
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Mol. Biosci. , 24 October 2019
Sec. Molecular Diagnostics and Therapeutics
Volume 6 - 2019 | https://doi.org/10.3389/fmolb.2019.00114
 Maria Rosa Conserva†
Maria Rosa Conserva† Immacolata Redavid†
Immacolata Redavid† Luisa Anelli
Luisa Anelli Antonella Zagaria
Antonella Zagaria Giorgina Specchia
Giorgina Specchia Francesco Albano*
Francesco Albano*Retinoic acid receptor γ (RARγ) belongs to the nuclear receptor superfamily and shares 90% homology with retinoic acid receptor α (RARα) and retinoic acid receptor β (RARβ). RARA rearrangements are well-known to be involved in acute promyelocytic leukemia (APL), but RARG rearrangements can also resemble this kind of leukemia. In this review we trace the role of RARγ, considering both its physiological and oncogenic contribution; from 2011 to date, nine cases of patients harboring RARG fusions have been reported. These patients showed typical APL features, including the clinical presentation, coagulation abnormalities and morphological features of bone marrow (BM), but are not responsive to APL standard therapy. We stress the urgent need for a better comprehension of the critical role of RARG dysregulation in the leukemogenesis process, since optimum therapy strategies have not yet been established.
Acute myeloid leukemia (AML) is a hematologic malignancy that may arise in patients with another hematological disorder already present or after previous therapy, for instance following previous exposure to alkylating agents or DNA topoisomerase inhibitors or radiation (Sill et al., 2011), but most frequently it appears as a de novo malignancy in previously healthy individuals (De Kouchkovsky and Abdul-Hay, 2016). The bases of the pathogenesis reside in an abnormal and malignant clonal expansion of specific progenitor myeloid cells that do not complete their physiological differentiation process (De Kouchkovsky and Abdul-Hay, 2016). The cause is to be found in a genetic alteration of the progenitor hematopoietic cell that gives life to a clonal population of blast cells, with the manifestation of an atypical hematopoiesis (Levine, 2013; Thomas and Majeti, 2017). Already in 1957, APL accounted for 5–15% of the total AML cases (Szotkowski et al., 2015). In fact, APL is a different subtype of AML characterized by an accumulation and expansion of leukemic cells that fail to go beyond the promyelocyte stage of myelopoiesis (de Thé et al., 1991). Before the discovery of all-trans retinoic acid (ATRA) and arsenic trioxide (ATO), patients with APL had a high risk of mortality, due to hemorrhagic complications before and during induction therapy (Choudhry and DeLoughery, 2012). Both ATRA and ATO behave as differentiating agents, since they promote the differentiation and maturation of leukemic promyelocytes to neutrophils (Asou et al., 1998; Karim et al., 2014; Daver et al., 2015). According to the current WHO criteria, the presence of t(15;17)(q22;q12), hence of the promyelocytic leukemia protein (PML)-RARA gene fusion, allows an AML to be classified as APL, but other variant RARA translocations with other partner genes are not only considered distinct, but not all have typical APL features and indeed, some patients show resistance to ATRA (Arber et al., 2016). In particular, in a subset of patients the PML-RARA fusion is not detected, but other rearrangements are identified that involve RARB and RARG, two distinct isoforms of RARA, that cause a pathological phenotype that resembles APL (Marinelli et al., 2007; Osumi et al., 2018). The aim of this review is to focus on the various emerging cases of APL without the RARA gene rearrangement and with the RARG gene rearrangement or dysregulation that presented abnormal promyelocytes fully in accordance with the APL phenotype.
RARγ belongs to the nuclear receptor superfamily and shares 90% homology with RARα and RARβ (Chambon, 1996). These three types of RARs have been discovered by cDNA cloning in both human and mouse and were identified as the only three members of the RAR family in mouse and man (Kastner et al., 1994). Moreover, all three RAR types have been characterized in the newt (Giguère et al., 1989; Ragsdale et al., 1989), RARβ cDNA has been cloned from chicken (Noji et al., 1991; Padanilam et al., 1991; Rowe et al., 1991; Smith and Eichele, 1991), RARα and RARγ cDNA have been found in Xenopus (Ellinger-Ziegelbauer and Dreyer, 1991; Blumberg et al., 1992) and have been isolated from zebrafish (Kastner et al., 1994), so these findings suggest that this RAR genes family exists in all vertebrates, mediating a series of effects, that may sometimes even be conflicting. RARγ is involved in several biological processes and cellular pathways, interacting with different proteins (Figure 1). The chromosomal mapping of RARG in humans is on 12q13, while RARA and RARB mapping is on 17q21.1 and 3p24, respectively (Mattei et al., 1988, 1991; Ishikawa et al., 1990). The protein structure of RARγ presents a DNA binding domain (DBD) that receives two “zinc fingers” that allow the protein to bind to specific DNA sequences in the promoter of their target genes, known as retinoic acid response elements (RAREs), and a ligand-binding domain (LBD) that confers the ligand binding specificity, assuming a ligand-inducible transactivation function (Kastner et al., 1990; Leroy et al., 1991; Zelent et al., 1991; Liu and Linney, 1993; Bastien and Rochette-Egly, 2004; Storlazzi et al., 2007; Al Tanoury et al., 2013). In detail, it is possible to recognize six different regions in the RAR primary sequence (A-F), which exhibit differential degrees of conservation (Kastner et al., 1994). The most conserved regions among the three RARs, within a given species, are the DBD (region C), the LBD (region E) and the region B which presents a promoter-specific transcription activation function (Nagpal et al., 1992). Although the D region, known as a hinge region, is well-conserved in its N-terminal portion, the central region appears to be more variable. Finally, the A and F regions differ notably among the three receptors (Kastner et al., 1994). For each RAR isotype it is possible to identify two or more isoforms; for regarding RARγ, two main isoforms (RARγ1 and RARγ2) have been identified, that show a different spatio-temporal expression. RARγ1 is found in the later phase of embryogenesis, as well as in the skin of newborns and adults, while RARγ2 is expressed in the early embryo (Kastner et al., 1990; Chambon, 1993). In the presence of a ligand, such as ATRA, RARs form heterodimers with the retinoid X receptors (RXRs) that operate as transcription factors, activating the RAREs regions in the target genes promoter (Mic et al., 2003; Bastien and Rochette-Egly, 2004). In detail, each RARs subtype presents a different sensitivity at different concentrations of ATRA: RARα activation occurs in the presence of high concentrations of ATRA, whereas RARγ activation requires the lowest amount of ligand (Beard et al., 2002).
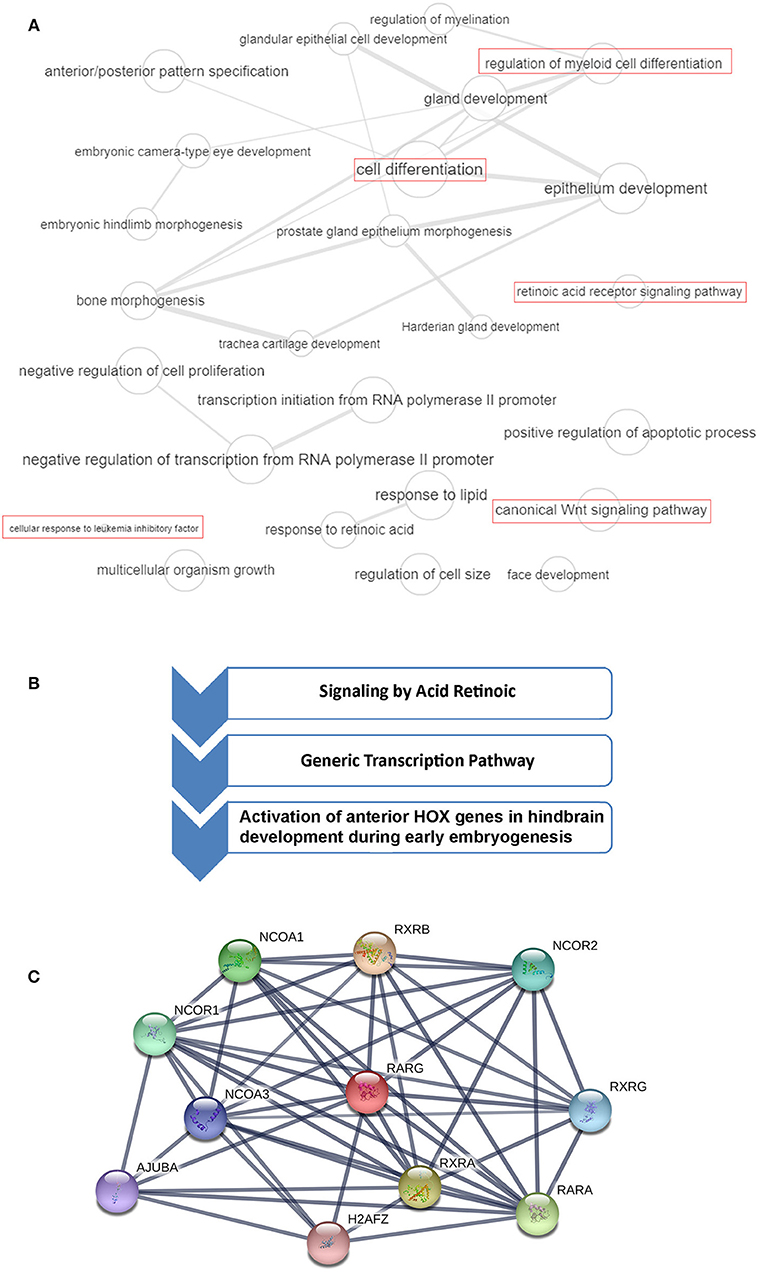
Figure 1. RARG gene ontology (GO), cellular pathways and protein-protein interaction analysis. (A) GO terms of RARG visualized by REVIGO (http://revigo.irb.hr/). Bubble size indicates the frequency of the GO term in the Gene Ontology Annotation database. Lines in the graph link highly similar GO terms, where their width indicates the degree of similarity. In red are highlighted RARG cellular processes that may be altered in leukemia. (B) RARG main cellular pathways derived from reactome database (https://reactome.org/). (C) RARG protein-protein interactions obtained by STRING database (https://string-db.org/). The interactions include direct (physical) and indirect (functional) associations.
Although knockout mice studies have shown some functional redundancy among the different RAR isotypes, in reality each of them has a unique role in the processes of development and differentiation that cannot be replaced by the action of the other isotypes (Taneja et al., 1995; Mark et al., 2006). RARG null mice display growth deficiency (Lohnes et al., 1993); indeed, RARγ is highly expressed in the growth plate and its absence is associated with a reduced chondrocyte proliferation and decreased expression and deposition of proteoglycans (Williams et al., 2009). In addition, RARγ is required for the correct formation of the axial skeleton including anteriorization of the cervical and thoracic vertebrae (Lohnes et al., 1993; Wendling et al., 2001). Other studies have demonstrated that the genetic ablation of RARG in mice induced a decrease in bone mass due to an increase in osteoclastogenesis and consequent loss of trabecular bone mass (Walkley et al., 2007; Green et al., 2015). In addition, RARγ is crucial for the formation of normal alveoli and alveoli elastic fibers in the lung (McGowan et al., 2000) and its genetic ablation is associated with male sterility and with alterations of the prostatic glandular epithelia (Lohnes et al., 1993). Moreover, RARγ signaling is critical for epigenetic changes induced by retinoic acid (RA) (Kashyap et al., 2013). RARγ, as well as RARα, are crucial for hematopoietic development (Purton, 2007). Indeed, while RARα induces granulocytic differentiation, RARγ plays a central role in maintaining the balance between the self-renewal state of hematopoietic stem cells (HSCs) and their differentiation (Purton et al., 2006; Purton, 2007). Purton et al. reported that RARG null mice show reduced numbers of long-term repopulating HSCs and increased numbers of more committed hematopoietic progenitors, therefore RARG−/− BM displayed significantly increased numbers of common myeloid progenitors and common lymphoid progenitors, unlike RARG+/+ BM. This finding suggests that loss of RARG induces an imbalance in HSC self-renewal decisions, supporting differentiation divisions. Moreover, Purton et al. have shown that RARγ signaling affected other genes known to elicit HSC self-renewal, such as NOTCH1, whose expression decreases severely in the absence of RARγ (Purton et al., 2006). Walkley et al., showed that RARG null mice exhibit a considerable increase in granulocytes in the peripheral blood (PB), in the BM and spleen, developing a myeloproliferative-like syndrome and displaying defects in both erythropoiesis and in B lymphopoiesis (Walkley et al., 2007; Dewamitta et al., 2014). Transplantation studies have confirmed that the expression of RARγ is essential in the BM microenvironment for a correct hematopoiesis to occur (Joseph et al., 2016). Indeed, Dewamitta et al. suggested that deficiencies observed in RARG−/− mice are due to an aberrant RARG−/− BM microenvironment, and therefore it is not an intrinsic cellular defect (Dewamitta et al., 2014). Green et al. also confirmed that RARγ is a crucial regulatory key for the presence of a healthy BM microenvironment, since mice with conditionally deleted RARG in more primitive limb bud-derived mesenchymal stem cells and their progeny, achieved through the use of Prrx1-Cre, showed alterations in the number of both osteoclasts and osteoblasts, with consequent modifications in the trabecular bone and an abnormal angiogenesis and B lymphopoiesis. These results seem to confirm the key role of RARγ in maintaining homeostasis in the various processes that occur in the BM microenvironment (Conserva et al., 2019), such as endochondral bone formation, angiogenesis, osteoclastogenesis and B lymphopoiesis (Green et al., 2018). Considering that vitamin A-derived retinoids play a central role in the growth and differentiation of a variety of cell types, RARγ in particular mediates various anti-proliferative and apoptotic effects of retinoids in certain tissues and cancer cells, such as melanoma and neuroblastoma cells (Spanjaard et al., 1997; Meister et al., 1998). Chen et al. also showed that the genetic ablation of RARG in a model of epidermal tumorigenesis enhanced the tumor incidence of Ras-transformed keratinocytes and was associated with retinoids resistance (Chen et al., 2004). Several studies, in recent years, have described various cases of patients carrying translocations involving RARG and showing a leukemic phenotype that resembles APL (Table 1). Therefore, it seems that RARG, just like RARA, can somehow rearrange and mediate oncogenic effects.
RARG rearranged leukemia is a rare specific subtype of AML, but its characterization is very ambiguous because its features are so strongly similar to APL. Indeed, patients presenting RARG rearrangements share typical APL clinical presentations, coagulation abnormalities and morphological and immunophenotypic features of BM (Luo et al., 2019). RARA rearrangements are known to generate APL, and rearrangements involving RARB or RARG can resemble APL. Typical APL is characterized by recurrent PML-RARA expression, but up to now, to our knowledge, eleven cases with RARG dysregulation have been identified (Such et al., 2011; Ha et al., 2017; Liu et al., 2018; Miller et al., 2018; Qin et al., 2018; Chen et al., 2019; Luo et al., 2019; Zhang et al., 2019b) (Figure 2). As previously reported by Conserva et al., the three RARs are highly homologous so it is not unexpected that rearrangements involving both RARB and RARG can generate similar diseases (Conserva et al., 2019). However, the involvement of RARG principal cellular pathways in AML needs to be further clarified (Figure 1A).
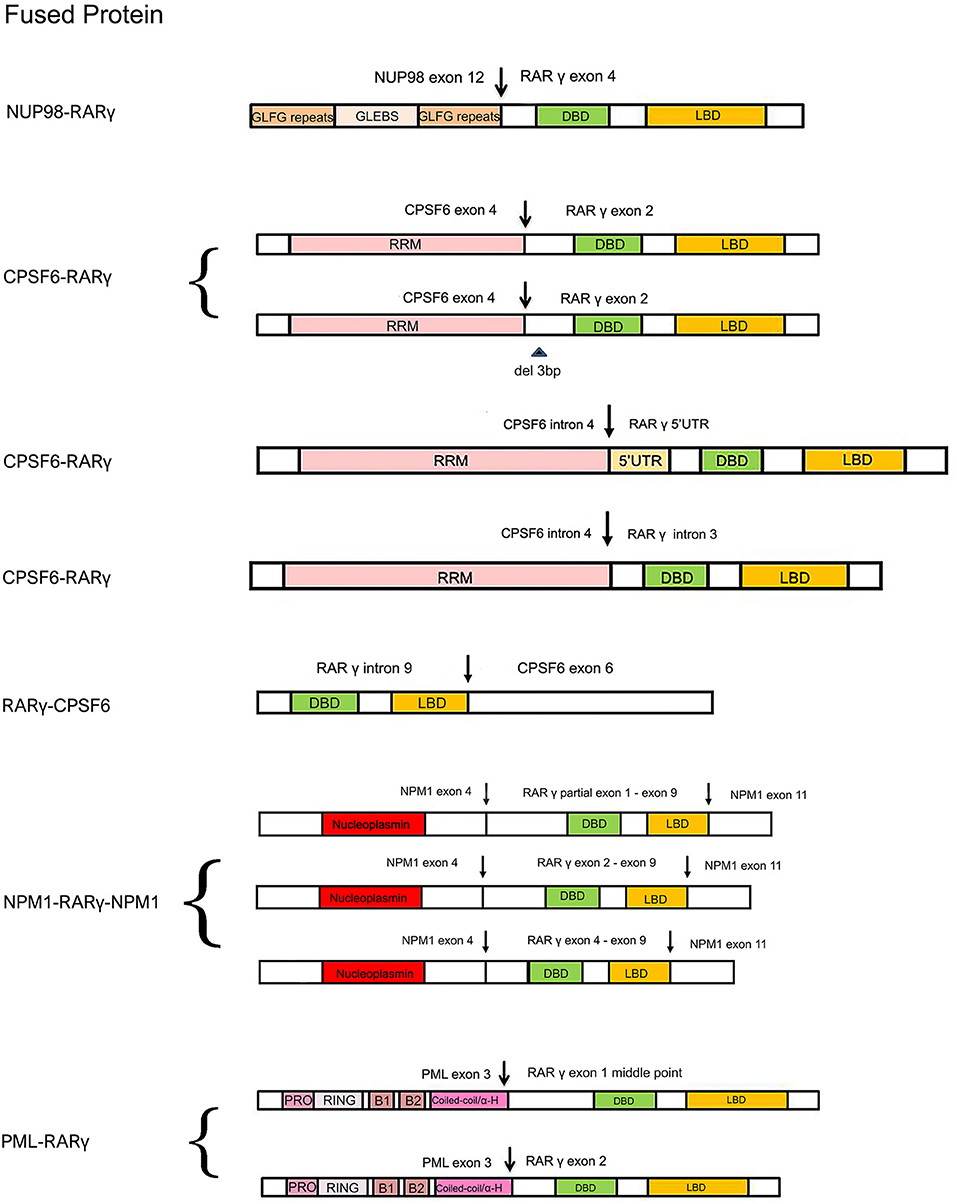
Figure 2. Schematic comparison of RARγ fusion proteins identified or expected in AML reported cases. The variably colored rectangles indicate all the important domains of RARγ and its partner proteins. The arrows indicate DNA break points. Curly brackets are used to group fusion proteins all found in one patient. GLFG, Gly-Leu-Phe-Gly; GLEBS, GLE2p-binding sequence; RRM, RNA recognition motif; PRO, proline-rich region; RING, RING finger; B1/B2, B-Box; Coiled-coil/α-H, coiled-coil/alpha-helical; NLS, nuclear localization signal.
The first AML patient with a RARG rearrangement, NUP98-RARG, was reported in 2011 (Such et al., 2011). NUP98 is the nucleoporin 98 gene mapped on chromosome 11p15, frequently involved in several rearrangements in both myeloid and lymphoid malignancies (Romana et al., 2006; La Starza et al., 2009). The patient (no. 1, Table 1) was a 35-years-old man exhibiting morphologic and clinical characteristics resembling APL. The genomic aberration found was translocation t(11;12)(p15;q13), plus a 1.0 Mb microdeletion in 11p15 and a 2.5 Mb microdeletion in 12q13. NUP98 exon 12 was fused in frame to RARG exon 4. Interestingly, the NUP98 5′ region encodes Gly-Leu-Phe-Gly-repeats (GLFG) and GLE2p-binding sequence (GLEBS)-like motifs, which act as transcription activators, providing docking sites for the nuclear transport of RNA and protein between the nucleus and the cytoplasm; while the 3′ region RARG involved in the fusion includes DBD and LBD. The mechanism underlying the transformation mediated by NUP98-RARG is unknown. The patient was started on ATRA treatment that was soon discontinued and switched to a standard 7 + 3 schedule (cytarabine and idarubicin) as induction therapy, then he achieved complete remission (CR). Two years later, the patient relapsed; he achieved second CR but then he died due to an infection complication (Such et al., 2011). Several studies were conducted by Qiu et al. focused on the RARγ involvement in retinoid signaling. Using green fluorescent protein-tagged versions of these proteins and analyzing their signal, they demonstrated that the novel NUP98-RARγ fusion protein acquires a different subcellular localization compared to NUP98 and wild-type RARγ. This localization is strongly dependent on the RARγ DBD domain, as shown by different experiments conducted using mutant variants of NUP98-RARγ. They also verified, through co-immunoprecipitation assays, the NUP98-RARγ ability to form aberrant homo-dimers and homo-oligomers, but this feature is granted by the NUP98 portion of the chimeric protein. The RARγ fraction is essential for interacting with RXRα: they also demonstrated that the RXR agonist can suppress the NUP98-RARγ/RXR connection. Luciferase reporter experimental results also suggested that NUP98-RARγ may have equivalent transcriptional properties to those of the RARα fusion protein, involved in APL, on its downstream targets. The oncogenic potential of NUP98-RARγ was confirmed by retroviral transduction/transformation assay in murine HSCs; furthermore, the cellular transformation ability requires both RARγ and NUP98 contributions and these transformed cells are sensitive to RXR agonist and ATRA treatment (Qiu et al., 2015). It was necessary to conduct several experiments to test ATRA responsivity since the first patient with RARG rearrangement received ATRA simultaneously to chemotherapy, so it was difficult to evaluate the effective in vivo responsivity to ATRA, even if it has been reported that fusion proteins generated with RARγ confer responsiveness to ATRA in vitro and in vivo (Marinelli et al., 2007, 2009). The in vitro studies showed that the NUP98/RARG rearrangement confers ATRA resistance (Such et al., 2014). Moreover, another patient (no. 2, Table 1) with NUP98/RARG exhibited a primary resistance to ATRA treatment (Luo et al., 2019). Sequencing of the fusion transcript showed also that NUP98 exon 12 was fused in frame to RARG exon 4, in the first patient described, and the Wilms' tumor (WT1) mutation was also identified. These studies point out an evident paradox of ATRA resistance/responsiveness in vitro and in vivo: it may depend on the culture assay condition or on the acquisition of additional mutations causing resistance (Such et al., 2014; Qiu et al., 2015). A third case of NUP98-RARG AML (no. 3, Table 1) was observed by Zhang et al. The break points were the same as in the other two cases reported in literature. In addition, two WT1 mutations were found. Given the two previous cases, the author established that NUP98-RARG AML is sensitive to the standard 3+7 regimen, and that allogeneic HSC transplantation (allo-HSCT) may be the best procedure to achieve CR. It is possible that additional genetic mutations can cause ATRA resistance (Zhang et al., 2019b).
Cleavage and polyadenylation specific factor 6 (CPSF6) is a subunit of cleavage factor I, which is an RNA binding protein complex involved in the alternative cleavage and polyadenylation process (Millevoi and Vagner, 2010). Although several studies have demonstrated the involvement of CPSF6 in HIV infection and breast cancer (Rasheedi et al., 2016; Binothman et al., 2017), the role of CPSF6 in myeloid leukemia has not yet been clarified. The first case of CPSF6-RARG rearrangement was found by Qin et al. in an AML 38-years-old male (no. 4, Table 1) presenting clinical and morphological features of classic APL. Both CPSF6 and RARG map on chromosome 12. In addition, a WT1 mutation was identified. The rearrangement within chromosome 12 led to the generation of two different fusion transcripts, depending on the breaking points: the major transcript made up by CPSF6 exon 4 fused to RARG exon 2 and the minor transcript by a deletion of 3 bp at the 5′ end of RARG exon 2. As regards therapy, the patient was treated with ATRA, but the ATRA treatment was interrupted because of the differentiation syndrome, so the best therapy has still to be established (Qin et al., 2018). Two cases of CPSF6-RARG rearrangements were then identified, with different genomic aberrations. In patient no. 5 (Table 1) a tetraploidy karyotype in two metaphases and a DNMT3A mutation were found; in patient no. 6 (Table 1) a del(12)(p12) and WT1 and K-RAS mutations were detected. Both patients showed ATRA resistance. The breakpoint in CPSF6 was located in intron 4 in both patients, while there were 2 breakpoints in intron 3 or 5′UTR and the telomeric region of exon 9 of RARG. The 3′ region of RARG (from exon 1 or exon 4 to exon 9) was reversed and fused in frame with the 5′ region of the CPSF6 gene (from exon 1 to exon 4). Even in these rearrangements, RARγ retained DBD and LBD domain, as well as in the NUP98-RARG rearrangements described before. Moreover, the RARG breakpoint in patient no. 5 is the same as in NUP98-RARG, patient no.1 (Table 1) (Such et al., 2011; Liu et al., 2018). These two cases were included in a study which enrolled 1,401 patients with suspected AML. Among these, 19 patients had alternative RARA or RARG rearrangements. The paper shows an interesting point about the prognostic impact of the presence of this kind of rearrangements: the overall survival and the leukemia-free survival were significantly lower in patients with alternative RARA or RARG fusions in comparison with patients with the PML-RARA classic gene fusion (Wen et al., 2019). Interestingly, the RARG-CPSF6 fusion was identified in a 26-years-old patient (no. 7, Table 1) with AML resembling APL, with a profoundly rearranged region on chromosome 12, where both RARG and CPSF6 are mapped, featuring breakpoints in these two genes and in the eukaryotic translation initiation factor 4B (EIF4B). RARG intron 9 was fused to EIF4B intron 8, but there was a deletion after this point and EIFB4 was then fused into the intron leading into CPSF6 exon 6. These rearrangements led to the formation of a novel RARG-CPSF6 transcript fusion, but there was no evidence of the corresponding protein (Miller et al., 2018).
Recently, another gene involved in the RARG rearrangement was identified. NPM1 is the nucleophosmin 1 gene mapped on chromosome 5 and it has been indicated as a partner of RARA in APL variant translocations (Kikuma et al., 2015). In this AML case (no. 8, Table 1), NPM1 is fused with RARG. There was a deletion of about 16 kb of NPM1 from intron 4 to intron 10 and an insertion of about 23 kb of RARG from 5′ UTR to intron 9. Sequencing analysis revealed that both NMP1 and RARG had two breakpoints each. The NPM1 breakpoints were located in intron 4 and intron 10; the RARG breakpoints were identified in 5′UTR and intron 9. The presence of three NPM1-RARG-NPM1 different chimeric transcripts was confirmed: from exon 1 to exon 4 of NPM1 fused with RARG, from partial exon 1 to exon 9, then fused with exon 11 of NPM1; exon 1 to exon 4 of NPM1 fused with RARG, from exon 2 to exon 9, then fused with exon 11 of NPM1; exon 1 to exon 4 of NPM1 fused with RARG, from exon 4 to exon 9, then fused with exon 11 of NPM1. In all three transcripts RARG maintained its DBD, while the deletion of RARG exon 10 resulted in the loss of LBD 25 amino acids. The patient, like others harboring RARG rearrangements, had an APL-like clinical presentation and morphological features, so he received ATRA treatment, but resulted resistant. This is a unique case of AML with NPM1-RARG-NMP1 chimeric fusion (Chen et al., 2019).
PML is well-known to be involved in translocation with RARA in APL, in which they produce the PML-RARα chimeric protein that behaves as an altered RARα, and represses the transcription of RA target genes (Kakizuka et al., 1991). In vitro and in vivo studies had already demonstrated that PML-RARγ has an oncogenic potential (Marinelli et al., 2007, 2009). Ha et al. identified a novel involvement of PML in a fusion with RARG in an AML patient (no. 9, Table 1) with morphologic and immunophenotypic features of classical APL. The translocation t(12;15)(q13;q22) was identified; the breakpoints were on intron 3 of PML and the 5′ upstream region of RARG. Two kinds of PML-RARG transcripts arise from this gene fusion: a longer transcript (PML exon3-RARG exon1 middle point) and a shorter one (PML exon3-RARG exon2). RARγ preserves its DBD and LBD, as in the other cases. It may be interesting to note that the fusion site of PML in this rearrangement is consistent with breakpoint cluster region 3 (bcr3) type of PML-RARA. The patient had intermittent ATRA treatment, so the sensitivity was not clarified (Ha et al., 2017).
Physiologically, the enhancer of zeste 2 polycomb repressive complex 2 subunit (EZH2) is involved in gene silencing thanks to its capability to recruit DNA methyltransferases (DNMTs) for gene repression and its interaction with two epigenetic repression systems (Viré et al., 2006; Sashida and Iwama, 2017). Epigenetic alterations of RARs genes can also lead to an AML resembling APL. In one patient (no. 10, Table 1) presenting similar features of APL an EZH2 mutation was found by our group, without any RARs rearrangement (Coccaro et al., 2018). The EZH2 mutation was in its DNMT binding region. Analysis showed a profound downregulation of RARA and RARG expression compared with APL, AML and normal control pools (Coccaro et al., 2018). Zhang et al. also reported that a patient harboring ELL-MLL (no. 11, Table 1) can seem to have APL; in particular RARG was upregulated (Zhang et al., 2019a).
Despite the diversity of gene partners, RARG rearrangements identified up to now have shown similar features, such as the retention of DBD and LBD RARγ domains; the presence of the chimeric transcript, but not always of the protein, and ATRA resistance. Probably, DBD/LBD maintenance affects the fusion protein different nuclear localization and is involved in the leukemogenesis process, leading to an alteration especially of the granulocyte lineage, as well as PML-RARA. Nevertheless, RARγ, like RARα, is critical for the physiological hemopoietic process, although its role is not comparable. In fact, RARγ is involved in maintaining a balance between the self-renewal and differentiation of HSCs rather than in the granulocytic differentiation. Surely, RARγ exhibits an indirect role in this, but it is not clear why RARG rearrangements are capable of resembling APL phenotype. Hence, this aspect must be more closely investigated, mainly because these patients seem to be ATRA-resistant. Indeed, ATRA responsivity also needs to be clarified since in vitro and in vivo studies have shown contrasting results. In addition, in most reported cases, several mutations in different genes have been identified, especially in the WT1 gene. This factor could be crucial in the oncogenic process and might also influence ATRA sensitivity. In our opinion, further studies are needed to better understand the oncogenic role of RARG rearrangements, and the molecular features of both the chimeric transcript and fusion protein, in particular to define the best therapy for these APL atypical patients.
MC and IR conceived and wrote the review. LA and AZ performed the literature analysis. FA and GS supervised and approved the final manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The authors would like to thank Mary Victoria Pragnell, B.A. for language revision of the manuscript. This work was supported by Associazione Italiana contro le Leucemie (AIL)-BARI.
RARγ, retinoic acid receptor γ; RARα, retinoic acid receptor α; RARβ, retinoic acid receptor β; APL, acute promyelocytic leukemia; BM, bone marrow; AML, acute myeloid leukemia; ATRA, all-trans retinoic acid; ATO, arsenic trioxide; PML, promyelocytic leukemia protein; DBD, DNA binding domain; RAREs, retinoic acid response elements; RXRs, retinoid X receptors; RA, retinoic acid; HSCs, hematopoietic stem cells; PB, peripheral blood; NUP98, nucleoporin 98; CR, complete remission; WT1, Wilms’ tumor; Allo-HSCT, allogeneic HSC transplantation; CPSF6, Cleavage and polyadenylation specific factor 6; OS, overall survival; LFS, leukemia free survival; EIF4B, eukaryotic translation initiation factor 4B; NPM1, nucleophosmin 1; bcr3, breakpoint cluster region 3; EZH2, enhancer of zeste 2 polycomb repressive complex 2 subunit; DNMTs, DNA methyltransferases.
Al Tanoury, Z., Piskunov, A., and Rochette-Egly, C. (2013). Vitamin A and retinoid signaling: genomic and nongenomic effects. J. Lipid Res. 54, 1761–1775. doi: 10.1194/jlr.R030833
Arber, D. A., Orazi, A., Hasserjian, R., Thiele, J., Borowitz, M. J., Le Beau, M. M., et al. (2016). The 2016 revision to the World Health Organization classification of myeloid neoplasms and acute leukemia. Blood 127, 2391–2405. doi: 10.1182/blood-2016-03-643544
Asou, N., Adachi, K., Tamura, J., Kanamaru, A., Kageyama, S., Hiraoka, A., et al. (1998). Analysis of prognostic factors in newly diagnosed acute promyelocytic leukemia treated with all-trans retinoic acid and chemotherapy. Japan Adult Leukemia Study Group. J. Clin. Oncol. 16, 78–85. doi: 10.1200/JCO.1998.16.1.78
Bastien, J., and Rochette-Egly, C. (2004). Nuclear retinoid receptors and the transcription of retinoid-target genes. Gene 328, 1–16. doi: 10.1016/J.GENE.2003.12.005
Beard, R. L., Duong, T. T., Teng, M., Klein, E. S., Standevan, A. M., and Chandraratna, R. A. S. (2002). Synthesis and biological activity of retinoic acid receptor-α specific amides. Bioorg. Med. Chem. Lett. 12, 3145–3148. doi: 10.1016/S0960-894X(02)00647-9
Binothman, N., Hachim, I. Y., Lebrun, J.-J., and Ali, S. (2017). CPSF6 is a clinically relevant breast cancer vulnerability target: role of CPSF6 in breast cancer. EBioMedicine 21, 65–78. doi: 10.1016/j.ebiom.2017.06.023
Blumberg, B., Mangelsdorf, D. J., Dyck, J. A., Bittner, D. A., Evans, R. M., and De Robertis, E. M. (1992). Multiple retinoid-responsive receptors in a single cell: families of retinoid ‘X' receptors and retinoic acid receptors in the Xenopus egg. Proc. Natl. Acad. Sci. U.S.A. 89, 2321–2325. doi: 10.1073/pnas.89.6.2321
Chambon, P. (1993). The molecular and genetic dissection of the retinoid signalling pathway. Gene 135, 223–228. doi: 10.1016/0378-1119(93)90069-F
Chambon, P. (1996). A decade of molecular biology of retinoic acid receptors. FASEB J. 10, 940–954. doi: 10.1096/FASEBJ.10.9.8801176
Chen, C. F., Goyette, P., and Lohnes, D. (2004). RARγ acts as a tumor suppressor in mouse keratinocytes. Oncogene 23, 5350–5359. doi: 10.1038/sj.onc.1207682
Chen, X., Wang, F., Zhang, Y., Teng, W., Cao, P., Ma, X., et al. (2019). A novel NPM1-RARG-NPM1 chimeric fusion in acute myeloid leukaemia resembling acute promyelocytic leukaemia but resistant to all-trans retinoic acid and arsenic trioxide. Br. J. Cancer 120, 1023–1025. doi: 10.1038/s41416-019-0456-z
Choudhry, A., and DeLoughery, T. G. (2012). Bleeding and thrombosis in acute promyelocytic leukemia. Am. J. Hematol. 87, 596–603. doi: 10.1002/ajh.23158
Coccaro, N., Zagaria, A., Orsini, P., Anelli, L., Tota, G., Casieri, P., et al. (2018). RARA and RARG gene downregulation associated with EZH2 mutation in acute promyelocytic-like morphology leukemia. Hum. Pathol. 80, 82–86. doi: 10.1016/j.humpath.2018.02.023
Conserva, M. R., Anelli, L., Zagaria, A., Specchia, G., and Albano, F. (2019). The pleiotropic role of retinoic acid/retinoic acid receptors signaling: from vitamin A metabolism to gene rearrangements in acute promyelocytic leukemia. Int. J. Mol. Sci. 20:2921. doi: 10.3390/ijms20122921
Daver, N., Kantarjian, H., Marcucci, G., Pierce, S., Brandt, M., Dinardo, C., et al. (2015). Clinical characteristics and outcomes in patients with acute promyelocytic leukaemia and hyperleucocytosis. Br. J. Haematol. 168, 646–653. doi: 10.1111/bjh.13189
De Kouchkovsky, I., and Abdul-Hay, M. (2016). Acute myeloid leukemia: a comprehensive review and 2016 update. Blood Cancer J. 6:e441. doi: 10.1038/bcj.2016.50
de Thé, H., Lavau, C., Marchio, A., Chomienne, C., Degos, L., and Dejean, A. (1991). The PML-RARα fusion mRNA generated by the t(15;17) translocation in acute promyelocytic leukemia encodes a functionally altered RAR. Cell 66, 675–684. doi: 10.1016/0092-8674(91)90113-D
Dewamitta, S. R., Joseph, C., Purton, L. E., and Walkley, C. R. (2014). Erythroid-extrinsic regulation of normal erythropoiesis by retinoic acid receptors. Br. J. Haematol. 164, 280–285. doi: 10.1111/bjh.12578
Ellinger-Ziegelbauer, H., and Dreyer, C. (1991). A retinoic acid receptor expressed in the early development of Xenopus laevis. Genes Dev. 5, 94–104. doi: 10.1101/gad.5.1.94
Giguère, V., Ong, E. S., Evans, R. M., and Tabin, C. J. (1989). Spatial and temporal expression of the retinoic acid receptor in the regenerating amphibian limb. Nature 337, 566–569. doi: 10.1038/337566a0
Green, A. C., Poulton, I. J., Vrahnas, C., Häusler, K. D., Walkley, C. R., Wu, J. Y., et al. (2015). RARγ is a negative regulator of osteoclastogenesis. J. Steroid Biochem. Mol. Biol. 150, 46–53. doi: 10.1016/j.jsbmb.2015.03.005
Green, A. C., Rudolph-Stringer, V., Straszkowski, L., Tjin, G., Crimeen-Irwin, B., Walia, M., et al. (2018). Retinoic acid receptor γ activity in mesenchymal stem cells regulates endochondral bone, angiogenesis, and B lymphopoiesis. J. Bone Miner. Res. 33, 2202–2213. doi: 10.1002/jbmr.3558
Ha, J.-S., Do, Y. R., Ki, C.-S., Lee, C., Kim, D.-H., Lee, W., et al. (2017). Identification of a novel PML-RARG fusion in acute promyelocytic leukemia. Leukemia 31, 1992–1995. doi: 10.1038/leu.2017.167
Ishikawa, T., Umesono, K., Mangelsdorf, D. J., Aburatani, H., Stanger, B. Z., Shibasaki, Y., et al. (1990). A functional retinoic acid receptor encoded by the gene on human chromosome 12. Mol. Endocrinol. 4, 837–844. doi: 10.1210/mend-4-6-837
Joseph, C., Nota, C., Fletcher, J. L., Maluenda, A. C., Green, A. C., and Purton, L. E. (2016). Retinoic acid receptor γ regulates B and T lymphopoiesis via nestin-expressing cells in the bone marrow and thymic microenvironments. J. Immunol. 196, 2132–2144. doi: 10.4049/jimmunol.1501246
Kakizuka, A., Miller, W. H., Umesono, K., Warrell, R. P., Frankel, S. R., Murty, V. V, et al. (1991). Chromosomal translocation t(15;17) in human acute promyelocytic leukemia fuses RAR alpha with a novel putative transcription factor, PML. Cell 66, 663–74.
Karim, F., Shaikh, U., Adil, S. N., and Khurshid, M. (2014). Clinical characteristics, outcome and early induction deaths in patients with acute promyelocytic leukaemia: a five-year experience at a tertiary care centre. Singapore Med. J. 55, 443–447. doi: 10.11622/SMEDJ.2014105
Kashyap, V., Laursen, K. B., Brenet, F., Viale, A. J., Scandura, J. M., and Gudas, L. J. (2013). RARγ is essential for retinoic acid induced chromatin remodeling and transcriptional activation in embryonic stem cells. J. Cell Sci. 126, 999–1008. doi: 10.1242/jcs.119701
Kastner, P., Krust, A., Mendelsohn, C., Garnier, J. M., Zelent, A., Leroy, P., et al. (1990). Murine isoforms of retinoic acid receptor gamma with specific patterns of expression. Proc. Natl. Acad. Sci. U.S.A. 87, 2700–4. doi: 10.1073/pnas.87.7.2700
Kastner, P., Leid, M., and Chambon, P. (1994). “Metabolism of vitamin A, the retinol-binding protein superfamily,” in Vitamin A in Health and Disease, ed. R. Blomhoff Dekker (New York, NY: Rune Blomhoff) 87–118.
Kikuma, T., Nakamachi, Y., Noguchi, Y., Okazaki, Y., Shimomura, D., Yakushijin, K., et al. (2015). A new transcriptional variant and small azurophilic granules in an acute promyelocytic leukemia case with NPM1/RARA fusion gene. Int. J. Hematol. 102, 713–718. doi: 10.1007/s12185-015-1857-2
La Starza, R., Brandimarte, L., Pierini, V., Nofrini, V., Gorello, P., Crescenzi, B., et al. (2009). A NUP98-positive acute myeloid leukemia with a t(11;12)(p15;q13) without HOXC cluster gene involvement. Cancer Genet. Cytogenet. 193, 109–111. doi: 10.1016/j.cancergencyto.2009.04.015
Leroy, P., Krust, A., Zelent, A., Mendelsohn, C., Garnier, J. M., Kastner, P., et al. (1991). Multiple isoforms of the mouse retinoic acid receptor alpha are generated by alternative splicing and differential induction by retinoic acid. EMBO J. 10, 59–69. doi: 10.1002/j.1460-2075.1991.tb07921.x
Levine, R. L. (2013). Molecular pathogenesis of AML: Translating insights to the clinic. Best Pract. Res. Clin. Haematol. 26, 245–248. doi: 10.1016/J.BEHA.2013.10.003
Liu, Q., and Linney, E. (1993). The mouse retinoid-X receptor-gamma gene: genomic organization and evidence for functional isoforms. Mol. Endocrinol. 7, 651–658. doi: 10.1210/mend.7.5.8391126
Liu, T., Wen, L., Yuan, H., Wang, Y., Yao, L., Xu, Y., et al. (2018). Identification of novel recurrent CPSF6-RARG fusions in acute myeloid leukemia resembling acute promyelocytic leukemia. Blood 131, 1870–1873. doi: 10.1182/blood-2017-11-818716
Lohnes, D., Kastner, P., Dierich, A., Mark, M., LeMeur, M., and Chambon, P. (1993). Function of retinoic acid receptor γ in the mouse. Cell 73, 643–658. doi: 10.1016/0092-8674(93)90246-M
Luo, H., Zhang, S., Li, K., Chen, X.-H., Li, Y.-C., Sun, Y., et al. (2019). A novel entity of acute myeloid leukaemia with recurrent RARG-rearrangement resembling acute promyelocytic leukaemia. Leuk. Res. 77, 14–16. doi: 10.1016/j.leukres.2018.12.009
Marinelli, A., Bossi, D., Pelicci, P. G., and Minucci, S. (2007). A redundant oncogenic potential of the retinoic receptor (RAR) α, β and γ isoforms in acute promyelocytic leukemia. Leukemia 21, 647–650. doi: 10.1038/sj.leu.2404572
Marinelli, A., Bossi, D., Pelicci, P. G., and Minucci, S. (2009). Redundant function of retinoic acid receptor isoforms in leukemogenesis unravels a prominent function of genome topology and architecture in the selection of mutagenic events in cancer. Leukemia 23, 417–419. doi: 10.1038/leu.2008.205
Mark, M., Ghyselinck, N. B., and Chambon, P. (2006). Function of retinoid nuclear receptors: lessons from genetic and pharmacological dissections of the retinoic acid signaling pathway during mouse embryogenesis. Annu. Rev. Pharmacol. Toxicol. 46, 451–480. doi: 10.1146/annurev.pharmtox.46.120604.141156
Mattei, M. G., Petkovich, M., Mattei, J. F., Brand, N., and Chambon, P. (1988). Mapping of the human retinoic acid receptor to the q21 band of chromosome 17. Hum. Genet. 80, 186–188.
Mattei, M. G., Rivière, M., Krust, A., Ingvarsson, S., Vennström, B., Islam, M. Q., et al. (1991). Chromosomal assignment of retinoic acid receptor (RAR) genes in the human, mouse, and rat genomes. Genomics 10, 1061–1069.
McGowan, S., Jackson, S. K., Jenkins-Moore, M., Dai, H.-H., Chambon, P., and Snyder, J. M. (2000). Mice bearing deletions of retinoic acid receptors demonstrate reduced lung elastin and alveolar numbers. Am. J. Respir. Cell Mol. Biol. 23, 162–167. doi: 10.1165/ajrcmb.23.2.3904
Meister, B., Fink, F. M., Hittmair, A., Marth, C., and Widschwendter, M. (1998). Antiproliferative activity and apoptosis induced by retinoic acid receptor-gamma selectively binding retinoids in neuroblastoma. Anticancer Res. 18, 1777–86.
Mic, F. A., Molotkov, A., Benbrook, D. M., and Duester, G. (2003). Retinoid activation of retinoic acid receptor but not retinoid X receptor is sufficient to rescue lethal defect in retinoic acid synthesis. Proc. Natl. Acad. Sci. U.S.A. 100, 7135–7140. doi: 10.1073/pnas.1231422100
Miller, C. A., Tricarico, C., Skidmore, Z. L., Uy, G. L., Lee, Y.-S., Hassan, A., et al. (2018). A case of acute myeloid leukemia with promyelocytic features characterized by expression of a novel RARG–CPSF6 fusion. Blood Adv. 2, 1295–1299. doi: 10.1182/bloodadvances.2017014183
Millevoi, S., and Vagner, S. (2010). Molecular mechanisms of eukaryotic pre-mRNA 3′ end processing regulation. Nucleic Acids Res. 38, 2757–2774. doi: 10.1093/nar/gkp1176
Nagpal, S., Saunders, M, Kastner, P, Durand, B, Nakshatri, H, and Chambon, P. (1992). Promoter context- and response element-dependent specificity of the transcriptional activation and modulating functions of retinoic acid receptors. Cell. 70, 1007–1019.
Noji, S., Nohno, T., Koyama, E., Muto, K., Ohyama, K., Aoki, Y., et al. (1991). Retinoic acid induces polarizing activity but is unlikely to be a morphogen in the chick limb bud. Nature 350, 83–86. doi: 10.1038/350083a0
Osumi, T., Tsujimoto, S., ichi, Tamura, M., Uchiyama, M., Nakabayashi, K., Okamura, K., et al. (2018). Recurrent RARB translocations in acute promyelocytic leukemia lacking RARA translocation. Cancer Res. 78, 4452–4458. doi: 10.1158/0008-5472.CAN-18-0840
Padanilam, B. J., Mcleod, L. B., Suzuki, H., and Solursh, M. (1991). Nucleotide sequence of an isoform of chicken retinoic acid binding protein-β varying in its A domain. Nucleic Acids Res. 19, 395–395. doi: 10.1093/nar/19.2.395
Purton, L. E. (2007). Roles of retinoids and retinoic Acid receptors in the regulation of hematopoietic stem cell self-renewal and differentiation. PPAR Res. 2007:87934. doi: 10.1155/2007/87934
Purton, L. E., Dworkin, S., Olsen, G. H., Walkley, C. R., Fabb, S. A., Collins, S. J., et al. (2006). RARgamma is critical for maintaining a balance between hematopoietic stem cell self-renewal and differentiation. J. Exp. Med. 203, 1283–1293. doi: 10.1084/jem.20052105
Qin, Y.-Z., Huang, X.-J., and Zhu, H.-H. (2018). Identification of a novel CPSF6-RARG fusion transcript in acute myeloid leukemia resembling acute promyelocytic leukemia. Leukemia 32, 2285–2287. doi: 10.1038/s41375-018-0095-z
Qiu, J. J., Zeisig, B. B., Li, S., Liu, W., Chu, H., Song, Y., et al. (2015). Critical role of retinoid/rexinoid signaling in mediating transformation and therapeutic response of NUP98-RARG leukemia. Leukemia 29, 1153–1162. doi: 10.1038/leu.2014.334
Ragsdale, C. W., Petkovich, M., Gates, P. B., Chambon, P., and Brockes, J. P. (1989). Identification of a novel retinoic acid receptor in regenerative tissues of the newt. Nature 341, 654–657. doi: 10.1038/341654a0
Rasheedi, S., Shun, M.-C., Serrao, E., Sowd, G. A., Qian, J., Hao, C., et al. (2016). The cleavage and polyadenylation specificity factor 6 (CPSF6) subunit of the capsid-recruited pre-messenger RNA cleavage factor I (CFIm) complex mediates HIV-1 integration into genes. J. Biol. Chem. 291, 11809–11819. doi: 10.1074/jbc.M116.721647
Romana, S., Radford-Weiss, I., Abdelali, R., Ben, Schluth, C., Petit, A., Dastugue, N., et al. (2006). NUP98 rearrangements in hematopoietic malignancies: a study of the Groupe Francophone de Cytogénétique Hématologique. Leukemia 20, 696–706. doi: 10.1038/sj.leu.2404130
Rowe, A., Richman, J. M., and Brickell, P. M. (1991). Retinoic acid treatment alters the distribution of retinoic acid receptor-beta transcripts in the embryonic chick face. Development 111, 1007–1016.
Sashida, G., and Iwama, A. (2017). Multifaceted role of the polycomb-group gene EZH2 in hematological malignancies. Int. J. Hematol. 105, 23–30. doi: 10.1007/s12185-016-2124-x
Sill, H., Olipitz, W., Zebisch, A., Schulz, E., and Wölfler, A. (2011). Therapy-related myeloid neoplasms: pathobiology and clinical characteristics. J. Pharmacol. 162, 792–805. doi: 10.1111/j.1476-5381.2010.01100.x
Smith, S. M., and Eichele, G. (1991). Temporal and regional differences in the expression pattern of distinct retinoic acid receptor-beta transcripts in the chick embryo. Development 111, 245–252.
Spanjaard, R. A., Ikeda, M., Lee, P. J., Charpentier, B., Chin, W. W., and Eberlein, T. J. (1997). Specific activation of retinoic acid receptors (RARs) and retinoid X receptors reveals a unique role for RARgamma in induction of differentiation and apoptosis of S91 melanoma cells. J. Biol. Chem. 272, 18990–18999. doi: 10.1074/jbc.272.30.18990
Storlazzi, C. T., Albano, F., Lo Cunsolo, C., Doglioni, C., Guastadisegni, M. C., Impera, L., et al. (2007). Upregulation of the SOX5 by promoter swapping with the P2RY8 gene in primary splenic follicular lymphoma. Leukemia 21, 2221–2225. doi: 10.1038/sj.leu.2404784
Such, E., Cervera, J., Valencia, A., Barragán, E., Ibañez, M., Luna, I., et al. (2011). A novel NUP98/RARG gene fusion in acute myeloid leukemia resembling acute promyelocytic leukemia. Blood 117, 242–245. doi: 10.1182/blood-2010-06-291658
Such, E., Cordón, L., Sempere, A., Villamón, E., Ibañez, M., Luna, I., et al. (2014). In vitro all-trans retinoic acid sensitivity of acute myeloid leukemia blasts with NUP98/RARG fusion gene. Ann. Hematol. 93, 1931–1933. doi: 10.1007/s00277-014-2073-5
Szotkowski, T., Faber, E., Hubacek, J., Raida, L., Rohon, P., Kuba, A., et al. (2015). Acute promyelocytic leukemia successfully treated also in elderly patients with significant comorbidities: a 20-year single-center experience. Neoplasma 62, 146–151. doi: 10.4149/neo_2015_019
Taneja, R., Bouillet, P., Boylan, J. F., Gaub, M. P., Roy, B., Gudas, L. J., et al. (1995). Reexpression of retinoic acid receptor (RAR) gamma or overexpression of RAR alpha or RAR beta in RAR gamma-null F9 cells reveals a partial functional redundancy between the three RAR types. Proc. Natl. Acad. Sci. U.S.A. 92, 7854–7858. doi: 10.1073/pnas.92.17.7854
Thomas, D., and Majeti, R. (2017). Biology and relevance of human acute myeloid leukemia stem cells. Blood 129, 1577–1585. doi: 10.1182/blood-2016-10-696054
Viré, E., Brenner, C., Deplus, R., Blanchon, L., Fraga, M., Didelot, C., et al. (2006). The Polycomb group protein EZH2 directly controls DNA methylation. Nature 439, 871–874. doi: 10.1038/nature04431
Walkley, C. R., Olsen, G. H., Dworkin, S., Fabb, S. A., Swann, J., McArthur, G. A., et al. (2007). A microenvironment-induced myeloproliferative syndrome caused by retinoic acid receptor gamma deficiency. Cell 129, 1097–1110. doi: 10.1016/j.cell.2007.05.014
Wen, L., Xu, Y., Yao, L., Wang, N., Wang, Q., Liu, T., et al. (2019). Clinical and molecular features of acute promyelocytic leukemia with variant retinoid acid receptor fusions. Haematologica 104, e195–e199. doi: 10.3324/haematol.2018.205369
Wendling, O., Ghyselinck, N. B., Chambon, P., and Mark, M. (2001). Roles of retinoic acid receptors in early embryonic morphogenesis and hindbrain patterning. Development 128, 2031–2038.
Williams, J. A., Kondo, N., Okabe, T., Takeshita, N., Pilchak, D. M., Koyama, E., et al. (2009). Retinoic acid receptors are required for skeletal growth, matrix homeostasis and growth plate function in postnatal mouse. Dev. Biol. 328, 315–327. doi: 10.1016/J.YDBIO.2009.01.031
Zelent, A., Mendelsohn, C., Kastner, P., Krust, A., Garnier, J. M., Ruffenach, F., et al. (1991). Differentially expressed isoforms of the mouse retinoic acid receptor beta generated by usage of two promoters and alternative splicing. EMBO J. 10, 71–81. doi: 10.1002/j.1460-2075.1991.tb07922.x
Zhang, X., Huang, X., Xu, H., Li, J., and Yu, W. (2019a). MLL-rearrangement can resemble acute promyelocytic leukemia. Leuk. Lymphoma 1–3. doi: 10.1080/10428194.2019.1607328
Keywords: retinoic acid receptor γ, acute promyelocytic leukemia, acute myeloid leukemia, gene fusions, protein fusions
Citation: Conserva MR, Redavid I, Anelli L, Zagaria A, Specchia G and Albano F (2019) RARG Gene Dysregulation in Acute Myeloid Leukemia. Front. Mol. Biosci. 6:114. doi: 10.3389/fmolb.2019.00114
Received: 08 July 2019; Accepted: 10 October 2019;
Published: 24 October 2019.
Edited by:
Anton A. Buzdin, I.M. Sechenov First Moscow State Medical University, RussiaReviewed by:
Nikolay Mikhaylovich Borisov, I.M. Sechenov First Moscow State Medical University, RussiaCopyright © 2019 Conserva, Redavid, Anelli, Zagaria, Specchia and Albano. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Francesco Albano, ZnJhbmNlc2NvLmFsYmFub0B1bmliYS5pdA==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.