
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol., 24 March 2025
Sec. Infectious Agents and Disease
Volume 16 - 2025 | https://doi.org/10.3389/fmicb.2025.1560340
This article is part of the Research TopicHerpesvirus: Transmission, Pathogenesis, Host-Pathogen Interaction, Prevention and TreatmentView all 4 articles
Introduction: The objective of this study was to investigate the effect of acyclovir (ACV) on the TLR9 signaling pathway after human immortalized epidermal (HaCaT) cell infection with herpes simplex virus type 2 (HSV-2).
Methods: In this study, an in vitro cell model of HSV-2 infection was successfully constructed by infecting HaCaT with HSV-2 virus. In order to explore the antiviral mechanism of acyclovir (ACV), high-throughput transcriptome sequencing (RNA-seq) was used to analyze the genome-wide expression profiling of infected cells before and after ACV treatment, and to systematically compare the change characteristics of differentially expressed genes (DEGs). Based on the sequencing results, the study further focused on Toll-like receptor (TLR) 9 signaling, using quantitative real-time reverse transcriptase chain reaction (qRT-PCR) to quantitatively detect the effect of ACV intervention on the mRNA expression level of key molecules of TLR 9 signaling pathway in HSV-2 infected HaCaT cells.
Results: A total of 896 significant changes in gene expression were identified by the transcriptome analysis, including 314 upregulated genes and 582 downregulated genes. GO enrichment analysis showed that the differentially expressed genes were mainly related to CC includes the ubiquitin ligase complex, mitochondrial protein-containing complex, DNA-binding transcription activator activity, exonuclease activity, catabolic process, nuclear-transcribed mRNA catabolic process nuclear-transcribed mRNA catabolic process; KEGG enrichment analysis showed that the differentially expressed genes were mainly related to Toll-like receptor signaling pathway, herpes simplex virus 1 infection, and TNF signaling pathway. The RT-PCR results were confirmed to be basically consistent with the sequencing results.
Conclusion: ACV altered the transcriptome level of HSV-2 infection in HaCaT cells. The RT-PCR results confirmed that ACV intervened in HSV-2 infection through the TLR9 signaling pathway.
Genital herpes (GH) is a sexually transmitted disease caused by herpes simplex virus 2 (HSV-2) infection, clinically manifested as erythema, papules, vesicles, erosion and exudation, associated with burning skin and mucous membranes with genital and anal (Parra-Sánchez, 2019). HSV infection is characterized by lifelong infection, recurrent recurrence, and neurotropic (Gatherer et al., 2021). The pathogenesis of genital herpes involves complex virus-host interactions.
Acyclovir (ACV) as a first-line treatment for genital herpes. Belongs to nucleoside analogues that exert antiviral effects through selective inhibition of HSV DNA polymerase (Bourne et al., 2019). It is phosphorylated in infected cells by the viral thymidine kinase in infected cells and converted to an active form, which competitively inhibits viral DNA synthesis (Lee and Ashkar, 2012). Recent studies suggest that acyclovir may enhance the antiviral effect by modulating the host innate immune response. The mechanism of action involves the regulation of the TLR 9 signaling pathway.
Recently, Toll-like receptor 9 (TLR 9) was found to play a key role in the immune response against HSV (Lv et al., 2018). Toll-like receptors (TLRs) are the pattern recognition receptor (Gill et al., 2008) for microbial pathogens. TLR 9 is a key pattern recognition receptor for recognizing viral DNA and is an important component of innate immunity (Feng et al., 2021), which is mainly expressed in epithelial cells and immune cells. After HSV-2 infection, TLR9 recognizes the DNA of viral HSV-2, initiates the innate immune response, activates NF-κB and IRF7 (Gill et al., 2005) through MyD 88-dependent signaling pathway (Gill et al., 2006), and induce production of type I interferon and pro-inflammatory cytokines (Tognarelli et al., 2019).
Transcriptome sequencing (RNA-seq) is a high-throughput sequencing technology that was developed in recent years, which can effectively evaluate the interactions between viruses and host cells (Kumar, 2025). Transcriptome sequencing has been used to analyze gene transcription levels and related mechanisms in different cells and in animal models of HSV infection (Rumpf and Schulz, 2025). In this study, high-throughput RNA sequencing technology was used to perform transcriptomic analysis of ACV-treated HaCaT cells infected with HSV-2, and the differentially expressed genes after ACV-infected HSV-2 infection were screened using virus-infected cells as controls. The functions of differentially expressed genes and the cellular processes and signaling pathways that may be involved in HSV-2 infection were further analyzed. In this study, we explored the mechanism of action of ACV against HSV-2 based on the TLR9 signaling pathway. And the replication efficiency of the virus was observed by determining HSV-2 gD levels, an advanced gene product of the virus.
The HaCaT (Procell CL-0090) cell line was purchased from Procell Life Science & Technology Co., Ltd. HSV-2 was purchased from the Hunan Prema Pharmaceutical Research Center as human herpesvirus 2/ATCC VR-1779. Both overexpression and knockdown viruses were constructed by Shanghai Genechem Co., Ltd.
Fetal bovine serum (FBS), MEM medium, penictomycin, and pancreatic enzymes were purchased from Wuhan Pricella Company. An RNA extraction kit was purchased from SIMGEN, and the cDNA reverse transcription and amplification kit was purchased from Novoprotein. Puromycin was purchased from Shanghai Jikai Gene Medical Technology Co., Ltd. Primers were purchased from Beijing Qingke Biotechnology Co., Ltd. Acyclovir tablets were produced by Sichuan Kelun Pharmaceutical Co., Ltd. (Approval Number: Guoyao Zhunzi H10983103).
Secondary biosafety cabinet (Thermo Fisher Scientific, Germany), high-speed refrigerated centrifuge (Thermo Fisher, Germany), carbon dioxide incubator (Thermo, Germany); inverted fluorescence microscope (CarelZeiss, Germany); real-time quantitative PCR instrument (Bio-Rad/GFX Connect); gene amplifier (Eppendorf/Mastercycler nexus GX2, Germany); vertical automatic pressure steam sterilizer (Xiamen Zhi Micro Instrument Company).
Twenty SPF-grade SD rats were purchased from Hunan SJA Laboratory Animal Co., Ltd. [License Number: SCKX (Xiang) 2019-0004]. There were 10 male and 10 female rats, 8 weeks old, weighing 200 ± 20 g. The rats were maintained in an isolated environment at a temperature of 22–26°C and a humidity of 40–60% and ate and drank freely. After 3 days of adaptive feeding, the animals were divided into a blank control group and an acyclovir group according to the random number table method, with 10 animals in each group. The rat administration dose was calculated based on the equivalent dose ratio of the human and rat body surface areas. Acyclovir was administered at 0.37 mg/kg 3 times each day for 7 days, and the blank control group was given the same volume of normal saline for 2 times each day for 7 days. After fasting for 24 h after the last dose, pentobarbital sodium was intraperitoneally injected. After the rats were completely anesthetized, abdominal aortic blood was collected, and after standing at room temperature for 2 h, the upper serum was collected by centrifugation for 15 min at 3,000 r/min and frozen in a −80°C freezer. The animal experiment operation was approved by the Experimental Animal Ethics Committee of Hunan University of Traditional Chinese Medicine (Approval Number: HNUCM21-2405-19).
Cells in logarithmic growth and displaying good growth and an appropriate multiplicity of infection were selected according to a pre-experiment, and an appropriate amount of lentiviral vector was added. The negative control group (CON335) of the lentivirus overexpression group (LV-TLR9-L) was transfected with CON335 virus. The negative control group (CON313) of the lentiviral knockdown group (LV-TLR9-H) was transfected with CON313 virus.
After 24 h of infection, the effect of the infection was preliminarily observed by fluorescence microscopy. After 72 h of infection, an appropriate concentration of puromycin and complete medium was added. The solution was changed regularly, and cells were screened for stable transfection. The cells were collected for RT-PCR analysis after 7 days of culture to identify the expression level of the target gene TLR9, and the cells with qualified identification results were cryopreserved and passaged for follow-up experiments.
HaCaT cells were cultured in 10% FBS with 1% penicillin/streptomycin and 89% MEM medium at 37°C and 5% CO2. After 1.25% trypsinization, cells were seeded at a density of 4 × 105/mL in 6-well plates to continue culturing.
Logarithmic cells in the growth phase were seeded in 6-well plates, and when the cell density reached 90%, the following treatments were applied: KBZ: only 2,000 μL complete medium; BLZ: add 50 μL HSV-2 virus solution and 1,950 μL of complete medium; ACV: add 50 μL HSV-2 virus solution, 100 μL ACV of drug-containing serum and 1,850 μL of complete medium; LV-TLR 9-H + H: Hacat cells overexpressing TLR 9 with 50 μL HSV-2 viral solution and 1,950 μL of complete medium; LV-TLR 9-H + HA: Hacat cells overexpressing TLR 9 with 50 μL HSV-2 viral solution, 100 μL ACV of drug-containing serum and 1,850 μL of complete medium; LV-TLR 9-L + H: Hacat cells expressing TLR 9 were supplemented with 50 μL HSV-2 viral solution and 1,950 μL of complete medium; LV-TLR 9-L + HA: Hacat cells with 50 μL HSV-2 virus solution 100 μL, ACV-containing serum and 1,850 μL of complete medium. After intervention, put into the incubator. This experiment was performed with the virus-related operations in the BSL-2 laboratory. Each experiment was repeated 3 times.
Three samples were selected from the BLZ group and the ACV group for biological replicates, and the RNA extraction kit was used to extract the total RNA. An Agilent 2100 bioanalyzer was used to determine the RNA integrity and total amount. After the quality inspection was confirmed, the cDNA library was sequenced using the Illumina platform for high-throughput sequencing.
DESeq2 software was used to analyze differentially expressed genes (DEGs) in BLZ group and ACV group. DEGs satisfied the criteria p < 0.05 and |log2(fold change)| >1. ClusterProfiler was used to perform GO and KEGG enrichment analysis of DEGs.
Transcriptomics was performed by the Shanghai Tanpu Biotechnology Co., Ltd. (Shanghai, China).
When the cell viability was about 60%, the total RNA was extracted with an RNA extraction kit. The RNA extraction kit and reverse transcription kit were used for mRNA extraction and reverse transcription of cDNA from HaCaT cells, respectively, and reverse transcription polymerase chain reaction (RT-PCR) was carried out with cDNA, SYBR master mix, and PCR primers in a 10-μL system. Relative mRNA expression was calculated by PCR using the method (see Table 1).
SPSS 26.0 and GraphPad Prism 8.0 software were used for statistical analysis. The experimental data were used as continuous data, expressed as , and the independent samples t-test was used for comparisons between the two groups. One-way ANOVA was used for comparing differences between multiple groups, and p < 0.05 was considered statistically significant.
According to the expression value of all genes in each sample: FPKM, the correlation coefficient of samples within and between groups is calculated and drawn into a heat map, which can directly show the sample difference between groups and the sample duplication within groups. The higher the correlation coefficient between samples, the closer the expression pattern is, and the sample correlation heat map is shown in Figure 1.

Figure 1. Differential gene cluster plots. According to the standardized expression values of the differentially expressed genes in the samples, the Pearson correlation was calculated, and the systematic clustering method (hierarchical cluster) was used to obtain the overall clustering results of the samples. In the figure, changes in expression are indicated by changes in color, where blue indicates lower expression and red indicates higher expression.
In sequencing of BLZ and ACV genes, DEGs 896 were select 582 downregulated genes and 314 upregulated genes (Figure 2).
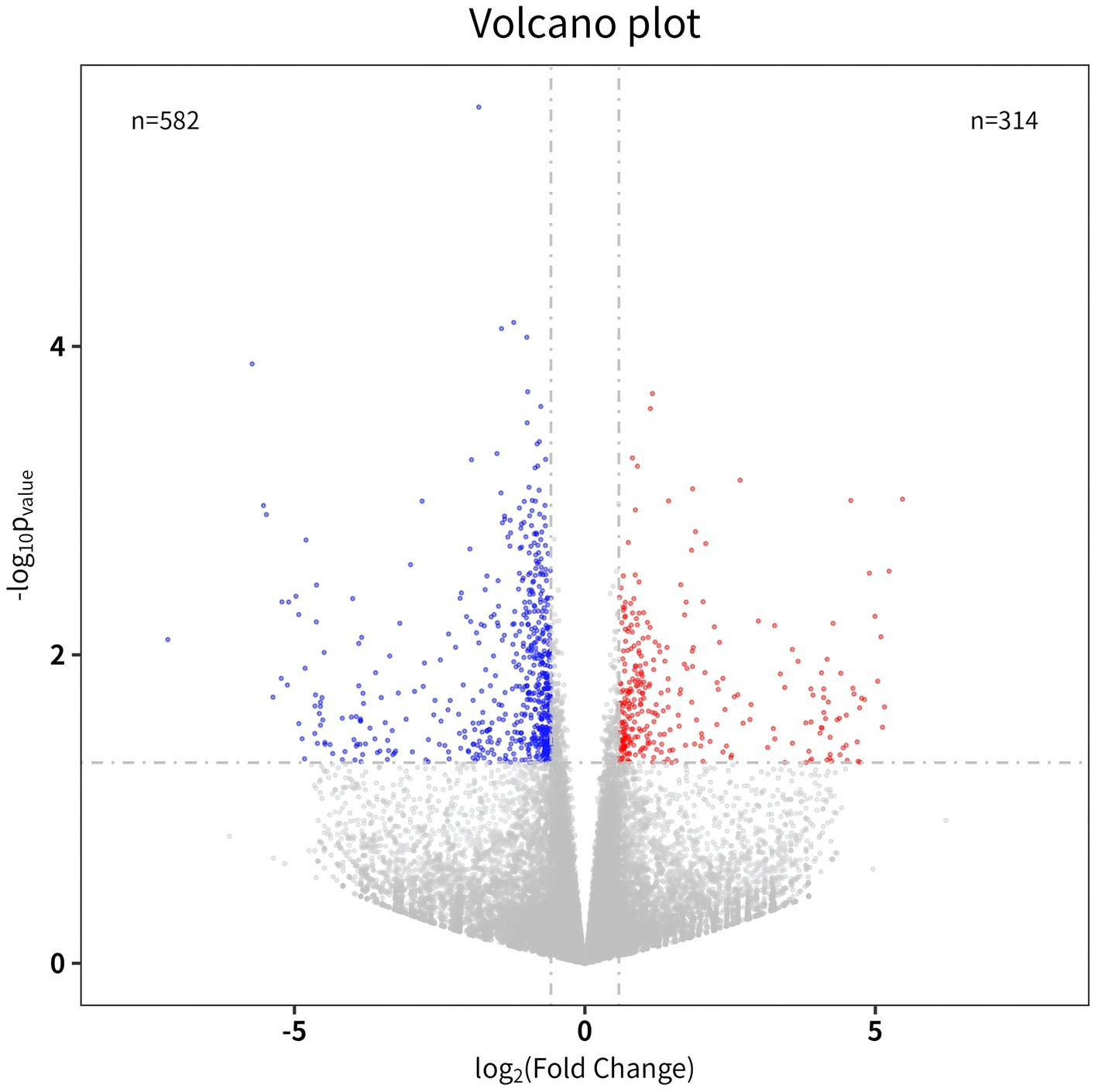
Figure 2. Volcano map. The abscissa is a multiple of the difference (logarithmic transformation with a base of 2), and the ordinate is the p-value (logarithmic transformation with a base of 10).
GO functional enrichment analysis was performed from biological process (BP), cellular component (CC) and molecular function (MF). The main enrichment of DEGs is shown in Figure 3. CC includes the ubiquitin ligase complex, mitochondrial protein-containing complex, cytoplasmic ribonucleoprotein granule, etc. MF includes the DNA-binding transcription activator activity, DNA-binding transcription activator activity, RNA polymerase II-specific, exonuclease activity, ubiquitin ligase-substrate adaptor activity, etc. BP includes proteasome-mediated ubiquitin-dependent protein, catabolic process, nuclear-transcribed mRNA catabolic process nuclear-transcribed mRNA catabolic process, cellular response to glucocorticoid stimulus cellular response to corticosteroid stimulus, etc.
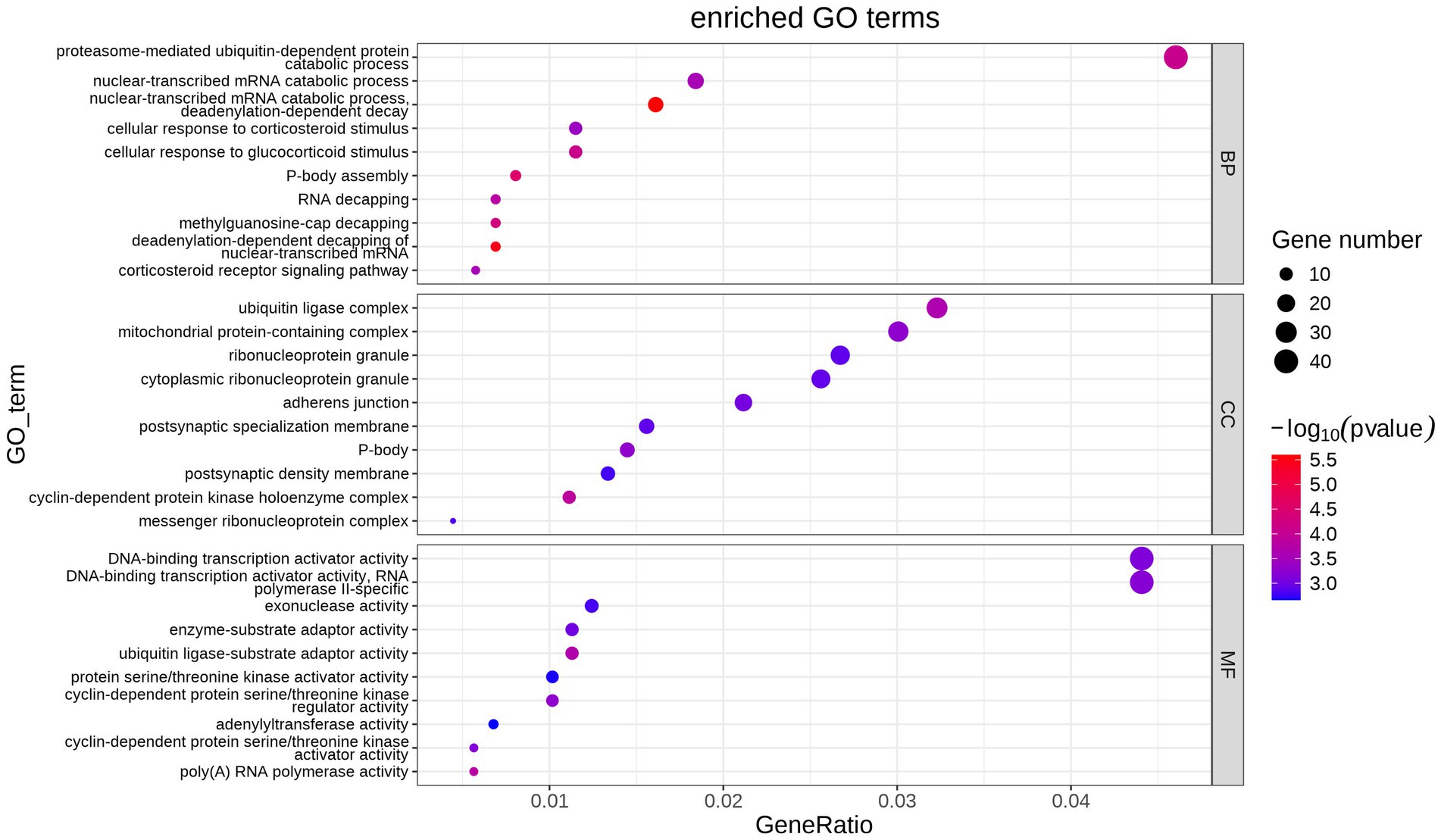
Figure 3. Bubble map of GO term enrichment statistics. Each point indicates the enrichment of the GO entry, and the closer the color is to the red, the higher the enrichment is. The size of each dot indicates the number of genes enriched to that GO entry, and larger dots indicate more genes enriched to that GO entry, and less vice versa.
The DEGs KEGG enrichment analysis obtained by screening showed that these DEGs were mainly distributed in 31 pathways, as shown in Figure 4, mainly include: Toll-like receptor signaling pathway, herpes simplex virus 1 infection, TNF signaling pathway, AGE-RAGE signaling pathway in diabetic complications, human cytomegalovirus infection, pertussis, HIF-1 signaling pathway, Kaposi sarcoma-associated herpesvirus infection, Epstein–Barr virus infection, IL-17 signaling pathway, malaria, hepatitis C, measles, pathways in cancer, prostate cancer.
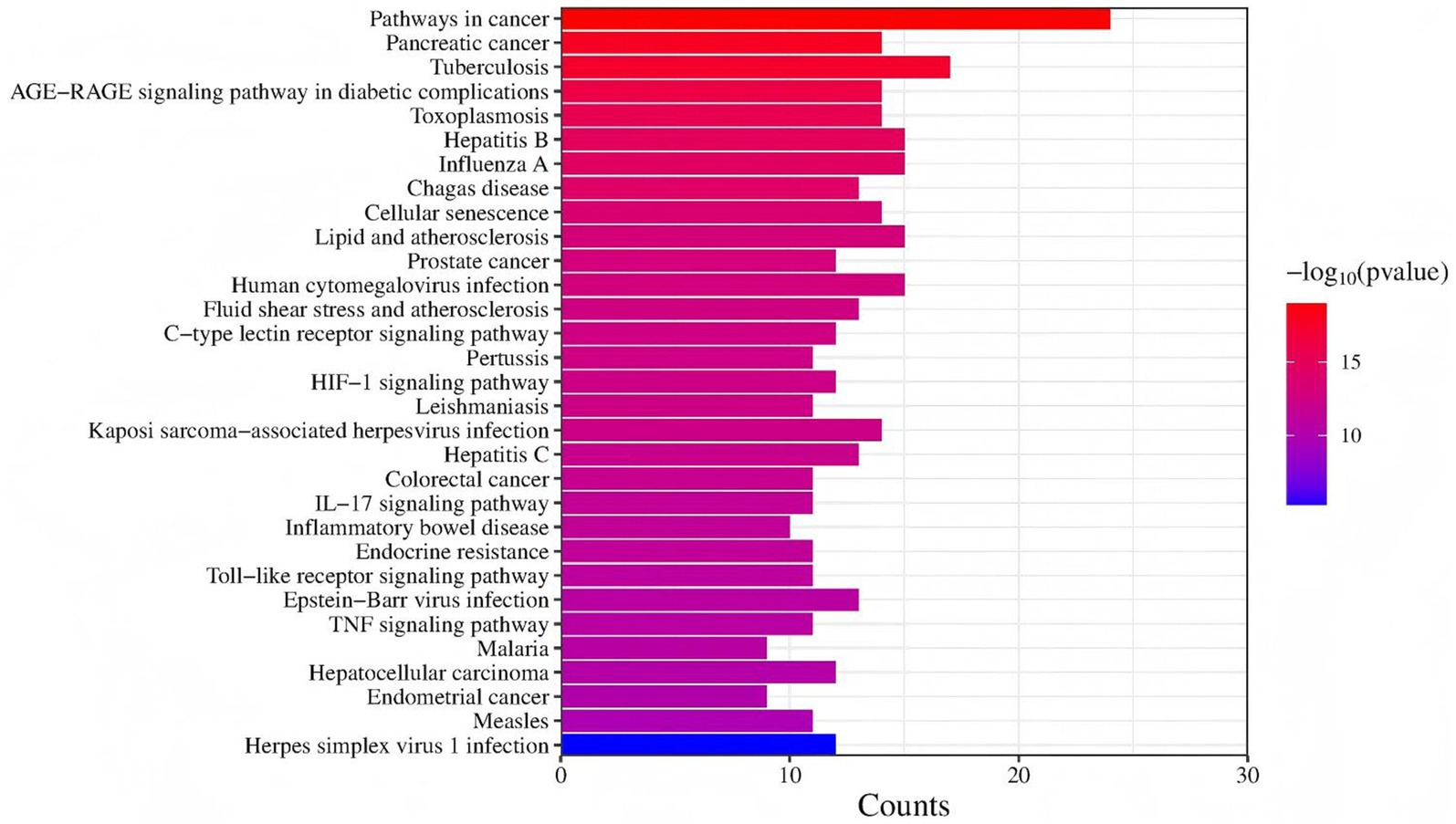
Figure 4. The KEGG pathway annotation results. The KEGG entries enriched in all samples were taken for analysis, the ordinate is KEGG entries, the abscissa is different sample names, and the closer the color is to red, the higher the enrichment is.
After transfection of HaCaT cells with lentivirus, models were made and cells of each group were observed by fluorescence microscope. The results showed that strong green fluorescence was observed in the transfection groups (Figure 5) and the transfection efficiency was higher. RT-PCR showed that TLR9 was significantly expressed in the LV-TLR9-H group compared with the CON335 group (Figure 6a). TLR9 expression was significantly reduced and statistically significantly different in the LV-TLR9-L group compared with the CON313 group (Figure 6b). These results indicated that TLR9 overexpression and knockdown in HaCaT cell lines had been successfully constructed after lentivirus transfection and screening.
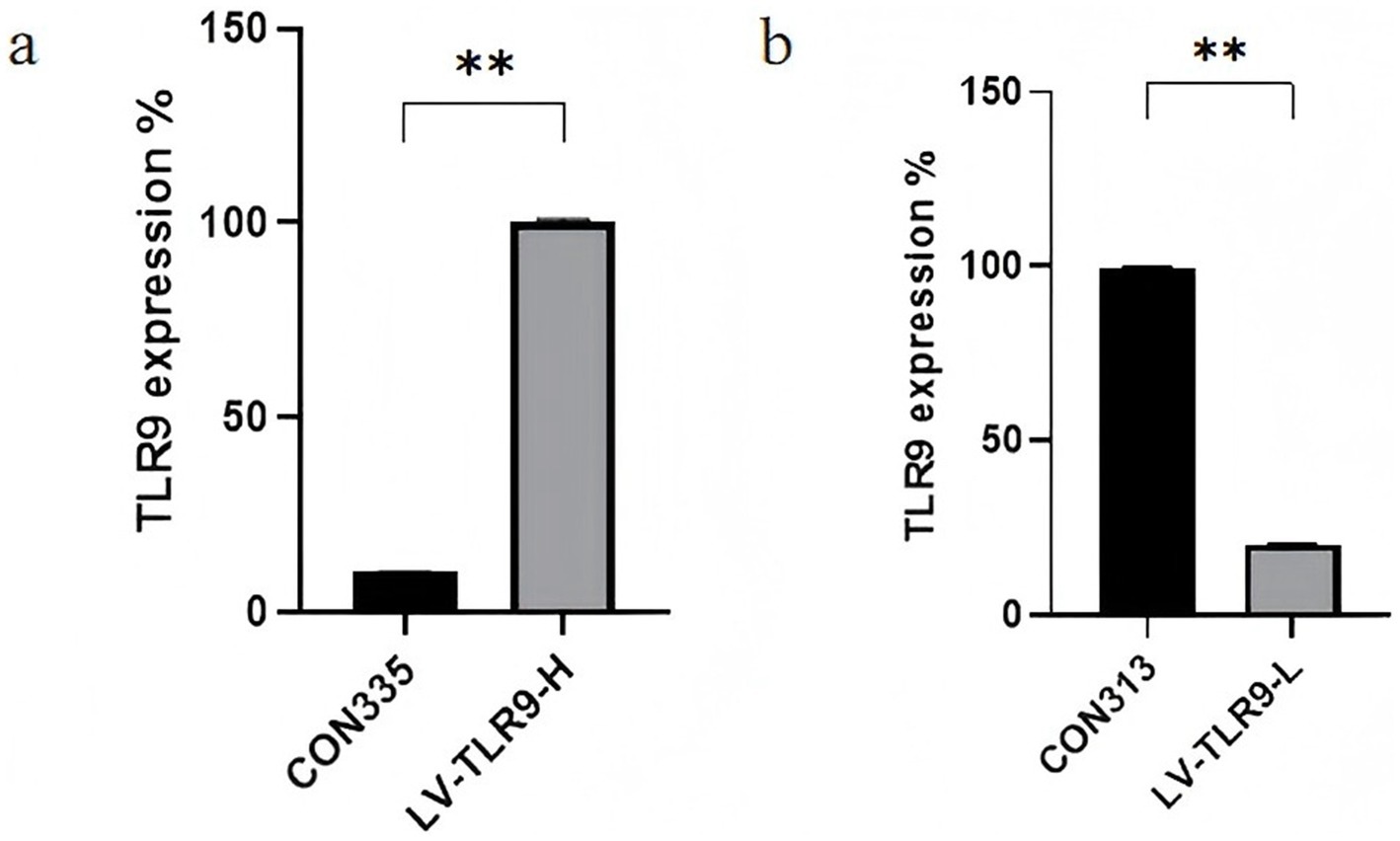
Figure 6. Identification of RNA expression levels in stable transgenic cell lines. (a) Construction of TLR 9-overexpressing cells. (b) Construction of TLR 9-knockdown cells.
The difference in expression of the target genes TLR9, MyD88, NF-κB, and gD in HSV-2-infected HaCaT cells after ACV intervention was verified by RT-PCR. The results showed that compared with the KBZ group, HSV-2 upregulated TLR9, MyD88, NF-κB, and gD, and there were statistically significant differences. Compared with the BLZ group, ACV downregulated the expression of TLR9, MyD88, NF-κB, and gD with statistical differences (Figure 7).
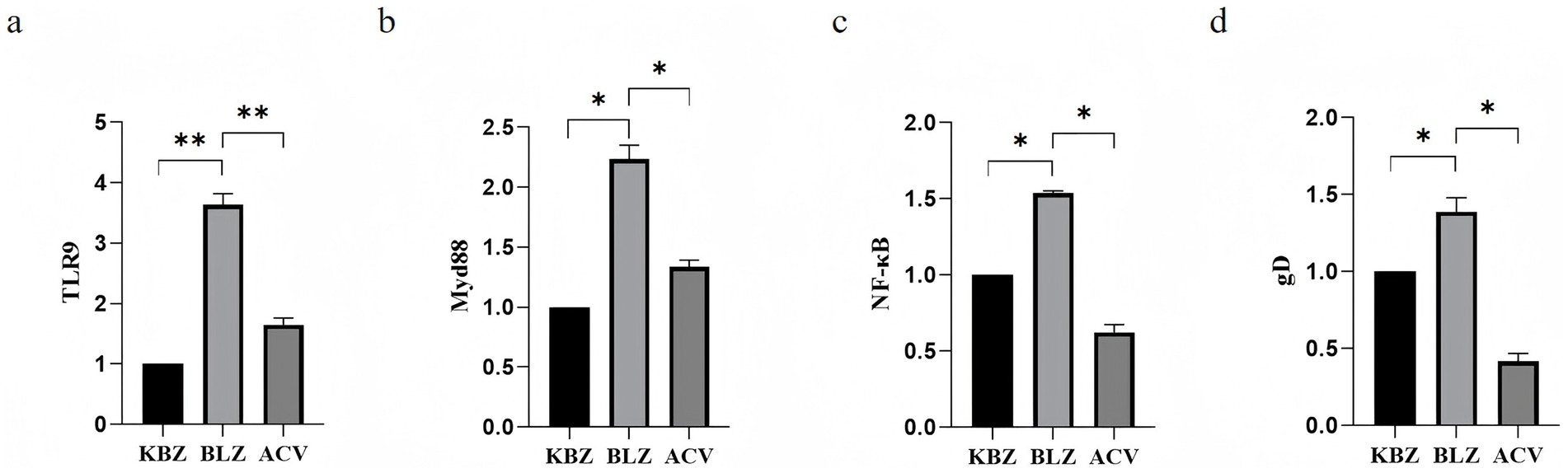
Figure 7. Expression of TLR9 signaling pathway-related genes after ACV intervention in HSV-2-infected HaCaT cells. (a) The expression status of TLR 9. (b) The expression status of MyD 88. (c) The expression status of NF-κB. (d) The expression status of gD.
After ACV intervention, the differential expression of mRNA TLR9, TLR 9, MyD 88, NF-κB, and gD in HSV-2-overexpressing Hacat cells infected with GAPDH as an internal reference was verified by RT-PCR. Results shown in Figure 8, ACV decreased the expression of TLR 9, MyD 88, NF-κB, gD compared to BLZ (p < 0.05). LV-TLR 9-H + H caused the up-regulation of MyD 88 and NF-κB compared to BLZ (p < 0.05). Compared with LV-TLR9-H + H group, LV-TLR9-H + HA group could down-regulate the expression of TLR 9, MyD 88, NF-κB, and gD.
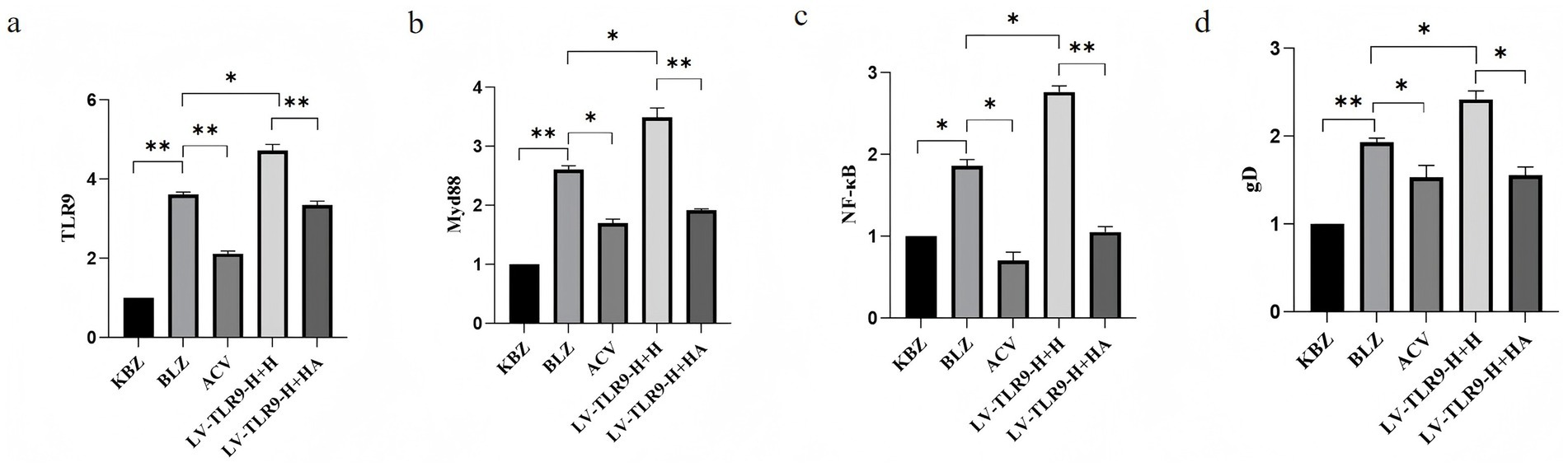
Figure 8. Expression of TLR9 signaling pathway-related genes in HSV-2-infected HaCaT cells overexpressing TLR9 after ACV intervention. *p < 0.05, **p < 0.01, and ns: p > 0.05. (a) The expression status of TLR 9. (b) The expression status of MyD 88. (c) The expression status of NF-κB. (d) The expression status of gD.
Using GAPDH as internal reference, RT-PCR verified the differential expression of mRNA TLR9, MyD 88, NF-κB, and gD in Hacat TLR 9 cells infected with HSV-2. As shown in Figure 9, ACV decreased the expression of TLR 9, MyD 88, NF-κB, and gD compared to the BLZ group (p < 0.05). Compared with BLZ, LV-TLR 9-L + H, caused the downregulation of TLR 9, MyD 88, NF-κB, and gD (p < 0.05). Compared with LV-TLR 9-L + H group, TLR 9, MyD 88, NF-κB, and gD in LV-TLR 9-L + H group (p < 0.05).
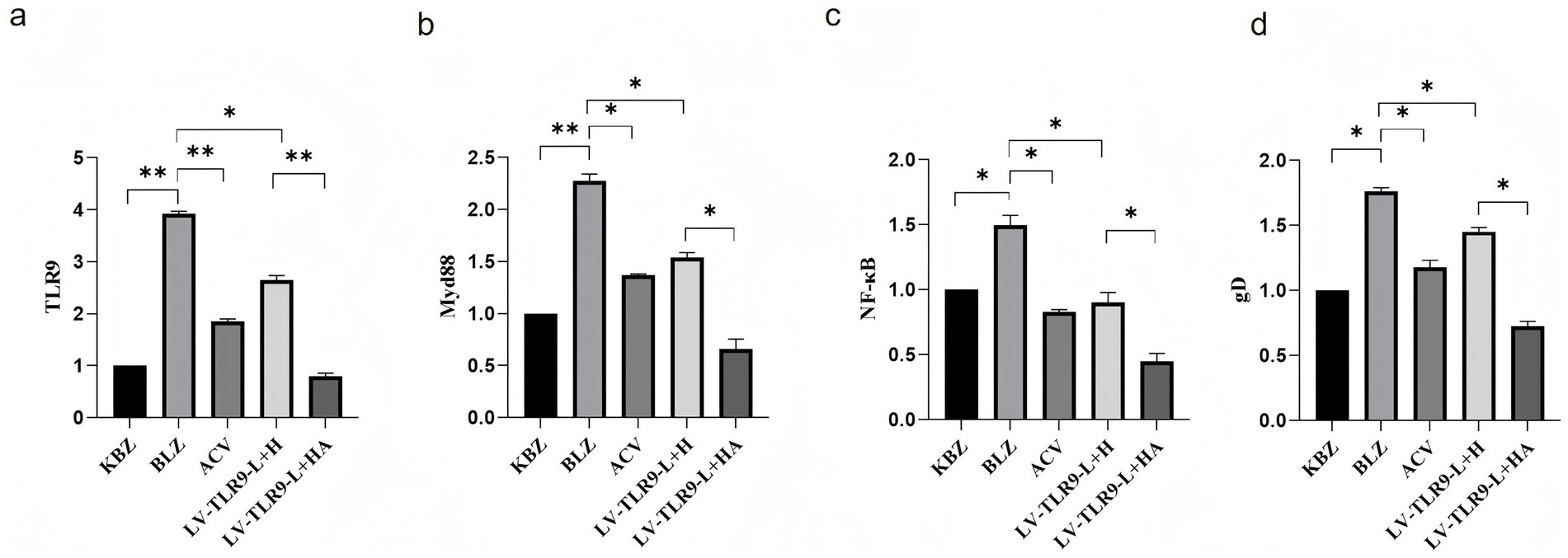
Figure 9. Expression of TLR9 signaling pathway-related genes in HSV-2-infected HaCaT cells knockdown TLR9 after ACV intervention. *p < 0.05, **p < 0.01, and ns: p > 0.05. (a) The expression status of TLR 9. (b) The expression status of MyD 88. (c) The expression status of NF-κB. (d) The expression status of gD.
The body’s immune response to HSV-2 is both innate and acquired (Kurt-Jones et al., 2017). Innate immunity is primarily mediated by leukocytes, such as neutrophils, macrophages, and dendritic cells, which are able to engulf pathogens and coordinate other responses with the synthesis of various inflammatory mediators and cytokines (Lucinda et al., 2017). A wide range of pattern recognition receptor families (PPRs) can help the immune system recognize pathogen-related molecular patterns, such as double-stranded RNA and lipopolysaccharides (Bauer and Wagner, 2002). TLR9 play an important role in this process.
TLR9 is involved in the recognition of DNA with unmethylated CpG structures in the genetic material of higher organisms or microorganisms or viruses (Gill et al., 2006). It was originally discovered in bacteria and has characteristic immunomodulatory effects based on the motifs present in bacterial DNA (Takeshita et al., 2004). However, it has since been found to be common in viruses and vertebrates and plays an important role in the recognition of Herpesviridae viruses, such as herpes simplex virus (HSV-1 and HSV-2), cytomegalovirus, and Epstein–Barr virus, and can help trigger a rapid innate immune response (Krug et al., 2004). The TLR9 receptor requires a non-canonical NF-κB pathway for pathogen recognition (Hochrein et al., 2004). Upon ligand binding, the receptor structure changes, and the MyD88 linker docks to the cytoplasmic domain of TLR9 and then forms a special complex with kinases of the IRAK family (Latz et al., 2004). This complex triggers a signaling cascade that causes NF-κB transcription factors to enter the nucleus, thereby initiating the expression of cytokines and chemokines (van Lint et al., 2010).
TLR9/MyD88/NF-κB signaling pathway is a classical pathway existing in epithelial cells (Zyzak et al., 2020), and its upregulation regulates the secretion of pro-inflammatory factors such as IL-6, TNF-α, and IL-1β in vivo, which plays an important role in the pathogenic process of GH (Svensson et al., 2007). MyD88 is involved in signaling in most tlr (Gaajetaan et al., 2012), and downregulation of MyD 88 is able to significantly inhibit HSV-2-mediated activation of AP-1 in reproductive epithelial cells, inhibiting viral replication (Wang et al., 2018). The mechanism of action may be that it suppresses the expression of related proteins on the TLR9/MyD88/NF-κB signaling pathway, and reduces the content of IL-6, TNF-α, and IL-1β inflammatory factors in vivo, thus achieving the effect of reducing skin inflammation (Zhang et al., 2013). The present experiment proved that TLR 9, NF-κB and MyD 88 were significantly lower in epithelial cells of ACV group than that in the model group (Barrow et al., 2024). This indicates that ACV may downregulate the content of proinflammatory factors and reduce skin inflammation by inhibiting the expression of TLR9/MyD88/NF-κB signaling pathway.
We propose that acyclovir not only has direct antiviral effects, but also enhances the host antiviral immune response by regulating the TLR 9 signaling pathway and improves the therapeutic efficacy on genital herpes (Bao and Liu, 2013). Acyclovir may also reduce the disease rate of recurrence by enhancing TLR 9-mediated immune memory. This dual mechanism of action provides a new idea for improving the therapeutic effect of genital herpes (Sauter et al., 2014).
In summary, we conclude that after HSV-2 infection, TLR9 effectively stimulates innate immune cells to control HSV-2 infection in HaCaT cells.
The original data in this study are included in the Supplementary material, further inqueries can be directed to the corresponding author.
The animal study was approved by Experimental Animal Ethics Committee of Hunan University of Traditional Chinese Medicine. The study was conducted in accordance with the local legislation and institutional requirements.
JS: Conceptualization, Data curation, Investigation, Methodology, Software, Validation, Writing – original draft, Writing – review & editing. LK: Conceptualization, Data curation, Funding acquisition, Methodology, Project administration, Resources, Supervision, Writing – review & editing. LQ: Data curation, Methodology, Software, Supervision, Validation, Writing – review & editing. RL: Methodology, Software, Supervision, Validation, Writing – review & editing. YW: Investigation, Project administration, Supervision, Writing – review & editing.
The author(s) declare that financial support was received for the research and/or publication of this article. This work was supported by National Natural Science Foundation of China (82174391), and Hunan Key Subject of Integrated Traditional Chinese and Western Medicine.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The authors declare that no Gen AI was used in the creation of this manuscript.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2025.1560340/full#supplementary-material
Bao, M., and Liu, Y. J. (2013). Regulation of TLR7/9 signaling in plasmacytoid dendritic cells. Protein Cell 4, 40–52. doi: 10.1007/s13238-012-2104-8
Barrow, A. D., Cella, M., Edeling, M. A., Khan, M. A. A. K., Cervantes-Barragan, L., Bugatti, M., et al. (2024). Cutting edge: PDGF-DD binding to NKp44 costimulates TLR9 signaling and proinflammatory cytokine secretion in human plasmacytoid dendritic cells. J. Immunol. 212, 369–374. doi: 10.4049/jimmunol.2200496
Bauer, S., and Wagner, H. (2002). Bacterial CpG-DNA licenses TLR9. Curr. Top. Microbiol. Immunol. 270, 145–154. doi: 10.1007/978-3-642-59430-4_9
Bourne, N., Banasik, B. N., Perry, C. L., Miller, A. L., White, M., Pyles, R. B., et al. (2019). Development of disease and immunity at the genital epithelium following intrarectal inoculation of male guinea pigs with herpes simplex virus type 2. Virology 526, 180–188. doi: 10.1016/j.virol.2018.10.023
Feng, E., Balint, E., Vahedi, F., and Ashkar, A. A. (2021). Immunoregulatory functions of interferons during genital HSV-2 infection. Front. Immunol. 12:724618. doi: 10.3389/fimmu.2021.724618
Gaajetaan, G. R., Geelen, T. H., Grauls, G. E., Bruggeman, C. A., and Stassen, F. R. (2012). CpG and poly(I:C) stimulation of dendritic cells and fibroblasts limits herpes simplex virus type 1 infection in an IFNβ-dependent- and independent way. Antivir. Res. 93, 39–47. doi: 10.1016/j.antiviral.2011.10.015
Gatherer, D., Depledge, D. P., Hartley, C. A., Szpara, M. L., Vaz, P. K., Benkő, M., et al. (2021). ICTV virus taxonomy profile: Herpesviridae 2021. J. Gen. Virol. 102:001673. doi: 10.1099/jgv.0.001673
Gill, N., Davies, E. J., and Ashkar, A. A. (2008). The role of Toll-like receptor ligands/agonists in protection against genital HSV-2 infection. Am. J. Reprod. Immunol. 59, 35–43. doi: 10.1111/j.1600-0897.2007.00558.x
Gill, N., Deacon, P. M., Lichty, B., Mossman, K. L., and Ashkar, A. A. (2006). Induction of innate immunity against herpes simplex virus type 2 infection via local delivery of Toll-like receptor ligands correlates with beta interferon production. J. Virol. 80, 9943–9950. doi: 10.1128/JVI.01036-06
Gill, N., Rosenthal, K. L., and Ashkar, A. A. (2005). NK and NKT cell-independent contribution of interleukin-15 to innate protection against mucosal viral infection. J. Virol. 79, 4470–4478. doi: 10.1128/JVI.79.7.4470-4478.2005
Hochrein, H., Schlatter, B., O'Keeffe, M., Wagner, C., Schmitz, F., Schiemann, M., et al. (2004). Herpes simplex virus type-1 induces IFN-alpha production via Toll-like receptor 9-dependent- and independent pathways. Proc. Natl. Acad. Sci. U.S.A. 101, 11416–11421. doi: 10.1073/pnas.0403555101
Krug, A., Luker, G. D., Barchet, W., Leib, D. A., Akira, S., and Colonna, M. (2004). Herpes simplex virus type 1 activates murine natural interferon-producing cells through Toll-like receptor 9. Blood 103, 1433–1437. doi: 10.1182/blood-2003-08-2674
Kumar, A. (2025). Genome Annotation. Methods Mol. Biol. 2859, 21–37. doi: 10.1007/978-1-0716-4152-1_2
Kurt-Jones, E. A., Orzalli, M. H., and Knipe, D. M. (2017). Innate immune mechanisms and herpes simplex virus infection and disease. Adv. Anat. Embryol. Cell Biol. 223, 49–75. doi: 10.1007/978-3-319-53168-7_3
Latz, E., Schoenemeyer, A., Visintin, A., Fitzgerald, K. A., Monks, B. G., Knetter, C. F., et al. (2004). TLR9 signals after translocating from the ER to CpG DNA in the lysosome. Nat. Immunol. 5, 190–198. doi: 10.1038/ni1028
Lee, A. J., and Ashkar, A. A. (2012). Herpes simplex virus-2 in the genital mucosa: insights into the mucosal host response and vaccine development. Curr. Opin. Infect. Dis. 25, 92–99. doi: 10.1097/QCO.0b013e32834e9a56
Lucinda, N., Figueiredo, M. M., Pessoa, N. L., Santos, B. S. Á. S., Lima, G. K., Freitas, A. M., et al. (2017). Dendritic cells, macrophages, NK and CD8+ T lymphocytes play pivotal roles in controlling HSV-1 in the trigeminal ganglia by producing IL1-beta, iNOS and granzyme B. Virol. J. 14:37. doi: 10.1186/s12985-017-0692-x
Lv, X., Wang, H., Su, A., Xu, S., and Chu, Y. (2018). Herpes simplex virus type 2 infection triggers AP-1 transcription activity through TLR4 signaling in genital epithelial cells. Virol. J. 15:173. doi: 10.1186/s12985-018-1087-3
Parra-Sánchez, M. (2019). Genital ulcers caused by herpes simplex virus. Úlceras genitales por virus herpes simplex. Enferm Infecc Microbiol Clin 37, 260–264. doi: 10.1016/j.eimc.2018.10.020
Rumpf, L., and Schulz, M. H. (2025). Learning enhancer-gene associations from bulk transcriptomic and epigenetic sequencing data with STITCHIT. Methods Mol. Biol. 2856, 341–356. doi: 10.1007/978-1-0716-4136-1_21
Sauter, M. M., Gauger, J. J., and Brandt, C. R. (2014). Oligonucleotides designed to inhibit TLR9 block herpes simplex virus type 1 infection at multiple steps. Antivir. Res. 109, 83–96. doi: 10.1016/j.antiviral.2014.06.015
Svensson, A., Bellner, L., Magnusson, M., and Eriksson, K. (2007). Role of IFN-alpha/beta signaling in the prevention of genital herpes virus type 2 infection. J. Reprod. Immunol. 74, 114–123. doi: 10.1016/j.jri.2006.09.002
Takeshita, F., Gursel, I., Ishii, K. J., Suzuki, K., Gursel, M., and Klinman, D. M. (2004). Signal transduction pathways mediated by the interaction of CpG DNA with Toll-like receptor 9. Semin. Immunol. 16, 17–22. doi: 10.1016/j.smim.2003.10.009
Tognarelli, E. I., Palomino, T. F., Corrales, N., Bueno, S. M., Kalergis, A. M., and González, P. A. (2019). Herpes simplex virus evasion of early host antiviral responses. Front. Cell. Infect. Microbiol. 9:127. doi: 10.3389/fcimb.2019.00127
van Lint, A. L., Murawski, M. R., Goodbody, R. E., Severa, M., Fitzgerald, K. A., Finberg, R. W., et al. (2010). Herpes simplex virus immediate-early ICP0 protein inhibits Toll-like receptor 2-dependent inflammatory responses and NF-kappaB signaling. J. Virol. 84, 10802–10811. doi: 10.1128/JVI.00063-10
Wang, S., Ljubimov, A. V., Jin, L., Pfeffer, K., Kronenberg, M., and Ghiasi, H. (2018). Herpes simplex virus 1 latency and the kinetics of reactivation are regulated by a complex network of interactions between the herpesvirus entry mediator, its ligands (gD, BTLA, LIGHT, and CD160), and the latency-associated transcript. J. Virol. 92:e01451. doi: 10.1128/JVI.01451-18
Zhang, S. Y., Herman, M., Ciancanelli, M. J., Pérez de Diego, R., Sancho-Shimizu, V., Abel, L., et al. (2013). TLR3 immunity to infection in mice and humans. Curr. Opin. Immunol. 25, 19–33. doi: 10.1016/j.coi.2012.11.001
Keywords: HSV-2, acyclovir, TLR9, RNA-seq, HaCaT
Citation: Shi J, Kuang L, Qi L, Li R and Wu Y (2025) Effect of the TLR9 signaling pathway on acyclovir infection with herpes simplex virus type 2 in HaCaT cells. Front. Microbiol. 16:1560340. doi: 10.3389/fmicb.2025.1560340
Received: 14 January 2025; Accepted: 04 March 2025;
Published: 24 March 2025.
Edited by:
Jinlin Li, Uppsala University, SwedenReviewed by:
Valentina Dell’Oste, University of Turin, ItalyCopyright © 2025 Shi, Kuang, Qi, Li and Wu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lin Kuang, eWFuZ21pbWkwNkAxNjMuY29t
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.