- 1Center for Molecular Genetics, Institute for Translational Medicine, The Affiliated Hospital of Qingdao University, College of Medicine, Qingdao University, Qingdao, China
- 2School of Basic Medicine, Qingdao University, Qingdao, China
- 3Jiangsu Provincial Engineering Research Center for Biomedical Materials and Advanced Medical Device, Huaiyin Institute of Technology, Huaian, China
Compared with the research on DNA damage, there are fewer studies on RNA damage, and the damage mechanism remains mostly unknown. Recent studies have shown that RNA is more vulnerable to damage than DNA when the cells are exposed to endogenous and exogenous insults. RNA injury may participate in a variety of disease occurrence and development. RNA not only has important catalytic functions and other housekeeping functions, it also plays a decisive role in the translation of genetic information and protein biosynthesis. Various kinds of stressors, such as ultraviolet, reactive oxygen species and nitrogen, can cause damage to RNA. It may involve in the development and progression of diseases. In this review, we focused on the relationship between the RNA damage and disease as well as the research progress on the mechanism of RNA damage, which is of great significance for the pathogenesis, diagnosis, and treatment of related diseases.
Introduction
As the main component of genetic material, DNA damage and repair mechanism has been paid close attention by researchers. However, RNA damage has not been paid enough attention resulting in an underestimated toxic effect of RNA damage on cells and the body. In a cell, RNAs are more abundant than DNAs, accounting for 80–90% of the cell’s total nucleic acid. Antisense RNA can bind to the target site by complementing the target site sequence, or directly prevent its function or change the conformation of the target site to affect its function. DNA post-transcriptional processing and modification usually involve some special RNA. RNA plays an important role in biological evolution. The discovery of ribozyme indicates that RNA is both an information molecule and a functional molecule, and RNA may be the first to appear in the early origin of life (Higgs and Lehman, 2015). Studies have shown that RNA damage exists in both physiological and pathological conditions (Ishii and Sekiguchi, 2019). A large number of experiments have shown that when RNA damage is severe, it may affect its normal physiological function and damage cells and organisms (Yan and Zaher, 2019; Giorgio et al., 2020). Since most DNA is double-stranded and the nucleotide bases on the two strands complement each other, bases are protected by hydrogen bonds and their structure is relatively stable and less subject to degradation (Vologodskii, 2016), while RNA is generally single-stranded and its structure is unstable (Sankaran, 2016). RNA instability may also be related to the half-life of the mRNA. Of the three common RNAs, the one with the shortest half-life is mRNA, which is immediately degraded after translation and is therefore the most unstable. tRNA and rRNA are relatively stable and generally exist for a long time (Peterson et al., 2016; Mauger et al., 2019). RNA is more abundant in cytoplasm and located near mitochondria at the subcellular level, and the bases are not protected by hydrogen bonds, which makes RNA more vulnerable to damage than DNA under the same nucleic acid damage pressure (Hofer et al., 2006; Nunomura et al., 2012; Fimognari, 2015).
Various kinds of stress, such as ultraviolet (UV), reactive oxygen species (ROS) and reactive nitrogen species (RNS), can cause damage to RNAs (Wurtmann and Wolin, 2009). RNA damage may have serious detrimental effects on the multifaceted functions of RNA and the viability of cells or organisms. Oxidative damage of ROS or RNS is a common damage in cells and can affect all macromolecules under physiological and pathological conditions (Li et al., 2014). Oxidized RNAs can be detected in rats, human urine and human plasma under normal physiological conditions. In particular, significant RNA oxidative damage can be detected in aging animal models and neurons of patients with neurodegenerative diseases (Nunomura, 2013). ROS are the main cause of RNA oxidative damage which are generated through the Fenton reaction (iron catalyzed oxidation) and are promoted by mitochondrial dysfunction in vivo (Poulsen et al., 2012). Ultraviolet radiation in the environment can cause photochemical modification and cross-linking of RNAs and can also cause RNA damage. Nitric oxide produced by the inflammatory response in vivo can also damage RNAs (Ishii et al., 2005).
At present, although there is an increasing number of studies on oxidative damage of RNA, the mechanism is still unclear. The purpose of this review was to describe the relationship between the RNA damage and disease as well as the research progress on the mechanism of RNA damage, which is of great significance for the pathogenesis, diagnosis, and treatment of related diseases.
Research Progress on Oxidative Damage Mechanism of RNA
At present, it is believed that cells have corresponding repair mechanisms for different forms of RNA oxidative damage. However, it is not still clear whether cells have corresponding repair mechanisms for oxidative damage of RNA, but some studies have shown that stress molecules involved in DNA damage may participate in oxidative damage stress response of RNAs (Mikolaskova et al., 2018). The mRNA monitoring and control mechanisms can remove the wrong mRNA transcripts, including nonsense codon mediated mRNA degradation (NMD), mRNA degradation without termination codon, and degradation of translation blocked mRNA, which may be involved in the treatment of mRNA damage. Protein truncations translated from oxidized mRNA may activate the NMD process (Fukuhara et al., 2005; Sheth and Parker, 2006). The repair mechanism of RNA methylation damage is the same as that of DNA damage. AlkB in E. coli and homologous molecule hABH3 in mammals can reverse the damage by direct hydroxylation of methyl groups on DNA or RNA bases damaged by methylation (Liu et al., 2010; Boniecki and Martinis, 2012). The most prevalent oxidized base in RNA is 8-hydroxyguanosine (8-OHG), which is oxidized by hydroxyl radicals to form C8-OH adjunct radicals. Different studies demonstrate that the oxidation of RNA can lead to the breaking of the stranded and the modification or removal of the bases (Nunomura et al., 2009; Jacobs et al., 2010).
8-OHG has been the focus of studies because it appears to be mutagenic (Castellani et al., 2008). For example, E. coli neutralized polynucleotide phosphorylase (hPNPase), human cell y-box binding protein 1 (Yb-1) and heterogeneous nuclear ribonucleoprotein (hnRNP) in human cells, which may lead to the silencing of damaged RNA and thus terminate its translation process after binding to damaged RNA (Maughan and Shuman, 2015). hPNPase is a human 3′-5′ RNA exonuclease that specifically degrades damaged RNA, update coding and non-coding RNA under the action of various damage recognition proteins. Overexpression of hPNPase can reduce the content of 8-OHG and improve the cell viability under oxidative stress. After hPNPase knockdown, the content of oxidative damaged RNA increases and the cell viability decreases (Wu and Li, 2008). Yb-1 is a component of intracellular mRNA degradation complex p-body. In the process of oxidative stress, Yb-1 may be involved in mediating the degradation of damaged RNAs (Figure 1). After interference with HNRNP, the sensitivity of cells to H2O2 stimulation was enhanced (Das et al., 2007; Hayakawa et al., 2010).
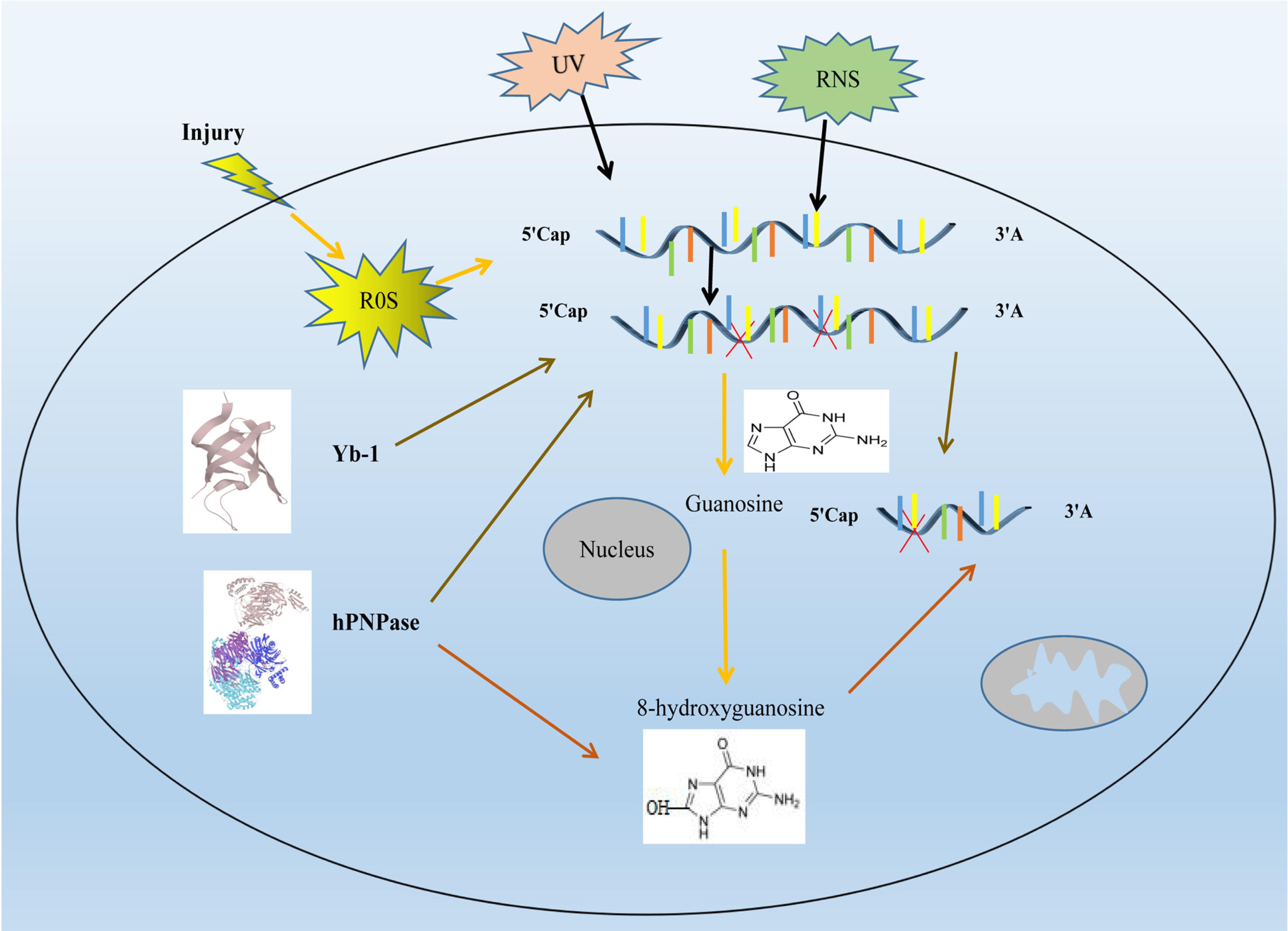
Figure 1. Research progress on oxidative damage mechanism of RNA. Ultraviolet (UV), reactive oxygen species (ROS) and reactive nitrogen species (RNS), will cause damage to RNAs RNA oxidative damage in cell. The increase of intracellular ROS leads to oxidative damage to RNA, which leads to an increase in intracellular 8-hydroxyguanosine (8-OHG), it is formed because the guanosine in RNA in RNA can be oxidized by highly reactive hydroxyl radicals to form a C8-OH adduct radical. hPNPase can degrades damaged RNA and reduces the content of 8-OHG, thereby reducing RNA damage. Yb-1 attenuates RNA damage by silencing damaged RNA. RNA molecules are shown as blue lines. red X represent the damaged RNA.
Some studies have shown that capillary dilated ataxia mutant protein (ATM), may be involved in RNA damage stress response. ATM is an important DNA damage stress molecule, and the autophosphorylation at serine is a marker of its activation. The activity and signaling pathway of ATM are disrupted after the mutation of the self-phosphorylation site (Bakkenist and Kastan, 2003). Recent studies reported that ATM participates in cellular RNA oxidative damage stress through autophosphorylation activation (Bhatia et al., 2018). However, the role of ATM in RNA damage stress, as well as its involvement in RNA damage recognition, repair, or degradation and renewal, are yet to be explored. Furthermore, studies of animal and human models suggest that the role of modified ribonucleotides in cells may be more profound. For example, m6A has been reported to be involved in transcriptional splicing and transposition in addition to regulating mRNA stability. Studies on leukemia cells have shown that complexes formed by m5C and specific proteins can regulate chromatin structure and its affinity to biomolecules (Chmielowska-Bąk et al., 2019).
Unlike AUF1, which recognizes an 8-OHG RNA, PCBP1 recognizes heavily oxidized RNA. This factor does not promote the degradation of target cells but induces cell death. A mutation occurs in one of the two RNA binding zinc finger domain and hnRNPK-homology domain (KH) domains cancel the binding of 8-OHG and inhibit the induced RNA binding activity of apoptosis-related reactions, which is necessary to induce programmed cell death. These observations underscore the ability of cells to use damaged mRNA as a signal of stress. Moreover, this PCBP1-mediated response is activated only in the presence of a large amount of modified RNAs (Ishii et al., 2018), which are activated only in the presence of excessive oxidative stress (Table 1).
Relationship Between Oxidative Damage of RNA and Diseases
The damage of genomic coding RNAs can affect gene expression and cause the disorder of proteome expression. About 98% of gene transcripts are non-coding RNAs. Studies have shown that non-coding RNAs have structural functions and catalytic activities. In recent years, a large number of non-coding RNAs have been discovered and identified in the central nervous system. Therefore, the physiological dysfunction caused by RNA damage has a close and complex relationship with the pathophysiology of neurons. Recently, more and more experiments have proved that oxidative RNA damage is related to the pathogenesis of neurodegenerative diseases. Obvious oxidative RNA damage can be detected in vulnerable neurons in the early stage of neurodegenerative disease, indicating that RNA oxidation may contribute to the occurrence and development of the disease (Butterfield et al., 2001). In addition, RNA damage may also be associated with atherosclerosis, type 2 diabetes, cancers, and other diseases (Figure 2).
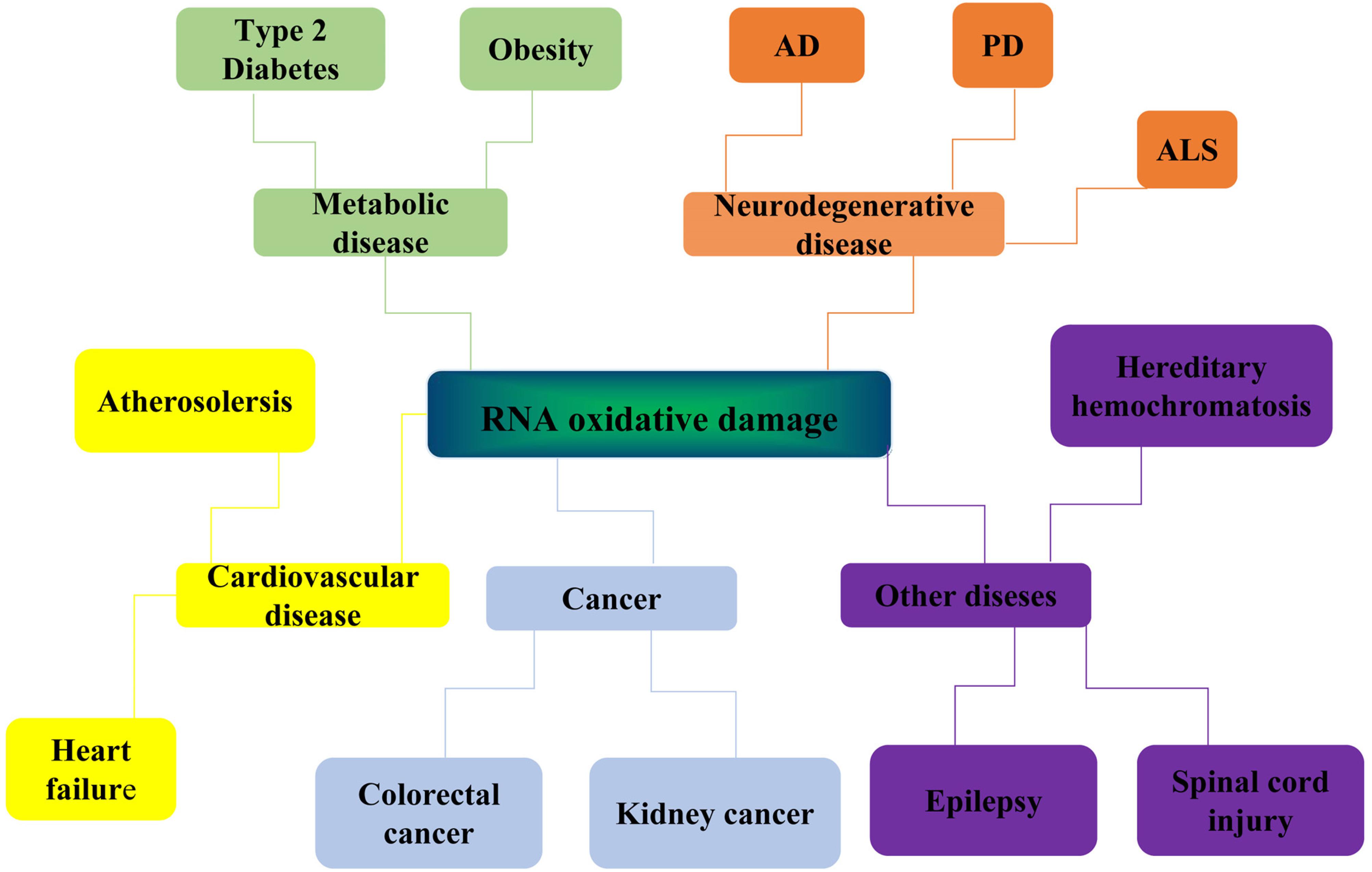
Figure 2. Diseases related to RNA oxidative damage. RNA oxidative damage is associated with the development of many diseases. It is mainly related to the occurrence and development of four kinds of diseases: neurodegenerative diseases, metabolic diseases, cardiovascular diseases, and cancer. Neurodegenerative diseases include Alzheimer’s disease (Alzheimer diseases, AD), Parkinson’s disease (PD) and Amyotrophic lateral sclerosis (ALS). Metabolic diseases include type 2 diabetes and obesity. Cardiovascular diseases include atherosclerosis and heart failure. Cancer include colorectal cancer and kidney cancer. It is also associated with hereditary hemochromatosis, spinal cord injury, epilepsy, and other diseases.
Neurodegenerative Disease
Studies found that patients at the early and middle phase of Alzheimer’s disease (Alzheimer diseases, AD) showed up to 50% of mRNA in their frontal cortex had oxidative damage, but in the control group mRNA damage degree is lower than 2%, and further combining with DNA chip analysis found that many mRNAs are associated with the AD (Shan et al., 2003; Ahmad et al., 2017). It is worth noting that the AD’s and mild cognitive impairment (MCI) patients, the rate of protein synthesis and ribosome complex ability in brain tissue specific areas are significantly drop. More interestingly, the ribosome concentration of rRNA and tRNA decreased in this areas, while the content of 8-OHG increased, suggesting the dysfunction of ribosome related to RNA oxidative damage (Ding et al., 2006).
In addition, several studies also detected a significant increase in RNA oxidative damage in Parkinson’s disease (PD) (Yoritaka et al., 1996; Ding et al., 2005; Annesley et al., 2016). Elevated RNA oxidation along with the oxidative damage to lipid, protein, and DNA were also observed in postmortem substantia nigra tissue and Cerebrospinal Fluid (CSF) from living PD patients (Zhang et al., 1999; Kikuchi et al., 2002; Abe et al., 2003). Investigations of the relationship between the levels of 8-OHG in the CSF and the duration of disease suggest that RNA oxidation may occur in the early stage of PD. However, studies are needed to determine if RNA oxidation contributes to the pathogenesis of PD.
Amyotrophic lateral sclerosis (ALS) is one of the most devastating neurological disorders (Kuzma-Kozakiewicz and Kwiecinski, 2011). Study suggest that oxidative stress can modulate the function of membrane related receptor protein tyrosine kinase c-ret, thereby reducing glial cell line-derived neurotrophic factor (GDNF) signaling in motor neurons (Ryu et al., 2011).
Cardiovascular Disease
It has also been reported that the intracellular contents of 18 s and 28 s rRNA in atherosclerotic patients decrease with the increase of 8-OHG (Li et al., 2019). Oxidative stress plays a key role in the progression of atherosclerosis. Elevated oxidative damage to DNA has been reported in both human and experimental atherosclerosis (Martinet et al., 2001; Tu et al., 2019). All RNA oxidation had been detected in smooth muscle cells and endothelial cells of the human atherosclerotic plaques (Martinet et al., 2004, 2005). The implications of RNA oxidation in atherosclerosis still remain unknown. Studies have found that RNA oxidation is associated with the occurrence and development of a variety of chronic diseases in the elderly, but whether RNA oxidation is related to the pathogenesis of HF remains unclear (Liu et al., 2019).
Metabolic Disease
Recent studies have shown that levels of RNA oxidative damage in the urine of patients with type 2 diabetes can predict long-term mortality. The urinary excretion of the RNA oxidation marker 8-OHG measured shortly after diagnosis of type 2 diabetes predicts the long-term mortality independently of other risk factors. Broedbaek et al. (2011) suggested that 8-OHG could serve as a new clinical biomarker in diabetes (Kjær et al., 2017). There are studies found high RNA oxidation is associated with an cause and cardiovascular mortality risk in patients with type 2 diabetes (Rafeh et al., 2020; Whipple et al., 2020; Yandrapalli et al., 2020). One study examined the urine content of 8-OHG in obese normal men, thin normal men and normal weight men, and found that the urine content of 8-OHG in obese normal men was higher than that in other groups, indicating that obesity in men is associated with increased RNA oxidative damage (Cejvanovic et al., 2016). These results suggest that RNA oxidation may be involved in the pathophysiological process of chronic diseases.
Cancer
Besides, oxidative damage to RNA may also be related to the development of cancers. Compared to normal cells, cancer cells exhibit an accelerated metabolism and demand high ROS concentrations to maintain their high proliferation rate. ROS is of high significance in cancer treatment and it influences tumor outcome (Andrisic et al., 2018). Recent research has found that 8-OHG as a possible RNA oxidative modification marker in urine from colorectal cancer patients associated lipid peroxidation might eventually help to defend adjacent non-malignant cells from cancer invasion (Guo et al., 2020). The number of kidney cancer is growing 3–5% each year due to unknown etiologies. Intra and inter-tumor mediators increase oxidative stress and drive tumor heterogeneity. Clinical analyses of large renal tumors reveal substantial heterogeneity within the tumors themselves, which are composed of low and high-grade tumor regions with areas of necrosis and vascularity. This heterogeneity likely explains drug resistance, despite targeted therapies. ROS play a major role in the development and progression of renal carcinogenesis (Shanmugasundaram and Block, 2016; Lichner et al., 2019). Studies have found that changes in RNA damage can promote the occurrence of tumors (Jorgensen et al., 2018; Janostiak and Wajapeyee, 2019).
Other Diseases
Investigated RNA oxidation after spinal cord contusion injury in rats. There are findings a significant increase in RNA oxidation in the lesion center immediately after the injury and the level remains high over 3 h, while in the cord segment rostral to the lesion center, RNA oxidation did not increase significantly until 3 h after the injury. Increased RNA oxidation primarily occurs in the neurons around the lesion center at an early stage and in the oligodendrocytes during secondary injury progression, and these cells die later. These results indicate a spreading pattern of the RNA oxidation from the lesion site to the distal areas, and this coincides with the developmental pattern of the secondary injury. These results suggest that RNA oxidation may play an important role in the development of secondary injury (Xu et al., 2005; Kong and Lin, 2010).
As a common chronic neurological disorder, epilepsy is characterized by recurrent unprovoked seizures. It afflicts more than 50 million people worldwide. Studies have demonstrated that seizure-induced mitochondrial dysfunction and excess free radical production cause oxidative damage to cellular components and initiate the mitochondrial apoptotic pathway (Henshall et al., 2001, 2002).
Oxidatively generated damage to nucleic acids is considered to play a significant role in carcinogenesis, and it has been shown that people with hereditary hemochromatosis are at increased risk of cancer. In this study, they used a new refined liquid chromatography-tandem mass spectrometry method to measure the level of 8-oxo-7 and 8-OHG in the urine of hereditary hemochromatosis patients, and they investigated the effect of treatment on the levels of these modifications. The study was carried out as a classical case-control study of 21 newly diagnosed, never treated hereditary hemochromatosis patients and 21 matched controls. They found that the levels of 8-OHG in the urine of the experimental group were much higher than those of the control group, with a similar level of 8-oxo-7, which returned to normal after treatment (Broedbaek et al., 2009; Lawrence et al., 2018). 8-OHG is the signature product of RNA oxidative damage, it can be seen that hereditary hemochromatosis is related to the oxidative damage of RNA.
Conclusion and Perspectives
At present, the research on the mechanisms of RNA damage oxidation is still scarce. RNA oxidative damage is involved in the pathogenesis of various pathological diseases, such as ischemia, cancer, obesity, diabetes, viral infections, chronic fatigue syndrome, amyotrophic lateral sclerosis (ALS), depression, kidney disease, heart failure, and atherosclerosis. RNA oxidation is a marker of many neurodegenerative diseases as well as an indicator of an increase in oxidative stress which can lead to an exacerbation of diseases. However, the molecular mechanism of RNA oxidative damage in these diseases remains to be explored, the direct cause-and-effect relationship between oxidized RNA and disease pathogenicity is not clear (Simms and Zaher, 2016). Although different forms of RNA damage are known to trigger different mechanisms of recognition and repair, whether there is a common pathway between multiple complex stress responses, whether RNA damage and DNA damage share some signaling pathways will be the focus of our future.
Author Contributions
ZhL, LA, and PL discussed the structure and content of the review. ZhL wrote the manuscript. LA and PL revised the manuscript. XC and ZiL helped to revise the manuscript. WY, LL, LQ, and HD did the final editing. All authors contributed to the article and approved the submitted version.
Funding
This project was supported by the National Natural Science Foundation of China Research Fund for International Young Scientists (81850410551), Natural Science Foundation of Shandong Province (ZR2019BH 014), and the Major Research Program of the National Natural Science Foundation of China (91849209).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Abe, T., Isobe, C., Murata, T., Sato, C., and Tohgi, H. (2003). Alteration of 8-hydroxyguanosine concentrations in the cerebrospinal fluid and serum from patients with Parkinson’s disease. Neurosci. Lett. 336, 105–108. doi: 10.1016/s0304-3940(02)01259-4
Ahmad, W., Ijaz, B., Shabbiri, K., Ahmed, F., and Rehman, S. (2017). Oxidative toxicity in diabetes and Alzheimer’s disease: mechanisms behind ROS/RNS generation. J. Biomed. Sci. 24:76. doi: 10.1186/s12929-017-0379-z
Andrisic, L., Dudzik, D., Barbas, C., Milkovic, L., Grune, T., and Zarkovic, N. (2018). Short overview on metabolomics approach to study pathophysiology of oxidative stress in cancer. Redox Bio. 14, 47–58. doi: 10.1016/j.redox.2017.08.009
Annesley, S. J., Lay, S. T., Piazza, S. W., Sanislav, O., Hammersley, E., Allan, C. Y., et al. (2016). Immortalized Parkinson’s disease lymphocytes have enhanced mitochondrial respiratory activity. Dis. Mod. Mech. 9, 1295–1305.
Bakkenist, C. J., and Kastan, M. B. (2003). DNA damage activates ATM through intermolecular autophosphorylation and dimer dissociation. Nature 421, 499–506. doi: 10.1038/nature01368
Bhatia, V., Valdés-Sánchez, L., Rodriguez-Martinez, D., and Bhattacharya, S. S. (2018). Formation of 53BP1 foci and ATM activation under oxidative stress is facilitated by RNA:DNA hybrids and loss of ATM-53BP1 expression promotes photoreceptor cell survival in mice. F1000res. 7:1233. doi: 10.12688/f1000research.15579.1
Boniecki, M. T., and Martinis, S. A. (2012). Coordination of tRNA synthetase active sites for chemical fidelity. J. Biol. Chem. 287, 11285–11289. doi: 10.1074/jbc.C111.325795
Broedbaek, K., Poulsen, H. E., Weimann, A., Kom, G. D., Schwedhelm, E., Nielsen, P., et al. (2009). Urinary excretion of biomarkers of oxidatively damaged DNA and RNA in hereditary hemochromatosis. Free Radical Biol. Med. 47, 1230–1233. doi: 10.1016/j.freeradbiomed.2009.08.004
Broedbaek, K., Siersma, V., Henriksen, T., Weimann, A., Petersen, M., Andersen, J. T., et al. (2011). Urinary markers of nucleic acid oxidation and long-term mortality of newly diagnosed type 2 diabetic patients. Diabetes Care 34, 2594–2596. doi: 10.2337/dc11-1620
Butterfield, D. A., Drake, J., Pocernich, C., and Castegna, A. (2001). Evidence of oxidative damage in Alzheimer’s disease brain: central role for amyloid beta-peptide. Trends Mol. Med. 7, 548–554.
Castellani, R. J., Nunomura, A., Rolston, R. K., Moreira, P. I., Takeda, A., and Perry, G. (2008). Sublethal RNA oxidation as a mechanism for neurodegenerative disease. Int. J. Mol. Sci. 9, 789–806. doi: 10.3390/ijms9050789
Cejvanovic, V., Asferg, C., Kjær, L. K., Andersen, U. B., Linneberg, A., Frystyk, J., et al. (2016). Markers of oxidative stress in obese men with and without hypertension. Scand. J. Clin. Labinv. 76, 620–625. doi: 10.1080/00365513.2016.1230776
Chmielowska-Bąk, J., Arasimowicz-Jelonek, M., and Deckert, J. (2019). In search of the mRNA modification landscape in plants. BMC Plant Biol. 19:421. doi: 10.1186/s12870-019-2033-2
Das, S., Chattopadhyay, R., Bhakat, K. K., Boldogh, I., Kohno, K., Prasad, R., et al. (2007). Stimulation of NEIL2-mediated oxidized base excision repair via YB-1 interaction during oxidative stress. J. Biol. Chem. 282, 28474–28484. doi: 10.1074/jbc.M704672200
Ding, Q., Markesbery, W. R., Cecarini, V., and Keller, J. N. (2006). Decreased RNA, and increased RNA oxidation, in ribosomes from early Alzheimer’s disease. Neurochem. Res. 31, 705–710. doi: 10.1007/s11064-006-9071-5
Ding, Q., Markesbery, W. R., Chen, Q., Li, F., and Keller, J. N. (2005). Ribosome dysfunction is an early event in Alzheimer’s disease. J. Neurosci. 25, 9171–9175. doi: 10.1523/jneurosci.3040-05.2005
Fimognari, C. (2015). Role of oxidative RNA damage in chronic-degenerative diseases. Oxid. Med. Cell Longev. 2015:358713. doi: 10.1155/2015/358713
Fukuhara, N., Ebert, J., Unterholzner, L., Lindner, D., Izaurralde, E., and Conti, E. (2005). SMG7 is a 14-3-3-like adaptor in the nonsense-mediated mRNA decay pathway. Mol. Cell 17, 537–547. doi: 10.1016/j.molcel.2005.01.010
Giorgio, M., Dellino, G. I., Gambino, V., Roda, N., and Pelicci, P. G. (2020). On the epigenetic role of guanosine oxidation. Redox Biol. 29:101398. doi: 10.1016/j.redox.2019.101398
Guo, C., Chen, Q., Chen, J., Yu, J., Hu, Y., Zhang, S., et al. (2020). 8-Hydroxyguanosine as a possible RNA oxidative modification marker in urine from colorectal cancer patients: evaluation by ultra performance liquid chromatography-tandem mass spectrometry. J. Chromatogr. B 1136:121931. doi: 10.1016/j.jchromb.2019.121931
Hayakawa, H., Fujikane, A., Ito, R., Matsumoto, M., Nakayama, K. I., and Sekiguchi, M. (2010). Human proteins that specifically bind to 8-oxoguanine-containing RNA and their responses to oxidative stress. Biochem. Biophys. Res. Commun. 403, 220–224. doi: 10.1016/j.bbrc.2010.11.011
Henshall, D. C., Araki, T., Schindler, C. K., Lan, J. Q., Tiekoter, K. L., Taki, W., et al. (2002). Activation of Bcl-2-associated death protein and counter-response of Akt within cell populations during seizure-induced neuronal death. J. Neurosci. 22, 8458–8465. doi: 10.1523/jneurosci.22-19-08458.2002
Henshall, D. C., Bonislawski, D. P., Skradski, S. L., Lan, J. Q., Meller, R., and Simon, R. P. (2001). Cleavage of bid may amplify caspase-8-induced neuronal death following focally evoked limbic seizures. Neurobil Dis. 8, 568–580. doi: 10.1006/nbdi.2001.0415
Higgs, P. G., and Lehman, N. (2015). The RNA World: molecular cooperation at the origins of life. Nat. Rev. Genet. 16, 7–17. doi: 10.1038/nrg3841
Hofer, T., Seo, A. Y., Prudencio, M., and Leeuwenburgh, C. (2006). A method to determine RNA and DNA oxidation simultaneously by HPLC-ECD: greater RNA than DNA oxidation in rat liver after doxorubicin administration. J. Biol. Chem. 387, 103–111. doi: 10.1515/bc.2006.014
Ishii, T., Hayakawa, H., Igawa, T., Sekiguchi, T., and Sekiguchi, M. (2018). Specific binding of PCBP1 to heavily oxidized RNA to induce cell death. Proc. Natl. Acad. Sci. U.S.A. 115, 6715–6720. doi: 10.1073/pnas.1806912115
Ishii, T., Sakurai, T., Usami, H., and Uchida, K. (2005). Oxidative modification of proteasome: identification of an oxidation-sensitive subunit in 26S proteasome. Biochemistry 44, 13893–13901. doi: 10.1021/bi051336u
Ishii, T., and Sekiguchi, M. (2019). Two ways of escaping from oxidative RNA damage: selective degradation and cell death. DNA Repair (Amst) 81, 102666. doi: 10.1016/j.dnarep.2019.102666
Jacobs, A. C., Resendiz, M. J., and Greenberg, M. M. (2010). Direct strand scission from a nucleobase radical in RNA. J. Am. Chem. Soc. 132, 3668–3669. doi: 10.1021/ja100281x
Janostiak, R., and Wajapeyee, N. (2019). RNA modification regulatory genes in DNA damage. Methods Mol. Biol. 1870, 249–262. doi: 10.1007/978-1-4939-8808-2_19
Jorgensen, A., Staalsoe, J. M., and Simonsen, A. H. (2018). Progressive DNA and RNA damage from oxidation after aneurysmal subarachnoid haemorrhage in humans. Free Radic. Res. 52, 51–56. doi: 10.1080/10715762.2017.1407413
Kikuchi, A., Takeda, A., Onodera, H., Kimpara, T., Hisanaga, K., Sato, N., et al. (2002). Systemic increase of oxidative nucleic acid damage in Parkinson’s disease and multiple system atrophy. Neurobiol. Dis. 9, 244–248. doi: 10.1006/nbdi.2002.0466
Kjær, L. K., Cejvanovic, V., Henriksen, T., Petersen, K. M., Hansen, T., Pedersen, O., et al. (2017). Cardiovascular and all-cause mortality risk associated with urinary excretion of 8-oxoGuo, a biomarker for rna oxidation, in patients with type 2 diabetes: a prospective cohort study. Diabetes Care 40, 1771–1778. doi: 10.2337/dc17-1150
Kong, Q., and Lin, C. L. (2010). Oxidative damage to RNA: mechanisms, consequences, and diseases. Cell Mol. Life Sci. CMLS 67, 1817–1829. doi: 10.1007/s00018-010-0277-y
Kuzma-Kozakiewicz, M., and Kwiecinski, H. (2011). New therapeutic targets for amyotrophic lateral sclerosis. Expert Opin Ther Tar. 15, 127–143. doi: 10.1517/14728222.2011.542152
Lawrence, E. M., Pooler, B. D., and Pickhardt, P. J. (2018). Opportunistic screening for hereditary hemochromatosis with unenhanced CT: determination of an optimal liver attenuation threshold. Am. J. Roentgenol. 211, 1206–1211. doi: 10.2214/ajr.18.19690
Li, S., Jin, Y., Zhao, H., Jiang, Y., and Cai, Z. (2019). Evaluation of bisphenol A exposure induced oxidative RNA damage by liquid chromatography-mass spectrometry. Chemosphere 222, 235–242. doi: 10.1016/j.chemosphere.2019.01.120
Li, Z., Malla, S., Shin, B., and Li, J. M. (2014). Battle against RNA oxidation: molecular mechanisms for reducing oxidized RNA to protect cells. Wires RNA 5, 335–346. doi: 10.1002/wrna.1214
Lichner, Z., Mac-Way, F., and Yousef, G. M. (2019). Obstacles in renal regenerative medicine: metabolic and epigenetic parallels between cellular reprogramming and kidney cancer oncogenesis. Eur. Urol. 5, 250–261. doi: 10.1016/j.euf.2017.08.003
Liu, R. M., Choi, J., Wu, J. H., Pravia, K. A., Lewis, K. M., Brand, J. D., et al. (2010). Oxidative modification of nuclear mitogen-activated protein kinase phosphatase 1 is involved in transforming growth factor beta1-induced expression of plasminogen activator inhibitor 1 in fibroblasts. J Biol Chem. 285, 16239–16247. doi: 10.1074/jbc.M110.111732
Liu, T., Cai, J. P., Zhang, L. Q., Sun, N., Cui, J., Wang, H., et al. (2019). The mechanism of RNA oxidation involved in the development of heart failure. Free Radical Res. 53, 910–921. doi: 10.1080/10715762.2019.1646424
Martinet, W., de Meyer, G. R., Herman, A. G., and Kockx, M. M. (2004). Reactive oxygen species induce RNA damage in human atherosclerosis. Eur. J. Clin. Invest. 34, 323–327. doi: 10.1111/j.1365-2362.2004.01343.x
Martinet, W., Knaapen, M. W., Meyer, G. R., Herman, A. G., and Kockx, M. M. (2001). Oxidative DNA damage and repair in experimental atherosclerosis are reversed by dietary lipid lowering. Circ. Res. 88, 733–739. doi: 10.1161/hh0701.088684
Martinet, W., Meyer, G. R., Herman, A. G., and Kockx, M. M. (2005). Amino acid deprivation induces both apoptosis and autophagy in murine C2C12 muscle cells. Biotechnol. Lett. 27, 1157–1163. doi: 10.1007/s10529-005-0007-y
Mauger, D. M., Cabral, B. J., Presnyak, V., Su, S. V., Reid, D. W., Goodman, B., et al. (2019). mRNA structure regulates protein expression through changes in functional half-life. Proc. Natl. Acad. Sci. U.S.A. 116, 24075–24083. doi: 10.1073/pnas.1908052116
Maughan, W. P., and Shuman, S. (2015). Characterization of 3’-phosphate RNA ligase paralogs RtcB1, RtcB2, and RtcB3 from Myxococcus xanthus highlights DNA and RNA 5’-phosphate capping activity of RtcB3. J. Bacteriol. 197, 3616–3624. doi: 10.1128/jb.00631-15
Mikolaskova, B., Jurcik, M., Cipakova, I., Kretova, M., Chovanec, M., and Cipak, L. (2018). Maintenance of genome stability: the unifying role of interconnections between the DNA damage response and RNA-processing pathways. Curr. Genet. 64, 971–983. doi: 10.1007/s00294-018-0819-7
Nunomura, A. (2013). Role of oxidative RNA damage in aging and neurodegenerative disorders. Brain Nerve 65, 179–194.
Nunomura, A., Hofer, T., Moreira, P. I., Castellani, R. J., Smith, M. A., and Perry, G. (2009). RNA oxidation in Alzheimer disease and related neurodegenerative disorders. Acta Neuropathol. 118, 151–166. doi: 10.1007/s00401-009-0508-1
Nunomura, A., Moreira, P. I., Castellani, R. J., Lee, H. G., Zhu, X., Smith, M. A., et al. (2012). Oxidative damage to RNA in aging and neurodegenerative disorders. Neurotox. Res. 22, 231–248. doi: 10.1007/s12640-012-9331-x
Peterson, J. R., Thor, S., Kohler, L., Kohler, P. R., Metcalf, W. W., and Luthey-Schulten, Z. (2016). Genome-wide gene expression and RNA half-life measurements allow predictions of regulation and metabolic behavior in Methanosarcina acetivorans. BMC Genomics 17:924. doi: 10.1186/s12864-016-3219-8
Poulsen, H. E., Specht, E., Broedbaek, K., Henriksen, T., Ellervik, C., Mandrup-Poulsen, T., et al. (2012). RNA modifications by oxidation: a novel disease mechanism? Free Radic. Biol. Med. 52, 1353–1361. doi: 10.1016/j.freeradbiomed.2012.01.009
Rafeh, R., Viveiros, A., Oudit, G. Y., and El-Yazbi, A. F. (2020). Targeting perivascular and epicardial adipose tissue inflammation: therapeutic opportunities for cardiovascular disease. Clin. Sci. 134, 827–851. doi: 10.1042/cs20190227
Ryu, H., Jeon, G. S., Cashman, N. R., Kowall, N. W., and Lee, J. (2011). Differential expression of c-Ret in motor neurons versus non-neuronal cells is linked to the pathogenesis of ALS. Lab. Invest. 91, 342–352. doi: 10.1038/labinvest.2010.203
Sankaran, N. (2016). The RNA world at thirty: a look back with its author. J. Mol. Evol. 83, 169–175. doi: 10.1007/s00239-016-9767-3
Shan, X., Tashiro, H., and Lin, C. L. (2003). The identification and characterization of oxidized RNAs in Alzheimer’s disease. J. Neurosci. 23, 4913–4921. doi: 10.1523/jneurosci.23-12-04913.2003
Shanmugasundaram, K., and Block, K. (2016). Renal carcinogenesis, tumor heterogeneity, and reactive oxygen species: tactics evolved. Antioxid. Redox Sig. 25, 685–701. doi: 10.1089/ars.2015.6569
Sheth, U., and Parker, R. (2006). Targeting of aberrant mRNAs to cytoplasmic processing bodies. Cell 125, 1095–1109. doi: 10.1016/j.cell.2006.04.037
Simms, C. L., and Zaher, H. S. (2016). Quality control of chemically damaged RNA. CMLS 73, 3639–3653. doi: 10.1007/s00018-016-2261-7
Tu, X., Qin, B., Zhang, Y., Zhang, C., Kahila, M., Nowsheen, S., et al. (2019). PD-L1 (B7-H1) competes with the RNA exosome to regulate the DNA damage response and can be targeted to sensitize to radiation or chemotherapy. Mol. Cell. 74, 1215–1226.e1214. doi: 10.1016/j.molcel.2019.04.005
Vologodskii, A. (2016). Disentangling DNA molecules. Phys. Life Rev. 18, 118–134. doi: 10.1016/j.plrev.2016.05.001
Whipple, M. O., Schorr, E. N., Talley, K. M. C., Wolfson, J., Lindquist, R., Bronas, U. G., et al. (2020). Influence of changes in sedentary time on outcomes of supervised exercise therapy in individuals with comorbid peripheral artery disease and type 2 diabetes. Ann. Vasc. Surg. 5096, 30312–30315. doi: 10.1016/j.avsg.2020.03.040
Wu, J., and Li, Z. (2008). Human polynucleotide phosphorylase reduces oxidative RNA damage and protects HeLa cell against oxidative stress. Biochem. Biophys. Res. Commun. 372, 288–292. doi: 10.1016/j.bbrc.2008.05.058
Wurtmann, E. J., and Wolin, S. L. (2009). RNA under attack: cellular handling of RNA damage. Crit. Rev. Biochem. Mol. 44, 34–49. doi: 10.1080/10409230802594043
Xu, M., Yip, G. W., Gan, L. T., and Ng, Y. K. (2005). Distinct roles of oxidative stress and antioxidants in the nucleus dorsalis and red nucleus following spinal cord hemisection. Brain Res. 1055, 137–142. doi: 10.1016/j.brainres.2005.07.003
Yan, L. L., and Zaher, H. S. (2019). How do cells cope with RNA damage and its consequences? J. Biol. Chem. 294, 15158–15171. doi: 10.1074/jbc.REV119.006513
Yandrapalli, S., Malik, A., Horblitt, A., Pemmasani, G., Aronow, W. S., and Frishman, W. H. (2020). Cardiovascular safety and benefits of non-insulin antihyperglycemic drugs for the treatment of type 2 diabetes mellitus-part 2. Cardiol. Rev. doi: 10.1097/crd.0000000000000311 [Epub ahead of print].
Yoritaka, A., Hattori, N., Uchida, K., Tanaka, M., Stadtman, E. R., and Mizuno, Y. (1996). Immunohistochemical detection of 4-hydroxynonenal protein adducts in Parkinson disease. Proc. Natl. Acad. Sci. U.S.A. 93, 2696–2701. doi: 10.1073/pnas.93.7.2696
Keywords: RNA oxidation, oxidative damage mechanism, 8-hydroxyguanosine (8-OHG), disease, research progress
Citation: Li Z, Chen X, Liu Z, Ye W, Li L, Qian L, Ding H, Li P and Aung LHH (2020) Recent Advances: Molecular Mechanism of RNA Oxidation and Its Role in Various Diseases. Front. Mol. Biosci. 7:184. doi: 10.3389/fmolb.2020.00184
Received: 06 May 2020; Accepted: 13 July 2020;
Published: 31 July 2020.
Edited by:
Ulf Andersson Vang Ørom, Aarhus University, DenmarkReviewed by:
Edoardo Milanetti, Sapienza University of Rome, ItalyDario Balestra, University of Ferrara, Italy
Copyright © 2020 Li, Chen, Liu, Ye, Li, Qian, Ding, Li and Aung. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lynn Htet Htet Aung, bHlubmF1bmdAcWR1LmVkdS5jbg==; Peifeng Li, cGVpZmxpQHFkdS5lZHUuY24=; Hongyan Ding, ZGh5QGh5aXQuZWR1LmNu
 Zhe Li
Zhe Li Xiatian Chen
Xiatian Chen Ziqian Liu1,2
Ziqian Liu1,2 Lynn Htet Htet Aung
Lynn Htet Htet Aung