
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 17 March 2025
Sec. Antimicrobials, Resistance and Chemotherapy
Volume 16 - 2025 | https://doi.org/10.3389/fmicb.2025.1561624
This study isolated a Klebsiella pasteurii strain, K1134, from the sputum of an ICU patient, revealing its resistance to the carbapenem antibiotics meropenem and imipenem. Whole-genome sequencing identified a plasmid pK1134-KPC, which carries the carbapenem resistance gene blaKPC-2. pK1134-KPC, belonging to the IncFIIpCP020359 plasmid group, exhibits a modular structure with blaKPC-2 embedded in a 32.09 kb accessory region containing multiple accessory genetic elements (AGEs). Comparative genomic analysis of 48 K. pasteurii isolates from 12 countries showed high genetic diversity, with strains clustered into three clades. Notably, K. pasteurii harbors extensive antimicrobial resistance genes across diverse AGEs, classifying it as multidrug-resistant. Twelve blaKPC-carrying AGEs were identified from the sequences of the isolates, classified into two groups: Tn7551 and Tn6296-related elements. The gene clusters for enterotoxins tilimycin and tilivalline, encompassing key regulators and operons, were present in nearly all strains, with incomplete clusters exclusively observed in clade 3 isolates. This study underscores the global dissemination and genetic adaptability of K. pasteurii, highlighting its potential role as a reservoir for resistance genes and emphasizing the need for robust surveillance to mitigate its public health impact.
The Klebsiella oxytoca species complex (KoSC) is part of the human microbiome (Wan et al., 2023), but also an opportunistic pathogen causing various infections, including antibiotic-associated hemorrhagic colitis (AAHC) (Yang et al., 2022; Long et al., 2022). Members of KoSC are highly versatile pathogens, isolated from a variety of infections including wound, intra-abdominal, urinary, and lower respiratory tract infections. Outbreaks often stem from environmental sources, and the subsequent spread of phylogroup members within healthcare settings presents a significant challenge—particularly in neonatal intensive care units (Cosic et al., 2021). The KoSC has been found to comprise nine species, and each species has been assigned based on average nucleotide identity (ANI) and in silico DNA–DNA hybridization (isDDH). K. pasteurii is a genospecies of KoSC defined by genomic, phenotypic and proteomic characteristics (Merla et al., 2019). Until now, this potential emerging pathogen has been reported in clinical settings in many countries. However, there is still a lack of comprehensive and large-scale genomic investigation to reveal the genetic heterogeneity in K. pasteurii.
The carbapenems, the latest generation of β-lactam antibiotics, are commonly used to treat infections that are resistant to nearly all other antibiotics, including those resistant to extended-spectrum β-lactams (Ma et al., 2021). In the Ambler classification, proteins responsible for carbapenem resistance are categorized into several classes: class A (KPC and certain GES variants), class D (OXA enzymes), and class B (metallo-β-lactamases, MBL) (Nordmann et al., 2012). A study of carbapenem-resistant KoSC strains collected from several hospitals in China showed that K. pasteurii strains have been found to carry blaKPC, blaNDM and blaIMP (Wan et al., 2023). One study indicated that Klebsiella sp. was the most common carbapenem-resistant Enterobacteriaceae (CRE) in samples isolated from patients (Stefaniak et al., 2024), and the most common carbapenem resistance gene (CRG) was blaKPC, including subtypes blaKPC-2 and blaKPC-3. In addition, another analysis of the KoSC genome showed that blaKPC-2 was the most prevalent acquired resistance gene (Long et al., 2022).
CRGs are usually located on plasmids or transposons and spread rapidly among bacteria by horizontal gene transfer (HGT), such as conjugation, transformation, or transduction (Botelho et al., 2019). Plasmids carry multiple drug resistance genes, often leading to multiple drug resistance. Currently, the most commonly used plasmid typing scheme is called Inc/rep typing. This classification method largely aligns with the conjugation-based scheme (Rozwandowicz et al., 2018). Plasmid-mediated transmission of CRGs among KoSC has been widely reported, such as the IncA plasmid carrying blaVIM (Biedrzycka et al., 2023), the IncX3 plasmid carrying blaNDM-1 (Jiang et al., 2023), the IncHI2 plasmid carrying blaSIM-1 (Li et al., 2022). In addition, there are still many sequenced CRG-carrying plasmids from KoSC that could not be assigned to any known Inc groups.
The dissemination of KoSC in healthcare settings, coupled with its ability to colonize the skin, respiratory tract, and urogenital tract, as well as to invade the bloodstream, drives outbreaks of both community-acquired and nosocomial infections globally (Glabonjat et al., 2021). KoSC causes AAHC, which is linked to the ability of these strains to produce enterotoxins, namely tilimycin (TV) and tilivalline (TM), and these cytotoxins also contribute to colonization resistance (Osbelt et al., 2024). In an animal model, K. oxytoca carry a secondary metabolite biosynthetic gene cluster that encodes a nonribosomal peptide assembly pathway (Schneditz et al., 2014), and it comprises 12 genes.
Here, we reported a carbapenem drug resistant K. pasteurii isolate K1134 harboring carbapenemase-encoding gene blaKPC-2, and its genetic characteristics were comprehensively analyzed. Together with the 47 K. pasteurii genomes available in GenBank, we used a total collection of the 48 global K. pasteurii sequenced isolates for further genomic epidemiology analyses, aiming to reveal the population structure, disclose the prevalence of antimicrobial resistance and virulence, and dissect the genetic environment of blaKPC in K. pasteurii, contributing to better understanding and management of its role in antimicrobial resistance and public health threats.
Strain K1134 was isolated in 2020 from the sputum specimen of a patient from a tertiary hospital located in Beijing, China. Activity of Ambler class A/B/D carbapenemases in bacterial cell extracts was determined by a modified CarbaNP test (Feng et al., 2016). The presence of the blaKPC gene was investigated using PCR assays. Bacterial species identification was done using genome sequence-based ANI analysis (Jain et al., 2018). Bacterial antimicrobial susceptibility was tested by BioMérieux VITEK 2, and interpreted as per the 2020 Clinical and Laboratory Standards Institute (CLSI) guidelines (Humphries et al., 2021).
Bacterial genomic DNA was isolated using the UltraClean Microbial Kit (Qiagen, NW, Germany) and sequenced from a paired-end library with an average insert size of 350 bp (range: 150–600 bp) on a HiSeq sequencer (Illumina, CA, United States), as well as a shared DNA library with an average size of 15 kb (range: 10–20 kb) on a PacBio RSII sequencer (Pacific Biosciences, CA, United States). The paired-end short Illumina reads were used to correct the long PacBio reads with the software proovread (Hackl et al., 2014), and then corrected PacBio reads were assembled de novo using SMARTdenovo.1 The sequencing data were checked using NanoPack29 and FastQC.2
Open reading frames (ORFs) and pseudogenes were predicted using RAST 2.03 combined with BLASTP/BLASTN searches against the UniProtKB/Swiss-Prot database4 and the RefSeq database.5 Annotation of resistance genes, mobile elements, and other features were carried out using the online databases including CARD,6 ResFinder,7 ISfinder,8 INTEGRALL9 and Tn Number Registry.10 Multiple and pairwise sequence comparisons were performed using MUSCLE 3.8.31 and BLASTN, respectively. Gene organization diagrams were drawn in Inkscape 1.0.11 Heatmaps were plotted with MeV 4.9.0.
All the publicly available genomes of K. pasteurii isolates in GenBank (n = 47, updated on September 9, 2024) were downloaded. The pairwise ANI values of the 48 global K. pasteurii genomes were calculated using FastANI. The K. pasteurii genome sequences were aligned to the complete chromosome sequence (GenBank accession number NZ_CP073236) of the reference strain Sb-24, and the core single nucleotide polymorphisms (SNPs) were identified by Mummer v3.2.12 All the SNPs in the repetitive DNA regions were identified and filtered by RepeatMasker.13 Based on the final recombination-free core SNPs, a maximum-likelihood phylogenetic tree was constructed using RAxML with a bootstrap iteration of 1,000 and displayed using ChiPlot (Xie et al., 2023).
A K. pasteurii strain named K1134, was isolated from the sputum specimen of a male patient admitted to the ICU surgery unit during routine sampling and culturing. Antimicrobial susceptibility testing revealed that strain K1134 was resistant to the carbapenem drugs meropenem and imipenem (Table 1). Whole-genome sequencing of K1134 demonstrated the presence of a ~ 6.2 Mb circular chromosome and six plasmids (Table 2). These plasmids were identified as pK1134-KPC, pK1134-aadA, pK1134-qnrS, pK1134-NR1, pK1134-NR2, and pK1134-NR3. Comparative genomic analysis showed that strain K1134 shared an ANI of 99.55% with the reference K. pasteurii strain Sb-24, highlighting its close genetic relationship.
We identified one blaKPC-2-carrying plasmid pK1134-KPC from the isolate we collected, which had a circular DNA sequence of 137.21 kb in length, with an average GC content of 52.98%. The plasmid (Figure 1) was identified as belonging to the IncFIIpCP020359 group because it and the reference plasmid unitig_2 (accession number CP020359) had identical repAIncpCP020359 genes sharing ≥95% nucleotide identity. The plasmid unitig_2 was found in the K. oxytoca strain AR_0147 in 2017. The modular structure of pK1134-KPC was divided into the backbone and two separated accessory regions, including a blaKPC-2 region. The blaKPC-2 region (Figure 2) was 32.09 kb in length with insertion of five accessory modules: IS5, ISKpn19, ISEc22, a truncated Tn1696, and a truncated Tn6296. Tn6296, a 14.49-kb Tn21 family unit transposon initially found in K. pneumoniae strain KP048 (Shen et al., 2009) and composed of the core backbone structure IRL (inverted repeat left)–tnpA (transposase)–tnpR (resolvase)–res–IRR-1 (inverted repeat right)–Δmcp (truncated methyl-accepting chemotaxis protein)–IRR-2, with insertion of the local blaKPC genetic environment, which consisted of the following: ΔISKpn6, blaKPC-2, and Tn6376.
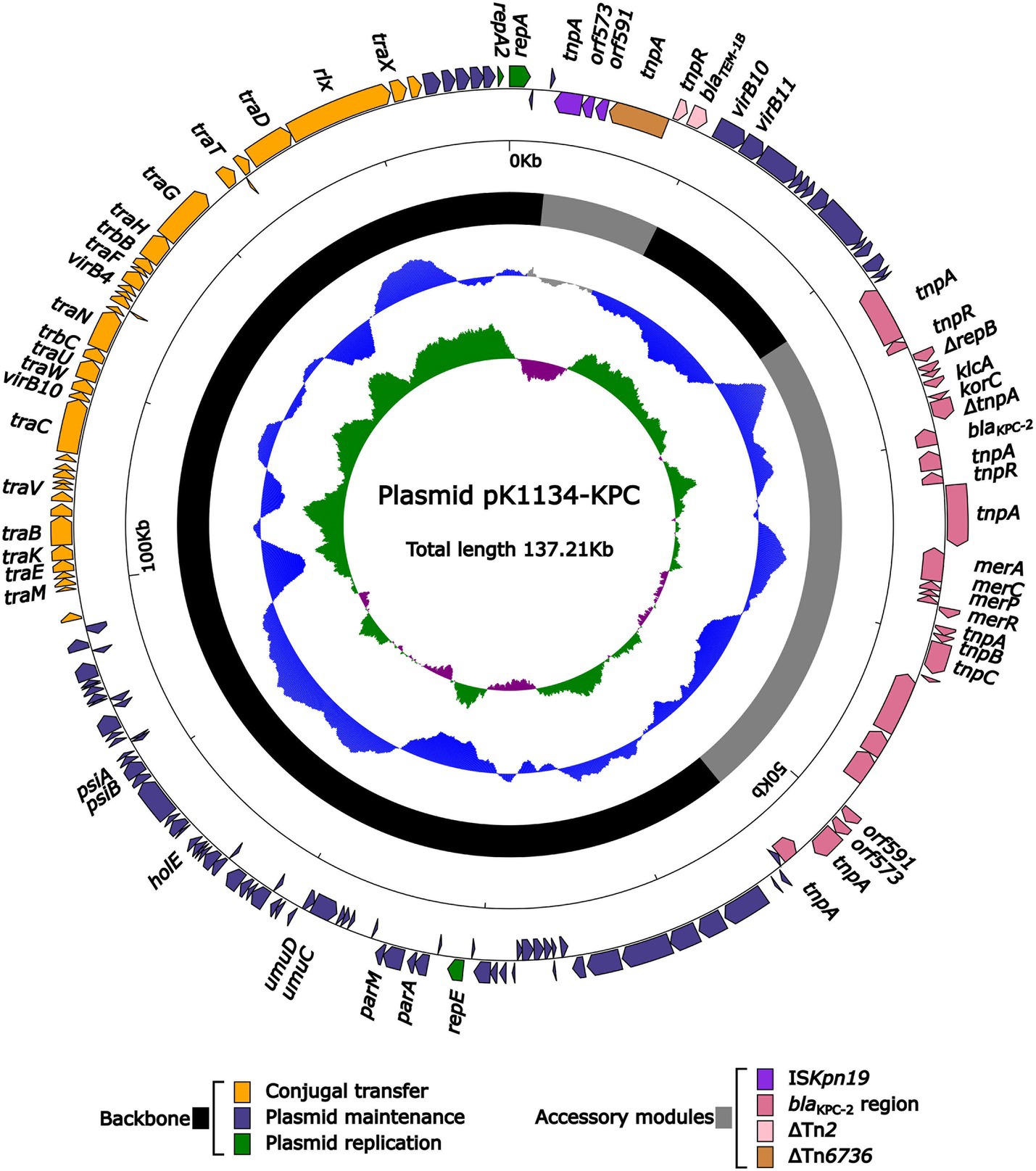
Figure 1. Schematic maps of the sequenced plasmid pK1134-KPC. Genes are denoted by arrows, and the backbone and accessory module regions are highlighted in black and gray, respectively. The next-to-innermost circle presents GC content.
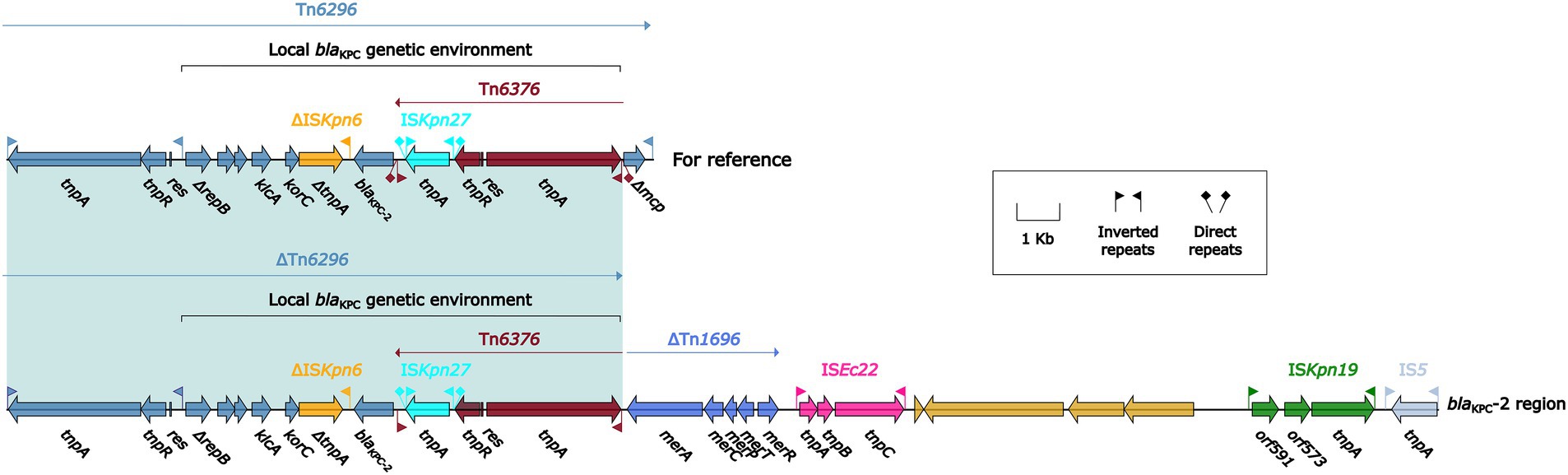
Figure 2. Organization of blaKPC-2 region and comparison to related regions. Genes are denoted by arrows. Genes, mobile elements and other features are colored based on their functional classification. Shading denotes regions of homology (nucleotide identity >95%). The accession number of Tn6296 used as reference is FJ628167.
To further investigate the population structure and antibiotic resistance of K. pasteurii, we then performed the species classification and phylogenomic analysis on a collection of 48 global sequenced K. pasteurii isolates from 12 countries (Figure 3 and Supplementary Table S1), including the above one together with the other 47 from GenBank (last accessed July 1 2024). By comparison with the reference, all strains belonged to K. pasteurii based on the threshold of 95% ANI for genospecies delineation. Through genomic analysis, we identified three unique clades within the species: Clade1 (n = 1), Clade2 (n = 12) and Clade3 (n = 35). 35.45% (n = 17) of the isolates were from China (Supplementary Figure S1), and 15 isolates, including K1134 in this study, belonged to Clade3. Most of the other strains came from Western Europe, East Asia and North America (Supplementary Table S1). In addition, we performed MLST analysis on these K. pasteurii strains. The results showed that ST416 (n = 10, 20.83%) was the most common ST, and this ST type was only found in Clade2. Through MLST analysis, 10 novel STs, designated as ST628 to ST637 were identified, which had not been previously reported in the ST database, including the only strain in Clade1, whose identification result was ST629. The analysis revealed a high level of genetic diversity among the strains.
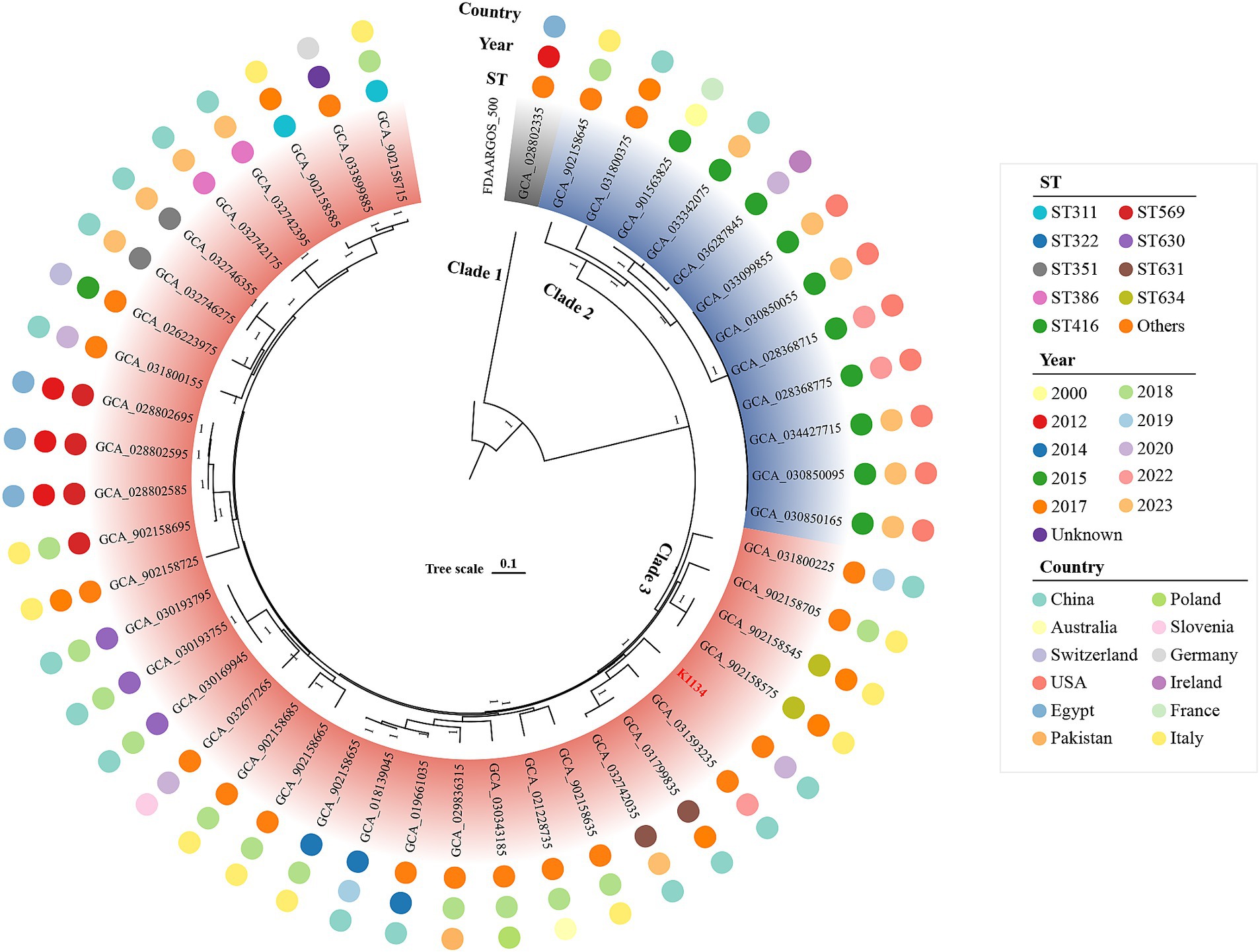
Figure 3. A maximum-likelihood phylogenetic tree of K. pasteurii isolates. Bar corresponds to scale of sequence divergence. K. oxytoca strain FDAARGOS_500 (accession number CP033844) is used as the outgroup. The strain K1134 is marked with red.
At least 43 resistance genes, involved in resistance to 11 different categories of antibiotic, were identified in these 48 global K. pasteurii genomes (Figure 4). KoSC strains are in general intrinsically resistant to β-lactams (Dabernig-Heinz et al., 2024), in addition to blaOXY, efflux pump-encoding oqxA/oqxB and the fosfomycin resistance gene fosA were also found in all isolates. Of these 48 K. pasteurii isolates, 20 were identified as containing no other resistance genes, and the remaining 28 isolates all contained one or more acquired antimicrobial resistance genes. Other drugs, such as trimethoprim resistance gene dfrA (n = 22), sulphonamide resistance gene sul1 (n = 21), and β-lactamase resistance gene blaTEM (n = 18) were found in most isolates. sul1 genes are often situated within class 1 integrons and usually co-exist in the integrons with other resistance genes, such as aminoglycoside resistance genes and quinolone resistance genes (Jacoby et al., 2014). All in all, K. pasteurii has a large number of acquired antimicrobial resistance genes, which can carried by diverse AGEs such as plasmids.
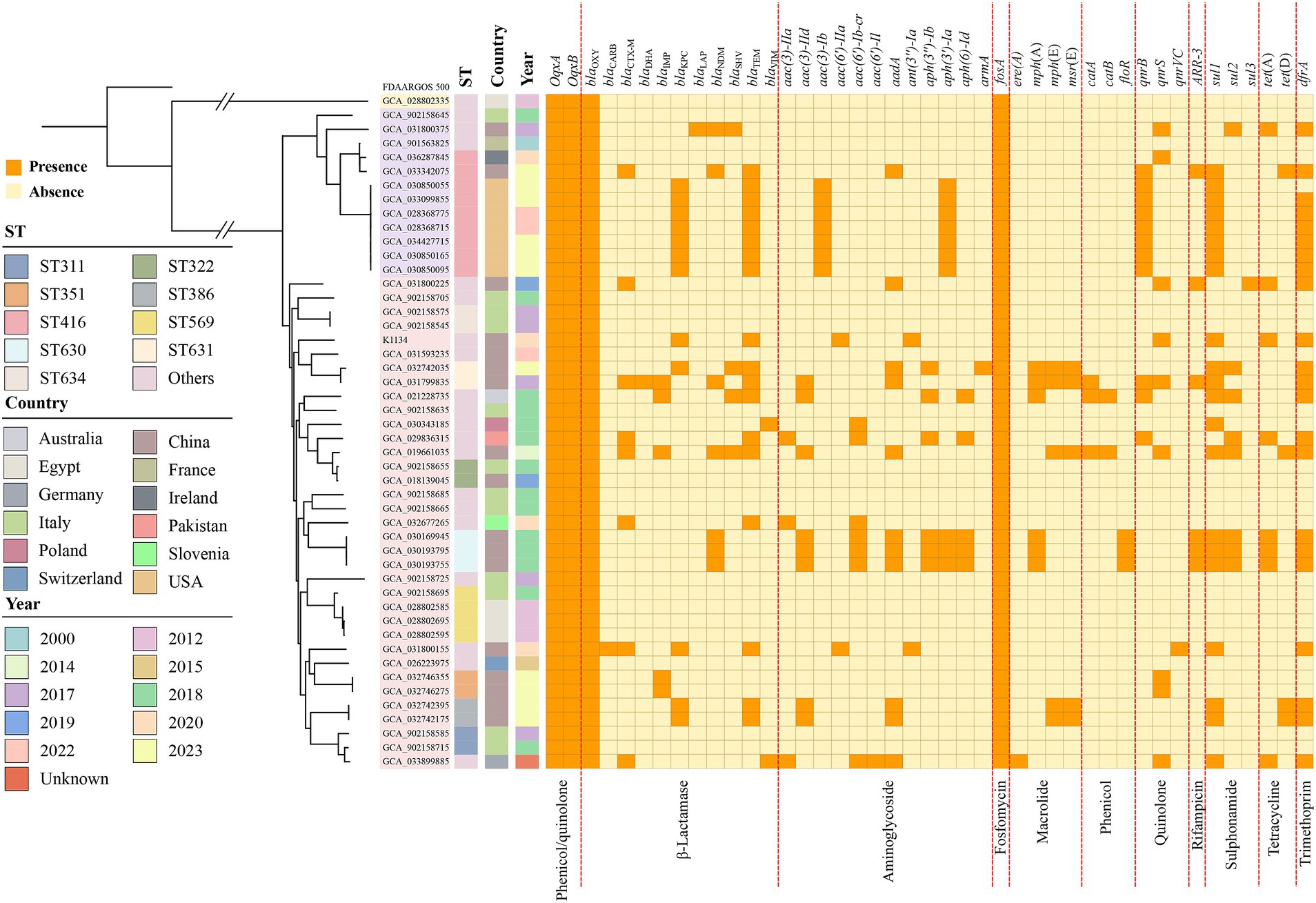
Figure 4. The maximum-likelihood tree and ARGs distributions in K. pasteurii. The matrix of ARGs is present for all K. pasteurii genomes against the dendrogram. The top of the matrix indicates the names of ARGs, while the bottom of that represents the groups of ARGs. The colored cells represent the “presence” of the ARGs.
blaKPC gene was detected in 12 isolates, and 12 blaKPC-carrying AGEs were identified from the sequences of the isolates, which belonging to two groups: Tn7551 and Tn6296-related elements. Seven isolates contained the blaKPC-4 carrying Tn7551 (Figure 5), which was a Tn3-family unit transposon and composed of the core backbone structure IRL–tnpA–res–tnpR–blaTEM-1–IRR, with a Tn4401-family unit transposon Tn4401b inserted into tnpA, and it had terminal 32-bp IRL/IRR pairs and were further bracketed by 5-bp direct repeats (DRs; target site duplication signals for transposition). The remaining four strains were detected to carry blaKPC-202 and their genetic environment was related to Tn6296. However, due to the inability to obtain the complete genome sequences, it is not possible to describe the specific genetic environment of blaKPC-202 from the four isolates. In addition, according to previous research, these genes are commonly carried by plasmids (Long et al., 2022). repAIncFIB and repAIncHI5 were also identified from the genome sequences.
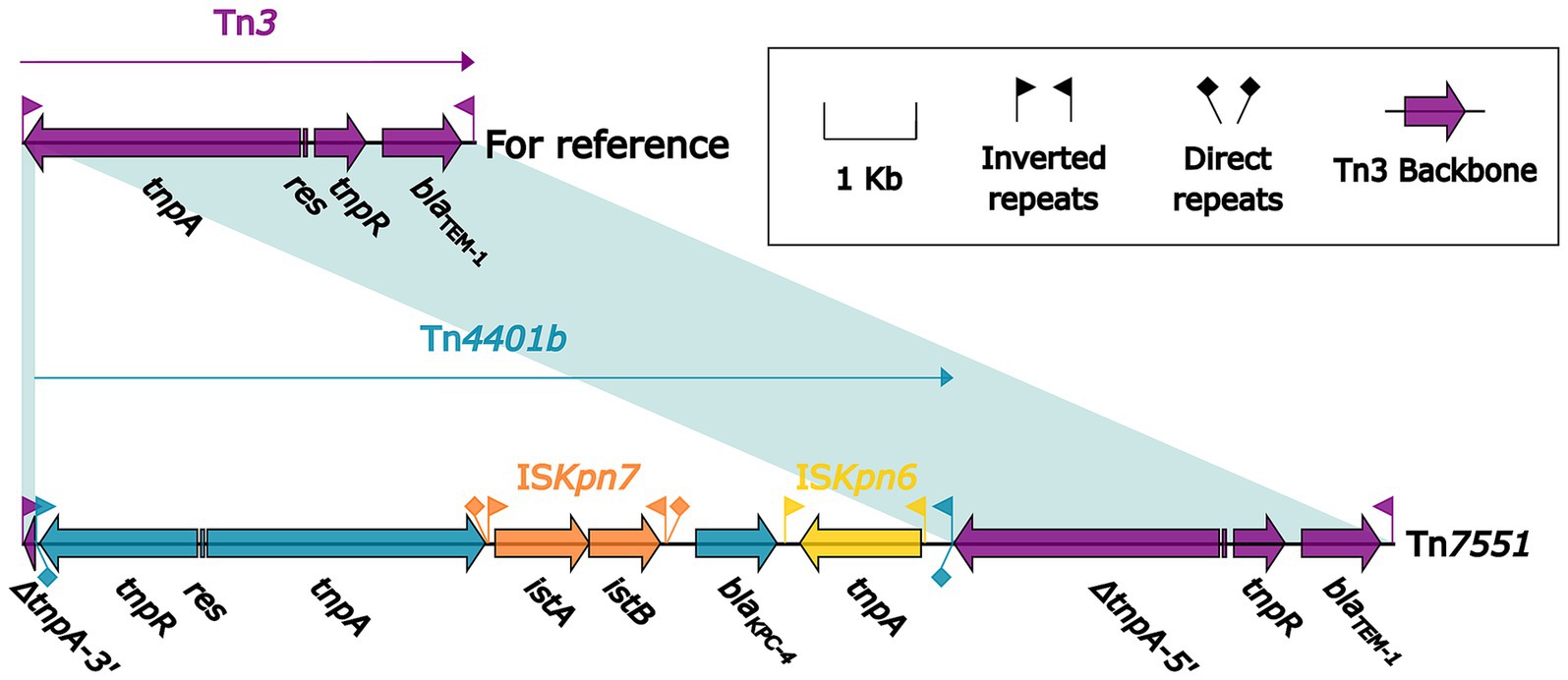
Figure 5. Comparison of Tn3 and Tn7551. Genes are denoted by arrows. Genes, mobile elements and other features are colored based on their functional classification. Shading denotes regions of homology (nucleotide identity >95%). The accession number of Tn3 used as reference is HM749966.
The gene cluster for tilimycin and tilivalline contains the regulators npsC and marR, mfsX (multidrug efflux MFS transporter), uvrX (excinuclease ABC subunit UvrA), an NRPS operon including npsA (amino acid adenylation domain-containing protein), thdA (acyl carrier protein), and npsB (nonribosomal peptide synthetase), and an aroX operon including aroX (3-deoxy-7-phosphoheptulonate synthase), dhbX (2,3-dihydro-2,3-dihydroxybenzoate dehydrogenase), icmX (isochorismatase), adsX (2-amino-2-deoxy-isochorismate synthase), and hmoX (4 hydroxyphenyl acetate-3-monooxygenase) (Yang et al., 2022; von Tesmar et al., 2018). Our results showed that these virulence factors were present in almost all strains (Supplementary Figure S2). The isolates with incomplete gene clusters were all derived from Clade3. Among them, one strain lacked all the genes, two strains were missing both the NRPS and aroX operons, and one strain exhibited a partial deletion within the aroX operon.
Infections caused by KoSC are the second most common cause of Klebsiella infections in humans (Stewart et al., 2022). The KoSC consists of at least nine species, including K. grimontii, K. huaxiensis, K. michiganensis, K. oxytoca, K. pasteurii, K. spallanzanii, and three unnamed novel species. Phylogroups (Ko) has been established based on differences in the intrinsic β-lactamase gene blaOXY encoded by KoSC, and K. pasteurii represents the phylogroup Ko4 (Shibu et al., 2021). In this study, all K. pasteurii strains contained the gene blaOXY-4. Moreover, the identity of all 48 genomes included in this work was confirmed by ANI analysis, which shared 98.10–100.00% ANI with each other. Therefore, all strains were confirmed as belonging to the species K. pasteurii.
As shown in this study, three unique clades were identified of the species: Clade1, Clade2 and Clade3, and most of which were identified as Clade3 (72.92%), including the reference strain Sb-24, and the K1134 sequenced in this study. The only Clade1 strain was from Egypt in 2012. Geographic or temporal patterns also reveal genetic diversity within the species, and it was found that the strains with the same spatial and temporal distribution also had genomic differences. The MLST is a powerful tool for epidemiological investigation of bacterial pathogens. A total of 28 distinct STs were identified, including 10 that were novel to this study. ST416 (20.83%) was the most common ST in these strains, and this ST type was only found in Clade2 and has become prevalent worldwide. Except for ST416, ST569 and ST630, which were slightly common (n ≥ 3), other STs were mostly detected in a single strain.
Acquired antimicrobial resistance is an emerging concern for K. pasteurii. Whole-genome sequencing revealed K1134 includes a blaKPC-2-carrying plasmid pK1134-KPC, which belongs to the IncFIIpCP020359 group. Comparative genomic analysis showed that Tn6296 is associated with the local blaKPC genetic environment. blaKPC gene was detected in other 11 strains, and the 12 blaKPC-carrying AGEs belong to two groups: Tn7551 and Tn6296-related elements. The seven strains carrying Tn7551 transposons showed high homology and were isolated from the United States in 2022 and 2023 with ST type ST416, suggesting clonal spread of these strains. The strains carrying Tn6296-related elements exhibit a dispersed distribution across the phylogenetic tree, indicating a lack of clear clonal clustering. This suggests substantial genetic diversity within the group of drug-resistant strains.
In the 48 global K. pasteurii genomes, at least 43 resistance genes were identified, conferring resistance to 11 classes of antibiotics and heavy metals, with strains generally being intrinsically resistant to β-lactams. These genomes also harbored a variety of acquired resistance genes. KoSC carries intrinsic β-lactamase-encoding blaOXY and fosfomycin resistance gene fosA (Merla et al., 2019). Many resistance genes in addition to these co-exist in these starins, suggest that these strains are multidrug-resistant, with the ability to resist a wide range of antibiotics. In addition, coexistence of two carbapenem genes was found in several strains, which will further complicates the treatment of K. pasteurii infections. Analysis of KoSC collected at a hospital in Australia showed that one K. pasteurii strain carried blaIMP-4 IncL/M plasmids, which indicates that resistance plasmids similar to those found in other Enterobacterales, were found in KoSC, so it is possible for KoSC to develop multidrug resistance (Stewart et al., 2022).
KoSC is rising as a significant opportunistic pathogen causing nosocomial infections (Neog et al., 2021). The pathogenicity of the microbe has been attributed to the production of cytotoxins, namely tilivalline and tilimycin, which are implicated in certain intestinal disorders (Minami et al., 1992). Our results showed that the related virulence factors were present in almost all K. pasteurii strains, which raises concerns about the widespread potential for these strains to cause intestinal infections in hospital settings, particularly in high-risk areas such as intensive care units and neonatal care units.
This study reveals the molecular epidemiology, antimicrobial resistance profiles, and pathogenicity of K. pasteurii. First, K. pasteurii is part of the KoSC and is classified into three major clades, with Clade3 being the most common. MLST analysis identified 28 distinct STs, with ST416 being the most prevalent and widely distributed globally. Additionally, K. pasteurii exhibits significant antimicrobial resistance, particularly to β-lactams. Various resistance genes including blaKPC were identified in these strains, further complicating treatment options. The study underscores the role of plasmids and other genetic elements in conferring resistance. The presence of plasmids and other genetic elements, such as transposons plays a crucial role in the emergence of multidrug-resistant K. pasteurii strains. This underscores the need for careful monitoring and control measures in healthcare settings. Almost all K. pasteurii strains in the study carried these virulence factors related to tilivalline and tilimycin, raising concerns about their potential to cause infections in hospital settings. In conclusion, K. pasteurii is an emerging opportunistic pathogen with broad antimicrobial resistance and significant virulence potential. These factors warrant increased clinical attention to manage its spread and infection risk, particularly in hospital environments.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.
The specimens were obtained with the patient’s consent. The use of human specimens and all related experimental protocols were reviewed and approved by the Ethics Committee of Taizhou Municipal Hospital, Zhejiang, China, in accordance with the medical research regulations of the Ministry of Health, China. Research and all related procedures involving biohazardous materials were approved by the Biosafety Committee of Taizhou Municipal Hospital affiliated with Taizhou University. This research was conducted in China.
XLi: Methodology, Supervision, Writing – original draft. ZS: Data curation, Investigation, Writing – review & editing. JL: Formal analysis, Writing – review & editing. JJ: Conceptualization, Writing – review & editing. HW: Validation, Writing – review & editing. HC: Project administration, Writing – review & editing. XLu: Funding acquisition, Writing – review & editing, Writing – original draft.
The author(s) declare that financial support was received for the research and/or publication of this article. This work was supported by the Natural Science Foundation of Zhejiang Province (LTGY23H190003) and the Medical and Health Science and Technology Project of Zhejiang Province (2023KY1323 and 2025KY461).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The authors declare that no Gen AI was used in the creation of this manuscript.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2025.1561624/full#supplementary-material
1. ^https://github.com/ruanjue/smartdenovo
2. ^https://www.bioinformatics.babraham.ac.uk/projects/fastqc
4. ^https://web.expasy.org/docs/swiss-prot_guideline.html
5. ^https://www.ncbi.nlm.nih.gov/refseq/
6. ^https://card.mcmaster.ca/browse
7. ^https://cge.cbs.dtu.dk/services/ResFinder/
8. ^https://www-is.biotoul.fr/
9. ^http://integrall.bio.ua.pt/?glossary
10. ^https://www.ucl.ac.uk/eastman/tn-number-registry
Biedrzycka, M., Urbanowicz, P., Żabicka, D., Hryniewicz, W., Gniadkowski, M., and Izdebski, R. (2023). Country-wide expansion of a VIM-1 carbapenemase-producing Klebsiella oxytoca ST145 lineage in Poland, 2009–2019. Eur. J. Clin. Microbiol. Infect. Dis. 42, 1449–1457. doi: 10.1007/s10096-023-04682-x
Botelho, J., Lood, C., Partridge, S. R., van Noort, V., Lavigne, R., Grosso, F., et al. (2019). Combining sequencing approaches to fully resolve a carbapenemase-encoding megaplasmid in a Pseudomonas shirazica clinical strain. Emerg. Microbes Infect. 8, 1186–1194. doi: 10.1080/22221751.2019.1648182
Cosic, A., Leitner, E., Petternel, C., Galler, H., Reinthaler, F. F., Herzog-Obereder, K. A., et al. (2021). Variation in accessory genes within the Klebsiella oxytoca species complex delineates monophyletic members and simplifies coherent genotyping. Front. Microbiol. 12:692453. doi: 10.3389/fmicb.2021.692453
Dabernig-Heinz, J., Wagner, G. E., Prior, K., Lipp, M., Kienesberger, S., Ruppitsch, W., et al. (2024). Core genome multilocus sequence typing (cgMLST) applicable to the monophyletic Klebsiella oxytoca species complex. J. Clin. Microbiol. 62:e0172523. doi: 10.1128/jcm.01725-23
Feng, W., Zhou, D., Wang, Q., Luo, W., Zhang, D., Sun, Q., et al. (2016). Dissemination of IMP-4-encoding pIMP-HZ1-related plasmids among Klebsiella pneumoniae and Pseudomonas aeruginosa in a Chinese teaching hospital. Sci. Rep. 6:33419. doi: 10.1038/srep33419
Glabonjat, R. A., Kitsera, M., Unterhauser, K., Lembacher-Fadum, C., Högenauer, C., Raber, G., et al. (2021). Simultaneous quantification of enterotoxins tilimycin and tilivalline in biological matrices using HPLC high resolution ESMS2 based on isotopically 15N-labeled internal standards. Talanta 222:121677. doi: 10.1016/j.talanta.2020.121677
Hackl, T., Hedrich, R., Schultz, J., and Förster, F. (2014). proovread: large-scale high-accuracy PacBio correction through iterative short read consensus. Bioinformatics 30, 3004–3011. doi: 10.1093/bioinformatics/btu392
Humphries, R., Bobenchik, A. M., Hindler, J. A., and Schuetz, A. N. (2021). Overview of changes to the Clinical and Laboratory Standards Institute Performance Standards for Antimicrobial Susceptibility Testing, M100, 31st Edition. J. Clin. Microbiol. 59:e0021321. doi: 10.1128/JCM.00213-21
Jacoby, G. A., Strahilevitz, J., and Hooper, D. C. (2014). Plasmid-mediated quinolone resistance. Microbiol. Spectr. 2. doi: 10.1128/microbiolspec.PLAS-0006-2013
Jain, C., Rodriguez, R. L., Phillippy, A. M., Konstantinidis, K. T., and Aluru, S. (2018). High throughput ANI analysis of 90K prokaryotic genomes reveals clear species boundaries. Nat. Commun. 9:5114. doi: 10.1038/s41467-018-07641-9
Jiang, T., Li, G., Huang, L., Ding, D., Ruan, Z., and Yan, J. (2023). Genomic and phylogenetic analysis of a multidrug-resistant blaNDM-carrying Klebsiella michiganensis in China. Infect. Drug Resist. 16, 3109–3116. doi: 10.2147/IDR.S409544
Li, S., Jiang, X., Li, C., Ju, Y., Yue, L., Chen, F., et al. (2022). A blaSIM-1 and mcr-9.2 harboring Klebsiella michiganensis strain reported and genomic characteristics of Klebsiella michiganensis. Front. Cell. Infect. Microbiol. 12:973901. doi: 10.3389/fcimb.2022.973901
Long, H., Hu, Y., Feng, Y., and Zong, Z. (2022). Genome analysis of Klebsiella oxytoca complex for antimicrobial resistance and virulence genes. Antimicrob. Agents Chemother. 66:e0218321. doi: 10.1128/aac.02183-21
Ma, P., He, L. L., Pironti, A., Laibinis, H. H., Ernst, C. M., Manson, A. L., et al. (2021). Genetic determinants facilitating the evolution of resistance to carbapenem antibiotics. eLife 10:e67310. doi: 10.7554/eLife.67310
Merla, C., Rodrigues, C., Passet, V., Corbella, M., Thorpe, H. A., Kallonen, T. V. S., et al. (2019). Description of Klebsiella spallanzanii sp. nov. and of Klebsiella pasteurii sp. nov. Front. Microbiol. 10:2360. doi: 10.3389/fmicb.2019.02360
Minami, J., Saito, S., Yoshida, T., Uemura, T., and Okabe, A. (1992). Biological activities and chemical composition of a cytotoxin of Klebsiella oxytoca. J. Gen. Microbiol. 138, 1921–1927. doi: 10.1099/00221287-138-9-1921
Neog, N., Phukan, U., Puzari, M., Sharma, M., and Chetia, P. (2021). Klebsiella oxytoca and emerging nosocomial infections. Curr. Microbiol. 78, 1115–1123. doi: 10.1007/s00284-021-02402-2
Nordmann, P., Dortet, L., and Poirel, L. (2012). Carbapenem resistance in Enterobacteriaceae: here is the storm! Trends Mol. Med. 18, 263–272. doi: 10.1016/j.molmed.2012.03.003
Osbelt, L., Almási, É. D. H., Wende, M., Kienesberger, S., Voltz, A., Lesker, T. R., et al. (2024). Klebsiella oxytoca inhibits Salmonella infection through multiple microbiota-context-dependent mechanisms. Nat. Microbiol. 9, 1792–1811. doi: 10.1038/s41564-024-01710-0
Rozwandowicz, M., Brouwer, M. S. M., Fischer, J., Wagenaar, J. A., Gonzalez-Zorn, B., Guerra, B., et al. (2018). Plasmids carrying antimicrobial resistance genes in Enterobacteriaceae. J. Antimicrob. Chemother. 73, 1121–1137. doi: 10.1093/jac/dkx488
Schneditz, G., Rentner, J., Roier, S., Pletz, J., Herzog, K. A., Bücker, R., et al. (2014). Enterotoxicity of a nonribosomal peptide causes antibiotic-associated colitis. Proc. Natl. Acad. Sci. U.S.A. 111, 13181–13186. doi: 10.1073/pnas.1403274111
Shen, P., Wei, Z., Jiang, Y., Du, X., Ji, S., Yu, Y., et al. (2009). Novel genetic environment of the carbapenem-hydrolyzing beta-lactamase KPC-2 among Enterobacteriaceae in China. Antimicrob. Agents Chemother. 53, 4333–4338. doi: 10.1128/AAC.00260-09
Shibu, P., McCuaig, F., McCartney, A. L., Kujawska, M., Hall, L. J., and Hoyles, L. (2021). Improved molecular characterization of the Klebsiella oxytoca complex reveals the prevalence of the kleboxymycin biosynthetic gene cluster. Microb. Genom. 7:000592. doi: 10.1099/mgen.0.000592
Stefaniak, K., Kiedrzyński, M., Korzeniewska, E., Kiedrzyńska, E., and Harnisz, M. (2024). Preliminary insights on carbapenem resistance in Enterobacteriaceae in high-income and low-/middle-income countries. Sci. Total Environ. 957:177593. doi: 10.1016/j.scitotenv.2024.177593
Stewart, J., Judd, L. M., Jenney, A., Holt, K. E., Wyres, K. L., and Hawkey, J. (2022). Epidemiology and genomic analysis of Klebsiella oxytoca from a single hospital network in Australia. BMC Infect. Dis. 22:704. doi: 10.1186/s12879-022-07687-7
von Tesmar, A., Hoffmann, M., Abou Fayad, A., Hüttel, S., Schmitt, V., Herrmann, J., et al. (2018). Biosynthesis of the Klebsiella oxytoca pathogenicity factor tilivalline: heterologous expression, in vitro biosynthesis, and inhibitor development. ACS Chem. Biol. 13, 812–819. doi: 10.1021/acschembio.7b00990
Wan, W., Yang, X., Yu, H., Wang, M., Jia, W., Huang, B., et al. (2023). Genomic characterization of carbapenem-resistant Klebsiella oxytoca complex in China: a multi-center study. Front. Microbiol. 14:1153781. doi: 10.3389/fmicb.2023.1153781
Xie, J., Chen, Y., Cai, G., Cai, R., Hu, Z., and Wang, H. (2023). Tree Visualization By One Table (tvBOT): a web application for visualizing, modifying and annotating phylogenetic trees. Nucleic Acids Res. 51, W587–w592. doi: 10.1093/nar/gkad359
Keywords: Klebsiella pasteurii, carbapenem resistance, plasmid, blaKPC, mobile genetic element, tilimycin and tilivalline
Citation: Li X, Song Z, Liu J, Jin J, Wan H, Chen H and Luo X (2025) Unraveling the genomic epidemiology and plasmid-mediated carbapenem resistance of Klebsiella pasteurii. Front. Microbiol. 16:1561624. doi: 10.3389/fmicb.2025.1561624
Received: 16 January 2025; Accepted: 03 March 2025;
Published: 17 March 2025.
Edited by:
Magdalena Popowska, University of Warsaw, PolandReviewed by:
Costas C. Papagiannitsis, University of Thessaly, GreeceCopyright © 2025 Li, Song, Liu, Jin, Wan, Chen and Luo. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xinhua Luo, c2x5eV8wMTU0MUB0emMuZWR1LmNu
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.