- Department of Genetics, Microbiology and Statistics, University of Barcelona, Barcelona, Spain
Conjugation of R27 plasmid is thermoregulated, being promoted at 25°C and repressed at 37°C. Previous studies identified plasmid-encoded regulators, HtdA, TrhR and TrhY, that control expression of conjugation-related genes (tra). Moreover, the nucleoid-associated protein H-NS represses conjugation at non-permissive temperature. A transcriptomic approach has been used to characterize the effect of temperature on the expression of the 205 R27 genes. Many of the 35 tra genes, directly involved in plasmid-conjugation, were upregulated at 25°C. However, the majority of the non-tra R27 genes—many of them with unknown function—were more actively expressed at 37°C. The role of HtdA, a regulator that causes repression of the R27 conjugation by counteracting TrhR/TrhY mediated activation of tra genes, has been investigated. Most of the R27 genes are severely derepressed at 25°C in an htdA mutant, suggesting that HtdA is involved also in the repression of R27 genes other than the tra genes. Interestingly, the effect of htdA mutation was abolished at non-permissive temperature, indicating that the HtdA-TrhR/TrhY regulatory circuit mediates the environmental regulation of R27 gene expression. The role of H-NS in the proposed model is discussed.
Introduction
Horizontal gene transfer (HGT) is a process of genetic exchange that highly contributes to evolution and adaptation of bacteria to new niches by promoting acquisition of genes coding for different metabolic pathways, toxins, adhesins, or antimicrobial resistances. Plasmid conjugation is one of the main HGT mechanisms responsible for the rapid dissemination of antibiotic resistances between pathogenic strains (Bennett, 2008; Smillie et al., 2010). In order to develop strategies to avoid multidrug spread, a detailed knowledge of the conjugation process and the mechanisms regulating that transfer process is needed.
In our research group we have focused in the study of the IncHI conjugative plasmids, which are associated with multidrug resistance in several pathogens, like Salmonella enterica and Escherichia coli (Holt et al., 2011). The conjugation of this incompatibility group plasmid is thermosensitive, showing higher conjugation frequencies at low temperatures, between 22 and 30°C (Taylor and Levine, 1980). This feature suggests that the dissemination of resistances among pathogenic species by IncHI plasmids is enhanced in water and soil environments (Maher and Taylor, 1993). In addition to temperature, the physiological state of the cell is critical to define the conjugative potential of the donor cells during the transfer process of R27 plasmid, the prototype of IncHI1 plasmids (Gibert et al., 2016). The possible role of other environmental factors in the conjugation of R27, such as osmolarity, anaerobiosis, quorum sensing and acidity, has been tested (Alonso et al., 2005). No significant influence of these environmental parameters was found on R27 transfer frequency.
The genes involved in the conjugation process of the R27 plasmid, denominated tra (transfer) genes, are clustered in two separated regions, Tra1 and Tra2 (Sherburne et al., 2000). Each region contains three operons: R, F and H operons in the Tra1 region (Lawley et al., 2002) and AC, Z and AN operons in the Tra2 region (Rooker et al., 1999; Lawley et al., 2003). We described a regulatory circuit, highly conserved among the IncHI plasmids, composed of three R27-encoded elements, HtdA, TrhR and TrhY, which is crucial in the temperature control of conjugation of R27 plasmid (Gibert et al., 2013, 2014). TrhR and TrhY are both required simultaneously to activate transcriptional expression of the F, H, AC and Z operons and consequently to promote R27 conjugation. HtdA has an overall negative role by counteracting the stimulatory activity of TrhR and TrhY (Gibert et al., 2013, 2014, 2016).
Most regulatory studies of R27 conjugation focused on the effect of different environmental or genetic factors on the expression of one or several tra genes by using gene fusions and/or mRNA quantification. In a recent report, we demonstrated that, other genetic loci located outside the tra regions seem to be coregulated together with the tra genes and, by their putative functions, might be also involved in promoting an efficient R27 conjugation (Gibert et al., 2016).
To further characterize the thermoregulation of R27 conjugation, the variations on the expression pattern of the totality of genes of the R27 plasmid at permissive (25°C) and non-permissive (37°C) temperatures was determined. Having in consideration that the HtdA-TrhR/TrhY regulatory circuit described seems to play a crucial role in the environmental control of the R27 conjugation, the transcriptomic studies have been extended to an htdA derivative of the R27 plasmid.
Materials and Methods
Bacterial Strains, Plasmids, and Growth Conditions
The strains used are MG1655 (Guyer et al., 1981) and its Δlac derivative AAG1 (Aberg et al., 2008) harboring either R27 (TcR, Grindley et al., 1972) or drR27 (R27htdA− TcR, Gibert et al., 2013) plasmids. Strain AAG1-F contains a fusion between the F operon promoter and the lacZ gene, located at the attB chromosomal locus (Gibert et al., 2013). Plasmid pBADtrhRY (Gibert et al., 2014) was used to overexpress the TrhR/TrhY proteins. Bacteria were routinely grown in LB (10 g/L NaCl, 10 g/L tryptone, 5 g/L yeast extract). For conjugation experiments strains were grown in PB medium (1.5 g/L meat extract, 1.5 g/L yeast extract, 5 g/L peptone, 1 g/L glucose, 3.5 g/L NaCl, 1.32 g/L KH2PO4, 4.82 g/L K2HP4·3H2O). M9 minimal media plates with the following composition: 1 x M9 salts, 0.2 % lactose, 10 μM thiamine and 1.5 % bactoagar, were used to differentiate donor from transconjugant cells in conjugation experiments, as described previously (Gibert et al., 2014). When needed, tetracycline (Tc) and arabinose were added at the concentration of 15 μg/mL and 0.02%, respectively. Unless indicated, all cultures were grown at either 25 or 37°C and under shaking (200 rpm) conditions.
Strain Constructions
lacZ fusions with the intergenic regions containing putative promoter sequences were constructed (fragments a, b and c in Figure 4B). An intragenic region within R0009 gene was used as a negative control (fragment d in Figure 4B). PCR amplification of those regions was performed using primers pairs described in Table S1. Primers were designed to incorporate either EcoRI (forward primers) or BamHI (reverse primers) restriction sites. The PCR-amplified fragments were cloned in plasmid pGEM-T and subsequently in pRS551, either in BamHI or EcoRI-BamHI sites. The resulting constructs were transferred to the attB chromosomal locus of the AAG1 strain using previously described protocols (Simons et al., 1987). Controls to confirm single gene fusion in the attB locus were performed for all fusions. All genetic constructions were confirmed by DNA sequencing.
Conjugation Experiments
Mating experiments, using strains AGG1 R27 and MG1655 as donor and recipient strains, respectively, were performed as described previously (Gibert et al., 2014).
β-Galactosidase Assay
β-Galactosidase assays were performed as described previously (Miller, 1992). Data are given as mean values from at least three independent experiments, plotted with standard deviations.
Total RNA Isolation
The RNA used in microarray experiments was purified from three independent cultures grown in LB under shaking conditions. The temperature of incubation was either 25 or 37°C and samples were taken at mid logarithmic phase (log, OD600nm of 0.4). The RNA was purified using an SV Total RNA Isolation System (Promega) according to the manufacturer's instructions. The RNA was DNase treated with TURBO DNAse (Ambion). After concentration, using a RNeasy Minielute Clean-up kit (Qiagen), purity and quality of the purified RNA was tested by Bionalyzer 2100 (Agilent Technologies). All samples show a RNA integrity number (RIN) over 8.0.
Microarray Analysis
Transcriptomic analysis was performed on a custom-designed DNA microarray engineered by NimbleGen, containing two replicates of seven selected probes for each of the 205 annotated genes of the R27 plasmid (NC_002305), as previously described (Paytubi et al., 2014). In this work we compare the microarray data obtained from cultures of strains harboring R27 or drR27 plasmid, grown at 25 or 37°C to mid log phase of growth. Microarray data of R27 genes expression at 25°C have been previously described in a comparative study between logarithmic and stationary phase cultures (Gibert et al., 2016). The complete dataset has been deposited under the accession number E-MTAB-9150 at http://www.ebi.ac.uk/arrayexpress.
Characterization of Transcripts
To describe the transcriptional organization of the AN operon, total RNA was isolated from cultures of strain AAG1(R27) grown in LB at 25°C to an OD600nm of 2.0. To characterize the 5′ and 3′ end of the transcripts generated from the AN operon, circularized RNA was obtained as follows. CircRNA samples containing processed transcripts (monophosphated in the original RNA samples) were obtained after direct ligation of 10 μg of total RNA with CircLigase RNA Ligase (Epicentre). To obtain circRNA samples carrying primary transcripts (triphosphated in the original RNA samples), prior ligation, 10 μg of total RNA was treated with phosphatase (CIAP, Invitrogen) and subsequent RNA 5′ polyphosphatase (Epicentre). CircRNA samples were retrotranscribed to cDNA using the AMV RT (Promega) with specific primers of the AN operon (Table S2). cDNA sequences generated from the junction between the 5′- and 3′- ends were PCR amplified, using two subsequent rounds of PCR with the indicated primers (Table S2). The PCR products were further purified and sequenced with the primers used in the second round PCR by a Sanger approach using the BigDye™ Terminator v3.1 Cycle Sequencing Kit.
The 5′RACE was performed using the FirstChoice RLM-RACE kit (Ambion) and following manufacturer's instructions. After cDNA synthesis, two rounds of PCR were performed using the primer pairs outer primer/R1-trhU and inner primer/R2-trhU. The outer and inner primers are supplied by the manufacturer. The amplicons generated were purified and sequenced.
To determine cotranscription among the different transcripts generated from the AN operon, cDNA was obtained from total RNA samples using primers R1-0009, R1-trhP, and R1-trhU and the reverse transcriptase AMV RT (Promega). PCR amplification using the following primer pairs was used to detect cotranscription: F1-htdK/R1-0009, F1-0009/R1-trhP, and F1-trhW/R1-trhU. As control of DNA contamination, not retrotranscribed samples were used.
Results and Discussion
Transcription Profile of the R27 Plasmid at Permissive and Non-permissive Temperatures
The conjugation of the R27 plasmid is tightly regulated by temperature (Figure 1A) (Taylor and Levine, 1980; Forns et al., 2005). Moreover, R27 transfer is promoted at temperatures lower than 30°C whereas is strongly repressed at 37°C. In order to characterize the effect of temperature on the expression of the R27 genes, the transcriptional expression of the 205 genes was monitored by using specific microarrays.
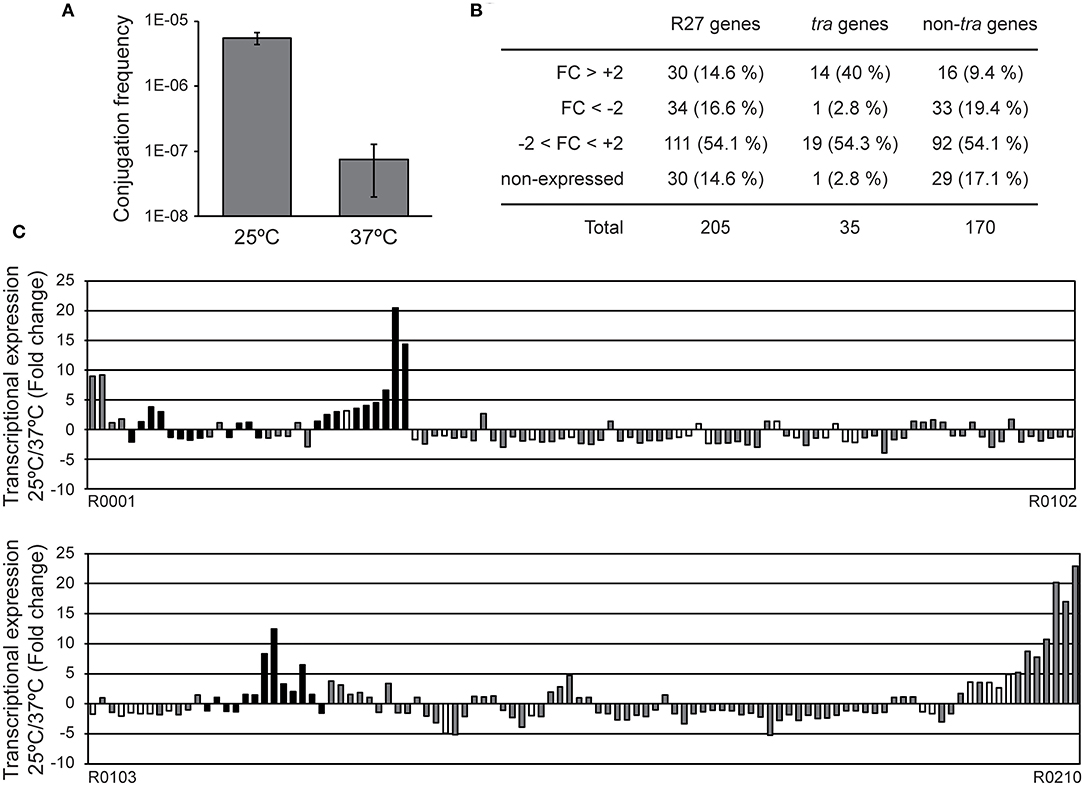
Figure 1. Expression profile of the R27 genes at permissive and non-permissive temperatures. (A) Conjugation rates were calculated using cultures of both donor [AAG1(R27)] and recipient (MG1655) strains grown at either 25 or 37°C. Average values and standard deviation of three independent experiments is shown. (B) Summary of the genes with an altered transcriptional expression in cultures of the strain AAG1(R27) grown at either 25 or 37°C. R27 genes are also divided in tra and non-tra genes. Attending to the fold change (FC), genes are classified in derepressed at 25°C (FC>+2), repressed at 25°C (FC < -2), no thermoregulated (-2 < FC < +2) and genes with signal values lower than 100 fluorescence units in both culture conditions were arbitrarily considered as non-expressed. (C) Fold change expression of the 205 ORFs encoded in R27 plasmid between AAG1(R27) cells grown at either 25 or 37°C. Expressed genes were classified as tra genes (black bars) and non-tra genes (gray bars). Non-expressed genes are indicated as white bars.
RNA samples extracted from AAG1 (a Δlac derivative of MG1655) carrying the R27 plasmid were analyzed. Cultures were grown in LB at permissive (25°C) and non-permissive (37°C) temperatures. Previous studies showed that R27 conjugation occurs more efficiently in cells growing exponentially as compared with cells in stationary phase of growth (Gibert et al., 2016). For this reason, cultures were grown up to mid logarithmic phase (OD600nm 0.4). Only expression values higher than 100 (arbitrary units of intensity of fluorescence), in at least one of the two conditions compared, were considered as significant expression and thus, the fold-change expression between conditions was calculated. We arbitrarily defined the fold-change threshold for the differential expression of a gene to 2-fold or higher. Therefore, genes with a fold-change ≥ +2 are induced at 25°C whereas genes with a fold-change ≤ −2 are induced at 37°C. A summary of the number of genes with a temperature-dependent expression (Figure 1B) and the fold change of all R27 genes (Figure 1C and Table S3) are shown. Overall, 64 genes have the expression altered by the temperature under the experimental conditions used, representing a 31.2% over the total number of R27 genes. Amongst these genes, approximately half (30) are induced at low temperature whereas the other half (34) are repressed at 25°C. No expression was detected in 30 genes, representing the 14.6% of all R27 genes. When discriminating between tra and non-tra genes, the distribution of affected genes showed different patterns. Up to 42.8% of the tra genes are affected by temperature. Most of them, 14 of the 15 genes affected, were stimulated at 25°C consistent with the clear increase in the conjugation ratio detected at this temperature. On the other hand, only 28.8% of the non-tra genes were affected, being most of them (33 of 49) repressed at 25°C. In Table 1, all genes with altered expression at low temperature are detailed. Genes from all tra operons, except for the Z operon coding for the entry-exclusion system, were found significantly induced at 25°C. Consistent with the increased conjugation frequency detected at low temperature, several genes from the AC operon, coding mostly for proteins required for mating pair formation, were found among the genes with the greatest induction. For instance, the gene trhA, coding for the major subunit of the putative conjugative pilus, is induced more than 14-fold. The expression of the R operon, coding for the regulators TrhR and TrhY, that are required for activation of tra operons expression, was induced at 25°C (3.2 and 1.9-fold for trhR and trhY, respectively) consistent with previous transcriptional studies (Gibert et al., 2014). Most of the non-tra R27 genes have not been characterized and their predicted function has been assigned attending to protein homology studies. Among the non-tra genes induced at 25°C, some of them might contribute to promote plasmid transfer, such as the muramidase (R0130) and a protein involved in the turnover of disulphide bonds (R0135) (Elton et al., 2005; Zahrl et al., 2005). Interestingly, all those non-tra genes were also found to be induced under physiological conditions promoting R27 conjugation (Gibert et al., 2016). A cluster of 9 genes, from ORF R0204 to ORF R0002, are importantly induced at low temperature. The function of the predicted proteins is not known.
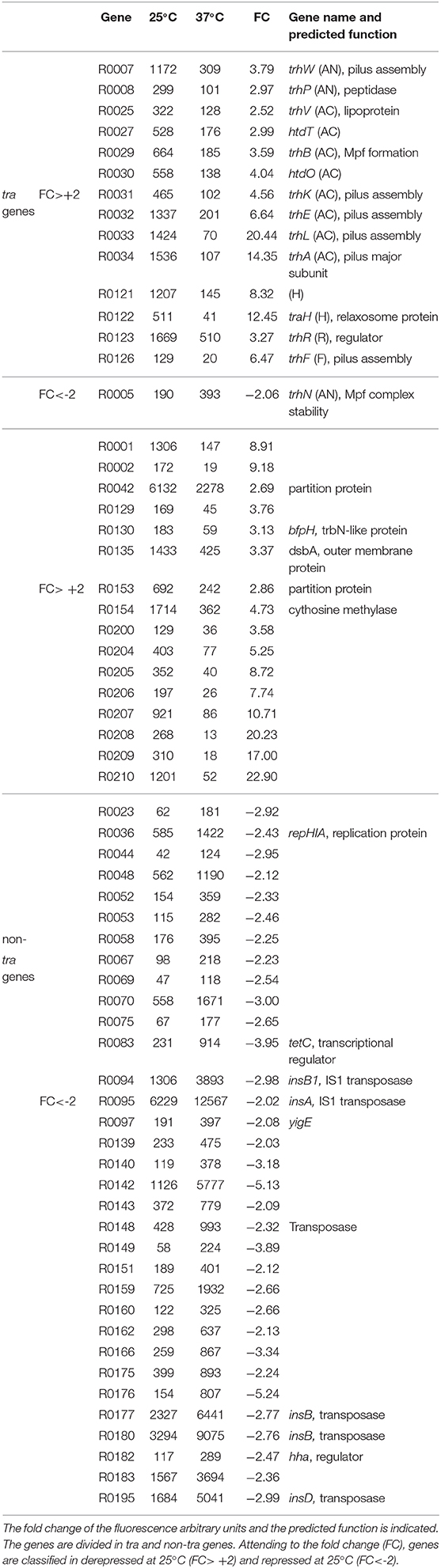
Table 1. Expression level of the R27 genes with altered expression at either 25 or 37°C in cultures grown up to mid-logarithmic phase.
Regarding the genes overexpressed at 37°C, only one belongs to the tra group of genes, trhN, coding for a predicted protein involved in mating pair formation complex stability (Lawley et al., 2003). The trhN gene was over 2-fold overexpressed at 37°C. Only a few genes with a higher expression at non-permissive temperature encoded predicted proteins with homologies to an annotated gene (Table 1). We found the repHIA gene (R0036), involved in plasmid replication; tetC (R0083), the tetracycline repressor; insA (R0095) and three insB genes (R0094, R0177 and R0180) that encode putative transposases of the insertion element IS1; the ORF R0195, encoding a IS2 transposase; and another putative transposase, R0148. Interestingly, the hha gene, coding for a transcriptional regulator that acts in combination with H-NS, is more expressed at 37°C, in concordance with its repressor role in the regulation of the conjugation process at non-permissive temperature described for H-NS/Hha proteins (Forns et al., 2005).
Altogether, our data clearly indicate that at 25°C there is a higher expression of the tra genes, consistent with the promoted R27 conjugation at low temperatures, whereas at 37°C the expression of mobile elements and transposase genes is induced. The results at high temperature might suggest that within the host (37°C), the mobilization of IS elements is induced promoting the transfer of material from the plasmid to the chromosome.
Evidently, many R27 genes were not thermoregulated. Some of R27 genes were highly expressed (higher than 600 arbitrary units of intensity of fluorescence) at both temperatures. These results could be expected, since many of these genes are involved in global processes that may take place at any temperature. Among these genes we found the tetracycline operon (tetR, tetA, and tetD), genes involved in partitioning (parA/R0020 and parB/R0019), replication (repHIB) or transposition (R0046, R0076, and R0085).
HtdA Causes Repression of R27 Genes Expression Only at Permissive Temperature
HtdA is a R27 encoded regulator that represses conjugation since R27 plasmid transfer is strongly promoted in an htdA mutant (Whelan et al., 1994; Gibert et al., 2013). HtdA is involved in the repression of the transcriptional regulation of four tra operons (F, Z, AC, and H) by counteracting the activation mediated by TrhR/TrhY (Gibert et al., 2014). Moreover, variations in the cellular levels of HtdA seem to play a crucial role in the growth phase dependency of R27 conjugation (Gibert et al., 2016). Microarray analyses using a strain carrying a drR27 plasmid (htdA mutant) were performed to monitor the effect of HtdA in the transcriptional expression of the R27 genes at both permissive and non-permissive temperatures (Figure 2, Table S4). At permissive temperature (Figures 2A,C), the htdA mutation causes a generalized derepression of R27 plasmid genes. Under the applied experimental conditions, 139 out of 181 expressed genes were upregulated in the htdA mutant strain whereas only one was downregulated (R0176). The downregulated gene is a non-tra gene with unpredicted function. Among the tra genes, 33 out of the 35 genes are upregulated in the htdA mutant. Only the R operon, containing the trhR and trhY genes, is not upregulated more than two-fold. This is consistent with previous transcriptional data from a lacZ fusion with the R operon promoter, indicating that the R operon was not regulated by the HtdA protein (Gibert et al., 2013). Remarkably, TrhR and TrhY are defined as activators of the tra genes whose activity is counteracted at the protein level by HtdA (Gibert et al., 2014). The fact that most non-tra genes (106 of 170) are derepressed in an htdA mutant strain clearly indicates that HtdA, initially described as a regulator of the tra genes, controls directly or indirectly the expression of most R27 genes. Interestingly, among the genes derepressed in the htdA mutant, we found hns and hha which are also involved in the temperature regulation of R27 conjugation (Forns et al., 2005). An earlier study showed that H-NS silences trhR transcriptional expression at non-permissive temperature whereas no relevant repression was detected at 25°C (Gibert et al., 2014). The HtdA-mediated repression of H-NS/Hha at 25°C might contribute to promote expression of trhR and trhY at low temperature.
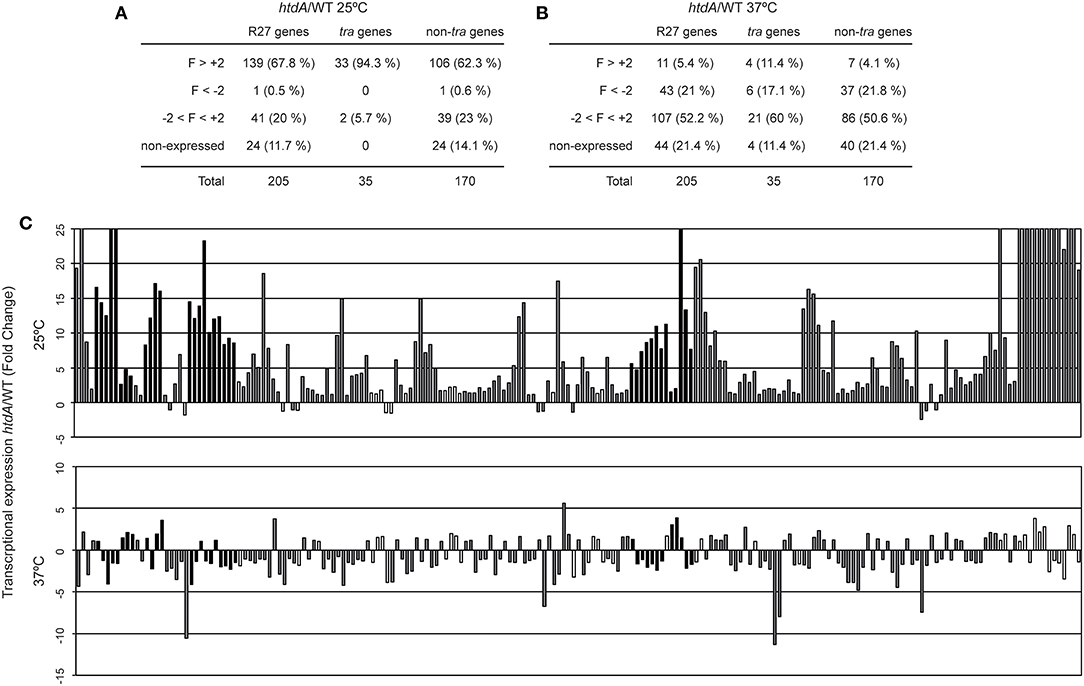
Figure 2. HtdA represses expression of R27 genes primarily at 25°C. Summary of the genes showing an altered transcriptional expression in cultures of the strains AAG1(drR27) and AAG1(R27) grown at either 25°C (A) or 37°C (B). R27 genes are also divided in tra and non-tra genes. Attending to the fold change (FC), genes are classified in derepressed at (FC>+2), repressed (FC < -2), no thermoregulated (-2 < FC < +2) and genes with signal values lower than 100 fluorescence units in both culture conditions were arbitrarily considered as non-expressed. (C) Fold change expression of the 205 ORFs encoded in R27 plasmid between AAG1(drR27) cells and AAG1(R27) cells grown at either 25°C (upper panel) or 37°C (lower panel). Expressed genes were classified as tra genes (black bars) and non-tra genes (gray bars). Non-expressed genes are indicated as white bars.
Interestingly, among the non-tra genes induced in the htdA mutant we found genes bfph, dsbA and two partition genes (R0042 and R00153), which have been found induced in R27 (htdA+) under all conditions known to promote R27 conjugation such as low temperature (Table S3) and log phase (Gibert et al., 2016).
Remarkably, all the 39 non-tra genes HtdA-independent were not induced at permissive temperature as compared to 37°C in the R27 plasmid (htdA+) (Tables S3, S4). Among these genes, 8 are involved in transposition (R0046, R0076, R0085, R0094, R0095, R0177, R0180, and R0181). We also found 2 partition genes (parA/R0020 and parB/R0019), 2 genes involved in citrate transport (citA and citB), part of the Tc operon (tetR, tetA and tetC) and 2 genes involved in UV protection (mucA and mucB) (Tables S3, S4). All these genes are involved in processes that presumably are independent of the conjugation process of the R27 plasmid.
The effect of the htdA mutation at non-permissive temperature was tested (Figures 2B,C) and a completely different pattern was observed. Most of the expressed genes did not show altered expression in the htdA mutant strain (107 of 161). Only 11 genes were derepressed by the htdA mutation as compared with the 139 genes detected at permissive temperature. On the other hand, at 37°C, 43 genes have lower expression in the htdA mutant than in the strain harboring the wt plasmid whereas at 25°C only one gene showed a decrease in the expression. Among the tra genes, 4 genes are derepressed in the htdA mutant, htdF from AN operon, trhZ from the Z operon and the 2 genes from the R operon; whereas 6 genes are repressed trhH (F operon), traI and R0118 (H operon), trhW (AN operon), R0016 (Z operon) and trhC (AC operon). Among the non-tra genes, only 7 genes were depressed in the htdA mutant, all of them were also repressed by HtdA at permissive temperature.
Overall our data suggest that the repressor role of HtdA at permissive temperature is not limited to the tra genes and its regulatory role vanishes at 37°C.
Overexpression of TrhR/TrhY Induces Expression of the F Operon Even at Non-permissive Temperature
Our data demonstrate a powerful repressor role of HtdA on the expression of R27 genes, especially among the tra genes, which is temperature dependent. Having in consideration that HtdA acts by counteracting the activation mediated by TrhR/TrhY, the role of the activators in the temperature mediated regulation was studied. F operon expression has been systematically used as a reporter to study HtdA-mediated regulation of the tra genes (Gibert et al., 2013, 2014, 2016). F operon is repressed by HtdA at permissive temperature but not at non-permissive temperature (Table S4). The effect of overexpressing TrhR/TrhY on F operon expression was monitored at permissive and non-permissive temperature in both htdA+ and htdA− genetic backgrounds. Overexpression of trhR/trhY using the arabinose inducible promoter from pBAD18 causes a robust upregulation of F operon independently of the temperature (Figure 3). These results suggest that TrhR/TrhY activity is crucial for the temperature-dependent regulation of R27 conjugation and provides a rational for the mechanisms of HtdA in the thermoregulation.
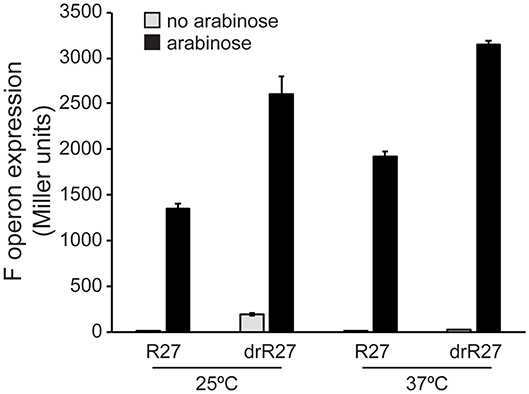
Figure 3. TrhR and TrhY overexpression causes induction of the F operon even at non-permissive temperature. Transcriptional expression of the F operon of R27 was monitored in cultures of the strain AAG1-F, carrying a chromosomal lacZ fusion with the promoter sequence of the F operon. The effect of the pBADtrhRY plasmid in the presence of either R27 or its htdA derivative drR27 was assessed. Cultures were grown at either 25 or 37°C to mid-logarithmic phase in the absence (gray bars) or presence (black bars) of arabinose (0.02%). The β-galactosidase activity (Miller units) was determined in three independent cultures and mean values with standard deviations are plotted.
New Insights in the Transcriptional Expression of the an Operon
In Figure 4A, the expression pattern of the six tra operons is depicted using the M value (log2FC) between 25 and 37°C of the different genes. Genes with no altered expression are defined by having a M value between +1 and−1, equivalent to−2 > FC > +2. A closer look to the gene expression pattern of the tra operon in response to the temperature reveals three different patterns. Pattern 1, unresponsiveness, as shown by the Z operon, where the expression of the three genes is not altered by temperature. Pattern 2, operons with induced expression at 25°C and the temperature responsiveness is greater among the proximal genes than among the distal ones. This is the most common pattern, shared by the operons AC, H, R, and F. The decrease in the expression of downstream genes in the same operon is defined as transcriptional polarity and it is a common feature among polycistronic operons. Pattern 3, the unusual pattern shown by the AN operon. The 4 proximal genes (htdA, htdF, htdK and R0009) do not respond to temperature, the 4 following genes show a pattern similar to those described in pattern 2. trhP and trhW, are induced at 25°C (2.9 and 3.7-fold) and the 2 distal genes (trhU and trhN) are again unresponsive to temperature. This expression profile within a polycistronic operon suggests the presence of complex regulatory mechanisms. Previous studies on the transcriptional organization of the tra genes revealed that the AN operon contains 8 genes, spanning from htdA to trhN which were apparently cotranscribed (Alonso et al., 2005). However, those studies did not rule out the occurrence of several transcripts arising from different events such as partial termination, mRNA processing and/or the possible presence of internal promoters. Interestingly, we found 3′ intergenic regions in the AN operon, located between htdK and R0009 (197 bp), R0009 and trhP (233 bp), and trhW and trhU (249 bp). Promoter search with the BPROM software (Solovyev and Salamov, 2011) predicts two putative promoters located upstream of the R009 gene and the trhP gene. Transcriptional terminators searches, using the ARNold software (Naville et al., 2011) identify a putative Rho-independent terminator downstream of the htdK gene. To characterize the transcripts generated from the AN operon, total RNA was isolated from cultures of strain AAG1(R27) grown at 25°C. Circularized RNA was obtained from either the isolated RNA (monophosphated mRNA, processed transcripts) or after 5′polyphosphatase RNA treatment (originally triphosphated, real transcriptional starts). The circRNA samples were used as template to obtain cDNA using specific primers (Table S2). Sequencing of the cDNA allowed us to clearly identify three transcripts, labeled as #2, #3 and #4 in Figures 4B,C. The fact that these three transcripts have been detected provides a feasible explanation to the particular expression profile of the AN operon independently of the mechanisms involved in its generation. Interestingly, transcript #2 (htdA-htdK) was the only transcript detected after polyphosphatase treatment, indicating a triphosphated mRNA and thus, a transcript with a real transcriptional start. Transcript #3 was detected from RNA untreated (processed transcript). Remarkably, its 5′ end overlaps with the 3′ end of transcript #2, by 46 bp. A possible explanation of this result is that transcript #3 was generated from a secondary promoter different from the promoter upstream of htdA and once is generated this transcript would be further processed to the final transcript #3 detected. Transcript #4 has a 5′ end located downstream of the 3′ end of transcript #3, therefore it could be generated from processing of a pre-existing polycistronic operon. After several trials, a transcript containing the sequences of the distal genes trhU-trhN was not identified by circRNA. However, 5′RACE experiments identified several transcripts trhU-trhN with distinctive 5′ ends (transcript #5). Further studies will be required to identify the exact location of the 3′ end of this transcript.
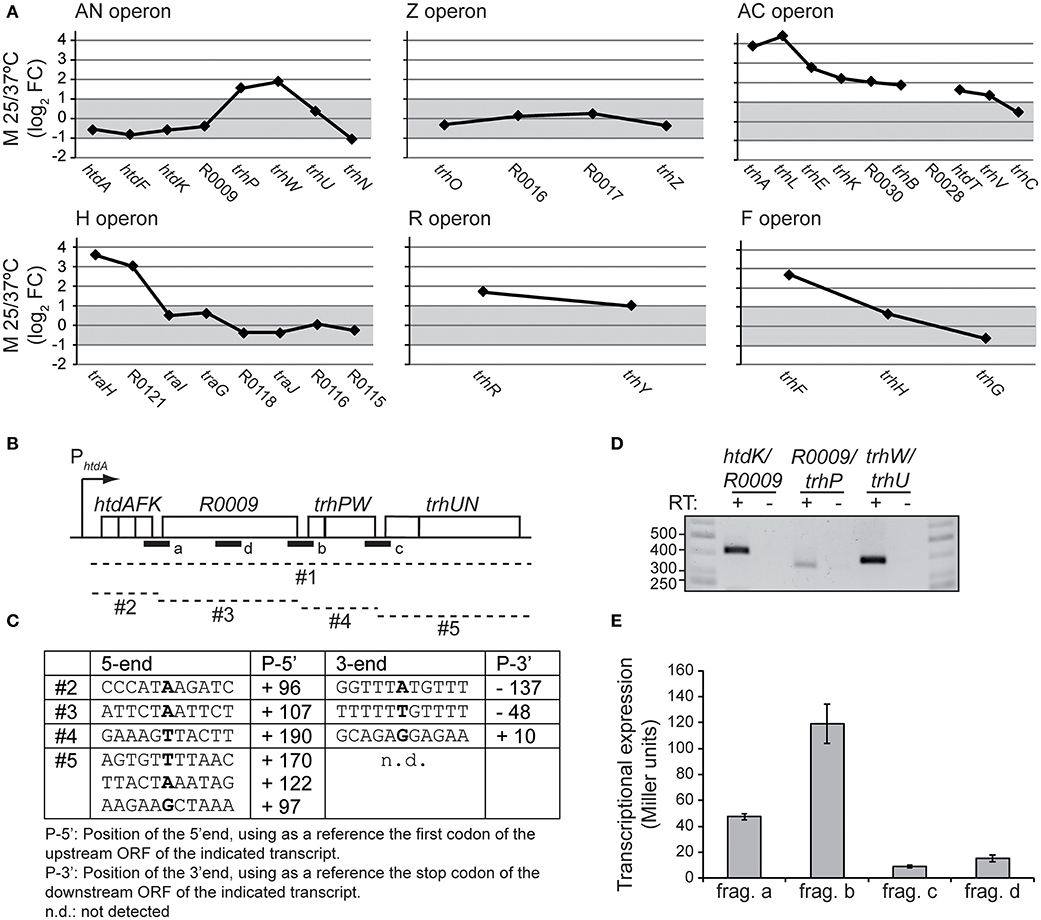
Figure 4. Transcriptional organization of the AN operon. (A) M-plots of the temperature dependent expression of the tra operons. The M value (log2FC 25/37°C) for the expressed genes of the different tra operons is depicted. An M value between +1 and−1, equivalent to a FC of +2 and−2, is considered no alteration in the expression and is shadowed in gray. (B) Representation of the AN operon. Dashed lines indicate the different putative transcripts (#1 to #5) derived from the AN operon. A thick line indicates the fragments (a to d) cloned in pRS551 to construct the lacZ fusions used. (C) The positions of 5′- and 3′ ends of the transcripts #2, #3 and #4, labeled in bold, were determined by circRNA. The putative 5′ ends of transcript #5 identified by 5′RACE assays. (D) RT-PCR using primers within the ORFs flanking the intergenic regions indicated. In all cases RNA not-RT AMV-treated was used as negative control. (E) Transcriptional expression of the lacZ fusions with the indicated fragments of AN operon was determined in cultures grown at 25°C to mid-logarithmic phase. The β-galactosidase activity (Miller units) was determined in three independent cultures and mean values with standard deviations are plotted.
RT-PCR assays (Figure 4D) using primers from the internal sequences of the transcripts #2, #3, #4, and #5 let us detect cotranscription among the different transcripts as previously described in Alonso et al. (2005) suggesting the presence of a polycistronic transcript, named #1.
The presence of putative promoter sequences in the intergenic regions (IR) upstream of R0009, trhP and trhU was assessed. The three IR and a R0009 intragenic region used as a negative control (named a, b, c, and d in Figure 4B) were cloned in pRS551 vector, upstream of the promoterless lacZ gene. The lacZ_fusion constructs generated were transferred to the attB site present in the E. coli chromosome. Transcriptional expression was measured by a β-galactosidase assay. The results (Figure 4E) suggest the presence of putative active promoter sequences upstream the R0009 and trhP genes. No activity was detected in the third IR between trhW and trhU.
Further extensive experiments will be required to fully describe the transcripts generated from the AN operon and the mechanisms involved in its generation. We would like to highlight that our data identify different mRNA species that provide a feasible explanation to the differential expression from the AN operon genes as revealed by the microarray data. The results obtained suggest that different transcriptional and posttranscriptional events may participate in the control of AN operon expression, such as partial termination, processing and transcription from secondary promoters. As reported by Conway et al. (2014), a large proportion (36%) of operons in E. coli are complex, with internal promoters or terminators that generate multiple transcription units. For 43% of operons, differential expression of polycistronic genes was observed, despite being in the same operons, indicating that E. coli operon architecture allows fine-tuning of gene expression.
Concluding Remarks
The transcriptional expression of R27 genes involved in plasmid transfer was induced at 25°C, consistent with R27 conjugation being optimal at this temperature. R27 conjugation is modulated by a regulator circuit composed by HtdA and TrhR/TrhY. Our data indicate that HtdA is not thermoregulated, while TrhR and TrhY are slightly more expressed at 25°C. Moreover, TrhR and TrhY expression is not regulated by HtdA at permissive temperature. These two proteins are responsible, at least partially, of the induction of the tra genes observed under these conditions (Figure 3).
A common expression profile shared among many tra and non-tra genes is exemplified by the trhA gene, coding for the pilin. A dramatic upregulation at 25°C in the absence of HtdA, but similar expression either in the presence or in the absence of the repressor at 37°C was observed (Table S4). This profile suggests that HtdA represses primarily at low temperature whereas at non-permissive temperature the presence or the absence of some specific regulators might avoid upregulation even in the absence of HtdA. HtdA acts by counteracting the activity of TrhR/TrhY and it has previously been described that H-NS silences tra genes expression at 37°C, by repressing trhRY expression (Figure 5). Having that in mind, we suggest that HtdA can exert its regulatory function only when H-NS is not blocking the expression of TrhR/TrhY. The TrhR/TrhY-HtdA mediated regulation is not restricted to the tra operon genes since many other R27 genes were also thermoregulated in a HtdA-dependent manner. These results also suggest that the effect of the H-NS/Hha proteins is more general as regulators of the tra operons than previously reported.
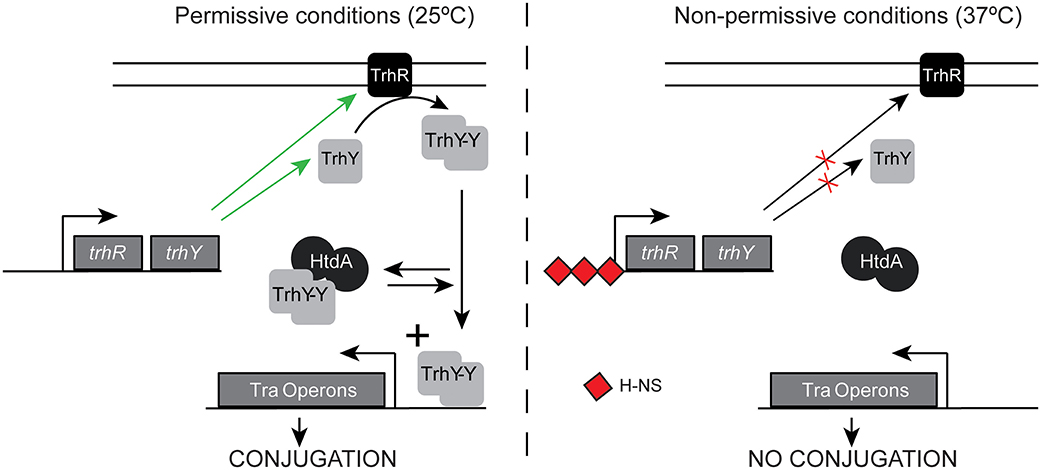
Figure 5. Schematic model of regulation of the tra operons at permissive and non-permissive temperatures. Green arrows indicate expression is promoted, whereas red crosses indicate expression is repressed.
Data Availability Statement
The datasets generated for this study can be found in the ArrayExpress repository, under the accession number E-MTAB-9150 (http://www.ebi.ac.uk/arrayexpress).
Author Contributions
MG contributed in the investigation. SP contributed in the investigation and writing the manuscript. CM and CB contributed in the conceptualization, investigation, formal analysis, and writing the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the Spanish Ministry of Economy and Competitiveness (grant AGL 2013-45339R), the Spanish Ministry of Science, Innovation and Universities (MCIU), State Bureau of Investigation (AIE), and European Regional Development Fund (FEDER) (grant PGC 2018-096958-B-I00), and the Catalonian government (grant 2017SGR499). MG was recipient of a FPU grant from the Spanish Ministry of Science and Innovation.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmolb.2020.00124/full#supplementary-material
References
Aberg, A., Shingler, V., and Balsalobre, C. (2008). Regulation of the fimB promoter: a case of differential regulation by ppGpp and DksA in vivo. Mol. Microbiol. 67, 1223–1241. doi: 10.1111/j.1365-2958.2008.06115.x
Alonso, G., Baptista, K., Ngo, T., and Taylor, D. E. (2005). Transcriptional organization of the temperature-sensitive transfer system from the IncHI1 plasmid R27. Microbiology 151, 3563–3573. doi: 10.1099/mic.0.28256-0
Bennett, P. M. (2008). Plasmid encoded antibiotic resistance: acquisition and transfer of antibiotic resistance. Br. J. Pharmacol. 153, S347–357. doi: 10.1038/sj.bjp.0707607
Conway, T., Creecy, J. P., Maddox, S. M., Grissom, J. E., Conkle, T. L., Shadid, T. M., et al. (2014). Unprecedented high-resolution view of bacterial operon architecture revealed by RNA sequencing. MBio 5:e01442-14. doi: 10.1128/mBio.01442-14
Elton, T. C., Holland, S. J., Frost, L. S., and Hazes, B. (2005). F-like type IV secretion systems encode proteins with thioredoxin folds that are putative DsbC homologues. J. Bacteriol. 187, 8267–8277. doi: 10.1128/JB.187.24.8267-8277.2005
Forns, N., Baños, R. C., Balsalobre, C., Juárez, A., and Madrid, C. (2005). Temperature-dependent conjugative transfer of R27: role of chromosome- and plasmid-encoded Hha and H-NS proteins. J. Bacteriol. 187, 3950–3959. doi: 10.1128/JB.187.12.3950-3959.2005
Gibert, M., Juárez, A., Madrid, C., and Balsalobre, C. (2013). New insights in the role of HtdA in the regulation of R27 conjugation. Plasmid 70, 61–68. doi: 10.1016/j.plasmid.2013.01.009
Gibert, M., Juárez, A., Zechner, E. L., Madrid, C., and Balsalobre, C. (2014). TrhR, TrhY and HtdA, a novel regulatory circuit that modulates conjugation of the IncHI plasmids. Mol. Microbiol. 94, 1146–1161. doi: 10.1111/mmi.12823
Gibert, M., Paytubi, S., Beltrán, S., Juárez, A., Balsalobre, C., and Madrid, C. (2016). Growth phase-dependent control of R27 conjugation is mediated by the interplay between the plasmid-encoded regulatory circuit TrhR/TrhY-HtdA and the cAMP regulon. Environ. Microbiol. 18, 5277–5287. doi: 10.1111/1462-2920.13579
Grindley, N. D., Grindley, J. N., and Anderson, E. S. (1972). R factor compatibility groups. Mol. Gen. Genet. 119, 287–297. doi: 10.1007/BF00272087
Guyer, M. S., Reed, R. R., Steitz, J. A., and Low, K. B. (1981). Identification of a sex-factor-affinity site in E. coli as gamma delta. Cold Spring Harb. Symp. Quant. Biol. 45(Pt 1), 135–140. doi: 10.1101/SQB.1981.045.01.022
Holt, K. E., Phan, M. D., Baker, S., Duy, P. T., Nga, T. V. T., Nair, S., et al. (2011). Emergence of a globally dominant IncHI1 plasmid type associated with multiple drug resistant typhoid. PLoS Negl. Trop. Dis. 5:e1245. doi: 10.1371/journal.pntd.0001245
Lawley, T. D., Gilmour, M. W., Gunton, J. E., Standeven, L. J., and Taylor, D. E. (2002). Functional and mutational analysis of conjugative transfer region 1 (Tra1) from the IncHI1 plasmid R27. J. Bacteriol. 184, 2173–2180. doi: 10.1128/JB.184.8.2173-2180.2002
Lawley, T. D., Gilmour, M. W., Gunton, J. E., Tracz, D. M., and Taylor, D. E. (2003). Functional and mutational analysis of conjugative transfer region 2 (Tra2) from the IncHI1 plasmid R27. J. Bacteriol. 185, 581–591. doi: 10.1128/JB.185.2.581-591.2003
Maher, D., and Taylor, D. E. (1993). Host range and transfer efficiency of incompatibility group HI plasmids. Can. J. Microbiol. 39, 581–587. doi: 10.1139/m93-084
Miller, J. H. (1992). A Short Course in Bacterial Genetics : A Laboratory Manual and Handbook for Escherichia coli and Related Bacteria. New York, NY: Cold Spring Harbor Laboratory Press.
Naville, M., Ghuillot-Gaudeffroy, A., Marchais, A., and Gautheret, D. (2011). ARNold: a web tool for the prediction of Rho-independent transcription terminators. RNA Biol. 8, 11–13. doi: 10.4161/rna.8.1.13346
Paytubi, S., Aznar, S., Madrid, C., Balsalobre, C., Dillon, S. C., Dorman, C. J., et al. (2014). A novel role for antibiotic resistance plasmids in facilitating Salmonella adaptation to non-host environments. Environ. Microbiol. 16, 950–962. doi: 10.1111/1462-2920.12244
Rooker, M. M., Sherburne, C., Lawley, T. D., and Taylor, D. E. (1999). Characterization of the Tra2 region of the IncHI1 plasmid R27. Plasmid 41, 226–239. doi: 10.1006/plas.1999.1396
Sherburne, C. K., Lawley, T. D., Gilmour, M. W., Blattner, F. R., Burland, V., Grotbeck, E., et al. (2000). The complete DNA sequence and analysis of R27, a large IncHI plasmid from Salmonella typhi that is temperature sensitive for transfer. Nucleic Acids Res. 28, 2177–2186. doi: 10.1093/nar/28.10.2177
Simons, R. W., Houman, F., and Kleckner, N. (1987). Improved single and multicopy lac-based cloning vectors for protein and operon fusions. Gene 53, 85–96. doi: 10.1016/0378-1119(87)90095-3
Smillie, C., Garcillán-Barcia, M. P., Francia, M. V., Rocha, E. P. C., and de la Cruz, F. (2010). Mobility of plasmids. Microbiol. Mol. Biol. Rev. 74, 434–452. doi: 10.1128/MMBR.00020-10
Solovyev, V., and Salamov, A. (2011). “Automatic annotation of microbial genomes and metagenomics sequences,” in Metagenomics and its Applications in Agriculture, Biomedicine and Environmental Studies, ed R. W. Li (New York, NY: Nova Science Publishers), 61–78.
Taylor, D. E., and Levine, J. G. (1980). Studies of temperature-sensitive transfer and maintenance of H incompatibility group plasmids. J. Gen. Microbiol. 116, 475–484. doi: 10.1099/00221287-116-2-475
Whelan, K. F., Maher, D., Colleran, E., and Taylor, D. E. (1994). Genetic and nucleotide sequence analysis of the gene htdA, which regulates conjugal transfer of IncHI plasmids. J. Bacteriol. 176, 2242–2251. doi: 10.1128/JB.176.8.2242-2251.1994
Keywords: plasmid conjugation, temperature-dependent control, TrhR/TrhY-HtdA, transcriptional regulation, R27
Citation: Gibert M, Paytubi S, Madrid C and Balsalobre C (2020) Temperature Dependent Control of the R27 Conjugative Plasmid Genes. Front. Mol. Biosci. 7:124. doi: 10.3389/fmolb.2020.00124
Received: 30 April 2020; Accepted: 02 June 2020;
Published: 10 July 2020.
Edited by:
Manuel Espinosa, Margarita Salas Center for Biological Research (CSIC), SpainReviewed by:
Elisabeth Grohmann, Beuth Hochschule für Technik Berlin, GermanyFabian Lorenzo-Diaz, University of La Laguna, Spain
Copyright © 2020 Gibert, Paytubi, Madrid and Balsalobre. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Cristina Madrid, Y21hZHJpZEB1Yi5lZHU=; Carlos Balsalobre, Y2JhbHNhbG9icmVAdWIuZWR1
†Present address: Sonia Paytubi, Infections and Cancer Laboratory, Cancer Epidemiology Research Program (CERP), Institut Català d'Oncologia (ICO)–IDIBELL, Barcelona, Spain
 Marta Gibert
Marta Gibert Sonia Paytubi
Sonia Paytubi Cristina Madrid
Cristina Madrid Carlos Balsalobre
Carlos Balsalobre