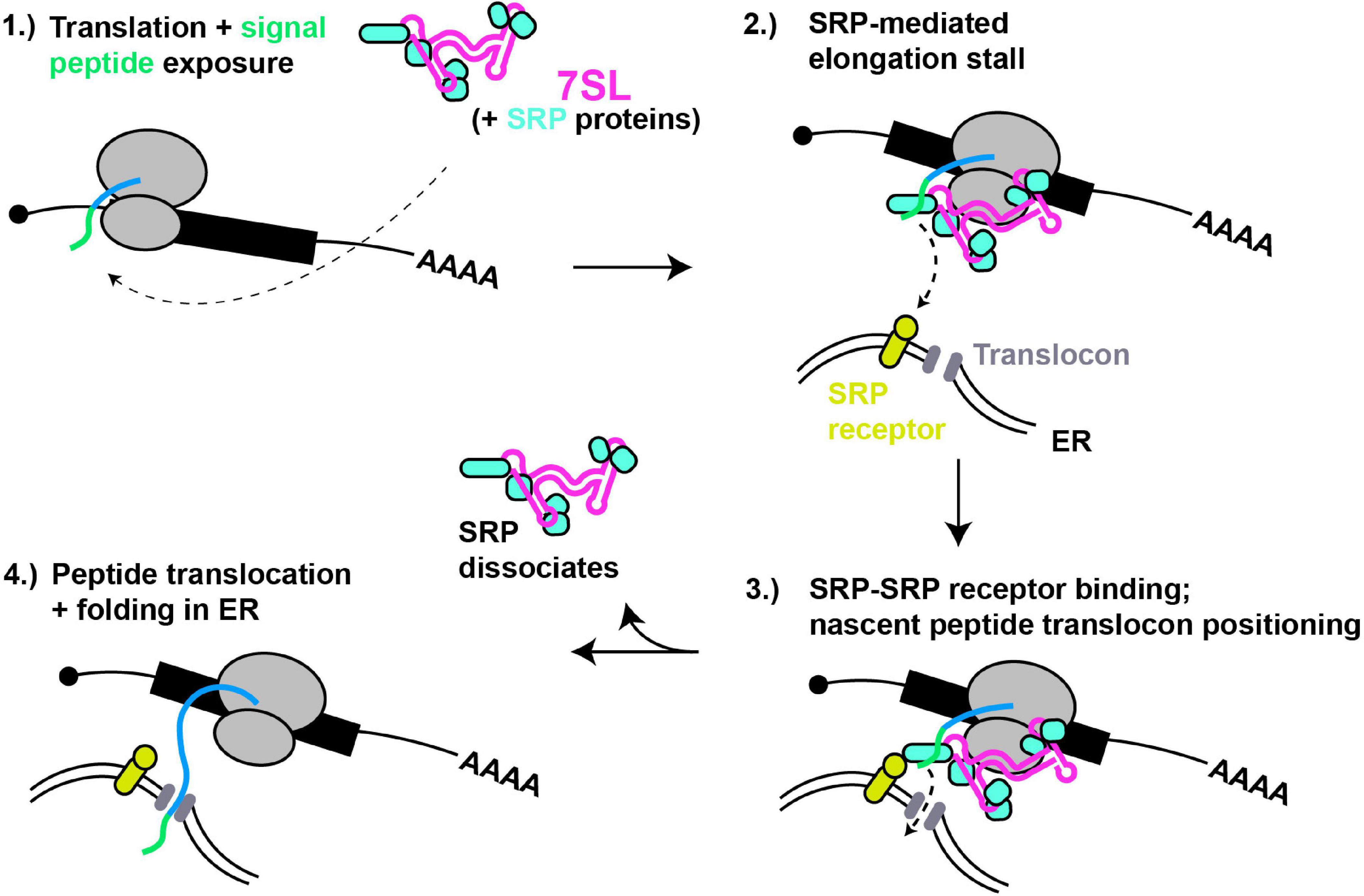
Figure 2. 7SL – a paradigm of cytoplasmic RNA scaffolding. (1) Translation of proteins with N-terminal signal peptides (green) are bound by SRP complex, scaffolded by 7SL. (2) Signal peptide binding induces SRP conformational change and tighter binding, thus block ribosomal A-site and stalling translation elongation. (3) SRP interaction with SRP receptor positions nascent peptide for entry into ER translocon. (4) Dissociation of SRP relieves elongation stall, and nascent peptide extends into and folds within ER lumen.
The signal recognition particle (SRP) is a well-studied RNP complex that is scaffolded by an RNA Polymerase III-derived RNA (7SL; Figure 2). SRP recognizes and binds to N-terminal signal peptides present on proteins destined for secretion or membrane localization as they emerge from a ribosome. SRP binding to signal peptides causes a stall in translation elongation, and targets the ribosome to a translocon pore in the ER membrane where translation then resumes, resulting in the unstructured nascent peptide being threaded into the ER lumen, where folding and later trafficking can ultimately occur (Keenan et al., 2001).
The human 7SL lncRNA is a ∼300 nt long structured RNA characterized by helical regions forming 2 domains separated by a flexible linker, a smaller domain (Alu) and a larger domain (S), with each part recruiting specific proteins (6 in total) that form the SRP (Pool, 2005). The Alu domain of 7SL preferentially binds the SRP14/SRP9 heterodimer whose function involves binding the ribosome and inhibiting translation elongation after signal recognition by obscuring access to the ribosomal A-site, while the S domain binds the heterodimer SRP68/SRP72, SRP19 and SRP54, and binds in proximity to the nascent peptide tunnel (Siegel and Walter, 1988; Wild et al., 2001). Besides signal peptide binding (via SRP54), the S domain also mediates SRP interactions with the SRP receptor at the ER membrane (Gowda et al., 1997; Pool et al., 2002). Crystal structure analyses suggests the SRP68/SRP72 heterodimer binding of 7SL drives an initial conformation change enabling proper interaction of SRP with the ribosome (Grotwinkel et al., 2014). Furthermore, Cryo-EM studies indicate that SRP bound to the elongating ribosome exists in at least two distinct states. In the “scanning” state, SRP samples nascent peptides for signal peptide sequences. If none are detected, elongation factor binding readily displaces the Alu domain away from the ribosomal A-site, whereas in the “engaged state,” with SRP54 bound to the nascent peptide, the Alu domain remains robustly bound near the A-site, thus inhibiting translation (Voorhees and Hegde, 2015). Thus, dynamic flexibility of the 7SL RNA scaffold is critical to SRP function.
While most lncRNAs exhibit a significant bias toward nuclear localization, many also localize in the cytoplasm and thus unsurprisingly, are increasingly being linked to an array of diverse scaffolding and decoy roles.
lncRNAs can bind to mRNA in trans, and scaffold complexes that impact mRNA function. For example, numerous lncRNAs harboring Alu elements (“1/2-sbsRNAs”) were identified that form partially complementary duplexes in trans with Alu elements present in various mRNA 3′UTRs (Gong and Maquat, 2011; Figure 3). Upon lncRNA-mRNA binding, the resulting double stranded RNA serves as a binding site for the RBP Staufen, which in turn recruits the RNA helicase Upf1, resulting in degradation of many target mRNAs by a process termed Staufen mediated decay (SMD) (Kim et al., 2005). SMD regulation by lncRNAs is complex, given that a) several different Alu-containing lncRNAs can promote decay of the same mRNA; b) a given Alu-containing lncRNA can regulate several mRNAs; and c) not all mRNA targets with complementarity to a given Alu-containing lncRNA necessarily undergo decay, possibly owing to other structural aspects of the mRNA-protein complex (mRNP) (Gong and Maquat, 2011; Gong et al., 2012; Park and Maquat, 2013). In another case, p21, a highly studied lncRNA transcriptional target of p53 with many nuclear functions, also forms hybrids with target cytoplasmic mRNAs (Figure 3). Specifically, p21 exhibits partial base pair complementarity and affinity purifies with CTNNB1 and JUNB mRNAs, which encode oncogenic proteins β-catenin and JunB (Yoon et al., 2012). Based on knockdown studies, p21 exerts an inhibitory effect on translation of these mRNAs, rather than mRNA decay. Consistent with this, p21 also affinity purifies with the translational repressors RCK and FMRP, and fractionates in polysomes, suggesting that p21 pairs to and recruits translation repressors to mRNA targets (Yoon et al., 2012).
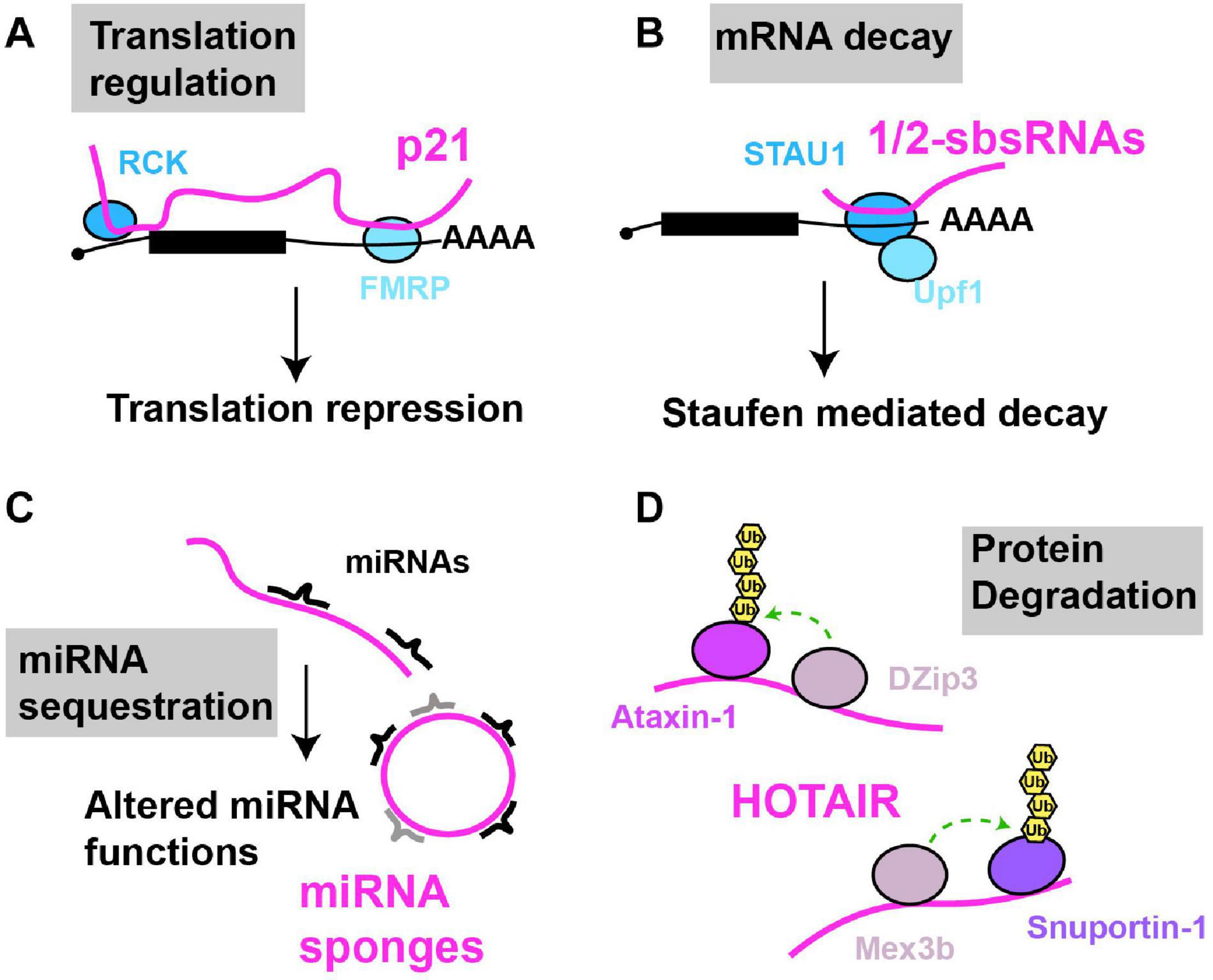
Figure 3. Examples of cytoplasmic RNA scaffolds and decoys. Scaffold and decoy RNAs are depicted in pink (A) mRNA translation: lncRNA-p21 partially base pairs with target mRNAs leading to the recruitment of translation repression factors like RCK and FMRP, thus inhibiting mRNA translation. (B) mRNA decay: 1/2sbsRNAs base pair with target mRNAs; the resulting dsRNA recruits Staufen, leading to staufen-mediated mRNA decay. (C) miRNA sequestration: various lncRNAs (linear and circular) in multiple species, regulated in a tissue, developmental or environmentally sensitive manner, can base-pair with and sequester miRNAs, preventing their regulation of mRNA translation or decay. (D) Protein degradation: HOTAIR lncRNA binds to two ubiquitin ligases and their substrates causing their ubiquitination and degradation.
A second example of 3′UTR co-translationally scaffolding assembly of protein complexes involves the Baculoviral IAP repeat-containing protein 3 (BIRC3) gene. BIRC3 encodes a ubiquitin ligase, and alternative cleavage and polyadenylation generates short (BIRC3-SU) and long (BIRC3-LU) 3′UTR mRNA isoforms, the latter of which is significantly upregulated in chronic lymphocytic leukemia (CLL) cells (Lee and Mayr, 2019). While not affecting BIRC3 mRNA or protein localization, or protein levels, BIRC3-LU is specifically bound by HuR and Staufen, which in turn recruits trafficking-regulating proteins (IQ motif containing GTPase IQGAP1, and the Ras GTPase RALA) (Lee and Mayr, 2019). These interact with nascently translated BIRC3 such that the 3′UTR scaffolds formation of a BIRC3-RALA-IQGAP1 complex. This complex in turn binds and promotes plasma membrane recycling of a receptor protein called CXCR-4, which promotes CLL progression by aiding malignant B cell migration and survival via targeting to protective bone marrow niches (Burger et al., 2005). Finally, the BIRC3-LU 3′UTR, distinct from the BIRC3-SU isoform, uniquely binds proteins involved in mitochondrial biology and chromatin remodeling, suggesting that multiple functionally distinct protein complexes may be scaffolded by this particular 3′UTR (Lee and Mayr, 2019).
While best known for its nuclear function HOTAIR is also found in the cytoplasm, particularly under cellular conditions such as senescence (Yoon et al., 2013). Cytoplasmic HOTAIR serves as a binding site for two ubiquitin ligases DZIP3 and MEX3B, and their substrates Snurportin 1 (SNUPN) and Ataxin-1 (ATXN1); this binding leads to increased SNUPN and ATXN1 ubiquitination and their degradation. HOTAIR accumulation is necessary for eliciting proper cellular senescence phenotypes in response to inducing stimuli, possibly in part through SNUPN and ATXN1 degradation (Yoon et al., 2013). Both HOTAIR and p21 are downregulated by binding of the RBP HuR, which normally stabilizes mRNAs, but in the case of these lncRNAs, stimulates assembly and recruitment of a miRNA silencing complex (Let-7/Ago2) to elicit their decay (Yoon et al., 2012). Thus, cytoplasmic lncRNAs can regulate translation and protein turnover events and can be subject to miRNA-mediated regulation of their stability.
Conversely, a well appreciated mode of lncRNA function in the cytoplasm is to function as decoys for miRNAs (aka sponges; Figure 3). First described in plants where the lncRNA IPS1 sequesters miR-399 as part of the regulation of phosphate homeostasis (Franco-Zorrilla et al., 2007), several lncRNA miRNA decoys are now known, some of which harbor multiple sites for distinct miRNAs and can exist as splicing-intermediate derived circular lncRNAs (Hansen et al., 2013; Memczak et al., 2013). Many miRNA decoys appear highly regulated, with distinct tissue, developmental and environmentally responsive expression patterns, and have the potential to exhibit large-scale effects on mRNA translation and decay, owing to the diversity of miRNA molecules whose function they may impede. We guide readers to other detailed reviews on these topics (Ebert and Sharp, 2010a, b; Yoon et al., 2014; Rong et al., 2017).
Increasing evidence indicates that mRNAs serve as scaffolding platforms for co-translational protein complex assembly (Natan et al., 2017). In prokaryotes, subunits of a given protein complex are often encoded in operons (a cluster of genes of common function under a single promoter), which are transcribed into polycistronic mRNAs. Translation of polycistronic mRNAs in turn enhances the likelihood of the encoded protein complex subunits interacting. Such interactions can occur on polycistronic mRNAs even when nascently synthesized proteins are still associated with ribosomes, owing in part to their close spatial proximity upon synthesis (Shieh et al., 2015). In this way, the mRNA serves as a scaffolding molecule facilitating nascent protein interactions. Furthermore, adjacent genes in operons tend to exhibit larger protein interaction interfaces (Koonin and Mushegian, 1996; Dandekar et al., 1998; Shieh et al., 2015; Wells et al., 2016). These findings, combined with additional bioinformatic and structural analyses of protein complex assembly suggest that operon gene order often matches (and thus may help direct) the order in which protein subunits assemble (Wells et al., 2016). Besides spatial proximity of subunits, lower stoichiometric variation in protein complex subunit expression is an additional advantage of protein complex assembly on polycistronic mRNAs (Swain, 2004; Sneppen et al., 2010; Shieh et al., 2015).
Eukaryotes lack operons, meaning any co-translational interactions must involve an in trans event such as recruitment of an interacting protein to a translating mRNP, or juxtaposition of translating mRNPs. Nonetheless, co-translational assembly of protein complexes in eukaryotes has been reported (Natan et al., 2017). Notably, recent affinity purification studies of well characterized protein complexes in yeast (Shiber et al., 2018) and human cells (Kamenova et al., 2019) demonstrate in many cases that nascently translating proteins interact strongly with proteins with which they are destined to form complexes with. Co-translation interactions were identified via careful interaction domain mapping, mRNA-protein co-localization microscopy and critically by detection of both protein interactors, and their mRNA template molecules, following purification of a protein complex subunit of interest. Combining nascent peptide purification with ribosome profiling (an RNA sequencing based method to map ribosome mRNA footprints) allowed determination of precisely where ribosomes were located on a given mRNA when their nascent peptides interacted with a protein of interest (Shiber et al., 2018). Co-translational protein interactions could be unidirectional (meaning Protein A and its mRNA are purified by Protein B isolation, but not vice versa) or bidirectional. This depended largely on how close a given protein’s interaction domain was to the C-terminus, as this affects the degree and timing of exposure of the interaction domain from within the translating ribosome (Shiber et al., 2018). Co-translational assembly of protein complexes was important in preventing protein misfolding and increasing the efficiency of protein complex assembly (Shiber et al., 2018; Kamenova et al., 2019). Thus, co-translational assembly of proteins in eukaryotes may be a common phenomenon.
Two primary models for eukaryotic co-translational protein interaction are generally postulated. The first states that a fully synthesized protein interacts with a nascently synthesizing protein, though whether this simply involves random diffusion, co-localization or an active chaperone driven recruiting process is unclear. A second model is that, particularly in the case of bidirectional co-translational protein interactions, translating mRNPs may be tethered to one another in trans by their nascent peptides (Natan et al., 2017; Schwarz and Beck, 2019). Though the above discussed examples do not distinguish between these models, recent data, discussed below, adds weight and new insight to both ideas and focuses attention on the importance of mRNA as a scaffolding molecule.
mRNAs possess three primary domains: a 5′ UTR, an open reading frame and a 3′UTR. While all are implicated in translation, ribosomes ordinarily only transit through 5′UTRs and ORFs, leaving 3′UTRs as regions where more stable interactions with proteins and other RNA molecules can be sustained. 3′UTR sequences are unconstrained by selection for a given protein sequence, are significantly longer than 5′UTRs, and increase in length with organismal complexity (Pesole, 2002; Mayr, 2017). Thus, it is not surprising that while 3′UTR localized interactions clearly govern individual mRNA functions, such as translation, decay and localization (Wilkie et al., 2003; Andreassi and Riccio, 2009), new functions for 3′UTRs are being uncovered.
Pioneering work by the Mayr lab demonstrated that specific mRNA 3′UTRs can scaffold protein-protein interactions in which a 3′UTR bound protein (or proteins) interacts with nascently synthesized protein translated on the mRNA itself, with important physiological consequences (Figure 4). The first report of this phenomenon (Berkovits and Mayr, 2015) concerned the CD47 gene, which encodes a protein capable of both ER and cell surface localization; at this latter site, CD47 protects cells from phagocytosis by macrophages (Oldenborg et al., 2000; Jaiswal et al., 2009). Alternative cleavage and polyadenylation usage, a highly regulated means by which alternate 3′UTR isoforms are generated (Sandberg et al., 2008; Mayr and Bartel, 2009; Lianoglou et al., 2013), results in two primary CD47 3′UTR mRNA isoforms - a short (CD47-SU) and long (CD47-LU) isoform. Knockdown of CD47-LU inhibits CD47 protein cell surface localization, independently of any mRNA localization effects, as both short and long CD47 mRNA reporters localized to the ER. Subsequent analysis of 3′UTR binding sites, gene knockdown, RNA-immunoprecipitation and mRNA-protein tethering approaches determined that HuR likely binds the CD47-LU 3′UTR in association with SET. In turn, SET interacts with nascently translated CD47 protein, and the membrane-localizing small GTPase RAC1, which then aids translocation of CD47 (and SET) to the plasma membrane (Ten Klooster et al., 2007). Underlying the importance of this 3′UTR scaffolding of a protein-localizing complex, cells forced to express either the CD47-LU or CD47-SU in isolation showed significant differences in phagocytic susceptibility (CD47-LU cells more resistant) and induction of apoptosis following γ-irradiation (only CD47-SU cells induce apoptosis) (Berkovits and Mayr, 2015).
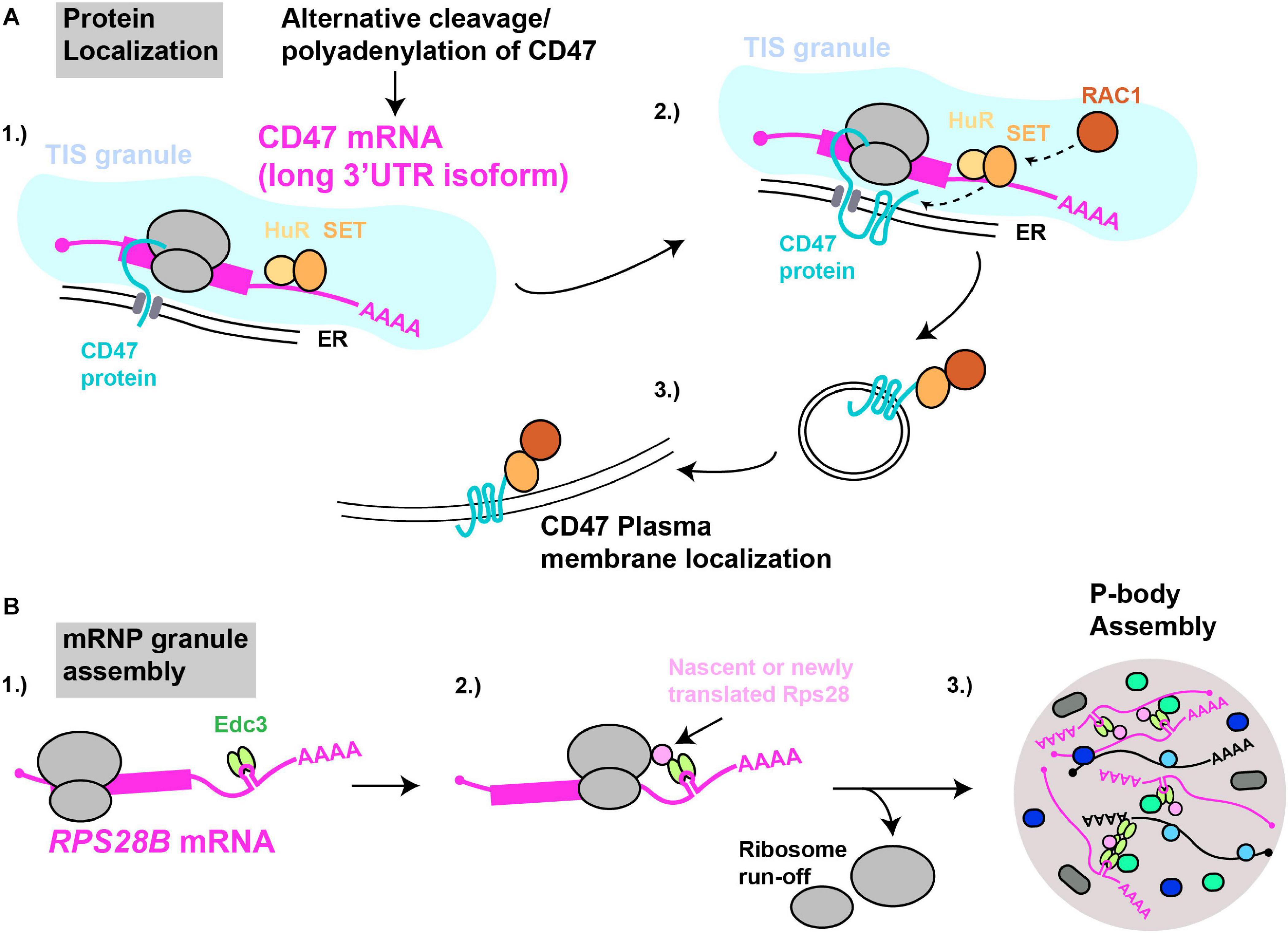
Figure 4. mRNAs as cytoplasmic RNA scaffolds. (A) Protein localization: (1) The CD47-LU mRNA 3′UTR recruits HuR and SET. (2) Within TIS11B-ER membrane compartments (TIS granules), nascently translated CD47’s interaction with SET is facilitated. (3) Recruitment of RAC1 by SET results in subsequent translocation of CD47 to the plasma membrane. (B) mRNP granule assembly: (1) The RPS28B 3′UTR is presumed to recruit Edc3 prior to its subsequent interaction, (2) with either nascently or newly translated Rps28. (3) Since translating mRNAs are excluded from mRNP granules, ribosome run-off is likely required for an RPS28B-Edc3-Rps28 RNP complex to help nucleate yeast P-body assembly.
3′UTR driven co-translational assembly of protein complexes likely occurs in all eukaryotes. We recently demonstrated in yeast that a specific mRNA 3′UTR acts as an mRNA scaffold promoting the assembly of P-bodies (Fernandes and Buchan, 2020), which are cytoplasmic membrane-less organelles enriched in non-translating mRNPs. The mRNA in question is RPS28B, one of two RPS28 paralogs (which code for ribosomal protein S28) that has an unusually long 3′UTR (643 nts) and which harbors a stem-loop structural element which binds Edc3 (He et al., 2014), a protein involved in P-body assembly (Decker et al., 2007). Using genetics, microscopy, and protein interaction assays, we demonstrated that Edc3 recruited by the RPS28B 3′UTR binds nascently or newly translated Rps28b in a manner dependent on the RPS28 ORF and 3′UTR being in cis (Figure 4). The resulting Edc3-Rps28 interaction in turn is key to normal P-body assembly.
To our knowledge, RPS28B is the first case of a specific mRNA implicated in scaffolding an mRNP granule, and it is likely that other RNA molecules (like NEAT1 in paraspeckles) will possess potent scaffolding potential for RNP granules, based on their potentially high interaction valency (either with proteins or other RNA molecules (Van Treeck and Parker, 2018; Van Treeck et al., 2018)) and ability to exist in a non-translating state. Indeed, recent data argues that poorly translated, long RNAs preferentially accumulate within RNP granules such as P-bodies and stress granules (Khong et al., 2017). RPS28B also meets these criteria (Ingolia, 2009), though questions remain as to the nature of the Rps28-Edc3 protein interaction, and whether regulation of RPS28B translation rates or abundance may also occur to influence P-body assembly. Finally, our work demonstrates that mRNAs from gene paralogs, in addition to transcript isoforms from a single gene, represents another means to allow mRNA-based regulation of macromolecular complex assembly. Given that 50–80% of all human genes generate mRNAs with alternative 3′UTRs (Lianoglou et al., 2013) a broad role for 3′UTRs in scaffolding co-translational protein complex assembly seems feasible (Mayr, 2019).
While mRNA localization clearly facilitates protein localization (Holt and Bullock, 2009), co-localization of mRNAs in specific cellular compartments may also aid co-translational protein interactions and/or protein complex assembly. Such a mechanism is an appealing explanation for co-translation protein assembly mechanisms discussed above. Interestingly, a few studies suggest the existence of “translation factories,” where functionally related mRNAs are spatially concentrated in cellular compartments.
In human cells, several membrane protein-encoding mRNAs, including CD47-LU (but not CD47-SU), harbor binding sites for an RBP called TIS11B. These mRNAs, as well as SET and HuR, coalesce with TIS11B in an ER-intertwined reticular structure termed TIS granules, that rely on TIS11B expression for their assembly (Ma and Mayr, 2018). Importantly, TIS11 expression, and formation of TIS granules promotes interaction of SET with CD47, and localization of CD47 to the plasma membrane. This may reflect increased retention of CD47 mRNA and SET within the TIS granule based on fluorescence recovery after photobleaching data, thus increasing the likelihood of co-translational CD47 interaction with SET (Ma and Mayr, 2018).
In yeast, specific glycolytic mRNAs (Lui et al., 2014) and select translation factor mRNAs (Pizzinga et al., 2019) co-localize in distinct translationally active granules, at least some of which encode proteins that collectively form multi-subunit complexes. Finally, several mRNAs encoding components of the Arp2/Arp3 complex, an actin cytoskeleton-regulating complex, co-localize and at least in some cases are translated at the leading edge of fibroblasts (Mingle et al., 2005; Willett et al., 2013). However, direct testing of whether mRNA co-localization effectively serves as a eukaryotic analog of a prokaryotic operon-based protein complex assembly mechanism remains a question for future study.
Estimates in the human genome suggest between 20 and 60k loci in the human genome transcribe lncRNA (Iyer et al., 2015; Hon et al., 2017), with over 170k distinct lncRNA transcripts now reported (Zhao et al., 2021). A considerable challenge in the field lies in discriminating transcriptional noise from functional lncRNA molecules that may act as scaffolds or decoys. However, several methodologies now exist that promise more widespread detection of functional RNA scaffold and decoy molecules, including those that may exhibit conservation.
Assessing reproducibility and condition-specific changes in expression levels, as well as conservation of RNA sequence, exon number, genomic position and transcriptional level is a generally utilized approach for identifying putative functional lncRNAs, for which various amenable bioinformatic tools are now available (Derrien et al., 2012; Ulitsky, 2016; Nelson et al., 2017). LncRNAs typically exhibit a lower degree of sequence conservation and expression level than seen for most protein coding genes, making comparative genomic studies alone more challenging, though the presence of certain sequence features (transposable element-derived sequences (Wang et al., 2017), miRNA binding sites (Furió-Tarí et al., 2016)) can help predict potential scaffolding or decoy potential.
Known scaffold or decoy RNAs often harbor structured domains and typically interact with proteins. RNA-seq technologies that assess RNA secondary structure (e.g., SHAPE (Lucks et al., 2011), Structure-Seq/DMS-Seq (Ding et al., 2014)), structured RNA-regions bound by proteins (e.g., CLASH (Kudla et al., 2011)) or both (e.g., PIP-SEQ) (Foley and Gregory, 2016) may therefore provide suggestive data on a broad transcriptome-wide scale. Comparative analyses of RNA secondary or tertiary structures obtained either via structural means (e.g., NMR, X-ray crystallography), RNA-seq methods or solely via bioinformatic sequence prediction can also help identify functional RNA scaffolds or decoys. While structural methods are clearly limited in throughput, they represent the gold standard in reliably defining the presence of an RNA structural element and identifying sequence targets for subsequent genetic perturbation and functional analysis.
Genetic screening remains a powerful initial tool to identify functional lncRNAs with possible scaffolding or decoy functions. Genome wide studies directly targeting lncRNA loci via CRISPR-Cas9 means (Joung et al., 2017; Liu et al., 2017), and a study of 285 lncRNAs degraded in the nucleus by Antisense oligonucleotide (ASO) mediated RNase-H degradation (Ramilowski et al., 2020) have revealed growth phenotypes in about 0.1–7% of cases. Cell-line and context specific phenotypes may explain this relatively low percentage (Joung et al., 2017; Liu et al., 2017). Indeed a larger fraction of ASO-depleted lncRNAs revealed significant alterations in transcriptome composition, often in the absence of growth phenotypes (Ramilowski et al., 2020). Though not commonly utilized, discrimination of nuclear versus cytoplasmic lncRNA functions may also be achieved in principle by siRNA knockdown, which only targets cytoplasmic RNA species for degradation.
Another means of identifying RNA scaffolds or decoys is to biochemically isolate their interacting biomolecular partners and co-purify and sequence the RNA. For example, isolation of the telomerase complex (via chromatography, TERT enzyme pulldown) has been used to identify or validate prediction of TR RNAs in several species (Greider and Blackburn, 1989; Feng et al., 1995; Dew-Budd et al., 2020). Cross-linking and Immunoprecipitation methods (Lee and Ule, 2018) can also be applied not only to detect putative interacting RNA scaffolds or decoys, but also the specific location on the RNA where the protein binds. This in turn may help identify key RNA sequence/structural elements to base comparative genomic analyses upon when determining if an RNA scaffold or decoy may be functionally conserved.
Finally, microscopy-based analyses using FISH, in combination with immunohistochemistry of other cellular organelles or proteins, can provide insight as to functional and regulatory mechanisms of lncRNAs. A study of 61 lncRNAs in various human cell types revealed that while most were nuclear, highly diverse patterns of localization were observed (Cabili et al., 2015). Notably, nuclear lncRNA foci dissolve during mitosis and become generally dispersed; what the functional impacts of this dispersal process are, and how nuclear localization is re-established remain unclear.
The above methods can also be applied to identifying mRNA scaffold and decoy molecules, though separating 3′UTR-based regulation of nascent protein-interaction from 3′UTR based regulation of the mRNA (i.e., mRNA translation, stability, localization) provides significant additional challenges. Thus, perturbation or forced expression of a mRNA 3′UTR isoform of interest, coupled to assessment of protein interactions (e.g., Co-immunoprecipitation, unbiased mass spectrometry) should include control experiments to verify no alteration of mRNA function or overall protein expression.
It is clear there is no one single path to identification of functional RNA scaffolds and decoys. While sequencing, bioinformatic and comparative analyses studies generally offer the most rapid means to identify putative RNA scaffolds and decoys, genetic, biochemical and microscopy methods can all be brought to bear to enhance mechanistic understanding.
The evidence is mounting that RNA molecules ubiquitously function as scaffolds and decoys bringing together or sequestering specific biomolecules to regulate macromolecular complex assembly. LncRNAs primarily accumulate in the nucleus (Derrien et al., 2012; Cabili et al., 2015), which has led to a historical focus on their roles as RNA scaffolds and decoys in this compartment. However, lncRNAs also accumulate, and in some cases are preferentially enriched, in the cytoplasm (Gudenas and Wang, 2018). This, together with emerging data that cytoplasmic mRNAs can also play scaffolding roles suggests that many new cytoplasmic scaffolding and decoy functions for RNA molecules await discovery, aided by the paradigms provided by their nuclear counterparts.
RNA scaffolds and decoys enable dynamic and condition-specific responses to a variety of cellular stimuli (Table 1), with lncRNAs notably exhibiting significant developmental and tissue-specific control of expression (Derrien et al., 2012; Sarropoulos et al., 2019). However, detailed understanding of how synthesis, decay, and localization (when acting in trans) of RNA scaffolds and decoys is regulated in response to changing cellular environments is lacking in most cases. Additionally, while several RNA scaffolds and decoys, particularly within the category of lncRNAs, have been identified and functionally characterized in mammalian model systems, fewer characterized examples exist in other model systems. How, or indeed if, RNA-scaffolded complexes are actively assembled and disassembled is also unclear, though we speculate that RNA helicases may play a prominent role given their RNP-remodeling roles in other RNA biology processes and at least one prior example of precedent (Wongtrakoongate et al., 2015).
A unique feature of mRNAs is their potential to scaffold nascent protein interactions by virtue of the fact they also serve as protein synthesis templates. To date, characterized cytoplasmic mRNA scaffolds are driven by the presence of unique 3′UTR sequences generated by alternative cleavage and polyadenylation or paralogous gene transcription. Distinct mRNA 3′UTR sequences, with binding sites for nascent protein interaction partners can then help define a nascent protein’s interaction fate and function. How widespread this mode of facilitating protein-protein interaction is remains unknown, though it offers several potential advantages. For instance, protein interaction partners and their respective template mRNAs do not have to be as carefully orchestrated for simultaneously translation and co-localization to avoid competing off-target interactions. A nascent protein’s interaction fate could also be pre-determined by loading of its mRNA’s 3′UTR, possibly in the nucleus, with suitable nascent protein interaction partners. Additionally, this approach may facilitate generation of several distinct nascent protein subcomplexes, as occurs with BIRC3 (Burger et al., 2005; Holt and Bullock, 2009). A caveat of this mechanism might be a limited capacity of any 3′UTR to store (or reacquire) a supply of nascent protein interactors. Thus, this mechanism may favor assembly of low-frequency, regulatory protein interactions, or be more prevalent on poorly translated mRNAs.
Identifying new RNA scaffolds and decoys remains a significant challenge. Nonetheless, new advances in sequencing, bioinformatics and protein-RNA interaction methods promise progress toward this goal. Given the intrinsic advantages of RNA as a scaffolding molecule, and that many RNA scaffolds incorporate other functions (e.g., catalysis, modifying protein activities, guide and template functions; Table 1), we anticipate that intriguing and impactful new modes of RNA-scaffolded interactions await discovery.
NF and JRB conceived and wrote the review and generated the figures and table. Both authors contributed to the article and approved the submitted version.
This work was supported by funding from NIGMS (NIH R01-GM1145664, JRB).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We thank Keith Aspinall for feedback and useful suggestions.
Abdelmohsen, K., Panda, A. C., Kang, M.-J., Guo, R., Kim, J., Grammatikakis, I., et al. (2014). NAR breakthrough article 7SL RNA represses p53 translation by competing with HuR. Nucleic Acids Res. 42, 10099–10111. doi: 10.1093/nar/gku686
Achsel, T., Ahrens, K., Brahms, H., Teigelkamp, S., and Lührmann, R. (1998). The human U5-220kD protein (hPrp8) forms a stable RNA-free complex with several U5-specific proteins, including an RNA unwindase, a homologue of ribosomal elongation factor EF-2, and a Novel WD-40 protein. Mol. Cell. Biol. 18, 6756–6766. doi: 10.1128/mcb.18.11.6756
Almeida, M., Pintacuda, G., Masui, O., Koseki, Y., Gdula, M., Cerase, A., et al. (2017). PCGF3/5-PRC1 initiates Polycomb recruitment in X chromosome inactivation. Science 356, 1081–1084. doi: 10.1126/science.aal2512
Amente, S., Bertoni, A., Morano, A., Lania, L., Avvedimento, E. V., Majello, B., et al. (2010a). LSD1-mediated demethylation of histone H3 lysine 4 triggers Myc-induced transcription. Oncogene 29, 3691–3702. doi: 10.1038/onc.2010.120
Amente, S., Lania, L., Avvedimento, E. V., and Majello, B. (2010b). DNA oxidation drives Myc mediated transcription. Cell Cycle 9, 3074–3076. doi: 10.4161/cc.9.15.12499
Andreassi, C., and Riccio, A. (2009). To localize or not to localize: mRNA fate is in 3′UTR ends. Trends Cell Biol. 19, 465–474. doi: 10.1016/j.tcb.2009.06.001
Andrulis, E. D., Neiman, A. M., Zappulla, D. C., and Sternglanz, R. (1998). Perinuclear localization of chromatin facilitates transcriptional silencing. Nature 394, 592–595. doi: 10.1038/29100
Athanasiadis, A., Rich, A., and Maas, S. (2004). Widespread A-to-I RNA editing of Alu-containing mRNAs in the human transcriptome. PLoS Biol. 2:e391. doi: 10.1371/journal.pbio.0020391
Audas, T. E., Jacob, M. D., and Lee, S. (2012). Immobilization of proteins in the nucleolus by ribosomal intergenic spacer noncoding RNA. Mol. Cell 45, 147–157. doi: 10.1016/j.molcel.2011.12.012
Balas, M. M., and Johnson, A. M. (2018). Exploring the mechanisms behind long noncoding RNAs and cancer. Non Cod. RNA Res. 3, 108–117. doi: 10.1016/j.ncrna.2018.03.001
Berkovits, B. D., and Mayr, C. (2015). Alternative 3′ UTRs act as scaffolds to regulate membrane protein localization. Nature 522, 363–367. doi: 10.1038/nature14321
Berry, J., Weber, S. C., Vaidya, N., Haataja, M., and Brangwynne, C. P. (2015). RNA transcription modulates phase transition-driven nuclear body assembly. Proc. Natl. Acad. Sci. U.S.A. 112, E5237–E5245.
Bhan, A., and Mandal, S. S. (2015). LncRNA HOTAIR: a master regulator of chromatin dynamics and cancer. Biochim. Biophys. Acta Rev. Cancer 1856, 151–164. doi: 10.1016/j.bbcan.2015.07.001
Boeynaems, S., Alberti, S., Fawzi, N. L., Mittag, T., Polymenidou, M., Rousseau, F., et al. (2018). Protein phase separation: a new phase in cell biology steven. Trends Cell Biol. 28, 420–435. doi: 10.1016/j.tcb.2018.02.004
Bohmdorfer, G., Sethuraman, S., Rowley, M. J., Krzyszton, M., Hafiz Rothi, M., Bouzit, L., et al. (2016). Long non-coding RNA produced by RNA polymerase V determines boundaries of heterochromatin. eLife. 4, 1–24. doi: 10.1002/9783527678679.dg06939
Bond, C. S., and Fox, A. H. (2009). Paraspeckles: nuclear bodies built on long noncoding RNA. J. Cell Biol. 186, 637–644. doi: 10.1083/jcb.200906113
Brockdorff, N. (2017). Polycomb complexes in X chromosome inactivation. Phil. Trans. R. Soc. B 372:20170021. doi: 10.1098/rstb.2017.0021
Brodersen, D. E., Clemons, W. M., Carter, A. P., Wimberly, B. T., and Ramakrishnan, V. (2002). Crystal structure of the 30 S ribosomal subunit from Thermus thermophilus: structure of the proteins and their interactions with 16 S RNA. J. Mol. Biol. 316, 725–768.
Bronicki, L. M., and Jasmin, B. J. (2013). Emerging complexity of the HuD/ELAVl4 gene; Implications for neuronal development, function, and dysfunction. RNA 19, 1019–1037. doi: 10.1261/rna.039164.113
Burger, M., Hartmann, T., Krome, M., Rawluk, J., Tamamura, H., Fujii, N., et al. (2005). Small peptide inhibitors of the CXCR4 chemokine receptor (CD184) antagonize the activation, migration, and antiapoptotic responses of CXCL12 in chronic lymphocytic leukemia B cells. Blood 106, 1824–1830. doi: 10.1182/blood-2004-12-4918
Cabili, M. N., Dunagin, M. C., McClanahan, P. D., Biaesch, A., Padovan-Merhar, O., Regev, A., et al. (2015). Localization and abundance analysis of human lncRNAs at single-cell and single-molecule resolution. Genome Biol. 16:20.
Carmo-Fonseca, M. (2015). Assembly of the nucleolus: in need of revision. EMBO J. 34, 2731–2732. doi: 10.15252/embj.201593185
Caudron-Herger, M., Pankert, T., and Rippe, K. (2016). Regulation of nucleolus assembly by non-coding RNA polymerase II transcripts. Nucleus 7, 308–318. doi: 10.1080/19491034.2016.1190890
Caudron-Herger, M., Pankert, T., Seiler, J., Németh, A., Voit, R., Grummt, I., et al. (2015). Alu element-containing RNA s maintain nucleolar structure and function. EMBO J. 34, 2758–2774. doi: 10.15252/embj.201591458
Cerase, A., Pintacuda, G., Tattermusch, A., and Avner, P. (2015). Xist localization and function: new insights from multiple levels. Genome Biol. 16:166. doi: 10.1186/s13059-015-0733-y
Chan, S. R. W. L., and Blackburn, E. H. (2004). Telomeres and telomerase. Philos. Trans. R. Soc. B Biol. Sci. 359, 109–121.
Chaudhary, R., Gryder, B., Woods, W. S., Subramanian, M., Jones, M. F., Li, X. L., et al. (2017). Prosurvival long noncoding RNA PINCR regulates a subset of p53 targets in human colorectal cancer cells by binding to Matrin 3. Elife 6:e23244. doi: 10.7554/eLife.23244
Chen, C.-K., Blanco, M., Jackson, C., Aznauryan, E., Ollikainen, N., Surka, C., et al. (2016). Xist recruits the X chromosome to the nuclear lamina to enable chromosome-wide silencing. Science 354, 468–473. doi: 10.1126/science.aae0047
Chen, J. L., and Greider, C. W. (2005). Functional analysis of the pseudoknot structure in human telomerase RNA. Proc. Natl. Acad. Sci. U.S.A. 102, 8080–8085. doi: 10.1073/pnas.0502259102
Chen, L. L., and Carmichael, G. G. (2009). Altered nuclear retention of mRNAs containing inverted repeats in human embryonic stem cells: functional role of a nuclear noncoding RNA. Mol. Cell 35, 467–478. doi: 10.1016/j.molcel.2009.06.027
Chen, L. L., DeCerbo, J. N., and Carmichael, G. G. (2008). Alu element-mediated gene silencing. EMBO J. 27, 1694–1705. doi: 10.1038/emboj.2008.94
Chow, J. C., Yen, Z., Ziesche, S. M., and Brown, C. J. (2005). Silencing of the mammalian X chromosome. Annu. Rev. Genomics Hum. Genet. 6, 69–92. doi: 10.1146/annurev.genom.6.080604.162350
Chu, C., Qu, K., Zhong, F. L., Artandi, S. E., and Chang, H. Y. (2011). Genomic maps of long noncoding RNA occupancy reveal principles of RNA-chromatin interactions. Mol. Cell 44, 667–678. doi: 10.1016/j.molcel.2011.08.027
Chu, C., Zhang, Q. C., Da Rocha, S. T., Flynn, R. A., Bharadwaj, M., Calabrese, J. M., et al. (2015). Systematic discovery of Xist RNA binding proteins. Cell 161, 404–416. doi: 10.1016/j.cell.2015.03.025
Chujo, T., Yamazaki, T., and Hirose, T. (2016). Architectural RNAs (arcRNAs): a class of long noncoding RNAs that function as the scaffold of nuclear bodies. Biochim. Biophys. Acta Gene Regul. Mech. 1859, 139–146. doi: 10.1016/j.bbagrm.2015.05.007
Clemson, C. M., Hutchinson, J. N., Sara, S. A., Ensminger, A. W., Fox, A. H., Chess, A., et al. (2009). An architectural role for a nuclear noncoding RNA: NEAT1 RNA is essential for the structure of paraspeckles. Mol. Cell 33, 717–726. doi: 10.1016/j.molcel.2009.01.026
Clemson, C. M., McNeil, J. A., Willard, H. F., and Lawrence, J. B. (1996). XIST RNA paints the inactive X chromosome at interphase: evidence for a novel RNA involved in nuclear/chromosome structure. J. Cell Biol. 132, 259–275. doi: 10.1083/jcb.132.3.259
Collins, K. (2008). Physiological assembly and activity of human telomerase complexes. Mech. Ageing Dev. 129, 91–98.
Dandekar, T., Snel, B., Huynen, M., and Bork, P. (1998). Conservation of gene order: a fingerprint of proteins that physically interact. Trends Biochem. Sci. 23, 324–328. doi: 10.1016/s0968-0004(98)01274-2
Davis, B. M., Mccurrach, M. E., Taneja, K. L., Singer, R. H., and Housman, D. E. (1997). Expansion of a CUG trinucleotide repeat in the 3′ untranslated region of myotonic dystrophy protein kinase transcripts results in nuclear retention of transcripts. Proc. Natl. Acad. Sci. U.S.A. 94, 7388–7393. doi: 10.1073/pnas.94.14.7388
Decker, C. J., Teixeira, D., and Parker, R. (2007). Edc3p and a glutamine/asparagine-rich domain of Lsm4p function in processing body assembly in Saccharomyces cerevisiae. J. Cell Biol. 179, 437–449. doi: 10.1083/jcb.200704147
Deckert, J., Hartmuth, K., Boehringer, D., Behzadnia, N., Will, C. L., Kastner, B., et al. (2006). Protein composition and electron microscopy structure of affinity-purified human spliceosomal B complexes isolated under physiological conditions. Mol. Cell. Biol. 26, 5528–5543. doi: 10.1128/mcb.00582-06
Deininger, P. (2011). Alu elements: know the SINEs. Genome Biol. 12, 1–12. doi: 10.1002/9781118584538.ieba0500
Deng, X., Ma, W., Ramani, V., Hill, A., Yang, F., Ay, F., et al. (2015). Bipartite structure of the inactive mouse X chromosome. Genome Biol. 16, 1–21.
Derrien, T., Johnson, R., Bussotti, G., Tanzer, A., Djebali, S., Tilgner, H., et al. (2012). The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res. 22, 1775–1789. doi: 10.1101/gr.132159.111
Dew-Budd, K., Cheung, J., Palos, K., Forsythe, E. S., and Beilstein, M. A. (2020). Evolutionary and biochemical analyses reveal conservation of the Brassicaceae telomerase ribonucleoprotein complex. PLoS One 15:e0222687. doi: 10.1371/journal.pone.0222687
Ding, Y., Tang, Y., Kwok, C. K., Zhang, Y., Bevilacqua, P. C., and Assmann, S. M. (2014). In vivo genome-wide profiling of RNA secondary structure reveals novel regulatory features. Nature 505, 696–700. doi: 10.1038/nature12756
Duszczyk, M. M., Wutz, A., Rybin, V., and Sattler, M. (2011). The Xist RNA A-repeat comprises a novel AUCG tetraloop fold and a platform for multimerization. RNA 17, 1973–1982. doi: 10.1261/rna.2747411
Ebert, M. S., and Sharp, P. A. (2010a). Emerging role of natural microrna sponge. Curr. Biol. 20, R858–R861.
Ebert, M. S., and Sharp, P. A. (2010b). MicroRNA sponges: progress and possibilities. RNA 16, 2043–2050. doi: 10.1261/rna.2414110
Falahati, H., Blythe, S., Falahati, H., Pelham-webb, B., Blythe, S., Wieschaus, E., et al. (2016). Nucleation by rRNA dictates the precision of nucleolus assembly article nucleation by rRNA dictates the precision of nucleolus assembly. Curr. Biol. 26, 277–285. doi: 10.1016/j.cub.2015.11.065
Fang, H., Bonora, G., Lewandowski, J. P., Thakur, J., Filippova, G. N., Henikoff, S., et al. (2020). Trans- and cis-acting effects of the lncRNA Firre on epigenetic and structural features of the inactive X chromosome. Nat. Commun. 11:6053.
Feng, J., Funk, W. D., Wang, S. S., Weinrich, S. L., Avilion, A. A., Chiu, C. P., et al. (1995). The RNA component of human telomerase. Science 269, 1236–1241.
Fernandes, N., and Buchan, J. R. (2020). RPS28B mRNA acts as a scaffold promoting Cis-translational interaction of proteins driving P-body assembly. Nucleic Acids Res. 48, 6265–6279. doi: 10.1093/nar/gkaa352
Foley, S. W., and Gregory, B. D. (2016). Protein interaction profile sequencing (PIP-seq). Curr. Protoc. Mol. Biol. 116:27.5.1.
Franco-Zorrilla, J. M., Valli, A., Todesco, M., Mateos, I., Puga, M. I., Rubio-Somoza, I., et al. (2007). Target mimicry provides a new mechanism for regulation of microRNA activity. Nat. Genet. 39, 1033–1037. doi: 10.1038/ng2079
Fu, D., and Collins, K. (2007). Purification of human telomerase complexes identifies factors involved in telomerase biogenesis and telomere length regulation. Mol. Cell 28, 773–785. doi: 10.1016/j.molcel.2007.09.023
Furió-Tarí, P., Tarazona, S., Gabaldón, T., Enright, A. J., and Conesa, A. (2016). spongeScan: a web for detecting microRNA binding elements in lncRNA sequences. Nucleic Acids Res. 44, W176–W180.
Galej, W. P. (2018). Structural studies of the spliceosome: past, present and future perspectives. Biochem. Soc. Trans. 46, 1407–1422. doi: 10.1042/bst20170240
Gao, S., Guo, J., Li, F., Zhang, K., Zhang, Y., Zhang, Y., et al. (2017). Long non-coding RNA lncTCF7 predicts poor prognosis and promotes tumor metastasis in osteosarcoma. Int. J. Clin. Exp. Pathol. 10, 10918–10925.
Genuth, N. R., and Barna, M. (2018). Heterogeneity and specialized functions of translation machinery: from genes to organisms. Nat. Rev. Genet. 19, 431–452. doi: 10.1038/s41576-018-0008-z
Gloss, B. S., and Dinger, M. E. (2016). The specificity of long noncoding RNA expression. Biochim. Biophys. Acta Gene Regul. Mech. 1859, 16–22. doi: 10.1016/j.bbagrm.2015.08.005
Gong, C., and Maquat, L. E. (2011). lncRNAs transactivate STAU1-mediated mRNA decay by duplexing with 3′UTRs via Alu elements. Nature 470, 284–288. doi: 10.1038/nature09701
Gong, C., Popp, M. W. L., and Maquat, L. E. (2012). Biochemical analysis of long non-coding RNA-containing ribonucleoprotein complexes. Methods 58, 88–93. doi: 10.1016/j.ymeth.2012.06.020
Gowda, K., Chittenden, K., and Zwieb, C. (1997). Binding site of the M-domain of human protein SRP54 determined by systematic site-directed mutagenesis of signal recognition particle RNA. Nucleic Acids Res. 25, 388–394. doi: 10.1093/nar/25.2.388
Greider, C. W., and Blackburn, E. H. (1989). A telomeric sequence in the RNA of Tetrahymena telomerase required for telomere repeat synthesis. Nature 337, 331–337. doi: 10.1038/337331a0
Grob, A., Colleran, C., and McStay, B. (2014). Construction of synthetic nucleoli in human cells reveals how a major functional nuclear domain is formed and propagated through cell division. Genes Dev. 28, 220–230. doi: 10.1101/gad.234591.113
Grotwinkel, J. T., Wild, K., Segnitz, B., and Sinning, I. (2014). ISRP RNA remodeling by SRP68 explains its role in protein translocation. Science 344, 101–105. doi: 10.1126/science.1249094
Gudenas, B. L., and Wang, L. (2018). Prediction of LncRNA subcellular localization with deep learning from sequence features. Sci. Rep. 8:16385.
Hacisuleyman, E., Goff, L. A., Trapnell, C., Williams, A., Henao-Mejia, J., Sun, L., et al. (2014). Topological organization of multichromosomal regions by the long intergenic noncoding RNA Firre. Nat. Struct. Mol. Biol. 21, 198–206. doi: 10.1038/nsmb.2764
Hajjari, M., and Salavaty, A. (2015). HOTAIR: an oncogenic long non-coding RNA in different cancers. Cancer Biol. Med. 12, 1–9.
Han, P., Li, W., Lin, C. H., Yang, J., Shang, C., Nurnberg, S. T., et al. (2014). A long noncoding RNA protects the heart from pathological hypertrophy. Nature 514, 102–106. doi: 10.1038/nature13596
Hansen, T. B., Jensen, T. I., Clausen, B. H., Bramsen, J. B., Finsen, B., Damgaard, C. K., et al. (2013). Natural RNA circles function as efficient microRNA sponges. Nature 495, 384–388. doi: 10.1038/nature11993
Hasegawa, Y., Brockdorff, N., Kawano, S., Tsutui, K., Tsutui, K., and Nakagawa, S. (2010). The matrix protein hnRNP U is required for chromosomal localization of xist RNA. Dev. Cell 19, 469–476. doi: 10.1016/j.devcel.2010.08.006
He, F., Li, C., Roy, B., and Jacobson, A. (2014). Yeast Edc3 targets RPS28B mRNA for decapping by binding to a 3′ -UTR decay-inducing regulatory element. Mol. Cell. Biol. 34, 1438–1451. doi: 10.1128/mcb.01584-13
Hernandez-verdun, D. (2011). Assembly and disassembly of the nucleolus during the cell cycle Assembly and disassembly of the nucleolus during the cell cycle. Nucleus 1034, 189–194. doi: 10.4161/nucl.2.3.16246
Hirose, T., Virnicchi, G., Tanigawa, A., Naganuma, T., Li, R., Kimura, H., et al. (2014). NEAT1 long noncoding RNA regulates transcription via protein sequestration within subnuclear bodies. Mol. Biol. Cell 25, 169–183. doi: 10.1091/mbc.e13-09-0558
Hisaoka, M., Ueshima, S., Murano, K., Nagata, K., and Okuwaki, M. (2010). Regulation of nucleolar chromatin by B23/nucleophosmin jointly depends upon its RNA binding activity and transcription factor UBF. Mol. Cell. Biol. 30, 4952–4964. doi: 10.1128/mcb.00299-10
Holt, C. E., and Bullock, S. L. (2009). Subcellular mRNA localization in animal cells and why it matters. Science 326, 1212–1216. doi: 10.1126/science.1176488
Hon, C. C., Ramilowski, J. A., Harshbarger, J., Bertin, N., Rackham, O. J. L., Gough, J., et al. (2017). An atlas of human long non-coding RNAs with accurate 5′ ends. Nature 543, 199–204.
Huarte, M., Guttman, M., Feldser, D., Garber, M., Koziol, M. J., Kenzelmann-Broz, D., et al. (2010). A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response. Cell 142, 409–419. doi: 10.1016/j.cell.2010.06.040
Iarovaia, O. V., Minina, E. P., Sheval, E. V., Onichtchouk, D., Dokudovskaya, S., Razin, S. V., et al. (2019). Nucleolus: a central hub for nuclear functions. Trends Cell Biol. 29, 647–659. doi: 10.1016/j.tcb.2019.04.003
Imamura, K., Imamachi, N., Akizuki, G., Kumakura, M., Kawaguchi, A., Nagata, K., et al. (2014). Long noncoding RNA NEAT1-dependent SFPQ relocation from promoter region to paraspeckle mediates IL8 expression upon immune stimuli. Mol. Cell 53, 393–406. doi: 10.1016/j.molcel.2014.01.009
Imashimizu, M., Shimamoto, N., Oshima, T., and Kashlev, M. (2014). Transcription elongation. Heterogeneous tracking of RNA polymerase and its biological implications. Transcription 5:e28285. doi: 10.4161/trns.28285
Ingolia, N. (2009). Genome-Wide Analysis in Vivo of Translation with nucleotide resolution using ribosome profiling. Science 324, 218–223. doi: 10.1126/science.1168978
Iyer, M. K., Niknafs, Y. S., Malik, R., Singhal, U., Sahu, A., and Hosono, Y. (2015). The landscape of long noncoding RNAs in the human transcriptome. Nat. Genet. 47, 199–208.
Jaiswal, S., Jamieson, C. H. M., Pang, W. W., Park, C. Y., Chao, M. P., Majeti, R., et al. (2009). CD47 is upregulated on circulating hematopoietic stem cells and leukemia cells to avoid phagocytosis. Cell 138, 271–285. doi: 10.1016/j.cell.2009.05.046
Jalali, S., Bhartiya, D., Lalwani, M. K., Sivasubbu, S., and Scaria, V. (2013). Systematic transcriptome wide analysis of lncRNA-miRNA interactions. PLoS One 8:e53823. doi: 10.1371/journal.pone.0053823
Jiang, L., Shao, C., Wu, Q. J., Chen, G., Zhou, J., Yang, B., et al. (2017). NEAT1 scaffolds RNA-binding proteins and the microprocessor to globally enhance pri-miRNA processing. Nat. Struct. Mol. Biol. 24, 816–824. doi: 10.1038/nsmb.3455
Johnsson, P., Lipovich, L., Grandér, D., and Morris, K. V. (2014). Biochimica et Biophysica Acta Evolutionary conservation of long non-coding RNAs; sequence, structure, function. BBA Gen. Subj. 1840, 1063–1071. doi: 10.1016/j.bbagen.2013.10.035
Joung, J., Engreitz, J. M., Konermann, S., Abudayyeh, O. O., Verdine, V. K., and Aguet, F. (2017). Genome-scale activation screen identifies a lncRNA locus regulating a gene neighbourhood. Nature 548, 343–346. doi: 10.1038/nature23451
Kamenova, I., Mukherjee, P., Conic, S., Mueller, F., El-Saafin, F., Bardot, P., et al. (2019). Co-translational assembly of mammalian nuclear multisubunit complexes. Nat. Commun. 10, 25–28.
Kaneko, S., Li, G., Son, J., Xu, C. F., Margueron, R., Neubert, T. A., et al. (2010). Phosphorylation of the PRC2 component Ezh2 is cell cycle-regulated and up-regulates its binding to ncRNA. Genes Dev. 24, 2615–2620. doi: 10.1101/gad.1983810
Kawaguchi, T., Tanigawa, A., Naganuma, T., Ohkawa, Y., Souquere, S., Pierron, G., et al. (2015). SWI/SNF chromatin-remodeling complexes function in noncoding RNA-dependent assembly of nuclear bodies. Proc. Natl. Acad. Sci. U.S.A. 112, 4304–4309. doi: 10.1073/pnas.1423819112
Keenan, R. J., Freymann, D. M., Stroud, R. M., and Walter, P. (2001). The signal recognition particle. Annu. Rev. Biochem. 70, 755–775.
Khandelwal, A., Bacolla, A., Vasquez, K. M., and Jain, A. (2015). Long non-coding RNA: a new paradigm for lung cancer. Mol. Carcinog. 54, 1235–1251. doi: 10.1002/mc.22362
Khong, A., Matheny, T., Jain, S., Mitchell, S. F., Wheeler, J. R., and Parker, R. (2017). The stress granule transcriptome reveals principles of mRNA accumulation in stress granules. Mol. Cell 68, 808–820. doi: 10.1016/j.molcel.2017.10.015
Kim, Y. K., Furic, L., DesGroseillers, L., and Maquat, L. E. (2005). Mammalian Staufen1 recruits Upf1 to specific mRNA 3′UTRs so as to elicit mRNA decay. Cell 120, 195–208. doi: 10.1016/j.cell.2004.11.050
Klein, D. J., Moore, P. B., and Steitz, T. A. (2004). The roles of ribosomal proteins in the structure assembly, and evolution of the large ribosomal subunit. J. Mol. Biol. 340, 141–177. doi: 10.1016/j.jmb.2004.03.076
Knott, G. J., Bond, C. S., and Fox, A. H. (2016). The DBHS proteins SFPQ, NONO and PSPC1: a multipurpose molecular scaffold. Nucleic Acids Res. 44, 3989–4004. doi: 10.1093/nar/gkw271
Köhn, M., Ihling, C., Sinz, A., Krohn, K., and Hüttelmaier, S. (2015). The Y3 ncRNA promotes the 3′ end processing of histone mRNAs. Genes Dev. 29, 1998–2003. doi: 10.1101/gad.266486.115
Kong, Y., Hsieh, C.-H., and Alonso, L. C. (2018). ANRIL: a lncRNA at the CDKN2A/B locus with roles in cancer and metabolic disease. Front. Endocrinol. (Lausanne) 9:405. doi: 10.3389/fendo.2018.00405
Koonin, E. V., and Mushegian, A. R. (1996). Complete genome sequences of cellular life forms: glimpses of theoretical evolutionary genomics. Curr. Opin. Genet. Dev. 6, 757–762. doi: 10.1016/s0959-437x(96)80032-3
Kopp, F., and Mendell, J. T. (2018). Functional classification and experimental dissection of long noncoding RNAs. Cell 172, 393–407. doi: 10.1016/j.cell.2018.01.011
Kudla, G., Granneman, S., Hahn, D., Beggs, J. D., and Tollervey, D. (2011). Cross-linking, ligation, and sequencing of hybrids reveals RNA-RNA interactions in yeast. Proc. Natl. Acad. Sci. U.S.A. 108, 10010–10015. doi: 10.1073/pnas.1017386108
Lecompte, O., Ripp, R., Thierry, J. C., Moras, D., and Poch, O. (2002). Comparative analysis of ribosomal proteins in complete genomes: an example of reductive evolution at the domain scale. Nucleic Acids Res. 30, 5382–5390. doi: 10.1093/nar/gkf693
Lee, F. C. Y., and Ule, J. (2018). Advances in CLIP technologies for studies of protein-RNA interactions. Mol. Cell 69, 354–369. doi: 10.1016/j.molcel.2018.01.005
Lee, S.-H., and Mayr, C. (2019). Gain of additional BIRC3 protein functions through 3′-UTR-mediated protein complex formation. Mol. Cell 74, 701.e9–712.e9.
Leisegang, M. S., Fork, C., Josipovic, I., Richter, F. M., Preussner, J., Hu, J., et al. (2017). Long noncoding RNA MANTIS facilitates endothelial angiogenic function. Circulation 136, 65–79. doi: 10.1161/CIRCULATIONAHA.116.026991
Lewandowski, J. P., Lee, J. C., Hwang, T., Sunwoo, H., Goldstein, J. M., Groff, A. F., et al. (2019). The Firre locus produces a trans-acting RNA molecule that functions in hematopoiesis. Nat. Commun. 10, 1–13. doi: 10.1002/9783527678679.dg13413
Li, B., Chen, P., Qu, J., Shi, L., Zhuang, W., Fu, J., et al. (2014). Activation of LTBP3 gene by a long noncoding RNA (IncRNA) MALAT1 transcript in mesenchymal stem cells from multiple myeloma. J. Biol. Chem. 289, 29365–29375. doi: 10.1074/jbc.M114.572693
Li, W., Notani, D., and Rosenfeld, M. G. (2016). Enhancers as non-coding RNA transcription units: recent insights and future perspectives. Nat. Rev. Genet. 17, 207–223. doi: 10.1038/nrg.2016.4
Li, Y., Wang, Z., Shi, H., Li, H., Li, L., Fang, R., et al. (2016). HBXIP and LSD1 scaffolded by lncRNA hotair mediate transcriptional activation by c-Myc. Cancer Res. 76, 293–305. doi: 10.1158/0008-5472.CAN-14-3607
Li, Y. P., Busch, R. K., Valdez, B. C., and Busch, H. (1996). C23 interacts with B23, a putative nucleolar-localization-signal-binding protein. Eur. J. Biochem. 237, 153–158. doi: 10.1111/j.1432-1033.1996.0153n.x
Li, Z., Chao, T. C., Chang, K. Y., Lin, N., Patil, V. S., Shimizu, C., et al. (2014). The long noncoding RNA THRIL regulates TNFα expression through its interaction with hnRNPL. Proc. Natl. Acad. Sci. U.S.A. 111, 1002–1007. doi: 10.1073/pnas.1313768111
Lianoglou, S., Garg, V., Yang, J. L., Leslie, C. S., and Mayr, C. (2013). Ubiquitously transcribed genes use alternative polyadenylation to achieve tissue-specific expression. Genes Dev. 27, 2380–2396. doi: 10.1101/gad.229328.113
Liu, J., Wang, L., Wang, Z., and Liu, J.-P. (2019). Roles of telomere biology in cell senescence, replicative and chronological ageing. Cells 8:54. doi: 10.3390/cells8010054
Liu, S. J., Horlbeck, M. A., Cho, S. W., Birk, H. S., Malatesta, M., He, D., et al. (2017). CRISPRi-based genome-scale identification of functional long noncoding RNA loci in human cells. Science 355:aah7111.
Livengood, A. J., Zaug, A. J., and Cech, T. R. (2002). Essential regions of Saccharomyces cerevisiae telomerase RNA: separate elements for Est1p and Est2p interaction. Mol. Cell. Biol. 22, 2366–2374. doi: 10.1128/mcb.22.7.2366-2374.2002
Long, Y., Wang, X., Youmans, D. T., and Cech, T. R. (2017). How do lncRNAs regulate transcription? Sci. Adv. 3:eaao2110. doi: 10.1126/sciadv.aao2110
Lowenstein, C. J., and Padalko, E. (2004). iNOS (NOS2) at a glance. J. Cell Sci. 117, 2865–2867. doi: 10.1242/jcs.01166
Lu, Y., Liu, X., Xie, M., Liu, M., Ye, M., Li, M., et al. (2017). The NF-κB–responsive long noncoding RNA FIRRE regulates posttranscriptional regulation of inflammatory gene expression through interacting with hnRNPU. J. Immunol. 199, 3571–3582. doi: 10.4049/jimmunol.1700091
Lu, Z., Carter, A. C., and Chang, H. Y. (2017). Mechanistic insights in X-chromosome inactivation. Phil. Trans. R. Soc. B 372:20160356. doi: 10.1098/rstb.2016.0356
Luan, W., Li, L., Shi, Y., Bu, X., Xia, Y., Wang, J., et al. (2016). Long non-coding RNA MALAT1 acts as a competing endogenous RNA to promote malignant melanoma growth and metastasis by sponging miR-22. Oncotarget 7, 63901–63912. doi: 10.18632/oncotarget.11564
Lucks, J. B., Mortimer, S. A., Trapnell, C., Luo, S., Aviran, S., Schroth, G. P., et al. (2011). Multiplexed RNA structure characterization with selective 2’-hydroxyl acylation analyzed by primer extension sequencing (SHAPE-Seq). Proc. Natl. Acad. Sci. U.S.A. 108, 11063–11068. doi: 10.1073/pnas.1106501108
Lui, J., Castelli, L. M., Pizzinga, M., Simpson, C. E., Hoyle, N. P., Bailey, K. L., et al. (2014). Granules harboring translationally active mRNAs provide a platform for P-body formation following stress. Cell Rep. 9, 944–954. doi: 10.1016/j.celrep.2014.09.040
Lunde, B. M., Moore, C., Varani, G., and Avenue, H. (2017). RNA-binding proteins: modular design for efficient function. Nat. Rev. Mol. Cell Biol. 8, 479–490. doi: 10.1038/nrm2178
Lynch, M., and Marinov, G. K. (2015). The bioenergetic costs of a gene. Proc. Natl. Acad. Sci. U.S.A. 112, 15690–15695. doi: 10.1073/pnas.1514974112
Ma, W., and Mayr, C. A. (2018). Membraneless organelle associated with the endoplasmic reticulum enables 3′UTR-mediated protein-protein interactions. Cell 175, 1492–1506. doi: 10.1016/j.cell.2018.10.007
Mallik, M., and Lakhotia, S. C. (2009). The developmentally active and stress-inducible noncoding hsrω gene is a novel regulator of apoptosis in Drosophila. Genetics 183, 831–852. doi: 10.1534/genetics.109.108571
Marijan, D., Tse, R., Elliott, K., Chandhok, S., Luo, M., Lacroix, E., et al. (2019). Stress- specific aggregation of proteins in the amyloid bodies. FEBS Lett. 593, 3162–3172. doi: 10.1002/1873-3468.13597
Mankodi, A., Urbinati, C. R., Yuan, Q. P., Moxley, R. T., Sansone, V., Krym, M., et al. (2001). Muscleblind localizes to nuclear foci of aberrant RNA in myotonic dystrophy types 1 and 2. Hum. Mol. Genet. 10, 2165–2170. doi: 10.1093/hmg/10.19.2165
Mao, Y. S., Zhang, B., and Spector, D. L. (2011). Biogenesis and function of nuclear bodies. Trends Genet. 27, 295–306. doi: 10.1016/j.tig.2011.05.006
Massenet, S. (2019). In vivo assembly of eukaryotic signal recognition particle: a still enigmatic process involving the SMN complex. Biochimie 164, 99–104. doi: 10.1016/j.biochi.2019.04.007
Mastroyiannopoulos, N. P., Shammas, C., and Phylactou, L. A. (2010). Tackling the pathogenesis of RNA nuclear retention in myotonic dystrophy. Biol. Cell 102, 515–523. doi: 10.1042/bc20100040
Matera, A. G., and Wang, Z. (2014). A day in the life of the spliceosome. Nat. Rev. Mol. Cell Biol. 15, 108–121. doi: 10.1038/nrm3742
Mathis, A. D., Naylor, B. C., Carson, R. H., Evans, E., Harwell, J., Knecht, J., et al. (2017). Mechanisms of in vivo ribosome maintenance change in response to nutrient signals. Mol. Cell. Proteomics 16, 243–254. doi: 10.1074/mcp.m116.063255
Mayr, C. (2017). Regulation by 3 -untranslated regions. Annu. Rev. Genet. 51, 171–194. doi: 10.1146/annurev-genet-120116-024704
Mayr, C. (2019). What are 3′ utrs doing? Cold Spring Harb. Perspect. Biol. 11:a034728. doi: 10.1101/cshperspect.a034728
Mayr, C., and Bartel, D. P. (2009). Widespread shortening of 3′UTRs by alternative cleavage and polyadenylation activates oncogenes in cancer cells. Cell 138, 673–684. doi: 10.1016/j.cell.2009.06.016
McHugh, C. A., Chen, C. K., Chow, A., Surka, C. F., Tran, C., McDonel, P., et al. (2015). The Xist lncRNA interacts directly with SHARP to silence transcription through HDAC3. Nature 521, 232–236. doi: 10.1038/nature14443
Melé, M., Mattioli, K., Mallard, W., Shechner, D. M., Gerhardinger, C., and Rinn, J. L. (2017). Chromatin environment, transcriptional regulation, and splicing distinguish lincRNAs and mRNAs. Genome Res. 27, 27–37. doi: 10.1101/gr.214205.116
Memczak, S., Jens, M., Elefsinioti, A., Torti, F., Krueger, J., Rybak, A., et al. (2013). Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 495, 333–338. doi: 10.1038/nature11928
Miller, J. W. (2000). Recruitment of human muscleblind proteins to (CUG)n expansions associated with myotonic dystrophy. EMBO J. 19, 4439–4448. doi: 10.1093/emboj/19.17.4439
Mingle, L. A., Okuhama, N. N., Shi, J., Singer, R. H., Condeelis, J., and Liu, G. (2005). Localization of all seven messenger RNAs for the actin-polymerization nucleator Arp2/3 complex in the protrusions of fibroblasts. J. Cell Sci. 118, 2425–2433. doi: 10.1242/jcs.02371
Mukherjee, N., Calviello, L., Hirsekorn, A., De Pretis, S., Pelizzola, M., and Ohler, U. (2017). Integrative classification of human coding and noncoding genes through RNA metabolism profiles. Nat. Struct. Mol. Biol. 24, 86–96. doi: 10.1038/nsmb.3325
Naganuma, T., Nakagawa, S., Tanigawa, A., Sasaki, Y. F., Goshima, N., Hirose, T., et al. (2012). Alternative 3′-end processing of long noncoding RNA initiates construction of nuclear paraspeckles. EMBO J. 31, 4020–4034. doi: 10.1038/emboj.2012.251
Nakagawa, S., and Hirano, T. (2014). Gathering around Firre. Nat. Struct. Mol. Biol. 21, 207–208. doi: 10.1038/nsmb.2782
Natan, E., Wells, J. N., Teichmann, S. A., and Marsh, J. A. (2017). Regulation, evolution and consequences of cotranslational protein complex assembly. Curr. Opin. Struct. Biol. 42, 90–97. doi: 10.1016/j.sbi.2016.11.023
Nelson, A. D. L., Devisetty, U. K., Palos, K., Haug-Baltzell, A. K., Lyons, E., and Beilstein, M. A. (2017). Evolinc: a tool for the identification and evolutionary comparison of long intergenic non-coding RNAs. Front. Genet. 8:52. doi: 10.3389/fgene.2017.00052
Noller, H. F., Hoffarth, V., and Zimniak, L. (1992). Unusual resistance of peptidyl transferase to protein extraction procedures. Science 256, 1416–1419. doi: 10.1126/science.1604315
Oldenborg, P. A., Zheleznyak, A., Fang, Y. F., Lagenaur, C. F., Gresham, H. D., and Lindberg, F. P. (2000). Role of CD47 as a marker of self on red blood cells. Science 288, 2051–2054. doi: 10.1126/science.288.5473.2051
O’Leary, V. B., Hain, S., Maugg, D., Smida, J., Azimzadeh, O., Tapio, S., et al. (2017). Long non-coding RNA PARTICLE bridges histone and DNA methylation. Sci. Rep. 7:1790. doi: 10.1038/s41598-017-01875-1
Osenberg, S., Dominissini, D., Rechavi, G., and Eisenberg, E. (2009). Widespread cleavage of A-to-I hyperediting substrates. RNA 15, 1632–1639. doi: 10.1261/rna.1581809
Padeken, J., and Heun, P. (2014). Nucleolus and nuclear periphery: velcro for heterochromatin. Curr. Opin. Cell Biol. 28, 54–60. doi: 10.1016/j.ceb.2014.03.001
Park, E., and Maquat, L. E. (2013). Staufen-mediated mRNA decay eonyoung. Wiley Interdiscip. Rev. RNA 4, 423–435. doi: 10.1002/wrna.1168
Pesole, G. (2002). UTRdb and UTRsite: specialized databases of sequences and functional elements of 5′ and 3′ untranslated regions of eukaryotic mRNAs. Update 2002. Nucleic Acids Res. 30, 335–340. doi: 10.1093/nar/30.1.335
Pintacuda, G., Wei, G., Roustan, C., Kirmizitas, B. A., Solcan, N., Cerase, A., et al. (2017a). hnRNPK Recruits PCGF3/5-PRC1 to the Xist RNA B-repeat to establish polycomb-mediated chromosomal silencing. Mol. Cell 68, 955.e10–969.e10.
Pintacuda, G., Young, A. N., and Cerase, A. (2017b). Function by structure: spotlights on xist long non-coding RNA. Front. Mol. Biosci. 4:90. doi: 10.3389/fmolb.2017.00090
Pisani, G., and Baron, B. (2019). Nuclear paraspeckles function in mediating gene regulatory and apoptotic pathways. Non Coding RNA Res. 4, 128–134. doi: 10.1016/j.ncrna.2019.11.002
Pizzinga, M., Bates, C., Lui, J., Forte, G., Morales-Polanco, F., Linney, E., et al. (2019). Translation factor mRNA granules direct protein synthetic capacity to regions of polarized growth. J. Cell Biol. 218, 1564–1581. doi: 10.1083/jcb.201704019
Pool, M. R. (2005). Signal recognition particles in chloroplasts, bacteria, yeast and mammals. Mol. Membr. Biol. 22, 3–15. doi: 10.1080/09687860400026348
Pool, M. R., Stumm, J., Fulga, T. A., Sinning, I., and Dobberstein, B. (2002). Distinct modes of signal recognition particle interaction with the ribosome. Science 297, 1345–1348. doi: 10.1126/science.1072366
Prasanth, K. V., Prasanth, S. G., Xuan, Z., Hearn, S., Freier, S. M., Bennett, C. F., et al. (2005). Regulating gene expression through RNA nuclear retention. Cell 123, 249–263.
Prikryl, J., Rojas, M., Schuster, G., and Barkan, A. (2011). Mechanism of RNA stabilization and translational activation by a pentatricopeptide repeat protein. PNAS 108, 415–420.
Qiao, F., and Cech, T. R. (2008). Triple-helix structure in telomerase RNA contributes to catalysis. Nat. Struct. Mol. Biol. 15, 634–640.
Ramilowski, J. A., Yip, C. W., Agrawal, S., Chang, J. C., Ciani, Y., Kulakovskiy, I. V., et al. (2020). Functional annotation of human long noncoding RNAs via molecular phenotyping. Genome Res. 30, 1060–1072.
Rashid, R., Liang, B., Baker, D. L., Youssef, O. A., He, Y., Phipps, K., et al. (2006). Crystal structure of a Cbf5-Nop10-Gar1 complex and implications in RNA-guided pseudouridylation and dyskeratosis congenita. Mol. Cell 21, 249–260.
Rego, A., Sinclair, P. B., Tao, W., Kireev, I., and Belmont, A. S. (2008). The facultative heterochromatin of the inactive X chromosome has a distinctive condensed ultrastructure. J. Cell Sci. 121, 1119–1127.
Rinn, J. L., and Chang, H. Y. (2012). Genome regulation by long noncoding RNAs. Annu. Rev. Biochem. 81, 145–166.
Rinn, J. L., and Chang, H. Y. (2020). Long noncoding RNAs: molecular modalities to organismal functions. Annu. Rev. Biochem. 89, 283–308.
Rinn, J. L., Kertesz, M., Wang, J. K., Squazzo, S. L., Xu, X., Brugmann, S. A., et al. (2007). Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell 129, 1311–1323.
Rong, D., Sun, H., Li, Z., Liu, S., Dong, C., Fu, K., et al. (2017). An emerging function of circRNA-miRNAs-mRNA axis in human diseases. Oncotarget 8, 73271–73281.
Rosenblad, M. A., Larsen, N., Samuelsson, T., and Zwieb, C. (2009). Kinship in the SRP RNA family. RNA Biol. 6, 508–516. doi: 10.4161/rna.6.5.9753
Rutenberg-Schoenberg, M., Sexton, A. N., and Simon, M. D. (2016). The properties of long noncoding RNAs that regulate chromatin. Annu. Rev. Genomics Hum. Genet. 17, 69–94.
Salmena, L., Poliseno, L., Tay, Y., Kats, L., and Pandolfi, P. P. (2011). A ceRNA hypothesis: the rosetta stone of a hidden RNA language? Cell 146, 353–358. doi: 10.1016/j.cell.2011.07.014
Sandberg, R., Neilson, J. R., Sarma, A., Sharp, P. A., and Burge, C. B. (2008). Proliferating cells express mRNAs with shortened 3′ untranslated regions and fewer microRNA target sites. Science 320, 1643–1647.
Sarropoulos, I., Marin, R., Cardoso-Moreira, M., and Kaessmann, H. (2019). Developmental dynamics of lncRNAs across mammalian organs and species. Nature 571, 510–514.
Scarola, M., Comisso, E., Pascolo, R., Chiaradia, R., Maria Marion, R., Schneider, C., et al. (2015). Epigenetic silencing of Oct4 by a complex containing SUV39H1 and Oct4 pseudogene lncRNA. Nat. Commun. 6:7631. doi: 10.1038/ncomms8631
Savino, T. M., Gébrane-Younès, J., De Mey, J., Sibarita, J. B., and Hernandez-Verdun, D. (2001). Nucleolar assembly of the rRNA processing machinery in living cells. J. Cell Biol. 153, 1097–1110.
Schmidt, J. C., Zaug, A. J., and Cech, T. R. (2016). Live cell imaging reveals the dynamics of telomerase recruitment to telomeres. Cell 166, 1188–1197.
Schwarz, A., and Beck, M. (2019). The benefits of cotranslational assembly: a structural perspective. Trends Cell Biol. 29, 791–803.
Selleri, L., Bartolomei, M. S., Bickmore, W. A., He, L., Stubbs, L., Reik, W., et al. (2016). A hox-embedded long noncoding RNA: is it all hot air? PLoS Genet. 12:e0222687.
Shay, J. W., and Wright, W. E. (2019). Telomeres and telomerase: three decades of progress. Nat. Rev. Genet. 20, 299–309. doi: 10.1038/s41576-019-0099-1
Shiber, A., Döring, K., Friedrich, U., Klann, K., Merker, D., Zedan, M., et al. (2018). Cotranslational assembly of protein complexes in eukaryotes revealed by ribosome profiling. Nature 561, 268–272.
Shieh, Y. W., Minguez, P., Bork, P., Auburger, J. J., Guilbride, D. L., Kramer, G., et al. (2015). Operon structure and cotranslational subunit association direct protein assembly in bacteria. Science 350, 678–680.
Shukla, C. J., McCorkindale, A. L., Gerhardinger, C., Korthauer, K. D., Cabili, M. N., Shechner, D. M., et al. (2018). High-throughput identification of RNA nuclear enrichment sequences. EMBO J. 37, 1–11.
Siegel, V., and Walter, P. (1988). Each of the activities of signal recognition particle (SRP) is contained within a distinct domain: analysis of biochemical mutants of SRP. Cell 52, 39–49.
Slavoff, S. A., Mitchell, A. J., Schwaid, A. G., Cabili, M. N., Ma, J., Levin, J. Z., et al. (2013). Peptidomic discovery of short open reading frame-encoded peptides in human cells. Nat. Chem. Biol. 9, 59–64.
Smith, K. P., Byron, M., Johnson, C., Xing, Y., and Lawrence, J. B. (2007). Defining early steps in mRNA transport: mutant mRNA in myotonic dystrophy type I is blocked at entry into SC-35 domains. J. Cell Biol. 178, 951–964.
Smola, M. J., Christy, T. W., Inoue, K., Nicholson, C. O., Friedersdorf, M., Keene, J. D., et al. (2016). SHAPE reveals transcript-wide interactions, complex structural domains, and protein interactions across the Xist lncRNA in living cells. Proc. Natl. Acad. Sci. U.S.A. 113, 10322–10327.
Sneppen, K., Pedersen, S., Krishna, S., Dodd, I., and Semsey, S. (2010). Economy of operon formation: cotranscription minimizes shortfall in protein complexes. mBio 1, 3–5.
Spitale, R. C., Tsai, M. C., and Chang, H. Y. (2011). RNA templating the epigenome: long noncoding RNAs as molecular scaffolds. Epigenetics 6, 539–543. doi: 10.4161/epi.6.5.15221
Souquere, S., and Pierron, G. (2015). Ultrastructural analysis of nuclear bodies using electron microscopy. Methods Mol. Biol. 1262, 105–118.
Stuckenholz, C., Meller, V. H., and Kuroda, M. I. (2003). Functional redundancy within roX1, a noncoding RNA involved in dosage compensation in Drosophila melanogaster. Genetics 164, 1003–1014.
Sun, Q., Hao, Q., and Prasanth, K. V. (2018). Nuclear long noncoding RNAs: key regulators of gene expression. Trends Genet. 34, 142–157.
Swain, P. S. (2004). Efficient attenuation of stochasticity in gene expression through post-transcriptional control. J. Mol. Biol. 344, 965–976.
Swinnen, B., Robberecht, W., and Van Den Bosch, L. (2020). RNA toxicity in non-coding repeat expansion disorders. EMBO J. 39, 1–23.
Taneja, K. L., McCurrach, M., Schalling, M., Housman, D., and Singer, R. H. (1995). Foci of trinucleotide repeat transcripts in nuclei of myotonic dystrophy cells and tissues. J. Cell Biol. 128, 995–1002.
Ten Klooster, J. P., Leeuwen, I. V., Scheres, N., Anthony, E. C., and Hordijk, P. L. (2007). Rac1-induced cell migration requires membrane recruitment of the nuclear oncogene SET. EMBO J. 26, 336–345.
Theimer, C. A., Blois, C. A., and Feigon, J. (2005). Structure of the human telomerase RNA pseudoknot reveals conserved tertiary interactions essential for function. Mol. Cell 17, 671–682.
Tian, B., White, R. J., Xia, T., Welle, S., Turner, D. H., Mathews, M. B., et al. (2000). Expanded CUG repeat RNAs form hairpins that activate the double-stranded RNA-dependent protein kinase PKR. RNA 6, 79–87.
Topisirovic, I., and Sonenberg, N. (2011). mRNA translation and energy metabolism in cancer: the role of the MAPK and mTORC1 Pathways. Cold Spring Harb. Symp. Quant. Biol. 76, 355–367.
Tsai, M., Manor, O., Wan, Y., Mosammaparast, N., Wang, J. K., Lan, F., et al. (2010). Modification complexes. Science 329, 689–693.
Tripathi, V., Ellis, J. D., Shen, Z., Song, D. Y., Pan, Q., Watt, A. T., et al. (2010). The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol. Cell 39, 925–938. doi: 10.1016/j.molcel.2010.08.011
Tucey, T. M., and Lundblad, V. (2014). Regulated assembly and disassembly of the yeast telomerase quaternary complex. Genes Dev. 28, 2077–2089.
Ulitsky, I. (2016). Evolution to the rescue: using comparative genomics to understand long non-coding RNAs. Nat. Rev. Genet. 17, 601–614.
Valgardsdottir, R., Chiodi, I., Giordano, M., Cobianchi, F., Riva, S., and Biamonti, G. (2005). Structural and functional characterization of noncoding repetitive RNAs transcribed in stressed human cells. Mol. Biol. Cell 16, 2597–2604. doi: 10.1091/mbc.e04-12-1078
Van Treeck, B., and Parker, R. (2018). Emerging roles for intermolecular RNA-RNA interactions in RNP assemblies. Cell 174, 791–802.
Van Treeck, B., Protter, D. S. W., Matheny, T., Khong, A., Link, C. D., and Parker, R. (2018). RNA self-assembly contributes to stress granule formation and defining the stress granule transcriptome. Proc. Natl. Acad. Sci. U.S.A. 115, 2734–2739.
Vasianovich, Y., and Wellinger, R. J. (2017). Life and death of yeast telomerase RNA. J. Mol. Biol. 429, 3242–3254.
Villegas, V. E., and Zaphiropoulos, P. G. (2015). Neighboring gene regulation by antisense long non-coding RNAs. Int. J. Mol. Sci. 16, 3251–3266.
Voorhees, R. M., and Hegde, R. S. (2015). Structures of the scanning and engaged states of the mammalian srp-ribosome complex. eLife 4, 1–21.
Wang, D., Qu, Z., Yang, L., Zhang, Q., Liu, Z. H., Do, T., et al. (2017). Transposable elements (TEs) contribute to stress-related long intergenic noncoding RNAs in plants. Plant J. 90, 133–146.
Wang, Y., He, L., Du, Y., Zhu, P., Huang, G., Luo, J., et al. (2015). The long noncoding RNA lncTCF7 promotes self-renewal of human liver cancer stem cells through activation of Wnt signaling. Cell Stem Cell 16, 413–425. doi: 10.1016/j.stem.2015.03.003
Wang, Y., Yesselman, J. D., Zhang, Q., Kang, M., and Feigon, J. (2016). Structural conservation in the template/pseudoknot domain of vertebrate telomerase RNA from teleost fish to human. Proc. Natl. Acad. Sci. U.S.A. 113, E5125– E5134.
Wells, J. N., Bergendahl, L. T., and Marsh, J. A. (2016). Operon gene order is optimized for ordered protein complex assembly. Cell Rep. 14, 679–685.
Werner, M. S., and Ruthenburg, A. J. (2015). Nuclear fractionation reveals thousands of chromatin-tethered noncoding RNAs adjacent to active genes. Cell Rep. 12, 1089–1098.
West, J. A., Mito, M., Kurosaka, S., Takumi, T., Tanegashima, C., Chujo, T., et al. (2016). Structural, super-resolution microscopy analysis of paraspeckle nuclear body organization. J. Cell Biol. 214, 817–830.
Wild, K., Sinning, I., and Cusack, S. (2001). Crystal structure of an early protein-RNA assembty complex of the signal recognition particle. Science 294, 598–601.
Wilkie, G. S., Dickson, K. S., and Gray, N. K. (2003). Regulation of mRNA translation by 5′- and 3′-UTR-binding factors. Trends Biochem. Sci. 28, 182–188.
Willett, M., Brocard, M., Pollard, H. J., and Morley, S. J. (2013). MRNA encoding WAVE-Arp2/3-Associated proteins is co-localized with foci of active protein synthesis at the leading edge of MRC5 fibroblasts during cell migration. Biochem. J. 452, 45–55.
Wojciechowska, M., and Krzyzosiak, W. J. (2011). Cellular toxicity of expanded RNA repeats: focus on RNA foci. Hum. Mol. Genet. 20, 3811–3821.
Wongtrakoongate, P., Riddick, G., Fucharoen, S., and Felsenfeld, G. (2015). Association of the long non-coding RNA steroid receptor RNA activator (SRA) with TrxG and PRC2 complexes. PLoS Genet. 11:e1005615. doi: 10.1371/journal.pgen.1006485
Wu, R. A., Upton, H. E., Vogan, J. M., and Collins, K. (2017). Telomerase mechanism of telomere synthesis. Annu. Rev. Biochem. 86, 439–460.
Wutz, A. (2011). Gene silencing in X-chromosome inactivation: advances in understanding facultative heterochromatin formation. Nat. Rev. Genet. 12, 542–553.
Wutz, A., Rasmussen, T. P., and Jaenisch, R. (2002). Chromosomal silencing and localization are mediated by different domains of Xist RNA. Nat. Genet. 30, 167–174.
Yang, F., Deng, X., Ma, W., Berletch, J. B., Rabaia, N., Wei, G., et al. (2015). The lncRNA Firre anchors the inactive X chromosome to the nucleolus by binding CTCF and maintains H3K27me3 methylation. Genome Biol. 16, 1–17.
Yang, F., Zhang, H., Mei, Y., and Wu, M. (2014). Reciprocal regulation of HIF-1α and LincRNA-p21 modulates the warburg effect. Mol. Cell 53, 88–100. doi: 10.1016/j.molcel.2013.11.004
Yang, L., Lin, C., Liu, W., Zhang, J., Ohgi, K. A., Grinstein, J. D., et al. (2011). NcRNA- and Pc2 methylation-dependent gene relocation between nuclear structures mediates gene activation programs. Cell 147, 773–788. doi: 10.1016/j.cell.2011.08.054
Yamashita, A. (2019). meiRNA, A polyvalent player in fission yeast meiosis. Non Cod. RNA 5:45. doi: 10.3390/ncrna5030045
Yamashita, A., Watanabe, Y., Nukina, N., and Yamamoto, M. (1998). RNA-assisted nuclear transport of the meiotic regulator Mei2p in fission yeast. Cell 95, 115–123. doi: 10.1007/978-3-662-10360-9_19
Yamazaki, T., Souquere, S., Chujo, T., Kobelke, S., Chong, Y. S., Fox, A. H., et al. (2018). Functional domains of NEAT1 architectural lncRNA induce paraspeckle assembly through phase separation. Mol. Cell 70, 1038.e7– 1053.e7.
Yoon, J. H., Abdelmohsen, K., and Gorospe, M. (2014). Functional interactions among microRNAs and long noncoding RNAs. Semin. Cell Dev. Biol. 34, 9–14.
Yoon, J. H., Abdelmohsen, K., Kim, J., Yang, X., Martindale, J. L., Tominaga-Yamanaka, K., et al. (2013). Scaffold function of long non-coding RNA HOTAIR in protein ubiquitination. Nat. Commun. 4:2939.
Yoon, J. H., Abdelmohsen, K., Srikantan, S., Yang, X., Martindale, J. L., De, S., et al. (2012). LincRNA-p21 Suppresses Target mRNA Translation. Mol. Cell 47, 648–655.
Zappulla, D. C. (2020). Yeast telomerase RNA flexibly scaffolds protein subunits: results and repercussions. Molecules 25, 1–13.
Zappulla, D. C., and Cech, T. R. (2006). RNA as a flexible scaffold for proteins: yeast telomerase and beyond. Cold Spring Harb. Symp. Quant. Biol. 71, 217–224.
Zappulla, D. C., Goodrich, K., and Cech, T. R. (2005). A miniature yeast telomerase RNA functions in vivo and reconstitutes activity in vitro. Nat. Struct. Mol. Biol. 12, 1072–1077.
Zhang, B., Arun, G., Mao, Y. S., Lazar, Z., Hung, G., Bhattacharjee, G., et al. (2012). The lncRNA Malat1 is dispensable for mouse development but its transcription plays a cis-regulatory role in the adult. Cell Rep. 2, 111–123. doi: 10.1016/j.celrep.2012.06.003
Zhang, B., Gunawardane, L., Niazi, F., Jahanbani, F., Chen, X., and Valadkhan, S. A. (2014). Novel RNA Motif Mediates the Strict Nuclear Localization of a Long Noncoding RNA. Mol. Cell. Biol. 34, 2318–2329.
Zhang, C., Ge, S., Gong, W., Xu, J., Guo, Z., Liu, Z., et al. (2020). LncRNA ANRIL acts as a modular scaffold of WDR5 and HDAC3 complexes and promotes alteration of the vascular smooth muscle cell phenotype. Cell Death Dis. 11:435. doi: 10.1038/s41419-020-2645-3
Zhang, L. F., Huynh, K. D., and Lee, J. T. (2007). Perinucleolar targeting of the inactive X during S phase: evidence for a role in the maintenance of silencing. Cell 129, 693–706.
Zhang, Q., Kim, N. K., and Feigon, J. (2011). Architecture of human telomerase RNA. Proc. Natl. Acad. Sci. U.S.A. 108, 20325–20332.
Zhang, Y., Yang, L., and Chen, L. L. (2014). Life without A tail: new formats of long noncoding RNAs. Int. J. Biochem. Cell Biol. 54, 338–349.
Zhang, Z., and Carmichael, G. G. (2001). The fate of dsRNA in the Nucleus: a p54nrb-containing complex mediates the nuclear retention of promiscuously A-to-I edited RNAs. Cell 106, 465–476.
Keywords: RNA scaffolds, RNA decoys, 3′UTR, nascent protein interactions, lncRNA, mRNA
Citation: Fernandes N and Buchan JR (2021) RNAs as Regulators of Cellular Matchmaking. Front. Mol. Biosci. 8:634146. doi: 10.3389/fmolb.2021.634146
Received: 27 November 2020; Accepted: 22 February 2021;
Published: 09 April 2021.
Edited by:
Purusharth I. Rajyaguru, Indian Institute of Science (IISc), IndiaReviewed by:
Alfredo Berzal-Herranz, Instituto de Parasitología y Biomedicina López-Neyra (IPBLN), SpainCopyright © 2021 Fernandes and Buchan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: J. Ross Buchan, cmJ1Y2hhbkBlbWFpbC5hcml6b25hLmVkdQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.