
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 04 April 2025
Sec. Food Microbiology
Volume 16 - 2025 | https://doi.org/10.3389/fmicb.2025.1575725
 Qiuqian Zeng1,2,3
Qiuqian Zeng1,2,3 Xiaomei Lyu1,2,3*
Xiaomei Lyu1,2,3*Introduction: Cellobiose 2-epimerase (CE) catalyzes the interconversion of glucosyl and mannosyl groups at the reducing end of β-1,4-linked disaccharides. This enzyme is pivotal for converting lactose into prebiotics like epilactose, offering a potential solution for lactose-intolerant-friendly dairy products. However, current CEs are hindered by pH and thermal instability in milk processing, as their neutral-to-alkaline pH optima clash with milk’s mildly acidic conditions (pH 6.5–6.7), and their poor thermolability requires costly post-processing enzyme removal.
Methods: We identified a novel CE from the acidophilic Acidobacteriota bacterium (Acba-CE) and characterized its properties. Its enzymatic activity was assessed under varying pH and temperature conditions, including milk-processing environments.
Results: Acba-CE exhibits an acidic pH optimum (6.0), retaining 95% activity at milk pH (6.5). Notably, it undergoes rapid thermal inactivation at pasteurization temperatures, enabling complete enzyme deactivation during standard pasteurization without additional steps. In milk systems, Acba-CE achieves 28.5% lactose-to-epilactose conversion at refrigeration temperatures (10°C), demonstrating strong cold adaptability.
Discussion: To our knowledge, this is the first reported CE from the Acidobacteriota phylum, combining acidophilic activity with low-temperature adaptability. Acba-CE represents a breakthrough for in situ dairy modification, eliminating key bottlenecks in milk processing and enabling next-generation functional milk production.
Cellobiose 2-epimerase (CE, EC 5.1.3.11) is a class of enzymes that plays a pivotal role in carbohydrate metabolism. Initially identified in Ruminococcus albus (Kim et al., 2012; Ito et al., 2007), CE belongs to the N-acetyl-glucosamine 2-epimerase (AGE) superfamily. These enzymes are widely distributed in nature, particularly among microorganisms, and demonstrate notable catalytic diversity with significant potential for industrial applications. The primary function of CE is to catalyze the interconversion of glucosyl and mannosyl groups at the reducing end of disaccharides linked by β-1,4-glycosidic bonds (Fujiwara et al., 2014). Common substrates catalyzed by CE include cellobiose, lactose, and 4-O-β-D-mannosyl-D-glucose. When lactose serves as the substrate, the epimerization activity of CE catalyzes its conversion into epilactose. As a derivative of lactose, epilactose is a non-digestible disaccharide with prebiotic properties, exhibiting various health-promoting bioactivities (Xiong et al., 2024), such as regulating intestinal flora (Watanabe et al., 2008) and promoting calcium absorption (Nishimukai et al., 2008; Suzuki et al., 2010). In recent years, research on the bioconversion of this sugar has garnered widespread attention, with CE playing a crucial role in this process (Pang et al., 2025).
Beyond its application in the industrial production of epilactose, CE is now being considered for use in the processing of milk (Eat et al., 2024; Krewinkel et al., 2014; Rentschler et al., 2015). Milk is abundant in lactose, but some individuals lack the enzyme lactase, which is primarily produced by cells in the small intestine, to digest it. When these individuals consume milk containing lactose, due to insufficient secretion or reduced activity of lactase, a significant amount of lactose remains undigested and accumulates in the intestine, potentially leading to adverse symptoms such as bloating, abdominal pain, diarrhea, nausea, and vomiting (Szilagyi and Ishayek, 2018; Angima et al., 2024).
The addition of CE to milk could enable in situ conversion of lactose into prebiotic epilactose. However, this strategy faces a critical challenge: milk’s mildly acidic pH (6.5–6.7) drastically reduces the activity of conventional CEs, which typically exhibit neutral-to-alkaline pH optima. Consequently, these enzymes experience varying degrees of activity loss in acidic environments. Table 1 summarizes the enzymatic properties of the main CE enzymes currently used for epilactose production. Figure 1 illustrates the relative activity of CE enzymes from different sources at various pH levels. Although the CE enzyme derived from Rhodothermus marinus (Ojima et al., 2014) is suitable for acidic environments, its optimal temperature is too high, making it difficult to inactivate during processing at high temperatures and unsuitable for use in milk processing environments.
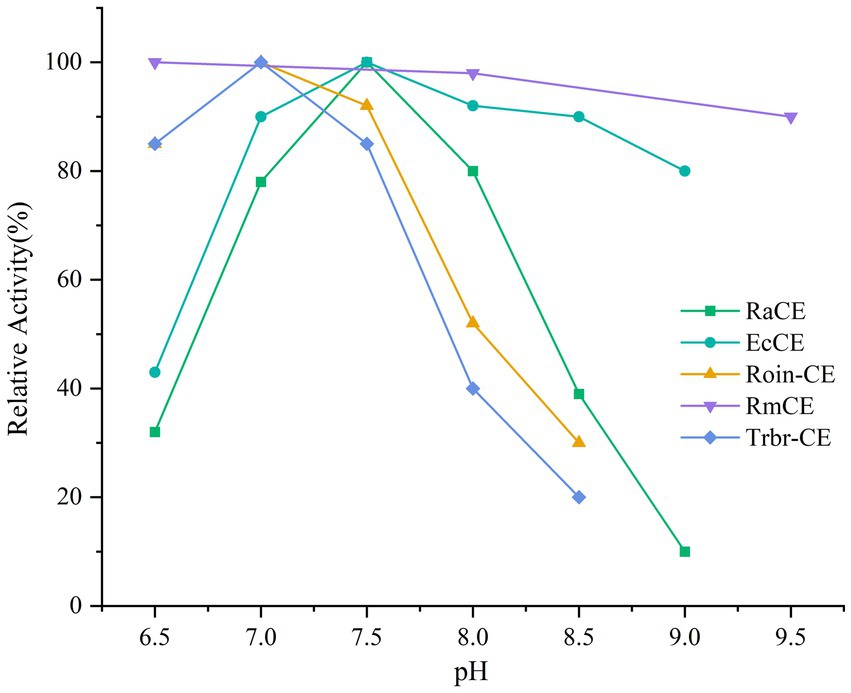
Figure 1. Relative activity of CEs from different sources at various pH levels. RaCE, Ruminococcus albus; EcCE, Eubacterium cellulosomes; Roin-CE, Roseburia intestinalis; RmCE, Rhodothermus marinus; Trbr-CE, Treponema brennaborense.
To identify enzymes that maintain high activity in slightly acidic environments, the exploration of novel CEs sourced from microorganisms adapted to acidic conditions is considered. This study focuses on the CE derived from Acidobacteriota bacterium (Acba-CE), examining its role in catalyzing the conversion of lactose to epilactose. As a recently classified bacterial phylum, Acidobacteriota bacterium is categorized as acidophiles, capable of surviving and reproducing in acidic environments (Ludwig et al., 2006). This unique physiological characteristic enables enzymes derived from Acidobacteriota bacterium to exhibit a degree of acidophilia, thus better adapting to and being applicable in slightly acidic industrial production environments.
In this research, our objective was to gain insights into the catalytic properties and application potential of Acba-CE, particularly its advantages in acidic environments. We conducted extensive expression and purification of this enzyme in Escherichia coli host, conducted in-depth investigations into its enzymatic properties and activity, and achieved efficient conversion of lactose to epilactose within a short period. The ultimate goal is to facilitate its widespread application in fields such as food, pharmaceuticals, and health supplements.
Isopropyl β-D-1-thiogalactopyranoside (IPTG), Kanamycin (Kana), Piperazine-N,N′-bis(2-ethanesulfonic acid) (PIPES), N-[2-Hydroxyethyl]piperazine-N′-[2-ethanesulfonic acid] (HEPES), Ni-NTA Sepharose 6FF (for His-Tag purification), and associated reagents for enzyme purification were procured from Sangon Biotech. Epilactose was sourced from Aladdin. Lactose and sodium chloride were obtained from Sinopharm Chemical Reagent Co., Ltd. Tryptone and yeast extract were acquired from Oxoid. The gene of Acba-CE (National Center for Biotechnology Information: NCBI accession number is RPJ56071.1) was synthesized by Dynegene Technologies and ligated between the NdeI and XhoI sites of the pET-28a(+) vector. Subsequently, the plasmid was transformed into Escherichia coli BL21(DE3) host cells through heat shock at 42°C, facilitating subsequent protein expression.
For recombinant protein production, a nutrient-rich culture solution was prepared containing 1% peptone, 1% NaCl, and 0.5% yeast extract, supplemented with 50 μg/mL of the antibiotic kanamycin. The engineered bacterial strains harboring the target enzyme were initially cultured in this medium for 12–16 h at 37°C. Subsequently, 1 mL of the pre-culture was introduced into 100 mL of fresh medium for large-scale cultivation. When the bacterial density, as measured by spectrophotometric analysis at 600 nm, reached 0.6–0.8 absorbance units, protein expression was initiated by adding IPTG to a concentration of 200 μM. The induction process was carried out at a reduced temperature of 16°C for an extended period of 20 h to facilitate proper protein folding.
Following cultivation, bacterial cells were harvested by centrifugation at 6,500 × g for 10 min at 4°C. The pellet was washed twice with ice-cold PBS and resuspended in lysis buffer (50 mM phosphate buffer, 200 mM NaCl, 10 mM imidazole [pH 7.5]) using a 50 mL beaker. Cell lysis was performed with a φ10 mm ultrasonic probe under pulsed conditions (5 s ON/9 s OFF per cycle) at 500 W output power for 15 min total duration, with the suspension maintained below 4°C using an ice bath. The lysate was clarified by centrifugation at 9,500 × g for 10 min at 4°C.
Protein isolation was performed using immobilized metal affinity chromatography with Ni-NTA Sepharose 6FF resin (His-Tag specific, Sangon Biotech) through gravity-flow purification according to the manufacturer’s protocol. The resin (5 mL bed volume) was equilibrated with 10 column volumes (CV) of lysis buffer (50 mM phosphate buffer, 200 mM NaCl, 10 mM imidazole, pH 7.5) at a natural flow rate controlled by column height adjustment. After loading the crude enzyme lysate, the column was sequentially washed with 5 CV of low-imidazole buffer (10 mM imidazole in lysis buffer) followed by 5 CV of intermediate-imidazole buffer (35 mM imidazole in lysis buffer) to remove nonspecifically bound proteins. His-tagged target proteins were eluted with 3 CV of high-imidazole buffer (250 mM imidazole in lysis buffer), and the eluate was collected in 2 mL microcentrifuge tubes. The collected fractions were immediately subjected to buffer exchange using dialysis against 50 mM PIPES (pH 6.0) at 4°C.
Recombinant cell lysate and purified protein was analyzed by 12% SDS-PAGE (150 V for 90 min, Bio-Rad Mini-PROTEAN) with Coomassie Blue staining. Quantitative analysis used a BCA assay with BSA standards, measured at 562 nm.
The enzymatic activity of Acba-CE toward lactose was assessed by monitoring substrate conversion through high performance liquid chromatography (HPLC). Reactions were conducted in 0.4 mL systems containing 200 mM lactose dissolved in 50 mM PIPES buffer (pH 6.0, adjusted with 5 M NaOH) with 0.02 mg/mL enzyme. After incubation at 60°C for 15 min in a temperature-controlled thermal cycler, reactions were terminated by boiling at 100°C for 3 min. Terminated samples were centrifuged at 7,000 × g for 3 min. To remove residual proteins, trichloroacetic acid (TCA) was added to a final concentration of 15% (w/v). The clarified supernatants were filtered through 0.22 μm hydrophilic nylon membranes prior to HPLC analysis as described in subsequent sections.
The enzymatic activity of Acba-CE was determined by quantifying epilactose production via HPLC. Enzyme activity units (U) were defined as the amount of enzyme required to generate 1 μmol of epilactose per minute under standard conditions (60°C, pH 6.0), calculated based on epilactose concentrations derived from a 0.1–20 mM calibration curve of purified standard. Negative controls with heat-inactivated enzyme (100°C, 10 min pretreatment) demonstrated negligible background conversion (<2%). All assays included triplicate technical replicates, with inter-experimental variation controlled to <5% coefficient of variation through standardized protocols.
The optimal temperature and pH of Acba-CE were determined by quantifying epilactose production via HPLC across temperature (4–90°C) and pH gradients. Temperature profiling was performed using a calibrated thermal block, with 5 min pre-incubation at each temperature. For pH optimization, three buffer systems were employed: 50 mM sodium acetate–acetic acid (pH 4.0–6.0), 50 mM PIPES (pH 6.0–7.5), and 50 mM HEPES (pH 7.5–8.5). All activity values were calculated based on epilactose generation, quantified against a 0.1–20 mM epilactose calibration curve.
Thermal stability was evaluated by pre-incubating the enzyme at 50–70°C for a period of time in assay buffer. Residual activity was defined as the percentage of epilactose produced relative to untreated controls (100% activity), measured under standard conditions (60°C, pH 6.0).
Michaelis–Menten kinetics were analyzed through nonlinear regression (Origin 2021) using epilactose production rates. Reactions containing 0.02 mg/mL purified enzyme were conducted at pH 6.0 and 60°C with lactose concentrations spanning 30–800 mM (8 gradient points).
The time-dependent catalytic conversion of lactose by Acba-CE was evaluated through parallel reaction systems. For lactose solution studies, individual 1.5 mL microcentrifuge tubes containing 0.4 mL reaction mixtures (0.5 mg/mL enzyme, 200 mM lactose in 50 mM PIPES pH 6.0) were incubated at 50°C. Triplicate samples were independently prepared for each time point (2, 6, 12, 24, 36, and 48 h). At designated intervals, entire reaction volumes (0.4 mL) were heat-inactivated (100°C, 3 min), followed by centrifugation (8,000 × g, 3 min), TCA precipitation, and filtration as per standard protocols.
Milk-based reactions were conducted in 5 mL microcentrifuge tubes with 1 mL systems, comprising 850 μL pasteurized milk (50 g/L carbohydrates) and 150 μL enzyme solution (final 1 mg/mL Acba-CE). Triplicate tubes were incubated at 10°C for 12 h. Reactions were terminated by heating (100°C, 3 min), followed by centrifugation (8,000 × g, 10 min), TCA precipitation, and filtration as per standard protocols. Lactose concentrations were corrected for initial 15% dilution during data normalization.
Quantitative analysis of epilactose, lactulose, and lactose was performed using an HPLC system (Waters Alliance e2695, USA) equipped with a differential refractive index detector (RID, Waters 2,414, USA), following chromatographic conditions from previously reported methodologies (Chen et al., 2016). Separation was achieved on a Shodex VG50-4E carbohydrate column (4.6 mm × 250 mm, 5 μm particle size) maintained at 40°C. The mobile phase consisted of acetonitrile/methanol/water (75:20:5, v/v/v) delivered isocratically at 1.0 mL/min. Sample injections of 20 μL were performed using an autosampler after filtration through 0.22 μm nylon membranes. Analyte identification was confirmed by retention time alignment with authentic standards, and quantification utilized external calibration curves.
Collect currently reported 19 kinds of CE sequence using the basic local alignment search tool1 of Acba-CE and the CE sequence similarity comparison, and use the ClustalW neighborhood connection method to construct phylogenetic tree.
Multiple sequence alignments and phylogenetic trees were used to analyze the Acba-CE sequences mined from the UniProtKB TrEMBL database. The Acba-CE sequence was identified through a targeted screening strategy using the UniProtKB query system: initial retrieval of all unreviewed entries annotated as “cellobiose 2-epimerase” (2,265 candidates) followed by taxonomy filtering for Acidobacteriota phylum organisms inhabiting acidic environments (5 candidates). The final prioritization is based on structural homology with characterized CE enzymes, preferentially selecting candidates that exhibit higher homology to CE enzymes with lower optimal temperatures and lower optimal pH.
The phylogenetic tree in Figure 2 shows that the Acba-CE clusters within a clade containing experimentally validated epimerases, despite moderate global sequence similarity (35–50% identity range). Among the compared CEs, Acba-CE exhibits the highest sequence similarity (46.61%) with the CE enzyme derived from Pedobacter heparinus (Krewinkel et al., 2014). According to previously reported literature, PhCE is a mesophilic enzyme with an optimal pH in the mildly acidic range, showing properties closely aligned with the target enzyme in this study (adapted to mildly acidic and low-temperature catalytic environments). Key catalytic residues were fully conserved across aligned sequences (Figure 3), while variable loop regions (e.g., Acba-CE_loop 175–186 and loop 236–248) exhibited less than 20% similarity, potentially explaining substrate specificity differences. Quantitative similarity metrics across functional domains are presented in Table 2.
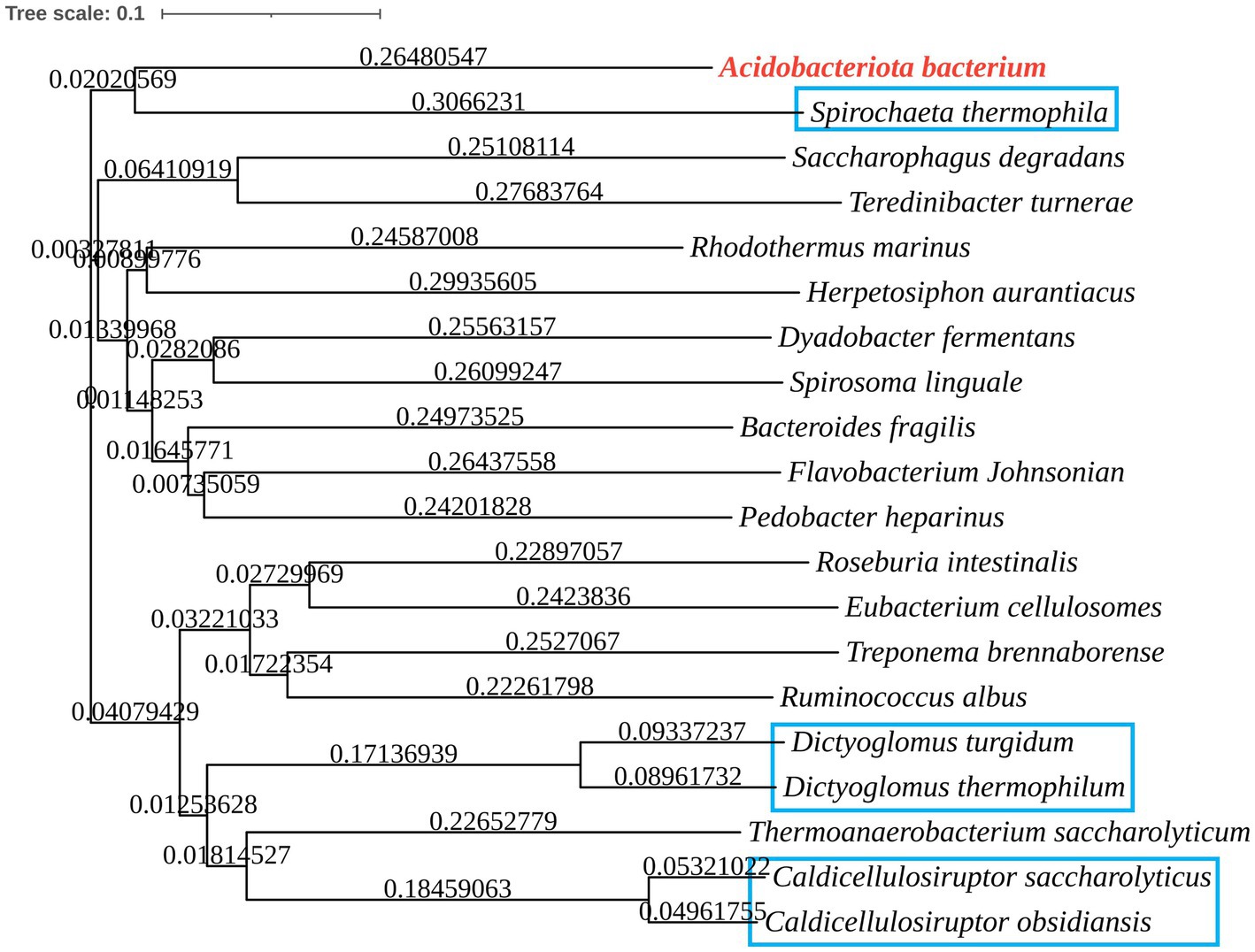
Figure 2. Phylogenetic tree of CEs from various microorganisms, comparing some reported CE enzymes with Acba-CE (blue box for CE producing lactulose).
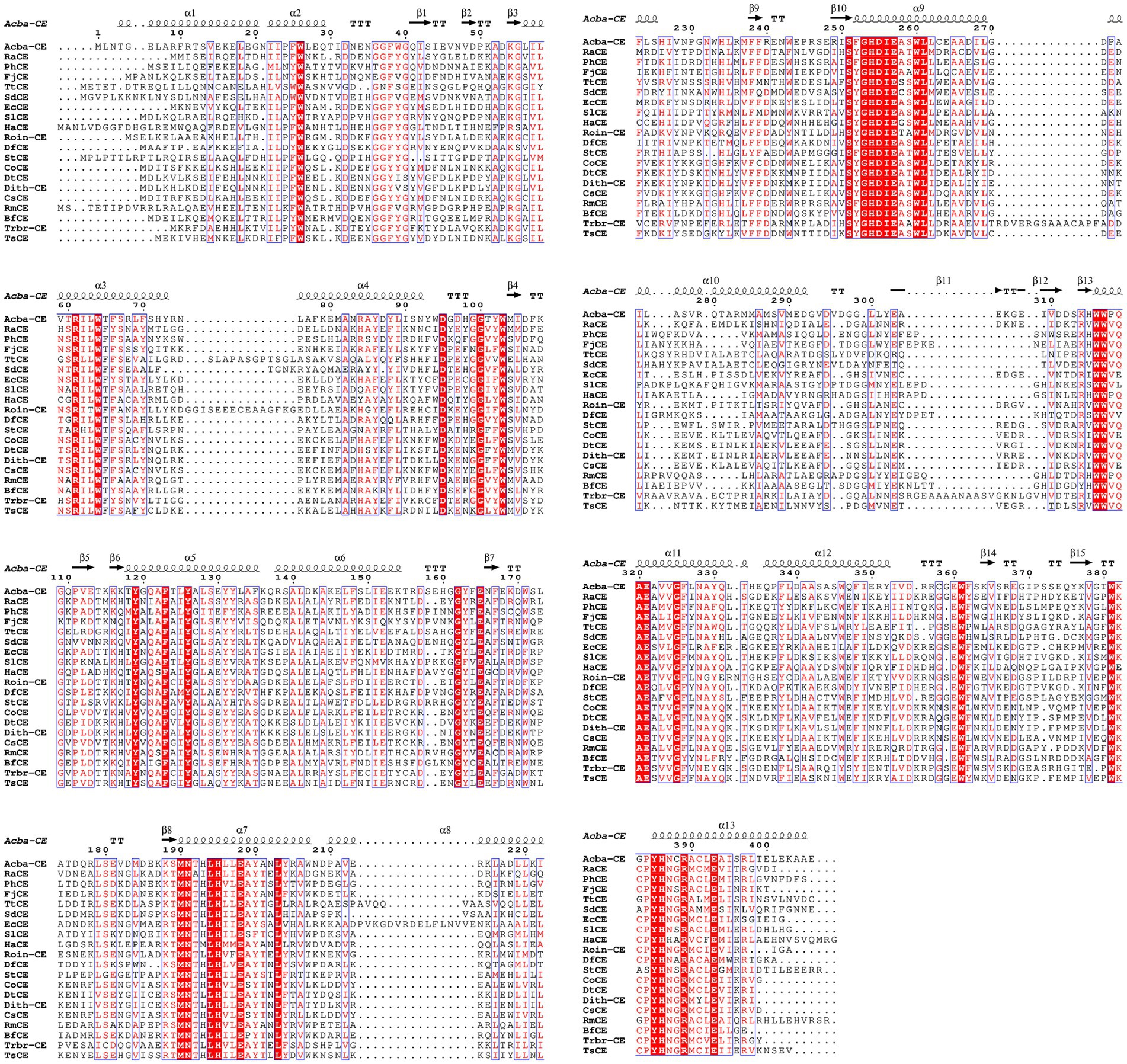
Figure 3. Multiple sequence alignment of the selected CEs. The alignment and its secondary structure analysis were performed using ClustalOmega and Espript 3.0. Three histidines involved in each catalytic center of CEs were symbolized according to the crystal structure of Acba-CE. The strict conserved residues are demonstrated by white characters with red boxes, red characters denote the residues with high similarity in each group. The residues with high similarity across the groups are boxed in blue frames.
To validate the expression and solubility of the recombinant protein, we have conducted SDS-PAGE analyses of both the supernatant and pellet fractions of the lysed recombinant cells (as shown in Figure 4a). The electrophoretic results clearly demonstrate successful protein expression, with distinct bands corresponding to the expected molecular weight of the target protein. This experiment confirms the effective expression and partial solubility of the recombinant enzyme, providing critical support for the subsequent functional characterization described in the study.
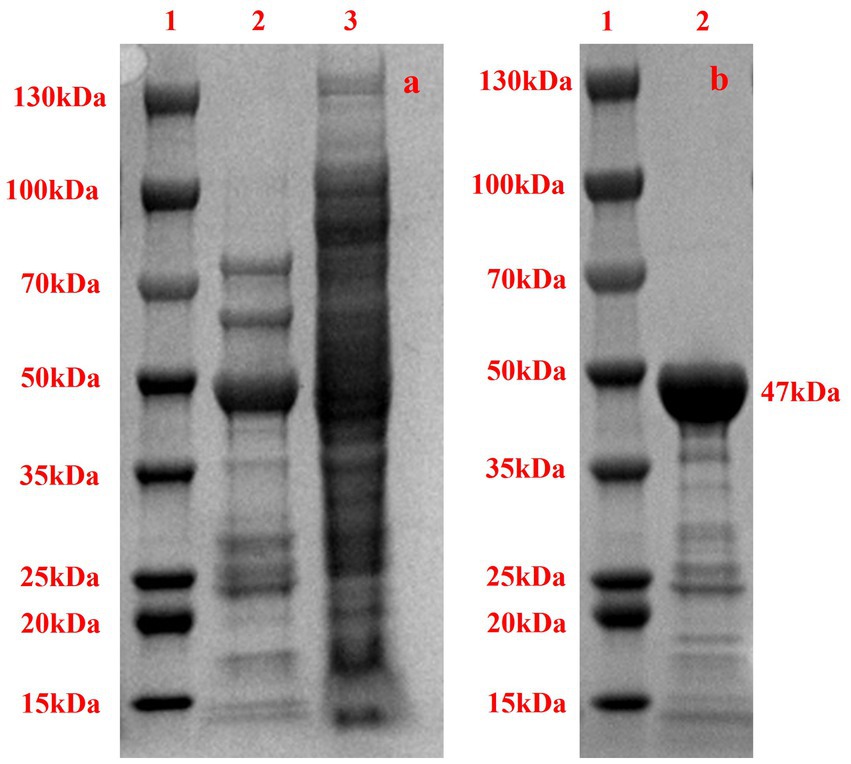
Figure 4. SDS-PAGE of Acba-CE. (a) SDS-PAGE analysis of recombinant cell lysate. Lane 1: Protein molecular weight marker. Lane 2: Pellet fraction after cell lysis. Lane 3: Supernatant fraction after centrifugation. (b) Recombinant Acba-CE, purified from E. coli transformant, were analyzed on SDS-PAGE. Lane 1 indicates molecular mass standard, with the size of each marker indicated in the figure. Lane 2 indicates purified Acba-CE protein.
To obtain pure Acba-CE enzyme for subsequent assays, the recombinant protein with a His tag, induced by IPTG, was purified using a Ni2+ column. SDS-PAGE analysis of the purified protein (Figure 4b) revealed that the size of the purified protein was approximately 47 kDa, which is generally consistent with the theoretical size of 47 kDa. Additionally, the electrophoresis diagram demonstrated that the purified solution contained only a small amount of other impurity proteins, making it suitable for further research.
The Acba-CE produces epilactose when catalyzing lactose and does not form lactulose, demonstrating the typical characteristics of a mesophilic CE enzyme. The activity of catalyzing lactose to epilactose is 37.3 ± 1.3 U/mg. Under standard assay conditions, the optimal temperature for Acba-CE was found to be 60°C (Figure 5a), which aligns with the general characteristics of mesophilic CE enzymes. While the enzyme exhibits residual activity at 70°C-90°C(~30–40%), its significant activity loss after prolonged incubation indicates limited thermal stability. Additionally, Acba-CE exhibits stronger cold adaptability compared to heat adaptability, with a lesser decrease in enzyme activity when the temperature drops than when it rises, as also reflected in the thermostability assay of Acba-CE (Figures 5c,d). When Acba-CE is incubated at different temperatures, the enzyme activity decreases significantly after incubation at 60°C for a period of time, with a more pronounced decrease than at 50°C, and the enzyme is inactivated after incubation at 70°C for half an hour.
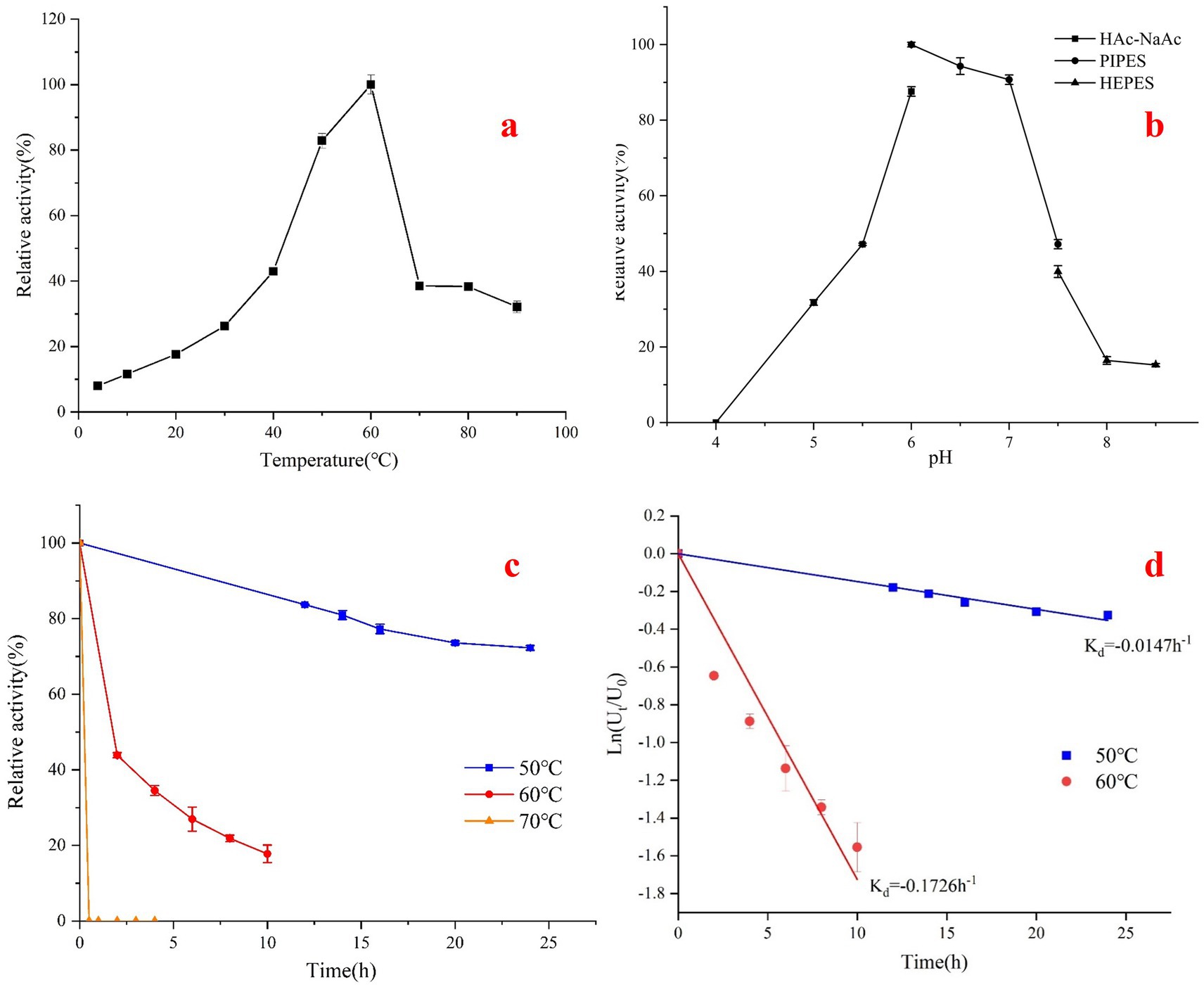
Figure 5. The optimum temperature, pH and thermal stability of Acba-CE were determined. (a) Optimal temperature. The reaction was carried out in 50 mM PIPES buffer (pH 6.0) at 4–90°C, supplemented with lactose substrate at a final concentration of 200 mM. (b) Optimal pH. The reaction was carried out in 50 mM PIPES buffer (pH 4.0–8.5) at 60°C with a final concentration of 200 mM lactose substrate. (c) Thermal stability and (d) half-life assays were performed in PIPES buffer (pH 6.0) at 50 mM after treatment at temperatures 50 and 60°C, followed by addition of lactose substrate at a final concentration of 200 mM at 60°C.
Under standard assay conditions, the optimal pH for Acba-CE was determined to be 6.0 (Figure 5b). Additionally, Acba-CE exhibited high activity within the pH range of 6.0–7.0, with relative activity consistently maintained above 90%. Outside this range, both an increase and a decrease in pH resulted in a significant reduction in enzyme activity. As Acidobacteriota bacterium is an acidophile, the enzyme isolated from it also demonstrated a certain degree of acidophilia, making it more suitable for application in slightly acidic environments. The stability of Acba-CE under acidic conditions may be related to the distribution of surface acidic amino acids. According to calculations of the solvent-accessible surface area, the proportion of acidic amino acids on the surface of Acba-CE reaches 27%, whereas the proportion on the commonly used mesophilic RaCE surface is only 22%. The elevated density of surface-exposed acidic amino acids potentially enhances the protein’s capacity for proton binding in acidic environments.
The kinetic parameters of Acba-CE were determined within the range of 30 mM to 800 mM, with a Km value of 147.4 ± 10.6 mM and a kcat of 35.02 ± 0.94 s−1 when lactose was used as the substrate.
Compared to the CE enzymes from other sources listed in Table 1, only two CEs exhibit lower optimal pH values. Most CEs have optimal pH ranges in the neutral to alkaline region, and their residual activity decreases to varying degrees when the pH drops to mildly acidic conditions. The most significant activity loss is observed in the enzyme from Ruminococcus albus (Ito et al., 2007), which retains only 25% activity at pH 6.5. Although the CE from Rhodothermus marinus (Ojima et al., 2014) can catalyze reactions under mildly acidic conditions, it is a thermophilic enzyme unsuitable for low-temperature processing environments and difficult to inactivate by heat. While the CE from Pedobacter heparinus (Krewinkel et al., 2014) meets the requirements for mildly acidic and low-temperature processing, Acba-CE demonstrates higher kcat and kcat/Km values, indicating superior catalytic efficiency.
The catalytic efficiency of Acba-CE was systematically evaluated across distinct operational conditions to assess its adaptability and industrial potential. Due to the poor thermal stability of Acba-CE, the protein was easy to precipitate at 60°C for a long time. The conversion rate of Acba-CE catalyzed lactose to epilactose was determined at 50°C. In lactose solution systems (100 mM, pH 6.0) at 50°C, the enzyme exhibited remarkably rapid kinetics, achieving 28% lactose-to-epilactose conversion within the first 2 h—a value approaching the final equilibrium conversion rate of 30.6% observed after 48 h. Subsequent sampling intervals (6, 12, 24, 36, and 48 h) revealed only marginal increases in conversion (29.1% at 6 h; 30.6% at 48 h), indicating that the majority of substrate turnover occurred within the initial reaction phase. This accelerated equilibration likely stems from conformational flexibility in Acba-CE’s substrate-binding loops.
In contrast, when applied to milk systems (50 g/L lactose, pH 6.7) under refrigeration conditions (10°C), Acba-CE demonstrated robust cold adaptability, attaining 28.5 ± 1.5% conversion after 12 h. Despite a significant decrease in temperature, the enzyme still retained a high conversion rate compared to its near-optimal performance at 50°C. Notably, despite a slight decrease in the equilibrium conversion rate between purified lactose (30.6%) and the complex milk matrix (28.5%), the enzyme maintained a high conversion rate, confirming its adaptability to inhibitory milk components (e.g., casein and calcium ions)—a critical advantage for dairy applications.
The temperature-dependent divergence in reaction kinetics underscores Acba-CE’s operational versatility. While elevated temperatures (50°C) enable rapid substrate turnover for time-sensitive processes, the enzyme’s sustained activity at 10°C aligns with industrial cold-chain requirements. This dual functionality bridges the gap between catalytic efficiency and practical feasibility, allowing seamless integration into both batch processing (e.g., rapid enzymatic modification prior to pasteurization) and prolonged cold storage workflows. The enzyme’s rapid thermal inactivation at 70°C further ensures compatibility with existing dairy sterilization protocols, eliminating the need for post-reaction purification—a persistent challenge for thermostable epimerases.
This study establishes Acba-CE as a novel cellobiose 2-epimerase with unique functional attributes used for dairy biotechnology applications. By mining the UniProtKB TrEMBL database and prioritizing Acidobacteriota-derived sequences, we identified a CE enzyme exhibiting dual advantages of acidophilic activity and temperature-responsive inactivation. Heterologous expression in E. coli BL21(DE3) yielded a highly pure enzyme (>95%) with robust catalytic efficiency (Km value of 147.4 ± 10.6 mM and kcat of 35.02 ± 0.94 s−1).
Structurally, Acba-CE’s elevated surface density of acidic residues (27% vs. 22% in R. marinus CE) correlates with enhanced proton retention in acidic environments, while its flexible loop regions (e.g., residues 175–186) likely contribute to cold adaptability. These traits collectively position Acba-CE as a biocatalyst uniquely suited for minimally processed dairy products, where preserving native flavor profiles and nutritional integrity is paramount.
Acba-CE’s optimal pH of 6.0 and sustained activity (>90%) within the pH 6.0–7.0 range align precisely with the natural acidity of milk (pH 6.5–6.8), eliminating the need for pH adjustment during processing. The enzyme’s 29 ± 1.5% lactose-to-epilactose conversion in raw milk at 10°C—demonstrates its potential for energy-efficient cold-processing workflows. Meanwhile, the heat-labile nature of Acba-CE under high-temperature processing ensures minimal post-process enzymatic activity, thereby eliminating the need for downstream purification to remove residual enzymes—a critical bottleneck in current dairy enzyme modification practices.
Looking forward, Acba-CE’s operational stability in milk matrices and compatibility with refrigeration temperatures open avenues for sustainable dairy processing. Future efforts should focus on loop engineering to enhance substrate affinity and immobilization strategies to extend catalytic longevity, ultimately bridging the gap between laboratory-scale efficacy and industrial-scale implementation.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/supplementary material.
QZ: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Supervision, Validation, Visualization, Writing – original draft, Writing – review & editing. XL: Conceptualization, Funding acquisition, Investigation, Project administration, Resources, Supervision, Writing – review & editing.
The author(s) declare that financial support was received for the research and/or publication of this article. This work was funded by National Key Research and Development Program of China (2022YFF1100300), Fundamental Research Funds for the Central Universities (JUSRP622024), Taihu Innovation-Leading Talent of Wuxi City (1026010241230040), and Cross-integration innovation funding of SFST (SFST2023-KY-10).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The authors declare that no Gen AI was used in the creation of this manuscript.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Angima, G., Qu, Y., Park, S. H., and Dallas, D. C. (2024). Prebiotic strategies to manage lactose intolerance symptoms. Nutrients 16:1002. doi: 10.3390/nu16071002
Chen, Q., Levin, R., Zhang, W., Zhang, T., Jiang, B., Stressler, T., et al. (2016). Characterisation of a novel cellobiose 2-epimerase from thermophilic Caldicellulosiruptor obsidiansis for lactulose production. J. Sci. Food Agric. 97, 3095–3105. doi: 10.1002/jsfa.8148
Chen, Q., Wu, Y., Huang, Z., Zhang, W., and Mu, W. (2021). Molecular characterization of a mesophilic cellobiose 2-epimerase that maintains a high catalytic efficiency at low temperatures. J. Agric. Food Chem. 69, 8268–8275. doi: 10.1021/acs.jafc.1c02025
Chen, Q., Xiao, Y., Zhang, W., Stressler, T., Fischer, L., Jiang, B., et al. (2020). Computer-aided search for a cold-active cellobiose 2-epimerase. J. Dairy Sci. 103, 7730–7741. doi: 10.3168/jds.2020-18153
Chen, Q., Zhang, W., Zhang, T., Jiang, B., and Mu, W. (2015). Characterization of an epilactose-producing cellobiose 2-epimerase from Thermoanaerobacterium saccharolyticum. J. Mol. Catal. B Enzym. 116, 39–44. doi: 10.1016/j.molcatb.2015.03.005
Eat, S., Wulansari, S., Ketbot, P., Waeonukul, R., Pason, P., Uke, A., et al. (2024). A novel cellobiose 2-epimerase from anaerobic halophilic Iocasia fonsfrigidae and its ability to convert lactose in fresh goat milk into epilactose. J. Sci. Food Agric. 104, 8529–8540. doi: 10.1002/jsfa.13680
Fujiwara, T., Saburi, W., Matsui, H., Mori, H., and Yao, M. (2014). Structural insights into the epimerization of β-1,4-linked oligosaccharides catalyzed by cellobiose 2-epimerase, the sole enzyme epimerizing non-anomeric hydroxyl groups of unmodified sugars. J. Biol. Chem. 289, 3405–3415. doi: 10.1074/jbc.M113.531251
Ito, S., Hamada, S., Yamaguchi, K., Umene, S., Ito, H., Matsui, H., et al. (2007). Cloning and sequencing of the cellobiose 2-epimerase gene from an obligatory anaerobe, Ruminococcus albus. Biochem. Biophys. Res. Commun. 360, 640–645. doi: 10.1016/j.bbrc.2007.06.091
Ito, S., Taguchi, H., Hamada, S., Kawauchi, S., Ito, H., Senoura, T., et al. (2008). Enzymatic properties of cellobiose 2-epimerase from Ruminococcus albus and the synthesis of rare oligosaccharides by the enzyme. Appl. Microbiol. Biotechnol. 79, 433–441. doi: 10.1007/s00253-008-1449-7
Kim, J.-E., Kim, Y.-S., Kang, L.-W., and Oh, D.-K. (2012). Characterization of a recombinant cellobiose 2-epimerase from Dictyoglomus turgidum that epimerizes and isomerizes β-1,4- and α-1,4-gluco-oligosaccharides. Biotechnol. Lett. 34, 2061–2068. doi: 10.1007/s10529-012-0999-z
Kim, Y.-S., and Oh, D.-K. (2012). Lactulose production from lactose as a single substrate by a thermostable cellobiose 2-epimerase from Caldicellulosiruptor saccharolyticus. Bioresour. Technol. 104, 668–672. doi: 10.1016/j.biortech.2011.11.016
Krewinkel, M., Gosch, M., Rentschler, E., and Fischer, L. (2014). Epilactose production by 2 cellobiose 2-epimerases in natural milk. J. Dairy Sci. 97, 155–161. doi: 10.3168/jds.2013-7389
Ludwig, W., Bauer, S. H., Bauer, M., Held, I., Kirchhof, G., Schulze, R., et al. (2006). Detection and in situ identification of representatives of a widely distributed new bacterial phylum. FEMS Microbiol. Lett. 153, 181–190. doi: 10.1111/j.1574-6968.1997.tb10480.x
Nishimukai, M., Watanabe, J., Taguchi, H., Senoura, T., Hamada, S., Matsui, H., et al. (2008). Effects of epilactose on calcium absorption and serum lipid metabolism in rats. J. Agric. Food Chem. 56, 10340–10345. doi: 10.1021/jf801556m
Ojima, T., Saburi, W., Sato, H., Yamamoto, T., Mori, H., and Matsui, H. (2014). Biochemical characterization of a thermophilic cellobiose 2-epimerase from a Thermohalophilic bacterium, Rhodothermus marinusjcm9785. Biosci. Biotechnol. Biochem. 75, 2162–2168. doi: 10.1271/bbb.110456
Pang, B., Yang, J., Song, M., Zhang, W., Qian, S., Xu, M., et al. (2025). Advances and prospects on production of lactulose and epilactose by cellobiose 2-epimerases: a review. Int. J. Biol. Macromol. 305:141283. doi: 10.1016/j.ijbiomac.2025.141283
Park, C.-S., Kim, J.-E., Lee, S.-H., Kim, Y.-S., Kang, L.-W., and Oh, D.-K. (2013). Characterization of a recombinant mannobiose 2-epimerase from Spirochaeta thermophila that is suggested to be a cellobiose 2-epimerase. Biotechnol. Lett. 35, 1873–1880. doi: 10.1007/s10529-013-1267-6
Rentschler, E., Schuh, K., Krewinkel, M., Baur, C., Claaßen, W., Meyer, S., et al. (2015). Enzymatic production of lactulose and epilactose in milk. J. Dairy Sci. 98, 6767–6775. doi: 10.3168/jds.2015-9900
Senoura, T., Taguchi, H., Ito, S., Hamada, S., Matsui, H., Fukiya, S., et al. (2014). Identification of the cellobiose 2-epimerase gene in the genome of Bacteroides fragilis nctc 9343. Biosci. Biotechnol. Biochem. 73, 400–406. doi: 10.1271/bbb.80691
Suzuki, T., Nishimukai, M., Takechi, M., Taguchi, H., Hamada, S., Yokota, A., et al. (2010). The nondigestible disaccharide Epilactose increases Paracellular ca absorption via rho-associated kinase- and myosin light chain kinase-dependent mechanisms in rat small intestines. J. Agric. Food Chem. 58, 1927–1932. doi: 10.1021/jf9035063
Szilagyi, A., and Ishayek, N. (2018). Lactose intolerance, dairy avoidance, and treatment options. Nutrients 10:1994. doi: 10.3390/nu10121994
Taguchi, H., Senoura, T., Hamada, S., Matsui, H., Kobayashi, Y., Watanabe, J., et al. (2008). Cloning and sequencing of the gene for cellobiose 2-epimerase from a ruminal strain of Eubacterium cellulosolvens. FEMS Microbiol. Lett. 287, 34–40. doi: 10.1111/j.1574-6968.2008.01281.x
Watanabe, J., Nishimukai, M., Taguchi, H., Senoura, T., Hamada, S., Matsui, H., et al. (2008). Prebiotic properties of epilactose. J. Dairy Sci. 91, 4518–4526. doi: 10.3168/jds.2008-1367
Xiao, Y., Chen, Q., Shakhnovich, E. I., Zhang, W., and Mu, W. (2019). Simulation-guided enzyme discovery: a new microbial source of cellobiose 2-epimerase. Int. J. Biol. Macromol. 139, 1002–1008. doi: 10.1016/j.ijbiomac.2019.08.075
Keywords: cellobiose 2-epimerase, acidophilic enzyme, milk processing, low-lactose milk, epilactose, prebiotics
Citation: Zeng Q and Lyu X (2025) Identification of a novel cellobiose 2-epimerase from Acidobacteriota bacterium and its application for in-situ milk catalysis. Front. Microbiol. 16:1575725. doi: 10.3389/fmicb.2025.1575725
Received: 12 February 2025; Accepted: 20 March 2025;
Published: 04 April 2025.
Edited by:
Haifeng Zhao, South China University of Technology, ChinaReviewed by:
Sazzad Hossen Toushik, North South University, BangladeshCopyright © 2025 Zeng and Lyu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiaomei Lyu, eG1seXVAamlhbmduYW4uZWR1LmNu
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.