- Department of Biological Sciences, The University of Illinois at Chicago, Chicago, IL, United States
The alternative streptococcal σ-factor and master competence regulator, σX, stimulates transcription from competence promoters, in vitro. As the only known alternative σ-factor in streptococci, σX expression is tightly controlled in each species and has a specific physiological role. Pneumococcal transformation also requires the DNA binding activity of ComW, a known σX activator and stabilizer. Mutations to the housekeeping σ factor, σA, partially alleviate the ComW requirement, suggesting that ComW is a key player in the σ factor swap during the pneumococcal competence response. However, there is no evidence of a direct ComW – RNA polymerase interaction. Furthermore, if and how ComW functions directly at combox promoters is still unknown. Here we report that a DNA-binding ComW variant, ComΔ6, can stimulate transcription from σX promoters in vitro.
Introduction
Streptococci are Gram-positive commensal cocci for a wide range of animal hosts, including humans. Streptococcus pneumoniae (pneumococcus), an inhabitant of the human nasopharynx, is a causative agent of pneumonia and meningitis in children, the elderly, and immune-compromised individuals. Natural genetic transformation (NGT) was discovered in the pneumococcus (Griffith, 1928; Avery and MacLeod, 1944), and subsequently the genes required for transformation have been found in the genomes of streptococci from all species groups (Håvarstein, 2010). Their genomes encode transcriptional regulators and apparatuses that enable exogenous DNA uptake and genetic recombination. This ability yields highly diverse genomes, making streptococci well equipped for adaptation, as seen with frequent capsular switching and the rapid spread of genes that mediate antibiotic resistance (Andam and Hanage, 2015).
Pneumococci primed for transformation must develop competence, a transient state marked by a shift in both transcriptomic and proteomic profiles, including two waves of new gene transcription, yielding early and late competence gene groups (Peterson et al., 2000; Fontaine et al., 2010; Gao et al., 2014; Khan et al., 2016, 2017). The link between early gene expression and late gene expression is the production of the alternative σ-factor, SigX (σX), a member of the σ70 family of proteins (Lee and Morrison, 1999, 2000; Luo and Morrison, 2003). All bacteria produce the principal σ factor σA, responsible for most gene transcription, and many produce multiple alternative σ factors that promote specific cellular responses, however, σX is the only alternative σ factor produced in pneumococcus (Håvarstein, 2010), and sigX expression is strictly linked to competence development.
Streptococci utilize tightly regulated quorum-sensing systems to coordinate σX- mediated competence, as well as other group behaviors (Cheng et al., 1997; Peterson et al., 2004; Mashburn-Warren et al., 2010, 2012). Species of the angionsus and mitis groups use the ComCDE pathway (Cheng et al., 1997), an auto-regulated two-component signal transduction system (TCSTS), that responds to the Competence Stimulating Peptide (CSP) (Håvarstein et al., 1995). Much of our understanding of streptococcal transformation comes from initial work done with the ComCDE system in pneumococcus.
During exponential growth, the σA-RNA polymerase complex (σA holo-enzyme) basally transcribes comC leading to production of the 41 aa pro-peptide, ComC, 41 (Håvarstein et al., 1995). The ComC pro-peptide is simultaneously cleaved and exported to form mature CSP by the ABC-transporter, ComAB (comAB). Extracellular CSP is sensed by the histidine-kinase receptor, ComD, resulting in auto-phosphorylation and activation of its cognate response regulator, ComE, via a phospho-relay event (Håvarstein et al., 1996; Martin et al., 2010, 2012). Activated ComE promotes transcription of comCDE and other early competence genes (Ween et al., 1999). ComE-mediated transcription triggers robust competence among the cell population in a positive-feedback auto-regulatory loop. This response culminates in ComE dependent production of σX (Ween et al., 1999; Peterson et al., 2004). During competence, the σX – RNA polymerase (σX holo-enzyme) transcribes from the competence specific combox promoter, directly linking ComCDE quorum sensing to the expression of the transformation regulon (Peterson et al., 2004). Genes under control of the σX holo-enzyme include all genes that are required for DNA uptake and recombination.
Although σX is the master competence regulator required for late gene expression in pneumococcus, it is not sufficient for high transformation efficiencies. An additional ComE dependent early gene product, ComW, a 9.5 kDa DNA-binding protein (Luo et al., 2004; Inniss et al., 2019), is required for the strongest transformation phenotype possible. sigX is expressed independently of ComW (Luo et al., 2004; Piotrowski et al., 2009) and σX activity is dependent on the presence of ComW (Sung and Morrison, 2005). SigX’s requirement for ComW is supported by observations that mutants lacking ComW (ΔcomW) transcribe late genes and transform at levels 10- and 10,000-fold below that of wild type cells, respectively (Tovpeko and Morrison, 2014). Furthermore, ΔcomW cells have decreased σX levels (Sung and Morrison, 2005), and ComW and σX weakly interact in yeast-2-hybrid assays (Tovpeko et al., 2016), suggesting that they physically interact as protein partners in the cell. Interestingly, mutations to σA that presumably decrease its affinity for RNA polymerase (RNAP) alleviate the ComW requirement, indicating that ComW functions on or with the σX holo-enzyme. Beyond this, there is currently no evidence of a physical interaction between ComW and RNAP.
A structural model of ComW suggests that is structurally similar to known σ factors, and ComW binds to DNA, non-specifically (Inniss et al., 2019). DNA binding does not require residues 73RGFISC78, as demonstrated with the truncated variant, ComWΔ6, although ComWΔ6 producing cells transform at levels slightly below wild type cells (Inniss et al., 2019). Instead, the conserved 38LxYYLxR44 motif is important for DNA binding, and L42A and R44A mutations in the 38LxYYLxR44 motif decrease transformation efficiency in pneumococci (Inniss et al., 2019).
Although much is known about how σX functions in streptococcal species the details of how it facilitates combox promoter recognition and melting are not known. Moreover, how and why σX-mediated transcription depends on ComW in unknown. Here we show that ComW stimulates transcription of competence genes, in vitro.
Materials and Methods
Bacterial Strains, Culture Media, and Plasmids
Bacterial strains and plasmids used are listed in Table 1. Plasmids pNLI60, pNLI115, and pNLI116 were synthesized by GenScript. E. coli strains DH5α and BL21De3 were hosts for plasmid isolation and protein expression, respectively. For plasmid introduction, E. Coli strains were chemically transformed according to (Mandel and Higa, 1970). E. coli strains were cultured in lysogeny broth (LB) (Bertani, 1951). LB was prepared from 5 g bacto tryptone (Difco), 5 g NaCl (Fisher) 2.5 g of yeast extract (Difco) in 1 L H2O, sterilized for 20 min at 121°C, and supplemented with appropriate antibiotics and 1.5% agar, as needed. Ampicillin was used at 100 μg/mL, for growth of E. coli strains. Antibiotics were purchased from Sigma-Aldrich.
Expression and Purification of Pneumococcal Proteins From E. coli
Pneumococcal ComWΔ6 was expressed from pNLI60 and purified according to (Inniss et al., 2019). Pneumococcal σX and σA were expressed from pNLI94 and pNLI114, respectively. E. coli transformed with pNLI94 (SigX-V5H6, pI ∼8.42) or with pNLI114 (σA-V5H6, pI ∼5.02) were cultured in 3 L of LB medium at 37°C, 200 rpm to an OD600 0.5. To prepare a sample for SDS-PAGE, 2 mL of uninduced cells were collected and pelleted for 10 min at room temperature, and then boiled at 95°C for 2 min in a 3:1 mixture of E. coli resuspension buffer (50 mM Tris-HCl, pH 8.0, 500 mM NaCl) and Laemmli’s buffer (Bio Rad), respectively, then stored at 4°C. The large culture was induced for protein expression by addition of [1 mM]f isopropyl β-D-1-thiogalactopyranoside (IPTG) (Gold Biotechnology). Induced cultures were incubated overnight at 20°C, 200 rpm. The next day, 2 mL of induced culture was prepped for analysis by SDS PAGE, and the large cultured collected at 5,000 rpm, 4°C, for 30 min. Cells were resuspended in 50 mL of E. coli resuspension buffer supplemented with 25 mM imidazole, 50 mM MgCl2, 100 μg/mL DNase, and a protease inhibitor tablet (Roche). The cell resuspension was lysed using an Emulsiflex-C3 (Avestin). The soluble fraction was separated by centrifugation at 30,000 × g for 30 min at 4°C.
For σX purification, the soluble fraction was discarded. The pellet was washed in 15 mL of cold, filtered 20 mM L-arginine, 1 mM EDTA, 5% glycerol (RGE buffer, final pH, 10.95) supplemented with 1 mM βME and 2% w/v sodium deoxycholate (NaDOC) (Sigma Aldrich) with a homogenizer, followed by sonication for 2 min at 65% amplitude (0.8/0.2, on/off cycles). The washed pellet was spun was 30,000 × g for 30 min at 4°C. Washing, sonication, and spinning were repeated. Then the pellet was resuspended and dissolved in 15 mL of RGE supplemented with 1 mM βME and 0.6% N-lauroyl sarcosine sodium salt (Sigma-Aldrich) at 4°C with slow stirring for up to 2 days. The dissolved inclusion body was collected by centrifugation at 30,000 × g for 30 min at 4°C. The supernatant was saved and dialyzed for 8 h at room temperature in 2 L of 25 mM Tris-HCl, pH 8.3, 500 mM NaCl, 30 mM L-arginine, 10% glycerol, and 5 mM βME (CB4), and then overnight at 4°C in 2 L of CB4. The soluble supernatant was passed through a 5 mL Ni-NTA resin (Thermo Fisher Scientific) on a column equilibrated with CB4 (no βME) via gravity flow. The column was washed with 500 mL of CB4 supplemented with 5 mM βME and 40 mM imidazole. SigX-V5H6 was eluted from the column with 10 mL of CB4 supplemented with 5 mM βME and 275 mM imidazole. Subsequently, the flow through was saved and concentrated using a Vivaspin 15R 2,000 MWCO column concentrator (Sartorius). The concentrated solution was applied to a pre-packed Superdex 200 column (GE) on an AKTA (Amersham Biosciences). SigX-V5H6 was eluted at 0.3 mL/min in CB4 supplemented with 1 mM βME and collected in 1 ml fractions for analysis on SDS-PAGE gels. The fractions were pooled and concentrated again using a VivaSpin column concentrator. Protein concentration was measured at 5.6 mg/mL using a Nanodrop. Purified SigX-V5H6 was diluted to 1.2 mg/mL in CB4 supplemented with 1 mM EDTA, 1 mM βME and 20% glycerol for storage at −20°C until use.
For purification of σA, the soluble fraction was separated by centrifugation at 30,000 × g for 30 min at 4°C. The soluble supernatant was passed through a 5 mL Ni-NTA resin (Thermo Fisher Scientific) on a column equilibrated with Column Buffer (50 mM Tris-HCl, pH 8.0, 500 mM NaCl, 10% glycerol) via gravity. The column was washed with 500 mL of Column Buffer supplemented with 5 mM βME and 40 mM imidazole. σA was eluted from the column with 10 mL of column buffer supplemented with 5 mM βME and 275 mM imidazole. The eluate was concentrated using a Vivaspin 15R concentrator column into 50 mM Tris-HCl pH 8.0, 20 mM L-arginine, 500 mM NaCl, 1 mM EDTA, 10% glycerol, and 1 mM βME (DBR buffer). Protein concentration was measured as 1.85 mg/mL using a Nanodrop. SigA-V5H6 was diluted to 1 mg/mL in DBR supplemented with 20% glycerol and stored at −20°C until use.
Amplification of Linear Templates for in vitro Transcription
Primers used in this study are listed in Table 1. Gene sequences for amiA, comEA, and ssbB were taken from the genome of S. pneumoniae R6 (NC_003098.1). Oligonucleotides were synthesized by IDT (Coralville, Iowa). Equal amounts (1 μM) of forward and reverse primers were used to amplify amiA (NL299/NL300), comEA (NL295/NL296), and ssbB (NL228/NL229) from 50 ng of CP2137 genomic DNA with 0.2 mM dNTP (Thermo Fisher Scientific) and 1 μL of Phire Hot Start DNA Polymerase II (Thermo Fisher Scientific) in 50 μL reactions in a thermocycler. PCR products were purified with Zymo kits, eluted in diethyl pyrocarbonate (DEPC) H2O and visualized on a 1% agarose gel prior to storage at −20°C.
In vitro Transcription From DNA Templates
In vitro transcription from linear templates was carried out using 100 nM of E. coli RNAP obtained from NEB, pneumococcal σX and σA purified from E. coli, and ComWΔ6 purified from E. coli, when needed. To determine the minimum concentration of each σ-factor required to stimulate RNAP, initiation complexes were formed with 100 nM of E. coli RNAP, 20 nM of ssbB or amiA, and 0–200 nM of σX or σA, in 50 mM Tris-acetate, 50 mM sodium-acetate, 12.5% glycerol, 0.25 mM EDTA, 15 mM magnesium-acetate, and 35 mM βME (4X TGA buffer). Transcription from plasmid templates was carried out using 0–400 nM of E. coli RNAP and 0–320 nM of σA or σX. Solutions were incubated at 37°C for 15 min in a thermocycler, followed by addition of 1 μL of a master mix containing (120 nM) ATP, CTP, and GTP (Thermo Fisher Scientific) with α P32UTP at 0.5 μCi/μL (3000 ci/mmol, Perkin Elmer). Runoff transcription was allowed to proceed for 6 min at 37°C in a thermocycler, followed by addition of 20 nM cold UTP (0.5 μL of a 200 nM stock) and a final elongation step at 37°C for 10 min in a final volume of 5.5 μL. Reactions were stopped by addition of one volume of formamide dye (80% formamide, 10 mM EDTA pH 8.0, 1 μg/mL xylene cyonal, and 1 μg/mL bromophenol blue) and incubation at 65°C for 2 min in a thermocycler. The samples were placed on ice until gel loading.
To measure transcription stimulation by ComWΔ6 on PCR templates, 100 nM of E. coli RNAP, 80 nM σX or σA, 20 nM of comEA, ssbB, or amiA, and 0–160 nM of ComWΔ6 were mixed in 50 mM Tris-acetate, 50 mM sodium-acetate, 12.5% glycerol, 0.25 mM EDTA, 15 mM magnesium-acetate, and 35 mM βME (4X TGA buffer). Solutions were incubated at 37°C for 15 min in a thermocycler, followed by addition of 1 μL of a master mix containing (120 nM) ATP, CTP, and GTP (Thermo Fisher Scientific) with α P32UTP at 0.5 μCi/μL (3000 ci/mmol, Perkin Elmer). Runoff transcription was allowed to proceed for 6 min at 37°C in a thermocycler, followed by addition of 20 nM cold UTP (0.5 μL of a 200 nM stock) and a final elongation step at 37°C for 10 min in a final volume of 5.5 μL. Reactions were stopped by addition of one volume of formamide dye (80% formamide, 10 mM EDTA pH 8.0, 1 μg/mL xylene cyonal, and 1 μg/mL bromophenol blue) and incubation at 65°C for 2 min in a thermocycler. The samples were placed on ice until gel loading.
To visualize mRNA transcripts, 2 μL of each reaction was run on a 20 × 20, 0.375 mm thick, 6% polyacrylamide urea gel at 20 watts for 45 min. Gels were dried for 40 min at 80°C and incubated on a phosphorimager and imaged on a Typhoon 3000 (GE). The expected mRNA transcript sizes were determined using the transcriptional start sites (TSS) for each gene as determined by Aprianto et al. (2018). Experimental transcripts were compared to markers generated by in vitro transcription using the MAXIscript T7 Transcription Kit (Invirtogen) and the RNA Century-Plus Marker Templates (Invitrogen).
Results
Pneumococcal σ-Factors Stimulate Escherichia coli Core RNA Polymerase, in vitro
In vitro transcription assays confirmed that σX, in molar excess to RNAP, promotes transcription from combox promoters (Luo et al., 2003). However σX-mediated late gene transcription and transformation requires ComW in vivo (Sung and Morrison, 2005). Previous work concluded that pneumococcal RNAP exists at ∼2000 molecules per cell, and that σX and ComW peak at ∼3000 and ∼500 molecules per cell, respectively (Luo and Morrison, 2003; Piotrowski, 2010). Although not confirmed, σX might have a lower affinity for RNAP than σA, a trait shared among multiple alternative σ-factors (Gruber and Gross, 2003), so excess σX could increase the amount of σX holo-enzyme formation. Interestingly, levels of σA decrease 20–40% during the pneumococcal competence response (Luo and Morrison, 2003; Luo et al., 2003), and mutations to σA, that presumably decrease its affinity for RNAP, can suppress ΔcomW phenotypes (Tovpeko and Morrison, 2014; Tovpeko et al., 2016). These data hint that σX access to RNAP and/or competence promoters is an important step in efficient competence gene expression. In addition, we have shown that ComW, a likely σX binding partner (Tovpeko et al., 2016), interacts with DNA and this interaction is important for transformation (Inniss et al., 2019). We hypothesize that ComW functions at or near competence promoters to boost σX transcriptional activity. We tested this theory with in vitro transcription (IVT) assays.
Luo and Morrison used a ∼1.7 molar excess of σX to pneumococcal RNAP (RNAPSp) in their IVT assays, making the number of molecules of σX higher than that of RNAPSp (roughly 5.12 × 1012 vs. 3.13 × 1012 molecules, respectively, and based on the average molar ratio of σX:RNAPSp in cells as determined by Luo and Morrison, 2003). Thus, IVT from every competence template tested was successful and did not require ComW. As RNAP is conserved in prokaryotes (Murakami and Darst, 2003), we titrated pneumococcal σX and σA to determine the minimal amount of each σ-factor required to target 100 nM of E. coli RNAP (RNAPEcoli) to the pneumococcal combox and housekeeping promoters, in vitro.
The promoter regions of pneumococcal ssbB and amiA were PCR amplified for use in in vitro transcription assays (Figure 1A). At 40 nM σX and 100 nM RNAPEcoli, a 121-base (b) mRNA product from 20 nM of an ssbB linear template was produced, although the amount of mRNA produced (Figure 1B). The amount of ssbB transcript produced increased as σX was titrated up to 160 nM. At 80 nM, there are approximately 1.93 × 1011 molecules of σX versus 2.41 × 1011 molecules of RNAPEcoli at 100 nM in the reaction. This molar ratio (80 nM σX: 100 nM RNAPEcoli) allowed for observable, yet sub-optimal transcription from 20 nM of linear ssbB DNA template containing a combox promoter, without ComW. In addition, 80 nM of σX + 100 nM RNAPEcoli did not produce an mRNA product when incubated with 20 nM of linear amiA template, a gene under control of the primary σ-factor, σA (not shown), demonstrating that σX transcription specifically initiates from the combox promoter.
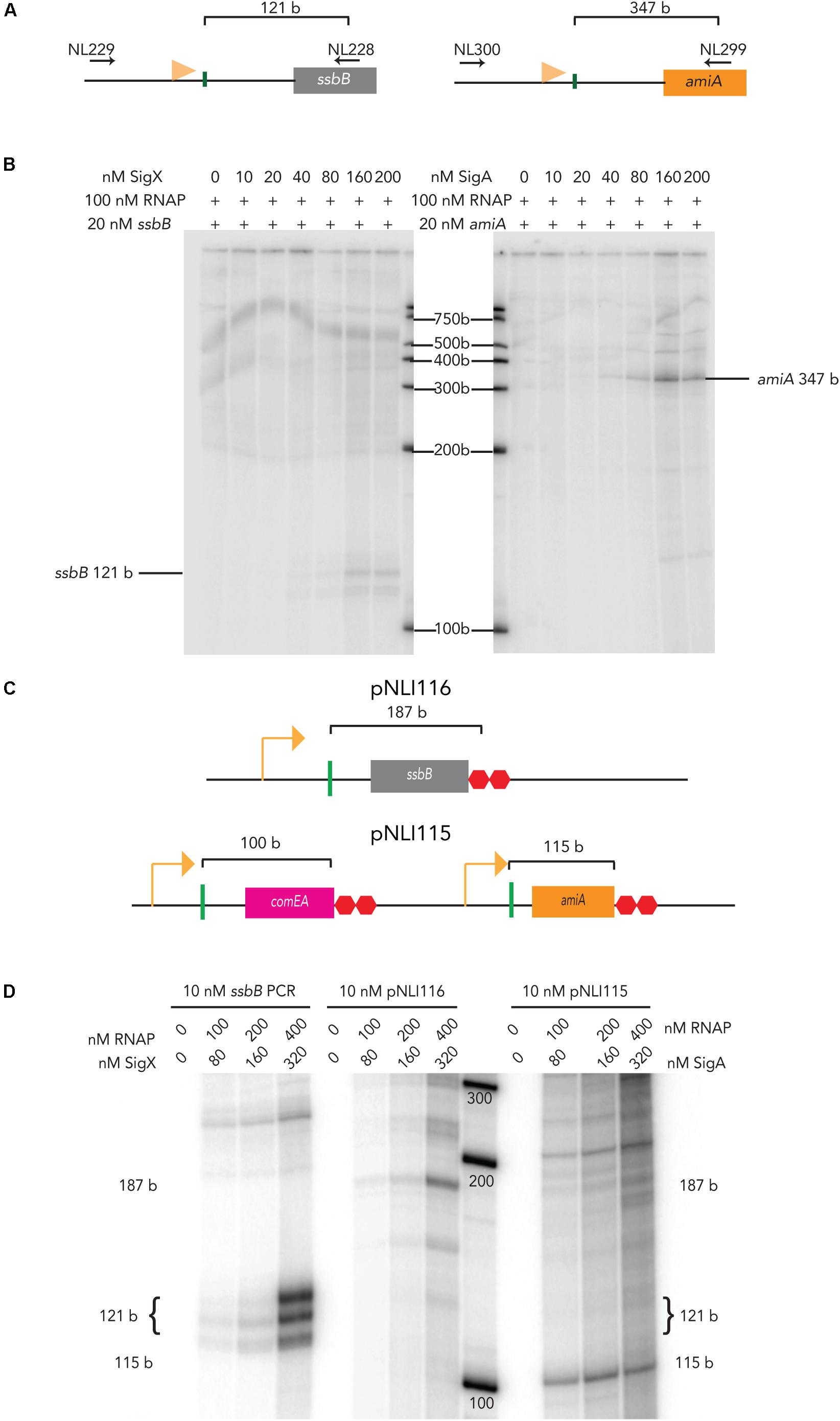
Figure 1. E. coli RNAP activation by pneumococcal sigma factors on linear and plasmid templates. (A) Schematic of ssbB (left) and amiA (right) PCR templates amplified from CP2137 genomic DNA using gene specific primers (their positions are indicated by the horizontal black arrows). The horizontal orange flags mark the positions of the combox promoter for ssbB and the Pribnow promoter box for amiA. The vertical green lines mark the transcription start sites for each gene, and the brackets indicate the size of the expected mRNA product (Aprianto et al., 2018). (B) The mRNA products from titration of pneumococcal σX (left blot) or σA with (right blot) with 100 nM of E. coli RNA polymerase and 20 nM of ssbB (left) or amiA (right) PCR templates. Pneumococcal σ-factors were titrated up to 200 nM. (C) (Top) Schematic of relevant region of plasmid pNLI116 containing pneumococcal combox promoter upstream of an ssbB gene fragment. (Bottom) Schematic of the relevant region of plasmid pNLI115, containing pneumococcal combox promoter upstream of comEA, followed the promoter upstream of pneumococcal amiA. The promoter upstream of each gene fragment is indicated by an orange arrow, and the vertical green lines mark the transcription start sites, with brackets that indicate the size of the expected mRNA product (Aprianto et al., 2018), and two red octagons indicate one copy of a pneumococcal termination signal (Aprianto et al., 2018) and a ltR2 terminator. (D) mRNA products that result from in vitro transcription with pneumococcal σX or σA and E. coli RNA polymerase from 10 nM of PCR or plasmid template with ssB or amiA gene fragments, respectively. The sizes of the mRNA products produced are indicated on the side of the gel, and were estimate based on Aprianto et al. (2018).
In contrast at 20 nM, pneumococcal σA produced detectable levels of a 347-base mRNA product from the housekeeping promoter upstream of amiA when incubated with RNAPEcoli (Figure 1B). The amount of amiA transcript produced increased as σA was titrated up to 160 nM, with a detectable amiA transcript at 80 nM. Interestingly, we did observe production of an ssbB transcript with 80 nM of σA (not shown). This might result from an orphan -10 promoter region σA binding site that is present 29 bp downstream of the ssbB transcription start site, according to Aprianto et al. (2018). The nature of this mRNA product must be examined more thoroughly. Nonetheless, these results demonstrate that both pneumococcal σ-factors direct RNAPEcoli to pneumococcal promoters, in vitro. In addition, as it appeared that more σX was required to stimulate RNAPEcoli activity at combox promoters than σA at housekeeping promoters, these results might indicate that more σX is needed to stimulate RNAPSp. As 80 nM of σX and σA are below saturating concentrations but still produced visible ssbB and amiA transcripts, respectively, we used 80 nM of each σ-factor in subsequent reactions.
To further explore in vitro transcription with pneumococcal σ factor, we tested transcription from plasmid templates. Supercoiling can affect bacterial gene transcription (Pruss and Drlica, 1989; Zhi et al., 2017), thus we asked whether or not our chimeric holo-enzymes could transcribe the ssbB and amiA genes from these plasmid templates. We designed two plasmids that carry pneumococcal promoters upstream of pneumococcal gene fragments, pNLI115 and pNLI116 (Figure 1C). pNLI116 contained only the combox promoter followed by an ssbB gene fragment, similar to our original PCR ssbB IVT template. pNLI115 contained the combox promoter region upstream of competence gene fragment comEA, and the housekeeping promoter region upstream of amiA, to serve as an internal control for promoter specific IVT from plasmid assays. After isolation from E. coli, each plasmid preparation contained predominantly supercoiled species, as observed on DNA agarose gels.
We titrated holo-enzymes into IVT reactions with 10 nM of either PCR DNA templates or plasmid templates. As expected, the amount of the 121 b mRNA transcript produced from linear DNA increased as the amount of σX holo-enzyme increased. Similar results were observed from pNLI116, as increasing amounts of a 187 b mRNA transcript was produced as the amount of σX holo-enzyme was titrated onto the plasmid (Figure 1D, left). In addition, a 115 b mRNA transcript was produced from pNLI115 when σA-RNAPE. coli complex was titrated onto the plasmid (Figure 1D, right). Together these data show that our chimeric holo-enzymes can be targeted to specific promoters on either linear or plasmid DNA templates, in vitro.
ComWΔ6 Stimulates Transcription From σX Promoters, in vitro
Both ssbB and comEA are induced by CSP and require ComW for robust transcription (Sung and Morrison, 2005). To determine the effect of ComW on transcription from combox promoters upstream of these two genes, we titrated ComWΔ6 into IVT reactions with σX holo-enzyme. Transcripts from both linear ssbB and comEA templates (Figure 2) were produced when ComWΔ6 was added to IVT reactions. Specifically, the σX holo-enzyme incubated with ComWΔ6 at 160 nM (2X molar excess of ComW to σX) produced 2X as much mRNA signal from the sbbB template compared to reactions with 0 nM ComWΔ6, an increase that was statistically significant (Figure 2). Similarly, the σX holo-enzyme incubated with ComWΔ6 at 160 nM produced 4.5X as much mRNA signal from the comEA template compared to σX holo-enzyme with no ComWΔ6, also an increase that was statistically significantly (Figure 2). The 4.5X increase in comEA mRNA signal production versus the 2X increase in ssbB signal production also represented a statistically significant difference. These data demonstrate that ComW is required for robust transcription when the number of σX molecules is below the number of core RNAP molecules, in vitro. Furthermore, the difference in transcription stimulation suggests that the requirement for ComW at combox promoters differs. Lastly, as ComWΔ6 can stimulate σX-mediate transcription, in vitro, it is possible that residues 73RGFISC78 are dispensable for transcription activation in pneumococci.
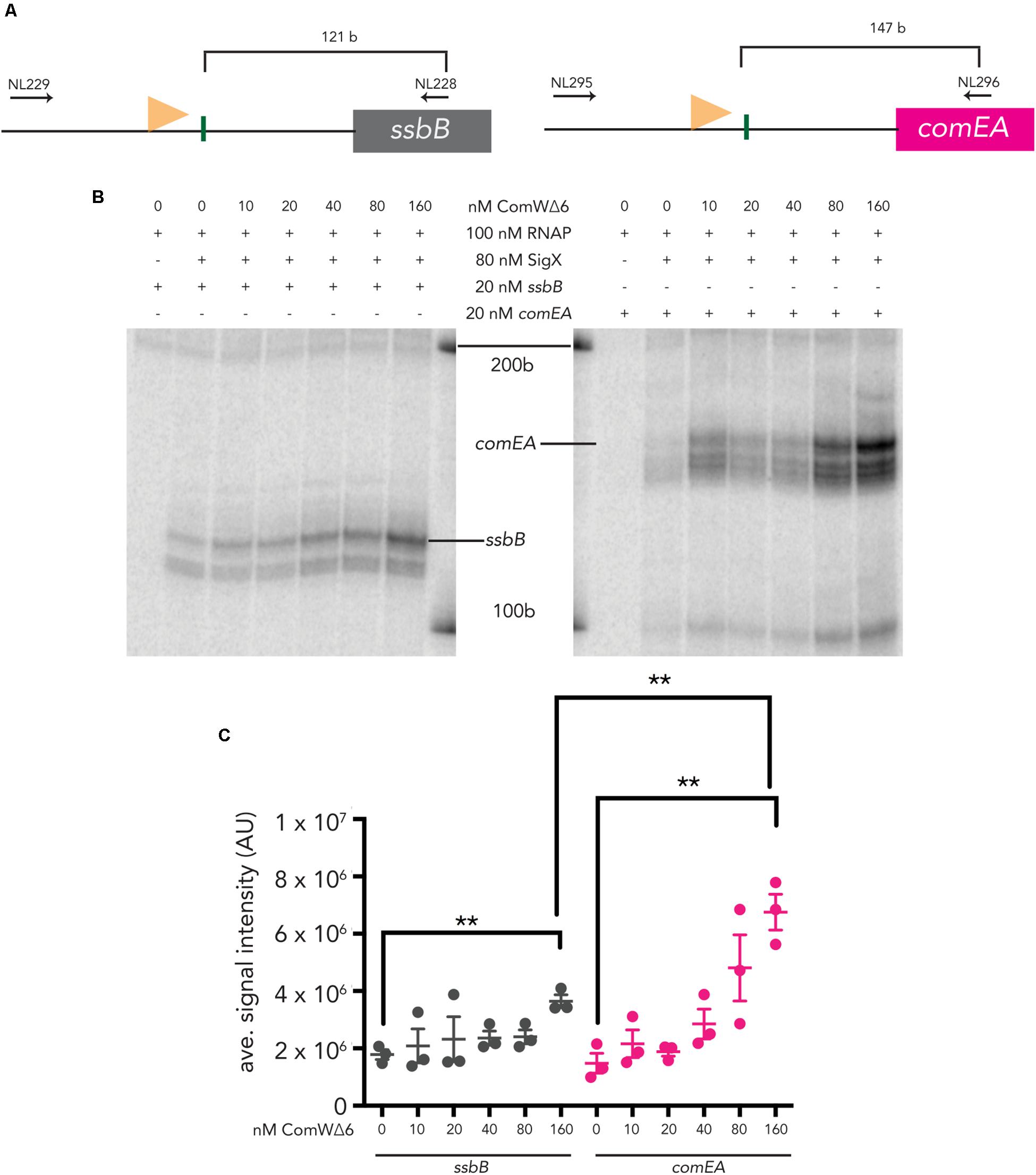
Figure 2. In vitro transcription from two pneumococcal late competence promoters using ComWΔ6. (A) A schematic of pneumococcal ssbB (left) and comEA (right) templates. A light orange flag represents each promoter and vertical green lines mark the transcription start sites. Horizontal arrows mark the positions of the primers used to amplify each template. (B) An image of ssbB (121 b, left) and comEA (147 b, right) mRNA products produced by σX holoenzymes with increasing amounts of ComWΔ6. The positions of the 100 b and 200 b standard bands are indicated. (C) Quantification of the signal intensities of mRNA transcripts from three in vitro transcription experiments. Asterisks mark statistically significant differences between samples with 0 nM ComWΔ6 and 160 nM ComWΔ6, and between samples with ssbB and comEA templates.
Discussion
Anginosus and mitis group streptococci like the pneumococcus use the DNA binding protein, ComW, to regulate σX-mediated transcription. However, previous data show that σX transcribes late genes in the absence of ComW, in vitro (Luo and Morrison, 2003). As we hypothesized that ComW is an active and important member of the competence-specific holo-enzyme, we examined how ComWΔ6 affected IVT from σX promoters. Remarkably, two σX targets, ssbB and comEA appeared to respond differently to the addition of ComWΔ6 to reactions; although transcription stimulation was observed for both genes, the level of stimulation appears greater for comEA. This is common in other naturally transformable species. For example, the V. cholerae competence regulator, QstR, binds upstream of some competence genes, but not others (Jaskólska et al., 2018), and the number of AT boxes that lie upstream of competence promoters in B. subtilis can differ from gene to gene, likely reflecting the difference in ComK activity at these promoters (Hamoen et al., 2003). Thus, at this moment, we cannot rule out that ComW is specific for transcription of some pneumococcal late competence genes, and might be dispensable for the transcription of others.
Interestingly, it appears that σA better stimulates E. coli RNAP than σX. This might suggest that pneumococcal RNAP responds similarly to these two σ factors, and the specifics of pneumococcal σ – RNAP interactions must be determined. This could be another reason as to why σX – mediated transcription requires ComW in the pneumococcus. Furthermore, the σX – ComW pair appear similar to a two-part σ factor, σO – RsoA, identified in B. subtilis (Maclellan et al., 2009). In this system RsoA aids in σO – RNAP open complex formation to promote transcription of genes required during growth in acidic conditions. Like σO, σX likely provides promoter recognition, and ComW, like RsoA, might aid in stabilizing the σX complex at the promoter (Maclellan et al., 2009). However, the specifics of a probable σX – ComW or ComW – RNAP interaction are yet to be determined.
The DNA – binding protein, ComW does stimulate σX – mediated transcription, in vitro. Although more effort is required to fully work out the details, we posit that there are likely two possible roles for how ComW functions (Figure 3). (A) As ComW interacts non-specifically with DNA, it is likely brought to competence promoters by σX, and increases promoter melting. (B) ComW might function as a component of a two-part σ factor with σX and RNAP as an active member of the holo-enzyme.
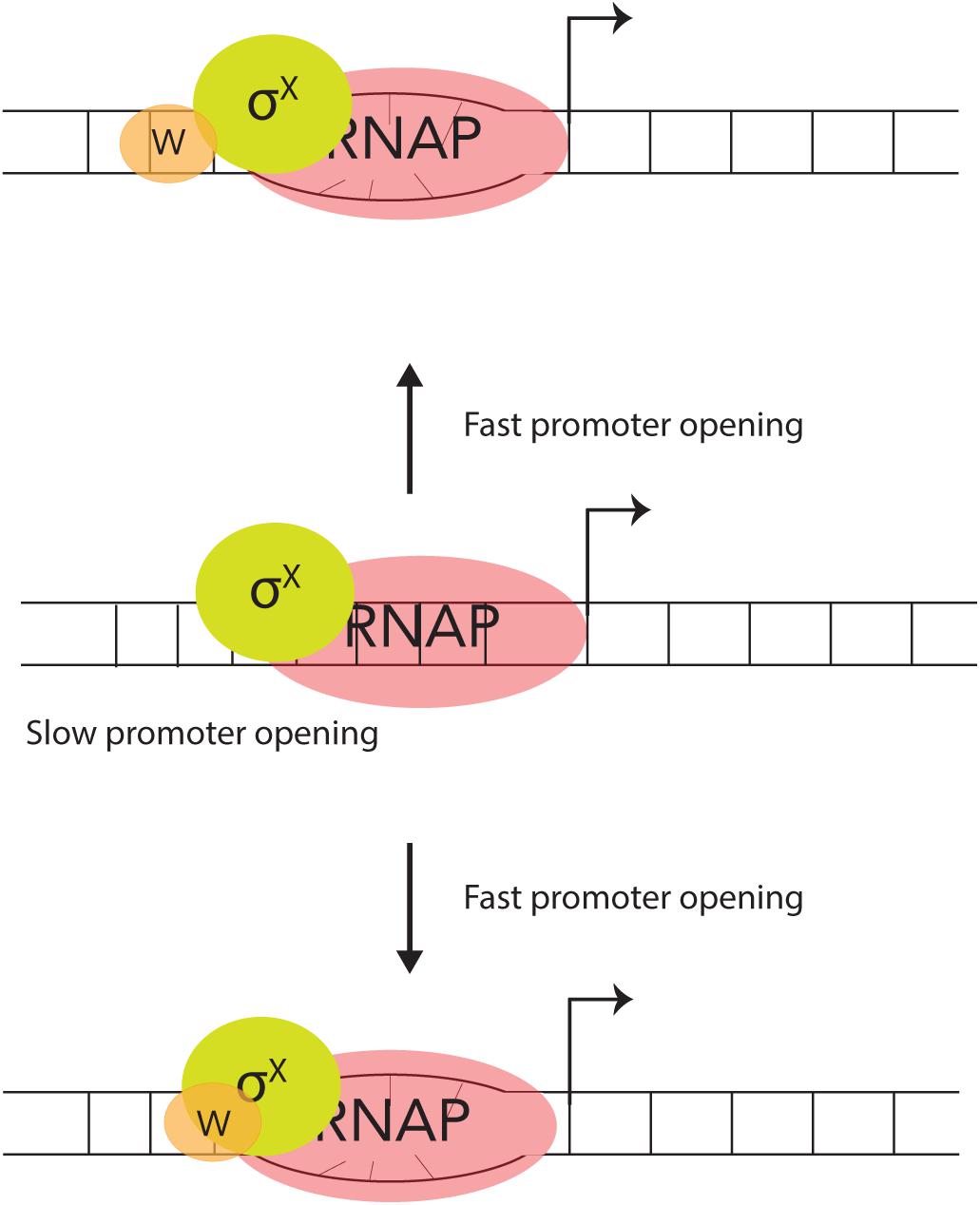
Figure 3. Two proposed mechanisms for ComW-dependent promoter melting. In the absence of ComW the pneumococcal σX-holoenzyme can recognize and bind to combox containing DNA, but is slow to melt the promoter (middle). ComW is brought to combox promoters via interaction with σX (top) and/or RNAP (bottom). ComW uses its non-specific DNA binding function to aid in melting promoter DNA.
Data Availability Statement
The datasets generated for this study are available on request to the corresponding author.
Author Contributions
NI conceived, conducted experiments and wrote the manuscript. DM edited the manuscript.
Funding
Funding was provided by The University of Illinois at Chicago through the Abraham Lincoln Fellowship, as well as an R03 grant from the National Institute of Allergy and Infectious Diseases of the National Institutes of Health (1R03AI128228).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Andam, C. P., and Hanage, W. P. (2015). Mechanisms of genome evoluation of Streptococcus. Infect. Genet. Evol. 33, 334–342. doi: 10.1016/j.meegid.2014.11.007
Aprianto, R., Slager, J., Holsappel, S., and Veening, J.-W. (2018). High-resolution analysis of the pneumococcal transcriptome under a wide range of infection-relevant conditions. Nucleic Acids Res. 17:1133. doi: 10.1093/nar/gky750
Avery, O. T., and MacLeod, C. M. (1944). Studies on the chemical nature of the substance inducing transformation of pneumococcal types. J. Exp. Med. 79, 137–158. doi: 10.1084/jem.149.2.297
Bertani, G. (1951). Studies on lysogenesis. J. Bacteriol. 62, 293–300. doi: 10.1128/jb.62.3.293-300.1951
Cheng, Q., Campbell, E. A., Naughton, A. M., Johnson, S., and Masure, H. R. (1997). The com locus controls genetic transformation in Streptococcus pneumoniae. Mol. Microbiol. 23, 683–692. doi: 10.1046/j.1365-2958.1997.2481617.x
Fontaine, L., Boutry, C., de Frahan, M. H., Delplace, B., Fremaux, C., Horvath, P., et al. (2010). A novel pheromone quorum-sensing system controls the development of natural competence in Streptococcus thermophilus and Streptococcus salivarius. J. Bacteriol. 192, 1444–1454. doi: 10.1128/JB.01251-09
Gao, X.-Y., Zhi, X.-Y., Li, H.-W., Klenk, H.-P., and Li, W.-J. (2014). Comparative genomics of the bacterial genus Streptococcus illuminates evolutionary implications of species groups. PLoS One 9:e101229. doi: 10.1371/journal.pone.0101229.s005
Gruber, T. M., and Gross, C. A. (2003). Multiple sigma subunits and the partitioning of bacterial transcription space. Annu. Rev. Microbiol. 57, 441–466. doi: 10.1146/annurev.micro.57.030502.090913
Hamoen, L. W., Venema, G., and Kuipers, O. P. (2003). Controlling competence in Bacillus subtilis: shared use of regulators. Microbiology 149, 9–17. doi: 10.1099/mic.0.26003-0
Håvarstein, L. S. (2010). Increasing competence in the genus Streptococcus. Mol. Microbiol. 78, 541–544. doi: 10.1111/j.1365-2958.2010.07380.x
Håvarstein, L. S., Coomaraswamy, G., and Morrison, D. A. (1995). An unmodified heptadecapeptide pheromone induced competence for genetic transformation in Streptococcus pneumoniae. Proc. Natl. Acad. Sci. U.S.A. 92, 11140–11144. doi: 10.1073/pnas.92.24.11140
Håvarstein, L. S., Gaustad, P., Nes, I. F., and Morrison, D. A. (1996). Identification of the streptococcal competence- pheromone receptor. Mol. Microbiol. 21, 863–869. doi: 10.1046/j.1365-2958.1996.521416.x
Inniss, N. L., Prehna, G., and Morrison, D. A. (2019). The pneumococcal σX activator, ComW, is a DNA-binding protein critical for natural transformation. J. Biol. Chem. 294, 11101–11118. doi: 10.1074/jbc.RA119.007571
Jaskólska, M., Stutzmann, S., Stoudmann, C., and Blokesch, M. (2018). QstR-dependent regulation of natural competence and Type VI secretion in Vibrio cholerae. Nucleic Acid Res. 46, 10619–10634. doi: 10.1093/nar/gky717
Khan, R., Junges, R., Mdal, H. A. A. X., Chen, T., Morrison, D. A., and Petersen, F. C. (2017). A positive feedback loop mediated by Sigma X enhances expression of the streptococcal regulator ComR. Sci. Rep. 7:5984. doi: 10.1038/s41598-017-04768-5
Khan, R., Rukke, H. V., Høvik, H., Åmdal, H. A., Chen, T., Morrison, D. A., et al. (2016). Comprehensive transcriptome profiles of Streptococcus mutans UA159 map core streptococcal compentence genes. mSystems 1:e00038-15. doi: 10.1128/mSystems.00038-15
Lee, M. S., and Morrison, D. A. (1999). Identification of a new regulator in Streptococcus pneumoniae linking qurum sensing to competence for genetic transformation. J. Bacteriol. 181, 5004–5016. doi: 10.1128/jb.181.16.5004-5016.1999
Lee, M. S., and Morrison, D. A. (2000). Regulation of competence for genetic transformation in Streptococcus pneumoniae: a link between quorum sensing and DNA processing genes. Res. Microbiol. 151, 445–451. doi: 10.1016/s0923-2508(00)00171-6
Luo, P., and Morrison, D. A. (2003). Transient association of an alternative sigma factor, ComX, with RNA polymerase during the period of competence for genetic transformation in Streptococcus pneumoniae. J. Bacteriol. 185, 349–358. doi: 10.1128/JB.185.1.349-358.2003
Luo, P., Li, H., and Morrison, D. A. (2003). ComX is a unique link between multiple quorum sensing outputs and competence in Streptococcus pneumoniae. Mol. Microbiol. 50, 623–633. doi: 10.1046/j.1365-2958.1999.01528.x
Luo, P., Li, H., and Morrison, D. A. (2004). Identification of ComW as a new component in the regulation of genetic transformation in Streptococcus pneumoniae. Mol. Microbiol. 54, 172–183. doi: 10.1111/j.1365-2958.2004.04254.x
Maclellan, S. R., Guariglia-Oropeza, V., Gaballa, A., and Helmann, J. D. (2009). A two-subunit bacterial σ-factor activates transcription in Bacillus subtilis. Proc. Natl. Acad. Sci. 106, 21232–21328.
Mandel, M., and Higa, A. (1970). Calcium-dependent bacteriophage DNA infection. J. Mol. Biol. 53, 159–162. doi: 10.1016/0022-2836(70)90051-3
Martin, B., Granadel, C., Campo, N., Hénard, V., Prudhomme, M., and Claverys, J.-P. (2010). Expression and maintenance of ComD-ComE, the two-component signal-transduction system that controls competence of Streptococcus pneumoniae. Mol. Microbiol. 75, 1513–1528. doi: 10.1111/j.1365-2958.2010.07071.x
Martin, B., Soulet, A.-L., Mirouze, N., Prudhomme, M., Mortier-Barrière, I., Granadel, C., et al. (2012). ComE/ComE P interplay dictates activation or extinction status of pneumococcal X-state (competence). Mol. Microbiol. 87, 394–411. doi: 10.1046/j.1365-2958.1999.01528.x
Mashburn-Warren, L., Morrison, D. A., and Federle, M. J. (2010). A novel double-tryptophan peptide pheromone controls competence in Streptococcus spp. via an Rgg regulator. Mol. Microbiol. 78, 589–606. doi: 10.1111/j.1365-2958.2010.07361.x
Mashburn-Warren, L., Morrison, D. A., and Federle, M. J. (2012). The cryptic competence pathway in Streptococcus pyogenes is controlled by a peptide pheromone. J. Bacteriol. 194, 4589–4600. doi: 10.1128/JB.00830-12
Murakami, K. S., and Darst, S. A. (2003). Bacterial RNA polymerases: the wholo story. Curr. Opin. Struct. Biol. 13, 31–39. doi: 10.1016/S0959-440X(02)00005-2
Peterson, S., Cline, R. T., Tettelin, H., Sharov, V., and Morrison, D. A. (2000). Gene expression analysis of the Streptococcus pneumoniae competence regulons by use of DNA microarrays. J. Bacteriol. 182, 6192–6202. doi: 10.1128/jb.182.21.6192-6202.2000
Peterson, S. N., Sung, C. K., Cline, R., Desai, B. V., Snesrud, E. C., Luo, P., et al. (2004). Identification of competence pheromone responsive genes in Streptococcus pneumoniae by use of DNA microarrays. Mol. Microbiol. 51, 1051–1070. doi: 10.1046/j.1365-2958.2003.03907.x
Piotrowski, A. (2010). Regulation of the Activities of ComX and ComW During Competence Development in Streptococcus Pneumoniae. Ph.D. thesis, Univerisity of Illinois at Chicago, Chicago. 1–169.
Piotrowski, A., Luo, P., and Morrison, D. A. (2009). Competence for genetic transformation in Streptococcus pneumoniae: termination of activity of the alternative sigma factor ComX is independent of proteolysis of ComX and ComW. J. Bacteriol. 191, 3359–3366. doi: 10.1128/JB.01750-08
Pruss, G. J., and Drlica, K. (1989). DNA supercoiling and prokaryotic transcription. Cell 56, 521–523. doi: 10.1016/0092-8674(89)90574-6
Sung, C. K., and Morrison, D. A. (2005). Two distinct functions of ComW in stabilization and activation of the alternative sigma factor ComX in Streptococcus pneumoniae. J. Bacteriol. 187, 3052–3061. doi: 10.1128/JB.187.9.3052-3061.2005
Tovpeko, Y., Bai, J., and Morrison, D. A. (2016). Competence for genetic transformation in Streptococcus pneumoniae: mutations in σA bypass the ComW requirement for late gene expression. J. Bacteriol. 198, 2370–2378. doi: 10.1128/JB.00354-16
Tovpeko, Y., and Morrison, D. A. (2014). Competence for genetic transformation in Streptococcus pneumoniae: mutations in a bypass the comW requirement. J. Bacteriol. 196, 3724–3734. doi: 10.1128/JB.01933-14
Ween, O., Gaustad, P., and Håvarstein, L. S. (1999). Identification of DNA binding sites for ComE, a key regulator of natural competence in Streptococcus pneumoniae. Mol. Microbiol. 33, 817–827. doi: 10.1046/j.1365-2958.1999.01528.x
Keywords: alternative sigma factor, S. pneumoniae competence, genetic transformation, ComW, transcription
Citation: Inniss NL and Morrison DA (2020) ComWΔ6 Stimulates Transcription of Pneumococcal Competence Genes in vitro. Front. Mol. Biosci. 7:61. doi: 10.3389/fmolb.2020.00061
Received: 05 February 2020; Accepted: 24 March 2020;
Published: 06 May 2020.
Edited by:
Manuel Espinosa, Margarita Salas Center for Biological Research (CSIC), SpainReviewed by:
Marco Rinaldo Oggioni, University of Leicester, United KingdomGee W. Lau, University of Illinois at Urbana-Champaign, United States
Copyright © 2020 Inniss and Morrison. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Donald A. Morrison, ZGFtb3JyaXNAdWljLmVkdQ==
 Nicole L. Inniss
Nicole L. Inniss Donald A. Morrison
Donald A. Morrison