
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
SYSTEMATIC REVIEW article
Front. Microbiomes , 12 September 2023
Sec. Nutrition, Metabolism and the Microbiome
Volume 2 - 2023 | https://doi.org/10.3389/frmbi.2023.1208166
 Linda Simon Paulo1,2*
Linda Simon Paulo1,2* George Msema Bwire2,3
George Msema Bwire2,3 K. Klipstein-Grobusch1,4,5
K. Klipstein-Grobusch1,4,5 Appolinary Kamuhabwa2,6
Appolinary Kamuhabwa2,6 Gideon Kwesigabo2,7
Gideon Kwesigabo2,7 Pilly Chillo2,8
Pilly Chillo2,8 Folkert W. Asselbergs9,10
Folkert W. Asselbergs9,10 Virissa C. Lenters1,11
Virissa C. Lenters1,11Introduction: As Sub-Saharan Africa (SSA) undergoes rapid urbanization changes in diet and lifestyle have contributed to a rise in non-communicable diseases (NCDs) across the region. Changes in gut microbiota which play an important role in human health may be an underlying driving factor. While evidence suggests that the gut microbiota differs between the extreme levels of economic development (least vs highly developed), it is not well-established which factors along the urbanization gradient are most influential, especially for SSA. This systematic review analyzed published articles from SSA countries that examined the differences in the composition and diversity of gut microbiota along the urbanization gradient. The findings of this review have important implications for understanding the impact of urbanization on human health in the SSA.
Methods: Peer-reviewed articles that examined the link between the urbanization gradient, dietary patterns, and gut microbiota using culture-independent techniques were included in the review.
Results: A total of 3,265 studies were identified and screened. Eighty-nine (89) studies underwent full-text review, and 23 studies were extracted and included for final analysis. Among these studies, it was observed that hunter-gatherers had high alpha diversity (within-person variation) and beta diversity (between-person variation) in their gut microbiota compared to rural and urban residents in SSA. However, there were inconsistent differences between rural and urban at the individual taxa levels, potentially due to limited statistical power and large variability in the study techniques and designs. Similarly, there were no clear differences in the relative abundance of genera across the urbanization gradient. Additionally, both diet and intestinal parasites were associated with the composition and diversity of the gut microbiota.
Conclusion: The review revealed there are variations in both alpha and beta diversity of the gut microbiota across the urbanization gradient with a higher diversity observed in rural areas. However, we did not observe significant differences in the relative abundance at phyla or genus levels consistently across the urbanization gradient. Moreover, our findings suggest that the mode of subsistence, diet, and intestinal parasites play a role in shaping the composition and diversity of the gut microbiota in SSA.
Systematic review registration: https://www.crd.york.ac.uk/prospero/display_record.php?ID=CRD42021251006, identifier CRD42021251006.
Frequent contact with the natural environment including soil and vegetation is postulated to enrich the diversity and composition of the gut microbiota (Mancabelli et al., 2017; Fragiadakis et al., 2019). High interaction with the natural environment has remained a major characteristic of rural/traditional communities giving rise to the term “rural microbiome” (Du et al., 2021). The rural microbiome is hypothesized to enhance health and protect against chronic illnesses (Blaser, 2017; Zuo et al., 2018). The number of rural/traditional communities continues to decline globally due to rapid urbanization, population growth, and technological advances (Ilbery, 2014; Li et al., 2019). The SSA is among the remaining geographical regions with transitioning communities at different levels of interaction with the natural environment such as; pre-agriculture, agro-pastoralists, nomadic pastoralists, and communities in rural and urban areas (Njoh, 2003; Kassouri and Okunlola, 2022). Overall, SSA is undergoing rapid urbanization, and population growth and sees an increasing burden of NCDs (Popkin, 1998; Steyn and McHiza, 2014). As such, the role of altered microbiomes in driving the increase in NCDs deserves attention.
Urbanization and modernization have several features that are linked to the changes in the composition and diversity of the gut microbiota (Yatsunenko et al., 2012; Morton et al., 2015; Mancabelli et al., 2017; Jha et al., 2018). For example, the frequency of using antimicrobials, exposure to cleaning chemicals, less interaction with the natural environment, population density, and excessive cleaning increases as one moves from traditional/hunter-gatherers to urbanized communities (Yatsunenko et al., 2012; Jha et al., 2018; Fragiadakis et al., 2019; McCall et al., 2020; Gacesa et al., 2022; Rosas-Plaza et al., 2022). The difference between urban and rural living environments is also evident in dietary patterns as urban residents more often eat processed foods with lower contents of fibers compared to traditional or other rural communities (David et al., 2014; Cockx et al., 2018). They also have access to a wider variety of foods and dishes compared to their counterparts (Cockx et al., 2018).
The gut microbiota plays a complex key role in human metabolism through different biochemical activities (Bäckhed et al., 2012; Doumatey et al., 2020). While no single optimal healthy composition and diversity of the gut microbiota in humans are known, there is increasing evidence for various sub-optimal microbiota compositions. And several probiotics, prebiotics, and post-biotic supplements to balance the gut microbiota are already in use (Fan and Pedersen, 2021; Oniszczuk et al., 2021; Zhang et al., 2021). Different healthy populations around the world have been found to possess unique core gut microbiota compositions and diversity (Sekirov et al., 2010; King et al., 2019; Lu et al., 2021). Similar to diets, there is no singular best gut microbiota, but it is essential to maintain important functions and resilience.
Evidence exists on the differences in the composition and diversity of the gut microbiota between highly and least industrialized communities (Gupta et al., 2017; Mancabelli et al., 2017). Generally, residing in industrialized communities is linked with less diverse gut microbiota and altered composition at different taxa levels (Gupta et al., 2017; Mancabelli et al., 2017). However, this may not reflect the situation in transitioning communities for example in SSA where there is a co-existence of traditional hunter-gatherers, pastoralists, agro-pastoralists, and rural and urban communities (Morton et al., 2015; Lokmer et al., 2020; Even et al., 2021). Single studies done in other geographical regions (China, India, South America, and Europe) in rural/urban communities have shown mixed results (Tyakht et al., 2014; Ramadass et al., 2017; Das et al., 2018; Li et al., 2018; McCall et al., 2020; Cui et al., 2021). Hence synthesized evidence on the gut microbiota across the urbanization gradient is still needed to expand the knowledge of the drivers of NCDs.
This systematic review analyzed published articles from SSA countries that examined the differences in the composition and diversity of gut microbiota of healthy people along the urbanization gradient. It aims at examining the differences in the gut microbiota and determining the potential explanatory factors that influence the gut microbiota of the residents in Sub-Saharan African countries.
We conducted a PROSPERO-registered systematic review (CRD42021251006) of peer-reviewed articles that examined the link between the urbanization gradient, dietary patterns, and gut microbiota. We searched PubMed/Medline, Scopus, Web of Science, and Embase databases. Three main concepts were used in search studies: gut microbiota, urbanization, and Sub-Saharan African countries (refer to Supplementary Material Appendix 1 for the search strings). Studies were included if they met the inclusion criteria: observational and interventional studies, restricting to control groups of case-control studies or baseline data of randomized controlled trials; published in English from 2000 to 2022; and used a culture-independent identification (i.e., next-generation sequencing) of gut microbiota and studied adult populations. Searches were run up to 1 March 2022. Two independent reviewers (LSP and GMB) performed title and abstract screening using Rayyan (Ouzzani et al., 2016). Quality assessment and characteristics of the included studies were extracted, and a full-text review was conducted using a structured online form (https://app.covidence.org/).
We used a modified Newcastle–Ottawa Scale (NOS) to assess the quality of cross-sectional and case-control studies that consider the selection of study participants, comparability, and outcome features related to the microbiome assessment (Table 1 and Supplementary Material Appendix 2), as previously used (van den Munckhof et al., 2018; Gong et al., 2021). No intervention studies met the inclusion criteria. As no study was critically biased, none was excluded from the review.
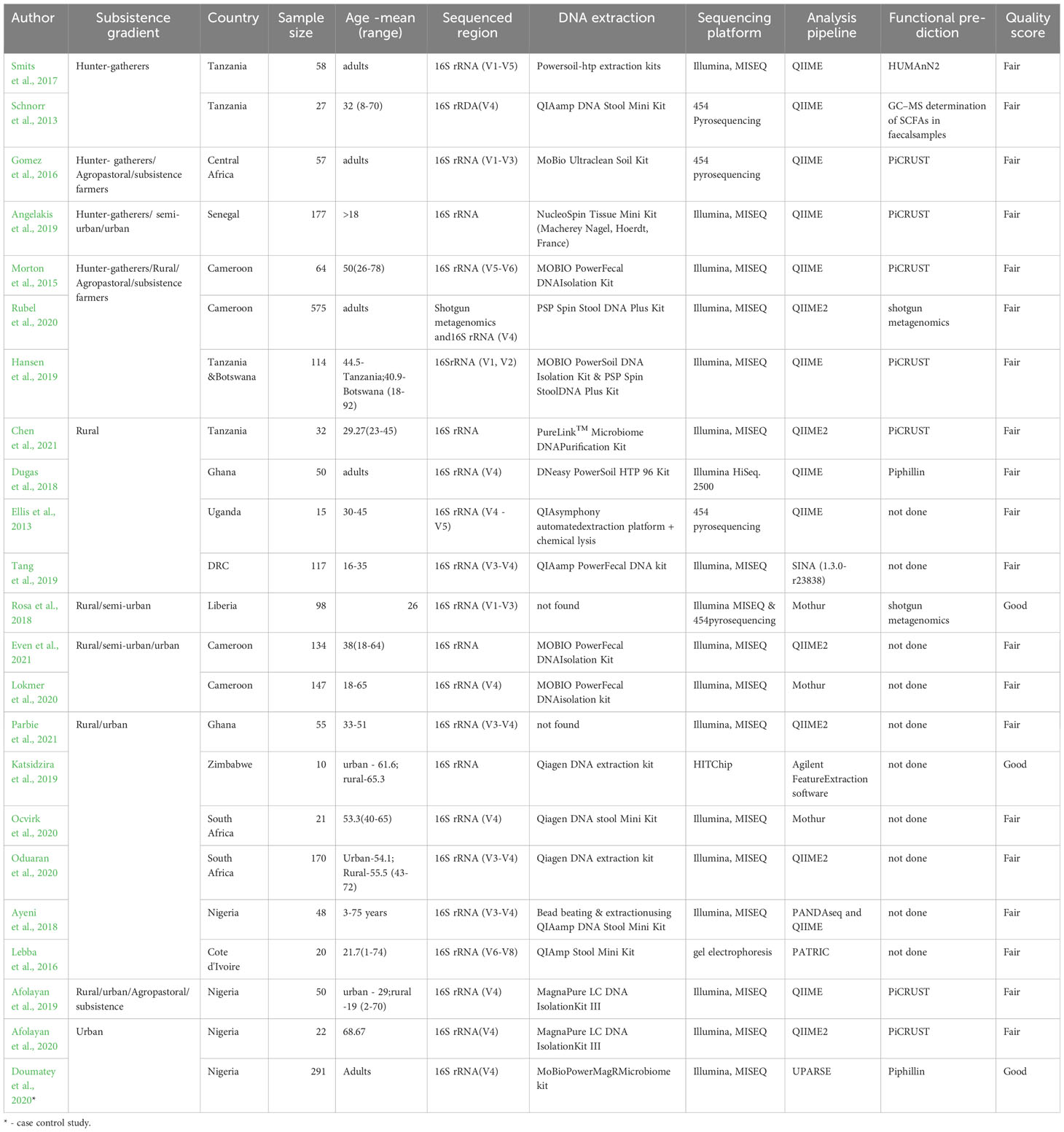
Table 1 A summary of included studies, sample characteristics, and methodologies of the included studies
A total of 3,265 studies were identified, 89 studies were eligible for full-text review, and 23 studies met the inclusion criteria as shown in Figure 1.
The characteristics of the studies are summarized in Supplementary Material Table 1. The studies included 22 cross-sectional studies (Ellis et al., 2013; Schnorr et al., 2014; Morton et al., 2015; Gomez et al., 2016; Iebba et al., 2016; Smits et al., 2017; Ayeni et al., 2018; Dugas et al., 2018; Rosa et al., 2018; Afolayan et al., 2019; Angelakis et al., 2019; Hansen et al., 2019; Katsidzira et al., 2019; Tang et al., 2019; Afolayan et al., 2020; Lokmer et al., 2020; Ocvirk et al., 2020; Oduaran et al., 2020; Rubel et al., 2020; Chen et al., 2021; Even et al., 2021; Parbie et al., 2021) and 1 case-control study (Doumatey et al., 2020), representing a total of 2,372 individuals. For the case-control study, we considered only the control group that assessed gut microbiota in the adult population with and without diabetes (Doumatey et al., 2020). Most studies (Morton et al., 2015) originated from countries in West Africa (n=13 studies; (Nigeria (Ayeni et al., 2018; Afolayan et al., 2019; Afolayan et al., 2020; Doumatey et al., 2020), Ghana (Dugas et al., 2018; Parbie et al., 2021), Cameroon (Morton et al., 2015; Lokmer et al., 2020; Rubel et al., 2020; Even et al., 2021), Cote d’Ivoire (Iebba et al., 2016), Senegal (Angelakis et al., 2019), Liberia (Rosa et al., 2018)) followed by East Africa (n=5 studies; (Tanzania (Schnorr et al., 2014; Smits et al., 2017; Hansen et al., 2019; Chen et al., 2021) and Uganda (Ellis et al., 2013)), Southern Africa (n=3 studies; (South Africa (Ocvirk et al., 2020; Oduaran et al., 2020) and Zimbabwe (Katsidzira et al., 2019)) and Central Africa (n=2 studies; (Central Africa Republic (Gomez et al., 2016) and DRC (Tang et al., 2019)). Seven studies included participants practicing hunting-gathering, while others combined rural, subsistence farmers, or urban-based populations. The age of adult participants ranged from 18-78 years although it was not defined in four studies (Gomez et al., 2016; Smits et al., 2017; Dugas et al., 2018; Rubel et al., 2020).
Of the 23 included studies, three were classified as having good quality (Rosa et al., 2018; Katsidzira et al., 2019; Doumatey et al., 2020) while the rest were ranked as fair quality. Studies ranked as good described the sampling strategies and justified the sample size used (Rosa et al., 2018; Katsidzira et al., 2019; Doumatey et al., 2020). These features were missing in other studies that scored fair. All studies scored high in the use and description of the measurement tools, ascertainment of outcomes (i.e., gut microbiota), and measurement of association between the exposure and outcome factors. However, the heterogeneity in comparison groups and microbiota measures precluded doing a meta-analysis.
Several approaches have been used to define urbanization and quantify the urban-rural gradient considering different features such as population density, administrative status, land cover, and socioeconomic activities (Urbanization - Overview | US EPA; Gibbs, 1966; Li and Lu, 2021). In examining the relationship between the different environmental factors and gut microbiota scientists have used the words urban, rural, and subsistence patterns in isolation or as an index measure by combining different attributes (Obregon-Tito et al., 2015; Mancabelli et al., 2017).
In the included studies (refer to Table 1), urban was defined as highly urbanized (Oduaran et al., 2020) or based on the consumption of industrially processed foods or a Westernized lifestyle (Ayeni et al., 2018; Afolayan et al., 2019; Afolayan et al., 2020). Rural was classified in terms of subsistence patterns such as transitioning rural (Oduaran et al., 2020), cattle keepers for pastoralists and nomadic pastoralists or agro-pastoralists described as cattle keepers who cultivate crops (Afolayan et al., 2019; Rubel et al., 2020), agriculturalists with western-like subsistence pattern (Gomez et al., 2016), agrarian community (Ayeni et al., 2018), and rural farming and fishing communities (Morton et al., 2015). Hunter-gatherers were referred to as small-scale farmers who hunt and gather food (Rubel et al., 2020) or simply hunter-gatherers (Morton et al., 2015; Gomez et al., 2016; Ayeni et al., 2018). Lokmer et al. created an index to assess the urbanization gradient based on the availability of electricity, use of tap water, housing floor, in-house animal rearing, use of antibiotics, and the level of education (Lokmer et al., 2020). Other included studies used urban or rural without further characterization.
The procedures and kits used to extract DNA from stool samples were reported in 21 studies but were not explicitly described in 2 studies (Rosa et al., 2018; Parbie et al., 2021). The most used extraction kit was from Qiagen (8 studies) followed by the MOBIO isolation kit (6 studies). Approaches used for DNA lysis varied between studies as some used mechanical bead beating (Dugas et al., 2018; Hansen et al., 2019; Katsidzira et al., 2019; Rubel et al., 2020) while others used both mechanical and chemical lysis (Tang et al., 2019) or chemical lysis alone (Iebba et al., 2016). It was not clearly stated in the rest of the studies whether mechanical/chemical lysis was applied in addition to the extraction protocol used.
All studies sequenced the 16S rRNA gene using at least one of the variable regions (V1-V9), although some studies did not report the exact region that was sequenced (Angelakis et al., 2019; Katsidzira et al., 2019; Chen et al., 2021; Even et al., 2021). V4 was the most sequenced region reported in 13 studies. In addition to 16S rRNA, Rubel et al. used shotgun metagenomics (Rubel et al., 2020). Sequencing the entire 16S rRNA gene is associated with a higher bacteria diversity although a higher yield is also seen with the V4 region compared to other regions. Most studies used Illumina, Miseq as a sequencing platform and QIIME and QIIME2 for sequence analysis except a few (GS FLX Titanium platform (Ellis et al., 2013), 454 pyrosequencing (Schnorr et al., 2014; Gomez et al., 2016; Rosa et al., 2018), HITChip (Katsidzira et al., 2019) and gel electrophoresis (Iebba et al., 2016).
The total reads ranged from 50,000 – 51,655,653 while the average reads per sample ranged from 6,000 - 93,171.06. Overall, the reporting of the reads varied and was not reported in half of the studies identified.
Various methods were used to assess the diversity of the gut microbiota. Beta diversity (variation between samples, reflecting (dis)similarity in community composition of groups) was assessed in 17 studies, and measures including Bray-Curtis, Unweighted UniFrac distance, and Weighted UniFrac distance were reported in 17 studies. Alpha diversity (within-sample diversity) was assessed in 20 studies and measures including Shannon, Chao1, Simpson, and OTU richness. Predictive functional profiling was most frequently performed using “Phylogenetic Investigation of Communities by Reconstruction of Unobserved States” (PiCRUST) in 7 studies followed by shotgun metagenomics (Rosa et al., 2018; Rubel et al., 2020), Piphilin (Dugas et al., 2018; Doumatey et al., 2020) and HUMAnN2 (Smits et al., 2017). Other approaches include the use of gas-chromatography–mass spectrometry (GC–MS) to determine short-chain fatty acids (SCFAs) in fecal samples (Schnorr et al., 2014). Ten studies did not perform functional analysis.
Three studies reported alpha diversity across an urbanization gradient whereby one study compared urban and rural (Oduaran et al., 2020), the second compared urban, semi-urban, and rural (Lokmer et al., 2020), and the third one compared hunter-gatherers versus rural communities-pastoralists and subsistence farmers (Rubel et al., 2020). Lokmer et al. showed an overall decrease in alpha diversity along the urbanization gradient (Lokmer et al., 2020). These findings were echoed by Oduaran et al. where alpha diversity was higher in rural compared to urban communities (Oduaran et al., 2020). The hunter-gatherers in Rubel et al. (2020) showed higher alpha diversity compared to other communities while the pastoralists had the lowest alpha diversity measures (Rubel et al., 2020). Overall, the alpha diversity was higher in hunter-gatherers/rural compared to urban and lower in pastoralists compared to other rural communities.
Beta diversity was presented in five studies (Gomez et al., 2016; Ayeni et al., 2018; Afolayan et al., 2019; Angelakis et al., 2019; Oduaran et al., 2020). Two studies compared hunter-gatherers versus urban communities (Gomez et al., 2016; Angelakis et al., 2019), and both showed a significant difference in beta diversity revealing greater diversity in hunter-gatherer populations. Differences in the diversity measures between urban and rural were also shown in Oduaran et al. (p=0.001 for Bray-Curtis dissimilarity measure) and Ayeni et al. (p<0.05 for Unweighted and Weighted UNIFRAC distances) with higher diversity in rural communities (Ayeni et al., 2018; Oduaran et al., 2020). This is similarly reported in Afolayan et al. where the pastoralists had a more diverse gut microbiome compared to the urban residents’ (Afolayan et al., 2019).
Reporting of the composition of gut microbiota varied between studies. Therefore, complete mapping of the gut microbiota across the hunter-gatherers, rural and urban gradients is limited. Out of the included studies, 5 (4 studies included rural and 3 in hunter-gatherers’ settings) reported 6 phyla in common (higher taxonomic level); Firmicutes, Bacteroidetes, Cyanobacteria, Spirochaetes, Actinobacteria and Proteobacteria (Schnorr et al., 2014; Gomez et al., 2016; Afolayan et al., 2019; Tang et al., 2019; Chen et al., 2021). Firmicutes and Bacteroidetes were the most abundant phyla in both settings. The range for Firmicutes in rural vs hunter-gatherers areas was (46.9-77.63%) vs (49.5-72%) respectively; the range for Bacteroidetes also among rural and hunter-gatherers areas was (13.8-33%) vs (17-44.4%) respectively (Schnorr et al., 2014; Gomez et al., 2016; Afolayan et al., 2019; Tang et al., 2019; Chen et al., 2021). This was followed by Proteobacteria which was higher in rural settings compared to hunter-gatherers, with a range of (5.5 -17.73%) in rural vs (6-8.49%) in hunter-gatherers. The presence of Actinobacteria was relatively low in both rural (1.27-1.79%) and hunter-gatherer populations (less than 0.54%). Spirochaetes were reported in all 3 studies focusing on hunter-gatherer populations, but their abundance was relatively low. In contrast, only 50% of the studies done in rural areas reported the presence of Spirochaetes, with their abundance ranging from low to high. Cyanobacteria were the least commonly reported in 1 study in each setting (0.29% rural) vs (0.7% Hunter-gatherers). Most phyla did not differ between hunter-gatherers and rural in Morton et al. except for Proteobacteria where the relative abundance was higher among hunter-gatherers. The details of the direction of the relative abundance at the phyla level reported across the gradient are shown in Figure 2.
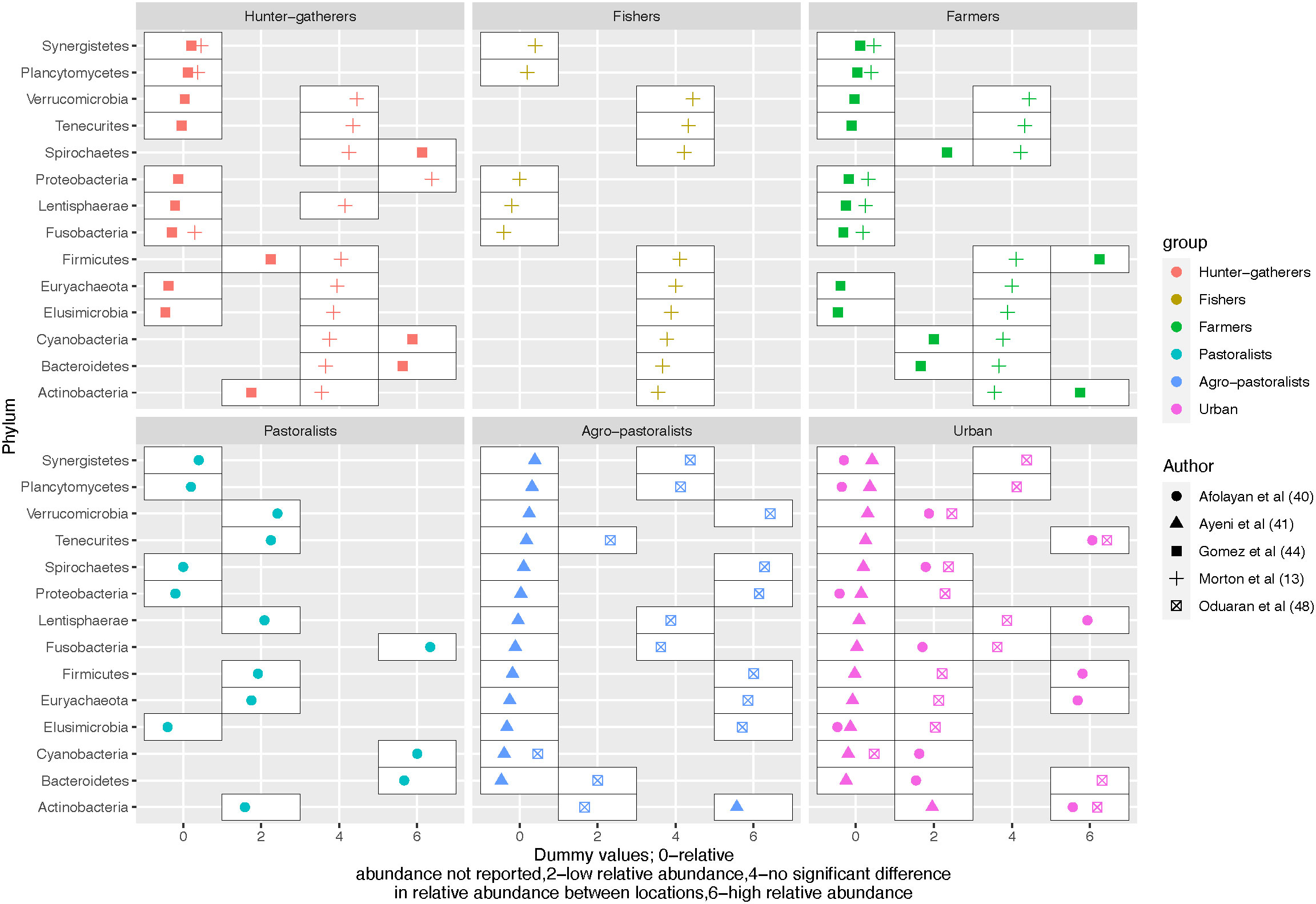
Figure 2 An overview of the differences in relative abundance at phylum level across the urbanization gradient.
Eight studies reported gut microbiota at the family level (Dugas et al., 2018; Afolayan et al., 2019; Hansen et al., 2019; Tang et al., 2019; Afolayan et al., 2020; Doumatey et al., 2020; Even et al., 2021; Parbie et al., 2021). The Prevotellacea family was the most frequently mentioned in 5 studies (Dugas et al., 2018; Afolayan et al., 2019; Hansen et al., 2019; Tang et al., 2019; Parbie et al., 2021) and is also mentioned to be the most prevalent in the African populations by Hansen et al. (2019).
Eight studies compared the relative abundance across hunter-gatherers, rural, semi-urban, or urban areas at the genus level as shown in Figure 3. Complex carbohydrate digesters Bacteroides were the most frequently reported and appeared in five studies (Morton et al., 2015; Afolayan et al., 2019; Lokmer et al., 2020; Oduaran et al., 2020; Rubel et al., 2020). The relative abundance of Bacteroides according to the urbanization level of the assessed population varied across studies. They had a higher relative abundance among hunter-gatherers and rural farmers (Morton et al., 2015), pastoralists (Rubel et al., 2020), and urban (Afolayan et al., 2019). In contrast, their relative abundances were low in hunter-gatherers (Rubel et al., 2020), rural fishers (Morton et al., 2015), pastoralists (Afolayan et al., 2019), and agro-pastoralists (Oduaran et al., 2020; Rubel et al., 2020) and decreased over the urbanization gradient from urban to rural (Lokmer et al., 2020). Hence, the direction of the relative abundance of Bacteroides does not follow a clear gradient. Similar to Bacteroides, there is no clear difference between urban and rural residents for Bifidobacterium, microorganisms known to digest carbohydrates and regulate gut health (Naumova et al., 2020). This component of the microbiota was compared across the urbanization gradient where it had a high relative abundance among rural fishers (Morton et al., 2015) and urban residents (Oduaran et al., 2020) but a low relative abundance among hunter-gatherers (Morton et al., 2015), and farmers and agro-pastoralists (Oduaran et al., 2020).
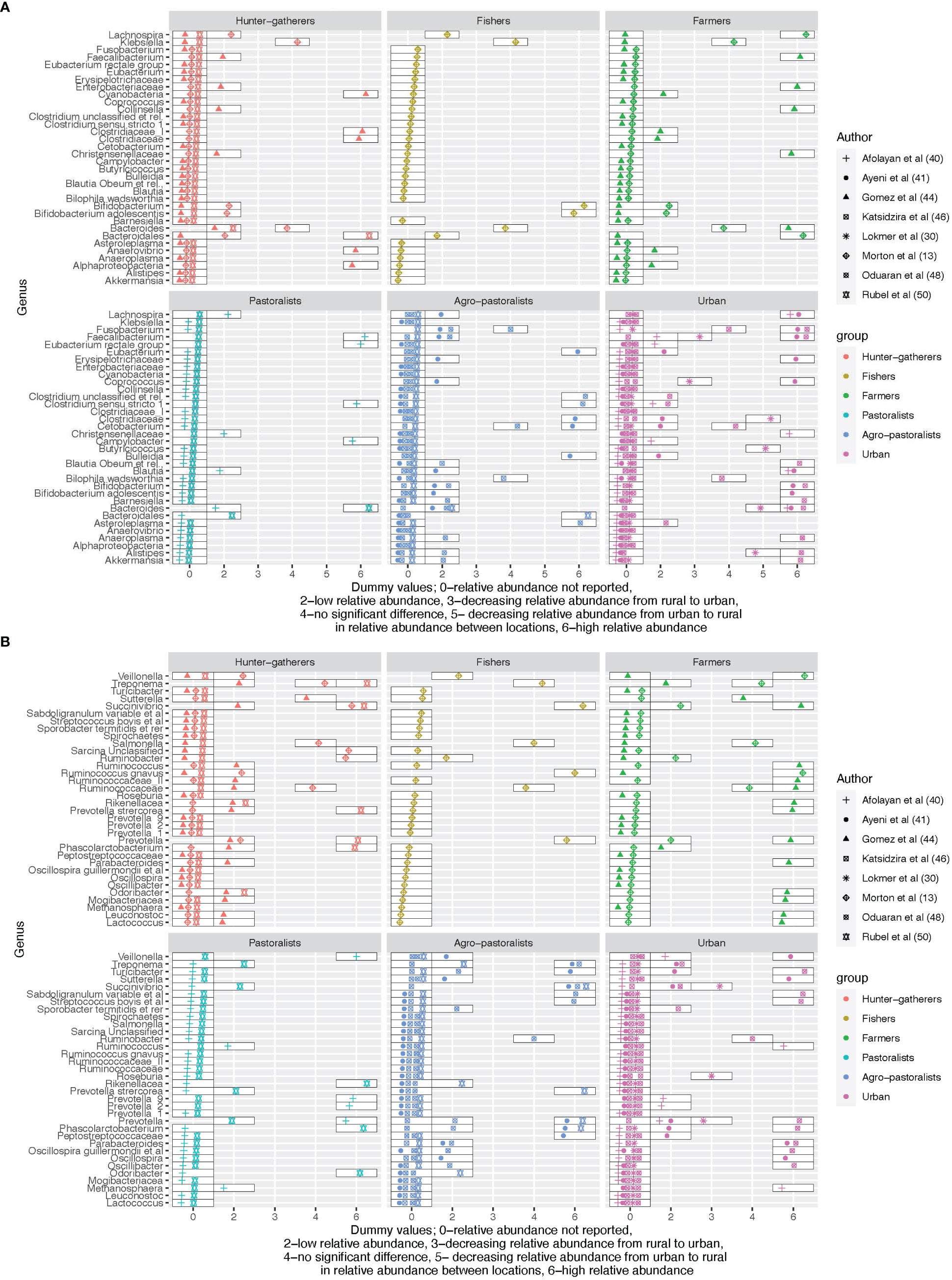
Figure 3 (A) An overview of the differences in relative abundance at genus level across the urbanization gradient for the first 34 reported genera. (B) An overview of the differences in relative abundance at genus level across the urbanization gradient for the last 34 reported genera.
Blautia was more enriched among rural pastoralists and agro-pastoralists but low in urban (Ayeni et al., 2018; Afolayan et al., 2019) while Blautia (species - obeum) was high in urban and low among agro-pastoralists (Katsidzira et al., 2019). The relative abundance of Bilophila (species - wadsworthia) (linked with disrupted glucose metabolism in mice) (Natividad et al., 2018), Cetobacterium, Ruminobacter, and Turicibacter did not differ between rural agropastoralists and urban residents (Katsidzira et al., 2019; Oduaran et al., 2020).
The relative abundance of the Treponema genus was high in hunter-gatherers compared to rural and was also detected in urban (Oduaran et al., 2020) contrary to the findings in (Angelakis et al., 2019) where no Treponema was detected in urban areas. Out of the six named core genera, Faecalibacterium was the most frequently reported in the comparison (4 out of 8 studies), followed by Blautia (3 studies), Clostridium (2 studies), Ruminococcus (2 studies) and Eubacterium (1 study) (Dehingia et al., 2015). Roseburia was also reported in one study only where its relative abundance increased from rural to urban (Lokmer et al., 2020).
Similar to previously mentioned genera, Faecalibacterium did not follow a consistent pattern. It showed a positive correlation with the urbanization gradient from rural to urban (Lokmer et al., 2020), and displayed a high abundance among pastoralists (Afolayan et al., 2019) and urban (Ayeni et al., 2018; Oduaran et al., 2020), but a low abundance in agro-pastoralists (Ayeni et al., 2018; Oduaran et al., 2020) and urban residents (Afolayan et al., 2019). The relative abundance of Fusobacterium was similar for rural agro-pastoralists and urban residents (Katsidzira et al., 2019), low in agro-pastoralists but high in urban (Oduaran et al., 2020) and vice versa (Ayeni et al., 2018).
Carbohydrate fermenters and producers of the short-chain fatty acid butyrate (Ezaki, 2015), Coprococcus were more abundant in urban residents and low in agro-pastoralists (Ayeni et al., 2018) and increased with the urbanization gradient from rural to urban (Lokmer et al., 2020).
The genus Prevotella was reported in several studies including Prevotella, Prevotella_1, Prevotella_2, Prevotella_9, and Prevotella stercorea. In this review, Prevotella was more abundant in Hunter-gatherers (Gomez et al., 2016; Rubel et al., 2020), agro-pastoralists (Rubel et al., 2020), and urban (Oduaran et al., 2020). The genus decreased in relative abundance across the urbanization gradient from rural to urban (Lokmer et al., 2020) and was less abundant in rural farmers, pastoralists, and agro-pastoralists (Gomez et al., 2016; Oduaran et al., 2020; Rubel et al., 2020). Prevotella_1 was more abundant among hunter-gatherers (Gomez et al., 2016) and urban (Oduaran et al., 2020) and less abundant among farmers (Gomez et al., 2016) and agro-pastoralists (Oduaran et al., 2020). Prevotella_2 and Prevotella_9 were both abundant among pastoralists compared to urbanites (Afolayan et al., 2019) while Prevotella stercorea were more abundant among hunter-gatherers and low among pastoralists (Rubel et al., 2020). Refer to Figure 3 for further details on the distribution of the relative abundance of the genera across the gradient.
Diet was assessed directly or indirectly in 11 studies using 24-hour recalls (Tang et al., 2019; Lokmer et al., 2020), food frequency questionnaires (FFQ) (Schnorr et al., 2014; Morton et al., 2015; Lokmer et al., 2020), narrative reports (Afolayan et al., 2019; Ocvirk et al., 2020), and nutritional questionnaires (Dugas et al., 2018; Rubel et al., 2020). Diets of urban residents consisted of dairy products, meat, sweets, and soft drinks (Lokmer et al., 2020) (Cameroon), low fiber-diet, processed foods, high-fiber diet, fermented drinks (Afolayan et al., 2019) (Nigeria), cereals, refined grains, fruits, and tubers, moderate use of antibiotics, use of treated water with a high level of hygiene (Ayeni et al., 2018) (Nigeria). In contrast, the rural residents reported consumption of tubers, leafy vegetables, peanuts, and palm wine (Lokmer et al., 2020) (Cameroon), grains, tubers, fruits and soups (okra, melon) and traditional soups, regular fish intake with less meat intake, a variety of fermented foods, use river water and rely on local herbs (Ayeni et al., 2018) and cassava, fish and meat (Morton et al., 2015) (Cameroon). Pastoralists reported eating high-fibre-based foods and fermented drinks while agro-pastoralists reported grain and vegetable consumption. Their livestock was mostly for selling purposes (Afolayan et al., 2019). Hunter-gatherers showed high-fiber diets and other food items from hunting and foraging (Rubel et al., 2020) (Cameroon), and high meat and nut consumption (Gomez et al., 2016) (Central Africa) but also low meat consumption (Morton et al., 2015) (Cameroon). Carbohydrate and fiber intake were observed to be similar in urban and rural settings with a high intake of protein in urban (Zimbabwe) (Katsidzira et al., 2019).
The relationship between diet and gut microbiota was assessed in 4 studies only (Morton et al., 2015; Smits et al., 2017; Tang et al., 2019; Rubel et al., 2020). For the Hadza community (Hunter-gatherers) there was a strong link between seasonal changes in diet and gut microbiota composition (Smits et al., 2017). During the wet season, Hadza consumes a plant-based diet (berries, honey, baobab, and tubers) and has access to more meat during dry seasons. Consequently, there was a decline in the population of Bacteroidetes (Prevotellaceae family) were depleted during the wet season and reappeared in the dry season. In contrast, Firmicutes remained stable throughout the seasons.
Second, after adjusting for the presence of Entamoeba spp, Morton et al. (2015) found a persistent low gut microbiota diversity among the fishing community compared to hunter-gatherers and subsistence farmers. Lastly, the presence of genus Ruminoccocus was associated with meat and iron intake while a high relative abundance of Faecalibacterium and Succininvibrio was positively correlated with fish and/or insect intake and vitamin A-rich green vegetables and fruits (Tang et al., 2019). Further details are given in Supplementary Material Appendix 3.
The relationship between the gut microbiota and intestinal parasites was reported in seven studies (Morton et al., 2015; Iebba et al., 2016; Rosa et al., 2018; Lokmer et al., 2020; Rubel et al., 2020; Chen et al., 2021; Even et al., 2021). Details are given in Figure 4 below. The presence of the intestinal parasites was assessed using microscopy in three studies (Morton et al., 2015; Lokmer et al., 2020; Chen et al., 2021), a combination of microscopy and quantitative PCR (qPCR) in two studies (Rosa et al., 2018; Rubel et al., 2020), qPCR in one study (Iebba et al., 2016) and 18S rRNA gene sequencing in one study (Even et al., 2021). The techniques used for the diagnosis of intestinal parasites complement each other as there is no gold standard (Maurelli et al., 2022). All studies that examined the relationship between intestinal parasites and gut microbiota showed that the presence of parasites is correlated with the diversity of the gut microbiota. Their presence was associated with a higher diversity of gut microbiota in two studies (Rubel et al., 2020; Even et al., 2021) but the direction of the association was not explicitly shown in the remaining 5 studies. In a longitudinal study, Rosa et al. (2018) found that gut microbiota alpha diversity was not fully restored two years after the administration of oral anti-helminths (Figure 4).
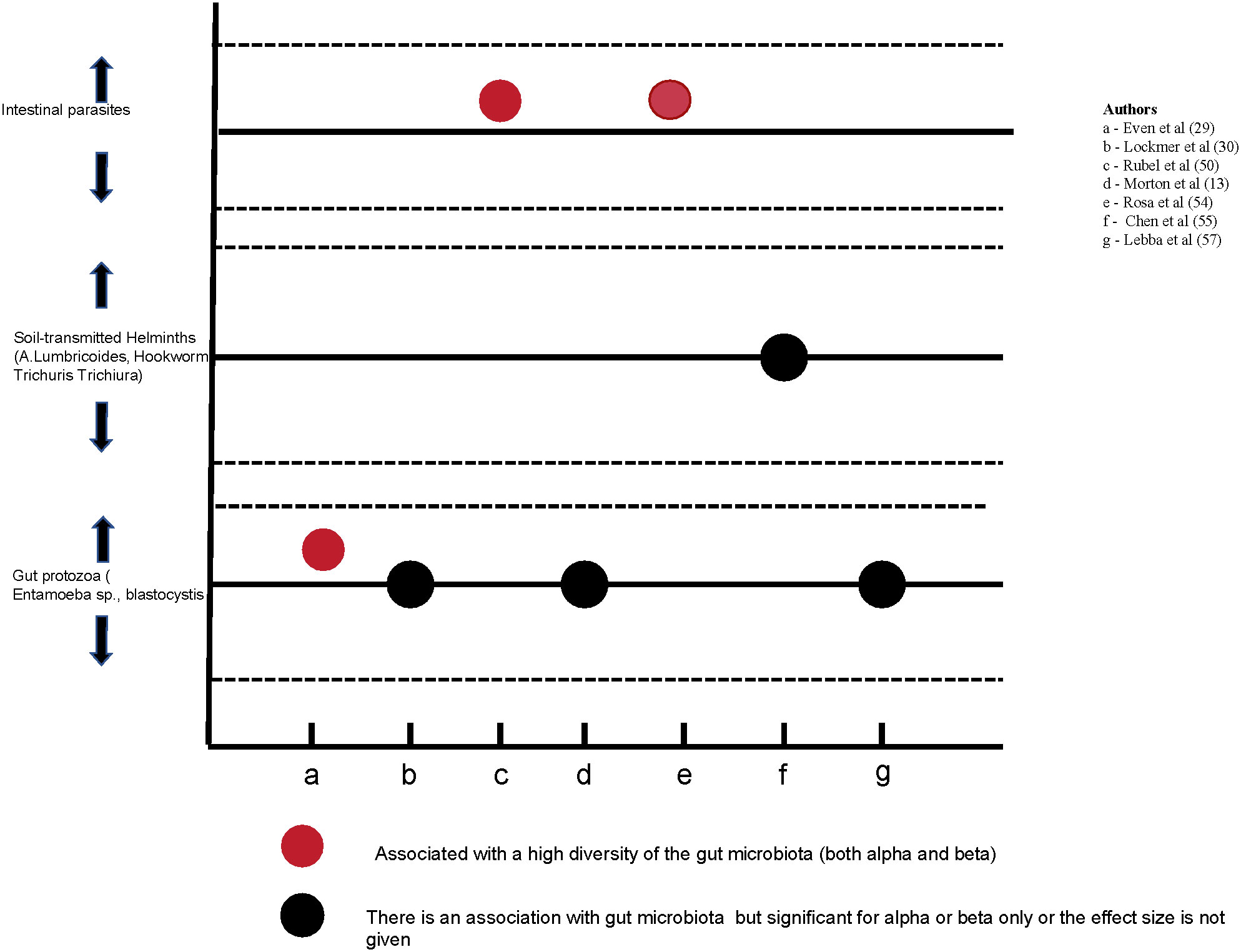
Figure 4 The relationship between the gut microbiota and intestinal parasites as reported in 7 studies.
To identify pathways and hierarchy, information obtained from the pipelines was collapsed using the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways and metabolites classification. Metabolic pathways were reported in 13 studies and were reported across the gradient in 5 studies (refer to Supplementary Material Appendix 4). Although most pathways are shared across the urbanization gradient differences in enrichment were reported in two studies (Afolayan et al., 2019; Hansen et al., 2019). In a study comparing urban and rural, methane metabolism, arginine and proline metabolism, pyruvate metabolism, valine, leucine, and isoleucine biosynthesis, and fructose and mannose metabolism were more enriched in urban residents (Afolayan et al., 2019). The same pathways are reported among hunter-gatherers and rural residents with no details on the differences in enrichment (Morton et al., 2015; Gomez et al., 2016; Dugas et al., 2018). Similarly, in a study conducted by Afolayan et al. as well as in other studies (Supplementary Material Appendix 4), lipopolysaccharide biosynthesis, ubiquinone and other terpenoid quinone synthesis, glycosyltransferases, arachidonic acid metabolism, riboflavin, vitamin B6 metabolism, protein digestion, and absorption were shown to be more enriched among rural residents (Afolayan et al., 2019). Finally, amino acid metabolism was reported throughout the urbanization gradient and no differences were reported in the pathway enrichment.
We reviewed 23 peer-reviewed publications that assessed the gut microbiota profile of healthy residents of 13 SSA countries across the urban-rural gradient. In addition to geographical differences the studies reported on the diet, the use of anti-helminths, intestinal parasites, and the functional potential of the identified pathways. All the included studies used Next Generation Sequencing techniques although the heterogeneity was high in sample collection, data processing including sequencing depth and platform, regions of the 16S gene sequenced, and downstream analysis like diversity and differential abundance analysis. Ten studies reported taxonomic levels and other differences related to the gut microbiota between geographical locations along the urbanization gradient (Schnorr et al., 2014; Gomez et al., 2016; Ayeni et al., 2018; Afolayan et al., 2019; Angelakis et al., 2019; Hansen et al., 2019; Katsidzira et al., 2019; Lokmer et al., 2020; Oduaran et al., 2020; Rubel et al., 2020).
We observed a difference in both alpha and beta diversity across the urbanization gradient with a higher diversity in hunter-gatherers/rural compared to urban communities. However, the taxa level and diversity metric analysis used differed between studies. The results we found are similar to a meta-analysis on the impact of urbanization on the gut microbiota globally (including a subset of the same studies from SSA included in this review) that showed a lower alpha diversity in urban populations compared to rural/hunter-gatherers settings (Rosas-Plaza et al., 2022). In this meta-analysis beta diversity at family and genera, levels showed similar trends, with the urban and Hunter-gatherers communities at the extremes (lowest vs highest diversity) (Rosas-Plaza et al., 2022). Several studies have evaluated this in other parts of the world with varying results (Clemente et al., 2015; Das et al., 2018; Jha et al., 2018; McCall et al., 2020; Rosas-Plaza et al., 2022). For example, a study in Tibet China showed differences in beta diversity and not alpha diversity across the urbanization gradient, contrary to the studies included here with differences in both measures (Li et al., 2018). Likewise, a study conducted among populations transitioning from foragers to farmers in the Himalayan region did not find any variations in alpha diversity (Jha et al., 2018). Conversely, a study done in South America, which explored the overall and chemical variation across the spectrum from traditional villages to the city center, did not find any differences in diversity along the urbanization gradient (McCall et al., 2020). In general, evidence indicates that gut microbiota in higher-income/industrialized exhibit lower diversity compared to lower-income/non-industrialized/rural/traditional communities (Obregon-Tito et al., 2015; Mancabelli et al., 2017; Rosas-Plaza et al., 2022). Nevertheless, there is a need for additional studies to fully understand the distinctions in gut microbiota among individuals residing in geographically close areas.
Bacteroidetes and Firmicutes are known to dominate the gut microbiota. Six other phyla are proposed to form the 8 core gut microbiota by the healthy gut microbiota (GutFeelingKB data and metadata); Proteobacteria, Actinobacteria, Spirochaetes, Plancytomycetes, Euryarchaeota and Cyanobacteria (King et al., 2019). Similar, to the above-established core gut microbiota, Firmicutes and Bacteroidetes were the most abundant phyla in the included studies. Further, there are differences in the relative abundance of gut microbiota across the urbanization gradient although not uniform for all the phyla/genera as some were relatively more abundant while others were less abundant, as shown in Figure 2.
The microbiome of the rural (Hunter-gatherers/traditional communities and non-urban) is associated with a low prevalence of diseases including inflammatory bowel diseases, colorectal cancer, and metabolic diseases (Zuo et al., 2018; Cui et al., 2021). Perturbations in a few phyla have been linked to health outcomes such as the Firmicutes: Bacteroidetes ratio where higher Bacteroidetes levels are seen in lean compared to obese individuals (Johnson et al., 2017). Also, a higher relative abundance of Bacteroidetes and Proteobacteria compared to Firmicutes was reported among patients with chronic kidney disease (Lun et al., 2019). In this review, the ratio (Firmicutes: Bacteroidetes) was reported in one study only where it was 5:1 in rural and 1:1 in the Hunter-gatherers’ community (Gomez et al., 2016).
A study in South America from the jungle to the city center by McCall et al. showed a high proportion of Proteobacteria with increasing urbanization (McCall et al., 2020). Similarly, in this review, the relative abundance of Proteobacteria was higher in rural compared to hunter-gatherers’ settings. In contrast to the findings in South America, where the relative abundance of Actinobacteria decreased with urbanization (higher in hunter-gatherers), our review did not identify any clear trend for this phylum (McCall et al., 2020).
Most studies reported the gut microbiota differences at the genus level. Existing evidence has linked individual genera to disease conditions and health outcomes. In this review, there are no clear differences in the relative abundance of identified genera across the urbanization gradient except for Coprococcus where the relative abundance was consistently low in rural and higher in urban (Ayeni et al., 2018; Lokmer et al., 2020). Bacteroides were the most frequently reported in the studies that compared the relative abundance across the gradient. They are known to maintain gut health, digest complex carbohydrates, metabolize bile, maintain the immune system and gut barrier, and produce Vitamins K and B (Xu and Gordon, 2003). This genera together with Faecalibacterium and Blautia were more abundant in urban herdsmen in Tibet contrary to the findings of this review where their relative abundance did not follow a clear gradient (Li et al., 2018).
The relative abundance of the Lactobacillus genus was reported to be high among the Mongolian pastoralists compared to the urban Mongolians possibly due to the high consumption of dairy products (Iebba et al., 2016). However, in our review, Lactobacillus was reported in two rural communities (Iebba et al., 2016; Tang et al., 2019) and there was no difference between pastoral and non-pastoral communities (Afolayan et al., 2019). The biological importance of Lactobacillus is not fully understood as their relative abundance is both negatively and positively correlated with disease outcomes (Heeney et al., 2018).
A study assessing the gut microbiota based on diet, ethnicity, and urbanization in China identified four types of enterotypes including one dominated by the Escherichia genus (Lu et al., 2021). This genus was also reported in a study in Korea contrasting gut microbiota between rural villages and urban (Wu et al., 2011). However, in this review, the Escherichia genus was not reported in any of the studies that compared gut microbiota across the gradient.
The relative abundance of the genus Prevotella varied across the urbanization gradient with a high and low relative abundance of the genera across the gradient without clear direction. Prevotella and its sub-genera were reported across the gradient with alternating relative abundances. The abundance of the Prevotella genus has been shown to correlate with carbohydrate consumption, which is documented in the included studies across the urbanization gradient (Sonnenburg et al. ). A higher relative abundance of Prevotella is often associated with plant-based diets and improved glucose metabolism (Kovatcheva-Datchary et al., 2015; Ley, 2016). The presence of Prevotella throughout the urbanization gradient indicates a potential trend between urbanization and industrialization, as the abundance of Prevotella is nearly absent in industrialized societies (Ley, 2016). This observation suggests that industrialization may have a greater effect on the abundance of this genus compared to the process of urbanization seen in SSA.
The composition and relative abundance of Bifidobacterium in mice fed with fish oil were higher compared to those eating lard (David et al., 2014). In line with this finding, this review found Bifidobacterium to have a high relative abundance among the fishing and urban communities compared to hunter-gatherers and agro-pastoralists (Oduaran et al., 2020). Bifidobacterium has been used as an important probiotic in several food products including yogurt and supplements and is believed to enhance a healthy gut (Naumova et al., 2020).
Bilophila wadsworthia is known to digest taurine a naturally occurring compound in meat and milk fat and produce hydrogen sulfide an inflammatory agent (Yazici et al., 2017; Hanson et al., 2021). Also, their ability to produce secondary bile is linked with the occurrence of colorectal cancer (Turnbaugh, 2012). The relative abundance of Bilophila wadsworthia did not differ between rural agro-pastoralists and urban residents (Katsidzira et al., 2019; Oduaran et al., 2020). This may signify a similarity in animal-based diet consumption among rural agro-pastoralists and urban residents (Mehta et al., 2017).
Blautia was reported to be low among rural pastoralists and agro-pastoralists but high in urban (Ayeni et al., 2018; Afolayan et al., 2019) while Blautia Obeum was high in urban and low among agro-pastoralists (Katsidzira et al., 2019). In Kim et al, the relative abundance of Blautia was low in urban Korea compared to rural villages (Wu et al., 2011). This genus is associated with better host health and reduced risk of metabolic syndrome (Liu et al., 2021).
Coprococcus, a butyrate producer, is positively correlated with a high-quality diet and negatively correlated with hypertension (Mancabelli et al., 2017; Laitinen and Mokkala, 2019). Two studies found Coprococcus were more abundant in urban residents and low in agro-pastoralists (Ayeni et al., 2018) and increased with the urbanization gradient from rural to urban (Lokmer et al., 2020). This is contrary to a study in Japan where Coprococcus were least abundant in urban (Naito et al., 2019). Further, without following the urbanization gradient, the Coprococcus genus was found to dominate the Brazilian population compared to Cameroonians (Schaan et al., 2021).
Fusobacterium is an important gut commensal organism that plays a key role in protecting the body from pathogens (Brennan and Garrett, 2019; Stokowa-Sołtys et al., 2021). However, its isolation from disease-related samples (clinical) has indicated a potential role in disease causation and progression questioning the direction of its effect on health (Brennan and Garrett, 2019). Fusobacterium is linked to an increased risk of colorectal cancer and its high relative abundance is associated with the consumption of Westernized diets (Mehta et al., 2017). In this review, the distribution of Fusobacterium was mixed with some studies reporting high relative abundance in urban areas while others reported a low abundance or no difference between urban and rural residents (Katsidzira et al., 2019; Afolayan et al., 2020; Oduaran et al., 2020).
Similar to Fusobacterium, Faecalibacterium strengthens the gut mucosal integrity and maintains the health of the gut and organism physiology through the production of different anti-inflammatory metabolites such as butyrate and salicylic acid (He et al., 2021). It is recognized as the most ubiquitous species due to its presence in the gut of most mammals including humans and was similarly the most frequently reported genus in this review (He et al., 2021). Faecalibacterium was more abundant in the mountainous rural compared to urbanizing and urbanized areas in Ningxia China (Du et al., 2021). We did not find a clear gradient for Faecalibacterium in the included studies. Like Fusobacterium, Fecalibacterium also exhibits a bidirectional impact on health, with a low relative abundance being associated with irritable bowel syndrome and a high relative abundance being linked to colorectal cancer (Rajilić-Stojanović et al., 2011; Keku et al., 2013).
Diets and individual foods influence the diversity and composition of the gut microbiota as shown in several reviews (Frame et al., 2019; Willis and Slavin, 2020; Kimble et al., 2022; Wang et al., 2022). A scoping review of randomized controlled trials on the effects of meat on the gut microbiota showed an increase in Ruminococcus, Roseburia, Bacteroides, and Anaerostipes genera with a high intake of meat while Faecalibacterium decreased (Wang et al., 2022). A review of the adherence to the Mediterranean diet and gut microbiota did not show any relationship at the higher taxonomic level (phyla) but at the genera level with Ruminococcus, Bacteroides, and Faecalibacterium (Kimble et al., 2022). Another systematic review of the effect of fiber supplementation on gut microbiota showed a significantly higher relative abundance of Bifidobacterium and Lactobacillus compared to other genera (So et al., 2018).
In this review, we could not conclude on the differences in diets across the urbanization gradient and its effect on the gut microbiota as only a few studies used standard methods of dietary assessment. One study reported the link between diet and gut microbiota where similar to (Wang et al., 2022) Ruminococcus was linked to meat and iron intake and Faecalibacterium to fish, Vitamin A-rich green vegetables and fruits similar to (Kimble et al., 2022). Also, there was evidence of changes in the gut microbiota in response to changing diets and seasons. This phenomenon was described in one study only and not across the gradient. Hence, it is evident that the effects of diet on the diversity and composition of the gut microbiota are still under-researched in the SSA. Included studies have mostly named the macronutrients without further quantification of the consumed diets, the micronutrient contents, or the analysis of dietary patterns. For example, the high-fiber diets reported in both rural and urban communities may differ significantly in quantity and quality hence their effect on the gut microbiota.
Intestinal parasites are associated with the composition and diversity of the gut microbiota. Their low prevalence in industrialized communities is thought to contribute to the lower diversity of the gut microbiota, although the mechanism underlying this association is poorly understood (Chabé et al., 2017). A study in Malaysia showed a higher microbial diversity among helminths positive compared to negative colonized individuals (Lee et al., 2014). The evidence from this review partly affirms this finding as it shows a relationship between gut microbiota and intestinal parasites. The presence of intestinal parasites was associated with a high diversity of the gut microbiota in two studies (Rubel et al., 2020; Even et al., 2021) but the effect size was not shown in some studies. Further, the use of anti-helminths was linked to a long-term loss of gut microbiota diversity (Rosa et al., 2018). As the relationship between gut microbiota and intestinal parasites is increasingly seen further studies to inform the public health efforts of mass deworming are needed. The absence of intestinal parasites has also been linked to an increased risk of inflammatory bowel diseases and Crohn’s disease (Panelli et al., 2020).
Evidence links metabolites and enriched metabolic pathways of the gut microbiota with several negative and positive health outcomes. Metabolites and pathways, such as the production of short-chain fatty acids in the colon, amino acid metabolism, carbohydrate metabolism, and secondary metabolite biosynthesis pathways, have been shown to positively influence health outcomes (Lin et al., 2017). Gut microbiome metabolites such as trimethylamine N-oxide (TMAO) and hydrogen sulfide are thought to affect gut and cardiovascular health in both positive and negative ways (Wu et al., 2021). However, evidence is still building on other gut microbiome by-products such as “p-cresol” being a potential neurotoxic (Tevzadze et al., 2017; Tevzadze et al., 2018). In this review, differences in pathway enrichment between rural and urban were reported in two studies where amino acid and carbohydrate metabolism pathways were enriched in the urban population (Afolayan et al., 2019) while carbohydrate metabolism gene pathways were depleted in the hunter-gatherers population (Gomez et al., 2016). The differences in the enrichment were not stated in most studies, however, we noted a uniform reporting of the amino acid metabolism pathways.
Low dietary protein intake is linked with the body using essential amino acids produced by the gut microbiome (Wu et al., 2021). Some bacteria species (e.g. Clostridium acetobutylicum) are known to have a complete gene set for essential amino acids biosynthesis (Nolling et al., 2001). Hence, the wide reporting of the amino acid metabolism pathways may signify a diet lacking all the essential amino acids by the included population. However, given the complex relationship and interpretation of metabolic pathways other factors may explain the pattern of the reported amino acid metabolism pathways. A study done in China in two adjacent rural and urban provinces reported 266 out of 1108 differences in metabolites between the two sites (Wang et al., 2020). Six metabolites were twice the amount in urban compared to rural for Xanthine metabolism (derived from caffeine) (Wang et al., 2020). Other metabolites and pathways enriched in urban included those causing inflammation and metabolic derangements (Wang et al., 2020). Characterization of the gut microbiome metabolic pathways and endogenous and exogenous metabolites is still a growing field. Thus, we did not find a large body of evidence comparing the differences along the urbanization gradient to draw robust inferences.
In addition to external perturbation, the gut microbiota is sensitive to several individual factors including physical activity levels, household, and lifestyle factors such as smoking, alcohol intake, occupation, economic levels, and cultural beliefs (Xu and Gordon, 2003). The assessment of these factors with the gut microbiota was not explicitly featured in the included studies. Evidence from McCall et al. shows the differences in the gut microbiota diversity are partly explained by the individual’s socioeconomic levels and not their location along the urbanization gradient (McCall et al., 2020). Further, we noted a variation in the definition of urbanization gradient as disparate approaches were used across studies which limited inter-study comparison. In some studies, the definition was based on subsistence modes while others considered geographical locations or developed indices. Multiple factors related to urbanization are known to impact gut microbiota, therefore, it is important to develop and use standard approaches for unified assessments. In this instance, indices to measure the urbanization gradient as done by Lokmer et al. and adapting standard approaches in the dietary assessment such as the 24-hour recall and Food frequency questionnaires (FFQ) should be considered (Lokmer et al., 2020).
The methods for sample collection, DNA extraction, and analysis varied significantly among the studies. Although alpha and beta diversities were often reported, the reporting was not consistent. In general, the results suggested the diversity of the gut microbiota differed across the urbanization gradient. Thus, the comparability of the studies was limited by heterogeneity in sequencing, analytical approaches, and reporting. Several initiatives propose approaches to standardizing methodologies and reporting microbiome studies (Amos et al., 2020; Mirzayi et al., 2021). Hence, upcoming studies should strive to incorporate these standards for robust comparisons across different studies and datasets. Standardization approaches, similar to those used in the study of microbiome should be considered in the investigation of the determinants of the gut microbiota.
The included studies had several limitations which affected the inter-study comparisons. Statistically, most studies did not give the sample selection strategy and rationale. Secondly, estimated sample sizes were justified in two studies only and there was a remarkable variation in the sample sizes that ranged from 10 to 575. Most studies were insufficiently powered to detect statistical differences. In this context, we assume most study samples were influenced by high sequencing costs although not evident in all studies. As the costs of sequencing decrease and the quality of sequencing technologies improve, larger sample sizes and studies beyond cross-sectional studies should be considered. These may include longitudinal studies to document population changes in gut microbiota including case-crossover studies that follow people at the time when they move or undergo diet transitions.
This review aimed to examine the dissimilarities in the composition and diversity of gut microbiota in the transitioning SSA region. Generally, the findings indicate that there are variations in the gut microbiota diversity between urban and rural/traditional communities, although the evidence regarding specific taxa levels for transitioning communities is inconsistent. In comparison to traditional/rural communities, urban communities exhibited lower diversity in their gut microbiota, while certain rural communities (pastoralists) show decreased diversity. Therefore, the urbanization process in the SSA is associated with the loss of diversity, but there is insufficient evidence to indicate changes in the composition of the gut microbiota. The lower diversity of the gut microbiota may be a driving factor in increasing NCDs in transitioning communities in SSA.
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s.
The research question was conceptualized by VL, FA, and LP. LP and GB independently screened abstracts and extracted study findings. LP analyzed and synthesized extracted data, supported by VL. LP wrote and revised the manuscript, incorporating comments from VL and other authors KK-G, FA, PC, AK, GK, and GB. All authors contributed to the article and approved the submitted version.
LP is supported by the University Medical Center Utrecht Global Health Support PhD programme, and the Cardiac Center of Excellence of the Muhimbili University of Health and Allied Sciences, Tanzania.
Special thanks to the Utrecht University library, especially Dr Paulien H. Wiersma for her guidance and support in the development of the search terms and strategies.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/frmbi.2023.1208166/full#supplementary-material
Afolayan A. O., Adebusoye L. A., Cadmus E. O., Ayeni F. A. (2020). Insights into the gut microbiota of Nigerian elderly with type 2 diabetes and non-diabetic elderly persons. Heliyon 6 (5), e03971. doi: 10.1016/j.heliyon.2020.e03971
Afolayan A. O., Ayeni F. A., Moissl-Eichinger C., Gorkiewicz G., Halwachs B., Högenauer C. (2019). Impact of a nomadic pastoral lifestyle on the gut microbiome in the fulani living in Nigeria. Front. Microbiol. 10, 2138. doi: 10.3389/fmicb.2019.02138
Amos G. C. A., Logan A., Anwar S., Fritzsche M., Mate R., Bleazard T., et al. (2020). Developing standards for the microbiome field. Microbiome 8 (1), 98. doi: 10.1186/s40168-020-00856-3
Angelakis E., Bachar D., Yasir M., Musso D., Djossou F., Gaborit B., et al. (2019). Treponema species enrich the gut microbiota of traditional rural populations but are absent from urban individuals. New Microbes New Infect. 27, 14–21. doi: 10.1016/j.nmni.2018.10.009
Ayeni F. A., Biagi E., Rampelli S., Fiori J., Soverini M., Audu H. J., et al. (2018). Infant and adult gut microbiome and metabolome in rural bassa and urban settlers from Nigeria. Cell Rep. 23 (10), 3056–3067. doi: 10.1016/j.celrep.2018.05.018
Bäckhed F., Fraser C. M., Ringel Y., Sanders M. E., Sartor R. B., Sherman P. M., et al. (2012). Defining a healthy human gut microbiome: Current concepts, future directions, and clinical applications. Cell Host Microbe 12 (5), 611–622. doi: 10.1016/j.chom.2012.10.012
Blaser M. J. (2017). The theory of disappearing microbiota and the epidemics of chronic diseases. Nat. Rev. Immunol. 17 (8), 461–463. doi: 10.1038/nri.2017.77
Brennan C. A., Garrett W. S. (2019). Fusobacterium nucleatum—symbiont, opportunist and oncobacterium. Nat. Rev. Microbiol. 17 (3), 156–166. doi: 10.1038/s41579-018-0129-6
Chabé M., Lokmer A., Ségurel L. (2017). Gut protozoa: friends or foes of the human gut microbiota? Trends Parasitol.: Elsevier Ltd. p, 925–934. doi: 10.1016/j.pt.2017.08.005
Chen H., Mozzicafreddo M., Pierella E., Carletti V., Piersanti A., Ali S. M., et al. (2021). Dissection of the gut microbiota in mothers and children with chronic Trichuris trichiura infection in Pemba Island, Tanzania. Parasites Vectors 14 (1), 62. doi: 10.1186/s13071-021-04580-1
Clemente J. C., Pehrsson E. C., Blaser M. J., Sandhu K., Gao Z., Wang B., et al. (2015). The microbiome of uncontacted AmerIndians. Sci. Adv. 1 (3), e1500183. doi: 10.1126/sciadv.1500183
Cockx L., Colen L., De Weerdt J. (2018). From corn to popcorn? Urbanization and dietary change: Evidence from rural-urban migrants in Tanzania. World Dev. 110, 140–159. doi: 10.1016/j.worlddev.2018.04.018
Cui G., Liu H., Xu G., Laugsand J.-B., Pang Z. (2021). Exploring links between industrialization, urbanization, and Chinese inflammatory bowel disease. Front. Med. 2047. doi: 10.3389/fmed.2021.757025
Das B., Ghosh T. S., Kedia S., Rampal R., Saxena S., Bag S., et al. (2018). Analysis of the gut microbiome of rural and urban healthy Indians living in sea level and high altitude areas. Sci. Rep. 8 (1), 1–15. doi: 10.1038/s41598-018-28550-3
David L. A., Maurice C. F., Carmody R. N., Gootenberg D. B., Button J. E., Wolfe B. E., et al. (2014). Diet rapidly and reproducibly alters the human gut microbiome. Nature 505 (7484), 559–563. doi: 10.1038/nature12820
Dehingia M., Devi K. T., Talukdar N. C., Talukdar R., Reddy N., Mande S. S., et al. (2015). Gut bacterial diversity of the tribes of India and comparison with the worldwide data. Sci. Rep. 5 (1), 1–12. doi: 10.1038/srep18563
Doumatey A. P., Adeyemo A., Zhou J., Lei L., Adebamowo S. N., Adebamowo C., et al. (2020). Gut microbiome profiles are associated with type 2 diabetes in urban Africans. Front. Cell. Infect. Microbiol. 10. doi: 10.3389/fcimb.2020.00063
Du Y., Ding L., Na L., Sun T., Sun X., Wang L., et al. (2021). Prevalence of chronic diseases and alterations of gut microbiome in people of Ningxia China during urbanization: an epidemiological survey. Front. Cell. Infect. Microbiol. 1067. doi: 10.3389/fcimb.2021.707402
Dugas L. R., Bernabé B. P., Priyadarshini M., Fei N., Park S. J., Brown L., et al. (2018). Decreased microbial co-occurrence network stability and SCFA receptor level correlates with obesity in African-origin women. Sci. Rep. 8 (1), 1–17. doi: 10.1038/s41598-018-35230-9
Ellis R. J., Bruce K. D., Jenkins C., Stothard J. R., Ajarova L., Mugisha L., et al. (2013). Comparison of the distal gut microbiota from people and animals in Africa. PloS One 8 (1), e54783. doi: 10.1371/journal.pone.0054783
Even G., Lokmer A., Rodrigues J., Audebert C., Viscogliosi E., Ségurel L., et al. (2021). Changes in the Human Gut Microbiota Associated With Colonization by Blastocystis sp. and Entamoeba spp. in Non-Industrialized Populations. Front. Cell. Infect. Microbiol. 11. doi: 10.3389/fcimb.2021.533528
Ezaki T. (2015). Coprococcus. Bergey’s Manual of Systematics of Archaea and Bacteria (John Wiley & Sons, Inc.), 1–3. doi: 10.1002/9781118960608.gbm00642
Fan Y., Pedersen O. (2021). Gut microbiota in human metabolic health and disease. Nat. Rev. Microbiol.: Nat. Res. p, 55–71. doi: 10.1038/s41579-020-0433-9
Fragiadakis G. K., Smits S. A., Sonnenburg E. D., Van Treuren W., Reid G., Knight R., et al. (2019). Links between environment, diet, and the hunter-gatherer microbiome. Gut. Microbes 10 (2), 216–227. doi: 10.1080/19490976.2018.1494103
Frame L., Costa E., Jackson S. (2019). Current explorations of nutrition and the gut microbiome: A systematic review (P20-032-19). Curr. Developments Nutr. 3 (Supplement_1), nzz040. doi: 10.1093/cdn/nzz040.P20-032-19
Gacesa R., Kurilshikov A., Vich Vila A., Sinha T., Klaassen M. A. Y., Bolte L. A., et al. (2022). Environmental factors shaping the gut microbiome in a Dutch population. Nature 604 (7907), 732–739. doi: 10.1038/s41586-022-04567-7
Gomez A., Petrzelkova K. J., Burns M. B., YeOman C. J., Amato K. R., Vlckova K., et al. (2016). Gut microbiome of coexisting BaAka pygmies and Bantu reflects gradients of traditional subsistence patterns. Cell Rep. 14 (9), 2142–2153. doi: 10.1016/j.celrep.2016.02.013
Gong B., Wang C., Meng F., Wang H., Song B., Yang Y., et al. (2021). Association between gut microbiota and autoimmune thyroid disease: A systematic review and meta-analysis. Front. Endocrinol. 12, 1544. doi: 10.3389/fendo.2021.774362
Gupta V. K., Paul S., Dutta C. (2017). Geography, ethnicity or subsistence-specific variations in human microbiome composition and diversity. Front. Microbiol.: Front. Media S. A. p, 1162– doi: 10.3389/fmicb.2017.01162
Hansen M. E. B., Rubel M. A., Bailey A. G., Ranciaro A., Thompson S. R., Campbell M. C., et al. (2019). Population structure of human gut bacteria in a diverse cohort from rural Tanzania and Botswana. Genome Biol. 20 (1), 16. doi: 10.1186/s13059-018-1616-9
Hanson B. T., Dimitri Kits K., Löffler J., Burrichter A. G., Fiedler A., Denger K., et al. (2021). Sulfoquinovose is a select nutrient of prominent bacteria and a source of hydrogen sulfide in the human gut. ISME J. 15 (9), 2779–2791. doi: 10.1038/s41396-021-00968-0
He X., Zhao S., Li Y. (2021). Faecalibacterium prausnitzii: A next-generation probiotic in gut disease improvement. Can. J. Infect. Dis. Med. Microbiol. 2021, 1–10. doi: 10.1155/2021/6666114
Heeney D. D., Gareau M. G., Marco M. L. (2018). Intestinal Lactobacillus in health and disease, a driver or just along for the ride? Curr. Opin. Biotechnol. 49, 140–147. doi: 10.1016/j.copbio.2017.08.004
Iebba V., Santangelo F., Totino V., Pantanella F., Monsia A., Di Cristanziano V., et al. (2016). Gut microbiota related to Giardia duodenalis, Entamoeba spp. and Blastocystis hominis infections in humans from Côte d’Ivoire. J. Infect. Developing Countries 10 (09), 1035–1041. doi: 10.3855/jidc.8179
Jha A. R., Davenport E. R., Gautam Y., Bhandari D., Tandukar S., Ng K. M., et al. (2018). Gut microbiome transition across a lifestyle gradient in Himalaya. PloS Biol. 16 (11), e2005396–e. doi: 10.1371/journal.pbio.2005396
Johnson E. L., Heaver S. L., Walters W. A., Ley R. E. (2017). Microbiome and metabolic disease: revisiting the bacterial phylum Bacteroidetes. J. Mol. Med. 95, 1–8. doi: 10.1007/s00109-016-1492-2
Kassouri Y., Okunlola O. A. (2022). Analysis of spatio-temporal drivers and convergence characteristics of urban development in Africa. Land Use Policy 112, 105868. doi: 10.1016/j.landusepol.2021.105868
Katsidzira L., Ocvirk S., Wilson A., Li J., Mahachi C. B., Soni D., et al. (2019). Differences in fecal gut microbiota, short-chain fatty acids and bile acids link colorectal cancer risk to dietary changes associated with urbanization among Zimbabweans. Nutr. Cancer 71 (8), 1313–1324. doi: 10.1080/01635581.2019.1602659
Keku T. O., McCoy A. N., Azcarate-Peril A. M. (2013). Fusobacterium spp. And colorectal cancer: cause or consequence? Trends Microbiol.: Elsevier p, 506–508. doi: 10.1016/j.tim.2013.08.004
Kimble R., Gouinguenet P., Ashor A., Stewart C., Deighton K., Matu J., et al. (2022). Effects of a mediterranean diet on the gut microbiota and microbial metabolites: A systematic review of randomized controlled trials and observational studies. Crit. Rev. Food Sci. Nutr., 1–22. doi: 10.1080/10408398.2022.2057416
King C. H., Desai H., Sylvetsky A. C., LoTempio J., Ayanyan S., Carrie J., et al. (2019). Baseline human gut microbiota profile in healthy people and standard reporting template. PloS One 14 (9), e0206484. doi: 10.1371/journal.pone.0206484
Kovatcheva-Datchary P., Nilsson A., Akrami R., Lee Y. S., De Vadder F., Arora T., et al. (2015). Dietary fiber-induced improvement in glucose metabolism is associated with increased abundance of Prevotella. Cell Metab. 22 (6), 971–982. doi: 10.1016/j.cmet.2015.10.001
Laitinen K., Mokkala K. (2019). Overall dietary quality relates to gut microbiota diversity and abundance. Int. J. Mol. Sci. 20 (8), 1835. doi: 10.3390/ijms20081835
Lee S. C., Tang M. S., Lim Y. A., Choy S. H., Kurtz Z. D., Cox L. M., et al. (2014). Helminth colonization is associated with increased diversity of the gut microbiota. PloS Negl. Trop. Dis. 8 (5), e2880. doi: 10.1371/journal.pntd.0002880
Ley R. E. (2016). Gut microbiota in 2015: Prevotella in the gut: Choose carefully. Nat. Rev. Gastroenterol. Hepatol.: Nat. Publishing Group p, 69. doi: 10.1038/nrgastro.2016.4
Li H., Li T., Li X., Wang G., Lin Q., Qu J. (2018). Gut microbiota in Tibetan herdsmen reflects the degree of urbanization. Front. Microbiol. 9, 1745. doi: 10.3389/fmicb.2018.01745
Li X., Lu Z. (2021). Quantitative measurement on urbanization development level in urban Agglomerations: A case of JJJ urban agglomeration. Ecol. Indicators 133, 108375. doi: 10.1016/j.ecolind.2021.108375
Li Y., Westlund H., Liu Y. (2019). Why some rural areas decline while some others not: An overview of rural evolution in the world. J. Rural Stud. 68, 135–143. doi: 10.1016/j.jrurstud.2019.03.003
Lin R., Liu W., Piao M., Zhu H. (2017). A review of the relationship between the gut microbiota and amino acid metabolism. Amino Acids 49, 2083–2090. doi: 10.1007/s00726-017-2493-3
Liu X., Mao B., Gu J., Wu J., Cui S., Wang G., et al. (2021). Blautia—a new functional genus with potential probiotic properties? Gut. Microbes: Bellwether Publishing Ltd. p, 1–21. doi: 10.1080/19490976.2021.1875796
Lokmer A., Aflalo S., Amougou N., Lafosse S., Froment A., Tabe F. E., et al. (2020). Response of the human gut and saliva microbiome to urbanization in Cameroon. Sci. Rep. 10 (1), 2856. doi: 10.1038/s41598-020-59849-9
Lu J., Zhang L., Zhai Q., Zhao J., Zhang H., Lee Y. K., et al. (2021). Chinese gut microbiota and its associations with staple food type, ethnicity, and urbanization. NPJ Biofilms Microbiomes 7 (1), 1–12. doi: 10.1038/s41522-021-00245-0
Lun H., Yang W., Zhao S., Jiang M., Xu M., Liu F., et al. (2019). Altered gut microbiota and microbial biomarkers associated with chronic kidney disease. Microbiologyopen 8 (4), e00678. doi: 10.1002/mbo3.678
Mancabelli L., Milani C., Lugli G. A., Turroni F., Ferrario C., van Sinderen D., et al. (2017). Meta-analysis of the human gut microbiome from urbanized and pre-agricultural populations. Environ. Microbiol. 19 (4), 1379–1390. doi: 10.1111/1462-2920.13692
Maurelli M., Pepe P., Gualdieri L., Bosco A., Cringoli G., Rinaldi L. (2022). Improving diagnosis of intestinal parasites towards a migrant-friendly health system. Curr. Trop. Med. Rep. 10, 17–25. doi: 10.1007/s40475-022-00280-7
McCall L. I., Callewaert C., Zhu Q., Song S. J., Bouslimani A., Minich J. J., et al. (2020). Home chemical and microbial transitions across urbanization. Nat. Microbiol.: Nat. Res. p, 108–115. doi: 10.1038/s41564-019-0593-4
Mehta R. S., Nishihara R., Cao Y., Song M., Mima K., Qian Z. R., et al. (2017). Association of dietary patterns with risk of colorectal cancer subtypes classified by Fusobacterium nucleatum in tumor tissue. JAMA Oncol. 3 (7), 921–927. doi: 10.1001/jamaoncol.2016.6374
Mirzayi C., Renson A., Consortium G. S., Analysis M., QCSFCSS-A, Zohra F., et al. (2021). Reporting guidelines for human microbiome research: the STORMS checklist. Nat. Med. 27 (11), 1885–1892. doi: 10.1038/s41591-021-01552-x
Morton E. R., Lynch J., Froment A., Lafosse S., Heyer E., Przeworski M., et al. (2015). Variation in rural African gut microbiota is strongly correlated with colonization by Entamoeba and subsistence. PloS Genet. 11 (11), e1005658–e. doi: 10.1371/journal.pgen.1005658
Naito Y., Takagi T., Inoue R., Kashiwagi S., Mizushima K., Tsuchiya S., et al. (2019). Gut microbiota differences in elderly subjects between rural city Kyotango and urban city Kyoto: an age-gender-matched study. J. Clin. Biochem. Nutr. 65 (2), 125–131. doi: 10.3164/jcbn.19-26
Natividad J. M., Lamas B., Pham H. P., Michel M. L., Rainteau D., Bridonneau C., et al. (2018). Bilophila wadsworthia aggravates high fat diet induced metabolic dysfunctions in mice. Nat. Commun. 9 (1), 1–15. doi: 10.1038/s41467-018-05249-7
Naumova N., Alikina T., Tupikin A., Kalmykova A., Soldatova G., Vlassov V., et al. (2020). Human gut microbiome response to short-term bifidobacterium-based probiotic treatment. Indian J. Microbiol. 60 (4), 451–457. doi: 10.1007/s12088-020-00888-1
Njoh A. J. (2003). Urbanization and development in sub-Saharan Africa. Cities 20 (3), 167–174. doi: 10.1016/S0264-2751(03)00010-6
Nolling J., Breton G., Omelchenko M. V., Makarova K. S., Zeng Q., Gibson R., et al. (2001). Genome sequence and comparative analysis of the solvent-producing bacterium Clostridium acetobutylicum. J. Bacteriol. 183 (16), 4823–4838. doi: 10.1128/JB.183.16.4823-4838.2001
Obregon-Tito A. J., Tito R. Y., Metcalf J., Sankaranarayanan K., Clemente J. C., Ursell L. K., et al. (2015). Subsistence strategies in traditional societies distinguish gut microbiomes. Nat. Commun. 6 (1), 1–9. doi: 10.1038/ncomms7505
Ocvirk S., Wilson A. S., Posma J. M., Li J. V., Koller K. R., Day G. M., et al. (2020). A prospective cohort analysis of gut microbial co-metabolism in Alaska Native and rural African people at high and low risk of colorectal cancer. Am. J. Clin. Nutr. 111 (2), 406–419. doi: 10.1093/ajcn/nqz301
Oduaran O. H., Tamburini F. B., Sahibdeen V., Brewster R., Gómez-Olivé F. X., Kahn K., et al. (2020). Gut microbiome profiling of a rural and urban South African cohort reveals biomarkers of a population in lifestyle transition. BMC Microbiol. 20 (1), 1–17. doi: 10.1186/s12866-020-02017-w
Oniszczuk A., Oniszczuk T., Gancarz M., Szymańska J. (2021). Role of gut microbiota, probiotics and prebiotics in the cardiovascular diseases. Molecules 26 (4), 1172. doi: 10.3390/molecules26041172
Ouzzani M., Hammady H., Fedorowicz Z., Elmagarmid A. (2016). Rayyan—a web and mobile app for systematic reviews. Systematic Rev. 5, 1–10. doi: 10.1186/s13643-016-0384-4
Panelli S., Epis S., Cococcioni L., Perini M., Paroni M., Bandi C., et al. (2020). Inflammatory bowel diseases, the hygiene hypothesis and the other side of the microbiota: parasites and fungi. Pharmacol. Res. 159, 104962. doi: 10.1016/j.phrs.2020.104962
Parbie P. K., Mizutani T., Ishizaka A., Kawana-Tachikawa A., Runtuwene L. R., Seki S., et al. (2021). Fecal microbiome composition in healthy adults in Ghana. Japanese J. Infect. Dis. 74 (1), 42–47. doi: 10.7883/yoken.JJID.2020.469
Popkin B. M. (1998). The nutrition transition and its health implications in lower-income countries. Public Health Nutr. 1 (1), 5–21. doi: 10.1079/PHN19980004
Rajilić-Stojanović M., Biagi E., Heilig H. G. H. J., Kajander K., Kekkonen R. A., Tims S., et al. (2011). Global and deep molecular analysis of microbiota signatures in fecal samples from patients with irritable bowel syndrome. Gastroenterology 141 (5), 1792–1801. doi: 10.1053/j.gastro.2011.07.043
Ramadass B., Rani B. S., Pugazhendhi S., John K., Ramakrishna B. S. (2017). Faecal microbiota of healthy adults in south India: comparison of a tribal & a rural population. Indian J. Med. Res. 145 (2), 237. doi: 10.4103/ijmr.IJMR_639_14
Rosa B. A., Supali T., Gankpala L., Djuardi Y., Sartono E., Zhou Y., et al. (2018). Differential human gut microbiome assemblages during soil-transmitted helminth infections in Indonesia and Liberia. Microbiome 6 (1), 33. doi: 10.1186/s40168-018-0416-5
Rosas-Plaza S., Hernández-Terán A., Navarro-Díaz M., Escalante A. E., Morales-Espinosa R., Cerritos R. (2022). Human gut microbiome across different lifestyles: from hunter-gatherers to urban populations. Front. Microbiol. 13, 843170. doi: 10.3389/fmicb.2022.843170
Rubel M. A., Abbas A., Taylor L. J., Connell A., Tanes C., Bittinger K., et al. (2020). Lifestyle and the presence of helminths is associated with gut microbiome composition in Cameroonians. Genome Biol. 21 (1), 1–32. doi: 10.1186/s13059-020-02020-4
Schaan A. P., Sarquis D., Cavalcante G. C., Magalhães L., Sacuena E. R., Costa J., et al. (2021). The structure of Brazilian Amazonian gut microbiomes in the process of urbanisation. NPJ Biofilms Microbiomes 7 (1), 65. doi: 10.1038/s41522-021-00237-0
Schnorr S. L., Candela M., Rampelli S., Centanni M., Consolandi C., Basaglia G., et al. (2014). Gut microbiome of the Hadza hunter-gatherers. Nat. Commun. 5 (1), 1–12. doi: 10.1038/ncomms4654
Sekirov I., Russell S. L., Antunes L. C., Finlay B. B. (2010). Gut microbiota in health and disease. Physiol. Rev. 90 (3), 859–904. doi: 10.1152/physrev.00045.2009
Smits S. A., Leach J., Sonnenburg E. D., Gonzalez C. G., Lichtman J. S., Reid G., et al. (2017). Seasonal cycling in the gut microbiome of the Hadza hunter-gatherers of Tanzania. Science 357 (6353), 802–805. doi: 10.1126/science.aan4834
So D., Whelan K., Rossi M., Morrison M., Holtmann G., Kelly J. T., et al. (2018). Dietary fiber intervention on gut microbiota composition in healthy adults: a systematic review and meta-analysis. Am. J. Clin. Nutr. 107 (6), 965–983. doi: 10.1093/ajcn/nqy041
Sonnenburg J. L., Bäckhed F. (2016). Diet-microbiota interactions as moderators of human metabolism. Nature 535 (7610), 56–64. doi: 10.1038/nature18846
Steyn N. P., McHiza Z. J. (2014). Obesity and the nutrition transition in Sub-Saharan Africa. Ann. New York Acad. Sci. 1311 (1), 88–101. doi: 10.1111/nyas.12433
Stokowa-Sołtys K., Wojtkowiak K., Jagiełło K. (2021). Fusobacterium nucleatum–Friend or foe? J. Inorg. Biochem. 224, 111586. doi: 10.1016/j.jinorgbio.2021.111586
Tang M., Frank D. N., Tshefu A., Lokangaka A., Goudar S. S., Dhaded S. M., et al. (2019). Different gut microbial profiles in sub-saharan african and south asian women of childbearing age are primarily associated with dietary intakes. Front. Microbiol. 10, 1848. doi: 10.3389/fmicb.2019.01848
Tevzadze G., Oniani N., Zhuravliova E., Darchia N., Eliozishvili M., GogiChadze M., et al. (2018). Effects of a gut microbiome toxin, p-cresol, on the indices of social behavior in rats. Neurophysiology 50, 372–377. doi: 10.1007/s11062-019-09764-1
Tevzadze G., Shanshiashvili L., Mikeladze D. (2017). Children with epilepsy and autistic spectrum disorders show similarly high levels of urinary p-cresol. JBPC 17 (1), 77–80. doi: 10.4024/05TE17A.jbpc.17.02
Tyakht A. V., Alexeev D. G., Popenko A. S., Kostryukova E. S., Govorun V. M. (2014). Rural and urban microbiota: to be or not to be? Gut. Microbes 5 (3), 351–356. doi: 10.4161/gmic.28685
van den Munckhof I. C. L., Kurilshikov A., ter Horst R., Riksen N. P., Joosten L. A. B., Zhernakova A., et al. (2018). Role of gut microbiota in chronic low-grade inflammation as potential driver for atherosclerotic cardiovascular disease: a systematic review of human studies. Obes. Rev. 19 (12), 1719–1734. doi: 10.1111/obr.12750
Wang Y., Sha W., Wang H., Howard A. G., Tsilimigras M. C., Zhang J., et al. (2020). Urbanization in China is associated with pronounced perturbation of plasma metabolites. Metabolomics 16, 1–13. doi: 10.1007/s11306-020-01724-9
Wang Y., Uffelman C. N., Bergia R. E., Clark C. M., Reed J. B., Tzu-Wen L., et al. (2022). Meat consumption and gut microbiota: A scoping review of literature and systematic review of randomized controlled trials in adults. Adv. Nutr. 14 (2), 215–237. doi: 10.1016/j.advnut.2022.10.005
Willis H. J., Slavin J. L. (2020). The influence of diet interventions using whole, plant food on the gut microbiome: a narrative review. J. Acad. Nutr. Dietetics 120 (4), 608–623. doi: 10.1016/j.jand.2019.09.017
Wu G. D., Chen J., Hoffmann C., Bittinger K., Chen Y. Y., Keilbaugh S. A., et al. (2011). Linking long-term dietary patterns with gut microbial enterotypes. Science 334 (6052), 105–108. doi: 10.1126/science.1208344
Wu L., Tang Z., Chen H., Ren Z., Ding Q., Liang K., et al. (2021). Mutual interaction between gut microbiota and protein/amino acid metabolism for host mucosal immunity and health. Anim. Nutr. 7 (1), 11–16. doi: 10.1016/j.aninu.2020.11.003
Xu J., Gordon J. I. (2003). Honor thy symbionts. Proc. Natl. Acad. Sci. 100 (18), 10452–10459. doi: 10.1073/pnas.1734063100
Yatsunenko T., Rey F. E., Manary M. J., Trehan I., Dominguez-Bello M. G., Contreras M., et al. (2012). Human gut microbiome viewed across age and geography. Nature: Nat. Publishing Group p, 222–227. doi: 10.1038/nature11053
Yazici C., Wolf P. G., Kim H., Cross T.-W. L., Vermillion K., Carroll T., et al. (2017). Race-dependent association of sulfidogenic bacteria with colorectal cancer. Gut 66 (11), 1983–1994. doi: 10.1136/gutjnl-2016-313321
Zhang X. F., Guan X. X., Tang Y. J., Sun J. F., Wang X. K., Wang W. D., et al. (2021). Clinical effects and gut microbiota changes of using probiotics, prebiotics or synbiotics in inflammatory bowel disease: a systematic review and meta-analysis. Eur. J. Nutr. 60 (5), 2855–2875. doi: 10.1007/s00394-021-02503-5
Keywords: gut microbiota, human microbiome, urbanization, Sub-Saharan Africa, diet, nutrition transition, intestinal parasites
Citation: Paulo LS, Msema Bwire G, Klipstein-Grobusch K, Kamuhabwa A, Kwesigabo G, Chillo P, Asselbergs FW and Lenters VC (2023) Urbanization gradient, diet, and gut microbiota in Sub-Saharan Africa: a systematic review. Front. Microbiomes 2:1208166. doi: 10.3389/frmbi.2023.1208166
Received: 18 April 2023; Accepted: 22 August 2023;
Published: 12 September 2023.
Edited by:
David C. B. Taras, Boehringer Ingelheim, GermanyReviewed by:
Laura-Isobel McCall, University of Oklahoma, United StatesCopyright © 2023 Paulo, Msema Bwire, Klipstein-Grobusch, Kamuhabwa, Kwesigabo, Chillo, Asselbergs and Lenters. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Linda Simon Paulo, TGluZGEucC5zaW1vbkBnbWFpbC5jb20=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.