
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol., 10 April 2025
Sec. Virology
Volume 16 - 2025 | https://doi.org/10.3389/fmicb.2025.1581330
 Ayako Sedohara1*
Ayako Sedohara1* Tomohiko Koibuchi2
Tomohiko Koibuchi2 Makoto Yamagishi3
Makoto Yamagishi3 Michiko Koga1
Michiko Koga1 Kotaro Arizono2
Kotaro Arizono2 Kazuhiko Ikeuchi4
Kazuhiko Ikeuchi4 Tadashi Kikuchi2
Tadashi Kikuchi2 Makoto Saito1
Makoto Saito1 Eisuke Adachi2
Eisuke Adachi2 Takeya Tsutsumi4
Takeya Tsutsumi4 Daisuke Honma5
Daisuke Honma5 Kazushi Araki6
Kazushi Araki6 Kaoru Uchimaru2,3
Kaoru Uchimaru2,3 Hiroshi Yotsuyanagi1,2*
Hiroshi Yotsuyanagi1,2*For the eradication of human immunodeficiency virus type 1 (HIV-1) provirus from people living with HIV-1, reactivation of latently HIV-1-infected cells is essential. Although several latency reversing agents have been identified, eradication of HIV-infected cells has been a challenge. Here, we investigated whether the novel enhancer of zeste homolog 1/2 (EZH1/2) dual inhibitor valemetostat/DS-3201/(R)-OR-S2 could efficiently reactivate latently HIV-1-infected cells in vitro and ex vivo. People living with HIV-1 who were on suppressive combined antiretroviral therapy and with plasma HIV-1 virus levels consistently below 50 copies/mL were enrolled in this study. ACH2 cells were treated with valemetostat for 7–14 days and with suberoylanilide hydroxamic acid (SAHA). CD4+ T cells were treated with valemetostat or the EZH2-selective inhibitors GSK126 and E7438 for 22 days alone or in combination with SAHA. HIV-1 expression in CD4+ T cells was determined. Valemetostat more effectively induced HIV-1 mRNA expression in ACH-2 cells when administered for 14 days than when administered for 7 days. Valemetostat reversed latently HIV-l-infected CD4+ T cells isolated from patients with HIV-1 and induced HIV-1 mRNA expression more potently than GSK126 and E7438. In addition, valemetostat induced HIV-1 mRNA expression more strongly when used in combination with SAHA compared with GSK126 and E7438. Expression levels of 21 hub genes were markedly increased after treatment with valemetostat. Gene Ontology analysis revealed that proteins encoded by these 21 genes were localized to the cell membrane and involved in the immune response. Kyoto Encyclopedia of Genes and Genomes enrichment pathway analysis showed that these 21 hub genes contributed to various signaling pathways, including the JAK–STAT signaling pathway. This study provides novel insights for the development of treatments to reactivate latently HIV-1-infected cells.
Human immunodeficiency virus-1 (HIV-1) is an RNA virus targeting T CD4+ lymphocytes. The HIV-1 genome is converted into HIV-1 DNA fragments by reverse transcriptase, and these fragments, referred to as proviruses, are subsequently integrated into the host genomic DNA (Sadowski and Hashemi, 2019; Shan et al., 2017). Antiretroviral therapy prevents viral replication and new infections of CD4+ T cells; however, it cannot eliminate HIV-1-infected cells. CD4+ T cells possess self-renewal capability and have a long lifespan (Chahroudi et al., 2015). They carry the HIV-1 provirus over an extended period, complicating the eradication of HIV-1-infected cells (Chomont et al., 2009; Hosmane et al., 2017; Kwon et al., 2020). In addition, because the HIV-1 provirus is silenced by epigenetic modifications (Sadowski and Hashemi, 2019), infected CD4+ T cells do not express the HIV-1 antigen on the cell surface and evade cell-mediated immunity. These features render CD4+ T cells as latent HIV-1 reservoirs, the eradication of which in patients with HIV-1 infection is a crucial and long-standing therapeutic challenge.
The “kick and kill” strategy aims to eradicate latently HIV-1-infected CD4+ T cells by reactivating them with a latency reversing agent (LRA) and eliminating them by the host immune system activation (Sadowski and Hashemi, 2019; Thorlund et al., 2017). Various LRAs have been identified but eliminating HIV-infected cells in vivo remains challenging (Ait-Ammar et al., 2019; Gay et al., 2024; Kim et al., 2018; Ta et al., 2022). Enhancer of zeste homolog 2 (EZH2), a component of polycomb repressive complex 2, is a histone methyltransferase that methylates H3K27 (Holoch and Margueron, 2017). EZH2 contributes to chromatin compaction and prevents RNA polymerase II elongation, limiting access of both transcription factors and ATP-dependent chromatin-remodeling machinery to chromatin (Di Croce and Helin, 2013; Khan et al., 2018). EZH2 is involved in the silencing of intracellular HIV-1 expression and maintenance of HIV-1 proviral latency (Friedman et al., 2011; Matsuda et al., 2015; Nguyen et al., 2017; Sharma et al., 2020). Therefore, EZH2-selective inhibitors reactivate intracellular HIV-1 expression (Friedman et al., 2011; Matsuda et al., 2015; Nguyen et al., 2017; Turner et al., 2020). EZH1, an EZH2 homolog, also functions as a catalytic domain of PRC2 methyltransferase and compensates for the loss of EZH2 activity (Shen et al., 2008). To effectively suppress PRC2 methyltransferase activity, EZH1 and EZH2 should be inhibited simultaneously.
In this study, we investigated whether the novel EZH1/2 dual inhibitor valemetostat/DS-3201/(R)-OR-S2 (Honma et al., 2017; Yamagishi et al., 2019), which was approved in Japan for the treatment of relapsed or refractory adult T-cell leukemia/lymphoma in September 2022 (Izutsu et al., 2023; Keam, 2022) and peripheral T-cell lymphoma in June 2024 (Zinzani et al., 2024), could reactivate latently HIV-1-infected cells in vitro and ex vivo more effectively than the EZH2-selective inhibitors GSK126 (Gulati et al., 2018) and E7438 (Izutsu et al., 2021; Morschhauser et al., 2020). Furthermore, we used transcriptome analysis to compare differences in gene expression changes in CD4+ T cells treated with valemetostat, GSK126, and E7438.
Patients infected with HIV-1 were enrolled in this study if they were on suppressive combined antiretroviral therapy for >3 years and with plasma HIV-1 virus levels consistently below 50 copies/mL for at least 2 years (Table 1). This study was reviewed and approved by the Research Ethics Committee of the University of Tokyo (approval No. 30-96-B20190402). All participants provided written informed consent to participate in the study and for their data to be published.
ACH2 cells (1.0 × 106 cells) were cultured at 37°C in the atmosphere of 95% air and 5% CO2 in 12-well plates (Sumitomo Bakelite Co. Ltd., Tokyo, Japan) with 1 mL of Roswell Park Memorial Institute (RPMI) 1,640 medium (plus L-glutamine, without phenol red) (Gibco, New York, NY, USA) supplemented with 10% 0.22 μM filtered fetal bovine serum (Thermo Fisher Scientific, Waltham, MA, USA) and 0.6% of penicillin–streptomycin solution (Merck, Darmstadt, Germany).
Peripheral blood mononuclear cells were isolated using SepMate™ (STEMCELL Technologies, Vancouver, BC, Canada) according to the manufacturer’s protocol. CD4+ T cells were isolated by negative selection using an EasySep™ Human CD4+ T Cell Isolation Kit (STEMCELL Technologies) and cultured at 37°C in the atmosphere of 95% air and 5% CO2 in a 48-well flat-bottomed plate (Sumitomo Bakelite Co.) filled with 500 μL of RPMI-1640 medium (plus L-glutamine, without phenol red; Gibco) supplemented with 10% 0.22 μM filtered fetal bovine serum (Thermo Fisher Scientific), 0.6% of penicillin–streptomycin solution (Merck), and 300 nM efavirenz (Merck), which prevented new HIV-1 infection. To maintain primary CD4+ T cells ex vivo for >7 days, they were treated with beads coated with antibodies against CD3 and CD28 (STEMCELL Technologies) for 4 days (Kuzmichev et al., 2017). After the removal of the beads, 5 mg/mL interleukin 7 (IL-7) was added every 3–4 days (Murera et al., 2018). Half of the culture medium was renewed every 7 days.
EZH2-selective inhibitors E7438 and GSK126 (Duan et al., 2020), and EZH1/2 dual inhibitor valemetostat/DS-3201/(R)-OR-S2 were synthesized in-house (Honma et al., 2017). Histone deacetylase inhibitor vorinostat/suberoylanilide hydroxamic acid (SAHA) (Merck) and reverse transcriptase inhibitor efavirenz (Merck) were dissolved in dimethyl sulfoxide (FUJIFILM Wako Pure Chemical Corporation, Osaka, Japan). Recombinant human IL-7 (Thermo Fisher Scientific) was dissolved in phosphate-buffered saline containing 0.1% bovine serum albumin (FUJIFILM Wako Pure Chemical Corporation).
ACH2 cells (1.0 × 106) were treated with 100 nM valemetostat for 7–14 days and with the HDAC inhibitor SAHA (100 nM, 500 nM, or 1 μM), an LRA, for 24 h, 1 day before the last day of valemetostat treatment. CD4+ T cells (1.0 × 106) were treated with 1,000 nM valemetostat, GSK126, or E7438 for 22 days alone or in combination with 500 nM SAHA for 6 h on the final day of treatment.
Total RNA and genomic DNA were extracted from cells using ISOGEN II and ISOGENOME (Nippon Gene Co. Ltd., Tokyo, Japan), respectively. The total RNA precipitate was then dissolved in ddH2O. The DNA pellet was resuspended in 8 mM NaOH and adjusted to pH 8.0 with a 0.1 M HEPES buffer (Nippon Gene Co. Ltd.).
cDNA was synthesized using SuperScript IV Reverse Transcriptase (Thermo Fisher Scientific). The THUNDERBIRD™ Probe qRT-PCR kit (TOYOBO, Osaka, Japan) was used for qPCR analysis. The targets were amplified using CFX Connect (Bio-Rad, Hercules, CA, USA) at 95°C for 10 s (95°C for 5 s, 60°C for 20 s) × 50 cycles. Primers and probes used in this study are listed in Table 2.
In ACH2 cells, cellular HIV-1 expression was normalized by that of importin 8 (IPO8) (Ledderose et al., 2011). HIV-1 expression in CD4+ T cells was calculated as HIV-1 per HIV-1 provirus. The number of genomic HIV-1 DNA (HIV provirus) copies was normalized by the number of genomic IPO8 copies. Next, the normalized number of cellular HIV-1 DNA copies was divided by the normalized HIV-1 provirus. Expression of H1 histone 1 family, member 0 (H1F0) was also normalized by that of IPO8.
qRT-PCR data in ACH2 cells sampled at 7 and 14 days were analyzed using the two-sample t-test (BellCurve for Excel ver. 4.07; Social Survey Research Information Co., Ltd., Tokyo, Japan). The variance of the population was estimated via the unbiased variance. The Kruskal–Wallis test was used to perform multiple comparisons of three or more experimental groups using the Bell Curve for Excel. The Steel–Dwass test was used to compare each experimental group individually. Differences were considered significant at p < 0.05.
Total RNA was quantified using a Bioanalyzer 2,100 system (Agilent Technologies, Santa Clara, CA, USA) (Supplementary Figure 1). The Clariom™ S Assay for human samples (Thermo Fisher Scientific) was used for transcriptome analysis. Biotinylated sense-strand DNA targets were amplified from total RNA and hybridized using a GeneChip™ WT Plus Reagent Kit (Applied Biosystems™, Waltham, MA, USA) according to the manufacturer’s protocol. The cartridge was scanned using a GnenChip™ Scanner 3,000 7G system (Applied Biosistems™). Microarray images were analyzed using GeneChip™ Command Console Software ver. 3.2 and GeneChip™ Expression Console Software ver. 1.3.0. (Applied Biosistems™). Microarray data were analyzed using Transcriptome Analysis Console ver. 4.0 (Thermo Fisher Scientific). DAVID1 was used for the functional analysis of miRNAs (Sherman et al., 2022), Gene Ontology (GO) analysis of miRNA target mRNAs (Young et al., 2010), and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis (Huang et al., 2009). The protein–protein interaction (PPI) network of proteins encoded by differentially expressed RNAs was analyzed using STRING2 (Szklarczyk et al., 2015). Hub genes in the completed PPI network were screened using CytoHubba Ver. 0.1 (Chin et al., 2014).
We investigated the effects of 7–14-day treatment with valemetostat on the ACH2 cells, a model of latent HIV-1 infection (Ishida et al., 2006). The HDAC inhibitor SAHA (Matsuda et al., 2015) was used as the positive control. The qRT-PCR results showed that valemetostat reactivated cellular HIV-1 expression in ACH2 cells, similar to the action of SAHA (Figure 1). A co-treatment of valemetostat and SAHA increased cellular HIV-1 mRNA expression to a greater extent than treatment with either agent alone. In addition, the level of cellular HIV-1 mRNA expression was higher in ACH2 cells treated with valemetostat for 14 days than in those treated for 7 days.
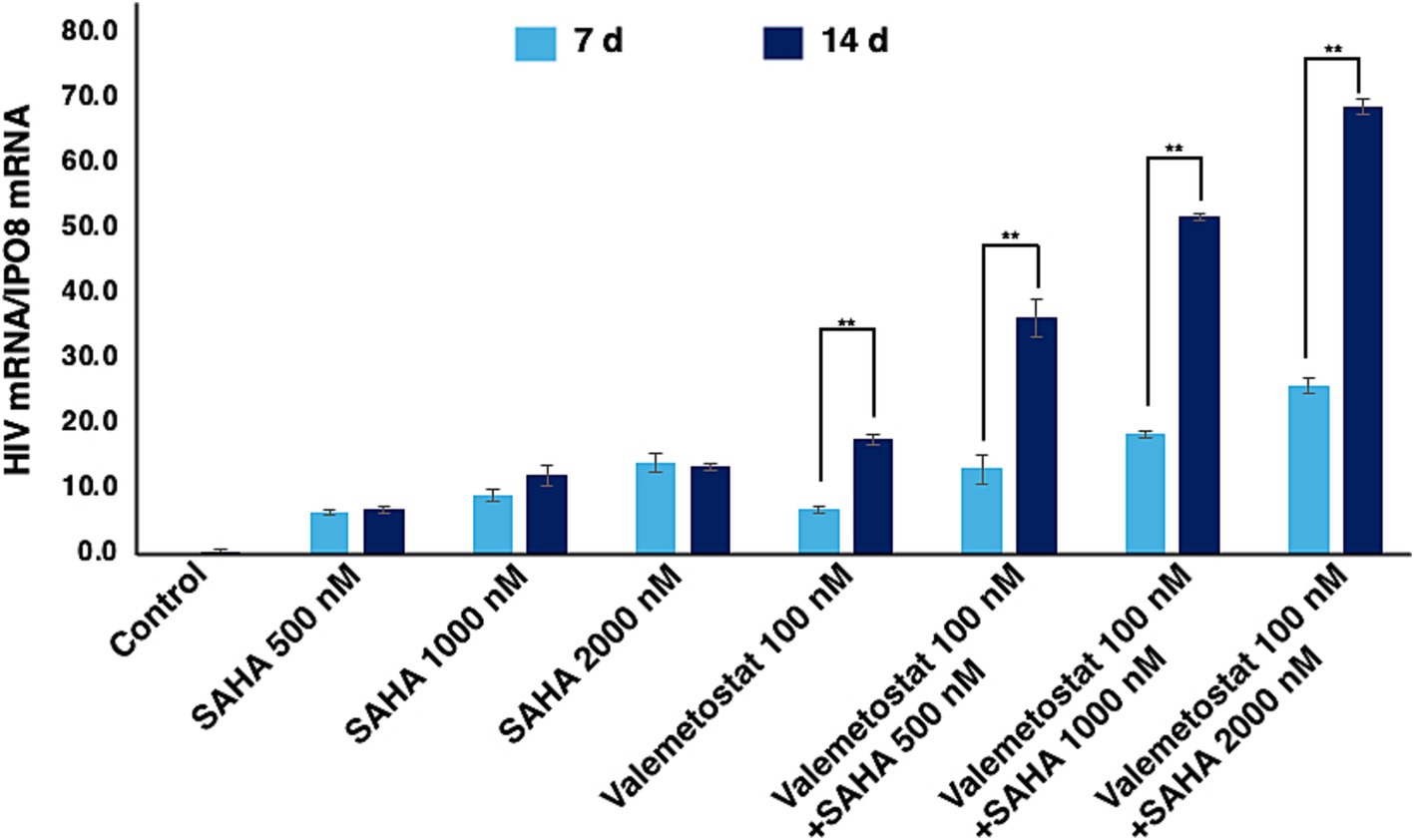
Figure 1. Reactivation of latently HIV-1-infected cells in vitro by valemetostat. qRT-PCR analysis of HIV-1 mRNA expression in ACH2 cells that were treated with 100 nM valemetostat for 7–14 days; 100 nM, 500 nM, or 1 μM SAHA for 24 h; or a combination of valemetostat and SAHA with the latter being added 1 day before the last day of treatment with valemetostat.
Next, we investigated whether long-term treatment with valemetostat could efficiently induce cellular HIV-1 expression in primary CD4+ T cells ex vivo. Although such cells cannot be maintained ex vivo for >7 days without immunostimulation, murine non-polarized primary CD4+ T cells can be maintained for a longer period following stimulation with anti-CD3/CD28 antibodies and IL-7 treatment (Kuzmichev et al., 2017; Murera et al., 2018). We successfully maintained primary CD4+ T cells derived from a patient with HIV-1 ex vivo for over 50 days (Figure 2A). These cells had high viability (over 80%) and normal cell diameter (over 7.5 μm) (Figure 2B), whereas primary CD4+ T cells cultured without beads coated with anti-CD3/CD28 antibodies had low viability (approximately 20%) and shrunken appearance (approximately 4–5 μm in diameter). The valemetostat-treated CD4+ T cells also maintained high viability for over 50 days, similar to that of the untreated CD4+ T cells.
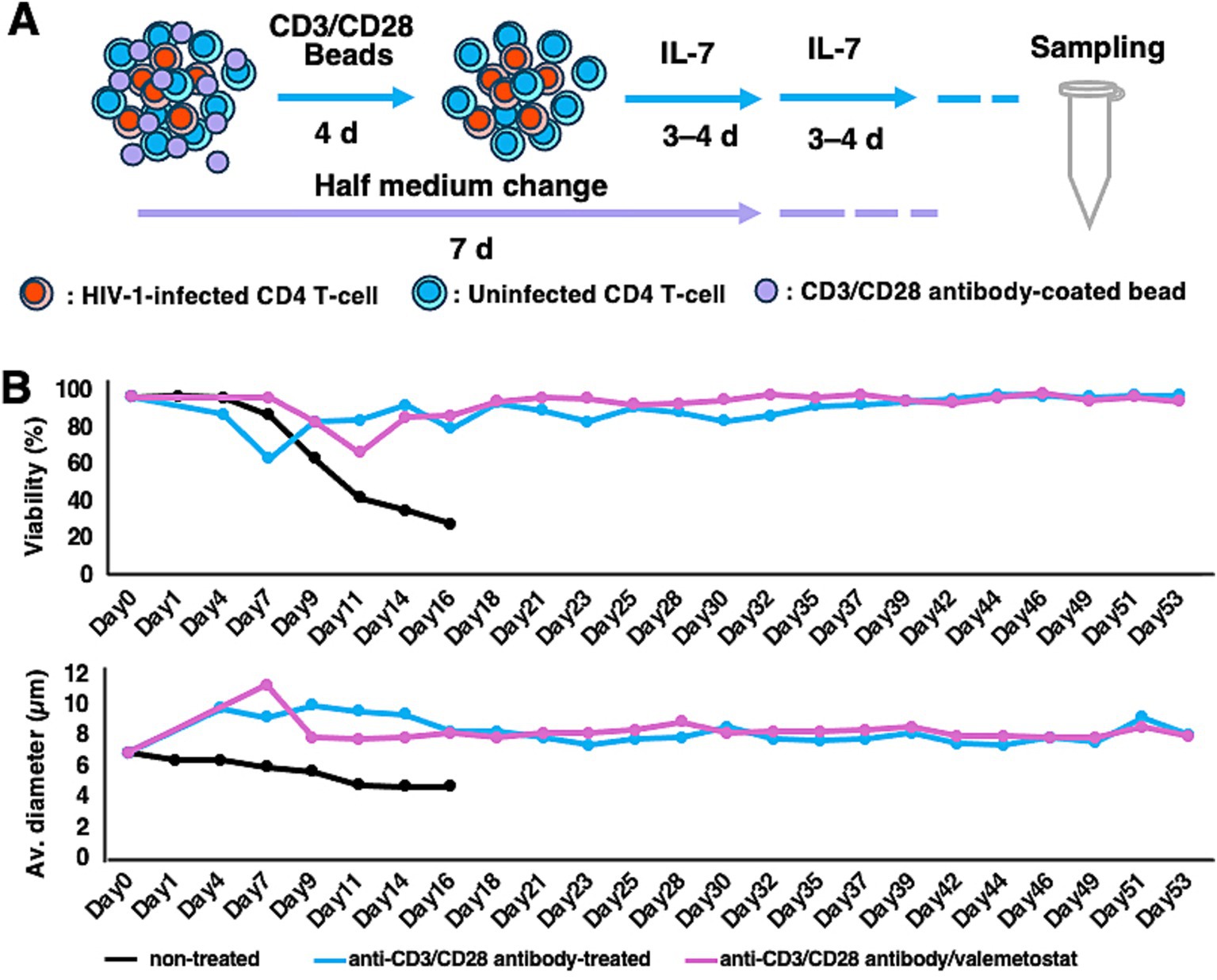
Figure 2. Maintenance of CD4+ T cells for more than 50 days ex vivo. (A) Long-term culture method for primary CD4+ T cells. Primary CD4+ T cells were treated with magnetic beads coated with anti-human CD3 and anti-human CD28 antibodies for 4 days and with IL-7 (5 μg/mL) every 3–4 days. Half of the medium was replaced with new culture medium every 7 days. (B) Viability and cell diameter of long-term cultured primary CD4+ T cells. These graphs show the results of one of the 12 cases. Black bar, non-treated CD4+ T cells; blue bar, CD4+ T cells treated with anti-human CD3 and anti-human CD28 antibodies; pink bar, CD4+ T cells treated with anti-human CD3 and anti-human CD28 antibodies and valemetostat.
We investigated whether valemetostat acts as an LRA on CD4+ T cells derived from patients with HIV-1 (n = 12). CD4+ T cells were treated with 1 mM valemetostat for 22 days. SAHA (500 nM) was added for 6 h on day 22 of culture. qRT-PCR results showed that valemetostat reactivated latently HIV-1-infected cells ex vivo (Figures 3A, P < 0.05). The level of HIV-1 mRNA expression induced by valemetostat was similar to that induced by SAHA (p > 0.05). When CD4+ T cells were treated with both valemetostat and SAHA, latently HIV-1-infected cells were more effectively reactivated than those treated with valemetostat alone (p < 0.05). Expression of the SAHA response gene H1F0 was induced only in the SAHA-treated groups (Figures 3B, P < 0.05). H1F0 expression was not induced by valemetostat, and the level of H1F0 expression after co-treatment with valemetostat and SAHA was the same as that following treatment with SAHA alone (p > 0.05).
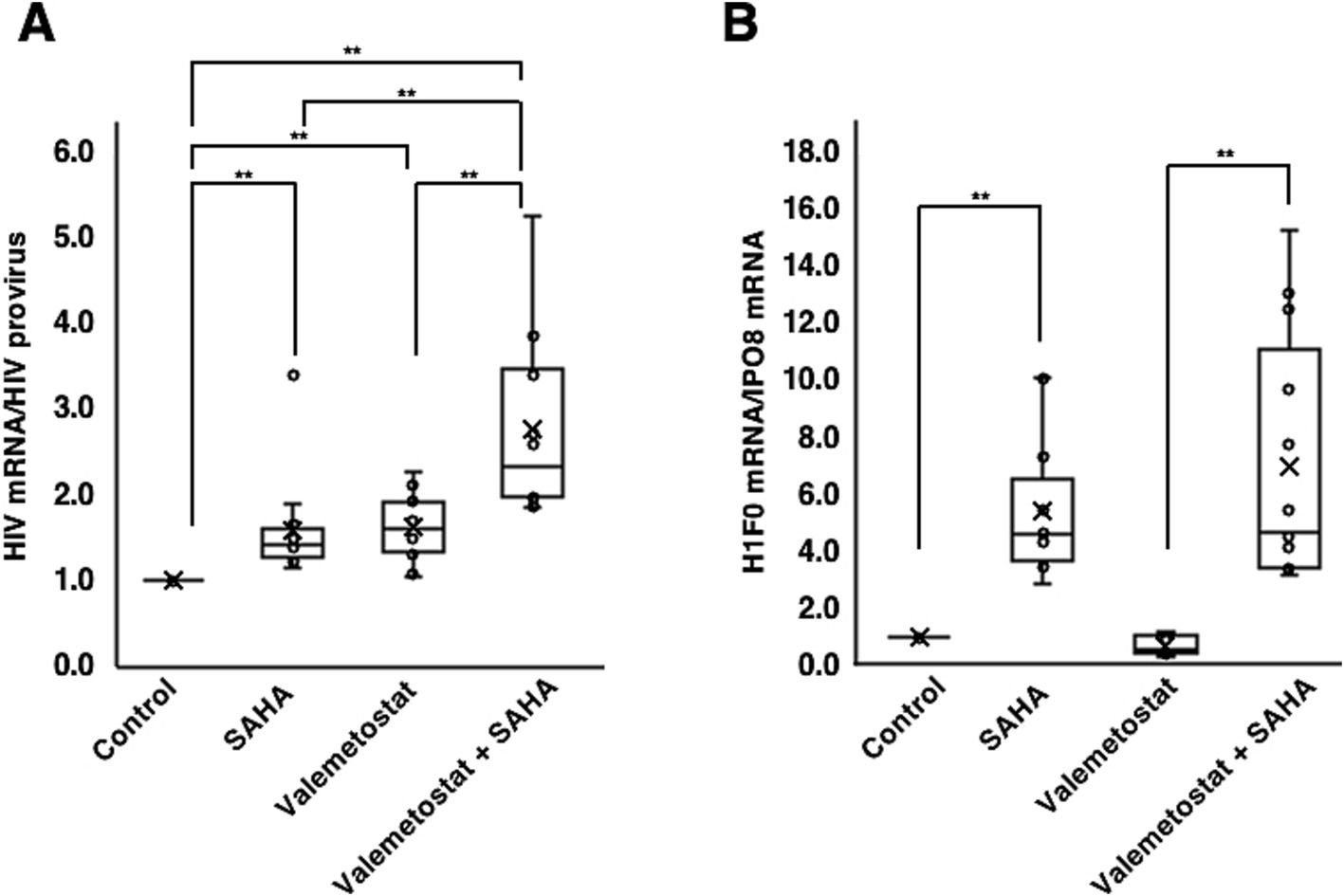
Figure 3. Reactivation by valemetostat of latently HIV-1-infected cells ex vivo. (A) qRT-PCR analysis of cellular HIV-1 expression ex vivo. CD4+ T cells were treated with 1,000 nM valemetostat for 22 days and treated with 500 nM SAHA for 6 h on the last day of treatment with valemetostat (n = 12). (B) qRT-PCR analysis of the expression of SAHA-responsive gene H1F0 (n = 12). **p < 0.01.
We investigated whether EZH1/2 dual inhibitor valemetostat could reverse HIV-1 latency ex vivo more effectively than EZH2-selective inhibitors GSK126 and E7438 (Duan et al., 2020). qRT-PCR revealed that valemetostat treatment induced HIV-1 mRNA expression more effectively than treatment with GSK126 or E7438 (Figures 4A, P < 0.05). The effect of valemetostat on the activation of latently HIV-1-infected CD4+ T cells was increased by co-treatment with SAHA. Notably, the additive effect of co-treatment using SAHA with either GSK126 or E7438 was weaker than that of the combination of SAHA and valemetostat. H1F0 expression was also induced by co-treatment with SAHA (Figures 4B, P < 0.05).
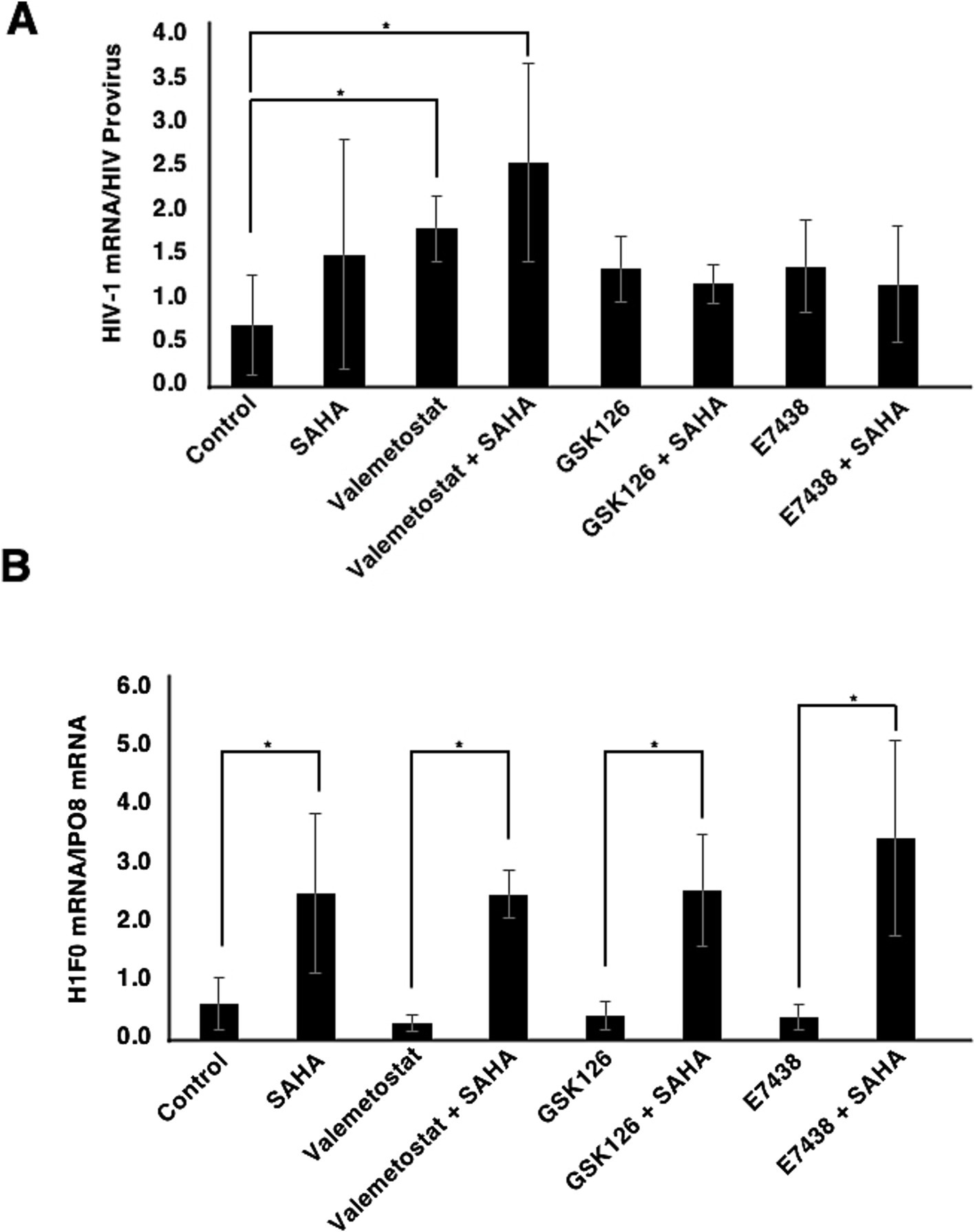
Figure 4. Comparison of the effects of valemetostat, GSK126, and E7538 on the reactivation of latently HIV-1-infected CD4+ T cells. (A) qRT-PCR analysis of cellular HIV-1 mRNA expression ex vivo. CD4+ T cells were treated for 22 days with 1,000 nM valemetostat, 1,000 nM GSK126, or 1,000 nM E7438 alone or in combination with 500 nM SAHA added for 6 h on the last day of treatment with one of these drugs (n = 3). (B) qRT-PCR analysis of cellular H1F0 expression (n = 3). *p < 0.05.
To evaluate the differences in the effects of valemetostat, GSK126, and E7438 on latently HIV-1-infected cells, the transcriptomes of CD4+ T cells were analyzed upon treatment with drugs for 95 days. The quality of the extracted total RNA was examined (Supplementary Figure 1), and the transcriptomes were analyzed using a microarray. To investigate the similarity of the transcriptomes between each sample, we performed principal component analysis on each transcriptome. The principal component analysis plot shown in Figure 5A indicates that the closer the samples are plotted, the greater the similarity between each sample, and it visually shows the overall relationship between samples. When the gene expression patterns after valemetostat treatment were compared with the gene expression patterns after E7438 and GSK126 treatment, the gene expression patterns after valemetostat treatment were more similar to the gene expression patterns after E7438 treatment than to the gene expression patterns after GSK126 treatment (Figure 5A). Figure 5B shows a heat map of genes whose expression levels changed >3-fold after treatment with valemetostat, GSK126, or E7438 (Supplementary Table S1). Expression levels of 177 genes increased >3-fold after valemetostat treatment compared with the effect of GSK126 (Supplementary Table S2). Similarly, for 17 genes, expression levels increased by >3-fold in cells treated with valemetostat compared to those in cells treated with E7438 (Supplementary Table S3). Among the genes whose expression levels increased >3-fold after treatment with valemetostat, 11 genes were common to both GSK126 and E7438. GO analysis of these 11 genes revealed that their protein products localize to the cell membrane and are involved in integrin-mediated signal transduction, cell division, and immune responses (Figure 5C and Table 3). In a similar comparison, only two genes showed a > 3-fold decrease in expression (Supplementary Table 4).
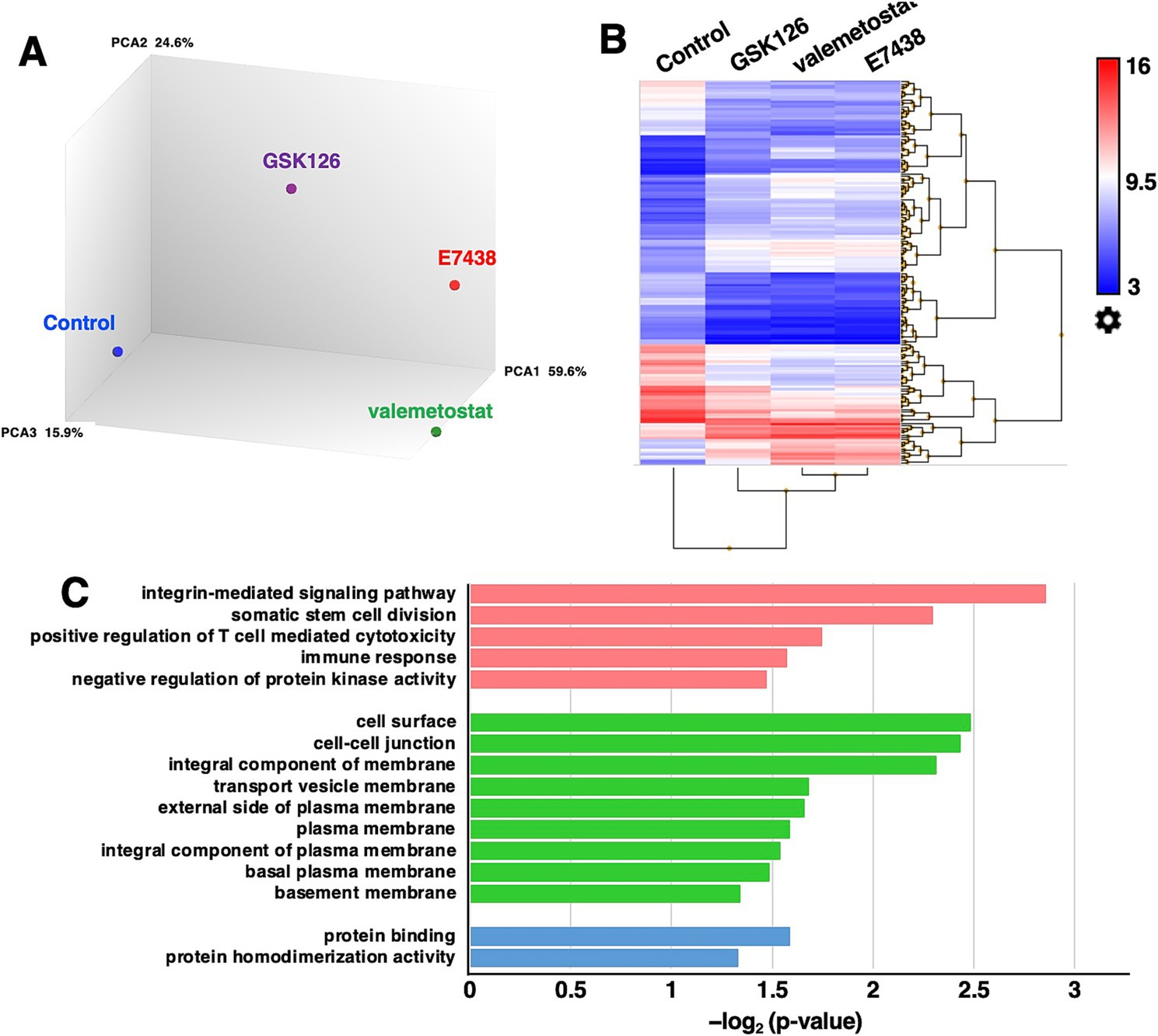
Figure 5. Microarray transcriptome analysis. (A) Principal component analysis of CD4+ T cells treated with 1,000 nM valemetostat, 1,000 nM GSK126, or 1,000 nM E7438 for 95 days (n = 1). Blue circle, control; purple circle, GSK126; green circle, valemetostat; red circle, E7438. The numbers on the right of PCA1, PCA2, and PCA3 indicate the contribution rate. (B) Heat map of genes whose expression levels changed more than threefold in CD4+ T cells treated with valemetostat, GSK126, or E7438 compared to the respective levels in untreated control cells. (C) Gene ontology analysis of the 11 genes that were three times more highly expressed in CD4+ T cells treated with valemetostat compared to their levels in GSK126- or E7438-treated cells. Pink, biological process; green, cellular component; blue, molecular function.
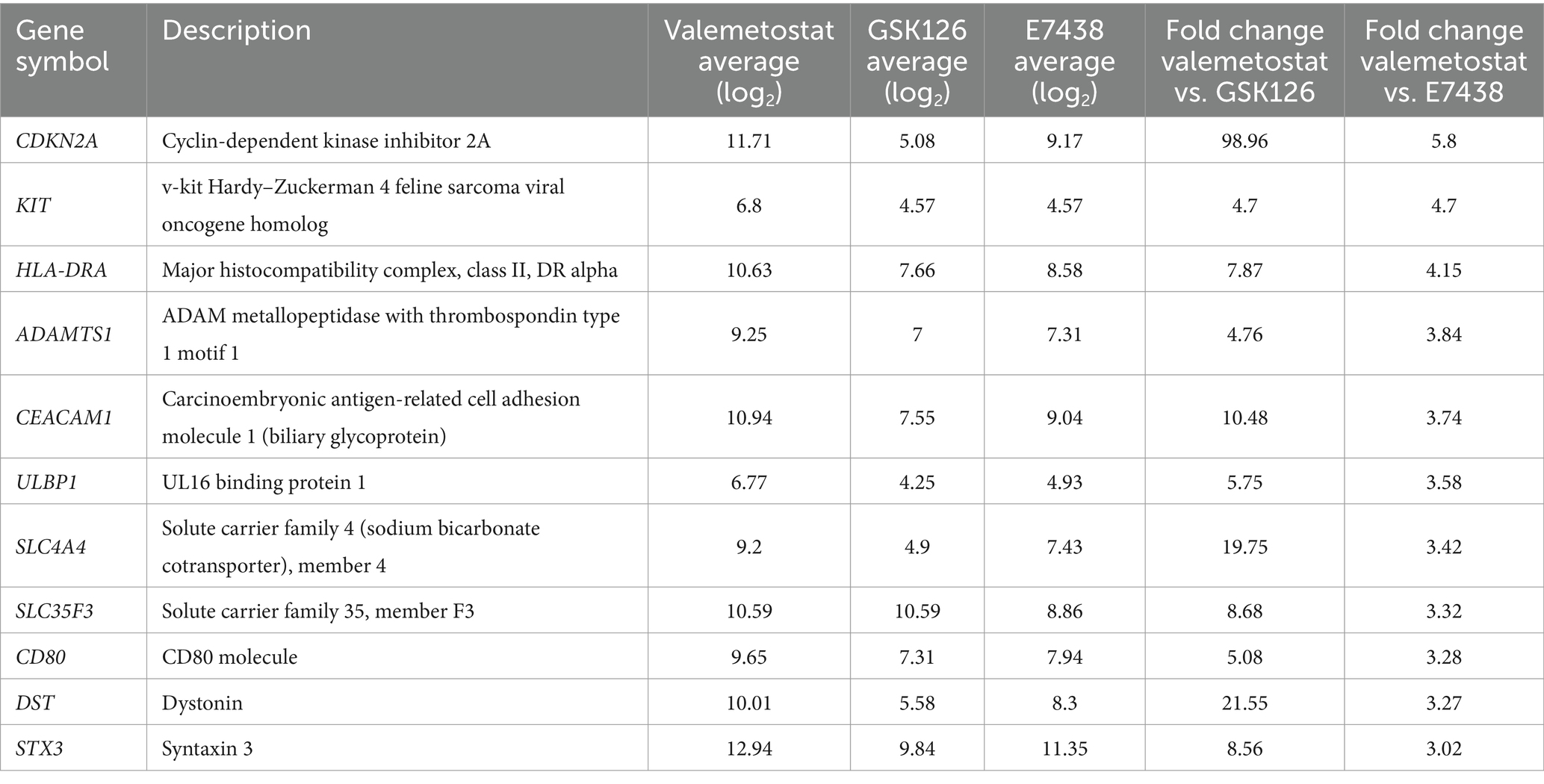
Table 3. Eleven genes with a 3-fold higher expression in valemetostat-treated CD4+ T cells than in GSK126- or E7438-treated cells.
Transcriptomic analysis showed that expression levels of 227 genes in valemetostat-treated cells changed compared with those in GSK126-treated CD4+ T cells (3-fold change, Supplementary Table 5). The PPI network revealed that proteins encoded by these 227 genes formed 212 nodes, 68 isolates, and 387 edges (Figure 6A and Supplementary Table 5). The 21 hub genes were identified from the PPI network (Table 4). The network of these 21 hub genes is shown in Figure 6B (Supplementary Table 6). GO analysis revealed that protein products of the 21 hub genes were localized in the cell membrane, cytoplasmic vesicles, and extracellular space (Supplementary Figure 2). The 21 hub genes were involved in immune response, T-cell proliferation, and B-cell differentiation. KEGG enrichment pathway analysis showed that the 21 hub genes contributed remarkably to the cytokine–cytokine receptor interactions and were involved in the JAK–STAT signaling pathway (Figure 6C). The same analysis, including PPI network analysis, was conducted for valemetostat and E7438, but only seven genes had expression levels altered by >3-fold (Supplementary Figure 3 and Supplementary Table 7). Expression levels of the 21 hub genes extracted from the transcriptomes modulated by valemetostat and GSK126 were also examined in the E7438-modulated transcriptome (Figure 6D). Expression levels of the 21 hub genes in the analysis of valemetostat vs. E7438 did not increase or decrease by >3-fold; however, they showed a similar increase/decrease pattern as observed in the comparison of valemetostat with GSK126.
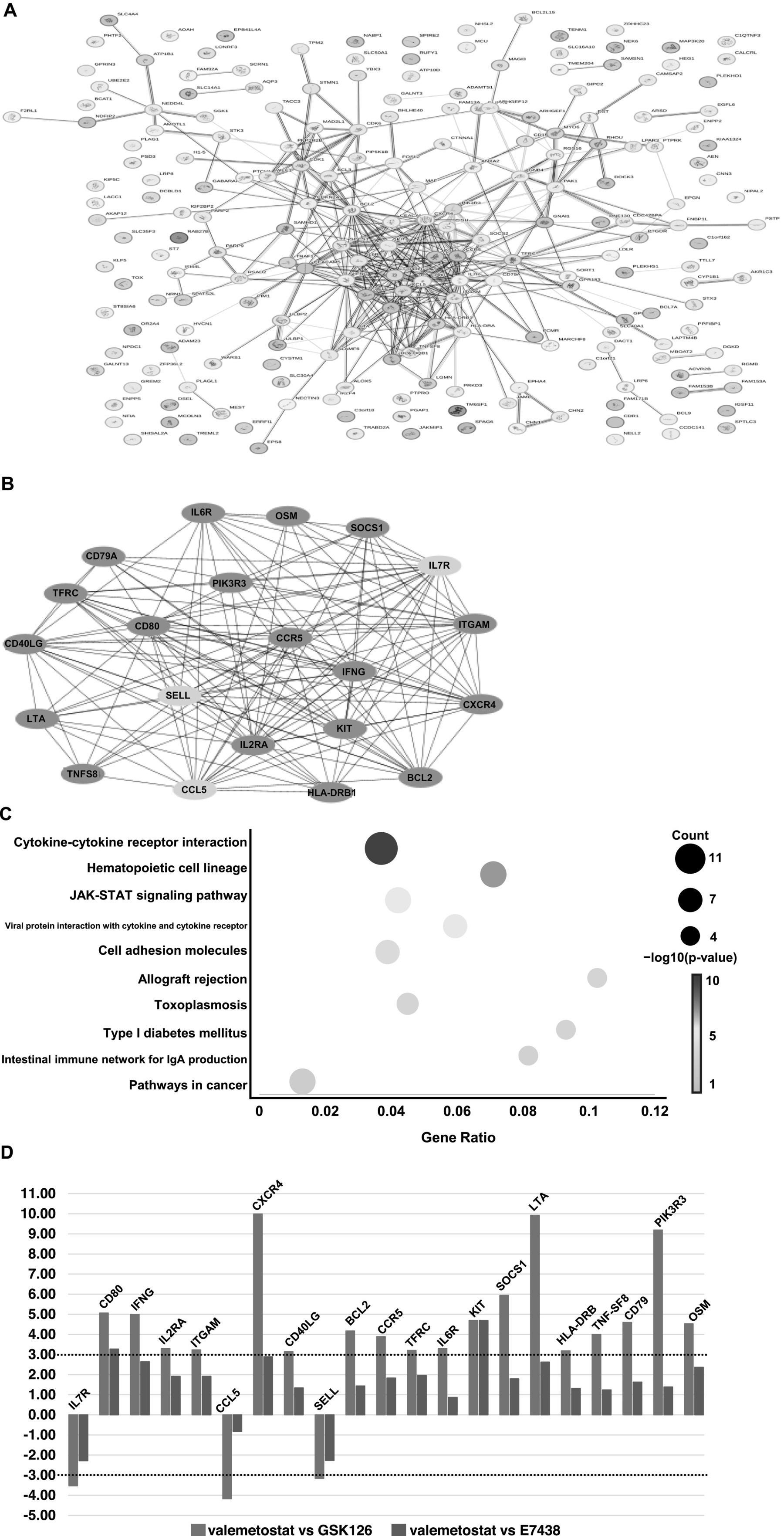
Figure 6. Analysis of genes with more than threefold changes in expression levels following treatment with valemetostat compared to the changes caused by the treatment with the EZH2-selective inhibitor GSK126. (A) PPI network of 227 genes whose expression levels changed more than threefold in CD4+ T cells upon treatment with valemetostat, compared to the changes caused by the treatment with GSK126. The colors of the edge lines indicate the following: light blue, database-supported associations; purple, experimentally demonstrated associations; red, fusion genes; yellow–green, associations supported by previous studies; green, gene neighborhoods; blue, cooccurring genes; black, coexpressing genes. (B) A network of 21 hub genes extracted from the 227 genes whose expression levels changed in valemetostat-treated cells, compared to their expression in GSK126-treated cells. The color of the nodes indicates that red indicates genes that were highly expressed in valemetostat, and blue indicates genes that were expressed at low levels in valemetostat. (C) KEGG enrichment pathway analysis of the 21 hub genes. (D) Twenty-one hub genes with more than threefold changes in expression levels changed in valemetostat-treated cells compared to their expression levels in GSK126- and E7438-treated cells. The dotted lines show positive and negative threefold change levels.
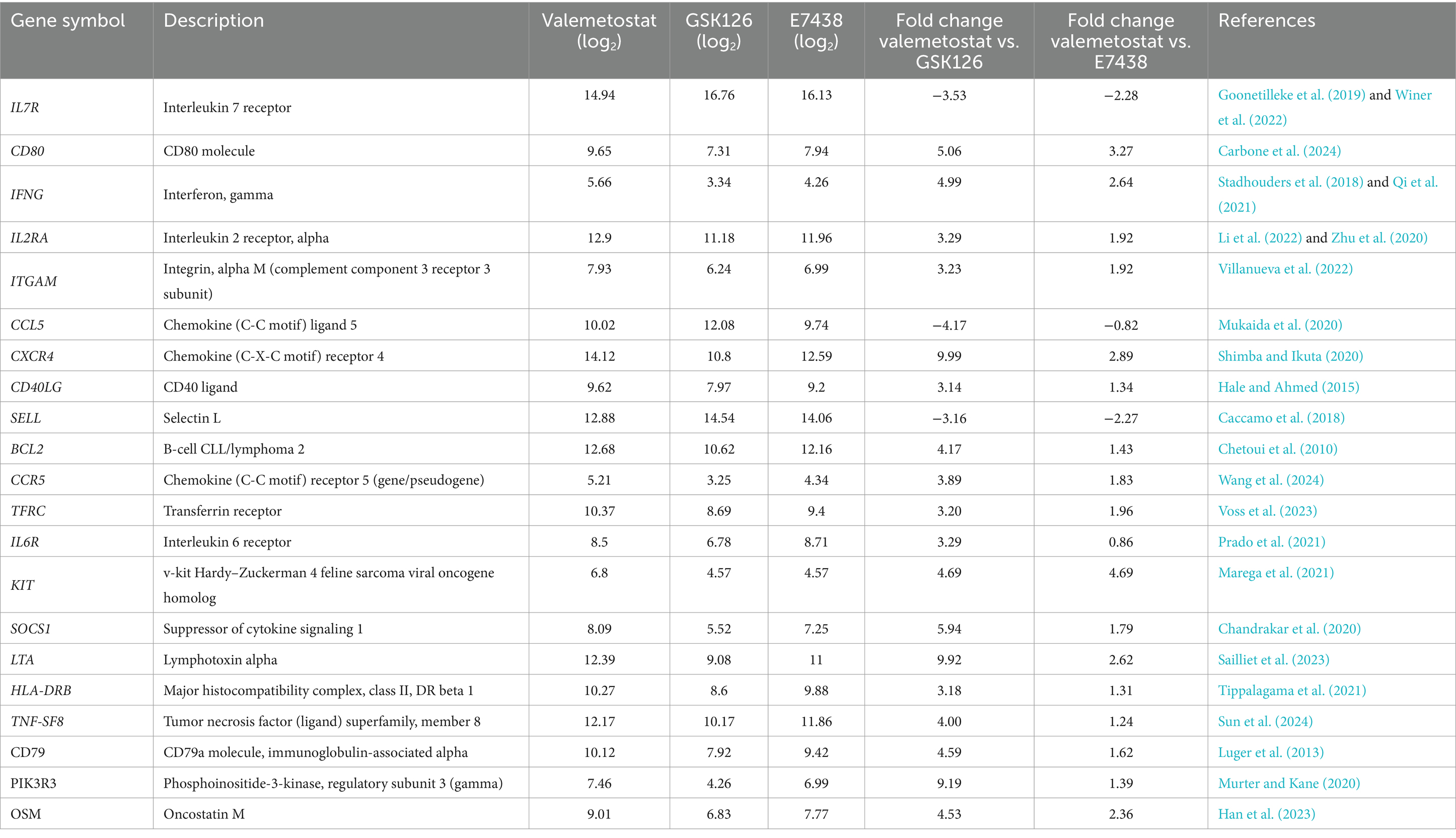
Table 4. Twenty-one hub genes extracted from the list of 227 genes exhibiting a > 3-fold change in expression levels in valemetostat-treated CD4+ T cells.
In this study, we demonstrated that the EZH1/2 dual inhibitor valemetostat demonstrated higher efficacy in reactivating latently HIV-1-infected cells than the EZH2 selective inhibitors GSK126 and E7438. Valemetostat was more effective in reactivating latently HIV-1-infected cells upon treatment over a longer period or in combination with SAHA. Our results support those of previous studies that co-treatment with an EZH2-selective inhibitor and SAHA enhanced cellular HIV-1 expression stronger than either agent alone (Tripathy et al., 2015). In addition, in the present study, we demonstrated that SAHA enhanced valemetostat-induced reactivation of latent HIV-1 infection ex vivo. However, the expression of the SAHA-responsive gene H1F0 was not enhanced by valemetostat, suggesting that this drug regulates HIV-1 expression by a mechanism not involving H1F0 upregulation. In future experiments, a detailed analysis of epigenetic changes caused by valemetostat may be warranted.
Valemetostat afforded similar reactivation of latently HIV-1-infected cells compared with GSK126 and E7438, although valemetostat tended to induce higher HIV-1 expression than GSK126 and E7438. Yamagishi et al. (2019) reported that EZH1 and EZH2 play complementary roles in regulating gene expression and that each regulates the expression of their own target genes, suggesting that the inhibition of EZH2 alone may not be sufficient (Yamagishi et al., 2019). Although the similarity between valemetostat and E7438 was high in the transcriptome analysis, valemetostat induced stronger gene expression changes compared to E7438. This is thought to be because valemetostat can inhibit EZH1 and EZH2 simultaneously and therefore has a greater effect than E7438 does. For this reason, it is thought that valemetostat shows a higher HIV-1 reactivation efficiency than E7438. SAHA more strongly enhanced the action of valemetostat compared with GSK126 and E7438. The induction of the SAHA-responsive gene H1F0 expression confirmed the efficacy of SAHA treatment. The additive effects of GSK126 and E7438 when co-treated with SAHA will be a topic for future studies.
The similarity of transcription profiles of valemetostat- and E7438-treated cells precluded comparison of these two drugs. In contrast, transcriptomes of valemetostat and GSK126 were different, and 21 distinct hub genes were identified. As valemetostat plays a role in regulating transcription, we expected that the genes with expression levels regulated by valemetostat would primarily consist of transcription factors. However, protein products of the genes that were differentially regulated by valemetostat and GSK126 were involved in the signal transduction systems localized on the cell membrane. The mechanism by which changes in the expression of these genes mediate the effects of valemetostat remains unclear. To elucidate the efficacy of valemetostat, it will be essential to investigate the relationships between these 21 genes.
This study has some limitations. The transcriptome analysis showed that valemetostat increased the expression of genes related to the JAK–STAT signaling pathway. In contrast, as the proportion of HIV-infected CD4+ T cells isolated from HIV-infected individuals is low (Woldemeskel et al., 2020), it was difficult to extract only HIV-1-infected cells and establish the mechanism of valemetostat action precisely. To solve this problem, it may be necessary to examine the direct effect of valemetostat on HIV-infected cells using in vitro experiments in cell lines, such as ACH2, which was used in this study.
The EZH1/2 dual inhibitor valemetostat reactivated latently HIV-l-infected cells ex vivo and tended to induce cellular HIV-1 mRNA expression more effectively than the existing drugs, EZH2-selective inhibitors GSK126 and E7438. A co-treatment with SAHA and valemetostat generated a stable additive effect that surpassed the effects of the combined treatment of SAHA with GSK126 or E7438. The transcriptomes of valemetostat- and E7438-treated cells were highly similar but differed from the transcriptome of GSK126-treated cells. We identified 21 hub genes that showed marked increases in expression levels upon valemetostat treatment. The products of these 21 genes are located on the cell membrane and involved in immune responses. This study provides new information relevant to the development of future treatments that would effectively reactivate latently HIV-1-infected cells and eliminate HIV-1-infected cells from patients.
All data supporting this study are available in the article 502 and supplementary data. The microarray data analyzed in this study are available in the DDBJ Genomic Expression Archive (GEA accession number : E-GEAD-876, https://ddbj.nig.ac.jp/public/ddbj_database/gea/experiment/E-GEAD-000/EGEAD-876/) and NCBI database (BioProject accession number : PRJDB19056, https://www.ncbi.nlm.nih.gov/bioproject/?term=PRJDB19056).
The studies involving humans were approved by the Research Ethics Committee of the University of Tokyo. The studies were conducted in accordance with the local legislation and institutional requirements. The participants provided their written informed consent to participate in this study.
AS: Formal Analysis, Investigation, Project administration, Supervision, Validation, Visualization, Writing – original draft, Writing – review & editing. ToK: Conceptualization, Resources, Writing – review & editing. MY: Conceptualization, Writing – review & editing. MK: Resources, Writing – review & editing. KoA: Data curation, Formal Analysis, Investigation, Writing – review & editing. KI: Writing – review & editing, Resources. TaK: Writing – review & editing, Resources. MS: Writing – review & editing, Resources. EA: Writing – review & editing, Resources. TT: Writing – review & editing, Resources. DH: Writing – review & editing, Resources. KaA: Writing – review & editing, Resources. KU: Writing – review & editing, Supervision. HY: Writing – review & editing, Funding acquisition, Supervision.
The author(s) declare that financial support was received for the research and/or publication of this article. This study was conducted under a joint research agreement with Daiichi Sankyo, Co. Ltd. Funding was provided by the company.
We thank Editage (http://www.editage.com) for editing and reviewing this manuscript. The microarray transcriptome analysis was conducted by Kurabo Industries, Ltd. (Osaka, Japan).
DH and KaA were employed by Daiichi Sankyo Co., Ltd.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The authors declare that no Generative AI was used in the creation of this manuscript.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2025.1581330/full#supplementary-material
Ait-Ammar, A., Kula, A., Darcis, G., Verdikt, R., De Wit, S., Gautier, V., et al. (2019). Current status of latency reversing agents facing the heterogeneity of HIV-1 cellular and tissue reservoirs. Front. Microbiol. 10:3060. doi: 10.3389/fmicb.2019.03060
Caccamo, N., Joosten, S. A., Ottenhoff, T. H. M., and Dieli, F. (2018). Cal human effector/memory CD4+ T cells with a naive-like phenotype. Front. Immunol. Atypi. 9:2832. doi: 10.3389/fimmu.2018.02832
Carbone, F., Russo, C., Colamatteo, A., La Rocca, C., Fusco, C., Matarese, A., et al. (2024). Cellular and molecular signaling towards T cell immunological self-tolerance. J. Biol. Chem. 300:107134. doi: 10.1016/j.jbc.2024.107134
Chahroudi, A., Silvestri, G., and Lichterfeld, M. (2015). T memory stem cells and HIV: a long-term relationship. Curr. HIV/AIDS Rep. 12, 33–40. doi: 10.1007/s11904-014-0246-4
Chandrakar, P., Parmar, N., Descoteaux, A., and Kar, S. (2020). Differential induction of SOCS isoforms by Leishmania donovani impairs macrophage-T cell cross-talk and host defense. J. Immunol. 204, 596–610. doi: 10.4049/jimmunol.1900412
Chetoui, N., Boisvert, M., Gendron, S., and Aoudjit, F. (2010). Interleukin-7 promotes the survival of human CD4+ effector/memory T cells by up-regulating Bcl-2 proteins and activating the JAK/STAT signalling pathway. Immunology 130, 418–426. doi: 10.1111/j.1365-2567.2009.03244.x
Chin, C. H., Chen, S. H., Wu, H. H., Ho, C. W., Ko, M. T., and Lin, C. Y. (2014). cytoHubba: identifying hub objects and sub-networks from complex interactome. BMC Syst. Biol. 8:S11. doi: 10.1186/1752-0509-8-S4-S11
Chomont, N., El-Far, M., Ancuta, P., Trautmann, L., Procopio, F. A., Yassine-Diab, B., et al. (2009). HIV reservoir size and persistence are driven by T cell survival and homeostatic proliferation. Nat. Med. 15, 893–900. doi: 10.1038/nm.1972
Di Croce, L., and Helin, K. (2013). Transcriptional regulation by Polycomb group proteins. Nat. Struct. Mol. Biol. 20, 1147–1155. doi: 10.1038/nsmb.2669
Duan, R., Du, W., and Guo, W. (2020). EZH2: a novel target for cancer treatment. J. Hematol. Oncol. 13:104. doi: 10.1186/s13045-020-00937-8
Friedman, J., Cho, W. K., Chu, C. K., Keedy, K. S., Archin, N. M., Margolis, D. M., et al. (2011). Epigenetic silencing of HIV-1 by the histone H3 lysine 27 methyltransferase enhancer of Zeste 2. J. Virol. 85, 9078–9089. doi: 10.1128/JVI.00836-11
Gay, C. L., Hanley, P. J., Falcinelli, S. D., Kuruc, J. D., Pedersen, S. M., Kirchherr, J., et al. (2024). The effects of human immunodeficiency virus type 1 (HIV-1) antigen-expanded specific T-cell therapy and vorinostat on persistent HIV-1 infection in people with HIV on antiretroviral therapy. J. Infect. Dis. 229, 743–752. doi: 10.1093/infdis/jiad423
Goonetilleke, N., Clutton, G., Swanstrom, R., and Joseph, S. B. (2019). Blocking formation of the stable HIV reservoir: a new perspective for HIV-1 cure. Front. Immunol. 10:1966. doi: 10.3389/fimmu.2019.01966
Gulati, N., Béguelin, W., and Giulino-Roth, L. (2018). Enhancer of zeste homolog 2 (EZH2) inhibitors. Leuk. Lymphoma 59, 1574–1585. doi: 10.1080/10428194.2018.1430795
Hale, J. S., and Ahmed, R. (2015). Memory T follicular helper CD4 T cells. Front. Immunol. 6:16. doi: 10.3389/fimmu.2015.00016
Han, L., Yan, J., Li, T., Lin, W., Huang, Y., Shen, P., et al. (2023). Multifaceted oncostatin M: novel roles and therapeutic potential of the oncostatin M signaling in rheumatoid arthritis. Front. Immunol. 14:1258765. doi: 10.3389/fimmu.2023.1258765
Holoch, D., and Margueron, R. (2017). Mechanisms regulating PRC2 recruitment and enzymatic activity. Trends Biochem. Sci. 42, 531–542. doi: 10.1016/j.tibs.2017.04.003
Honma, D., Kanno, O., Watanabe, J., Kinoshita, J., Hirasawa, M., Nosaka, E., et al. (2017). Novel orally bioavailable EZH1/2 dual inhibitors with greater antitumor efficacy than an EZH2 selective inhibitor. Cancer Sci. 108, 2069–2078. doi: 10.1111/cas.13326
Hosmane, N. N., Kwon, K. J., Bruner, K. M., Capoferri, A. A., Beg, S., Rosenbloom, D. I. S., et al. (2017). Proliferation of latently infected CD4+ T cells carrying replication-competent HIV-1: potential role in latent reservoir dynamics. J. Exp. Med. 214, 959–972. doi: 10.1084/jem.20170193
Huang, W., Sherman, B. T., and Lempicki, R. A. (2009). Systematic and integrative analysis of large gene lists using David bioinformatics resources. Nat. Protoc. 4, 44–57. doi: 10.1038/nprot.2008.211
Ishida, T., Hamano, A., Koiwa, T., and Watanabe, T. (2006). 5′ long terminal repeat (LTR)-selective methylation of latently infected HIV-1 provirus that is demethylated by reactivation signals. Retrovirology 3:69. doi: 10.1186/1742-4690-3-69
Izutsu, K., Ando, K., Nishikori, M., Shibayama, H., Teshima, T., Kuroda, J., et al. (2021). Phase II study of tazemetostat for relapsed or refractory B-cell non-Hodgkin lymphoma with EZH2 mutation in Japan. Cancer Sci. 112, 3627–3635. doi: 10.1111/cas.15040
Izutsu, K., Makita, S., Nosaka, K., Yoshimitsu, M., Utsunomiya, A., Kusumoto, S., et al. (2023). An open-label, single-arm phase 2 trial of valemetostat for relapsed or refractory adult T-cell leukemia/lymphoma. Blood 141, 1159–1168. doi: 10.1182/blood.2022016862
Keam, S. J. (2022). Valemetostat tosilate: first approval. Drugs 82, 1621–1627. doi: 10.1007/s40265-022-01800-5
Khan, S., Iqbal, M., Tariq, M., Baig, S. M., and Abbas, W. (2018). Epigenetic regulation of HIV-1 latency: focus on polycomb group (PcG) proteins. Clin. Epigenetics 10:14. doi: 10.1186/s13148-018-0441-z
Kim, Y., Anderson, J. L., and Lewin, S. R. (2018). Getting the “kill” into “shock and kill”: strategies to eliminate latent HIV. Cell Host Microbe 23, 14–26. doi: 10.1016/j.chom.2017.12.004
Kuzmichev, Y. V., Veenhuis, R. T., Pohlmeyer, C. W., Garliss, C. C., Walker-Sperling, V. E., and Blankson, J. N. (2017). A CD3/CD28 microbead-based HIV-1 viral outgrowth assay. J. Virus Erad. 3, 85–89. doi: 10.1016/S2055-6640(20)30292-2
Kwon, K. J., Timmons, A. E., SenGupta, S., Simonetti, F. R., Zhang, H., Hoh, R., et al. (2020). Different human resting memory CD4+ T cell subsets show similar low inducibility of latent HIV-1 proviruses. Sci. Transl. Med. 12:eaax6795. doi: 10.1126/scitranslmed.aax6795
Ledderose, C., Heyn, J., Limbeck, E., and Kreth, S. (2011). Selection of reliable reference genes for quantitative real-time PCR in human T cells and neutrophils. BMC. Res. Notes 4:427. doi: 10.1186/1756-0500-4-427
Li, Y., Li, X., Geng, X., and Zhao, H. (2022). The IL-2A receptor pathway and its role in lymphocyte differentiation and function. Cytokine Growth Factor Rev. 67, 66–79. doi: 10.1016/j.cytogfr.2022.06.004
Luger, D., Yang, Y. A., Raviv, A., Weinberg, D., Banerjee, S., Lee, M. J., et al. (2013). Expression of the B-cell receptor component CD79a on immature myeloid cells contributes to their tumor promoting effects. PLoS One 8:e76115. doi: 10.1371/journal.pone.0076115
Marega, L. F., Sabino, J. S., Pedroni, M. V., Teocchi, M., Lanaro, C., de Albuquerque, D. M., et al. (2021). Phenotypes of STAT3 gain-of-function variant related to disruptive regulation of CXCL8/STAT3, KIT/STAT3, and IL-2/CD25/Treg axes. Immunol. Res. 69, 445–456. doi: 10.1007/s12026-021-09225-0
Matsuda, Y., Kobayashi-Ishihara, M., Fujikawa, D., Ishida, T., Watanabe, T., and Yamagishi, M. (2015). Epigenetic heterogeneity in HIV-1 latency establishment. Sci. Rep. 5:7701. doi: 10.1038/srep07701
Morschhauser, F., Tilly, H., Chaidos, A., McKay, P., Phillips, T., Assouline, S., et al. (2020). Tazemetostat for patients with relapsed or refractory follicular lymphoma: an open-label, single-arm, multicentre, phase 2 trial. Lancet Oncol. 21, 1433–1442. doi: 10.1016/S1470-2045(20)30441-1
Mukaida, N., Sasaki, S. I., and Baba, T. (2020). CCL4 signaling in the tumor microenvironment. Adv. Exp. Med. Biol. 1231, 23–32. doi: 10.1007/978-3-030-36667-4_3
Murera, D., Arbogast, F., Arnold, J., Bouis, D., Muller, S., and Gros, F. (2018). CD4 T cell autophagy is integral to memory maintenance. Sci. Rep. 8:5951. doi: 10.1038/s41598-018-23993-0
Murter, B., and Kane, L. P. (2020). Control of T lymphocyte fate decisions by PI3K signaling. F1000Res 9:1171. doi: 10.12688/f1000research.26928.1
Nguyen, K., Das, B., Dobrowolski, C., and Karn, J. (2017). Multiple histone lysine methyltransferases are required for the establishment and maintenance of HIV-1 latency. MBio 8, e00133–e00117. doi: 10.1128/mBio.00133-17
Prado, D. S., Cattley, R. T., Shipman, C. W., Happe, C., Lee, M., Boggess, W. C., et al. (2021). Synergistic and additive interactions between receptor signaling networks drive the regulatory T cell versus T helper 17 cell fate choice. J. Biol. Chem. 297:101330. doi: 10.1016/j.jbc.2021.101330
Qi, F., Liu, F., and Gao, L. (2021). Janus kinase inhibitors in the treatment of vitiligo: a review. Front. Immunol. 12:790125. doi: 10.3389/fimmu.2021.790125
Sadowski, I., and Hashemi, F. B. (2019). Strategies to eradicate HIV from infected patients: elimination of latent provirus reservoirs. Cell. Mol. Life Sci. 76, 3583–3600. doi: 10.1007/s00018-019-03156-8
Sailliet, N., Mai, H. L., Dupuy, A., Tilly, G., Fourgeux, C., Braud, M., et al. (2023). Human granzyme B regulatory B cells prevent effector CD4+CD25− T cell proliferation through a mechanism dependent from lymphotoxin alpha. Front. Immunol. 14:1183714. doi: 10.3389/fimmu.2023.1183714
Shan, L., Deng, K., Gao, H., Xing, S., Capoferri, A. A., Durand, C. M., et al. (2017). Transcriptional reprogramming during effector-to-memory transition renders CD4+ T cells permissive for latent HIV-1. Infect. Immun. 47:e763, 766–775.e3. doi: 10.1016/j.immuni.2017.09.014
Sharma, A. L., Hokello, J., Sonti, S., Zicari, S., Sun, L., Alqatawni, A., et al. (2020). CBF-1 promotes the establishment and maintenance of HIV latency by recruiting polycomb repressive complexes, PRC1 and PRC2, at HIV LTR. Viruses 12:1040. doi: 10.3390/v12091040
Shen, X., Liu, Y., Hsu, Y. J., Fujiwara, Y., Kim, J., Mao, X., et al. (2008). EZH1 mediates methylation on histone H3 lysine 27 and complements EZH2 in maintaining stem cell identity and executing pluripotency. Mol. Cell 32, 491–502. doi: 10.1016/j.molcel.2008.10.016
Sherman, B. T., Hao, M., Qiu, J., Jiao, X., Baseler, M. W., Lane, H. C., et al. (2022). David: a web server for functional enrichment analysis and functional annotation of gene lists (2021 update). Nucleic Acids Res. 50, W216–W221. doi: 10.1093/nar/gkac194
Shimba, A., and Ikuta, K. (2020). Control of immunity by glucocorticoids in health and disease. Semin. Immunopathol. 42, 669–680. doi: 10.1007/s00281-020-00827-8
Stadhouders, R., Lubberts, E., and Hendriks, R. W. (2018). A cellular and molecular view of T helper 17 cell plasticity in autoimmunity. J. Autoimmun. 87, 1–15. doi: 10.1016/j.jaut.2017.12.007
Sun, N., Zhang, M., Kong, J., Li, J., Dong, Y., Wang, X., et al. (2024). Dysregulated T-cell homeostasis and decreased CD30+ Treg proliferating in aplastic anemia. Heliyon 10:e35775. doi: 10.1016/j.heliyon.2024.e35775
Szklarczyk, D., Franceschini, A., Wyder, S., Forslund, K., Heller, D., Huerta-Cepas, J., et al. (2015). STRING v10: protein–protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 43, D447–D452. doi: 10.1093/nar/gku1003
Ta, T. M., Malik, S., Anderson, E. M., Jones, A. D., Perchik, J., Freylikh, M., et al. (2022). Insights into persistent HIV-1 infection and functional cure: novel capabilities and strategies. Front. Microbiol. 13:862270. doi: 10.3389/fmicb.2022.862270
Thorlund, K., Horwitz, M. S., Fife, B. T., Lester, R., and Cameron, D. W. (2017). Landscape review of current HIV “kick and kill” cure research – some kicking, not enough killing. BMC Infect. Dis. 17:595. doi: 10.1186/s12879-017-2683-3
Tippalagama, R., Singhania, A., Dubelko, P., Lindestam Arlehamn, C. S., Crinklaw, A., Pomaznoy, M., et al. (2021). HLA-DR marks recently divided antigen-specific effector CD4 T cells in active tuberculosis patients. J. Immunol. 207, 523–533. doi: 10.4049/jimmunol.2100011
Tripathy, M. K., McManamy, M. E. M., Burch, B. D., Archin, N. M., and Margolis, D. M. (2015). H3K27 demethylation at the proviral promoter sensitizes latent HIV to the effects of vorinostat in ex vivo cultures of resting CD4+ T cells. J. Virol. 89, 8392–8405. doi: 10.1128/JVI.00572-15
Turner, A. W., Dronamraju, R., Potjewyd, F., James, K. S., Winecoff, D. K., Kirchherr, J. L., et al. (2020). Evaluation of EED inhibitors as a class of PRC2-targeted small molecules for HIV latency reversal. ACS Infect. Dis. 6, 1719–1733. doi: 10.1021/acsinfecdis.9b00514
Villanueva, V., Li, X., Jimenez, V., Faridi, H. M., and Gupta, V. (2022). CD11b agonists offer a novel approach for treating lupus nephritis. Transl. Res. 245, 41–54. doi: 10.1016/j.trsl.2022.03.001
Voss, K., Sewell, A. E., Krystofiak, E. S., Gibson-Corley, K. N., Young, A. C., Basham, J. H., et al. (2023). Elevated transferrin receptor impairs T cell metabolism and function in systemic lupus erythematosus. Sci. Immunol. 8:eabq0178. doi: 10.1126/sciimmunol.abq0178
Wang, L., Yukselten, Y., Nuwagaba, J., and Sutton, R. E. (2024). JAK/STAT signaling pathway affects CCR5 expression in human CD4+ T cells. Sci. Adv. 10:eadl0368. doi: 10.1126/sciadv.adl0368
Winer, H., Rodrigues, G. O. L., Hixon, J. A., Aiello, F. B., Hsu, T. C., Wachter, B. T., et al. (2022). IL-7: comprehensive review. Cytokine 160:156049. doi: 10.1016/j.cyto.2022.156049
Woldemeskel, B. A., Kwaa, A. K., and Blankson, J. N. (2020). Viral reservoirs in elite controllers of HIV-1 infection: implications for HIV cure strategies. EBioMedicine 62:103118. doi: 10.1016/j.ebiom.2020.103118
Yamagishi, M., Hori, M., Fujikawa, D., Ohsugi, T., Honma, D., Adachi, N., et al. (2019). Targeting excessive EZH1 and EZH2 activities for abnormal histone methylation and transcription network in malignant lymphomas. Cell Rep. 29, 2321–2337.e7. doi: 10.1016/j.celrep.2019.10.083
Young, M. D., Wakefield, M. J., Smyth, G. K., and Oshlack, A. (2010). Gene ontology analysis for RNA-seq: accounting for selection bias. Genome Biol. 11:R14. doi: 10.1186/gb-2010-11-2-r14
Zhu, M., Yang, L., Shi, X., Gong, Z., Yu, R., Zhang, D., et al. (2020). TPD7 inhibits the growth of cutaneous T cell lymphoma H9 cell through regulating IL-2R signalling pathway. J. Cell. Mol. Med. 24, 984–995. doi: 10.1111/jcmm.14810
Zinzani, P. L., Izutsu, K., Mehta-Shah, N., Barta, S. K., Ishitsuka, K., Córdoba, R., et al. (2024). Valemetostat for patients with relapsed or refractory peripheral T-cell lymphoma (valentine-PTCL01): a multicentre, open-label, single-arm, phase 2 study. Lancet Oncol. 25, 1602–1613. doi: 10.1016/S1470-2045(24)00503-5
Keywords: HIV-1 reservoir, latency reversing agents, EZH1/2 dual inhibitor, valemetostat, suberoylanilide hydroxamic acid, GSK126, E7438
Citation: Sedohara A, Koibuchi T, Yamagishi M, Koga M, Arizono K, Ikeuchi K, Kikuchi T, Saito M, Adachi E, Tsutsumi T, Honma D, Araki K, Uchimaru K and Yotsuyanagi H (2025) Enhancer of zeste homolog 1/2 dual inhibitor valemetostat outperforms enhancer of zeste homolog 2-selective inhibitors in reactivating latent HIV-1 reservoirs ex vivo. Front. Microbiol. 16:1581330. doi: 10.3389/fmicb.2025.1581330
Received: 22 February 2025; Accepted: 13 March 2025;
Published: 10 April 2025.
Edited by:
Yasuko Tsunetsugu Yokota, Tokyo University of Technology, JapanReviewed by:
Masaru Yokoyama, National Institute of Infectious Diseases (NIID), JapanCopyright © 2025 Sedohara, Koibuchi, Yamagishi, Koga, Arizono, Ikeuchi, Kikuchi, Saito, Adachi, Tsutsumi, Honma, Araki, Uchimaru and Yotsuyanagi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ayako Sedohara, c2Vkb2hhcmFAaW1zLnUtdG9reW8uYWMuanA=; Hiroshi Yotsuyanagi, eW90c3VkaWRAaW1zLnUtdG9reW8uYWMuanA=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.