
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 27 March 2025
Sec. Microbe and Virus Interactions with Plants
Volume 16 - 2025 | https://doi.org/10.3389/fmicb.2025.1572568
 Yunqing Cheng
Yunqing Cheng Xingtong Jiang
Xingtong Jiang Xinyi He
Xinyi He Zhaoyang Wu
Zhaoyang Wu Qiang Lv
Qiang Lv Shuang Zhao
Shuang Zhao Xinyue Zhang
Xinyue Zhang Shuai Wang
Shuai Wang Hongli He
Hongli He Jianfeng Liu*†
Jianfeng Liu*†Introduction: While co-inoculation with rhizobia and plant growth-promoting rhizobacteria (PGPR) can enhance soybean growth and nodulation, the interaction mechanisms between Bacillus velezensis 20507 and Bradyrhizobium japonicum USDA110 under varying nitrogen (N) supply levels (0–10 mmol/L) remain unclear. This study investigates how their synergistic interactions influence soybean nitrogen content per plant and molecular pathways.
Methods: Soybean plants were co-inoculated with B. velezensis and B. japonicum across four N levels. Nodulation, plant growth, physiology, and N content were quantified. Transcriptome sequencing of soybean roots under N deficiency compared single and co-inoculation treatments. Flavonoids in B. velezensis fermentation broth were identified via mass spectrometry, and rutin’s regulatory effects on B. japonicum nodulation genes (NodD1/NodD2) were tested in coculture.
Results: Co-inoculation significantly increased nodulation, biomass, and N content per plant compared to single inoculations across all N levels. Under N deficiency, co-inoculation induced 5,367 differentially expressed genes (DEGs), with Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment in phenylpropanoid (ko00940) and flavonoid biosynthesis (ko00941). B. velezensis produced 29 flavonoids and 4 isoflavonoids (including rutin). Rutin (5–10 mg/L) upregulated NodD1 and suppressed NodD2 in B. japonicum.
Discussion: B. velezensis enhances B. japonicum-soybean symbiosis via flavonoid secretion, particularly rutin, which modulates nodulation gene expression. This metabiotic interaction improves soybean N assimilation and growth, even under low N conditions. The findings provide a foundation for designing composite inoculants to optimize soybean yield and nitrogen-use efficiency.
The excessive use of chemical fertilizers in agricultural production has led to increasingly prominent problems such as soil fertility decline and environmental pollution. Microbial fertilizers have attracted widespread attention because they can enhance plant growth, crop yield, and quality, while reducing fertilizer use. Bradyrhizobium japonicum is a common microbial fertilizer that solubilizes potassium and phosphorus and promotes growth and symbiotic nitrogen fixation in soybeans (Nguyen et al., 2012; Afzal et al., 2010). B. japonicum provides nitrogen nutrition to its hosts and increases soybean yield through symbiotic nitrogen fixation in leguminous plants. B. japonicum secretes metabolites, such as organic acids, which can also chelate minerals, such as iron, phosphorus, and magnesium, making them one of the most valuable microbial resources in natural and agricultural systems (Argaw, 2012). Soybeans are important symbiotic nitrogen-fixing crops, with rhizobial nitrogen fixation supplying 50–90% of the nitrogen required for soybean growth (Bai et al., 2014). Rhizobial inoculation is widely used in many soybean-producing countries, including the United States, Brazil, and Argentina (Wu et al., 2017). However, the low survival rate of B. japonicum under environmental stress conditions, which requires repeated inoculation, limits its practical application (Armendariz et al., 2019). The unstable effects of inoculation have led to the development of optimized single strains (Melissa et al., 2022; Melchiorre et al., 2011), as well as multi-strain and multi-functional inoculants (Alemneh et al., 2020; Juge et al., 2012).
Combining B. japonicum with plant growth-promoting rhizobacterial (PGPR) strains is conducive to symbiotic nitrogen fixation and growth promotion (Alemneh et al., 2020; Mary et al., 2012), and soybean nodulation and nitrogen fixation can be enhanced by co-inoculation with other PGPR via various mechanisms (Zeffa et al., 2020; Majeed et al., 2015; Fox et al., 2011; Vessey, 2003). The potential mechanism of PGPR co-inoculation promoting the growth of leguminous plants mainly focuses on the following aspects: (1) PGPRs produce auxin, which can promote the growth and development of plant roots and improve the absorption capacity of nutrients and water (Kuan et al., 2016; Majeed et al., 2015; Shao et al., 2015); (2) PGPRs themself are capable of biological nitrogen fixing and provide essential ammonia for plant growth (Adedayo and Babalola, 2023; Sharma et al., 2022; Sun et al., 2022; Alemneh et al., 2020; Kuan et al., 2016; Majeed et al., 2015; Babu et al., 2015); (3) PGPRs can dissolve inorganic phosphate, providing more phosphorus for plant growth (Majeed et al., 2015); (4) PGPR can promote symbiosis between leguminous plants and nitrogen-fixing microorganisms, which is beneficial for the formation and development of plant nodules and enhances the ability of plants to fix nitrogen (Zeffa et al., 2020; Alemneh et al., 2020; Miransari, 2014; Mary et al., 2012; Fox et al., 2011; Vessey, 2003). Though synergistic effects of dual microbial inoculation involving arbuscular mycorrhizal (AM) fungi and free-living diazotrophs demonstrate significant enhancement in plant productivity through improved biomass accumulation and agricultural yield (Kasanke et al., 2024), To date, the mechanism by which PGPRs promote symbiosis between leguminous plants and nitrogen-fixing microorganisms has not yet been fully elucidated.
Bacillus velezensis, a spore-forming strain, is widely distributed in nature and used in agricultural production. Its ease of isolation and cultivation, rapid growth, and stress resistance make it a suitable inoculum (Cheng et al., 2024). It also promotes the growth of wheat, beets, carrots, peppers, potatoes, radishes (Meng et al., 2016), cucumbers (Sun et al., 2022), and soybean (Kondo et al., 2023). B. velezensis BAC03 promotes the growth of numerous plants under greenhouse conditions. Some studies have suggested that the mechanism by which this strain promotes plant growth may be related to its ability to synthesize auxins and ACC deaminase (1-aminocyclopropane-1-carboxylate deaminase) (Meng et al., 2016). B. velezensis SQR9 can directly promote the growth of cucumbers by producing auxins, volatile compounds acetoin and 2,3-butanediol, and extracellular phytases (Shao et al., 2015). There was a syntrophic cooperative relationship between the PGPR B. velezensis SQR9 and the indigenous Pseudomonas stutzeri in the cucumber rhizosphere. These two stains exchange metabolites and there was also a phenomenon of metabolic facilitation. Biofilm matrix components from Bacillus play an important role in this interaction, and the formation of a consortium is beneficial for promoting plant growth (Sun et al., 2022). B. velezensis S141 can promote the growth and nodulation of soybean plants, as well as the symbiosis between soybean and B. diazoefficiency USDA110. It is speculated that the main mechanism of action is that S141 can produce β-glucosidase, efficiently hydrolyzed isoflavone glucosides produced by soybean roots into isoflavone aglycones, and promote soybean nodulation. However, it is not clear which β-glucosidases are responsible for hydrolyzing isoflavone glucosides (Kondo et al., 2023).
B. velezensis exerts significant modulatory effects on indigenous soil microbial communities through competitive colonization dynamics and bioactive metabolite exchange. Spatial–temporal analyses reveal that B. velezensis FH-1 enhances early-stage seedling vigor in rice by reprogramming microbial community structure and functional profiles at critical developmental interfaces (Dai et al., 2024). Notably, B. velezensis SQR9 demonstrates dual functional capacities: direct plant growth promotion and metabolic mediation that selectively enriches native P. stutzeri populations. This interspecies synergy facilitates stable polymicrobial biofilm formation in the rhizosphere niche, establishing a persistent plant-beneficial consortium that significantly outperforms monoculture systems in cucumber cultivation trials (Sun et al., 2022; Chen et al., 2016). Mechanistic investigations of this syntrophic relationship reveal coordinated cellular protection mechanisms and enhanced Na+ homeostasis regulation under saline stress, providing the scientific basis for developing next-generation biofertilizers combining these microbial partners.
While extensive studies have documented individual applications of B. velezensis and B. japonicum in agricultural systems, critical knowledge gaps persist regarding their combinatorial effects on legume symbiosis regulation. Particularly, the mechanistic interplay between these microorganisms in modulating soybean growth-nodulation dynamics under varying nitrogen regimes remains poorly characterized—a significant oversight given the well-documented nitrogen-mediated suppression of diazotrophic activity (Saito et al., 2014). Our experimental design systematically evaluates single and co-inoculation strategies across nitrogen gradients, employing multi-omics approaches to: (1) quantify phenotypic responses in seedling growth and nodulation efficiency, (2) map root transcriptome reprogramming under nitrogen limitation, and (3) characterize metabolic cross-talk through bacterial exometabolite profiling. Crucially, we establish flavonoid-mediated regulation of nod gene expression as a key mechanism underlying microbial synergism. These findings collectively provide a robust theoretical framework for engineering advanced soybean inoculants and optimizing microbial consortia in sustainable fertilization practices.
The experimental strain B. velezensis 20507 (CGMCC deposit No. 20507) was isolated and maintained in axenic culture in our laboratory, while B. japonicum USDA110 was procured from Prof. Xuelu Wang (Zhengzhou University, China). Both strains were cultivated under standardized conditions (28°C, 200 rpm orbital shaking) using selective media: PDA (Potato Dextrose Agar) for B. velezensis and TY (Tryptone Yeast extract) agar for B. japonicum. Bacterial biomass was harvested via centrifugation (5,200 × g, 20 min, 4°C) followed by three sequential deionized water washing cycles. To delineate nitrogen-dependent microbial interactions, a factorial design encompassing four nitrogen regimes (N0: 0 mM, N2: 2 mM, N5: 5 mM, N10: 10 mM Hoagland’s solution) and three inoculation strategies (monocultures: 10 μL of B. velezensis [Bv] or B. japonicum [Bj] at 1 × 109 CFU/mL; co-culture [BB]: 5 μL Bv + 5 μL Bj) was implemented. Standardized cell suspensions (OD600 = 0.4) were inoculated into 400 μL culture systems (10 μL bacterial suspension +390 μL medium), with growth kinetics monitored using a Bioscreen C system (Lab Systems, Finland) under controlled parameters (28°C, 180 rpm, 7-day duration, OD600 measurements at 2-h intervals, n = 5 replicates). Concurrently, dual-culture assays on PDA plates inoculated with 20 μL aliquots of logarithmic-phase cultures (1 × 109 CFU/mL per strain, 2.0 cm separation distance) facilitated spatial interaction analysis, with phenotypic documentation at 24-h intervals under standard incubation (28°C).
Pot experiments were conducted in controlled-environment chambers (School of Life Sciences, Jilin Normal University) under optimized growth parameters: 28°C/22°C Day/night thermoperiod, 70% relative humidity, and 12-h photoperiod with 400 μmol·m−2·s−1 photosynthetic photon flux density (LED illumination). Glycine max cv. Williams82 seeds underwent surface sterilization through sequential washing protocols: 5-min tap water cleansing followed by three 3-min sterile water rinses, culminating in 12-h imbibition at 28–30°C for synchronized germination. Germinated propagules were transplanted into 4.4-L horticultural pots (12.7 cm top Ø × 11.4 cm height) containing vermiculite substrate irrigated with nitrogen-gradient Hoagland solutions (0, 2, 5, 10 mM NO₃−; designated N0-N10). A full-factorial design incorporating four nitrogen levels and four inoculation treatments [CK: autoclaved control; Bv: B. velezensis 20507 (3 mL, 1 × 109 CFU·mL−1); Bj: B. japonicum USDA110 (3 mL, 1 × 109 CFU·mL−1); BB: 1:1 Bv:Bj co-inoculation] generated 16 experimental groups, each comprising three biological replicates (n = 8 seedlings/replicate). For experimental controls, sterilized inoculum (121°C, 20 min) replaced live bacterial suspensions. Physiological assessments at 30 days post-inoculation included: (1) chlorophyll quantification via SPAD-502PLUS meter (Konica Minolta, Japan), (2) photosynthetic gas exchange parameters measured by LI-6400XT portable photosynthesis system (Li-Cor Biosciences, United States) under standardized conditions (28°C leaf temperature, 400 μmol·mol−1 CO₂, 400 μmol·m−2·s−1 irradiance) during morning hours (09:00–12:00), and (3) morphometric analyses encompassing plant height, stem diameter (digital caliper), leaf area (YMJ-B area meter, Top Yunnong Tech, China), and biomass partitioning (aboveground tissues, root systems excluding nodules, and nodular structures). Tissue nitrogen content was determined through sulfuric acid-hydrogen peroxide digestion followed by automated Kjeldahl analysis (K-360, Büchi Labortechnik, Switzerland).
After 30 days of cultivation, roots with seedling nodules were sampled and sent to Sangon Biotech (Shanghai) Co., Ltd. for RNA extraction, sequencing on an Illumina HiSeq4000 PE150 platform, and bioinformatic analysis. The obtained raw transcriptome sequencing data were stored in SRA1 under the accession number PRJNA1152285. Clean data were aligned to the soybean reference genome using HISAT2 v.2.1.0 (Kim et al., 2015), and FastQC v0.11.2 was used to visually evaluate the sequencing data. StringTie v1.3.3b (Pertea et al., 2015) was used to assemble the mapped sequences onto the genome and evaluate gene expression levels. DESeq2 v1.12.4 was used to identify differentially expressed genes (DEGs) using the criteria of |Log2Fold Change| > 1.00 and false discovery rate (FDR) < 0.05. Genegene ontology and Kyoto Encyclopedia of Genes and Genomes enrichment analyses were performed using topGO v2.24.0, clusterProfiler v3.0.5, and an adjusted Q-value < 0.05, which was used as the criterion to judge the significantly enriched GO terms and KEGG pathways.
B. velezensis 20507 was cultured for 3 days in PDA. The bacterial cells were then collected by centrifugation at 5,200 × g for 20 min at 4°C. The supernatant fermentation broth sample was freeze-dried, and 25 mg of solid sample was dissolved in 1,000 μL of extraction solution (methanol: acetonitrile: water = 2:2:1, (v/v/v)) containing an isotope-labeled internal standard. Vortex mixing was performed for 30 s, after which the solution was transferred to an ice-water bath and sonicated for 5 min. This step was repeated three times. After standing at −40°C for 1 h, the solution was filtered through a 0.22 μm filter membrane, and the filtrate was collected for detection. Vanquish (Thermo Fisher Scientific) ultra-high performance liquid chromatography was used to separate the target compound using a Phenomenex Kinetex C18 (2.1 mm × 50 mm, 2.6 μm) liquid chromatography column. The A phase of liquid chromatography was an aqueous phase containing 0.01% acetic acid and the B phase was isopropanol: acetonitrile (1:1, v/v). Column temperature: 25°C; sample tray temperature: 4°C; injection volume: 2 μL. The Orbitrap Exploris 120 mass spectrometer could collect primary and secondary mass spectrometry data under the control of a control software (Xcalibur, version 4.4, Thermo). The detailed parameters were as follows: sheath gas flow rate, 50 Arb; Aux gas flow rate, 15 Arb, Capillary temperature, 320°C; Sweep Gas, 1 arb; vaporizer temperature, 350°C. Full ms resolution: 60000, MS/MS resolution: 15000, Collision energy: SNCE 20/30/40, Spray Voltage: 3.8 kV (positive) or − 3.4 kV (negative). After converting the raw data into the mzXML format using ProteoWizard software, metabolite identification was performed using the R package (Zhou et al., 2022), and the database used was BiotreeDB (V3.0).
To investigate the regulatory effects of rutin (RU) and B. velezensis on nodD1/nodD2 expression in B. japonicum USDA110, six experimental treatments were established in nitrogen-free Hoagland’s solution: (i) blank control (CK); (ii) B. velezensis monoculture (Bv); (iii–vi) RU gradient concentrations (5, 10, 20, 40 mg/L designated RU5-RU40). Pharmaceutical-grade rutin (≥98% purity, Sigma-Aldrich, United States) was dissolved in DMSO (0.1% final concentration). Co-inoculation treatments (BB) received both B. velezensis (1 × 108 CFU/mL) and B. japonicum USDA110 (1 × 109 CFU/mL) in 400 mL culture systems. Bacterial cells were harvested after 48-h incubation via centrifugation (5,000 × g, 10 min), followed by RNA extraction using the EASYspin Plus Kit (Aidibo Biotechnology, China). Quantitative reverse transcription PCR (qRT-PCR) was conducted on an AriaMx system (Agilent Technologies, United States) with 10 μL reactions under standardized cycling parameters: initial denaturation at 95°C (2 min); 40 cycles of 95°C (15 s), 60°C (15 s), 72°C (20 s). The 16S rRNA (USDA110) and β-actin (plant samples) served as endogenous controls for microbial and plant gene quantification, respectively. Twelve differentially expressed genes (DEGs) from RNA-seq comparisons (BB vs. Bj) were validated through this protocol. Relative gene expression was calculated via the 2−ΔΔCt method (Schmittgen and Livak, 2008) with triplicate technical replicates. Primer sequences are cataloged in Supplementary Table S1.
Variance analysis was performed using two-way ANOVA (main factors of four N concentration treatments and four microbial inoculation treatments) in SPSS (Statistical Package for the Social Sciences) software version 26, with a t-test evaluating statistically significant differences at the 5% level.
The nitrogen concentration gradient in the culture medium and microbial inoculation significantly affected the growth of soybean seedlings (Figure 1). Compared with the control under the same nitrogen concentration, the seedling heights of the Bv, Bj, and BB treatments were higher under N0 conditions (Figure 1A). These results suggest that compared to the control, the Bv, Bj, and BB treatments enhanced soybean seedling growth. Moreover, among the different inoculation treatments, the BB treatment resulted in the most pronounced growth improvement. Similarly, under N2 (Figure 1B), N5 (Figure 1C), and N10 (Figure 1D) conditions, seedlings treated with Bv, Bj, or BB showed better growth than the control, with the BB treatment consistently achieving the highest growth performance. Collectively, these findings indicate that separate inoculation of B. velezensis or B. japonicum promotes soybean growth, while combined inoculation (BB) synergistically enhances seedling growth compared to single-strain applications.

Figure 1. Phenotypic responses of soybean seedlings to nitrogen gradients and bacterial inoculation treatments. (A–D) Representative images under 0, 2, 5, and 10 mM nitrogen concentrations with four treatments: CK (control), Bv (Bacillus velezensis), Bj (Bradyrhizobium japonicum USDA110), and BB (Bv + Bj co-inoculation). Scale bars: 8.0 cm. Plants were cultivated in vermiculite for 30 days.
Soybean seedlings were nourished with Hoagland’s solution containing graded nitrogen levels (N0, N2, N5, N10). B. velezensis and B. japonicum were inoculated either singly or in combination in the respective nitrogen media, and their growth curves were compared. For B. velezensis, the OD values in N2, N5, and N10 treatments exhibited an initial increase followed by a decline, whereas in N0, the OD value rose steadily, showing divergent trends among treatments (Figure 2A). After 168 h, the highest OD value for B. velezensis was observed in N0, followed by N10, while N2 and N5 had the lowest values (Figure 2A). B. japonicum displayed a similar pattern, with the highest OD value in N0 and the lowest in N5 and N10 after 168 h (Figure 2B). In co-inoculation experiments, the OD value in N0 was markedly higher than in other treatments at 168 h (Figure 2C). Specifically, under nitrogen-deficient conditions (N0), the OD value of the mixed inoculation reached 0.25 (Figure 2C), exceeding the values for B. velezensis (0.18; Figure 2A) and B. japonicum (0.13; Figure 2B) in monoculture. Plate confrontation assays (Figures 2D–F) revealed no antagonism between the two strains. Based on the observed metabolic interdependence, we propose that their interaction represents metabiosis—a relationship where organisms benefit mutually through metabolic activities. In summary, both strains exhibited higher proliferation rates in nitrogen-deficient media than in nitrogen-supplemented media, and their interaction was characterized by metabiosis.
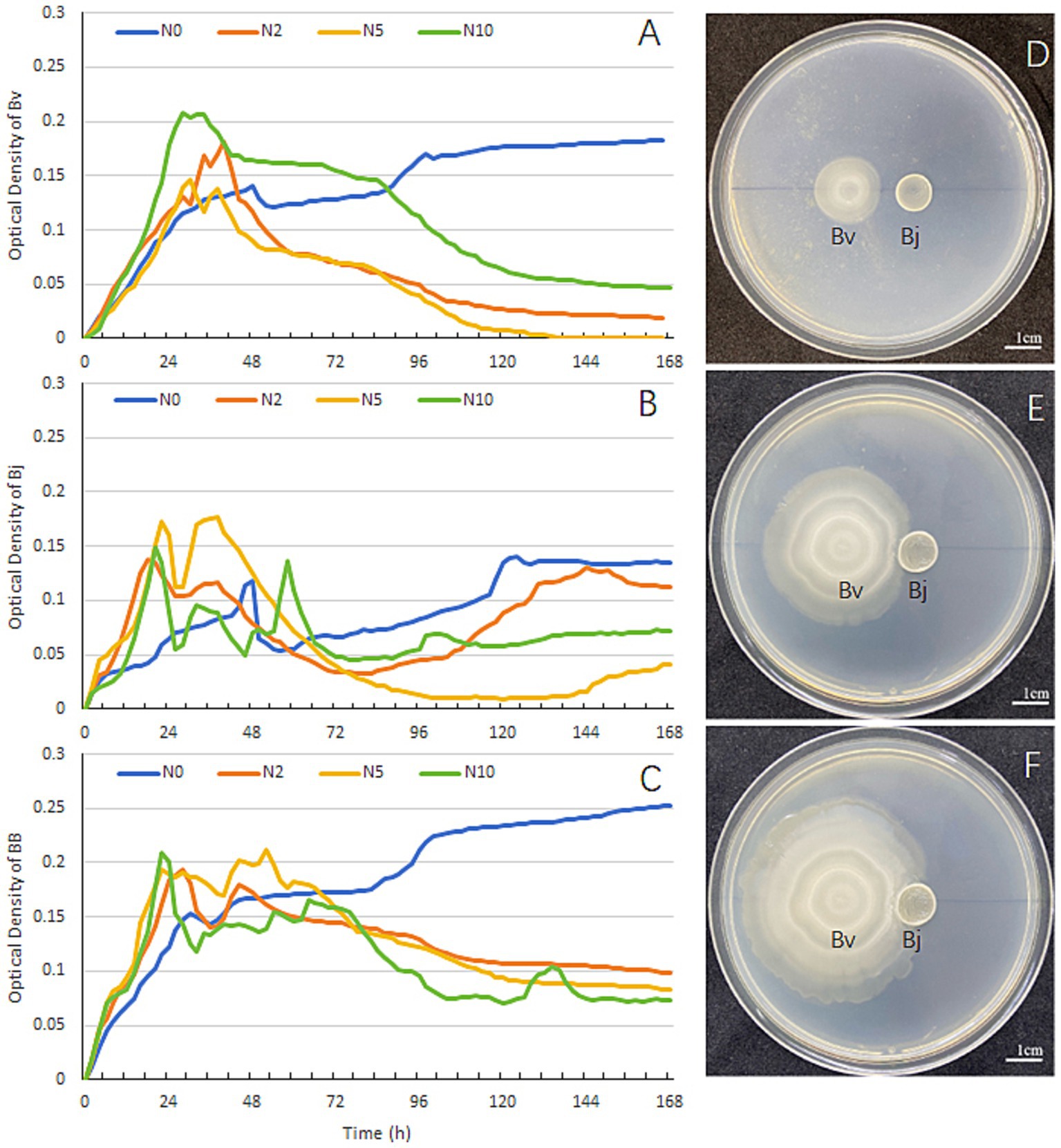
Figure 2. In vitro growth dynamics and spatial interactions between B. velezensis and B. japonicum USDA110. Growth curves of (A) B. velezensis, (B) B. japonicum, and (C) co-culture under four nitrogen regimes (N0-N10: 0–10 mM). (D–F) Plate confrontation assay at 24, 48, and 72 h post-inoculation. Cultures were maintained for 7 days at 28°C with continuous shaking (200 rpm).
Nitrogen concentration (N), strain inoculation (S), and N × S interaction significantly influenced plant height, stem diameter, leaf area, and SPAD (Figure 3). Increasing nitrogen levels led to reduced plant height (Figure 3A) but increased stem diameter (Figure 3B), resulting in more robust seedlings. Under the same nitrogen concentration, Bj and Bv treatments increased both plant height and stem diameter, while BB further enhanced these parameters compared to CK, Bj, and Bv (Figures 3A,B). Similarly, leaf area declined with increasing nitrogen levels under N2, N5, and N10 (Figure 3C), yet BB treatment maximized leaf area across all nitrogen conditions (Figure 3C). SPAD values were significantly higher in Bj and BB treatments than in CK under the same nitrogen levels (Figure 3D). Overall, the BB treatment achieved the highest values for all measured traits (plant height, stem diameter, leaf area, SPAD) and most effectively promoted soybean seedling growth.
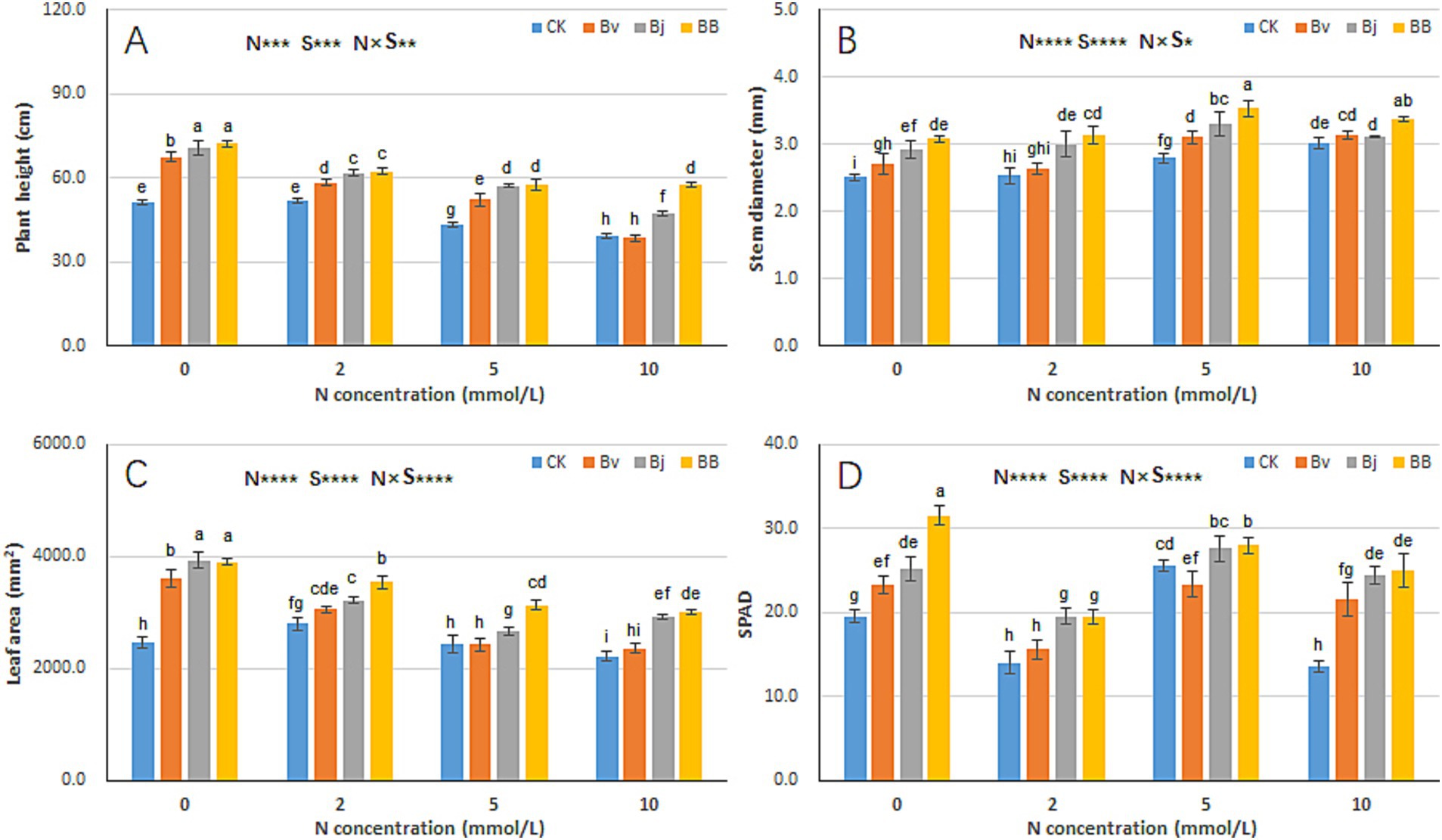
Figure 3. Nitrogen-inoculation interactions on soybean growth parameters. (A) Plant height, (B) stem diameter, (C) leaf area, and (D) SPAD values (Soil Plant Analysis Development). N: nitrogen levels (0–10 mM); S: treatments (CK, Bv, Bj, BB). Data represent mean ± SD (n = 12). Distinct lowercase letters denote significant differences (t-test, p < 0.05; ** < 0.01; *** < 0.001; **** < 0.0001).
Nitrogen (N), strain inoculation (S), and N × S interaction significantly affected total plant dry weight (Figure 4A). Additionally, N and S influenced root nodule dry weight (Figure 4B). Under the same nitrogen concentration, Bj and BB treatments yielded higher total plant weights than CK, with BB consistently outperforming other treatments (Figure 4A). In pot experiments, root nodules formed exclusively in Bj and BB treatments (inoculated with B. japonicum), whereas CK and Bv (lacking B. japonicum) produced no nodules. Root nodule weight was inversely correlated with medium nitrogen content. At N0, N2, and N5, BB treatment generated significantly heavier nodules than Bj (Figure 4B), suggesting that B. velezensis facilitates B. japonicum infection and nodulation in soybean roots.
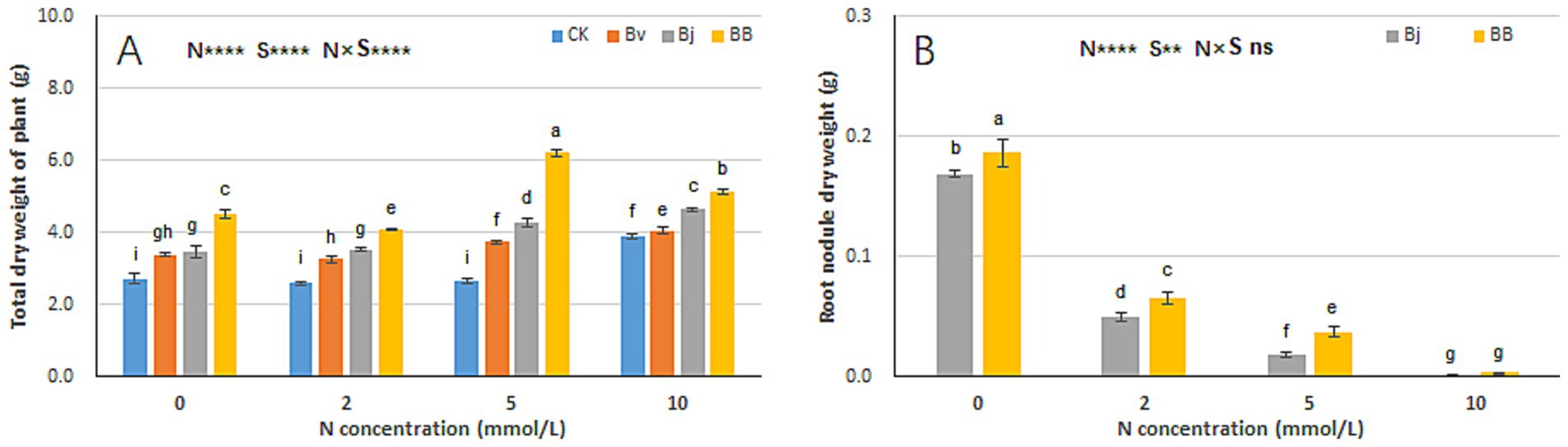
Figure 4. Biomass allocation patterns under experimental treatments. (A) Whole-plant dry weight, (B) nodule dry weight. Statistical conventions as in Figure 3.
The photosynthetic characteristics of soybean seedlings under different nitrogen concentrations and inoculation conditions were analyzed. Nitrogen (N), strain inoculation (S), and N × S interaction significantly affected photosynthetic parameters and water use efficiency (WUE) (Figure 5). Under the same nitrogen concentration, net photosynthetic rate (Pn) values of Bj (B. japonicum), Bv (B. velezensis), and BB (B. japonicum + B. velezensis) treatments were higher than those of CK (control). Notably, BB treatment exhibited the highest Pn values, demonstrating superior enhancement of photosynthetic efficiency compared to single-strain treatments (Figure 5A). Under N0, N2, and N5 conditions, intercellular CO₂ concentration (Ci) values of BB were lower than those of CK, Bj, and Bv (Figure 5C), while stomatal conductance (Cond) and WUE of BB were consistently higher than other treatments (Figures 5B,D). These findings suggest that co-inoculation (BB) synergistically optimizes photosynthetic performance and water utilization in soybean seedlings.
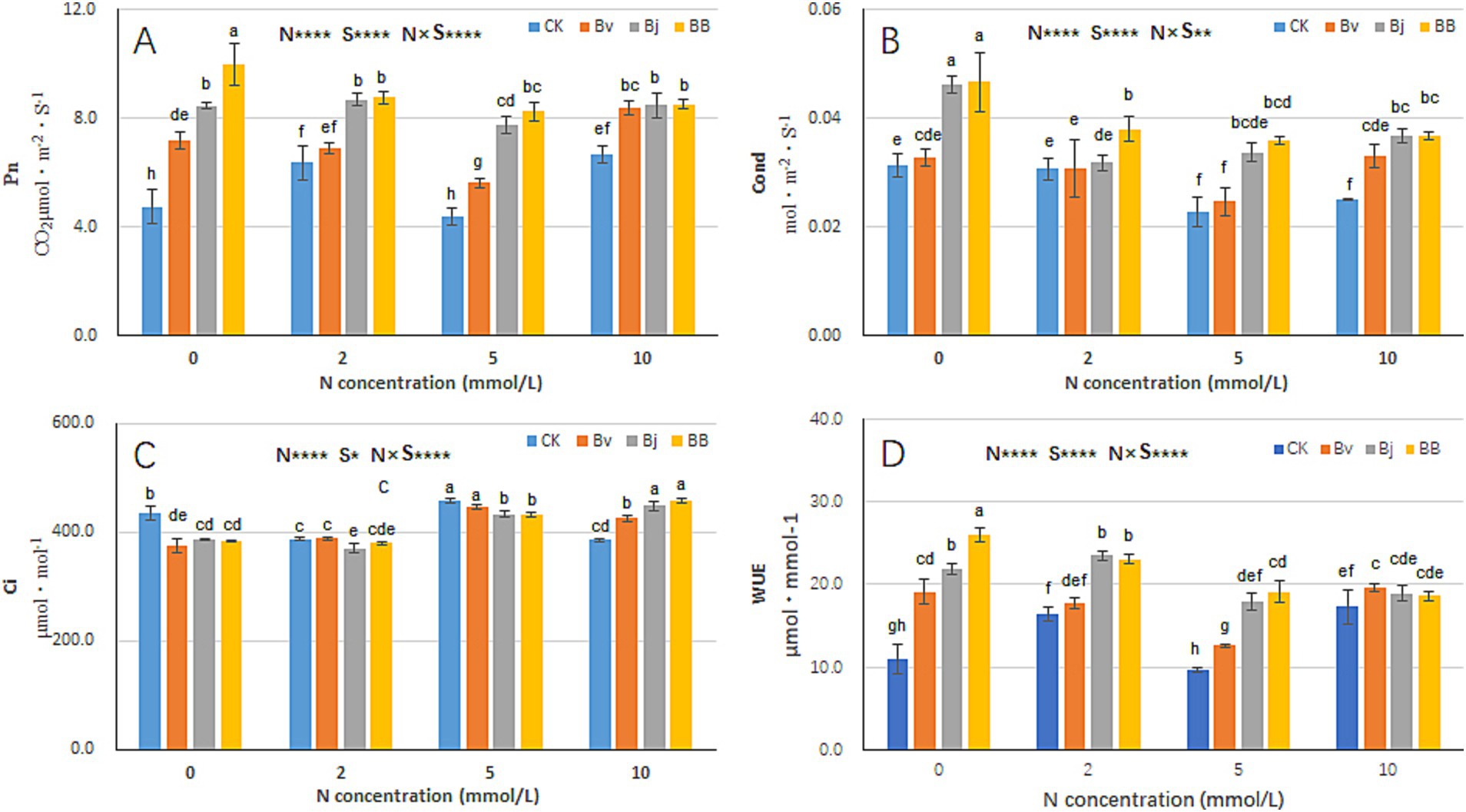
Figure 5. Photosynthetic performance under combinatorial treatments. (A) Net photosynthetic rate (Pn), (B) stomatal conductance (Cond), (C) intercellular CO2 concentration (Ci), (D) water-use efficiency (WUE). Data represent mean ± SD (n = 6). Statistical analysis as in Figure 3.
Nitrogen (N), strain inoculation (S), and N × S interaction significantly influenced nitrogen content in roots, stems, leaves, and total plant nitrogen (Figure 6). Increasing nitrogen levels correlated with elevated stem and leaf nitrogen content (Figures 6A,B). Under the same nitrogen concentration, Bj and BB treatments accumulated more nitrogen in leaves and stems than CK. Specifically, under N0 conditions, BB treatment achieved the highest nitrogen content, significantly surpassing CK and Bj (Figures 6A–C). Total plant nitrogen content increased with nitrogen concentration (Figure 6D), with the following efficacy ranking across treatments: BB > Bj > Bv > CK under N0, N2, and N5 conditions. At N10, BB, and Bj maintained higher total nitrogen than CK and Bv, highlighting BB as the most effective strategy for enhancing nitrogen uptake efficiency.
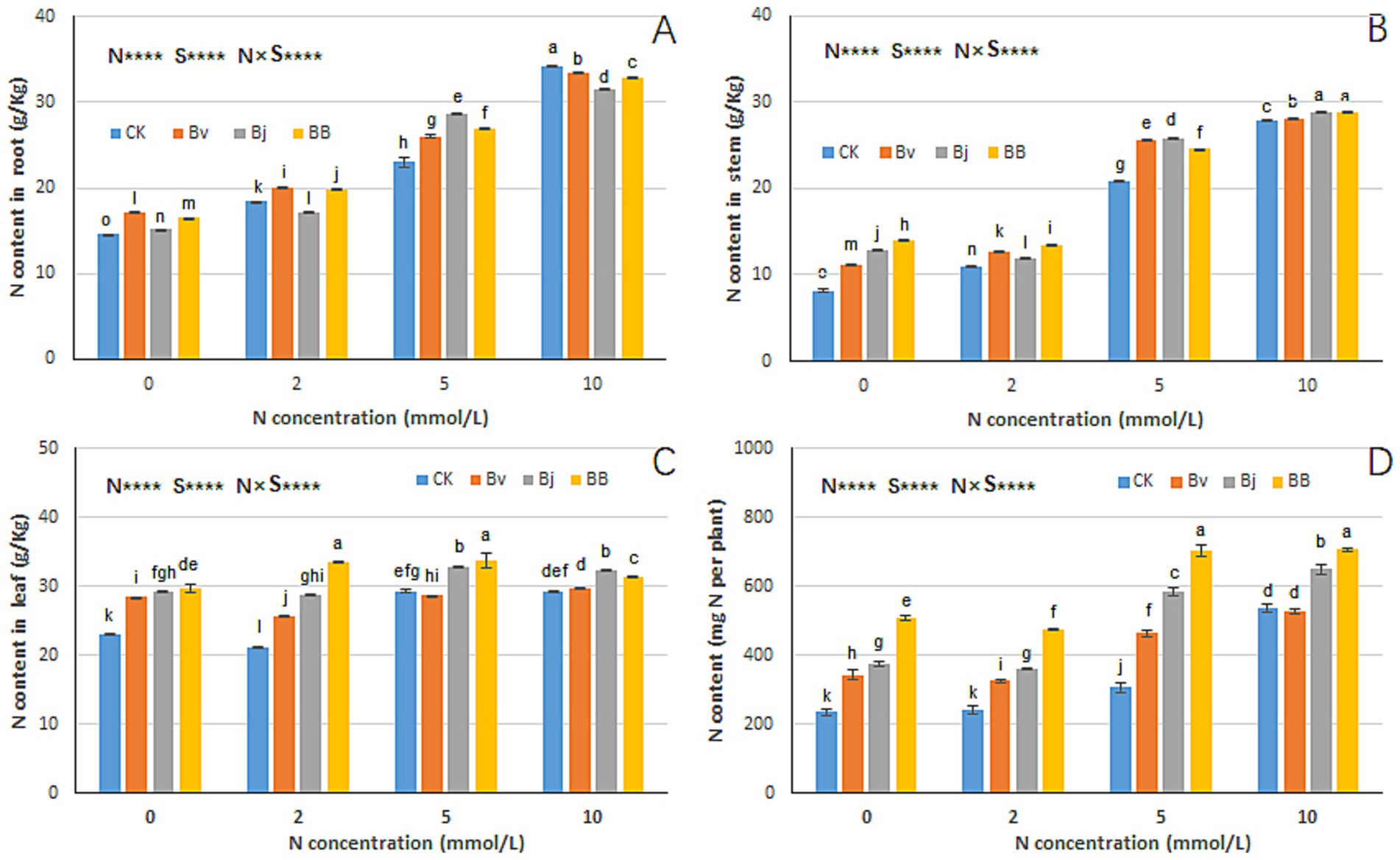
Figure 6. Nitrogen partitioning in plant tissues. Tissue-specific N content in (A) roots, (B) stems, (C) leaves, and (D) whole plants. Experimental conditions and statistical methods as in Figure 3.
Twelve root samples from the soybean seedlings were collected and sequenced, generating 603,274,676 raw reads. These reads covered a length of 90,491,201,400 bp (Supplementary Table S2). Low-quality reads were filtered, and 562, 043, and 316 clean reads were generated. Clean reads were mapped to the soybean reference genome, and 546,957,198 clean reads were aligned to the reference genome, accounting for 97.31% of the obtained clean reads (Supplementary Table S3). All DEGs in paired comparison of BB vs. Bj are listed in Supplementary Table S4.
There were 10,051 DEGs in the paired comparison of Bv vs. Ck, and the number of downregulated genes was 2.37 times that of upregulated genes. There were 4,697 DEGs between Bj and Ck, and the number of downregulated genes was relatively close to that of upregulated genes. There were 13,960 DEGs in Bj vs. Bv, and the number of downregulated genes was 2.11 times that of upregulated genes (Figure 7A). There were 18,098 DEGs in BB vs. Bv, and the number of upregulated genes far exceeded that of downregulated genes. There were 5,367 DEGs in the BB vs. Bj group, which was much fewer than the number of DEGs in the BB vs. Bv. There were 11,528 DEGs between BB and CK, and the numbers of upregulated and downregulated genes were similar (Figure 7B). The three comparisons generated 21,765 DEGs, and the common DEGs shared by the three groups accounted for 15.74% of all DEGs (Figure 7B). Among the three paired comparisons, BB vs. Bv produced the most DEGs, whereas BB vs. Bv produced fewer DEGs. In pairwise comparisons of Bv vs. CK and Bj vs. CK, there were more DEGs in Bv vs. CK. These results indicate that Bv significantly impacted the expression of soybean root genes, whereas Bj had a relatively small impact on the expression of root genes. In the three paired comparisons of Bv vs. CK, Bj vs. CK, and Bv vs. Bj, 17,294 DEGs were generated, with only 759 DEGs shared among the three, accounting for only 4.39% of all the DEGs (Figure 7C). Thus, there was a significant difference between the DEGs induced by Bv and Bj treatments.
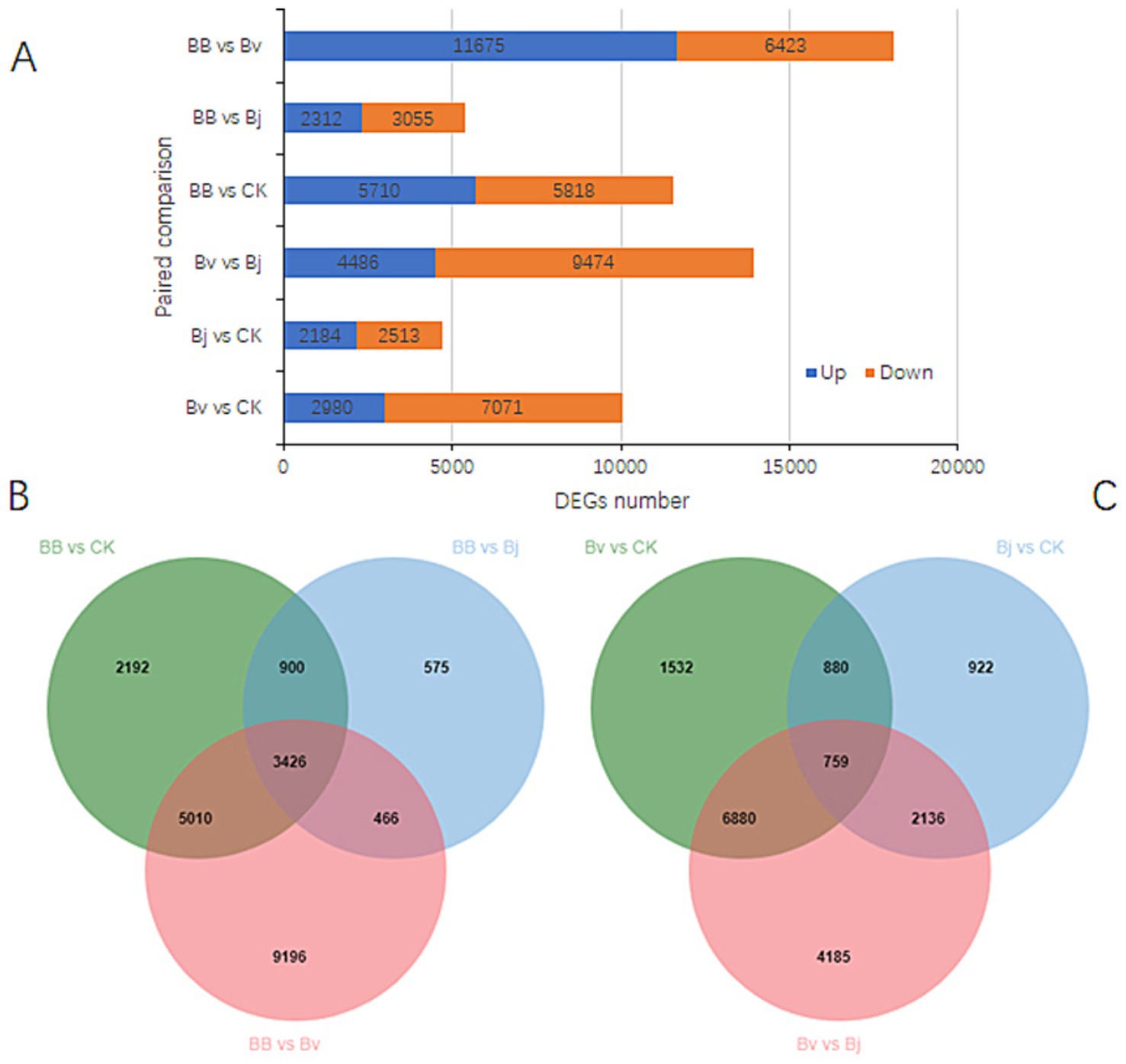
Figure 7. Differential gene expression profiling. (A) DEG counts across six comparisons. Venn diagrams of shared DEGs in (B) BB-centric comparisons and (C) pairwise monoculture contrasts. CK: control; BB: Bv + Bj co-inoculation.
The GO enrichment of all DEGs in BB vs. Bj comparison are listed in Supplementary Table S5. GO (Gene Ontology) clustering analysis of DEGs in BB vs. Bj was performed, and the results are shown in Figure 8. All DEGs were clustered into 63 GO Terms annotated to 25 biological processes, 20 cellular components, and 18 molecular functions. Catalytic activity (GO: 0003824) was the GO term with the most members in the molecular function category, with 1,150 DEGs, followed by binding (GO: 0005488) and transporter activity (GO: 0005215) with 711 and 239 DEGs, respectively. The GO term with the most members in the cellular component category was cell part (GO: 0044464), with 1,952 DEGs, followed by organelle (GO: 0043226) and membrane (GO: 0044425), with 1,289 and 851 DEGs, respectively. The GO term with the most members in the biological process category was cellular process (GO: 0009987), followed by metabolic process (GO: 0008152), and response to stimulus (GO:0050896). Based on these results, it is believed that co-inoculation with BB and Bj significantly impacted the expression of genes related to metabolism, response to stimulus, and substance transport in soybean seedling root cells compared with Bj treatment alone.
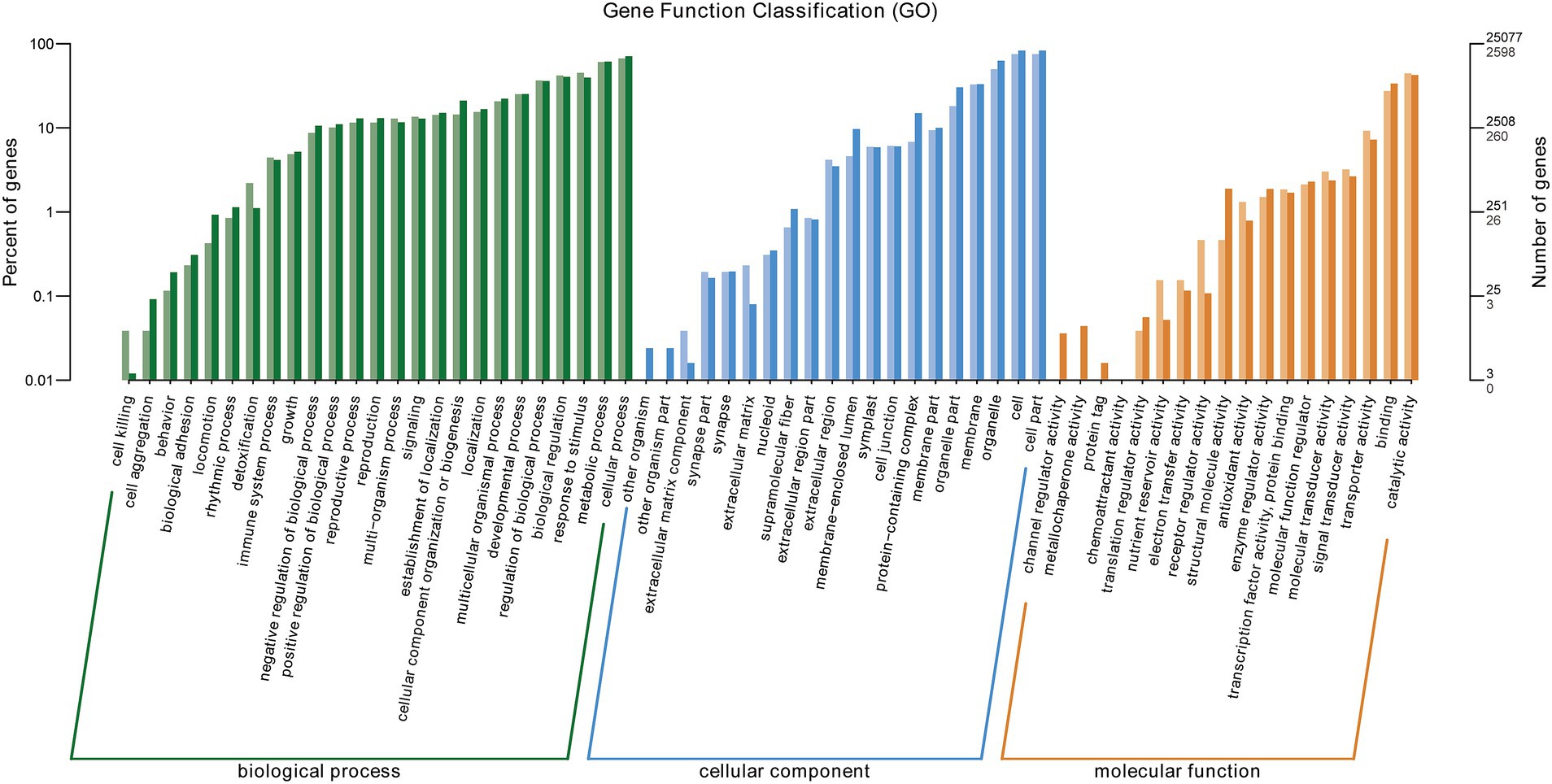
Figure 8. Gene ontology (GO) enrichment of BB vs. Bj DEGs. X-axis: GO terms; Y-axis: gene count/percentage.
KEGG enrichment analysis revealed 46 significantly enriched pathways in the BB vs. Bj comparison (Supplementary Table S6; Figure 9), despite no pathways being enriched in Bv vs. Bj (13,960 DEGs), indicating divergent molecular mechanisms between B. velezensis (Bv) and B. japonicum (Bj). Top enriched pathways included: phenylpropanoid biosynthesis (ko00940), flavonoid biosynthesis (ko00941), MAPK signaling (ko04016), photosynthesis (ko00195), plant hormone transduction (ko04075), and nitrogen metabolism (ko00910), suggesting co-inoculation (BB) broadly regulates secondary metabolism, stress adaptation, and nutrient utilization compared to Bj alone.
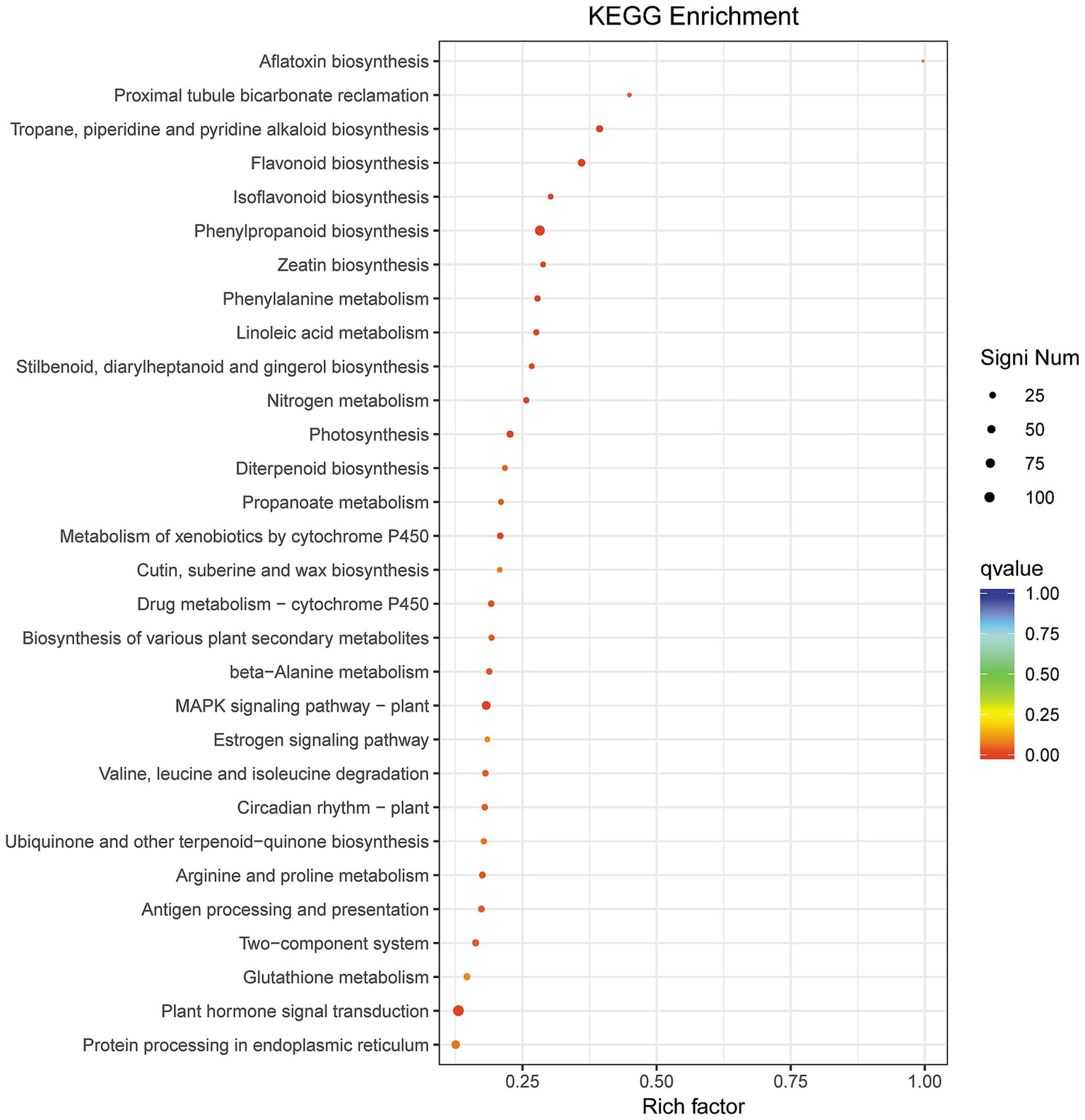
Figure 9. KEGG pathway analysis of BB vs. Bj DEGs. Dot plot visualization with rich factor (DEG proportion per pathway), Q-value (color gradient), and gene count (dot size). Red coloration indicates higher significance.
We sequenced and assembled the genome of B. velezensis 20507 and identified the chalcone synthase gene (tig00000001_pilon_281) (Supplementary Table S7), which is considered a key enzyme in flavonoid synthesis. Furthermore, to determine whether B. velezensis 20507 secretes flavonoids to promote the growth of USDA110 and soybean seedlings, metabolites were identified in the fermentation broth of B. velezensis 20507, and the identified compounds are listed in Supplementary Table S8. There were 390 metabolites in the broth of B. velezensis 20507, of which 360 belonged to seven superclasses (Figure 10A). The fatty acid superclass had the largest number of members, accounting for 29.49% of the total identified compounds, followed by shikimates and phenylpropanoids with 91 compounds accounting for 23.33% of the total. Further classification of shikimates and phenylpropanoids revealed that there were 29 flavonoid classes and four isoflavonoid classes, accounting for 31.87 and 4.40% of the total compounds in shikimates and phenylpropanoids, respectively (Figure 10B). The 29 flavonoid compounds were further divided into eight subclasses, including chalcones (e.g., 4-Hydroxydericin and Droloxifene), flavanones (e.g., Bavachinin and Glabrol), flavones (e.g., Hosloppin and Oroxindin), and flavonols (e.g., Rutin and Kaempferol) contained numerous compounds, totaling 24, accounting for 83% of flavonoid compounds (Figure 10C). In summary, total flavonoid/isoflavonoid content reached 8% of identified metabolites, supporting the role of B. velezensis in flavonoid-mediated symbiosis enhancement.
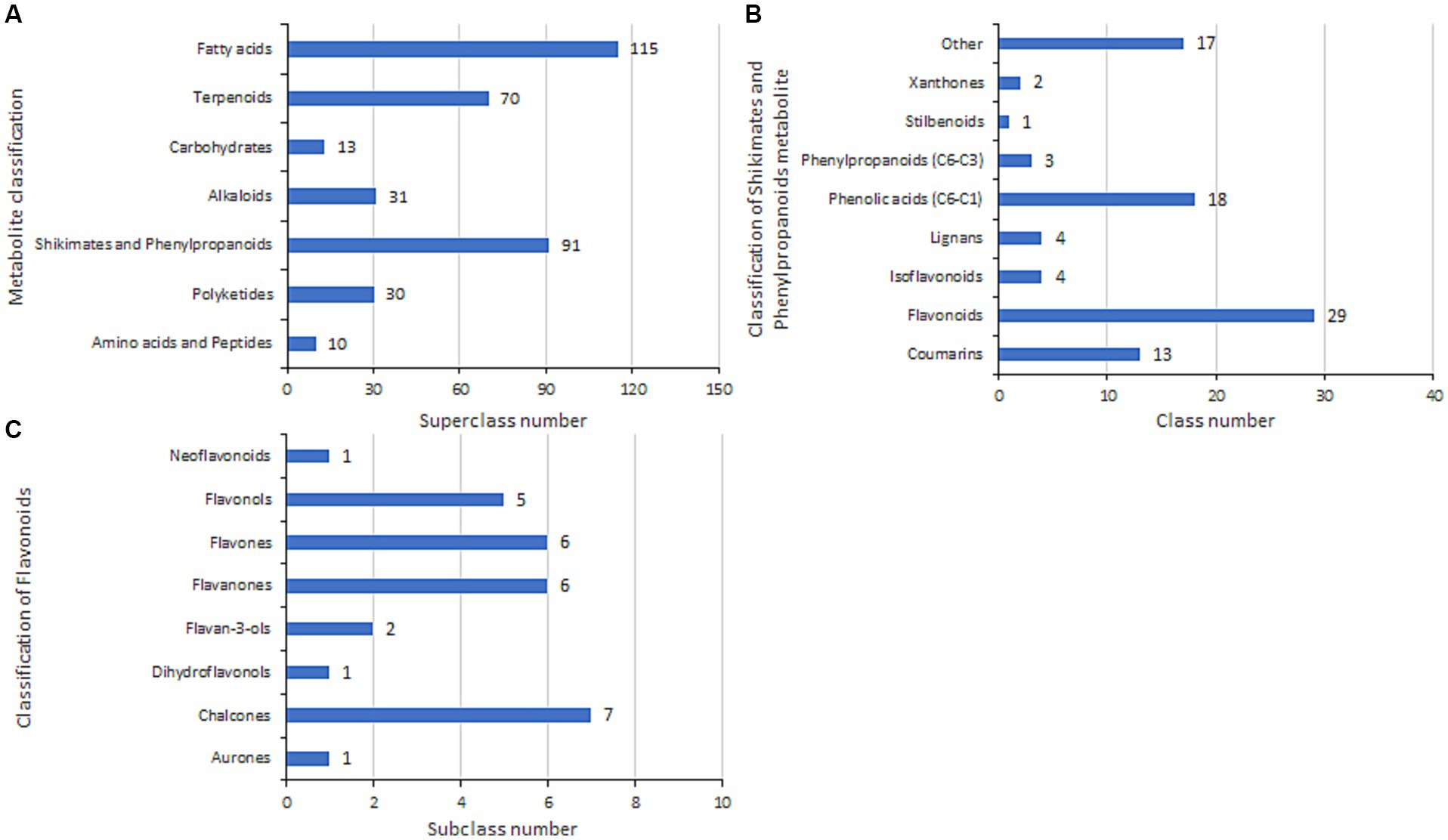
Figure 10. Metabolomic profiling of B. velezensis 20507 fermentation products. (A) Metabolite superclass distribution. (B) Shikimate-phenylpropanoid derivatives. (C) Flavonoid subclasses.
The addition of rutin to the culture medium and co-inoculation with B. velezensis had vital effects on the expression of NodD1 and NodD2. Within the range of 0–10 mg/L rutin, the expression level of NodD1 increased with increasing rutin concentrations. Within the range of 10–40 mg/L, the expression level of NodD1 decreased with increasing rutin concentration (Figure 11A). Compared with the control, the three treatments (RU5, RU10, and BB) significantly increased the expression levels of NodD1 (Figure 11A). Compared with the control, the other five treatments significantly reduced NodD2 expression levels (Figure 11B). Thus, the trend of NodD1 was opposite to that of NodD2 (Figures 11A,B). For paired comparisons of BB and Bj, a set of DEGs was selected for qRT-PCR analysis. RNA-seq and qRT-PCR results showed that DEGs related to flavonoid synthesis, such as PAL (phenylalanine ammonia lyase), CHS (chalcone synthase), CHI (chalcone isomerase), 4CL (4-coumaric coenzyme A ligase), and CHR6 (chalcone reductase6), were downregulated; nitrogen transport-related genes NSP2 (nodulation signaling pathway2) and NPF6.2 (nitrate transporter1/ peptide transporter family6.2) were upregulated, and DEGs related to auxin signal transduction, GH3 (GRETCHEN HAGEN3) and SAUR (small auxin-up RNA), were upregulated (Figure 11C). These results suggest that BB treatment may inhibit flavonoid synthesis, promote nitrogen transport, and alter auxin signaling in soybean seedlings. The trends in gene expression changes in RNA-seq and qRT-PCR were similar, indicating that the RNA-seq analysis results in this study were reliable.
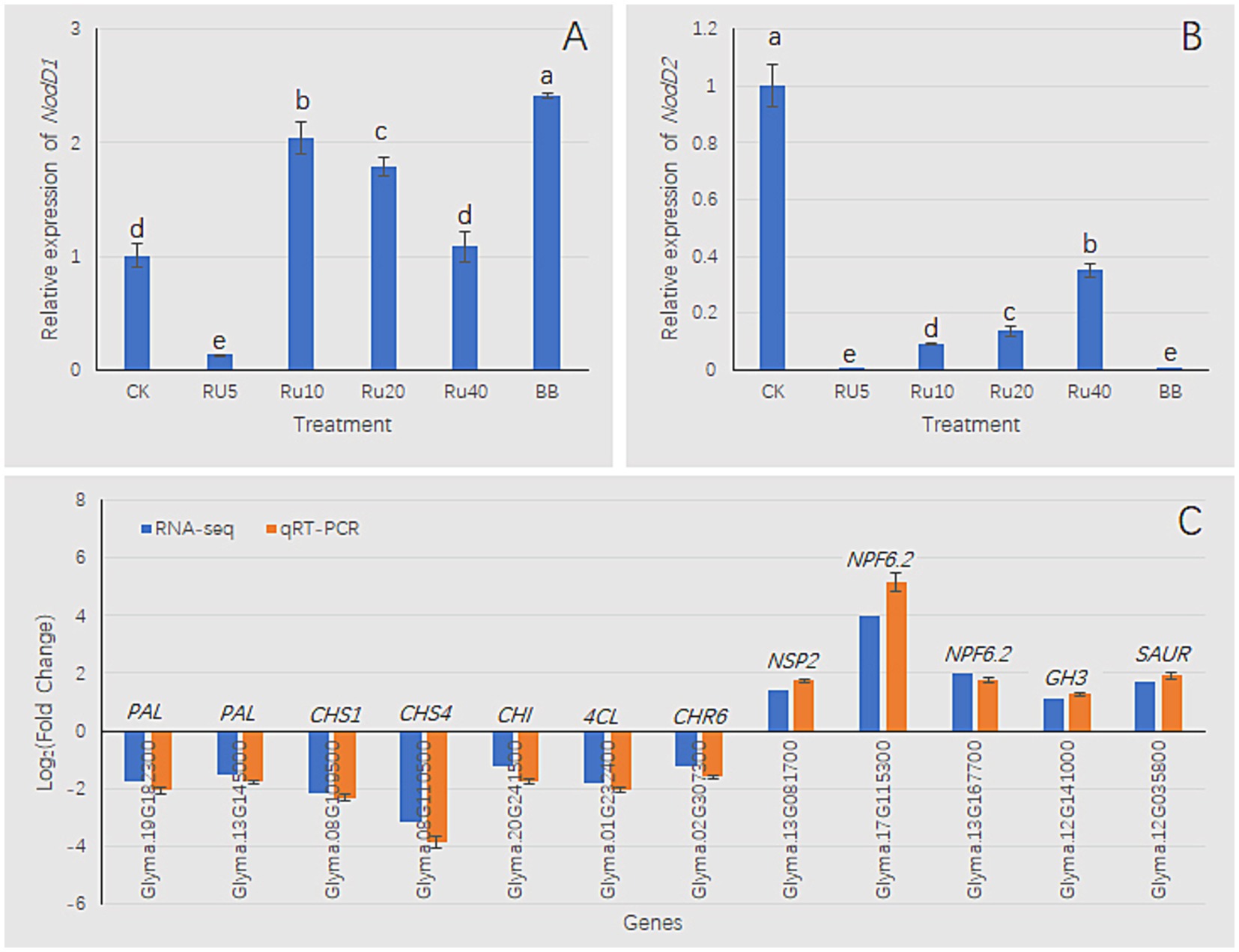
Figure 11. qRT-PCR analysis. (A,B) nodD1/nodD2 expression in B. japonicum USDA110 under rutin (RU) gradients (5–40 mg/L) with/without Bv co-inoculation. (C) RNA-seq verification using 16S rRNA as endogenous control. Fold change calculated as BB/Bj ratio. Bars: mean ± SD (n = 3); lowercase letters indicate significance (LSD, p < 0.05). Enzyme abbreviations: PAL, phenylalanine ammonia lyase; CHS, chalcone synthase; CHI, chalcone isomerase; 4CL, 4-coumarate-CoA ligase; CHR6, chalcone reductase 6; NSP2, nodulation signaling pathway2; NPF6.2, nitrate transporter1/ peptide transporter family6.2; GH3, GRETCHEN HAGEN3; SAUR, small auxin-up RNA.
Existing research has shown that PGPR are widely used in agriculture and have promising application prospects. PGPR can be used alone or combined with other microbial species to improve their application (Alemneh et al., 2020). Besides its own disease resistance and growth promotion, PGPR co-inoculated with rhizobia can also promote nodulation and nitrogen fixation in rhizobia and improve soil fertility and nutrient utilization efficiency through the synergistic effect of the two (Zeffa et al., 2020; Majeed et al., 2015; Fox et al., 2011; Vessey, 2003). The PGPR strains with these include Azotobacter (Abdiev et al., 2019), Azospirillum (Chibeba et al., 2015; Remans et al., 2008a), Pseudomonas (Nascimento et al., 2019), Streptomyces (Htwe et al., 2018), Bacillus (Korir et al., 2017), Staphylococcus (Prakamhang et al., 2015), and Serratia (Zahir et al., 2011). Juge et al. (2012) studied the effects of different inoculation methods of Bradyrhizobium, Azospirillum, and arbuscular mycorrhizae on soybean growth and found that all microbial combinations improved soybean root biomass. Argaw (2012) studied the effects of co-inoculation of B. japonicum and phosphate-solubilizing Pseudomonas spp. and traditional fertilizers on soybean nodulation, yield, and yield components, and found that co-inoculation could maximize soybean yield and quality. Mary et al. (2012) studied the effect of co-inoculation with B. japonicum and Bacillus subtilis on soybeans and found that co-inoculation could increase the number of nodules, dry weight of nodules, and biomass of soybeans. Co-inoculation with P. mucilaginosus and B. japonicum not only improved soybean quality and increased yield, but also increased soil fertility (Ma et al., 2015). Compared with single inoculation, co-inoculation with PGPR and USDA110 resulted in a significant increase in the number of root nodules, root nodule dry weight, and seed yield (Aung et al., 2013), and co-inoculation of B. subtilis or Staphylococcus aureus with USDA110 increased the nodule number and biological yield (Prakamhang et al., 2015). In this study, the plant height, stem diameter, biomass, and other indicators of soybean seedlings inoculated with B. velezensis and B. japonicum were significantly higher than those inoculated with B. velezensis and B. japonicum alone, indicating that the co-inoculation of B. velezensis and B. japonicum improved the growth of soybean seedlings, consistent with previous reports (Korir et al., 2017; Prakamhang et al., 2015). While the current study demonstrates B. velezensis’ rhizospheric competence, GFP-based spatial tracking coupled with confocal microscopy could further elucidate its microhabitat preferences and definitively rule out transient endophytic colonization. This study provides a new approach for the development of nitrogen-fixing bacterial fertilizers for soybean.
During the process of mutual recognition between soybeans and rhizobia, transduction of biochemical signals is a prerequisite for establishing a symbiotic system (Nakei et al., 2022). Leguminous plant roots secrete flavonoids in N-deficient environments, such as flavonoids, which have chemotactic effects on rhizobia and can attract rhizobia to the soil and bind them to the roots of leguminous plants, stimulating their massive reproduction (Sharma et al., 2021). Host-specific flavonoids interact with NodD proteins in rhizobia and induce Nod expression (Liu and Murray, 2016; Mario et al., 2015; Mandal et al., 2010). Subsequently, rhizobia produce structurally conserved lipochitooligosaccharide Nod factors, which are recognized by the NOD-ACTOR RECEPTOR Complex of leguminous plants. Finally, leguminous plants undergo physiological and biochemical changes to prepare for the formation of rhizobial symbionts (Roy et al., 2020; Mario et al., 2015; Geurts et al., 2005). To date, the underlying potential promotion mechanism of soybean growth and nitrogen fixation by co-inoculation with B. velezensis and USDA110. Among the various rhizobia, the NodD1 gene is the main regulatory factor for Nod factor biosynthesis and the symbiotic phenotype (del Cerro et al., 2015; Appelbaum et al., 1998; Gillette and Elkan, 1996). In contrast, NodD2 inhibits NodD1 expression (Loh and Stacey, 2003; Machado and Krishnan, 2003; Fellay et al., 1998; Machado et al., 1998). A previous study observed that co-inoculation of B. velezensis S141 with USDA110 promoted the growth, nodulation, and nitrogen fixation of soybeans. It is believed to S141 promotes nodulation and nitrogen fixation by secreting IAA and cytokinins. In addition, it is possible that the S141 strain secretes other substances that are beneficial for nodulation (Sibponkrung et al., 2020). In this study, a metabolomic analysis was conducted on the fermentation broth of the PGPR B. velezensis 20507, and it was found that the metabolites of B. velezensis contained 29 flavonoids and four isoflavonoids, including rutin, but not IAA and cytokinin. This may be related to the genetic background differences between the two strains. However, the observed promotion of soybean seedling growth and nodulation is consistent with previous reports (Sibponkrung et al., 2020). Appropriate concentrations of rutin (5 and 10 mg/L) in the culture medium significantly increased the expression of NodD1 and inhibited the expression of NodD2 in USDA110 cells. Consistent with this, when co-cultured with B. velezensis 20507, the expression of NodD1 was significantly upregulated, whereas that of NodD2 was significantly downregulated (Figures 11A,B). Therefore, the effects of adding rutin (5 and 10 mg/L) to the culture medium and co-culturing with B. velezensis on NodD1 and NodD2 are similar. It is inferred that B. velezensis can enhance the expression of NodD1 and inhibit the expression of NodD2 by secreting flavonoids, such as rutin. In the BB treatment in this study, it was inferred that the flavonoids produced by B. velezensis were beneficial for symbiosis between the nitrogen-fixing bacteria B. japonicum and soybean roots, and the growth and nitrogen content levels of soybeans in the BB treatment were significantly higher than those in the Bj treatment. Although the limited root nodule biomass (<0.1 g) under 2–5 mM nitrogen precluded quantitative nitrogen analysis via K-360 detection, the observed nodulation frequency (Figure 4) provides preliminary evidence for differential symbiotic efficiency. Future studies employing large-scale field trials with enhanced sampling protocols may overcome biomass constraints of nitrogen quantification.
Most studies suggest a negative correlation between flavonoid biosynthesis and auxin transport. Reduced flavonoid biosynthesis promotes auxin transport, whereas increased flavonoid synthesis hinders auxin transport (Santelia et al., 2008; Besseau et al., 2007; Peer et al., 2004). Some studies have suggested that flavonoid accumulation interferes with auxin distribution and turnover, inhibiting plant growth (Kuhn et al., 2016; Buer et al., 2013; Brown et al., 2001). By comparing the growth of different branches of flavonoid pathway mutants, Jeon et al. (2022) suggested that inhibition of flavonoid biosynthesis promotes plant growth, whereas promotion of flavonoid biosynthesis leads to growth inhibition. Studies have also been conducted from various perspectives. Li et al. (2010) suggested that the reduced growth associated with lignin biosynthesis in Arabidopsis was not related to flavonoids. Flavonoids are secondary metabolites with antioxidant and radical-scavenging activities. Nakabayashi et al. (2014) suggested that the accumulation of flavonoids in plants is beneficial for improving their ability to scavenge reactive oxygen species under drought stress conditions and that the accumulation of flavonoids does not inhibit plant growth. Therefore, the intrinsic relationship between plant growth and flavonoid biosynthesis remains unclear.
The phenylpropane pathway is an important metabolic pathway in plants that mainly synthesizes phenylpropane compounds such as flavonoids, soluble phenolic esters, and biopolymer precursors (Tohge et al., 2018). Flavonoids are widely distributed in plants, and are the main source of pigments that accumulate in various plant parts. Flavonoid synthesis begins with the phenylpropanoid metabolic pathway, which generates chalcones through a series of enzyme-catalyzed reactions and converts them into various flavonoid compounds. Several enzymes play crucial roles in flavonoid biosynthesis, including PAL, 4-coumaric coenzyme A ligase (4CL), chalcone synthase (CHS), and chalcone isomerase (CHI) (Wang et al., 2018; Wang et al., 2019). Compared with a single inoculation of rhizobia, co-inoculation with Rhizobium spp. and Azospirillum spp. can increase the number of root nodules by increasing the number of root hairs and the amount of flavonoids secreted by roots (Remans et al., 2008a, 2008b). Previous studies have found that when the key enzyme involved in the synthesis of flavonoids in Medicago truncatula, the chalcone synthase (CHS) gene, is disrupted, the inhibitory effect of rhizobia inoculation on local auxin transport in Medicago truncatula roots disappears and the plant cannot form nodules (Wasson et al., 2006, 2009). In paired comparisons of BB and Bj, 5,367 DEGs were identified. Among these, the number of downregulated genes was much higher than upregulated genes, with a quantity 1.32 times that of upregulated genes. The significant enrichment results of the KEGG indicated that the downregulated genes were significantly enriched in KEGG pathways, such as phenylpropanoid biosynthesis (ko00940), flavonoid biosynthesis (ko00941), and isoflavonoid biosynthesis (ko00943). Among the significantly enriched KEGG pathways, most of the key DEGs involved in flavonoid biosynthesis were downregulated (Supplementary Table S9), including PAL, CHS, CHI, 4CL, CHR2, and CHR6. Therefore, it is believed that inoculation with the PGPR B. velezensis inhibited the biosynthesis of flavonoids in soybeans, which may be related to the feedback inhibition caused by the production of flavonoids by B. velezensis. Nitrogen is the most essential mineral nutrient for plant growth and development, and NO3− is an important form of nitrogen absorption and utilization in plants. NPF genes are involved in the absorption, regulation, transport, and distribution of nitrate and nitrogen in plants (Sakuraba et al., 2021). Therefore, NPF genes play an important role and have practical value in improving and enhancing crop nitrogen utilization efficiency and yield-related traits. In this study, six DEGs encoding NPF were identified, which were upregulated 1.49–4.01-fold (Supplementary Table S9). In paired comparisons, DEGs were significantly enriched in the KEGG pathway ko04075 (Plant hormone signal transduction), in which most DEGs encoding GH3 and SAUR were upregulated. GH3 catalyzes the binding of auxin to amino acids, forming a complex that serves as an auxin storage reservoir. When the concentration of auxin is too low, the auxin-amino acid complex is hydrolyzed by proteolytic enzymes into auxin, which participates in the auxin signaling pathway and regulates the dynamic balance of auxin in plants (Bao et al., 2024). SAUR plays an important regulatory role in plant cell elongation and growth (Xu et al., 2023; Xu et al., 2017). In this study, a set of DEGs encoding GH3 and SAUR were upregulated by 1.13–3.02-fold (Supplementary Table S9). B. velezensis has been suggested to promote the growth of soybean seedlings by inhibiting the synthesis of soybean flavonoids, regulating auxin homeostasis and root elongation.
The enhanced growth performance of soybean seedlings in the BB treatment compared to Bj stems from three interrelated mechanisms. First, despite B. velezensis’ broad antimicrobial activity, it establishes a metabiotic relationship with B. japonicum, enhancing rhizobial proliferation through non-antagonistic interactions. Second, B. velezensis-secreted flavonoids (notably rutin) orchestrate nodulation in B. japonicum USDA110 via dual regulatory effects: activating NodD1 while suppressing NodD2 expression. This transcriptional reprogramming elevates NSP2 levels, triggering NOD factor signaling that initiates symbiosis, coupled with upregulated nitrate transporter (NPF) genes that enhance nitrogen assimilation efficiency, thereby stimulating chlorophyll biosynthesis and photosynthetic capacity. Third, bacterial flavonoids induce feedback inhibition of root flavonoid biosynthesis through downregulated phenylpropanoid genes (PAL, CHS, CHI), concomitant with upregulated auxin-responsive genes (GH3s, SAURs), potentially synergizing plant growth through phytohormone modulation. While these findings from 30-day-old seedlings in vermiculite systems provide mechanistic insights, field validation with mature plants under agricultural conditions remains essential. While bacterial transcriptomics would be informative, our experimental design prioritized host plant responses, and further full transcriptomic analysis of bacterial responses is planned for our next study. Ongoing CRISPR-Cas9 engineering of B. velezensis flavonoid mutants will conclusively map microbial metabolic contributions. This study advances multi-strain biofertilizer design by demonstrating how tailored microbial consortia can synergistically enhance legume productivity through complementary biochemical pathways.
The raw transcriptome sequencing data of soybean have been deposited in the national center for biotechnology information (NCBI) database under the accession number PRJNA1152285. The genome sequence of B. velezensis have been deposited in NCBI under the accession number PRJNA981422. Other relevant data supporting the findings of this study are available in this article and its associated Supplementary material.
YC: Conceptualization, Data curation, Writing – original draft, Writing – review & editing. XJ: Data curation, Investigation, Writing – review & editing, Writing – original draft. XH: Methodology, Writing – review & editing, Writing – original draft. ZW: Methodology, Writing – review & editing, Writing – original draft. QL: Methodology, Writing – review & editing, Writing – original draft. SZ: Methodology, Writing – review & editing, Writing – original draft. XZ: Methodology, Writing – review & editing, Writing – original draft. SW: Methodology, Writing – review & editing. HH: Data curation, Methodology, Writing – review & editing, Writing – original draft. JL: Writing – original draft, Writing – review & editing, Methodology.
The author(s) declare that financial support was received for the research and/or publication of this article. This work was financially supported by the National Natural Science Foundation of China (No. 32172079) and Outstanding Talents Team Project of Department of Science and Technology of Jilin Province (No. 20240601063RC).
We would like to thank all donors who supported this research.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The authors declare that no Gen AI was used in the creation of this manuscript.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2025.1572568/full#supplementary-material
Supplementary Table S1 | PCR primer sequences.
Supplementary Table S2 | Statistics of raw data information for each sample.
Supplementary Table S3 | Statics of clean reads mapping.
Supplementary Table S4 | DEGs in paired comparison of BB vs Bj.
Supplementary Table S5 | GO enrichment of DEGs in BB vs Bj.
Supplementary Table S6 | KEGG enrichment of DEGs in BB vs Bj.
Supplementary Table S7 | Gene annotation of B. velezensis 20507.
Supplementary Table S8 | Mass spectrometry identification results of fermentation broth of B. velezensis 20507.
Supplementary Table S9 | Interested genes and their expression in paired comparison of BB vs Bj.
Abdiev, A., Khaitov, B., Toderich, K., and Park, K. (2019). Growth, nutrient uptake and yield parameters of chickpea (Cicer arietinum L.) enhance by Rhizobium and Azotobacter inoculations in saline soil. J. Plant Nutr. 42, 2703–2714. doi: 10.1080/01904167.2019.1655038
Adedayo, A. A., and Babalola, O. O. (2023). Fungi that promote plant growth in the rhizosphere boost crop growth. J. Fungi 9:239. doi: 10.3390/jof9020239
Afzal, A., Bano, A., and Fatima, M. (2010). Higher soybean yield by inoculation with N-fixing and P-solubilizing bacteria. Agron. Sustain. Dev. 30, 487–495. doi: 10.1051/agro/2009041
Alemneh, A. A., Zhou, Y., Ryder, M. H., and Denton, M. D. (2020). Mechanisms in plant growth - promoting rhizobacteria that enhance legume–rhizobial symbioses. J. Appl. Microbiol. 129, 1133–1156. doi: 10.1111/jam.14754
Appelbaum, E. R., Thompson, D. V., Idler, K., and Chartrain, N. (1998). Rhizobium japonicum USDA 191 has two nodD genes that differ in primary structure and function. J. Bacteriol. 170, 12–20. doi: 10.1128/jb.170.1.12-20.1988
Argaw, A. (2012). Evaluation of co-inoculation of Bradyrhizobium japonicum and phosphate solubilizing Pseudomonas spp. effect on soybean (Glycine max L. (Merr.)) in Assossa area. J. Agric. Sci. Technol. 14, 213–224.
Armendariz, A. L., Talano, M. A., Olmos Nicotra, M. F., Escudero, L., Breser, M. L., Porporatto, C., et al. (2019). Impact of double inoculation with Bradyrhizobium japonicum E109 and Azospirillum brasilense Az39 on soybean plants grown under arsenic stress. Plant Physiol. Biochem. 138, 26–35. doi: 10.1016/j.plaphy.2019.02.018
Aung, T. T., Tittabutr, P., Boonkerd, N., Herridge, D., and Teaumroong, N. (2013). Co-inoculation effects of Bradyrhizobium japonicum and Azospirillum sp. on competitive nodulation and rhizosphere eubacterial community structures of soybean under rhizobia-established soil conditions. Afr. J. 12, 2850–2862. doi: 10.5897/AJB12.2557
Babu, S., Prasanna, R., Bidyarani, N., and Singh, R. (2015). Analysing the colonisation of inoculated cyanobacteria in wheat plants using biochemical and molecular tools. J. Appl. Phycol. 27, 327–338. doi: 10.1007/s10811-014-0322-6
Bai, Y., Guan, D., Li, L., Jiang, X., Ma, M., and Li, J. (2014). Screening and characterization of superior nitrogen-tolerance soybean rhizobia. Soybean Sci. 33, 861–864. doi: 10.11861/j.issn.1000-9841.2014.06.0861
Bao, D., Chang, S., Li, X., and Qi, Y. (2024). Advances in the study of auxin early response genes: aux/IAA, GH3, and SAUR. Crop J. 12, 964–978. doi: 10.1016/j.cj.2024.06.011
Besseau, S., Hoffmann, L., Geoffroy, P., Lapierre, C., Pollet, B., and Legrand, M. (2007). Flavonoid accumulation in Arabidopsis repressed in lignin synthesis affects auxin transport and plant growth. Plant Cell 19, 148–162. doi: 10.1105/tpc.106.044495
Brown, D. E., Rashotte, A. M., Murphy, A. S., Normanly, J., Tague, B. W., Peer, W. A., et al. (2001). Flavonoids act as negative regulators of auxin transport in vivo in Arabidopsis. Plant Physiol. 126, 524–535. doi: 10.1104/pp.126.2.524
Buer, C. S., Kordbacheh, F., Truong, T. T., Hocart, C. H., and Djordjevic, M. A. (2013). Alteration of flavonoid accumulation patterns in transparent testa mutants disturbs auxin transport, gravity responses, and imparts long-term effects on root and shoot architecture. Planta 238, 171–189. doi: 10.1007/s00425-013-1883-3
Chen, L., Liu, Y., Wu, G., Veronican Njeri, K., Shen, Q., Zhang, N., et al. (2016). Induced maize salt tolerance by rhizosphere inoculation of Bacillus amyloliquefaciens SQR9. Physiol. Plant. 158, 34–44. doi: 10.1111/ppl.12441
Cheng, Y., Lou, H., He, H., He, X., Wang, Z., Gao, X., et al. (2024). Genomic and biological control of Sclerotinia sclerotiorum using an extracellular extract from Bacillus velezensis 20507. Front. Microbiol. 15:1385067. doi: 10.3389/fmicb.2024.1385067
Chibeba, A. M., Guimarães, M. F., Brito, O. R., Nogueira, M. A., Araujo, R. S., and Hungria, M. (2015). Co-inoculation of soybean with Bradyrhizobium and Azospirillum promotes early nodulation. Am. J. Plant Sci. 6, 1641–1649. doi: 10.4236/ajps.2015.610164
Dai, S., Wang, J., Wei, M., Pang, Y., Yang, Y., Xu, S., et al. (2024). Bacillus velezensis FH-1 promotes rice seedlings growth by regulating specific spatiotemporal soil microbiome. Microbiol. China 51, 2381–2410. doi: 10.13344/j.microbiol.china.230799
del Cerro, P., Rolla-Santos, A. A. P., Gomes, D. F., Marks, B. B., Pérez-Montaño, F., Rodríguez-Carvajal, M. Á., et al. (2015). Regulatory nodD1 and nodD2 genes of Rhizobium tropici strain CIAT 899 and their roles in the early stages of molecular signaling and host-legume nodulation. BMC Genomics 16:251. doi: 10.1186/s12864-015-1458-8
Fellay, R., Hanin, M., Montorzi, G., Frey, J., Freiberg, C., Golinowski, W., et al. (1998). nodD2 of Rhizobium sp. NGR234 is involved in the repression of the nodABC operon. Mol. Microbiol. 27, 1039–1050. doi: 10.1046/j.1365-2958.1998.00761.x
Fox, S. L., O’Hara, G. W., and Bräu, L. (2011). Enhanced nodulation and symbiotic effectiveness of Medicago truncatula when co-inoculated with Pseudomonas fluorescens WSM3457 and Ensifer (Sinorhizobium) medicae WSM419. Plant Soil 348, 245–254. doi: 10.1007/s11104-011-0959-8
Geurts, R., Federova, E., and Bisseling, T. (2005). Nod factor signaling genes and their function in the early stages of rhizobium infection. Curr. Opin. Plant Biol. 8, 346–352. doi: 10.1016/j.pbi.2005.05.013
Gillette, W. K., and Elkan, G. H. (1996). Bradyrhizobium (Arachis) sp. strain NC92 contains two nodD genes involved in the repression of nodA and a nolA gene required for the efficient nodulation of host plants. J. Bacteriol. 178, 2757–2766. doi: 10.1128/jb.178.10.2757-2766.1996
Htwe, A. Z., Moh, S. M., Moe, K., and Yamakawa, T. (2018). Effects of co-inoculation of Bradyrhizobium japonicum SAY3-7 and Streptomyces griseoflavus P4 on plant growth, nodulation, nitrogen fixation, nutrient uptake, and yield of soybean in a field condition. Soil Sci. Plant Nutr. 64, 222–229. doi: 10.1080/00380768.2017.1421436
Jeon, J. S., Rybka, D., Carreno-Quintero, N., Vos, R. D., Raaijmakers, J. M., and Etalo, D. (2022). Metabolic signatures of rhizobacteria-induced plant growth promotion. Plant Cell Environ. 45, 3086–3099. doi: 10.1111/pce.14385
Juge, C., Prévost, D., Bertrand, A., Bipfubusa, M., and Chalifour, F. P. (2012). Growth and biochemical responses of soybean to double and triple microbial associations with Bradyrhizobium, Azospirillum and arbuscular mycorrhizae. Appl. Soil Ecol. 61, 147–157. doi: 10.1016/j.apsoil.2012.05.006
Kasanke, S. A., Cheeke, T. E., Moran, J. J., and Roley, S. S. (2024). Tripartite interactions among free-living, N-fixing bacteria, arbuscular mycorrhizal fungi, and plants: mutualistic benefits and community response to co-inoculation. Soil Sci. Soc. Am. 88, 1000–1013. doi: 10.1002/saj2.20679
Kim, D., Langmead, B., and Salzberg, S. L. (2015). HISAT: a fast spliced aligner with low memory requirements. Nat. Methods 12, 357–360. doi: 10.1038/nmeth.3317
Kondo, T., Sibponkrung, S., Hironao, K. Y., Tittabutr, P., Boonkerd, N., Ishikawa, S., et al. (2023). Bacillus velezensis S141, a soybean growth-promoting bacterium, hydrolyzes isoflavone glycosides into aglycones. J. Gen. Appl. Microbiol. 69, 175–183. doi: 10.2323/jgam.2023.02.002
Korir, H., Mungai, N. W., Thuita, M., Hamba, Y., and Masso, C. (2017). Co-inoculation effect of rhizobia and plant growth promoting Rhizobacteria on common bean growth in a low phosphorus soil. Front. Plant Sci. 8:141. doi: 10.3389/fpls.2017.00141
Kuan, K. B., Othman, R., Abdul Rahim, K., and Shamsuddin, Z. H. (2016). Plant growth-promoting Rhizobacteria inoculation to enhance vegetative growth, nitrogen fixation and nitrogen remobilisation of maize under greenhouse conditions. PLoS One 11:e0152478. doi: 10.1371/journal.pone.0152478
Kuhn, B. M., Errafi, S., Bucher, R., Dobrev, P., Geisler, M., Bigler, L., et al. (2016). 7-Rhamnosylated flavonols modulate homeostasis of the plant hormone auxin and affect plant development. J. Biol. Chem. 291, 5385–5395. doi: 10.1074/jbc.M115.701565
Li, X., Bonawitz, N. D., Weng, J.-K., and Chapple, C. (2010). The growth reduction associated with repressed lignin biosynthesis in Arabidopsis thaliana is independent of flavonoids. Plant Cell 22, 1620–1632. doi: 10.1105/tpc.110.074161
Liu, C., and Murray, J. D. (2016). The role of flavonoids in nodulation host-range specificity: an update. Plants (Basel) 5:33. doi: 10.3390/plants5030033
Loh, J., and Stacey, G. (2003). Nodulation gene regulation in Bradyrhizobium japonicum: a unique integration of global regulatory circuit. Appl. Environ. Microbiol. 69, 10–17. doi: 10.1128/AEM.69.1.10-17.2003
Ma, M., Liu, L., Jiang, X., Guan, D., and Li, J. (2015). Evaluation of the effect of co-inoculant of Paenibacillus mucilaginosus and Bradyrhizobium japonicum in application. Sci. Agric. Sin. 48, 3600–3611. doi: 10.3864/j.issn.0578-1752.2015.18.004
Machado, D., and Krishnan, H. B. (2003). nodD alleles of Sinorhizobium fredii USDA 191 differentially influence soybean nodulation, nodC expression, and production of exopolysaccharides. Curr. Microbiol. 7, 134–137. doi: 10.1007/s00284-002-3972-6
Machado, D., Pueppke, S. G., Vinardel, J. M., Ruiz-Sainz, J. E., and Krishnan, H. B. (1998). Expression of nodD1 and nodD2 in Sinorhizobium fredii, a nitrogen-fixing symbiont of soybean and other legumes. Mol. Plant-Microbe Interact. 11, 375–382. doi: 10.1094/MPMI.1998.11.5.375
Majeed, A., Abbasi, M. K., Hameed, S., Swelim, M. A., and Aty, A. A. A. E. (2015). Isolation and characterization of plant growth-promoting rhizobacteria from wheat rhizosphere and their effect on plant growth promotion. Front. Microbiol. 6:198. doi: 10.3389/fmicb.2015.00198
Mandal, S. M., Chakraborty, D., Dey, S., Mandal, S. M., Chakraborty, D., and Dey, S. (2010). Phenolic acids act as signaling molecules in plant-microbe symbioses phenolic acids act as signaling molecules in plant-microbe symbioses. Plant Signal. Behav. 5, 359–368. doi: 10.4161/psb.5.4.10871
Mario, J. R., Lira, A., Nascimento, L. R. S., and Fracetto, G. G. M. (2015). Legume-rhizobia signal exchange: promiscuity and environmental effects. Front. Microbiol. 6, 1–9. doi: 10.3389/fmicb.2015.00945
Mary, A., Laetitia, H., Robert, O., and Didier, L. (2012). Efficiency of different formulations of Bradyrhizobium japonicum and effect of co-inoculation of Bacillus subtilis with two different strains of Bradyrhizobium japonicum. World J. Microbiol. Biotechnol. 28, 2541–2550. doi: 10.1007/s11274-012-1062-x
Melchiorre, M., Luca, M. J. D., Anta, G. G., Suarez, P., Lopez, C., Lascano, R., et al. (2011). Evaluation of bradyrhizobia strains isolated from field-grown soybean plants in Argentina as improved inoculants. Biol. Fertil. Soils 47, 81–89. doi: 10.1007/s00374-010-0503-7
Melissa, O., Cristian, A., Sheila, C., Vanina, M., and Fabricio, C. (2022). Evaluation of nitrous oxide emission by soybean inoculated with Bradyrhizobium strains commonly used as inoculants in South America. Plant Soil 472, 311–328. doi: 10.1007/s11104-021-05242-y
Meng, Q., Jiang, H., and Hao, J. J. (2016). Effects of Bacillus velezensis strain BAC03 in promoting plant growth. Biol. Control 98, 18–26. doi: 10.1016/j.biocontrol.2016.03.010
Miransari, M. (2014). Plant growth promoting rhizobacteria. J. Plant Nutr. 37, 2227–2235. doi: 10.1080/01904167.2014.920384
Nakabayashi, R., Yonekura-Sakakibara, K., Urano, K., Suzuki, M., Yamada, Y., Nishizawa, T., et al. (2014). Enhancement of oxidative and drought tolerance in Arabidopsis by overaccumulation of antioxidant flavonoids. Plant J. 77, 367–379. doi: 10.1111/tpj.12388
Nakei, M. D., Venkataramana, P. B., and Ndakidemi, P. A. (2022). Soybean-Nodulating rhizobia: ecology, characterization, diversity, and growth promoting functions. Front. Sustain. Food Syst. 6:824444. doi: 10.3389/fsufs.2022.824444
Nascimento, F. X., Tavares, M. J., Franck, J., Ali, S., Glick, B. R., and Rossi, M. J. (2019). ACC deaminase plays a major role in Pseudomonas fluorescens YsS6 ability to promote the nodulation of alpha- and Betaproteobacteria rhizobial strains. Arch. Microbiol. 201, 817–822. doi: 10.1007/s00203-019-01649-5
Nguyen, T. H. N., Brechenmacher, L., Aldrich, J. T., Clauss, T. R., Gritsenko, M. A., Hixson, K. K., et al. (2012). Quantitative phosphoproteomic analysis of soybean root hairs inoculated with Bradyrhizobium japonicum. Mol. Cell. Proteomics 11, 1140–1155. doi: 10.1074/mcp.M112.018028
Peer, W. A., Bandyopadhyay, A., Blakeslee, J. J., Makam, S. N., Chen, R. J., Masson, P. H., et al. (2004). Variation in expression and protein localization of the PIN family of auxin efflux facilitator proteins in flavonoid mutants with altered auxin transport in Arabidopsis thaliana. Plant Cell 16, 1898–1911. doi: 10.1105/tpc.021501
Pertea, M., Pertea, G. M., Antonescu, C. M., Chang, T. C., Mendell, J. T., and Salzberg, S. L. (2015). StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 33, 290–295. doi: 10.1038/nbt.3122
Prakamhang, J., Tittabutr, P., Boonkerd, N., Teamtisong, K., Uchiumi, T., Abe, M., et al. (2015). Proposed some interactions at molecular level of PGPR coinoculated with Bradyrhizobium diazoefficiens USDA110 and B. japonicum THA6 on soybean symbiosis and its potential of field application. Appl. Soil Ecol. 85, 38–49. doi: 10.1016/j.apsoil.2014.08.009
Remans, R., Beebe, S., Blair, M., Manrique, G., Tovar, E., Rao, I., et al. (2008a). Physiological and genetic analysis of root responsiveness to auxin-producing plant growth-promoting bacteria in common bean (Phaseolus vulgaris L.). Plant Soil 302, 149–161. doi: 10.1007/s11104-007-9462-7
Remans, R., Ramaekers, L., Schelkens, S., Hernandez, G., Garcia, A., Reyes, J.-L., et al. (2008b). Effect of Rhizobium-Azospirillum coinoculation on nitrogen fixation and yield of two contrasting Phaseolus vulgaris L. genotypes cultivated across different environments in Cuba. Plant Soil 312, 25–37. doi: 10.1007/s11104-008-9606-4
Roy, S., Liu, W., Nandety, R. S., Crook, A., Mysore, K. S., Pislariu, C. I., et al. (2020). Celebrating 20 years of genetic discoveries in legume nodulation and symbiotic nitrogen fixation. Plant Cell 32, 15–41. doi: 10.1105/tpc.19.00279
Saito, A., Tanabata, S., Tanabata, T., Tajima, S., Ueno, M., Ishikawa, S., et al. (2014). Effect of nitrate on nodule and root growth of soybean (Glycine max (L.) Merr.). Int. J. Mol. Sci. 15, 4464–4480. doi: 10.3390/ijms15034464
Sakuraba, Y., Chaganzhana, M. A., Iba, K., and Yanagisawa, S. (2021). Enhanced NRT1.1/NPF6.3 expression in shoots improves growth under nitrogen deficiency stress in Arabidopsis. Commun. Biol. 4:256. doi: 10.1038/s42003-021-01775-1
Santelia, D., Henrichs, S., Vincenzetti, V., Sauer, M., Bigler, L., Klein, M., et al. (2008). Flavonoids redirect PIN-mediated polar auxin fluxes during root gravitropic responses. J. Biol. Chem. 283, 31218–31226. doi: 10.1074/jbc.M710122200
Schmittgen, T. D., and Livak, K. J. (2008). Analyzing real-time PCR data by the comparative CT method. PRO 3, 1101–1108. doi: 10.1038/nprot.2008.73
Shao, J., Xu, Z., Zhang, N., Shen, Q., and Zhang, R. (2015). Contribution of indole-3-acetic acid in the plant growth promotion by the rhizospheric strain Bacillus amyloliquefaciens SQR9. Biol. Fertil. Soils 51, 321–330. doi: 10.1007/s00374-014-0978-8
Sharma, K., Chaturvedi, U., Sharma, S., Vaishnav, A., and Singh, S. V. (2021). “Fenugreek-rhizobium symbiosis and flavonoids under stress condition” in Antioxidants in Plant-Microbe Interaction, 449–459.
Sharma, K., Sharma, S., Vaishnav, A., Jain, R., Singh, D., Singh, H. B., et al. (2022). Salt-tolerant PGPR strain Priestia endophytica SK1 promotes fenugreek growth under salt stress by inducing nitrogen assimilation and secondary metabolites. J. Appl. Microbiol. 133, 2802–2813. doi: 10.1111/jam.15735
Sibponkrung, S., Kondo, T., Tanaka, K., Tittabutr, P., Boonkerd, N., Yoshida, K., et al. (2020). Co-inoculation of Bacillus velezensis strain S141 and Bradyrhizobium strains promotes nodule growth and nitrogen fixation. Microorganisms 8:678. doi: 10.3390/microorganisms8050678
Sun, X., Xu, Z., Xie, J., Hesselberg-Thomsen, V., Tan, T., Zheng, D., et al. (2022). Bacillus velezensis stimulates resident rhizosphere Pseudomonas stutzeri for plant health through metabolic interactions. ISME J. 16, 774–787. doi: 10.1038/s41396-021-01125-3
Tohge, T., de Souza, L. P., and Fernie, A. R. (2018). Corrigendum: current understanding of the pathways of flavonoid biosynthesis in model and crop plants. J. Exp. Bot. 69:4497. doi: 10.1093/jxb/ery260
Vessey, J. K. (2003). Plant growth promoting rhizobacteria as biofertilizers. Plant Soil 255, 571–586. doi: 10.1023/A:1026037216893
Wang, S., Alseekh, S., Fernie, A. R., and Luo, J. (2019). The structure and function of major plant metabolite modifications. Mol. Plant 12, 899–919. doi: 10.1016/j.molp.2019.06.001
Wang, Q., Liu, J., and Zhu, H. (2018). Genetic and molecular mechanisms underlying symbiotic specificity in legume-Rhizobium interactions. Front. Plant Sci. 9:313. doi: 10.3389/fpls.2018.00313
Wasson, A. P., Pellerone, F. I., and Mathesius, U. (2006). Silencing the flavonoid pathway in Medicago truncatula inhibits root nodule formation and prevents auxin transport regulation by rhizobia. Plant Cell 18, 1617–1629. doi: 10.1105/tpc.105.038232
Wasson, A. P., Ramsay, K., Jones, M. G. K., and Mathesius, U. (2009). Differing requirements for flavonoids during the formation of lateral roots, nodules and root knot nematode galls in Medicago truncatula. New Phytol. 183, 167–179. doi: 10.1111/j.1469-8137.2009.02850.x
Wu, Z., Zhong, W., Fan, Y., Peng, S., Du, D., Chen, X., et al. (2017). Symbiotic compatibility among eight elite soybean rhizobia strains and twenty-seven soybean cultivars from different planting regions. Soybean Sci 36, 405–418. doi: 10.11861/j.issn.1000-9841.2017.03.0405
Xu, K., Lou, Q., Wang, D., Li, T., Chen, S., Li, T., et al. (2023). Overexpression of a novel small auxin-up RNA gene, OsSAUR11, enhances rice deep rootedness. BMC plant biology 23:5385. doi: 10.1186/s12870-023-04320-w
Xu, Y. X., Xiao, M. Z., Liu, Y., Fu, J. L., He, Y., and Jiang, D. A. (2017). The small auxin-up RNA OsSAUR45 affects auxin synthesis and transport in rice. Plant molecular biology. 94, 97–107. doi: 10.1007/s11103-017-0595-7
Zahir, Z. A., Zafar-Ul-Hye, M., Sajjad, S., and Naveed, M. (2011). Comparative effectiveness of Pseudomonas and Serratia sp. containing ACC-deaminase for coinoculation with Rhizobium leguminosarum to improve growth, nodulation, and yield of lentil. Biol. Fertil. Soils 47, 457–465. doi: 10.1007/s00374-011-0551-7
Zeffa, D. M., Fantin, L. H., Koltun, A., de Oliveira, A. L. M., Nunes, M. P. B. A., Canteri, M. G., et al. (2020). Effects of plant growth-promoting rhizobacteria on co-inoculation with Bradyrhizobium in soybean crop: a meta-analysis of studies from 1987 to 2018. PeerJ 8:e7905. doi: 10.7717/peerj.7905
Keywords: soybean, transcriptome, growth, nitrogen fixation, nodulation
Citation: Cheng Y, Jiang X, He X, Wu Z, Lv Q, Zhao S, Zhang X, Wang S, He H and Liu J (2025) Bacillus velezensis 20507 promotes symbiosis between Bradyrhizobium japonicum USDA110 and soybean by secreting flavonoids. Front. Microbiol. 16:1572568. doi: 10.3389/fmicb.2025.1572568
Received: 07 February 2025; Accepted: 18 March 2025;
Published: 27 March 2025.
Edited by:
Md. Motaher Hossain, Bangabandhu Sheikh Mujibur Rahman Agricultural University, BangladeshCopyright © 2025 Cheng, Jiang, He, Wu, Lv, Zhao, Zhang, Wang, He and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jianfeng Liu, amlhbmZlbmdsaXUxOTc2QDE2My5jb20=
†ORCID: Jianfeng Liu, orcid.org/0000-0003-3220-8941
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.