- 1Department of Medical Genetics, Shahid Beheshti University of Medical Sciences, Tehran, Iran
- 2Clinical Research Development Center (CRDU), Qom University of Medical Sciences, Qom, Iran
- 3Isfahan Neuroscience Research Center, Isfahan University of Medical Sciences, Isfahan, Iran
- 4Urogenital Stem Cell Research Center, Shahid Beheshti University of Medical Sciences, Tehran, Iran
Background: Long non-coding RNAs (lncRNAs) are a group of functional transcripts that are not translated to proteins. Recent investigations have underscored their role in the pathogenesis of neurodevelopmental disorders.
Methods: In the current investigation, we quantified expression levels of four lncRNAs (HOXA-AS2, SPRY4-IT1, MEG3, and LINC-ROR) in peripheral blood of epileptic patients and normal controls.
Results: Expression of HOXA-AS2 was significantly higher in patients compared with controls (Posterior beta = 1.982, P = 0.001). We detected interaction effects of gender on expression of HOXA-AS2 (P = 0.012). Further analyses showed over-expression of HOXA-AS2 in male patients compared with male controls (P = 0.003), in spite of similar levels of expression between female cases and female controls (P = 0.77). Expression of SPRY4-IT1 was higher in total patients compared with total controls (Posterior beta = 1.27, P = 0.02). Such difference was only observed between male patients and male controls when dividing study participants based on their gender (P = 0.012). There was no significant difference in expression of MEG3 and LINC-ROR between patients and controls.
Conclusion: Expression levels of all lncRNAs were correlated with each other with r values ranging from 0.61 to 0.76 (P < 0.0001). However, expressions of none of lncRNAs were correlated with age of study participants. The current data implies a putative role for two lncRNAs in the pathogenesis of epilepsy and warrants future functional studies to verify the observed association.
Introduction
The role of long non-coding RNAs (lncRNAs) has been recently assessed in human diseases such as cancer and neurodevelopmental disorders (Shao and Chen, 2017; Sanchez Calle et al., 2018; Mazdeh et al., 2019). Epilepsy is among neurodevelopmental disorders in which lncRNAs might be involved (Shao and Chen, 2017). This chronic disorder is characterized by the presence of periodic unprovoked seizures and affects over 50 million individuals globally (de Boer et al., 2008). Although a number of anti-epileptic drugs (AED) have been approved for clinical use, none of them have attempted to influence the pathologic process of epilepsy (Shao and Chen, 2017). Thus, new methods for treatment of this disorder are highly appreciated. A prerequisite for development of such modalities is clarification of the pathogenesis of epilepsy. A previous microarray study in animal models has identified hundreds of aberrantly expressed lncRNAs in epileptic animals (Lee et al., 2015). Neurogenesis, production of neurotransmitter, passage of ions through cellular membranes, and synaptic plasticity are among mechanisms which are influenced by lncRNAs (Ng et al., 2013). Moreover, lncRNAs regulate several steps of neurogenesis during embryonic development whose abnormalities might be involved in the epilepsy (Mercer et al., 2010). Based on the above mentioned studies, we focused on a number of lncRNAs with putative roles in the epileptogenic process. The lncRNA HOXA Cluster Antisense RNA 2 (HOXA-AS2) modulates the expression of SCN3A (Wu et al., 2019), an acknowledged gene in infantile epileptic encephalopathy (Zaman et al., 2018). Moreover, this antisense RNA is transcribed from HOXA cluster which contains genes being expressed in several regions of the developing brain (Nolte and Krumlauf, 2007). A recent study has also highlighted the role of aberrant DNA methylation of the HOXA gene cluster in the pathogenesis of Alzheimer's disease (Smith et al., 2018). SPRY4 intronic transcript 1 (SPRY4-IT1) has a role in regulation of expression of estrogen-related receptor α (ERRα) (Yu et al., 2017), an orphan receptor with extensive expression in brain (Saito and Cui, 2018). The maternally expressed gene 3 (MEG3) in highly expressed in brain and contributes in neuronal cell damage caused by subarachnoid hemorrhage through suppression of the PI3K/Akt pathway (Liang et al., 2018). The PI3K/Akt pathway has a protective role against epileptic seizure through enhancement of astrocyte proliferation and survival (Cao et al., 2018). Long Intergenic Non-Protein Coding RNA, Regulator Of Reprogramming (LINC-ROR) contributes in stem cell pluripotency (Loewer et al., 2010). In the current study, we measured expression of HOXA-AS2, SPRY4-IT1, MEG3, and LINC-ROR in the peripheral blood of epileptic patients and normal individuals to appraise their contribution in epileptogenic processes.
Materials and Methods
Study Participants
Totally, 40 epileptic patients and 40 normal individual were recruited. All patients had juvenile myoclonic epilepsy. All of them were taking valproic acid (between 3 and 18 months). Patients had no seizure attack throughout 6 months beforehand. None of them experienced febrile seizures. Diagnosis was based on electroencephalogram (EEG) and brain magnetic resonance imaging (MRI) [diffusion weighted (DW), T1, T2, and gradient eco images]. The study was approved by the ethics committee of Shahid Beheshti University of Medical Sciences. Informed consent forms were signed by all participants and the parents/Legally Authorized Representative of participants who were included in the study. Control group was chosen from individual who had no neurological, psychiatric or systemic disorder.
Expression Assays
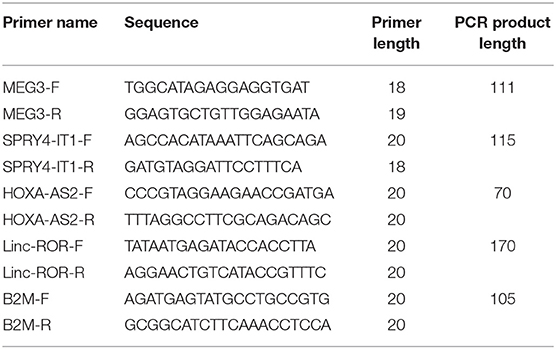
Five milliliters of peripheral blood was gathered from patients and control individuals in tubes containing 5 mM EDTA. Total RNA was extracted from blood specimens using Hybrid-RTM blood RNA extraction Kit (GeneAll, Seoul, South Korea). Next, the appropriateness of extracted RNA for additional phases of expression assay was judged using NanoDrop equipment (Thermo Scientific, MA, USA). First strand cDNA was produced using the OneStep RT-PCR Series Kit (BioFact™, Seoul, South Korea) according to company guidelines. Transcript levels of lncRNAs were quantified by using RealQ Plus 2 × PCR Master Mix Green Without ROX™ PCR Master Mix (Ampliqon, Odense, Denmark) in StepOnePlus™ RealTime PCR System (Applied Biosystems, Foster city, CA, USA). B2M gene was used as normalizer. Detailed information of primers is demonstrated in Table 1.
Statistical Analyses
Analyses were performed in R software version 3.3.2. The differences in expression of lncRNAs between epileptic patients and normal individuals were assessed using Bayesian estimation. Normal distribution was assumed for parameters with 200,000 iterations. Spearman correlation test was applied for assessment correlation between lncRNAs expression amount as well as lncRNA expression and age. P <0.05 were regarded as significant.
Results
Detailed Data of Study Participants
The available information on study participants is presented in Table 2.
Expression Assays
Expressions of HOXA-AS2 and SPRY4-IT1 were significantly different between male patients and male controls (Figure 1). However, there was no significant difference in expression of MEG3 and LINC-ROR between patients and controls.
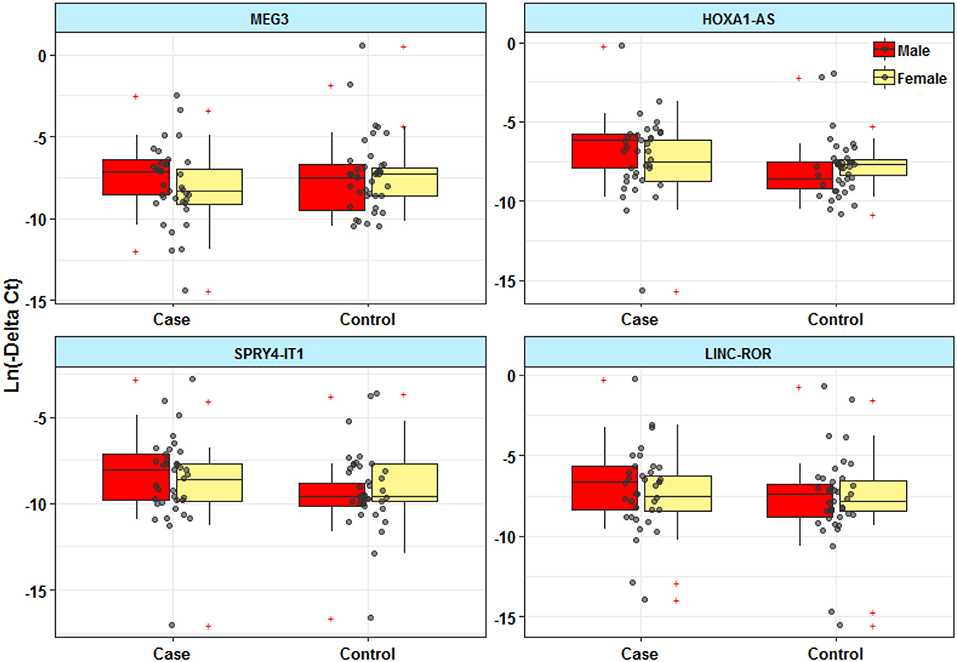
Figure 1. Expression levels of lncRNAs in peripheral blood of epileptic patients and healthy subjects.
Table 3 shows the detailed results of Bayesian Regression model for comparison of expression levels of lncRNAs between epileptic patients and healthy subjects. Expression of HOXA-AS2 was significantly higher in patients compared with controls (Posterior beta = 1.982, P = 0.001). We detected interaction effects of gender on expression of HOXA-AS2 (P = 0.012). Further analyses showed over-expression of HOXA-AS2 in male patients compared with male controls (P = 0.003), in spite of similar levels of expression between female cases and female controls (P = 0.77).
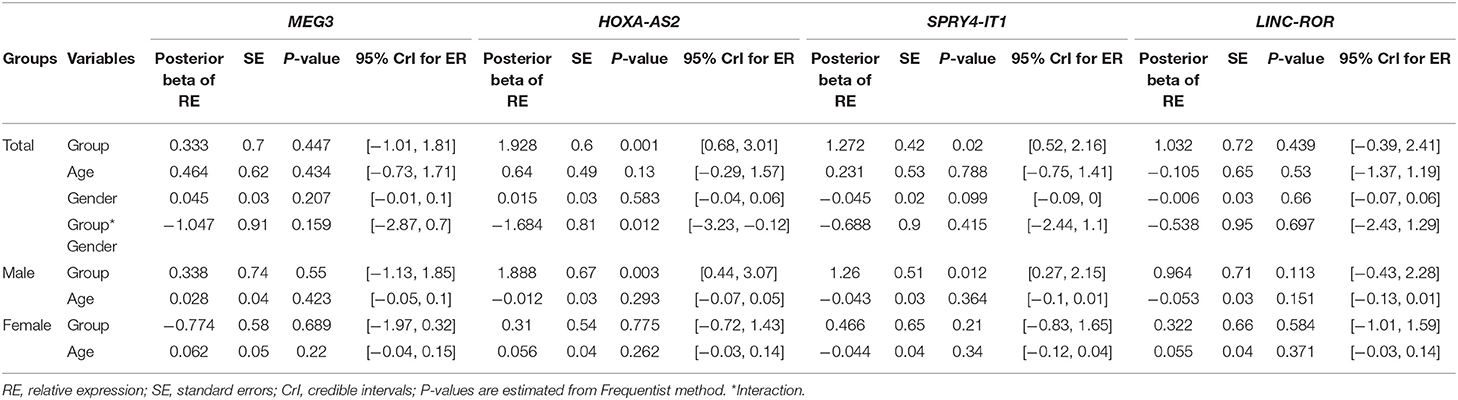
Table 3. Results of Bayesian Regression model for comparison of expression levels of lncRNAs between epileptic patients and healthy subjects with adjusting the effects of age and gender.
Expression of SPRY4-IT1 was higher in total patients compared with total controls (Posterior beta = 1.27, P = 0.02). Such difference was only observed between male patients and male controls when dividing study participants based on their gender (P = 0.012).
Correlations Between Expression Levels of lncRNAs
Expression levels of all lncRNAs were correlated with each other with r values ranging from 0.61 to 0.76 (P <0.0001). However, expressions of none of lncRNAs were correlated with age of study participants (Figure 2).
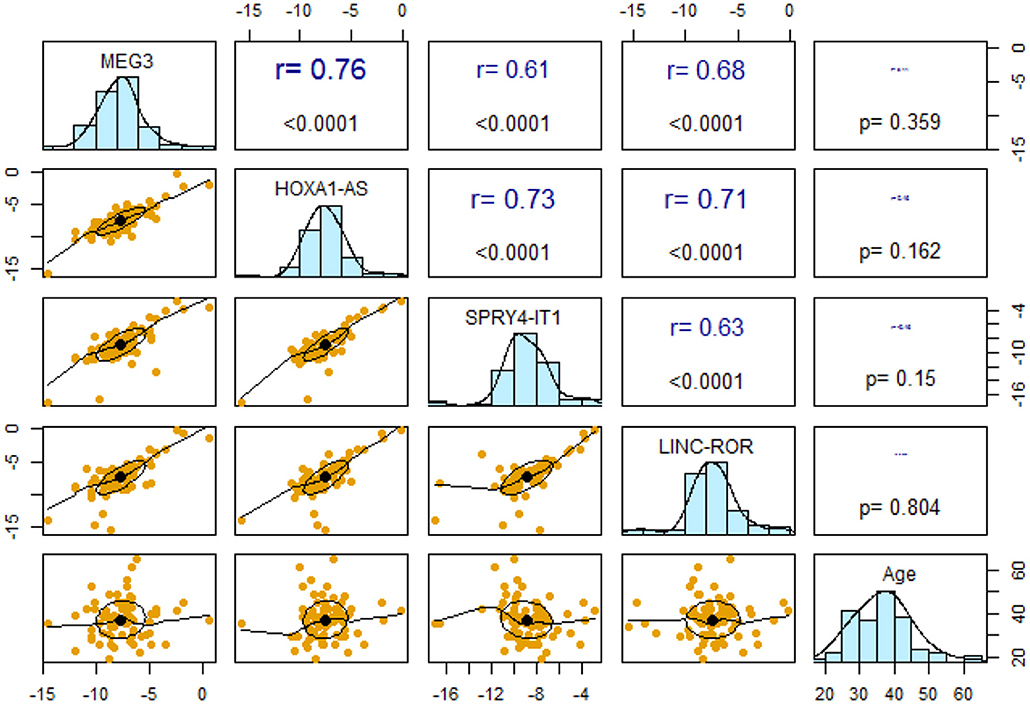
Figure 2. Pairwise correlations between expression levels of lncRNAs and their correlation with age.
Subsequently, we repeated correlation analyses in distinct gender-based groups of cases and controls (Table 4). The SPRY4-IT1/LINC-ROR and SPRY4-IT1/MEG3 pairwise correlations were insignificant in female controls.
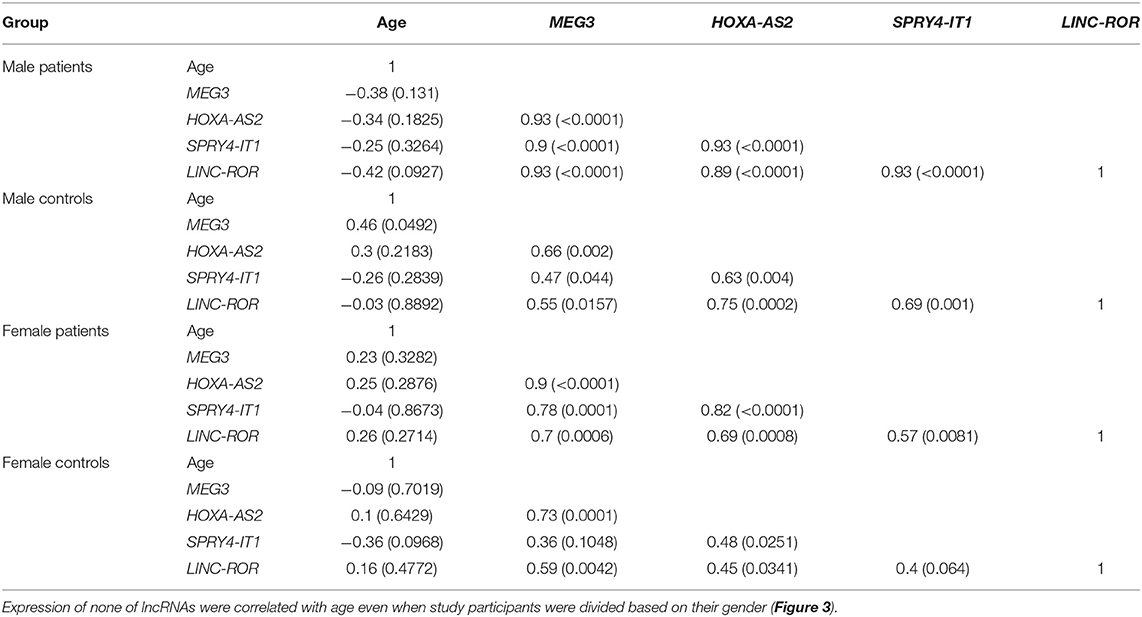
Table 4. Correlations between expression levels of lncRNAs in distinct groups [Spearman correlation coefficients (P-values) are presented].
Expression of none of lncRNAs were correlated with age even when study participants were divided based on their gender (Figure 3).
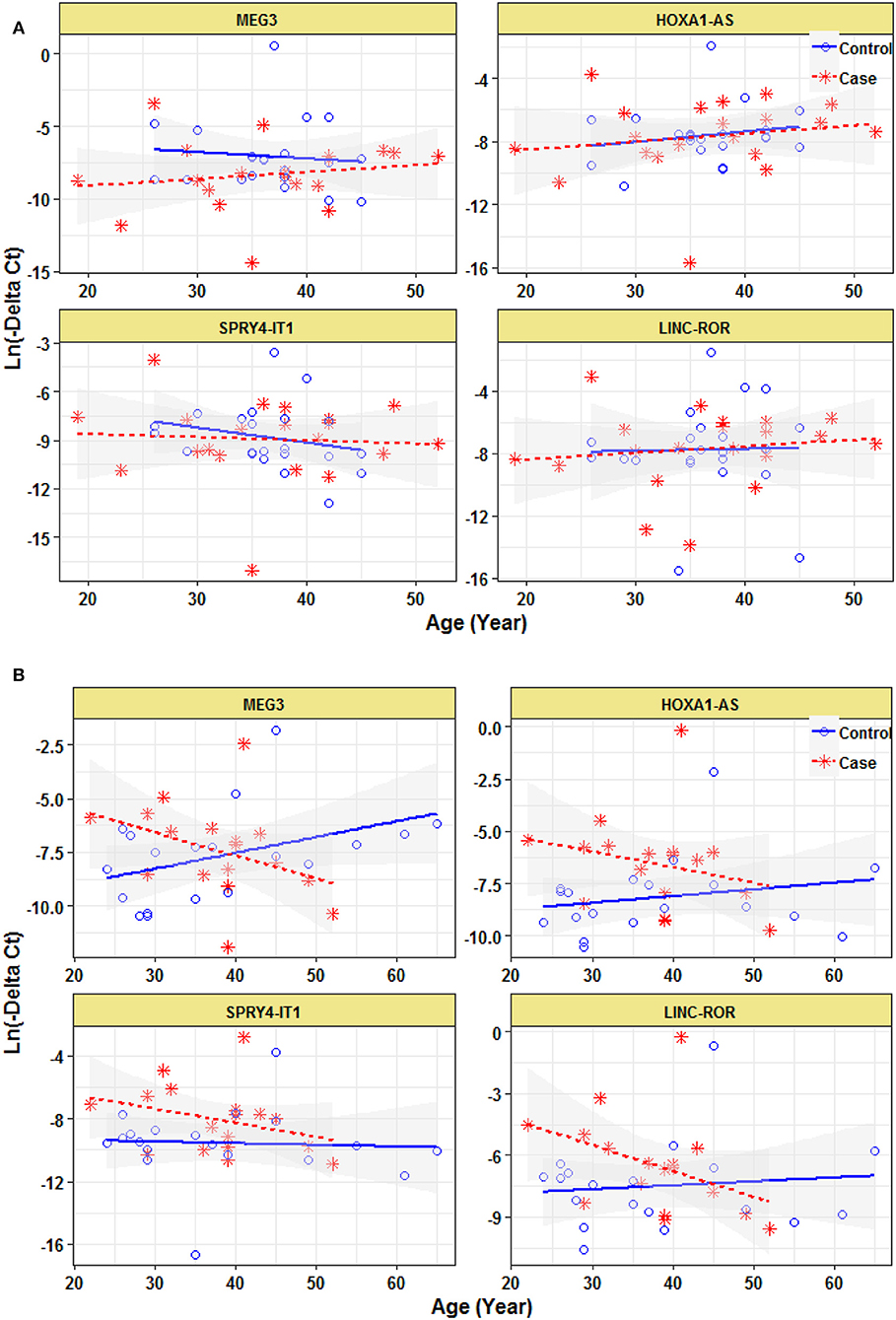
Figure 3. Correlation between expression levels of lncRNAs and age in female epileptic patients and healthy subjects (A) and male epileptic patients and healthy subjects (B).
Discussion
Several lncRNAs have been formerly introduced as regulators of epilepsy-related processes such as neurogenesis, production of neurotransmitter, passage of ions through cellular membranes, and synaptic plasticity (Ng et al., 2013). In the current study, we quantified expression of four lncRNAs in epileptic patients. The functions of these selected lncRNAs were possibly related with epileptogenesis but their expressions have not been evaluated in epileptic patients before. We reported higher expressions of HOXA-AS2 and SPRY4-IT1 in male patients compared with male controls. However, expressions of these two lncRNAs were not different between female cases and female controls. A recent study has reported higher expression of both HOXA-AS2 and SPRY4-IT1 in patients with schizophrenia compared with controls. Contradictory to the present study, when authors assessed expression of these genes in gender-based manner, the differences in the expression of these lncRNAs were remarkable only among females (Fallah et al., 2019). These findings suggest different roles of these lncRNAs based on the gender of patients.
HOXA-AS2 modulates the expression of SCN3A via interaction with hsa-miR-106a-5p (Wu et al., 2019). SCN3A gene has an acknowledged role in the epilepsy (Zaman et al., 2018) and hsa-miR-106a-5p participates in the pathogenesis of Alzheimer's disease (Patel et al., 2008). Notably, both gain- and loss-of-function mutations in SCN3A gene have been associated with seizure vulnerability (Chen et al., 2015; Lamar et al., 2017). So, the HOXA-AS2/ hsa-miR-106a-5p/ SCN3A axis is a putative target for future functional studies in epilepsy and a possible therapeutic target in this regard. The putative role of SPRY4-IT1 in epileptogenesis might be exerted through its role in regulation of expression of ERRα (Yu et al., 2017). ERRα is an orphan nuclear receptor which has similar sequences with ERα. These two kinds of nuclear receptors have common transcriptional networks. Most notably, brain is a tissue with high expression of both ERα and ERRs (Saito and Cui, 2018). Although estrogen is not the intrinsic ligand for ERRα, the interaction between estrogen-signaling and ERRα might be facilitated through transcriptional regulation, or mutual binding on responsive elements, or via the regulation of estrogen production by aromatase (Saito and Cui, 2018). The observed gender-based differences in expressions of HOXA-AS2 and SPRY4-IT1 are in accordance with the results of former studies which highlighted the role of gender in modulation of the evolution of epilepsy. Some underlying mechanisms have also been clarified in the developing brain. Notably, gender-based dissimilarities in neuronal excitability, reaction to environmental stimulants, and epigenetic regulation of gene expression are among these mechanisms (Kight and McCarthy, 2014; Surguchov et al., 2017). In the case of SPRY4-IT1, the differences in relative abundance of ERRs in normal male and female brain might explain the observed gender-based differences as well. Alternatively, the presence of gender-specific transcription factors and epigenetic regulators might explain such differences between males and females.
Based on the high expression of MEG3 in brain and its contribution in neuronal cell damage suppression of a brain-protective signaling pathway (Liang et al., 2018), we expected dysregulation of this lncRNA in epileptic patients. However, we did not detect any significant differences between study groups. So, we suggest that this lncRNA is not a probable contributor in epileptogenesis. Assessment of expression of this lncRNA in postmortem brain tissues might be needed for validation of this hypothesis.
In the current study, we did not have drug-naïve patients to assess the effects of valproic acid on expression of lncRNAs. A recent study has shown the effects of this AED on expression of certain genes associated with neuroprotection and neurotoxicity pathways in epileptic patients (Floriano-Sánchez et al., 2018). Both short and long-term treatment with this drug modulated mRNA levels of a several genes, and notably, returning most of the genes changed by the epileptic condition to a normal state (Floriano-Sánchez et al., 2018). Consequently, lack of difference in expression of MEG3 and LINC-ROR between patients and controls in the current study might be explained by the effects of valproic acid on reverting their levels to normal state.
Notably, expression levels of all lncRNAs were correlated with each other in all subgroups except for lack of correlations between SPRY4-IT1/LINC-ROR and SPRY4-IT1/MEG3 pairs in female controls. Such observation implies the presence of putative interaction network between them which might be affected by gender and disease status in some cases. However, expressions of none of lncRNAs were correlated with age of study participants which implies their independency from age or disease duration in patients. Taken together, the current data implies a putative role for two lncRNAs in the pathogenesis of epilepsy and warrants future functional studies to verify the observed association.
Our study has some limitations. The main weakness is that we assessed expression of a limited number of lncRNAs and limited amount of patients and, therefore, the conclusion about the significance of detected correlations is putative. Another limitation of our study is unavailability of drug-naïve patients.
Data Availability Statement
This manuscript contains previously unpublished data. The name of the repository and accession number(s) are not available.
Ethics Statement
The studies involving human participants were reviewed and approved by the ethics committee of Shahid Beheshti University of Medical Sciences. Informed consent forms were signed by all participants and the parents/Legally Authorized Representative of participants who were included in the study. The patients/participants provided their written informed consent to participate in this study.
Author Contributions
FH and HF performed the experiment. AS and MT designed and supervised the study. SG-F wrote the manuscript and revised the draft. JM and SA-J analyzed the data. SM contributed in experiments setup. All the authors contributed equally and fully aware of the submission.
Funding
The current study was supported by a grant fromShahid Beheshti University of Medical Sciences.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Cao, Q., Liu, X., Yang, F., and Wang, H. (2018). CB2R induces a protective response for epileptic seizure via the PI3K 110alpha-AKT signaling pathway. Exp. Ther. Med. 16, 4784–4790. doi: 10.3892/etm.2018.6788
Chen, Y. J., Shi, Y. W., Xu, H. Q., Chen, M. L., Gao, M. M., Sun, W. W., et al. (2015). Electrophysiological differences between the same pore region mutation in SCN1A and SCN3A. Mol. Neurobiol.51, 1263–1270. doi: 10.1007/s12035-014-8802-x
de Boer, H. M., Mula, M., and Sander, J. W. (2008). The global burden and stigma of epilepsy. Epilepsy Behav. 12, 540–546. doi: 10.1016/j.yebeh.2007.12.019
Fallah, H., Azari, I., Neishabouri, S. M., Oskooei, V. K., Taheri, M., and Ghafouri-Fard, S. (2019). Sex-specific up-regulation of lncRNAs in peripheral blood of patients with schizophrenia. Sci. Rep. 9:12737. doi: 10.1038/s41598-019-49265-z
Floriano-Sánchez, E., Brindis, F., Ortega-Cuellar, D., Ignacio-Mejía, I., Moreno-Arriola, E., Romero-Morelos, P., et al. (2018). Differential gene expression profile induced by valproic acid (VPA) in pediatric epileptic patients. Genes 9:E328. doi: 10.3390/genes9070328
Kight, K. E., and McCarthy, M. M. (2014). Using sex differences in the developing brain to identify nodes of influence for seizure susceptibility and epileptogenesis. Neurobiol. Dis. 72(Pt B), 136–143. doi: 10.1016/j.nbd.2014.05.027
Lamar, T., Vanoye, C. G., Calhoun, J., Wong, J. C., Dutton, S. B. B., Jorge, B. S., et al. (2017). SCN3A deficiency associated with increased seizure susceptibility. Neurobiol. Dis. 102, 38–48. doi: 10.1016/j.nbd.2017.02.006
Lee, D. Y., Moon, J., Lee, S. T., Jung, K. H., Park, D. K., Yoo, J. S., et al. (2015). Dysregulation of long non-coding RNAs in mouse models of localization-related epilepsy. Biochem. Biophys. Res. Commun. 462, 433–440. doi: 10.1016/j.bbrc.2015.04.149
Liang, Z., Chi, Y. J., Lin, G. Q., Xiao, L. F., Su, G. L., and Yang, L. M. (2018). LncRNA MEG3 participates in neuronal cell injury induced by subarachnoid hemorrhage via inhibiting the Pi3k/Akt pathway. Eur. Rev. Med. Pharmacol. Sci. 22, 2824–2831. doi: 10.26355/eurrev_201805_14983
Loewer, S., Cabili, M. N., Guttman, M., Loh, Y. H., Thomas, K., Park, I. H., et al. (2010). Large intergenic non-coding RNA-RoR modulates reprogramming of human induced pluripotent stem cells. Nat. Genet. 42, 1113–1117. doi: 10.1038/ng.710
Mazdeh, M., Zamani, M., Eftekharian, M. M., Komaki, A., Arsang-Jang, S., Taheri, M., et al. (2019). Expression analysis of vitamin D receptor-associated lncRNAs in epileptic patients. Metab. Brain Dis. 34, 1457–1465. doi: 10.1007/s11011-019-00446-9
Mercer, T. R., Qureshi, I. A., Gokhan, S., Dinger, M. E., Li, G., Mattick, J. S., et al. (2010). Long noncoding RNAs in neuronal-glial fate specification and oligodendrocyte lineage maturation. BMC Neurosci. 11:14. doi: 10.1186/1471-2202-11-14
Ng, S. Y., Lin, L., Soh, B. S., and Stanton, L. W. (2013). Long noncoding RNAs in development and disease of the central nervous system. Trends Genetics 29, 461–468. doi: 10.1016/j.tig.2013.03.002
Nolte, C., and Krumlauf, R. (2007). “Expression of Hox genes in the nervous system of vertebrates,” in HOX Gene Expression, ed Papageorgiou, S. (New York, NY: Springer, 14–41.
Patel, N., Hoang, D., Miller, N., Ansaloni, S., Huang, Q., Rogers, J. T., et al. (2008). MicroRNAs can regulate human APP levels. Mol. Neurodegener. 3:10. doi: 10.1186/1750-1326-3-10
Saito, K., and Cui, H. (2018). Emerging roles of estrogen-related receptors in the brain: potential interactions with estrogen signaling. Int. J. Mol. Sci. 19:E1091. doi: 10.3390/ijms19041091
Sanchez Calle, A., Kawamura, Y., Yamamoto, Y., Takeshita, F., and Ochiya, T. (2018). Emerging roles of long non-coding RNA in cancer. Cancer Sci. 109, 2093–2100. doi: 10.1111/cas.13642
Shao, Y., and Chen, Y. (2017). Pathophysiology and clinical utility of non-coding RNAs in epilepsy. Front. Mol. Neurosci. 10:249. doi: 10.3389/fnmol.2017.00249
Smith, R. G., Hannon, E., De Jager, P. L., Chibnik, L., Lott, S. J., Condliffe, D., et al. (2018). Elevated DNA methylation across a 48-kb region spanning the HOXA gene cluster is associated with Alzheimer's disease neuropathology. Alzheimer's Dement. 14, 1580–1588. doi: 10.1016/j.jalz.2018.01.017
Surguchov, A., Surgucheva, I., Sharma, M., Sharma, R., and Singh, V. (2017). Pore-forming proteins as mediators of novel epigenetic mechanism of epilepsy. Front. Neurol. 8:3. doi: 10.3389/fneur.2017.00003
Wu, J., Li, M., and Zhang, Y. (2019). Long noncoding RNA HOXA-AS2 regulates the expression of SCN3A by sponging miR-106a in breast cancer. J. Cell. Biochem. 120, 14465–14475. doi: 10.1002/jcb.28706
Yu, G., Lin, J., Liu, C., Hou, K., Liang, M., and Shi, B. (2017). Long non-coding RNA SPRY4-IT1 promotes development of hepatic cellular carcinoma by interacting with ERRalpha and predicts poor prognosis. Sci. Rep. 7:17176. doi: 10.1038/s41598-017-16781-9
Keywords: epilepsy, lncRNA, HOXA-AS2, SPRY4-IT1, MEG3, LINC-ROR
Citation: Hashemian F, Ghafouri-Fard S, Arsang-Jang S, Mirzajani S, Fallah H, Mehvari Habibabadi J, Sayad A and Taheri M (2019) Epilepsy Is Associated With Dysregulation of Long Non-coding RNAs in the Peripheral Blood. Front. Mol. Biosci. 6:113. doi: 10.3389/fmolb.2019.00113
Received: 27 August 2019; Accepted: 10 October 2019;
Published: 23 October 2019.
Edited by:
Andrei Surguchov, University of Kansas Medical Center, United StatesReviewed by:
Irina G. Sourgoutcheva, University of Kansas Medical Center, United StatesGenny Orso, University of Padova, Italy
Copyright © 2019 Hashemian, Ghafouri-Fard, Arsang-Jang, Mirzajani, Fallah, Mehvari Habibabadi, Sayad and Taheri. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Arezou Sayad, YXIuc2F5YWRAeWFob28uY29t; Mohammad Taheri, bW9oYW1tYWRfODIzQHlhaG9vLmNvbQ==
†These authors have contributed equally to this work
 Fatemeh Hashemian
Fatemeh Hashemian Soudeh Ghafouri-Fard1†
Soudeh Ghafouri-Fard1† Mohammad Taheri
Mohammad Taheri