- 1Sichuan Province Engineering Technology Research Center of Liquor-Making Grains, School of Bioengineering, Sichuan University of Science and Engineering, Yibin, China
- 2Liquor Brewing Biotechnology and Application Key Laboratory of Sichuan Province, Yibin, China
- 3National Engineering Research Center of Solid-State Brewing, Luzhou Laojiao Co., Ltd., Luzhou, China
- 4Sichuan Yibin Hengshengfu Liquor Industry Group Co., Ltd., Yibin, China
Introduction: Pichia kudriavzevii is a prevalent non-Saccharomyces cerevisiae yeast in baijiu brewing. The aim of this study was to isolate a high temperature resistant Pichia kudriavzevii strain from the daqu of strong flavor baijiu and to elucidate its molecular mechanism.
Methods: Growth activity was assessed at temperatures of 37°C, 40°C, 45°C, and 50°C. Morphological changes were observed by scanning electron microscopy at 37°C, 45°C, and 50°C. Subsequent analysis of the transcriptomics and metabolomics was undertaken to elucidate the molecular mechanism of heat tolerance.
Results: The strain was able to tolerate high temperature of 50°C, undergoing substantial morphological alterations. Gene ontology (GO) analysis of the transcriptomics revealed that differentially expressed genes (DEGs) were enriched in pathways such as ATP biosynthesis process and mitochondrion; Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis showed that DEGs were up regulated in oxidative phosphorylation. Utilising liquid chromatograph-mass spectrometer, a total of 463 cationic differential metabolites and 352 anionic differential metabolites were detected and screened for differential substances that were closely related to heat tolerance (NAD+ and ADP); KEGG analysis showed that metabolites were up regulated in purine metabolism. Furthermore, correlation analyses of transcriptomics-metabolomics demonstrated a strong positive correlation between the metabolites NAD+ and ADP, and multiple DEGs of the oxidative phosphorylation pathway.
Discussion: These results suggest that the heat tolerant strain can be able to counteract high temperature environment by up regulating energy metabolism (especially oxidative phosphorylation) to increase ATP production.
1 Introduction
Baijiu is an ancient distilled spirit that originated in China and has a history of more than 2,000 years. In comparison with foreign distilled spirits, there is a significant disparity in the raw materials and technology utilised in its production (Liu and Sun, 2018). It is typically produced through the fermentation of a blend of sorghum, rice, corn, barley, wheat, and other grain crops, following specific combinations and ratios (Kang et al., 2023). This fermentation process entails a complex microbial activity (Tu et al., 2022), wherein the microbiota, through its own succession and growth patterns, and metabolic processes, generates a multitude of fermentation metabolites that collectively contribute to the formation of the distinctive flavor profile of baijiu (Zhang et al., 2024). Daqu is a saccharification fermenter employed in the production of baijiu, and it is distinguished by its rich microbial communities, functional enzyme systems, and flavor precursors (Li et al., 2023). The classification of daqu utilised in the context of baijiu fermentation can be predicated on the processing temperature range: high temperature (60–70°C), medium-high temperature (55–60°C), medium-temperature (50–55°C), and low-temperature (40–50°C) (Zhang et al., 2024; Zhou et al., 2022). Therefore, the temperature of the microorganism growth environment frequently exceeds 40°C, particularly in high temperature daqu, and the microbial population decreases as the temperature rises (Gou et al., 2015; Wang et al., 2008). Indeed, in the process of producing baijiu, the microbiological system is frequently exposed to extreme conditions, including high temperature, high acidity, and high ethanol level (Wang N. et al., 2023). Notably, high temperature has been identified as a pivotal factor influencing the activity of brewing microorganisms, the rate of microbial growth, and the production of metabolites (Amillastre et al., 2012; Mejía-Barajas et al., 2017). It has been established that high temperature can induce hazards such as diminished cell viability, inhibition of the growth rate, disruption of cellular and mitochondrial membranes (Richter et al., 2010).
For many years, Saccharomyces cerevisiae has been considered the most prevalent yeast in the fermentation process of baijiu, and it is the microorganism that exerts a substantial influence on the fermentation process (Hu et al., 2021). Indeed, non-Saccharomyces cerevisiae yeast has been shown to play an essential role in the brewing process (Gou et al., 2015). It has been established that certain non-Saccharomyces cerevisiae yeasts possess remarkable heat tolerance and are capable of adapting to the high temperature encountered during baijiu fermentation (Xiong et al., 2014). Consequently, the isolation and utilisation of heat tolerant non-Saccharomyces cerevisiae yeast during baijiu brewing can contribute to the stabilisation of the ecological balance of brewing microorganisms under extreme high temperature conditions. Pichia kudriavzevii is a yeast that is found in nature and is frequently employed in the fermentation of foodstuffs due to its ability to withstand high temperature, acid, and ethanol (Chu et al., 2023; Zhou et al., 2022). Pichia kudriavzevii is the dominant non-Saccharomyces cerevisiae yeast in a wide range of aroma types of baijiu, including strong flavor, sauce flavor, and light flavor (Wang et al., 2019). During the initial phase of baijiu fermentation, a greater diversity of yeast species is present. However, as the fermentation process continues at high temperature, there is a notable decline in the number and species of yeast, but Pichia kudriavzevii maintains predominant at high temperature (You et al., 2021). The majority of Pichia kudriavzevii are capable of growth within the temperature range of 25–40°C, and it has been demonstrated that Pichia kudriavzevii exhibits greater heat tolerance than Saccharomyces cerevisiae (Wang et al., 2024).
In this study, a Pichia kudriavzevii strain was isolated from the daqu of strong flavor baijiu, which was found to have good heat tolerance by measuring growth activity. Utilising transcriptomics and metabolomics, we sought to elucidate the molecular underpinnings of the strain’s heat tolerance, with the objective of providing an idea for the investigation of heat tolerance mechanisms in Pichia kudriavzevii, as well as to develop methodologies for the control and optimisation of the baijiu brewing process.
2 Materials and methods
2.1 Screening of heat tolerant yeast strain
We isolated some heat tolerant strains from daqu of strong flavor baijiu, and the daqu sample was obtained from a baijiu distillery in Yibin, China. Subsequently, the most heat tolerant strain was selected and identified as a strain of Pichia kudriavzevii. The isolate has been registered with the China General Microbiological Culture Collection Center (CGMCC), and the accession number (CGMCC No. 33254). To evaluate the heat tolerance potential of the strain, a YPD liquid medium (Sangon Biotech, Shanghai, China) was prepared. The strain growth temperature was adjusted by setting different incubator temperatures, including 37°C, 40°C, 45°C, and 50°C. The 37°C was set as the control. Then, the samples were inoculated in a 48-well plate at 5% (v/v) inoculum, and the well plates were placed in a MicroScreen-HT real-time microbial growth analysis system (Gering, Tianjin, China). The plate was incubated at 180 rpm with shaking, and the OD600 was measured.
2.2 Observation on morphological changes of heat tolerant yeast
The methods are based on the previous report with some modifications (Wang et al., 2024). The strains were incubated at various temperatures (37°C, 45°C, and 50°C), with 37°C serving as the control group. Following a 48-h incubation period, the samples were washed 3 times with 0.1 M PBS (pH 7.0), fixed with 5.4% (v/v) glutaraldehyde at 4°C for 24 h. The fixative was discarded and the sample was rinsed with 0.1 M PBS (pH 7.0) for 3 times (15 min each time). Subsequently, the samples were dehydrated with different concentrations of ethanol (50, 60, 70, 80, 90, and 100%) for 15–20 min. The dehydrated samples were transferred to a mixture of ethanol and isoamyl acetate (v:v = 1:1) for about 30 min, then to pure isoamyl acetate for about 1 h. Finally, the samples were dried at critical point with liquid CO2, coated with palladium and observed under scanning electron microscopy (SEM; VEGA3 TESCAN, Tescan, Brno, Czech Republic).
2.3 RNA samples preparation, extraction, and sequencing
The strain was inoculated in YPD liquid medium (50 mL). Growth temperatures were adjusted to 37°C (C group) and 45°C (HT group). Then, the samples were inoculated at 5% (v/v) and shaken at 180 rpm. As indicated by the growth curves, the samples were collected from each group at the midpoint of the logarithmic growth period. The samples were then subjected to a centrifugal process at 3,000 rpm for 3 min, after which the resultant supernantants were discarded. The samples were then immediately placed into liquid nitrogen for a period of 10 min, after which they were stored at a temperature of −80°C.
Samples from the high temperature group (HT) and the control group (C) were collected for RNA extraction and three replicates were set up for each group. Total RNA was extracted using a Trizol kit (Invitrogen, Carlsbad, CA, United States). RNA quality and quantity were then determined, libraries were constructed and sequenced using the Illumina Novaseq platform to obtain paired-end reads of 150 bp.
2.4 Transcriptomics analysis
The transcriptomics analysis part was conducted by Novogene Co. (Beijing, China). The DESeq2 R package (1.20.0) was used to compare the differential expression analyses between groups, and the clusterProfiler R package (3.8.1) was used to perform Gene Ontology (GO) enrichment analyses of the differentially expressed genes (DEGs) and to correct for gene length bias. The Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis of DEGs was performed using clusterProfiler R package (3.8.1). In order to address the issue of the false positive rate resulting from multiple comparisons, the Benjamini–Hochberg method was employed to recalibrate the p-value and obtain p-adj, thereby ensuring the control of the false discovery rate (FDR). The screening of DEGs was conducted based on the absolute values of |log2 (FoldChange)| >1 and p-adj <0.05. DEGs were undertaken for GO enrichment and KEGG pathway enrichment, with the screening condition of p-adj <0.05.
2.5 Untargeted metabolomics analysis of heat tolerant yeast
Untargeted metabolomics analysis was performed using liquid chromatograph-tandem mass spectrometer (LC-MS/MS). Metabolomics analyses were performed on a Vanquish UHPLC (Thermo Fisher, Germany) equipped with a Hypersil Gold column (C18) (100 × 2.1 mm, 1.9 μm, Thermo Fisher, United States) coupled to Q Exactive™ HF/Q Exactive™ HF-X (Thermo Fisher, Germany) in Novogene Co., Ltd. (Beijing, China). Mobile phase A was 0.1% FA in water, while mobile phase B was methanol. The flow rate was 0.2 mL/min, and the column temperature was 40°C. The gradient elution program was as follows: 0–1.5 min, 2% B; 1.5–3 min, 2 to 85% B; 3–10 min, 85 to 100% B; 10–10.1 min, 100 to 2% B; 10.1–12 min, 2% B. Q ExactiveTM HF mass spectrometer was operated in positive/negative polarity mode with spray voltage of 3.5 kV, capillary temperature of 320°C, sheath gas flow rate of 35 psi and aux gas flow rate of 10 L/min, S-lens RF level of 60, Aux gas heater temperature of 350°C. Peak detection, molecular formula fitting, and peak area extraction were performed on the raw data using Compound Discoverer 3.3 (CD 3.3, Thermo Fisher). Principal component analysis (PCA) and partial least squares discriminant analysis (PLS-DA) were performed using the metaX (a flexible and comprehensive software for processing metabolomics data). The differential metabolites were screened according to the criteria of VIP >1.0, FC >1.5 or FC <0.667, and p-value <0.05. The differential metabolites were undertaken for KEGG pathway enrichment, with the screening condition of p-value <0.05.
2.6 Statistical analysis
The analysis of the data was conducted using GraphPad Prism 9.0 (San Diego, CA, United States), and the resulting data sets were visualised through the creation of graphs. In consideration of the divergent requirements of sequencing platforms for the screening of differential genes and differential metabolites, it was determined that p-adj would serve as a metric of significant difference in transcriptomics, and p-value in metabolomics. The specific screening criteria are shown above.
3 Results
3.1 Analysis of high temperature resistance of heat tolerant yeast
The strains were incubated at different temperatures, including 37°C (control), 40°C, 45°C, and 50°C. It was found that the strains proliferated rapidly during the logarithmic growth period at 37°C and 40°C. The results showed that varying incubation temperatures exerted a substantial influence on the OD600 of the strain (Figure 1). As the temperature continued to increase, the strain was still able to grow, although the growth rate and state decreased, indicating that the strain was able to tolerate the high temperature of 50°C, but the growth state of the strain at 45°C was better than that at 50°C. In order to successfully carry out the subsequent molecular mechanism investigation, we chose 45°C as the high temperature treatment condition.
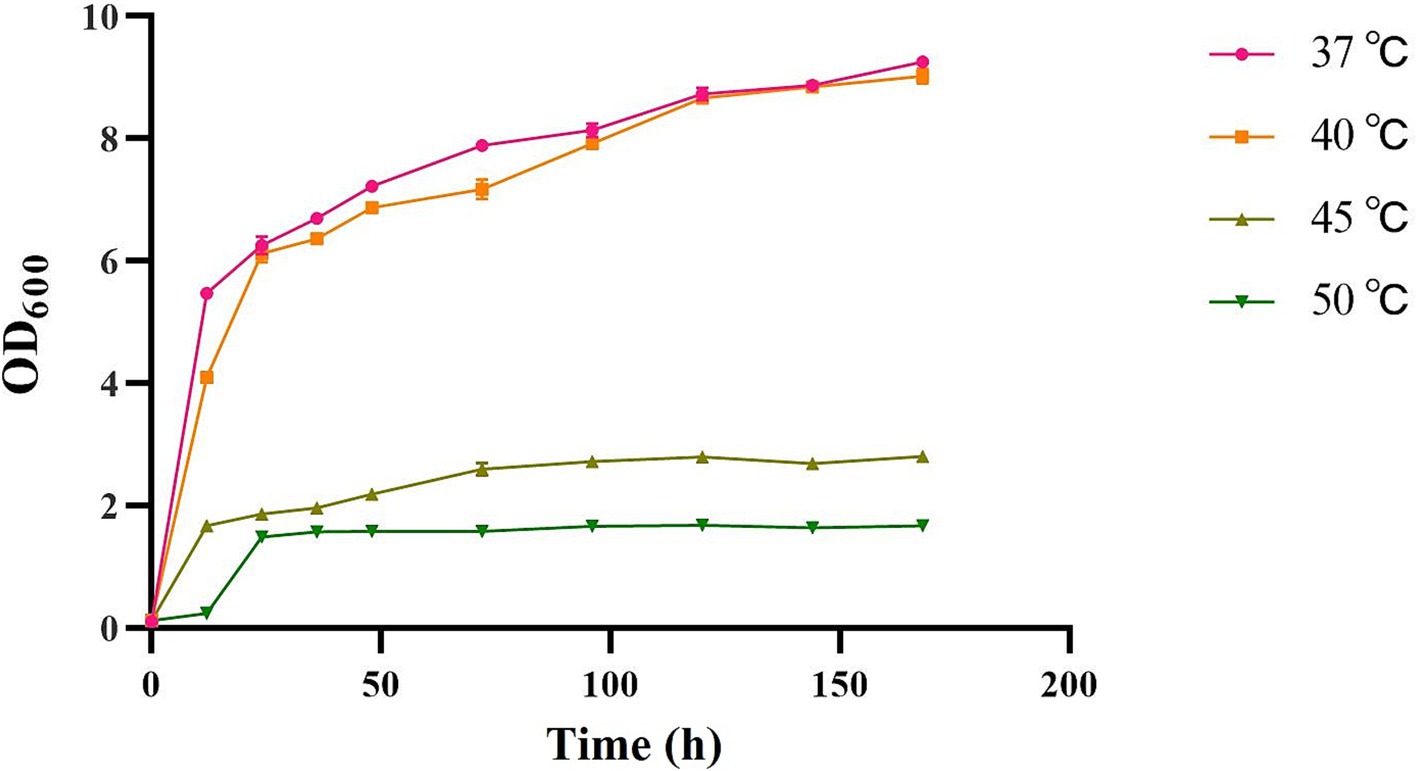
Figure 1. The OD600 of heat tolerant yeasts at different incubation temperatures (37°C, 40°C, 45°C, and 50°C).
3.2 Morphological and structural changes of heat tolerant yeast
Changes in the morphology and structure of the strains cultivated at varying temperatures for 48 h were observed using scanning electron microscopy (Figures 2A–C). The results showed that sample A (37°C, control group), the body was fuller; sample B (45°C) compared to A, the morphology changed significantly, the body became significantly longer, in the form of a long rod; sample C (50°C) compared to A, the body became significantly smaller, in the form of round ball.
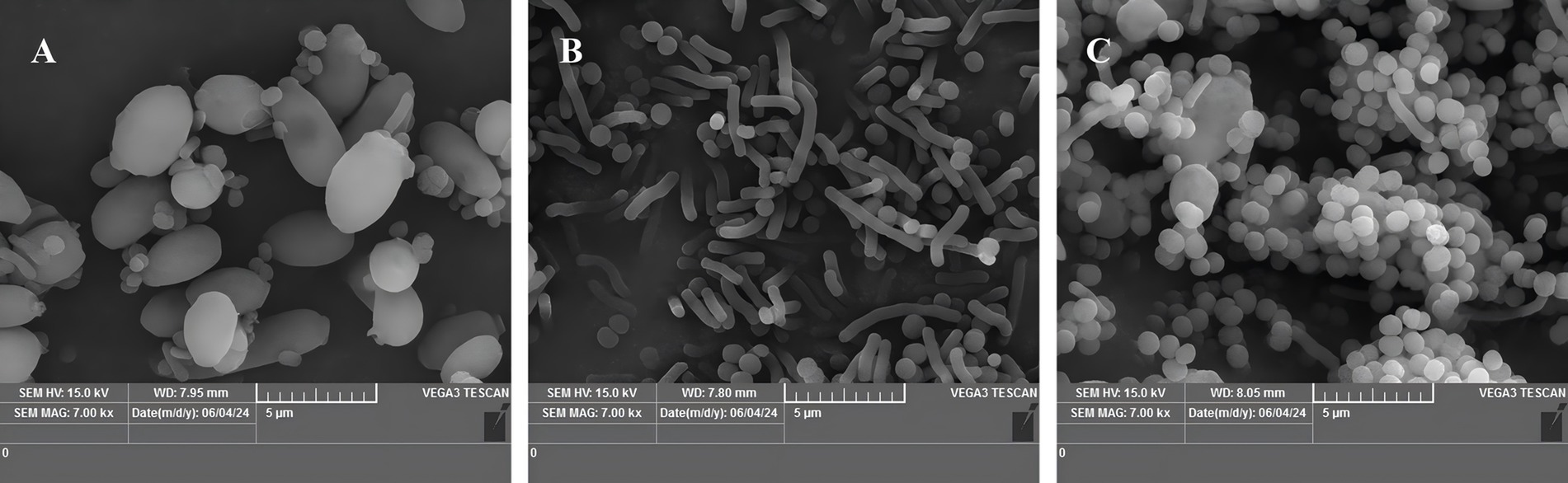
Figure 2. Scanning electron micrographs of strains at different incubation temperatures (7.00 k×). (A) Incubation at 37°C (control), (B) incubation at 45°C, (C) incubation at 50°C.
3.3 Transcriptomics analysis of heat tolerant yeast under high temperature stress
According to the results of the previous analysis, the strain exhibits better growth at a high temperature of 45°C. Therefore, this temperature was selected as the high temperature culture conditions of the strain. In total, 280,839,674 raw reads were generated, and 276,634,950 clean reads were obtained after quality control filtering. Principal component analysis (PCA) is performed on the gene expression values (FPKM) of all samples in order to assess the differences between groups and the reproducibility of samples within groups. The results show that the samples are more dispersed between the C and HT groups, but more concentrated within the same group, suggesting a significant difference in FPKM between the two groups (Figure 3A). In addition, the combined change in the first and second principal components was 97.5% (PC1: 67.61%, PC2: 29.89%), indicating that there were differences between the C and HT groups. Compared with group C, a total of 1716 DEGs were identified under 45°C high temperature stress (HT group), of which 732 genes were down regulated and 984 genes were up regulated (Figure 3B). In addition, 99 and 94 genes were unique to the C and HT groups, respectively, while 4,835 genes were shared (Figure 3C).
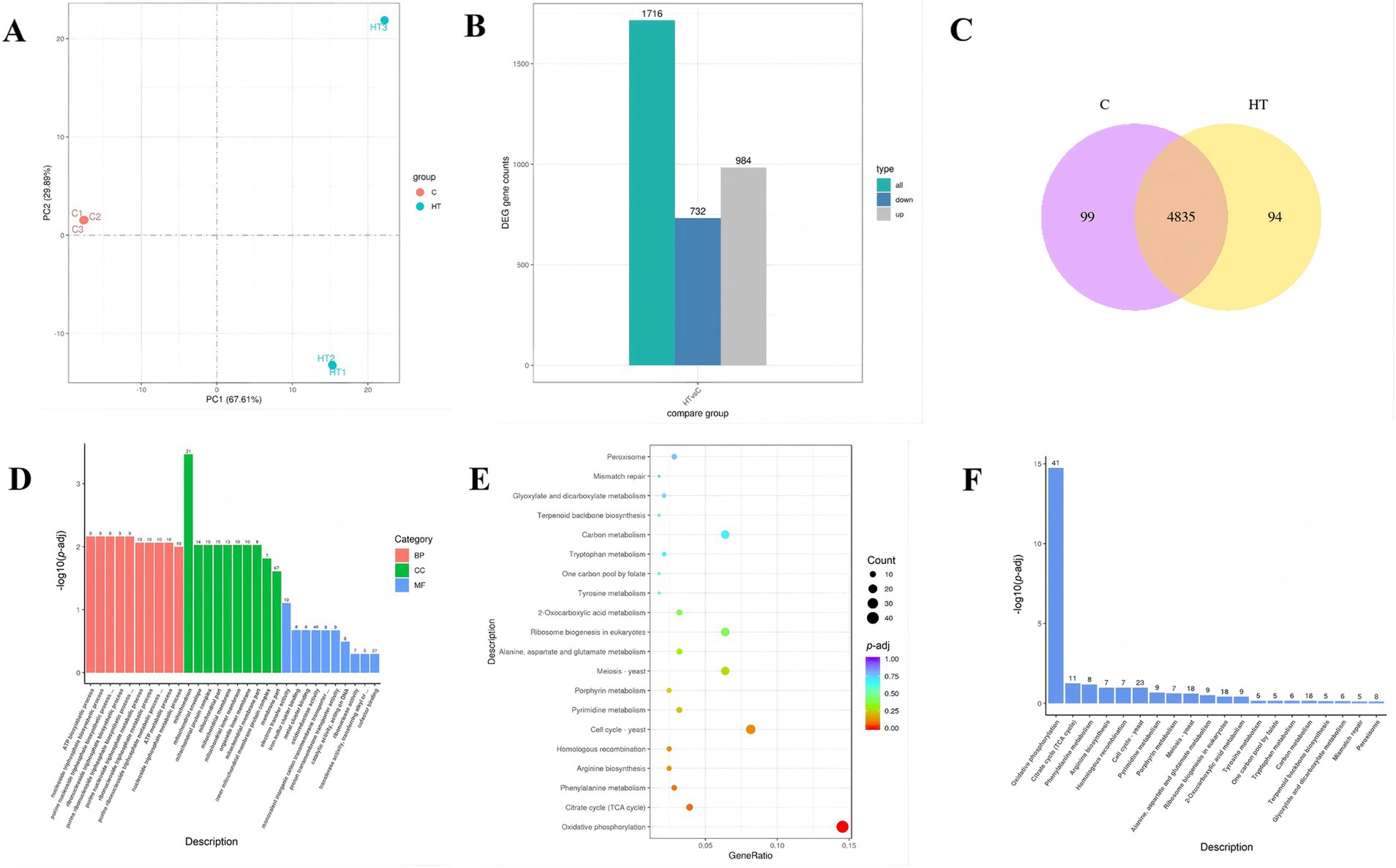
Figure 3. Transcriptomics analysis of heat tolerant yeast under high temperature stress (HT vs. C). C: control group (37°C), HT: high temperature group (45°C). (A) Principal component analysis (PCA) results. (B) Statistical histogram of the number of differential genes (DEG) in the differential comparison. (C) Co-expression Venn diagram. (D) Histogram of the GO up regulated enrichment analysis. The horizontal coordinate in the graph is the GO term and the vertical coordinate is the significance level of the GO term enrichment, expressed as −log10 (p-adj). −log10 (p-adj) represents the negative log10 transformation of the p-adj, highlighting the statistical significance of differential expression. Higher values indicate greater significance. The numbers on top of the bars denote the count of differentially expressed genes enriched in each GO term. (E) Scatter plot of the KEGG up regulated enrichment. The horizontal coordinate of the graph is defined as the ratio of the number of DEGs annotated to the KEGG pathway to the total number, and the vertical coordinate is represented by the KEGG pathway. (F) Bar graph of the KEGG up regulated enrichment analysis. The horizontal coordinate of the graph represents the KEGG pathway, while the vertical coordinate denotes the significance level of pathway enrichment. The numbers on top of the bars denote the count of differentially expressed genes enriched in each KEGG pathway.
DEGs were analysed for Gene Ontology (GO) enrichment. The results of the HT vs. C showed that the DEGs significantly up-regulated in biological processes (BP) were involved in ATP biosynthetic process (GO:0006754), nucleoside triphosphate biosynthetic process (GO:0009142), and purine nucleoside triphosphate biosynthetic process (GO:0009145), whereas DEGs significantly enriched in cellular component processes (CC) were involved in mitochondrion (GO:0005739), mitochondrial envelope (GO:0005740), and mitochondrial protein complex (GO:0098798) (Figure 3D and Supplementary Table S1). Consequently, the strain can enhance tolerance to high temperature by increasing the synthesis of energetic substances such as ATP.
As demonstrated in Figures 3E,F, KEGG pathway enrichment analysis showed that the pathways that were up regulated in the HT vs. C group predominantly associated with oxidative phosphorylation (ppa00190) and citrate cycle (TCA cycle) (ppa00020). Among these, the oxidative phosphorylation pathway exhibited a significant up regulation. This result lends further credence to the hypothesis that the thermotolerance mechanism of this strain is predominantly associated with the process of energy substance (e.g., ATP) production.
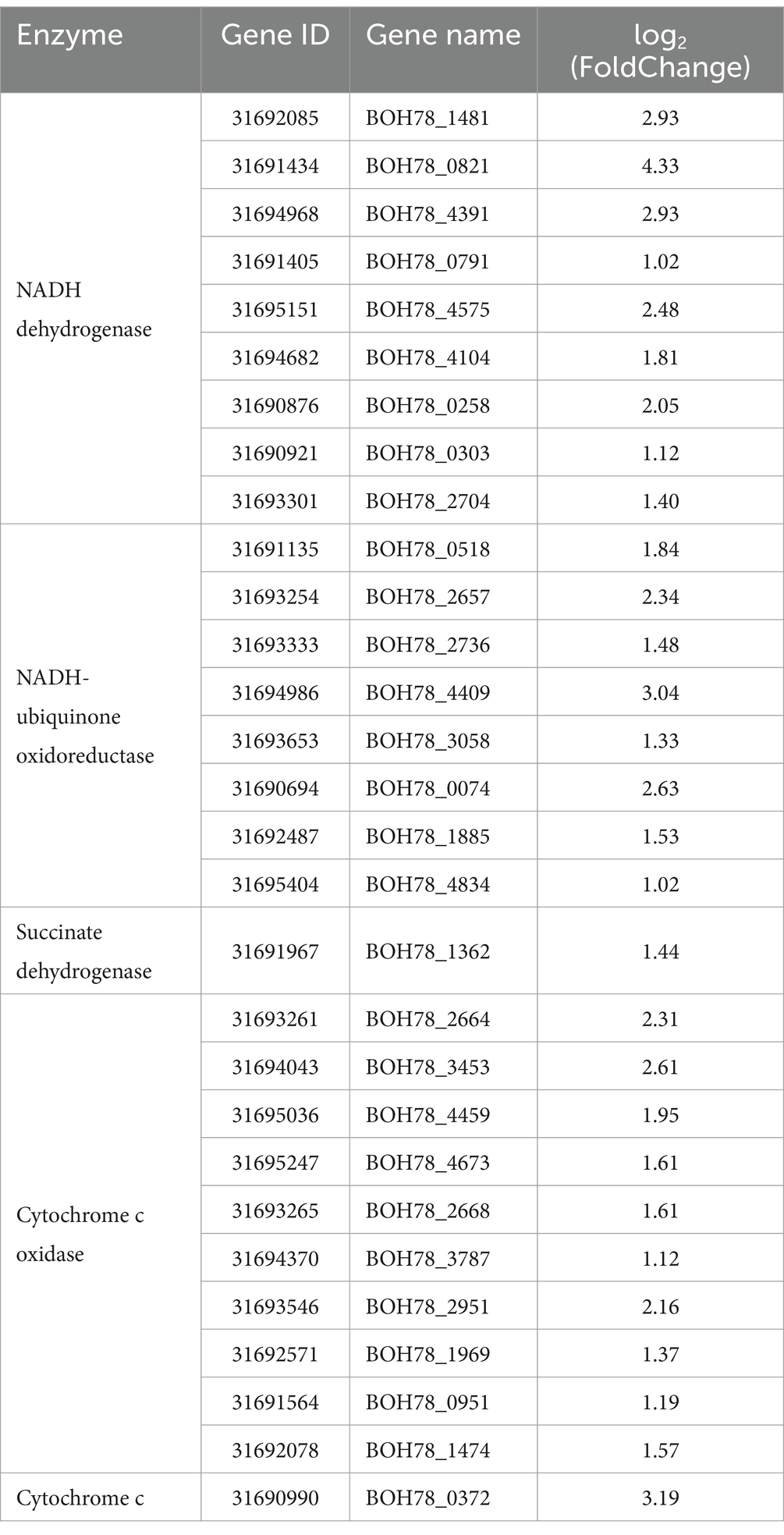
A detailed investigation into the DEGs associated with oxidative phosphorylation revealed that genes related to NADH dehydrogenase, complex I (NADH-ubiquinone oxidoreductase), complex II (succinate dehydrogenase), complex IV (cytochrome c oxidase), and cytochrome c were significantly up-regulated in the HT group (Table 1). In conditions of high temperature, the heat tolerant yeast predominantly regulates the oxidative phosphorylation pathway through the up regulation of the gene expression levels of key enzymes.
3.4 Metabolomics analysis of heat tolerant yeast under high temperature stress
In order to identify the differential metabolites, the partial least squares discrimination analysis (PLS-DA) was utilised to compare the metabolite changes in this heat tolerant yeast following exposure to high temperature stress. The results of the PLS-DA analysis show that the two sample groups are more dispersed between groups, while clustering is called within groups. And both R2Y and Q2Y are equal to 1, which shows the reliability of the model. A significant separation between the samples from the two groups was observed in the horizontal direction, indicating a substantial difference in their metabolite compositions (Figures 4A,B). The R2 (0.93) and Q2 (−0.68) values for the positive polarity mode, and the R2 (0.91) and Q2 (−0.69) values for the negative polarity mode. It has been determined that the R2 data exceeds the Q2 data in both detection modes, and the intercept between the Q2 regression line and the Y-axis is less than 0. This indicates that there is no “overfitting” of the model, and the model is stable and feasible (Figures 4C,D).
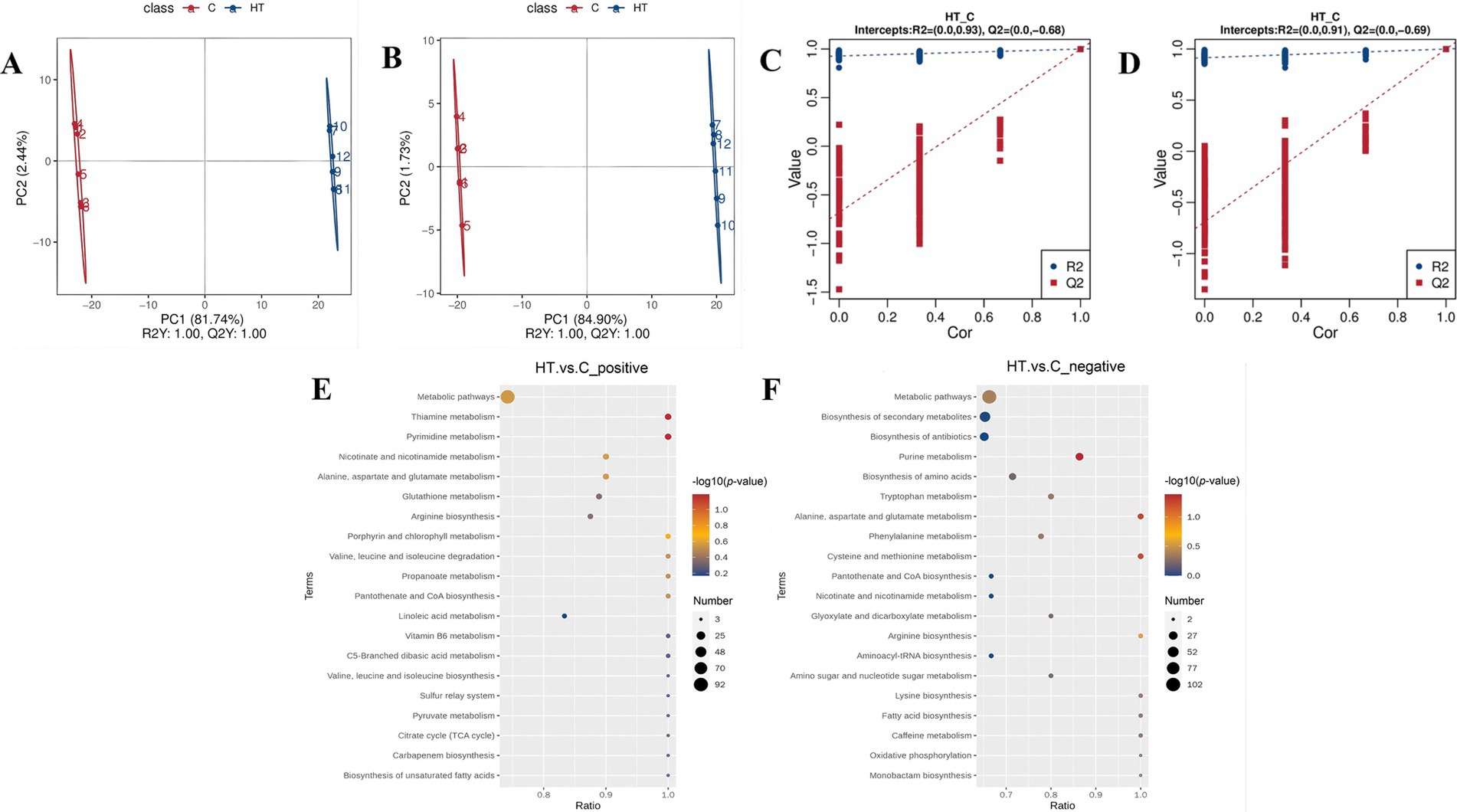
Figure 4. Metabolomics analysis of heat tolerant yeast under high temperature stress (HT vs. C). C: control group (37°C), HT: high temperature group (45°C). (A) PLS-DA score scatterplot_positive. (B) PLS-DA score scatterplot_negative. (C) PLS-DA ranked validation plot_positive. (D) PLS-DA ranked validation plot_negative. (E) Bubble plot of KEGG pathway enrichment analysis (HT vs. C_positive). (F) Bubble plot of KEGG pathway enrichment analysis (HT vs. C_negative). In the PLS-DA score scatterplot (A,B), the horizontal coordinates represent the scores of the samples on the first principal component, while the vertical coordinates denote the scores of the samples on the second principal component. R2Y signifies the explanatory rate of the model, and Q2Y is employed to evaluate the predictive ability of the PLS-DA model. When the values of R2Y and Q2Y are close to 1, it indicates that the model is more stable and reliable. In the PLS-DA ranked validation plot (C,D), the horizontal coordinate represents the correlation between the sample labeling information Y of the random grouping and the sample labeling information Y of the original grouping, and the vertical coordinate represents the scores of R2 and Q2. When the R2 data is greater than the Q2 data and the intercept of the Q2 regression line with the Y-axis is less than 0, it can be shown that the model is not “overfitted,” which means the model is feasible.
The results demonstrated that there were 463 cationic metabolites with significant differences, of which 214 metabolites were up regulated and 249 metabolites were down regulated, and there were 352 anionic metabolites with significant differences, of which 116 metabolites were up regulated and 236 metabolites were down regulated. Among the metabolites that were found to be significantly up-regulated were those associated with heat tolerance, including nicotinamide adenine dinucleotide (NAD+), adenosine diphosphate (ADP), betaine, and adenosine 3′5′-cyclic monophosphate (Supplementary Table S2). Of particular note are NAD+ and ADP, which are closely related to oxidative phosphorylation and energy metabolism, thus suggesting a possible regulatory pathway for this heat tolerant yeast confronting high temperature stress.
The annotation of differently expressed cationic metabolites revealed their association with 44 metabolic pathways, including pyrimidine metabolism and thiamine metabolism (Figure 4E). In the case of differentially expressed anionic metabolites, their annotation pointed to 45 metabolic pathways, encompassing purine metabolism, cysteine, and methionine metabolism, alanine, aspartate and glutamate metabolism (Figure 4F). Of particular note was the significant enrichment of purine metabolism (p-value <0.05).
3.5 Transcriptomics-metabolomics association analysis of heat tolerant yeast under high temperature stress
In order to provide a more accurate reflection of the relationship between genes and metabolites in this heat tolerant yeast, based on the results of the analyses above, the focus here was on the role of energy metabolism in the regulation of heat tolerance in the strain. Nicotinamide adenine dinucleotide (NAD+) and adenosine diphosphate (ADP) were consequently selected as the focal metabolites, and correlation analyses were conducted with oxidative phosphorylation-related DEGs in Table 1. A transcriptomics-metabolomics correlation network was then constructed (Figure 5). The DEGs associated with NADH dehydrogenase, NADH-ubiquinone oxidoreductase, cytochrome c oxidase, and cytochrome c in the oxidative phosphorylation pathway demonstrated a significant positive correlation with NAD+ and ADP. Collectively, these differential metabolites, associated with ATP production and energy metabolism in the metabolomics, exhibited a positive correlation with oxidative phosphorylated DEGs in the transcriptomics. This finding further supports the hypothesis that this heat tolerant yeast possesses the capacity to counteract high temperature by up regulating energy metabolism (especially oxidative phosphorylation). This process can constitute a pivotal mechanism through which the strain exhibits heat tolerance.
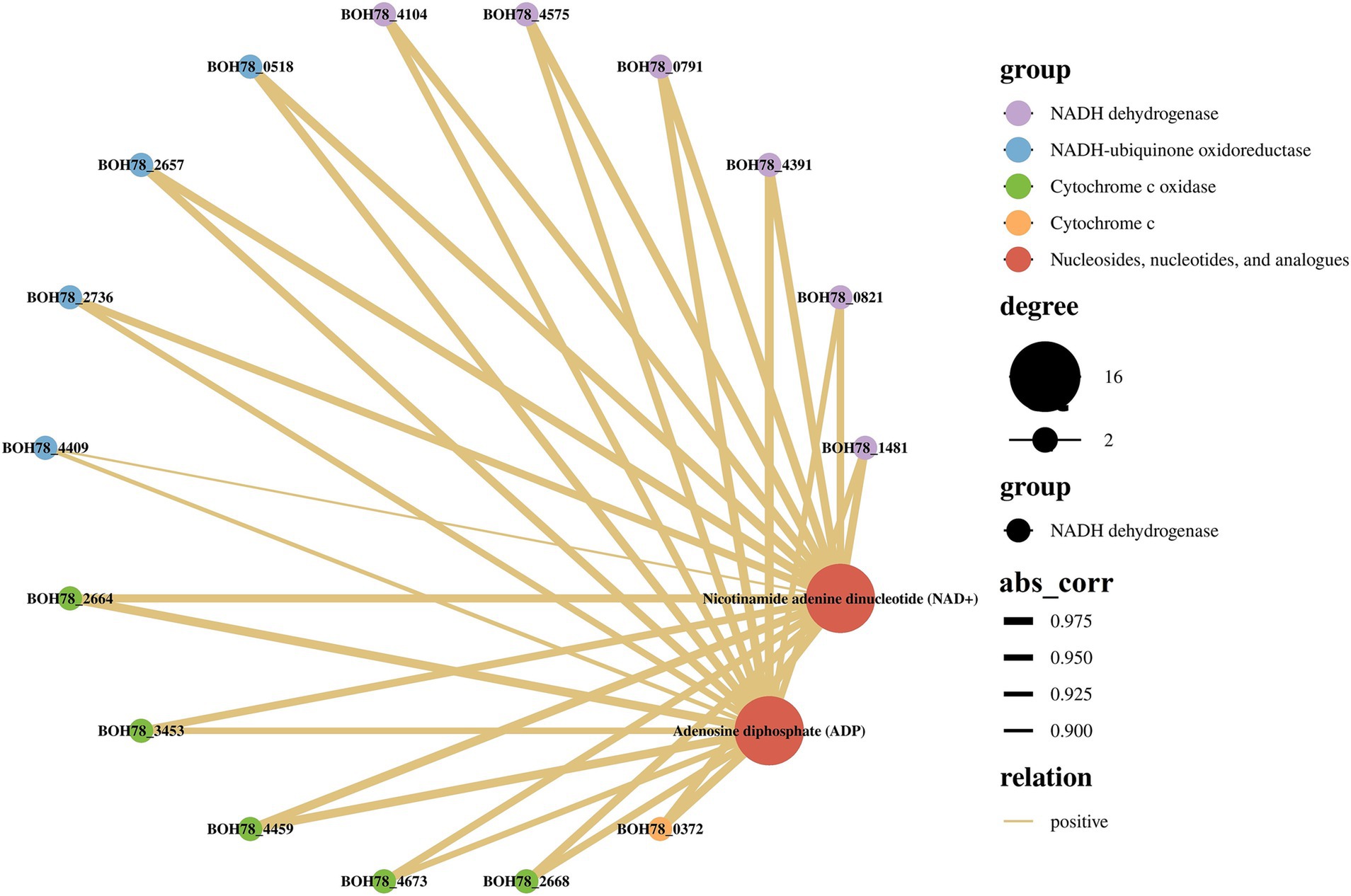
Figure 5. Transcriptomics-metabolomics association analysis in oxidative phosphorylation and energy metabolism (HT vs. C). C: control group (37°C), HT: high temperature group (45°C). The abs_corr refers to the absolute value of the correlation coefficient, which is a quantitative measure of the strength of the relationship between gene expression and metabolite levels. A higher abs_corr value (closer to 1) indicates a stronger correlation, while a lower value (closer to 0) suggests a weaker relationship. The larger the abs_corr coefficient, the thicker the line.
4 Discussion
The process of baijiu brewing frequently occurs in conditions characterised by high temperature, high acidity, and significant ethanol content. It is widely acknowledged that temperature is a pivotal factor in the fermentation process (Liu et al., 2023). The impact of fermentation temperature extends beyond the categorisation and quality of baijiu, also influencing the composition of the microbial community within the brewing process (Tu et al., 2022; Zhang et al., 2024). Pichia kudriavzevii plays a pivotal role in the brewing process of baijiu and is a dominant functional yeast in the fermentation process of typical baijiu (Chu et al., 2023; Li et al., 2013). Not only does it contribute to the establishment of the baijiu fermentation microbial community by influencing its dynamics during the fermentation process (Zhang et al., 2021), but it also enhances the ester content of baijiu and improves its flavor quality (Li et al., 2022). In addition, Pichia kudriavzevii was observed to be highly heat resistant, with existing studies demonstrating that temperatures ranging from 37 to 42°C significantly stimulated its growth during fermentation of sauce flavor baijiu (Zhang et al., 2021), and the yeast rapidly dominated the fermentation process at these temperatures. Furthermore, it was determined that Pichia kudriavzevii MBY1358 exhibited the capacity to withstand temperature of 44°C (Choi et al., 2017).
In this paper, a Pichia kudriavzevii strain that exhibited excellent heat tolerance was isolated from a strong flavor baijiu macroquat. The strain was cultivated at varying temperatures in order to ascertain its growth activity and to observe its morphological characteristics. In order to explore the molecular mechanism of heat resistance, transcriptomics, and metabolomics were employed. The findings of the present study demonstrated that the strain exhibited a high degree of temperature tolerance, with the capacity to withstand temperatures of up to 50°C. Furthermore, the incubation of the strain at elevated temperatures resulted in a substantial alteration of its morphological characteristics. It has been demonstrated by several studies that the morphology of strains frequently undergoes alterations in order to cope with high temperature damage subsequent to exposure to high temperature (Shen and Chou, 2016; Xiao et al., 2022). A study reported that heat stress (42°C) led to the development of a circular structure in Saccharomyces cerevisiae that differed from bud scars, with a twofold increase in cell wall stiffness (Pillet et al., 2014), which is related to heat stress-induced thickening of the yeast cell wall and alteration of cell membrane fluidity and the development of cell membrane fluidity (Lin et al., 2022). Furthermore, elevated temperatures have been observed to induce substantial internal structural changes in yeast, including a reduction in the size of the inner core (Webster and Watson, 1993) and alterations in yeast morphology due to changes in organelle size (Keuenhof et al., 2021). These results provide further evidence to support the hypothesis that yeast possess the capacity to counteract deleterious effects of elevated temperatures by undergoing alterations to their morphological characteristics. It is therefore hypothesised that the examined this heat tolerant Pichia kudriavzevii can exhibit resistance to thermal damage caused by heat stress through alterations in their morphology.
The transcriptomics analysis of the strain RNA samples revealed that the KEGG pathway enrichment analysis indicated up regulation of oxidative phosphorylation (p-adj <0.05) and citrate cycle (TCA cycle). These results suggested that the strain could counteract high temperature stress by enhancing oxidative phosphorylation and increasing energy metabolism, which might be the heat tolerance mechanism of the strain. Oxidative phosphorylation is considered to be one of the key activities of the protein complexes in the inner membrane of mitochondria, which is also viewed as the primary pathway for ATP synthesis within the mitochondria and a major source of energy in cellular pathways (Cooper and Hausman, 2007; van der Bliek et al., 2017). It has been established that during oxidative phosphorylation, electrons from NADH and FADH2 bind to O2. The energy released from these oxidation/reduction reactions is used to drive the synthesis of ATP from ADP (Cooper and Hausman, 2007). In response to altered environmental conditions, fungi have been observed to modify their energy metabolism pathways, thereby acclimatising to the novel conditions (Marcet-Houben et al., 2009). A multi-omics analysis of the mechanism of high temperature tolerance in Kluyveromyces marxianus was conducted, and oxidative phosphorylation was identified as a significant enrichment pathway. These findings suggest that this yeast possesses the capacity to counteract high temperature damage by regulating the oxidative phosphorylation pathway (Kosaka et al., 2022; Li et al., 2019). In addition to yeast, a proteomic analysis of Aspergillus niger under 50°C treatment conditions revealed significant changes in various proteins within the oxidative phosphorylation KEGG pathway (Deng et al., 2020). In addition, the TCA cycle has been demonstrated to be associated with microbial heat tolerance, as well as oxidative phosphorylation. The TCA cycle represents a pivotal energy metabolic pathway, serving as the central pathway of cellular oxidative phosphorylation, which generates NADH and FADH2 (Anderson et al., 2018; Shih and Pan, 2011). A study was conducted to perform a transcriptomics analysis for the purposes of investigating the heat tolerance exhibited by Pichia kudriavzevii LC375240, and the results obtained revealed that there was a significant up regulation of genes belonging to the TCA cycle pathway in the strain that had been subjected to heat treatment (Qi et al., 2025).
In this paper, LC-MS/MS was utilised to analyse the metabolite changes in heat tolerant yeast following exposure to high temperature stress. The study identified a total of 463 differential metabolites in the positive polarity mode and 352 differential metabolites in the negative polarity mode. Among these, the differential metabolites related to heat tolerance were found to be significant, including a significant up regulation of nicotinamide adenine dinucleotide (NAD+) and adenosine diphosphate (ADP). These substances are closely related to oxidative phosphorylation and energy metabolism. NAD+ is a vital pyridine nucleotide cofactor that plays a pivotal role in various aspects of cellular respiration and energy production. It is employed in the synthesis of ATP and the maintenance of membrane potential in mitochondria (Massudi et al., 2012). NAD+ is a cofactor that plays an essential role in a variety of significant biochemical processes within cells, including oxidative phosphorylation, ATP production, DNA repair, calcium-dependent secondary messengers (Braidy et al., 2019; Gindri et al., 2024). In the context of aerobic eukaryotic cells, the predominant pathway for the production of the energy metabolite ATP is through mitochondrial oxidative phosphorylation. The mitochondrial ADP/ATP carrier plays a pivotal role in this process by facilitating the input of ADP into the cytoplasmic lysosome and the export of ATP from the mitochondrial matrix. This process represents a pivotal step in the broader mechanism of oxidative phosphorylation within the context of eukaryotic cells (Clémençon et al., 2013; Kunji et al., 2016). In addition to substances associated with energy metabolism, a variety of other substances have been linked to increased heat resistance, including betaine and adenosine 3′5′-cyclic monophosphate. Betaine is a compound that is found in a wide variety of organisms, including bacteria, fungi, higher plants and animals. Microorganisms have been observed to utilise betaine as a stress protector against a range of environmental challenges, such as temperature stress, drought and other forms of stress (Mo et al., 2022). As a second messenger, cAMP has been demonstrated to exert a pivotal function in a broad spectrum of physiological responses in bacteria, fungi, and plants and animals (Liang et al., 2022), and can modulate the response of organisms to extreme environments such as heat stress and osmotic stress (Zhu et al., 2016).
Further KEGG pathway enrichment analysis of the differential metabolites revealed that the metabolites were enriched in purine metabolism (p-value <0.05), pyrimidine metabolism, and multiple amino acid metabolism (cysteine, methionine, alanine, aspartate, and glutamate, etc.) pathways. Purine metabolites, most notably purine nucleotides, function not only as building blocks for DNA and RNA, but also as vital sources of cellular energy (e.g., ATP) and cofactors that promote cell survival and proliferation (Tran et al., 2024). Furthermore, they play a crucial role in intracellular and intercellular signalling pathways (Gessner et al., 2023; Pedley and Benkovic, 2017; Wang L. et al., 2023). It has been demonstrated that the purine metabolism pathway in yeast exerts a dual regulatory effect on ATP levels and NAD+ synthesis. When purine metabolism is up regulated, there is an increase in intracellular ATP levels, and the levels of key metabolites in the NAD+ synthesis pathway are also elevated (Pinson et al., 2019). The extant research suggests a robust correlation between the aforementioned pathways and the heat tolerance of yeasts. Li et al. (2021) found that differential metabolites of Kluyveromyces marxianus were enriched in purine metabolism and pyrimidine metabolism under high temperature stress. Pan et al. (2019) performed metabolomics analysis of Saccharomyces cerevisiae after a high temperature incubation, and found that the differential metabolites were associated with a variety of amino acid metabolism (e.g., arginine), energy metabolism, and purine and pyrimidine metabolism.
In order to analyse and explore the effect of oxidative phosphorylation and energy metabolism on the heat tolerance of the strain, a correlation analysis was performed between transcriptomics and metabolomics. The results demonstrated that a variety of metabolites (NAD+ and ADP) linked to ATP production and metabolism in the metabolomics exhibited a positive correlation with oxidative phosphorylation DEGs in the transcriptomics. Overall, oxidative phosphorylation provides the cell with a substantial amount of ATP, while NAD+ and ADP are closely associated with oxidative phosphorylation and energy metabolism. Consequently, this suggests that the strain can withstand high temperature stress by up regulated energy metabolism (especially oxidative phosphorylation) and increasing ATP production.
5 Conclusion
In this study, we investigated a heat tolerant Pichia kudriavzevii strain isolated from strong flavor daqu of baijiu. The strain demonstrated considerable heat tolerance at temperatures up to 50°C, accompanied by significant morphological and structural alterations. To elucidate the molecular mechanisms underlying the heat resistance of this strain, transcriptomics and metabolomics analyses were employed. The results indicated that the strain primarily up-regulated energy metabolism, particularly oxidative phosphorylation, in order to cope with high temperature stress. Furthermore, metabolomics data demonstrated elevated levels of NAD+ and ADP, which are closely associated with energy metabolism. The correlation analysis between transcriptomics and metabolomics confirmed a strong positive relationship between these metabolites and differentially expressed genes (DEGs) involved in oxidative phosphorylation. This study provides a molecular basis for understanding the heat resistance mechanism in baijiu yeast and offers insights for optimizing production and processing control.
Data availability statement
The raw sequence data presented in this paper have been deposited in the Genome Sequence Archive in the National Genomics Data Center, China National Center for Bio information/Beijing Institute of Genomics, Chinese Academy of Sciences, under accession number CRA022684 and are publicly accessible at https://bigd.big.ac.cn/gsa (accessed on 1 February 2025).
Ethics statement
The manuscript presents research on animals that do not require ethical approval for their study.
Author contributions
NW: Conceptualization, Data curation, Resources, Writing – review & editing. LL: Formal analysis, Software, Visualization, Writing – original draft. YM: Formal analysis, Writing – original draft. CSh: Conceptualization, Writing – review & editing. ZA: Methodology, Writing – review & editing. CSo: Investigation, Writing – review & editing. MM: Validation, Writing – review & editing. PZ: Visualization, Writing – original draft. YL: Software, Writing – review & editing. XS: Methodology, Writing – review & editing. HZ: Funding acquisition, Project administration, Validation, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This work was supported by Luzhou Laojiao Postgraduate Innovation Fund (Grant No. LJCX2023-3), Luzhou Laojiao Postdoctoral Programme (Grant No. 2024HX08), 2024 Provincial Financial Rural Revitalization Transfer Payment Fund Park Construction Project, Yibin Agricultural Science and Technology Innovation Project (Grant No. H72313004).
Acknowledgments
The authors would like to thank Ms. Weiwei Li, Ms. Yuanlin Xiang, and Mr. Qiang Li for their timely assistance in sample data visualization and article conceptualization.
Conflict of interest
NW, CSh, ZA, and CSo were employed by Luzhou Laojiao Co., Ltd. YL and XS were employed by Sichuan Yibin Hengshengfu Liquor Industry Group Co., Ltd.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The authors declare that no Gen AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2025.1572004/full#supplementary-material
References
Amillastre, E., Aceves-Lara, C. A., Uribelarrea, J. L., Alfenore, S., and Guillouet, S. E. (2012). Dynamic model of temperature impact on cell viability and major product formation during fed-batch and continuous ethanolic fermentation in Saccharomyces cerevisiae. Bioresour. Technol. 117, 242–250. doi: 10.1016/j.biortech.2012.04.013
Anderson, N. M., Mucka, P., Kern, J. G., and Feng, H. (2018). The emerging role and targetability of the TCA cycle in cancer metabolism. Protein Cell 9, 216–237. doi: 10.1007/s13238-017-0451-1
Braidy, N., Berg, J., Clement, J., Khorshidi, F., Poljak, A., Jayasena, T., et al. (2019). Role of nicotinamide adenine dinucleotide and related precursors as therapeutic targets for age-related degenerative diseases: rationale, biochemistry, pharmacokinetics, and outcomes. Antioxid. Redox Signal. 30, 251–294. doi: 10.1089/ars.2017.7269
Choi, D., Park, E., and Kim, M. (2017). Isolation of thermotolerant yeast Pichia kudriavzevii from nuruk. Food Sci. Biotechnol. 26, 1357–1362. doi: 10.1007/s10068-017-0155-6
Chu, Y., Li, M., Jin, J., Dong, X., Xu, K., Jin, L., et al. (2023). Advances in the application of the non-conventional yeast Pichia kudriavzevii in food and biotechnology industries. J. Fungi 9:170. doi: 10.3390/jof9020170
Clémençon, B., Babot, M., and Trézéguet, V. (2013). The mitochondrial ADP/ATP carrier (SLC25 family): pathological implications of its dysfunction. Mol. Asp. Med. 34, 485–493. doi: 10.1016/j.mam.2012.05.006
Cooper, G. M., and Hausman, R. E. (2007). The cell: a molecular approach. 2nd Edn. Sunderland, MA: Sinauer Associates.
Deng, X., Du, B., Zhu, F., Gao, Y., and Li, J. (2020). Proteomic analysis of Aspergillus niger 3.316 under heat stress. Microbiology 9:e1012. doi: 10.1002/mbo3.1012
Gessner, P., Lum, J., and Frenguelli, B. G. (2023). The mammalian purine salvage pathway as an exploitable route for cerebral bioenergetic support after brain injury. Neuropharmacology 224:109370. doi: 10.1016/j.neuropharm.2022.109370
Gindri, I. M., Ferrari, G., Pinto, L. P. S., Bicca, J., Dos Santos, I. K., Dallacosta, D., et al. (2024). Evaluation of safety and effectiveness of NAD in different clinical conditions: a systematic review. Am. J. Physiol. Endocrinol. Metab. 326, E417–E427. doi: 10.1152/ajpendo.00242.2023
Gou, M., Wang, H., Yuan, H., Zhang, W., Tang, Y., and Kida, K. (2015). Characterization of the microbial community in three types of fermentation starters used for Chinese liquor production. J. Inst. Brew. 121, 620–627. doi: 10.1002/jib.272
Hu, Y., Yang, Q., Chen, D., Fu, B., Zhang, Y., Zhang, Y., et al. (2021). Study on microbial communities and higher alcohol formations in the fermentation of Chinese Xiaoqu Baijiu produced by traditional and new mechanical technologies. Food Res. Int. 140:109876. doi: 10.1016/j.foodres.2020.109876
Kang, Q., Sun, J., Wang, B., and Sun, B. (2023). Wine, beer and Chinese Baijiu in relation to cardiovascular health: the impact of moderate drinking. Food Sci. Human Wellness 12, 1–13. doi: 10.1016/j.fshw.2022.07.013
Keuenhof, K. S., Larsson Berglund, L., Malmgren Hill, S., Schneider, K. L., Widlund, P. O., Nyström, T., et al. (2021). Large organellar changes occur during mild heat shock in yeast. J. Cell Sci. 135:jcs258325. doi: 10.1242/jcs.258325
Kosaka, T., Tsuzuno, T., Nishida, S., Pattanakittivorakul, S., Murata, M., Miyakawa, I., et al. (2022). Distinct metabolic flow in response to temperature in thermotolerant Kluyveromyces marxianus. Appl. Environ. Microbiol. 88:e0200621. doi: 10.1128/aem.02006-21
Kunji, E. R., Aleksandrova, A., King, M. S., Majd, H., Ashton, V. L., Cerson, E., et al. (2016). The transport mechanism of the mitochondrial ADP/ATP carrier. Biochim. Biophys. Acta 1863, 2379–2393. doi: 10.1016/j.bbamcr.2016.03.015
Li, P., Fu, X., Chen, M., Zhang, L., and Li, S. (2019). Proteomic profiling and integrated analysis with transcriptomic data bring new insights in the stress responses of Kluyveromyces marxianus after an arrest during high-temperature ethanol fermentation. Biotechnol. Biofuels 12:49. doi: 10.1186/s13068-019-1390-2
Li, H., Liu, S., Liu, Y., Hui, M., and Pan, C. (2023). Functional microorganisms in Baijiu Daqu: research progress and fortification strategy for application. Front. Microbiol. 14:1119675. doi: 10.3389/fmicb.2023.1119675
Li, X., Ma, E., Yan, L., Meng, H., Du, X., and Quan, Z. (2013). Bacterial and fungal diversity in the starter production process of Fen liquor, a traditional Chinese liquor. J. Microbiol. 51, 430–438. doi: 10.1007/s12275-013-2640-9
Li, P., Tan, X., Fu, X., Dang, Y., and Li, S. (2021). Metabolomic analysis reveals Kluyveromyces marxianus’s stress responses during high-temperature ethanol fermentation. Process Biochem. 102, 386–392. doi: 10.1016/j.procbio.2021.01.024
Li, R., Xu, M., Zheng, J., Liu, Y., Sun, C., Wang, H., et al. (2022). Application potential of baijiu non-Saccharomyces yeast in winemaking through sequential fermentation with Saccharomyces cerevisiae. Front. Microbiol. 13:902597. doi: 10.3389/fmicb.2022.902597
Liang, S., Sun, J., Luo, Y., Lv, S., Chen, J., Liu, Y., et al. (2022). cAMP is a promising regulatory molecule for plant adaptation to heat stress. Life 12:885. doi: 10.3390/life12060885
Lin, N., Xu, Y., and Yu, X. (2022). Overview of yeast environmental stress response pathways and the development of tolerant yeasts. Syst. Microbiol. Biomanuf. 2, 232–245. doi: 10.1007/s43393-021-00058-4
Liu, X., Ma, D., Yang, C., Yin, Q., Liu, S., Shen, C., et al. (2023). Microbial community succession patterns and drivers of Luxiang-flavor Jiupei during long fermentation. Front. Microbiol. 14:1109719. doi: 10.3389/fmicb.2023.1109719
Liu, H., and Sun, B. (2018). Effect of fermentation processing on the flavor of baijiu. J. Agric. Food Chem. 66, 5425–5432. doi: 10.1021/acs.jafc.8b00692
Marcet-Houben, M., Marceddu, G., and Gabaldón, T. (2009). Phylogenomics of the oxidative phosphorylation in fungi reveals extensive gene duplication followed by functional divergence. BMC Evol. Biol. 9:295. doi: 10.1186/1471-2148-9-295
Massudi, H., Grant, R., Guillemin, G. J., and Braidy, N. (2012). NAD+ metabolism and oxidative stress: the golden nucleotide on a crown of thorns. Redox Rep. 17, 28–46. doi: 10.1179/1351000212y.0000000001
Mejía-Barajas, J. A., Montoya-Pérez, R., Salgado-Garciglia, R., Aguilera-Aguirre, L., Cortés-Rojo, C., Mejía-Zepeda, R., et al. (2017). Oxidative stress and antioxidant response in a thermotolerant yeast. Braz. J. Microbiol. 48, 326–332. doi: 10.1016/j.bjm.2016.11.005
Mo, X., Qian, J., Liu, P., Zeng, H., Chen, G., and Wang, Y. (2022). Exogenous betaine enhances the protrusion vigor of rice seeds under heat stress by regulating plant hormone signal transduction and its interaction network. Antioxidants 11:1792. doi: 10.3390/antiox11091792
Pan, D., Wiedemann, N., and Kammerer, B. (2019). Heat stress-induced metabolic remodeling in Saccharomyces cerevisiae. Metabolites 9:266. doi: 10.3390/metabo9110266
Pedley, A. M., and Benkovic, S. J. (2017). A new view into the regulation of purine metabolism: the purinosome. Trends Biochem. Sci. 42, 141–154. doi: 10.1016/j.tibs.2016.09.009
Pillet, F., Lemonier, S., Schiavone, M., Formosa, C., Martin-Yken, H., Francois, J. M., et al. (2014). Uncovering by atomic force microscopy of an original circular structure at the yeast cell surface in response to heat shock. BMC Biol. 12:6. doi: 10.1186/1741-7007-12-6
Pinson, B., Ceschin, J., Saint-Marc, C., and Daignan-Fornier, B. (2019). Dual control of NAD+ synthesis by purine metabolites in yeast. eLife 8:e43808. doi: 10.7554/eLife.43808
Qi, Y., Qin, Q., Ma, J., Wang, B., Jin, C., and Fang, W. (2025). Unveiling the thermotolerance mechanism of Pichia kudriavzevii LC375240 through transcriptomic and genetic analyses. BMC Biol. 23:55. doi: 10.1186/s12915-025-02159-1
Richter, K., Haslbeck, M., and Buchner, J. (2010). The heat shock response: life on the verge of death. Mol. Cell 40, 253–266. doi: 10.1016/j.molcel.2010.10.006
Shen, J., and Chou, C. (2016). Morphological plasticity of bacteria-open questions. Biomicrofluidics 10:031501. doi: 10.1063/1.4953660
Shih, T. W., and Pan, T. M. (2011). Stress responses of thermophilic Geobacillus sp. NTU 03 caused by heat and heat-induced stress. Microbiol. Res. 166, 346–359. doi: 10.1016/j.micres.2010.08.001
Tran, D. H., Kim, D., Kesavan, R., Brown, H., Dey, T., Soflaee, M. H., et al. (2024). De novo and salvage purine synthesis pathways across tissues and tumors. Cell 187, 3602–3618.e20. doi: 10.1016/j.cell.2024.05.011
Tu, W., Cao, X., Cheng, J., Li, L., Zhang, T., Wu, Q., et al. (2022). Chinese baijiu: the perfect works of microorganisms. Front. Microbiol. 13:919044. doi: 10.3389/fmicb.2022.919044
van der Bliek, A. M., Sedensky, M. M., and Morgan, P. G. (2017). Cell biology of the mitochondrion. Genetics 207, 843–871. doi: 10.1534/genetics.117.300262
Wang, D., Chen, L., Yang, F., Wang, H., and Wang, L. (2019). Yeasts and their importance to the flavour of traditional Chinese liquor: a review. J. Inst. Brew. 125, 214–221. doi: 10.1002/jib.552
Wang, L., Jiang, S., Li, D., and Sun, C. (2024). Combined transcriptomics and metabolomics analysis reveals the antifungal activity and mechanism of Enterococcus faecium cell-free supernatant against Alternaria alternata. Food Biosci. 57:103442. doi: 10.1016/j.fbio.2023.103442
Wang, L., Qiao, L., Li, A., Chen, L., He, B., Liu, G., et al. (2023). Integrative multiomics analysis of the heat stress response of Enterococcus faecium. Biomol. Ther. 13:437. doi: 10.3390/biom13030437
Wang, C., Shi, D., and Gong, G. (2008). Microorganisms in Daqu: a starter culture of Chinese Maotai-flavor liquor. World J. Microbiol. Biotechnol. 24, 2183–2190. doi: 10.1007/s11274-008-9728-0
Wang, N., Zhang, P., Zhou, X., Zheng, J., Ma, Y., Liu, C., et al. (2023). Isolation, identification, and characterization of an acid-tolerant Pichia kudriavzevii and exploration of its acetic acid tolerance mechanism. Fermentation 9:540. doi: 10.3390/fermentation9060540
Webster, D. L., and Watson, K. (1993). Ultrastructural changes in yeast following heat shock and recovery. Yeast 9, 1165–1175. doi: 10.1002/yea.320091103
Xiao, W., Zhang, J., Huang, J., Xin, C., Li, M. J., and Song, Z. (2022). Response and regulatory mechanisms of heat resistance in pathogenic fungi. Appl. Microbiol. Biotechnol. 106, 5415–5431. doi: 10.1007/s00253-022-12119-2
Xiong, X., Hu, Y., Yan, N., Huang, Y., Peng, N., Liang, Y., et al. (2014). PCR-DGGE analysis of the microbial communities in three different Chinese “Baiyunbian” liquor fermentation starters. J. Microbiol. Biotechnol. 24, 1088–1095. doi: 10.4014/jmb.1401.01043
You, L., Zhao, D., Zhou, R., Tan, Y., Wang, T., and Zheng, J. (2021). Distribution and function of dominant yeast species in the fermentation of strong-flavor baijiu. World J. Microbiol. Biotechnol. 37:26. doi: 10.1007/s11274-020-02988-y
Zhang, P., Liu, Y., Li, H., Hui, M., and Pan, C. (2024). Strategies and challenges of microbiota regulation in baijiu brewing. Foods 13:1954. doi: 10.3390/foods13121954
Zhang, H., Wang, L., Tan, Y., Wang, H., Yang, F., Chen, L., et al. (2021). Effect of Pichia on shaping the fermentation microbial community of sauce-flavor baijiu. Int. J. Food Microbiol. 336:108898. doi: 10.1016/j.ijfoodmicro.2020.108898
Zhou, Q., Ma, K., Song, Y., Wang, Z., Fu, Z., Wang, Y., et al. (2022). Exploring the diversity of the fungal community in Chinese traditional Baijiu daqu starters made at low-, medium- and high-temperatures. LWT 162:113408. doi: 10.1016/j.lwt.2022.113408
Keywords: Pichia kudriavzevii, transcriptomics, metabolomics, high temperature stress, heat tolerance mechanism
Citation: Wang N, Li L, Ma Y, Shen C, Ao Z, Song C, Mehmood MA, Zhang P, Liu Y, Sun X and Zhu H (2025) Combined transcriptomics and metabolomics analyses reveal the molecular mechanism of heat tolerance in Pichia kudriavzevii. Front. Microbiol. 16:1572004. doi: 10.3389/fmicb.2025.1572004
Edited by:
Jiong Hong, University of Science and Technology of China, ChinaReviewed by:
Pongsanat Pongcharoen, Naresuan University, ThailandDania Sandoval, University of Guadalajara, Mexico
Copyright © 2025 Wang, Li, Ma, Shen, Ao, Song, Mehmood, Zhang, Liu, Sun and Zhu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hui Zhu, emh1aHVpd25Ab3V0bG9vay5jb20=
†These authors have contributed equally to this work and share first authorship
 Ning Wang
Ning Wang Lu Li
Lu Li Yi Ma1,2
Yi Ma1,2 Muhammad Aamer Mehmood
Muhammad Aamer Mehmood Hui Zhu
Hui Zhu