
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Mol. Biosci. , 24 April 2020
Sec. Molecular Diagnostics and Therapeutics
Volume 7 - 2020 | https://doi.org/10.3389/fmolb.2020.00070
This article is part of the Research Topic Application of Systems Biology in Molecular Characterization and Diagnosis of Cancer View all 13 articles
Gallbladder cancer is a relatively uncommon human malignant tumor with an extremely poor prognosis. Currently, no biomarkers can accurately diagnose gallbladder cancer and predict patients’ prognosis. XRCC1 is involved in tumorigenesis, progression, and chemo-resistance of several human cancers, but the role of XRCC1 in gallbladder cancer is never reported. In this study, we investigated the expression of XRCC1 and its clinicopathological and prognostic significance in gallbladder cancer, and explored the biological role of XRCC1 in gallbladder cancer cells. We found that XRCC1 was significantly up-regulated in gallbladder cancer in protein and mRNA levels. Positive XRCC1 expression was correlated with aggressive clinicopathological features and was an independent poor prognostic factor in gallbladder cancer. The ROC curves suggested that XRCC1 expression had potential clinicopathological diagnostic value in gallbladder cancer. In vitro, XRCC1 was overexpression in CD133+GBC-SD cells compared to GBC-SD cells. In functional experiment, XRCC1 knockdown had a non-significant impact on proliferation, migration, invasion, and apoptosis of CD133+GBC-SD cells. But, XRCC1 knockdown could significantly improve the sensitivity of CD133+GBC-SD cells to 5-Fluorouracil via promoting cell necrosis and apoptosis. Thus, this study indicates that XRCC1 may be a promising predictive biomarker of gallbladder cancer and a potential therapeutic target for gallbladder cancer.
Gallbladder cancer (GBC) is a relatively uncommon human malignant tumor with an extremely poor prognosis. Histologically, GBC mainly consists of gallbladder adenocarcinoma (AC) (about 90%) and squamous cell/adenosquamous carcinoma (SC/ASC) (accounting for 1–12%) (Roa et al., 2011; Samuel et al., 2018). Although various studies about GBC were performed, GBC clinical outcome remains extremely poor. Currently, radical resection remains the only way to cure GBC, although adjuvant treatments (chemotherapy and radiotherapy) are available. Because GBC patients at early stages present asymptomatic and are difficult to be diagnosed, most patients are diagnosed at late stages when patients lost the chance to receive radical surgery (Reid et al., 2007; Henley et al., 2015). Additionally, GBC is often resistant to chemotherapy and radiotherapy (Horgan et al., 2012). These above reasons make the prognosis of GBC unsatisfactory. Although many biological marks have been studied, no one can accurately diagnose GBC and predict patients’ survival (Sicklick et al., 2016; Sharma et al., 2017). Therefore, discovering reliable early diagnostic biomarkers and exploring the mechanism of treatment resistance are critically important to improve the prognosis of GBC.
DNA repair pathways are related to tumorigenesis and treatment resistance, and base excision repair (BER) is one pathway of DNA repair systems (Wood et al., 2001). BER functions an essential role in protecting the genome against chemical carcinogens and ionizing radiations (Tudek, 2007). Chemotherapy and radiotherapy usually kill tumor cells by causing DNA damage which could be repaired via BER. As a BER protein, x-ray repair cross-complementing group 1 (XRCC1), a 70-kDa protein, is encoded by the gene located on chromosome 19q13.2–13.3 (Thompson and West, 2000). XRCC1 functions as a scaffold protein in the BER and its aberrant expression is associated with carcinogenesis of multiply human malignant tumors (Hanssen-Bauer et al., 2012; Meng et al., 2017; Mei et al., 2019). Currently, researches on XRCC1 mainly concentrate on the relationship between gene polymorphism and cancer susceptibility. Recently, several studies have investigated the clinicopathological and prognostic significance of XRCC1 in human cancers including gastric cancer, ovarian cancer, and non-small cell lung cancer (Wang et al., 2012; Abdel-Fatah et al., 2013; Liu et al., 2015). Moreover, XRCC1 affects the effectiveness of chemotherapy and the role of XRCC1 in chemosensitivity varies in different types of cancer. For example, in gastric and ovarian cancer, patients with low XRCC1 expression exhibited favorable response to platinum-based chemotherapy (Wang et al., 2012; Abdel-Fatah et al., 2013). However, bladder cancer patients with high XRCC1 expression had a favorable chemosensitivity to platinum-based chemotherapy (Sakano et al., 2013). As we know, the role of XRCC1 in GBC is never reported.
Therefore, in this study, we investigated the expression of XRCC1 and its clinicopathological and prognostic significance in gallbladder SC/ASC and AC. Furthermore, the biological function of XRCC1 in CD133+GBC cells was evaluated.
This study was approved by the Ethics Committee for Human Research, Central South University and performed in accordance with the Declaration of Helsinki. The included patients were histologically diagnosed by two pathologists. These patients never received chemotherapy or radiotherapy preoperatively and postoperatively. We collected 69 SC/ASC samples from January 2001 to December 2013 (16 from Xiangya Hospital, 31 from Second Xiangya Hospital, 10 from Third Xiangya Hospital, 5 from Hunan Provincial People Hospital, 5 from Hunan Provincial Tumor Hospital, and 1 each from Changde Central Hospital and Loudi Central Hospital). According to the recommendations of the American Joint Committee on Cancer, tumors with a squamous component ≥10% were considered as ASC. The 69 SC/ASCs accounted for 5.5% of 1248 GBCs. We collected 146 AC samples from January 2008 and December 2013 at Second Xiangya Hospital and Third Xiangya Hospital. Survival data for these patients was obtained through letters and telephone calls. The follow-up time was 2 years, and patients who survived over 2 years were considered as censored cases.
The rabbit anti-human XRCC1 primary antibody and HRP-conjugated anti-rabbit second antibody were purchased from Santa Cruz Biotechnology (CA, United States). EnVisionTM Detection Kit was purchased from Dako Laboratories (CA, United States). Immunohistochemistry was performed as previously described (Wu et al., 2017). Briefly, four-micrometer-thick sections were cut from routinely paraffin-embedded tissues. The sections were deparaffinized and then incubated with peroxidase inhibitor (3% H2O2) in the dark for 15 min, followed by EDTA-trypsin digestion for 15 min. Then, the sections were incubated with primary antibody for 60 min at 37°C. Next, the sections were incubated with the second antibody for 30 min at 37°C after being soaked with PBS for 3 × 5 min. Then, solution A was added to the sections for 30 min, followed by DAB staining and hematoxylin counter-staining. The slides were dehydrated with different concentrations (70%–100%) of alcohol, and soaked in xylene for 3 × 5 min and finally mounted with neutral balsam.
Ten random fields were examined per section by two independent pathologists. The percent of positively stained cells was determined. Strength of staining was rated on a scale of 1 to 3 (1: little to no positive staining or uncertainly weak staining; 2: weak to moderate staining; 3: moderate to strong staining). A section was determined as positive expression when the percent of positively stained cells was ≥10% and staining strength was ≥2. The few sections where percent positive staining was 5% to 10% and staining strength was 3 were also regarded as positive.
Total protein was extracted from frozen tissues or cell samples. Protein concentrations were tested via a BCA protein-assay. Protein samples were separated on 10% SDS-PAGE gel. The separated proteins were transferred to Immun-Blot PVDF membrane (Bio-Rad) using a wet transfer system (Bio-Rad). The membrane was blocked with 5% skimmed milk and then incubated with primary antibody (XRCC1, 1:500, proteintech, China) at 4°C overnight, followed by incubation with HRP-linked anti-rabbit IgG (Merck Millipore) in a dilution of 1: 10000 for 1 h at room temperature.
Trizol reagent (Beijing Dingguo Changsheng Biotech, Co., Ltd., China) was applied to extract total RNA. The RNA was reverse-transcribed to cDNA by the PrimeScript RT reagent Kit (Takara Biomedical Tech, Co., Ltd., China). The cDNA was subjected to qRT-PCR using SYBR Premix Ex Taq II (Takara, Co., Ltd., China) and the assay was performed on the CFX connect system (Bio-Rad Co., Ltd., United States). GAPDH was used as an internal control. The primers were synthesized from Tsingke Biological Technology Co., (Changsha, Hunan, China), and sequences of primers were listed as followed: XRCC1: Forward 5′-CCTTTGGCTTGAGTTTTGTACG-3′, Reverse 5′-CCTCCTTCACACGGAACTGG-3′; GAPDH: Forward 5′-ATGACCACAGTCCATGCCATCA-3′, Reverse 5′-TTACTCCTTGGAGGCCATGTAG-3′.
The human gallbladder cancer cell line GBC-SD was obtained from the Cell Bank of the Chinese Academy of Sciences (Shanghai, China). Cells were cultured in RPMI-1640 (Hyclone, United States) supplemented with 10% fetal bovine serum (Gibco, Grand Island, NY, United States), Penicillin 100 U/ml and Streptomycin 100 ug/ml (Beyotime, China) in humidified atmosphere at 37°C and 5% CO2.
For magnetic cell sorting, cells were labeled with CD133 microbeads and sorted using the Miltenyi Biotec CD133 Cell Isolation Kit according to the manufacturer’s protocols (Miltenyi Biotec, Germany). Magnetic separation was performed twice to obtain high purity of CD133+cells. The purity of sorted cells was evaluated by flow cytometry with a FACS Calibur machine after labeling with phycoerythrin (PE)-conjugated anti-human CD133 antibody (Biolegend, United States).
XRCC1 shRNA and negative control shRNA were purchased from GeneChem (Shanghai, China). XRCC1 shRNA or negative control shRNA were mixed with RPIM-1640 (Hyclone, United States) and Lip2000 (Invitrogen, United States), and then incubated at room temperature for 20 min. Approximately 2 × 105 CD133+cells were plated in 6-well plates, followed by treating them with the transfection mixture and incubated at 37°C with 5% CO2. Cells were harvested at 6 h post-transfection for further studies.
The proliferation of CD133+cells transfected with control shRNA or XRCC1 shRNA was detected by use of Cell Counting Kit-8 (CCK8) (DOJINDO, Japan). Cells were seeded into 96-well culture plates at a density of 1 × 104 cells/100 ul. Four wells of each group were detected every day. At the end of each experiment, CCK-8 solution was added to each well, and the cultures were incubated at 37°C for 4 h. Then, the cultures were detected by use of a microplate reader.
Cell migration assays were performed in a 24-well Transwell plate (Corning, United States). Cells in serum-free medium (1 × 105 cells) was added to the upper chamber. Complete medium was added to the bottom wells of the chamber. After 48 h of incubation at 37°C, the cells that did not migrate were removed from the upper face of the filters. The number of cells migrating to the lower face was counted after fixed with 4% formaldehyde and stained with 0.5% crystal violet. The number of cells was counted under a microscope. The cell invasion assay was essentially the same as the migration assays, except that the membrane filters were coated with Matrigel (Becton, Dickinson and Company, United States).
Cells transfected with control shRNA or XRCC1 shRNA were cultured in a 6-well plate. After 48 h, cells were harvested by trypsinization, washed twice with PBS, and then stained with annexin V-APC (APC) (NanJing KeyGen Biotech, Co., Ltd., China) and propidium iodide (PI) (NanJing KeyGen Biotech, Co., Ltd., China) to detect cell apoptosis. Samples were immediately detected in the flow cytometer. This method can distinguish the cells in early (APC+/PI-) and late (APC+/PI+) apoptosis.
CD133+cells transfected with XRCC1 shRNA or negative control shRNA were cultured in 96 well plates (1 × 104 cells/well) overnight. On the second day, the cells were treated with 5-Fluorouracil (5-FU, final concentration of 0.1 mg/L) (APExBIO, United States) (Paschall et al., 2016). After 72 h, CCK-8 solution was added to each well, and the cultures were incubated at 37°C for 4 h. Then, the cultures were detected by use of a microplate reader.
CD133+cells transfected with XRCC1 shRNA or negative control shRNA were cultured in 6 well plates (2 × 105 cells/well) overnight. On the second day, the cells were treated with 5-Fluorouracil (5-FU, final concentration of 0.1 mg/L). After 72 h, cells were harvested to assess cell apoptosis by flow cytometry as above.
Data was analyzed using SPSS 13.0. The relationship between XRCC1 expression and clinicopathological factors was analyzed using χ2 or Fisher’s exact test. Kaplan-Meier and Log-rank test were used for univariate survival analysis. Cox proportional hazards model was used for univariate and multivariate analysis. A P < 0.05 was considered as statistical significance.
Among the 69 SC/ASC samples, 44 were collected from female patients and patient ages ranged from 35 to 80 (53.8 ± 10.2) years. Among the 146 AC patients, 85 were female with an age range of 33 to 78 (52.4 ± 9.6) years. The detail clinicopathological information of the 146 SC/ASC patients and the 69 AC patients was presented in Table 1. Briefly, among the 69 SC/ASCs, the squamous cell component presented well-differentiated in 19 (27.5%), moderately differentiated in 33 (47.8%), and poorly differentiated in 17 (24.6%). The 146 ACs consisted of 51 well-differentiated types (34.9%), 54 moderately differentiated types (37.0%) and 41 poorly differentiated types (28.1%). Among the SC/ASC patients, invasion to surrounding tissues and organs was observed in 45 patients (65.2%); 42 (60.7%) occurred regional lymph node metastasis; and 38 (55.1%) existed gallstones. Among the 146 AC patients, 74 (50.7%) occurred invasion; 66 (45.2%) presented regional lymph node metastasis; and 68 (46.6%) had gallstones. According to tumor-node-metastasis (TNM) staging, 29 SC/ASCs and 40 SC/ASCs stage I + II and stage III + IV, respectively. Among the 146 ACs, 77 were in a stage of I or II and 69 were in a stage of III or IV. Among all patients, 27 SC/ASC patients and 75 AC patients received radical surgery; 28 SC/ASC patients and 50 AC patients received palliative surgery; 14 SC/ASC patients and 21 AC patients only underwent biops.
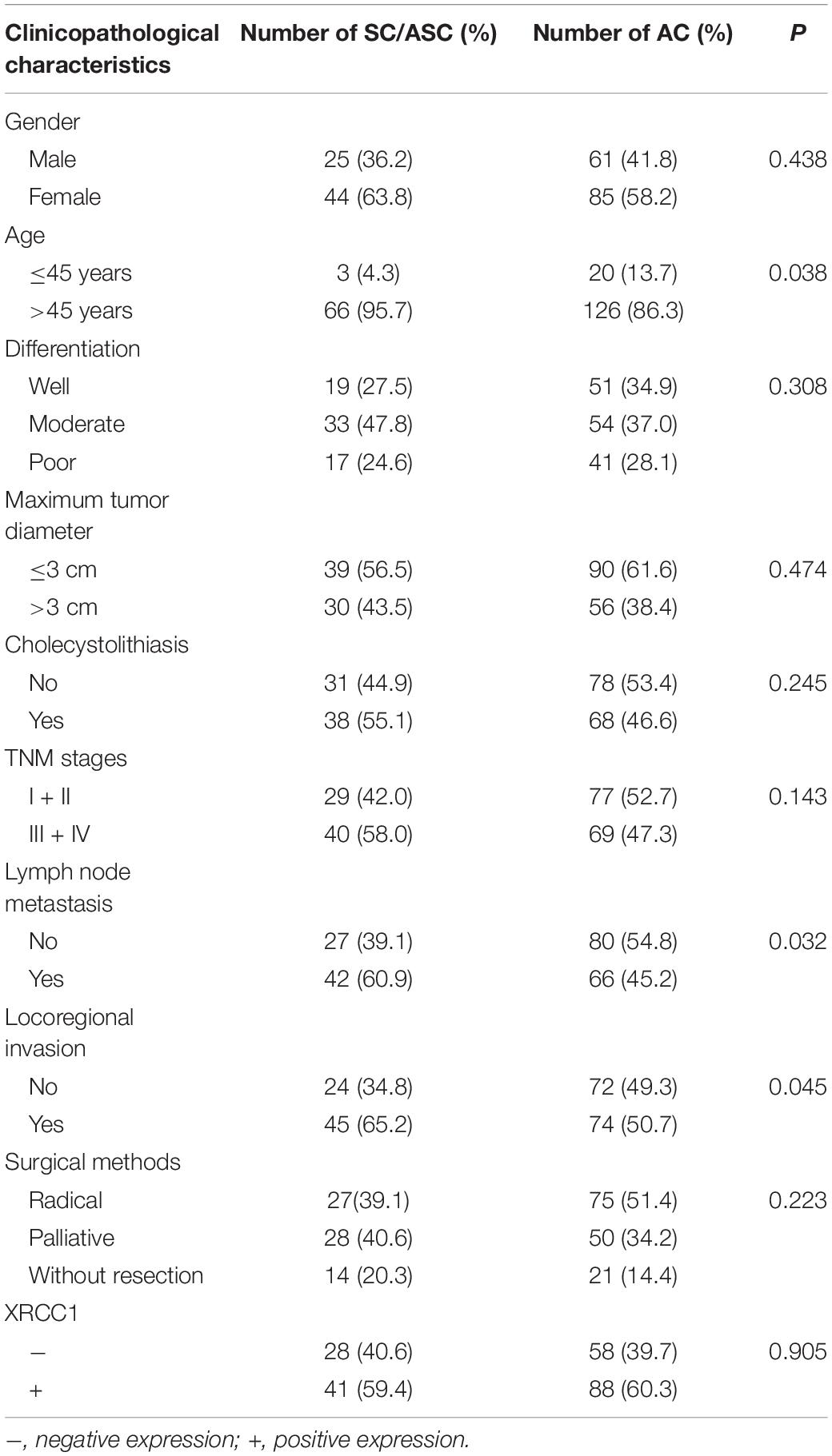
Table 1. Comparison of gallbladder SC/ASC and AC clinicopathological characteristics and XRCC1 expression status.
To evaluate the expression of XRCC1 in GBC tissues and corresponding adjacent non-tumor tissues, qRT-PCR and western blot were performed. The results demonstrated that XRCC1 expression in GBC tissues was significantly higher than adjacent non-tumor tissues both in mRNA and protein levels (Figures 1A,B).
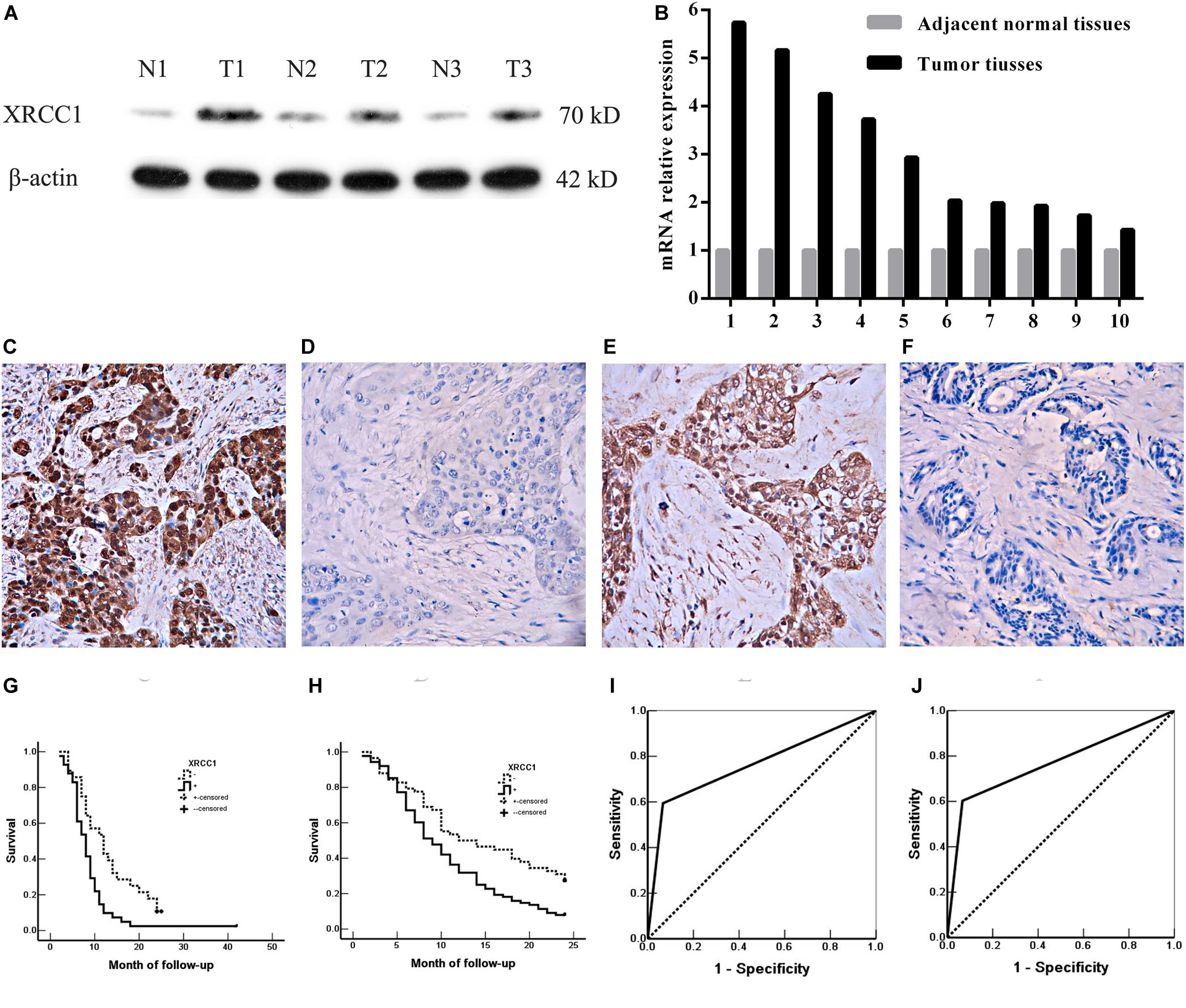
Figure 1. XRCC1 expression is up-regulated in GBC and associated with poor prognosis. (A) XRCC1 expression was analyzed by western blot. (B) XRCC1 expression was assessed by qRT-PCR. (C) Positive expression of XRCC1 in moderately differentiated ASC, ×200. (D) Negative expression of XRCC1 in moderately differentiated SC, ×200. (E) Positive expression of XRCC1 in moderately differentiated AC, ×200. (F) Negative expression of XRCC1 in well differentiated AC, ×200. (G) Kaplan-Meier curves for SC/ASC patients with positive and negative XRCC1 expression. (H) Kaplan-Meier curves for AC patients with positive and negative XRCC1 expression. (I) ROC of diagonal segments was produced by ties of XRCC1 in SC/ASC. (J) ROC of diagonal segments was produced by ties of XRCC1 in AC.
We then assessed XRCC1 expression in gallbladder cancer tissues (including 69 SC/ASCs and 146 ACs) and gallbladder epithelium with chronic cholecystitis by immunohistochemistry. The majority of XRCC1 positive-reaction was localized in the cytoplasm of the SC/ASC (Figure 1C) and AC (Figure 1E). The representative images of XRCC1 negative expression in SC/ASC and AC were seen in Figure 1D and Figure 1F, respectively. The staining positive rate was significantly higher in SC/ASC (59.4%) and AC (60.3%) than gallbladder epithelium with chronic cholecystitis (6.7%, P < 0.01). The epithelium of chronic cholecystitis with high XRCC1 expression showed moderate to severe dysplasia. This suggested that XRCC1 may be a biomarker to evaluate the pre-malignant changes.
As showed in Table 1, the percentage of cases with a patient age over 45 years, lymph node metastasis and invasion was significantly higher in SC/ASC compared with AC (all P < 0.05). However, there was a non-significant difference between SC/ASC and AC in other clinicopathological features including tumor differentiated degree, tumor size, TNM stages, receiving surgical methods, and XRCC1 positive expression (all P > 0.05, Table1).
We further evaluated the clinicopathological significance of XRCC1 expression in SC/ASC and AC patients. We found that XRCC1 positive expression was associated with several poor clinicopathological features of gallbladder cancer. In SC/ASC, XRCC1 positive expression was positively correlated with lymph node metastasis, invasion, and only receiving biopsy (all P < 0.05, Table 2). Similarly, XRCC1 positive expression was positively associated with large tumor size (>3 cm), lymph node metastasis, invasion, late TNM stages (III + IV), only receiving biopsy in AC (all P < 0.05, Table 2).
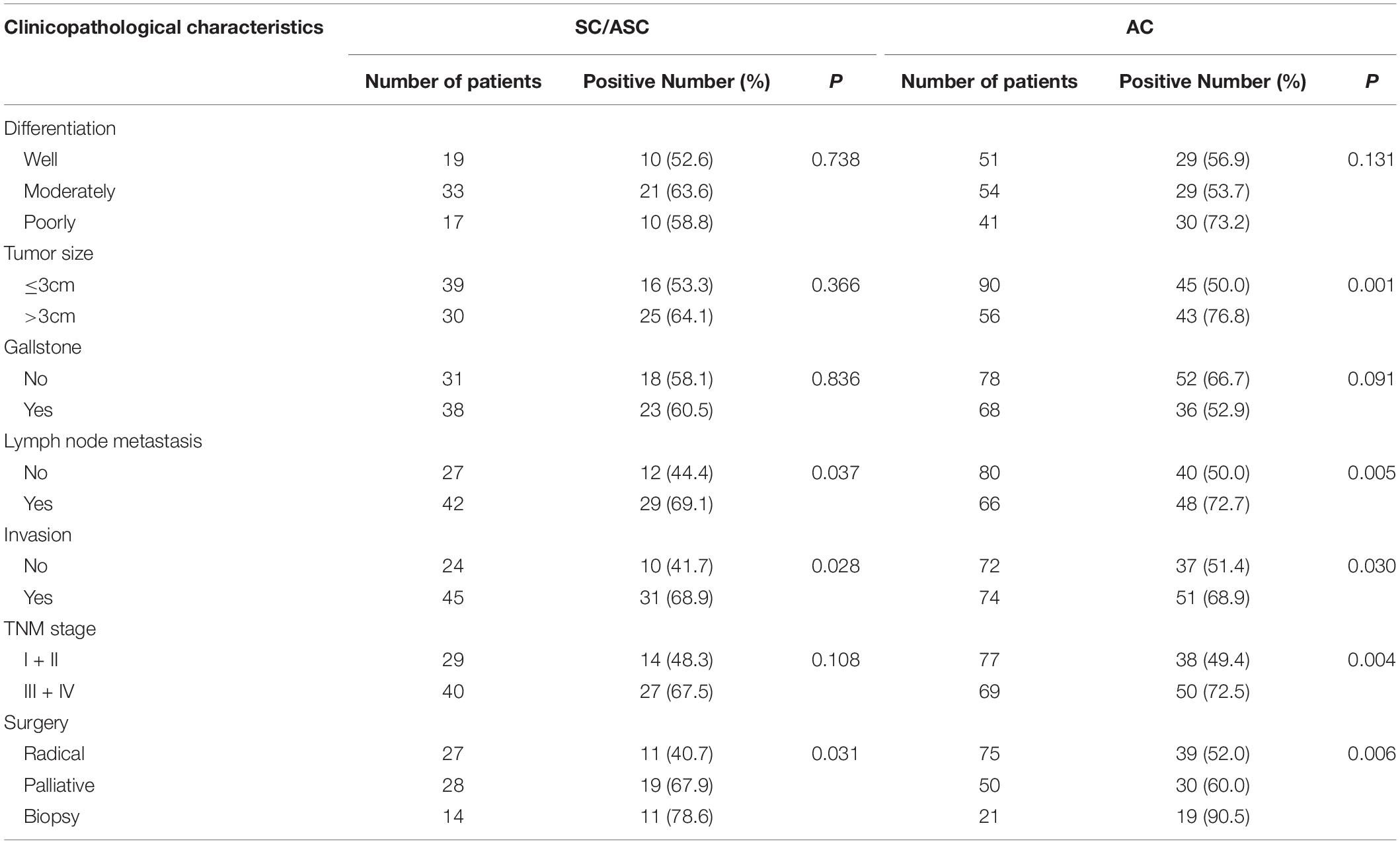
Table 2. Correlations of XRCC1 expression with the clinicopathological characteristics of gallbladder SC/ASC and AC.
Gallbladder cancer patients (both AC/ASC and AC) in XRCC1 positive expression group had significantly shorter average survival time than patients in the negative expression group (all P < 0.01, Table 3). The Kaplan-Meier survival curves demonstrated that patients with XRCC1 positive expression had a poor overall survival than patients with XRCC1 negative expression (Figures 1G,H). Moreover, univariate and multivariate analysis showed that XRCC1 positive expression was an independent risk factor for the overall survival of gallbladder SC/ASC and AC patients (Tables 4, 5). Finally, the receiver operating characteristic (ROC) curve was depicted to assess the diagnostic efficacy of XRCC1 expression in SC/ASC and AC. The AUC of XRCC1 expression in SC/ASC and AC was 0.764 (95%CI: 0.669–0.859) and 0.768 (95%CI: 0.689–0.847) respectively (Figures 1I,J). These results fully revealed that XRCC1 was closely related to poor survival and might be a novel independent prognosis biomarker for gallbladder SC/ASC and AC patients.
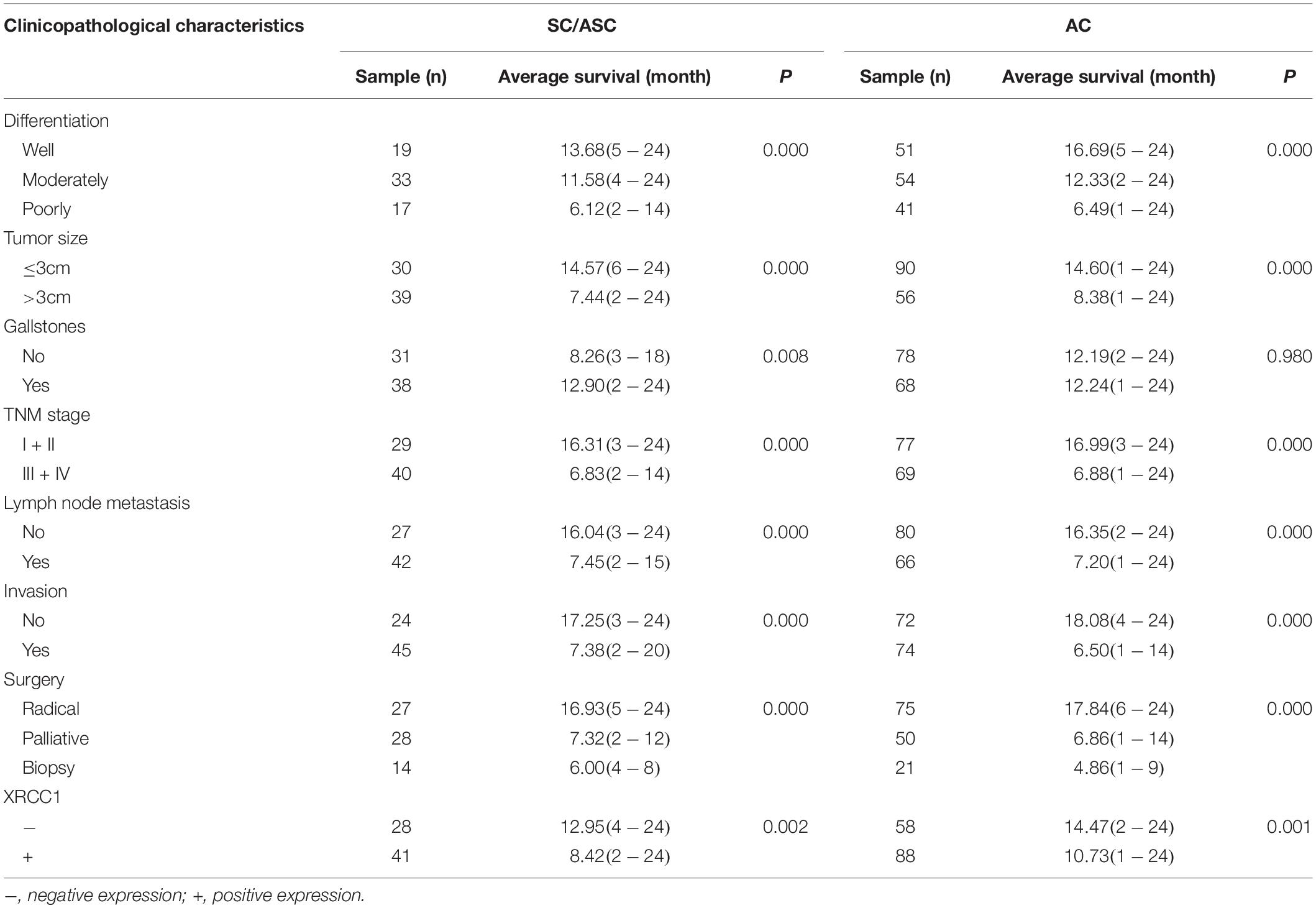
Table 3. Relationship between XRCC1 expression, clinicopathological characteristics and average survival of SC/ASC and AC patients.
Previous studies reported that both XRCC1 and CD133+cancer cells are related to tumor drug resistance so that we studied the role of XRCC1 in CD133+GBC-SD cells. CD133+GBC-SD cells were obtained from GBC-SD cells by CD133 magnetic bead sorting. We applied qRT-PCR and western blot to research XRCC1 expression in normal GBC-SD cells and CD133+GBC-SD cells. Compared with GBC-SD cells, XRCC1 mRNA and protein were overexpressed in CD133+GBC-SD cells (Figure 2). Based on previous studies, these results indicated that XRCC1 might affect the unique biological features of CD133+GBC-SD cells compared to normal GBC-SD cells, such as chemo-resistance.
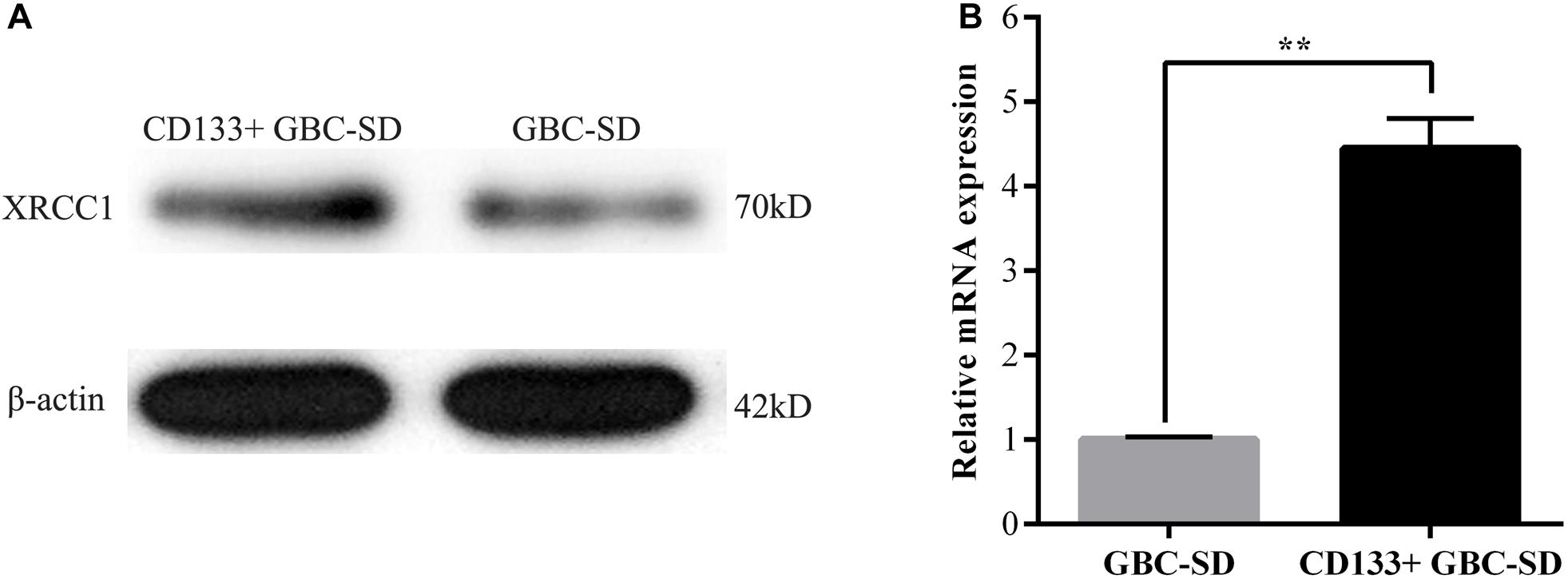
Figure 2. XRCC1 is up-regulated in CD133+ GBC-SD cells. (A) XRCC1 expression in GBC-SD cells and CD133+GBC-SD cells was analyzed by western blotting. (B) XRCC1 expression in GBC-SD cells and CD133+GBC-SD cells was assessed by qRT-PCR (**P < 0.01).
To further study the function of XRCC1 in CD133+GBC-SD cells, XRCC1 expression in cells was manipulated via short hairpin RNA (shRNA) knockdown. Three shRNAs (shRNA1, shRNA2, and shRNA3) were designed to knockdown XRCC1 expression in CD133+GBC-SD cells. After CD133+GBC-SD cells were infected with XRCC1-shRNA, the expression level of XRCC1 was tested by western blotting to evaluate the efficacy of shRNA knockdown. Among the three XRCC1-shRNAs, shRNA3 was the most effective one (Figure 3A) and was selected for further studies. To study the effect of XRCC1 knockdown on the proliferation, migration, invasion, and apoptosis of CD133+GBC-SD cells, CCK8 assay, transwell assay, and flow cytometry were performed. Our results showed that XRCC1 knockdown in CD133+GBC-SD cells had a non-significant impact on the ability of proliferation, migration, invasion, and apoptosis, compared with control-shRNA/CD133+GBC-SD cells (Figure 3).
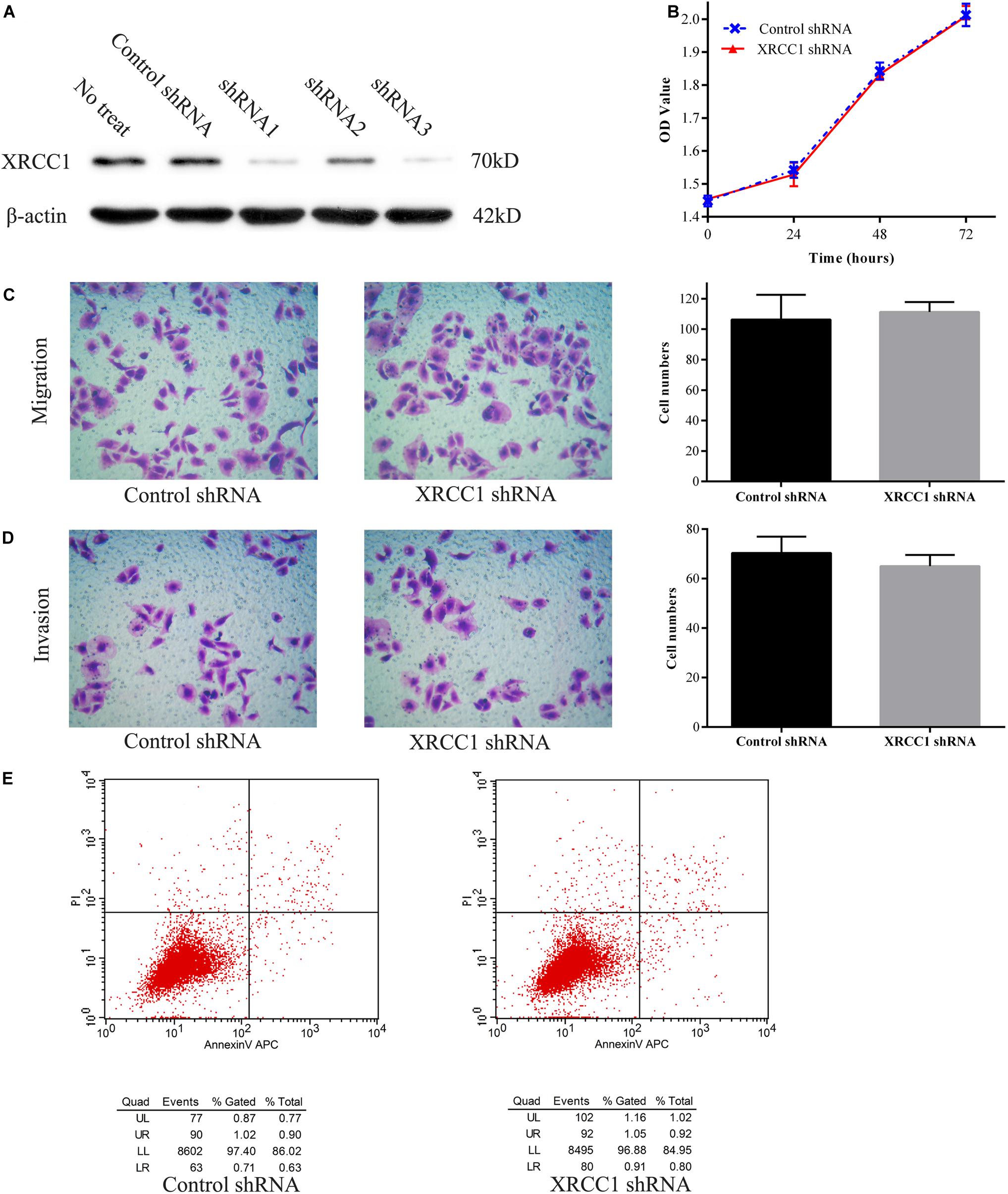
Figure 3. XRCC1 has no effect on proliferation, migration, invasion, and apoptosis of CD133+GBC-SD cells. (A) Western blot detected XRCC1 expression in CD133+GBC-SD cells transfected with different shRNAs. (B) Proliferation of XRCC1-shRNA/CD133+GBC-SD cells and control-shRNA/CD133+GBC-SD cells was examined by CCK8. (C) Transwell migration assay detected the migration capacity of XRCC1-shRNA/CD133+GBC-SD cells and control-shRNA/CD133+GBC-SD cells. (D) Transwell invasion assay detected the invasion capacity of XRCC1-shRNA/CD133+GBC-SD cells and control-shRNA/CD133+GBC-SD cells. (E) Flow cytometer detected the apoptosis capacity of XRCC1-shRNA/CD133+GBC-SD cells and control-shRNA/CD133+GBC-SD cells; UL: necrosis, UR: late apoptosis, LL: normal cells, LR: early apoptosis.
To explore the role of XRCC1 in CD133+GBC-SD cells drug resistance, 5-FU was used to treat control-shRNA/CD133+GBC-SD cells and XRCC1-shRNA/CD133+GBC-SD cells. After treated with 5-FU (0.1 mg/L) for 72 h, CCK8 assay was performed to assess the cell totality of every group. Results showed that in CCK8 assay, XRCC1-shRNA/CD133+GBC-SD had a lower absorbance compared to control-shRNA/CD133+GBC-SD, which suggested that XRCC1 could promote CD133+GBC-SD cell resistance to 5-FU (Figure 4A). To further validate the results of CCK8 assay, flow cytometry was performed. Under 5-FU treated 72 h, flow cytometry revealed that cell necrosis and apoptosis were significantly increased in XRCC1-shRNA/CD133+GBC-SD compared to control-shRNA/CD133+GBC-SD (Figure 4B). Thus, our results indicated that XRCC1 might promote CD133+GBC-SD cells resistance to 5-FU through inhibiting cell necrosis and apoptosis.
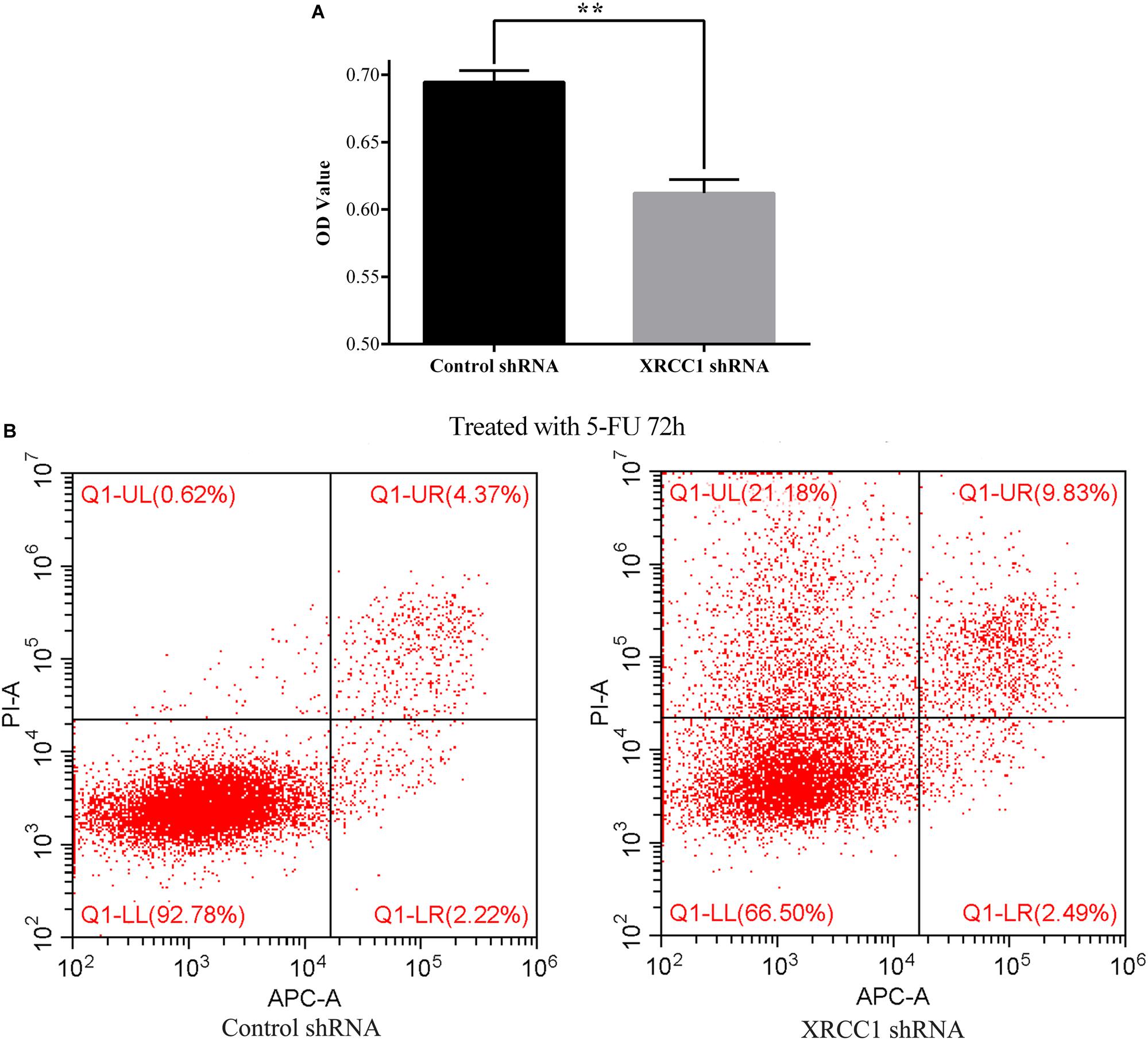
Figure 4. XRCC1 is related to CD133+GBC-SD cells resistance to 5-FU. (A) Cell viability of cells treated with 5-FU 72 h was assessed by CCK8 (**P < 0.01). (B) Apoptosis of cells treated with 5-FU 72 h was examined by Flow cytometer; Q1-UL: necrosis cells, Q1-UR: late apoptosis cells, Q1-LL: normal cells, Q1-LR: early apoptosis cells.
GBC is an aggressive malignant of the biliary tree and consists of several pathological subtypes including AC and SC/ASC. In comparison of AC, the incidence rate of gallbladder SC/ASC is relatively rare and its clinicopathological features remain to be further elucidated. Currently, most reports investigated SC/ASC based on individual cases or small case samples. As far as we know, the 69 SC/ASC cases that we included in this study are relatively large samples in current clinical studies on gallbladder SC/ASC, which could provide more detail clinicopathological knowledge about SC/ASC. In the present study, we found that SC/ASC accounted for 5.5% of GBC and the occurring rate of lymph node metastasis and invasion was significantly higher in SC/ASC than AC, which was consistent with previous reports (Kim et al., 2011; Roa et al., 2011; Samuel et al., 2018). In agreement with previous researches (Chan et al., 2007; Kim et al., 2011), our results also showed that gallbladder SC/ASC and AC had similar clinicopathological features such as tumor differentiated degree, tumor size, the existence of gallstone, TNM stage, and XRCC1 expression.
Nowadays, the prognosis of GBCs remains extremely poor. In this study, our data revealed that lymph node metastasis, invasion, large tumor size, and advanced TNM stages were independent risk factors for patient’s survival, and radical surgery could significantly prolong the mean survival time of patients in SC/ASC and AC. These results suggested that early diagnosis was very important for improving the clinical prognosis of GBC. Thus, it is extremely vital to discover early specific diagnostic biomarkers and explore the reason why GBC resists to chemotherapy. Previous works have demonstrated that XRCC1 is associated with tumor resistance to chemotherapy and radiotherapy, carcinogenesis, and tumor progression (Sak et al., 2005; Hanssen-Bauer et al., 2012; Xu et al., 2014; Li et al., 2018). CD133+cancer cells are a small subgroup of tumor cells and related to tumor resistance to chemotherapy and radiotherapy (Zhang et al., 2010; Desai et al., 2014; Vincent et al., 2014; Kanwal et al., 2018). Thus, we further studied the clinicopathological and prognostic significance of XRCC1 in gallbladder SC/ASC and AC, and evaluated the biological role of XRCC1 in CD133+ GBC-SD cells.
As a DNA repair gene, XRCC1 is involved in tumorigenesis, progression, and poor prognosis of many human cancer types. In this study, we observed that XRCC1 expression was up-regulated in GBC compared with non-tumor tissues, which was consistent with previous studies where XRCC1 was overexpression in ovarian cancer and head and neck squamous cell cancer (Ang et al., 2011; Abdel-Fatah et al., 2013). On the contrary, several reports showed that XRCC1 was down-regulated in glioma, bladder cancer, pancreatic cancer, and gastric cancer (Crnogorac-Jurcevic et al., 2002; Sak et al., 2005; Wang et al., 2012; Mei et al., 2019). This contradiction may be owed to the organ specificity. Furthermore, we found that the epithelium of chronic cholecystitis with high XRCC1 expression showed moderate to severe dysplasia, suggesting that XRCC1 may be involved in the processes that benign lesions evolve into GBC. Thus, we further evaluated the clinicopathological and prognostic significance of XRCC1 in gallbladder SC/ASC and AC. Our data demonstrated that XRCC1 positive expression was significantly related to lymph node metastasis, invasion, and poor prognosis, which was consistent with previous studies (Ang et al., 2011; Abdel-Fatah et al., 2013; Mian et al., 2016). Moreover, cox univariate and multivariate analysis further showed that XRCC1 positive expression was an independent risk factor for the overall survival of SC/ASC and AC. The AUC of XRCC1 indicated that the expression of XRCC1 might have potential clinicopathological diagnostic significance in SC/ASC and AC. These results suggested that XRCC1 might be involved in carcinogenesis and development of GBC.
Previous studies have demonstrated that XRCC1 plays a role in regulating cell biological features such as proliferation, migration, invasion, and drug resistance in several human cancer cell lines (Xu et al., 2014; Meng et al., 2017; Li et al., 2018; Mei et al., 2019). However, there is no study reporting the function of XRCC1 in gallbladder cancer cells. Herein, we firstly studied the biological role of XRCC1 in CD133+GBC-SD cells. Unexpectedly, the functional experiments revealed that knockdown of XRCC1 had no significant effect on the ability of proliferation, migration, invasion, and apoptosis in CD133+GBC-SD cells, which was inconsistent with previous researches (Li et al., 2018; Mei et al., 2019). This inconsistency may be caused by cell specificity. Additionally, we found that XRCC1 was up-regulated in CD133+GBC-SD cells compared with GBC-SD cells, indicating that XRCC1 might be associated with unique biological features of CD133+cancer cells, such as chemo-resistance. Therefore, we further investigated the impact of XRCC1 on CD133+GBC-SD cells resistance to 5-FU. As we suspected, our results showed that XRCC1 were contributed to the resistance of CD133+GBC-SD cells to 5-FU via inhibiting cell necrosis and apoptosis, which was in accordance with previous studies (Abdel-Fatah et al., 2013; Xu et al., 2014). Thus, XRCC1 may promote GBC resistance to chemotherapy, which needs further studies to validate and explore potential molecular mechanism. Thus, we speculated that XRCC1 might be a promising target to improve the sensitivity of GBC to chemotherapy.
In conclusion, this study demonstrated that XRCC1 was overexpression in gallbladder cancer tissues. XRCC1 positive expression was associated with aggressive clinicopathological features and poor prognosis of gallbladder SC/ASC and AC. Moreover, XRCC1 was related to the chemo-resistance of CD133+GBC-SD cells to 5-FU. Thus, XRCC1 may be a promising predictive biomarker and a potential therapeutic target for GBC.
The datasets generated for this study are available on request to the corresponding author.
This study was approved by the Ethics Committee for Human Research, Central South University and was carried out in accordance with Declaration of Helsinki.
ZW, RL, and YZ carried out studies and wrote the manuscript. ZY, XM, and RL designed the study and revised the manuscript. ZY and XM performed the statistical analysis. DL, YY, and QZ collected specimens and experimental materials. All authors read and approved the final manuscript.
This work was supported by The National Natural Science Foundation of China (81472738); Natural Science Foundation of Hunan Province, China (2019JJ10002); and Hunan Provincial Key Research and Development Program (2019SK2042).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Abdel-Fatah, T., Sultana, R., Abbotts, R., Hawkes, C., Seedhouse, C., Chan, S., et al. (2013). Clinicopathological and functional significance of XRCC1 expression in ovarian cancer. Int. J. Cancer 132, 2778–2786. doi: 10.1002/ijc.27980
Ang, M. K., Patel, M. R., Yin, X. Y., Sundaram, S., Fritchie, K., Zhao, N., et al. (2011). High XRCC1 protein expression is associated with poorer survival in patients with head and neck squamous cell carcinoma. Clin. Cancer Res. 17, 6542–6552.
Chan, K. M., Yu, M. C., Lee, W. C., Jan, Y. Y., and Chen, M. F. (2007). Adenosquamous/squamous cell carcinoma of the gallbladder. J. Surg. Oncol. 95, 129–134. doi: 10.1002/jso.20576
Crnogorac-Jurcevic, T., Efthimiou, E., Nielsen, T., Loader, J., Terris, B., Stamp, G., et al. (2002). Expression profiling of microdissected pancreatic adenocarcinomas. Oncogene 21, 4587–4594. doi: 10.1038/sj.onc.1205570
Desai, A., Webb, B., and Gerson, S. L. (2014). CD133+ cells contribute to radioresistance via altered regulation of DNA repair genes in human lung cancer cells. Radiother. Oncol. 110, 538–545. doi: 10.1016/j.radonc.2013.10.040
Hanssen-Bauer, A., Solvang-Garten, K., Akbari, M., and Otterlei, M. (2012). X-ray repair cross complementing protein 1 in base excision repair. Int. J. Mol. Sci. 13, 17210–17229. doi: 10.3390/ijms131217210
Henley, S. J., Weir, H. K., Jim, M. A., Watson, M., and Richardson, L. C. (2015). Gallbladder cancer incidence and mortality, United States 1999-2011. Cancer Epidemiol. Biomarkers. Prev. 24, 1319–1326. doi: 10.1158/1055-9965.EPI-15-0199
Horgan, A. M., Amir, E., Walter, T., and Knox, J. J. (2012). Adjuvant therapy in the treatment of biliary tract cancer: a systematic review and meta-analysis. J. Clin. Oncol. 30, 1934–1940. doi: 10.1200/JCO.2011.40.5381
Kanwal, R., Shukla, S., Walker, E., and Gupta, S. (2018). Acquisition of tumorigenic potential and therapeutic resistance in CD133+ subpopulation of prostate cancer cells exhibiting stem-cell like characteristics. Cancer Lett. 430, 25–33. doi: 10.1016/j.canlet.2018.05.014
Kim, W. S., Jang, K. T., Choi, D. W., Choi, S. H., Heo, J. S., You, D. D., et al. (2011). Clinicopathologic analysis of adenosquamous/squamous cell carcinoma of the gallbladder. J. Surg. Oncol. 103, 239–242. doi: 10.1002/jso.21813
Li, Q., Ma, R., and Zhang, M. (2018). XRCC1 rs1799782 (C194T) polymorphism correlated with tumor metastasis and molecular subtypes in breast cancer. Onco. Targets Ther. 11, 8435–8444. doi: 10.2147/OTT.S154746
Liu, J. Y., Liu, Q. M., and Li, L. R. (2015). Association of GSTP1 and XRCC1 gene polymorphisms with clinical outcomes of patients with advanced non-small cell lung cancer. Genet. Mol. Res. 14, 10331–10337. doi: 10.4238/2015.August.28.19
Mei, P. J., Bai, J., Miao, F. A., Li, Z. L., Chen, C., Zheng, J. N., et al. (2019). Relationship between expression of XRCC1 and tumor proliferation, migration, invasion, and angiogenesis in glioma. Invest. New Drugs 37, 646–657. doi: 10.1007/s10637-018-0667-669
Meng, Q., Wang, S., Tang, W., Wu, S., Gao, N., Zhang, C., et al. (2017). XRCC1 mediated the development of cervival cancer through a novel Sp1/Krox-20 swich. Oncotarget 8, 86217–86226. doi: 10.18632/oncotarget.21040
Mian, M., McNamara, M. G., Doherty, M., Hedley, D., Knox, J. J., and Serra, S. (2016). Predictive and prognostic values of ERCC1 and XRCC1 in biliary tract cancers. J. Clin. Pathol. 69, 695–701. doi: 10.1136/jclinpath-2015-203397
Paschall, A. V., Yang, D., Lu, C., Redd, P. S., Choi, J. H., Heaton, C. M., et al. (2016). CD133+CD24lo defines a 5-Fluorouracil-resistant colon cancer stem cell-like phenotype. Oncotarget 7, 78698–78712. doi: 10.18632/oncotarget.12168
Reid, K. M., Ramos-De, L. M. A., and Donohue, J. H. (2007). Diagnosis and surgical management of gallbladder cancer: a review. J. Gastrointest. Surg. 11, 671–681. doi: 10.1007/s11605-006-0075-x
Roa, J. C., Tapia, O., Cakir, A., Basturk, O., Dursun, N., Akdemir, D., et al. (2011). Squamous cell and adenosquamous carcinomas of the gallbladder: clinicopathological analysis of 34 cases identified in 606 carcinomas. Mod. Pathol. 24, 1069–1078. doi: 10.1038/modpathol.2011.68
Sak, S. C., Harnden, P., Johnston, C. F., Paul, A. B., and Kiltie, A. E. (2005). APE1 and XRCC1 protein expression levels predict cancer-specific survival following radical radiotherapy in bladder cancer. Clin. Cancer Res. 11, 6205–6211. doi: 10.1158/1078-0432.CCR-05-0045
Sakano, S., Ogawa, S., Yamamoto, Y., Nishijima, J., Miyachika, Y., Matsumoto, H., et al. (2013). ERCC1 and XRCC1 expression predicts survival in bladder cancer patients receiving combined trimodality therapy. Mol. Clin. Oncol. 1, 403–410. doi: 10.3892/mco.2013.85
Samuel, S., Mukherjee, S., Ammannagari, N., Pokuri, V. K., Kuvshinoff, B., Groman, A., et al. (2018). Clinicopathological characteristics and outcomes of rare histologic subtypes of gallbladder cancer over two decades: a population-based study. PLoS One 13:e0198809. doi: 10.1371/journal.pone.0198809
Sharma, A., Sharma, K. L., Gupta, A., Yadav, A., and Kumar, A. (2017). Gallbladder cancer epidemiology, pathogenesis and molecular genetics: recent update. World J. Gastroenterol. 23, 3978–3998. doi: 10.3748/wjg.v23.i22.3978
Sicklick, J. K., Fanta, P. T., Shimabukuro, K., and Kurzrock, R. (2016). Genomics of gallbladder cancer: the case for biomarker-driven clinical trial design. Cancer Metastasis Rev. 35, 263–275. doi: 10.1007/s10555-016-9602-9608
Thompson, L. H., and West, M. G. (2000). XRCC1 keeps DNA from getting stranded. Mutat. Res. 459, 1–18. doi: 10.1016/s0921-8777(99)00058-0
Tudek, B. (2007). Base excision repair modulation as a risk factor for human cancers. Mol. Aspects Med. 28, 258–275. doi: 10.1016/j.mam.2007.05.003
Vincent, Z., Urakami, K., Maruyama, K., Yamaguchi, K., and Kusuhara, M. (2014). CD133-positive cancer stem cells from Colo205 human colon adenocarcinoma cell line show resistance to chemotherapy and display a specific metabolomic profile. Genes Cancer 5, 250–260. doi: 10.18632/genesandcancer.23
Wang, S., Wu, X., Chen, Y., Zhang, J., Ding, J., Zhou, Y., et al. (2012). Prognostic and predictive role of JWA and XRCC1 expressions in gastric cancer. Clin. Cancer Res. 18, 2987–2996. doi: 10.1158/1078-0432.CCR-11-2863
Wood, R. D., Mitchell, M., Sgouros, J., and Lindahl, T. (2001). Human DNA repair genes. Science 291, 1284–1289. doi: 10.1126/science.1056154
Wu, Z. C., Xiong, L., Wang, L. X., Miao, X. Y., Liu, Z. R., Li, D. Q., et al. (2017). Comparative study of ROR2 and WNT5a expression in squamous/adenosquamous carcinoma and adenocarcinoma of the gallbladder. World J. Gastroenterol. 23, 2601–2612. doi: 10.3748/wjg.v23.i14.2601
Xu, W., Wang, S., Chen, Q., Zhang, Y., Ni, P., Wu, X., et al. (2014). TXNL1-XRCC1 pathway regulates cisplatin-induced cell death and contributes to resistance in human gastric cancer. Cell Death Dis. 5:e1055. doi: 10.1038/cddis.2014.27
Keywords: XRCC1, gallbladder cancer, prognosis, clinicopathological significance, chemo-resistance
Citation: Wu Z, Miao X, Zhang Y, Li D, Zou Q, Yuan Y, Liu R and Yang Z (2020) XRCC1 Is a Promising Predictive Biomarker and Facilitates Chemo-Resistance in Gallbladder Cancer. Front. Mol. Biosci. 7:70. doi: 10.3389/fmolb.2020.00070
Received: 19 February 2020; Accepted: 30 March 2020;
Published: 24 April 2020.
Edited by:
Yongjun Wei, Zhengzhou University, ChinaCopyright © 2020 Wu, Miao, Zhang, Li, Zou, Yuan, Liu and Yang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Rushi Liu, bGl1cnVzaGlAaHVubnUuZWR1LmNu; Zhulin Yang, eWFuZ3podWxpbjhAY3N1LmVkdS5jbg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.