
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 02 April 2025
Sec. Terrestrial Microbiology
Volume 16 - 2025 | https://doi.org/10.3389/fmicb.2025.1572497
This article is part of the Research Topic Monitoring, Modeling, and Mitigation in Terrestrial Ecosystems: Microbial Response to Climate Change View all 3 articles
In light of the challenges posed by contemporary global warming and soil acidification, the respective effects of pH and temperature on soil microbiome and functions have been explored. However, the combined influence of acidification and warming on soil denitrification and active microbial communities are still unclear. Here, we conducted a microcosm experiment to investigate the influences of increasing temperature and acidification on active microbes such as bacteria and eukaryotic microbes. Denitrification rate in soil were detected using a C2H2 inhibition method. The results showed that the Shannon index of bacterial communities exhibited significant enhancement in response to warming and acidification, whereas their community patterns were predominantly shaped by pH. For the micro-eukaryotic community, temperature emerged as the main driver of variations in the α-diversity, with the MT group exhibiting significantly lower Shannon indices compared to LT and HT groups. Both pH and temperature exerted a combined effect on their community patterns. Additionally, pH was detected as a crucial factor influencing denitrification rates, with a significant negative correlation between pH and denitrification rate within the pH range of 4.32–7.46 across all temperatures in this study. Our findings highlighted the significant impacts of acidification on soil denitrification rates and active microbes under global warming, which provided an important scientific basis for agricultural production management and environmental protection in the context of global climate warming.
Nitrous oxide (N2O) is a major greenhouse gas and is regarded as the most significant contributor to ozone depletion in the 21st century (Ravishankara et al., 2009; Montzka et al., 2011). As an essential component of the global biogeochemical cycle, soil serves as both the source and sink of atmospheric N2O (Ryden, 1981). Consequently, N2O released by soils, especially farmland soils, has received much attention in previous studies (Bhattarai et al., 2021), which have shown that microbial processes are critical to nitrous acid emissions from agricultural soils.
Denitrification is an important microbial process converting nitrate (NO3–) and nitrite (NO2–) into N2O and N2 in different ecosystems, for example, soil, sediment, and water (Long et al., 2013; Margalef-Marti et al., 2024). This process has been extensively studied in bacteria, showing as multiple reduction steps catalyzed by various enzymes, including nitrate reductase (NAR), nitrite reductase (NIR), nitric oxide reductase (NOR), and N2O reductase (NOS) (Sennett et al., 2024). In addition to bacteria, eukaryotic microbes represent a significant component of the soil microbial community, playing vital ecological roles in nitrogen processes (Mothapo et al., 2013, 2015; Maeda et al., 2015; Higgins et al., 2016). Differed from bacteria, several denitrifying eukaryotic microbes, such as Fusarium oxysporum, Bolivina plicata, and Stainforthia sp. (Kobayashi et al., 1996; Kamp et al., 2015), lack the nosZ gene, resulting in N2O as the final denitrification product (Laughlin and Stevens, 2002; Crenshaw et al., 2008). Soil environments would affect the relative contributions of bacteria and eukaryotic microbes to denitrification, especially N2O emission rates in soil. Eukaryotic microbes derived N2O emissions are comparable to, or even exceeded, those from bacterial denitrification (Chen et al., 2014).
Complex relationships between soil pH and denitrification processes have been determined in a previous study (Firestone et al., 1980). The ratios of denitrification products was strongly influenced by pH (ŠImek and Cooper, 2002), specifically, the N2O/(N2O + N2) ratio seemed to have a negative correlation with soil pH in agricultural settings pH 5–8 (Bakken et al., 2012). Additionally, there was an increase in the N2O/(N2O + N2) ratio during denitrification under acidic conditions (pH < 5.0). Raising soil pH to near-neutral levels (pH > 6.5) through liming can reduce N2O emissions; However, increasing the pH of acidic soils (pH < 5.6) to moderately acidic levels (pH 5.6–6.0) generally led to higher N2O emissions. A hump-shaped relationship existed between soil pH and N2O, leading to peak N2O emissions at moderate soil acidity (Qiu et al., 2024). Furthermore, variations in pH over both short and long terms affect soil N2O emissions differently, as the dominant microbial communities can shift due to pH-induced changes in the microbial source of N2O (Baggs et al., 2010). Additionally, temperature has also been indicated to be important factor affecting the distribution of soil denitrifying bacterial communities (Braker et al., 2010; Taylor et al., 2019). A previous study has demonstrated that ammonia oxidizers and bacterial denitrifiers were significantly inhibited at high temperatures, whereas micro-eukaryotic denitrifiers are well-adapted and may be the primary contributors to N2O emissions in acidic soils (Xu et al., 2017). Whereas, most of previous studies investigating pH or temperature impacts on microbes determined the community and abundance of microbes at DNA level, but not RNA level, which reflects active microbes in different ecosystems. Previous studies have determined the correlation between denitrification rate and microbial community including microbial abundance and diversity at DNA level (Yao et al., 2013; Wang et al., 2014; Chunyi et al., 2024). However, active microbes are the drivers of nutrient transformations in different ecosystems. The effects of pH and temperature on active microbial community are still limited, especially the combined impacts of pH and temperature.
To explore the combined effects of warming and acidification on soil denitrification rates and the active microbial communities, we conducted a microcosm experiment with a gradient of pH (4.9–7.7) and soil temperature (20°C–30°C). Additionally, we employed transcriptomic methods to analyze RNA levels, allowing us to investigate the relationship between denitrification rates and the dynamics of active microbial communities. This study will shed light on how active microorganisms, alongside soil nutrients, influence soil denitrification, offering insights into the combined effects of soil acidification and rising temperature on greenhouse gas production.
Soil samples with an initial pH of 4.95 were collected from a tea garden in Ningbo, China (121.86°E, 29.75°N). Soil was homogenized by sieving through a 2 mm mesh. The soil samples were maintained at room temperature with a 20% moisture content for 30 days to stabilize the soil properties and microbial community. Limestone (CaCO3, 99.0%, AR) was added to adjust the soil pH values. During the soil pH adjustment period, three pH levels were established as high pH 7.7 (HP), medium pH 6.4 (MP) and low pH 4.9 (LP), respectively. The soil with stabilized pH were incubated at three different soil temperature gradients (soil temperature was measured by using a thermometer) for 30 days, including low temperature 20°C (LT), medium temperature 25°C (MT), and high temperature 30°C (HT), respectively. During the adjustment of pH and temperature, the moisture content was maintained at 20%, consistent with level during previous stable incubation. Finally, a total of nine experimental treatments were prepared: LTHP, MTHP, HTHP, LTMP, MTMP, HTMP, LTLP, MTLP, and HTLP. Each treatment consisted of three replicates, with each replicate containing 200 g of soil. pH decreased slightly after 1 month of incubation (Supplementary Table 1).
The concentration of ammonia (NH4+) and nitrate (NO3–) were determined using a continuous flow analyzer (AA3 analyzer, German) after extracted by 1 mol/L KCl solution (soil:KCl solution = 1:10) (Li et al., 2025). The pH of each soil was measured using an XL60 pH meter (Fisher Scientific, United States) after being suspended in deionized water (ddH2O) with 1:2.5 soil-to-water ration (Li et al., 2016). Soil moisture was calculated after dried in an oven at 105°C for 16 h. Total carbon, nitrogen, and sulfur contents were analyzed using a Vario MAX CNS elemental analyzer (ELEMENTAR, German) (Xu et al., 2014).
Denitrification rate was measured using an acetylene (C2H2) inhibition method in accordance with a previous study (Xu et al., 2019). In brief, 10 g of fresh soil was placed in a 120 mL serum bottle with 5 mL 2.4 mM NaNO3 and 5 mL 0.06 M glucose. The serum bottles were sealed with rubber stoppers and were alternately vacuumed and flushed with helium (He) gas to establish anaerobic conditions. For determination of potential N2O production rate, 10% (v/v) of C2H2 was added to inhibit the reduction of N2O to N2. The rates of N2O production in the treatments without C2H2 were calculated as the real denitrification rates (Philippot et al., 2011). The concentration of N2O in the headspace was measured at 1 and 5 h (Supplementary Table 2) using a gas chromatograph (7890A; Agilent Technologies, Santa Clara, CA, United States) (Molstad et al., 2007; Xu et al., 2019).
Total RNA was extracted from 2 g fresh soil using a RNeasy PowerSoil Total RNA Kit (Qiagen) according to the manufacturer’s instructions, and the purified RNA without DNA was stored at −80°C until used. Complementary DNA (cDNA) was synthesized through reverse transcription using an ABKscript RT MasterMix (OneStep gDNA Removal) Kit, and the resulting cDNA was used as template for target gene amplification. To evaluate the communities of bacteria and eukaryotic microbes in different treatments, we performed PCR amplification for bacterial 16S rRNA gene and micro-eukaryotic 18S rRNA gene using primer set of 338F/806R (Yang et al., 2020) and 565F/981R (Salmaso et al., 2020), respectively. The amplicons were purified using a E.Z.N.A.® Gel Extraction Kit (Omega, United States) and sent to Magigene Biotechnology Co. (GuangZhou, China) for high-throughput sequencing on a Novaseq 6000 PE250 platform.
We utilized Quantitative Insights Into Microbial Ecology version 2 (QIIME2) (Bolyen et al., 2019) to analyze the sequences, and employed the DADA2 plugin (Callahan et al., 2016) to denoise sequences and generate Amplicon Sequence Variants (ASV) approximately 250 base pairs in length. For both bacteria and eukaryotic microbes, ASVs with only one sequence were discarded in the following analysis (Li et al., 2023). SILVA 138 SSU Ref NR99 and RDP 18S v4.1 database (Pruesse et al., 2007; Wang et al., 2007) were used for the classification of the bacterial and micro-eukaryotic taxonomy, respectively. ASVs identified as mitochondria and chloroplast sequences were removed. Alpha diversity indices (Shannon and Chao1) were calculated based on species richness to assess the biodiversity of microbial communities (Supplementary Table 3). Principal Co-ordinates Analysis (PCoA) were constructed to exhibit the distribution patterns of microbial (i.e., bacteria and eukaryotic microbes), bacterial, and micro-eukaryotic communities based on Bray-Curtis distances. Then, redundancy analysis (RDA) was selected to distinguish the soil properties affecting microbial communities. The relative abundances of bacterial and micro-eukaryotic species were displayed using heatmap plot using R with “pheatmap” package (version 1.0.12). We used LDA Effect Size (LEfSe) to identify species with significant differences between treatment via the website1, with an LDA threshold of four and a p-value threshold of 0.05. Additionally, classes differences between bacterial and micro-eukaryotic active microorganisms under different treatments were assessed using two-way ANOVA to extract F values for evaluating the impact magnitude (Package “vegan” v2.6–6.1). After filtering for ASVs with relative abundance greater than 0.01%, a heatmap was generated using the top 30 species by relative abundance at the class level. Mantel test and Pearson’s correlation analyses were conducted using R with “LinkET” package (version 0.0.7.4). The p-values of mantel test were adjusted using FDR. The partial least squares path modeling (PLSPM) was constructed to explore the mechanism of effects of microbial alpha and community pattern, total nutrient (including TS, TC, and TN), inorganic nitrogen (including NOx-N, and NH4+-N), pH, and temperature on soil denitrification rates using the “plspm” package (version 0.5.1) in R software. Multiple goodness of fit criteria was tested for the model as follows: Goodness of Fit (GoF > 0.6), Dillon-Goldstein’s rho (DG.rho > 0.7), Average Variance Extracted (AVE > 0.5). Random forest analyses were conducted using the “rfPermute” package (version 2.5.2) in R with 1,000 permutations and 500 decision trees. The data for Random Forest analysis incorporated actual denitrification rates (measured without C2H2 inhibition), soil physicochemical parameters, microbial diversity and community patterns (PCoA 1 axis). All data used R software (R4.3.1) for statistical testing and correlation analysis. Tukey-HSD was used for post hoc pairwise comparisons. The date for bacterial 16S rRNA genes and micro-eukaryotic 18S rRNA genes could be downloaded from ScienceDB using https://doi.org/10.57760/sciencedb.17932.
Significant differences in denitrification rates were observed among the various treatment groups, with rates ranging from 0.018 to 0.55 μg⋅g–1⋅h–1 in the absence of acetylene (C2H2) and N2O production rates ranging from 0.17 to 0.87 μg⋅g–1⋅h–1 in treatments with C2H2 (Figure 1A). In the treatment with C2H2, denitrification rates were significantly higher (p < 0.05) in the medium temperature and medium pH (MTMP) treatment than in other treatments. Conversely, high pH significantly decreased the denitrification rates in soils, especially treated with low and high temperature (p < 0.05). In the treatment without C2H2, the highest N2O production rate was observed in the LTMP treatment, while the lowest rate was found in the MTLP treatment. Additionally, N2O production rates in middle pH groups were the highest, followed by in high pH and low pH (p < 0.05). Calculating based on N2O emission in treatments with and without C2H2, the rates of N2O reduction, i.e., conversion of N2O to N2, were the lowest in low pH treatments compared in high and middle treatment groups (Supplementary Table 2).
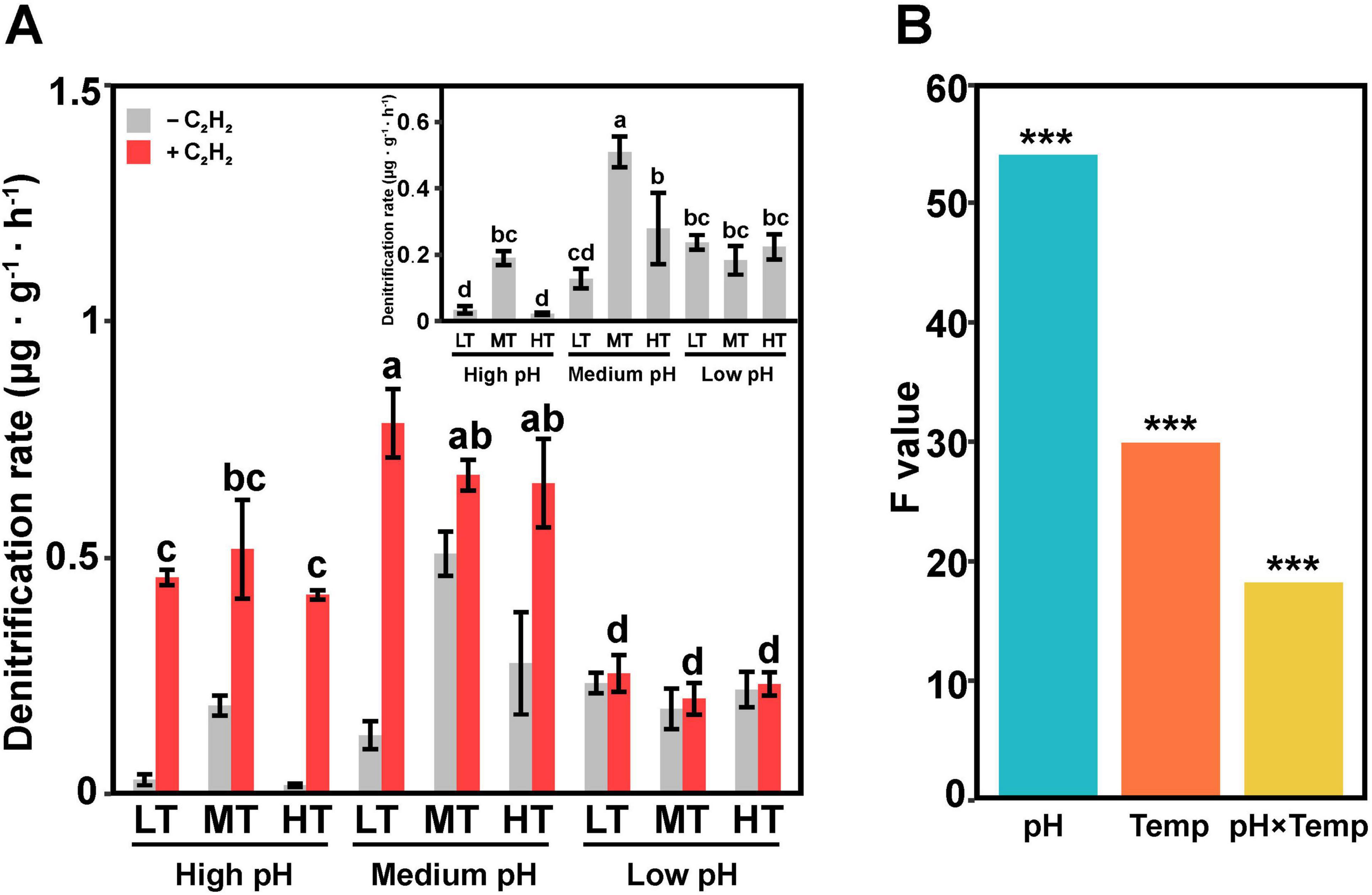
Figure 1. N2O emission from soil samples with different treatments. (A) N2O emission rates under different conditions: with (red bars) and without (gray bars) C2H2. (B) Factors, i.e., pH and Temp (temperature), influencing denitrification rates. The treatment groups g low temperature (LT), medium temperature (MT) and high temperature (HT) in the figure represented low temperature, medium temperature and high temperature, respectively. These letters employ the alphabet mark method to indicate statistically significant differences in multiple comparisons. The symbol ‘***’ represents asterisks for significance in figures.
Soil pH, temperature, and combination of pH and temperature significantly (two-way ANOVA test, p < 0.001) affected denitrification rates in soil. Notably, soil pH played a more important role in regulating soil denitrification rate in comparison with temperature (Figure 1B).
A total of 50,610 bacterial ASVs and 6,181 micro-eukaryotic ASVs were obtained in soil samples based on high-throughput sequencing of bacterial 16S rRNA genes and micro-eukaryotic 18S rRNA genes at RNA-level. The coverage of 99.20% and 99.64%, for bacterial and micro-eukaryotic communities, respectively, indicated sufficient sampling depth to capture overall microbial diversity across all 27 samples. Significant acidic and thermal variations (p <0.05) were observed in the Shannon index (Supplementary Figures 1A, C, E), with HT and LP treatments exhibiting higher values than LT and HP treatments (Supplementary Figure 1), indicating that pH and temperature changes played a pivotal role in shaping the alpha diversity of bacteria. While temperature mainly affected the alpha diversity of soil eukaryotic microbes (Supplementary Figures 2A, E), with HT and LT treatment showing higher values than MT treatment (Supplementary Figure 2). From the perspective of overall active microorganisms, temperature primarily had a significant effect on alpha diversity (Supplementary Figures 3A–E).
PCoA plots showed that significant differences were observed in community patterns of microbes including bacteria and eukaryotic microbes (Figure 2A) among various treatments (adonis R2 = 0.80, p < 0.001). Further, we found that microbial profiles in low pH (LP) treatments differed from those in middle (MP) and high (HP) groups. Moreover, pH changed the distribution patterns of microbes in soil samples across different pH (Figure 2A). Similarly, bacterial patterns in soil samples treated with low pH were separated from both MP and HP treatments at axis one, which explained 32.51% variation in bacterial communities (Figure 2B). In contrast, the micro-eukaryotic patterns were separated at axis 1 based on temperature, and three groups of micro-eukaryotic communities in soils with different temperatures were clearly separated (Figure 2C).
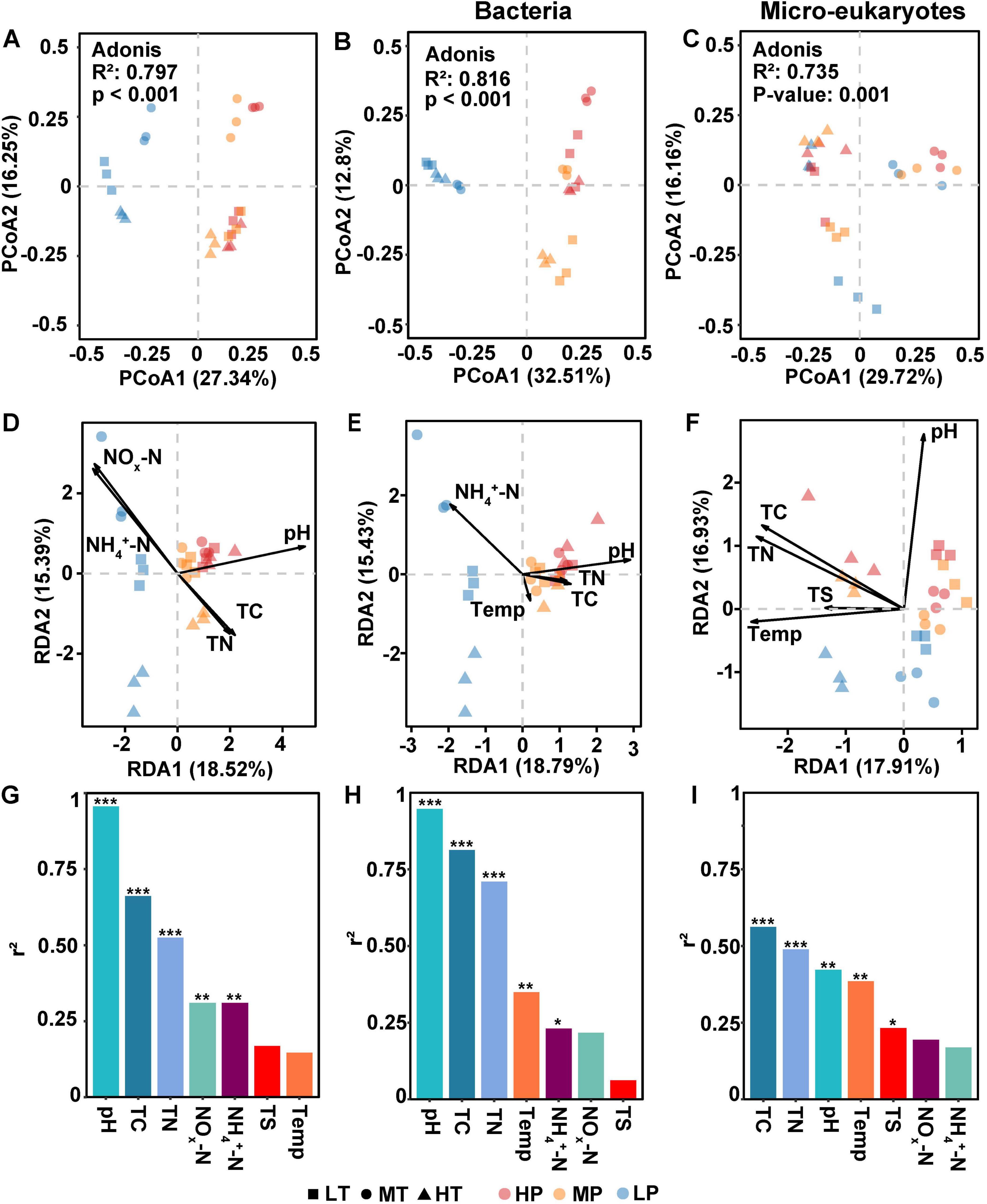
Figure 2. Distribution patterns of microbial communities and factors influencing microbial communities in soil samples. (A–C) Principal Co-ordinates Analysis (PCoA) of microbial communities, bacterial and micro-eukaryotic communities, respectively. (D–F) Environmental factors affecting microbial communities, bacterial communities, and micro-eukaryotic communities, respectively. (G–I) Correlation coefficient (r2) of environmental factors to microbial communities, bacterial communities, micro-eukaryotic communities, respectively, calculated by redundancy analysis (RDA) analysis. The environmental factors included pH, temperature (Temp), total carbon (TC), total nitrogen (TN), total sulfur (TS), nitrate and nitrite nitrogen (NOx-N), and ammonium nitrogen (NH4+-N). The symbol ‘*’, ‘**’, ‘***’ represents asterisks for significance in figures.
RDA analysis determined the factors influencing microbial communities (Figure 2D), bacterial communities (Figure 2E), and micro-eukaryotic communities (Figure 2F) in soil samples, respectively. pH, TC, TN, NOx-N, and NH4+-N were determined to significantly affect microbial communities (p < 0.01). Further, we found that pH played the most important role on regulating microbial community patterns in soil samples (Figure 2G). For bacterial community, pH, TC, TN, temperature, and NH4+-N concentration were detected as environmental factors significantly influencing their community structures (p <0.05). Correlation coefficient analysis further indicated that pH acted as the most important factor in changing bacterial communities, followed by TC, TN, and temperature (Figure 2H). Finally, TC, TN, TS, pH and temperature significantly influenced micro-eukaryotic communities, with pH and temperature showing equal contributions (Figure 2I).
The genus-level classification plots were shown in Supplementary Figures 4A, 5A. LEfSe analysis revealed that 62 bacterial and 77 micro-eukaryotic species exhibited significant differences (Supplementary Figures 4B, C, 5B, C) among treatments (p < 0.05, standardized scaling factor: 1000000). Among the top 30 bacterial classes, most showed significant differences (Figure 3A), with 18 classes significantly affected by both pH and temperature, including Alphaproteobacteria, Holophagae, and Vicinamibacteria. The acidification process enriched several bacteria, such as Acidimicrobiia (HP: 0.49%, MP: 0.51%, LP: 0.90%), Dehalococcoidia (HP: 0.072%, MP: 0.084%, LP: 0.15%) and Bdellovibrionia (HP: 0.37%, MP: 0.55%, LP: 2.56%). In addition, the warming process enriched several bacteria, such as Acidobacteriae (HP: 4.60%, MP: 4.77%, LP: 5.01%) and Bacteroidia (LT: 1.76%, MT: 5.11%, HT: 8.29%).
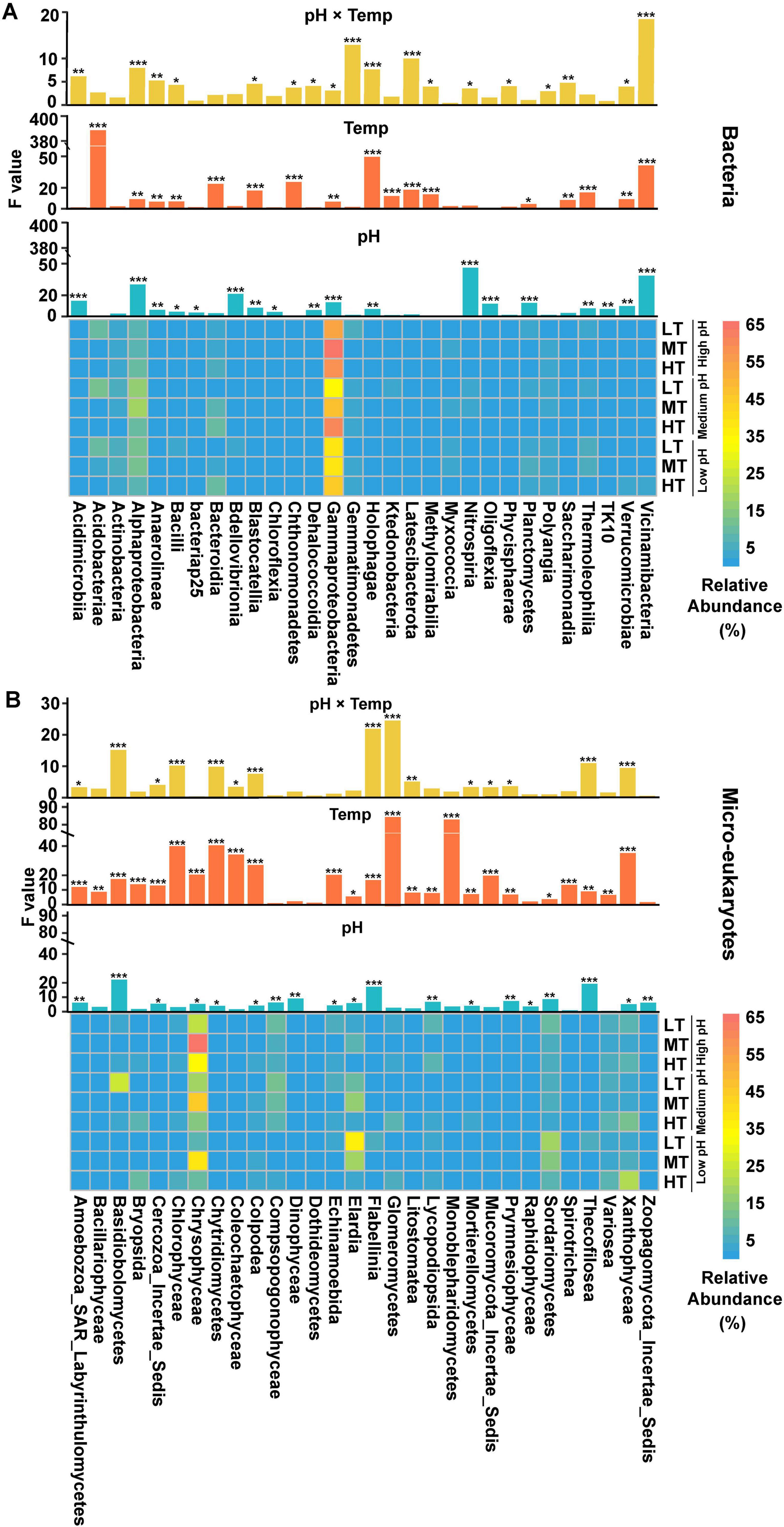
Figure 3. Factors influenced the relative abundance of variant taxa among different treatments, including (A) active bacterial microorganisms and (B) eukaryotic active microorganisms. The symbol ‘*’, ‘**’, ‘***’ represents asterisks for significance in figures.
Among the top 30 micro-eukaryotic classes, results showed temperature played a more important role in regulating the abundance of eukaryotic microbes in comparison with pH (Figure 3B). There were 25 classes of eukaryotic microbes significantly affected by temperature, such as Glomeromycetes (LT: 2.25%, MT: 1.35%, HT: 5.98%), Monoblepharidomycetes (LT: 0.22%, MT: 0.11%, HT: 0.73%) and Coleochaetophyceae (LT: 0.71%, MT: 0.21%, HT: 1.16%). In addition, Elardia reached its peak under LT groups (LT: 16.28%, MT: 14.18%, HT: 2.68%) and Chrysophyceae exhibited the highest relative abundance under MT treatment (LT: 16.18%, MT: 50.11%, HT: 19.88%).
Mantel tests were employed to detect the biotic and abiotic factors affecting denitrification and N2O production rates in soil and to analyze the relationships between biotic and abiotic factors (Figure 4). The results showed that pH (r = 0.68, p < 0.05), active bacterial diversity (Chao1 and Shannon: r = 0.22, p < 0.05 and r = 0.18, p < 0.05, respectively), community pattern (microbes and bacteria: r = 0.71, p < 0.05 and r = 0.74, p < 0.05, respectively), inorganic nitrogen (NH4+ and NOx–: r = 0.33, p < 0.05 and r = 0.32, p < 0.05, respectively) significantly influenced the rates of N2O production. Differing from N2O production, denitrification rate was significantly affected by pH (r = 0.26, p < 0.05), total carbon (TC, r = 0.25, p < 0.05), and total nitrogen (TN, r = 0.29, p < 0.05). Notably, no significant correlation between temperature and denitrification rate, and between temperature and N2O production rate was observed, while the significant correlations between temperature and microbial alpha diversity, and soil nutrients (e.g., TC. TN, and TS) were observed in this study.
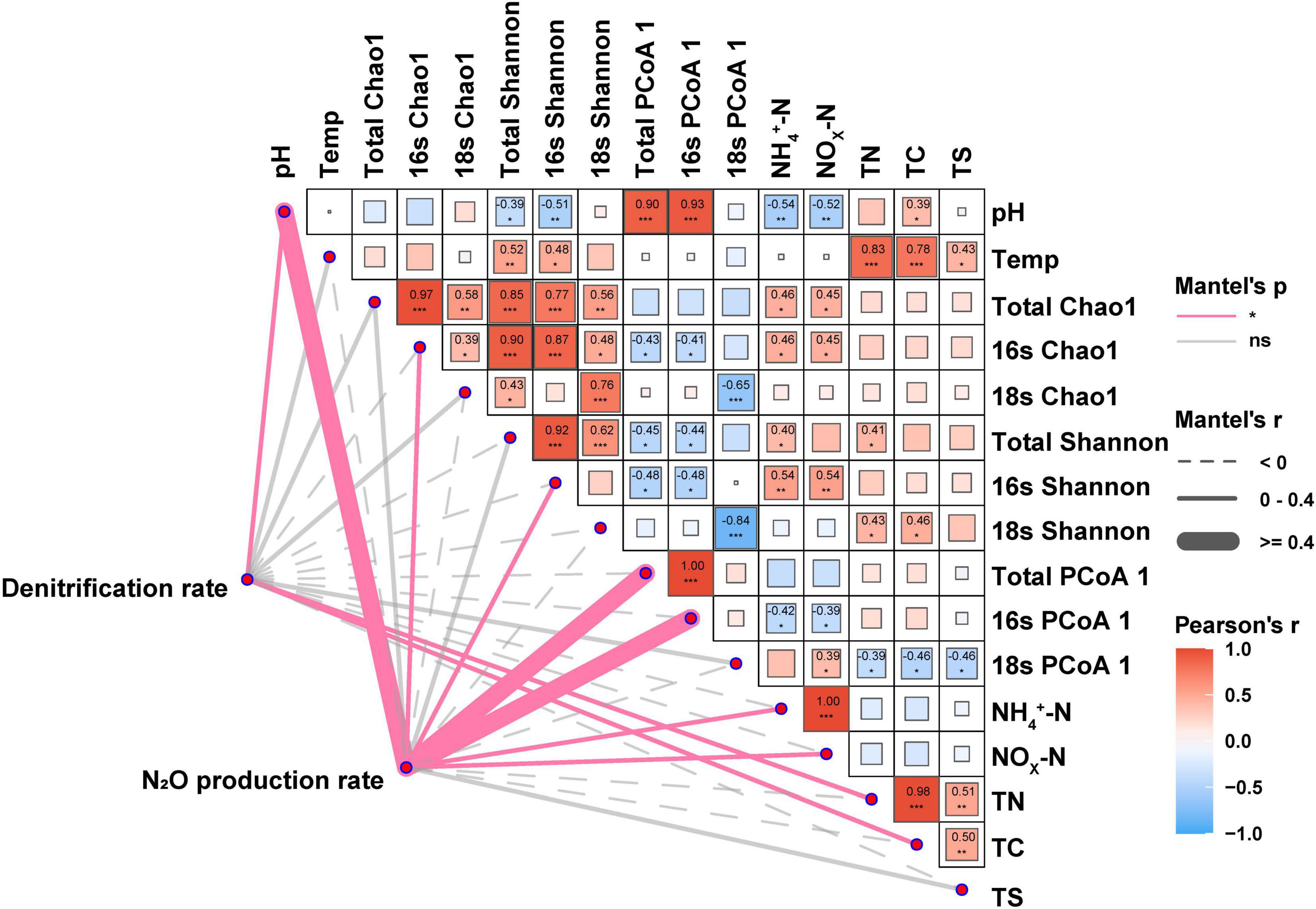
Figure 4. Mantel test analysis identified factors influencing both denitrification and N2O production rate. The Mantel’s r and p-values indicated the correlation and significance, respectively. Dashed lines and solid lines indicated negative and positive correlations, respectively, while red and gray color meant significant (p < 0.05) and non-significant, respectively. Total Chao1: Chao1 index of microbial including bacterial and micro-eukaryotic community. 16S Chao1 and 18S Chao1: Chao1 index of bacterial and micro-eukaryotic community, respectively. Total Shannon: Shannon index of microbial including bacterial and micro-eukaryotic community. 16S Shannon and 18S Shannon: Shannon index of bacterial and micro-eukaryotic communities, respectively. Total β diversity: PCoA1 axis index of microbial including bacterial and micro-eukaryotic community. 16S β diversity and 18S β diversity were used to represent bacterial and micro-eukaryotic PCoA1 axis, respectively. The symbol ‘*’, ‘**’, ‘***’ represents asterisks for significance in figures.
PLSPM was employed to further analyze the mechanisms of the effects of pH and temperature on soil denitrification rate and model explained 44%, 45%, and 50% of the variation in denitrification rate for overall active microbial, bacterial and micro-eukaryotic groups, respectively (Figures 5A, C, E). The alpha diversity of active microorganisms exerts the strongest positive effect on denitrification rates. Furthermore, inorganic nitrogen has the opposite effect on denitrification rates in both bacterial and micro-eukaryotic models. Notably, pH has a greater influence on denitrification rates than temperature (Supplementary Figures 6A–C). Consistent with the Mantel test results, pH exerted a stronger influence on denitrification rates than temperature (Figures 5B, D, F). Alpha diversity had the pH directly and indirectly affected the rates of denitrification through altering soil properties, bacterial alpha diversity (Figure 5C), and micro-eukaryotic community pattern (Figure 5E). Temperature, conversely, only indirectly affected denitrification rates by influencing inorganic nitrogen (NH4+ and NOx–) concentrations and microbial community pattern (Figures 5A, C, E). Additionally, increasing temperature significantly and directly affected the community compositions of eukaryotic microbes but not bacteria in soil ecosystems (Figures 5C, E). Further, a Random Forest analysis corroborated these findings, indicating that pH was the most significant factor influencing denitrification rates in both micro-eukaryotic and bacterial groups, while the importance of other physicochemical properties, including TC and TN, ranked secondary in comparison, consistent with the Mantel test results (Figures 5B, D, F).
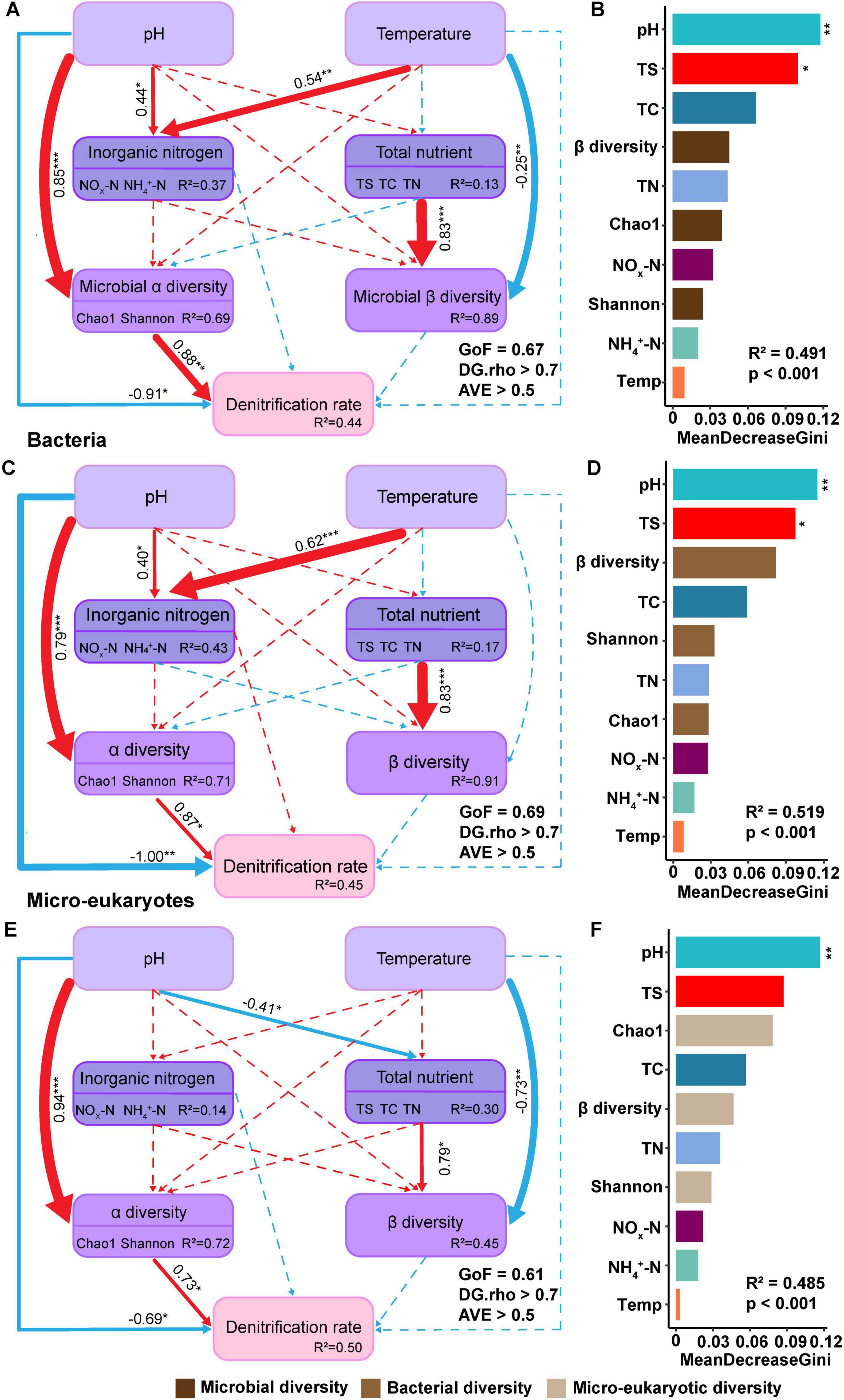
Figure 5. Partial least squares path modeling (PLSPM) model elucidated the influence of microbial and soil properties on denitrification rate. The model assessed the impacts of active soil microorganisms (bacteria and eukaryotic microbes) and physicochemical properties on denitrification rate. (A,C,E) The fitness of PLSPM model was acceptable (GoF > 0.6, DG.rho > 0.7, AVE > 0.5). Microbial alpha and beta diversity represented the overall diversity of bacterial and eukaryotic active microbes. Positive correlations were depicted by red solid lines, and negative correlations by blue solid lines. Dashed lines indicated non-significant correlations, with the color corresponding to the correlation status. Path coefficients and coefficients of determination (R2) were calculated after 999 bootstrapping iterations, and all path coefficients shown were statistically significant (p < 0.05). (B,D,F) Random Forest modeling with permutation tests (n = 999, *p < 0.05, **p < 0.01) indicated that the MeanDecreaseGini (MDG) values showed the significant importance of environmental variables and microbial biodiversity indices. (A,B) Represented overall microorganisms. The index of β diversity, Chao1 and Shannon indicated microbial communities including bacterial and micro-eukaryotic community diversity. (C,D) Denoted bacteria. Similarly, the index of β diversity, Chao1 and Shannon indicated bacterial community diversity. (E,F) Referred to eukaryotic microbes and their indexes of β diversity, Chao1 and Shannon indicated micro-eukaryotic community diversity. The environmental factors included pH, temperature (Temp), total carbon (TC), total nitrogen (TN), total sulfur (TS), nitrate and nitrite nitrogen (NOx-N), and ammonium nitrogen (NH4+-N). The symbol ‘***’ represents asterisks for significance in figures.
In the present study, we observed that the relative abundance of denitrifying bacteria, such as Pseudoxanthomonas, Bacillus, and Pseudomonas (Hartmann and Six, 2023), exhibited relatively high abundances under MTMP treatment, and this treatment had the highest denitrification rate. Denitrifying bacteria are abundant in various soil types, including farmland, parks, and tea gardens, where they enhance N2O emissions and play a crucial role in the nitrogen cycle (Crenshaw et al., 2008; Hiis et al., 2024; Tan et al., 2024). Additionally, there was a significant positive correlation between inorganic nitrogen levels and bacterial diversity (Figure 4), which may be attributed to the nitrification processes performed by nitrifying bacteria. Nitrifying bacteria, such as Nitrosospira and Nitrospira (Zhang et al., 2022; Deng et al., 2024), exhibited the highest abundance under MTMP treatment. This condition also led to the highest denitrification rates by providing substrates for denitrification and enhancing the activity of associated microorganisms (Su et al., 2021). These findings highlight the significant role of active bacteria in mediating biochemical processes and influencing soil multifunctionality.
Micro-eukaryotic denitrifiers play an important role in N2O emission (Figures 4, 5E). Although the richness of micro-eukaryotic denitrifiers was relatively rare compared to their bacterial counterparts (Bösch et al., 2023), they exhibited biome-specific differences in both relative abundance and species distributions (Figure 2C and Supplementary Figure 5A). Studies indicate that fungi contribute more to soil N2O emissions than bacteria in acidic soils (Laughlin and Stevens, 2002; Herold et al., 2012; Zheng et al., 2020; Xiong et al., 2024), especially those belonging to the genus Fusarium, can perform denitrification sensu stricto (Keuschnig et al., 2020), resulting in an increase in N2O emissions (Shoun et al., 1989; Zheng et al., 2020). As studies have shown that Fusarium is an important fungus in N2O emission (Maeda et al., 2015; Zheng et al., 2020; Shao et al., 2024), under acidic conditions (e.g., in LTLP treatment), the relative abundance of Fusarium is highest, which may explain the phenomenon that soil still retains denitrification capacity (Figure 1A) even in strongly acidic environments (pH 4.3–4.7) (Chen et al., 2014). In addition to fungi, other eukaryotic microorganisms, mainly including foraminifers such as Globobulimina pseudospinescens, Bolivina plicata and Stainforthia sp. (Kamp et al., 2015; Risgaard-Petersen et al., 2006), are also involved in denitrification processes that release N2O. Unlike active bacteria, eukaryotic microbes exhibit similar responses to pH and temperature (Figures 2I, 5E), demonstrating their ability to thrive under extreme climatic conditions with high temperatures (Xu et al., 2017). The relatively low sensitivity of micro-eukaryotic richness to pH and temperature changes compared to bacteria may explain their stability in denitrification processes (Huang et al., 2017; Banerjee et al., 2024). These results imply potential micro-eukaryotic contributions to nitrogen cycling processes, laying the foundation for future use of eukaryotic microbes to improve soil health.
Temperature is a critical factor influencing denitrification processes (Barnard et al., 2005). Elevated temperatures (typically 20°C–25°C) enhance N2O emissions (Dai et al., 2020). This phenomenon can be attributed to multiple mechanisms. Higher temperatures accelerate the mineralization process (Dai et al., 2020), which elevates soil inorganic nitrogen levels (Figure 5A) and supplies additional substrates for microbial nitrification and denitrification, thereby promoting N2O production. Furthermore, rising temperatures stimulate the activity and growth of denitrifying microorganisms in soil (Schulz et al., 2017). For instance, warmer conditions promote the proliferation of Bacillus (Choma et al., 2000), directly contributing to increased N2O emission.
This study reveals a negative correlation between soil pH and denitrification rate, aligning with earlier report (Čuhel and Šimek, 2011). Previous research demonstrated that pH dominates the ratio of denitrification products (Figures 2G–I, 4, 5B, D, F), with N2O being the primary product at pH 4.6–5.4 (Koskinen and Keeney, 1982). This phenomenon may be attributed to the significantly negative correlation between pH and inorganic nitrogen concentrations (Figure 4). Specifically, soil acidity enhances the effect of nitrate on the composition denitrification-derived gaseous products, driving higher N2O production at pH 4.9 than at pH 6.5 (Firestone et al., 1980). However, reduced pH hinders the decomposition of soil organic nitrogen (Li et al., 2018), partially suppressing denitrification rates. pH further indirectly inhibits denitrification (Krichels et al., 2025) by altering active microbial communities (Figures 2G–I). The limited N2O reduction in LP groups (Figure 1A) implies that acidic conditions may inhibit bacterial N2O reductase activity, potentially due to the sensitivity of reductase translation and assembly to pH value (ŠImek and Cooper, 2002). Active bacteria responses to pH fluctuations are rapid, as their lack of a cytoskeleton contrasts with the structural rigidity conferred by fungal chitinous cell walls (Wang and Kuzyakov, 2024). Additionally, fungal spore formation further reinforces resistance to stressors such as drought and acidification (Yang et al., 2024).
Soil pH exerts a stronger impact than temperature on N2O emission and active microorganisms (Figures 2–5), particularly in shaping the denitrifying bacterial community patterns (Lauber et al., 2009; Bakken et al., 2012). This likely arises because pH more robustly governs nitrate utilization by denitrifiers (Blackmer and Bremner, 1978; Senbayram et al., 2019). Our analysis demonstrated a significant negative correlation between nitrate and pH, but no such relationship with temperature (Figure 4). Elevated nitrate levels, as a substrate, promote the proliferation of nirK- and nirS- type denitrifiers (Hao et al., 2022). Moreover, pH exerts a marked effect on microbial diversity (Figures 4, 5) and community patterns (Figure 2), particularly on certain bacterial denitrifiers (Pan et al., 2023). Researchers revealed that significant changes in the proteome of denitrifier (e.g., P. denitrificans) were identified when comparing pH 6.5 and 7.2, exhibiting significant downregulation of functional proteins (Olaya-Abril et al., 2021). These findings suggest that pH stability, compared to temperature, exerts a stronger influence on the survival of denitrifiers, ultimately shaping microbial diversity and community patterns. This may account for the reduced relative abundance of Pseudomonas and Pseudoxanthomonas at low pH, as observed in genus-level bacterial community structure analyses.
While this study demonstrated that the effects of soil acidification and warming on denitrification processes through RNA-seq approaches, it should be noted that the current conclusions were exclusively based on correlation analyses, such as RDA, Mantel test and PLSPM model. To clarify the mechanisms on how active microorganisms impacts soil nitrogen cycling processes, future studies should consider combining quantitative PCR and metatranscriptomics to systematically assess the functional contributions of microbial communities (e.g., nirK, nirS, and nosZ genes) in soil denitrification processes.
In summary, the results of this study demonstrated a stronger impact of pH on denitrification rates than temperature. Micro-eukaryotic and bacterial communities exhibited distinct responses to soil acidification and warming. Bacterial communities were predominantly shaped by pH, while micro-eukaryotic communities were influenced similarly by pH and temperature. Additionally, we found that micro-eukaryotic active microbes also contributed to the denitrification and N2O emission in the used soil of this study. These findings highlighted the important role of pH on nitrogen cycling in soil through changing active bacterial and micro-eukaryotic communities and provided scientific basis for agricultural management during the global warming.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found here: https://www.scidb.cn/en, https://doi.org/10.57760/sciencedb.17932.
PX: Data curation, Formal Analysis, Investigation, Visualization, Writing – original draft, Writing – review and editing. MG: Investigation, Writing – original draft, Writing – review and editing, Formal Analysis. YL: Writing – original draft, Writing – review and editing, Data curation, Investigation. JY: Writing – original draft, Writing – review and editing, Supervision. JS: Writing – original draft, Writing – review and editing, Conceptualization, Supervision. HL: Funding acquisition, Investigation, Methodology, Validation, Writing – original draft, Writing – review and editing, Project administration, Conceptualization, Supervision.
The author(s) declare that financial support was received for the research and/or publication of this article. This work was financially supported by Fujian Provincial Department of Science and Technology (2023J06047) and National Natural Science Foundation of China (42177097).
We wish to thank and acknowledge the staffs at the Institute of Urban Environment, Chinese Academy of Sciences (IUE) for advice and assistance with laboratory instruments and labs.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The authors declare that no Generative AI was used in the creation of this manuscript.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2025.1572497/full#supplementary-material
Baggs, E. M., Smales, C. L., and Bateman, E. J. (2010). Changing pH shifts the microbial sourceas well as the magnitude of N2O emission from soil. Biol. Fertil. Soils 46, 793–805. doi: 10.1007/s00374-010-0484-6
Bakken, L. R., Bergaust, L., Liu, B., and Frostegård, Å (2012). Regulation of denitrification at the cellular level: A clue to the understanding of N2O emissions from soils. Philos. Trans. R. Soc. B Biol. Sci. 367, 1226–1234. doi: 10.1098/rstb.2011.0321
Banerjee, S., Zhao, C., Garland, G., Edlinger, A., García-Palacios, P., Romdhane, S., et al. (2024). Biotic homogenization, lower soil fungal diversity and fewer rare taxa in arable soils across Europe. Nat. Commun. 15:327. doi: 10.1038/s41467-023-44073-6
Barnard, R., Leadley, P. W., and Hungate, B. A. (2005). Global change, nitrification, and denitrification: A review. Glob. Biogeochem. Cycles 19:GB1007. doi: 10.1029/2004GB002282
Bhattarai, H. R., Wanek, W., Siljanen, H. M. P., Ronkainen, J. G., Liimatainen, M., Hu, Y., et al. (2021). Denitrification is the major nitrous acid production pathway in boreal agricultural soils. Commun. Earth Environ. 2:54. doi: 10.1038/s43247-021-00125-7
Blackmer, A. M., and Bremner, J. M. (1978). Inhibitory effect of nitrate on reduction of N2O to N2 by soil microorganisms. Soil Biol. Biochem. 10, 187–191. doi: 10.1016/0038-0717(78)90095-0
Bolyen, E., Rideout, J. R., Dillon, M. R., Bokulich, N. A., Abnet, C. C., Al-Ghalith, G. A., et al. (2019). Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 37, 852–857. doi: 10.1038/s41587-019-0209-9
Bösch, Y., Pold, G., Saghaï, A., Karlsson, M., Jones, C. M., and Hallin, S. (2023). Distribution and environmental drivers of fungal denitrifiers in global soils. Microbiol. Spectr. 11:e00061-23. doi: 10.1128/spectrum.00061-23
Braker, G., Schwarz, J., and Conrad, R. (2010). Influence of temperature on the composition and activity of denitrifying soil communities. FEMS Microbiol. Ecol. 73, 134–148. doi: 10.1111/j.1574-6941.2010.00884.x
Callahan, B. J., McMurdie, P. J., Rosen, M. J., Han, A. W., Johnson, A. J., and Holmes, S. P. (2016). DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 13, 581–583. doi: 10.1038/nmeth.3869
Chen, H., Mothapo, N. V., and Shi, W. (2014). The significant contribution of fungi to soil N2O production across diverse ecosystems. Appl. Soil Ecol. 73, 70–77. doi: 10.1016/j.apsoil.2013.08.011
Choma, C., Clavel, T., Dominguez, H., Razafindramboa, N., Soumille, H., Nguyen-the, C., et al. (2000). Effect of temperature on growth characteristics of Bacillus cereus TZ415. Int. J. Food Microbiol. 55, 73–77. doi: 10.1016/S0168-1605(00)00197-5
Chunyi, K., Wei, S., Mingken, W., Chunyu, X., and Changxiu, L. (2024). Diversity, community structure, and abundance of nirS-type denitrifying bacteria on suspended particulate matter in coastal high-altitude aquaculture pond water. Sci. Rep. 14:5594. doi: 10.1038/s41598-024-56196-x
Crenshaw, C. L., Lauber, C., Sinsabaugh, R. L., and Stavely, L. K. (2008). Fungal control of nitrous oxide production in semiarid grassland. Biogeochemistry 87, 17–27. doi: 10.1007/s10533-007-9165-4
Čuhel, J., and Šimek, M. (2011). Proximal and distal control by pH of denitrification rate in a pasture soil. Agric. Ecosyst. Environ. 141, 230–233. doi: 10.1016/j.agee.2011.02.016
Dai, Z., Yu, M., Chen, H., Zhao, H., Huang, Y., Su, W., et al. (2020). Elevated temperature shifts soil N cycling from microbial immobilization to enhanced mineralization, nitrification and denitrification across global terrestrial ecosystems. Glob. Change Biol. 26, 5267–5276. doi: 10.1111/gcb.15211
Deng, N., Gubry-Rangin, C., Song, X.-T., Ju, X.-T., Liu, S.-Y., Shen, J.-P., et al. (2024). AOB Nitrosospira cluster 3a.2 (D11) dominates N2O emissions in fertilised agricultural soils. J. Environ. Manage. 355:120504. doi: 10.1016/j.jenvman.2024.120504
Firestone, M. K., Firestone, R. B., and Tiedje, J. M. (1980). Nitrous oxide from soil denitrification: Factors controlling its biological production. Science 208, 749–751. doi: 10.1126/science.208.4445.749
Hao, J., Feng, Y., Wang, X., Yu, Q., Zhang, F., Yang, G., et al. (2022). Soil microbial nitrogen-cycling gene abundances in response to crop diversification: A meta-analysis. Sci. Total Environ. 838:156621. doi: 10.1016/j.scitotenv.2022.156621
Hartmann, M., and Six, J. (2023). Soil structure and microbiome functions in agroecosystems. Nat. Rev. Earth Environ. 4, 4–18. doi: 10.1038/s43017-022-00366-w
Herold, M. B., Baggs, E. M., and Daniell, T. J. (2012). Fungal and bacterial denitrification are differently affected by long-term pH amendment and cultivation of arable soil. Soil Biol. Biochem. 54, 25–35. doi: 10.1016/j.soilbio.2012.04.031
Higgins, S. A., Welsh, A., Orellana Luis, H., Konstantinidis Konstantinos, T., Chee-Sanford Joanne, C., Sanford Robert, A., et al. (2016). Detection and diversity of fungal nitric oxide reductase genes (p450nor) in agricultural soils. Appl. Environ. Microbiol. 82, 2919–2928. doi: 10.1128/AEM.00243-16
Hiis, E. G., Vick, S. H. W., Molstad, L., Røsdal, K., Jonassen, K. R., Winiwarter, W., et al. (2024). Unlocking bacterial potential to reduce farmland N2O emissions. Nature 630, 421–428. doi: 10.1038/s41586-024-07464-3
Huang, Y., Xiao, X., and Long, X. (2017). Fungal denitrification contributes significantly to N2O production in a highly acidic tea soil. J. Soils Sediments 17, 1599–1606. doi: 10.1007/s11368-017-1655-y
Kamp, A., Høgslund, S., Risgaard-Petersen, N., and Stief, P. (2015). Nitrate storage and dissimilatory nitrate reduction by eukaryotic microbes. Front. Microbiol. 6:1492. doi: 10.3389/fmicb.2015.01492
Keuschnig, C., Gorfer, M., Li, G., Mania, D., Frostegard, A., Bakken, L., et al. (2020). NO and N2O transformations of diverse fungi in hypoxia: Evidence for anaerobic respiration only in Fusarium strains. Environ. Microbiol. 22, 2182–2195. doi: 10.1111/1462-2920.14980
Kobayashi, M., Matsuo, Y., Takimoto, A., Suzuki, S., Maruo, F., and Shoun, H. (1996). Denitrification, a novel type of respiratory metabolism in fungal mitochondrion*. J. Biol. Chem. 271, 16263–16267. doi: 10.1074/jbc.271.27.16263
Koskinen, W. C., and Keeney, D. R. (1982). Effect of pH on the rate of gaseous products of denitrification in a silt loam soil. Soil Sci. Soc. Am. J. 46, 1165–1167. doi: 10.2136/sssaj1982.03615995004600060009x
Krichels, A. H., Sanford, R. A., Chee-Sanford, J. C., Connor, L., Van Allen, R., Kent, A. D., et al. (2025). Distinct N-cycling microbial communities contribute to microtopographic variation in soil N2O emissions from denitrification. Soil Biol. Biochem. 202:109683. doi: 10.1016/j.soilbio.2024.109683
Lauber, C. L., Hamady, M., Knight, R., and Fierer, N. (2009). Pyrosequencing-based assessment of soil pH as a predictor of soil bacterial community structure at the continental scale. Appl. Environ. Microbiol. 75, 5111–5120. doi: 10.1128/AEM.00335-09
Laughlin, R., and Stevens, R. J. (2002). Evidence for fungal dominance of denitrification and codenitrification in a grassland soil. Soil Sci. Soc. Am. J. 66, 1540–1548. doi: 10.2136/sssaj2002.1540
Li, H., Hong, Y. W., Gao, M. K., An, X. L., Yang, X. R., Zhu, Y. G., et al. (2023). Distinct responses of airborne abundant and rare microbial communities to atmospheric changes associated with Chinese New Year. Imeta 2:e140. doi: 10.1002/imt2.140
Li, H., Yang, X., Weng, B., Su, J., Nie, S., Gilbert, J. A., et al. (2016). The phenological stage of rice growth determines anaerobic ammonium oxidation activity in rhizosphere soil. Soil Biol. Biochem. 100, 59–65. doi: 10.1016/j.soilbio.2016.05.015
Li, H., Zhao, S., Gao, M.-K., Zhou, Y., Xu, B., Yang, L.-Y., et al. (2025). Experimental evidence for viral impact on microbial community, nitrification, and denitrification in an agriculture soil. J. Hazard. Mater. 489:137532. doi: 10.1016/j.jhazmat.2025.137532
Li, Y., Sun, J., Tian, D., Wang, J., Ha, D., Qu, Y., et al. (2018). Soil acid cations induced reduction in soil respiration under nitrogen enrichment and soil acidification. Sci. Total Environ. 615, 1535–1546. doi: 10.1016/j.scitotenv.2017.09.131
Long, A., Heitman, J., Tobias, C., Philips, R., and Song, B. (2013). Co-occurring anammox, denitrification, and codenitrification in agricultural soils. Appl. Environ. Microbiol. 79, 168–176. doi: 10.1128/AEM.02520-12
Maeda, K., Spor, A., Edel-Hermann, V., Heraud, C., Breuil, M.-C., Bizouard, F., et al. (2015). N2O production, a widespread trait in fungi. Sci. Rep. 5:9697. doi: 10.1038/srep09697
Margalef-Marti, R., Thibault de Chanvalon, A., Anschutz, P., Amouroux, D., and Sebilo, M. (2024). Synergies of chemodenitrification and denitrification in a saline inland lake. Chemosphere 359:142292. doi: 10.1016/j.chemosphere.2024.142292
Molstad, L., Dörsch, P., and Bakken, L. R. (2007). Robotized incubation system for monitoring gases (O2, NO, N2O N2O) in denitrifying cultures. J. Microbiol. Methods 71, 202–211. doi: 10.1016/j.mimet.2007.08.011
Montzka, S. A., Dlugokencky, E. J., and Butler, J. H. (2011). Non-CO2 greenhouse gases and climate change. Nature 476, 43–50. doi: 10.1038/nature10322
Mothapo, N. V., Chen, H., Cubeta, M. A., and Shi, W. (2013). Nitrous oxide producing activity of diverse fungi from distinct agroecosystems. Soil Biol. Biochem. 66, 94–101. doi: 10.1016/j.soilbio.2013.07.004
Mothapo, N., Chen, H., Cubeta, M. A., Grossman, J. M., Fuller, F., and Shi, W. (2015). Phylogenetic, taxonomic and functional diversity of fungal denitrifiers and associated N2O production efficacy. Soil Biol. Biochem. 83, 160–175. doi: 10.1016/j.soilbio.2015.02.001
Olaya-Abril, A., Hidalgo-Carrillo, J., Luque-Almagro, V. M., Fuentes-Almagro, C., Urbano, F. J., Moreno-Vivián, C., et al. (2021). Effect of pH on the denitrification proteome of the soil bacterium Paracoccus denitrificans PD1222. Sci. Rep. 11:17276. doi: 10.1038/s41598-021-96559-2
Pan, Y., She, D., Shi, Z., Cao, T., Xia, Y., and Shan, J. (2023). Salinity and high pH reduce denitrification rates by inhibiting denitrifying gene abundance in a saline-alkali soil. Sci. Rep. 13:2155. doi: 10.1038/s41598-023-29311-7
Philippot, L., Andert, J., Jones, C. M., Bru, D., and Hallin, S. (2011). Importance of denitrifiers lacking the genes encoding the nitrous oxide reductase for N2O emissions from soil. Global Change Biol. 17, 1497–1504. doi: 10.1111/j.1365-2486.2010.02334.x
Pruesse, E., Quast, C., Knittel, K., Fuchs, B. M., Ludwig, W., Peplies, J., et al. (2007). SILVA: A comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res. 35, 7188–7196. doi: 10.1093/nar/gkm864
Qiu, Y., Zhang, Y., Zhang, K., Xu, X., Zhao, Y., Bai, T., et al. (2024). Intermediate soil acidification induces highest nitrous oxide emissions. Nat. Commun. 15:2695. doi: 10.1038/s41467-024-46931-3
Ravishankara, A. R., Daniel, J. S., and Portmann, R. W. (2009). Nitrous Oxide (N2O): The dominant ozone-depleting substance emitted in the 21st century. Science 326, 123–125. doi: 10.1126/science.1176985
Risgaard-Petersen, N., Langezaal, A., Ingvardsen, S., et al. (2006). Evidence for complete denitrification in a benthic foraminifer. Nature 443, 93–96. doi: 10.1038/nature05070
Ryden, J. C. (1981). N2O exchange between a grassland soil and the atmosphere. Nature 292, 235–237. doi: 10.1038/292235a0
Salmaso, N., Boscaini, A., and Pindo, M. (2020). Unraveling the diversity of eukaryotic microplankton in a large and deep perialpine lake using a high throughput sequencing approach. Front. Microbiol. 11:168. doi: 10.3389/fmicb.2020.00789
Schulz, S., Kölbl, A., Ebli, M., Buegger, F., Schloter, M., and Fiedler, S. (2017). Field-scale pattern of denitrifying microorganisms and N2O emission rates indicate a high potential for complete denitrification in an agriculturally used organic soil. Microb. Ecol. 74, 765–770. doi: 10.1007/s00248-017-0991-1
Senbayram, M., Budai, A., Bol, R., Chadwick, D., Marton, L., Gündogan, R., et al. (2019). Soil NO3– level and O2 availability are key factors in controlling N2O reduction to N2 following long-term liming of an acidic sandy soil. Soil Biol. Biochem. 132, 165–173. doi: 10.1016/j.soilbio.2019.02.009
Sennett, L. B., Roco, C. A., Lim, N. Y. N., Yavitt, J. B., Dörsch, P., Bakken, L. R., et al. (2024). Determining how oxygen legacy affects trajectories of soil denitrifier community dynamics and N2O emissions. Nat. Commun. 15:7298. doi: 10.1038/s41467-024-51688-w
Shao, S., Li, Y., Li, Z., Ma, X., Zhu, Y., Luo, Y., et al. (2024). Impact of tea tree cultivation on soil microbiota, soil organic matter, and nitrogen cycling in mountainous plantations. Agronomy 14:638. doi: 10.3390/agronomy14030638
Shoun, H., Suyama, W., and Yasui, T. (1989). Soluble, nitrate/nitrite-inducible cytochrome P-450 of the fungus, Fusarium oxysporum. FEBS Lett. 244, 11–14. doi: 10.1016/0014-5793(89)81151-2
ŠImek, M., and Cooper, J. E. (2002). The influence of soil pH on denitrification: Progress towards the understanding of this interaction over the last 50 years. Eur. J. Soil Sci. 53, 345–354. doi: 10.1046/j.1365-2389.2002.00461.x
Su, X., Li, G., Cotner, J. B., Wei, L., Wang, Y., Pan, T., et al. (2021). Long-term organic fertilization changes soil active bacterial composition and multifunctionality: RNA-based bacterial community and qPCR-based SmartChip analysis. J. Soils Sediments 21, 799–809. doi: 10.1007/s11368-020-02854-2
Tan, Y., Chen, Z., Liu, W., Yang, M., Du, Z., Wang, Y., et al. (2024). Grazing exclusion alters denitrification N2O/(N2O + N2) ratio in alpine meadow of Qinghai–Tibet Plateau. Sci. Total Environ. 912:169358. doi: 10.1016/j.scitotenv.2023.169358
Taylor, A. E., Myrold, D. D., and Bottomley, P. J. (2019). Temperature affects the kinetics of nitrite oxidation and nitrification coupling in four agricultural soils. Soil Biol. Biochem. 136:107523. doi: 10.1016/j.soilbio.2019.107523
Wang, C., and Kuzyakov, Y. (2024). Mechanisms and implications of bacterial–fungal competition for soil resources. ISME J. 18:wrae073. doi: 10.1093/ismejo/wrae073
Wang, L., Zheng, B., Nan, B., and Hu, P. (2014). Diversity of bacterial community and detection of nirS- and nirK-encoding denitrifying bacteria in sandy intertidal sediments along Laizhou Bay of Bohai Sea, China. Mar. Pollut. Bull. 88, 215–223. doi: 10.1016/j.marpolbul.2014.09.002
Wang, Q., Garrity, G. M., Tiedje, J. M., and Cole, J. R. (2007). Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 73, 5261–5267. doi: 10.1128/aem.00062-07
Xiong, R., He, X., Gao, N., Li, Q., Qiu, Z., Hou, Y., et al. (2024). Soil pH amendment alters the abundance, diversity, and composition of microbial communities in two contrasting agricultural soils. Microbiol. Spectr. 12:e04165-23. doi: 10.1128/spectrum.04165-23
Xu, H.-J., Wang, X.-H., Li, H., Yao, H.-Y., Su, J.-Q., and Zhu, Y.-G. (2014). Biochar impacts soil microbial community composition and nitrogen cycling in an acidic soil planted with rape. Environ. Sci. Technol. 48, 9391–9399. doi: 10.1021/es5021058
Xu, H.-J., Yang, X.-R., Li, S., Xue, X.-M., Chang, S., Li, H., et al. (2019). Nitrogen inputs are more important than denitrifier abundances in controlling denitrification-derived N2O emission from both urban and agricultural soils. Sci. Total Environ. 650, 2807–2817. doi: 10.1016/j.scitotenv.2018.10.001
Xu, X., Liu, X., Li, Y., Ran, Y., Liu, Y., Zhang, Q., et al. (2017). High temperatures inhibited the growth of soil bacteria and archaea but not that of fungi and altered nitrous oxide production mechanisms from different nitrogen sources in an acidic soil. Soil Biol. Biochem. 107, 168–179. doi: 10.1016/j.soilbio.2017.01.003
Yang, H., Lyu, W., Lu, L., Shi, X., Li, N., Wang, W., et al. (2020). Biogeography of microbiome and short-chain fatty acids in the gastrointestinal tract of duck. Poult. Sci. 99, 4016–4027. doi: 10.1016/j.psj.2020.03.040
Yang, W., Zhang, L., Yang, Y., Xiang, H., and Yang, P. (2024). Plant secondary metabolites-mediated plant defense against bacteria and fungi pathogens. Plant Physiol. Biochem. 217:109224. doi: 10.1016/j.plaphy.2024.109224
Yao, S., Ni, J., Chen, Q., and Borthwick, A. G. L. (2013). Enrichment and characterization of a bacteria consortium capable of heterotrophic nitrification and aerobic denitrification at low temperature. Bioresour. Technol. 127, 151–157. doi: 10.1016/j.biortech.2012.09.098
Zhang, Q., Han, P., Xu, H., Wang, Q., and Xu, G. (2022). Survival strategies of Nitrospira in a stable nitritation-denitritation system treating low-strength fermented wastewater. Biochem. Eng. J. 187:08674. doi: 10.1016/j.bej.2022.108674
Keywords: acidification, warming, RNA level, microbial community, denitrification rate
Citation: Xu P, Gao M, Li Y, Ye J, Su J and Li H (2025) Combined effects of acidification and warming on soil denitrification and microbial community. Front. Microbiol. 16:1572497. doi: 10.3389/fmicb.2025.1572497
Received: 07 February 2025; Accepted: 17 March 2025;
Published: 02 April 2025.
Edited by:
Xiaolong Liang, Key Laboratory of Pollution Ecology and Environmental Engineering, Institute of Applied Ecology (CAS), ChinaReviewed by:
Pengfei Liu, Lanzhou University, ChinaCopyright © 2025 Xu, Gao, Li, Ye, Su and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hu Li, aGxpQGl1ZS5hYy5jbg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.