
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Mol. Biosci., 18 February 2021
Sec. Cellular Biochemistry
Volume 7 - 2020 | https://doi.org/10.3389/fmolb.2020.598419
This article is part of the Research TopicTransport of Nutrients, Metabolites and Ions linked to Bioenergetics: Relevance to Human PathologyView all 16 articles
Human GLUT2 and GLUT3, members of the GLUT/SLC2 gene family, facilitate glucose transport in specific tissues. Their malfunction or misregulation is associated with serious diseases, including diabetes, metabolic syndrome, and cancer. Despite being promising drug targets, GLUTs have only a few specific inhibitors. To identify and characterize potential GLUT2 and GLUT3 ligands, we developed a whole-cell system based on a yeast strain deficient in hexose uptake, whose growth defect on glucose can be rescued by the functional expression of human transporters. The simplicity of handling yeast cells makes this platform convenient for screening potential GLUT2 and GLUT3 inhibitors in a growth-based manner, amenable to high-throughput approaches. Moreover, our expression system is less laborious for detailed kinetic characterization of inhibitors than alternative methods such as the preparation of proteoliposomes or uptake assays in Xenopus oocytes. We show that functional expression of GLUT2 in yeast requires the deletion of the extended extracellular loop connecting transmembrane domains TM1 and TM2, which appears to negatively affect the trafficking of the transporter in the heterologous expression system. Furthermore, single amino acid substitutions at specific positions of the transporter sequence appear to positively affect the functionality of both GLUT2 and GLUT3 in yeast. We show that these variants are sensitive to known inhibitors phloretin and quercetin, demonstrating the potential of our expression systems to significantly accelerate the discovery of compounds that modulate the hexose transport activity of GLUT2 and GLUT3.
Transport of hexoses across plasma membranes marks the first and rate-limiting step of energy metabolism in cells of all domains of life. In humans, 14 glucose transporter family members (GLUTs, SLC2 gene family), with differing tissue distributions, mediate the facilitative diffusion of sugar along a concentration gradient. Despite a high sequence similarity, GLUTs differ in substrate specificity and affinity (Mueckler and Thorens, 2013), matching the demands for complex, tissue-dependent hexose uptake. Abnormal expression, localization or function of GLUTs are related to the pathogenesis of several diseases including cancer (Barron et al., 2016), diabetes (Ohtsubo et al., 2005; Hajiaghaalipour et al., 2015), and other severe metabolic disorders (Santer et al., 1997; Brockmann, 2009), making these transporters important drug targets.
GLUT2 is the primary GLUT isoform found in the liver. It also mediates glucose transport in the kidney, intestine, pancreatic β-cells and the central nervous system (Fukumoto et al., 1988; Thorens, 2015). It exhibits a low affinity for glucose (KM = ∼17 mM (Uldry et al., 2002)) and even lower for fructose, galactose, and mannose (KM = ∼76, ∼92, and ∼125 mM, respectively (Mueckler and Thorens, 2013)). For glucosamine, however, GLUT2 shows a very high substrate affinity (KM = ∼0.8 mM) (Uldry et al., 2002). Under normal physiological conditions, the glucose concentration in human blood is ∼5.5 mM (Jung et al., 2013); the glucose uptake by GLUT2 would be inefficient, indicating that it is not the primary function of this transporter. More likely, glucose sensing (Ohtsubo et al., 2005) and/or signaling (Guillemain et al., 2000) are the main functions of GLUT2, consistent with its expression in tissues with high glucose fluxes. In murine pancreatic β-cells, GLUT2 mediates glucose stimulated insulin secretion, thereby regulating blood glucose levels. The absence of this function impairs glucose homeostasis leading to diabetes (Ohtsubo et al., 2005). Furthermore, GLUT2 may also be involved in mediating transcriptional glucose signaling (Guillemain et al., 2000). Loss of GLUT2 function causes the Fanconi Bickel Syndrome (Santer et al., 1997), a rare autosomal disease with various symptoms like hepatomegaly, tubular nephropathy, glucose and galactose intolerance, fasting hypoglycemia, rickets, and retarded growth (Santer et al., 2002).
In contrast to GLUT2, GLUT3 exhibits a high affinity for glucose (KM = 1.4 mM (Colville et al., 1993)). Other substrates of GLUT3 are mannose, galactose, and xylose (Simpson et al., 2008). GLUT3 shares a high sequence identity (66%) with GLUT1 (Deng et al., 2015), and together they are predominantly responsible for glucose uptake in the brain - GLUT1 in the blood-brain barrier and GLUT3 in neurons (Simpson et al., 2007). Accordant with its high affinity for glucose, GLUT3 plays a pivotal role in the glucose uptake of cell types with a high demand for energy, such as sperm, circulating white blood cells, and preimplantation embryos (Simpson et al., 2007). Consistently, as tumor tissues have an increased need for carbon sources to sustain uncontrolled proliferation, GLUT3 is upregulated in many cancers, including gliomas, lung, laryngeal and bladder tumors (Ancey et al., 2018; Barron et al., 2016). In general, high expression of GLUT3 (and GLUT1) is associated with severe pathogenesis and poor survival in most cancer tissues (Ancey et al., 2018). Recently, an association between diverse diseases and variations of copy numbers of the GLUT3 gene has been hypothesized (Ziegler et al., 2020). Hence, the discovery of activating or inhibiting drugs specific for GLUT2 or GLUT3 is highly desirable.
Both transporters GLUT2 and GLUT3 belong to the Class I GLUT family, therefore sharing certain structure similarities as, for example, the position of the glycosylation site in the extracellular loop between the transmembrane helices (TM) 1 and 2 (Joost and Thorens, 2009). For GLUT2, it has been proposed that its glycosylation is essential for proper anchoring of the transporter to the plasma membrane of β-cells and its stability in these cells (Ohtsubo et al., 2005). However, the loop itself differs in the two transporters, with GLUT2 exhibiting a significantly larger loop size than GLUT3. The role of the cytoplasmic and extracellular loops between transmembrane domains of GLUTs is still dramatically understudied and putatively underestimated. The C-terminal intracellular domain of GLUT2 has been implicated in the low glucose affinity of this transporter (Katagiri et al., 1992). Furthermore, it has been shown that conformational changes in facilitators like GLUTs, during the rocker switch mechanism, are the primary force for the translocation of sugar (Qureshi et al., 2020). Therefore, residues distant from the sugar-binding sites likely influence transport dynamics significantly, including the role of hydrophilic regions. Investigation of the extra-membrane domains will further elucidate the origins of the transporter’s different properties.
The need for a convenient platform to investigate GLUTs has been recognized, and among different systems (Gould and Lienhard, 1989; Zamora-Leon et al., 1996; Kraft et al., 2015), the yeast cell-based investigation system has many benefits (Schmidl et al., 2018). The yeast Saccharomyces cerevisiae is a widely used GRAS organism and a model organism for diverse research applications. Cells are easily manipulated and maintained and exhibit a short generation time. By deleting all endogenous hexose transporter genes (HXT1-17, GAL2) and the genes of hexose transporting maltose transporters (AGT1, MPH2, MPH3) with the loxP-Cre recombinase system in a CEN.PK2-1C strain background (Entian and Kötter, 2007), a hexose transporter-deficient (hxt0) strain, incapable of growing on glucose or related monosaccharides, was constructed and named EBY.VW4000 (Wieczorke et al., 1999; Solis-Escalante et al., 2015). The strain is maintained on maltose, a disaccharide taken up by specialized maltose symporters (Chow et al., 1989), and cleaved inside the cell into two glucose molecules. The selective uptake of the respective monosaccharide by a heterologously expressed transporter can, therefore, be examined by simple growth tests or uptake assays with the radiolabeled sugar (Boles and Oreb, 2018; Schmidl et al., 2018). Moreover, a radiolabel-free assay to determine transport kinetics in the hxt0 yeast system has been recently developed (Schmidl et al., 2021).
However, the functional, heterologous expression of GLUTs in this strain is a challenging task. In previous studies, the transformation of EBY.VW4000 cells with native rat GLUTs did not yield cell growth on glucose (for GLUT1 and GLUT4) (Kasahara and Kasahara, 1996; Kasahara and Kasahara, 1997) or fructose (for GLUT5) (Tripp et al., 2017). Nevertheless, single point mutations in the TM2 of GLUT1 and GLUT5 enabled their activity in EBY.VW4000 (Wieczorke et al., 2002; Tripp et al., 2017). Also, wild-type GLUT1 was functionally expressed in a hxt0 strain harboring the additional fgy1 (“functional expression of GLUT1 in yeast”) mutation (Wieczorke et al., 2002) that affects the scaffold protein Efr3 (Wieczorke and Boles, personal communication). Efr3 is essential for recruiting the Stt4 phosphatidylinositol-4-kinase to the plasma membrane and, consequently, builds a prerequisite for normal membrane phosphatidylinositol-4-phosphate levels (Wu et al., 2014). The corresponding strain was named EBY.S7 (Wieczorke et al., 2002). Murine GLUT4 was only active in a strain named SDY.022 that, besides the fgy1 mutation, had a mutation in the ERG4 gene, coding for the terminal enzyme of the ergosterol biosynthesis pathway (Boles et al., 2004). The latter mutation putatively leads to a different sterol composition in the yeast plasma membrane, which seems beneficial for GLUT4 activity.
Here, we report the functional expression of human GLUT2 and GLUT3 in the hxt0 yeast system. Thereby, we complete the accessibility of the well-characterized Class I GLUTs (GLUTs1-4, categorized according to their sequence similarities (Joost and Thorens, 2009)) in a convenient system that enables detailed characterizations of these transporters and the screening for small molecules affecting their activity in a high-throughput manner. These new platforms facilitate the rapid discovery of drugs addressing severe diseases associated with these essential human transporters.
The construction of the strains CEN.PK2-1C (Entian and Kötter, 2007), EBY.VW4000 (Wieczorke et al., 1999), EBY.S7 (Wieczorke et al., 2002) and SDY.022 (Boles et al., 2004) used in this study was reported previously and their genotypes are listed in Supplementary Table S1. For maintenance and preparation of competent cells, plasmid-free cells were grown in standard YEP-media (1% (w/v) yeast extract, 2% (w/v) peptone) supplemented with 1% (w/v) maltose. Frozen competent cells were prepared and transformed according to Gietz and Schiestl (Gietz and Schiestl, 2007). The transformants were plated on solid, selective synthetic complete (SC) medium with 1% (w/v) maltose (M) in which uracil was omitted (-URA) to maintain the selection pressure. For experiments with envyGFP constructs at the microscope, EBY.S7 or CEN.PK2-1C cells were grown in filter-sterilized, low fluorescent, synthetic complete medium (lf-SC) containing 6.9 g/l YNB with ammonium sulfate, without amino acids, without folic acid and without riboflavin (MP Biomedicals), containing 1% (w/v) maltose (for EBY.S7) or 2% (w/v) glucose (for CEN.PK2-1C) and amino acids as stated in Bruder et al. (Bruder et al., 2016), in which uracil was omitted. For subcloning of plasmids, E. coli strain DH10B (Gibco BRL, Gaithersburg, MD) was used.
DNA Sequences of GLUT2 and GLUT3 are listed in Supplementary Table S2. PCRs were performed with Phusion polymerase (New England Biolabs GmbH) and the respective primers, according to the intended modifications, which are listed in Supplementary Table S3. The resulting fragments were transformed together with the EcoR1/BamH1 linearized p426H7 vector into EBY.VW4000, EBY.S7 or SDY.022 frozen competent cells, respectively, to allow for plasmid assembly via homologous recombination (Oldenburg et al., 1997). Cells were plated on SCM (1% (w/v)) -URA agar plates and incubated for 3 days at 30°C. The grown colonies were then replica plated onto solid SC -URA medium with 0.2% (w/v) glucose (SCD (0.2% (w/v))). If growth occurred on glucose medium, single colonies from these plates were picked, sub-cultivated and plasmids were recovered by the standard alkaline lysis protocol. If no growth on glucose was observed, colonies from the maltose plates were picked and treated accordingly. For propagation and amplification, plasmids were transformed via electroporation in E. coli. Plasmid isolation from overnight E. coli cultures was carried out using a GeneJET Plasmid Miniprep Kit (Thermo Scientific) according to the manufacturer’s instructions and sequenced at GATC Biotech (Konstanz, Germany). For subcloning into a vector with a dominant marker, the respective transporter sequences were amplified from the p426H7 plasmids as described, with primers exhibiting overhangs to the HXT7 promotor or CYC1 terminator, respectively, and PCR products were transformed together with the linearized pRS62K plasmid, which contains the same promotor and terminator regions, into EBY.VW4000 cells to allow for homologous recombination. To check the in vivo localization, envyGFP (Slubowski et al., 2015) was fused to the C-terminus of transporter constructs via homologous recombination of PCR fragments presenting the specific overhangs, and the whole construct which was flanked by the HXT7 promotor and the CYC1 terminator was inserted into the low-copy CEN6/ARS4 vector pUCPY1. All plasmids used in this study are listed in Supplementary Table S4.
For growth tests on solid medium, drop tests were performed on minimal SC -URA medium, containing the respective sugar, with cells expressing transporter constructs in the p426H7 vector backbone. Pre-cultures were grown overnight in 10 ml SCM (1% (w/v)) -URA medium at 30°C and 180 rpm, centrifuged (3,000 g, 3 min, 20°C) and washed twice in double-distilled, sterile water (ddH2O). Cells were resuspended in ddH2O and OD600nm was adjusted to 1. Dilutions of OD600nm 0.1, 0.01 and 0.001 were prepared and 4 µl of each dilution was dropped onto the agar plate. Plates were incubated at 30°C for 5 days.
Cell growth in liquid YEP medium was measured with the Cell Growth Quantifier (Aquila Biolabs) (Bruder et al., 2016). Pre-cultures of cells expressing the transporter constructs in the pRS62K vector backbone were grown overnight in 10 ml YEPM (1% (w/v)) medium with 200 µg/ml G418 for plasmid selection, harvested by centrifugation (3,000 g, 3 min, 20°C) and washed twice with ddH2O. Washed cells were used to inoculate 30 ml YEP G418 (200 µg/ml) medium with the indicated sugar to an OD600nm of 0.2 in 300 ml Erlenmeyer flasks, which were mounted onto the sensor plate. Quantification of cell growth and calculation of apparent maximal growth rates were performed with the CGQuant software (Aquila Biolabs) as previously described (Bruder et al., 2016).
To investigate the in vivo localization of the (modified) transporters, CEN.PK2-1C and EBY.S7 cells expressing the envyGFP-tagged constructs on the low-copy CEN.ARS plasmid pUCPY1 were grown overnight in filter-sterilized, low fluorescent SC medium in which uracil was omitted (lf-SC -URA) and 1% (w/v) maltose (for EBY.S7 cells) or 2% (w/v) glucose (for CEN.PK2-1C) was added. 500 µl cell suspension of an OD600nm between 1.5 and 3 was mixed with 500 µl lf-SC -URA medium with the respective sugar containing 1.2% (w/v) low melting agarose (Roth) to reach a suspension with 0.6% (w/v) low melting agarose for immobilization. Six microliters were applied to an object plate, sealed with a cover slip and GFP fluorescence was located with the Confocal Laser Scanning Microscope (Zeiss LSM 780, Jena, Germany).
To confirm the activity of envyGFP-tagged transporter constructs, EBY.S7 cells expressing the respective construct were streaked out on solid SC -URA medium with 0.2% (w/v) glucose and growth was recorded after 5 days of incubation at 30°C.
The crystal structure of GLUT2 is unknown and only the outward-facing GLUT3 structures are available. The homology models of the inward-facing GLUT2 and GLUT3 were generated with the ‘Homology Model’ function of the program package Molecular Operating Environment (MOE; Chemical Computing Group, https://www.chemcomp.com), using as a template the crystal structure of GLUT1 (PDB ID 4PYP). The homology models for the outward-facing GLUT2 were generated with MOE from the crystal structure of GLUT3 (PDB ID 5C65 or 4ZWC). The amino acid sequence identity and similarity between GLUT2 and GLUT3 are 50% and 68%, between GLUT1 and GLUT2 are 52% and 68%, and between GLUT1 and GLUT3 are 63% and 78%, respectively, as determined with the alignment function from MOE. The homology models generated were scored with GB/VI. The mutations were performed in MOE Protein Designing function and subject to energy minimization with the Forcefield Amber10.
The culturing of yeast cells was done at 30°C with shaking (180 rpm). EBY.S7 yeast cells expressing GLUT2∆loopS_Q455R were grown for a day in YEPM (1% (w/v)) media containing 200 µg/ml G418. Cells were washed once in YEP media containing 0.2% (w/v) glucose (YEPG) and 200 µg/ml G418 and transferred in the same media so that OD600nm ∼ 0.5 and grown further for 1 to 2 days. GLUT3S66Y expressed in EBY.S7 were grown for two days in SC-URA with 1% (w/v) maltose, then washed and transferred in SC-URA with 0.2% (w/v) glucose, followed by further growth for 1-2 days. For transport activity assay, cells were centrifuged (1000 g, 5 min), washed once with PBS solution (10 mM Na2HPO4, 1.8 mM KH2PO4, 2.7 mM KCl, 137 mM NaCl, pH 7.4), and resuspended in PBS buffer at an OD600nm ∼ 10; each assay determination contained 100 µl of this cell solution. Transport activity assay was started by adding C14-glucose (10 mM for GLUT2∆loopS_Q455R or 1 mM for GLUT3S66Y). Transport activity assay was halted after 10 min by adding 3 ml ice-chilled Quench buffer (0.1 M KPi, 0.1 M LiCl, pH 5.5), followed by filtration through a glass fiber channel (GC50; Advantec, Tokyo, Japan) under vacuum, and another two washes with 3 ml Quench buffer and filtration. The filtration membranes were transferred into scintillation vials with 10 ml of Scintillation Solution (BioSafeII; Research Products International, Mount Prospect, IL, United States), and vortexed briefly. The radioactivity was determined with a scintillation counter (Tri-carb 2900TR, Perkin Elmer, Waltham, MA, USA). Phloretin and quercetin were dissolved in DMSO at 20 mM stock concentration, and inhibitor concentrations used for IC50 determination were 100x stocks so that the final concentration of DMSO in the assay was 1%. Controls for determining the relative transport activity included 1% (v/v) DMSO to account for DMSO presence due to inhibitor, and the cells transformed with the empty vector. We found that the background activity with the empty vector was comparable to the transport activity of the transporters at 200 µM phloretin. Data were analyzed with the nonlinear fit analysis of GraphPad Prism (San Diego, CA, United States).
A plasmid for the expression of GLUT3 in yeast was generated using either the native human coding sequence or a codon-optimized sequence for the expression in insect cells. Amplification of the open reading frame (ORF) with oligonucleotides having 30-40 base pair overhangs to the applied promoter (HXT7-1–329) or terminator (CYC1) region, respectively, and co-transformation with the linearized host plasmid (p426H7) in EBY.VW4000, EBY.S7, and SDY.022 cells allowed for the plasmid assembly via homologous recombination (Oldenburg et al., 1997; Boles and Oreb, 2018). The strong, truncated HXT7 promoter region and the CYC1 terminator were chosen to achieve high expression levels. Transformants were plated on selective SC -URA medium, containing 1% (w/v) maltose, and incubated for three days at 30°C, resulting in the growth of approximately 1000 colonies per plate. Subsequently, cells were replica plated on SC -URA medium, containing 0.2% (w/v) glucose, to screen for cells that regained the ability to use glucose as a carbon source. EBY.VW4000 transformants did not show any growth on the solid glucose medium, even after a prolonged incubation of seven days. Most of the EBY.S7 and SDY.022 transformants grew after five days of incubation on glucose. To investigate if wild-type GLUT3 is active in these strain backgrounds, we isolated and sequenced plasmids of several colonies with various sizes. In bigger colonies, one single mutation (S66Y) was found, located in TM2. However, also unmodified GLUT3 mediated the growth of EBY.S7 and SDY.022 transformants on glucose. Re-transformation of the native GLUT3 and the modified GLUT3S66Y construct in EBY.S7 cells resulted in growth on glucose medium, proving that no further mutations, except for the fgy1 mutation, in the yeast strain were essential for this phenotype. However, while wild-type GLUT3 was incapable of mediating growth of EBY.VW4000 cells on solid glucose medium, slight growth was observed for GLUT3S66Y-expressing EBY.VW4000 cells (Supplementary Figure S7A). Furthermore, the drop test of the EBY.S7 and SDY.022 cells expressing the different constructs (GLUT3 and GLUT3S66Y) on a plasmid, revealed larger colonies for those expressing the mutated version (Supplementary Figures S7B,C), indicating that the S66Y mutation is beneficial for the functionality of the transporter in the heterologous system. Growth tests with hxt0 cells expressing GLUT3 or GLUT3S66Y were performed in liquid YEP medium with 0.2% (w/v) glucose, for which the GLUT3 ORFs were transferred to pRS62K plasmid backbones. Consistent with the results observed in the drop tests, both constructs enabled the growth of EBY.S7 cells, but GLUT3-expressing cells showed a significantly longer lag phase and a 1.8 times lower apparent maximal growth rate than those expressing GLUT3S66Y (Figure 1A; Supplementary Table S5). For EBY.VW4000 cells, only GLUT3S66Y mediated growth, while the unmodified GLUT3 did not (Figure 1B). Positive controls were GLUT1, as a representative of a heterologous transporter active in EBY.S7 cells (Wieczorke et al., 2002), and Hxt1, an endogenous low-affinity transporter for glucose and fructose (Leandro et al., 2009).
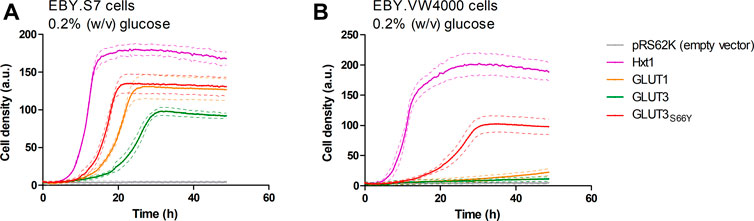
FIGURE 1. Growth of hxt0 cells expressing GLUT3 or GLUT3S66Y on glucose. Cell growth of EBY.S7 (A) or EBY.VW4000 (B) cells in the YEP medium containing 200 µg/ml G418 and 0.2% (w/v) glucose was recorded with the Cell Growth Quantifier (Aquila Biolabs). The cells were transformed with the empty pRS62K vector, pRS62K vector or with plasmids encoding the endogenous Hxt1 the endogenous Hxt1 transporter, GLUT1, GLUT3, or GLUT3S66Y. The average of three biological replicates (continuous lines) and the error (dashed lines) are shown. Apparent maximal growth rates for the growing cells were calculated with the CGQuant software and are listed together with the duration of lag phases in Supplementary Table S5.
Expression plasmids carrying either the native or a codon-optimized (for insect cells) human GLUT2 sequence were generated as described above for GLUT3. However, after replica plating, EBY.VW4000, EBY.S7, or SDY.022 transformants, of which more than 1000 colonies grew on solid maltose medium, showed no growth on glucose-containing agar plates, even after prolonged incubation at 30°C for one week.
Like the S66Y mutation in GLUT3, previous studies found single point mutations in the second transmembrane region of GLUT1 (e.g., V69M) (Wieczorke et al., 2002) and GLUT5 (e.g., S72Y and S76I) (Tripp et al., 2017) that enabled or improved the functional expression of these GLUTs in yeast. Therefore, we focused on this critical region in GLUT2 as well. Comparison of the amino acids 96 – 104 in GLUT2 with the corresponding area in GLUT1, GLUT3, and GLUT5 (Supplementary Figure S2) revealed that GLUT2 primary sequence has two consecutive serines at positions 102-103 vs. hydrophobic amino acids in the other GLUTs successfully expressed in yeast (Supplementary Figure S2).
Interestingly, in GLUT3 and GLUT5, a single serine mutation to a more hydrophobic amino acid enabled or improved the functional expression of these two transporters in yeast (as shown in this study and by Tripp et al. (Tripp et al., 2017)). This observation prompted us to direct mutagenesis to the amino acids 101-103, including the valine at position 101, considering the effect of the V69M mutation in GLUT1 (Wieczorke et al., 2002). By using degenerate primers that allow for the introduction of different nucleotides at a particular position (Kwok et al., 1994) (see Supplementary Table S3), different codons were inserted to mutate the chosen amino acids. The following degenerate codons were used to insert the desired range of substitutions: at position 101, RTK (encoding Ile, Met and Val); at position 102, RYT (encoding Ala, Ile, Thr and Val); at position 103, ATK (encoding Ile and Met). Considering all permutations, a library encoding 24 different protein sequence variants of the critical region resulted from this approach. The sequencing of the isolated plasmids from random clones confirmed the successful introduction of the expected range of substitutions. Still, screening of more than 1000 colonies for growth on glucose-containing agar plates yielded no positive clones. In addition to these mutations in TM2, we mutated the N glycosylation site at position 62 (GLUT2N62Q) within the first extracellular loop, just in case the glycosylation disturbed the activity of the transporter in yeast cells. This modification also failed to restore hxt0 yeast cell growth on glucose.
A conspicuous structural difference between GLUT2 and the other Class I GLUTs, which are active in yeast, is the extension of the extracellular loop connecting the TM helices H1 and H2 (Figure 2).
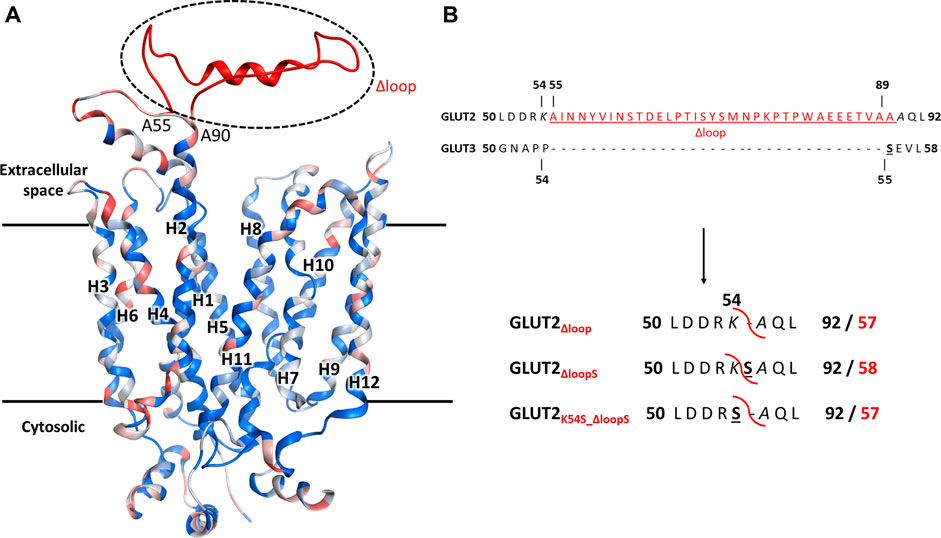
FIGURE 2. The structural model for GLUT2, GLUT3, and GLUT2 constructs. (A) The structural model was generated in MOE, based on the crystal structure of the outward-facing conformation of human GLUT3 (PDB ID 4ZWC). The color code represents the sequence homology between GLUT2 and GLUT3, ranging from identical residues (dark blue) to very dissimilar substitutions (red). Unique to GLUT2 is the extension in the loop between transmembrane helices H1 and H2, in the extracellular space (shown in red). (B) GLUT2 and GLUT3 sequence alignment in the region flanking the extended extracellular loop of GLUT2 (in red, underlined). The loop deletion constructs of GLUT2 are: GLUT2Δloop, which lacks residues 55-89, GLUT2ΔloopS, which lacks residues 55-88 and has the mutation A89S, and GLUT2K54S_Δloop, which lacks residues 55-89 and has the mutation K54S. For the two latter constructs, the single substitutions to serine are in analogy to S55 of GLUT3 (black bold underlined). The red curved line in the constructs shows the loop excision site. For the GLUT2 constructs, the number for both the original (bold black) and post-deletion (bold red) sequences are shown.
Since minor sequence modifications have not led to an active expression of GLUT2 in hxt0 yeast strains, we reasoned that shortening the extension, thereby introducing a GLUT3-like-loop, could promote GLUT2 activity in the heterologous system.
In addition to just deleting the amino acids 55-89 (GLUT2∆loop), we also constructed one chimera in which those amino acids were replaced by one serine (GLUT2∆loopS) and one in which the lysine preceding the deleted region was mutated to serine (GLUT2K54S_∆loop) (Figure 2B). Compared to GLUT3, the loop of GLUT2∆loop is one residue shorter, lacking the residue corresponding to S55 of GLUT3, while that of GLUT2∆loopS is the same length and includes a residue in analogy to S55 of GLUT3. GLUT2K54S_∆loop contains the mutation corresponding to S55 of GLUT3 but at the beginning instead of the end of the deleted loop. The modifications were introduced into the ORFs via PCR (primers are listed in Supplementary Table S3), and the individual fragments were transformed together with a linearized p426H7 vector in EBY.VW4000, EBY.S7, or SDY.022 cells. Transformants that grew on solid maltose medium (more than 1000 colonies per plate) were then replica plated onto glucose containing plates. A substantial number (approximately 60%) of colonies expressing one of the three loop-modified constructs, respectively, grew on glucose. Multiple plasmids from each strain background and loop-construct combination were isolated and sequenced. Most of these plasmids did not show mutations other than those intentionally introduced. Strikingly, in 2 out of 3 sequenced plasmids harboring the GLUT2∆loopS construct that were isolated from EBY.VW4000 cells, a point mutation led to the substitution of the amino acid glutamine at position 455 with either lysine (Q455K) or arginine (Q455R). Also, Q455R mutation was found in one out of three GLUT2∆loopS constructs isolated from SDY.022 cells, indicating an additional beneficial effect of this mutation on the glucose uptake via GLUT2. The isolated loop constructs GLUT2∆loop, GLUT2K55S_∆loop, GLUT2∆loopS, and GLUT2∆loopS_Q455R were re-transformed in all three hxt0 strains, and their growth was analyzed by a drop test on solid glucose medium (Supplementary Figure S9). The ability to grow on glucose was regained by EBY.S7 and SDY.022 cells with all loop constructs, whereas cells expressing GLUT2∆loopS and especially GLUT2∆loopS_Q455R, respectively, showed bigger colonies in comparison to GLUT2∆loop or GLUT2K55S_∆loop expressing cells (Supplementary Figures S9B,C). EBY.VW4000 cells grew significantly only when expressing GLUT2∆loopS_Q455R (Supplementary Figure S9), providing further evidence for a crucial effect of this additional point mutation, when combined with the ∆loopS modification, on GLUT2 activity.
As with GLUT3, the ORFs of different GLUT2 variants were transferred into the pRS62K vector and tested for growth in liquid YEP media, using the Cell Growth Quantifier (Aquila Biolabs) (Bruder et al., 2016). In agreement with the results from the drop test, only GLUT2∆loopS and GLUT2∆loopS_Q455R, but not wild-type GLUT2, conferred growth of EBY.S7 cells on 0.2% (w/v) glucose (Figure 3A). For EBY.VW4000 cells, the additional point mutation in GLUT2∆loopS_Q455R was essential for growth in this medium (Figure 3B). Interestingly, no growth of EBY.S7 cells expressing either of the loop constructs was observed on 0.2% (w/v) fructose medium, while the positive controls expressing Hxt1 showed normal growth (data not shown). This is likely due to the very low affinity of GLUT2 for fructose (KM = ∼76 mM) (Mueckler and Thorens, 2013). However, on 2% (w/v) fructose, both loop constructs conferred robust growth (Figure 3C). Analysis of the apparent maximal growth rates with the CGQuant Software (Bruder et al., 2016) showed a significantly faster doubling rate for EBY.S7 cells expressing GLUT2∆loopS_Q455R compared to GLUT2∆loopS-expressing cells on 0.2% (w/v) glucose medium (1.7 times) as well as on 2% (w/v) fructose medium (1.5 times) (Supplementary Table S5).
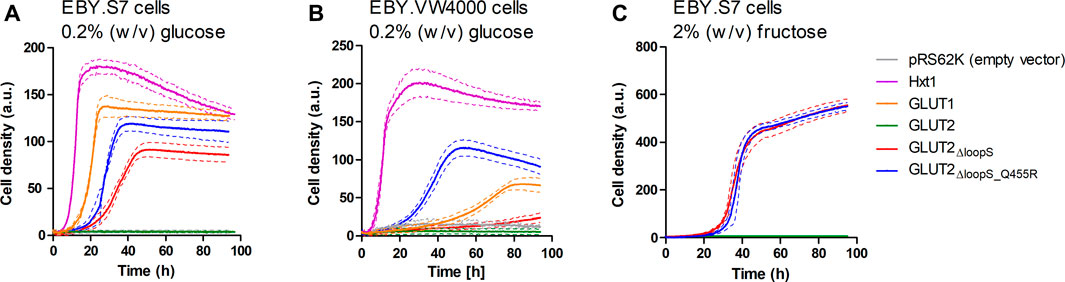
FIGURE 3. Growth of hxt0 cells expressing GLUT2 variants on hexoses. Cell growth of EBY.S7 (A,C) or EBY.VW4000 (B) cells containing the empty pRS62K vector, the endogenous Hxt1 transporter (only (A),(B)), GLUT1 (only (A),(B)), GLUT2, GLUT2ΔloopS or GLUT2ΔloopS_Q455R, was recorded with the Cell Growth Quantifier (Aquila Biolabs). The YEP medium was supplemented with 200 μg/ml G418 for plasmid selection and 0.2% (w/v) glucose (A,B) or 2% (w/v) fructose (C). The average of three biological replicates (continuous lines) and the error (dashed lines) are shown. Apparent maximal growth rates for the growing cells were calculated with the CGQuant software and are listed together with the duration of lag phases in Supplementary Table S5.
One hypothesis to explain the adverse effect of the extended loop on GLUT2 activity in yeast is possible interference with the transporter’s targeting to the plasma membrane. To test if our loop modifications influence the transporter trafficking, we fused the strong GFP variant envyGFP (Slubowski et al., 2015) to the C-terminal ends of the two most active constructs GLUT2∆loopS and GLUT2∆loopS_Q455R, and of the inactive wild-type GLUT2. The activity of these envyGFP-tagged transporters was confirmed by the growth of GLUT2∆loopS_Q455R-envyGFP and GLUT2∆loopS-envyGFP-expressing EBY.S7 cells on solid SC -URA 0.2% (w/v) glucose medium (Supplementary Figure S11). Fluorescence microscopy of CEN.PK2-1C cells indeed revealed significant differences between the localization of the wild-type GLUT2 and the two truncated constructs. While cells expressing the latter showed a patchy distribution at the plasma membrane and partial retention in the endomembrane system, unmodified GLUT2 does not reach the plasma membrane and is located in vacuoles (Figure 4), thereby being destined for degradation.
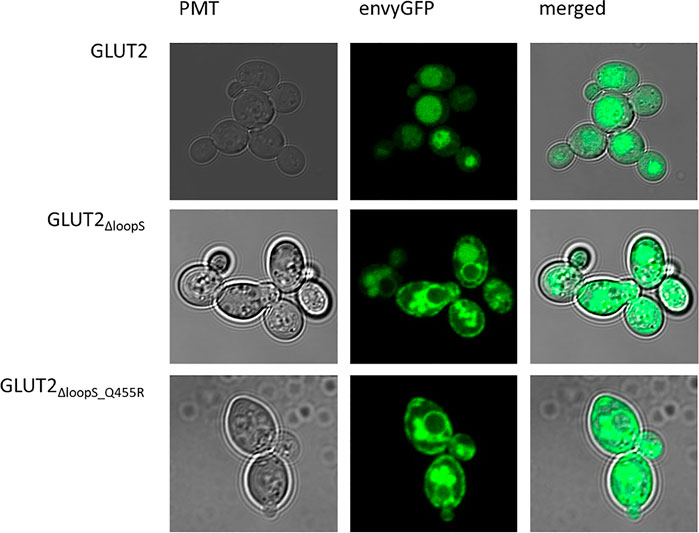
FIGURE 4. Subcellular localization of GLUT2 variants. envyGFP was fused to the C-termini of GLUT2 (wild-type), GLUT2ΔloopS, and GLUT2ΔloopS_Q455R. CEN.PK2-1C cells, expressing the respective envyGFP construct, were grown in low fluorescent SC -URA medium containing 0.2% (w/v) glucose. For immobilization, 0.6% (w/v) low melt agarose was added to the suspension. Localization was analyzed with the Confocal Laser Scanning Microscope (Zeiss LSM 780, Jena, Germany).
The same pattern was observed in EBY.S7 cells (Supplementary Figure S10), demonstrating that the effect of the loop is not dependent on the strain background. Therefore, we conclude that the implemented loop modifications rescue the trafficking defect of GLUT2 in yeast cells. The additional point mutation appears not to influence the subcellular localization of the transporter and putatively improves the transporter’s substrate-gating activity by other effects (see below).
Since the best activities of GLUT2 and GLUT3 constructs were observed in the EBY.S7 cells, this strain background was chosen to establish a yeast-based platform for screening and characterizing GLUT inhibitors. Phloretin (Kwon et al., 2007) and quercetin (Lee et al., 2015) are well-known inhibitors of GLUT2 and GLUT3 (Kwon et al., 2007; Augustin, 2010; Lee et al., 2015), and, therefore, we determined their half maximal inhibitory concentration (IC50 values) for GLUT2∆loopS_Q455R expressed in EBY.S7 (Figures 5A,B).
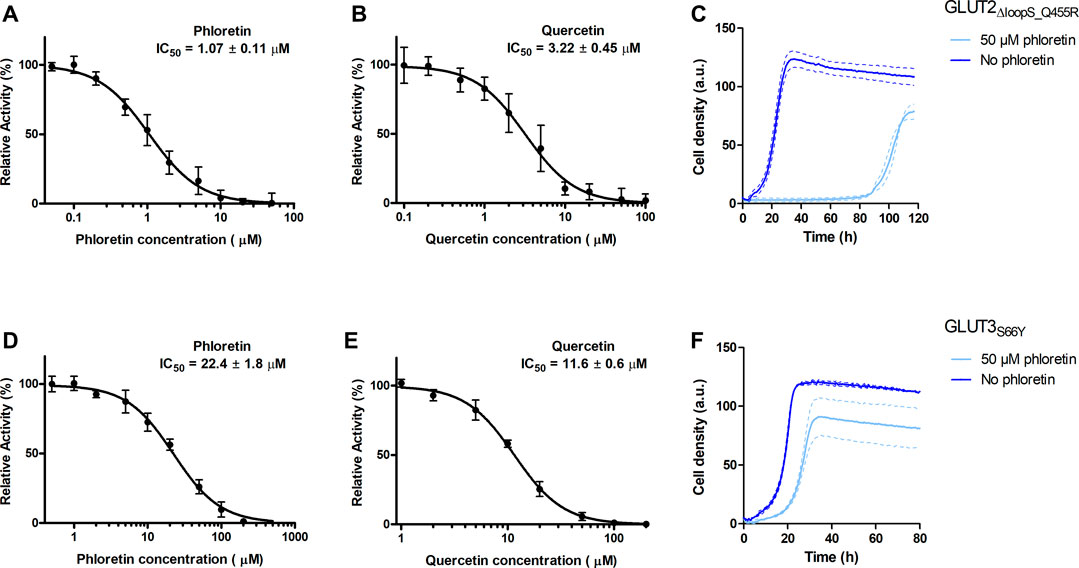
FIGURE 5. Effect of phloretin and quercetin on GLUT2∆loopS_Q455R and GLUT3S66Y expressed in hxt0 EBY.S7 cells. The relative transport activity (expressed as % of the transport in the absence of the inhibitor) of GLUT2∆loopS_Q455R(A,B) and GLUT3S66Y(D,E) as a function of phloretin (A,D) or quercetin (B,E) concentrations at 10 mM (for GLUT2∆loopS_Q455R, (A,B)) or 1 mM (for GLUT3S66Y, (D,E)) glucose is shown. Error bars represent the standard deviation from three independent measurements. Growth assays with GLUT2∆loopS_Q455R(C) or GLUT3S66Y(F) expressing EBY.S7 cells on YEP medium supplemented with 0.2% (w/v) glucose and G418 (200 μg/ml) in the presence of 50 μM phloretin (light blue) or its absence (dark blue) were recorded with the Cell Growth Quantifier (Aquila Biolabs). The average of two biological replicates (continuous lines) and the error (dashed lines) are shown. Apparent maximal growth rates of the here-depicted experiments, and EBY.S7 cells expressing Hxt1 with and without phloretin as a control, were calculated with the CGQuant software and are listed together with the duration of lag phases in Supplementary Table S6.
The values obtained for GLUT2∆loopS_Q455R (IC50 of 1.07 ± 0.11 and 3.22 ± 0.45 µM for phloretin and quercetin, respectively) indicate a strong inhibition and confirm that yeast-expressed GLUT2 can be reliably used to identify and characterize transport inhibitors despite the deletion of the extended extracellular loop. This is supported by growth tests in the presence of phloretin. We found that 50 µM phloretin inhibited the growth of EBY.S7 cells expressing GLUT2∆loopS_Q455R significantly, leading to an extensive long lag phase of 87.4 hours, a diminished apparent maximal growth rate (Supplementary Table S6) and a lower maximal cell density (Figure 5C).
Inhibition of GLUT3S66Y with phloretin and quercetin is less potent than for GLUT2∆loopS_Q455R but still effective, as demonstrated by the IC50 values (22.4 ± 1.8 µM for phloretin and 11.6 ± 0.6 µM for quercetin) (Figures 5D,E). Moreover, the phloretin IC50 values are in the same range as those reported for the related GLUT1 and GLUT4 (Kasahara and Kasahara, 1997). Reflecting the higher IC50 values of phloretin compared to those measured with GLUT2∆loopS_Q455R, the growth delay of GLUT3S66Y expressing EBY.S7 cells in glucose containing medium in the presence of 50 µM phloretin is less pronounced but still significant (Figure 5F; Supplementary Table S6). In contrast, the apparent maximal growth rates of Hxt1-expressing EBY.S7 cells do not show significant differences when grown in the absence or presence of 50 μM phloretin (Supplementary Table S6). As all tested transporters were expressed under the control of the truncated Hxt7 promotor, the Hxt1 control shows that the diminished growth phenotypes of GLUT2∆loopS_Q455R and GLUT3S66Y-expressing EBY.S7 cells upon phloretin addition do not arise from lowered expression levels. Together, these data demonstrate that the yeast system can be conveniently used to screen large compound libraries for potential inhibitors of GLUT2 and GLUT3 in a simple, growth-based manner, amenable to high throughput screening.
The prevailing transport mechanism for the Major Facilitator Superfamily (MFS) proteins, including GLUTs, postulates that the substrate cavity of the transporter is alternately open to either side of the membrane with the transporter cycling between the so-called outward- and inward-facing conformations, through a relative rocking of the transporter N- and C- halves (domains) around the active site (Abramson et al., 2003; Yan, 2015). The structural models for GLUT2ΔloopS, GLUT2ΔloopS_Q455R, GLUT3, and GLUT3S66Y were based on the crystal structures of the inward-facing conformation for GLUT1 (PDB ID 4PYP) and the outward-facing conformation of GLUT3 (PDB ID 5C65 or 4ZWC) (Figure 6).
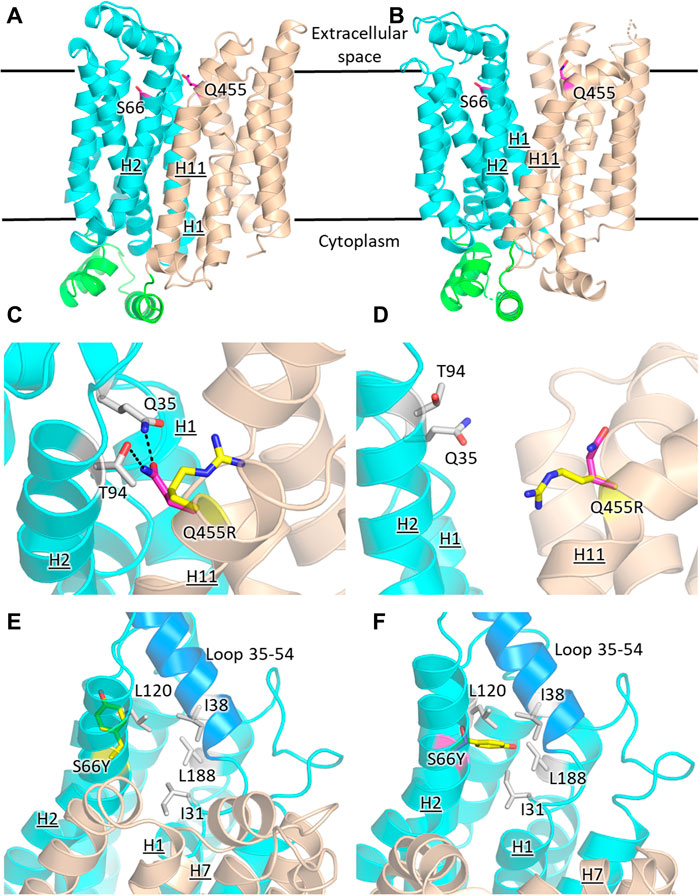
FIGURE 6. Structural models of GLUT2 and GLUT3 variants with functional transporter expression in hxt0 cells. Inward-facing (A,C,E) and outward-facing (B,D,F) conformations of GLUT2 and GLUT3 based on the crystal structures for the corresponding conformations of GLUT1 (PDB ID 4PYP, inward-facing) or GLUT3 (PDB ID 5C65, outward-facing), showing the locations of the single-site mutations that increased the transport activity of GLUT2loop and GLUT3 expressed in hxt0 yeast cells: S66 in H2 of GLUT3 and Q455 in H11 of GLUT2loop. The transporter N- and C-halves are in magenta and gold, respectively, and the transmembrane helices are labeled as H1-H12. (C,D) Close-up views of (A,B), respectively, focused on Q455R in GLUT2loop, showing the original side-chain (magenta), and the mutant (yellow). (E,F) Close-up views of (A,B), respectively, focused on S66Y in GLUT3. The mutant side chain is shown in yellow. The extracellular loop between helices H1 and H2 (residues 35-54, shown in blue) is close to the mutated side chain Y66 in the outward-facing conformation (F). Compared with the inward-facing conformation, in the outward-facing conformation, the loop spanning residues 35-54 moves at least 1 Å in the direction indicated by the red arrow. Figures were drawn using Open-Source PyMol Version 2.3.0 (The PyMOL Molecular Graphics System, Version 2.3.0 Schrödinger, LLC).
GLUT2 and GLUT3 share 50% and 68% amino acid sequence identity and homology, respectively (determined with MOE, Chemical Computing Group). Cell growth of GLUT-expressing EBY.S7 cells showed that additional single mutations in GLUT2ΔloopS and GLUT3 improved the transport activities (Figures 1, 3): Q455R in GLUT2ΔloopS and S66Y in GLUT3. Q455 of GLUT2 is in the outward edge of the TM helix H11, close to the interface between the N- and C-domains (Figure 6A). Its substitution with arginine is inconsequential for the outward-facing conformation (Figures 6B,D), but disruptive for the inward-facing conformation of the transporter; the bulkier guanidinium moiety can no longer participate in the hydrogen bond interactions with Q35 of H1 and T94 of H2 (Figure 6C), sterically crowding its vicinity. In GLUT3, S66 is in the outer periphery of H2, part of a hydrophobic pocket composed of N-domain residues including, I31, I38, L120, and L188, close to the soluble loop linking H1 and H2 (Figures 6A,E,F). In the outward-facing conformation of GLUT3, S66 substitution with the larger side-chain of a tyrosine integrates well, as there is ample space around the loop connecting H1 and H2 (loop 35-54) (Figure 6F). Conversely, in the inward-facing conformation, loop 35-54 packs near H7, and the tyrosine side-chain is pushed away from the hydrophobic pocket (Figure 6E).
The work here completes the generation of GLUT-specific assay systems for class I GLUTs in the versatile and simple platform provided by the engineered yeast cells, hxt0. These systems enable high-throughput ligand screening for a particular GLUT and determination of the potency and selectivity of the ligand candidates relative to closely related GLUTs within class I, and also class II member, GLUT5. The co-existence of several GLUT isoforms within one tissue or cancer cell line complicates efforts to understand the role of a particular GLUT in the cell pathology or physiology. Thus, besides their medical importance as potential drugs, GLUT-selective inhibitors can also serve as investigative tools to unravel how healthy and abnormal cells adjust their energetic needs through GLUT expression, regulation or function (Carreño et al., 2019). Additionally, GLUT-specific substrate analogs can be optimized as diagnostic tools, especially in cancer detection (Barron et al., 2016; George Thompson et al., 2016; Wuest et al., 2018). Finally, discovery of GLUT-selective activators would be of particular interest in the case of GLUT4 or GLUT2, as strategies to ameliorate diabetes.
Having a GLUT2 assay system is particularly useful. The low affinity for glucose and fructose of this transporter makes it difficult to study its activity when other GLUT isoforms are present, even when GLUT2 is overexpressed. Deleting the loop extension between TM1 and TM2 helices was necessary for the functional expression of GLUT2, and the two transporters showed differences in their inhibition by phloretin and quercetin, with GLUT2 being significantly more sensitive than GLUT3 (Figure 5).
Besides their value for GLUT-specific ligand discovery and characterization, the GLUT expressing yeast systems point to intriguing clues regarding the regions important for transporters activity. As shown here for GLUT2 and GLUT3 and previously reported for GLUT1 (Wieczorke et al., 2002) and GLUT5 (Tripp et al., 2017), the functional expression of these transporters is facilitated or improved by single amino acid substitutions in the transporter sequence (summarized in Table 1, Figure 7). Although the elucidation of the underlying mechanism is complex and not in the scope of this study, structural modeling can help derive hypotheses to explain the observations.
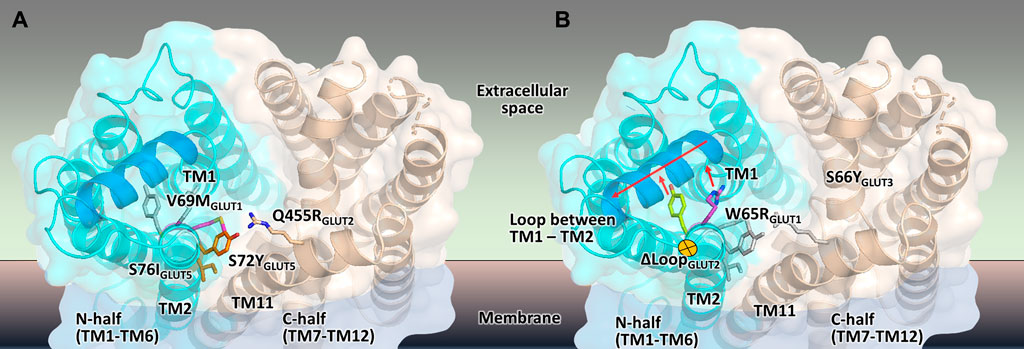
FIGURE 7. Mapping the mutations that render different GLUTs active in hxt0 yeast cells. (A,B) The structural model of the outward-facing conformation for GLUT1-3 and GLUT5 showing the single-site mutations that provide or increase transport activity of the respective human GLUT upon expression in hxt0 yeast cells, viewed from the extracellular side. The mutants are generally substitutions to bulkier side chains and fall into two categories. (A) Mutants that disrupt the packing between the N- and C-domains (colored as cyan and wheat, respectively), such as S72I and S76Y of GLUT5, Q455R of GLUT2, and V69M of GLUT1. (B) Mutants that dislocate the loop between TM helices TM1 and TM2 (the part of the loop that needs to move to accommodate the mutations is shown in blue), causing it to move away from the central cavity, such as S66Y of GLUT3 and W65R of GLUT1.
Wild-type transporters and GLUT2ΔloopS, although localized at the membrane, could not compensate for the absence of the endogenous hexose transporters when EBY.VW4000 cells were grown in media with glucose or fructose as the carbon source. W65R and V69M of GLUT1 (Wieczorke et al., 2002), S66Y of GLUT3 (this study), S72Y and S76I of GLUT5 (Tripp et al., 2017) are all residues in the TM2, near the loop connecting TM helices 1 and 2, which in turn either packs closely to TM helices 7 and 11 (in the inward-facing conformation) or relaxes by moving at least 1 Å away from these helices (in the outward-facing conformation). Intriguingly, Q455R of GLUT2ΔloopS is at the end of TM11, which also approaches the same region: TM1, TM2, and their connecting loop (residues 35-54) (Figures 6, 7). The convergence in the location of the mutations that functionalize the transporters (Figure 7), whether they are part of the Class I GLUTs (like GLUT1-3) or Class II (like GLUT5), suggests that this region is critical for transport activity of human GLUTs in the yeast system. Consistent with the proposed role of the mutations in GLUT5 (Tripp et al., 2017), the ones described here for GLUT3 and GLUT2ΔloopS (S66Y and Q455R, respectively) also seem to destabilize the inward-facing conformation while leaving the outward-facing one unaffected. More generally, the substitutions that functionalize GLUTs in yeast cells favor an open substrate cavity towards the extracellular space (i.e., outward-facing conformation), either by destabilizing the N- and C-domain interface of the inward-facing conformation (Figure 7A) or by pushing the TM1-TM2 extracellular loop away, leading to a more accessible substrate cavity from the extracellular side (Figure 7B).
Evidently, the lipid composition of the yeast’s plasma membrane also impacts the heterologous expression of some human GLUTs (Wieczorke et al., 2002; Schmidl et al., 2018).
In EBY.S7, a strain in which the phosphatidylinositol-4-phosphate composition is putatively altered due to the fgy1 mutation (Schmidl et al., 2018), wild-type GLUT1 and GLUT3, and GLUT2ΔloopS, are actively expressed without any further point mutations. Conceivably, the cycling between the transporter conformations depends on the lipid environment; the relative stabilization of the outward-facing conformation vs. the inward-facing one may be a feature of functional expression of human GLUTs in yeast cells. Activating transport by stabilizing the outward-facing conformation has been documented for another classic MFS protein, LacY, for which interaction of the transporter periplasmic loops with a nanobody against the outward-facing conformation increased the substrate affinity 50-fold (Smirnova et al., 2014). Strikingly, the critical role of lipids in yeast for the functionality of transporters that undergo conformational changes during the transport cycle has been recently demonstrated, supporting our hypothesis (van’t Klooster et al., 2020a; van’t Klooster et al., 2020b).
For GLUT2, introducing point mutations or screening a combinatorial mini-library of 24 possible aminoacid combinations in the critical TM2 region was not sufficient for activating the transporter, even in strain backgrounds EBY.S7 and SDY.022 exhibiting an altered lipid composition. We demonstrated that truncating the large extension of the second extracellular loop, which is a distinctive property of GLUT2 compared to other GLUTs functionally expressed in yeast, was a successful strategy based on a rational approach. As shown by fluorescence microscopy, this modification promotes the trafficking of the transporter to the plasma membrane, whereas the full-length protein is largely retained in the vacuole. In general, the role of the soluble loops in MFS transporters is not well understood, but it is believed that they might affect the gating and conformational dynamics of the transporters (Qureshi et al., 2020). Our results suggest that the GLUT2-specific extension of the second loop is not essential for glucose transport per se. It might rather have a regulatory function, which is probably relevant only within its native environment in the human cells. Similar to our observations, the soluble loops were dispensable for the function of the human nucleoside transporters (belonging to the MFS superfamily) expressed in Xenopus oocytes (Aseervatham et al., 2015).
In summary, despite the necessary modifications of their sequence, our transporter variants retain their native function in facilitating glucose (GLUT2, GLUT3) and fructose (GLUT2) uptake. Importantly, they are sensitive to the known inhibitors phloretin and quercetin, which validates our system as a platform for screening and characterizing potential transport inhibitors.
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Experimental work was performed by SS, SR, and CI. All authors contributed to experimental design and manuscript preparation. J-YC and MO guided the project.
Financial support by NIH, grant number R01-GM123103 (to J-YC and MO) and the Boehringer Ingelheim Fonds (travel grant to SS) is gratefully acknowledged.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We thank Christine Essl for assistance and Eckhard Boles for stimulating discussions.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmolb.2020.598419/full#supplementary-material.
Abramson, J., Smirnova, I., Kasho, V., Verner, G., Iwata, S., and Kaback, H.R. (2003). The lactose permease of Escherichia coli: overall structure, the sugar-binding site and the alternating access model for transport. FEBS Lett. 555, 96–101. doi:10.1016/s0014-5793(03)01087-1
Ancey, P.-B., Contat, C., and Meylan, E. (2018). Glucose transporters in cancer-from tumor cells to the tumor microenvironment. FEBS J. 285, 2926–2943. doi:10.1111/febs.14577
Aseervatham, J., Tran, L., Machaca, K., and Boudker, O. (2015). The role of flexible loops in folding, trafficking and activity of equilibrative nucleoside transporters. PLoS One 10, e0136779. doi:10.1371/journal.pone.0136779
Augustin, R. (2010). The protein family of glucose transport facilitators: It’s not only about glucose after all. IUBMB life 62, 315–333. doi:10.1002/iub.315
Barron, C. C., Bilan, P. J., Tsakiridis, T., and Tsiani, E. (2016). Facilitative glucose transporters: Implications for cancer detection, prognosis and treatment. Metabolism 65, 124–139. doi:10.1016/j.metabol.2015.10.007
Boles, E., Dlugai, S., Mueller, G., and Voss, D. (2004). Use of Saccharomyces cerevisiae ERG4 mutants for the expression of glucose transporters from mammals. Geneva, Switzerland: World Intellectual Property Organization. WO002004026907A3.
Boles, E., and Oreb, M. (2018). A growth-based screening system for hexose transporters in yeast. Methods Mol. Biol. 1713, 123–135. doi:10.1007/978-1-4939-7507-5_10
Brockmann, K. (2009). The expanding phenotype of GLUT1-deficiency syndrome. Brain Dev. 31, 545–552. doi:10.1016/j.braindev.2009.02.008
Bruder, S., Reifenrath, M., Thomik, T., Boles, E., and Herzog, K. (2016). Parallelised online biomass monitoring in shake flasks enables efficient strain and carbon source dependent growth characterisation of Saccharomyces cerevisiae. Microb. Cell Fact. 15, 127. doi:10.1186/s12934-016-0526-3
Carreño, D., Corro, N., Torres-Estay, V., Véliz, L. P., Jaimovich, R., Cisternas, P., et al. (2019). Fructose and prostate cancer: toward an integrated view of cancer cell metabolism. Prostate Cancer Prostatic Dis. 22, 49–58. doi:10.1038/s41391-018-0072-7
Chow, T. H. C., Sollitti, P., and Marmur, J. (1989). Structure of the multigene family of MAL loci in Saccharomyces. Mol. Gen. Genet. 217, 60–69. doi:10.1007/BF00330943
Colville, C. A., Seatter, M. J., Jess, T. J., Gould, G. W., and Thomas, H. M. (1993). Kinetic analysis of the liver-type (GLUT2) and brain-type (GLUT3) glucose transporters in Xenopus oocytes: substrate specificities and effects of transport inhibitors. Biochem. J. 290, 701–706. doi:10.1042/bj2900701
Deng, D., Sun, P., Yan, C., Ke, M., Jiang, X., Xiong, L., et al. (2015). Molecular basis of ligand recognition and transport by glucose transporters. Nature 526, 391–396. doi:10.1038/nature14655
Entian, K.-D., and Kötter, P. (2007). 25 yeast genetic strain and plasmid collections. Methods Microbiol. 36, 629–666. doi:10.1016/S0580-9517(06)36025-4
Fukumoto, H., Seino, S., Imura, H., Seino, Y., Eddy, R. L., Fukushima, Y., et al. (1988). Sequence, tissue distribution, and chromosomal localization of mRNA encoding a human glucose transporter-like protein. Proc. Natl. Acad. Sci. U.S.A. 85, 5434–5438. doi:10.1073/pnas.85.15.5434
George Thompson, A. M., Ursu, O., Babkin, P., Iancu, C. V., Whang, A., Oprea, T. I., et al. (2016). Discovery of a specific inhibitor of human GLUT5 by virtual screening and in vitro transport evaluation. Sci. Rep. 6, 24240. doi:10.1038/srep24240
Gietz, R. D., and Schiestl, R. H. (2007). Frozen competent yeast cells that can be transformed with high efficiency using the LiAc/SS carrier DNA/PEG method. Nat. protoc. 2, 1–4. doi:10.1038/nprot.2007.17
Gould, G. W., and Lienhard, G. E. (1989). Expression of a functional glucose transporter in Xenopus oocytes. Biochemistry 28, 9447–9452. doi:10.1021/bi00450a030
Guillemain, G., Loizeau, M., Pinnçon-Raymond, M., Girard, J., and Leturque, A. (2000). The large intracytoplasmic loop of the glucose transporter GLUT2 is involved in glucose signaling in hepatic cells. J. Cell Sci. 113, 841–847.
Hajiaghaalipour, F., Khalilpourfarshbafi, M., and Arya, A. (2015). Modulation of glucose transporter protein by dietary flavonoids in type 2 diabetes mellitus. Int. J. Biol. Sci. 11, 508–524. doi:10.7150/ijbs.11241
Joost, H.-G., and Thorens, B. (2009). The extended GLUT-family of sugar/polyol transport facilitators: Nomenclature, sequence characteristics, and potential function of its novel members. Mol. Membr. Biol. 18, 247–256. doi:10.1080/09687680110090456
Jung, J. H., Wang, X. D., and Loeken, M. R. (2013). Mouse embryonic stem cells established in physiological-glucose media express the high Km Glut2 glucose transporter expressed by normal embryos. Stem Cell Transl. Med. 2, 929–934. doi:10.5966/sctm.2013-0093
Kasahara, T., and Kasahara, M. (1996). Expression of the rat GLUT1 glucose transporter in the yeast Saccharomyces cerevisiae. Biochem. J. 315, 177–182. doi:10.1042/bj3150177
Kasahara, T., and Kasahara, M. (1997). Characterization of rat Glut4 glucose transporter expressed in the yeast Saccharomyces cerevisiae: comparison with Glut1 glucose transporter. Biochim. Biophys. Acta 1324, 111–119. doi:10.1016/S0005-2736(96)00217-9
Katagiri, H., Asano, T., Ishihara, H., Tsukuda, K., Lin, J.-L., Inukai, K., et al. (1992). Replacement of intracellular C-terminal domain of GLUT1 glucose transporter with that of GLUT2 increases Vmax and Km of transport activity. J. Biol. Chem. 267, 22550–22555.
Kraft, T. E., Hresko, R. C., and Hruz, P. W. (2015). Expression, purification, and functional characterization of the insulin-responsive facilitative glucose transporter GLUT4. Protein Sci. 24, 2008. doi:10.1002/pro.2812
Kwok, S., Chang, S.-Y., Sninsky, J. J., and Wang, A. (1994). A guide to the design and use of mismatched and degenerate primers. PCR Meth. Appl. 3, 39–47. doi:10.1101/gr.3.4.s39
Kwon, O., Eck, P., Chen, S., Corpe, C. P., Lee, J.-H., Kruhlak, M., et al. (2007). Inhibition of the intestinal glucose transporter GLUT2 by flavonoids. FASEB J. 21, 366–377. doi:10.1096/fj.06-6620com
Leandro, M. J., Fonseca, C., and Gonçalves, P. (2009). Hexose and pentose transport in ascomycetous yeasts: An overview. FEMS Yeast Res. 9, 511–525. doi:10.1111/j.1567-1364.2009.00509.x
Lee, Y., Lim, Y., and Kwon, O. (2015). Selected phytochemicals and culinary plant extracts inhibit fructose uptake in Caco-2 cells. Molecules 20, 17393–17404. doi:10.3390/molecules200917393
Mueckler, M., and Thorens, B. (2013). The SLC2 (GLUT) family of membrane transporters. Mol. Aspects Med. 34, 121–138. doi:10.1016/j.mam.2012.07.001
Ohtsubo, K., Takamatsu, S., Minowa, M. T., Yoshida, A., Takeuchi, M., and Marth, J. D. (2005). Dietary and genetic control of glucose transporter 2 glycosylation promotes insulin secretion in suppressing diabetes. Cell 123, 1307–1321. doi:10.1016/j.cell.2005.09.041
Oldenburg, K., Vo, K. T., Michaelis, S., and Paddon, C. (1997). Recombination-mediated PCR-directed plasmid construction in vivo in yeast. Nucleic Acids Res. 25, 451–452. doi:10.1093/nar/25.2.451
Qureshi, A. A., Suades, A., Matsuoka, R., Brock, J., McComas, S. E., Nji, E., et al. (2020). The molecular basis for sugar import in malaria parasites. Nature 578, 321–325. doi:10.1038/s41586-020-1963-z
Santer, R., Groth, S., Kinner, M., Dombrowski, A., Berry, G. T., Brodehl, J., et al. (2002). The mutation spectrum of the facilitative glucose transporter gene SLC2A2 (GLUT2) in patients with Fanconi-Bickel syndrome. Hum. Genet. 110, 21–29. doi:10.1007/s00439-001-0638-6
Santer, R., Schneppenheim, R., Dombrowski, A., Götze, H., Steinmann, B., and Schaub, J. (1997). Mutations in GLUT2, the gene for the liver-type glucose transporter, in patients with Fanconi-Bickel syndrome. Nat. Genet. 17, 324–326. doi:10.1038/ng1197-324
Schmidl, S., Iancu, C. V., Choe, J.-Y., and Oreb, M. (2018). Ligand screening systems for human glucose transporters as tools in drug discovery. Front. Chem. 6, 183. doi:10.3389/fchem.2018.00183
Schmidl, S., Iancu, C. V., Reifenrath, M., Choe, J.-Y., and Oreb, M. (2021). A label-free real-time method for measuring glucose uptake kinetics in yeast. FEMS Yeast Res. 21, foaa069. doi:10.1093/femsyr/foaa069
Simpson, I. A., Carruthers, A., and Vannucci, S. J. (2007). Supply and demand in cerebral energy metabolism: The role of nutrient transporters. J Cereb. Blood. Flow. Metab. 27, 1766–1791. doi:10.1038/sj.jcbfm.9600521
Simpson, I. A., Dwyer, D., Malide, D., Moley, K. H., Travis, A., and Vannucci, S. J. (2008). The facilitative glucose transporter GLUT3: 20 years of distinction. Am. J. Physiol. Endocrinol. Metab. 295, E242–E253. doi:10.1152/ajpendo.90388.2008
Slubowski, C. J., Funk, A. D., Roesner, J. M., Paulissen, S. M., and Huang, L. S. (2015). Plasmids for C-terminal tagging in Saccharomyces cerevisiae that contain improved GFP proteins. Envy and Ivy. Yeast 32, 379–387. doi:10.1002/yea.3065
Smirnova, I., Kasho, V., Jiang, X., Pardon, E., Steyaert, J., and Kaback, H. R. (2014). Outward-facing conformers of LacY stabilized by nanobodies. Proc. Natl. Acad. Sci. U.S.A. 111, 18548–18553. doi:10.1073/pnas.1422265112
Solis-Escalante, D., van den Broek, M., Kuijpers, N. G. A., Pronk, J. T., Boles, E., Daran, J.-M., et al. (2015). The genome sequence of the popular hexose-transport-deficient Saccharomyces cerevisiae strain EBY.VW4000 reveals LoxP/Cre-induced translocations and gene loss. FEMS Yeast Res. 15, fou004. doi:10.1093/femsyr/fou004
Thorens, B. (2015). GLUT2, glucose sensing and glucose homeostasis. Diabetologia 58, 221–232. doi:10.1007/s00125-014-3451-1
Tripp, J., Essl, C., Iancu, C. V., Boles, E., Choe, J.-Y., and Oreb, M. (2017). Establishing a yeast-based screening system for discovery of human GLUT5 inhibitors and activators. Sci. Rep. 7, 124. doi:10.1038/s41598-017-06262-4
Uldry, M., Ibberson, M., Hosokawa, M., and Thorens, B. (2002). GLUT2 is a high affinity glucosamine transporter. FEBS Lett. 524, 199–203. doi:10.1016/s0014-5793(02)03058-2
van’t Klooster, J. S., Cheng, T.-Y., Sikkema, H. R., Jeucken, A., Moody, B., and Poolman, B. (2020a). Periprotein lipidomes of Saccharomyces cerevisiae provide a flexible environment for conformational changes of membrane proteins. eLife 9, e57003. doi:10.7554/eLife.57003
van’t Klooster, J. S., Cheng, T.-Y., Sikkema, H. R., Jeucken, A., Moody, D. B., and Poolman, B. (2020b). Membrane lipid requirements of the lysine transporter Lyp1 from Saccharomyces cerevisiae. J. Mol. Biol. 432, 4023–4031. doi:10.1016/j.jmb.2020.04.029
Wieczorke, R., Dlugai, S., Krampe, S., and Boles, E. (2002). Characterisation of mammalian GLUT glucose transporters in a heterologous yeast expression system. Cell. Physiol. Biochem. 13, 123–134. doi:10.1159/000071863
Wieczorke, R., Krampe, S., Weierstall, T., Freidel, K., Hollenberg, C. P., and Boles, E. (1999). Concurrent knock-out of at least 20 transporter genes is required to block uptake of hexoses in Saccharomyces cerevisiae. FEBS Lett. 464, 123–128. doi:10.1016/S0014-5793(99)01698-1
Wu, X., Chi, R. J., Baskin, J. M., Lucast, L., Burd, C. G., De Camilli, P., et al. (2014). Structural insights into assembly and regulation of the plasma membrane phosphatidylinositol 4-kinase complex. Dev. Cell 28, 19–29. doi:10.1016/j.devcel.2013.11.012
Wuest, M., Hamann, I., Bouvet, V., Glubrecht, D., Marshall, A., Trayner, B., et al. (2018). Molecular imaging of GLUT1 and GLUT5 in breast cancer: a multitracer positron emission tomography imaging study in mice. Mol. Pharmacol. 93, 79–89. doi:10.1124/mol.117.110007
Yan, N. (2015). Structural biology of the major facilitator superfamily transporters. Annu. Rev. Biophys. 44, 257–283. doi:10.1146/annurev-biophys-060414-033901
Zamora-Leon, S. P., Golde, D. W., Concha, I. I., Rivas, C. I., Delgado-Lopez, F., Baselga, J., et al. (1996). Expression of the fructose transporter GLUT5 in human breast cancer. Proc. Natl. Acad. Sci. U.S.A. 93, 1847–1852. doi:10.1073/pnas.93.5.1847
Keywords: GLUT2, GLUT3, Glucose transport inhibitor, drug screening system, hxt0 yeast strain
Citation: Schmidl S, Tamayo Rojas SA, Iancu CV, Choe J-Y and Oreb M (2021) Functional Expression of the Human Glucose Transporters GLUT2 and GLUT3 in Yeast Offers Novel Screening Systems for GLUT-Targeting Drugs. Front. Mol. Biosci. 7:598419. doi: 10.3389/fmolb.2020.598419
Received: 24 August 2020; Accepted: 23 December 2020;
Published: 18 February 2021.
Edited by:
Cesare Indiveri, University of Calabria, ItalyReviewed by:
Sergi Puig, Institute of Agrochemistry and Food Technology (IATA), SpainCopyright © 2021 Schmidl, Tamayo Rojas, Iancu, Choe and Oreb. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Mislav Oreb, bS5vcmViQGJpby51bmktZnJhbmtmdXJ0LmRl; Jun-Yong Choe, Q2hvZWoxOEBlY3UuZWR1
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.