- 1College of Food Science and Engineering, Tarim University, Alar, Xinjiang, China
- 2State Key Laboratory of Mycology, Institute of Microbiology, Chinese Academy of Sciences, Beijing, China
- 3Production & Construction Group Key Laboratory of Special Agricultural Products Further Processing in Southern Xinjiang, Alar, China
Domesticated Saccharomyces cerevisiae is one of the most significant microbial populations in human civilization due to its remarkable diversity and high adaptability to human environments. However, the adaptability mechanisms underlying this population ecological behavior remain unclear. This study explored the adaptive behaviors of S. cerevisiae strains from the Wine and Mantou genetic lineages under both artificial stress conditions and natural or near-natural environments. A total of 307 diploid S. cerevisiae strains were analyzed, including 169 strains derived from grape sources and 138 from sourdough sources. Various stress factors, including sodium chloride, tannins, ethanol, pH, temperature, and sulfur dioxide (SO2), as well as different substrates (natural grape juice, simulated grape juice, and simulated dough), were applied to evaluate adaptability. The results demonstrated that Wine population exhibited superior performance in grape juice environments, characterized by higher CO2 production. The biomass of both the Wine and Mantou populations in the simulated dough was significantly higher than that in the simulated grape juice. In the simulated grape juice environment, the adaptability of the Wine population was significantly superior to that of the Mantou population. In contrast, in the simulated dough environment, the Mantou population exhibited better adaptability than the Wine population. Furthermore, Wine population displayed higher tolerance to ethanol, extreme temperatures, tannins, and sodium chloride in YPD medium compared to Mantou population. Diploid strains also exhibited greater stress tolerance than haploid strains. These findings offer valuable insights into the distinct adaptive mechanisms of domesticated S. cerevisiae lineages.
1 Introduction
Recent research on the evolutionary phylogeny of S. cerevisiae populations has identified two major groups: wild and domesticated (Duan et al., 2018). The domesticated group is further classified into sub-lineages specifically adapted to liquid fermentation environments (e.g., the Wine lineage) and those adapted to solid fermentation environments (e.g., the Mantou lineage) (Duan et al., 2018; Han et al., 2021; Tellini et al., 2024). Domesticated S. cerevisiae is one of the most important microbial populations for human civilization. The domesticated population plays a crucial role in the production of bread and wine, exhibiting extensive diversity and strong adaptability to environments such as grape juice and wet dough. Grape juice introduces a range of stress factors that challenge S. cerevisiae survival, including high polyphenol content, acidity, osmotic pressure, and sugar levels, as well as low pH, added sulfur dioxide (SO2), controlled low temperatures, elevated temperatures during natural fermentation, and high alcohol content (Gao et al., 2022; García-Ríos and Guillamón, 2019; Gobert et al., 2019; Lin et al., 2024). Despite these challenges, S. cerevisiae remains the dominant microorganism in fermentation processes due to its exceptional adaptability. In starch-rich environments, such as with a nearly neutral pH, S. cerevisiae thrives by efficiently metabolizing key nutrients like maltose (Aydın et al., 2022; Lahue et al., 2020). This suggests that S. cerevisiae has developed distinct adaptive strategies, enabling it to thrive in various ecological niches, which has resulted in the formation of specialized lineages (Han et al., 2021; Tellini et al., 2024; Wang et al., 2012). Based on the excellent fermentation properties of indigenous S. cerevisiae in grape juice and dough fermentation, many researchers have selected potential strains from natural habitats for industrial applications (Mudoor Sooresh et al., 2023; Parapouli et al., 2020; Zhang et al., 2021). The application of individual strain fermentation and limited strain combinations has significantly advanced the industrial production of wine and bread (de Gioia et al., 2022; Ojeda-Linares et al., 2022; Rădoi-Encea et al., 2023). However, this approach has limitations to rich the quality of fermented foods, and to balance between tradition and innovation in contemporary winemaking (Gardner et al., 2023; Zampi and Ranfagni, 2024). Microbial populations, compared to individual strains, demonstrate a greater capacity to endure intense selective pressures (Swamy and Zhou, 2019). They undergo adaptive selection, characterized by mutations that confer a survival advantage in specific new environments (Heidenreich, 2007; Massoud and Zoghi, 2022; Tellini et al., 2024). These mutations reflect stable evolutionary adaptations to environmental changes. For example, in grape juice and dough, S. cerevisiae relies on the collective behavior of its population to carry out enological functions. Effectively managing this population during natural fermentation has increasingly become a central focus of research (Yi et al., 2024; Zhang et al., 2021).
Currently, research on the adaptability of S. cerevisiae populations derived from limited ancestors’ strains is primarily conducted in laboratory evolution (Caspeta and Nielsen, 2015; Godara and Kao, 2021; Randez-Gil et al., 2020). Through serial propagation, the resulting populations exhibit genetic variations, phenotypic changes, and alterations in survival capabilities as they adapt to the predefined environment (Betlej et al., 2020; Godara and Kao, 2021; Zheng and Wang, 2015). There are also many studies that focus on single strains or a limited number of strains cultivated under specific artificial conditions (Lázari et al., 2022; Mudoor Sooresh et al., 2023). This approach seeks to elucidate the mechanisms by which microorganisms adapt to specific natural environments. However, it may inadequately represent the adaptability of natural S. cerevisiae populations under actual conditions, potentially leading to discrepancies.
The complexity of natural environments, such as grape juice ecosystems, has led many scholars to propose that S. cerevisiae populations from different regions display distinct “terroir characteristics” (Alexandre, 2020; Pretorius, 2020; Rădoi-Encea et al., 2023). However, some researchers contend that this assumption is unfounded (Marsit and Dequin, 2015; Šuranská et al., 2016). In nature, S. cerevisiae exhibits both haploid and diploid life forms (i.e., MAT-a and MAT-α mating types, with diploid forms being predominant) (Zhang et al., 2017). Although the molecular mechanisms driving the adaptive evolution of various evolutionary lineages and ploidy types of S. cerevisiae are well studied, our understanding of population-level adaptive behaviors under real or near-real environmental conditions remains limited.
This study examined the population-level adaptive behaviors of S. cerevisiae in 169 Wine strains and 138 Mantou strains collected from regions along the Silk Road (within China) under multi-stress conditions in both native and non-native habitats. The findings reveal the adaptability of domesticated populations, shedding light on potential intrinsic patterns and providing novel strategies and perspectives for industrial applications.
2 Materials and methods
2.1 Experimental strains
This study utilized a total of 310 yeast strains, including 307 wild strains. Among these, 169 strains of S. cerevisiae were from the Wine population of grapes (SCP), and 138 strains were from the Mantou population of sourdough (SCJ). Specifically, 149 strains originated from Xinjiang (XJ), while 158 strains were sourced from regions outside Xinjiang (NXJ). The phylogenetic analysis revealed that these strains belonged to various lineages, including CHN-VI/VII, Daqu/Baijiu, CHN-VIII, Mantou 3, West African cocoa, Milk/Cheese Milk, European Wine, Mantou 5, Huangjiu, Mantou 7, and Mosaic lineages (e.g., Ecuador Beer 8. Mixed origin, Belgium Human/Clinical 8. Mixed origin, France Human/Clinical 8. Mixed origin, Ethiopia Honey Wine ADY/Mixed, China Plant 8. Mixed origin, China Commercial ADY, Slovakia Water 19. Mixed origin, China Mantou ADY). All wild strains were confirmed to be diploid through ploidy analysis (Supplementary Table 1) (Xiao et al., 2004; Xue et al., 2012). From each lineage, 2–3 S. cerevisiae strains with distinct geographical origins were selected, resulting in a total of 25 strains for the construction of wild-type haploid strains (Xiao et al., 2004; Xue et al., 2012). Additionally, haploid strains BY4741 (MAT-a) and BY4742 (MAT-α) (obtained from the Institute of Microbiology, Chinese Academy of Sciences), as well as the reference strain S288C (purchased from Biosune Biotechnology), were used as reference strains.
2.2 Reagents and medium
Reagents: Sterile 30% glycerol, sodium chloride, tannic acid, sodium thiosulfate, and snail enzyme (100 mg/mL).
YPD liquid medium was prepared by dissolving 10 g of yeast extract, 20 g of glucose, and 20 g of peptone in distilled water. The solution was sterilized at 121°C for 30 min.
Grape juice was freshly pressed and stored at −20°C for subsequent use.
Synthetic grape juice (SG) medium composition:
The SG medium was prepared with the following components (per liter): glucose (60 g), fructose (60 g), dipotassium hydrogen phosphate (1.14 g), magnesium sulfate heptahydrate (MgSO4⋅7H2O, 1.23 g), calcium chloride dihydrate (CaCl2⋅2H2O, 0.44 g), potassium bitartrate (cream of tartar, 2.5 g), malonic acid (3 g), citric acid (0.2 g), ammonium hydrogen phosphate [(NH4)2HPO4, 0.4 g], vitamin solution (10 mL), amino acid solution (10 mL), trace element solution (1 mL), and ergosterol (12.5 mg).
Vitamin solution (mg/L): the vitamin solution contained inositol (100 mg), pyridoxine hydrochloride (Vitamin B6, 2 mg), niacin (2 mg), calcium pantothenate (1 mg), thiamine hydrochloride (Vitamin B1, 0.5 mg), para-aminobenzoic acid (0.2 mg), riboflavin (Vitamin B2, 0.2 mg), biotin (0.125 mg), and folic acid (0.2 mg).
Amino acid solution (mg/L): the amino acid solution included alanine (5.9 mg), arginine (137.3 mg), asparagine (36.5 mg), aspartic acid (23.1 mg), glutamine (48.7 mg), glutamic acid (30.8 mg), glycine (4.1 mg), histidine (45.8 mg), isoleucine (24.1 mg), lysine (61.5 mg), methionine (20 mg), phenylalanine (11.6 mg), serine (48.2 mg), threonine (42.2 mg), tryptophan (12.1 mg), tyrosine (2.4 mg), and valine (24.1 mg).
Trace element solution (μg/L): the trace element solution comprised manganese chloride tetrahydrate (MnCl2⋅4H2O, 200 μg), zinc chloride (ZnCl2, 135 μg), iron chloride tetrahydrate (FeCl2⋅4H2O, 30 μg), copper chloride dihydrate (CuCl2⋅2H2O, 15 μg), boric acid (H3BO3, 5 μg), cobalt nitrate hexahydrate [Co(NO3)2⋅6H2O, 30 μg], sodium molybdate dihydrate (Na2MoO4⋅2H2O, 25 μg), and potassium iodate (KIO3, 10 μg).
The medium was prepared by adding 1,000 mL of distilled water, thoroughly mixing all components, and sterilizing at 121°C for 30 min (Camesasca et al., 2018).
Synthetic flour juice (SF) composition:
The SF medium was composed of the following components (per liter): wheat oligopeptides (12 g), magnesium sulfate heptahydrate (MgSO4⋅7H2O, 0.2 g), manganese sulfate monohydrate (MnSO4⋅H2O, 0.05 g), potassium dihydrogen phosphate (KH2PO4, 4 g), dipotassium hydrogen phosphate (K2HPO4, 4 g), Tween 80 (1 mL), and a vitamin solution (1 mL).
Vitamin solution (mg/L): the vitamin solution contained cobalamin (200 mg), folic acid (200 mg), niacinamide (200 mg), pantothenic acid (200 mg), pyridoxal phosphate (200 mg), and thiamine (200 mg).
Carbohydrate solution (g/L): the carbohydrate solution included glucose (0.5 g), maltose (10 g), fructose (0.5 g), and sucrose (2 g).
The final medium was prepared by adding distilled water to reach a total volume of 1,000 mL, thoroughly mixing all components, and sterilizing at 121°C for 30 min (Vrancken et al., 2008).
2.3 Experimental methods
Fresh YPD cultures (1%) were inoculated into 10 mL of SG and SF media. The cultures were incubated at 28°C for 7 days. On days 0 and 7, 200 μL of the cultures were transferred to a 96-well microplate, and absorbance at 600 nm was measured using a Synergy H1 multifunctional microplate reader (BioTek). The initial absorbance (OD1) and post-incubation absorbance (OD2) were recorded. Calculate fungal biomass ΔOD = (OD2–OD1)/3 (Zhu et al., 2017).
Tolerance behavior under stressful conditions:
Fresh YPD cultures were inoculated (1% v/v) into YPD media under various stress conditions, including sodium chloride (NaCl), tannin (TA), alcohol (AL), pH, temperature, and sulfur dioxide (SO2). The specific stress conditions were as follows:
Sodium chloride (NaCl): 6, 8, and 10% in YPD.
Tannin (TA): 1 g/L, 1.5 g/L, and 2 g/L in YPD.
Alcohol (AL): 12, 14, and 16% in YPD.
pH levels: 3.0, 3.5, 4.0, 5.0, and 7.0 in YPD.
Sodium thiosulfate (S): 0.632 g/L (S-4), 0.1264 g/L (S-8), and 0.1896 g/L (S-12) in YPD.
Cultures were incubated at 28°C for 48 h after inoculation into YPD liquid medium. At both 0 h and 48 h, 200 μL of the culture was transferred into a 96-well plate. Absorbance was measured at 600 nm using a multifunctional plate reader, recording the initial optical density (OD1) and the optical density after 48 h (OD2). The change in microbial density (ΔOD) was calculated using the formula: (OD2–OD1)/3.
All experiments were performed in triplicate, and data analysis was conducted using SPSS 26.0 software. One-way ANOVA was employed to assess differences across various sources of isolation, regions, lineages, and substrates, with statistical significance defined as P < 0.05. Graphs were created using Origin 2021, while data processing and heatmap generation were performed with MetaboAnalyst.1
3 Results
3.1 Adaptability of diploid S. cerevisiae populations
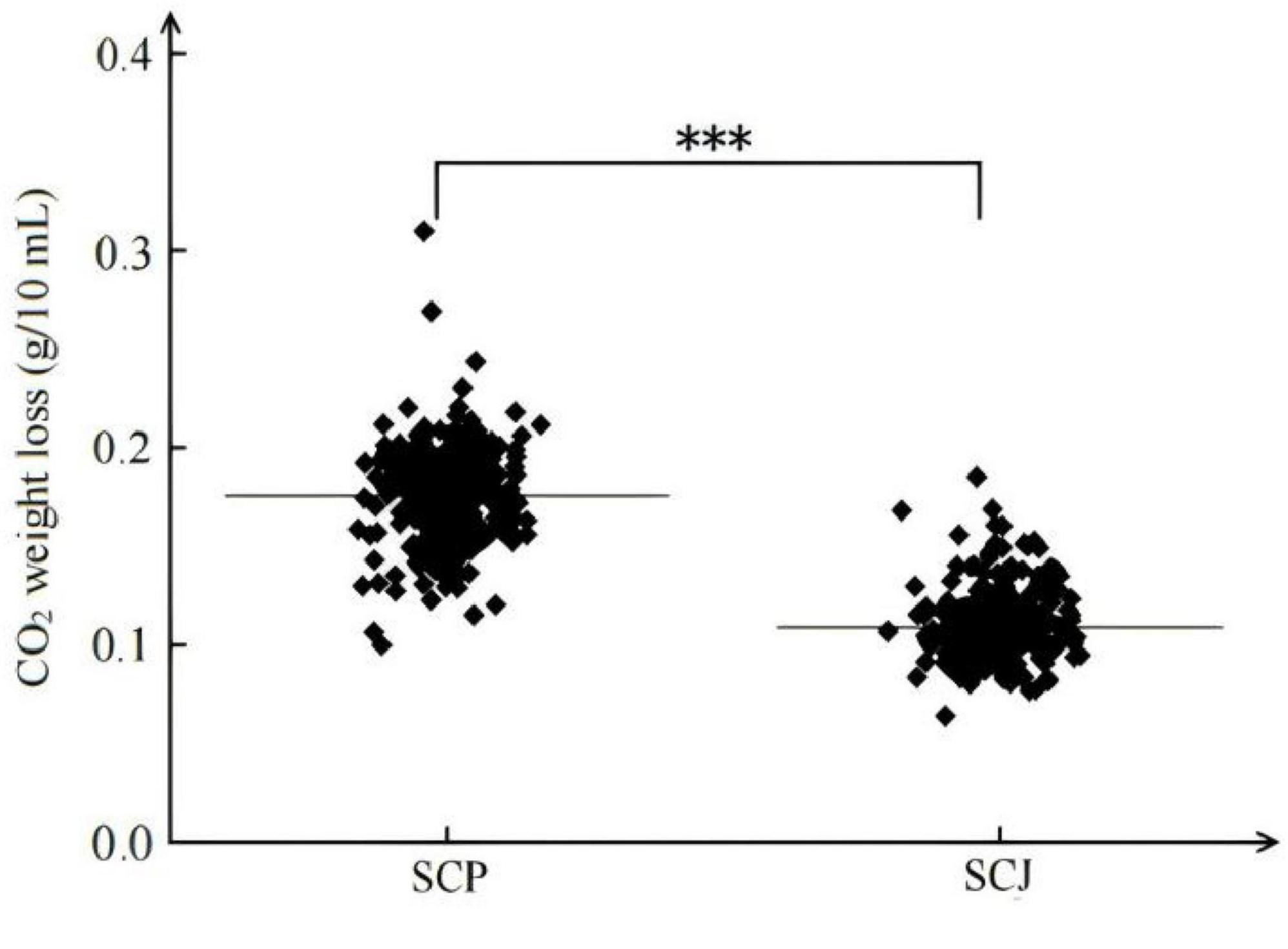
3.1.1 CO2 weight loss in SCP and SCJ populations in grape juice
As illustrated in Figure 1, In grape juice, S. cerevisiae populations exhibited relatively similar CO2 release patterns for SCP or SCJ subpopulation. However, a significant difference was observed between the two subpopulations. The CO2 release from the 169-strain SCP population was significantly higher than that from the 138-strain SCJ population (P < 0.001).
3.1.2 Adaptation behaviors of SCP and SCJ populations in SG and SF medium
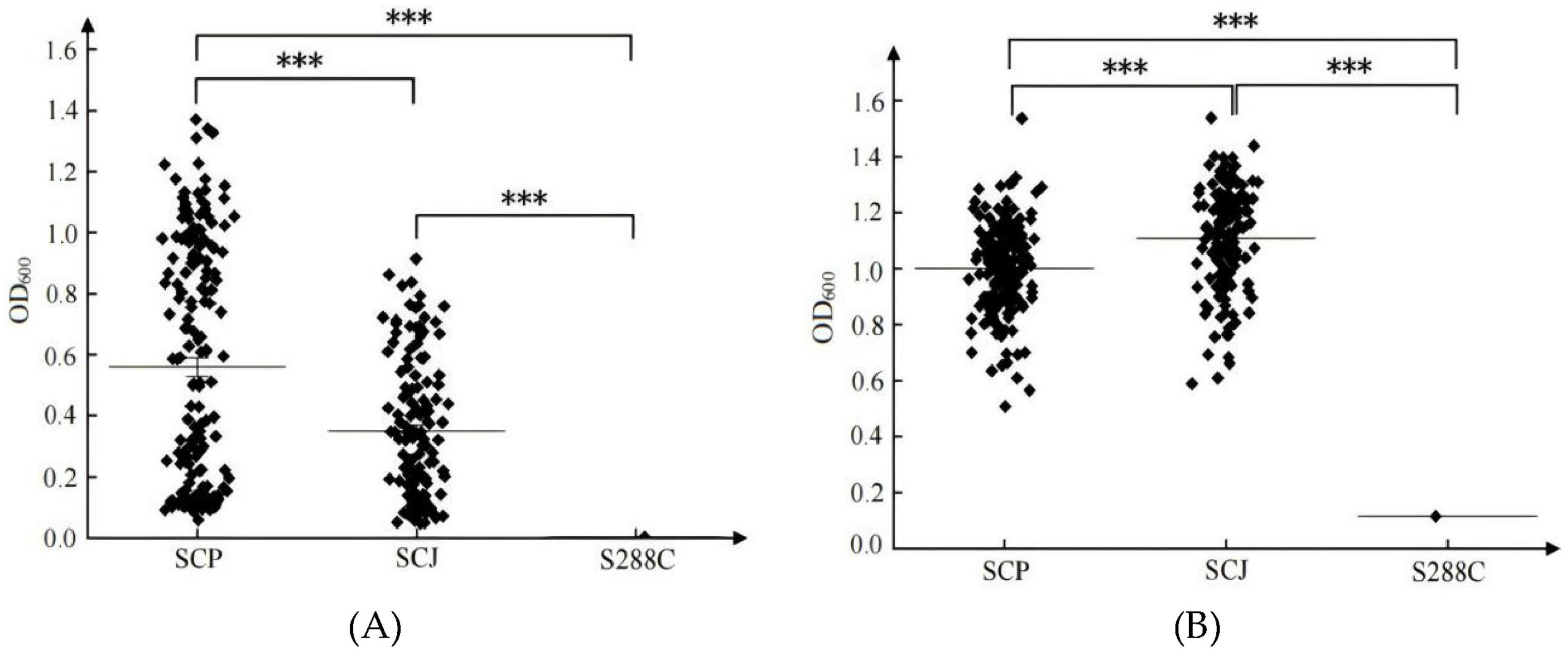
The adaptation behaviors of SCP and SCJ S. cerevisiae populations differed significantly between the SG and SF media (Figure 2). In SG medium, both populations exhibited highly dispersed biomass distributions with lower mean values (mean < 0.6), whereas in SF medium, the biomass was more concentrated and displayed higher mean values (> 1.0). Specifically, in SG medium, the SCP population demonstrated significantly better growth compared to the SCJ population, while in SF medium, the SCJ population outperformed the SCP population (P < 0.001). The S288C model strain, isolated from a spontaneous fermentation environment (Liu and Huang, 2022), exhibited superior growth in SF medium compared to SG medium. These findings indicate that SF medium provides a more favorable environment for the growth of S. cerevisiae, whereas the simulated grape juice (SG) medium does not adequately support its growth. Under native or near-native conditions, the SCJ and SCP populations display distinct growth advantages within their respective ecological niches.
3.1.3 Adaptive analysis of XJ and NXJ populations in SG and SF medium
A total of 149 S. cerevisiae strains from the Xinjiang region (XJ) and 158 strains from outside the Xinjiang region (NXJ) were cultured in SG medium (Figures 3A, B). The SCP population demonstrated significantly better growth in NXJ strains compared to XJ strains (Figure 3A), with the average OD600 of the NXJ SCP group (0.8) being markedly higher than that of the SCJ groups from both regions (0.3–0.4) (Figure 3B). In SF medium, the SCP population within Xinjiang exhibited significantly higher growth compared to the SCP population outside Xinjiang (Figure 3C). However, no significant difference was observed in the growth characteristics of the SCJ population between the two regions (Figure 3D). The average OD600 value of the SCP population within Xinjiang was slightly higher than that of the SCP population outside Xinjiang (Figure 3C).
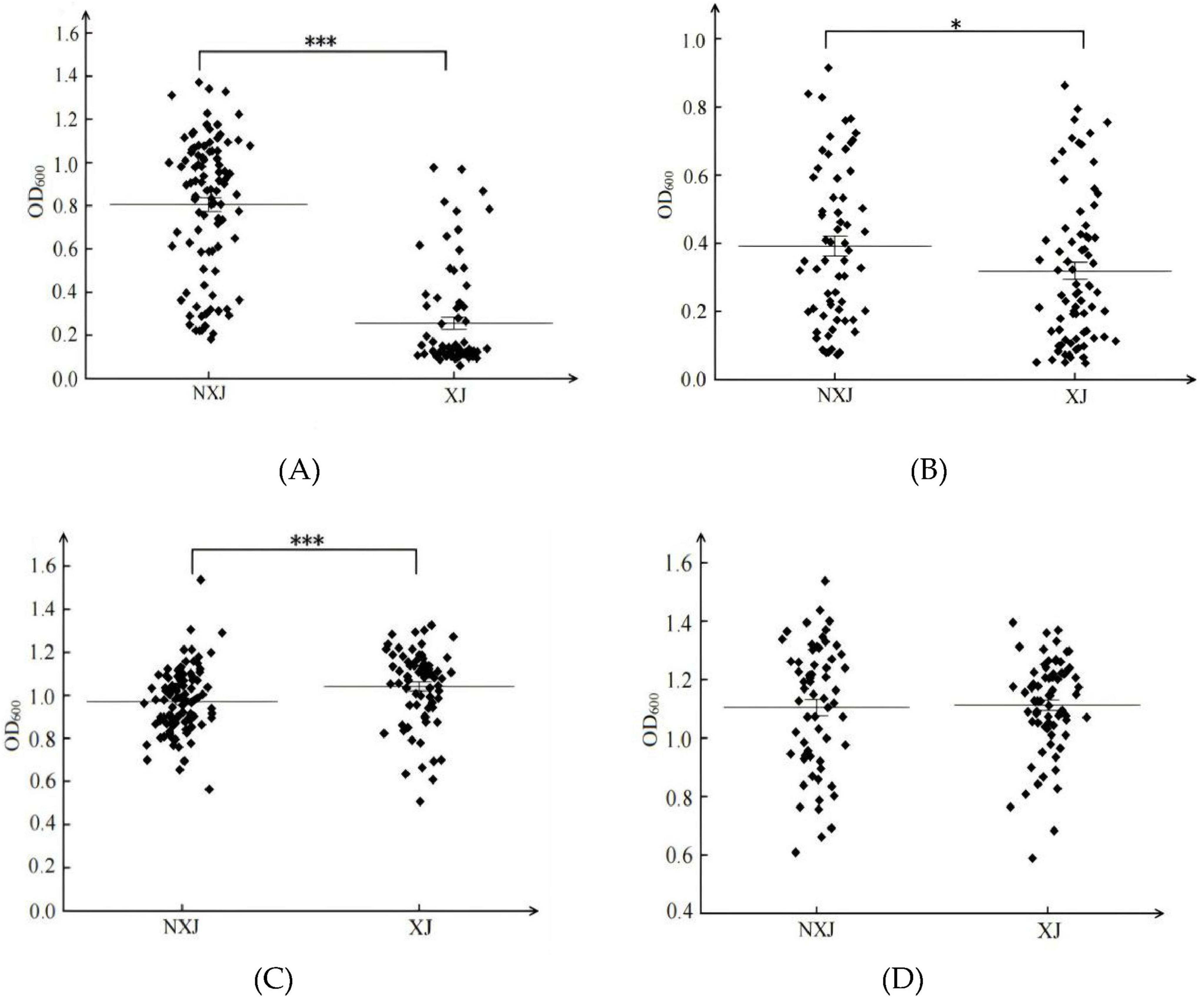
Figure 3. Biomass (OD600) of XJ (Xinjiang) and NXJ (no Xinjiang) S. cerevisiae populations. SCP populations from XJ and NXJ in SG medium (A); SCJ populations from XJ and NXJ in SG medium (B); SCP populations from XJ and NXJ in SF medium (C); SCJ populations from XJ and NXJ in SF medium (D). *p < 0.05, ***p < 0.001.
3.1.4 Adaptation of different lineages of S. cerevisiae in SG and SF media
In SG medium, significant differences in growth abilities were observed among different lineages within the SCP population (Figure 4A) and the SCJ population (Figure 4B) (P < 0.05). The CHN-VIII lineage exhibited the highest growth ability, whereas the CHN-VI/VII lineage showed the weakest adaptation. Within the SCJ population, strains from the Mosaic lineage demonstrated the strongest adaptation capacity.
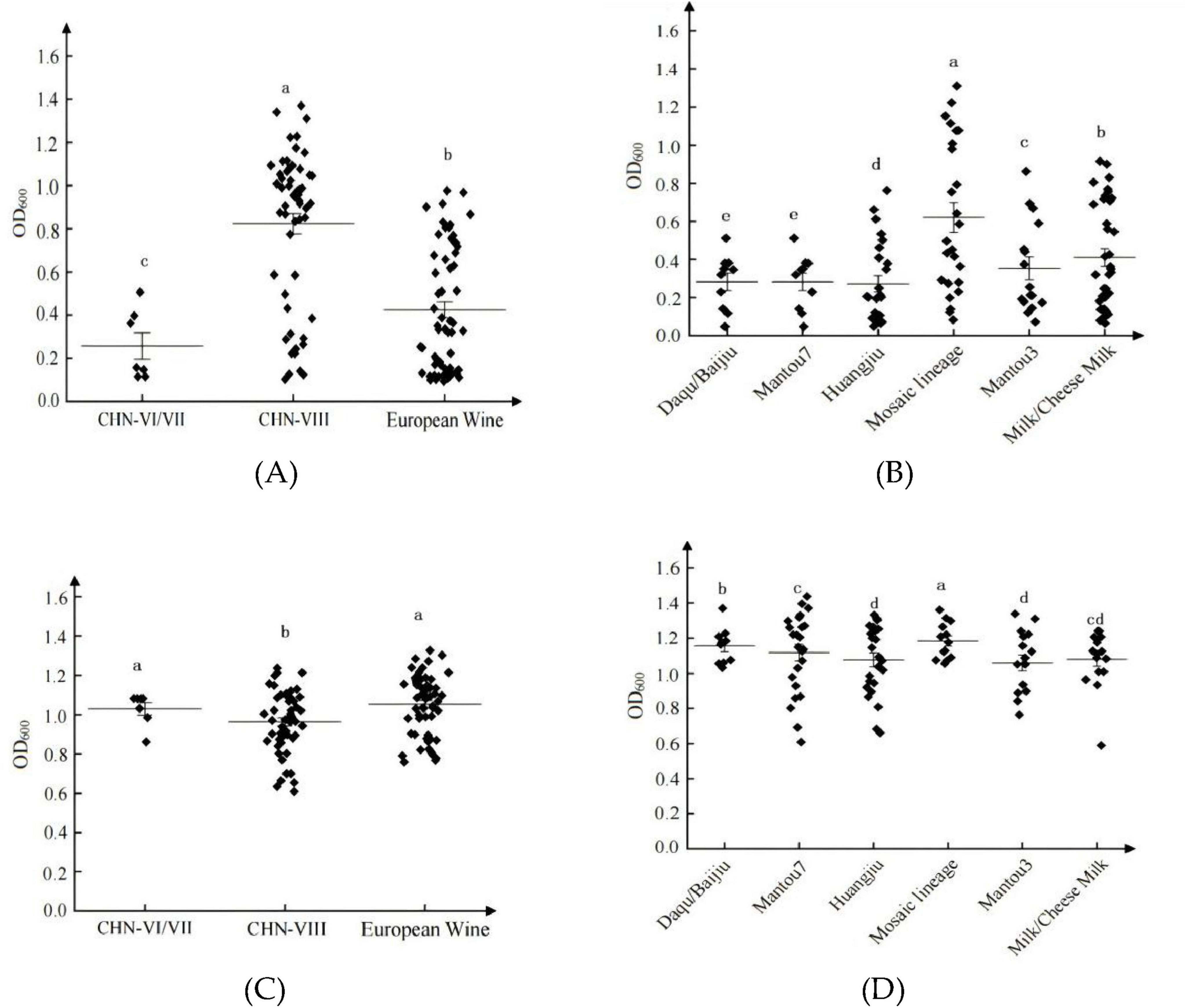
Figure 4. Growth (OD600) of different lineages of S. cerevisiae. SCP population in SG medium (A); SCJ population in SG medium (B); SCP population in SF medium (C); SCJ population in SF medium (D). a, b, c, d, and e indicate significant differences between groups, the same letters indicate insignificant differences, and different letters indicate significant differences.
In SF medium, notable differences in biomass (P < 0.05) were also detected among lineages within the SCP population (Figure 4C) and the SCJ population (Figure 4D). Overall, lineages within the SCJ population showed stronger adaptation abilities, with the Mosaic lineage achieving the highest performance.
3.1.5 Tolerance of SCP and SCJ populations under various stresses
Under stress conditions such as 6%–10% NaCl (Figure 5A), 1–1.5 g/L tannin (Figure 5B), 14%–16% ethanol (Figure 5C), extreme temperatures (15°C and 42°C, Figure 5E), and 40–80 mg/L SO2 (Figure 5F), the SCP population demonstrated significantly higher adaptability compared to the SCJ population (P < 0.05). In contrast, under high pH conditions (pH 5–7), the biomass of the SCJ population was significantly higher than that of the SCP population. However, under low pH conditions (pH 3–4, Figure 5D) and high SO2 stress (120 mg/L, Figure 5F), the SCJ population exhibited stronger inhibition. No significant differences in adaptability were observed between the SCP and SCJ populations under these conditions, nor under optimal growth conditions at 30°C (Figure 5E).
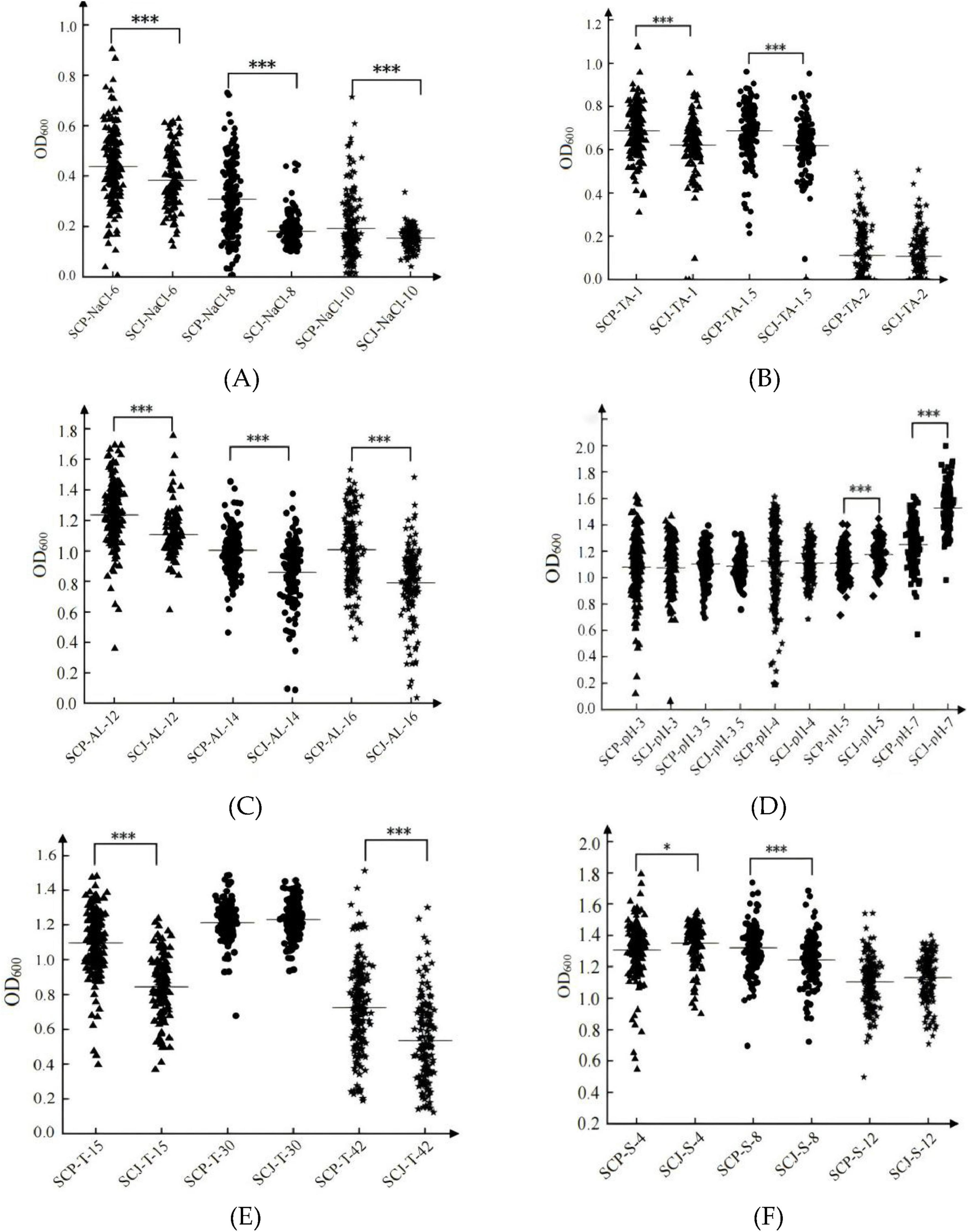
Figure 5. Biomass (OD600) of the SCP and SCJ populations under NaCl stress. Biomass of the two groups under NaCl stress (A); Biomass of the two groups under tannin stress (B); Biomass of the two groups under ethanol stress (C); Biomass of the two groups under pH stress (D); Biomass of the two groups under temperature stress (E); Biomass of the two groups under SO2 stress (F). *p < 0.05, ***p < 0.001.
3.2 Adaptability of representative haploid strains
3.2.1 Adaptability of haploid SCP and SCJ populations
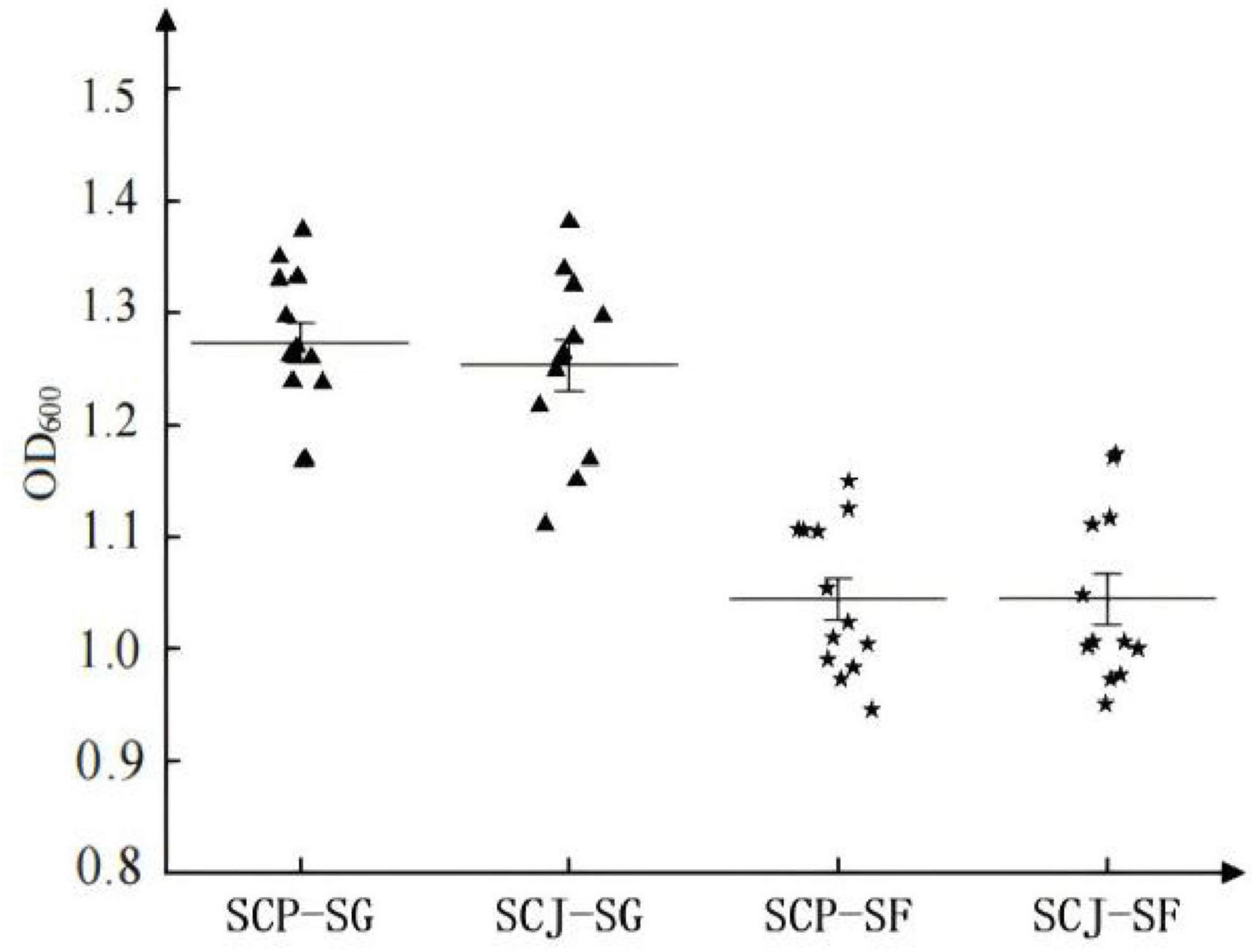
In a comparison between SG and SF media, the biomass accumulation of haploid SCP and SCJ populations was significantly higher in SG medium than in SF medium (Figure 6). However, within the same medium, no significant difference in biomass accumulation was observed between the haploid SCP and SCJ populations (Figure 6).
3.2.2 Comparison of the tolerance characteristics of haploid and diploid S. cerevisiae populations
Under stress conditions, including 6–10 g/L NaCl (Figure 7A), 1–2 g/L tannins (Figure 7B), 12%–14% ethanol (Figure 7C), pH 4–5 (Figure 7D), and low temperatures at 15°C (Figure 7E), diploid S. cerevisiae populations demonstrated significantly higher tolerance compared to haploid populations (P < 0.05). In contrast, under high-temperature stress at 42°C, haploid populations exhibited significantly greater tolerance than diploid populations (P < 0.05) (Figure 7E). For SO2 tolerance (40 mg/L–120 mg/L), haploid populations showed slightly higher tolerance than diploid populations (Figure 7F).
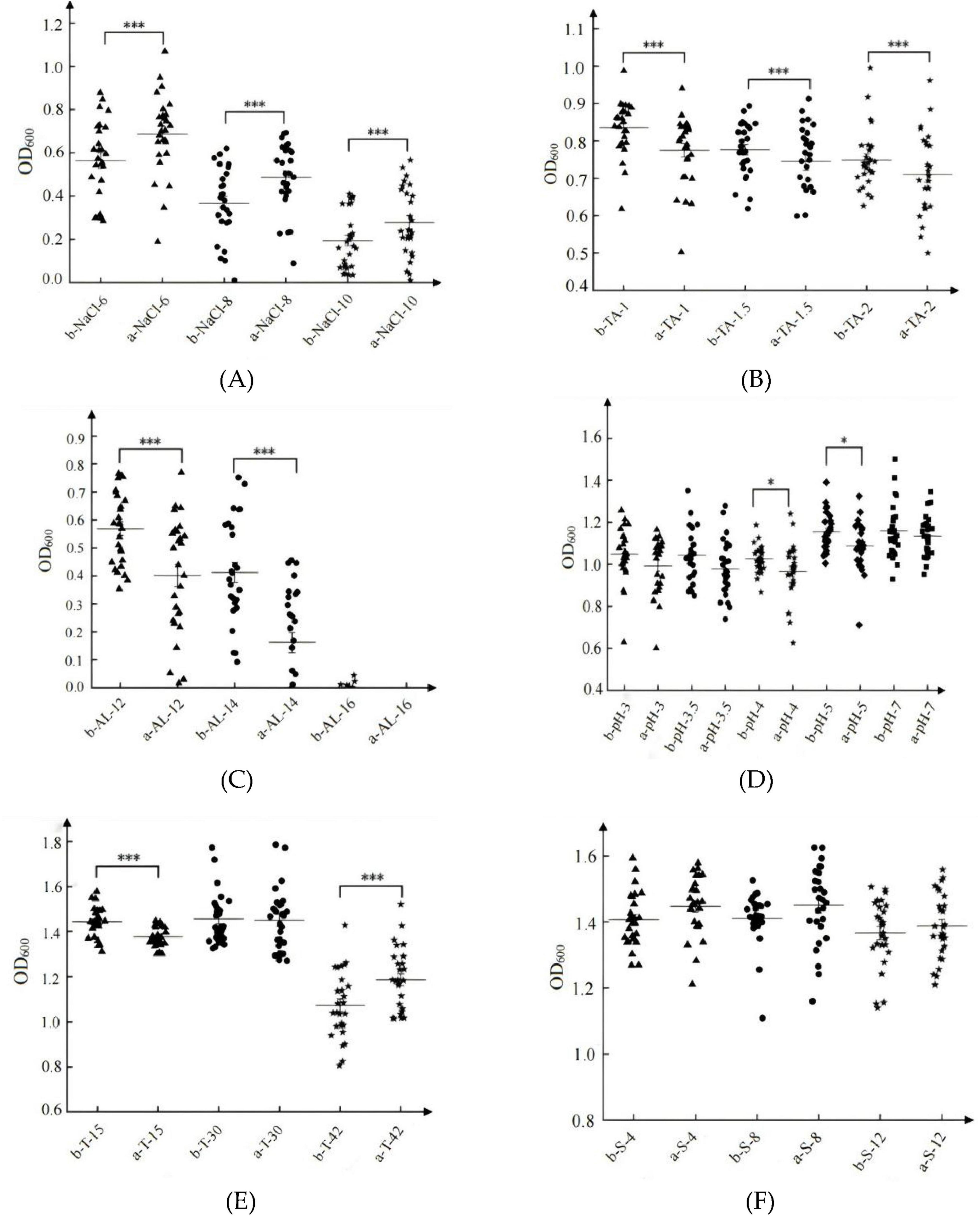
Figure 7. Tolerances of representative haploid and diploid S. cerevisiae strains. Biomass of S. cerevisiae under NaCl stress (A); Biomass of S. cerevisiae under tannin stress (B); Biomass of S. cerevisiae under ethanol stress (C); Biomass of S. cerevisiae under pH stress (D); Biomass of S. cerevisiae under temperature stress (E); Biomass of S. cerevisiae under SO2 stress (F). In each graph, “a” represents haploid S. cerevisiae, and “b” represents diploid S. cerevisiae. *p < 0.05, ***p < 0.001.
The adaptive responses of S. cerevisiae populations varied depending on the type of stress, irrespective of ploidy. Among the tested stress factors, ethanol stress exerted the most pronounced impact, with biomass values dropping below 1.0 (OD600) and mean values falling below 0.6 (OD600) (Figure 7C). Under NaCl stress, while some individual strains achieved biomass values exceeding 1.0, the population mean remained below 0.6 (Figure 7A). Tannin stress resulted in biomass values below 1.0 (OD600), with mean values ranging from 0.7 to 0.9 (Figure 7B). Under pH stress, the population biomass mean was approximately 1.0, with relatively low strain variation (Figure 7D). Compared to these stressors, both temperature extremes (Figure 7E) and SO2 stress (Figure 7F) had relatively minor effects on the growth of S. cerevisiae populations.
3.2.3 Tolerance of haploid and diploid SCP and SCJ S. cerevisiae populations
A comparison of 25 representative haploid strains of S. cerevisiae (Figure 8A) and 307 diploid strains (Figure 8B) revealed significant differences in stress tolerance between the SCP and SCJ populations under various stress conditions. Most haploid SCP strains exhibited lower tolerance compared to haploid SCJ strains (Figure 8A). In contrast, the diploid SCP population demonstrated superior tolerance relative to the diploid SCJ population under conditions of high ethanol, low tannins, extreme temperatures (both high and low), high osmotic pressure, and low pH (< 4) (Figure 8B). These results suggest that the diploid SCP population exhibits significantly enhanced tolerance characteristics compared to its haploid counterpart.
4 Discussion and conclusion
A comparison of biomass production between the two populations in SG and SF media revealed significantly lower biomass production in SG compared to SF. This suggests that the simulated grape juice medium inhibits S. cerevisiae growth and fails to provide an optimal environment for its development (Bagheri et al., 2017; Raas and Dutheil, 2024). In both grape juice and SG media, the SCP population exhibited significantly better growth than the SCJ population. In contrast, the SCJ population demonstrated significantly better adaptability in SF medium than the SCP population. The model strain S288C displayed markedly better growth characteristics in SF medium compared to SG medium, suggesting that S. cerevisiae strains from the Wine and Mantou lineages possess distinct growth advantages in their respective native environments, reflecting clear ecological adaptation (Bai et al., 2022). The differences in adaptability between the two populations are associated with specific functional genes and metabolic pathways (Chen and Zhang, 2024). In SG medium, the carbon sources primarily come from glucose and fructose. S. cerevisiae may adapt better to the SG medium by upregulating genes related to sugar metabolism, such as the glucose transporter HXT (Karri et al., 2024), the glucose kinase gene GLK1 (Zhang et al., 2024), and the phosphofructokinase gene PFK1 (Zheng et al., 2024), thereby enabling the SCP population to better thrive in this medium. The simulated dough medium typically contains rich and complex carbon sources. The SCP population may demonstrate enhanced adaptability in such an environment by upregulating genes associated with starch hydrolysis and sugar transport, such as the STA gene, which is responsible for breaking down starch into smaller fermentable sugars (Krogerus et al., 2019). genes such as GPA1, SAG1, and MAL32, which are involved in starch degradation and conversion, contribute to enhanced adaptability in starch-rich environments (Dietvorst et al., 2007; Schlarmann et al., 2024; Tamaki, 2007). S. cerevisiae adapts to varying environmental pH levels by regulating genes such as the phosphate transporter gene PHO84 and the proton pump gene PMA1 (Antunes and Sá-Correia, 2024; Eskes et al., 2018). Grape juice is typically weakly acidic, whereas the pH of dough is closer to neutral or slightly acidic (Bovo et al., 2018; Jayaram et al., 2013). The SCP population has already adapted to this low-pH environment.
In diploid populations, variations in adaptive behavior are observed between different regions and lineages, suggesting that both genetic and environmental factors play a significant role in shaping the adaptive behaviors of S. cerevisiae populations (Bai et al., 2022; Legras et al., 2018). Brewing yeasts are classified into distinct phylogenetic lineages based on their geographical origins, including Malaysian, North American, West African, and Wine/European lineages. This classification highlights the combined effects of genetic and geographical factors on the adaptive behavior of brewing yeast populations (Bai et al., 2023). S. cerevisiae strains with higher ploidy exhibit improved adaptability (Lahue et al., 2020). Diploid S. cerevisiae exhibited greater tolerance and stability compared to haploid populations, consistent with findings from numerous related studies (Harari et al., 2018; Liang and Wang, 2022; Yona et al., 2012). Diploid strains of brewing yeast possess a repertoire of stress tolerance genes, such as Th2CysPrx, which significantly enhance their ability to tolerate multiple stressors (Wang et al., 2022), The stress tolerance of diploid S. cerevisiae populations under multiple stress factors was found to be stable in the study. Moreover, in the same habitat, diploid yeast populations from SCP habitats demonstrated superior tolerance traits compared to haploid populations. In contrast, haploid populations exhibited instability in their tolerance characteristics, this highlights the key rationale for the widespread distribution of diploid S. cerevisiae populations in natural ecosystems.
In stressful environments, the SCP population demonstrates greater tolerance to NaCl, tannin, alcohol, and both low and high temperatures, while the SCJ population exhibits significantly higher biomass in near-neutral environments compared to the SCP population. This is associated with stress acclimatization in their native habitats, driving adaptive evolution in gene expression, metabolic pathways, and stress response mechanisms (Bai et al., 2022). When S. cerevisiae is exposed to high osmotic pressure environments (e.g., high salt, high alcohol), the expression of the GPD1 gene is typically upregulated to help the yeast synthesize more glycerol (Eriksson et al., 2000), maintaining osmotic balance inside and outside the cell. The SCP population enhances its tolerance to low temperatures by upregulating cold-response genes (e.g., HSP30) (Sahana et al., 2024), which helps maintain the fluidity and activity of the yeast cell membrane. Additionally, S. cerevisiae may upregulate genes such as alcohol dehydrogenase (ADH1) and boost lipid metabolism and membrane stability to mitigate alcohol-induced damage to the cell membrane and enzymes (Gutiérrez-Lomelí et al., 2008). In response to tannin stress, the expression of the antioxidant gene SOD1 may be regulated to mitigate oxidative damage caused by tannin in near-neutral environments (Subramaniyan et al., 2019), the SCJ population outperforms the SCP population, which is related to the slightly acidic nature of the dough, further demonstrating that, under favorable environmental conditions, the best growth environment for S. cerevisiae is still its native habitat.
From a laboratory perspective, the phenomena described above clearly indicate that S. cerevisiae populations from different habitats exhibit similar population characteristics and demonstrate distinct growth advantages in their native environments. This observation provides theoretical support for the selection of superior strains within populations and offers valuable insights into the development of applications utilizing populations composed of different strains of the same species. Further research is necessary to investigate the relationship between multidimensional stress testing at the strain level and the metabolic activities of these populations, with the goal of optimizing the potential of limited strains in ecological environments.
Current assessments of S. cerevisiae adaptability primarily focus on strain selection and practical applications, such as evaluating the fermentation tolerance of S. cerevisiae strains isolated from orchards to identify those with superior fermentation performance. Furthermore, individual strains are often evaluated for their adaptability and differentiation potential through laboratory passages. In contrast, this paper adopts a population-level perspective, investigating the ecological adaptive behavior of S. cerevisiae. The objective is to provide direct evidence of the species’ remarkable adaptability in ecological environments, while also offering insights from an ecological population perspective to inform the development and application of S. cerevisiae strains.
Data availability statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Author contributions
CS: Formal Analysis, Methodology, Validation, Visualization, Writing – original draft. HW: Formal Analysis, Methodology, Validation, Writing – original draft. YJ: Resources, Writing – original draft. WW: Formal Analysis, Resources, Writing – original draft. XM: Data curation, Formal Analysis, Methodology, Validation, Visualization, Writing – original draft. PH: Methodology, Writing – original draft. LZ: Conceptualization, Data curation, Methodology, Writing – review and editing.
Funding
The authors declare that financial support was received for the research and/or publication of this article. This research was supported by the National Natural Science Foundation of China (Grant Nos. 32272457 and 31871777).
Acknowledgments
We extend our sincere gratitude to the Key Laboratory of Southern Xinjiang Agricultural Product Processing, Tarim University, of the Xinjiang Production and Construction Corps, for providing an excellent research platform. We also acknowledge the generous support of the Institute of Microbiology, Chinese Academy of Sciences, and Biosun Biotechnology Co., Ltd., for supplying the strains utilized in this study.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The authors declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2025.1581370/full#supplementary-material
Footnotes
References
Alexandre, H. (2020). Wine yeast terroir: Separating the wheat from the chaff-for an open debate. Microorganisms 8:787. doi: 10.3390/microorganisms8050787
Antunes, M., and Sá-Correia, I. (2024). The role of ion homeostasis in adaptation and tolerance to acetic acid stress in yeasts. FEMS Yeast Res. 24:foae016. doi: 10.1093/femsyr/foae016
Aydın, F., Özer, G., Alkan, M., and Çakır, İ (2022). Genetic diversity and population structure of Saccharomyces cerevisiae isolated from Turkish sourdough by iPBS-retrotransposons markers. Arch. Microbiol. 204:693. doi: 10.1007/s00203-022-03313-x
Bagheri, B., Bauer, F. F., and Setati, M. E. (2017). The impact of Saccharomyces cerevisiae on a wine yeast consortium in natural and inoculated fermentations. Front. Microbiol. 8:1988. doi: 10.3389/fmicb.2017.01988
Bai, F., Han, D., Duan, S., and Wang, Q. (2022). The ecology and evolution of the Baker’s yeast saccharomyces cerevisiae. Genes 13:230. doi: 10.3390/genes13020230
Bai, F., Han, D., Duan, S., and Wang, Q. (2023). The ecology and evolution of the Baker’s yeast Saccharomyces cerevisiae. Genes 13, 1748–1770. doi: 10.3390/genes13020230
Betlej, G., Bator, E., Oklejewicz, B., Potocki, L., Górka, A., Slowik-Borowiec, M., et al. (2020). Long-term adaption to high osmotic stress as a tool for improving enological characteristics in industrial wine yeast. Genes 11:576. doi: 10.3390/genes11050576
Bovo, B., Nadai, C., Lemos Junior, W., Carlot, M., Giacomini, A., and Corich, V. (2018). The different physical and chemical composition of grape juice and marc influence Saccharomyces cerevisiae strains distribution during fermentation. J. Food Sci. 83, 2191–2196. doi: 10.1111/1750-3841.14274
Camesasca, L., Minteguiaga, M., Fariña, L., Salzman, V., Aguilar, P., Gaggero, C., et al. (2018). Overproduction of isoprenoids by Saccharomyces cerevisiae in a synthetic grape juice medium in the absence of plant genes. Int. J. Food Microbiol. 282, 42–48. doi: 10.1016/j.ijfoodmicro.2018.05.025
Caspeta, L., and Nielsen, J. (2015). Thermotolerant yeast strains adapted by laboratory evolution show trade-off at ancestral temperatures and preadaptation to other stresses. mBio 6:e00431. doi: 10.1128/mBio.00431-15
Chen, P., and Zhang, J. (2024). The loci of environmental adaptation in a model eukaryote. Nat. Commun. 15:5672. doi: 10.1038/s41467-024-50002-y
de Gioia, M., Russo, P., and de Simone, N. (2022). Interactions among relevant non-saccharomyces, Saccharomyces, and lactic acid bacteria species of the wine microbial consortium: Towards advances in antagonistic phenomena and biocontrol potential. Appl. Sci. 12, 12760–12760. doi: 10.3390/APP122412760
Dietvorst, J., Blieck, L., Brandt, R., Van Dijck, P., and Steensma, H. (2007). Attachment of MAL32-encoded maltase on the outside of yeast cells improves maltotriose utilization. Yeast 24, 27–38. doi: 10.1002/yea.1436
Duan, S., Han, P., Wang, Q., Liu, W., Shi, J., Li, K., et al. (2018). The origin and adaptive evolution of domesticated populations of yeast from Far East Asia. Nat. Commun. 9, 1–13. doi: 10.1038/s41467-018-05106-7
Eriksson, P., Alipour, H., Adler, L., and Blomberg, A. (2000). Rap1p-binding sites in the saccharomyces cerevisiae GPD1 promoter are involved in its response to NaCl. J. Biol. Chem. 275, 29368–29376. doi: 10.1074/jbc.M001663200
Eskes, E., Deprez, M., Wilms, T., and Winderickx, J. (2018). pH homeostasis in yeast; the phosphate perspective. Curr. Genet. 64, 155–161. doi: 10.1007/s00294-017-0743-2
Gao, J., Wang, M., Huang, W., You, Y., and Zhan, J. (2022). Indigenous Saccharomyces cerevisiae could better adapt to the physicochemical conditions and natural microbial ecology of prince grape must compared with commercial Saccharomyces cerevisiae FX10. Molecules 27, 6892–6892. doi: 10.3390/molecules27206892
García-Ríos, E., and Guillamón, J. M. (2019). Mechanisms of yeast adaptation to wine fermentations. Prog. Mol. Subcell. Biol. 58, 5837–5859. doi: 10.1007/978-3-030-13035-0_2
Gardner, J., Alperstein, L., Walker, M., Zhang, J., and Jiranek, V. (2023). Modern yeast development: finding the balance between tradition and innovation in contemporary winemaking. FEMS Yeast Res. 23:foac049. doi: 10.1093/femsyr/foac049
Gobert, A., Tourdot-Maréchal, R., Sparrow, C., Morge, C., and Alexandre, H. (2019). Influence of nitrogen status in wine alcoholic fermentation. Food Microbiol. 83, 71–85. doi: 10.1016/j.fm.2019.04.008
Godara, A., and Kao, K. (2021). Adaptive laboratory evolution of β-caryophyllene producing Saccharomyces cerevisiae. Microb. Cell Fact. 20:106. doi: 10.1186/s12934-021-01598-z
Gutiérrez-Lomelí, M., Torres-Guzmán, J., González-Hernández, G., Cira-Chávez, L., Pelayo-Ortiz, C., and Ramírez-Córdova Jde, J. (2008). Overexpression of ADH1 and HXT1 genes in the yeast Saccharomyces cerevisiae improves the fermentative efficiency during tequila elaboration. Antonie Van Leeuwenhoek 93, 363–371. doi: 10.1007/s10482-007-9213-z
Han, D., Han, P., Rumbold, K., Koricha, A., Duan, S., Song, L., et al. (2021). Adaptive gene content and allele distribution variations in the wild and domesticated populations of Saccharomyces cerevisiae. Front. Microbiol. 12:631250. doi: 10.3389/fmicb.2021.631250
Harari, Y., Ram, Y., and Kupiec, M. (2018). Frequent ploidy changes in growing yeast cultures. Curr. Genet. 64, 1001–1004. doi: 10.1007/s00294-018-0823-y
Heidenreich, E. (2007). Adaptive mutation in Saccharomyces cerevisiae. Crit. Rev. Biochem. Mol. Biol. 42, 285–311. doi: 10.1080/10409230701507773
Jayaram, V., Cuyvers, S., Lagrain, B., Verstrepen, K., Delcour, J., and Courtin, C. (2013). Mapping of Saccharomyces cerevisiae metabolites in fermenting wheat straight-dough reveals succinic acid as pH-determining factor. Food Chem. 136, 301–308. doi: 10.1016/j.foodchem.2012.08.039
Karri, S., Dickinson, Q., Jia, J., Yang, Y., Gan, H., Wang, Z., et al. (2024). The role of hexokinases in epigenetic regulation: altered hexokinase expression and chromatin stability in yeast. Epigenetics Chromatin 17:27. doi: 10.1186/s13072-024-00551-9
Krogerus, K., Magalhães, F., Kuivanen, J., and Gibson, B. A. (2019). deletion in the STA1 promoter determines maltotriose and starch utilization in STA1+ Saccharomyces cerevisiae strains. Appl. Microbiol. Biotechnol. 103, 7597–7615. doi: 10.1007/s00253-019-10021-y
Lahue, C., Madden, A., Dunn, R., and Smukowski Heil, C. (2020). History and domestication of Saccharomyces cerevisiae in bread baking. Front. Genet. 11:584718. doi: 10.3389/fgene.2020.584718
Lázari, L., Wolf, I., Schnepper, A., and Valente, G. (2022). LncRNAs of Saccharomyces cerevisiae bypass the cell cycle arrest imposed by ethanol stress. PLoS Comput. Biol. 18:e1010081. doi: 10.1371/journal.pcbi.1010081
Legras, J., Galeote, V., Bigey, F., Camarasa, C., Marsit, S., Nidelet, T., et al. (2018). Adaptation of S. cerevisiae to fermented food environments reveals remarkable genome plasticity and the footprints of domestication. Mol. Biol. Evol. 35, 1712–1727. doi: 10.1093/molbev/msy066
Liang, S., and Wang, G. (2022). Research progress in wine-making characteristic of wine-related yeast based on ploidy. Food Fermentation Industries 58, 121–130. doi: 10.3969/j.issn.1674-506X.2022.01-017
Lin, Y., Zhang, N., Lin, Y., Gao, Y., Li, H., Zhou, C., et al. (2024). Transcriptomic and metabolomic correlation analysis: effect of initial SO2 addition on higher alcohol synthesis in Saccharomyces cerevisiae and identification of key regulatory genes. Front. Microbiol. 15:1394880. doi: 10.3389/fmicb.2024.1394880
Liu, Z., and Huang, X. (2022). Copy number variants impact phenotype-genotype relationships for adaptation of industrial yeast Saccharomyces cerevisiae. Appl. Microbiol. Biotechnol. 106, 6611–6623. doi: 10.1007/s00253-022-12137-0
Marsit, S., and Dequin, S. (2015). Diversity and adaptive evolution of Saccharomyces wine yeast: A review. FEMS Yeast Res. 15:fov067. doi: 10.1093/femsyr/fov067
Massoud, R., and Zoghi, A. (2022). Potential probiotic strains with heavy metals and mycotoxins bioremoval capacity for application in foodstuffs. J. Appl. Microbiol. 133, 1288–1307. doi: 10.1111/jam.15685
Mudoor Sooresh, M., Willing, B., and Bourrie, B. (2023). Opportunities and challenges of understanding community assembly in spontaneous food fermentation. Foods 12:673. doi: 10.3390/foods12030673
Ojeda-Linares, C., Solís-García, I. A., and Casas, A. (2022). Constructing micro-landscapes: Management and selection practices on microbial communities in a traditional fermented beverage. Front. Ecol. Evol. 10:821268. doi: 10.3389/fevo.2022.821268
Parapouli, M., Vasileiadis, A., Afendra, A., and Hatziloukas, E. (2020). Saccharomyces cerevisiae and its industrial applications. AIMS Microbiol. 6, 1–31. doi: 10.3934/microbiol.2020001
Pretorius, I. (2020). Tasting the terroir of wine yeast innovation. FEMS Yeast Res. 20:foz084. doi: 10.1093/femsyr/foz084
Raas, M., and Dutheil, J. (2024). The rate of adaptive molecular evolution in wild and domesticated Saccharomyces cerevisiae populations. Mol. Ecol. 33:e16980. doi: 10.1111/mec.16980
Rădoi-Encea, R. -Ş, Pădureanu, V., and Diguţă, C. F. (2023). Achievements of autochthonous wine yeast isolation and selection in romania—A review. Fermentation 9:407. doi: 10.3390/fermentation9050407
Randez-Gil, F., Prieto, J., Rodríguez-Puchades, A., Casas, J., Sentandreu, V., and Estruch, F. (2020). Myriocin-induced adaptive laboratory evolution of an industrial strain of Saccharomyces cerevisiae reveals its potential to remodel lipid composition and heat tolerance. Microb. Biotechnol. 13, 1066–1081. doi: 10.1111/1751-7915.13555
Sahana, G. R., Balasubramanian, B., and Joseph, K. S. A. (2024). review on ethanol tolerance mechanisms in yeast: Current knowledge in biotechnological applications and future directions. Process Biochem. 138, 1–13. doi: 10.1016/j.procbio.2023.12.024
Schlarmann, P., Sakuragi, K., Ikeda, A., Yang, Y., Sasaki, S., Hanaoka, K., et al. (2024). The tricalbin family of membrane contact site tethers is involved in the transcriptional responses of Saccharomyces cerevisiae to glucose. J. Biol. Chem. 300:107665. doi: 10.1016/j.jbc.2024.107665
Subramaniyan, S., Alugoju, P., Sj, S., Veerabhadrappa, B., and Dyavaiah, M. (2019). Magnolol protects Saccharomyces cerevisiae antioxidant-deficient mutants from oxidative stress and extends yeast chronological life span. FEMS Microbiol. Lett. 366:fnz065. doi: 10.1093/femsle/fnz065
Šuranská, H., Vránová, D., and Omelková, J. (2016). Isolation, identification and characterization of regional indigenous Saccharomyces cerevisiae strains. Braz. J. Microbiol. 47, 181–190. doi: 10.1016/j.bjm.2015.11.010
Swamy, K., and Zhou, N. (2019). Experimental evolution: Its principles and applications in developing stress-tolerant yeasts. Appl. Microbiol. Biotechnol. 103, 2067–2077. doi: 10.1007/s00253-019-09616-2
Tamaki, H. (2007). Glucose-stimulated cAMP-protein kinase A pathway in yeast Saccharomyces cerevisiae. J. Biosci. Bioeng. 104, 245–250. doi: 10.1263/jbb.104.245
Tellini, N., De Chiara, M., Mozzachiodi, S., Tattini, L., Vischioni, C., Naumova, E., et al. (2024). Ancient and recent origins of shared polymorphisms in yeast. Nat. Ecol. Evol. 8, 761–776. doi: 10.1038/s41559-024-02352-5
Vrancken, G., Rimaux, T., De Vuyst, L., and Leroy, F. (2008). Kinetic analysis of growth and sugar consumption by Lactobacillus fermentum IMDO 130101 reveals adaptation to the acidic sourdough ecosystem. Int. J. Food Microbiol. 128, 58–66. doi: 10.1016/j.ijfoodmicro.2008.08.001
Wang, Q., Liu, W., Liti, G., Wang, S., and Bai, F. (2012). Surprisingly diverged populations of Saccharomyces cerevisiae in natural environments remote from human activity. Mol. Ecol. 21, 5404–5417. doi: 10.1111/j.1365-294X.2012.05732.x
Wang, Y., Liu, B., and Jiang, B. (2022). Th2CysPrx gene enhanced abiotic stress tolerances of Saccharomyces cerevisiae. J. Nanjing For. Univer. 46:87. doi: 10.12302/j.issn.1000-2006.202105023
Xiao, G., Liu, Q., and Li, J. (2004). Optimization of Sporulation and Haploidization of S.Cerevisiae BY-6. Liquor-Making Sci. Technol. 4, 21–22. doi: 10.3969/j.issn.1001-9286.2004.04.002
Xue, R., Li, L., and Wang, L. (2012). Methods for the preparation of haploid Pichia pastoris cells. China Brewing 31, 128–130. doi: 10.3969/j.issn.1001-6678.2015.03.006
Yi, X., Xia, H., Huang, P., Ma, S., and Wu, C. (2024). Exploring community succession, assembly patterns, and metabolic functions of ester-producing-related microbiota during the production of nongxiangxing baijiu. Foods 13, 3169–3169. doi: 10.3390/foods13193169
Yona, A., Manor, Y., Herbst, R., Romano, G., Mitchell, A., Kupiec, M., et al. (2012). Chromosomal duplication is a transient evolutionary solution to stress. Proc. Natl. Acad. Sci. U S A. 109, 21010–21015. doi: 10.1073/pnas.1211150109
Zampi, V., and Ranfagni, S. A. (2024). question of style: an analysis of the use of the concept of style in the wine industry and research. J. Wine Res. 35, 218–234. doi: 10.1080/09571264.2024.2394033
Zhang, B., Ivanova-Petropulos, V., and Duan, C. (2021). Distinctive chemical and aromatic composition of red wines produced by Saccharomyces cerevisiae co-fermentation with indigenous and commercial non-Saccharomyces strains. Food Biosci. 41:100925. doi: 10.1016/j.fbio.2021.100925
Zhang, C., Tian, J., Zhang, J., Liu, R., Zhao, X., and Lu, W. (2024). Engineering and transcriptome study of Saccharomyces cerevisiae to produce ginsenoside compound K by glycerol. Biotechnol. J. 19:e2300383. doi: 10.1002/biot.202300383
Zhang, K., Fang, Y., Gao, K., Sui, Y., Zheng, D., and Wu, X. (2017). Effects of genome duplication on phenotypes and industrial applications of Saccharomyces cerevisiae strains. Appl. Microbiol. Biotechnol. 101, 5405–5414. doi: 10.1007/s00253-017-8284-7
Zheng, C., Hou, S., Zhou, Y., Yu, C., and Li, H. (2024). Regulation of the PFK1 gene on the interspecies microbial competition behavior of Saccharomyces cerevisiae. Appl. Microbiol. Biotechnol. 108:272. doi: 10.1007/s00253-024-13091-9
Zheng, Y., and Wang, S. (2015). Stress tolerance variations in Saccharomyces cerevisiae strains from diverse ecological sources and geographical locations. PLoS One 10:e0133889. doi: 10.1371/journal.pone.0133889
Zhu, L., Wang, G., Xue, J., Gou, D., and Duan, C. (2017). Direct stamp of technology or origin on the genotypic and phenotypic variation of indigenous Saccharomyces cerevisiae population in a natural model of boiled grape juice fermentation into traditional Msalais wine in China. FEMS Yeast Res. 17:fow108. doi: 10.1093/femsyr/fow108
Keywords: Saccharomyces cerevisiae, population adaptability, dough, grape juice, stress tolerance
Citation: Su C, Wang H, Jia Y, Wang W, Ma X, Han P and Zhu L (2025) Comparative adaptability of 307 Saccharomyces cerevisiae strains from winemaking and Mantou fermentation. Front. Microbiol. 16:1581370. doi: 10.3389/fmicb.2025.1581370
Received: 22 February 2025; Accepted: 28 March 2025;
Published: 11 April 2025.
Edited by:
Weiwei Dong, Hubei Normal University, ChinaReviewed by:
Wenchao Cai, Shihezi University, ChinaDongqing Ye, Guangxi Academy of Agricultural Science, China
Copyright © 2025 Su, Wang, Jia, Wang, Ma, Han and Zhu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lixia Zhu, MTIwMDUwMDY4QHRhcnUuZWR1LmNu
 Cairong Su1
Cairong Su1 Lixia Zhu
Lixia Zhu