
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol., 21 March 2025
Sec. Microbe and Virus Interactions with Plants
Volume 16 - 2025 | https://doi.org/10.3389/fmicb.2025.1573724
Introduction: The pomegranate (Punica granatum) is a significant economic tree species. In recent years, the root collar rot has severely affected pomegranates in the dry-hot valley regions of Yunnan Province, China. The rhizosphere microbiome plays a crucial role in plant growth, development, and disease resistance.
Methods: This study utilized Illumina MiSeq sequencing to analyze the fungal communities in the roots and rhizosphere soils of healthy and diseased pomegranates, focusing on the impact of root collar rot disease on the diversity and structural composition of these communities.
Results: The results indicated that in the unique fungal communities of healthy plant roots, the relative abundance of ectomycorrhizal and arbuscular mycorrhizal functional (AMF) groups was 53.77%, including genera such as Glomus and Septoglomus. After infection with root collar rot disease, the rhizosphere fungal communities became more monotonous, with increased differentiation within sample groups. Fungal groups associated with plant diseases and soil nutrient structures underwent significant changes. The disease altered the composition and functional group proportions of rhizosphere fungal communities, a process linked to soil nutrient structures. And the balance between plant-pathogen-related and saprotrophic functional groups in the rhizosphere was disrupted. Through Koch’s postulates verification, the pathogen was identified as Lauriomyces bellulus.
Discussion: This is the first report of collar rot of pomegranate caused by L. bellulus in China. Studying the differences in rhizosphere fungal community structures and quantities in response to new diseases aids in the rapid prediction of pathogens, effectively saving diagnostic time, and provides theoretical support for disease prediction, diagnosis, and control.
Pomegranate (Punica granatum L.) has been widely cultivated for its nutritional and medicinal value, strong environmental adaptability, ease of management, and high economic benefits. In recent years, root collar rot has become increasingly prevalent in the dry-hot valley regions of Yunnan Province, China, causing significant economic losses for local fruit growers. This soilborne disease occurs when the base of the plant stem and the root collar are infected by pathogens. Internationally, Phytophthora species have been identified as common pathogens (Basım and Basım, 2013; Kurbetli et al., 2020; Ghaderi and Habibi, 2021). However, in China, there is currently no documentation of these pathogens or the associated changes in fungal community structure in the rhizosphere under disease-inducing conditions.
The rhizosphere encompasses the plant roots and surrounding soil. It is a highly dynamic region where material and information exchange occur between plants, soil, and microorganisms. Rhizosphere microorganisms are crucial for plant growth and development, and they are often the primary source of pathogens for soilborne diseases like root collar rot (Sasse et al., 2018). For instance, Li et al. (2021) utilized amplicon sequencing to compare fungal community structures in healthy and root rot infected Astragalus roots, identifying Fusarium oxysporum as the probable root rot pathogen, which was later confirmed through pathogenicity assays. Long-term monoculture and excessive fertilizer use disrupt soil physicochemical properties, leading to soil eutrophication and changes in microbial communities. This reduces rhizosphere fungal diversity and richness, creating conditions conducive to outbreaks of harmful microorganisms (Zuppinger-Dingley et al., 2014; Liu et al., 2015; Wang et al., 2022). Pathogen invasion or environmental changes can alter plant physiological and metabolic pathways, thereby modifying rhizodeposit composition. This, in turn, attracts specific soil microorganisms to the rhizosphere, either recruiting or reducing beneficial or pathogenic microbes, ultimately influencing plant growth and disease resistance (Berendsen et al., 2018; Yuan et al., 2018; Qi et al., 2019; You et al., 2024). For example, Berendsen et al. (2018) found that under stimulation from Hyaloperonospora arabidopsidis, the downy mildew pathogen, Arabidopsis selectively enriched three rhizosphere-specific bacteria that collaboratively induced systemic resistance against H. arabidopsidis and promoted plant growth. The abundance of key species is one of the factors reflecting microbial community functionality. In amplicon sequencing studies, dominant taxa with relative abundances greater than 1% often constitute a minority within the community but perform vital ecological roles. Rare taxa, with relative abundances below 0.1%, serve as a seed bank and contribute to nutrient cycling (Kurm et al., 2017), redox reactions (Hausmann et al., 2016), plant growth promotion, and resistance induction (Zhang et al., 2019).
This study analyzes the fungal diversity and functionality in the roots and rhizosphere soils of healthy and root collar rot-infected pomegranates. By comparing changes in fungal community structures and diversity, we identify differential species and preliminarily explore their potential ecological roles. These findings provide a foundation for identifying pathogens, understanding pathogenic mechanisms, and screening biocontrol strains for managing root collar rot in pomegranates.
Over the past 4 years, investigations on pomegranate root collar rot have been conducted in the dry-hot valley regions of Yunnan Province, China, and samples were collected in September 2023. Following preliminary investigations across multiple pomegranate-producing areas, a representative orchard (26°43′ N, 103°14′ E) with a certain scale was selected as the sampling site. Sampling sites were selected based on the following criteria: Consistent soil type; Representative dry-hot valley climate; Perennial pomegranate cultivation history; Uniform fertilization practices (type, rate, frequency); Root collar rot prevalence (>70% incidence); Randomized sampling design accounting for management variations.
Nine healthy pomegranate with uniform growth and nine pomegranate with consistent severity of root collar rot were selected. Roots and rhizosphere soil were collected from each plant. Samples from three plants were mixed to create one composite sample as one replicate, resulting in three composite samples per group. Healthy root samples were labeled as J1, J2, and J3, with corresponding rhizosphere soil samples labeled as K1, K2, and K3. Diseased root samples were labeled as B1, B2, and B3, with corresponding rhizosphere soil samples labeled as M1, M2, and M3. The sample types were categorized as follows: HR (healthy root), DR (diseased root), HS (healthy rhizosphere soil), and DS (diseased rhizosphere soil). The sampling method for individual plants as follow: Litter and surface soil layers were removed, and roots were excavated in a circle around the plant trunk at 20–50 cm from the trunk and 10–20 cm below the surface. Roots were extracted, and large soil clumps (over 15 cm in diameter) attached to the roots were broken apart. Fine roots and their adhering soil were collected into resealable bags and thoroughly mixed. Roots were shaken free and separated, mixed uniformly, and sealed in separate bags. Both soil and root samples were transported back to the laboratory in dry ice containers. Samples were categorized and labeled by location and replicate, then weighed, subsampled, and stored at −80°C for further analysis. Root surface sterilization was performed sequentially with: (1) 75% ethanol (40-s immersion); (2) 2.5% sodium cyclamate +0.01% Tween 80 (10-min immersion); Followed by 2–3 sterile water rinses.
Total DNA was extracted from root and soil samples using the E.Z.N.A.™ Mag-Bind Soil DNA Kit (OMEGA) following the manufacturer’s protocol. The integrity of the extracted DNA was assessed using 1% agarose gel electrophoresis, and genomic DNA was accurately quantified using the Qubit 3.0 DNA Assay Kit before proceeding to PCR amplification. The primers used for PCR included sequencing platform-compatible ITS1 (5′-CTTGGTCATTTAGAGGAAGTAA-3′) and ITS2 (5′-GCTGCGTTCTTCATCGATGC-3′) (White et al., 1990). First-round PCR: The amplification was conducted in a 30 μL reaction system containing: 15 μL of 2× Hieff® Robust PCR Master Mix, 1 μL of Bar-PCR primer F, 1 μL of primer R, 10–20 ng of DNA template, ddH₂O to make up to 30 μL. The thermal cycling conditions were as follows: 94°C for 3 min (initial denaturation); 5 cycles of 94°C for 30 s, 45°C for 20 s, and 65°C for 30 s; 20 cycles of 94°C for 20 s, 55°C for 20 s, and 72°C for 30 s; Final extension at 72°C for 5 min, and storage at 10°C. Second-round PCR: To incorporate Illumina bridge PCR-compatible primers, a second round of amplification was performed in a 30 μL reaction system containing: 15 μL of 2× Hieff® Robust PCR Master Mix, 1 μL of primer F, 1 μL of Index-PCR primer R, 20–30 ng of first-round PCR product, ddH₂O to make up to 30 μL. The thermal cycling conditions were as follows: 95°C for 3 min (initial denaturation); 5 cycles of 94°C for 20 s, 55°C for 20 s, and 72°C for 30 s; Final extension at 72°C for 5 min, and storage at 10°C.
For arbuscular mycorrhizal functional (AMF), the polymerase chain reaction was performed in a 50 μL mixture as follow: 16.2 μL of 2× Hieff® Robust PCR Master Mix, 2.5 μL of each primer (AMV4.5NF 5’-AAGCTCGTAGTTGAATTTCG-3′ and AMDGR 5’-CCCAACTATCCCTATTAATCAT-3′), 4 μL of DNA template. The following thermal profile was used: 95°C for 10 min; 94°C for 30 s, 55°C for 30s, 72°C for 1 min for 35 cycles; fnally 74°C for 9 min (Sato et al., 2005).
The PCR products were analyzed for library size using 2% agarose gel electrophoresis and quantified using a Qubit 3.0 fluorometer (Hu et al., 2017). Sequencing was performed by Bioengineering (Shanghai) Co., Ltd. on the Illumina MiSeq™/HiSeq™ Illumina MiSeq sequencing platform.
The physicochemical properties of soil were determined following the methods described in the Agricultural Industry Standards of the People’s Republic of China, the Forestry Industry Standards of the People’s Republic of China.
• Soil pH: Measured using the potentiometric method.
• Soil Organic Matter (OM): Determined by the potassium dichromate dilution method.
• Available Phosphorus (AP): Measured using the bicarbonate extraction-molybdenum-antimony anti-colorimetric method.
• Available and Slow-Release Potassium (AK): Determined by ammonium acetate extraction-flame photometry.
• Total Nitrogen (TN): Measured using an automatic nitrogen analyzer.
• Total Phosphorus (TP): Determined by the sodium hydroxide fusion-molybdenum-antimony anti-colorimetric method.
• Total Potassium (TK): Measured using the hydrofluoric acid digestion method.
• Nitrate Nitrogen (NO₃−-N) and Ammonium Nitrogen (NH₄+-N): Measured using the phenol disulfonic acid colorimetric method and indophenol blue colorimetric method, respectively.
Each group consisted of three replicates, with three tests conducted for each sample. Differences among groups were assessed using one-way analysis of variance (One-Way ANOVA) to determine statistical significance.
The DNA samples were sequenced on the Illumina MiSeq™/HiSeq™ platform. The raw sequencing data were processed using tools such as Cutadapt 1.18, PEAR 0.9.8, and PRINSEQ 0.20.4 for base recognition, sequence assembly, quality control, and filtering. The processed data were uploaded to the NCBI database (accession number: PRJNA1170523).
All sequences were clustered into operational taxonomic units (OTUs) using Usearch 11.0.667 at varying similarity levels. Subsequent analyses focused on OTUs clustered at a 97% similarity threshold for in-depth bioinformatic and statistical evaluations (Nguyen et al., 2016). Taxonomic annotation of OTU representative sequences was performed using the RDP Classifier Bayesian algorithm (version 2.12) and SINTAX, with database selection based on amplification types (Zhou et al., 2020). For ITS regions, UNITE was used for Blast-based annotation to identify closely related species. Community composition at phylum, class, order, family, and genus levels was summarized, and tables of relative abundance were created for fungal taxa across these taxonomic levels (Edgar, 2013; Bolger et al., 2014).
α-diversity indices were calculated by randomly subsampling a fixed number of sequences from each sample. Rarefaction curves were generated using R (4.2.1) to assess whether sequencing depth was sufficient, based on curve saturation. OTU clustering results were used to calculate sample library coverage and α-diversity indices such as the Simpson and Shannon indices using Mothur software. The t-test was employed to assess the significance of differences in diversity indices between groups.
Beta diversity was analyzed based on weighted normalized UniFrac distances. Principal coordinate analysis (PCoA) was conducted using the vegan package in R to visualize differences in fungal community composition between samples. Parametric tests were used to identify taxa with significant abundance differences across groups, and intergroup comparisons were performed using the Wilcoxon rank-sum test. Linear discriminant analysis (LDA) was applied to reduce data dimensionality and assess the influence of significantly different taxa, with an LDA threshold of 3. The all-against-all strategy was employed for LEfSe analysis. Redundancy analysis (RDA) was used to evaluate the relationships between environmental factors and microbial community distribution.
Analyses were performed in SPSS 26.0.0.0, and the differences among samples were assessed using a one-way ANOVA analysis, and the calculated means were subjected to Duncan’s multiple range test at p < 0.05.
Plants showing aerial symptoms were categorized by disease severity. Bark tissue from root collar lesions was excised, with samples collected from both advanced infection zones (with sporulating structures) and disease fronts (symptomatic tissue without sporulation). Samples were separately bagged, labeled, and transported to the laboratory for isolation.
Symptoms on the collars of diseased pomegranate were meticulously examined using Zeiss Discovery-V20, Leica M166FC, and Leica DM2500 microscopes. Conidiophore clusters from the affected tissue were collected into sterile water. The suspension in the centrifuge tubes was gently agitated to ensure an even distribution of conidia.
Pathogenicity assays were conducted on four pomegranate seedlings. The collars of these seedlings were sequentially washed with 75% ethanol, 1% sodium hypochlorite solution, and sterile water. Five small incisions were made on the young stems using a sterile needle at each of the five designated inoculation sites per seedling. Each site was then inoculated with the prepared conidium suspension. Three seedlings were treated with the suspension, while the fourth served as a control, receiving only sterile distilled water. The inoculated areas were covered with sterile, moistened absorbent cotton and sealed with a preservative film.
Genomic DNA was extracted from freshly harvested conidiophore clusters on pomegranate tissues using the CTAB method. Three ribosomal DNA regions were amplified with specific primers: NS1 and NS4 for SSU, LROR and LR7 for LSU, and ITS1 and ITS2 for ITS regions (White et al., 1990; Bunyard et al., 1994; Somrithipol et al., 2017). The resulting amplicons were sequenced by TsingKe Biotechnology. Sequence alignments and comparisons were performed using the NCBI database. Phylogenetic relationships were elucidated through Maximum Parsimony (MP) and Maximum Likelihood (ML) analyses.
A total of 12 samples, including both healthy and diseased pomegranate roots and their corresponding rhizosphere soils, were sequenced. The Sequencing information and α-diversity index at genus level of fungi and AMF were showed in Table 1.
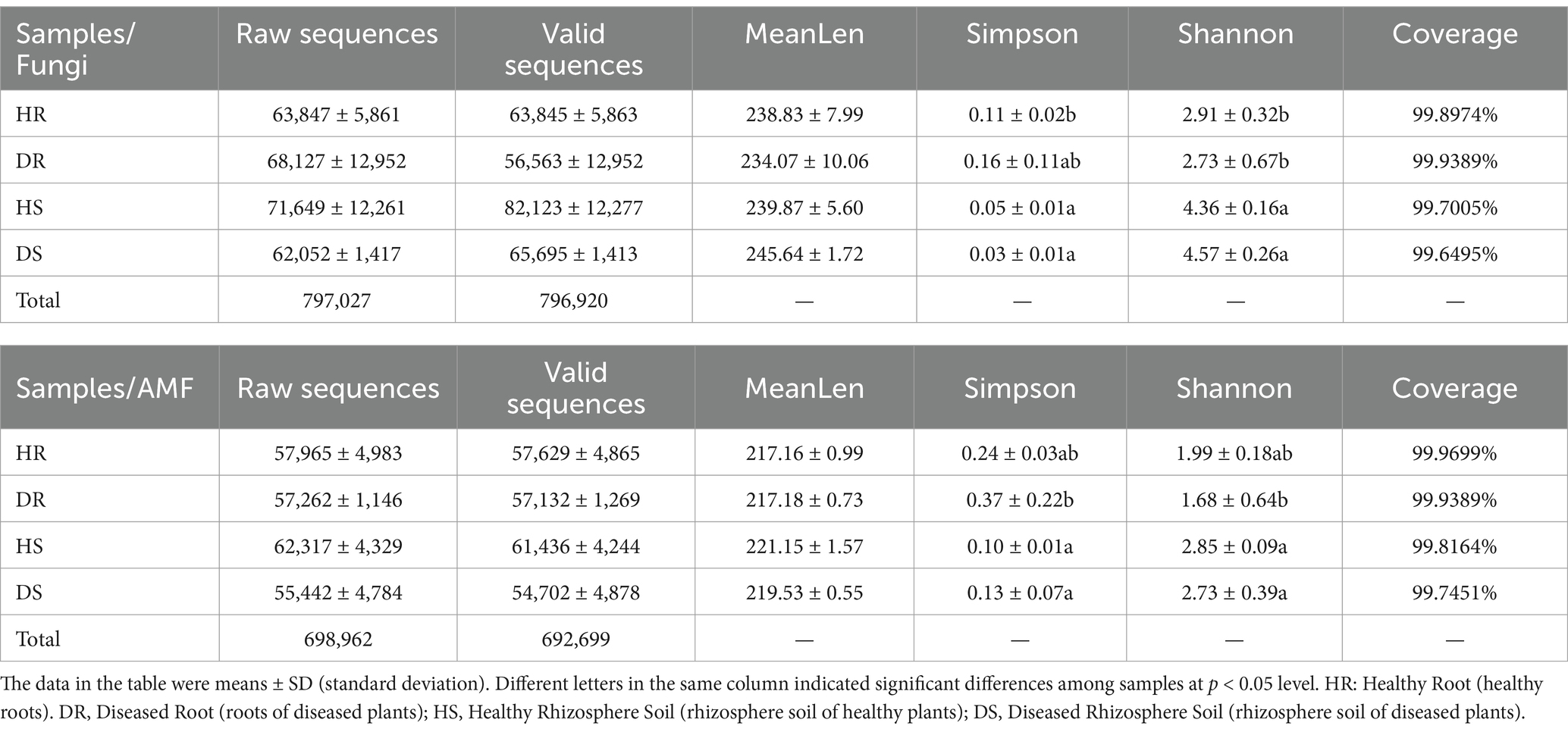
Table 1. Sequencing information and α-diversity index at genus level of root and rhizosphere soil of heathy and diseased pomegranate.
At the genus level, no significant α-diversity differences was observed between diseased and healthy roots. For fungal communities, the Simpson index for healthy root samples (HR) was significantly different from that of soil samples (p < 0.05). In contrast, Shannon indices at the genus level were significantly higher in soil samples compared to root samples (p < 0.05). The similar situation was appeared of AMF communities. Fungal abundance and community diversity were greater in soil samples than in root samples, with more evenly distributed species in soil. Pomegranate roots demonstrated strong selectivity for fungal colonization. The occurrence of root collar rot did not significantly affect fungal diversity or total fungal abundance in roots and rhizosphere soils. However, based on the Simpson index, species richness in diseased root and soil samples was lower, and community composition tended to become more homogeneous. The proportion of non-dominant species decreased, while the relative abundance of potential dominant species—likely pathogens of root collar rot—increased.
From the Beta diversity principal coordinate analysis (PCoA) plot, the fungal community (Figures 1A,B) composition within each group—HR, DR, HS, and DS—was relatively consistent. The contribution rates of PCoA1 and PCoA2 were 45.67 and 25.67%, respectively. Regardless of whether the samples were from roots or rhizosphere soil, there was significant clustering differentiation between diseased and healthy samples. This indicates that root collar rot in pomegranate had a significant impact on the fungal communities in both roots and the surrounding rhizosphere soil. Principal coordinates analysis of AMF β-diversity revealed disease-induced community shifts in both roots and rhizosphere soil (Figures 1C,D).
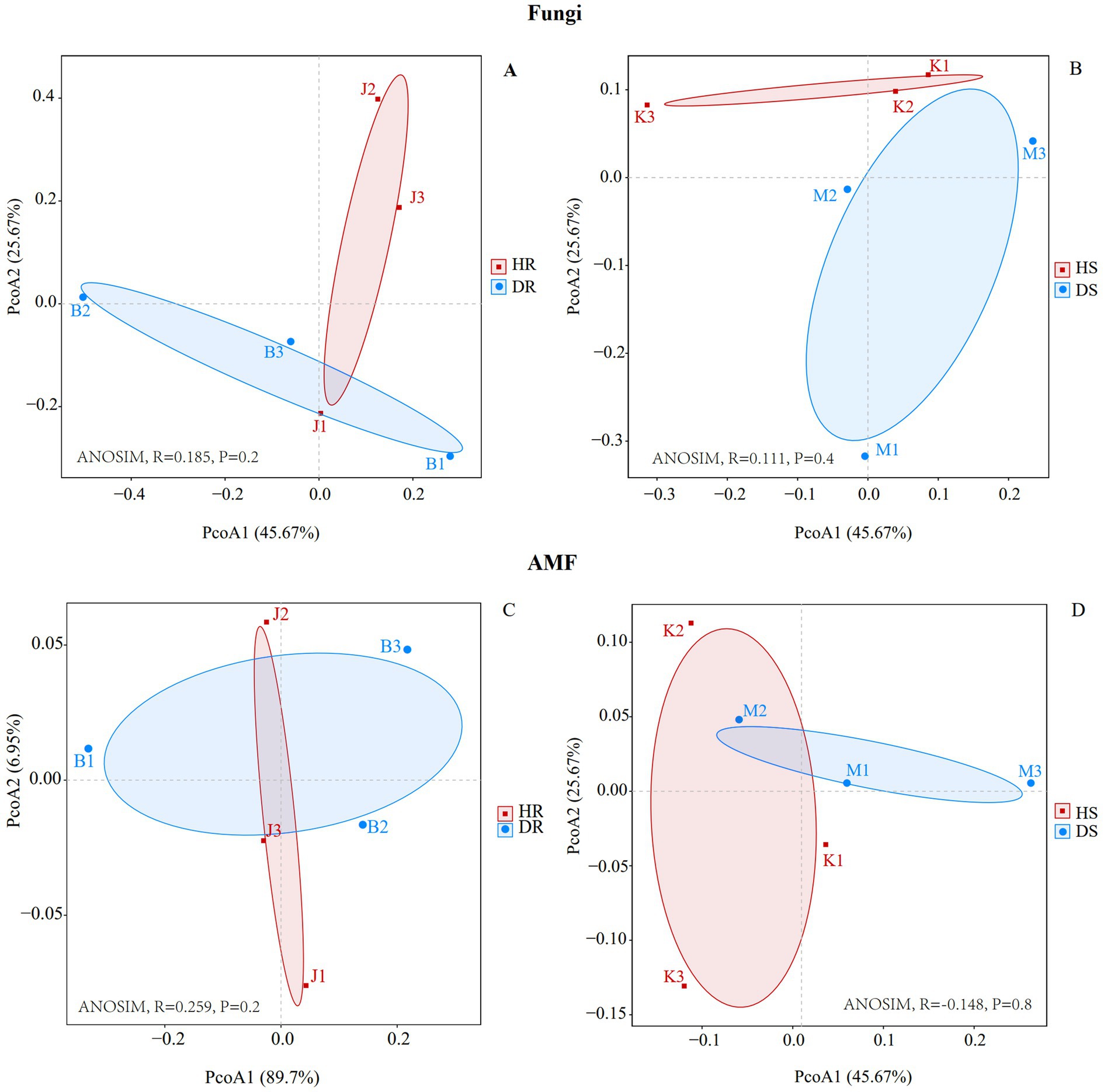
Figure 1. PCoA analysis of root (A), rhizosphere soil (B) fungi communities and root (C), rhizosphere soil (D) AMF communities at genus level. Healthy root (HR): J1/J2/J3. Diseased root (DR): B1/B2/B3. Healthy rhizosphere soil (HS): K1/K2/K3. Diseased rhizosphere soil (DS): M1/M2/M3.
A total of 1,472 fungal OTUs were identified from the diseased and healthy roots and rhizosphere soils. Based on a 97% sequence similarity threshold, fungal sequences were classified into 196–961 OTUs, primarily belonging to 4 phyla: Ascomycota, Basidiomycota, Mortierellomycota, and Chytridiomycota. These were further categorized into 10 classes, 23 orders, 36 families, and 41 genera.
A Venn diagram analysis (Figure 2A) of fungal OTUs in the four sample groups [healthy root (HR), diseased root (DR), healthy rhizosphere soil (HS), and diseased rhizosphere soil (DS)] revealed that 19.36% of OTUs were shared among all groups. The healthy root samples contained 10 unique fungal OTUs, annotated to genera such as Pezicula (a genus of inoperculate Discomycetes) and arbuscular mycorrhizal fungi including Septoglomus. In soil samples, a total of 1,436 OTUs were identified, with 14.55 and 8.29% of OTUs unique to healthy and rhizosphere soil, respectively, contributing to the overall richness. The dominant fungal taxa varied across different sample types. In healthy roots, the dominant genera were Oliveonia (15.44%) and Dactylonectria (12.25%), while in diseased roots, Lauriomyces (20.36%) and Fusidium (16.65%) were predominant. The diseased root samples contain 18 unique fungal OTUs, among which 5 genera of them annotated as Amphobotrys, Paraconiothyrium, Phaeomoniella, Thyronectria and Truncatella have been reported to cause plant diseases (Guarnaccia et al., 2022; Sokolova et al., 2022; Ji et al., 2023; Greeshma et al., 2024; Li et al., 2024). Venn analysis identified only one unique OTU of AMF (Figure 2B) present in diseased roots was assigned to Paraglomus (Supplementary Tables 1, 2).
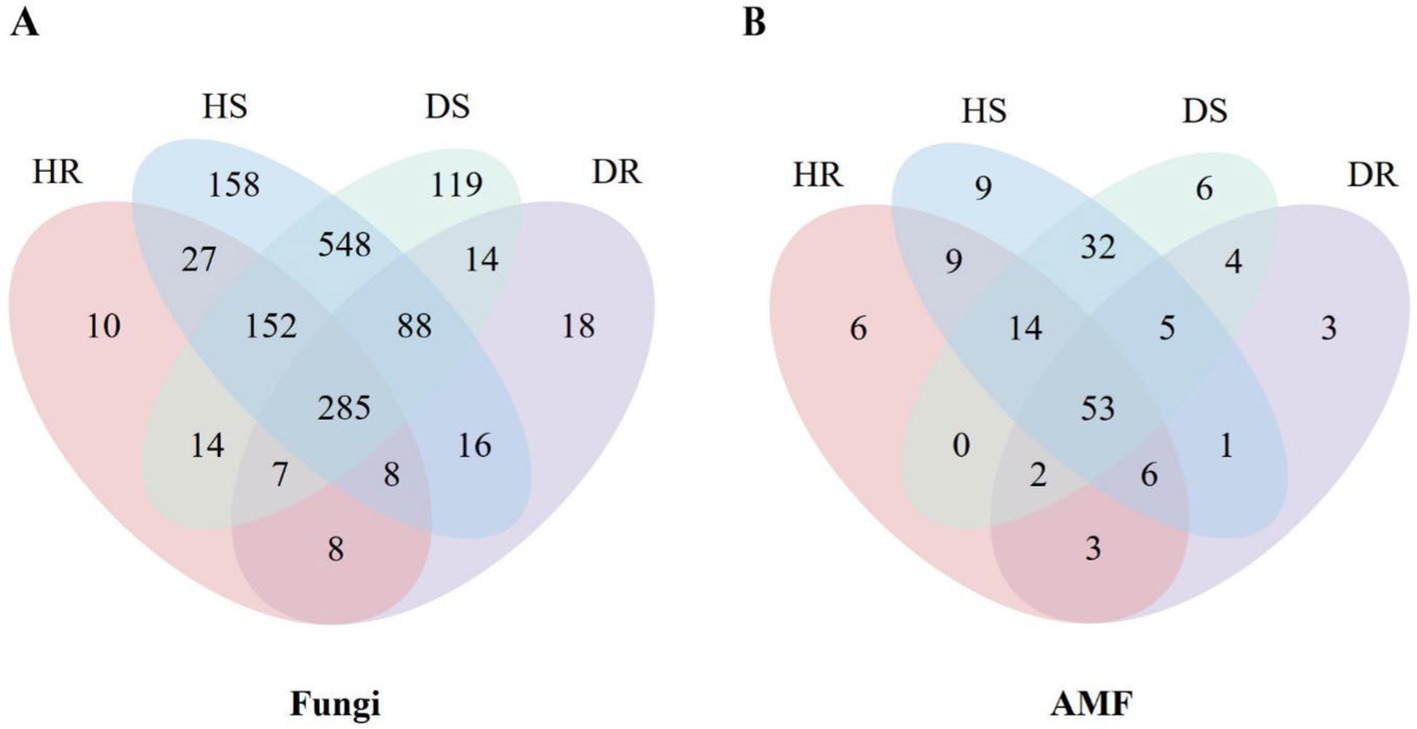
Figure 2. Venn diagram of fungi (A) and AMF (B) communities of root and rhizosphere soil at the OTU level.
During the transition of pomegranate from healthy to diseased states, fungal distribution in the roots and rhizosphere soils of healthy and diseased plants was compared. Distinct biomarker fungal taxa were identified in the rhizosphere soils of healthy (HS) and diseased (DS) plants (Figure 3). At the class level, Dothideomycetes, Microbotryomycetes, Chytridiomycetes, and Mortierellomycetes were significantly enriched in diseased rhizosphere soil (DS). At the order, family, and genus levels, the number of differential fungal taxa in rhizosphere soils was significantly higher than that in root samples. At the genus level, fungal taxa with significant intergroup abundance differences in healthy soil (HS) included: Aureobasidium, Coniothyrium, Alternaria, Arthrobotrys, Orbilia, Trichoderma, Paramyrothecium, Arxiella, Phaeoacremonium, and Geastrum. In diseased soil (DS), taxa with significant intergroup differences included: Preussia, Epiphyte, Coniella, Alfaria, Humicola, Cercophora, Psathyrella, Pseudohyphozyma, Curvibasidium, and Mortierella. In the roots, Aspergillus, Dactylonectria, and Oliveonia were significantly enriched in healthy roots, while Fusidium was significantly enriched in diseased roots. Among these taxa, Aureobasidium, Alternaria, Paramyrothecium, Coniella, Dactylonectria, and Fusidium were identified as potential plant pathogenic fungi. These taxa may play a critical role in the progression and severity of root collar rot in pomegranate.
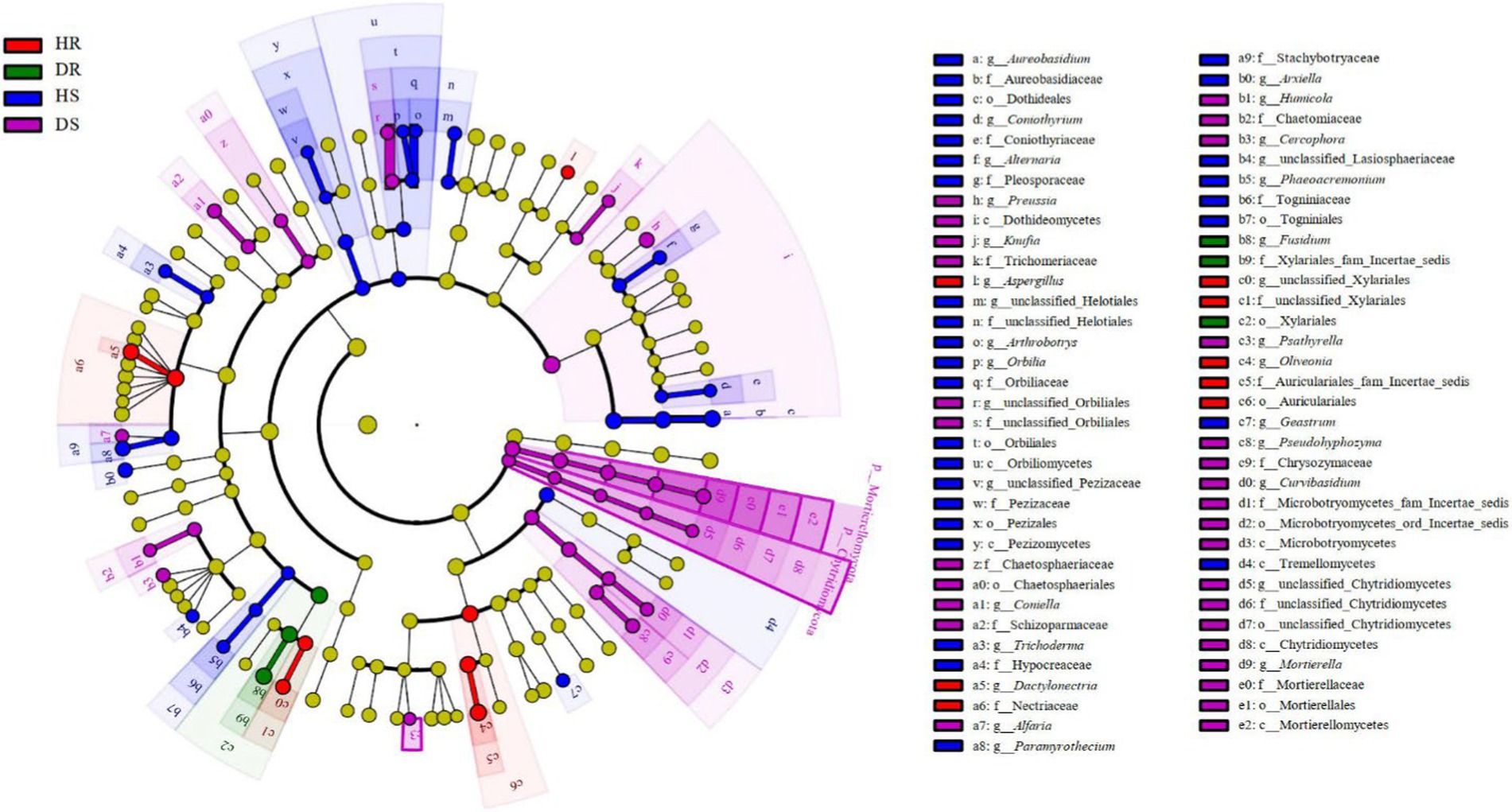
Figure 3. LEfSe analysis of root and rhizosphere soil fungi communities. The red, green, blue and purple nodes in the figure represented fungal taxa that were significantly enriched in HR, DR, HS and DS respectively, and significantly influenced the differences between groups at the same time. The yellow nodes represented nondifferentiated fungal taxa. The c, o, f and g represented class, order, family and genus, respectively.
In addition, a heatmap was created to study fungi with higher relative abundances at the genus level in each sample (Figure 4). In the diseased and healthy roots of pomegranate, there are also several other distinct fungal groups. For instance, pathogenic fungi such as Rhizoctonia sp., Gibberella sp., Diaporthe sp., Plectosphaerella sp., etc. (Morin et al., 2022; da Silva et al., 2024); biocontrol fungi including Aureobasidium sp., Talaromyces sp., Metacordyceps sp., etc.; and probiotic fungi like Cladorrhinum sp., Papiliotrema sp., etc. (Ali et al., 2022). These fungi play a role during the disease development process. Most AMF genera (beneficial symbionts) showed reduced abundance in diseased roots and rhizosphere soil. Interestingly, Glomus displayed contrasting trends: decreasing in roots but increasing in rhizosphere soil. This divergence warrants further investigation.
The functional roles of fungal communities in the pomegranate rhizosphere were predicted using FUNGuild 1.0 software. In healthy roots, the dominant functional group was ectomycorrhizal-undetermined saprotrophic fungi, with a relative abundance of 50.08%. After root collar rot infection, the relative abundance of this group decreased sharply to 3.69%. Similarly, the relative abundance of the arbuscular mycorrhizal functional group declined from 3.94% in healthy roots to 0.79% in diseased roots, with a similar decreasing trend observed in soil samples. Conversely, in diseased roots, the total relative abundance of plant-pathogen-related functional groups increased significantly to 68.36%, compared to 24.74% in healthy roots (Figure 5).
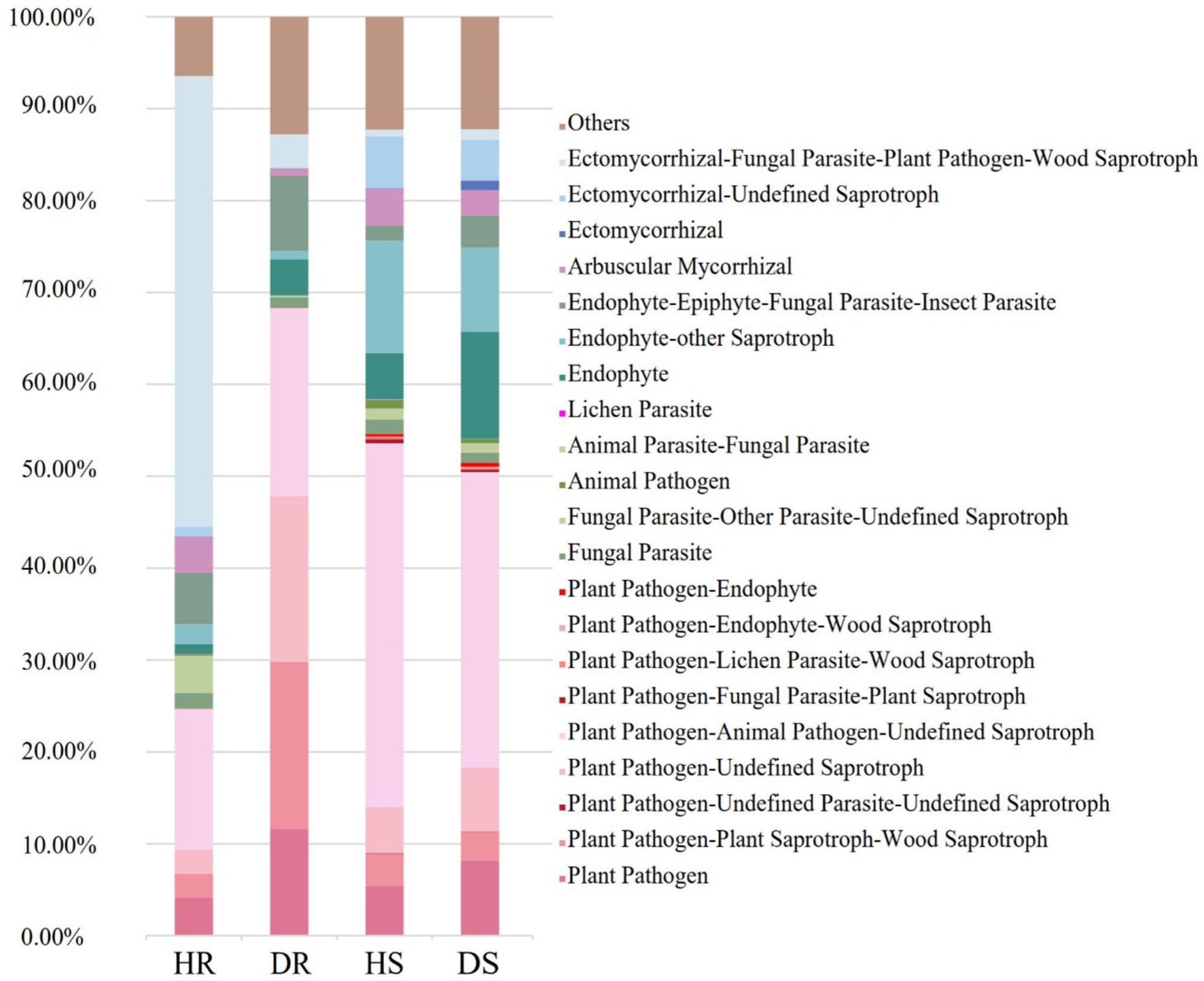
Figure 5. The nutritional structure of fungi in root and rhizosphere soil fungi communities. The ‘Others’ represented the sum of fungal taxa with relative abundance of less than 1%.
No significant differences were observed in the physicochemical properties of rhizosphere soil between healthy and diseased pomegranate. The rhizosphere soil had a neutral pH. Based on the soil nutrient grading standards from the Second National Soil Survey of China, all tested nutrient levels in the sampling site soil were categorized as Level 5 or Level 6, indicating high to extremely high nutrient availability (Table 2). These findings suggest that the sampling site’s soil is nutrient-rich and unaffected by root collar rot, potentially due to long-term artificial fertilization.
Redundancy Analysis (RDA) of Soil Physicochemical Properties and Fungal Community Composition. Redundancy analysis was conducted by integrating data on the physicochemical properties of pomegranate rhizosphere soils and fungal communities. The cumulative contribution rates of RDA1 and RDA2 were 28.37 and 23.72%, respectively. Five soil physicochemical properties played significant roles in shaping the fungal community structure, with their influence ranked as follows: AK > pH > AP > TN > OM. Compared to healthy soils, diseased soil samples exhibited closer intra-group distances, indicating that the fungal community structure within diseased soils was more consistently influenced by soil physicochemical properties (Figure 6). The effects of AK and TN were more pronounced on the fungal community structure of diseased soils (e.g., M1, M3) compared to healthy soils (e.g., K1). In contrast, pH and OM primarily influenced fungal community composition in healthy soil samples.
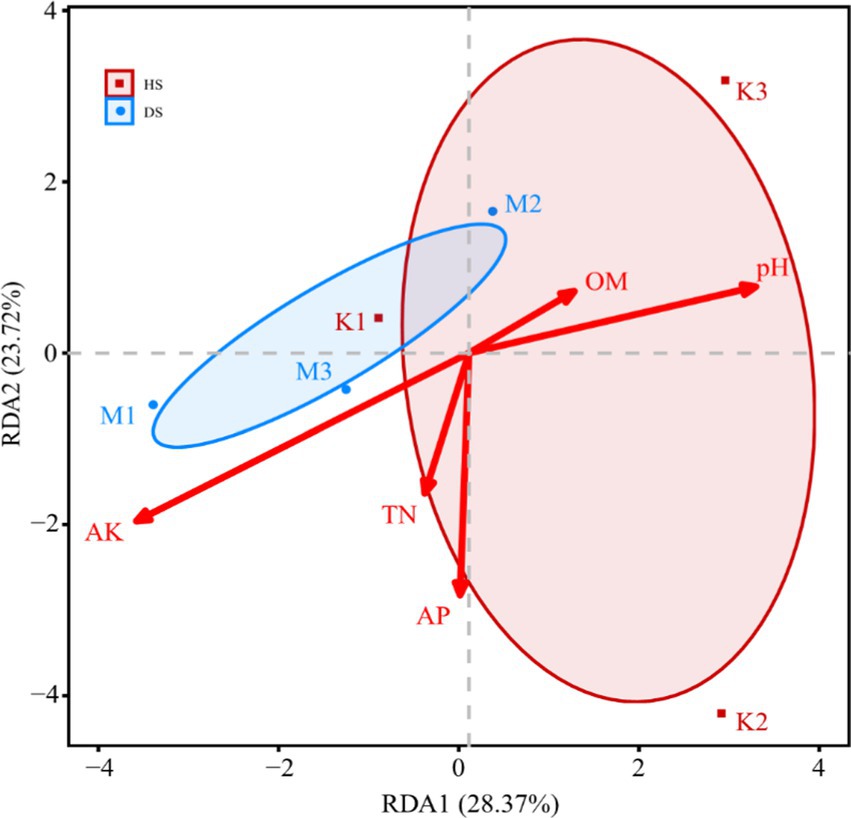
Figure 6. Effects of soil physicochemical properties on relative abundance at genus level. Healthy root (HR): J1/J2/J3. Diseased root (DR): B1/B2/B3. Healthy rhizosphere soil (HS): K1/K2/K3. Diseased rhizosphere soil (DS): M1/M2/M3.
Spearman correlation analysis based on soil physicochemical properties and Illumina MiSeq sequencing results was conducted to further investigate the relationships between soil physicochemical properties and dominant soil fungi at the genus level (relative abundance >1%) (Figure 7). The results indicated that pH was significantly negatively correlated with the genus Plectosphaerella (p < 0.05). Organic matter (OM) was significantly negatively correlated with Alfaria and Psathyrella (p < 0.05) but significantly positively correlated with Arxiella (p < 0.05). Ammonium nitrogen (NH₄+-N) was significantly negatively correlated with Knufia (p < 0.05). Available potassium (AK) was significantly positively correlated with Lauriomyces (p < 0.05) and highly significantly negatively correlated with Trichoderma (p < 0.01). Nitrate nitrogen (NO₃−-N) was significantly positively correlated with Cercophora (p < 0.05). Available phosphorus (AP) was significantly positively correlated with Gibberella (p < 0.05). These correlations directly reflect the structural composition of the fungal community.
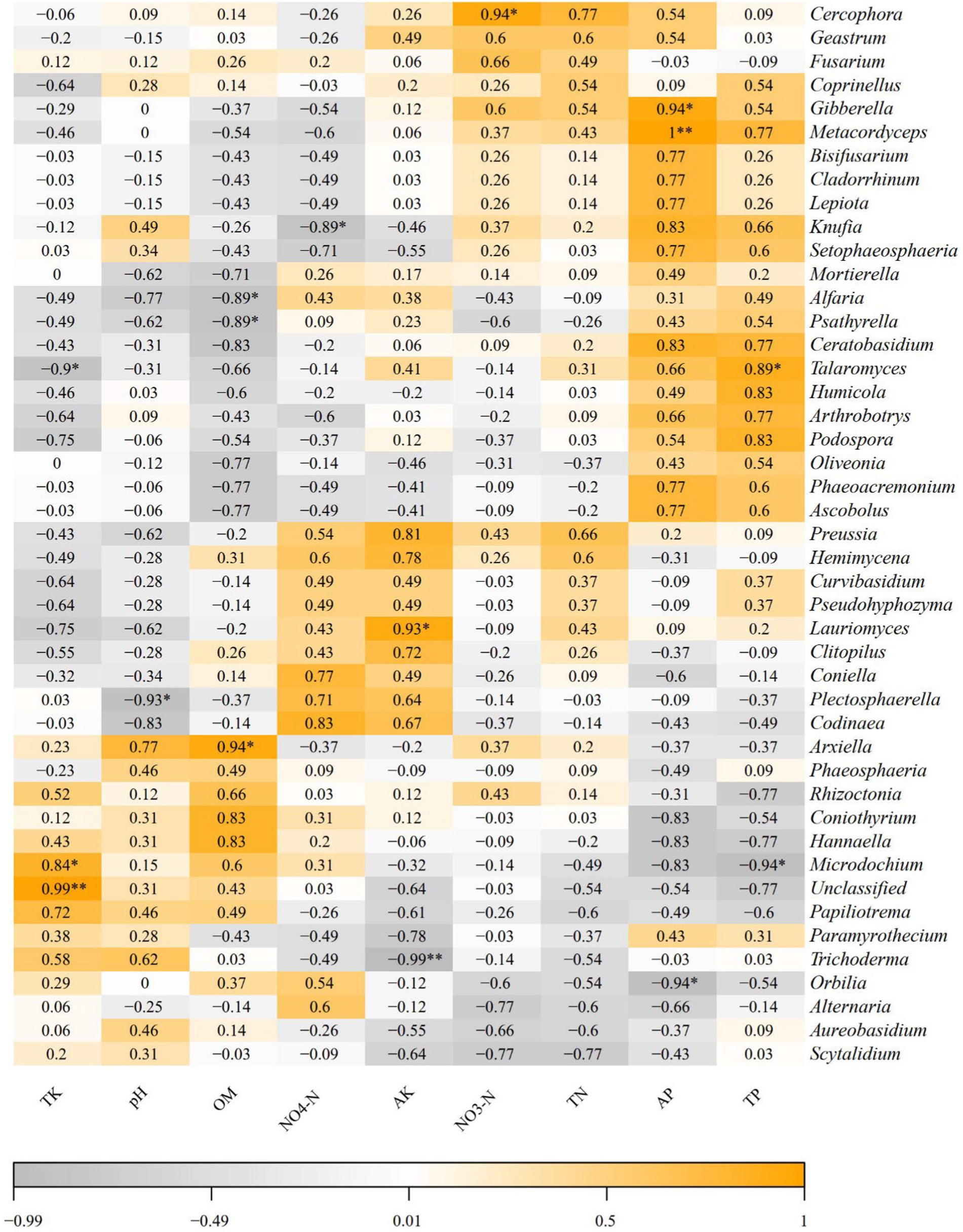
Figure 7. Spearman corrlation between rhizosphere soil fungi communities at genus level and soil physicochemical properties. The color represented the correlation. * and ** indicate significant correlation at p < 0.05, p < 0.01, respectively.
Approximately one-month post-inoculation, the inoculated seedlings exhibited necrotic lesions and decay symptoms. In contrast, the control seedlings remained symptom-free. To verify the identity of the pathogen, hyphae, and conidiophores were collected from the affected seedlings and subjected to reidentification through morphological and molecular biological techniques. These findings confirmed the presence of the initially inoculated pathogen, consistent with Koch’s postulates (Figures 8O–P).

Figure 8. (A) Diseased pomegranate; (B–D) Symptom on pomegranate collar associated with Lauriomyces bellulus. (E–J) Conidiophore groups observed with stereomicroscope (Zeiss Discovery-V20, Leica M166FC); (K,L) Conidiophore groups observed with optical microscope (Leica DM2500); (M,N) Conidium; (O,P): Inoculated seedings: O, control, P, symptoms caused by L. bellulus. Scale bars: E = 0.15 mm; F, H, J = 0.05 mm; G, I = 0.02 mm; K = 25 μm; L–N = 10 μm.
The symptoms observed on the diseased collar of a pomegranate are depicted in Figures 8A–D. For morphological identification, Lauriomyces bellulus (P.W Crous & M. J. Wingfield anam., Figures 8E–N): hyphae were septate, branched, smooth and hyaline, becoming brown near the conidiophores. The conidiophore was macronematous, mononematous, simple, solitary, erect, smooth, septate, dark brownand thick-walled at the base, becoming thin-walled and light brown toward the apex, about 600 μm long, 4–7 μm wide. Ramoconidia blastic-acropetal, catenate, hyaline, smooth, cylindrical to ellipsoidal, rounding towards flattened, subtruncate ends, 6–9 × 1.5–3 μm, with up to 10 conidia in the main branches conidial showing thickened eonidial dehiscence scars (Crous and Wingfield, 1994).
Molecular identification was performed using BLASTn searches against the ITS sequences. The sequence NGSL-P5 exhibited the highest similarity to Lauriomyces bellulus, with 99.39% identity (673/bp) compared to the type culture CBS 517.93 (GenBank Accession No. NR137151). The sequences have been deposited in GenBank with the following accession numbers: ITS: ON495704; LSU: ON495742; SSU: ON495748. Phylogenetic analyses utilizing the combined dataset (Table 3, Somrithipol et al., 2017) were conducted using Maximum Parsimony (MP) and Maximum Likelihood (ML) methods, as presented in Figure 9. Both morphological analysis and molecular characteristics confirmed the identification of NGSL-P5 as Lauriomyces bellulus.
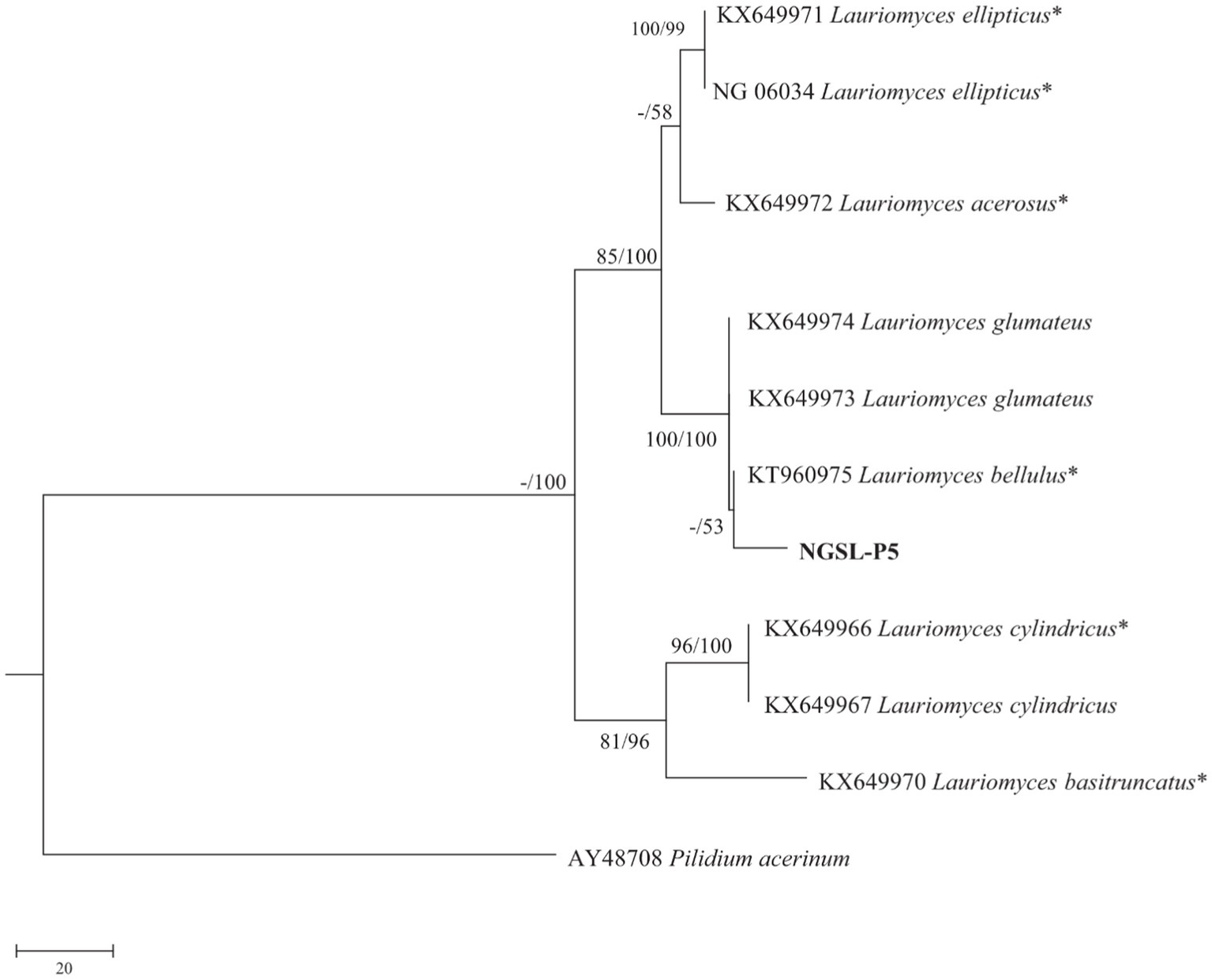
Figure 9. Maximum likelihood strict consensus tree illustrating the phylogeny of Lauriomyces bellulus and related species based on ITS, LSU and SSU sequences. Branches are labeled with maximum likelihood bootstrap higher than 70%, parsimony bootstrap proportions higher than 50%.
The dynamics of soil microbial core communities, including their growth, decline, and their capability to suppress root rot disease, are influenced by environmental factors. These factors also regulate the interactions between plant roots and soil microorganisms (Dasgupta and Brahmaprakash, 2021). In this study, the physicochemical properties of the soil associated with pomegranate did not show significant changes and exhibited eutrophic characteristics, which were associated with the anthropogenic application of chemical fertilizers.
Research has indicated that the long-term overapplication of chemical fertilizers severely threatens the health of soil ecosystems, alters soil nutrient dynamics, and consequently modifies microbial community structures, ultimately driving the soil ecosystem toward a pathological state (Wang et al., 2019). The accumulation of potassium (K), phosphorus (P), and nitrogen (particularly ammonium nitrogen, NH₄+-N, and nitrate nitrogen, NO₃−-N) can reduce microbial activity and diversity, disrupt nutrient absorption by microorganisms, and induce microbial cell apoptosis (Sahab et al., 2021; Shen et al., 2021; Yang et al., 2011). (1) The redundancy analysis results in this study revealed that the top five soil physicochemical properties influencing fungal community composition were, in descending order, available potassium (AK) > pH > available phosphorus (AP) > total nitrogen (TN) > organic matter (OM). In the rhizosphere soil of diseased plants, AK was the most influential soil physicochemical factor. The dominant genus Lauriomyces in the roots of diseased pomegranates showed a significant positive correlation with AK (p < 0.05). Pathogen invasion may cause root exudate changes that modulate rhizosphere microbiota, give rise to potassium-increasing, to trigger plant defense responses (Ansari et al., 2024; Chen et al., 2024). (2) AP was also a critical factor influencing fungal communities. A global study by Ma et al. (2023) found that the colonization rate and abundance of arbuscular mycorrhizal fungi (AMF) decreased significantly with increasing soil AP (p < 0.001). As an essential nutrient for plant growth, variations in AP can lead to changes in plant biomass, which may, in turn, alter fungal groups and nutritional conditions, causing competition among bacteria, soil animals, and fungi (He et al., 2016; Huang et al., 2016). Furthermore, AP can influence soil microorganisms by altering other soil physical and chemical properties, such as pH (Cai et al., 2018). (3) Compared to the healthy group, NH₄+-N content in diseased samples increased significantly (by 28.69%). NH₄+-N was significantly negatively correlated with the genus Knufia (p < 0.05). Knufia petricola is associated with ammonia oxidation processes and may influence soil physicochemical properties (Erdmann et al., 2022). High concentrations of NH₄+ can induce stress symptoms in some plants, including ionic imbalance, increased oxidative stress, and reduced biomass accumulation. Nitrogen forms in the soil can elicit differential responses between plants and microorganisms. For instance, under NO₃− nutrient conditions, the endophytic fungus Phomopsis liquidambaris promotes Arabidopsis seedling growth, whereas under NH₄+ nutrient conditions, it inhibits seedling growth (Sun et al., 2022). (4) Field investigations revealed that orchards primarily rely on pumped tap water from mountain foothills for irrigation. Due to high elevation, water supply remains highly unstable. Combined with precipitation patterns in dry-hot valley climates, this leads to long-term alternation between drought and waterlogging in orchard soils. Concentrated rainfall or irrigation causes abrupt surges in soil moisture, potentially breaking pathogen dormancy and triggering disease outbreaks. Moreover, irrigation water may carry pathogens for field dispersal, exacerbating disease spread (Zhang and Li, 2021).
In summary, the long-term application of chemical fertilizers has led to soil eutrophication, preventing plants from adjusting to the rhizosphere fungal community structure to meet their growth requirements. This disruption ultimately destroys fungal community homeostasis, resulting in the degradation or even deterioration of microbial communities (Lu et al., 2022; Lei et al., 2024). This may be one of the primary reasons for the severe occurrence of pomegranate collar rot disease in the study region. This study represents the first systematic analysis of microbial community structure differences in pomegranate root collar rot within dry-hot valley regions. It should be noted that this study does not cover regions with different climatic conditions or soil types, which may limit the universality of the conclusions. Given that this pomegranate variety is primarily cultivated in dry-hot valley areas of Yunnan Province, China, we maintain that these limitations do not diminish the reference value of current conclusions for similar ecological zones. However, special attention should be given to necessary adaptive modifications during practical applications based on regional characteristics.
The structural integrity and diversity of soil microorganisms are pivotal in maintaining soil health and quality. Pathogenic microorganisms, capable of surviving in soil for extended periods, can propagate through soil particles, roots, and water. This propagation may initiate new diseases, trigger regional epidemics, and cause chain reactions that heighten the risk of crop diseases. Rhizosphere microorganisms, in particular, are significant reservoirs of soil-borne pathogens, which include those responsible for seedling damping off (Philippot et al., 2013; Zhalnina et al., 2018).
ITS-based fungal identification has limitations and cannot rely solely on Illumina MiSeq sequencing to determine the pathogen of a certain disease. Therefore, this study still adopted the Koch’s postulates to verify the pathogen of pomegranate rot collar rot. The combination of Illumina MiSeq sequencing results helped narrow down the range of fungi to be tested. For complex and difficult to identify plant diseases, amplicon sequencing can effectively identify potential pathogenic regions by analyzing the differences between microbial communities in diseased and healthy plants, thereby saving time in formulating pathogen-specific control measures (Mukuma et al., 2020; He et al., 2022). In this study, less than 20% of microbial taxa were shared between healthy and diseased pomegranate root samples, with significant changes in dominant genera. The healthy roots were significantly enriched with genera such as Aspergillus and Oliveonia, while the relative abundance of Lauriomyces and Fusidium (the sexual stage of which belongs to Nectria) increased significantly in diseased roots (p < 0.01).
Among these genera, Aspergillus and Oliveonia are primarily saprotrophic. Lauriomyces sakaeratensis has been identified as a pathogen causing fruit rot in Dipterocarpus (Somrithipol et al., 2006). Some fungi in the genus Nectria are common plant pathogens, capable of causing a variety of stem and root diseases (Keith et al., 2010; Li, 2017; Račko et al., 2020). The pathogen responsible for pomegranate root collar rot was confirmed to be Lauriomyces bellulus through pathogenicity tests, demonstrating that amplicon sequencing can significantly expedite the identification of diseases, especially for pathogens that are challenging to cultivate on artificial media. In addition, this is the first report of collar rot of pomegranate caused by L. bellulus in China.
In healthy roots, various pathogenic fungi maintain a dynamic equilibrium. However, when Lauriomyces bellulus dominates a specific ecological niche and produces Haplofungins—compounds with potent inhibitory effects on fungal inositol phosphoceramide synthase (IPC)—it suppresses other pathogenic fungi. This leads to an increased abundance of single colonies, which in turn causes plant disease (Ohnuki et al., 2009). Furthermore, there was a notable increase in the abundance of the Plectosphaerella in the roots of diseased plants. This genus includes several fungi known to cause root diseases, particularly P. melonis, which is associated with root rot in various crops. Lauriomyces bellulus, may in conjunction with Plectosphaerella sp., plays a role in accelerating the development of pomegranate root collar rot. But this inference still needs to be verified through later experiments.
Rhizosphere microorganisms represent a crucial reservoir of beneficial microbes. Probiotic organisms in the rhizosphere can synergistically extract nutrients from the soil, bolster plant growth, and fortify plant resistance against pathogens (Das et al., 2021; Huang et al., 2025). Shifts in microbial community structures are known to directly influence the prevalence and severity of soil-borne diseases (Jayaraman et al., 2021; Chauhan et al., 2023). In response to threats such as Lauriomyces bellulus infection, pomegranate roots can recruit antagonistic fungal communities that not only combat this pathogen but also enhance overall tree vigor by fostering the aggregation of probiotic communities.
Although constrained by data acquisition dimensions such as sample time series, this study has not yet established direct correlation models with continuous succession processes of soil health status. Nevertheless, it reveals that pomegranate plants exhibit reduced abundance of beneficial biological communities after pathogen infection, while showing a trend of recruiting beneficial microorganisms through pathogen interaction. This aligns with the theory proposed by van der Heijden et al. (2016). FUNGuild analysis revealed that the healthy rhizosphere fungal community was predominantly composed of ectomycorrhizal-undefined saprotrophic functional groups and arbuscular mycorrhizal (AM) functional groups. Mycorrhizal fungi not only benefit plant growth and development but also enhance host resistance, improve soil fertility, and maintain soil health (Liu and Chen, 2007). Specific genera in healthy roots included Pezicula and AM fungi such as Glomus and Septoglomus. Secondary metabolites of Pezicula fungi, such as Pezicumin, exhibit inhibitory activity against nine plant pathogens, including Botrytis cinerea and Passalora fulva (Wang et al., 2014). Aureobasidium is a genus with a relatively high abundance in healthy roots, where A. Pullulans is well-recognized as a mature biocontrol agent. However, a notable decline in the abundance of this genus has been observed in samples from diseased roots. This reduction could impact on the natural biocontrol mechanisms within the rhizosphere, potentially diminishing the soil’s disease-suppressive capabilities.
Following the onset of collar rot disease, the relative abundance of ectomycorrhizal-undefined saprotrophic and AM functional groups declined, while the abundance of pathology-related functional groups increased. Despite the decline in certain beneficial fungi like Aureobasidium, there are still other beneficial fungal groups that have undergone a degree of enrichment. Research indicates that in the early stages of plant disease, changes in root exudates may recruit or enrich beneficial microorganisms as a form of self-rescue (Bakker et al., 2018; Lyu and Smith, 2022; Feng et al., 2024). These fungi play various roles, including biocontrol, promoting plant growth, and improving soil conditions, which collectively contribute to the plant’s ability to combat disease and stress. Dominant genera in diseased root samples were mostly saprotrophic Basidiomycota fungi, including Pluteus, Lepiota, Tremella, and Thyronectria. Saprotrophic fungi contribute to organic matter degradation, facilitating carbon and nitrogen cycling in soil, which can enhance or improve soil organic matter and nutrient composition (Clocchiatti et al., 2020). Additionally, unique fungal taxa in diseased pomegranate roots included Fusidium and Mortierella. Secondary metabolites of Fusidium fungi, such as fusidic acid, act as antibiotics against certain fungal pathogens (Huang et al., 2021). Mortierella fungi contribute to soil nutrient improvement through phosphate solubilization (Osorio and Habte, 2013) and the degradation of herbicides such as diuron (Ellegaard-Jensen et al., 2013) and endosulfan (Kataoka et al., 2010; Li et al., 2018). In the diseased root samples, there is an observed increase in the abundance of non-endemic beneficial fungi, encompassing five genera. Notably, the relative abundance of Talaromyces has increased by 1.05%. The ready-to-use dry-powder formulation of T. flavus Bodhi001 demonstrates an effective inhibitory effect against rice brown leaf spot disease, highlighting its potential as a biocontrol agent (López-Moral et al., 2021; Volynchikova and Kim, 2022; Jantasorn et al., 2025). Other fungi such as Acremonium alternatum and Metacordyceps chlamydosporia have been effectively reported to prevent and control plant diseases in both greenhouse and field conditions (Auer et al., 2024). Additionally, Aureobasidium pullulans has been developed into a relatively mature biocontrol agent (Cignola et al., 2024). Some fungi compete with plant pathogens for livable space and nutrients, such as Papiliotrema flavescens, which exerts antifungal effects through potential competition for nutrition, space, and parasitism and parasitism. Another species, P. huenov, improves soil conditions by decomposing heavy metals (Nguyen et al., 2020; Nguyen Van et al., 2021). In pot experiments, P. laurentii showed significant effects on Brinjal growth promotion and biocontrol of Fusarium wilt, indicating a beneficial role in plant health management (Das et al., 2023). Fungal communities play critical roles in disease suppression not only in green plants but also in mushroom crops. For example, Yu et al. (2025) indicated that inoculating the Morchella importuna mycosphere with Pseudomonas chlororaphis increased the α-diversity of the soil fungal community and suppressed the abundance of the pathogen Paecilomyces penicillatus, leading to reduced white mould disease incidence and increased morel yield. Thus, there is a discernible relationship between the differential changes in these fungal groups and pomegranate root and neck rot disease, and that manipulation of beneficial microorganisms could be a broadly applicable strategy in disease management.
The colonization of microbial strains has long been considered a major limiting factor affecting the effectiveness of biocontrol strains. Extensive research suggests that microbial communities possess resilience and resistance, capable of quickly reverting to their original structure following external disturbances or the introduction of new species (Beisner et al., 2003; Toju et al., 2018; Hayashi et al., 2024). Consequently, changing established microbial community types is often challenging, especially when a single or a few strains are introduced into an existing microbial system without continuous supplementation, leading to their rapid replacement by other existing microorganisms (Castro-Sowinski et al., 2007; Luo et al., 2019; Quiroz, 2022). Reports suggest plants can “perceive and respond to their environment” through genetic experience, emitting specific chemical signals to attract mobile organisms that meet their needs, thereby achieving self-protection (Spoel and Dong, 2012; Leopold, 2014; Das et al., 2021). The enrichment behavior of beneficial fungi during the “healthy-diseased plant” transition in pomegranates may relate to this mechanism. During this process, organic matter released by rhizosphere microbes becomes incorporated into soil organic matter, and the soil develops memory of these dynamic changes (Lapsansky et al., 2016). Rhizosphere microbes from plants successfully adapted to environments can persist and influence subsequent plants even after original plant removal. If more comprehensive experiments could validate this hypothesis, selected microbes might be used to construct beneficial microbial communities within the “ecological memory of plants and rhizosphere soil fungi,” potentially circumventing challenges caused by microbial communities’ inherent resilience and resistance. Therefore, utilizing Illumina MiSeq sequencing to compare the differences in rhizosphere microorganisms between diseased and healthy plants, and based on ecological memory of plants and rhizosphere soil fungi, and selecting core microbial communities that are beneficial for plant growth and disease resistance from the inherent microbial community of the environment, constructing a rhizosphere microbial community assembly (MCA), and transforming soil from a susceptible state to a disease inhibiting state, can be an effective strategy for preventing this disease (Tian et al., 2024).
This research investigated the roots and rhizosphere soils of healthy and collar rot-affected pomegranate, analyzing the relationship between collar rot disease and the diversity and structural composition of rhizosphere fungal communities. The rhizosphere of healthy plants exhibited high species richness, with no dominant potential pathogenic fungi, indicating a relatively stable community structure. However, after infection, the distribution of rhizosphere fungal communities became more homogenized, the balance of the rhizosphere fungal community structure was disrupted, and soil nutrient composition shifted, transitioning from a probiotic to a pathological state.
The prevention and control of soil-borne diseases present significant challenges. Traditional chemical pesticide methods offer limited efficacy against soil pathogens and can contribute to environmental pollution. As a result, integrated management strategies are typically employed to address soil-borne diseases. The findings clarify the impact of collar rot disease on the structure of pomegranate rhizosphere fungal communities, providing a basis for further research into the disease’s pathogenic mechanisms. Meanwhile, this work also offers theoretical support for identifying biocontrol microorganisms to manage pomegranate collar rot.
All raw data for this study is available on the NCBI database (accession number: PRJNA1170523): https://www.ncbi.nlm.nih.gov/search/all/?term=PRJNA1170523.
ZW: Data curation, Writing – original draft, Writing – review & editing, Funding acquisition, Investigation, Resources, Validation, Visualization, Formal analysis. JiaC: Conceptualization, Methodology, Writing – review & editing. JieC: Formal analysis, Supervision, Visualization, Writing – review & editing. YY: Formal analysis, Writing – original draft. AZ: Formal analysis, Writing – original draft. JW: Conceptualization, Funding acquisition, Project administration, Supervision, Writing – review & editing, Writing – original draft.
The author(s) declare that financial support was received for the research and/or publication of this article. This research was funded by the National Natural Science Foundation of China (31860208); the National Key Research and Development Program of China (2019YFD100200X); Yunnan Provincial Department of Education Science Research Fund Project (2023J0706); the Open Foundation of Key Laboratory of Forest Disaster Warning and Control of Yunnan Province (ZKJS-T-202202); Yunnan Fundamental Research Projects (202401AU070137).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The authors declare that no Gen AI was used in the creation of this manuscript.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2025.1573724/full#supplementary-material
SUPPLEMENTARY Table 1 | Otu_core_spec.groups.
Supplementary Table 2 | Otu_taxonomy.
Ali, A., Elrys, A. S., Liu, L., Iqbal, M., Zhao, J., Huang, X. -Q., et al. (2022). Cover plants-mediated suppression of fusarium wilt and root-knot incidence of cucumber is associated with the changes of rhizosphere fungal microbiome structure-under plastic shed system of North China. Front. Microbiol. 13:697815. doi: 10.3389/fmicb.2022.697815
Ansari, M. M., Bisht, N., Singh, T., and Chauhan, P. S. (2024). Symphony of survival: insights into cross-talk mechanisms in plants, bacteria, and fungi for strengthening plant immune responses. Microbiol. Res. 285:127762. doi: 10.1016/j.micres.2024.127762
Auer, S., Zamani-Noor, N., Mahfoud, Y., and Ludwig-Müller, J. (2024). Exploring the influence of rapeseed cultivar and pathogen isolate on Acremonium alternatum's efficacy in clubroot disease control. Eur. J. Plant Pathol. 170, 519–534. doi: 10.1007/s10658-024-02916-y
Bakker, P. A., Pieterse, C. M., de Jonge, R., and Berendsen, R. L. (2018). The soil-borne legacy. Cell 172, 1178–1180. doi: 10.1016/j.cell.2018.02.024
Basım, E., and Basım, H. (2013). Determination of root and crown rot diseases in pomogranate (Punica granatum L.) orchards in Antalya province. Fruit Sci. 1, 23–26.
Beisner, B. E., Haydon, D. T., and Cuddington, K. (2003). Alternative stable states in ecology. Front. Ecol. Environ. 1, 376–382. doi: 10.1890/1540-9295(2003)001[0376:ASSIE]2.0.CO;2
Berendsen, R. L., Vismans, G., Yu, K., Song, Y., de Jonge, R., Burgman, W. P., et al. (2018). Disease-induced assemblage of a plant-beneficial bacterial consortium. ISME J. 12, 1496–1507. doi: 10.1038/s41396-018-0093-1
Bolger, A. M., Lohse, M., and Usadel, B. (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. doi: 10.1093/bioinformatics/btu170
Bunyard, B. A., Nicholson, M. S., and Royse, D. J. (1994). A systematic assessment of Morchella using RFLP analysis of the 28S ribosomal RNA gene. Mycologia 86, 762–772. doi: 10.1080/00275514.1994.12026481
Cai, Z. -Q., Zhang, Y. -H., Yang, C., and Wang, S. (2018). Land-use type strongly shapes community composition, but not always diversity of soil microbes in tropical China. Catena 165, 369–380. doi: 10.1016/j.catena.2018.02.018
Castro-Sowinski, S., Herschkovitz, Y., Okon, Y., and Jurkevitch, E. (2007). Effects of inoculation with plant growth-promoting rhizobacteria on resident rhizosphere microorganisms. FEMS Microbiol. Lett. 276, 1–11. doi: 10.1111/j.1574-6968.2007.00878.x
Chauhan, P., Sharma, N., Tapwal, A., Kumar, A., Verma, G. S., Meena, M, et al. (2023). Soil microbiome: Diversity, benefits and interactions with plants. Sustainability 15:14643. doi: 10.3390/su151914643
Chen, J., Liu, Z., Liu, Y., Ji, X., Li, X., Wei, Y., et al. (2024). Differences in autotoxic substances and microbial community in the root space of Panax notoginseng coinducing the occurrence of root rot. Appl. Environ. Microb. 90:e0228723. doi: 10.1128/aem.02287-23
Cignola, R., Carminati, G., Natolino, A., and Di Francesco, A. (2024). Effects of bioformulation prototype and bioactive extracts from Agaricus bisporus spent mushroom substrate on controlling Rhizoctonia solani of Lactuca sativa L. Front. Plant Sci. 15:1466956. doi: 10.3389/fpls.2024.1466956
Clocchiatti, A., Hannula, S. E., van den Berg, M., Korthals, G., and De Boer, W. (2020). The hidden potential of saprotrophic fungi in arable soil: patterns of short-term stimulation by organic amendments. Appl. Soil Ecol. 147:103434. doi: 10.1016/j.apsoil.2019.103434
Crous, P. W., and Wingfield, M. J. (1994). Sporendocladia fumosa and Lauriomyces bellulus sp. nov. from Castanea cupules in Switzerland. Sydowia 46, 193–203.
da Silva, J. S. A., Alves, V. C. S., da Silva, S. F., Do Nascimento Barbosa, R., De Souza, C. A. F., Da Costa, D. P., et al. (2024). Diaporthe ueckeri causing cassava root rot in Pernambuco, Brazil. Crop Prot. 184:106811. doi: 10.1016/j.cropro.2024.106811
Das, N., Mishra, S. K., Bishayee, A., Ali, E. S., and Bishayee, A. (2021). The phytochemical, biological, and medicinal attributes of phytoecdysteroids: an updated review. Acta Pharm. Sin. B 11, 1740–1766. doi: 10.1016/j.apsb.2020.10.012
Das, S., Rabha, J., and Narzary, D. (2023). Assessment of soil yeasts Papiliotrema laurentii S-08 and Saitozyma podzolica S-77 for plant growth promotion and biocontrol of fusarium wilt of brinjal. J. Appl. Microbiol. 134:lxad252. doi: 10.1093/jambio/lxad252
Dasgupta, D., and Brahmaprakash, G. P. (2021). Soil microbes are shaped by soil physico-chemical properties: a brief review of existing literature. Int. J. Plant Soil Sci. 33, 59–71. doi: 10.9734/IJPSS/2021/V33I130409
Edgar, R. C. (2013). UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 10, 996–998. doi: 10.1038/nmeth.2604
Ellegaard-Jensen, L., Aamand, J., Kragelund, B. B., Johnsen, A. H., and Rosendahl, S. (2013). Strains of the soil fungus Mortierella show different degradation potentials for the phenylurea herbicide diuron. Biodegradation 24, 765–774. doi: 10.1007/s10532-013-9624-7
Erdmann, E. A., Nitsche, S., Gorbushina, A. A., and Schumacher, J. (2022). Genetic engineering of the rock inhabitant Knufia petricola provides insight into the biology of extremotolerant black fungi. Front. Fungal Biol. 3:862429. doi: 10.3389/ffunb.2022.862429
Feng, Z. -W., Liang, Q. -H., Yao, Q., Bai, Y., and Zhu, H. -H. (2024). The role of the rhizobiome recruited by root exudates in plant disease resistance: current status and future directions. Environ. Microbiome 19:91. doi: 10.1186/s40793-024-00638-6
Ghaderi, F., and Habibi, A. (2021). Morphological and molecular characterization of Phytophthora species associated with root and crown rot of pomegranate in Iran. Plant Pathol. 70, 615–629. doi: 10.1111/ppa.13320
Greeshma, K., Uma Devi, G., Senthilvel, S., Dinesh Kumar, V., Gandhi, B., Shiva Shanker, K., et al. (2024). Lupeol a predisposing factor in grey mould (Amphobotrys ricini [N. F. Buchw.]) Hennebert disease pathogenesis in castor (Ricinus communis L.). J. Phytopathol. 172:e13357. doi: 10.1111/jph.13357
Guarnaccia, V., Martino, I., Brondino, L., and Gullino, M. L. (2022). Paraconiothyrium fuckelii, Diaporthe eres and Neocosmospora parceramosa causing cane blight of red raspberry in northern Italy. J. Plant Pathol. 104, 683–698. doi: 10.1007/s42161-022-01068-4
Hausmann, B., Knorr, K. H., Schreck, K., Tringe, S. G., Glavina del Rio, T., Loy, A., et al. (2016). Consortia of low-abundance bacteria drive sulfate reduction-dependent degradation of fermentation products in peat soil microcosms. ISME J. 10, 2365–2375. doi: 10.1038/ismej.2016.42
Hayashi, I., Fujita, H., and Toju, H. (2024). Deterministic and stochastic processes generating alternative states of microbiomes. ISME Commun. 4:ycae007. doi: 10.1093/ismeco/ycae007
He, Y., Li, Y., Song, Y., Hu, X., Liang, J., Shafik, K., et al. (2022). Amplicon sequencing reveals novel fungal species responsible for a controversial tea disease. J. Fungi 8:782. doi: 10.3390/jof8080782
He, D., Xiang, X. -J., He, J. -S., Wang, C., Cao, G. -M., Adams, J., et al. (2016). Composition of the soil fungal community is more sensitive to phosphorus than nitrogen addition in the alpine meadow on the Qinghai-Tibetan plateau. Biol. Fertil. Soils 52, 1059–1072. doi: 10.1007/s00374-016-1142-4
Hu, H., Chen, X., Hou, F., Wu, Y., and Cheng, Y. (2017). Bacterial and fungal community structures in loess plateau grasslands with different grazing intensities. Front. Microbiol. 8:606. doi: 10.3389/fmicb.2017.00606
Huang, W. -W., Ge, X. -Y., Huang, Y., Chai, X. -T., Zhang, L., Zhang, Y. -X., et al. (2021). High-yield strain of fusidic acid obtained by atmospheric and room temperature plasma mutagenesis and the transcriptional changes involved in improving its production in fungus Fusidium coccineum. J. Appl. Microbiol. 130, 405–415. doi: 10.1111/jam.14797
Huang, J., Hu, B., Qi, K., Chen, W., Pang, X., Bao, W., et al. (2016). Effects of phosphorus addition on soil microbial biomass and community composition in a subalpine spruce plantation. Eur. J. Soil Biol. 72, 35–41. doi: 10.1016/j.ejsobi.2015.12.007
Huang, P., Shi, H., Jiang, L., Zhu, D., Zhou, Z., Hou, Z., et al. (2025). Soil microbial community and influencing factors of different vegetation restoration types in a typical agricultural pastoral ecotone. Front. Microbiol. 15:1514234. doi: 10.3389/fmicb.2024.1514234
Jantasorn, A., Oiuphisittraiwat, T., Wangsawang, S., and Cha-Aim, K. (2025). Application of ready-to-use dry-powder formulation of Talaromyces flavus Bodhi001 against rice brown leaf spot disease and to promote the yield components of rice in saline-alkaline soils. Eur. J Plant Pathol. 1–12. doi: 10.1007/s10658-024-02995-x
Jayaraman, S., Naorem, A. K., Lal, R., Dalal, R. C., Sinha, N. K., Patra, A. K., et al. (2021). Disease-suppressive soils—beyond food production: a critical review. J Soil Sci. Plant Nut. 21, 1437–1465. doi: 10.1007/s42729-021-00451-x
Ji, T., Altieri, V., Salotti, I., and Rossi, V. (2023). Effects of temperature and moisture duration on spore germination of four fungi that cause grapevine trunk diseases. Plant Dis. 107, 1005–1008. doi: 10.1094/PDIS-08-22-1802-SC
Kataoka, R., Takagi, K., and Sakakibara, F. (2010). A new endosulfan-degrading fungus, Mortierella species, isolated from a soil contaminated with organochlorine pesticides. J. Pestic. Sci. 35, 326–332. doi: 10.1584/jpestics.G10-10
Keith, L., Sugiyama, L., and Nagao, M. (2010). Macadamia quick decline caused by Phytophthora tropicalis is associated with sap bleeding, frass, and Nectria in Hawaii. Plant Dis. 94:128. doi: 10.1094/PDIS-94-1-0128B
Kurbetli, İ., Karaca, G., Aydoğdu, M., and Sülü, G. (2020). Phytophthora species causing root and collar rot of pomegranate in Turkey. Eur. J. Plant Pathol. 157, 485–496. doi: 10.1007/s10658-020-02007-8
Kurm, V., Van Der Putten, W. H., De Boer, W., Naus-Wiezer, S., and Hol, W. G. (2017). Low abundant soil bacteria can be metabolically versatile and fast growing. Ecology 98, 555–564. doi: 10.1002/ecy.1670
Lapsansky, E. R., Milroy, A. M., Andales, M. J., and Vivanco, J. M. (2016). Soil memory as a potential mechanism for encouraging sustainable plant health and productivity. Pol. J. Environ. Stud. 38, 137–142. doi: 10.1016/j.copbio.2016.01.014
Lei, Y., Ding, D., Duan, J. -H., Luo, Y., Huang, F. -Y., Kang, Y. -K., et al. (2024). Soil microbial community characteristics and their effect on tea quality under different fertilization treatments in two tea plantations. Genes 15:610. doi: 10.3390/genes15050610
Leopold, A. C. (2014). Smart plants: memory and communication without brains. Plant Signal. Behav. 9:e972268. doi: 10.4161/15592316.2014.972268
Li, X. -P. (2017). Naked barley root rot diseases and influence on its rhizosphere microecology in Qinghai-Tibet plateau. China, Gansu: GansuAgricultural University (Doctoral dissertation).
Li, Z., Bai, X., Jiao, S., Li, Y., Li, P., Yang, Y., et al. (2021). A simplified synthetic community rescues Astragalus mongholicus from root rot disease by activating plant-induced systemic resistance. Microbiome 9, 217–220. doi: 10.1186/s40168-021-01169-9
Li, F., Chen, L., Redmile-Gordon, M., Zhang, J. -B., Zhang, C. -Z., Ning, Q., et al. (2018). Mortierella elongata's roles in organic agriculture and crop growth promotion in a mineral soil. Land Degrad. Dev. 29, 1642–1651. doi: 10.1002/ldr.2965
Li, P., Zhao, X., Chai, Q., Hou, D., Hou, X., Luo, Q., et al. (2024). A new canker on Gleditsia sinensis caused by Thyronectria austroamericana in China. Crop Prot. 182:106740. doi: 10.1016/j.cropro.2024.106740
Liu, W., Wang, Q., Wang, B., Wang, X., Franks, A. E., Teng, Y., et al. (2015). Changes in the abundance and structure of bacterial communities under long-term fertilization treatments in a peanut monocropping system. Plant Soil 395, 415–427. doi: 10.1007/s11104-015-2569-3
López-Moral, A., Agustí-Brisach, C., and Trapero, A. (2021). Plant biostimulants: new insights into the biological control of Verticillium wilt of olive. Front. Plant Sci. 12:662178. doi: 10.3389/fpls.2021.662178
Lu, Y., Cong, P., Kuang, S., Tang, L., Li, Y. -Y., Dong, J. -X., et al. (2022). Long-term excessive application of K2SO4 fertilizer alters bacterial community and functional pathway of tobacco-planting soil. Front. Plant Sci. 13:1005303. doi: 10.3389/fpls.2022.1005303
Luo, L. -F., Guo, C. -W., Wang, L. -T., Zhang, J. -X., Deng, L. -M., Luo, K. -F., et al. (2019). Negative plant-soil feedback driven by re-assemblage of the rhizosphere microbiome with the growth of Panax notoginseng. Front. Microbiol. 10:1597. doi: 10.3389/fmicb.2019.01597
Lyu, D., and Smith, D. L. (2022). The root signals in rhizospheric inter-organismal communications. Front. Plant Sci. 13:1064058. doi: 10.3389/fpls.2022.1064058
Ma, X. -C., Xu, X., Geng, Q. -H., Luo, Y. -Q., Ju, C. -H., Li, Q., et al. (2023). Global arbuscular mycorrhizal fungal diversity and abundance decreases with soil available phosphorus. Glob. Ecol. Biogeogr. 32, 1423–1434. doi: 10.1111/geb.13704
Morin, L., Bissett, A. B., and van Klinken, R. D. (2022). Exploring the role of fungal endophytes in the sudden death syndrome of the invasive shrub Chrysanthemoides monilifera subsp. rotundata in Australia. Phytobiomes J. 6, 13–25. doi: 10.1094/PBIOMES-04-21-0027-R
Mukuma, C., Godoy-Lutz, G., Eskridge, K., Steadman, J., Urrea, C., and Muimui, K. (2020). Use of culture and molecular methods for identification and characterization of dry bean fungal root rot pathogens in Zambia. Trop. Plant Pathol. 45, 385–396. doi: 10.1007/s40858-020-00336-x
Nguyen, K. C. T., Nguyen, P. V., and Truong, H. T. H. (2020). Heavy metal tolerance of novel Papiliotrema yeast isolated from Vietnamese mangosteen. Mycobiology 48, 296–303. doi: 10.1080/12298093.2020.1767020
Nguyen Van, P., Thi Hong Truong, H., Pham, T. A., Le Cong, T., Le, T., and Thi Nguyen, K. C. (2021). Removal of manganese and copper from aqueous solution by yeast Papiliotrema huenov. Mycobiology 49, 507–520. doi: 10.1080/12298093.2021.1968624
Nguyen, N. P., Warnow, T., Pop, M., and White, B. (2016). A perspective on 16S rRNA operational taxonomic unit clustering using sequence similarity. NPJ Biofilms Microbiomes 2, 16004–16008. doi: 10.1038/npjbiofilms.2016.4
Ohnuki, T., Yano, T., Ono, Y., Kozuma, S., Suzuki, T., Ogawa, Y., et al. (2009). Haplofungins, novel inositol phosphorylceramide synthase inhibitors, from Lauriomyces bellulus SANK 26899 I. Taxonomy, fermentation, isolation and biological activities. J. Antibiot. 62, 545–549. doi: 10.1038/ja.2009.72
Osorio, N. W., and Habte, M. (2013). Phosphate desorption from the surface of soil mineral particles by a phosphate-solubilizing fungus. Biol. Fertil. Soils 49, 481–486. doi: 10.1007/s00374-012-0763-5
Philippot, L., Raaijmakers, J. M., Lemanceau, P., and Van Der Putten, W. H. (2013). Going back to the roots: the microbial ecology of the rhizosphere. Nat. Rev. Microbiol. 11, 789–799. doi: 10.1038/nrmicro3109
Qi, L., Ge, Y., Xia, T., He, J. Z., Shen, C., Wang, J., et al. (2019). Rare earth oxide nanoparticles promote soil microbial antibiotic resistance by selectively enriching antibiotic resistance genes. Environ. Sci. 6, 456–466. doi: 10.1039/C8EN01129J
Quiroz, R. N. (2022). Antarctic soil microbiomes as a promising strategy to counteract the effects of climate change by natural microbiome engineering in tomato plants growing under water deficit stress Chile, Temuco: Universidad de la Frontera (Doctoral dissertation).
Račko, V., Mihál, I., and Mišíková, O. (2020). Beech bark disease in Slovakia related to fungi of the genus Nectria S.L. and the anatomy of necrotised bark and wood: a brief review. Folia Oecol. 47, 16–22. doi: 10.2478/foecol-2020-0003
Sahab, S., Suhani, I., Srivastava, V., Chauhan, P. S., Singh, R. P., and Prasad, V. (2021). Potential risk assessment of soil salinity to agroecosystem sustainability: current status and management strategies. Sci. Total Environ. 764:144164. doi: 10.1016/j.scitotenv.2020.144164
Sasse, J., Martinoia, E., and Northen, T. (2018). Feed your friends: do plant exudates shape the root microbiome? Trends Plant sci. 23, 25–41. doi: 10.1016/j.tplants.2017.09.003
Sato, K., Suyama, Y., Saito, M., and Sugawara, K. (2005). A new primer for discrimination of arbuscular mycorrhizal fungi with polymerase chain reaction-denature gradient gel electrophoresis. Grassl. Sci. 51, 179–181. doi: 10.1111/J.1744-697X.2005.00023.X
Shen, W. -S., Hu, M. -C., Qian, D., Xue, H. -W., Gao, N., and Lin, X. -H. (2021). Microbial deterioration and restoration in greenhouse-based intensive vegetable production systems. Plant Soil 463, 1–18. doi: 10.1007/s11104-021-04933-w
Sokolova, O., Sivicka, I., Krivmane, B., and Kārkliņa, K. (2022). First report of Truncatella angustata causing leaf spot on oregano (Origanum vulgare) in Latvia. J. Phytopathol. 170, 167–175. doi: 10.1111/jph.13064
Somrithipol, S., Jones, E. G., Bahkali, A. H., Suetrong, S., Sommai, S., Chamoi, C., et al. (2017). Lauriomyces, a new lineage in the Leotiomycetes with three new species. Cryptogam. Mycol. 38, 259–273. doi: 10.7872/crym/v38.iss2.2017.259
Somrithipol, S., Kosol, S., and Jones, E. G. (2006). Lauriomyces sakaeratensis sp. nov., a new hyphomycete on decaying Dipterocarpus costatus fruits from Sakaerat biosphere reserve, Thailand. Nova Hedwig. 82, 209–215. doi: 10.1127/0029-5035/2006/0082-0209
Spoel, S. H., and Dong, X. (2012). How do plants achieve immunity? Defence without specialized immune cells. Nat. Rev. Immunol. 12, 89–100. doi: 10.1038/nri3141
Sun, K., Lu, F., Huang, P. -W., Tang, M. -J., Xu, F. -J., Zhang, W., et al. (2022). Root endophyte differentially regulates plant response to NO3− and NH4+ nutrition by modulating N fluxes at the plant–fungal interface. Plant Cell Environ. 45, 1813–1828. doi: 10.1111/pce.14304
Tian, J. -N., Zhang, M. -H., Huang, Y., Luo, X. -N., Shao, F. -X., Lei, W. -Q., et al. (2024). Biocontrol potential of an artificial synthetic bacterial consortium against peony root rot disease. Biol. Control 195:105563. doi: 10.1016/j.biocontrol.2024.105563
Toju, H., Peay, K. G., Yamamichi, M., Narisawa, K., Hiruma, K., Naito, K., et al. (2018). Core microbiomes for sustainable agroecosystems. Nat. Plants 4, 247–257. doi: 10.1038/s41477-018-0139-4
Van Der Heijden, M. G., De Bruin, S., Luckerhoff, L., Van Logtestijn, R. S., and Schlaeppi, K. (2016). A widespread plant-fungal-bacterial symbiosis promotes plant biodiversity, plant nutrition and seedling recruitment. ISME J. 10, 389–399. doi: 10.1038/ismej.2015.120
Volynchikova, E., and Kim, K. D. (2022). Biological control of oomycete soilborne diseases caused by Phytophthora capsici, Phytophthora infestans, and Phytophthora nicotianae in solanaceous crops. Mycobiology 50, 269–293. doi: 10.1080/12298093.2022.2136333
Wang, X. -J., Peng, C. -Y., Liang, J. -S., Liang, Q. -D., Xu, C. -L., and Guo, W. (2019). The complete chloroplast genome of Paris polyphylla var. chinensis, an endemic medicinal herb in China. Mitochondrial DNA Part B Resour. 4, 3888–3889. doi: 10.1080/23802359.2019.1687351
Wang, X. -B., Wang, X. -L., Sheng, H. -J., Wang, X. -Z., Zhao, H. -T., and Feng, K. (2022). Excessive nitrogen fertilizer application causes rapid degradation of greenhouse soil in China. Pol. J. Environ. Stud. 31, 1527–1534. doi: 10.15244/pjoes/143293
Wang, J. -Y., Wang, G. -P., Zhang, Y. -L., Zheng, B. -Q., Zhang, C. -L., and Wang, L. -W. (2014). Isolation and identification of an endophytic fungus Pezicula sp. in Forsythia viridissima and its secondary metabolites. World J. Microbiol. Biotechnol. 30, 2639–2644. doi: 10.1007/s11274-014-1686-0
White, T. J., Bruns, T., Lee, S. J. W. T., and Taylor, J. (1990). “Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics”, in: PCR Protocols: A Guide to Methods and Applications, eds. Innis, M.A., D.H. Gelfand, J. J. Sninsky, and T. J. White. (New York: Academic Press), 315–322.
Yang, H. -S., Yuan, Y. -G., Zhang, Q., Tang, J. -J., Liu, Y., and Chen, X. (2011). Changes in soil organic carbon, total nitrogen, and abundance of arbuscular mycorrhizal fungi along a large-scale aridity gradient. Catena 87, 70–77. doi: 10.1016/j.catena.2011.05.009
You, C., Yang, T. J., Zhou, X. G., Wang, X. F., Xu, Y. C., Shen, Q. R., et al. (2024). Research advances on mechanisms and preventions of soil-borne diseases exacerbated by root exudates in continuous cropping systems. Acta Pedol. Sin. 6, 1201–1211. doi: 10.11766/trxb202307180281
Yu, Y., Kang, X., Liu, T., Wang, Y., Tang, J., Peng, W., et al. (2025). Inoculation of the Morchella importuna mycosphere with Pseudomonas chlororaphis alleviated a soil-borne disease caused by Paecilomyces penicillatus. Biol. Fertil. Soils 61, 141–161. doi: 10.1007/s00374-024-01874-1
Yuan, J., Zhao, J., Wen, T., Zhao, M., Li, R., Goossens, P., et al. (2018). Root exudates drive the soil-borne legacy of aboveground pathogen infection. Microbiome 6, 156–112. doi: 10.1186/s40168-018-0537-x
Zhalnina, K., Louie, K. B., Hao, Z., Mansoori, N., Da Rocha, U. N., Shi, S., et al. (2018). Dynamic root exudate chemistry and microbial substrate preferences drive patterns in rhizosphere microbial community assembly. Nat. Microbiol. 3, 470–480. doi: 10.1038/s41564-018-0129-3
Zhang, Y., Dong, S., Gao, Q., Ganjurjav, H., Wang, X., and Geng, W. (2019). “Rare biosphere” plays important roles in regulating soil available nitrogen and plant biomass in alpine grassland ecosystems under climate changes. Agric. Ecosyst. Environ. 279, 187–193. doi: 10.1016/j.agee.2018.11.025
Zhang, X., and Li, Y. (2021). Drought-flood alternation dynamics drive the resurgence of soil-borne pathogens in agroecosystems. Front. Microbiol. 12:742315. doi: 10.3389/fmicb.2021.742315
Zhou, H., Gao, Y., Jia, X., Wang, M., Ding, J., Cheng, L., et al. (2020). Network analysis reveals the strengthening of microbial interaction in biological soil crust development in the mu us Sandy land, northwestern China. Soil Biol. Biochem. 144:107782. doi: 10.1016/j.soilbio.2020.107782
Keywords: pomegranate, root collar rot, fungal in the pomegranate rhizosphere, the diversity and structural composition, the soil physicochemical properties
Citation: Wu Z, Chen J, Chen J, Yang Y, Zhou A and Wu J (2025) The relationship between pomegranate root collar rot and the diversity of fungal communities in its rhizosphere. Front. Microbiol. 16:1573724. doi: 10.3389/fmicb.2025.1573724
Received: 09 February 2025; Accepted: 06 March 2025;
Published: 21 March 2025.
Edited by:
Md. Motaher Hossain, Bangabandhu Sheikh Mujibur Rahman Agricultural University, BangladeshReviewed by:
Hao Tan, Sichuan Academy of Agricultural Sciences, ChinaCopyright © 2025 Wu, Chen, Chen, Yang, Zhou and Wu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jianrong Wu, MTE3NjI3OTA0NEBxcS5jb20=
†These authors have contributed equally to this work and share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.